Abstract
Advances in three-dimensional secondary ion mass spectrometry (SIMS) imaging have enabled visualizing the subcellular distributions of various lipid species within individual cells. However, the difficulty of locating organelles using SIMS limits efforts to study their lipid compositions. Here, the authors have assessed whether endoplasmic reticulum (ER)-Tracker Blue White DPX®, which is a commercially available stain for visualizing the endoplasmic reticulum using fluorescence microscopy, produces distinctive ions that can be used to locate the endoplasmic reticulum using SIMS. Time-of-flight-SIMS tandem mass spectrometry (MS2) imaging was used to identify positively and negatively charged ions produced by the ER-Tracker stain. Then, these ions were used to localize the stain and thus the endoplasmic reticulum, within individual human embryonic kidney cells that contained higher numbers of endoplasmic reticulum-plasma membrane junctions on their surfaces. By performing MS2 imaging of selected ions in parallel with the precursor ion (MS1) imaging, the authors detected a chemical interference native to the cell at the same nominal mass as the pentafluorophenyl fragment from the ER-Tracker stain. Nonetheless, the fluorine secondary ions produced by the ER-Tracker stain provided a distinctive signal that enabled locating the endoplasmic reticulum using SIMS. This simple strategy for visualizing the endoplasmic reticulum in individual cells using SIMS could be combined with existing SIMS methodologies for imaging intracellular lipid distribution and to study the lipid composition within the endoplasmic reticulum.
I. INTRODUCTION
In mammalian cells, lipids and cholesterol form the selectively permeable plasma membrane (PM) that separates the cell from its surroundings and the intracellular membranes that delineate the boundaries of organelles and transport vesicles. Cholesterol and distinct lipid species are unevenly distributed between the membranes of different organelles and are also hypothesized to be nonrandomly organized within them.1–3 For example, different phosphatidylinositol lipid subspecies are predominantly found in the Golgi apparatus, endosomes, and plasma membrane.3,4 Lipid exchange between the plasma membrane and endoplasmic reticulum (ER) may induce gradients in the concentrations of sterols and certain lipid species within the endoplasmic reticulum.4–6 Changes in the lipid compositions of certain organellar membranes are correlated with disease, but whether these changes are the cause or consequence of cellular dysfunction is not clear.7–9 Strategies that enable detecting both distinct lipid species and specific organelles with the same imaging modality could help to define the roles of organelle lipids in cellular function and disease.
The ability to image the distributions of distinct lipid species on the surfaces of cells using secondary ion mass spectrometry (SIMS) has significantly advanced understanding of lipid distribution within the plasma membrane.10–17 This progress was built upon results acquired using two different types of instrumentation, time-of-flight-SIMS (TOF-SIMS) for imaging unlabeled lipids and magnetic sector SIMS instruments for imaging isotope-labeled lipids with a high spatial resolution.11–18,48 These complementary SIMS approaches have been used to create three-dimensional (3D) renderings of the lipid distributions within cells,19–28 which may open the door to both discovery-based and target-focused intracellular lipid imaging. Nonetheless, mapping the 3D distributions of lipid species within individual cells is only one obstacle to assessing the lipid composition within specific organelles. The difficulty of detecting organelles of interest using SIMS remains.
To date, the routine imaging of organelles using SIMS is limited to the nucleus, which is localized by detecting rare stable isotope or non-native elemental labels on metabolically incorporated nucleoside analogs.20,21,27,29 Strategies that employ functionalized antibodies or genetic approaches for conjugating proteins of interest to distinctive mass tags14,28,30,31 could be adapted to permit organelle detection using SIMS. Alternatively, lipids and organelles could be imaged sequentially using correlated fluorescence and SIMS imaging.29,32 Unfortunately, these strategies are labor intensive.
In pursuit of a simpler approach for detecting additional organelles of interest using SIMS, we began to explore whether fluorophores that preferentially accumulate in specific organelles could be repurposed for SIMS imaging. We focused on ER-Tracker Blue-White DPX®, referred to as ER-Tracker herein, which is a commercially available fluorophore that is used to selectively label the endoplasmic reticulum for fluorescence microscopy imaging. We selected this organelle-specific fluorophore because it contains fluorine atoms, which are not native to mammalian cells, and the distributions of other fluorine-labeled probes in cells have been detected using SIMS according to their distinctive fluorine ions.14,30 However, the concentration of the ER-Tracker within the endoplasmic reticulum might not be sufficient to permit imaging it using SIMS.
