Abstract
N 6-methyladenosine (m6A) is the most prevalent modification in eukaryotic messenger RNAs (mRNAs) and is interpreted by its readers, such as YTH domain-containing proteins, to regulate mRNA fate. Here we report the insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs; including IGF2BP1/2/3) as a distinct family of m6A readers that target thousands of mRNA transcripts through recognizing the consensus GG(m6A)C sequence. In contrast to the mRNA-decay-promoting function of YTHDF2, IGF2BPs promote the stability and storage of their target mRNAs (e.g., MYC) in an m6A-depedent manner under normal and stress conditions and thus affect gene expression output. Moreover, the K homology (KH) domains of IGF2BPs are required for their recognition of m6A and are critical for their oncogenic functions. Our work therefore reveals a different facet of the m6A-reading process that promotes mRNA stability and translation, and highlights the functional importance of IGF2BPs as m6A readers in post-transcriptional gene regulation and cancer biology.
Keywords: IGF2BP, RNA N6-methyladenosine, readers, mRNA stability, translation, K homology domain, MYC
INTRODUCTION
As the most abundant mRNA modification, N6-methyladenosine (m6A) modification is reversible and plays critical roles in multiple fundamental biological processes (e.g., cell differentiation, tissue development, and tumorigenesis)1–13. High throughput sequencing revealed that m6A is especially enriched in the 3′ untranslated regions (UTRs) and near the stop codons of mRNAs with a consensus sequence of RRACH (R=G or A; H=A, C or U)2, 3. The biological importance of m6A modification relies on m6A-binding proteins (i.e., readers). Therefore, it is crucial to identify and characterize m6A readers that directly guide distinct bioprocesses. A group of YT521-B homology (YTH) domain-containing proteins have been identified as m6A readers that control messenger RNA (mRNA) fate by regulating pre-mRNA splicing, facilitating translation, or promoting mRNA decay2, 10, 14–16.
In the YTHDF2-mediated decay pathway10, mRNA levels are expected to increase when m6A abundance is reduced. However, our recent data showed that a large portion of mRNAs with reduced m6A abundance tended to be down-regulated due to decreased RNA stability12, suggesting the presence of alternative mechanism(s) to stabilize m6A-modified mRNAs. Here we report the insulin-like growth factor-2 (IGF2) mRNA-binding proteins 1, 2, and 3 (IGF2BP1/2/3) as a new family of m6A readers that guard m6A-modified mRNAs from decay. IGF2BPs, a conserved family of single-stranded RNA-binding proteins17, are composed of six canonical RNA-binding domains, including two RNA recognition motif (RRM) domains and four K homology (KH) domains17, 18. Besides IGF2, a few other well-known mRNAs (e.g., MYC, ACTIN and LIN28B) have been reported as targets of IGF2BPs19–21. However, the exact molecular mechanisms by which IGF2BPs recognize and regulate expression of their targets remain elusive. Here we provide compelling evidence showing that IGF2BPs preferentially recognize m6A-modified mRNAs and promote the stability (and likely also translation) of thousands of potential mRNA targets (including MYC) in an m6A-dependent manner, thereby globally affecting gene expression output. Furthermore, as m6A readers, IGF2BPs play oncogenic roles in cancer cells, likely by stabilizing methylated mRNAs of oncogenic targets (e.g., MYC).
RESULTS
Identification of IGF2BPs as m6A-binding proteins
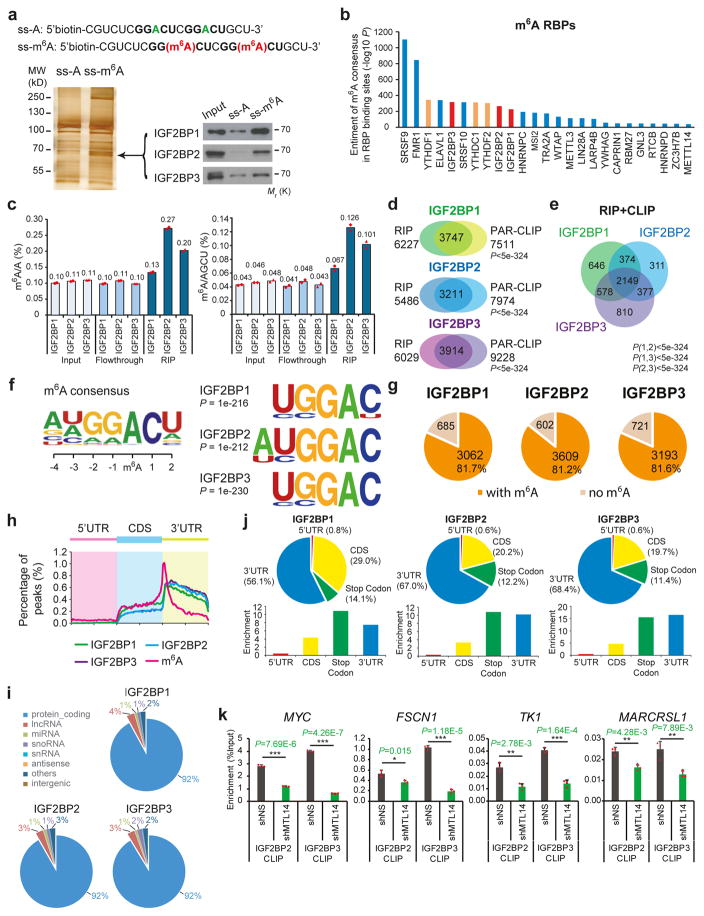
To identify m6A-binding proteins, we applied two independent methods: (i) using methylated single-stranded RNA bait (ss-m6A, with the consensus sequence GG(m6A)CU) or unmethylated control RNA (ss-A) for RNA pull-down (Fig. 1a and Supplementary Fig. 1a), followed by mass spectrometry analysis2; (ii) developing a computational pipeline for screening potential m6A binding proteins by using published RNA-binding protein (RBP) CLIP-seq datasets and known m6A modification sites3. All three IGF2BP proteins were identified by mass spectrum and confirmed to selectively bind the methylated bait (ss-m6A) with a 3–4 fold higher affinity than the unmethylated control (ss-A) (Fig. 1a and Supplementary Fig. 1b), and the binding seems to be independent of RNA secondary structure (Supplementary Fig. 1c). Similar to endogenous proteins, recombinant IGF2BP proteins purified from human cells also preferentially bound to methylated RNA probe over the unmethylated one (Supplementary Fig. 1d and 1e). In addition to ss-m6A, IGF2BPs also preferentially bound to methylated hairpin RNA (hp-m6A) probes over control (hp-A) probes (Supplementary Fig. 1f and 1g). Meanwhile, our computational pipeline revealed that all three IGF2BPs were among the top 15 of 112 RBPs in terms of both the significance (Fig. 1b) and the frequency (Supplementary Fig. 1h) of m6A motifs enriched in their RNA binding sites. Thus, IGF2BP proteins are potential direct m6A-binding proteins.
Figure 1. Selective binding of IGF2BPs to m6A-modified RNAs.
(a) Identification of m6A specific binding proteins by RNA affinity chromatography using ssRNA probes with methylated (red) or unmethylated (green) adenosine. Silver staining (lower left) and Western blotting (lower right) showed selective pulldown of ~68kDa IGF2BP proteins from HEK293T nuclear extract. Western blot images were representative of 3 independent experiments. (b) Enrichment of m6A consensus sequence “GGAC” in the binding sites of RBPs. The three IGF2BP paralogues were shown in red, while the YTH domain proteins were shown in orange. (c) Quantification of m6A/A and m6A/AGCU ratios by LC-MS/MS in RNAs bound by ectopically expressed IGF2BP1 (chicken ZBP1), IGF2BP2 (human), or IGF2BP3 (human). Values are mean of n =2 independent experiments and individual data points are showed. (d) Overlap of IGF2BP target genes identified by RIP-seq and published PAR-CLIP in HEK293T cells. RIP-seq was performed once. P value was calculated by Fisher’s test. (e) Venn diagram showing the numbers of shared high-confidence targets (i.e., CLIP+RIP targets) amongst IGF2BP paralogues. P value was calculated by Fisher’s test. (f) Top consensus sequences of IGF2BP binding sites and the m6A motif detected by HOMER Motif analysis with PAR-CLIP data. (g) Pie charts showing numbers and percentages of IGF2BP high-confidence target genes that contain m6A peaks. The m6A-seq data was reported in Ref. 3. (h) Metagene profiles of enrichment of IGF2BP binding sites and m6A modifications across mRNA transcriptome. (i) Percentages of various RNA species bound by IGF2BPs. (j) The distribution (upper) and enrichment (lower) of IGF2BPs binding peaks within different gene reions. The enrichment was determined by the proportion of IGF2BPs binding peaks normalized by the length of the region. Analyses in i and j were performed twice with similar results. (k) In vivo binding of Flag-IGF2BP2 to representative target genes in METTL14 knockdown or control HEK293T cells. Values are mean±s.d. of n =3 independent experiments. *, P<0.05; **, P <0.01; ***, P <0.001; two-tailed Student’s t-test. Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of c and k are in Supplementary Table 3.
We then overexpressed FLAG-tagged IGF2BPs in HEK293T cells and immunoprecipitated ribonucleoprotein complexes to evaluate the in cellulo binding. A significant enrichment of m6A modifications in FLAG-IGF2BPs-bound RNA was observed (Fig. 1c and Supplementary Fig. 1i), similar to that in RNA immunoprecipitates (RIP) of endogenous IGF2BPs (Supplementary Fig. 1j). Sequencing purified RNA from FLAG-RIP samples identified more than 5000 genes from each RIP sample; among them, over 50% overlapped with published PAR-CLIP-seq targets22 (P<5e-324, Fisher’s exact test; Fig. 1d). The 3747, 3211 and 3914 transcripts identified by both RIP and PAR-CLIP methods can be considered as high-confidence targets of IGF2BP1, IGF2BP2 and IGF2BP3, respectively (Fig. 1d and Supplementary Table 1). The three IGF2BP proteins shared 2149 (55%–70%) high-confidence RNA targets (Fig. 1e). All three IGF2BPs preferentially bind to the “UGGAC” consensus sequence containing the “GGAC” m6A core motif (Fig. 1f), and more than 80% of the high-confidence targets contain at least one m6A peak as detected by m6A-seq3 (Fig. 1g). Moreover, most of the IGF2BP binding sites (92%) are located in protein-coding transcripts (i.e., mRNAs) and highly enriched near stop codons and in 3′ UTRs, coinciding with the m6A distribution (Fig. 1h–1j). In addition, we analyzed ENCODE eCLIP-seq data in HepG2 cells and human embryonic stem cells (hESCs), and found that the “UGGAC” motif was also enriched in the targets of IGF2BPs in both cell types (Supplementary Fig. 1k and ll).
METTL3 and METTL14 are two critical components of the methyltransferase complex catalyzing methylation at N6-adenosine. We performed CLIP of Flag-tagged IGF2BP2 and IGF2BP3 in HEK293T cells with or without METTL14 knockdown. Four representative high confidence targets, including MYC, FSCN1, TK1, and MARCKSL1, exhibit strong binding with IGF2BPs around their m6A motifs in control cells (Fig. 1k). Such binding was largely impaired upon METTL14 knockdown (Fig. 1k), suggesting the requirement of cellular m6A modification for the binding. Taken together, these data demonstrated the role of IGF2BPs as direct m6A binding proteins in vitro and in cellulo.
