SUMMARY
Assembly of the endocytic machinery is a constitutively active process that is important for the organization of the plasma membrane, signal transduction, and membrane trafficking. Existing research has focused on the stochastic nature of endocytosis. Here, we report the emergence of the collective dynamics of endocytic proteins as periodic traveling waves on the cell surface. Coordinated clathrin assembly provides the earliest spatial cue for cortical waves and sets the direction of propagation. Surprisingly, the onset of clathrin waves, but not individual endocytic events, requires feedback from downstream factors, including FBP17, Cdc42, and N-WASP. In addition to the localized endocytic assembly at the plasma membrane, intracellular clathrin and phosphatidylinositol-3,4-bisphosphate (PI(3,4)P2) predict the excitability of the plasma membrane and modulate the geometry of traveling waves. Collectively, our data demonstrate the multiplicity of clathrin functions in cortical pattern formation and provide important insights regarding the nucleation and propagation of single cell patterns.
INTRODUCTION
Collective dynamics refers to the motion displayed by a group of subjects for which each subject’s motion is related to that of its neighbours. It is an emergent property readily observed in dynamical systems, such as social networks, bird flocks, bacterial colonies, cellular slime molds, and groups of migrating cells (Watts and Strogatz, 1998). Despite the differences in scales, this self-organization arises due to common physical principles and could lead to selective advantages for the organisms (Laan et al., 2014). Collective dynamics is also common in molecular and cellular systems. At the single cell level, the collective dynamics of cortical actin have been documented in many cellular systems and are referred to as cortical waves. Understanding the cortical dynamics and the emergence of the collective dynamics within the cortex are critical for our understanding of cellular behaviours, such as polarity (Howell et al., 2012), migration (Weiner et al., 2007), mitosis (Bement et al., 2015), and cell growth (Das et al., 2012).
Gaining a molecular understanding of pattern formation is the most promising at the single cell level, but doing so still poses great challenges due to the nonlinearity of regulatory feedback mechanisms and cross-talk between different molecular networks. Particularly, little is known about the mechanisms of wave initiation. For cortical actin waves, molecular players that act upstream of actin, including Rho family GTPases and their activating lipid second messengers have been heavily investigated. These include Rac and phosphatidylinositol-3,4,5-trisphosphate (PIP3) in HL-60 neutrophils (Weiner et al., 2007), Rho in Xenopus eggs and starfish oocytes (Bement et al., 2015), PIP3 and phosphoinositide-3-kinase (PI3K) in Dictyostelium discoideum (Arai et al., 2010), and Cdc42 and PI3K in RBL-2H3 mast cells (Xiong et al., 2016). Because waves of Rho GTPase activation usually precedes actin waves and inactivation of these Rho GTPases could abolish actin waves, activation of Rho GTPases has been suggested to be involved in wave nucleation. Yet, the mechanisms that mediate the spatiotemporal activation of Rho GTPases in the cortical waves remain obscure. Mathematically, the nucleation of a patch of activation of GTPases has been modeled as molecular “noise” amplified by positive feedback mechanisms (Bement et al., 2015; Dyer et al., 2013; Holmes et al., 2012; Ozbudak et al., 2005).
A more recently identified group of actin-regulatory proteins involved in cortical waves is Fer/CIP4, Bin, amphiphysin, and Rvs (F-BAR) proteins of the Toca family, including FBP17, CIP4, and Toca-1 (Wu et al., 2013; Xiong et al., 2016). In mast cells, receptor activation by multivalent antigen leads to pronounced dynamic accumulation of F-BAR proteins and the formation of traveling waves. F-BAR waves are temporally and spatially synchronized with Cdc42 waves and precede actin waves (Wu et al., 2013). Based on in vivo and in vitro evidence, F-BAR proteins can participate in endocytosis. Specifically, knockdown of FBP17 impairs the endocytosis of epidermal growth factor and transferrin (Tfn) (Itoh et al., 2005; Kamioka et al., 2004; Tsujita et al., 2006). Loss of the only two members of the Toca family F-BAR proteins, Toca-1 and Toca-2, in Caenorhabditis elegans leads to severe endocytic and growth defects (Giuliani et al., 2009). How F-BAR proteins of the Toca family participate in endocytosis is less clear. FBP17 and CIP4 could be recruited to the late stage of growing clathrin-coated pits (CCPs) in NIH-3T3 cells (Taylor et al., 2011), but the percentage of CCPs with such recruitment has been shown to be vanishingly low in BSC1 cells (Henne et al., 2010). Nevertheless, the connection between F-BAR proteins and the endocytic machinery raises the question as to whether the endocytic process itself could also be collective and whether clathrin could be involved in the nucleation of cortical waves, which specifically refers to waves composed of Rho GTPases Cdc42, F-BAR proteins FBP17 and CIP4, and actin in this work.
Due to the accessibility of the plasma membrane, sensitive imaging methods, such as total internal reflection fluorescence (TIRF) microscopy, have facilitated the study of endocytosis, making it one of the most well-characterized cellular events in terms of its dynamics. Yet, the assembly of endocytic machinery is not known to be collective even though collective behaviour is predicted to be a generic phenomenon for self-propelled or active particles and filaments (Schaller and Bausch, 2013). Observations of CCP dynamics have given the impression that endocytic assembly is stochastic. When distances between neighbouring CCPs were specifically quantified, it was concluded that the distribution of CCPs was largely random (Ehrlich et al., 2004). This is also reflected by almost all existing studies on clathrin-mediated endocytosis (CME), which typically treat the assembly and disassembly cycles of CCPs as completely autonomous events.
Strictly speaking, not all visualization studies of clathrin are consistent with the random nucleation model. Endocytic hotspots, as indicated by multiple vesicles emanating from the same location, have been frequently observed. Occasionally a new CCP appears at the same location within minutes of the disappearance of an old CCP (Gaidarov et al., 1999), but the majority of the reported hotspots refer to multiple CCPs appearing from a single clathrin-coated structure (CCS) that never fully disappears (Saffarian et al., 2009). These large CCSs likely represent flat clathrin lattices that are not diffraction-limited in size. Successful endocytic events from these clathrin plagues have been inferred based on the recruitment of late stage markers, such as actin and Dynamin (Merrifield et al., 2005). The prevalent existence of endocytic hotspots suggests that different endocytic events may be temporally coordinated, but whether endocytic events from different CCSs could be coordinated is largely unknown.
Using secretory mast cells as a model, we investigated the dynamics of endocytic proteins and discovered that CCP assembly can be collective in the absence of stimulation or when a stimulus is uniformly present. We systematically monitored the temporal profiles of various modules of the endocytic process. Based on our observations, we propose that the assembly of the endocytic machinery mediates the onset of cortical waves. In contrast to a purely upstream role for endocytosis, the collective dynamics of endocytic assembly requires feedback from downstream factors of the clathrin machinery. We also reveal an unexpected role for intracellular clathrin in organizing spiral waves, suggesting that clathrin plays an additional role in modulating cortical excitability and wave geometry.
RESULTS
Coordinated Clathrin Waves Precede Waves of Active Cdc42 and FBP17
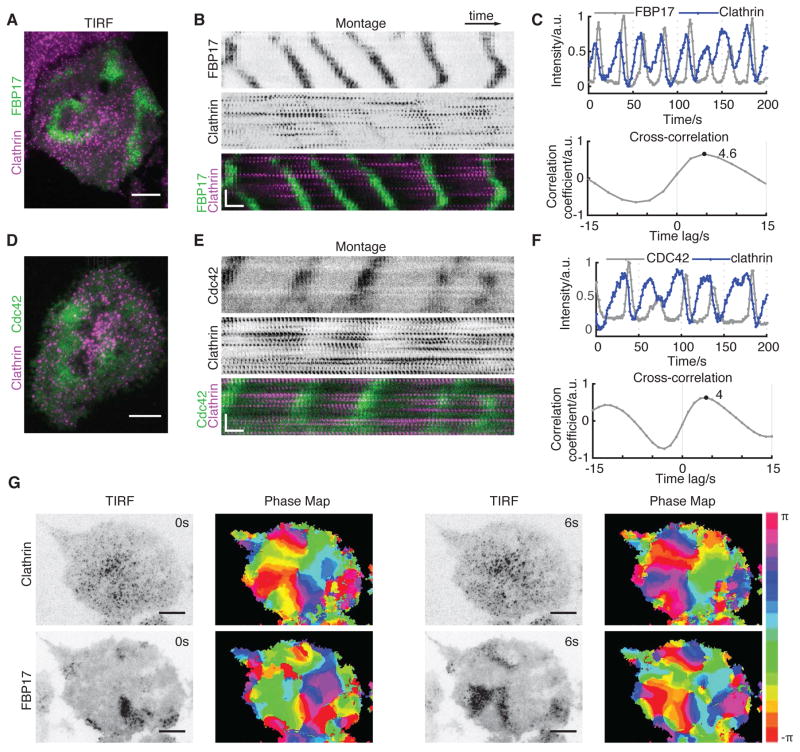
To determine whether traveling cortical waves are coordinated with cycles of endocytosis, we monitored the behavior of clathrin using TIRF microscopy. Because waves of FBP17, CIP4, and N-WASP are always coupled with each other and are present in all cortical waves, we interchangeably used them as generic markers of cortical waves. Individual CCSs displayed coordinated dynamics and appeared as traveling waves (Figure 1A and Movie S1). Although clathrin did not colocalize with FBP17 at any given time point, waves of clathrin and waves of FBP17 were temporally related (Figure 1B). Both shared the same frequency, and the peak of clathrin signal preceded that of FBP17 by 4.4 ± 0.6 sec, as determined by time correlation analysis using the intensity plots (Figure 1C). We further generated a phase map by extracting phase information using the time traces at individual pixels and a time-delay embedding technique (Gray et al., 1998). As shown by the phase map of clathrin and FBP17, their phase differences are consistent throughout the cell and independent of the region chosen. The phase map of clathrin at 0 sec closely matches the phase map of FBP17 at 6 sec (Figure 1G), providing a more clear demonstration of the correlation between the waves of clathrin and FBP17 despite their apparent lack of colocalization. Consistent with our previous findings that FBP17 and Cdc42 share the same phase in cortical waves, clathrin waves precede Cdc42 with a similar phase shift (Figures 1D–1F). Over-expression of clathrin light chain (CLC) in our experimental condition does not have obvious side effects on endocytosis as measured by Tfn uptake (Figure S1A). We also tagged endogenous CLC with CRISPR-cas9 system (Leonetti et al., 2016). These endogenously labeled clathrin displayed similar collective dynamics as the overexpressed clathrin (Figure S1B), confirming that clathrin waves are not artifacts of CLC overexpression.
Figure 1. Coordinated Clathrin Waves Precede Waves of Active Cdc42 and FBP17.
(A) Merged TIRF image of an RBL-2H3 cell stably expressing FBP17-EGFP (green) and transfected with mCherry-clathrin light chain (CLC) (magenta).
(B) Montages of FBP17 and clathrin waves in the cell shown in (A). A rectangular region of interest (ROI) of 7 ×102-pixel over 120 s is shown for montage.
(C) Representative intensity profile (top) of clathrin and FBP17 waves and cross correlation between the two intensity profiles (bottom) (n = 32 cells in 11 experiments).
(D) Merged TIRF image of an RBL-2H3 cell transfected with active CBD-EGFP (green) and mCherry-CLC (magenta).
(E) Montages of active Cdc42 and clathrin waves in the cell shown in (D). A rectangular ROI of 7 × 102-pixel over 120 s is shown for montage.
(F) Representative intensity profile (top) of clathrin and active Cdc42 waves and cross correlation (bottom) between the two intensity profiles (n = 13 cells from six experiments).
(G) Phase map of FBP17-EGFP and mCherry-CLC at two time-points with a 6 s interval. TIRF image and the corresponding phase map are side-by-side.
Inverted look-up table was used for single-channel TIRF images and montages. Scale bars represent 10 μm in all whole cell images and represent 10 s (horizontal), 5 μm (vertical) in all montages.