The structure and intracellular positioning of the endoplasmic reticulum further complicate the assessment of whether the ER-Tracker stain could be repurposed to permit imaging this organelle using SIMS. The endoplasmic reticulum is a network of membrane-enclosed sheets and tubules that is generally found throughout the cell interior. Sheets of endoplasmic reticulum surround the nucleus, and endoplasmic reticulum tubules extend toward the cell surface, often forming junctions with the plasma membrane33 which are called ER-PM junctions. To expose the endoplasmic reticulum sheets to the SIMS analysis beam, an indeterminate amount of material would need to be sputtered from the cell surface. Less sputtering would be required to uncover the endoplasmic reticulum tubules if these tubules formed junctions with the plasma membrane. Thus, to facilitate this study, we used human embryonic kidney (HEK) cells with ER-PM junctions covering up to 20% of the cell surface.34 These ER-PM junctions were induced by expression of a green fluorescent protein-labeled voltage-gated potassium channel, Kv2.1 (GFP-Kv2.1), that has the unusual property of inducing and stabilizing ER-PM junctions. GFP-Kv2.1 forms clusters in the plasma membrane which have a size in the range of 500 nm to over 1 μm.34–38 This clustering results from channel binding to the cortical ER and the subsequent induction in ER-PM junctions.31
To assess whether the ER-Tracker stain can be used to visualize the endoplasmic reticulum using SIMS, we used TOF-SIMS Parallel Imaging MS/MS (i.e., TOF-SIMS tandem MS imaging) to identify the ion signals produced by the ER-Tracker. TOF-SIMS tandem MS imaging enables the simultaneous detection of the precursor ion (MS1) spectrum and the product ion (MS2) spectrum arising from fragmentation of a selected precursor at every pixel in the analysis region.39,40 TOF-SIMS tandem MS imaging was able to detect and image ions characteristic of the ER-Tracker within a transfected HEK cell. These results demonstrate that the ER-Tracker stain can be repurposed to image the endoplasmic reticulum in transfected cells using TOF-SIMS.
II. EXPERIMENT
A. Materials
HEK cells [HEK 293] were obtained from ATCC and cultured in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS) (Gibco). Poly-l-lysine and glutaraldehyde were obtained from Electron Microscopy Sciences. Silicon wafer chips (5 × 5 mm) were purchased from Ted Pella, Inc. Lipofectamine 2000, OptiMEM, and ER-Tracker Blue-White DPX were all obtained from Thermo Fisher. A DNA plasmid for the expression of the GFP-labeled Kv2.1 ion channel (GFP-Kv2.1loopBAD) that stabilizes ER-PM junctions was used as previously described.34,37,41,42 HEK imaging saline is composed of 146 mM NaCl, 4.7 mM KCl, 2.5 mM CaCl2, 0.6 mM MgSO4, 1.6 mM NaHCO3, 0.15 mM NaH2PO4, 0.1 mM ascorbic acid, 8 mM glucose, and 20 mM 4–(2-hydroxyethyl)–1-piperazineethanesulfonic acid (HEPES) at pH 7.4.
B. Sample preparation
To prepare samples for TOF-SIMS analysis, HEK cells were seeded on poly-l-lysine coated silicon chips in DMEM containing 10% FBS and incubated at 37 °C in 5% CO2. A DNA plasmid solution was prepared by incubating ∼3 μg of DNA plasmid (GFP-Kv2.1loopBAD) and 8 μl of Lipofectamine 2000 in 400 μl of OptiMEM at room temperature for 30 min.34 Each silicon chip with adherent cells was treated with 100 μl of plasmid solution and incubated for 48 h at 37 °C in 5% CO2. Next, the cells were rinsed with HEK imaging saline and incubated for 30 min in 1 μM ER-Tracker in imaging saline at 37 °C in 5% CO2. Cells were fixed with glutaraldehyde as previously reported.17 Briefly, cells were rinsed twice with Hendry's phosphate buffer (HPB) for 3 min and fixed with 4% glutaraldehyde in HPB for 30 min. Cell substrates were then rinsed twice with HPB for 5 min and once with triple-distilled water for 5 min.
ER-Tracker standards were prepared by drying the pure compound dissolved in dimethyl sulfoxide (DMSO) on silicon substrates.
B. Fluorescence imaging
To assess the distribution of Kv2.1 and the endoplasmic reticulum at the bottom of the cell using total internal reflection fluorescence (TIRF) microscopy imaging, HEK cells were cultured and transfected as previously described.34 In short, cells were transfected via electroporation with DsRed2-ER, Kv2.1BAD, and BirA and plated on Matrigel coated 35 mm glass bottom dishes 24 h before experiments. The biotin acceptor domain (BAD) containing construct has been previously described.36,37,41,42 Prior to imaging, cells were labeled for ten min with CF640R Streptavidin (Biotium, Cat:29041) at 1:1000 dilution in HEK imaging saline. Imaging was performed on a Nikon Eclipse Ti Perfect-Focus equipped TIRF microscope as previously described.34,43
To image ER-Tracker stain and Kv2.1 localization using confocal fluorescence microscopy, cells cultured on a glass-bottom dish (Cellvis) for 24 h were transfected and after one day labeled with ER-Tracker as described above. A Zeiss laser scanning microscope 710 confocal fluorescence microscope at the Carl R. Woese Institute for Genomic Biology (University of Illinois, Urbana, IL) was used to acquire fluorescence images from live cells in HEK imaging saline at a magnification of 63× using oil immersion. GFP fluorescence was performed with maximum excitation and emission wavelengths of 488 and 510 nm, respectively, while ER-Tracker imaging was performed with maximum excitation and emission wavelengths of 405 and 460 nm, respectively. Images were processed using Zen lite software.
