SUMMARY
Typhoid fever caused by Salmonella enterica serovar (S.) Typhi differs in its clinical presentation from gastroenteritis caused by S. Typhimurium and other non-typhoidal Salmonella serovars. The different clinical presentations are attributed in part to the virulence-associated capsular polysaccharide (Vi antigen) of S. Typhi, which prevents phagocytes from triggering a respiratory burst by preventing antibody-mediated complement activation. Paradoxically, the Vi antigen is absent from S. Paratyphi A, which causes a disease that is indistinguishable from typhoid fever. Here, we show that evasion of the phagocyte respiratory burst by S. Paratyphi A required very long O antigen chains containing the O2 antigen to inhibit antibody binding. We conclude that the ability to avoid the phagocyte respiratory burst is a property distinguishing typhoidal from non-typhoidal Salmonella serovars that was acquired by S. Typhi and S. Paratyphi A independently through convergent evolution.
Graphical abstract
INTRODUCTION
Typhoid fever, caused by the human-adapted Salmonella enterica serovar (S.) Typhi, is a major human disease that is responsible for 21.6 million illnesses annually, whereas paratyphoid fever, associated predominantly with the human-adapted S. Paratyphi A, is responsible for 5.4 million cases per year (Crump et al., 2004). Typhoid and paratyphoid fever are indistinguishable in their clinical signs and symptoms (Nuccio et al., 2013), suggesting that typhoidal Salmonella serovars possess similar virulence strategies.
The ability to breach an intact mucosal barrier and cause an invasive bloodstream infection in humans is a property that sets typhoidal Salmonella serovars apart from non-typhoidal -Salmonella serovars associated with gastroenteritis (Keestra-Gounder et al., 2015; Raffatellu et al., 2006; Tsolis et al., 2008; Wangdi et al., 2012). Clues regarding the identities of the mucosal barrier functions limiting the dissemination of the non-typhoidal S. Typhimurium in individuals with an intact immune system can be gleaned by analyzing the immune defects that put patients at risk of developing bacteremia. Chronic granulomatous disease, which is caused by deficiencies in genes encoding subunits of the phagocyte NADPH oxidase (the enzyme that generates superoxide radicals during the respiratory burst of phagocytes [Hohn and Lehrer, 1975; McPhail et al., 1977; Moellering and Weinberg, 1970; Seger et al., 1983]), renders individuals susceptible to develop non-typhoidal Salmonella bacteremia (Winkelstein et al., 2000). Thus, the generation of reactive oxygen species (ROS) by phagocytes is essential for maintaining an intact mucosal barrier impeding S. Typhimurium dissemination beyond the mesenteric lymph node in humans. Whereas the phagocyte respiratory burst helps prevent S. Typhimurium dissemination in individuals with an intact immune system, S. Typhi and S. Paratyphi A are able to cross an intact mucosal barrier and spread from the mesenteric lymph node via the circulation to internal organs. A supposition from these observations is that S. Typhi and S. Paratytphi A must possess virulence mechanisms for averting the respiratory burst of human phagocytes, but these virulence mechanisms must be absent from S. Typhimurium.
A DNA region present in S. Typhi but absent from the S. Typhimurium genome is the viaB locus, which contains genes for the regulation (tviA), synthesis (tviBCDE), and export (vexABCDE) of the Vi antigen (Liston et al., 2016; Virlogeux et al., 1995; Wetter et al., 2012). TviA activates genes involved in Vi antigen synthesis and export while repressing genes encoding flagella and the invasion-associated type III secretion system (Winter et al., 2010, 2009). The latter reduces the generation of inflammatory responses during the invasion of the intestinal mucosa (Raffatellu et al., 2007; Winter et al., 2008, 2015, 2014). More important, acquisition of the viaB locus by horizontal gene transfer helps explain why S. Typhi is able to avert the respiratory burst of neutrophils (Kossack et al., 1981; Miller et al., 1972). When a neutrophil comes within close proximity of S. Typhimurium, it migrates toward the intruder by following a chemotactic gradient of complement component 5 fragment a (C5a) that emanates from the bacterial surface (Wangdi et al., 2014). C5a is a neutrophil chemoattractant that triggers Rac-dependent NADPH oxidase activation (Bokoch, 1995), thereby rendering the S. Typhimurium-induced neutrophil oxidative burst complement dependent. The Vi antigen encoded by the S. Typhi viaB locus prevents complement activation and C5a-mediated neutrophil chemotaxis (Wangdi et al., 2014).
However, the viaB locus is not present in S. Paratyphi A (McClelland et al., 2004), which is puzzling because host responses elicited by this pathogen are more similar to those triggered by S. Typhi than those observed during S. Typhimurium infection. Here, we resolve this apparent paradox by showing that typhoidal Salmonella serovars acquired virulence traits for averting the phagocyte respiratory burst through convergent evolution.
RESULTS
Typhoidal Salmonella Serovars Evade the Phagocyte Respiratory Burst
To test the hypothesis that unlike non-typhoidal Salmonella serovars, typhoidal Salmonella serovars possess virulence mechanisms for averting the phagocyte respiratory burst, we infected human neutrophil-like (HL-60) cells with typhoidal and non-typhoidal Salmonella serovars and monitored the generation of ROS using a luminol-based bioassay. Infection with non-typhoidal Salmonella serovars, including S. Typhimurium (strains from the American Type Culture Collection [ATCC]: ATCC14028 and ATCC7823) and S. Enteritidis (strain SSU7998), triggered a robust respiratory burst (Figures 1A and 1B; Figure S1A). In contrast, infection of HL-60 cells with typhoidal Salmonella serovars, including S. Typhi strain ATCC700931 (Ty2) or S. Paratyphi A strain ATCC9150, did not elicit this response (Figures 1A and 1B). Similar results were obtained using primary human neutrophils (Figures S1B and S1C).
Figure 1. S. Typhi Evades the Respiratory Burst in Neutrophil-like Cells Using the Vi Antigen.
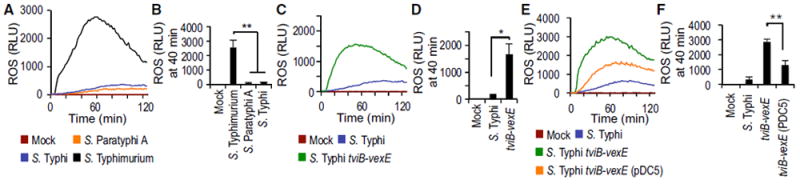
HL-60 cells were infected with the indicated opsonized bacterial strains and ROS production monitored over time using chemiluminescence.
(A and B) HL60 cells were infected with the indicated Salmonella serovars.
(C and D) HL60 cells were infected with capsulated and non-capsulated S. Typhi strains.
(E and F) HL60 cells were infected with S. Typhi wild type, a tviB-vexE mutant and a tviB-vexE mutant complemented by introducing the viaB locus on a plasmid (pDC5).
(A, C, and E) Representative experiment showing generation of chemiluminescence over time.
(B, D, and F) Quantification of chemiluminescence from three independent experiments at the indicated time point after infection. Bars represent means ± SE. Mock, mock infection; pDC5, XX; RLU, relative luminescence units. *p < 0.05; **p < 0.01.
The Vi Antigen Enables S. Typhi to Avert the Phagocyte Respiratory Burst
To assess whether averting the phagocyte respiratory burst is a feature that is conserved among S. Typhi isolates, HL-60 cells were infected with S. Typhi strains Ty2, ATCC19430, and ATCC33458. Similar to S. Typhi strain Ty2, S. Typhi strain ATCC19430 did not elicit marked ROS production in HL-60 cells. However, robust ROS production was observed with S. Typhi strain ATCC33458 (Figure S1A). The Vi antigen of S. Typhi has been implicated in averting the neutrophil respiratory burst (Kossack et al., 1981; Miller et al., 1972), and its biosynthesis is encoded by the viaB locus on Salmonella pathogenicity island (SPI) 7, a DNA region that can be lost upon laboratory passage of clinical isolates (Bueno et al., 2004; Nair et al., 2004). Interestingly, production of the Vi antigen could be detected by dot blot analysis in S. Typhi strains Ty2 and ATCC19430, but not in S. Typhi strain ATCC33458 (Figure S1D). These data suggested that robust ROS production elicited by S. Typhi ATCC33458 (Figure S1A) may result from the inability of the strain to produce the Vi antigen (Figure S1D). To test this idea, we compared ROS production elicited by S. Typhi strain Ty2 in HL-60 cells with that elicited by an isogenic mutant carrying a precise deletion of the Vi capsule biosynthesis genes (S. Typhi ΔtviB-vexE mutant). Unlike S. Typhi strain Ty2, the non-capsulated S. Typhi ΔtviB-vexE mutant elicited robust ROS production in HL-60 cells (Figures 1C and 1D). Similar results were obtained using primary human neutrophils (Figures S1E and S1F). Introduction of a plasmid (pDC5) carrying the cloned viaB region into the S. Typhi ΔtviB-vexE mutant restored synthesis of the Vi antigen (Figure S1G) and moderated ROS production in HL-60 cells (Figures 1E and 1F).
