Abstract
The clinical presentation of primary hyperparathyroidism (PHPT) varies widely, although the underlying mechanistic reasons for this disparity remain unknown. We recently reported that parathyroid tumors can be functionally segregated into two distinct groups on the basis of their relative responsiveness to ambient calcium, and that patients in these groups differ significantly in their likelihood of manifesting bone disability. To examine the molecular basis for this phenotypic variation in PHPT, we compared the global gene expression profiles of calcium-sensitive and calcium-resistant parathyroid tumors. RNAseq and proteomic analysis identified a candidate set of differentially expressed genes highly correlated with calcium-sensing capacity. Subsequent quantitative assessment of the expression levels of these genes in an independent cohort of parathyroid tumors confirmed that calcium-sensitive tumors cluster in a discrete transcriptional profile group. These data indicate that PHPT is not an etiologically monolithic disorder and suggest that divergent molecular mechanisms could drive the observed phenotypic differences in PHPT disease course, provenance, and outcome.
Keywords: Primary hyperparathyroidism, parathyroid adenoma, tumor profiling, gene expression
Introduction
Primary hyperparathyroidism (PHPT) is a common endocrine neoplastic disorder of calcium metabolism that occurs in 11 to 30 individuals per 100,000 in the U.S. (Wermers, et al. 2006). The disease can induce a broad spectrum of pathophysiological sequelae, including nephrolithiasis, neurologic symptoms, and reduced bone density (Bilezikian and Silverberg 2004; Hyperparathyroidism 2005). Despite the variable clinical presentation of the disease, efforts to define the molecular mechanisms underlying PHPT phenotypic heterogeneity have been limited to date (Lee, et al. 2016; Morrison, et al. 2004). To address this issue, we developed a comprehensive clinical registry of PHPT patient information and related these data to live-cell functional assessment of parathyroid tumor biochemical signaling capacity. To measure calcium sensing capacity, we employed an ex vivo system we developed for quantitating cellular responsiveness to changes in ambient calcium concentrations (Koh, et al. 2016a; Koh, et al. 2016b). In this system, parathyroid tumor cells loaded with an intracellular fluorescent indicator (Fluo-4-AM) are challenged with varying levels of extracellular calcium, and intracellular calcium flux is detected in real time as a dynamic readout of signal transduction mediated through the calcium sensing receptor (CASR). The proportions of responding cells are plotted as a function of extracellular calcium concentration to generate a sigmoid dose-response curve. The extracellular calcium concentration required to induce a half-maximal response (Ca EC50) is calculated from the dose-response curve as an indicator of tumor calcium sensitivity. We have found that parathyroid adenomas segregate into two functionally distinct groups with respect to calcium responsiveness. We classify tumors with a Ca EC50 greater than 3.0 mM as “calcium-resistant”, while those with a Ca EC50 of less than 3.0 mM are annotated as “calcium-sensitive.”
Using this approach, we showed that parathyroid tumors exhibiting calcium resistance were strongly associated with reduced bone density in patients with sporadic, non-familial PHPT (Weber, et al. 2017). The finding that attenuated calcium responsiveness in a subset of parathyroid tumors is selectively linked to clinically significant bone mineral density (BMD) loss suggests the existence of phenotypically distinct sub-classes of PHPT patients with differential disease courses and alternative underlying etiologies. To examine the molecular basis for variable calcium sensing in parathyroid tumor tissue that gives rise to these alternative phenotypes, we performed a transcriptional profiling study seeking to identify gene expression patterns that differentiate calcium-resistant and calcium-sensitive tumors.
Materials and Methods
Human Subjects Procedures
All procedures using materials derived from human subjects were reviewed and approved by the Duke University Institutional Review Board (IRB). Parathyroid tumor sections were prepared from surgically resected tissue obtained from patients with non-familial, sporadic PHPT undergoing parathyroidectomy at Duke University Medical Center. Patients pre-operatively diagnosed with PHPT who were referred to Duke University Medical Center for surgical management were eligible for participation and enrolled after providing informed consent as described under IRB-approved protocol #Pro00046210. Pertinent pre-operative clinical information was obtained through review of Duke Maestro Care, an EPIC-based electronic health record, as well as outside clinical documents from referring providers.
RNAseq
Total RNA was extracted from freshly acquired parathyroid tumor specimens using a commercial reagent kit (Promega ReliaPrep RNA Tissue System, Z6110). Prior to sequencing, RNA integrity was verified using an Agilent 2100 Bioanalyzer (RIN > 9). RNA-seq data were processed using the TrimGalore toolkit (http://www.bioinformatics.babraham.ac.uk/projects/trim_galore), which employs Cutadapt (Martin 2011) to trim low quality bases and Illumina sequencing adapters from the 3′ end of the reads. Only reads that were 20nt or longer after trimming were kept for further analysis. Reads were mapped to the GRCh37v75 version of the human genome and transcriptome (Kersey, et al. 2012) using the STAR RNA-seq alignment tool (Dobin, et al. 2013). Reads were kept for subsequent analysis if they mapped to a single genomic location. Gene counts were compiled using the HTSeq tool (https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu638). Only genes that had at least 10 reads in any given library were used in subsequent analyses. Normalization and differential expression were carried out using the DESeq2 (Love, et al. 2014) Bioconductor (Huber, et al. 2015) package with the R statistical programming environment (www.r-project.org). The false discovery rate (FDR) was calculated using the Benjamini-Hochberg approach (Benjamini and Hochberg 1995; Benjamini and Yekutieli 2001) to control for multiple hypothesis testing. The significance threshold for adjusted p-values was set at 0.01. Gene Set Enrichment Analysis (Mootha, et al. 2003) was performed to identify differentially regulated pathways and gene ontology terms for each of the comparisons performed.
Proteomics
Formalin-fixed, paraffin-embedded 10 micron sections were prepared from each specimen as sample input sources. 250 ng of tryptic protein digest from each sample were analyzed using a nanoAcquity UPLC system (Waters) coupled to a Q Exactive Plus Orbitrap high resolution accurate mass tandem mass spectrometer (Thermo) via a nanoelectrospray ionization source. Raw data were imported into Rosetta Elucidator v4.0 software and aligned based on the accurate mass and retention time of detected ions. Relative peptide abundance was calculated based on area under the curve (AUC) measurements of aligned peak features across all runs. The dataset had 271,196 quantified features and 396,039 MS/MS spectra for sequencing. Peptides were matched against a custom Swiss-Prot database of unique human peptide sequences seeded with internal standard and contaminant protein controls along with an equivalent number of reverse-sequence decoy entries for false positive rate determination (40,546 total entries).
Nanostring Data Acquisition
Tumor sections were selected following histological review to confirm parathyroid adenoma tissue identity and adequate cross-sectional area for RNA extraction. RNA was prepared from 5 micron sections of formalin-fixed, paraffin-embedded parathyroid tumor sections using the Qiagen RNeasy commercial reagent kit (Qiagen, 73504), substituting the included proteinase K with a proteinase K solution acquired from Roche (Roche 03115836001) as recommended by Nanostring. RNA was evaluated using an Agilent 2100 Bioanalyzer G2939A instrument and an RNA Nano Chip for quantitation and quality control assessment. DV200 smear analysis was performed to verify that all samples met the quality control criteria of >50% of the total RNA content being greater than 200 nt in length. Gene expression was evaluated using the Nanostring nCounter platform (Geiss, et al. 2008). 200 ng of isolated total RNA was hybridized to capture and reporter probes for a minimum of 16 hours at 65°C, according to the standardized Nanostring protocol. Data were normalized to the mean of assay positive controls as well as the mean of housekeeping gene internal standards (B2M, G6PD, GAPDH, PGK1) incorporated into the codeset design.
Nanostring Data Analysis
The raw NanoString data were processed, underwent strict quality control, and were normalized using the NanoStringNorm package from the R statistical programming environment (Waggott, et al. 2012). In brief, samples were first removed if they had more than 90% missing data. The data were then normalized to eliminate systematic differences across the samples by using the geometric mean of the positive controls as a scaling factor. The samples then underwent background correction by removing the mean plus two standard deviations of the negative controls from the signal. The samples then were re-normalized for RNA-concentration by generating a scaling factor from the geometric mean of the housekeeping genes. Individual expression values for genes that were below 1 were set to a value of 1. Genes whose expression values were below 1 in all samples were removed prior to clustering. Expression values for each gene were then log2 transformed followed by z-score transformation across the samples. After transformation, both genes and samples were clustered using a correlation distance with complete linkage.