C. TOF-SIMS tandem MS analysis
The TOF-SIMS tandem MS imaging analyses were performed on a PHI nanoTOF II Parallel Imaging MS/MS instrument (Physical Electronics, MN), which has been previously described.39,40 The instrument was equipped with a 20 kV argon gas cluster ion beam for sputtering and a 30 kV Binq+ cluster liquid metal ion gun for analysis. Secondary ions were generated from the specimens using a mass pure and pulsed beam of Bi3+ primary ions at a typical unfiltered dc beam current of 3 nA. The pulse width and repetition rate were 16 ns, uncompressed, and ≈ 8300 Hz, respectively.
Cells were characterized using a <100 μm field-of-view (FOV) containing 512 × 512 pixels. A typical 2D parallel imaging MS/MS analysis was conducted with a 70 μm FOV to a primary ion dose density (PIDD) of 1.16 × 1013 < PIDD< 9.26 × 1013 Bi3+/cm2. Although the ion dose density often exceeded the static limit, the time profile for characteristic or molecular ion signals was not observed to degrade as a function of ion dose density. 3D imaging depth profile data were collected by sputtering with a 5 keV beam of Ar2,500+ ions (2.5 nA dc beam current) rastered over an 800 × 800 μm area (PIDD = 8.06 ×1014 Ar2500+/cm2). With these sputter beam conditions, the sputter rate was measured to be between 1 and 4 nm/s using poly(methyl methacrylate) (PMMA) thin film references from different sources. Although the sputter rate on cells is expected to be heterogeneous and is certain to differ from PMMA, the sputter rate of PMMA is used as a rough approximation for these studies. Because this study does not seek to identify the precise depth of the observed features, this rough approximation will not affect the conclusions drawn herein. The effective diameter of the primary ion beam that defines the analytical lateral resolution was measured to be 80 nm, which is smaller than the size of the individual pixels (137 × 137 nm) in the 512 × 512 pixel TOF-SIMS images acquired from a 70 μm FOV. Thus, the working lateral resolution of the ion images was defined by the pixel size. Raw data were collected using PHI SmartSoft-TOF software (Physical Electronics, MN), and all images and spectra were produced retrospectively from the raw data files using phi tof-dr software (Physical Electronics, MN).
At each image pixel, both TOF-SIMS (MS1) and tandem MS (MS2) imaging data are simultaneously collected. In other words, one primary ion beam pulse triggers one parallel cycle of the MS1 (TOF-SIMS) and MS2 (tandem MS) spectrometers. Monoisotopic precursor selection in a 1 Da window is achieved by the design for the TOF-SIMS tandem MS imaging. Collisional activation of the selected precursors is achieved at ≈1.5 keV in a cell of Ar gas. The percent duty cycle of the precursor selection is variable by the operator such that, if desired, a fractional portion of the precursor ions may remain in the MS1 imaging data for the purpose of normalization or quantification. For the analyses presented herein, the percent duty cycle was set at 100% such that all precursor ions generated from each primary ion pulse appear in the MS2 imaging data to maximize the sensitivity.
D. Data analysis
phi tof-dr software was used to visualize and evaluate MS1 and MS2 spectra and to create ion images. When the figure caption indicates binning was applied to an image, the 512 × 512 pixel images were binned to produce 256 × 256 pixel or 128 × 128 pixel images with a pixel size, and thus an effective lateral resolution, of approximately 273 × 273 or 547 × 547 nm, respectively. Intensity scaling was adjusted to maintain consistency across images of the same m/z value and throughout the 3D stack. For 3D visualization, data from 255 individual image cycles acquired at the same sample location were used to construct a stack of 17 separate ion images that span a depth of approximately 40 nm from the cell surface. Each image in this stack was compiled from the ion signals detected in 15 consecutive image cycles. These ion images were scaled so that the ion intensities encoded in their color scales were identical throughout the image stack. To account for any instrument or stage drift, features that were easily identified in the total ion count (TIC) images, such as the cell–substrate boundaries, were visually aligned in the x- and y-directions so that they appeared to be positioned at approximately the same x- and y-coordinates. This visual alignment produced images that appeared to be in registration when viewed through the stack (i.e., see Fig. 4). Then, the aligned images were stacked using the 3D viewer plugin of imagej.