Plasma Membrane Localizations and Endocytic Nature of the Clathrin Waves
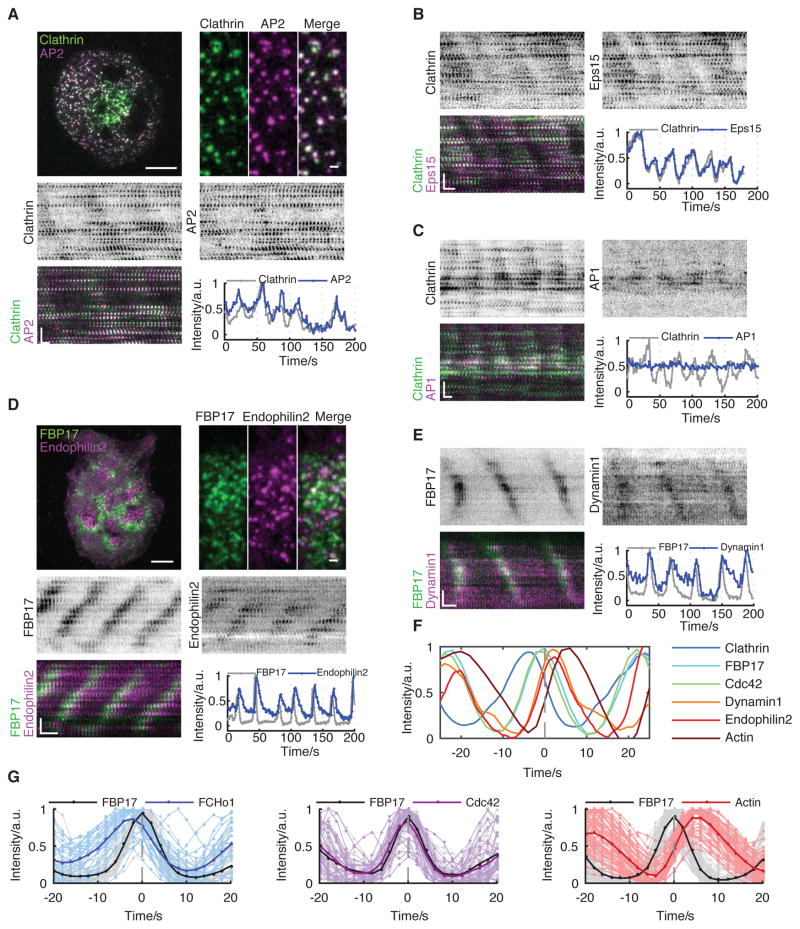
Clathrin is the most important protein in CME, but it is not exclusively localized to the plasma membrane. CCSs can also bud from the trans-Golgi network, late endosomes, and lysosomes. To test if clathrin waves are assembled on the plasma membrane, we monitored AP2, a known plasma membrane-specific adaptor for clathrin. AP2 shows wave dynamics and its phase of recruitment is identical to that of clathrin (Figure 2A). Such dynamics could be extended to other endocytic adaptor proteins, including FCHo1 (Figure S2B and Movie S2) and Eps15 (Figure 2B). Whereas clathrin and adaptor proteins colocalize at individual puncta in the traveling waves and share the same phase (Figures 2A, 2B), faster time-resolution could still reveal a phase shift between the recruitment of FCHo1 and clathrin in the waves, confirming that they are de novo nucleated pits instead of the membrane fluctuating in and out of the TIRF field (Figure S2A). In contrast, AP1, the intracellular adaptor of clathrin, did not form a wave pattern in cells with clathrin waves (Figure 2C), which is consistent with the plasma membrane localization of the waves.
Figure 2. Plasma Membrane Localizations of Clathrin Waves.
(A) Representative merged image, montage, and intensity profile of a cell co-transfected with CLC-GFP and AP2μ2-mCherry. Upper left, merged image of clathrin (green) and AP2 (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the colocalization. Scale bar represents 1 μm (n = 11 cells in four experiments).
(B) Representative montage and intensity profile of the cell co-transfected with CLC-GFP (green) and Eps15-mCherry (magenta) (n = 16 cells in four experiments).
(C) Representative montage and intensity profile of the cell co-transfected with AP1σ1-EGFP (magenta) and mCherry-CLC (green) (n = 18 cells in five experiments).
(D) Representative merged image, montage, and intensity profile of the cell co-transfected with mCherry-FBP17 and Endophilin2-GFP. Upper left, merged image of FBP17 (green) and Endophilin2 (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the localization. Scale bar represents 1 μm (n = 17 cells in three experiments).
(E) Representative montage and aligned profile of the cell co-transfected with mCherry-FBP17 and Dynamin1-GFP (n = 15 cells in four experiments).
(F) The average of the aligned intensity profiles of different endocytic modules. Each line is the average of multiple peaks from a single ROI from a representative cell. Each probe is aligned according to FBP17.
(G) Aligned intensity profile of FBP17 with that of proteins from early to late endocytic modules. Left, peaks of intensity profiles of FBP17 and FCHo1 (n = 251 ROIs from 20 cells). Middle, peaks of intensity profiles of FBP17 and Cdc42 (n = 5 ROIs from one cell). Right, peaks of intensity profiles of FBP17 and actin (n = 232 ROIs from 11 cells). For each probe, profiles of 5-10 ROIs from a representative cell over 5 min is aligned according to FBP17 and shown here.
Inverted look-up table was used in all single channel montages. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
It is theoretically possible that CCSs that participate in the waves are not productive CME, but abortive pits that disassembled without successful fission. Thus, we examined the late-stage markers of endocytosis, including Dynamin, Endophilin, and actin. The recruitment of Dynamin, the most well-known scission protein, and Endophilin, an N-BAR protein that favors the curvature of the CCP-neck and associates with Dynamin through its SH3 domain, implicates the fission step of CME. Endophilin2 (Figure 2D and Movie S3) and Dynamin1 (Figure 2E) were recruited around the same time as FBP17 but displayed longer lifetimes in the waves. In contrast, actin was recruited with a significant delay compared to FBP17 (Figure S2B). We use the recurring traveling waves to align multiple cycles of waves at their peaks and determine the phase difference. Using FBP17 as a common reference, we found that the phase progression of the endocytic protein waves was reminiscent of the previously characterized time course for CME (Figure 2F). These proteins could be classified into three groups: early module including AP2, FCHo1, Eps15, and clathrin; Cdc42 module including FBP17 and CIP4 acts after clathrin; and late module, including Endophilin2, Dynamin1, and actin (Figure 2G). The time delay was 3.8 ± 0.7 sec between Endophilin2 and FBP17 (n = 17 cells), 1.5 ± 1.2 sec between Dynamin1 and FBP17 (n = 15 cells), and 4.6 ± 0.6 sec between actin and FBP17 (n = 11 cells). FBP17 has the shortest lifetime, and a symmetrical assembly and disassembly profile. The early module had a slow assembly phase followed by a sharp decay phase, whereas the late module had the opposite profile: sharp onset and a slow decay phase. The phase-shifted recruitment of endocytic proteins confirms that these waves involve coordinated progression expected for endocytosis.
To further demonstrate the endocytic nature of the clathrin waves, we examined cargo loading in the waves. Transferrin receptor (TfR), a canonical CME cargo, and Glut4, which was reported to be internalized through CME pathway (Hartig et al., 2009), both exhibited coordinated waves colocalizing with clathrin waves at the level of individual puncta (Figure S2C).
Clathrin Waves Emerge from Coordinated Assembly and Act Upstream of Cortical Waves
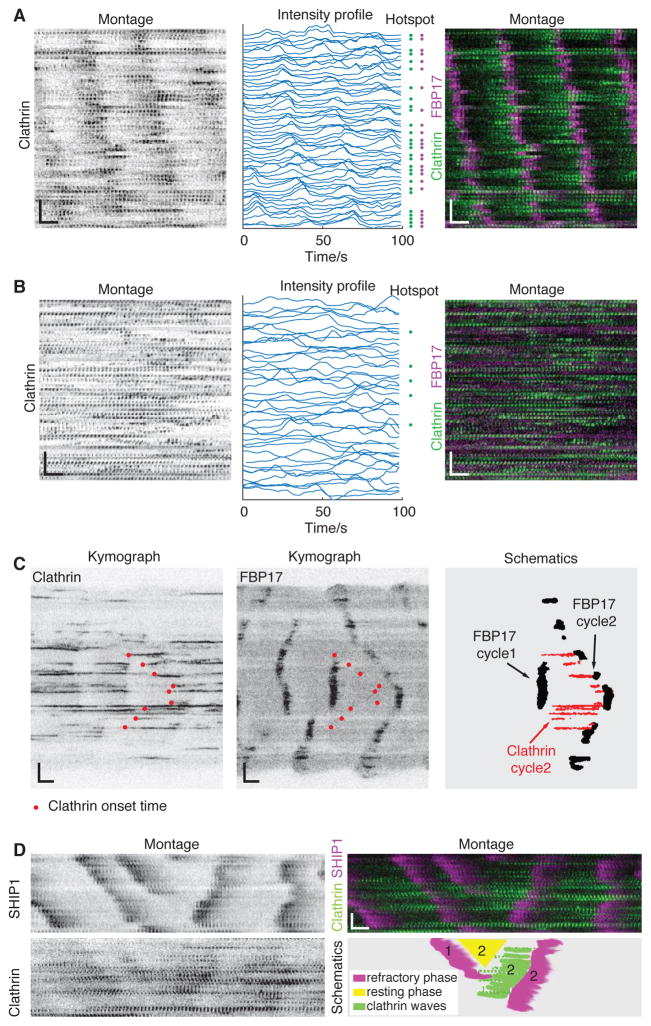
The sharp rise of the proteins involved in the late stage of endocytic assembly, including BAR proteins and actin, is synchronized with the onset of the decay phase for clathrin. The decay of the clathrin signal is consistent with the coordinated displacement of clathrin from the membrane due to its invagination. Since the decay phase of clathrin is visually more apparent in the movies, it raises the question whether the waves of clathrin displacement alone would generate the impression of clathrin waves. To determine whether coordinated assembly of early coat proteins is involved or if the apparent clathrin waves are passive, i.e., appear by random nucleation of CCSs that are synchronously displaced by actin waves, we carefully analyzed the single puncta that made up the clathrin waves. We sorted all the single CCSs according to their locations along the direction of wave propagation. We found that the majority of the clathrin that disassembled in waves also assembled in a coordinated fashion (Figure 3A). Notably, not all synchronized assemblies were de novo. A significant percentage of the CCSs (51.2 ± 7.2 %, n = 10 cells) in the waves originated from “hotspots” that support multiple, periodic budding events (Figure S3). A higher percentage of hotspots was observed in the clathrin waves (Figure 3A) than in cells without clathrin waves (22.0 ± 8.7 %, n = 6 cells) (Figure 3B).
Figure 3. Clathrin Waves Emerge from Coordinated Assembly and Act Upstream of Cortical Waves.
(A) CCPs in a single cell with clathrin waves sorted by the coordinates of the pits along the direction of wave propagation. Left, single channel CCP montage. Middle, corresponding line profiles of each single pit. Blue dots at the right side indicate the corresponding line is hotspot positive. Right, merged montage of FBP17 (magenta) and clathrin (green).
(B) CCPs in a single cell without clathrin waves sorted by the coordinates of the pits. Left, single channel CCP montage. Middle, corresponding line profiles of each single pit. Blue dots at the right side indicate the corresponding line is hotspot positive. Right, merged montage of FBP17 (magenta) and clathrin (green).
(C) Kymographs of diverted waves of mCherry-clathrin (left) and the corresponding FBP17-EGFP (middle) and a schematic illustration of the relationship between a cycle of clathrin waves (red) with two neighboring cycles of a FBP17 wave (black) (right). Red dots in kymographs indicate the initiation points of CCPs in the diverted waves.
(D) Montages of diverted waves of CLC-GFP (upper left), the corresponding mCherry-SHIP1 (lower left), and the merge of the two with CLC in green and SHIP1 in magenta (upper right), and a schematic illustration of the relationship between a cycle of clathrin waves (green) with two neighboring cycles(‘1’ marks the previous cycle and ‘2’ marks the current cycle) of refractory phase markers (magenta) and a resting phase (yellow) (lower right).
Inverted look-up table was used in all single channel montages. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
As further evidence arguing against the finding that apparently coordinated assembly can result passively from the synchronized disassembly in the previous cycle, in which case assembly restarts together due to a fixed refractory period, we examined the assembly trace of clathrin when cortical waves altered their direction of propagation. Changes in the direction of propagation are due to the difference in the oscillation period at neighboring loci. In kymographs of FBP17 waves, these events appear as stripes of different slopes (Figure 3C). We used these events to determine how the onset of coordinated clathrin assembly correlates with the consecutive pulses of FBP17 recruitment. With clearly divergent FBP17 waves, the corresponding clathrin assembly correlated with FBP17 in the subsequent cycle, but not the previous one, as illustrated in the simplified schematic picture (Figure 3C).