Results
Characteristics of the study population
The demographic, clinical, and biochemical characteristics of the 48 patients included in this study are summarized in Table 1. Patients with calcium-resistant tumors (n=27) were older and have a lower body mass index than those with calcium-sensitive tumors (n=21). Race, sex, and ethnicity distributions did not differ between the two patient groups. As we reported previously, reduced bone mineral density (BMD) was strongly associated with tumor calcium-sensing status; patients with osteoporosis or osteopenia were much more likely to have calcium-resistant tumors. The incidence of nephrolithiasis, pathological fracture, peptic ulcers, or multi-gland disease was not significantly different between the two patient groups. With respect to preoperative biochemical findings, patients with calcium-resistant tumors had higher serum calcium and ionized calcium levels, and significantly lower BMD as measured by skeletal T-score.
Table 1.
Study Population Characteristics (N=48)
| Patient Characteristics | All Patients (N=48) |
Calcium-Resistant (CaEC50 > 3.0 mM) (N=27) |
Calcium-Sensitive (CaEC50 < 3.0 mM) (N=21) |
|
|---|---|---|---|---|
| N (%) | N (%) | N (%) | p | |
| Demographics | ||||
| Age at Consent (Years) – Median (IQR) | 61 (50, 70) | 67 (60, 73) | 52 (40, 56) | 0.008 |
| Body Mass Index – Median (IQR) | 30.85 (24.28, 37.75) | 27 (39.5, 55.5) | 36.7 (30.25, 50.6) | 0.0003 |
| Sex | NS | |||
| Female | 40 (83.3%) | 22 (81.5%) | 18 (85.7%) | |
| Male | 8 (16.7%) | 5 (18.5%) | 3 (14.3%) | |
| Race | NS | |||
| White | 35 (72.9%) | 22 (81.5%) | 13 (61.9%) | |
| Black or African American | 8 (16.7%) | 3 (11.1%) | 5 (23.8%) | |
| Other/Unknown | 5 (10.4%) | 2 (7.4%) | 3 (14.3%) | |
| Ethnicity | NS | |||
| Hispanic or Latino | 3 (6.3%) | 3 (11.1%) | 0 (0%) | |
| Non-Hispanic or Latino | 45 (93.7%) | 24 (88.9%) | 21 (100%) | |
| Clinical History | ||||
| Nephrolithiasis | 13 (27.1%) | 7 (25.9%) | 6 (28.6%) | NS |
| Pathologic Fracture | 1 (2.1%) | 1 (3.7%) | 0 (0%) | NS |
| Peptic Ulcer Disease | 0 (0%) | 0 (0%) | 0 (0%) | NS |
| Osteoporosis | 13 (27.1%) | 13 (48.1%) | 0 (0%) | <0.0001 |
| Osteopenia | 11 (22.9%) | 9 (33.3%) | 2 (9.5%) | 0.0019 |
| Number of Glands Removed | NS | |||
| 1 | 43 (89.5%) | 24 (88.9%) | 19 (90.5%) | |
| 2 | 2 (4.2%) | 2 (7.4%) | 0 (0%) | |
| 3 | 3 (6.3%) | 1 (3.7%) | 2 (9.5%) | |
| 4 | 0 (0%) | 0 (0%) | 0 (0%) | |
| Preoperative Biochemical Characteristics | Median (IQR) | Median (IQR) | Median (IQR) | |
| Intact Parathyroid Hormone (14–72 pg/ml) | 134 (98, 187.3) | 143 (96, 230) | 119 (98, 157.5) | NS |
| Serum Calcium (8.7–10.2 mg/dl) | 10.9 (10.6, 11.5) | 11.0 (10.8, 11.7) | 10.8 (10.5, 11.2) | 0.0493 |
| Albumin (3.5–4.8 g/dl) | 4.1 (3.8, 4.3) | 4.1 (3.8, 4.3) | 4.0 (3.8, 4.2) | NS |
| Ionized Calcium (1.15–1.32 mM/L) | 1.40 (1.36, 1.57) | 1.54 (1.36, 1.74) | 1.38 (1.35, 1.54) | 0.031 |
| Serum Creatinine (0.4–1.0 mg/dl) | 0.8 (0.7, 1) | 0.9 (0.7, 1) | 0.80 (0.7, 0.9) | NS |
| MDRD calculated GFR (ml/min) | 77.98 (62.39, 93.47) | 73.04 (61.34, 86.01) | 79.98 (75.32, 94.68) | NS |
| Vitamin 25 D (30–100 ng/ml) | 22 (15, 32) | 24 (13, 34) | 22 (20, 32) | NS |
| Insufficient (<20 ng/ml) – N (%) | 14 (31.1%) | 10 (40%) | 4 (20%) | NS |
| Sufficient (≥20 ng/ml) – N (%) | 31 (68.9%) | 15 (60%) | 16 (80%) | NS |
| 24 Hour Urine Calcium (50–300 mg) | 257 (223, 448) | 245 (197, 424) | 298 (229, 454) | NS |
| Lowest T Score | −2.1 (−3.23, −0.9) | −3.1 (−3.75, −2.15) | −0.8 (−1.7, −0.2) | <0.0001 |
Data are presented as N (%) unless otherwise specified.
Percentages may not add up to 100% due to rounding or missing values.
The T-score metric is a comparative measure of an individual’s bone mineral density, indicating the number of standard deviations above (positive) or below (negative) the age-matched mean. By convention, an individual’s lowest T-score among multiple skeletal sites is used as the diagnostic criterion for reduced bone density. A T-score of <−2.5 indicates osteoporosis; a T-score of −1.0 to −2.0 indicates osteopenia.
Abbreviations: IQR=Interquartile Range. GFR=Glomerular Filtration Rate. MDRD=Modification of Diet in Renal Disease. NS=Not Significant
Comparative RNAseq analysis of calcium-resistant and calcium-sensitive parathyroid tumors
We initially performed a series of RNAseq and proteomics comparisons between calcium-resistant and calcium-sensitive tumors. The specimens utilized for the RNAseq and proteomics assays were drawn from a consecutive series of non-familial sporadic PHPT cases whose demographic and clinical characteristics are not significantly different from the overall PHPT patient population at our institution. Equal numbers of calcium-resistant and calcium-sensitive tumors were included in each sample group. Consistent with our earlier findings, each patient in the calcium-resistant sample group had reduced bone density. Six representative single gland parathyroid adenoma tumor specimens were selected for transcriptional profiling (Table 2). Using conservative estimates (false positive rate = 0.05; power=0.8; sequencing depth to produce a minimum count of 200 per transcript; a coefficient of variation of 0.5; and a target effect size of 3-fold expression change between groups), a widely accepted statistical power model designed for RNAseq comparative expression studies (Hart, et al. 2013) determined that the minimum required number of samples in each calcium response group is 3. All six specimens were pathologically confirmed as parathyroid adenomas, and all shared similar tumor architecture and histological appearance (Fig. S1). Three of the tumors displayed normal calcium sensitivity (calcium EC50<3.0) and were not associated with osteoporosis or osteopenia. Three other tumors were calcium-resistant (calcium EC50>3.0) and derived from patients with bone mineral density deficit (osteoporosis or osteopenia) demonstrated by dual-energy X-ray absorptiometry (DXA). Using a FDR threshold of 5%, 128 genes were found to be differentially expressed between calcium-sensitive and calcium-resistant tumors, with 65 genes meeting a FDR threshold of 1% (Table S1). Of these genes, 28 were found to be expressed from 4-fold to as much as 20-fold higher in calcium-sensitive tumors than in calcium-resistant tumors (Table S2). Ten genes were expressed from four-fold to 10-fold higher in calcium-resistant tumors than in calcium-sensitive tumors (Table S3). Parathyroid hormone (PTH) was highly expressed at equivalent levels in all six tumors, confirming parathyroid tissue identity. Notably, CASR also was highly expressed at equivalent levels in both calcium-sensitive and calcium-resistant tumors (Fig. S2).
Table 2.
