SUMMARY
Lysine succinylation was recently identified as a post-translational modification in cells. However, the molecular mechanism underlying lysine succinylation remains unclear. Here, we show that carnitine palmitoyltransferase 1A (CPT1A) has lysine succinyl-transferase (LSTase) activity in vivo and in vitro. Using a stable isotope labeling by amino acid in cell culture (SILAC)-based proteomics approach, we found that 101 proteins were more succinylated in cells expressing wild-type (WT) CPT1A compared with vector control cells. One of the most heavily succinylated proteins in this analysis was enolase 1. We found that CPT1A WT succinylated enolase 1 and reduced enolase enzymatic activity in cells and in vitro. Importantly, mutation of CPT1A Gly710 (G710E) selectively inactivated carnitine palmitoyltransferase (CPTase) activity but not the LSTase activity that decreased enolase activity in cells and promoted cell proliferation under glutamine depletion. These findings suggest that CPT1A acts as an LSTase that can regulate enzymatic activity of a substrate protein and metabolism independent of its classical CPTase activity.
In Brief
Kurmi et al. find that carnitine palmitoyltransferase (CPT) 1A has lysine succinyltransferase (LSTase) activity in vivo and in vitro. Mutation of CPT1A Gly710 (G710E) selectively inactivates canonical carnitine palmitoyltransferase (CPTase) activity but not LSTase activity.
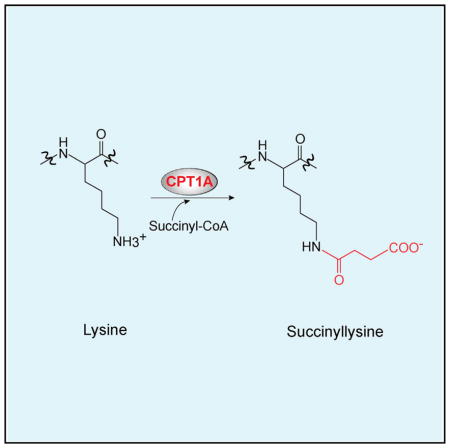
INTRODUCTION
Multiple post-translational modifications on the epsilon-amino group of lysine regulate protein functions. Recent studies have added lysine succinylation, which has been observed in many species (Colak et al., 2013; Park et al., 2013; Rardin et al., 2013; Weinert et al., 2013; Zhang et al., 2011), as an additional lysine modification. Sirtuin (SIRT) 5 can remove succinyl modifications from lysine (Du et al., 2011). Indeed, recent studies showed that increased accumulation of succinyl-coenzyme A (CoA) caused increased lysine succinylation, likely because of non-enzymatic lysine succinylation (Li et al., 2015; Wagner and Payne, 2013; Weinert et al., 2013). However, given that (1) lysine acetylation is catalyzed by acetyltransferases even though acetyl-CoA can non-enzymatically acetylate lysines (Choudhary et al., 2014) and (2) regulatory post-translational modifications are typically catalyzed by enzymes, it is likely that there is a lysine succinyltransferase (LSTase) that catalyzes the forward reaction of lysine succinylation using succinyl-CoA as a substrate.
Therefore, succinyl-CoA is considered to be a putative substrate for an LSTase. It was also shown, more than 30 years ago, that short-chain dicarboxylic-acyl-CoAs, including succinyl-CoA, bind to carnitine palmitoyltransferase 1A (CPT1A) and inhibit its carnitine palmitoyltransferase (CPTase) activity (McGarry et al., 1977; Mills et al., 1983). CPT1A catalyzes the formation of long-chain acylcarnitines from long-chain acyl-CoAs and carnitine, the rate-limiting step of mitochondrial fatty acid oxidation (FAO) that metabolizes fatty acids into acetyl-CoA to produce ATP in mitochondria. Although the inhibitory effect of succinyl-CoA on CPTase activity is well accepted in the field, how succinyl-CoA binds to CPT1A remains unclear because of the lack of the crystal structure of the CPT1A protein. In this study, we show that CPT1A is able to use succinyl-CoA as a substrate to function as an LSTase in vitro and in vivo. Our SILAC-based quantitative succinylation proteomics analysis identified 171 lysine sites on 101 proteins (out of 550 lysine sites on 247 proteins total) that were succinylated in a CPT1A expression-dependent manner in cells. Importantly, the CPTase activity and the LSTase activity of CPT1A can be separated by mutation of CPT1A Gly710 (G710E). Using this CPT1A G710E mutant, we show that the LSTase activity of CPT1A inhibited enolase 1 enzymatic activity and promoted cell proliferation under glutamine depletion. Thus, we propose that CPT1A regulates cellular metabolism by succinylating substrate proteins independent of its canonical CPTase activity.
RESULTS AND DISCUSSION
Identification of CPT1A as a Potential LSTase
Because succinyl-CoA is a short-chain acyl-CoA, we hypothesized that acyltransferases with CoA binding domains may have LSTase activity. To identify potential candidates, we first searched the Swiss-Prot database using “acyltransferase” and “CoA binding” as keywords and obtained 33 candidate proteins (Figure S1A). We next excluded acyltransferases expressed in yeast, because yeasts do not express desuccinylases (e.g., SIRT5) that would be expected to reverse succinylation catalyzed by an LSTase. After these screening steps, we obtained nine candidates, including all CPT family members (Figure S1A). Among the CPT family members, CPT1A is the most expressed isoform in the liver, where lysine succinylation occurs at highest levels compared with other tissues (Park et al., 2013; Rardin et al., 2013). Interestingly, short-chain dicarboxylic-acyl-CoAs, including succinyl-CoA, are known to inhibit the CPTase activity (McGarry et al., 1977; Mills et al., 1983). A structural modeling study of CPT1A proposed two potential CoA binding pockets within the active site (López-Vinãs et al., 2007). Also, lysine succinylation and carnitine acylation are quite different in terms of chemical reactivity and steric occlusion because lysine succinylation is linked to the epsilon-amino group of lysine, whereas carnitine acylation occurs on a secondary hydroxyl group. Thus, we hypothesize that succinyl-CoA may bind to a locus different from the palmitoyl-CoA-binding locus within the CPT1A active site, which triggers CPT1A to have an additional enzymatic activity: the ability to succinylate lysine residues. To examine this hypothesis, we generated human 293T cell lines stably expressing wild-type (WT) human CPT1A and vector control and measured the total lysine succinylation levels in lysates by western blotting (WB) with a pan anti-succinylated lysine antibody. Expression of CPT1A WT increased the total lysine succinylation levels in 293T cell lysates compared with expression of vector control (Figure 1A, left). We next measured intracellular succinyl-CoA levels in 293T cell lines expressing CPT1A WT and vector control by an isotope-ratio-based approach with gas chromatography (GC)/mass spectrometry (MS) as we have performed previously (Hitosugi et al., 2012). The detailed calculations of intracellular succinyl-CoA levels in the cells are described in Figure S1B. Using this isotope-ratio based approach, we found no significant increase in succinyl-CoA levels by expression of CPT1A WT compared with expression of vector control in 293T cells, indicating that the increase in lysine succinylation by CPT1A WT is not due to higher intracellular succinyl-CoA levels (Figure 1A, right). We also measured NAD+ concentration in vector and CPT1A WT-expressed 293T cells and found no significant change in NAD+ concentration by CPT1A expression (Figure S1C). These results suggest that CPT1A may function as an LSTase in cells. In addition, we sought to investigate whether CPT1A is the specific enzyme that has the observed LSTase activity. We examined lysine succinylation levels in cells expressing another isoform of CPT1 family member CPT1C, which was identified as a potential LSTase from Swiss-Prot search in addition to CPT1A (Figure S1A). We ectopically expressed CPT1C in 293T cells and measured the total lysine succinylation levels in lysates by WB with a pan anti-succinylated lysine antibody. Expression of CPT1C WT did not increase the total lysine succinylation levels in 293T cell lysates compared with expression of vector control (Figure S1D).