We previously showed that SHIP1, a PIP3 5-phosphatase is a molecular marker for the refractory phase of FBP17 waves (Xiong et al., 2016). To determine whether the difference in time of the clathrin onset was determined by the length of the refractory phase in the previous cycle, we imaged SHIP1 together with clathrin. As shown in Figure 3D, loci with longer oscillation periods displayed similar refractory phases compared to those with shorter periods. The difference in the timing in the subsequent cycle of FBP17 oscillation is mainly due to the length of the resting phase (Figure 3D). In other words, the relative timing of clathrin assembly appears to determine the relative phases of F-BAR waves; hence, the apparent change in the direction of wave propagation.
Clathrin Acts Upstream of Cortical Waves
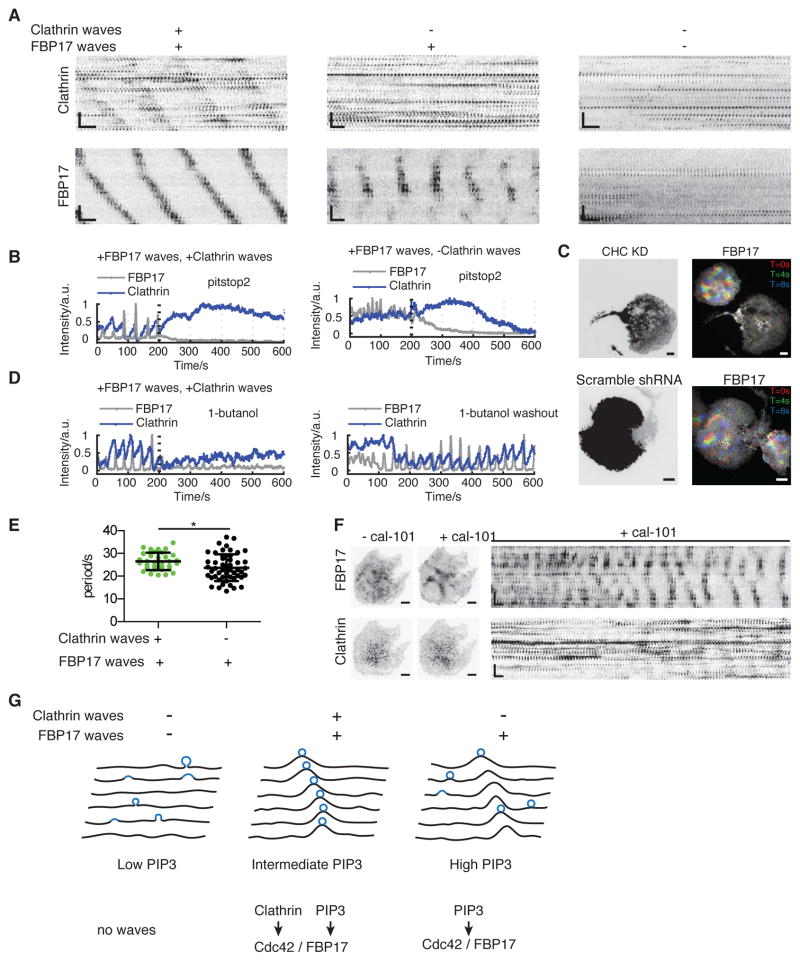
One observation that is inconsistent with the absolute requirement of clathrin waves in nucleating cortical waves, however, is the presence of cells with cortical waves but no apparent coordination of clathrin (Figure 4A). While the majority of cells shows FBP17 waves and actin waves (95.0%, n=161 cells), clathrin waves were only visible in about 25% of the cells with cortical waves (76/310 cells exhibiting FBP17 waves). One explanation for this observation is that there could be both clathrin-dependent and independent mechanisms of wave nucleation (Schroth-Diez et al., 2009). We therefore examined whether cortical waves in the absence of clathrin waves depend on clathrin. Pharmaceutically inhibiting clathrin function using the chemical inhibitor, Pitstop 2 (Kleist et al., 2011), abolished FBP17 waves instantly regardless of whether clathrin waves were visible in the same cell (Figure 4B, Movie S4). Genetically, knocking down clathrin-heavy-chain (CHC) greatly inhibited FBP17 waves formation (33.9% exhibiting waves, n=62 cells, compared to 89.3% in control cells, n=140 cells) (Figure 4C, S4A). Scramble shRNA had no effect (96.4% cells exhibiting FBP17 waves, n=56 cells) (Figure 4C). To confirm that not only clathrin but also successful endocytosis is required, we used Dynamin1-K44A, a dominant negative (DN) mutant of Dynamin1, to inhibit endocytosis. Cells expressing Dynamin1-K44A were greatly inhibited in forming waves of FBP17 (31.7% exhibiting waves, n=60 cells) and clathrin (1.7% exhibting waves, n=60 cells) (Figure S4B). To examine whether the dependence on endocytosis was acute and reversible, we used 1-butanol, which decreases the nucleation of CCPs (Boucrot et al., 2006). Waves of FBP17 that were coupled or uncoupled with clathrin waves were both abolished upon the addition of 2% (v/v) 1-butanol, and such effects were reversible when 1-butanol was washed out (Figure 4D). In order to determine whether the requirement of clathrin in cortical waves could be generalizable in other cortical waves, we examined whether clathrin function affected chemoattractant-induced mHem1 waves in HL-60 neutrophils. Inhibiting clathrin function with Pitstop 2, acutely abolished formation of mHem1 waves (9 out of 9 cells), suggesting that clathrin also functions upstream of the collective dynamics involving WAVE2 complex in neutrophils (Figure S4C,S4D).
Figure 4. Clathrin Acts Upstream of Cortical Waves, Independently of Whether Clathrin Has Waves.
(A) Representative montages of stably expressed FBP17 and transiently co-expressed clathrin in cells with different combinations of clathrin waves and FBP17 waves. Cells with no cortical waves (right) were rare after stimulation and were not considered in this study.
(B) Intensity profiles of the inhibitory effect of Pitstop 2 on waves of FBP17-EGFP in cells with (n = 6 cells in three experiments) or without (n = 13 cells in four experiments) waves of mCherry-CLC. 7 μM (final concentration) Pitstop 2 was added at the 200th second (dashed line) (n = 6 cells in three experiments).
(C) Images of representative cells with or without CHC knockdown (KD). Cells stably expressing FBP17-EGFP were transfected with CHC-shRNA in RFP expressing vector (upper row) or scramble shRNA in RFP expressing vector (lower row) as control. Left: Expression of the RFP. Right, corresponding time projection of FBP17 waves of 3 time points. Inverted look-up table was used in all montages and single time point images. Scale bars represent 10 μm in all whole cell images and represent 10 s (horizontal), 5 μm (vertical) in all montages.
(D) Intensity profiles of a movie in which waves of mCherry-CLC and FBP17-EGFP terminate upon the addition of 1-butanol and recover after washout of 1-butanol. 2% (v/v) 1-butanol was added at the 200th second (dashed line), and washout was performed right before the movie on the right (n = 15 cells in three experiments).
(E) Quantification of the average cycle time of FBP17 waves in cells with or without clathrin waves (mean ± STD., n = 33 and 51 for each category, *P = 0.0182, t-test).
(F) Before-and-after comparisons and montage of a cell with clathrin waves treated with 800 nM cal-101 (n = 9 cells from eight experiments).
Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical). Inverted look-up table was used.
(G) Cartoon illustrating formation of clathrin wave at the intermediate level of PIP3.
PIP3 Levels Determine the Visual Appearance of Clathrin Waves
To reconcile the requirement of clathrin in cortical wave formation but the apparent appearance of clathrin waves only in a subset of the cells, we hypothesized that the requirement of clathrin for cortical wave nucleation is indirect. The formation of CCPs could generate an intermediate species which favours the nucleation of cortical waves, leading to local coupling between clathrin and FBP17, but such spatial connections could be bypassed if the concentration of the intermediate species on the plasma membrane is sufficiently high. To explore the molecular link between CCP and FBP17, we compared FBP17 waves in the presence or absence of coupled clathrin waves. We found that the oscillation period is statistically longer in cells with clathrin waves (27.9 ± 3.5 sec, n = 34 cells) compared to cells without clathrin waves (20.7 ± 4.4 sec, n = 51 cells, p<0.05) (Figure 4E). Particularly, cortical waves with periods shorter than 20 sec (frequently observed within 15 min after antigen stimulation) typically do not have clathrin waves in the same cell (Figure 4E). Since PIP3 is involved in setting the periodicity of FBP17 waves and that PI3K inhibitor can decrease the wave frequency (Xiong et al., 2016), we reasoned that cells without clathrin waves could contain slightly higher levels of PIP3. To test whether PIP3 could link clathrin and FBP17, we lowered the concentration of PIP3 by adding the PI3K inhibitor, cal-101, to cells with no clathrin waves (high FBP17 oscillation frequency). As the PI3K inhibitor slowed down the frequency of the waves, clathrin waves emerged (Figure 4F, Movie S5). These results indicate that the onset of clathrin waves require an intermediate level of PIP3 (Figure 4G, Figure S5). This is reminiscent of the FBP17 waves (Xiong et al., 2016), but the range of PIP3 levels required for coordinated clathrin assembly appears to be narrower than that of FBP17.
Coordinated Clathrin Assembly Requires Feedback from Cdc42/FBP17/N-WASP Module
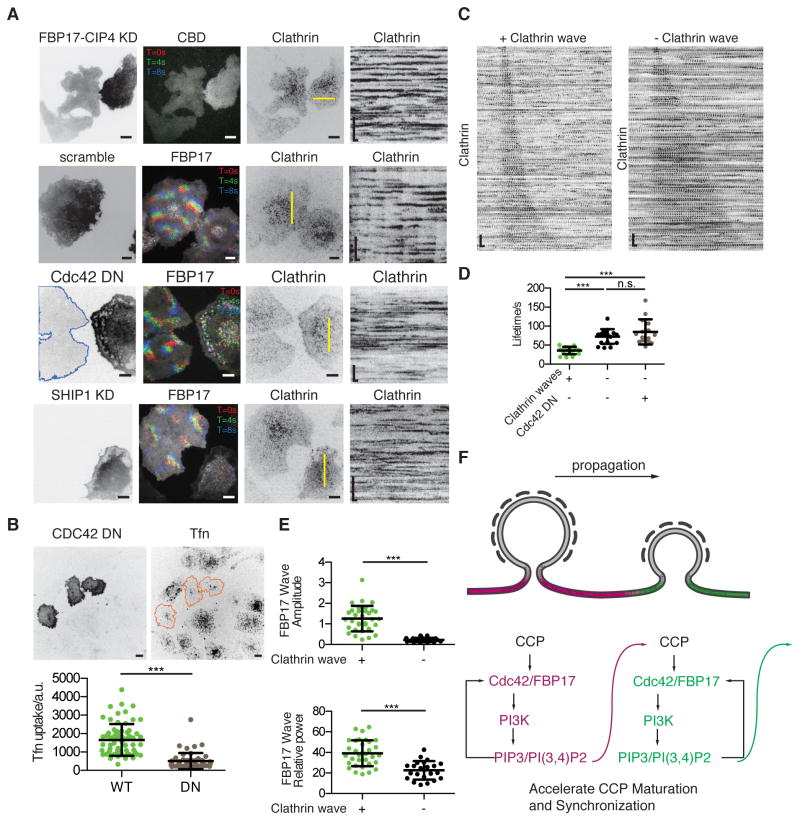
Next we examined the mechanisms through which clathrin becomes synchronized at the intermediate PIP3 level. Considering the greater percentage of hotspots in cells with clathrin waves than in cells without clathrin waves (Figure 3A, 3B, S5), we hypothesized that clathrin assembly functions as an oscillatory reaction on the plasma membrane. These local oscillations are weakly coupled and can be coordinated in the presence of positive feedback and mutual entrainment. Induction of clathrin waves in cells with FBP17 waves indicates that FBP17 waves may have a retrograde effect on clathrin. To test whether FBP17 represents part of the positive feedback for clathrin to be synchronized into waves, we knocked down FBP17 and CIP4, the only two Toca family F-BAR proteins expressed in RBL-2H3 cells according to our RNA sequencing data (Figure S6A). Double knockdown of FBP17 and CIP4, but not single knockdown of FBP17 alone, inhibited the formation of Cdc42 waves (15.4%, n=52 cells, compared to ~90% in controls) (Wu et al., 2017), and all of the clathrin waves (0%, n=52 cells) (Figure 5A). We also visualized clathrin dynamics in cells co-transfected with Cdc42-T17N, a dominant negative mutant of Cdc42. In all imaged cells with positive Cdc42-T17N signals, clathrin and FBP17 were less dynamic and no clathrin waves or FBP17 waves were visualized (n=19 cells) (Figure 5A). CCP formation was not abolished by Cdc42 inhibition, but they appeared random with Cdc42 inhibition. Furthermore, wave abolishment by overexpressing Cdc42-T17N greatly impaired CME as indicated by Tfn uptake (Figure 5B), suggesting that this positive feedback plays an important role in clathrin-mediated cargo uptake. Lastly, knocking down SHIP1, the delayed inhibitor important for the oscillatory dynamics of FBP17 waves, inhibited clathrin waves as well (90.7%, n=53 cells) (Figure 5A). To compare clathrin dynamics in the same cell, we used N-WASP inhibitor Wiskostatin since N-WASP is in the same phase as Cdc42 and FBP17. Clathrin waves stopped instantly upon the addition of Wiskostatin (Figure S6B). All these demonstrate that feedback from downstream FBP17/Cdc42 module is necessary for clathrin wave formation.