Silencing of IGF2BPs globally down-regulates target gene expression
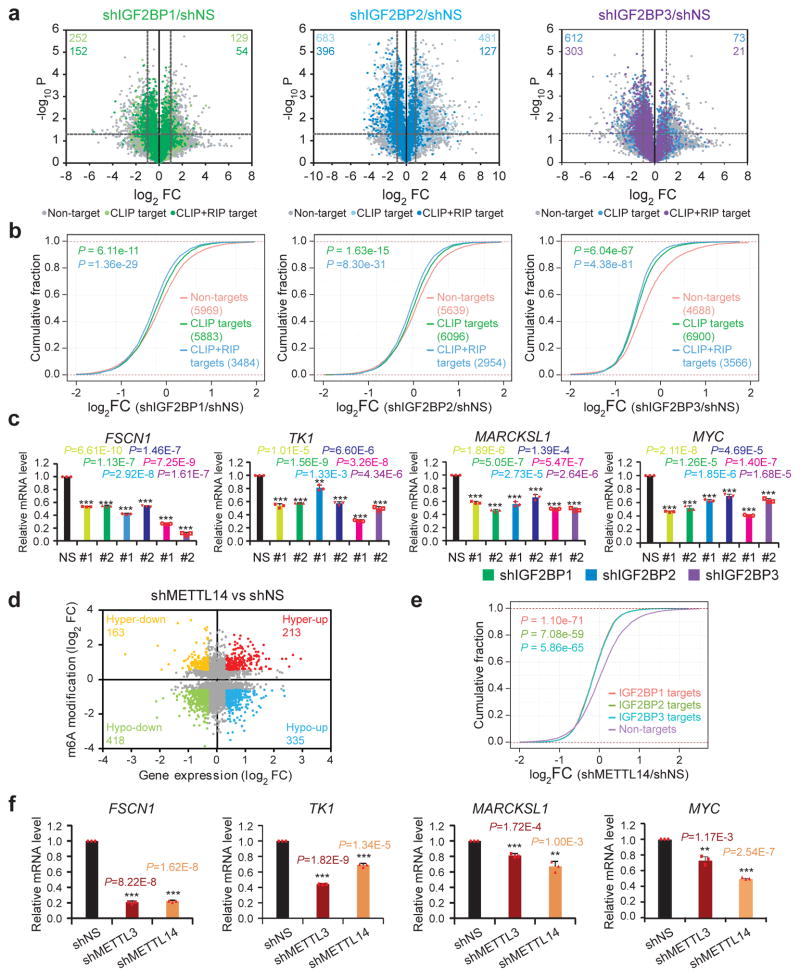
We next conducted RNA-seq in individual IGF2BP knockdown and control HepG2 cells (Supplementary Fig. 2a). The global transcripts were grouped into non-targets, CLIP targets and CLIP+RIP targets according to their binding by IGF2BPs in HEK293T cells (see Fig. 1d), considering the availability of CLIP data for all three IGF2BPs in this cell line. Knockdown of individual IGF2BPs globally and preferentially inhibited expression of CLIP targets and especially CLIP+RIP targets, with much more CLIP+RIP targets being down-regulated than up-regulated (Fig. 2a and 2b). Gene set enrichment analysis (GSEA) also showed that genes highly expressed in the control groups were enriched with the IGF2BP CLIP+RIP targets (FDR<0.05, Supplementary Fig. 2b). Functional annotation indicated that target genes with reduced expression were enriched in DNA replication, cell cycle, proliferation and cancer related biological processes and pathways (Supplementary Fig. 2c). In addition, an enrichment of cell cycle genes and MYC target genes was observed in control vs. shIGF2BPs (Supplementary Fig. 2d). The down-regulation of representative targets was confirmed by qPCR (Fig. 2c).
Figure 2. IGF2BPs regulate transcriptome-wide mRNA levels.
(a) Volcano plots displaying enrichment of dysregulated target genes in IGF2BP knockdown (shIGF2BP) vs. control (shNS) HepG2 cells. The numbers of significantly downregulated (log2FC<−1, P<0.05, two-tailed Student’s t-test) or upregulated genes (log2FC>1, P<0.05, two-tailed Student’s t-test) in the CLIP target group and CLIP+RIP target group are shown. FC, fold change. (b) Cumulative frequency of mRNA log2-fold change in non-target, CLIP target and CLIP+RIP target genes upon IGF2BP silencing. P values were calculated using two-sided Wilcoxon and Mann-Whitney test. (c) Relative changes in FSCN1, TK1, MARCKSL1, and MYC mRNA levels upon IGF2BP silencing. Results from 2 shRNAs for each IGF2BP are shown. Values are mean±s.d. of n = 3 independent experiments. Two-tailed Student’s t-tests were used (**, P <0.01; ***, P <0.001). (d) Distribution of genes with a significant change in both m6A level and gene expression level in METTL14 knockdown HepG2 cells compared to control cells. (e) Cumulative frequency of mRNA log2-fold change showing global reduction of IGF2BPs high-confidence target genes in shMETTL14 vs. shNS cells. P values were calculated using two-sided Wilcoxon and Mann-Whitney test. Exponential regression was used in d and e. Gene expression in a, b were analyzed for three times, while e were analyzed twice. (f) Relative changes in FSCN1, TK1, MARCKSL1, and MYC mRNA levels upon METTL3 or METL14 silencing. Values are mean±s.d. of n =3 independent experiments. Two-tailed Student’s t-tests were used (**, P <0.01; ***, P <0.001). Source data of c and f are in Supplementary Table 3.
To determine whether the expression of IGF2BP targets is also affected by cellular m6A level, we performed m6A-seq and RNA-seq in control or METTL14 knockdown HepG2 cells. Upon METTL14 knockdown, 1516 genes showed reduced m6A modifications (i.e., m6A-Hypo genes; fold change (FC)<0.667). Among these genes, 418 had reduced (i.e., m6A-Hypo-down genes; FC<0.8) mRNA levels whereas 335 had increased levels (i.e., m6A-Hypo-up genes; FC>1.2) (Fig. 2d). As expected, IGF2BP high-confidence targets showed a global and significant reduction in mRNA level upon METTL14 knockdown (Fig. 2e). Expression of individual targets, including FSCN1, TK1, MARCKSL1, and MYC, was confirmed by qPCR to be significantly down-regulated upon METTL3 or METTL14 knockdown (Fig. 2f). Similar changes were observed for FSCN1 and MYC, two shared targets of IGF2BPs and YTHDF2, in IGF2BP1/YTHDF2 double knockdown cells (Supplementary Fig. 2e). On the other hand, the m6A-Hypo-down genes were globally down-regulated upon IGF2BPs knockdown, as compared to their non-target counterparts (Supplementary Fig. 2f). The correlated regulation of gene expression by IGF2BPs and METTL14 indicates that IGF2BPs are responsible for the expression output of m6A-regulated genes.
Regulation of mRNA stability by IGF2BPs
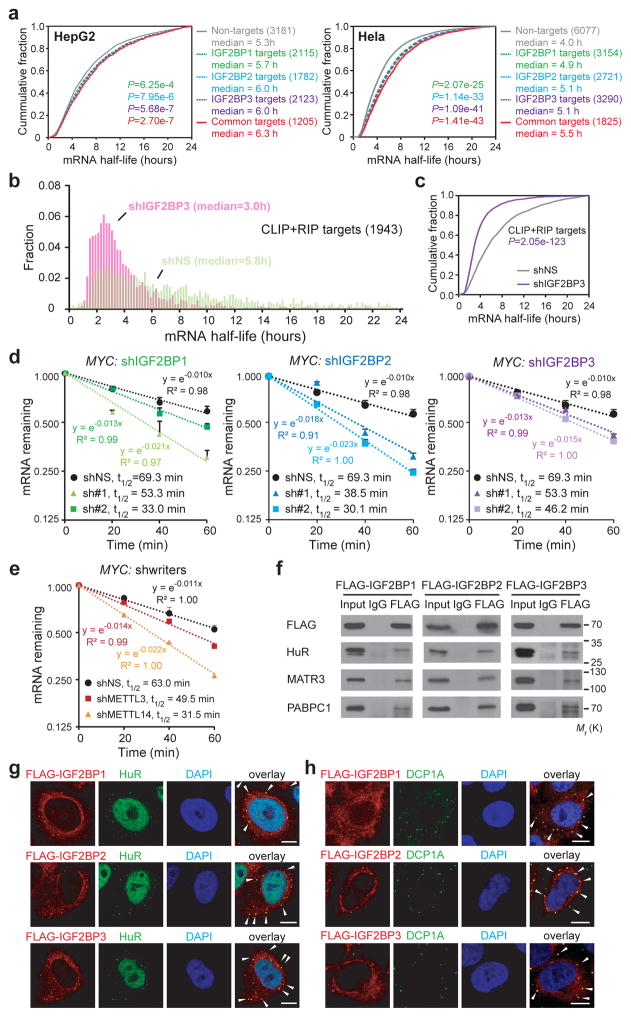
mRNA stability profiling in HepG2 cells revealed that high-confidence targets of IGF2BP1-3 tend to have longer half-lives (p<0.001) than their non-target counterparts (Fig. 3a), whereasYTHDF2 targets tend to have shorter half-lives (Supplementary Fig. 3a). Similar results were observed in Hela cells10 (Fig. 3a and Supplementary Fig. 3a). Since IGF2BP3 silencing affected target gene expression most potently (Fig. 2a and 2b), we performed mRNA stability profiling in IGF2BP3 knockdown cells. The median half-life of IGF2BP3 high-confidence targets as well as CLIP targets were significantly reduced in IGF2BP3-depleted cells to approximately 50% of that in control cells (Fig. 3b and 3c, and Supplementary Fig. 3b and 3c). Accelerated mRNA decay of MYC, FSCN1, TK1, and MARCKSL1 upon knockdown of each IGF2BP was confirmed in HepG2 cells (Fig. 3d and Supplementary Fig. 3d) and human cord blood CD34+ cells (Supplementary Fig. 3e). Furthermore, the stability of MYC, FSCN1, TK1, and MARCKSL1 mRNAs was also reduced when m6A writers were silenced (Fig. 3e and Supplementary Fig. 3f).
Figure 3. Knockdown of IGF2BPs decreases mRNA stability.
(a) Cumulative distribution of mRNA half-lives of non-target or IGF2BP high-confidence target genes in HepG2 (left) and Hela (right) cells. (b) Distribution of mRNA half-lives in IGF2BP3 high-confidence targets in HepG2 cells with shIGF2BP3 or shNS. (c) Cumulative distribution of mRNA half-life of IGF2BP3 high-confidence targets in shIGF2BP3 or shNS HepG2 cells. mRNA half-life analyses in a, b and c were repeated twice. (d and e) Reducing MYC mRNA half-life by silencing IGF2BPs (d) or m6A writers (e) in HepG2 cells. Values are mean±s.d. of n =3 independent experiments. (f) Co-Immunoprecipitation and Western blotting showing the binding of mRNA stabilizers with FLAG-tagged IGF2BPs in HEK293T cells, representative of 3 independent experiments. (g and h) Co-localization of IGF2BP proteins with HuR (g) or DCP1A (h) in Hela cells. Arrows indicate co-localization in cytoplasmic granules. Scale bar=10μm. Images were representative of 3 independent experiments. P values were calculated using two-sided Wilcoxon and Mann-Whitney test in a, b, c. Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of d and e are in Supplementary Table 3.
To identify cofactors of IGF2BPs that may enhance stability of mRNA targets, we pulled down the IGF2BP2 complexes and conducted mass spectrometry analysis. Notably, ELAV like RNA binding protein 1 (ELAVL1, also known as HuR), matrin 3 (MATR3), and poly(A) binding protein cytoplasmic 1 (PABPC1), three known mRNA stabilizers23–26, were identified. Western blotting confirmed their binding to ectopically expressed IGF2BPs in HEK293T cells (Fig. 3f), consistent with previous reports21, 27. Moreover, co-localization of HuR and IGF2BPs was observed in cytoplasmic granules (Fig. 3g), highly likely in P-bodies, as shown by DCP1A (i.e., P-body marker) staining (Fig. 3h). Thus, IGF2BPs likely recruit RNA stabilizers to promote the stability of their mRNA targets.