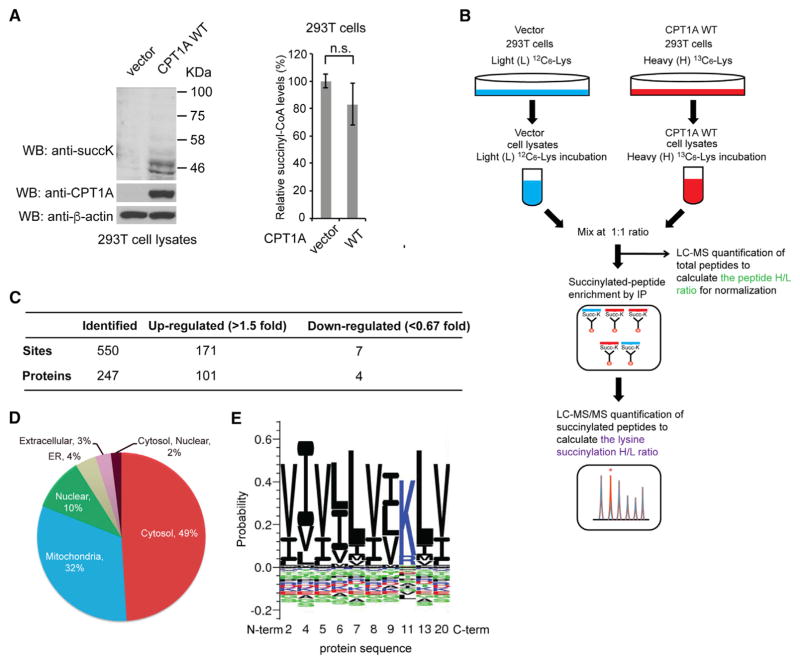
Figure 1. Identification of CPT1A-Dependent Lysine-Succinylated Proteins Using a SILAC-Based Quantitative Proteomics Approach.
(A) Left: WB of lysates from 293T cells stably expressing vector control and CPT1A WT with anti-pan-succinylated lysine, anti-CPT1A, and anti-β-actin antibodies. Right: GC-MS analysis of succinyl-CoA extracted from 293T cells expressing vector control and CPT1A WT. Error bars, ±SD of three independent measurements. P values were determined using a two-tailed Student’s t test. n.s., not significant.
(B) Schematic diagram of the SILAC-based quantitative proteomics approach for the identification of CPT1A-dependent lysine succinylation.
(C) Summary of the quantified lysine-succinylated sites and proteins.
(D) Subcellular distribution of CPT1A-dependent lysine-succinylated proteins.
(E) Consensus sequence motif logo for the amino acid sequences surrounding the 171 CPT1A-dependent succinylated lysine sites (>1.5-fold).
Identification of CPT1A-Dependent Lysine-Succinylated Proteins Using a SILAC-Based Quantitative Proteomics Approach
To identify CPT1A-dependent lysine-succinylated proteins and the sites of succinylation, we next performed quantitative succinylation analysis using stable isotope labeling by amino acid in cell culture (SILAC). 293T cells stably transfected with vector control (vector control cells) were incubated with light (L) [U-12C6]-lysine, while 293T CPT1A WT cells were incubated with heavy (H) [U-13C6]-lysine. Lysates from these two cell lines were mixed in a 1:1 ratio. One half of the mixed lysates was used for quantitative liquid chromatography (LC)-MS analysis for the total peptide normalization, and the other half was used for affinity enrichment by immunoprecipitation with pan anti-succinylated lysine antibody-conjugated beads followed by LC-MS/MS (Figure 1B). We found that succinylation levels of 171 unique lysine sites on 101 proteins (31% of the total 550 detected unique lysine sites on 247 proteins) were increased by more than 1.5-fold in CPT1A WT cells compared with vector control cells (Figure 1C; Tables S1 and S2). These results suggest that CPT1A regulates lysine succinylation in cells. We also investigated the possibility that the increased succinylation bands could be degradation products of expressed CPT1A (Figure 1A). We found no lysine-succinylated CPT1A peptide in our SILAC proteomics data from the CPT1A-expressed 293T cell lysates (Table S1), indicating that the increased succinylation bands by CPT1A expression would appear to be the lysine-succinylated downstream substrate proteins.
Using the SILAC proteomics data, we next examined the sub-cellular distribution of the CPT1A-dependent lysine-succinylated proteins and assessed whether there is a consensus sequence motif for CPT1A-dependent lysine succinylation. We found that almost 50% of the proteins with greater than a 1.5-fold increase in succinylation were cytosolic proteins (Figure 1D; Table S2). Analysis of the amino acids flanking the 171 succinylated lysines revealed that they were enriched in nonpolar hydrophobic amino acids such as leucine, valine, and isoleucine (Figure 1E; Table S2). These results are in contrast to recent studies on SIRT5 showing that most of the SIRT5-dependent lysine-succinylated proteins were mitochondrial proteins and that succinylated lysine sites targeted by SIRT5 tended to be near glycine, alanine, serine, or threonine residues (Park et al., 2013; Rardin et al., 2013). These differences may be because the substrate binding sites of CPT1A are exposed to the cytosolic side of the mitochondrial outer membrane, while SIRT5 localizes to the mitochondrial matrix. Supporting this reasoning, among 32 mitochondrial CPT1A-dependent lysine-succinylated proteins (32% of the total 101 CPT1A-dependent lysine-succinylated proteins), only 8 proteins were previously identified as potential SIRT5 substrates (Figure 1D; Table S2) (Park et al., 2013).