Tumor samples submitted for RNAseq analysis. The T-score metric is a comparative measure of an individual’s bone mineral density, indicating the number of standard deviations above (positive) or below (negative) the age-matched mean. By convention, an individual’s lowest T-score among multiple skeletal sites is used as the diagnostic criterion for reduced bone density. A T-score of <−2.5 indicates osteoporosis; a T-score of −1.0 to −2.0 indicates osteopenia.
| ID | EC50 | Lowest T-score | Gender | BMD diagnosis |
|---|---|---|---|---|
| 217 | 2.46 | −1.7 | F | Normal |
| 230 | 2.48 | −1.2 | F | Normal |
| 238 | 2.67 | −0.2 | F | Normal |
| 221 | 3.62 | −3.1 | F | Osteoporosis |
| 235 | 3.73 | −4.4 | F | Osteoporosis |
| 223 | 3.45 | −2.4 | F | Osteopenia |
A heat map of genes differentially expressed (FDR ≤0.05) in calcium-resistant (high EC50) vs calcium-sensitive (low EC50) tumors reveals distinct global transcriptional profiles associated with calcium sensing capacity, and by extension, with the clinical presentation of diminished BMD (Fig. 1). The emergence of consistent transcriptional profiles correlated with calcium sensing functional response in human parathyroid tumors strongly suggests the existence of discrete and definable programs of gene expression linked to tumor behavior and PHPT patient bone density outcome.
Figure 1.
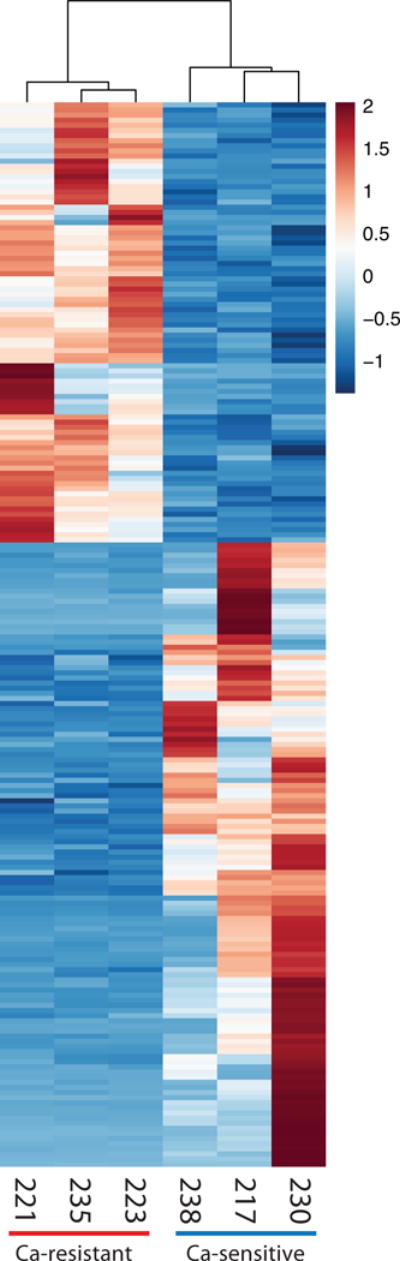
Heat map of 208 differentially expressed genes (FDR ≤ 0.05). Gene expression has been z-score normalized. Samples and genes are clustered by correlation distance with complete linkage. Sample ID numbers for each specimen are shown below their corresponding columns.
Gene Set Enrichment Analysis of RNAseq Data
The RNAseq data were subjected to Gene Set Enrichment Analysis (GSEA) to identify pathways or functional groups associated with the high and low EC50 transcriptional signatures (Mootha et al. 2003). The MSigDB database of manually curated pathways (including KEGG and Reactome) and the Gene Ontology terms (GO) database were queried in parallel against the list of differentially expressed genes arranged in rank order of significance of differential expression and direction of change. 67 gene sets from the GO database and 126 gene sets from the MSigDB database were significantly enriched (p-value adjusted for multiple testing <0.01) in calcium-resistant tumors (Table S4). Gene sets associated with mitochondrial function were significantly enriched (Normalized Enrichment Score ≥2.30) in calcium-resistant tumors and dominated both the MSigDB curated pathways and GO database results. Twelve of the 20 most significantly enriched MSigDB pathways, including the top five, were associated with mitochondrial function, as were 13 of 20 enriched GO terms, including the top 11. All 12 of the mitochondrial gene set enrichment plots displayed a similar pattern of association with calcium-resistant tumors. Enrichment Score (ES) plots for two of these mitochondrial gene sets are shown in Fig. 2. 124 GO terms gene sets and 126 MSigDB gene sets were significantly associated (p-value adjusted for multiple testing <0.01) with the calcium-sensitive phenotype (Table S5). Differentially expressed genes down-regulated in calcium-resistant tumors (NES ≤−2.1) fell into three groups: genes associated with the extracellular matrix (ECM; 6 of 20 enriched gene sets, including the top 4 among GO terms; 6 of 20 among the curated pathways gene sets); genes associated with receptor signaling (3 of 20 in GO; 5 of 20 curated pathways); and genes associated with cation homeostasis (3 of 20 in GO; 1 of 20 curated pathways). These results suggest that increased expression of mitochondrial components is a key characteristic of calcium-resistant (high EC50) tumors associated with compromised bone density. Conversely, reduced expression of receptor signaling and ECM genes may contribute to attenuation of calcium sensitivity in calcium-resistant tumors compared to calcium-sensitive tumors. For example, the mitochondrial genes NDUFV1 (NADH:Ubiquinone oxidoreductase core subunit V1), COX7B (cytochrome C oxidase subunit 7B), CYCS (cytochrome C, somatic), and ATP5G3 (ATP Synthase, H+ transporting, mitochondrial Fo complex subunit C3) are expressed at significantly higher levels in calcium-resistant tumors relative to calcium-sensitive tumors (Fig. 3). In contrast, membrane trafficking proteins such as LIN7A (lin-7 homolog A) and the ECM proteins such as TNXB (Tenascin) are preferentially expressed in calcium-sensitive (low EC50) tumors (Fig. 4).
Figure 2.
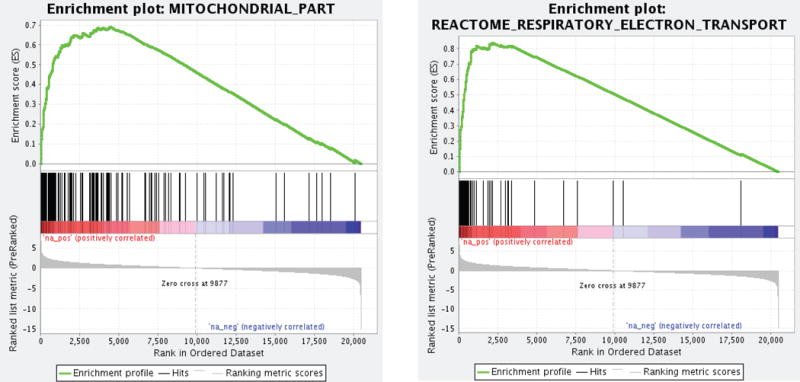
GSEA plots for GO (left) and MSigDB (right) mitochondrial gene sets in the rank ordered list of genes differentially expressed in calcium-resistant tumors. The red end of the GSEA spectrum indicates genes preferentially expressed in calcium-resistant tumors.
Figure 3.
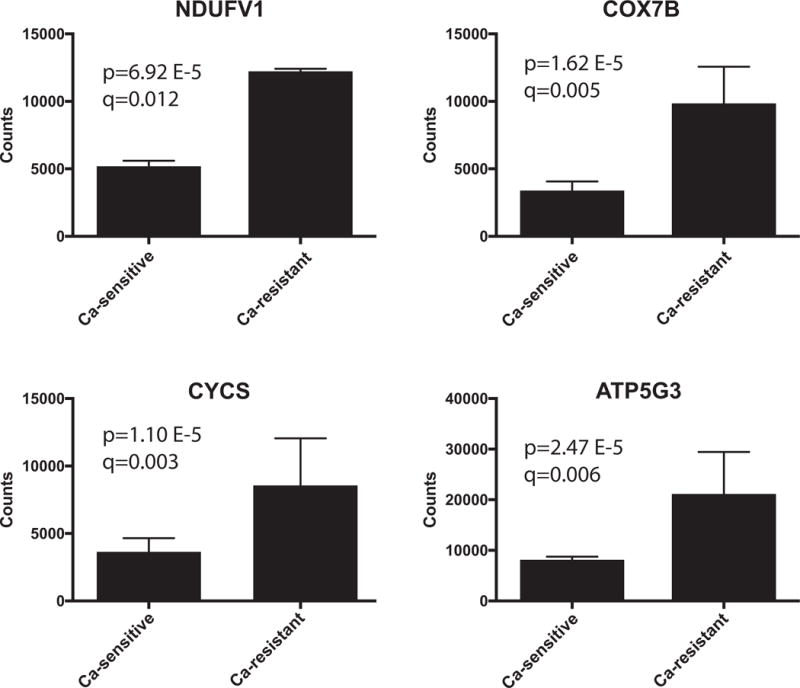
Elevated expression of mitochondrial genes in calcium-resistant (high EC50) parathyroid tumors. The y-axis displays the mean and standard deviation of normalized RNAseq counts for each of the four genes in three calcium-sensitive and three calcium-resistant tumors. The p- and q- values for differential expression between the two tumor types are shown for each gene. The q-values are the p-values adjusted for FDR.