Stress granules are a second type of cytoplasmic mRNP granule thought to protect mRNAs from harmful conditions, and the protected mRNAs could be returned for translation after stress is relieved. We found that IGF2BPs were well co-localized with the stress granule marker TIAR, but only partially overlapped with DCP1A under heat shock (Supplementary Fig. 3g and 3h), which is consistent with previous reports 28, 29 and further reveals the roles of IGF2BPs in mRNA stabilization and storage during stress. Thus, our data indicate that IGF2BPs can promote RNA stability and/or increase mRNA storage in a dynamic manner under different physiological conditions.
Polysome profiling showed that FLAG-IGF2BP1 and FLAG-IGF2BP2 were present in most of the sucrose gradient fractions, while FLAG-IGF2BP3 was accumulated in 60S and 40S (Supplementary Fig. 4a). A similar distribution of endogenous IGF2BP1/2/3 proteins was observed in HepG2 cells (Supplementary Fig. 4b). Interestingly, HuR was detected along with each FLAG-tagged IGF2BP (Supplementary Fig. 4a). Moreover, knockdown of IGF2BP1 in HEK293T cells significantly reduced MYC mRNA in the translating pool (Supplementary Fig. 4c, fractions 13–18), suggesting a role of IGF2BPs in the active translation of their target genes. In agreement with the localization of IGF2BPs to stress granules during heat shock, IGF2BP2 shifted to non-ribosome fractions, and was found to gradually return to ribosome fractions during recovery from heat shock (Supplementary Fig. 4d).
IGF2BPs regulate MYC expression in an m6A-dependent manner
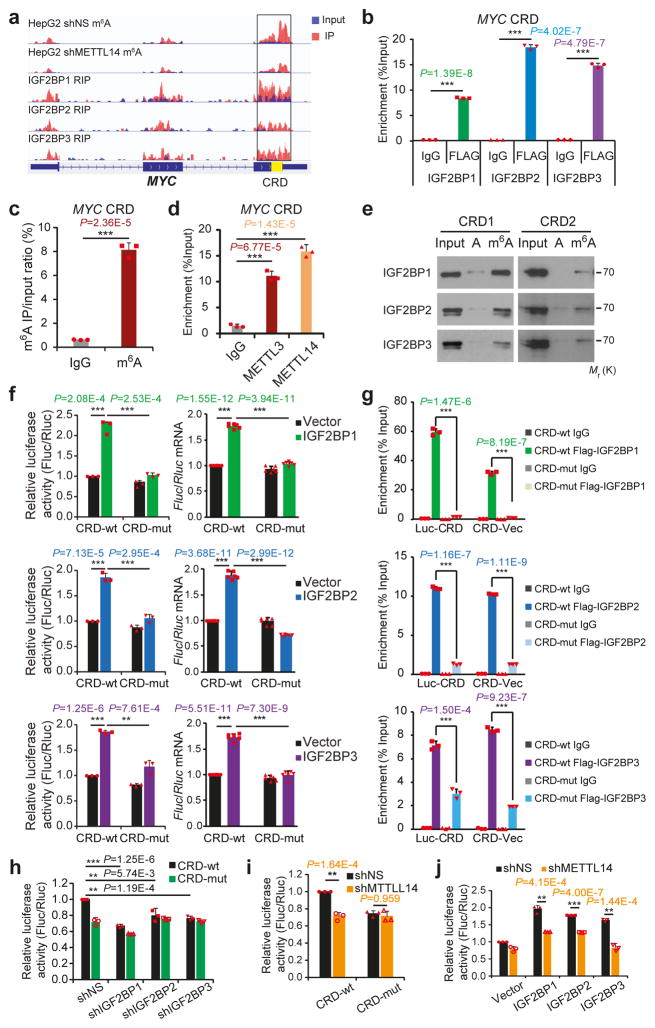
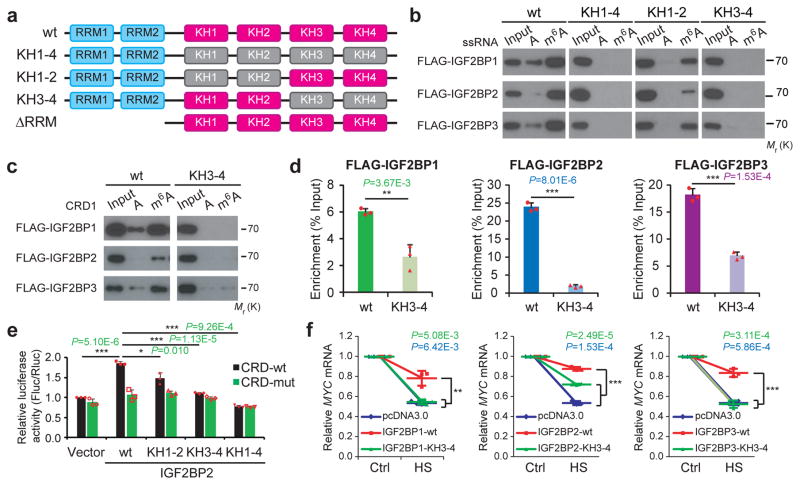
To determine whether IGF2BP-mediated gene regulation is m6A-dependent, we chose MYC, a well-known target of IGF2BP119, 21, 30, for a systematic study. A ~250 nucleotide (nt) cis-acting element called coding region instability determinant (CRD) resides in the 3′-terminus of the MYC coding region and has been proved critical for IGF2BP1 binding30. As shown in Figure 4a, m6A modifications are accumulated across MYC transcript, and the m6A peaks coincide well with IGF2BP binding sites. Notably, the CRD-containing region has a high abundance of m6A modifications that decreases remarkably upon METTL14 knockdown (Fig. 4a). By conducting RIP and gene-specific m6A assays, we confirmed the in cellulo IGF2BP binding (Fig. 4b and Supplementary Fig. 5a) and m6A modification (Fig. 4c), as well as METTL3 and METTL14 binding (Fig. 4d), in CRD. Moreover, the m6A modifications in the consensus sites of the synthetic CRD RNA oligos greatly facilitated their binding by endogenous IGF2BPs (Fig. 4e).
Figure 4. IGF2BPs regulate MYC expression through binding to methylated CRD.
(a) Distribution of m6A peaks across MYC mRNA transcript. The coding region instability determinant (CRD) region is highlighted in yellow. m6A-seq was repeated twice while RIP-seq was performed once.(b) RIP-qPCR showing the association of MYC CRD with FLAG-tagged IGF2BPs in HEK293T cells. (c) Enrichment of m6A modification in MYC CRD as detected by gene specific m6A qPCR assay. (d) RIP-qPCR showing the binding of METTL3 and METTL14 to the MYC CRD. (e) RNA pulldown of endogenous IGF2BP proteins from HEK293T nuclear extract using synthetic CRD RNA fragments, CRD1 and CRD2, with (m6A) or without (A) m6A modifications. Images are representative of 3 independent experiments. (f) Relative firefly luciferase (Fluc) activity (i.e., protein level; left) and Fluc mRNA level (right) of wild-type (CRD-wt) or mutated (CRD-mut) CRD reporters in HEK293T cells with ectopically expressed IGF2BP1, IGF2BP2, or IGF2BP3. (g) RIP-qPCR detecting the in vivo binding of Flag-IGF2BPs to the transcripts of CRD-wt or CRD-mut luciferase reporter in HEK293T cells. (h and i) Relative luciferase activity of CRD-wt or CRD-mut in Hela cells with or without stable knockdown of IGF2BPs (h) or METTL14 (i). (j) Relative luciferase activity of CRD-wt or CRD-mut in METTL14 stable knockdown or control Hela cells with ectopic expression of IGF2BPs. For all luciferase assays, the Fluc/Rluc ratio (representing luciferase activity) of CRD-wt with empty vector or shNS was used for normalization. Values are mean±s.d. of n =3 independent experiments, and two-tailed Student’s t-tests were used in b, c, d, f, g, h, i, j. (**, P <0.01; ***, P <0.001). Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of b, c, d, f, g, h, i, j are in Supplementary Table 3.
We next inserted the 249-nt wild-type or mutant CRD sequence into a firefly luciferase (Fluc) reporter (Supplementary Fig. 5b). Mutations in the m6A sites of CRD (CRD1 and CRD2 RNA oligos) dramatically abrogated the association with IGF2BP proteins in vitro (Supplementary Fig. 5c). As expected, ectopic IGF2BPs induced a significant increase in Fluc activity of wild-type reporter in a dose-dependent manner (Supplementary Fig. 5d). Such increase was largely impaired by mutations in the m6A consensus sites (Fig. 4f, left, and Supplementary Fig. 5b). Consistently, relative Fluc mRNA level of CRD-wt, but not CRD-mut, was increased by IGF2BP overexpression (Fig. 4f, right). RIP-qPCR demonstrated a strong binding of IGF2BPs with CRD-wt reporters and a much less or no binding with CRD-mut in cellulo (Fig. 4g). Conversely, knockdown of individual IGF2BPs, similar to knockdown of METTL14, caused inhibited Fluc activity, which also relies on the presence of wild-type m6A motifs within CRD (Fig. 4h and 4i). Noticeably, IGF2BPs-mediated increase of luciferase activity could be partially or completely blocked by METTL14 knockdown (Fig. 4j). Taken together, our data demonstrate that m6A modifications in CRD are required for the binding of IGF2BPs to MYC and for IGF2BP-mediated regulation of MYC expression.
Recognition of m6A by IGF2BPs’ KH domains
RRM and KH, the RNA binding domains of IGF2BPs, are different from YTH domain, the known m6A binding domain2, 10, 14, 16. We constructed IGF2BP mutants with truncation of the two RRM domains, or with mutations of GxxG to GEEG in the KH domains as reported20 (Fig. 5a), and showed KH domains (KH1-4) mutations, but not RRM truncation, completely abolished the interaction between IGF2BPs and ss-m6A probes (Fig. 5b and Supplementary Fig. 5e). Interaction of IGF2BPs with ss-m6A probes was only partially reduced by KH1-2 di-domain mutation but was almost completely abolished by KH3-4 mutation (Fig. 5b), indicating that the KH3-4 di-domain is indispensable for m6A recognition and binding while KH1-2 might play an accessory role. KH domain mutations including KH3-4 and KH1-4, but less likely KH1-2 mutation or RRM truncation, also impaired the binding of IGF2BP2 to hp-m6A probes (Supplementary Fig. 5f). Accordingly, mutation of KH3-4 interrupted the association of IGF2BPs with methylated CRD probes in vitro and with MYC CRD in cellulo (Fig. 5c and 5d). Moreover, IGF2BP2-promoted luciferase expression of CRD-wt reporter was partially impaired by KH1-2 mutation and completely diminished by KH3-4 or KH1-4 mutations (Fig. 5e).
Figure 5. The KH domains of IGF2BPs are critical for m6A recognition and binding.
(a) Schematic structures showing RNA binding domains within IGF2BP proteins and a summary of IGF2BP variants used in this study. Blue boxes are RRM domains, red boxes are wild-type KH domains with GxxG core, and grey boxes are inactive KH domain with GxxG to GEEG conversions. (b) RNA pulldown followed by Western blotting showed in vitro binding of ssRNA baits with wild-type (wt) or KH domain-mutated IGF2BP variants, representative of 3 independent experiments. (c) In vitro binding of CRD1 RNA probes with wild-type or KH3-4 mutated IGF2BPs, representative of 3 independent experiments. (d) The association of wild-type and KH3-4 mutated IGF2BPs with MYC CRD in HEK293T cells as assessed by RIP-qPCR. (e) Relative luciferase activity of CRD reporters in HEK293T cells with forced expression of wild-type or mutated IGF2BP2 variants. (f) Changes in MYC mRNA levels in Hela cells with empty vector or forced expression of wild-type or KH3-4 mutated IGF2BPs one hour post-heat shock (HS). Values are mean±s.d. of n =3 independent experiments, and two-tailed Student’s t-tests were used in d, e, f (*, P <0.05; **, P <0.01; ***, P <0.001). Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of d, e, f are in Supplementary Table 3.