CPT1A Is a Regulator of Lysine Succinylation In Vivo and Can Lysine-Succinylate Enolase 1 to Inhibit Enolase 1 Activity In Vitro
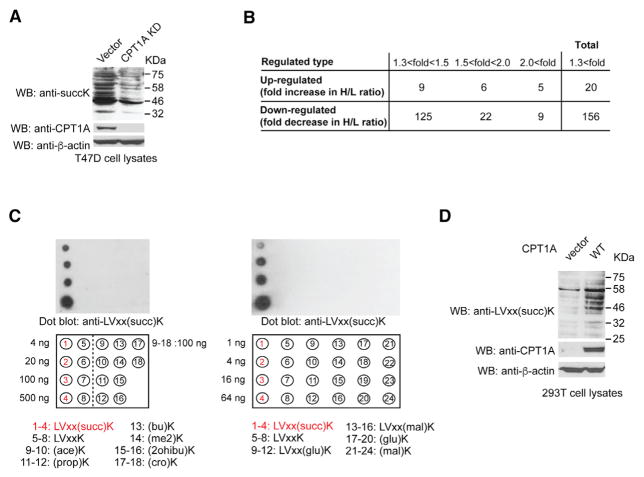
To further examine the role of CPT1A as an LSTase in cells, we knocked down endogenous CPT1A by short hairpin RNA (shRNA) in a breast cancer cell line, T47D cells, which express high levels of CPT1A. We found that CPT1A knockdown (KD) decreased total lysine succinylation levels in lysates (Figure 2A). We further performed SILAC succinyl proteomics analysis using T47D cells with or without stable KD of CPT1A, as similarly performed in 293T cells (Figure 1B). In this case, T47D cells stably transfected with vector control (vector control cells) were incubated with L [U-12C6]-lysine, while T47D cells stably expressing shRNA against CPT1A (CPT1A KD cells) were incubated with H [U-13C6]-lysine. The SILAC succinyl proteomics data showed that succinylation levels of 156 unique lysine sites were decreased by more than 1.3-fold in CPT1A KD cells compared with vector control cells (13% of the total 1,202 detected unique succinyl lysine sites) (Figure 2B; Table S3), suggesting that CPT1A KD caused the overall decrease in lysine succinylation in T47D cells. Furthermore, to verify CPT1A-dependent lysine succinylation in cells, we developed an anti-succinylated lysine motif antibody using a peptide library containing the succinylated motif sequence LVxx(succ)K that we identified using SILAC (Figure 1E). The specificity of this antibody was confirmed by dot blot assay, which showed that the antibody did not recognize acetyl, malonyl, glutaryl lysines, or other acyllysine modifications (Figure 2C). Similar to our findings in Figure 1A, WB with the anti-LVxx(succ)K antibody showed that CPT1A WT expression increased LVxxK succinylation in 293T cells (Figure 2D). Together, these results support the notion that CPT1A is a regulator of lysine succinylation in vivo.
Figure 2. CPT1A Is a Major Regulator of Lysine Succinylation In Vivo.
(A) WB of lysates from T47D cells stably expressing vector control and CPT1A shRNA with anti-succinylated lysine, anti-CPT1A, and anti-β-actin antibodies.
(B) Summary of the quantified lysine-succinylated sites in T47D cells with CPT1A KD from SILAC analysis.
(C) Left: dot blotting of unmodified and acylated lysine peptides including succinylated lysine motif LVxx(succ)K, acetyllysine (ace)K, propionyllysine (prop)K, butyryllysine (bu)K, 2-methyllysine (me2)K, 2-hydroxyisobutyryllysine (2ohibu)K, and crotonyllysine (cro)K peptides with anti-LVxx(succ)K antibody. Right: dot blotting of unmodified and succinylated lysine LVxx(succ)K, malonylated lysine LVxx(mal)K, (mal)K, and glutarylated lysine LVxx(glu)K, (glu)K peptides with anti-LVxx(succ)K antibody.
(D) WB of lysates from 293T cells stably expressing vector control and CPT1A WT with anti-LVxx(succ) K succinylated lysine motif, anti-CPT1A, and anti-β-actin antibodies.
We next sought to examine the effects of CPT1A as an LSTase on individual substrate proteins. We focused on enolase 1 because it contained the greatest number of succinylation sites of the 40 proteins identified in our SILAC analysis using CPT1A-expressed 293T cells (Figures S2A and S2B). We ectopically expressed Flag-tagged enolase 1 in 293T cells and showed that CPT1A WT expression increased LVxxK succinylation of immunoprecipitated Flag-tagged enolase 1 compared with expression of the vector control (Figure 3A), confirming our SILAC proteomics data. We further performed WB analysis of two more identified potential CPT1A substrate proteins (creatine kinase B [CKB] and 14-3-3 zeta), one non-target protein that was not detected by our SILAC analysis (glutathione S-transferase [GST]), and one identified protein in which lysine succinylation was decreased by CPT1A expression (histone 4 [H4]). We separately expressed Flag-tagged CKB, GST-tagged 14-3-3 zeta, and GST alone in 293T cells and showed that CPT1A WT expression increased LVxxK succinylation of immunoprecipitated Flag-tagged CKB and pulled down GST-tagged 14-3-3 zeta compared with vector control, but no LVxxK succinylation was observed in pulled-down GST protein alone from 293T cells expressing CPT1A (Figures S3A–S3C). We also showed that CPT1A WT expression decreased K32 succinylation of H4 in 293T cells, but the mechanism of the reduction in H4 K32 succinylation by CPT1A expression remains elusive (Figure S3D). These results together validate the accuracy of our SILAC succinyl proteomics data.