Figure 5. Coordinated Clathrin Assembly Requires Feedback from Cdc42/FBP17/N-WASP module.
(A) Images and kymographs of representative cells of clathrin waves under downstream purterbation. In each row, cells with or without expression of FBP17-shRNA and CIP4-shRNA in RFP expressing vectors, scramble shRNA in RFP expressing vectors, mCherry-Cdc42-DN, SHIP1-shRNA were co-expressed with iRFP-CLC in cells stably expressing FBP17-EGFP or GFP-CBD (for double knockdown (DKD)). Cells with very low (contoured with blue line) or no expression in the same field are controls. All kymographs were generated by projecting kymographes of 10 adjacent pixels at the yellow lines in the third column.
(B) Representative confocal images and quantification of Tfn uptake in CDC42 DN over-expressing cells. Upper row shows the Lyn-tailed mCherry-CDC42 T17N (added Lyn-tail for better membrane localization and wave inhibition) expression on the left and the corresponding 20min Tfn uptake image on the right. Three cells with positive CDC42-DN signal were circled out with orange line. Intensity of Tfn in background subtracted maximum projection image of whole cell were used for quantification shown in the bottom (mean ± STD, n = 67 and 52, respectively, for each category, ***P < 0.001, t-test).
(C) Representative CCP montages sorted by CCP lifetime in a cell with clathrin waves compared to a cell without clathrin waves (but with FBP17 waves). All CCSs in the movie of the cell over 5 min are identified and sorted, but only 100 were selected arithmetically from top to bottom (listed here).
(D) Quantification of the median of the CCP lifetime distribution in cells with and without clathrin waves (mean ± STD, n = 19 and 20, respectively, for each category, ***P < 0.001, t-test).
(E) Quantification of the average amplitude and relative power of FBP17 waves in cells with and without clathrin waves (mean ± STD, n = 33 and 22, respectively, for each category, ***P < 0.001, t-test).
(F) Cartoon illustrating the hypothesis of how retrograde effect from Cdc42/FBP17 module could mediate the coordination between CCPs.
Inverted look-up table was used in all single-channel and single-time point images, and singel channel kymographs/montages. Scale bars in all whole cell images represent 10 μm and in kymographs/montages represent 10 s (horizontal) and 5 μm (vertical) unless otherwise stated.
To understand the effect of the FBP17/Cdc42 module on clathrin that could mediate its synchronization, we quantified the lifetime of individual CCSs. For oscillatory clathrin dynamics, we found that the lifetime of clathrin matches well with the cycling time of the oscillations. The lifetime of CCSs in cells that exhibit clathrin waves (medium: 37.4 ± 11.1 s; mean: 32 + 42 s) is consistently shorter than in cells without clathrin waves (72.5 ± 18.3 s, mean 48 + 38 s) (Figures 5C, 5D). In any synchronization system, the frequency distribution of single oscillators needs to be sufficiently narrow in order for synchronization to emerge (Strogatz and Stewart, 2001). Consistent with this, the variations of the lifetime of clathrin in cells from the above two categories is also different, with synchronized clathrin exhibited less variation (Figure 5C, 5D). To examine whether feedbacks from FBP17/Cdc42 module could account for the differences in clathrin lifetime, we measured lifetime of clathrin in cells expressing Cdc42-T17N. Cells expressing Cdc42-T17N assembled CCPs with longer lifetime, which was not significantly different from cells without clathrin waves (Figures 5D). Cdc42 is known to stimulate PI3K activity, which could modulate the lifetime of CCPs (Nakatsu et al., 2010). To test whether activation of PI3K could change clathrin dynamics in this system, we used CRY2-CIBN based optogenetics to recruit PI3K to the plasma membrane. We found that activation of PI3K can increase the frequencies of clathrin waves (Figure S6C), similar to what has been reported for individual CCPs (Nakatsu et al., 2010).
If collective dynamics of clathrin appears because of the retrograde effects from the Cdc42/FBP17 module, why would FBP17 waves fail to synchronize clathrin at high PIP3 level? We found that the spatial connection between clathrin and FBP17 was reduced in cells without clathrin waves. For cells with high PIP3 levels, FBP17 could assemble away from clathrin and form waves. Quantification shows that 36.0±10.1% (n=10 cells) of the CCSs in cells exhibiting clathrin waves recruit FBP17 at the late stage (Figure S6D). Among them, a great majority of the clathrin hotspots in clathrin waves were also FBP17 hotspots (85.0 ± 7.2 %, n=10 cells) (Figure 3A). In contrast, in cells with FBP17 waves but no clathrin waves, only 10.3±6.8% (n=5 cells) of the CCSs recruited FBP17 at the late stage. Besides the reduced physical proximity, the amplitude and power of FBP17 waves were significantly lower when they were not coupled with clathrin waves (Figure 5E). These results suggest that clathrin wave formation, but not clathrin assembly itself, requires a minimum level of activation that is dependent on FBP17/Cdc42/N-WASP and their connection with clathrin. The retrograde effect of FBP17/Cdc42 module on clathrin therefore manifest as a local but non-autonomous effect that could mediate the communications of the neighboring CCPs (Figure 5F).
The Clusters of Long-lived Clathrin Organize Wave Geometry
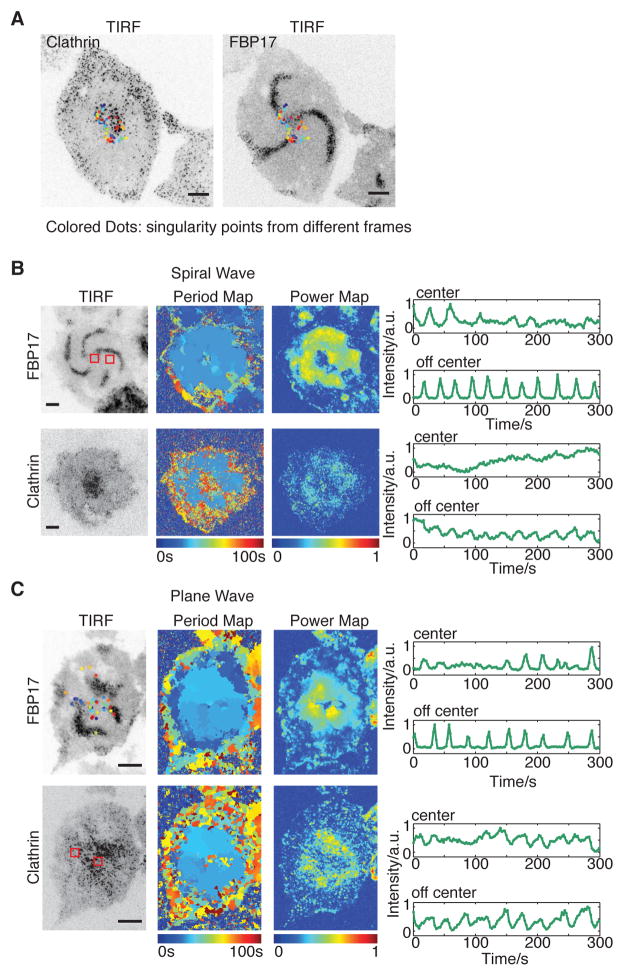
If clathrin could regulate the onset of cortical waves, we wondered whether it could also be involved in organizing cortical wave geometry. This involvement is the most striking in the case of spiral waves (Figure 6A, Movie S6) where there is a clearly identifiable “vortex core” in the neighborhood of the phase singularity that organizes the rotating waves (Winfree, 1989). In the frequency map and the power map generated from probes of both FBP17 and clathrin, the vortex core can be easily identified as the region with less power due to the reduced amplitude of the oscillation within the cortex core (Figure 6B). When the singularity points (identified in phase map as points whose surrounding phases add up to zero) of the spiral waves were projected into a single frame, they were concentrated in the vortex where clathrin formed clusters (Figure 6B). These long-lived clathrin signals are also visible in cells without striking spirals (Figure 6C). Although they do not participate in the waves, these smaller cluster had lower oscillation amplitude, and were highly enriched in singularity points too.
Figure 6. Clusters of Long-lived Clathrin Organize Cortical Wave Geometry.
(A) Singularities generated through out 5 min projected onto the same frame of clathrin/FBP17.
(B) Representative TIRF image, frequency map, power map, and intensity profiles of cells with armed waves. mCherry-CLC was transiently expressed in cells stably expressing FBP17-EGFP. Intensity profiles of the ROIs in and off center are separately plotted. ROIs are indicated as red squares in the power map of FBP17.
(C) Representative TIRF image, frequency map and power map of FBP17 waves in cells with plane clathrin waves. Colored dots on TIRF image indicates singularity points from different frames.
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm.
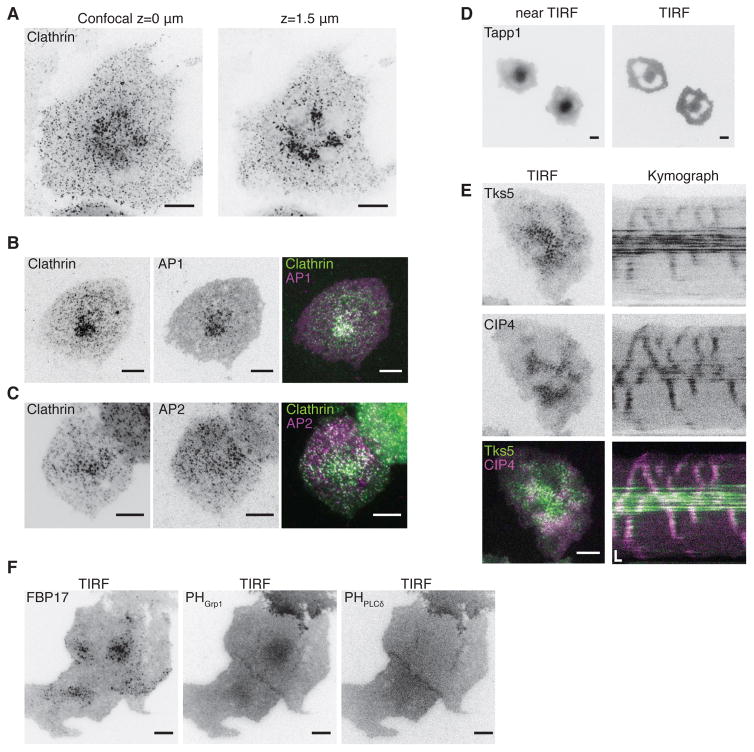
Clathrin clusters are different from clathrin waves that precede cortical waves. When imaged by spinning disk confocal fluorescence microscopy, the contrast of the cluster becomes higher when the distance from the substrate increases (Figure 7A), indicating that the cluster is localized intracellularly but that it could be in the vicinity of the plasma membrane. Consistent with its intracellular localization, AP1, but not plasma membrane-localized AP2, colocalizes with the clustered clathrin signals (Figure 7B–C). FBP17 also decorates this cluster of punctae (Figure S7A). Both FBP17 and clathrin puncta in the cluster are resistant to PI3K inhibition (Figure S7B, Movie S7).
Figure 7. The Intracellular Pool of Clathrin and PI(3,4)P2 defines a Non-excitable Vortex Core.
(A) Confocal view of the center cluster of mCherry-CLC. Left, bottom surface. Right, 1.5 μm above the surface.