IGF2BP1 was reported to play a role in stabilizing mRNA under stress conditions29, 31. We found that when Hela cells were exposed to heat shock at 42˚C for 1 hour, MYC mRNA was decreased to about 50% (Fig. 5f). Overexpression of wild-type IGF2BPs significantly diminished the reduction of MYC mRNA; however, this protective effect was largely or completely impaired by the mutation in KH3-4 (Fig. 5f). Collectively, these results demonstrate the essential roles of KH3-4 di-domains in IGF2BP binding to m6A-modified mRNAs and in regulation of target gene expression in both normal and stress conditions.
IGF2BPs play oncogenic roles in cancers as m6A readers
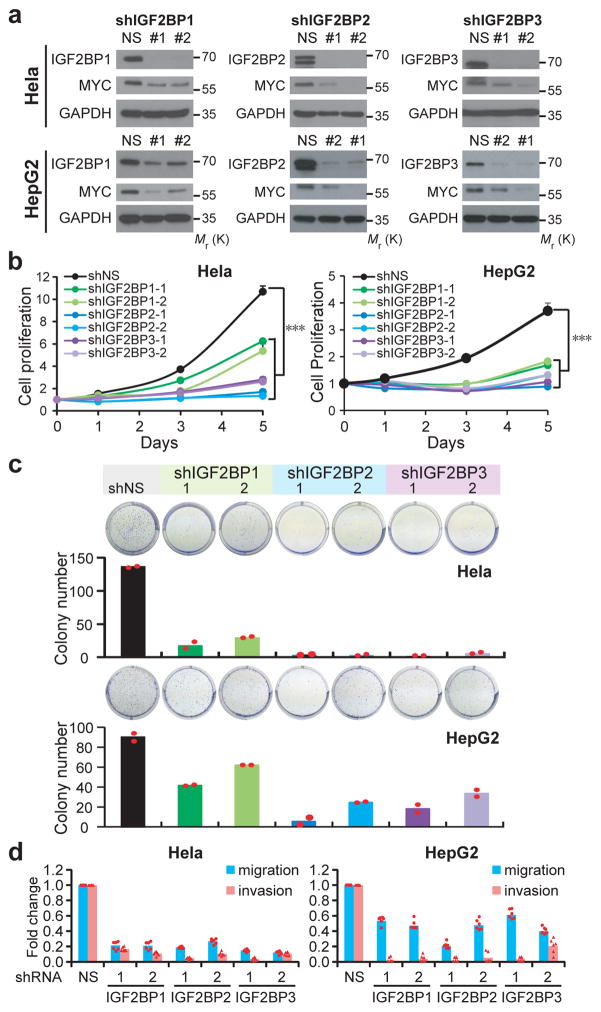
Detailed analysis of public databases from cBioPortal for Cancer Genomics (http://www.cbioportal.org) and The Cancer Genome Atlas (TCGA) (http://cancergenome.nih.gov) showed that all three IGF2BPs are frequently amplified or highly expressed in a variety of human cancers (Supplementary Fig. 6a and 6b). Given their similar effect on stabilizing oncogenic transcripts such as MYC, we assume that all three IGF2BPs could exert oncogenic functions in cancer. Indeed, knocking down of each individual IGF2BPs in Hela (cervical cancer) and HepG2 (liver cancer) cells significantly repressed MYC expression (Fig. 6a) and inhibited cancer cell proliferation, colony formation ability, and cell migration/invasion (Fig. 6b–6d and Supplementary Fig. 6c–6e), which mimics the effect of MYC silencing (Supplementary Fig. 6f–6j).
Figure 6. The oncogenic functions of IGF2BPs in human cancer cells.
(a) Western blot showing depletion of MYC protein in IGF2BPs-silenced Hela and HepG2 cells, representative of 3 independent experiments. (b) Inhibition of cell proliferation in IGF2BP-silenced cells compared to control cells as determined by MTT assays. Values are mean±s.d. of n =3 independent experiments. Two-tailed Student’s t-tests were used (***, P <0.001). (c) Effect of IGF2BP silencing on colony formation ability. Representative images of crystal violet staining of cells in 6-well plate are shown on top of the histograms. Colonies were counted from 3 replicate wells and 2 independent experiments were performed. The colony number of each experiment represents the total count of 3 replicate wells. (d) Repression of migration and invasion by shRNAs against IGF2BPs. Numbers of migrated or invaded cells were counted from 3 replicate wells and 2 independent experiments were performed. The numbers in the control (shNS) groups were set as 1. Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of b, c, d are in Supplementary Table 3.
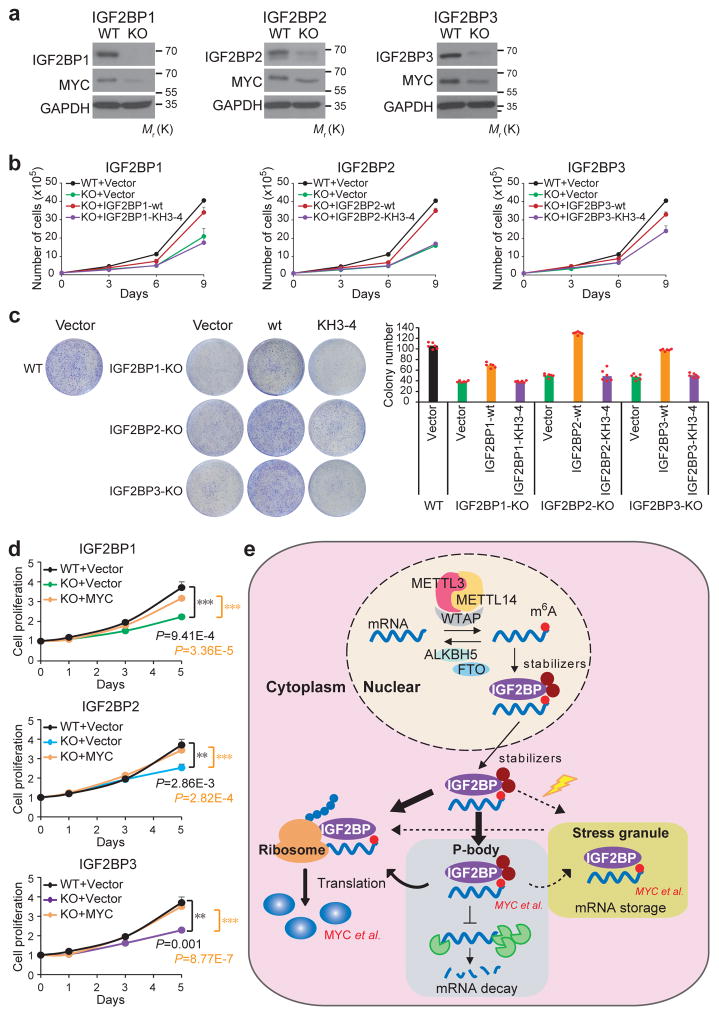
By utilizing the CRISPR-Cas9 system, we generated IGF2BPs-knockout (KO) cells (Fig. 7a) and performed rescue experiments. The decreased proliferation and colony formation ability of KO cells could be reversed by forced expression of wild-type IGF2BPs, but not the KH3-4-mutants (Fig. 7b and 7c), suggesting the oncogenic function of IGF2BPs relies on their role as m6A readers. Ectopic expression of MYC also restored the proliferative ability of IGF2BP-KO cells (Fig. 7d), further supporting MYC as a critical target of IGF2BPs.
Figure 7. IGF2BPs are oncogenic m6A readers.
(a) CRISPR-Cas9 mediated knockout (KO) of IGF2BPs and the subsequent depletion of MYC in HepG2 cells as detected by Western blotting. Images were representative of 3 independent experiments. (b) Effect of wild-type or KH3-4 mutated IGF2BPs on restoring cell proliferation in IGF2BP-KO cells. Data shown represent mean value of viable cell numbers of 2 independent experiments. (c) Colony formation assay using wild-type (WT) or IGF2BP KO (sgIGF2BP) HepG2 cells. Representative images of crystal violet staining of cells were shown on top of the histograms of colony numbers. Colonies were counted from 3 replicate wells of and 2 independent experiments were performed. The colony number of each experiment represents the average count of 3 replicate wells. (d) MTT assays displaying the effect of MYC on restoring cell proliferation in IGF2BP-KO cells. Values are mean±s.d. of n =3 independent experiments. Two-tailed student t-test were used (**, P <0.01; ***, P <0.001). (e) Working model of IGF2BP-mediated regulation of m6A modified mRNAs. mRNAs were methylated de novo by the methyltransferase complex composed of METTL3, METTL14 and a regulatory subunit WTAP. The naïve mRNA with m6A modifications were preferentially recognized by IGF2BP proteins. By recruiting mRNA stabilizers, such as HuR and MATR3, IGF2BPs protect target mRNAs from degradation in the P-body while facilitating translation after being exported to cytoplasm. Under stress conditions such as heat shock, IGF2BP-containing mRNPs are translocated to stress granules for the storage of their mRNA targets. Unprocessed scans of western blot analysis are available in Supplementary Figure 8. Source data of b, c, d are in Supplementary Table 3.
Discussion
The characterization of the YTH-domain-containing proteins as direct m6A readers2, 10, 14, 16 has provided profound insights into our understanding of effects of m6A modification on genetic information flow. Our findings add a distinct RNA binding protein family, IGF2BPs, into the catalogue of m6A readers, and reveal their roles in mRNA stabilization and translation (Fig. 7e). Notably, over 3,000 mRNA transcripts were identified as targets of each individual IGF2BP protein, while over 5,000 mRNAs being targeted by at least one protein and more than 2,000 mRNAs being co-targeted by all three IGF2BPs. The binding sites of IGF2BPs are enriched with m6A motif “GGAC”. Given that only around 7,000 mRNA transcripts are m6A modified in individual mammalian cells 2, 3 and that more than 80% of IGF2BP targets have at least one m6A peak (Fig. 1g), IGF2BPs likely have a broad impact on m6A-associated gene regulation. Accordingly, reduction of cellular m6A levels upon METTL14 knockdown impairs the in cellulo binding of Flag-IGF2BP2/3 to their RNA targets. Furthermore, we demonstrated that IGF2BPs bind directly to MYC CRD and promote MYC expression in an m6A-dependent manner.
The direct binding of IGF2BPs to m6A RNAs through their KH domains was demonstrated both in vitro and in cellulo. KH domain is an evolutionarily conserved RNA recognition element found in several proteins, such as FMR1, HNRPK, and PTBP1 32. KH domains in IGF2BPs were responsible for the recognition and binding of some specific mRNAs, including ACTIN, MYC, and IGF217, 33, 34. Our work reveals a new, m6A-dependent binding mode on top of the primary sequence. We show that KH domains, especially the KH3-4 di-domain, are critical for the binding of IGF2BPs to m6A-modified RNAs. Nonetheless, KH3-4 peptides alone showed poor selectivity for m6A RNA compared to full length proteins (data not shown), consistent with previous reports that post-translational modifications in the KH3-4 flanking regions may be important for IGF2BP selectivity17, 35. Future structural studies are warranted to understand how specific KH domains bind to m6A-modified RNAs. It will also be interesting to investigate other KH domain proteins as potential m6A-binding readers.