Next, we characterized the mechanism through which the Vi antigen inhibits the neutrophil respiratory burst. The Vi antigen inhibits complement activation on the bacterial surface (Looney and Steigbigel, 1986; Wilson et al., 2011), thereby conferring serum resistance on S. Typhi (Figures 2A and 2B; Figure S1H) (Hart et al., 2016). We investigated whether preventing complement activation would inhibit the phagocyte respiratory burst. ROS production triggered by infecting HL-60 cells with the non-capsulated S. Typhi ΔtviB-vexE mutant was complement dependent because it required opsonization with human serum (data not shown) and could be blunted by treatment with Futhan (6-amidino-2-naphthyl p-guanidinobenzoate dimethanesulfonate; BD Biosciences), a complement inhibitor (Figures 2C and 2D). The main conduit of lipopolysaccharide (LPS)-mediated complement activation in an immunologically naive host is mediated by binding of B-1 cell-derived natural immunoglobulin (Ig)M to the bacterial surface, which leads to complement activation through the classical pathway (Reid et al., 1997). We thus determined whether IgM in human serum binds to purified LPS or to purified Vi antigen. Human IgM bound to LPS purified from S. Typhimurium or an S. Typhi ΔtviB-vexE mutant, but it did not bind to purified Vi antigen (Figure 2E). We next investigated whether production of Vi antigen would prevent IgM binding to the bacterial surface. After incubation of bacteria in human serum, proteins bound to the surface were eluted using glycine and analyzed by western blot with anti-IgM antibody. Human IgM bound to the surface of S. Typhimurium (ATCC14028) and S. Enteritidis (SSU7998), but not to S. Typhi (Ty2). Inhibition of IgM binding to the surface of S. Typhi was mediated by the capsular polysaccharide because binding of IgM was detected at the surface of an S. Typhi ΔtviB-vexE mutant (Figure 2F), which was consistent with a previous report (Hart et al., 2016).
Figure 2. The Vi Antigen Averts the Phagocyte Respiratory Burst by Inhibiting Antibody-Dependent Complement Activation.
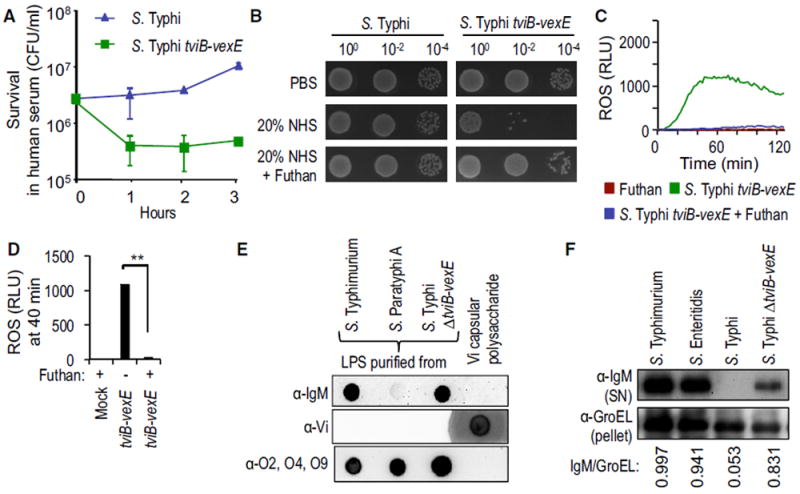
(A) Viability of the indicated bacterial strains incubated in 10% NHS was monitored over time.
(B) Viability of the indicated bacterial strains incubated in PBS, 20% NHS, or 20% NHS supplemented with Futhan was determined by spotting serial dilutions on agar plates.
(C and D) HL-60 cells were infected with the indicated opsonized bacterial strains and ROS production monitored over time using chemiluminescence.
(C) Representative experiment showing generation of chemiluminescence over time.
(D) Quantification of chemiluminescence from three independent experiments at the indicated time point after infection.
(E) Binding of IgM in human serum (α-IgM; top); binding of rabbit anti-Vi antigen serum (α-Vi; middle); or binding of rabbit anti-O2, -O4, -O9 serum (α-O2, O4, O9; bottom panel) to purified LPS of the indicated bacterial strains or to purified Vi antigen was detected by dot blot.
(F) The indicated bacterial strains were incubated in human serum and surface-bound proteins eluted with glycine. IgM eluted from the bacterial surface was detected by western blot (α-IgM; top). The bacterial protein GroEL was detected in the bacterial pellet (α-GroEL; bottom) and used as a loading control by measuring the IgM/GroEL using densitometry.
(A and D) Bars represent means ± SE.
Mock, mock infection; RLU, relative luminescence units. **p < 0.01.
S. Typhi carries a loss-of-function mutation in fepE (Bravo et al., 2008), encoding the length regulator of very long O antigen chains (Murray et al., 2003). Because human IgM bound to purified S. Typhi LPS (Figure 2E), we investigated whether restoring the expression of very long O antigen chains in S. Typhi would result in IgM binding to the bacterial surface. Interestingly, introduction of a plasmid (pTW134) carrying the cloned fepE gene from S. Paratyphi A into S. Typhi resulted in the expression of very long O antigen chains (Figure S2A) and the binding of human IgM to the bacterial surface (Figure S2B), suggesting that the Vi antigen cannot prevent the binding of IgM to very long LPS species. These data were consistent with a previous report, showing that lack of very long O antigen chains is required for Vi antigen-mediated evasion of complement activation in S. Typhi (Crawford et al., 2013).
Very Long O Antigen Chains of S. Paratyphi A Inhibit the Phagocyte Respiratory Burst
To investigate the mechanism by which S. Paratyphi A averts the neutrophil respiratory burst, we incubated S. Typhimurium (ATCC14028) and S. Paratyphi A (ATCC9150) in human serum and analyzed proteins eluted from the bacterial surface (Figure S3A) by mass spectrometry (MS) (Figure S3B). The results showed that increased amounts of IgM and complement components were eluted from the surface of S. Typhimurium compared to S. Paratyphi A (Figure S3). Similarly, S. Typhi bound less IgM and complement components compared to S. Typhimurium (Figure S3). However, S. Paratyphi A reduced binding of IgM and deposition of complement through a mechanism that did not require the Vi antigen, because the biosynthesis of this capsular polysaccharide is encoded by the viaB locus (Liston et al., 2016; Virlogeux et al., 1995; Wetter et al., 2012), a DNA region present in S. Typhi but absent from S. Paratyphi A (Figure 2E) (McClelland et al., 2004).
Next, we investigated whether evasion of the neutrophil respiratory burst was a general feature of S. Paratyphi A isolates. To this end, HL-60 cells were infected with S. Paratyphi A var. Durazzo strain ATCC11511 and S. Paratyphi A strains ATCC9150 and ATCC9281. ROS production was blunted in HL-60 cells infected with S. Paratyphi A strains ATCC9150 and ATCC9281, but S. Paratyphi A var. Durazzo strain ATCC11511 elicited a robust respiratory burst (Figures 3A and 3B). The latter isolate was first described in a 1934 article cautioning that laboratory passage may have begun to alter the O antigen of S. Paratyphi A var. Durazzo toward a rough form (Kauffmann and Silberstein, 1934). Dot blot analysis of purified LPS suggested that the O2 antigen, which is characteristic of S. Paratyphi A O antigen repeat units, was produced by all three isolates (Figure 3C). However, comparison of LPS chain lengths revealed that S. Paratyphi A var. Durazzo (ATCC: 11511) lacked very long O antigen chains, whereas very long O antigen chains were present in other S. Paratyphi A isolates (Figure 3D).
Figure 3. Very Long O Antigen Chains of S. Paratyphi A Are Necessary for Evasion of the Respiratory Burst in Neutrophil-like Cells.