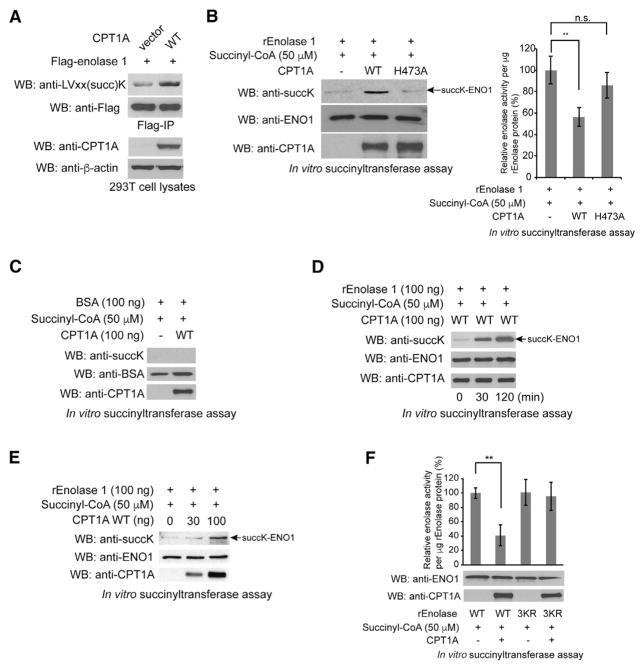
Figure 3. CPT1A Lysine Succinylates Enolase 1 to Inhibit Its Enzymatic Activity.
(A) Immunoprecipitated Flag-tagged enolase 1 was western blotted with anti-LVxx(succ)K succinylated lysine motif and anti-Flag antibodies. Lysates were western blotted for CPT1A and β-actin.
(B) Left: WB of the in vitro succinyltransferase assay samples using purified CPT1A WT or H473A and enolase 1 with anti-pan-succinylated lysine, anti-CPT1A, and anti-enolase 1 antibodies. Right: enolase activity of the in vitro succinyltransferase assay samples.
(C) WB of the in vitro succinyltransferase assay samples using purified CPT1A WT and BSA with anti-pan-succinylated lysine, anti-CPT1A, and anti-BSA antibodies.
(D) WB of the in vitro succinyltransferase assay samples at the indicated time points with anti-pan-succinylated lysine, anti-CPT1A, and anti-enolase 1 antibodies.
(E) WB of the in vitro succinyltransferase assay using the indicated amounts of purified CPT1A WT and enolase 1 with anti-pan-succinylated lysine, anti-CPT1A, and anti-enolase 1 antibodies.
(F) Enolase activity of the in vitro succinyltransferase assay samples using purified CPT1A WT with enolase 1 WT or 3KR.
Error bars, ±SD of three independent measurements. P values were determined using a two-tailed Student’s t test. **p < 0.01; n.s., not significant. See also Figure S3.
To examine whether CPT1A directly succinylates lysine residues in vitro, we purified recombinant CPT1A WT as well as CPT1A H473A, a catalytically inactive mutant that disrupts the putative binding pocket for the sulfur atom of the acyl-CoA thioester (López-Vinãs et al., 2007; Morillas et al., 2001, 2004) (Figure S3E). Next, to evaluate the structural properties of purified CPT1A WT and H473A proteins, we examined the relative stability of the CPT1A proteins to limited proteolytic digestion by trypsin as described previously (Kang et al., 2007). Silver staining results showed that there was no obvious difference in the digestion patterns of trypsin-treated CPT1A WT and H473A mutant proteins, suggesting that H473A mutation does not disrupt the overall structure of CPT1A protein (Figure S3F). It is of note that purified CPT1A WT protein, but not purified CPT1A H473A proteins, still possesses the canonical CPTase activity in vitro (Figure S3G). We next incubated the purified CPT1A proteins with purified enolase 1 in the presence of 50 μM succinyl-CoA, which is below the physiological concentration of succinyl-CoA in 293T cells (100 μM) (Figure S1B). WB analysis showed that CPT1A WT, but not CPT1A H473A, succinylated enolase 1 on lysines, thus demonstrating that purified CPT1A can directly succinylate enolase 1 in vitro and that the H473A mutant lacks LSTase activity (Figure 3B, left). To assess whether lysine succinylation affects enolase 1 catalytic activity, we incubated purified enolase 1 with CPT1A WT and CPT1A H473A and assayed enolase activity. CPT1A WT significantly inhibited enolase 1, whereas CPT1A H473A did not (Figure 3B, right). Together, these results suggest that enolase 1 is a substrate for CPT1A and lysine succinylation of enolase 1 by CPT1A alters enolase 1 catalytic activity. We further performed the in vitro succinyltransferase assay to show (1) that BSA, which is a protein that is not a target of CPT1A, is not succinylated (Figure 3C); (2) a time-dependent increase in succinylation on enolase 1 by CPT1A (Figure 3D); and (3) a dose-dependent increase in succinylation on enolase 1 by increased amount of CPT1A (Figure 3E). In addition, we did not observe the time-dependent increase in lysine succinylation of enolase 1 without CPT1A or without succinyl-CoA (Figure S3H) and demonstrated that no lysine succinylation on enolase 1 was induced by CPT1A with malonyl-CoA (Figure S3I).
To further investigate which succinylated lysine sites are responsible for enolase 1 inhibition by CPT1A, we first separately introduced succinylation mimic K-to-E mutations into enolase 1. These mutations included major succinylated lysine sites such as K5, K80, K89, and K233 identified from our SILAC succinyl proteomics data. Although K81 has lowest stoichiometry of succinylation within enolase 1, we introduced K80/81E mutations together because of the proximity of the two sites (Figure S3J). We also introduced another K335E mutation because K335 is proximal to the substrate binding region of enolase 1 (Larsen et al., 1996). We assayed enolase activity of WT, K5 K80/81E, K89E, K233E, and K335E proteins and found that K80/81E and K335E mutant exhibited lower enzymatic activity compared with WT, suggesting that K80/81 and K335 succinylation are likely important for the inhibition of enolase 1 by lysine succinylation (Figure S3K). On the basis of these results, we further made succinylation deficient 3KR mutant protein in which all the K80/81 and K335 sites were substituted to Arg (R) and incubated with purified CPT1A WT along with succinyl-CoA and assayed for enolase activity. Purified CPT1A WT did not inhibit the 3KR mutant protein, suggesting that succinylation on K80/81 and K335 is important for CPT1A-dependent inhibition of enolase 1 (Figure 3F). It is of note that the protein sequence surrounding K80/81 of enolase 1 is IAPA(LVSKK)LNVTEQ, which contains the LVxxK motif sequence. In addition, we performed the in vitro succinyltransferase assay using purified CPT1A WT and enolase 3KR mutant followed by WB analysis with anti-pan succinyl lysine, anti-CPT1A, and anti-enolase 1 antibodies. The WB results showed that CPT1A did not increase lysine succinylation on enolase 3KR mutant, suggesting that any or all of K80, K81, and K335 are major succinylation sites by CPT1A in the in vitro succinyltransferase assay (Figure S3L). Consistent with this finding, we observed that CPT1A KD decreased lysine succinylation of Flag-enolase 1 WT, but not Flag-enolase 1 3KR, compared with vector control in T47D cells (Figure S3M). These results support our notion that endogenous CPT1A can regulate lysine succinylation of enolase 1 in cells.