Our RNA stability profiling revealed that IGF2BPs stabilize target RNAs. The opposite role of IGF2BPs versus YTHDF2 imposes an additional layer of complexity on m6A function. It is possible that IGF2BPs and YTHDF2 recognize different targets, or compete for the same m6A sites to fine-tune expression of shared targets. In fact, YTHDF2 binding sites show a lower density in the 3′UTRs than in the coding regions10, which is distinct from IGF2BP binding sites (Fig. 1h). Analysis of the ENCODE PAR-CLIP data revealed only a very small proportion (0.85%–1.20%) of IGF2BP binding sites being shared by YTHDF2 (Supplementary Fig. 7a). Moreover, a consensus of “GAAC” besides “GGAC” was found from all significant YTHDF2 binding sites (Supplementary Fig. 7b). In addition, we found that IGF2BP binding sites have a significant lower guanine-cytosine (GC) content than YTHDF2 binding sites (Supplementary Fig. 7c and 7d), suggesting that local nucleotide composition may also contribute to the binding preference of different readers. Together, these data suggest that IGF2BPs and YTHDF2 have distinct pattern in target recognition and regulation.
The mRNA stabilizing function of IGF2BPs was also supported by its cofactors, HuR and MATR3. Particularly, HuR was previously identified as an indirect m6A binding protein, which increased stability of bound RNA and blocked microRNA targeting8, 25, 36. Interestingly, we showed here that HuR was colocalized with IGF2BPs in P-bodies, locations for mRNA fate decision37–39. Our findings suggest that HuR could be recruited by IGF2BPs to protect m6A-containing mRNAs from degradation and facilitate their translation (Fig. 7e). We found that IGF2BPs colocalize with stress granules and shuttle between ribosome and non-ribosome fractions during heat shock and recovery, suggesting a role of IGF2BPs in mRNA translation in stress response. Collectively, IGF2BPs can promote stability by inhibiting mRNA degradation or enhancing mRNA storage under stress, and facilitate their translation (Fig. 7e). We recently reported that METTL14 promotes the stability and translation of MYC mRNA and plays an essential oncogenic role in leukemia40, which is likely also attributed to IGF2BP-mediated, m6A-dependent regulation of MYC expression.
Dysregulation of IGF2BPs could result in abnormal accumulation of oncogenic products such as MYC, and therefore support the malignant state of cancer cells. Consistent with the frequent amplification of IGF2BP genes in various types of cancers, our finding that IGF2BPs exhibit oncogenic roles as m6A readers demonstrates the functional importance of IGF2BPs and their associated m6A reading processes in tumorigenesis, and highlights the therapeutic potential of targeting IGF2BPs in cancers.
METHODS
Plasmids and siRNAs
pcDNA3-based vectors encoding wild-type and KH domain mutant FLAG-tagged chicken ZBP1 (refer to as IGF2BP1), human IGF2BP2 and IGF2BP3 were kindly provided by Dr. Hüttelmaier (Martin-Luther-University, Germany). RRM domain truncation of IGF2BP2 (IGF2BP2-ΔRRM) was produced by PCR using the Q5 Site Directed Mutagenesis Kit (NEB) with Forward primer 5′-GAAGAGGTGAGCTCCCCT-3′ and Reverse primer 5′-GAATTCCTTGTCGTCGTCC-3′. The plasmid encoding human MYC (pCDNA3-HA-HA-humanCMYC) was obtained from Addgene. The TRC lentiviral vectors encoding shRNAs against IGF2BP1 (TRCN0000075149 and TRCN0000075152), IGF2BP2 (TRCN0000149002 and TRCN0000148565), IGF2BP3(TRCN0000074677 and TRCN0000074673), METTL3 (TRCN0000034715) and METTL14 (TRCN0000015933) and their non-specific control (shNS, RHS6848) were purchased from GE Dharmacon (Lafayette, CO), while the packing vectors, pMD2.G, pMDLg/pRRE and pRSV-Rev were obtained from Addgene. The MYC siRNA was purchased from Santa Cruz Biotechnology (Santa Cruz, CA), while the HuR siRNA was from GE Dharmacon.
Cell culture and transfection
The human hepatocellular carcinoma cell line HepG2 (ATCC HB-8065) was maintained in EMEM medium (ATCC) supplemented with 10% FBS (Invitrogen, Carlsbad, CA), 2 mM L-Glutamine and 1% penicillin-streptomycin. HEK293T and Hela cells were grown in DMEM medium (Invitrogen) containing 10% FBS, 2 mM L-Glutamine and 1% penicillin-streptomycin. All cell lines were purchased from the American Type Culture Collection (ATCC) and were not authenticated by ourselves. All cell lines were routinely tested for mycoplasma contamination. These cell lines were not listed in the database of commonly misidentified cell lines maintained by ICLAC. For heat shock treatment, Hela cells were incubated at 42 °C for 1 hour. Plasmids and siRNAs were transfected into cells with Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer.
Isolation and lentiviral infection of CD34+ cells
Cord blood of healthy donors was obtained from Cincinnati Children’s Hospital Medical Center (CCHMC, Cincinnati, USA) and subjected to isolation of mononuclear cells (MNCs) using Ficoll-Paque PLUS (GE Healthcare Life Sciences). Human CD34+ hematopoietic stem/progenitor cells (HSPCs) were then purified from MNCs by using human CD34 MicroBead Kit (Miltenyi Biotec, Auburn, CA). The CD34+ cells were cultured in StemSpan™ SFEM medium (StemCell Technologies, Vancouver, Canada) supplemented with 1% Lipid Mixture 1 (L0288, Sigma-Aldrich, St. Louis, MO), 2 mmol/L L-glutamine, 1% penicillin-streptomycin, 100 ng/mL SCF, and 2 ng/mL IL-3. Cells were infected with concentrated lentiviral particles through two rounds of “spinoculation”.
RNA affinity chromatography and nLC-ESI-MS/MS
Biotin-labelled RNA oligonucleotides containing adenosine or m6A were synthesized by GE Dharmacon. The hpRNA baits were denatured at 99 °C for 10 minutes and slowly cooled down to room temperature to allow the formation of stem-loop structure before use. The ssRNA baits were denatured at 99 °C for 10 minutes and put on ice immediately unless otherwise specified. Each RNA oligonucleotide (0.4 pmol, unless otherwise specified) was immobilized onto 50 μl streptavidin magnetic beads (Thermo Fisher Scientific, BufferRockford, IL) in binding buffer (20 mM Tris, 200 mM NaCl, 6 mM EDTA, 5 mM potassium fluoride, 5 mM β-glycerophosphate, 2 μg/ml aprotinin at pH 7.5) at 4 °C for 4 hours. RNA bait-conjugated streptavidin beads were then incubated with 200 μg of HEK293T nuclear extract or 0.5 μg to 1 ug of human recombinant proteins in binding buffer in a final volume of 400 μl overnight at 4 °C. After extensive washing, RNA–protein complexes were dissolved in 1x SDS buffer, separated under denaturing conditions on 10% SDS-polyacrylamide Bis-Tris gels and detected by silver staining (Thermo Fisher Scientific) or Western blot analysis. For mass spectrometry analysis, proteins in gel slices were digested with trypsin. The recovered peptides were introduced into an Eksigent (Dublin, CA) nanoLC.ultra nanoflow system attached to a TripleTOF 5600 plus (Sciex, Toronto, Canada) for nLC-ESI-MS/MS analyses.
Protein expression and purification
Flag-tagged IGF2BP1, IGF2BP2 and IGF2BP3 were expressed in HEK293T cells. For each protein, four 15 cm dishes of cells were prepared and lysed in 4 ml lysis buffer (50 mM Tris-HCl pH 7.5, 300 mM KCl, 0.5% NP-40, 5% Glycerol, 5 μg/ml DNase I, 1% RNase T1/A, 1% protease inhibitor, 1 mM DTT) at 4 °C for 1 hour and sonicated (5 seconds on, 25 seconds off, 24 cycles). The lysate was then cleared by centrifuge at 4 °C. The proteins were affinity purified using 40 μl anti-Flag M2 resin (Sigma-Aldrich) at 4 °C for 2 hour. After extensive wash with wash buffer (50 mM Tris-HCl, pH 7.5. 300 mM KCl, 5% Glycerol, 1 mM DTT), proteins were eluted in 500 μl 1x Flag elution solution (0.5 mg/ml Flag peptide in wash buffer) at 4 °C for 1 hour. Protein purity was verified with SDS-PAGE followed by coomassie staining.
Electrophoretic mobility shift assay/gel shift assay (EMSA)
The Cy5.5-labelled RNA oligonucleotides [ss-A: Cy5.5-CGUCUCGGACUCGGACUGCU; ss-m6A: Cy5.5-CGUCUCGG(m6A)CUCGG(m6A)CUGCU] with the same sequences of biotin-labelled ss-A and ss-m6A were synthesized by GE Dharmacon. The gel shift assay was performed as previously described10. Briefly, RNA probes were denatured by heating at 65 °C for 5 min and slowly cooling down to room temperature. For each reaction, 1 μl RNA probes (4 nM final concentration) and 1 μl protein (10x of the concentration gradient indicated in Fig. S1d) were incubated in 8 μl binding buffer (10 mM Tris-HCl pH7.5, 50 mM KCl, 1 mM EDTA, 0.05% Triton-X-100, 5% glycerol, 1 mM DTT and 40 U/ml RNasin) on ice for 30 min. The RNA-protein mixtures were separated in 5% native polyacrylamide gels (in 0.5x Tris-borate-EDTA buffer) at 4 °C for 60 min at 13 V/cm. The fluorescence signal was visualized by Odyssey Imaging Systems (LI-COR Biosciences) and quantified by ImageMaster TotalLab (GE Healthcare). The dissociation constant (Kd) was calculated with nonlinear curve fitting (Function one site-specific binding) using GraphPad Prism with y=Bmax*x/(Kd+x), where y is the ratio of [RNA-protein]/([free RNA]+[RNA-protein]), x is the input protein concentration, and Bmax is set to 1.
m6A dot blot
The m6A dot blot assay was conducted as previously described 12. Briefly, the indicated amount of total cellular RNA or synthesized RNA oligonucleotide was denatured in 3-fold volume of RNA incubation buffer (65.7% fomamide, 7.77% formaldehyde and 1.33x MOPS) at 65 °C for 5 min, followed by chilling on ice and mixing with 1 volume of 20x SSC. RNA samples were applied to Amersham Hybond-N+ membrane (GE Healthcare, Piscataway, NJ) with a Bio-Dot Apparatus (Bio-Rad, Hercules, CA). After UV crosslinking, the membrane was stained with 0.02% methylene blue (MB) in 0.3M sodium acetate. The membrane was then washed with 1x PBST buffer, blocked with 5% non-fat milk in PBST, and incubated with anti- m6A antibody (#202003, 1:1000; Synaptic Systems, Göttingen, Germany) overnight at 4 °C. After incubating with HRP-conjugated anti-rabbit IgG secondary antibody (Santa Cruz Biotechnology), the membrane was visualized using Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare).
Western blotting
Cells were lysed using 1x SDS buffer and sonicated. Equal amounts of proteins were loaded and separated by 10% SDS-PAGE, transferred to polyvinylidene fluoride membranes, and detected by immunoblotting with the Pierce ECL Western Blotting Substrate (Thermo Fisher Scientific) or Amersham ECL Prime Western Blotting Detection Reagent (GE Healthcare). Antibodies used for Western blotting were as follows unless otherwise specified: IGF2BP1(IMP1, clone D33A2, #8482), IGF2BP2 (IMP2, clone D4R2F, #14672), MYC (clone D3N8F, #13987), HuR (ELAVL1, clone D9W7E, #12582) were from Cell Signaling Technology (CST, Beverly, MA), IGF2BP3 (IMP3, A303-426A) and MATRIN3 (A300-591A-T) were from Bethyl Laboratories (Montgomery, TX), and FLAG (M2, F3165) was from Sigma-Aldrich (St. Louis, MO). GAPDH (sc-47724, Santa Cruz) was used as a loading control.