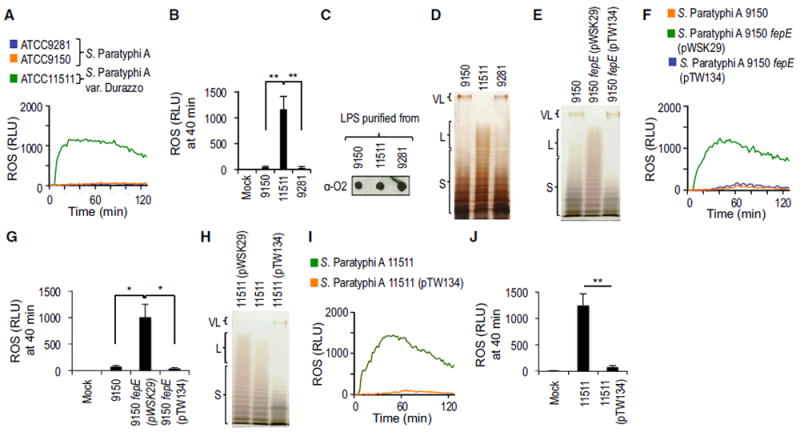
(A and B) HL-60 cells were infected with the indicated opsonized S. Paratyphi A or S. Paratyphi A var. Durazzo strains and ROS production monitored over time using chemiluminescence.
(C) Production of the O2-antigen was detected in LPS purified from the indicated bacterial strains using rabbit anti-O2 serum.
(D) LPS purified from the indicated S. Paratyphi A or S. Paratyphi A var. Durazzo strains was separated by SDS-PAGE and silver stained.
(E) LPS purified from S. Paratyphi A strain ATCC9150, an isogenic fepE mutant or a complemented fepE mutant was separated by SDS-PAGE and silver stained.
(F and G) S. Paratyphi A strain ATCC9150, an isogenic fepE mutant or a complemented fepE mutant were opsonized and ROS production elicited after infection of HL-60 cells monitored over time using chemiluminescence.
(H) LPS purified from the indicated S. Paratyphi A var. Durazzo strains was separated by SDS-PAGE and silver stained.
(I and J) HL-60 cells were infected with the indicated opsonized S. Paratyphi A var. Durazzo strains and ROS production monitored over time using chemiluminescence.
(A, B, F, G, I, and J) HL-60 cells were infected with the indicated opsonized bacterial strains and ROS production monitored over time using chemiluminescence.
(A, F, and I) Representative experiments showing generation of chemiluminescence over time.
(B, G, and J) Quantification of chemiluminescence from three independent experiments at the indicated time point after infection. Bars represent means ± SE. L, long O antigen chains; Mock, mock infection; pTW134, plasmid carrying the cloned fepE gene from S. Paratyphi A strain ATCC9150; pWSK29, cloning vector; RLU, relative luminescence units; S, short O antigen chains; VL, very long O antigen chains; ; 9150, S. Paratyphi A strain ATCC9150; 9281, S. Paratyphi A strain ATCC9281; 11511, S. Paratyphi A var. Durazzo strain ATCC11511. **p < 0.01.
O antigen chains of Salmonella LPS typically exhibit a trimodal length distribution, including short LPS species containing between 1 and 15 O antigen repeat units, long LPS species carrying between 16 and 35 O antigen repeat units, and very long LPS species with >100 O antigen repeat units (Batchelor et al., 1992, 1991; Murray et al., 2003) (Figure 3D). A length regulator encoded by the fepE gene controls the synthesis of very long O antigen chains (Murray et al., 2003). Inspection of the genomic sequence revealed that the fepE gene was disrupted in S. Paratyphi A var. Durazzo (ATCC11511) by a frameshift mutation (i.e., insertion of an adenine nucleotide) in position 33 of the open reading frame. In contrast, the fepE gene was intact in the other S. Paratyphi A isolates. These data raised the possibility that very long O antigen chains of S. Paratyphi A prevented the generation of the phagocyte respiratory burst. Remarkably, inactivation of the fepE gene in S. Paratyphi A strain ATCC9150 resulted in the loss of very long O antigen production (Figure 3E) and the generation of a robust respiratory burst (Figures 3F and 3G), whereas complementation of the mutation via the introduction of a plasmid (pTW134) carrying the cloned fepE gene restored the inhibition of ROS production in HL-60 cells. The S. Paratyphi A ATCC9150 fepE mutant also exhibited increased serum sensitivity compared to the wild-type or a complemented mutant (Figure S4). Introduction of a plasmid encoding an intact fepE gene (pTW134) into S. Paratyphi A var. Durazzo (ATCC11511) restored very long O antigen production (Figure 3H), restored serum resistance (Figure S4A), and abrogated ROS production in HL-60 cells (Figures 3I and 3J) and primary human neutrophils (Figures S4B and S4C). These data provided direct support for the idea that very long O antigen chains enable S. Paratyphi A to avert the phagocyte respiratory burst.
The O2 Antigen of S. Paratyphi A Reduces Antibody-Mediated ROS Production
Studies with normal human serum (NHS) from different individuals suggested that IgM binds to LPS purified from S. Typhimurium and S. Typhi, but binding to LPS purified from S. Paratyphi A was observed in only a fraction of individuals (Figure 2E; Figure S5). The occasional presence of IgM antibodies against S. Paratyphi A LPS in NHS may be the result of previous exposure to related antigens in the respective donors. However, the absence of IgM binding in a fraction of NHS samples suggested that natural antibodies did not recognize S. Paratyphi A LPS. Increased IgM binding to the surface of an S. Paratyphi A (ATCC9150) fepE mutant (Figure 4A) suggested that very long O antigen chains reduced IgM binding to the bacterial surface.
Figure 4. Heterologous Production of the O9 Antigen by S. Paratyphi A Activates the Respiratory Burst in Neutrophil-like Cells.
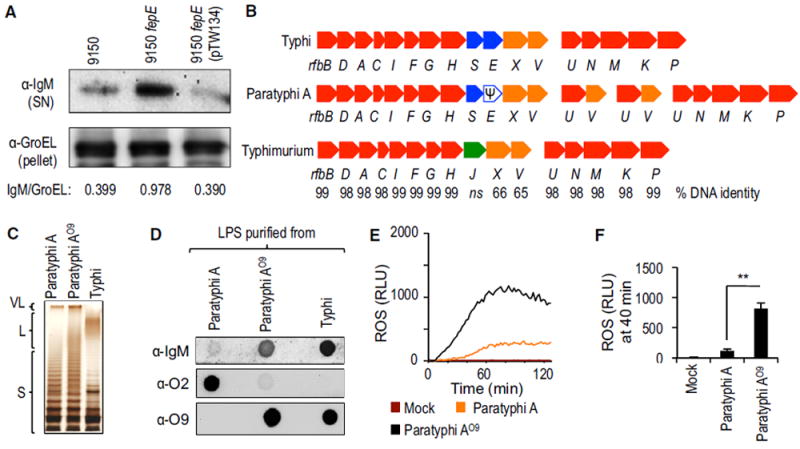
(A) The indicated S. Paratyphi A strains were incubated in human serum and surface-bound proteins eluted with glycine. IgM eluted from the bacterial surface was detected by western blot (α-IgM; top). The bacterial protein GroEL was detected in the bacterial pellet (α-GroEL; bottom) and used as a loading control by measuring the IgM/GroEL using densitometry.
(B) Schematic of the O antigen biosynthesis gene clusters in the indicated Salmonella serovars.
(C) LPS purified from the indicated bacterial strains was separated by SDS-PAGE and silver stained.
(D) LPS purified from the indicated bacterial strains was loaded on a dot blot to detect binding of IgM present in NHS (α-IgM; top), production of the O2 antigen (α-O2; middle), or production of the O9 antigen (α-O9; bottom).
(E and F) HL-60 cells were infected with the indicated opsonized bacterial strains and ROS production monitored over time using chemiluminescence.
(E) Representative experiment showing generation of chemiluminescence over time.
(F) Quantification of chemiluminescence from four independent experiments at the indicated time point after infection. Bars represent means ± SE.
Ψ, pseudogene; L, long O antigen chains; Mock, mock infection; RLU, relative luminescence units; S, short O antigen chains; VL, very long O antigen chains. **p < 0.01.
The O antigen of S. Paratyphi A is composed of an α-d-mannose-(1,4)-α-l-rhamnose-(1,3)-α-d-galactose backbone (the O12 antigen) and a branching paratose residue that is α-(1,3)-linked to d-mannose in the backbone (the O2 antigen). O antigens of S. Typhimurium and S. Typhi have an identical trisaccharide backbone (the O12 antigen), but they carry abequose (the O4 antigen) or tyvelose (the O9 antigen), respectively, as branching sugars. Interestingly, rfbE is a pseudogene (i.e., an open reading frame disrupted by a mutation) in S. Paratyphi A (Figure 4B) (McClelland et al., 2004), which prevents conversion of cytidine diphosphate (CDP)-paratose to CDP-tyvelose and accounts for the presence of paratose (the O2 antigen) in place of tyvelose (the O9 antigen), the only structural difference between the O antigens of S. Paratyphi A and S. Typhi.