Physiological Significance of CPT1A-Dependent Lysine Succinylation in Cells
Because CPT1A H473A lacks both LSTase and CPTase activities (Figures 3B and S3G), we sought a CPT1A mutation that separates LSTase and CPTase activities so that we could assess the impact of selectively disabling the LSTase activity in cells. We found that expression of the previously reported CPTase-deficient mutant CPT1A G710E (Prip-Buus et al., 2001) increased total LVxxK succinylation levels to a similar extent as expression of CPT1A WT in lysates of 293T cells (Figure 4A). We also confirmed that expression of CPT1A WT, but not G710E mutant, increased the CPTase activity in 293T cells compared with expression of vector control although G710E mutant lysine succinylates enolase 1 to a similar extent as CPT1A WT in the in vitro succinyltransferase assay (Figures 4B and S3I). G710E mutation was identified from patients with CPT1A deficiency and is a natural mutation that decreases the activity without changing the protein stability (Prip-Buus et al., 2001). Because no crystal structure of CPT1A has been reported yet, a previous study of structure modeling of CPT1A used the crystal structure of mouse carnitine acetyltransferase and showed that G710 is proximal to the catalytic H473 site, and the introduction of a negative charge may impair the catalytic process of the classical CPTase activity of CPT1A by inhibiting the binding of palmitoyl group to the putative substrate binding pocket (Morillas et al., 2004).
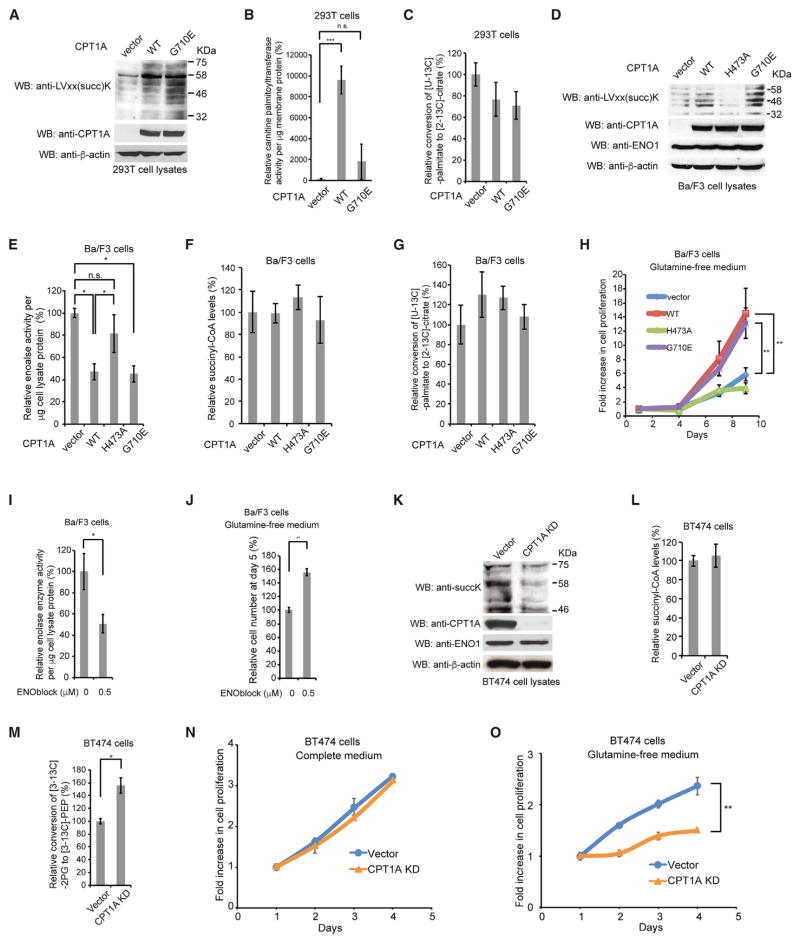
Figure 4. LSTase Activity of CPT1A Promotes Hematopoietic Cell Proliferation Under Glutamine Depletion.
(A) Lysates from 293T cells stably expressing vector control, CPT1A WT, and CPT1A G710E were western blotted with anti-LVxx(succ)K, anti-CPT1A, and anti-β-actin antibodies.
(B) CPTase activity of membrane proteins from 293T cells stably expressing vector control, CPT1A WT and G710E.
(C) FAO activity in 293T cells expressing vector control, CPT1A WT, and G710E.
(D) WB of lysates from Ba/F3 cells stably expressing vector control, CPT1A WT, H473A, and G710E with anti-LVxx(succ)K, anti-CPT1A, anti-enolase 1, and anti-β-actin antibodies.
(E) Enolase activity of lysates from Ba/F3 cells stably expressing vector control, CPT1A WT, H473A, and G710E.
(F) GC/MS analysis of succinyl-CoA extracted from Ba/F3 cells expressing vector control, CPT1A WT, H473A, and G710E.
(G) FAO activity in Ba/F3 cells expressing vector control, CPT1A WT, H473A, and G710E.
(H) Glutamine-independent proliferation of Ba/F3 cells stably expressing vector control, CPT1A WT, H473A, and G710E.
(I) Enolase activity of lysates from Ba/F3 cells treated with vehicle control and 0.5 μM ENOblock.
(J) Glutamine-independent proliferation of Ba/F3 cells treated with vehicle control and 0.5 μM ENOblock.
(K) WB of lysates from BT474 cells stably expressing vector control and CPT1A shRNA with anti-succinylated lysine, anti-CPT1A, anti-enolase 1, and anti-β-actin antibodies.
(L) GC/MS analysis of succinyl-CoA extracted from BT474 cells stably expressing vector control and CPT1A shRNA.
(M) GC/MS analysis of enolase activity by measuring the conversion rate of [3-13C]-2PG to [3-13C]-PEP in cells incubated with [U-13C6]-glucose.
(N) Proliferation of BT474 cells expressing vector control and CPT1A shRNA in complete medium.
(O) Proliferation of BT474 cells expressing vector control and CPT1A shRNA in glutamine-free medium.
Error bars, ±SD of three independent measurements. P values were determined by a two-tailed Student’s t test. *p < 0.05, **p < 0.01, ***p < 0.001; n.s., not significant. See also Figure S4 and Table S4.