Prediction of m6A binding proteins
We developed a computational pipeline to discover the potential m6A binding proteins from ENCODE and published RBP-CLIP data sets generated by various CLIP methods, including HITS-CLIP, PAR-CLIP, eCLIP, iCLIP and irCLIP. The CLIP-seq peaks of each RBP were intersected with known m6A sites 41 to calculate the ratio between the number of peaks overlapping with m6A-peak sites and total number of peaks (m6A-containing peak number/total peak number). These peaks were imported into HOMER software 42 for de novo motif identification. The potential m6A binding proteins should meet the following requirements: (i) the ratio (m6A-containing peak number/total peak number) should be larger than 10%; and (ii) for the identified motifs of each RBP, the p-value should be less than 1e-35. Based on this pipeline, all the top de novo motifs could be identified unbiasedly. Nevertheless, because we are interested in identification of m6A-binding proteins, we further focused on the proteins that have top motif(s) containing ‘GGAC’, which represents the most common m6A consensus sequence.
LC-MS/MS
Fifty to one hundred ng of mRNA were digested by nuclease P1 (1U, Wako Pure Chemical, Osaka, Japan) in 25 μl of buffer containing 20 mM of NH4OAc (pH = 5.3) at 42°C for 2 h, followed by the addition of NH4HCO3 (1 M, 3 μl, freshly made) and alkaline phosphatase (1 U, Sigma). After an additional incubation at 37°C for 2 h, the sample was diluted to 50 μl and filtered (0.22 μm pore size, 4 mm diameter, Millipore), and 5 μl of the solution was injected into LC-MS/MS. Nucleosides were separated by reverse phase ultra-performance liquid chromatography on a C18 column with on-line mass spectrometry detection using an Agilent 6410 QQQ triple-quadrupole LC mass spectrometer in positive electrospray ionization mode. The nucleosides were quantified by using retention time and the nucleoside to base ion mass transitions of 284 to 152 (G), 282.1 to 150.1 (m6A), 268 to 136 (A), 245 to 113.1 (U), and 244 to 112 (C). Quantification was performed in comparison with the standard curve obtained from pure nucleoside standards running with the same batch of samples. The m6A level was calculated as the ratio of m6A to A or the ratio of m6A to (A+U+C+G) based on the calibrated concentrations.
RIP-LC-MS/MS
The RNA immunoprecipitation (RIP) procedure was reported previously 10. Three 15 cm dishes of confluent HEK293T cells transiently overexpressing Flag tagged IGF2BP (1, or 2, or 3) were subjected to RIP procedure. Input, flow-through, and IGF2BP-bound RNA were purified with Trizol reagent. mRNA from the three portions was further purified by depleting rRNA with RiboMinus Eukaryote Kit v2 (Ambion, Austin, TX) followed by depleting tRNA with RNA Clean and Concentrator-5 (Zymo Research, Orange, CA). 50 ng purified mRNA of each sample was subjected to LC-MS/MS quantification of m6A levels as reported previously 10.
RNA immunoprecipitation (RIP)
RNA immunoprecipitation was performed as previously described 43 with some modifications. Briefly, cells seeded in 10 cm dish at 70–80% confluency were cross-linked by UV and harvested by trypsinization. Nuclear extraction was isolated and sonicated. One microgram of FLAG (F3165, Sigma-Aldrich), IGF2BP1 (#8482, CST), or IGF2BP2 (#14672, CST) antibody or a corresponding control IgG (mouse IgG (CS200621, Millipore) for Flag, rabbit IgG (#NI01, Millipore) for IGF2BP1 and IGF2BP2) was conjugated to Protein A/G Magnetic Beads (Thermo Fisher Scientific) by incubation for 4 hours at 4 °C, followed by 3x wash and incubation with pre-cleared nuclear extraction in RIP buffer [150 mM KCl, 25 mM Tris (pH 7.4), 5 mM EDTA, 0.5 mM DTT, 0.5% NP40, 1x protease inhibitor] at 4 °C overnight. After washing with RIP buffer for three times, beads were resuspended in 80 μl PBS, followed by DNA digestion at 37 °C for 15 min and incubation with 50 μg of Proteinase K (Thermo Fisher) at 37 °C for 15 min. Input and co-immunoprecipitated RNAs were recovered by TRIzol (Invitrogen) extraction and analyzed by qPCR or RNA-seq.
Crosslinking and Immunoprecipitation (CLIP)
CLIP was performed following previously reported protocol 10 with some modifications. HEK293T cells seeded in four 15-cm plates at approximately 80% confluency were treated with 200 μM 4-thiouridine (4-SU). After 16 hours, cells were washed with ice-cold PBS, crosslinked with 150 mJ/cm2 of 365 nm ultraviolet light and harvested by trypsinization. Nuclear extraction was isolated and sonicated. Two to three milligram of nuclear lysates were serially digested by 0.05U/μl DNase RQ1 at 37 °C for 5 min, and 0.2U/μl RNase T1 at 22 °C for 15 min. Ten microgram of FLAG (F3165, Sigma-Aldrich) or IGF2BP2 (#14672, CST) antibody was conjugated to protein A/G magnetic beads and incubated with nuclear lysates in RIP buffer at 4 °C overnight. After washing with RIP buffer for three times, a second round of RNase T1 digestion was conducted under 50U/μl at 22 °C for 15 min. Input and immunoprecipiated RNAs were recovered and analyzed by qPCR.
RNA-seq
Total RNA was isolated from IGF2BP knockdown or control HepG2 cells using miRNeasy Kit (Qiagen). PolyA RNA was subsequently purified from 50–100 ng total RNA using NEBNext Poly(A) mRNA Magnetic Isolation Module. NEBNext Ultra Directional RNA Library Prep Kit (New England BioLabs, Ipswich, MA) was used for library preparation. Each group was sequenced in triplicate.
RNA stability assay and sequencing for mRNA lifetime
HepG2 cells with stably expressed shRNAs against IGF2BPs or shNS were seeded into 6-well plates to get 50% confluency after 24 hours. Cells were treated with 5 μg/ml actinomycin D and collected at indicated time points. The total RNA was extracted by miRNeasy Kit (Qiagen) and analyzed by RT-PCR and RNA-seq. For RNA sequencing, equal amount of ERCC RNA spike-in control (Thermo Fisher Scientific) was added to the total RNA samples as internal controls before library construction. Sequencing libraries were prepared using NEBNext Ultra Directional RNA Library Prep Kit. RNA stability profiling was generated from two biological replicates.
The turnover rate and half-life of mRNA was estimated according to previously published paper44. Since actinomycin D treatment results in transcription stalling, the change of mRNA concentration at a given time (dC/dt) is proportional to the constant of mRNA decay (Kdecay) and mRNA concentration (C), leading to following equation:
Thus the mRNA degradation rate Kdecay was estimated by:
To calculate the mRNA half-life (t1/2), when 50% of mRNA is decayed (ie. C/C0=1/2), the equation was:
From where:
m6A-seq
Total RNA was extracted by homogenizing cells in TRIzol reagent and purifying with Direct-zol RNA MiniPrep kit (Zymo). mRNA was further purified using Dynabeads mRNA purification kit (Thermo). RNA fragmentation was performed by sonication at 10 ng/μl in 100 μl RNase-free water using Bioruptor Pico (Diagenode) with 30s on/30s off for 30 cycles. m6A-IP and library preparation were performed according to the published protocol 2. Sequencing was carried out on Illumina HiSeq 2000 according to the manufacturer’s instructions.
Sequencing data analysis
For RIP-seq data
Samples were sequenced by Illumine Hiseq 1000 with a single end 51-bp read length. The RIP-seq reads were mapped to human genome version hg19 by Tophat2 version 2.0.13 with default settings 45. Differential gene expression was calculated by Cuffdiff version v2.2.1 46. The RIP targets were defined as genes with RPKM >= 1, IP/input >= 2, and p-value < 0.05.
For PAR-CLIP data
The IGF2BPs PAR-CLIP data were obtained from the public database GEO (Accession No. GSE21918) 22. The adapters were trimmed by using cutadapt version 1.9.1 47. The processed reads were mapped to human genome version hg19 by bowtie version 1.1.2 with parameters: –v 3 –m 5 --best --strata 48. The mapped results were analyzed by PARalyzer v1.5 with default settings 49. The results were further filtered by ModeScore >= 0.6. For peaks that were larger than 50 nt, we extracted 50 nt centered on ModeLocation site. GENCODE v24 was used to annotate the filtered peaks, and finally the PAR-CLIP target genes of IGF2BP1 (7511), IGF2BP2 (7974) and IGF2BP3 (9228) were identified. We then chose peaks overlapping with RIP targets for de novo motif analysis using HOMER software with default RNA analysis parameters.
For eCLIP data
To identify IGF2BP footprints, all mapped reads to consecutive genomic peaks were first assembled. Significant peaks were calculated by determining read-number cutoffs using the Poisson distribution as described by Zisoulis et al 50. The Poisson distribution assumes all intervals are independent and have equal probability of an occurrence happening. Peaks with significant high read-number values (p<10e-5 and the minimum peak height >= 10) and occurring in both biological duplicates were defined as IGF2BP footprints. We performed de novo motif identifications on eCLIP peaks using the HOMER software 42 with default RNA analysis parameters.
For RNA-seq data
All RNA-seq samples were sequenced by Illumine Hiseq 1000 with single end 51-bp read length. All reads were mapped to human genome version hg19 by hisat2 version 2.0.4 with default settings 51. Reads count was calculated using HTSeq 52, and was converted to RPKM (reads per kilobase, per million reads) using our custom Perl script. The average gene expression values of three independent studies were used for following analysis.
For mRNA lifetime profiling
all RNA-seq samples for mRNA lifetime were sequenced by Illumine Hiseq1000 with single end 51-bp read length. All reads were mapped to human genome version hg19 by hisat2 version 2.0.4 with default settings 51. Reads count was calculated using HTSeq 52, and was converted to RPKM (reads per kilobase, per million reads) by our custom Perl script. RPKM was converted to attomole by linear-fitting of the RNA spike-in as described by Wang et al. 10. The degradation rate of RNA (k) and mRNA half-life (t1/2) were calculated according to the aforementioned formula. The final half-life was calculated by using the average value of 1h, 3h and 6h.
For m6A-seq
Samples were sequenced by Illumine Hiseq 2000 with single end 50-bp read length. All reads were mapped to human genome version hg19 by tophat version 2.0.13 with default settings 45. The m6A level changes for shMETTL14/shNS were calculated by using exomePeak 53. Gene expression level changes for input and treatment were analyzed using Cuffdiff.
Integrative data analysis and statistics
Three biological replicates of RNA-seq were conducted for shNS, shIGF2BP1, shIGF2BP2 and shIGF2BP3, respectively. The genes expression values used were the average of three replicates. The differential expression patterns of non-targets, PAR-CLIP targets and CLIP+RIP targets were compared for each IGF2BP isoform by log2 transformed fold change [i.e., log2(shIGF2BP1/shNS), log2(shIGF2BP2/shNS) and log2(shIGF2BP3/shNS)]. Nonparametric Mann–Whitney U-test (Wilcoxon rank-sum test, two sided, significance level = 0.05) was applied for calculating p-value 10. The same analysis was applied for mRNA half-life.
Gene-specific m6A qPCR
m6A modifications on individual genes were determined using Magna MeRIP m6A Kit (Millipore, Billerica, MA) following the manufacturer’s instructions. Briefly, 100 μg of total RNA was sheared to about 100 nt in length by metal-ion induced fragmentation, then purified and incubated with anti-m6A antibody (#202003, Synaptic Systems)- or mouse IgG (CS200621, Millipore)-conjugated beads in 500 μl 1x IP buffer supplemented with RNase inhibitors at 4 °C overnight. Methylated RNA was immunoprecipitated with beads, eluted by competition with free m6A, and recovered with RNeasy kit (Qiagen). One tenth of fragmented RNA was saved as input control, and further analyzed by qPCR along with MeRIPed RNA. The related enrichment of m6A in each sample was calculated by normalizing to ten-fold input.