(B) TIRF image of a cell transfected with mCherry-CLC and AP1σ1-EGFP. Right, merged image of CLC (green) and AP1 (magenta).
(C) TIRF image of a cell transfected with CLC-EGFP and AP2μ2-mCherry. Right, merged image of CLC (green) and AP2 (magenta).
(D) Near-TIRF (left) and TIRF (right) view of RFP-PH-Tapp1. Near-TIRF is achieved by elevating the incident angle.
(E) Representative TIRF images and kymograph of Tks5-GFP (top) and mCherry-CIP4 (middle) in the center cluster in a cell bearing waves. Bottom, merged image of Tks5 (green) and CIP4 (magenta).
(F) TIRF image of the cell co-expressing FBP17-EGFP (left) and mCherry-Grp1-PH (middle) and iRFP-PH-PLCδ (right).
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm. Scale bars in kymographs represent 10 s (horizontal) and 5 μm (vertical).
The appearance of non-excitable vortex cores indicates that there might be differences on the plasma membrane in terms of local excitability. The region on the plasma membrane opposing the intracellular cluster could have a longer refractory period than the rest of the membrane. Previously we showed that PI(3,4)P2 defines the refractory period of the oscillation (Xiong et al., 2016). Consistent with this finding, we found an aggregation of PI(3,4)P2 at the cluster (Figure 7D). Similarly, there is a strong enrichment of the PI(3,4)P2 binding proteins, Tks5 (Figure 7E) and PIP3, precursors of PI(3,4)P2 (Figure 7F). In contrast, there is a lack of enrichment of phosphoinositide-4,5-biphosphate (PI(4,5)P2) at the cluster (Figure 7F). While the PI(3,4)P2 cluster is clearly separated from the plasma membrane pool of PI(3,4)P2, its close proximity to the membrane suggests that a mechanism to dynamically regulate the local level of PI(3,4)P2 exists, which dictates local refractories and ultimately wave geometry.
DISCUSSION
Here, we report the existence of previously unrecognized collective dynamics of the endocytic machinery. Waves of endocytosis suggest that a non-autonomous mechanism for clathrin assembly exists that is different from the previously established model for endocytosis for which the assembly of endocytic pits is random and spatially uncoordinated. Our results suggest that CCPs can be coordinated with neighbouring pits. Such coordinated assembly, together with the mechanisms that rely on the spatially biased presentation of ligands for inducing endocytosis, could be important for controlling subcellular endocytic activity with spatial specificity. The relative timing of clathrin nucleation apparently set the propagation path for cortical waves, providing what is currently known as the earliest spatial cue for wave nucleation.
The coexistence of the collective dynamics of clathrin, F-BAR proteins, Cdc42, and actin raises the question as to which is the driving force for the synchronization of these activities. Intuitively, since clathrin is the most upstream factor in terms of its temporal recruitment within the periodic waves, the coordination of clathrin assembly may be the driving force, and one could speculate that dynamic gradients of ligands could be present that may affect the nucleation of clathrin. Surprisingly, although the assembly of clathrin precedes that of Cdc42/F-BAR in a given cycle, Cdc42/F-BAR waves could happen in the absence of coordinated clathrin assembly. The partial correlation of CCPs with the nucleation of actin waves has also been reported in Dictyostelium (Schroth-Diez et al., 2009), suggesting that the emergence of Cdc42/F-BAR waves could drive the synchronization of clathrin. Thus, we propose that the coordination of clathrin assembly stems from the positive feedback between clathrin assembly and its downstream factors, including Cdc42/F-BAR, and may not require pre-determined chemical gradients. The onset of clathrin synchronization could be analogous to how people on the Millennium Bridge could involuntarily walk in sync. Clathrin assembly precedes that of Cdc42/F-BAR (Figure 1A–F), and it takes place independently of Cdc42/F-BAR (Figure 5D), similar to how individuals could walk prior to, and independently of, the swinging of the bridge. Under normal conditions, there is no coordination between different clathrin assembly events, in the same way that individuals walk at their own pace. Synchronization could occur due to the positive feedback between the natural sway motion of people and that of the bridge.
Several positive feedback mechanisms between clathrin and F-BAR proteins could potentially promote clathrin synchronization. One possible scenario involves chemical coupling. Our results indicate that the Cdc42/F-BAR/N-WASP module is part of the feedback mechanism, which could be important for nucleating new clathrin assembly in the neighborhood of an existing CCP. This action could be facilitated by the local production of lipids that enhance CCP nucleation. One candidate lipid is PI(3,4)P2. If FBP17 recruits SHIP1, as we previously showed (Xiong et al., 2013), it will lead to the local production of PI(3,4)P2. Interestingly, PI(3,4)P2 has been shown to modulate the lifetime of CCPs in endocytosis (Nakatsu et al., 2010; Posor et al., 2013). The common rate-limiting lipid also reinforces the connection of cortical oscillations with the cycle of endocytosis and suggests that turnover of PI(3,4)P2 could serve as the mechanism that provides positive feedback between FBP17 and clathrin.
An equally attractive, but not mutually exclusive, possibility involves mechanical feedback. F-BAR proteins have been shown to localize to the tubular invagination connected to the CCPs in cell-free systems of endocytosis (Wu et al., 2010). The close proximity of F-BAR proteins with clathrin as well as their mutual segregation are consistent with the behaviour of F-BAR protein Bzz1 in Saccharomyces cerevisiae (Kishimoto et al., 2011), and F-BAR protein Bzz1 and Cdc15 in Schizosaccharomyces pombe (Arasada and Pollard, 2011). Although deletion of these F-BAR proteins in yeast has relatively subtle effects on endocytosis, genetic deletion of the F-BAR protein, Bzz1, together with the BAR protein, Rvs167, in S. cerevisiae leads to retraction of endocytic intermediates (Kishimoto et al., 2011), indicating that F-BAR proteins could play a role in the mechanical stability of CCPs. Besides the neck region, a second pool of diffusive FBP17 colocalizes with active Cdc42 in the region between different pits (Wu et al., 2013). In principle, these populations of FBP17 could modulate the regional mechanical property of the membrane, as recently proposed for the leading edge during cell migration (Tsujita et al., 2015). Positive feedback between membrane deformation and clathrin assembly during its budding, although the mechanism is unknown, seem plausible considering that curvature-mediated cooperativity has been suggested to function in virus budding (Reynwar et al., 2007).
Complete or partial synchronization by entrainment of a series of spatially arranged oscillators that are weakly coupled is a different mechanism instead of quorum sensing, where the system turns from a quiescence to an oscillatory state once a density threshold has been reached, that could explain the onset of synchronization (Gregor et al., 2010; Taylor et al., 2009). Here, prior to the formation of clathrin oscillations, clathrin also shows cycles of assembly and disassembly, which can be considered an excitable behaviour, but this excitability does not have a regular rhythm in time, nor are they coordinated in space. During oscillatory clathrin assembly, we observed a high percentage of hotspots where multiple clathrin budding cycles can occur at the same location. These hotspots have been observed in various cell systems, but their functions remain largely obscure. It is likely that they conveniently convert excitable clathrin assembly into localized oscillatory reactions and facilitate their mutual entrainments.
Theoretically, in order for the weakly coupled oscillator to synchronize, the distribution of their natural frequency needs to be sufficiently narrow (Strogatz and Stewart, 2001). If the frequency distribution is too broad, each oscillator will receive input from its neighbours, some of which some have higher frequencies than others. These mixed messages will prevent synchronization. Consistent with this notion, in oscillatory clathrin waves, we observed more uniform distribution of clathrin lifetimes compared to those in cells without clathrin waves (Figure 5C). Similarly, there is a requirement for the oscillation frequency and amplitude of FBP17 waves necesary to observe clathrin synchronization. There is an optimal range of frequency and minimal amplitude that is permissive for the onset of the clathrin pattern. We interpret these results as evidence that there is a minimal coupling strength of clathrin that is mediated by FBP17 and its interacting proteins, such as Cdc42 and N-WASP.
Our investigation of the dynamics of endocytic proteins also revealed a surprising role for the intracellular assembly of clathrin in defining the geometry of cortical waves, particularly the rotation vortex of spiral waves. Spiral waves, similar to plane waves or concentric target waves, are characteristic of the excitable and oscillatory systems. Compared to target waves, which require pacemakers for wave initiation, spiral waves are self-sustainable and are organized by the vortex core (Pálsson et al., 1997). Wave geometry could be biologically important. In the heart, the appearance of spiral waves has been linked to heart failure (Winfree, 1989). The conversion between target waves and spiral waves has been studied in the multicellular aggregates of Dictyostelium (Siegert and Weijer, 1995). Rotating spiral waves usually entrain the concentric waves with spontaneously firing centres. Ultimately, spiral cores serve as aggregation centres (Lee et al., 1996). In certain mutants, there are only spirals, and such mutants ultimately display developmental pathway defects (Lauzeral et al., 1997; Pálsson and Cox, 1996). In single cell patterns, wave geometry was proposed to be critical for cell morphodynamics (Taniguchi et al., 2013). Little is known about the molecular organization of the singularity points and vortex core. Our observations suggest that spatial features that define the spiral core do not have to come from the plasma membrane itself, rather it is due to the close proximity of the intracellular pools of clathrin and PI(3,4)P2. Collectively, our findings highlight the versatility of the endocytic machinery in regulating cortical dynamics.
EXPERIMENTAL PROCEDURES
Cell Culture and Transfection
RBL-2H3 cells(ATCC) were cultured in Minimum Essential Medium with Earles’ Salts (MEM) (Invitrogen 11095-098) with 20% Fetal Bovine Serum (FBS) (Sigma F4135-500ml) and 50 μg/ml Gentamicin (Invitrogen 15750-060) at 37°C and 5% CO2. Transfections were carried out with the Neon transfection system (Invitrogen) at 1200 pulse voltage, 20 ms pulse width, and 2 pulse number, with 1 μg plasmid. After transfection, cells (2 × 106) were seeded on 35 mm glass bottom dishes (MatTek) and sensitized with 0.4 μg/ml anti-Dinitrophenyl (DNP)-IgE (Sigma D8406-.5MG) and cultured overnight. For CHC knockdown, a total of 3 μg shRNA was used for each transfection and cells were cultured for 72 h before imaging. Stable RBL-2H3 cell lines of FBP17-EGFP were generated as previously described (Xiong et al., 2016). Endogenous clta gene (Clathrin light chain A) was tagged with GFP based on clustered regularly interspersed short palindromic repeat associated protein 9 (CRISPR/Cas9) method previously described (Leonetti et al., 2016). Briefly, Cas9/single-guide RNA (sgRNA) ribonucleoprotein complexes (RNPs) together with homologous-directed repair (HDR) coding GFP11(the 11th beta strand of GFP) were electroporated into single clonal RBL-2H3 cells stably expressing GFP1-10 (GFP without the 11th beta strand). Cells with positive GFP signal were selected with fluorescence-activated cell sorting (FACS) as cells with GFP tagging endogenous clta. The sequence of DNA oligo for clta specific sgRNA synthesis is TAATACGACTCACTATAGGTTGGGCCGTCATGGCTGAGTGTTTAAGAGCTATG CTGGAA. GFP11 HDR donor sequence was G GCG GGG TGT CGG CGG ATC GGT CGG TTG GTT TCG GCT TCG GTG TCG CTG CGT TTG AGC TGG GCT GTA ATG CGT GAC CAC ATG GTC CTT CAT GAG TAT GTA AAT GCT GCT GGG ATT ACA GGT GGC GGC GCA GAA TTA GAT CCA TTT GGC GCT CCC GCC GGT GCG CCC GGT GGC CCC GCG CTC GGG AAC GG. RNA extraction and RNA-seq library preparation were previously described (Xiong et al., 2016). HL-60 cells stably expressing mHem1 fused to mNeonGreen were grown in RPMI-1640 media supplemented with L-glutamine and 25 mM HEPES (Mediatech) and containing 10% (v/v) heat-inactivated fetal bovine serum (Gibco). Cultures were maintained at a density of 0.2–1.0 million cells/mL at 37°C/5% CO2. Differentiated HL-60 cells were obtained by adding 1.5% (v/v) DMSO (Sigma-Aldrich) to actively growing cells followed by incubation for an additional 5 days.