To determine whether the branching paratose residue (the O2 antigen) was necessary for inhibiting ROS production, we introduced the S. Typhi rfbE gene into S. Paratyphi A (ATCC9150). The resulting S. Paratyphi A strain, henceforth referred to as S. Paratyphi AO9, produced very long O antigen chains (Figure 4C) containing the O9 antigen instead of the O2 antigen (Figure 4D), which resulted in a robust induction of ROS production in HL-60 cells (Figures 4E and 4F) and primary human neutrophils (Figures S4D and S4E). Next, we replaced the S. Paratyphi A (ATCC9150) rfbS and rfbE genes, encoding paratose synthase and tyvelose epimerase, respectively, with the S. Typhimurium rfbJ gene, encoding abequose synthase (Figure 4B). The resulting S. Paratyphi A strain, henceforth referred to as S. Paratyphi AO4, produced very long O antigen chains (Figure 5A) containing the O4 antigen instead of the O2 antigen (Figure 5B); this resulted in robust ROS production in HL-60 cells (Figures 5C and 5D) and primary human neutrophils (Figures S4D and S4E). Collectively, these data supported the idea that the O2 antigen is necessary for averting the neutrophil respiratory burst.
Figure 5. Heterologous Production of the O4 Antigen by S. Paratyphi A Activates the Respiratory Burst in Neutrophil-like Cells.
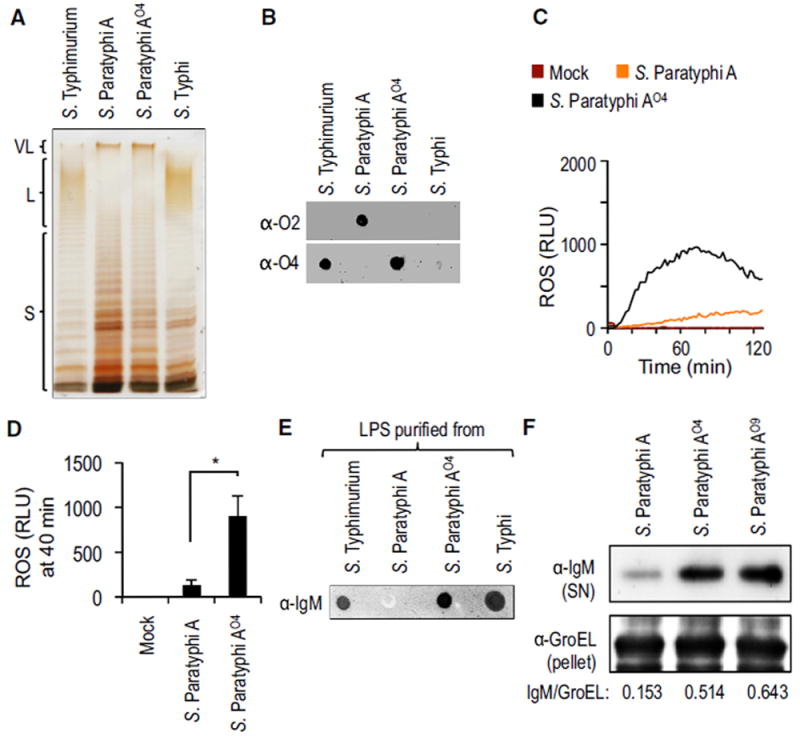
(A) LPS purified from the indicated bacterial strains was separated by SDS-PAGE and silver stained.
(B) LPS purified from the indicated bacterial strains was loaded on a dot blot to detect production of the O2 antigen (α-O2; top) or production of the O4 antigen (α-O4; bottom).
(C and D) HL-60 cells were infected with the indicated opsonized bacterial strains and ROS production monitored over time using chemiluminescence. (C) Representative experiment showing generation of chemiluminescence over time.
(D) Quantification of chemiluminescence from four independent experiments at the indicated time point after infection. Bars represent means ± SE.
(E) LPS purified from the indicated bacterial strains was loaded on a dot blot to detect binding of IgM present in NHS (α-IgM).
(F) The indicated bacterial strains were incubated in human serum and surface-bound proteins eluted with glycine. IgM eluted from the bacterial surface was detected by western blot (α-IgM; top). The bacterial protein GroEL was detected in the bacterial pellet (α-GroEL; bottom) and used as a loading control by measuring the IgM/GroEL using densitometry. L, long O antigen chains; Mock, mock infection; RLU, relative luminescence units; S, short O antigen chains; SN, supernatant; VL, very long O antigen chains. *p < 0.05.
Human IgM bound to LPS purified from S. Paratyphi AO4 but not to LPS purified from S. Paratyphi A (ATCC9150), suggesting that the O2 antigen was responsible for poor binding of human natural antibodies to LPS (Figure 5E). Interestingly, S. Paratyphi A opsonized with the serum of germ-free mice elicited robust ROS production in murine primary neutrophils, suggesting that unlike human natural antibodies, the natural antibody repertoire of mice recognizes the O2 antigen (Figures S4F and S4G). We then investigated the binding of human IgM to the bacterial surface. Human IgM bound to the surface of S. Paratyphi AO4 and Paratyphi AO9, but binding to the surface of S. Paratyphi A was reduced markedly in comparison (Figure 5F).
Next, we tested whether antibody-mediated complement activation through the classical pathway was responsible for the phagocyte respiratory burst triggered by S. Paratyphi AO4. Blocking the classical and the alternative pathways of complement activation with EGTA abrogated ROS production elicited by S. Paratyphi AO4 in HL-60 cells. Interestingly, selective inhibition of the classical pathway with EGTA and Mg2+ (Des Prez et al., 1975) resulted in a marked reduction of ROS production (Figure 6A). Similarly, the classical pathway was responsible for ROS production elicited by a fepE mutant of S. Paratyphi A strain ATCC9150 (Figure S6A), although IgM did not bind LPS purified from this strain (Figure S6B). To further investigate the contribution of antibodies to the triggering of the phagocyte respiratory burst, we depleted IgM or IgG from NHS. IgM depletion, but not IgG depletion, reduced the concentration of IgM in NHS (Figure 6B). Opsonization with IgM-depleted serum lowered ROS production triggered by S. Paratyphi AO4 in HL-60 cells to levels observed with its S. Paratyphi A parent strain (Figure 6C). In contrast, S. Paratyphi AO4 opsonized with IgG-depleted serum triggered significantly larger ROS production in HL-60 cells than the S. Paratyphi A parent strain. Collectively, these data provided compelling support for the idea that the O2 antigen averts the neutrophil respiratory burst in a fraction of individuals by preventing IgM binding.
Figure 6. Heterologous Production of the O4 Antigen by S. Paratyphi A Activates the Respiratory Burst in Neutrophil-like Cells through Antibody-Mediated Complement Activation.
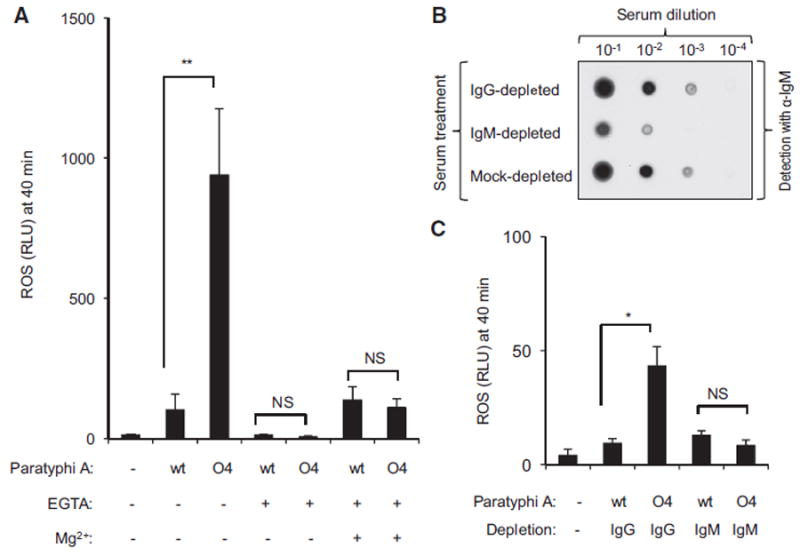
(A) HL60 cells were infected with the indicated opsonized bacterial strains and ROS production in the presence (+) or absence (-) of EGTA and magnesium (Mg2+) was monitored over time using chemiluminescence.