Immunofluorescence and microscopy
Hela cells were grown on cover slides, fixed, and stained with indicated antibodies as previously described 54. Antibodies used for immunofluorescence were as follows: FLAG (F3165, 1:200, Sigma-Aldrich), HuR (#12582, 1:200, CST), DCP1A (A303-590A-T, 1:200, Bethyl Laboratories), Alexa Fluor 488 anti-rabbit IgG (#4412, 1:500, CST), Alexa Fluor 594 anti-mouse IgG (#8890, 1:500, CST). Nuclei were stained by DAPI. Image acquisition was performed on a Zeiss LSM-710 confocal microscope under a 63x oil objective (Zeiss, Jena, Germany).
Polysome profiling
We followed the procedure reported previously 14 with the following modifications. We started with one 15 cm dish of confluent HEK293T cells transiently overexpressing FLAG tagged IGF2BP (1, or 2, or 3) or infected with lentiviral shRNA targeting IGF2BP1. Before collection, cycloheximide (CHX) was added to the media at 100 μg/ml for 7 min. The lysis buffer was formulated as 20 mM HEPES, pH 7.6, 100 mM KCl, 5 mM MgCl2, 100 μg/ml CHX, 1% Triton-X-100, freshly added 1:100 protease inhibitor (Roche), 40 U/ml SUPERasin (Ambion). The sample was then fractioned into 30 fractions, 0.5 ml per fraction, and analyzed by Gradient Station (BioCamp) equipped with ECONOUV monitor (BioRad, Hercules, CA) and Gilson FC203B fraction collector (Mandel Scientific, Guelph, Canada). Sample from each fraction was subjected to western blot analysis for Flag (A5892, Sigma-Aldrich), eIF3A (#3411, CST), eIF3B (sc-16377, Santa Cruz) and HuR (A-21277, Molecular Probes), or to qPCR analysis of MYC transcript.
For detection of endogenous IGF2BP proteins in ribosomal fractions, three 15 cm dishes of HepG2 cells at ~80% confluency were harvested as described above. Sample from each fraction was subjected to western blot analysis for IGF2BP1, IGF2BP2, and IGF2BP3.
Dual-Luciferase reporter assay
To generate the CRD firefly luciferase reporter construct, DNA fragments of wild-type and mutant CRD were synthesized by Integrated DNA Technologies (Coralville, Iowa) and cloned into the XhoI site of pMIR-REPORT vector (Ambion, Austin, TX). HEK293T or Hela cells were seeded in triplicate in 24-well plate to allow 70% confluency in the next day. 100 ng reporter plasmids with wild-type or mutant CRD (pMIR-CRD1-wt and pMIR-CRD-mut) and 20 ng renilla luciferase control plasmids (pRL-TK) were cotransfected with or without IGF2BPs expression vectors using Lipofactamine 2000 (Invitrogen). Twenty-four hours later, firefly luciferase (Fluc) and renilla luciferase (Rluc) activities were measured with Dual-Luciferase Reporter Assay System (Promega, Madison, WI) according to the supplier’s instructions. The relative luciferase activity was calculated by dividing Fluc by Rluc and normalized to individual control for each assay. For measuring Fluc mRNA changes, lysates left from luciferase activity measurement were lyzed with Qiazol reagent and total RNAs were extracted for qPCR analysis of Fluc and Rluc mRNA abundance. The relative luciferase mRNA was calculated as described above.
Protein co-immunoprecipitation and nLC-ESI-MS/MS
Cells grown in 10 cm dishes at 70–80% confluency were lysed with 500 μl NP40 buffer (150 mM NaCl, 1.5 mM MgCl2, 0.5% NP40, 50 mM Tris-HCl at pH8.0) and sonicated. Proteins were immunoprecipitated from 500 μg of cell lysates with FLAG or IGF2BP1 antibody and the corresponding IgG (as described above). After applying magnet, proteins associated with Protein A/G Magnetic Beads (Thermo Fisher Scientific) were washed 3 times and analyzed by Western blotting.
For mass spectrometry analysis, the protein complexes were eluted from beads by incubation with 0.2 mol/L glycine buffer (pH2.6), followed by neutralization with equal volume of 1 mol/LTris pH 8.0, and identified using nanoLC-MS/MS by the Proteomics Laboratory (University of Cincinnati, OH). Briefly, samples from immuno-enrichment with IgG and FLAG-antibodies were solubilized in Laemmli gel buffer and loaded onto separated lanes of a 4–12% MOPS mini SDS polyacrylamide gel, then electrophoresed for 15 min. The entire gel region containing the proteins was cut from the gel and subjected to in gel trypsin digestion and subsequent recovery of peptides as described previously 55. The recovered peptides were introduced into an Eksigent (Dublin, CA) nanoLC.ultra nanoflow system attached to a TripleTOF 5600 plus (Sciex, Toronto, Canada) for nLC-ESI-MS/MS analyses. Data were recorded using Analyst®-TF (v.1.7) software. Searches from the nLC-MS/MS were accomplished using ProteinPilot software (version 5.0, revision 4769) that utilizes Paragon algorithm (ver 5.0.0.0, 4769) against the complete database of Homo sapiens protein (20342 proteins) downloaded from ftp.uniprot.org on 1/21/16.
Lentiviral shRNA infection
Lentiviruses were produced in HEK293T cells by co-transfecting individual shRNA construct with packing vectors (pMD2.G, pMDLg/pRRE and pRSV-Rev) into HEK293T cells in 60 mm cell culture dish using X-tremeGENE HP DNA Transfection Reagent (Roche Diagnostics, Grenzach-Wyhlen, Germany). The lentivirus particles were harvested at 48 and 72 hours after transfection and directly added into target cells with 4 μg/ml polybrene. After two rounds of infection, cells were selected for at least two passages by adding 1 μg/ml puromycin into growth medium.
Cell proliferation, migration and invasion assays
Hela and HepG2 cells were seeded in 96-well plates at 1000 cells/well. Cell proliferation was evaluated by MTT Assay (Promega).
For colony formation assay, 2000 Hela cells or 10000 HepG2 cells were seeded in 6-well plates and stained with crystal violet 7–10 days later. Colonies were counted in three random fields under a 5x microscope.
For cell migration and invasion transwell assays, 25000 HepG2 cells or 50000 Hela cells in 500 μl starvation media were plated on the top chambers of Transwell Clear Polyester Membrane Inserts (for migration assay, Corning Costar, Vienna, Austria) and BioCoat Matrigel Invasion Chambers (for invasion assay, Corning Costar), while culture media with 20% FBS was applied on the bottom. After 48 to 72 hours, migrated or invaded cells were stained with crystal violet and counted under a 20x microscope.
For the wound healing assay, Hela and HepG2 cells were seeded in 24 well plates and allowed to grow to confluent monolayer in 24 hours. Cells were then scratched and allowed to continue to culture in complete culture media to allow for migration for 24–72 hours. Images were taken at indicated time under microscope. The scratched areas were quantified using Adobe Photoshop and used to calculate migration rate.
RNA isolation and quantitative RT-PCR
Total RNA was isolated from cultured cells using miRNeasy Kit (Qiagen, Valencia, CA) or Trizol reagent (Invitrogen). First strand cDNA was synthesized by reverse transcription of 500 ng RNA using QuantiTect Reverse Transcription Kit (Qiagen). Quantitative PCR (qPCR) was carried out using QuantiTect SYBR Green PCR Kit (Qiagen) and mRNA expression was normalized to reference genes, GAPDH and TATA-binding protein (TBP). The primers used in all qPCR assays were listed in Supplementary Table 2.
CRISPR-CSP9 KO
Human HepG2 cells were transiently transfected with a modified pSpCas9(BB)-2A-GFP plasmids (Addgene, #48138) containing IGF2BP1, IGF2BP2, or IGF2BP3 sgRNAs with optimized scaffold 56 and a high fidelity eSpCas9(1.1) 57 using Lipofectamine 2000 (Invitrogen) following the manufacturer’s instructions. Single cells were isolated 48 hours after transfection by FACS (BD FACS Aria III) into 96-well plates. Only cells with high GFP (top 4%) were selected. Independent clones were allowed to grow for three weeks. Knockout efficiency was tested by Western blotting with specific antibodies. The sgRNA sequences used are listed below: IGF2BP1, TATTCCACCCCAGCTCCGAT; IGF2BP2, GAGAAGTGCCCCCAGGGGCG; IGF2BP3, TGGCACCGACTGATAGAGCT.
Statistics and reproducibility
Statistical comparisons were performed by using t-tests (two tailed) or Wilcoxon and Mann-Whitney test as indicated in the figure legends. Data are presented as mean ± s.d.. P < 0.05 was considered significant. The number of biological (non-technical) replicates for each experiment is indicated in the figure legends. Three independent sets of RNA samples were used for RNA-seq, two independent sets of RNA samples were used for mRNA stability profiling, and two set of RNA samples was used for m6A-seq. All Western blot, dot blot, and immunofluorescence images are representative of three independent experiments. No statistical method was used to predetermine sample size.
Code availability
The custom Perl and R scripts used in this study are available on request to the corresponding authors.
Data availability
All sequencing data that support the findings of this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE90639 (for RIP-seq), GES90642 (for m6A-seq), and GSE90684 (for RNA-seq). Data for LC/MS/MS of IGF2BP2 coimmunoprecipitation have been deposited to Figshare (http://dx.doi.org/10.6084/m9.figshare.5693410). Previously published ENCODE PAR-CLIP and eCLIP data that were re-analyzed here are available under accession code: GSE21918 (for PAR-CLIP of IGF2BPs), GSE92021 (for IGF2BP1 eCLIP in HepG2),GSE92220 (for IGF2BP3 eCLIP in HepG2) and GSE78509 (for IGF2BPs eCLIP in hESCs).The human cancer data were derived from the TCGA Research Network (http://cancergenome.nih.gov/). The data-set derived from this resource that supports the findings of this study is available in cBioPortal for Cancer Genomics (http://www.cbioportal.org/, using TCGA pan-cancer studies).
Source data for Figures 1c, 1k, 2c, 2f, 3d, 3e, 4b, 4c, 4d, 4f, 4g, 4h, 4i, 4j, 5d, 5e, 5f, 6b, 6c, 6d, 7b, 7c, 7d and Supplementary Figures 2e, 3d, 3e, 3f, 4c, 5a, 5d, 6d, 6f, 6g, 6h, 6i, 6j are provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.
Supplementary Material
Acknowledgments
We thank Proteomics Laboratory at University of Cincinnati for mass spectrometry analysis; Transgenic Animal and Genome Editing Core at Cincinnati Children’s Hospital Medical Center for design and construction of sgRNA vectors; Genomics, Epigenomics and Sequencing Core at University of Cincinnati and the Genomic Facility at the University of Chicago for next generation sequencing. This work was supported in part by the National Institutes of Health (NIH) R01 Grants CA214965 (J.C.), CA211614 (J.C.), CA178454 (J.C.), CA182528 (J.C.), CA163493 (J.-L. G.), RM1 HG008935 (C.He), and grants 2017YFA0504400 (J.Y.), 91440110 (J.Y.) and 31671349 (L.Q.) from National Nature Science Foundation of China. J.C. is a Leukemia & Lymphoma Society (LLS) Scholar. C.He is an investigator of the Howard Hughes Medical Institute (HHMI). B.S.Z. is an HHMI International Student Research Fellow.
Footnotes
COMPETING FINANCIAL INTERESTS
C.He is a scientific founder of Accent Therapeutics, Inc.