Figure 4.
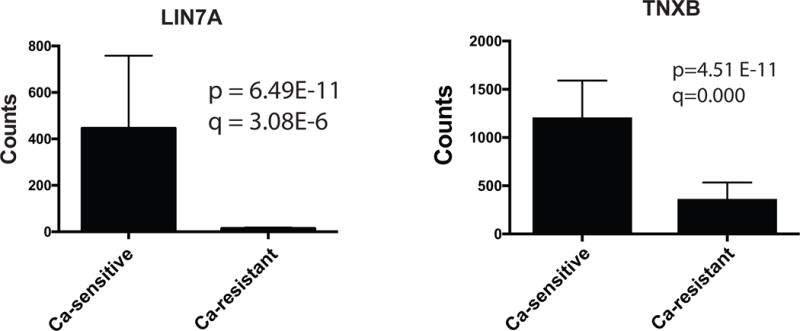
Elevated expression of ECM and trafficking genes LIN7A and TNXB in calcium-sensitive (low EC50) tumors. The y-axis displays the mean and standard deviation of normalized RNAseq counts for each gene in three calcium-sensitive and three calcium-resistant tumors. The p- and q- values for differential expression between the two tumor types are shown for each gene. The q-values are the p-values adjusted for FDR.
Global proteomics comparison of calcium-resistant and calcium sensitive tumors
In parallel with these transcriptome studies, we performed quantitative one-dimensional liquid-chromatography tandem mass spectroscopy (LC-MS/MS) proteomics analysis in an independent sample set of parathyroid adenomas to identify proteins whose relative abundance differed between calcium-resistant and calcium-sensitive tumors. A group of three calcium-resistant tumors derived from PHPT patients with osteoporosis and three calcium-sensitive tumors from patients with normal bone density were selected for comparison. The tumors all shared similar tumor architecture and histological appearance (Fig. S3). After annotation for a <1.0% peptide false discovery rate and filtering to remove low quality peptides, quantitative data were obtained for 18,844 peptides and 3,274 proteins. The mean percent coefficient of variation (%CV) among triplicate repeats of an all-sample pooled protein injection was 8.9% for all proteins and 5.3% for proteins quantified by two or more peptides. The %CV between individual tumor samples, a measure of biological variability, was 54% for all proteins and 48% for proteins quantified by 2 or more peptides. These data indicate that technical reproducibility was high and that quantitative differences in protein abundance observed in this experiment are predominantly reflective of biological rather than technical variability.
To identify proteins differentially expressed in calcium-resistant vs calcium-sensitive tumors, we calculated fold-change ratios of average protein intensities between the two sample groups;129 proteins were found to be differentially expressed (adjusted p-value for difference <0.05). The abundance of 43 proteins was found to be increased ≥2-fold in calcium-resistant tumors, with an average %CV of 8.8% across pooled technical replicates. 39 proteins were found to be ≥2-fold decreased in calcium-resistant tumors, with an average %CV of 8.0% across pooled technical replicates. In striking agreement with the RNAseq data, numerous mitochondrial proteins were significantly up-regulated in calcium-resistant (high EC50) tumors. Mitochondrial 28S and 39S ribosomal subunits were enriched in calcium-resistant tumors (Fig. 5A); in contrast, cytoplasmic ribosomal subunits (40S and 60S) were equally abundant in both EC50 groups (Fig. 5B). Multiple additional proteins with “mitochondria” in their gene ontology annotations or with known mitochondrial localization (e.g., sideroflexin-2, CDGSH iron sulfur domain-containing protein 1, and carnitine O-palmitoyltransferase 1) were preferentially expressed in calcium-resistant specimens (Fig. 5C), consistent with elevated mitochondrial mass in these tumors.
Figure 5.
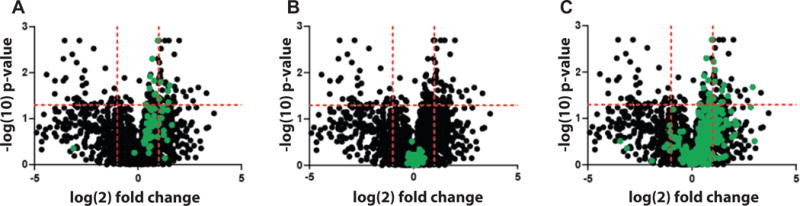
Proteins differentially expressed in calcium-resistant vs calcium-sensitive tumors are plotted with log-2-fold change on the x-axis and –log10(p-value) on the y-axis. The horizontal red line indicates p = 0.05 (proteins above the line are significantly different). Vertical red lines indicate -2-fold and +2-fold change in calcium-resistant vs calcium-sensitive groups. P-values are adjusted for multiple testing. (A) Mitochondrial ribosomal subunits are shown in green. (B) cytoplasmic ribosomal subunits in green. (C) proteins with “mitochondria” in their gene ontology descriptors are marked in green.
Verification of differential gene expression candidates in an independent cohort of parathyroid tumors
To confirm the emergent gene expression patterns observed in these studies in a larger, unselected cohort of PHPT cases, we assembled a retrospective sample set of 48 formalin-fixed, paraffin-embedded (FFPE) parathyroid tumor specimens for multiplexed gene expression quantitation using the Nanostring nCounter analysis system (Geiss et al. 2008). The Nanostring platform utilizes digital color-coded barcode technology for direct quantitation of individual transcripts without enzymatic amplification, making it possible to assay FFPE-derived RNA that would otherwise be too fragmented for analysis by sequencing-based methods. We designed a custom Nanostring chipset to query 33 genes from each sample (Table S6). The chipset included four internal positive controls known to be highly expressed in (PTH, CHGA, CASR) or restricted (GCM2) to parathyroid tissue, along with four housekeeping genes for signal normalization (B2M, G6PD, GAPDH, and PGK1). A total of 25 genes found in our RNAseq data to be differentially expressed between calcium-sensitive and calcium-resistant tumors were selected for the chipset. The primary inclusion criterion was strength of association with the calcium sensing phenotype as expressed by rank ordering each gene’s adjusted p-values for linkage to calcium sensing status. Twelve genes expressed more highly in calcium-sensitive tumors were selected. These genes included the top seven with the smallest adjusted p-values (RGS2, P2RX2, ADD2, HPSE, LIN7A, SAMD11, and TNXB), and five additional strongly associated genes, each chosen as representative of pathways from which multiple genes were found to be associated with the calcium-sensitive phenotype (SLC8A1, MME, SORL1, DDX43, VCAN). Similarly, 13 genes expressed more highly in calcium-resistant tumors were incorporated into the chipset. These genes included the top eight with the smallest adjusted p-values (DIRC3, SPSB4, ATP5A1, SEC14L4, HYAL1, FRK, MAP7D2, and EFHD2), and five additional strongly associated genes. Four of the five additional genes are mitochondrial pathway components (COX7B, CYCS, ATP5G3, and PCYT2). The fifth gene, RPH3AL, is a possible tumor suppressor protein known to play a regulatory role in calcium-dependent exocytosis in endocrine cells (Cheviet, et al. 2004; Smith, et al. 1999).
Validation of Nanostring chipset
We first evaluated the Nanostring chipset and methodology in a series of control experiments to determine (1) technical reproducibility; (2) consistency of results from intact vs FFPE-derived RNA from the same tumor; and (3) concordance between Nanostring data generated from FFPE-derived RNA and tumor-matched RNAseq data. To assess reproducibility, linear regression curves were plotted comparing log2-transformed Nanostring expression data for the 33-gene chipset from technical replicates of FFPE-derived parathyroid tumor RNA isolates. Technical replicates were highly consistent and linear over more than four orders of magnitude (R2 = 0.9966, p <0.0001) (Fig. S4). By paired two-sided t-test, mean difference between replicates is 0.1124 (95% confidence interval: −0.023 to 0.237). RNA source bias was not a factor, as Nanostring expression data from tumor-matched freshly prepared intact RNA and FFPE-extracted RNA were highly correlated (R2 = 0.9153, p < 0.0001) (Figure S5). By paired two-sided t-test, the mean difference between these two sources is −0.5531 (95% confidence interval: −0.836 to −0.270). Finally, we compared log2-transformed expression data from FFPE-extracted RNA evaluated on the Nanostring platform to the corresponding RNAseq gene counts from the same tumor. The data from the two platforms were strongly concordant (R2 = 0.8513, p < 0.0001) (Fig. S6). The mean of the differences between the two measures was 2.234 (95% confidence interval: 1.498 to 2.97).