(B) The indicated serum dilutions were applied to a dot blot to detect the presence of IgM using anti-human IgM antibody (α-IgM).
(C) HL60 cells were infected with the indicated bacterial strains opsonized with IgG-depleted serum or IgM-depleted serum and ROS production monitored over time using chemiluminescence.
(A and C) Chemiluminescence was quantified from three independent experiments at the indicated time point after infection. Bars represent means ± standard error.
Mock, mock infection; NS, not statistically significantly different; RLU, relative luminescence units; wt, wild-type. *p < 0.05; **p < 0.001.
DISCUSSION
The viaB locus is a DNA region acquired by horizontal gene transfer that is present exclusively in Salmonella serovars associated with extraintestinal disease, including two human-adapted typhoidal Salmonella serovars, S. Typhi and S. Paratyphi C (Selander et al., 1990), and the bovine-adapted S. Dublin, which causes bacteremia in cattle (Selander et al., 1992). The presence of the viaB locus confers some of the properties that distinguish S. Typhi from non-typhoidal Salmonella serovars associated with human gastroenteritis, such as S. Typhimurium (Keestra-Gounder et al., 2015; Raffatellu et al., 2006; Tsolis et al., 2008; Wangdi et al., 2012). However, the viaB locus is absent from S. Paratyphi A, which is puzzling because paratyphoid fever is indistinguishable in its symptoms from typhoid fever.
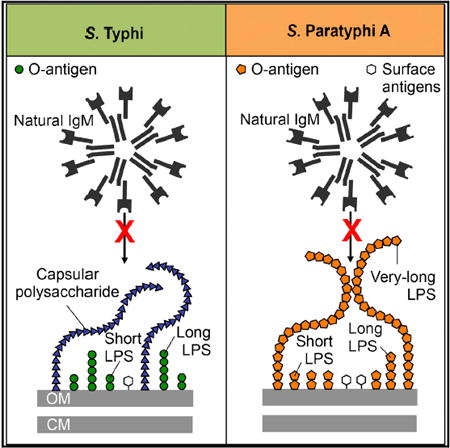
S. Paratyphi A evolved from an ancestral organism producing the O9 -antigen through a loss-of-function mutation in rfbE, resulting in expression of the O2 antigen instead of the O9 antigen (McClelland et al., 2004). Here, we show that very long O antigen chains carrying the O2 antigen do not trigger the neutrophil respiratory burst in humans because they prevent antibody binding, at least in a fraction of individuals. By analogy, S. Typhi covers its surface with the Vi antigen, which does not trigger the neutrophil respiratory burst by preventing antibody binding (Hart et al., 2016). In an immunologically naive host, binding of natural IgM to the bacterial surface activates complement through the classical pathway (Reid et al., 1997), which appeared to be the main pathway leading to ROS production in neutrophils infected with non-capsulated S. Typhi or S. Paratyphi A lacking very long O antigen chains. The picture emerging from this and previous work is that polysaccharide chains can prevent complement-dependent ROS production, if they (1) do not bind natural IgM and (2) are long enough to prevent pentameric IgM molecules from gaining access to other potential binding sites at the bacterial surface (Figure 7). In support of this idea, the presence of very long O antigen chains can interfere with immune evasion by the Vi antigen (Crawford et al., 2013), presumably because the former rival the capsule in length, thereby exposing IgM binding sites at the bacterial surface (Figures 7A–7C). Furthermore, short and long O antigen chains produced by an S. Paratyphi A fepE mutant did not bind IgM, but this was not sufficient for preventing ROS production, presumably because very long O antigen chains are the only surface structures long enough to cover up other IgM binding sites (Figures 7D and 7E). These results suggested that S. Typhi and S. Paratyphi A both used very long polysaccharide chains to prevent human IgM binding, complement activation, and ROS production, a shared virulence strategy that distinguishes them from non-typhoidal Salmonella serovars associated with human gastroenteritis. This remarkable example of convergent evolution was not apparent from previous work focusing on horizontal gene transfer of the viaB locus, but it becomes apparent only when the contribution of pseudogene formation is taken into account. This situation is reminiscent of that observed during the evolution of Yersinia pestis from a Yersinia pseudotuberculosis-like ancestor, which involved a combination horizontal gene transfer (ymt and pla) and pseudogene formation (ureD, rcsA, pde2, and pde3) to complete the transition from a gastrointestinal pathogen to an arthropod-borne extraintestinal pathogen (Hinnebusch et al., 2016). Recent work suggests that the transition from a gastrointestinal to an extraintestinal pathogen involved deletion of genes encoding a pathway for anaerobic b-oxidation of butyrate in S. Typhi and S. Paratyphi A, which moderates intestinal inflammation during typhoid and paratyphoid fever (Bronner et al., 2018).
Figure 7. Model for Evasion of the Phagocyte Respiratory Burst by Typhoidal Salmonella Serovars.
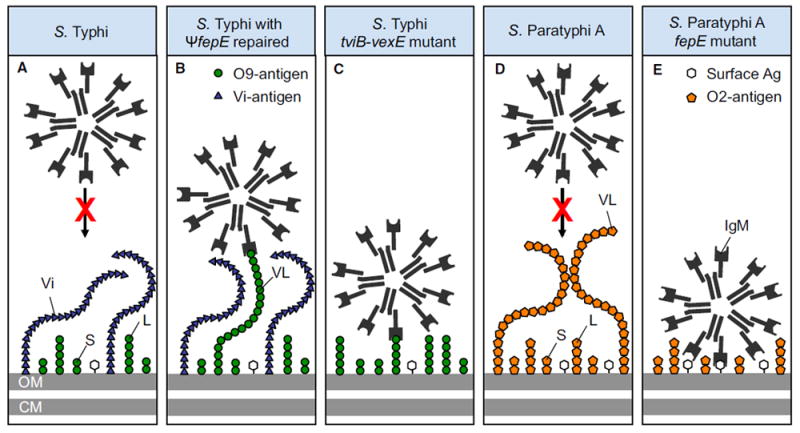
(A) The polysaccharide chains of the Vi antigen shield underlying surface structures from IgM binding.
(B) Restoration of the fepE pseudogene (ΨfepE) in S. Typhi results in production of very long O antigen chains that rival the Vi antigen in length, thereby elevating IgM binding to the bacterial surface.
(C) Deletion of the capsule biosynthesis genes in the S. Typhi tviB-vexE mutant exposes the short and long O antigen chains, thereby resulting in IgM binding.
(D) The very long O antigen chains of S. Paratyphi A contain the O2 antigen, which does not bind natural IgM, thereby shielding underlying surface structures.
(E) The short and long O antigen chains present in an S. Paratyphi A fepE mutant do not bind IgM, but are too short to prevent IgM binding to other surface antigens. Ag, antigens; CM, plasma membrane; L, long O antigen chains; OM, outer membrane; S, short O antigen chains; Vi, Vi antigen; VL, very long O antigen chains.
Interestingly, whereas the O2 antigen prevented binding of human natural antibodies (Figure 5), it did not prevent opsonization with serum from germ-free mice (Figures S4F and S4G). These data indicate that unlike the natural antibody reservoir of humans, the natural antibody reservoir of mice recognizes the O2 antigen. The O2 antigen also is present in a small number of non-typhoidal serovars, including S. Nitra, S. Kiel, and S. Koessen (Grimont and Weill, 2007), that are not commonly isolated from patients or animals bred as food (Edwards et al., 1948; Hargrett-Bean et al., 1988; Rigney et al., 2004). These epidemiological observations suggest that S. Nitra, S. Kiel, and S. Koessen are not adapted to humans, but they may circulate in an unknown wild animal reservoir.
EXPERIMENTAL PROCEDURES
Bacterial Strains, Plasmids, and Growth Conditions
Bacterial strains used in this study are presented in Table S1. Bacterial cultures were routinely incubated with aeration at 37°C in Luria-Bertani (LB) broth (10 g tryptone, 5 g yeast extract, and 10 g NaCl/L) or on LB agar plates unless indicated otherwise. Antibiotics were added as appropriate. To induce expression of Vi capsule polysaccharide, low sodium (0.25% NaCl) was used to preculture bacterial strains.
S. Paratyphi A strain TWS110 (ΔrfbSE∷rfbJSTM) was generated by replacing the rfbSE genes with the rfbJ gene from S. Typhimurium via homologous recombination. Briefly, the S. Typhimurium rfbHJ region was amplified with primers 83/84 (Table S2). S. Paratyphi A rfbX was amplified with primers 85/86. The two fragments were ligated in tandem and were cloned into plasmid pRDH10 using the Gibson Assembly kit (New England Biolabs). The construct was transformed into Escherichia coli S17λpir cells. The resulting donor strain (TWE76) was conjugated with wild-type S. Paratyphi A to generate strain TWS110 (ΔrfbSE∷rfbJSTM).