Plasmids and Chemicals
The sources of the plasmids used are as follows: Cltc (clathrin heavy chain) - Rat, 2 unique 29mer shRNA constructs in retroviral RFP vector (Origene Technologies, Inc, cat.no. FI741227: GCAGAAGTGGTTGCTCCTGACTGGCATAT, and FI741228: GGTGTTCCTGATTACCAAGTATGGTTATA), mCherry-CLC (Addgene #27680), AP2μ2-mCherry (Addgene #27672), Eps15-mCherry (Addgene #27696), EGFP-EPSIN1 (Addgene #22228), EGFP-Glut4 (cloned from Addgene #18087), TfR-pHuji (Addgene #61505), Dynamin1-K44A-EGFP (Addgene #22197) AP1σ1 (Lei Lu, Nanyang Technological University), mRFP-CBD (Addgene #26733), Dynamin1-GFP (Addgene #22163), Tks5-GFP (B. Diaz, Sanford-Burnham Medical Research Institute), GFP-FCHo1, GFP-CLC, Endophilin2-GFP, mCherry-actin, and mCherry-FBP17 (P. De Camilli, Yale University), Lyn-tailed mCherry Cdc42-T17N (cloned from Myc Cdc42-T17N from P. De Camili, Yale University), SHIP1 shRNA, CIBN-pmGFP, mCherry-CRY2-iSH2PI3K, mCherry-Grp1-PH, GFP-PH(TAPP1)-PH(TAPP1), iRFP-N-WASP, mCherry-SHIP1, and FBP17-EGFP were previously described (Xiong et al., 2016). CIP4 shRNA, FBP17 shRNA were previously described (Wu et al., 2017).
All chemicals were purchased from commercial sources: DNP-Albumin from Bovine Serum (BSA) (Invitrogen, A23018) as the antigen to stimulate the sensitized cells, clathrin inhibitor Pitstop 2 (Abcam, ab120687), PI3Kδ inhibitor CAL-101 (Cayman, 15279), PI3K inhibitor Wortmannin (Sigma, W1344-250ul), N-WASP inhibitor Wiskostatin (Sigma, W2270-5MG), and 1-butanol (Alfa Aesar L13171), Transferrin from human serum, Alexa Fluor 647 conjugated (Invitrogen T23366), clathrin light chain antibody (Millipore AB9884),.
Imaging
TIRF microscopy was performed with a Nikon Ti-E inverted microscope with motorized illuminator unit, equipped with a Multiplying Charge Coupled Device camera (Photometrics Evolve512 S/N: A11M105002), a 100X objective (CFI Apochromat, N.A. 1.49), and controlled by Metamorph software (Verison 7.8.6.0). Excitation light was provided by 491 nm (for GFP) Diode-pumped solid-state (DPSS) laser, 561 nm (for mRFP and mCherry) DPSS laser and 642 nm (for iRFP) Diode laser. The emission light is filtered with a quad-bandpass Dichroic mirror (Di01-R405/488/561/635, Semrock) and with single band filters as follows: 520/35 nm for GFP, 609/54 nm for RFP and mCherry, and 692/40 nm for iRFP(Semrock). Before imaging, cells were washed once in pre-warmed Tyrodes buffer (T buffer) containing: 135 mM NaCl, 5.0 mM KCl, 1.8 mM CaCl2, 1.0 mM MgCl2, 5.6 mM D-glucose, and 20 mM HEPES (pH 7.4). For stimulation, the final concentration of DNP-BSA added to the pre-sensitized cells was 80 ng/ml. Cells were kept at 37°C during imaging. To prep differentiated HL-60 cells for imaging, cell were 2x concentrated in imaging media (Leibovitz’s-15 medium with no phenol red [Gibco] and containing 0.5% [v/v] fetal bovine serum) and plated onto 0.2 μg/mL fibernectin-coated wells. Cells were incubated at 37°C/5% CO2 for 5 min and washed 2x with imaging media. To induce mHem1 waves with chemoattractant, a 2x stock of 20 nM fMLP was added. For the second acute perturbation, a 2x stock of 14 μM Pitstop 2 and 20 nM fMLP or 20 nM fMLP was added. All stocks were freshly prepared in imaging media. All imaging was performed at room temperature on a Nikon Eclipse Ti inverted microscope equipped with a CSU-X1 spinning disk confocal (Yokogawa) with a 60× 1.49NA Plan TIRF objective (Nikon).
Optogenetic experiments and analysis
For optogenetic experiments, the membrane-anchored probe CIBN-pmGFP was used to recruit its cytosolic binding partner CRY2 fused with iSH2PI3K and mCherry (mCherry-CRY2-iSH2PI3K) upon 491 nm laser activation. Due to the power dose-dependency of CRY2 recruitment, a train of 0.18mW 491-nm laser pulses with intervals of 1 min were used to achieve maximum CRY2 recruitment.
Tfn uptake assay
Cells transfected with Lyn-tailed mCherry Cdc42-DN, or mCherry-CLC after 24 hours were starved for 4h in starve medium (MEM, 0.3%BSA, 50mM HEPES). Then Cells were washed with T buffer and added DNP stimulation for 20min at 37°C. Then cells were incubated with 20 μg/μl Tfn-Alexa Fluor 647 in starve medium at 4°C for 1h. Cells were then washed with cold PBS and incubate with pre-warmed full medium for uptake at 37°C for 20min. Cells were then transferred to ice and wash the surface with acidic buffer (0.1M acetic acid, 0.25M NaCl) for 20min, and then fixed with 4% paraformaldehyde for confocal imaging analysis. Confocal image acquired whole cell z-stack with 1μm stepsize, and maximum intensity projection of the whole stack were used for analysis.
Image Analysis
Images acquired from the system above were 512 × 512 pixels, 0.16 μm/pixel, 16-bit. Displayed images and movies were generated with Fiji software. Line profiles and pseudo color maps were generated in MATLAB (MathWorks). Scatter plots were generated in Prism5 (GraphPad). All intensity profiles show the average intensity of a ROI of 21 × 21 pixel square over time, normalized by the difference between the local maximum and minimum intensity of the ROI. Dual and multi-colour peak alignments in Figure 2 were generated by aligning intensity profiles according to the peaks of FBP17 intensity. Phase lag between probes cross-correlation analysis between intensity profiles. Wave period/cycle time in Figure 1 were obtained using the major peak of the fast Fourier transformation (FFT) of intensity profile. Amplitude in Figure 5 was quantified as the difference between peaks and valleys identified from the intensity profile and normalized by the expression level of FBP17. Relative power in Figure 5 is defined as the percentage of the area under the major peak over all area from 0 to 100s in period spectrum generated from FFT. Phase, period, and relative power maps were constructed by calculating the phase (Figure 1)/period (Figure 5)/relative power (Figure 5) using the average intensity profile of a ROI of 11 × 11 pixels around each pixel. Singularity point is defined as pixels neighboring every phase of an oscillatory cycle (Figure 6). For WAVE2 complex quantification, background-subtracted mHem1-mNeonGreen intensity inside the cell was integrated with Fiji. All quantifications were normalized to the cell area at each time point using a cell mask. The cell mask was obtained after applying a 3×3 mean filter and thresholded in Fiji.
Single Particle Tracking Analysis
Tracking and lifetime determination of CCSs were performed in MATLAB. CCSs in image stacks were identified by detecting the local maxima after local background subtraction. Tracking was done by registration of puncta according to the nearest neighbour. Short tracks fewer than four frames were not quantified, and CCSs present in the first frame or the last frame were deleted as incomplete tracks. Lifetime, position, and the start and end frames of all tracks were recorded. Montages were aligned and organized according to either the direction of wave propagation (Figure 3A) or CCP lifetime (Figure 5C). For lifetime quantification, usually 2000 – 10000 tracks per cell were identified from a movie of 5 min. For late stage marker recruitment and hotspot quantification, usually 1000–6000 tracks per cell were identified from a movie of 200s. 100 dots selected from shortest lifetime to the longest lifetime arithmetically were quantified.
Statistics
P values were determined by two-tailed unpaired t-tests and noted in the legends. Mean ± STD was reported in the text and the figures for wave period, the median and the STD were reported for the CCP lifetime, wave amplitude, wave relative power, and phase delay between probes. For repeatability, the number of trials (n) was noted in legends. Generally, more than three cells from at least three independent experiments were quantified, and a mean of at least three ROIs in each cell were quantified for wave period, amplitude, relative power, and phase delay between probes. The lifetime of each CCP was identified in each cell, and the median and STD were quantified.
Supplementary Material
Figure S1. Clathrin waves is not due to overexpression of clathrin. Related to Figure 1.
(A) Comparison of Tfn uptake and CCP density between wild type cells and CLC overexpressing cells in the same field of view. Upper row: representative maximum intensity projection image of confocal z-stack of cells after 20 min of Tfn uptake. Cells expressing mCherry-CLC are shown on the left. Quantification of Tfn-Alexa-647 per cell area is shown on the right. Lower row: representative TIRF image of Immunofluoresence (IF) for CLC in cells with or without overexpressing of CLC. Cells expressing mCherry-CLC are shown on the left. Intensity per area of mCherry-CLC vs. intensity per area of Alexa fluor 488 IF-CLC is plotted on the right. R2 of the two is 0.0096.
(B) Endogeneous clathrin displays waves the same as overexpressed ones. Upper left: Representative TIRF image of an RBL-2H3 cell stably expressing GFP1-10 with GFP11 KI at endogeneous CLC N-terminus, and the same image (magenta) merged with transfected FBP17-GFP(green). Lower left: Montages of transfected FBP17-EGFP and endogenous clathrin waves in the cell shown above. A rectangular region of interest (ROI) of 7 ×102-pixel over 120 s is shown for montage. Right: representative intensity profile (top) of endoygenous clathrin and tranfected FBP17-EGFP waves and cross correlation between the two intensity profiles (bottom) (n = 3 cells in 3 experiments).
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm. Scale bars in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Figure S2. Additional components of early and late stage endocytic modules and endocytic cargoes in waves. Related to Figure 2.
(A) Representative merged image, montage, and intensity profile of the cell co-transfected with mCherry-CLC and GFP-FCHo1. Upper left, merged image of CLC (green) and FCHo1 (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the colocalization. Scale bar represents 1 μm. (n = 15 cells in five experiments). Bottom: Dot montage of transiently expressed GFP-FCHo1 and mCherry-CLC in a cell with clathrin waves. Time interval is 2 s. 0s marks the initial assembly of clathrin. ROI size is 11 × 11 pixels. Inverted look up table is used.
(B) Representative merged image, montage, and aligned profile of the cell stably expressing FBP17-EGFP transfected with mCherry-Actin. Upper left, merged image of FBP17 (green) and Actin (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the localization. Scale bar represents 1μm.(n = 11 cells in three experiments).
(C) Representative merged imgae, kymographs and intensity profiles of endocytic cargoes. Cell co-transfected with iRFP-CLC and TfR-pHuji (upper row), or with CLC-mCherry and EGFP-Glut4 (lower row). Scale bars 1 μm in snapshot images, and represent 20 s (horizontal) and 5 μm (vertical) in kymographs (n = 3 cells in 3 experiments for each group). Inverted look up table is used in all single channel montages. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
Figure S3. FBP17 recruitment and hotspots in clathrin waves at single puncta resolution. Related to Figure 3.
Representative zoomed in montages and corresponding intensity profiles of hotspots and non-hotspots. Three examples for each group. Crop ROI for montage is 9 × 9-pixel in size, and the intensity of the center 5 × 5 pixels are averaged and normalized for the intensity profiles.
Inverted look up table is used in all single channel montages.
Figure S4. Clathrin heavy chain knockdown efficiency; Requirement of Dynamin activity in wave formation; Inhibitory effect of Pitstop 2 on mHem1 waves in neutrophils. Related to Figure 4.
(A) TIRF images of cells with or without CHC knockdown after 72h. Left: shRNA-RFP transfected cells. Middle: immunostaining of CHC of the same field of view as the left image, with the knockdown cells outlined. Right: quantification of anti-CLC IF intensity per area in cells with or without CHC shRNA expression.
(B) Whole cell images and kymographs of representative cells with Dynamin1-shRNA in RFP expressing vectors. Cells stably expressing FBP17-EGFP were transfected with iRFP-CLC and Dynamin1-shRNA. Kymographs were generated by projecting kymographes of 10 adjacent pixels at the yellow lines in the third column.
(C) Confocal images of representative cells stably expressing mHem1 for control and Pitstop 2 treatments. The middle panels are 30 sec after induction of mHem1 waves with 10 nM (final) fMLP chemoattractant. The panels correspond to the time point marked by the circle in (D). The right panels are 30 sec after treatment without (control) or with 7 μM (final) Pitstop 2. The panels correspond to the time point marked by the triangle in (D).