We also measured FAO activity in 293T cell lines expressing CPT1A WT and mutants as well as vector control by monitoring the conversion rate of [U-13C]-palmitate to [2-13C]-citrate. We unexpectedly observed no significant alteration of FAO activity by expression of CPT1A WT compared with expression of vector control and CPT1A mutants in 293T cells (Figure 4C). These results may be explained by our finding from the global proteomics data that stable expression of CPT1A WT in 293T cells decreased expression levels of enzymes related to FAO pathway such as very long-chain specific acyl-CoA dehydrogenase (ACADVL), acyl-CoA dehydrogenase family member (ACAD) 9 and 11, 3-ketoacyl-CoA thiolase (ACAA), and 3-hydroxyacyl-CoA dehydrogenase type-2 (HSD17B10), which might decrease the activity of FAO to cancel out the effect of CPT1A WT expression while the mechanism underlying the decrease in expression levels of these proteins remains elusive (Figure S4A; Table S1).
To further ask whether the classical CPTase activity of endogenous CPT1A affects its LSTase activity, we treated breast cancer BT474 cells expressing high levels of endogenous CPT1A with an irreversible inhibitor of CPT1, etomoxir. We found that overnight etomoxir treatment reduced FAO activity to less than 10% but did not affect total LVxxK succinylation levels (Figure S4B). Because KD of CPT1A decreased total succinylation in BT474 cell lysates (Figure 4K), these data together suggest that the CPTase activity and LSTase activity are independently regulated by CPT1A.
The results from Figures 4A and 4B led us to use CPT1A G710E mutant for the following enolase activity assay in cell lysates as well as cell proliferation assay to examine the physiological functions of CPT1A-dependent lysine succinylation in cells. A previous proteomics study showed that among enolase isoforms, only enolase 1-derived peptides were detected in hematopoietic Ba/F3 cells (Pierce et al., 2008), suggesting that enolase 1 is predominantly expressed in Ba/F3 cells, and the total enolase activity in Ba/F3 cell lysates represents the activity of enolase 1. We therefore generated Ba/F3 cells stably expressing vector control, CPT1A WT, CPT1A H473A, and CPT1A G710E and examined the total enolase activity in lysates. Consistent with the results from Figures 3B and 4A, expression of CPT1A WT and CPT1A G710E, but not CPT1A H473A, increased total LVxxK succinylation and inhibited enolase enzyme activity in lysates without altering enolase 1 protein levels compared with vector control (Figures 4D and 4E). Furthermore, consistent with the results from Figures S1B and 4C, we did not observe any significant alteration in intracellular succinyl-CoA levels or FAO activity between these Ba/F3 cell lines (Figures 4F and 4G). Taken together, these results suggest that the LSTase activity, but not the CPTase activity, of CPT1A inhibits enolase in cells.
A recent study showed that shRNA-based silencing of enolase 1 expression induces senescence in breast cancer MDA-MB 231 cells but promotes cell survival under conditions in which glutamine metabolism is suppressed by the glutaminase inhibitor BPTES (bis-2-[5phenylacetamido-1,2,4- thia-diazol-2-yl] ethyl sulfide) (Capello et al., 2016). This led us to test whether the LSTase activity of CPT1A that inhibits enolase 1 may contribute to cell proliferation in glutamine-free medium. We found that CPT1A WT and G710E mutant-expressing Ba/F3 cells, but not H473A mutant-expressing Ba/F3 cells, exhibited increased cell proliferation in glutamine-free medium compared with vector control cells (Figure 4H). There was no significant difference in proliferation of these stable cell lines in complete medium (Figure S4C). Consistent with these results, CPT1A WT and G710E-expressed 293T cells exhibited increased cell proliferation in glutamine-free medium compared with vector control cells (Figure S4D). These results suggest that the LSTase activity, but not the CPTase activity, of CPT1A promotes cell proliferation under glutamine depletion.
To further examine whether the increase in glutamine-independent Ba/F3 cell proliferation by CPT1A WT and by G710E mutant is mediated through enolase inhibition, we treated Ba/F3 cells with an enolase-specific inhibitor, ENOblock, to mimic the inhibition effect of CPT1A on enolase 1 (Jung et al., 2013). We found that 0.5 μM ENOblock treatment reduced enolase 1 activity to a similar extent as in the lysates from CPT1A WT or G710E mutant-expressed Ba/F cells (50% decrease from control treatment or control vector cells) (Figures 4E and 4I). Consistent with our findings, 0.5 μM ENOblock treatment significantly increased cell proliferation in glutamine-free medium but not in complete medium compared with vehicle treatment (Figures 4J and S4E), suggesting that the LSTase activity of CPT1A promotes glutamine-independent Ba/F3 cell proliferation at least in part through the inhibition of enolase. We also performed cell proliferation assay of the CPT1A-expressed Ba/F3 cell lines treated with etomoxir in glutamine-free medium and found that expression of CPT1A WT and G710E, but not H73A, increased Ba/F3 cell proliferation compared with vector control even in the presence of etomoxir in glutamine-free medium (Figure S4F), suggesting that the growth enhancement observed in glutamine-free medium with expression of CPT1A was not due to the alteration of FAO. We also found no significant alteration in cell proliferation among these Ba/F3 cell lines when treated with AICAR as well as ENOblock and 2-DG in complete medium (Figures S4G and S4H).
To further study the physiological relevance of the LSTase activity of endogenous CPT1A, we sought to examine CPT1A KD BT474 cells of which lysine succinylation levels were decreased in lysates compared with vector control cells (Figure 4K). We confirmed that CPT1A KD did not cause any changes in enolase 1 expression levels and succinyl-CoA levels in BT474 cells (Figures 4K and 4L). It is difficult to assess enolase activity in lysates from BT474 cells with high expression of endogenous SIRT5 compared with other SIRT family members (Table S4), which may desuccinylate enolase to change its activity during cell lysis. To solve this problem, we incubated BT474 cells with [U-13C6]-glucose and measured the conversion rate of [3-13C]-2PG (enolase substrate) to [3-13C]-PEP (enolase product), which represents enolase activity in cells. We found that CPT1A KD cells displayed significantly higher conversion rate of [3-13C]-2PG to [3-13C]-PEP than vector control cells, suggesting that CPT1A KD increased enolase activity in BT474 cells (Figure 4M). To examine whether the observed increase in the total enolase activity by CPT1A KD in BT474 cells is due to changes in enolase 1, we performed total proteomics analysis of BT474 cell lysates. We found that signal intensities from enolase 1-derived peptides were 200 times higher than those from enolase 2-derived peptides, and no other enolase isoforms were detected (Table S4). These results suggest that enolase 1 is predominantly expressed in BT474 cells and the increase in enolase activity by CPT1A KD in BT474 cells is likely due to changes in enolase 1.