With the performance of the platform and chipset validated, we employed the Nanostring system to evaluate 36 additional unselected parathyroid tumors. Of these samples, 13 had previously been determined to be calcium-sensitive, with 23 calcium-resistant. RNA was prepared from FFPE sections from each tumor, and expression of the 33 genes in our chipset was assessed. The raw RCC output data were normalized and background subtracted as previously described (Waggott et al. 2012) to generate a heat map of relative gene expression values.
Cluster analysis of Nanostring data
Unsupervised cluster analysis of these data revealed three groups of tumors (Fig. 6). Cluster 1 consisted entirely of calcium-sensitive, low EC50 tumors. Of the 13 calcium-sensitive tumors in the sample set, 10 were grouped together in Cluster 1. Contingency analysis by two-sided Fisher’s exact test showed that the grouping of calcium-sensitive tumors within this cluster was highly significant (p = 0.0015); the exclusion of calcium-resistant tumors from Cluster 1 also was significant (Fisher’s exact test; p = 0.0035). Cluster 1 tumors featured increased levels of TNXB and LIN7A, consistent with the RNAseq data from calcium-sensitive tumors (Fig. 4), along with elevated expression of other RNAseq-based calcium-sensitive candidate markers, including DDX43, HPSE, and SLC8A. Expression of mitochondrial genes such as CYCS, COX7B, ATP5G3, and ATP5A1 was relatively low among Cluster 1 tumors, as expected from the RNAseq data.
Figure 6.
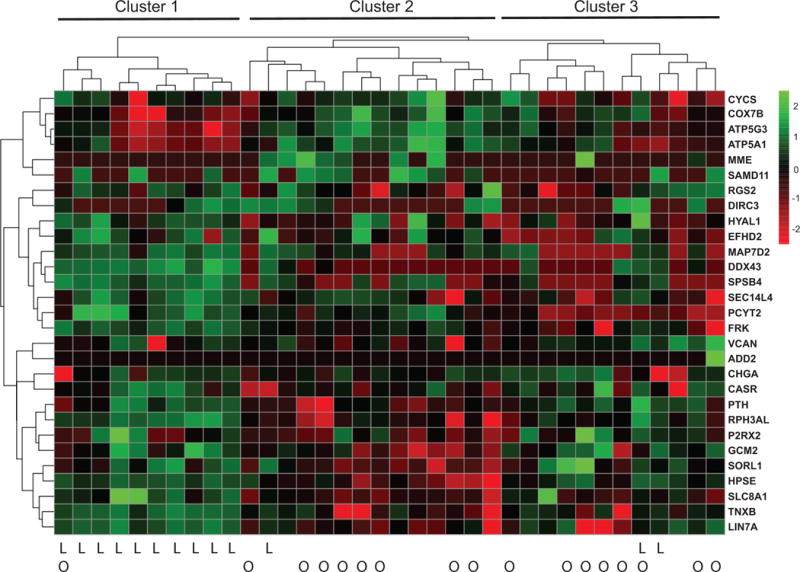
Unsupervised clustering of differentially expressed genes in calcium-sensitive vs calcium resistant tumors using a Nanogene candidate gene codeset based on RNAseq data. The heat map depicts log2-transformed normalized expression levels for 29 genes (rows) in 36 different parathyroid adenomas (columns). Hierarchical cluster relationships based on Euclidean distances are shown above the heat map. Calcium-sensitive (low EC50) tumors are indicated by “L” below the heat map; tumors associated with osteoporosis are marked with “O”.
Calcium-resistant tumors were distributed roughly equally between Clusters 2 and 3. Cluster 2 appeared to have the strongest mitochondrial signature, with elevated expression of CYCS, COX7B, ATP5G3, and ATP5A1, a result consistent with the prior RNAseq data (Fig. 3). Cluster 3 expressed generally lower amounts of these mitochondrial gene transcripts, although still at a somewhat higher level on average than the Cluster 1 tumors. In contrast to Cluster 2, Cluster 3 tumors expressed slightly higher levels of the protein trafficking gene SORL1 and the ECM protein HPSE, both of which were originally identified as candidate markers of calcium-sensitive tumors. Tumors derived from PHPT patients with osteoporosis fell almost exclusively within Clusters 2 and 3, with only a single instance found in Cluster 1. The association between calcium-resistant tumor gene expression profiles and bone mineral density loss observed in this sample set provides molecular evidence supporting our earlier report of a link between tumor biochemical behavior and PHPT clinical presentation (Weber et al. 2017).
Discussion
The data reported here, generated through three independent approaches analyzing non-overlapping sample sets, reveal that parathyroid tumors causative of PHPT can be subdivided into distinct classes based on molecular profiling. This finding is consistent with our earlier work demonstrating the segregation of parathyroid tumors into calcium-sensitive and calcium-resistant functional groups, and further refines the concept that diversity in PHPT clinical presentation may be reflective of differences in the underlying molecular mechanisms driving the disease. The emergence of a distinct molecular signature associated with calcium-sensitive tumors suggests that this subset of neoplasms shares a common etiology that may determine a disease course less likely to provoke bone density loss. The fact that calcium-resistant tumors appear to resolve into two transcriptional profile clusters suggests that the origin and behavior of these tumors may be more complex, perhaps reflective of alternative mechanistic pathways towards loss or attenuation of calcium responsiveness in incipient parathyroid tumor tissue. However, it is clear from our data that attenuation of calcium sensitivity in calcium-resistant tumors is not solely driven by silencing or loss of CASR expression as has been proposed (Cetani, et al. 2000; Corbetta, et al. 2000; Farnebo, et al. 1997). Since CASR mutations have been shown to be rare in sporadic PHPT adenomas (Arnold, et al. 2002; Brennan, et al. 2013; Cromer, et al. 2012), the underlying cause of failed calcium sensing in calcium-resistant tumors could involve additional components beyond CASR itself. Moreover, elevated expression of core mitochondrial components in calcium-resistant tumors suggests that diminished calcium sensing capacity or other physiological changes associated with the presence of this tumor class may create increased demand for electron transport chain capacity, calcium uptake, or other mitochondrial functions. Consistent with this idea, target genes induced by the transcriptional co-activator PPARGC1A, a master regulator of mitochondrial biogenesis, were found to be collectively enriched in calcium-resistant tumors. PPARGC1A, itself four-fold more abundant in calcium-resistant (high EC50) tumors (log2(fold change) = 2.21; p=0.0006) has been shown to induce mitochondrial biogenesis in response to a wide range of metabolic stresses in diverse tissues, including the kidney, cardiac muscle, neuronal tissue, skeletal muscle, and vascular endothelial cells (Benton, et al. 2010; Das and Sharma 2015; Lai, et al. 2014; Wang, et al. 2015; Xiong, et al. 2013; Yuan, et al. 2012). This central regulator could represent a key nodal point whose downstream targets collectively confer the calcium-resistant phenotype. Although proteomic analyses of parathyroid tumors have been limited to date, two prior studies (Giusti, et al. 2011; Varshney, et al. 2014) comparing extracts from adenomatous and normal parathyroid tissues using 2D gel electrophoresis found a number of mitochondrial components to be expressed more highly in tumors than in normal tissues. Moreover, recent paper (Akpinar, et al. 2017) describing a comparative proteomic study of parathyroid adenoma versus parathyroid hyperplasia tissue samples reported that mitochondrial components appeared to be more abundant in parathyroid hyperplasia samples relative to adenomas. This result is consistent with the idea that calcium-resistant (mitochondria-rich) and calcium-sensitive (mitochondria-poor) adenomas may arise via distinct etiological mechanisms, with calcium-resistant tumors perhaps originating from hyperplastic tissue. It is possible that heterogeneity with respect to the relative abundance of mitochondria-rich parathyroid oxyphil cells could contribute to the apparent enrichment of mitochondrial components in calcium-resistant tumors. However, we did not observe any consistent relationship between tumor oxyphilic content and calcium response category, suggesting that mitochondrial accumulation may be occurring in parathyroid adenomas prior to or independent of the appearance of morphologically identifiable oxyphil cells.