(D) Normalized intensity profiles of mHem1 waves. Left dashed line indicates time when the 10 nM (final) fMLP chemoattractant was added to induce mHem1 waves. Right dashed line indicates time of Pitstop 2 addition. By confocal microscopy, mHem1 waves correspond with a decrease in intensity as mHem1 is recruited to the membrane. Cytosolic signals of mHem1 was quantified and inverted here to indicate the recruitment of mHem1 in the waves. Control experiment: n=7, from 3 independent experiments. Pitstop 2 experiment: n=9, from 3 independent experiments. Error bars, SEM. *, p <= 0.05 by t-test.
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm and in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Figure S5. Cartoon of the model of clathrin synchronization. Related to Figure 5.
For each category, statistics and the cell numer quantified is as indicated. Error bar in statistics: STD. N.A: not applicable.
Figure S6. Retrograde effect of FBP17/N-WASP module on clathrin. Related to Figure 5.
(A) RNA-Seq analysis to assess expression of F-BAR family proteins in RBL-2H3. Gene expression levels are interpreted in read per kilobase per million mapped expression (RPKM) (n=2 total RNA samples; error bar: STD. N.D: not detected).
(B) Kymograph and intensity profile of the inhibitory effect of Wiskostatin on the waves of transiently expressed mCherry-CLC. Final concentration of 50 μM of Wiskostatin is added at 200 s (n = 15 cells in three experiments).
(C) Kymograph and time-series quantification showing that iRFP-clathrin waves cycle time decreased after the recruitment of mCherry-CRY2-iSH2PI3K to the plasma membrane. A train of five 57-ms, 491nm laser was used.
Fire look up table is used. Scale bare represents 10 s (horizontal) and 5 μm (vertical).
(D) Representative montage of CCSs and corresponding FBP17 channel in cells exhibiting clathrin waves. All CCSs in a cell was tracked and sorted with CCS lifetime. Middle: 100 of them selected from the top to the end are listed. Right: montage of the corresponding FBP17 channel. Left: color merge of the previous two. n=10 cells. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
Figure S7. PIP3 Sensitivity Reveals a Special Cluster of Endocytic Proteins in Cortical Waves. Related to Figure 7.
(A) TIRF images of a cell with FBP17 waves before and after the treatment of 10 μM wortmannin, a PI3K inhibitor (n = 31 cells in 19 experiments).
(B) Kymograph of a cell with clathrin waves treated with 10 μM wortmannin at 150 s. Above, transfected FBP17-EGFP. Below, transfected cherry-CLC. The intensity profiles of ROIs in the center and off center are laid out correspondingly.
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm and in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC(right) were imaged at 2.3 sec/frame, and played at speed 40 times faster than real time. Scale bars represent 10μm. Inverted look up table is used.
Cell cotransfected with GFP-FCHo1(left) and CLC-GFP (right) were imaged at 2 s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell cotransfected with mCherry-FBP17 (left) and Endophilin2-GFP (right) were imaged at 1s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Movie S4. TIRFM movie of Pitstop 2 inhibition on clathrin and FBP17 wave. Related to Figure 4
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC (right) were imaged at 1 s/frame for 10 min, 7 μM (final concentration) Pitstop 2 is added at the 200 second. Movie is played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Movie S5. TIRFM movie of cal-101 induced low frequency of FBP17 waves and emergence of clathrin waves. Related to Figure 4
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC (right) were imaged at 1s/frame for 15 min, 800 nM (final concentration) cal-101 is added at the 200 second. Movie is played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC(right) were imaged at 1 s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell stably expressing FBP17-EGFP were imaged at 2 s/frame for 16.4 min, 10 μM (final concentration) wortmannin is added at the 200th second. Movie is played at speed 40 times faster than real time. Scale bar represents 10 μm. Inverted look up table is used.
Acknowledgments
We thank Maohan Su in the lab for helping the Matlab code and generating the phase map. This work was supported by the National Research Foundation (NRF) of Singapore under its NRF Fellowship Program (M.W., NRF Award No. NRF-NRFF2011-09), the Mechanobiology Institute at National University of Singapore and American Heart Association Predoctoral Fellowship (AP), R35GM118167 (ODW).
Footnotes
AUTHOR CONTRIBUTIONS
Y.Y. and M.W. conceived and designed the experiments. Y.Y. and D.X. performed the experiments. Y.Y. analyzed the data. Y.Y. and M.W. wrote the paper. A.P. and O.D.W. performed and analyzed the experiments in neutrophils.
References
- Arai Y, Shibata T, Matsuoka S, Sato MJ, Yanagida T, Ueda M. Self-organization of the phosphatidylinositol lipids signaling system for random cell migration. Proceedings of the National Academy of Sciences. 2010;107:12399–12404. doi: 10.1073/pnas.0908278107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arasada R, Pollard TD. Distinct Roles for F-BAR Proteins Cdc15p and Bzz1p in Actin Polymerization at Sites of Endocytosis in Fission Yeast. Curr Biol. 2011;21:1450–1459. doi: 10.1016/j.cub.2011.07.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bement WM, Leda M, Moe AM, Kita AM, Larson ME, Golding AE, Pfeuti C, Su KC, Miller AL, Goryachev AB, et al. Activator–inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium. Nat Cell Biol. 2015;17:1471–1483. doi: 10.1038/ncb3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boucrot E, Saffarian S, Massol R, Kirchhausen T, Ehrlich M. Role of lipids and actin in the formation of clathrin-coated pits. Exp Cell Res. 2006;312:4036–4048. doi: 10.1016/j.yexcr.2006.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das M, Drake T, Wiley DJ, Buchwald P, Vavylonis D, Verde F. Oscillatory Dynamics of Cdc42 GTPase in the Control of Polarized Growth. Science. 2012 doi: 10.1126/science.1218377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer JM, Savage NS, Jin M, Zyla TR, Elston TC, Lew DJ. Tracking Shallow Chemical Gradients by Actin-Driven Wandering of the Polarization Site. Curr Biol. 2013;23:32–41. doi: 10.1016/j.cub.2012.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrlich M, Boll W, Van Oijen A, Hariharan R, Chandran K, Nibert ML, Kirchhausen T. Endocytosis by random initiation and stabilization of clathrin-coated pits. Cell. 2004;118:591–605. doi: 10.1016/j.cell.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Gaidarov I, Santini F, Warren RA, Keen JH. Spatial control of coated-pit dynamics in living cells. Nat Cell Biol. 1999;1:1–7. doi: 10.1038/8971. [DOI] [PubMed] [Google Scholar]
- Giuliani C, Troglio F, Bai Z, Patel FB, Zucconi A, Malabarba MG, Disanza A, Stradal TB, Cassata G, Confalonieri S, et al. Requirements for F-BAR proteins TOCA-1 and TOCA-2 in actin dynamics and membrane trafficking during Caenorhabditis elegans oocyte growth and embryonic epidermal morphogenesis. PLoS Genet. 2009;5:e1000675. doi: 10.1371/journal.pgen.1000675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray RA, Pertsov AM, Jalife J. Spatial and temporal organization during cardiac fibrillation. Nature. 1998;392:75–78. doi: 10.1038/32164. [DOI] [PubMed] [Google Scholar]
- Gregor T, Fujimoto K, Masaki N, Sawai S. The Onset of Collective Behavior in Social Amoebae. Science. 2010;328:1021–1025. doi: 10.1126/science.1183415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Boucrot E, Meinecke M, Evergren E, Vallis Y, Mittal R, Mcmahon HT. FCHo proteins are nucleators of clathrin-mediated endocytosis. Science. 2010;328:1281–1284. doi: 10.1126/science.1188462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes WR, Carlsson AE, Edelstein-Keshet L. Regimes of wave type patterning driven by refractory actin feedback: transition from static polarization to dynamic wave behaviour. Phys Biol. 2012;9:046005. doi: 10.1088/1478-3975/9/4/046005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell AS, Jin M, Wu CF, Zyla TR, Elston TC, Lew DJ. Negative Feedback Enhances Robustness in the Yeast Polarity Establishment Circuit. Cell. 2012;149:322–333. doi: 10.1016/j.cell.2012.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh T, Erdmann KS, Roux A, Habermann B, Werner H, De Camilli P. Dynamin and the actin cytoskeleton cooperatively regulate plasma membrane invagination by BAR and F-BAR proteins. Dev Cell. 2005;9:791–804. doi: 10.1016/j.devcel.2005.11.005. [DOI] [PubMed] [Google Scholar]
- Kamioka Y, Fukuhara S, Sawa H, Nagashima K, Masuda M, Matsuda M, Mochizuki N. A novel dynamin-associating molecule, formin-binding protein 17, induces tubular membrane invaginations and participates in endocytosis. J Biol Chem. 2004;279:40091–40099. doi: 10.1074/jbc.M404899200. [DOI] [PubMed] [Google Scholar]
- Kishimoto T, Sun Y, Buser C, Liu J, Michelot A, Drubin DG. Determinants of endocytic membrane geometry, stability, and scission. Proceedings of the National Academy of Sciences. 2011 doi: 10.1073/pnas.1113413108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Kleist L, Stahlschmidt W, Bulut H, Gromova K, Puchkov D, Robertson MJ, MacGregor KA, Tomlin N, Pechstein A, Chau N, et al. Role of the Clathrin Terminal Domain in Regulating Coated Pit Dynamics Revealed by Small Molecule Inhibition. Cell. 2011;146:471–484. doi: 10.1016/j.cell.2011.06.025. [DOI] [PubMed] [Google Scholar]
- Laan A, Gutnick T, Kuba MJ, Laurent G. Behavioral Analysisof Cuttlefish Traveling Wavesand Its Implications for Neural Control. Curr Biol. 2014;24:1737–1742. doi: 10.1016/j.cub.2014.06.027. [DOI] [PubMed] [Google Scholar]
- Lauzeral J, Halloy J, Goldbeter A. Desynchronization of cells on the developmental path triggers the formation of spiral waves of cAMP during Dictyostelium aggregation. Proc Natl Acad Sci USA. 1997;94:9153–9158. doi: 10.1073/pnas.94.17.9153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K, Cox E, Goldstein R. Competing patterns of signaling activity in dictyostelium discoideum. Phys Rev Lett. 1996;76:1174–1177. doi: 10.1103/PhysRevLett.76.1174. [DOI] [PubMed] [Google Scholar]
- Leonetti MD, Sekine S, Kamiyama D, Weissman JS, Huang B. A scalable strategy for high-throughput GFP tagging of endogenous human proteins. Proceedings of the National Academy of Sciences. 2016;113:E3501–E3508. doi: 10.1073/pnas.1606731113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrifield CJ, Perrais D, Zenisek D. Coupling between clathrin-coated-pit invagination, cortactin recruitment, and membrane scission observed in live cells. Cell. 2005;121:593–606. doi: 10.1016/j.cell.2005.03.015. [DOI] [PubMed] [Google Scholar]
- Nakatsu F, Perera RM, Lucast L, Zoncu R, Domin J, Gertler FB, Toomre D, de Camilli P. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J Cell Biol. 2010;190:307–315. doi: 10.1083/jcb.201005018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozbudak EM, Becskei A, van Oudenaarden A. A system of counteracting feedback loops regulates Cdc42p activity during spontaneous cell polarization. Dev Cell. 2005;9:565–571. doi: 10.1016/j.devcel.2005.08.014. [DOI] [PubMed] [Google Scholar]
- Pálsson E, Cox EC. Origin and evolution of circular waves and spirals in Dictyostelium discoideum territories. Proc Natl Acad Sci USA. 1996;93:1151–1155. doi: 10.1073/pnas.93.3.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pálsson E, Lee KJ, Goldstein RE, Franke J, Kessin RH, Cox EC. Selection for spiral waves in the social amoebae Dictyostelium. Proc Natl Acad Sci USA. 1997;94:13719–13723. doi: 10.1073/pnas.94.25.13719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posor Y, Eichhorn-Gruenig M, Puchkov D, Schöneberg J, Ullrich A, Lampe A, Müller R, Zarbakhsh S, Gulluni F, Hirsch E, et al. Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate. Nature. 2013;499:233–237. doi: 10.1038/nature12360. [DOI] [PubMed] [Google Scholar]
- Reynwar BJ, Illya G, Harmandaris VA, Müller MM, Kremer K, Deserno M. Aggregation and vesiculation of membrane proteins by curvature-mediated interactions. Nature. 2007;447:461–464. doi: 10.1038/nature05840. [DOI] [PubMed] [Google Scholar]
- Saffarian S, Cocucci E, Kirchhausen T. Distinct dynamics of endocytic clathrin-coated pits and coated plaques. PLoS Biol. 2009;7:e1000191. doi: 10.1371/journal.pbio.1000191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller V, Bausch AR. Topological defects and density fluctuations in collectively moving systems. Proc Natl Acad Sci USA. 2013;110:4488–4493. [Google Scholar]