Finally, we performed proliferation assays of vector control and CPT1A KD BT474 cells and demonstrated that CPT1A KD significantly decreased cell proliferation in glutamine-free medium but not in complete medium (Figures 4N and 4O). All of the data in Figures 4K–4O are consistent with the other data in Figure 4 and support the notion that the CPT1A increases total lysine succinylation levels without affecting intracellular succinyl-CoA levels to inhibit enolase 1 and enhances glutamine-independent cell proliferation.
Altogether we showed that CPT1A lysine succinylated its substrate proteins in vivo and in vitro, and such LSTase activity inhibits enolase 1 and promotes cell proliferation under glutamine depletion independent of its classical CPTase activity. These data provide evidence not only for the role of CPT1A as an LSTase but also a functional effect of CPT1A-dependent lysine succinylation on a substrate protein and cellular metabolism. Furthermore, we identified 101 potential CPT1A substrate proteins, expanding the understanding of lysine succinylation-dependent signal transduction in cells. However, it remains unclear how lysine succinylation affects the inhibition of CPT1A by malonyl-CoA, and it also is possible that any alteration in the succinyl-CoA level can regulate the levels of succinylation besides the LSTase activity of CPT1A. Hence, further work would be needed to resolve these issues.
EXPERIMENTAL PROCEDURES
Statistical Methods
Significance was tested using unpaired two-tailed Student’s t tests, assuming independent variables, normal distribution, and equal variance of samples. Data are presented as mean ± SD from three independent measurements. A p value < 0.05 was considered to indicate statistical significance.
In Vitro Succinyltransferase Assay
Purified recombinant CPT1A proteins (100 ng) were incubated with purified recombinant enolase 1 protein (100 ng) at 30°C for 2 hr in 50 μL of the buffer containing 10 mM HEPES (pH 7.5), 2 mM sodium orthovanadate, 5 mM sodium pyrophosphate, 300 mM NaCl, and 1% Nonidet P-40 plus 50 μM succinyl-CoA as a substrate and 50 ng of BSA as a protein stabilizer. The reaction was terminated by the addition of 6xSDS sample buffer and by boiling the samples for 5 min.
CPTase Activity Assay
CPTase activity was measured using 10 μCi L-[3H]carnitine as previously described (Dobrzyn et al., 2004).
Metabolic Flux Analysis of FAO
FAO activity was measured by monitoring the conversion rate of [U-13C]-palmitate to [2-13C]-citrate with GC/MS. In brief, the cells were incubated with 100 μM [U-13C]-palmitate-BSA conjugate for overnight. After the incubation, the metabolites were extracted and dried up and then derivatized with MSTFA. The FAO activity was calculated by dividing the citrate labeling rate by the palmitate uptake rate.
Cell Proliferation Assay
Fold increase in cell proliferation was determined by measuring the cell viability at the indicated days divided by the starting cell viability at day 1 using the CyQUANT Cell Proliferation Assay Kit (Thermo Fisher Scientific).
Supplementary Material
Highlights.
CPT1A succinylates proteins in vitro using succinyl-CoA as a substrate
CPT1A increases lysine succinylation in cells without altering succinyl-CoA levels
CPT1A inhibits enolase 1 by lysine succinylation in vivo and in vitro
About 100 CPT1A-dependent succinylated proteins were identified
Acknowledgments
We thank Dr. Scott Kaufmann for critical reading of the manuscript. We thank Drs. Yuichi Machida and Liewei Wang for providing 293T and T47D cell lines, respectively. We thank Thomas Larson for critical reading of the manuscript. We also thank the Mayo Clinic Cancer Center Pharmacology Shared Resource for providing tissue culture facilities and HPLC analytical expertise. SILAC succinyl proteomics analysis was performed by PTM-Bio. Research reported in this publication was supported by the Career Catalyst Research funding program from the Susan G. Komen Foundation (CCR14300798 to T.H.), the Eagles Cancer Research Fund (T.H.), the Team Science Platform Award from the Mayo Clinic Center for Biomedical Discovery (T.H. and Z.L.), the Developmental Therapeutics Program from the Mayo Clinic Cancer Center (T.H.), and the Mayo Clinic Breast SPORE (P50CA-116201-10 to T.H. and M.P.G.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the NIH. K.K. and E.K.W. were supported by predoctoral fellowships from the Mayo Foundation for Education and Research. F.B.-A. was supported by NIH Clinical Pharmacology Training Grant T32-GM08685. The Mayo Clinic Cancer Center Pharmacology Shared Resource was supported by NCI Cancer Center Support Grant P30-CA15083-40.
Footnotes
AUTHOR CONTRIBUTIONS
W.I.G., Z.L., L.M.K., and M.P.G. provided critical equipment, expertise, and reagents. F.B.-A. provided technical assistance for HPLC experiments. K.K., S.H., E.K.W., and T.H. designed the study and performed the experiments. K.K. and T.H. wrote the manuscript. All authors contributed to the discussion of the data.
DECLARATION OF INTERESTS
The authors declare no competing interests.
DATA AND SOFTWARE AVAILABILITY
The accession number for the proteomics data reported in this paper is PeptideAtlas: PASS01135 (Maxquant_Succinylome293T and Maxquant_SuccinylomeT47D).
Supplemental Information includes Supplemental Experimental Procedures, four figures, and four tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.030.