To generate strain TWS159 (S. Paratyphi A 9150 ΔfepE∷KanR), fepE flanking regions 1 and 2 were amplified from S. Paratyphi A 9150 with primers 133/134 and 135/136, respectively. The two fragments were ligated in tandem and were cloned into plasmid pRDH10 using the Gibson Assembly kit. The construct was transformed into E. coli S17λpir cells. The resulting plasmid (pTW93) was isolated and digested at the engineered BamHI restriction site located between flanking regions 1 and 2. A kanamycin-resistance cassette was inserted and the plasmid (pTW117) was transformed into E. coli S17λpir cells. E. coli S17λpir (pTW117) was conjugated with S. Paratyphi A 9150 carrying a temperature-sensitive plasmid pSW172 that conferred carbenicillin resistance for counter selection. An exconjugant was cured of pSW172 to generate strain TWS159. To complement the mutation, fepE was amplified from S. Paratyphi A 9150 with primers 179/180 and was inserted into pWSK29.
To generate an S. Paratyphi A strain producing the O9 antigen (S. Paratyphi A 9150 ΔrfbE∷rfbESTY), rfbE-flanking regions 1 and 2 were amplified from S. Paratyphi A 9150 with primers 171/172 and 173/174, respectively. The two fragments were ligated in tandem and were cloned into plasmid pRDH10 using the Gibson Assembly kit. The resulting plasmid (pTW126) was digested with BamHI to insert a kanamycin-resistance cassette to generate plasmid pTW131. E. coli S17λpir (pTW131) was conjugated with S. Paratyphi A 9150 (pSW172). An exconjugant was cured of pSW172 to generate S. Paratyphi A 9150 ΔrfbE (HH121). Next, rfbE and its two flanking regions were amplified from S. Typhi with primers 86/87. The fragment was inserted into pRDH10 to generate plasmid pTW131. E. coli S17λpir (pTW131) was conjugated with HH121 to generate strain HH122 (S. Paratyphi A 9150 ΔrfbE∷rfbESTY).
LPS Analysis
LPS was purified from overnight cultures of bacterial strains using hot-aqueous phenol extraction (Davis and Goldberg, 2012). A sensitive silverstaining protocol (Tsai and Frasch, 1982) was then used for detection of purified LPS separated by SDS-PAGE.
Animal Experiments
All of the experiments in this study were approved by the Institutional Animal Care and Use Committee at the University of California, Davis. Germ-free Swiss-Webster mice were bred in-house. Femurs collected from germ-free mice after euthanasia were used to isolate neutrophils.
Isolation of Primary Neutrophils
The University of California, Davis institutional review board approved the protocol for obtaining blood draws for this study. Human neutrophils were isolated from the peripheral blood of healthy adult donors using a procedure described previously (Lee et al., 2015).
To collect murine primary neutrophils, mice were euthanized and femurs were collected. Neutrophils were then isolated from bone marrow cell suspension using the EasySep mouse neutrophil enrichment kit (StemCell Technologies) in accordance with the manufacturer’s instructions.
Serum Sensitivity Test
Bacterial cultures were resuspended and adjusted to optical density (OD) 0.1 in PBS. A total of 180 μL of cultures were then dispensed into each well of a 96-well plate, containing 20 μL of undiluted human serum. Plates were incubated at room temperature with gentle agitation for 3 hr. At each time point, 20-μL samples were collected, and the bacterial numbers were enumerated. As an alternative method, bacteria were incubated with 20% serum for 3 hr. Then, 10 μL of undiluted or 10−2 and 10−4 diluted samples were spotted on an LB plate and incubated at 37°C overnight and images captured (BioSpectrum Imaging System, UVP).
Sequence Analysis
The following reference genomes were used in this study: S. Typhi Ty2 (GenBank: AE014613.1), S. Paratyphi A 9150 (GenBank: CP000026.1), S. Typhimurium LT2 (GenBank: AE006468.2), and S. Paratyphi A var. Durazzo 11511 gene (GenBank: CP019185.1). To verify the S. Paratyphi A 11511 fepE nucleotide sequence, the gene was amplified with primers 137/138 and cloned into plasmid PCR2.1 to yield pTW128. For amino acid sequence alignment, Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo) was used.
ROS Assay
ROS production by HL-60 cells or primary neutrophils was measured using chemiluminescence. HL-60 cells were differentiated into neutrophil-like cells by incubation with RPMI 1640 medium (Thermo Fisher Scientific) containing 1.3% DMSO (Sigma), 15% fetal bovine serum (FBS, Thermo Fisher Scientific) and 25 mM HEPES ((4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid), Thermo Fisher Scientific) for 5–6 days. Differentiated HL-60 cells or primary neutrophils were washed and resuspended in phenol red free RPMI 1640 containing 2% FBS and luminol (Sigma) was added (1 mM). Approximately 5 × 104 HL-60 cells in 90 μL were seeded into an opaque 96-well microplate (Perkin-Elmer) and the basal luminescence was measured every 2 min for 5 min on a FilterMax F3 plate reader (Molecular Devices). HL-60 cells were infected with 10 μL of bacteria (MOI = 10) opsonized with 20% human serum (30 min at room temperature) or were mock treated, and ROS production was measured every 2 min using a plate reader. In some experiments, human serum was treated with Futhan, 10 mM EGTA, or 10 mM Mg2+ plus 10 mM EGTA (MgEGTA) before opsonizing bacteria.
Dot Blot Assay
LPS was purified using an LPS extraction kit (iNtRON Biotechnology). Vi capsule polysaccharide was purified essentially as described for capsular polysaccharides from Bacteroides fragilis (Pantosti et al., 1991). Briefly, frozen cell pellets of S. Typhi strain Ty2 were lysed and extracted with 37% phenol (Sigma) in water at 68°C with vigorous agitation. The aqueous phase was collected and extracted with ethyl ether (Thermo Fisher Scientific) to remove the remaining phenol, and the ether was removed from the aqueous phase on a rotary evaporator (BUCHI) at 60°C for 2 hr. DNA and RNA were eliminated from the sample by DNase and RNase (both Worthington Biochemical) digestion, respectively, and protein removed by serial digests with Pronase (Calbiochem). Polysaccharide was then harvested by ethanol precipitation and separated from LPS on a Sephacryl S300 (GE Healthcare) column in 3% deoxycholate (Sigma).
Purified LPS at 5 μL or Vi capsule polysaccharide (100 μg/mL) were spotted onto a nitrocellulose membrane and air dried. The membrane was blocked with 5% skim milk in PBS-T (PBS containing 0.25% Tween 20). To detect specific O antigens or Vi capsule polysaccharide, we used anti-O2, -O4, -O9, or anti-Vi capsule polysaccharide polyclonal antibodies (BD Biosciences), respectively, and secondary horseradish peroxidase (HRP)-anti-rabbit Ig. To assess IgM binding to LPS or Vi capsule polysaccharide, the membrane was incubated with diluted (1/1000–1/5000) human serum overnight at 4°C. Bound IgM was detected using HRP-anti-human IgM (Jackson ImmunoResearch). Enhanced chemiluminescence (ECL; PerkinElmer) was used to develop, and images were obtained on an imager (BioSpectrum Imaging System, UVP).
IgM Binding
To assess human IgM binding to the surface of bacterial cells, 1 × 109 colony-forming units (CFUs) of the indicated bacterial strains were incubated with 500 μL of 20% NHS (heat inactivated) or PBS for 1 hr at room temperature. After washing the bacteria with PBS four times, bound serum proteins were eluted by resuspending the samples in 100 μL of 0.1 M glycine-HCl followed by centrifugation. A total of 80 μL supernatants were collected and subsequently neutralized with 20 μL of 1 M (tris(hydroxymethyl)aminomethane) (Tris)-HCl buffer (pH 8.0). Neutralized samples and bacterial pellets were precipitated with methanol-chloroform and resuspended with Laemmli sample buffer. Samples were subjected to 10% SDS-PAGE gel. After electrotransfer, the polyvinylidene fluoride (PVDF) membranes (Merck Millipore) were probed with HRP-conjugated anti-human IgM antibody (Jackson ImmunoResearch). As an internal control, bacterial pellets were separated by SDS-PAGE, blotted onto PVDF, and probed with anti-GroEL (Sigma). The blots were developed using ECL (PerkinElmer), and images were obtained on an imager (BioSpectrum Imaging System, UVP).