AUTHOR CONTRIBUTIONS
H.H., H. Weng, and J.C. conceived and designed the entire project. H.H., H. Weng, C. He, J.Y. and J.C. designed and supervised the research. H.H., H. Weng, X.Q., H.S., H. Wu, B.S.Z., A.M., C.L., C.Y., J.S., R.S., X.D., M.S., C.L., S.N., Y.W., C. Hu, K.F., and J.C. performed experiments and/or data analyses; H.H., H. Weng, W.S., L.D., and J.Y. performed the genome-wide or transcriptome-wide data analyses; Y.H., S.H., K.G., X.J., M.W., L.Q., J.G., C. He, J.Y., and J.C. contributed reagents/analytic tools and/or grant support; H.H., H. Weng, W.S., H.S., B.S.Z., A.M., S.N., C. He, J.Y., and J.C. wrote and revise the paper. All authors discussed the results and commented on the manuscript.
References
- 1.Jia G, et al. N6-methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO. Nat Chem Biol. 2011;7:885–887. doi: 10.1038/nchembio.687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dominissini D, et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature. 2012;485:201–206. doi: 10.1038/nature11112. [DOI] [PubMed] [Google Scholar]
- 3.Meyer KD, et al. Comprehensive analysis of mRNA methylation reveals enrichment in 3′ UTRs and near stop codons. Cell. 2012;149:1635–1646. doi: 10.1016/j.cell.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao X, et al. FTO-dependent demethylation of N6-methyladenosine regulates mRNA splicing and is required for adipogenesis. Cell Res. 2014;24:1403–1419. doi: 10.1038/cr.2014.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Su R, et al. R-2HG exhibits anti-tumor activity by targeting FTO/m6A/MYC/CEBPA signaling. Cell. 2017 doi: 10.1016/j.cell.2017.11.031. Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liu N, et al. N(6)-methyladenosine-dependent RNA structural switches regulate RNA-protein interactions. Nature. 2015;518:560–564. doi: 10.1038/nature14234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Geula S, et al. Stem cells. m6A mRNA methylation facilitates resolution of naive pluripotency toward differentiation. Science. 2015;347:1002–1006. doi: 10.1126/science.1261417. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, et al. N6-methyladenosine modification destabilizes developmental regulators in embryonic stem cells. Nat Cell Biol. 2014;16:191–198. doi: 10.1038/ncb2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen T, et al. m(6)A RNA methylation is regulated by microRNAs and promotes reprogramming to pluripotency. Cell Stem Cell. 2015;16:289–301. doi: 10.1016/j.stem.2015.01.016. [DOI] [PubMed] [Google Scholar]
- 10.Wang X, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature. 2014;505:117–120. doi: 10.1038/nature12730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao BS, Roundtree IA, He C. Post-transcriptional gene regulation by mRNA modifications. Nat Rev Mol Cell Biol. 2016 doi: 10.1038/nrm.2016.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Z, et al. FTO Plays an Oncogenic Role in Acute Myeloid Leukemia as a N6-Methyladenosine RNA Demethylase. Cancer Cell. 2017;31:127–141. doi: 10.1016/j.ccell.2016.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao BS, et al. m6A-dependent maternal mRNA clearance facilitates zebrafish maternal-to-zygotic transition. Nature. 2017;542:475–478. doi: 10.1038/nature21355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang X, et al. N-6-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell. 2015;161:1388–1399. doi: 10.1016/j.cell.2015.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Du H, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626. doi: 10.1038/ncomms12626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xiao W, et al. Nuclear m(6)A Reader YTHDC1 Regulates mRNA Splicing. Mol Cell. 2016;61:507–519. doi: 10.1016/j.molcel.2016.01.012. [DOI] [PubMed] [Google Scholar]
- 17.Bell JL, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cellular and Molecular Life Sciences. 2013;70:2657–2675. doi: 10.1007/s00018-012-1186-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen J, et al. A family of insulin-like growth factor II mRNA-binding proteins represses translation in late development. Molecular and Cellular Biology. 1999;19:1262–1270. doi: 10.1128/mcb.19.2.1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Noubissi FK, et al. CRD-BP mediates stabilization of beta TrCP1 and c-myc mRNA in response to beta-catenin signalling. Nature. 2006;441:898–901. doi: 10.1038/nature04839. [DOI] [PubMed] [Google Scholar]
- 20.Huttelmaier S, et al. Spatial regulation of beta-actin translation by Src-dependent phosphorylation of ZBP1. Nature. 2005;438:512–515. doi: 10.1038/nature04115. [DOI] [PubMed] [Google Scholar]
- 21.Weidensdorfer D, et al. Control of c-myc mRNA stability by IGF2BP1-associated cytoplasmic RNPs. RNA. 2009;15:104–115. doi: 10.1261/rna.1175909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M, et al. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 2010;141:129–141. doi: 10.1016/j.cell.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Behm-Ansmant I, Gatfield D, Rehwinkel J, Hilgers V, Izaurralde E. A conserved role for cytoplasmic poly(A)-binding protein 1 (PABPC1) in nonsense-mediated mRNA decay. The EMBO journal. 2007;26:1591–1601. doi: 10.1038/sj.emboj.7601588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mangus DA, Evans MC, Jacobson A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome biology. 2003;4:223. doi: 10.1186/gb-2003-4-7-223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–3460. doi: 10.1093/emboj/17.12.3448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Salton M, et al. Matrin 3 binds and stabilizes mRNA. PloS one. 2011;6:e23882. doi: 10.1371/journal.pone.0023882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boudoukha S, Cuvellier S, Polesskaya A. Role of the RNA-binding protein IMP-2 in muscle cell motility. Mol Cell Biol. 2010;30:5710–5725. doi: 10.1128/MCB.00665-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wachter K, Kohn M, Stohr N, Huttelmaier S. Subcellular localization and RNP formation of IGF2BPs (IGF2 mRNA-binding proteins) is modulated by distinct RNA-binding domains. Biol Chem. 2013;394:1077–1090. doi: 10.1515/hsz-2013-0111. [DOI] [PubMed] [Google Scholar]
- 29.Stohr N, et al. ZBP1 regulates mRNA stability during cellular stress. J Cell Biol. 2006;175:527–534. doi: 10.1083/jcb.200608071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Doyle GA, et al. The c-myc coding region determinant-binding protein: a member of a family of KH domain RNA-binding proteins. Nucleic acids research. 1998;26:5036–5044. doi: 10.1093/nar/26.22.5036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bley N, et al. Stress granules are dispensable for mRNA stabilization during cellular stress. Nucleic Acids Research. 2015;43 doi: 10.1093/nar/gku1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valverde R, Edwards L, Regan L. Structure and function of KH domains. Febs Journal. 2008;275:2712–2726. doi: 10.1111/j.1742-4658.2008.06411.x. [DOI] [PubMed] [Google Scholar]
- 33.Nielsen J, Kristensen MA, Willemoes M, Nielsen FC, Christiansen J. Sequential dimerization of human zipcode-binding protein IMP1 on RNA: a cooperative mechanism providing RNP stability. Nucleic acids research. 2004;32:4368–4376. doi: 10.1093/nar/gkh754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chao JA, et al. ZBP1 recognition of beta-actin zipcode induces RNA looping. Genes & development. 2010;24:148–158. doi: 10.1101/gad.1862910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dai N, et al. mTOR phosphorylates IMP2 to promote IGF2 mRNA translation by internal ribosomal entry. Genes & development. 2011;25:1159–1172. doi: 10.1101/gad.2042311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Peng SS, Chen CY, Xu N, Shyu AB. RNA stabilization by the AU-rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–3470. doi: 10.1093/emboj/17.12.3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brengues M, Teixeira D, Parker R. Movement of eukaryotic mRNAs between polysomes and cytoplasmic processing bodies. Science. 2005;310:486–489. doi: 10.1126/science.1115791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huch S, et al. The decapping activator Edc3 and the Q/N-rich domain of Lsm4 function together to enhance mRNA stability and alter mRNA decay pathway dependence in Saccharomyces cerevisiae. Biology open. 2016;5:1388–1399. doi: 10.1242/bio.020487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bett JS, et al. The P-body component USP52/PAN2 is a novel regulator of HIF1A mRNA stability. The Biochemical journal. 2013;451:185–194. doi: 10.1042/BJ20130026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Weng H, et al. METTL14 Inhibits Hematopoietic Stem/Progenitor Differentiation and Promotes Leukemogenesis via mRNA m6A Modification. Cell Stem Cell. 2018;22:1–15. doi: 10.1016/j.stem.2017.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sun WJ, et al. RMBase: a resource for decoding the landscape of RNA modifications from high-throughput sequencing data. Nucleic Acids Res. 2016;44:D259–265. doi: 10.1093/nar/gkv1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rinn JL, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell. 2007;129:1311–1323. doi: 10.1016/j.cell.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen CYA, Ezzeddine N, Shyu AB. Messenger Rna Half-Life Measurements in Mammalian Cells. Method Enzymol. 2008;448:335–357. doi: 10.1016/S0076-6879(08)02617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kim D, et al. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trapnell C, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. journal. 2011;17:10–12. [Google Scholar]
- 48.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome biology. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Corcoran DL, et al. PARalyzer: definition of RNA binding sites from PAR-CLIP short-read sequence data. Genome biology. 2011;12:R79. doi: 10.1186/gb-2011-12-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zisoulis DG, et al. Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nature structural & molecular biology. 2010;17:173–179. doi: 10.1038/nsmb.1745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nature methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anders S, Pyl PT, Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Meng J, et al. A protocol for RNA methylation differential analysis with MeRIP-Seq data and exomePeak R/Bioconductor package. Methods. 2014;69:274–281. doi: 10.1016/j.ymeth.2014.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Weng HY, et al. Inhibition of miR-17 and miR-20a by Oridonin Triggers Apoptosis and Reverses Chemoresistance by Derepressing BIM-S. Cancer Research. 2014;74:4409–4419. doi: 10.1158/0008-5472.CAN-13-1748. [DOI] [PubMed] [Google Scholar]
- 55.Eismann T, et al. Peroxiredoxin-6 protects against mitochondrial dysfunction and liver injury during ischemia-reperfusion in mice. Am J Physiol Gastrointest Liver Physiol. 2009;296:G266–274. doi: 10.1152/ajpgi.90583.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen B, et al. Dynamic imaging of genomic loci in living human cells by an optimized CRISPR/Cas system. Cell. 2013;155:1479–1491. doi: 10.1016/j.cell.2013.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slaymaker IM, et al. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351:84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All sequencing data that support the findings of this study have been deposited in NCBI’s Gene Expression Omnibus (GEO) under accession number GSE90639 (for RIP-seq), GES90642 (for m6A-seq), and GSE90684 (for RNA-seq). Data for LC/MS/MS of IGF2BP2 coimmunoprecipitation have been deposited to Figshare (http://dx.doi.org/10.6084/m9.figshare.5693410). Previously published ENCODE PAR-CLIP and eCLIP data that were re-analyzed here are available under accession code: GSE21918 (for PAR-CLIP of IGF2BPs), GSE92021 (for IGF2BP1 eCLIP in HepG2),GSE92220 (for IGF2BP3 eCLIP in HepG2) and GSE78509 (for IGF2BPs eCLIP in hESCs).The human cancer data were derived from the TCGA Research Network (http://cancergenome.nih.gov/). The data-set derived from this resource that supports the findings of this study is available in cBioPortal for Cancer Genomics (http://www.cbioportal.org/, using TCGA pan-cancer studies).
Source data for Figures 1c, 1k, 2c, 2f, 3d, 3e, 4b, 4c, 4d, 4f, 4g, 4h, 4i, 4j, 5d, 5e, 5f, 6b, 6c, 6d, 7b, 7c, 7d and Supplementary Figures 2e, 3d, 3e, 3f, 4c, 5a, 5d, 6d, 6f, 6g, 6h, 6i, 6j are provided as Supplementary Table 3. All other data supporting the findings of this study are available from the corresponding authors on reasonable request.