The functions of genes found to be differentially expressed between calcium-sensitive and calcium-resistant parathyroid adenomas suggest divergent potential mechanisms underlying the distinct biochemical phenotypes observed in these tumors (Fig. 7). For example, the preferential expression of ECM and protein trafficking genes in calcium-sensitive tumors could reflect a compensatory upregulation in components associated with CASR signal transduction, in response to chronically elevated serum calcium levels. ECM binding has been shown to play an important role in transmitting contextual cues required for appropriate CASR-mediated signal transduction (Tharmalingam and Hampson 2016). Maintained expression of ECM genes such as TNXB in calcium-sensitive tumors could thus contribute to retention of CASR signaling capacity. Because intracellular trafficking of CASR is widely understood to be essential for receptor desensitization and maintenance of functional responsiveness in the constant presence of its cognate ligand (Breitwieser 2013; Chakravarti, et al. 2012; Ray 2015), components such as LIN7A that regulate transmembrane receptor intracellular movement could directly influence relative calcium sensing capacity. LIN7A is involved in generating and maintaining a polarized distribution of channels and receptors at the plasma membrane, forming multiprotein complexes that regulate delivery and recycling of plasma membrane proteins to specific membrane domains (Rodriguez-Boulan and Macara 2014); diminution or loss of this key function could result in impaired CASR signaling in calcium-resistant tumors.
Figure 7.
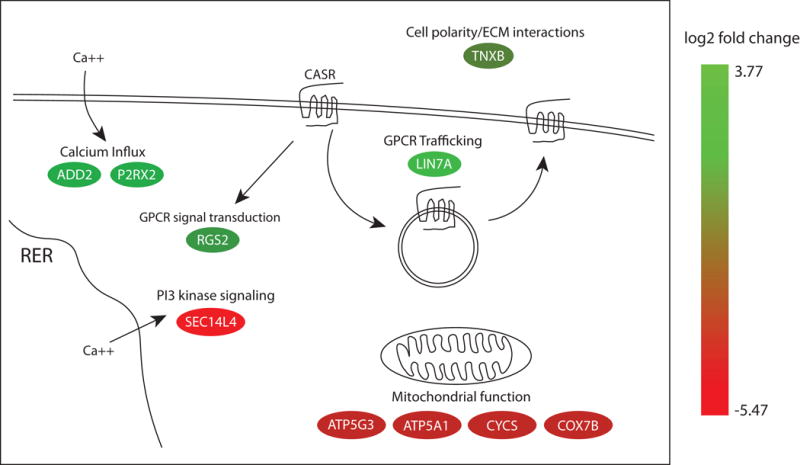
Functions of differentially expressed genes identified in the current study. The color of each oval indicates the log2-fold change in expression level in calcium-sensitive tumors relative to calcium-resistant tumors for each gene based on the color gradient index shown to the right. Genes expressed more highly in calcium-sensitive tumors are at the green end of the spectrum. Genes expressed more highly in calcium-resistant tumors are at the red end of the spectrum. Color gradients were generated using the Path Designer tool in the Ingenuity Pathway Analysis software package (12.18.2017). RER: rough endoplasmic reticulum; GPCR: G-protein coupled receptor; CASR: calcium-sensing receptor; ECM: extracellular matrix.
The known functions of additional differentially expressed genes could be envisioned to affect calcium responsiveness in parathyroid cells through other pathways (Fig. 7). RGS2 (Regulator of G-protein signaling 2) is a member of a protein family that modulates GPCR signaling and prevents receptor desensitization by deactivating Gi alpha subunits of the heterotrimeric G proteins (Cunningham, et al. 2001). We have shown previously that RGS5, a member of the same sub-family of RGS proteins, can modulate CASR signaling (Koh, et al. 2011). P2RX2 (Purinergic receptor, ligand-gated ion channel 2) is a broadly expressed, membrane bound ATP-gated ion channel permeable to Na+, K+, and Ca+ that has been found to activate transport of these cations in a broad range of tissues (North 2002). ADD2 (Adducin 2) is a member of a family of proteins that regulate calcium-dependent endothelial cell and epithelial junctional remodeling and other cellular activities by stimulating calcium influx through the plasma membrane (Kugelmann, et al. 2015; Rotzer, et al. 2014). Increased expression of these genes could help potentiate calcium signaling in calcium-sensitive tumors. SEC14L4 (SEC14-like lipid binding 4) is a member of a protein family that potentiates phosphoinositide signaling and intracellular calcium flux, a key downstream pathway utilized by CASR and many other GPCRs, as well as regulating membrane trafficking (Bankaitis, et al. 2010; Mousley, et al. 2007). The differential expression pattern of genes such as these that are linked to calcium responsiveness, membrane trafficking, and GPCR signal transduction provide an intriguing set of leads for mechanistic investigation.
The current study has limitations and strengths. While this report is the first to demonstrate differential gene expression patterns associated with alternative biochemical behaviors in parathyroid tumors, the cluster analysis groupings identified here do not yet represent definitive predictive signatures. For example, three of the calcium-sensitive tumors do not currently aggregate with the other ten sharing the same biochemical phenotype. This apparent mis-assignment can be rectified if we lower the threshold of called expression below our current, conservative setting, yielding three clusters but with a single cluster containing all 13 calcium-sensitive tumors (data not shown). The sensitivity of our provisional cluster analysis to threshold settings indicates that follow-up studies using larger sample sets will be required to extract robust standalone signatures capable of independent identification of tumor calcium-response capacity. Larger studies also could provide greater statistical power to refine phenotypic categorization by incorporating additional features with respect to clinical presentation, disease course, and outcome. Nonetheless, the primary finding of underlying molecular diversity among parathyroid tumors with differential calcium response characteristics represents a fundamental challenge to the current view of parathyroid tumors as essentially uniform entities. The visualization of distinct gene expression patterns among parathyroid tumors highlights the need for further investigation to define the mechanistic basis for these differences.
Collectively, the findings presented here indicate that phenotypic diversity in PHPT clinical presentation may be due to previously unrecognized subclasses of parathyroid tumors with different biochemical behaviors and distinct molecular profiles. The observation of differential gene expression patterns among parathyroid tumors suggests that PHPT could arise through alternative mechanisms with potentially divergent clinical outcomes. Developing a deeper understanding of the causes and consequences of these molecular profile patterns could provide important and much-needed insight for optimizing the clinical management of PHPT, as best treatment practices for this disease remain subject to debate. While parathyroidectomy is the only definitive cure for PHPT, not all patients are surgical candidates. Moreover, significant uncertainty exists with regard to how to address the growing cohort of asymptomatic PHPT patients identified incidentally through automated serum screening. Many patients undergo long-term surveillance or medical therapy with bisphosphonate agents or cinacalcet without referral for surgery, while others are sent for surgical consultation, a variation in practice that potentially implies variation in quality of care. Attaining a clearer, mechanistic grasp of underlying differences in the origins and behavior of parathyroid neoplasias, and development of the means for prospective identification of these differences, could provide an important new tool for personalizing the clinical management of PHPT to address the unique characteristics of each patient’s tumor. Refinement and validation of the molecular profiles described here into consolidated, predictive metagene signatures will be an important goal for future studies.
Supplementary Material
Figure S1. Tumor architecture of specimens subjected to RNAseq analysis. Images are hematoxylin/eosin stained 5 micron sections of formalin-fixed, paraffin-embedded (FFPE) tumor specimens. Magnification is 100X.
Figure S2. Mean RNAseq counts for CASR and PTH transcripts in calcium-sensitive and calcium-resistant tumors. Error bars represent standard deviations among three independent tumor specimens in each calcium response group. p-values are unadjusted; q-values represent the FDR-adjusted p-value.
Figure S3. Tumor architecture of specimens subjected to proteomics analysis. Images are hematoxylin/eosin stained 5 micron sections of FFPE tumor specimens. Magnification is 100X.
Figure S4. Technical reproducibility of transcript abundance from FFPE-extracted parathyroid tumor RNA. Log2-transformed values for the 33 transcripts in the custom Nanostring chipset are plotted for correlation between technical replicates relative to a linear regression line derived from the data.