MS
Proteins eluted from the bacterial surface were analyzed by MS. Briefly, 5 × 108 CFU of bacteria were incubated with 500 μL of 20% NHS, heat-inactivated 20% NHS, or PBS for 40 min at room temperature. Bacterial pellets were spun down and washed with PBS four times. Bound serum proteins and some bacterial proteins were eluted in 100 μL of 0.1 M glycine-HCl (pH 2.7). Supernatants (80 μL) were neutralized by adding 20 μL of 1 M Tris (pH 8.0). Samples were then precipitated using methanol-chloroform. The precipitated pellets were resuspended in 60 μL SDS-sample buffer and purified by PAGE followed by MS analysis by the Proteomics Core Facility at the University of California, Davis Genome Center.
Statistical Analysis
All of the data are expressed as the mean and standard error of the mean. Student’s t tests were used for statistical analyses. p < 0.05 was considered significant.
Supplementary Material
In Brief.
The clinical presentation of typhoid fever differs from gastroenteritis, which has been attributed to the Salmonella enterica serovar (S.) Typhi capsular polysaccharide. Paradoxically, S. Paratyphi A is not capsulated but causes typhoid-like disease. Hiyoshi et al. show that the very long O antigen of S. Paratyphi A functions as a capsule, an example of convergent evolution.
Highlights.
Typhoidal but not non-typhoidal Salmonella serovars foil the PMN respiratory burst
The Vi antigen averts binding of natural antibodies to foil the PMN respiratory burst
S. Paratyphi A very long O antigen chains foil the PMN respiratory burst
Virulence in typhoidal Salmonella serovars arose through convergent evolution
Acknowledgments
We acknowledge expert technical assistance by Brett S. Phinney and Michelle Salemi from the University of California, Davis Proteomics Core Facility. We thank Sumathi Sankaran and Briana Young for expert technical assistance. H.H. was supported by a fellowship from the Daiichi Sankyo Foundation of Life Sciences, Japan. B.A.C. was supported by Public Health Service grant GM082916. T.W. was supported by Public Health Service grant AI126080. Work in A.J.B.’s laboratory was supported by Public Health Service grants AI044170, AI096528, AI112445, and AI112949.
Footnotes
AUTHOR CONTRIBUTIONS
Conceptualization, H.H., T.W., and A.J.B.; Methodology, H.H., T.W., G.L., C.S., M.R., and B.A.C.; Investigation, H.H., T.W., G.L., C.S., M.R., B.A.C., and A.J.B.; Resources, M.R., B.A.C., and A.J.B.; Funding Acquisition, T.W., B.A.C., and A.J.B.; Writing–Original Draft, H.H., T.W., and A.J.B.; Writing–Review and Editing, H.H., T.W., and A.J.B.; Supervision, H.H., T.W., and A.J.B.
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and two tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.01.016.
References
- Batchelor RA, Haraguchi GE, Hull RA, Hull SI. Regulation by a novel protein of the bimodal distribution of lipopolysaccharide in the outer membrane of Escherichia coli. J Bacteriol. 1991;173:5699–5704. doi: 10.1128/jb.173.18.5699-5704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batchelor RA, Alifano P, Biffali E, Hull SI, Hull RA. Nucleotide sequences of the genes regulating O-polysaccharide antigen chain length (rol) from Escherichia coli and Salmonella typhimurium: protein homology and functional complementation. J Bacteriol. 1992;174:5228–5236. doi: 10.1128/jb.174.16.5228-5236.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokoch GM. Chemoattractant signaling and leukocyte activation. Blood. 1995;86:1649–1660. [PubMed] [Google Scholar]
- Bravo D, Silva C, Carter JA, Hoare A, Alvarez SA, Blondel CJ, Zaldívar M, Valvano MA, Contreras I. Growth-phase regulation of lipopolysaccharide O-antigen chain length influences serum resistance in serovars of Salmonella. J Med Microbiol. 2008;57:938–946. doi: 10.1099/jmm.0.47848-0. [DOI] [PubMed] [Google Scholar]
- Bronner DN, Faber F, Olsan EE, Byndloss MX, Sayed NA, Xu G, Yoo W, Kim D, Ryu S, Lebrilla CB, et al. Genetic ablation of butyrate utilization attenuates gastrointestinal Salmonella disease. Cell Host Microbe. 2018;23:266–273. doi: 10.1016/j.chom.2018.01.004. this issue. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bueno SM, Santiviago CA, Murillo AA, Fuentes JA, Trombert AN, Rodas PI, Youderian P, Mora GC. Precise excision of the large pathogenicity island, SPI7, in Salmonella enterica serovar Typhi. J Bacteriol. 2004;186:3202–3213. doi: 10.1128/JB.186.10.3202-3213.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crawford RW, Wangdi T, Spees AM, Xavier MN, Tsolis RM, Bäumler AJ. Loss of very-long O-antigen chains optimizes capsule-mediated immune evasion by Salmonella enterica serovar Typhi. MBio. 2013;4:e00232–13. doi: 10.1128/mBio.00232-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull World Health Organ. 2004;82:346–353. [PMC free article] [PubMed] [Google Scholar]
- Davis MR, Jr, Goldberg JB. Purification and visualization of lipopolysaccharide from Gram-negative bacteria by hot aqueous-phenol extraction. J Vis Exp. 2012;(63):3916. doi: 10.3791/3916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Prez RM, Bryan CS, Hawiger J, Colley DG. Function of the classical and alternate pathways of human complement in serum treated with ethylene glycol tetraacetic acid and MgCl2-ethylene glycol tetraacetic acid. Infect Immun. 1975;11:1235–1243. doi: 10.1128/iai.11.6.1235-1243.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards PR, Bruner DW, Moran AB. Further studies on the occurrence and distribution of Salmonella types in the United States. J Infect Dis. 1948;83:220–231. doi: 10.1093/infdis/83.3.220. [DOI] [PubMed] [Google Scholar]
- Grimont PAD, Weill F-X. WHO Collaborating Centre for Reference and Research on Salmonella. Ninth Edition. 2007. Antigenic Formulae of the Salmonella Serovars. [Google Scholar]
- Hargrett-Bean NT, Pavia AT, Tauxe RV. Salmonella isolates from humans in the United States, 1984-1986. MMWR CDC Surveill Summ. 1988;37:25–31. [PubMed] [Google Scholar]
- Hart PJ, O’Shaughnessy CM, Siggins MK, Bobat S, Kingsley RA, Goulding DA, Crump JA, Reyburn H, Micoli F, Dougan G, et al. Differential killing of Salmonella enterica serovar Typhi by antibodies targeting Vi and lipopolysaccharide O:9 antigen. PLoS One. 2016;11:e0145945. doi: 10.1371/journal.pone.0145945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch BJ, Chouikha I, Sun YC. Ecological opportunity, evolution, and the emergence of flea-borne plague. Infect Immun. 2016;84:1932–1940. doi: 10.1128/IAI.00188-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohn DC, Lehrer RI. NADPH oxidase deficiency in X-linked chronic granulomatous disease. J Clin Invest. 1975;55:707–713. doi: 10.1172/JCI107980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kauffmann F, Silberstein W. Untersuchungen über einige neue Salmonella-typen. Zentralbl Bakteriol Parasitenkd Infecktionskr. 1934;132:431–437. [Google Scholar]
- Keestra-Gounder AM, Tsolis RM, Bäumler AJ. Now you see me, now you don’t: the interaction of Salmonella with innate immune receptors. Nat Rev Microbiol. 2015;13:206–216. doi: 10.1038/nrmicro3428. [DOI] [PubMed] [Google Scholar]
- Kossack RE, Guerrant RL, Densen P, Schadelin J, Mandell GL. Diminished neutrophil oxidative metabolism after phagocytosis of virulent Salmonella typhi. Infect Immun. 1981;31:674–678. doi: 10.1128/iai.31.2.674-678.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee CY, Thompson GR, 3rd, Hastey CJ, Hodge GC, Lunetta JM, Pappagianis D, Heinrich V. Coccidioides endospores and spherules draw strong chemotactic, adhesive, and phagocytic responses by individual human neutrophils. PLoS One. 2015;10:e0129522. doi: 10.1371/journal.pone.0129522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liston SD, Ovchinnikova OG, Whitfield C. Unique lipid anchor attaches Vi antigen capsule to the surface of Salmonella enterica serovar Typhi. Proc Natl Acad Sci USA. 2016;113:6719–6724. doi: 10.1073/pnas.1524665113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Looney RJ, Steigbigel RT. Role of the Vi antigen of Salmonella typhi in resistance to host defense in vitro. J Lab Clin Med. 1986;108:506–516. [PubMed] [Google Scholar]
- McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M, et al. Comparison of genome degradation in Paratyphi A and Typhi, human-restricted serovars of Salmonella enterica that cause typhoid. Nat Genet. 2004;36:1268–1274. doi: 10.1038/ng1470. [DOI] [PubMed] [Google Scholar]
- McPhail LC, DeChatelet LR, Shirley PS, Wilfert C, Johnston RB, Jr, McCall CE. Deficiency of NADPH oxidase activity in chronic granulomatous disease. J Pediatr. 1977;90:213–217. doi: 10.1016/s0022-3476(77)80632-x. [DOI] [PubMed] [Google Scholar]
- Miller RM, Garbus J, Hornick RB. Lack of enhanced oxygen consumption by polymorphonuclear leukocytes on phagocytosis of virulent Salmonella typhi. Science. 1972;175:1010–1011. doi: 10.1126/science.175.4025.1010. [DOI] [PubMed] [Google Scholar]
- Moellering RC, Jr, Weinberg AN. Persistent Salmonella infection in a female carrier for chronic granulomatous disease. Ann Intern Med. 1970;73:595–601. doi: 10.7326/0003-4819-73-4-595. [DOI] [PubMed] [Google Scholar]
- Murray GL, Attridge SR, Morona R. Regulation of Salmonella typhimurium lipopolysaccharide O antigen chain length is required for virulence; identification of FepE as a second Wzz. Mol Microbiol. 2003;47:1395–1406. doi: 10.1046/j.1365-2958.2003.03383.x. [DOI] [PubMed] [Google Scholar]
- Nair S, Alokam S, Kothapalli S, Porwollik S, Proctor E, Choy C, McClelland M, Liu SL, Sanderson KE. Salmonella enterica serovar Typhi strains from which SPI7, a 134-kilobase island with genes for Vi exopolysaccharide and other functions, has been deleted. J Bacteriol. 2004;186:3214–3223. doi: 10.1128/JB.186.10.3214-3223.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nuccio S-P, Wangdi T, Winter SE, Baumler AJ. Typhoid. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F, editors. The Prokaryotes. Springer-Verlag; 2013. pp. 353–374. [Google Scholar]
- Pantosti A, Tzianabos AO, Onderdonk AB, Kasper DL. Immunochemical characterization of two surface polysaccharides of Bacteroides fragilis. Infect Immun. 1991;59:2075–2082. doi: 10.1128/iai.59.6.2075-2082.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Chessa D, Wilson RP, Tükel C, Akçelik M, Bäumler AJ. Capsule-mediated immune evasion: a new hypothesis explaining aspects of typhoid fever pathogenesis. Infect Immun. 2006;74:19–27. doi: 10.1128/IAI.74.1.19-27.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raffatellu M, Santos RL, Chessa D, Wilson RP, Winter SE, Rossetti CA, Lawhon SD, Chu H, Lau T, Bevins CL, et al. The capsule encoding the viaB locus reduces interleukin-17 expression and mucosal innate responses in the bovine intestinal mucosa during infection with Salmonella enterica serotype. Typhi Infect Immun. 2007;75:4342–4350. doi: 10.1128/IAI.01571-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid RR, Prodeus AP, Khan W, Hsu T, Rosen FS, Carroll MC. Endotoxin shock in antibody-deficient mice: unraveling the role of natural antibody and complement in the clearance of lipopolysaccharide. J Immunol. 1997;159:970–975. [PubMed] [Google Scholar]
- Rigney CP, Salamone BP, Anandaraman N, Rose BE, Umholtz RL, Ferris KE, Parham DR, James W. Salmonella serotypes in selected classes of food animal carcasses and raw ground products, January 1998 through December 2000. J Am Vet Med Assoc. 2004;224:524–530. doi: 10.2460/javma.2004.224.524. [DOI] [PubMed] [Google Scholar]
- Seger RA, Tiefenauer L, Matsunaga T, Wildfeuer A, Newburger PE. Chronic granulomatous disease due to granulocytes with abnormal NADPH oxidase activity and deficient cytochrome-b. Blood. 1983;61:423–428. [PubMed] [Google Scholar]
- Selander RK, Beltran P, Smith NH, Helmuth R, Rubin FA, Kopecko DJ, Ferris K, Tall BD, Cravioto A, Musser JM. Evolutionary genetic relationships of clones of Salmonella serovars that cause human typhoid and other enteric fevers. Infect Immun. 1990;58:2262–2275. doi: 10.1128/iai.58.7.2262-2275.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selander RK, Smith NH, Li J, Beltran P, Ferris KE, Kopecko DJ, Rubin FA. Molecular evolutionary genetics of the cattle-adapted serovar Salmonella dublin. J Bacteriol. 1992;174:3587–3592. doi: 10.1128/jb.174.11.3587-3592.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai CM, Frasch CE. A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal Biochem. 1982;119:115–119. doi: 10.1016/0003-2697(82)90673-x. [DOI] [PubMed] [Google Scholar]
- Tsolis RM, Young GM, Solnick JV, Bäumler AJ. From bench to bedside: stealth of enteroinvasive pathogens. Nat Rev Microbiol. 2008;6:883–892. doi: 10.1038/nrmicro2012. [DOI] [PubMed] [Google Scholar]
- Virlogeux I, Waxin H, Ecobichon C, Popoff MY. Role of the viaB locus in synthesis, transport and expression of Salmonella typhi Vi antigen. Microbiology. 1995;141:3039–3047. doi: 10.1099/13500872-141-12-3039. [DOI] [PubMed] [Google Scholar]
- Wangdi T, Winter SE, Bäumler AJ. Typhoid fever: “you can’t hit what you can’t see”. Gut Microbes. 2012;3:88–92. doi: 10.4161/gmic.18602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wangdi T, Lee C-Y, Spees AM, Yu C, Kingsbury DD, Winter SE, Hastey CJ, Wilson RP, Heinrich V, Bäumler AJ. The Vi capsular polysaccharide enables Salmonella enterica serovar typhi to evade microbeguided neutrophil chemotaxis. PLoS Pathog. 2014;10:e1004306. doi: 10.1371/journal.ppat.1004306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wetter M, Goulding D, Pickard D, Kowarik M, Waechter CJ, Dougan G, Wacker M. Molecular characterization of the viaB locus encoding the biosynthetic machinery for Vi capsule formation in Salmonella Typhi. PLoS One. 2012;7:e45609. doi: 10.1371/journal.pone.0045609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RP, Winter SE, Spees AM, Winter MG, Nishimori JH, Sanchez JF, Nuccio SP, Crawford RW, Tükel ç, Bäumler AJ. The Vi capsular polysaccharide prevents complement receptor 3-mediated clearance of Salmonella enterica serotype Typhi. Infect Immun. 2011;79:830–837. doi: 10.1128/IAI.00961-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelstein JA, Marino MC, Johnston RB, Jr, Boyle J, Curnutte J, Gallin JI, Malech HL, Holland SM, Ochs H, Quie P, et al. Chronic granulomatous disease. Report on a national registry of 368 patients. Medicine (Baltimore) 2000;79:155–169. doi: 10.1097/00005792-200005000-00003. [DOI] [PubMed] [Google Scholar]
- Winter SE, Raffatellu M, Wilson RP, Rüssmann H, Bäumler AJ. The Salmonella enterica serotype Typhi regulator TviA reduces interleukin-8 production in intestinal epithelial cells by repressing flagellin secretion. Cell Microbiol. 2008;10:247–261. doi: 10.1111/j.1462-5822.2007.01037.x. [DOI] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Thiennimitr P, Gerriets VA, Nuccio SP, Rüssmann H, Bäumler AJ. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol Microbiol. 2009;74:175–193. doi: 10.1111/j.1365-2958.2009.06859.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Godinez I, Yang H-J, Rüssmann H, Andrews-Polymenis HL, Bäumler AJ. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 2010;6:e1001060. doi: 10.1371/journal.ppat.1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Poon V, Keestra AM, Sterzenbach T, Faber F, Costa LF, Cassou F, Costa EA, Alves GE, et al. Salmonella enterica serovar Typhi conceals the invasion-associated type three secretion system from the innate immune system by gene regulation. PLoS Pathog. 2014;10:e1004207. doi: 10.1371/journal.ppat.1004207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Atluri V, Poon V, Romão EL, Tsolis RM, Bäumler AJ. The flagellar regulator TviA reduces pyroptosis by Salmonella enterica serovar Typhi. Infect Immun. 2015;83:1546–1555. doi: 10.1128/IAI.02803-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