- Schroth-Diez B, Gerwig S, Ecke M, Hegerl R, Diez S, Gerisch G. Propagating waves separate two states of actin organization in living cells. Hfsp J. 2009;3:412–427. doi: 10.2976/1.3239407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegert F, Weijer CJ. Spiral and concentric waves organize multicellular Dictyostelium mounds. Curr Biol. 1995;5:937–943. doi: 10.1016/s0960-9822(95)00184-9. [DOI] [PubMed] [Google Scholar]
- Strogatz SH, Stewart I. Coupled oscillators and biological synchronization. Scientific American. 2001:1–7. doi: 10.1038/scientificamerican1293-102. [DOI] [PubMed] [Google Scholar]
- Taniguchi D, Ishihara S, Oonuki T, Honda-Kitahara M, Kaneko K, Sawai S. Phase geometries of two-dimensional excitable waves govern self-organized morphodynamics of amoeboid cells. Proceedings of the National Academy of Sciences. 2013;110:5016–5021. doi: 10.1073/pnas.1218025110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AF, Tinsley MR, Wang F, Huang Z, Showalter K. Dynamical quorum sensing and synchronization in large populations of chemical oscillators. Science. 2009;323:614–617. doi: 10.1126/science.1166253. [DOI] [PubMed] [Google Scholar]
- Taylor MJ, Perrais D, Merrifield CJ. A high precision survey of the molecular dynamics of Mammalian clathrin-mediated endocytosis. PLoS Biol. 2011;9:e1000604. doi: 10.1371/journal.pbio.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Suetsugu S, Sasaki N, Furutani M, Oikawa T, Takenawa T. Coordination between the actin cytoskeleton and membrane deformation by a novel membrane tubulation domain of PCH proteins is involved in endocytosis. J Cell Biol. 2006;172:269–279. doi: 10.1083/jcb.200508091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsujita K, Takenawa T, Itoh T. Feedback regulation between plasma membrane tension and membrane-bending proteins organizes cell polarity during leading edge formation. Nat Cell Biol. 2015 doi: 10.1038/ncb3162. [DOI] [PubMed] [Google Scholar]
- Watts DJ, Strogatz SH. Collective dynamics of “small-world” networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW. An Actin-Based Wave Generator Organizes Cell Motility. PLoS Biol. 2007;5:e221. doi: 10.1371/journal.pbio.0050221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winfree AT. Electrical instability in cardiac muscle: phase singularities and rotors. J Theor Biol. 1989;138:353–405. doi: 10.1016/s0022-5193(89)80200-0. [DOI] [PubMed] [Google Scholar]
- Wu M, Huang B, Graham M, Raimondi A, Heuser JE, Zhuang X, De Camilli P. Coupling between clathrin-dependent endocytic budding and F-BAR-dependent tubulation in a cell-free system. Nat Cell Biol. 2010;12:902–908. doi: 10.1038/ncb2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M, Wu X, De Camilli P. Calcium oscillations-coupled conversion of actin travelling waves to standing oscillations. Proceedings of the National Academy of Sciences. 2013;110:1339–1344. doi: 10.1073/pnas.1221538110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Su M, Tong C, Wu M, Liu J. Collective membrane dynamics emerging from curvature-dependent spatial coupling. bioRxiv. 2017 Preprint. [Google Scholar]
- Xiong D, Xiao S, Guo S, Lin Q, Nakatsu F, Wu M. Frequency and amplitude control of cortical oscillations by phosphoinositide waves. Nat Chem Biol. 2016;12:159–166. doi: 10.1038/nchembio.2000. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Clathrin waves is not due to overexpression of clathrin. Related to Figure 1.
(A) Comparison of Tfn uptake and CCP density between wild type cells and CLC overexpressing cells in the same field of view. Upper row: representative maximum intensity projection image of confocal z-stack of cells after 20 min of Tfn uptake. Cells expressing mCherry-CLC are shown on the left. Quantification of Tfn-Alexa-647 per cell area is shown on the right. Lower row: representative TIRF image of Immunofluoresence (IF) for CLC in cells with or without overexpressing of CLC. Cells expressing mCherry-CLC are shown on the left. Intensity per area of mCherry-CLC vs. intensity per area of Alexa fluor 488 IF-CLC is plotted on the right. R2 of the two is 0.0096.
(B) Endogeneous clathrin displays waves the same as overexpressed ones. Upper left: Representative TIRF image of an RBL-2H3 cell stably expressing GFP1-10 with GFP11 KI at endogeneous CLC N-terminus, and the same image (magenta) merged with transfected FBP17-GFP(green). Lower left: Montages of transfected FBP17-EGFP and endogenous clathrin waves in the cell shown above. A rectangular region of interest (ROI) of 7 ×102-pixel over 120 s is shown for montage. Right: representative intensity profile (top) of endoygenous clathrin and tranfected FBP17-EGFP waves and cross correlation between the two intensity profiles (bottom) (n = 3 cells in 3 experiments).
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm. Scale bars in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Figure S2. Additional components of early and late stage endocytic modules and endocytic cargoes in waves. Related to Figure 2.
(A) Representative merged image, montage, and intensity profile of the cell co-transfected with mCherry-CLC and GFP-FCHo1. Upper left, merged image of CLC (green) and FCHo1 (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the colocalization. Scale bar represents 1 μm. (n = 15 cells in five experiments). Bottom: Dot montage of transiently expressed GFP-FCHo1 and mCherry-CLC in a cell with clathrin waves. Time interval is 2 s. 0s marks the initial assembly of clathrin. ROI size is 11 × 11 pixels. Inverted look up table is used.
(B) Representative merged image, montage, and aligned profile of the cell stably expressing FBP17-EGFP transfected with mCherry-Actin. Upper left, merged image of FBP17 (green) and Actin (magenta). Scale bar represents 10 μm. Upper right, zoomed in image to show the localization. Scale bar represents 1μm.(n = 11 cells in three experiments).
(C) Representative merged imgae, kymographs and intensity profiles of endocytic cargoes. Cell co-transfected with iRFP-CLC and TfR-pHuji (upper row), or with CLC-mCherry and EGFP-Glut4 (lower row). Scale bars 1 μm in snapshot images, and represent 20 s (horizontal) and 5 μm (vertical) in kymographs (n = 3 cells in 3 experiments for each group). Inverted look up table is used in all single channel montages. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
Figure S3. FBP17 recruitment and hotspots in clathrin waves at single puncta resolution. Related to Figure 3.
Representative zoomed in montages and corresponding intensity profiles of hotspots and non-hotspots. Three examples for each group. Crop ROI for montage is 9 × 9-pixel in size, and the intensity of the center 5 × 5 pixels are averaged and normalized for the intensity profiles.
Inverted look up table is used in all single channel montages.
Figure S4. Clathrin heavy chain knockdown efficiency; Requirement of Dynamin activity in wave formation; Inhibitory effect of Pitstop 2 on mHem1 waves in neutrophils. Related to Figure 4.
(A) TIRF images of cells with or without CHC knockdown after 72h. Left: shRNA-RFP transfected cells. Middle: immunostaining of CHC of the same field of view as the left image, with the knockdown cells outlined. Right: quantification of anti-CLC IF intensity per area in cells with or without CHC shRNA expression.
(B) Whole cell images and kymographs of representative cells with Dynamin1-shRNA in RFP expressing vectors. Cells stably expressing FBP17-EGFP were transfected with iRFP-CLC and Dynamin1-shRNA. Kymographs were generated by projecting kymographes of 10 adjacent pixels at the yellow lines in the third column.
(C) Confocal images of representative cells stably expressing mHem1 for control and Pitstop 2 treatments. The middle panels are 30 sec after induction of mHem1 waves with 10 nM (final) fMLP chemoattractant. The panels correspond to the time point marked by the circle in (D). The right panels are 30 sec after treatment without (control) or with 7 μM (final) Pitstop 2. The panels correspond to the time point marked by the triangle in (D).
(D) Normalized intensity profiles of mHem1 waves. Left dashed line indicates time when the 10 nM (final) fMLP chemoattractant was added to induce mHem1 waves. Right dashed line indicates time of Pitstop 2 addition. By confocal microscopy, mHem1 waves correspond with a decrease in intensity as mHem1 is recruited to the membrane. Cytosolic signals of mHem1 was quantified and inverted here to indicate the recruitment of mHem1 in the waves. Control experiment: n=7, from 3 independent experiments. Pitstop 2 experiment: n=9, from 3 independent experiments. Error bars, SEM. *, p <= 0.05 by t-test.
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm and in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Figure S5. Cartoon of the model of clathrin synchronization. Related to Figure 5.
For each category, statistics and the cell numer quantified is as indicated. Error bar in statistics: STD. N.A: not applicable.
Figure S6. Retrograde effect of FBP17/N-WASP module on clathrin. Related to Figure 5.
(A) RNA-Seq analysis to assess expression of F-BAR family proteins in RBL-2H3. Gene expression levels are interpreted in read per kilobase per million mapped expression (RPKM) (n=2 total RNA samples; error bar: STD. N.D: not detected).
(B) Kymograph and intensity profile of the inhibitory effect of Wiskostatin on the waves of transiently expressed mCherry-CLC. Final concentration of 50 μM of Wiskostatin is added at 200 s (n = 15 cells in three experiments).
(C) Kymograph and time-series quantification showing that iRFP-clathrin waves cycle time decreased after the recruitment of mCherry-CRY2-iSH2PI3K to the plasma membrane. A train of five 57-ms, 491nm laser was used.
Fire look up table is used. Scale bare represents 10 s (horizontal) and 5 μm (vertical).
(D) Representative montage of CCSs and corresponding FBP17 channel in cells exhibiting clathrin waves. All CCSs in a cell was tracked and sorted with CCS lifetime. Middle: 100 of them selected from the top to the end are listed. Right: montage of the corresponding FBP17 channel. Left: color merge of the previous two. n=10 cells. Scale bars in montages represent 10 s (horizontal) and 5 μm (vertical).
Figure S7. PIP3 Sensitivity Reveals a Special Cluster of Endocytic Proteins in Cortical Waves. Related to Figure 7.
(A) TIRF images of a cell with FBP17 waves before and after the treatment of 10 μM wortmannin, a PI3K inhibitor (n = 31 cells in 19 experiments).
(B) Kymograph of a cell with clathrin waves treated with 10 μM wortmannin at 150 s. Above, transfected FBP17-EGFP. Below, transfected cherry-CLC. The intensity profiles of ROIs in the center and off center are laid out correspondingly.
Inverted look-up table was used in all single channel images/montages. Scale bars in all whole cell images represent 10 μm and in kymographs represent 10 s (horizontal) and 5 μm (vertical).
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC(right) were imaged at 2.3 sec/frame, and played at speed 40 times faster than real time. Scale bars represent 10μm. Inverted look up table is used.
Cell cotransfected with GFP-FCHo1(left) and CLC-GFP (right) were imaged at 2 s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell cotransfected with mCherry-FBP17 (left) and Endophilin2-GFP (right) were imaged at 1s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Movie S4. TIRFM movie of Pitstop 2 inhibition on clathrin and FBP17 wave. Related to Figure 4
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC (right) were imaged at 1 s/frame for 10 min, 7 μM (final concentration) Pitstop 2 is added at the 200 second. Movie is played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Movie S5. TIRFM movie of cal-101 induced low frequency of FBP17 waves and emergence of clathrin waves. Related to Figure 4
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC (right) were imaged at 1s/frame for 15 min, 800 nM (final concentration) cal-101 is added at the 200 second. Movie is played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell stably expressing FBP17-EGFP (left) transfected with mCherry-CLC(right) were imaged at 1 s/frame, and played at speed 40 times faster than real time. Scale bars represent 10 μm. Inverted look up table is used.
Cell stably expressing FBP17-EGFP were imaged at 2 s/frame for 16.4 min, 10 μM (final concentration) wortmannin is added at the 200th second. Movie is played at speed 40 times faster than real time. Scale bar represents 10 μm. Inverted look up table is used.