Figure S5. Correlation between Nanostring quantitation of transcript abundance in FFPE-extracted RNA (x-axis) versus freshly isolated intact RNA (y-axis) from the same parathyroid tumor. Log2-transformed values for 33 transcripts are shown relative to a linear regression line derived from the data.
Figure S6. Correlation between Nanostring quantitation of transcript abundance from FFPE-extracted RNA (x-axis) relative to RNAseq quantitation of the same genes in intact freshly isolated RNA (y-axis) from the same tumor. Log2-transformed values for 33 transcripts are plotted relative to a linear regression line derived from the data.
Acknowledgments
The authors thank Kristen Lynam, R.N., for her outstanding efforts in clinical data management, informed consent procedures, and other essential tasks required for maintaining full compliance with institutional guidelines for human subjects research.
Funding: This work was supported by funds from the Duke Cancer Institute (J.A.S.) and by NIH grants 1R21CA192004–02 (J.K. and J.A.S.) and 2R24GM119888–02 (J.K. and J.A.S.)
Footnotes
Disclosures: JAS is a member of the Data Monitoring Committee of the Medullary Thyroid Cancer Consortium Registry supported by GlaxoSmithKline, Novo Nordisk, Astra Zeneca, and Eli Lilly.
Declaration of interest: JAS is a member of the Data Monitoring Committee of the Medullary Thyroid Cancer Consortium Registry supported by GlaxoSmithKline, Novo Nordisk, Astra Zeneca, and Eli Lilly. All of the other authors declare that there is no conflict of interest that could be perceived as prejudicing the impartiality of the research reported.
Literature Cited
- Akpinar G, Kasap M, Canturk NZ, Zulfigarova M, Islek EE, Guler SA, Simsek T, Canturk Z. Proteomics Analysis of Tissue Samples Reveals Changes in Mitochondrial Protein Levels in Parathyroid Hyperplasia over Adenoma. Cancer Genomics Proteomics. 2017;14:197–211. doi: 10.21873/cgp.20031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold A, Shattuck TM, Mallya SM, Krebs LJ, Costa J, Gallagher J, Wild Y, Saucier K. Molecular pathogenesis of primary hyperparathyroidism. J Bone Miner Res. 2002;17(Suppl 2):N30–36. [PubMed] [Google Scholar]
- Bankaitis VA, Mousley CJ, Schaaf G. The Sec14 superfamily and mechanisms for crosstalk between lipid metabolism and lipid signaling. Trends Biochem Sci. 2010;35:150–160. doi: 10.1016/j.tibs.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the False Discovery Rate - a Practical and Powerful Approach to Multiple Testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Annals of Statistics. 2001;29:1165–1188. [Google Scholar]
- Benton CR, Holloway GP, Han XX, Yoshida Y, Snook LA, Lally J, Glatz JF, Luiken JJ, Chabowski A, Bonen A. Increased levels of peroxisome proliferator-activated receptor gamma, coactivator 1 alpha (PGC-1alpha) improve lipid utilisation, insulin signalling and glucose transport in skeletal muscle of lean and insulin-resistant obese Zucker rats. Diabetologia. 2010;53:2008–2019. doi: 10.1007/s00125-010-1773-1. [DOI] [PubMed] [Google Scholar]
- Bilezikian JP, Silverberg SJ. Clinical practice. Asymptomatic primary hyperparathyroidism. N Engl J Med. 2004;350:1746–1751. doi: 10.1056/NEJMcp032200. [DOI] [PubMed] [Google Scholar]
- Breitwieser GE. The calcium sensing receptor life cycle: trafficking, cell surface expression, and degradation. Best Pract Res Clin Endocrinol Metab. 2013;27:303–313. doi: 10.1016/j.beem.2013.03.003. [DOI] [PubMed] [Google Scholar]
- Brennan SC, Thiem U, Roth S, Aggarwal A, Fetahu I, Tennakoon S, Gomes AR, Brandi ML, Bruggeman F, Mentaverri R, et al. Calcium sensing receptor signalling in physiology and cancer. Biochim Biophys Acta. 2013;1833:1732–1744. doi: 10.1016/j.bbamcr.2012.12.011. [DOI] [PubMed] [Google Scholar]
- Cetani F, Picone A, Cerrai P, Vignali E, Borsari S, Pardi E, Viacava P, Naccarato AG, Miccoli P, Kifor O, et al. Parathyroid expression of calcium-sensing receptor protein and in vivo parathyroid hormone-Ca(2+) set-point in patients with primary hyperparathyroidism. The Journal of clinical endocrinology and metabolism. 2000;85:4789–4794. doi: 10.1210/jcem.85.12.7028. [DOI] [PubMed] [Google Scholar]
- Chakravarti B, Chattopadhyay N, Brown EM. Signaling through the extracellular calcium-sensing receptor (CaSR) Adv Exp Med Biol. 2012;740:103–142. doi: 10.1007/978-94-007-2888-2_5. [DOI] [PubMed] [Google Scholar]
- Cheviet S, Coppola T, Haynes LP, Burgoyne RD, Regazzi R. The Rab-binding protein Noc2 is associated with insulin-containing secretory granules and is essential for pancreatic beta-cell exocytosis. Mol Endocrinol. 2004;18:117–126. doi: 10.1210/me.2003-0300. [DOI] [PubMed] [Google Scholar]
- Corbetta S, Mantovani G, Lania A, Borgato S, Vicentini L, Beretta E, Faglia G, Di Blasio AM, Spada A. Calcium-sensing receptor expression and signalling in human parathyroid adenomas and primary hyperplasia. Clin Endocrinol (Oxf) 2000;52:339–348. doi: 10.1046/j.1365-2265.2000.00933.x. [DOI] [PubMed] [Google Scholar]
- Cromer MK, Starker LF, Choi M, Udelsman R, Nelson-Williams C, Lifton RP, Carling T. Identification of somatic mutations in parathyroid tumors using whole-exome sequencing. J Clin Endocrinol Metab. 2012;97:E1774–1781. doi: 10.1210/jc.2012-1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cunningham ML, Waldo GL, Hollinger S, Hepler JR, Harden TK. Protein kinase C phosphorylates RGS2 and modulates its capacity for negative regulation of Galpha 11 signaling. J Biol Chem. 2001;276:5438–5444. doi: 10.1074/jbc.M007699200. [DOI] [PubMed] [Google Scholar]
- Das NR, Sharma SS. Peroxisome Proliferator Activated Receptor Gamma Coactivator 1 Alpha: An Emerging Target for Neuroprotection in Parkinson’s Disease. CNS Neurol Disord Drug Targets. 2015;14:1024–1030. doi: 10.2174/1871527314666150326000018. [DOI] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farnebo F, Enberg U, Grimelius L, Backdahl M, Schalling M, Larsson C, Farnebo LO. Tumor-specific decreased expression of calcium sensing receptor messenger ribonucleic acid in sporadic primary hyperparathyroidism. J Clin Endocrinol Metab. 1997;82:3481–3486. doi: 10.1210/jcem.82.10.4300. [DOI] [PubMed] [Google Scholar]
- Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T, et al. Direct multiplexed measurement of gene expression with color-coded probe pairs. Nature biotechnology. 2008;26:317–325. doi: 10.1038/nbt1385. [DOI] [PubMed] [Google Scholar]
- Giusti L, Cetani F, Ciregia F, Da Valle Y, Donadio E, Giannaccini G, Banti C, Pardi E, Saponaro F, Basolo F, et al. A proteomic approach to study parathyroid glands. Molecular bioSystems. 2011;7:687–699. doi: 10.1039/c0mb00191k. [DOI] [PubMed] [Google Scholar]
- Hart SN, Therneau TM, Zhang Y, Poland GA, Kocher JP. Calculating sample size estimates for RNA sequencing data. Journal of computational biology: a journal of computational molecular cell biology. 2013;20:970–978. doi: 10.1089/cmb.2012.0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- http://www.bioinformatics.babraham.ac.uk/projects/trim_galore.
- https://academic.oup.com/bioinformatics/article-lookup/doi/10.1093/bioinformatics/btu638.