References
- Capello M, Ferri-Borgogno S, Riganti C, Chattaragada MS, Principe M, Roux C, Zhou W, Petricoin EF, Cappello P, Novelli F. Targeting the Warburg effect in cancer cells through ENO1 knockdown rescues oxidative phosphorylation and induces growth arrest. Oncotarget. 2016;7:5598–5612. doi: 10.18632/oncotarget.6798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C, Weinert BT, Nishida Y, Verdin E, Mann M. The growing landscape of lysine acetylation links metabolism and cell signalling. Nat Rev Mol Cell Biol. 2014;15:536–550. doi: 10.1038/nrm3841. [DOI] [PubMed] [Google Scholar]
- Colak G, Xie Z, Zhu AY, Dai L, Lu Z, Zhang Y, Wan X, Chen Y, Cha YH, Lin H, et al. Identification of lysine succinylation substrates and the succinylation regulatory enzyme CobB in Escherichia coli. Mol Cell Proteomics. 2013;12:3509–3520. doi: 10.1074/mcp.M113.031567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobrzyn P, Dobrzyn A, Miyazaki M, Cohen P, Asilmaz E, Hardie DG, Friedman JM, Ntambi JM. Stearoyl-CoA desaturase 1 deficiency increases fatty acid oxidation by activating AMP-activated protein kinase in liver. Proc Natl Acad Sci U S A. 2004;101:6409–6414. doi: 10.1073/pnas.0401627101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du J, Zhou Y, Su X, Yu JJ, Khan S, Jiang H, Kim J, Woo J, Kim JH, Choi BH, et al. Sirt5 is a NAD-dependent protein lysine demalonylase and desuccinylase. Science. 2011;334:806–809. doi: 10.1126/science.1207861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hitosugi T, Zhou L, Elf S, Fan J, Kang HB, Seo JH, Shan C, Dai Q, Zhang L, Xie J, et al. Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth. Cancer Cell. 2012;22:585–600. doi: 10.1016/j.ccr.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung DW, Kim WH, Park SH, Lee J, Kim J, Su D, Ha HH, Chang YT, Williams DR. A unique small molecule inhibitor of enolase clarifies its role in fundamental biological processes. ACS Chem Biol. 2013;8:1271–1282. doi: 10.1021/cb300687k. [DOI] [PubMed] [Google Scholar]
- Kang S, Dong S, Gu TL, Guo A, Cohen MS, Lonial S, Khoury HJ, Fabbro D, Gilliland DG, Bergsagel PL, et al. FGFR3 activates RSK2 to mediate hematopoietic transformation through tyrosine phosphorylation of RSK2 and activation of the MEK/ERK pathway. Cancer Cell. 2007;12:201–214. doi: 10.1016/j.ccr.2007.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen TM, Wedekind JE, Rayment I, Reed GH. A carboxylate oxygen of the substrate bridges the magnesium ions at the active site of enolase: structure of the yeast enzyme complexed with the equilibrium mixture of 2-phosphoglycerate and phosphoenolpyruvate at 1.8 A resolution. Biochemistry. 1996;35:4349–4358. doi: 10.1021/bi952859c. [DOI] [PubMed] [Google Scholar]
- Li F, He X, Ye D, Lin Y, Yu H, Yao C, Huang L, Zhang J, Wang F, Xu S, et al. NADP(+)-IDH mutations promote hypersuccinylation that impairs mitochondria respiration and induces apoptosis resistance. Mol Cell. 2015;60:661–675. doi: 10.1016/j.molcel.2015.10.017. [DOI] [PubMed] [Google Scholar]
- López-Vinãs E, Bentebibel A, Gurunathan C, Morillas M, de Arriaga D, Serra D, Asins G, Hegardt FG, Gómez-Puertas P. Definition by functional and structural analysis of two malonyl-CoA sites in carnitine palmitoyltransferase 1A. J Biol Chem. 2007;282:18212–18224. doi: 10.1074/jbc.M700885200. [DOI] [PubMed] [Google Scholar]
- McGarry JD, Mannaerts GP, Foster DW. A possible role for malonyl-CoA in the regulation of hepatic fatty acid oxidation and ketogenesis. J Clin Invest. 1977;60:265–270. doi: 10.1172/JCI108764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mills SE, Foster DW, McGarry JD. Interaction of malonyl-CoA and related compounds with mitochondria from different rat tissues. Relationship between ligand binding and inhibition of carnitine palmitoyltransferase I. Biochem J. 1983;214:83–91. doi: 10.1042/bj2140083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morillas M, Gómez-Puertas P, Roca R, Serra D, Asins G, Valencia A, Hegardt FG. Structural model of the catalytic core of carnitine palmitoyltransferase I and carnitine octanoyltransferase (COT): mutation of CPT I histidine 473 and alanine 381 and COT alanine 238 impairs the catalytic activity. J Biol Chem. 2001;276:45001–45008. doi: 10.1074/jbc.M106920200. [DOI] [PubMed] [Google Scholar]
- Morillas M, López-Vinãs E, Valencia A, Serra D, Gómez-Puertas P, Hegardt FG, Asins G. Structural model of carnitine palmitoyltransferase I based on the carnitine acetyltransferase crystal. Biochem J. 2004;379:777–784. doi: 10.1042/BJ20031373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J, Chen Y, Tishkoff DX, Peng C, Tan M, Dai L, Xie Z, Zhang Y, Zwaans BM, Skinner ME, et al. SIRT5-mediated lysine desuccinylation impacts diverse metabolic pathways. Mol Cell. 2013;50:919–930. doi: 10.1016/j.molcel.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierce A, Unwin RD, Evans CA, Griffiths S, Carney L, Zhang L, Jaworska E, Lee CF, Blinco D, Okoniewski MJ, et al. Eight-channel iTRAQ enables comparison of the activity of six leukemogenic tyrosine kinases. Mol Cell Proteomics. 2008;7:853–863. doi: 10.1074/mcp.M700251-MCP200. [DOI] [PubMed] [Google Scholar]
- Prip-Buus C, Thuillier L, Abadi N, Prasad C, Dilling L, Klasing J, Demaugre F, Greenberg CR, Haworth JC, Droin V, et al. Molecular and enzymatic characterization of a unique carnitine palmitoyltransferase 1A mutation in the Hutterite community. Mol Genet Metab. 2001;73:46–54. doi: 10.1006/mgme.2001.3176. [DOI] [PubMed] [Google Scholar]
- Rardin MJ, He W, Nishida Y, Newman JC, Carrico C, Danielson SR, Guo A, Gut P, Sahu AK, Li B, et al. SIRT5 regulates the mitochondrial lysine succinylome and metabolic networks. Cell Metab. 2013;18:920–933. doi: 10.1016/j.cmet.2013.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner GR, Payne RM. Widespread and enzyme-independent Nε-acetylation and Nε-succinylation of proteins in the chemical conditions of the mitochondrial matrix. J Biol Chem. 2013;288:29036–29045. doi: 10.1074/jbc.M113.486753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinert BT, Schölz C, Wagner SA, Iesmantavicius V, Su D, Daniel JA, Choudhary C. Lysine succinylation is a frequently occurring modification in prokaryotes and eukaryotes and extensively overlaps with acetylation. Cell Rep. 2013;4:842–851. doi: 10.1016/j.celrep.2013.07.024. [DOI] [PubMed] [Google Scholar]
- Zhang Z, Tan M, Xie Z, Dai L, Chen Y, Zhao Y. Identification of lysine succinylation as a new post-translational modification. Nat Chem Biol. 2011;7:58–63. doi: 10.1038/nchembio.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.