- Huber W, Carey VJ, Gentleman R, Anders S, Carlson M, Carvalho BS, Bravo HC, Davis S, Gatto L, Girke T, et al. Orchestrating high-throughput genomic analysis with Bioconductor. Nature methods. 2015;12:115–121. doi: 10.1038/nmeth.3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- AACE/AAES Task Force on Primary Hyperparathyroidism. The American Association of Clinical Endocrinologists and the American Association of Endocrine Surgeons position statement on the diagnosis and management of primary hyperparathyroidism. Endocrine practice: official journal of the American College of Endocrinology and the American Association of Clinical Endocrinologists. 2005;11:49–54. doi: 10.4158/EP.11.1.49. [DOI] [PubMed] [Google Scholar]
- Kersey PJ, Staines DM, Lawson D, Kulesha E, Derwent P, Humphrey JC, Hughes DS, Keenan S, Kerhornou A, Koscielny G, et al. Ensembl Genomes: an integrative resource for genome-scale data from non-vertebrate species. Nucleic acids research. 2012;40:D91–97. doi: 10.1093/nar/gkr895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J, Dar M, Untch BR, Dixit D, Shi Y, Yang Z, Adam MA, Dressman H, Wang X, Gesty-Palmer D, et al. Regulator of G-protein signaling 5 is highly expressed in parathyroid tumors and inhibits signaling by the calcium-sensing receptor. Molecular Endo. 2011;25:867–876. doi: 10.1210/me.2010-0277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J, Hogue JA, Sosa JA. A Novel Ex Vivo Method for Visualizing Live-Cell Calcium Response Behavior in Intact Human Tumors. PloS one. 2016a;11:e0161134. doi: 10.1371/journal.pone.0161134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koh J, Hogue JA, Wang Y, DiSalvo M, Allbritton NL, Shi Y, Olson JA, Jr, Sosa JA. Single-cell functional analysis of parathyroid adenomas reveals distinct classes of calcium sensing behaviour in primary hyperparathyroidism. Journal of cellular and molecular medicine. 2016b;20:351–359. doi: 10.1111/jcmm.12732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kugelmann D, Waschke J, Radeva MY. Adducin is involved in endothelial barrier stabilization. PloS one. 2015;10:e0126213. doi: 10.1371/journal.pone.0126213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lai L, Wang M, Martin OJ, Leone TC, Vega RB, Han X, Kelly DP. A role for peroxisome proliferator-activated receptor gamma coactivator 1 (PGC-1) in the regulation of cardiac mitochondrial phospholipid biosynthesis. J Biol Chem. 2014;289:2250–2259. doi: 10.1074/jbc.M113.523654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee F, Lee JJ, Jan WC, Wu CJ, Chen HH, Cheng SP. Molecular pathways associated with transcriptional alterations in hyperparathyroidism. Oncol Lett. 2016;12:621–626. doi: 10.3892/ol.2016.4623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. Bioinformatics in action. 2011;17:10–12. [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nature genetics. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Morrison C, Farrar W, Kneile J, Williams N, Liu-Stratton Y, Bakaletz A, Aldred MA, Eng C. Molecular classification of parathyroid neoplasia by gene expression profiling. Am J Pathol. 2004;165:565–576. doi: 10.1016/S0002-9440(10)63321-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mousley CJ, Tyeryar KR, Vincent-Pope P, Bankaitis VA. The Sec14-superfamily and the regulatory interface between phospholipid metabolism and membrane trafficking. Biochim Biophys Acta. 2007;1771:727–736. doi: 10.1016/j.bbalip.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- North RA. Molecular physiology of P2Xreceptors. Physiol Rev. 2002;82:1013–1067. doi: 10.1152/physrev.00015.2002. [DOI] [PubMed] [Google Scholar]
- Ray K. Calcium-Sensing Receptor: Trafficking, Endocytosis, Recycling, and Importance of Interacting Proteins. Prog Mol Biol Transl Sci. 2015;132:127–150. doi: 10.1016/bs.pmbts.2015.02.006. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Boulan E, Macara IG. Organization and execution of the epithelial polarity programme. Nature reviews Molecular cell biology. 2014;15:225–242. doi: 10.1038/nrm3775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotzer V, Breit A, Waschke J, Spindler V. Adducin is required for desmosomal cohesion in keratinocytes. J Biol Chem. 2014;289:14925–14940. doi: 10.1074/jbc.M113.527127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JS, Tachibana I, Allen C, Chiappa SA, Lee HK, McIver B, Jenkins RB, Raffel C. Cloning of a human ortholog (RPH3AL) of (RNO)Rph3al from a candidate 17p13.3 medulloblastoma tumor suppressor locus. Genomics. 1999;59:97–101. doi: 10.1006/geno.1999.5864. [DOI] [PubMed] [Google Scholar]
- Tharmalingam S, Hampson DR. The Calcium-Sensing Receptor and Integrins in Cellular Differentiation and Migration. Front Physiol. 2016;7:190. doi: 10.3389/fphys.2016.00190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney S, Bhadada SK, Arya AK, Sharma S, Behera A, Bhansali A, Rao SD. Changes in parathyroid proteome in patients with primary hyperparathyroidism due to sporadic parathyroid adenomas. Clin Endocrinol (Oxf) 2014;81:614–620. doi: 10.1111/cen.12479. [DOI] [PubMed] [Google Scholar]
- Waggott D, Chu K, Yin S, Wouters BG, Liu FF, Boutros PC. NanoStringNorm: an extensible R package for the pre-processing of NanoString mRNA and miRNA data. Bioinformatics. 2012;28:1546–1548. doi: 10.1093/bioinformatics/bts188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Zhao X, Lotz M, Terkeltaub R, Liu-Bryan R. Mitochondrial biogenesis is impaired in osteoarthritis chondrocytes but reversible via peroxisome proliferator-activated receptor gamma coactivator 1alpha. Arthritis Rheumatol. 2015;67:2141–2153. doi: 10.1002/art.39182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber TJ, Koh J, Thomas SM, Hogue JA, Scheri RP, Roman SA, Sosa JA. Impaired calcium sensing distinguishes primary hyperparathyroidism (PHPT) patients with low bone mineral density. Metabolism. 2017;74:22–31. doi: 10.1016/j.metabol.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wermers RA, Khosla S, Atkinson EJ, Achenbach SJ, Oberg AL, Grant CS, Melton LJ., 3rd Incidence of primary hyperparathyroidism in Rochester, Minnesota, 1993–2001 an update on the changing epidemiology of the disease. J Bone Miner Res. 2006;21:171–177. doi: 10.1359/JBMR.050910. [DOI] [PubMed] [Google Scholar]
- www.r-project.org.
- Xiong S, Salazar G, Patrushev N, Ma M, Forouzandeh F, Hilenski L, Alexander RW. Peroxisome proliferator-activated receptor gamma coactivator-1alpha is a central negative regulator of vascular senescence. Arterioscler Thromb Vasc Biol. 2013;33:988–998. doi: 10.1161/ATVBAHA.112.301019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan Y, Huang S, Wang W, Wang Y, Zhang P, Zhu C, Ding G, Liu B, Yang T, Zhang A. Activation of peroxisome proliferator-activated receptor-gamma coactivator 1alpha ameliorates mitochondrial dysfunction and protects podocytes from aldosterone-induced injury. Kidney international. 2012;82:771–789. doi: 10.1038/ki.2012.188. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Tumor architecture of specimens subjected to RNAseq analysis. Images are hematoxylin/eosin stained 5 micron sections of formalin-fixed, paraffin-embedded (FFPE) tumor specimens. Magnification is 100X.
Figure S2. Mean RNAseq counts for CASR and PTH transcripts in calcium-sensitive and calcium-resistant tumors. Error bars represent standard deviations among three independent tumor specimens in each calcium response group. p-values are unadjusted; q-values represent the FDR-adjusted p-value.
Figure S3. Tumor architecture of specimens subjected to proteomics analysis. Images are hematoxylin/eosin stained 5 micron sections of FFPE tumor specimens. Magnification is 100X.
Figure S4. Technical reproducibility of transcript abundance from FFPE-extracted parathyroid tumor RNA. Log2-transformed values for the 33 transcripts in the custom Nanostring chipset are plotted for correlation between technical replicates relative to a linear regression line derived from the data.
Figure S5. Correlation between Nanostring quantitation of transcript abundance in FFPE-extracted RNA (x-axis) versus freshly isolated intact RNA (y-axis) from the same parathyroid tumor. Log2-transformed values for 33 transcripts are shown relative to a linear regression line derived from the data.
Figure S6. Correlation between Nanostring quantitation of transcript abundance from FFPE-extracted RNA (x-axis) relative to RNAseq quantitation of the same genes in intact freshly isolated RNA (y-axis) from the same tumor. Log2-transformed values for 33 transcripts are plotted relative to a linear regression line derived from the data.