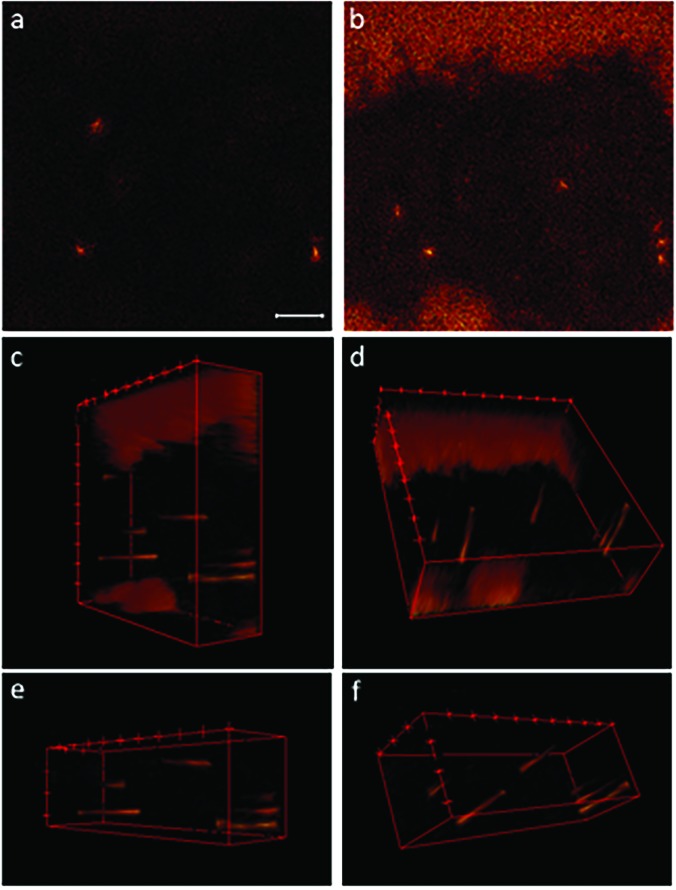
Fig. 4.
Individual images and 3D image stacks compiled from 3D depth profile data show the m/z 19 signals at the surface of a transfected HEK cell and at distances up to approximately 40 nm below the surface. Pixel binning was not applied to these images. The image that corresponds to the top (a) of the 3D stack shows the m/z 19 fluoride ions that locate the ER-Tracker on the cell surface. The last m/z 19 ion image acquired at approximately 40 nm below the cell surface (b) reveals additional hotspots inside the cell. 3D image stacks of the full FOV [(c) and (d)] and a smaller region within the cell [(e) and (f)] show that the elevated m/z 19 signals were detected in cylindrical regions that appear at different distances from the cell surface. These cylindrical morphologies are consistent with endoplasmic reticulum tubules that occasionally form ER-PM junctions. The scale bar is 10 μm.
III. RESULTS AND DISCUSSION
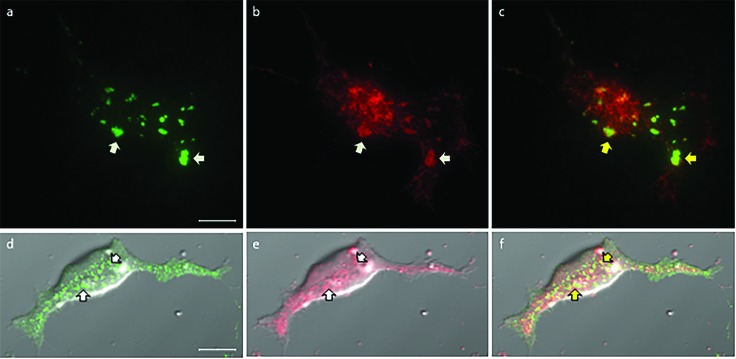
To show that Kv2.1 clusters localize at ER-PM junctions, HEK cells were transfected to express biotinylated Kv2.1 channels that reside in the plasma membrane, and a red fluorescent protein that resides in the endoplasmic reticulum (DsRed2-ER).44 Then the cells were treated with CF640-conjugated streptavidin, which selectively binds to the extracellular biotin, allowing only the biotinylated Kv2.1 on the surface of the cell to be detected with fluorescence microscopy. Imaging using TIRF microscopy revealed patches of green fluorescence from Kv2.1 clusters in the flat portion of the plasma membrane that was adjacent to the glass surface beneath the cell [Fig. 1(a)]. DsRed2-ER within the total internal reflection (TIR) illumination field appeared as bright red fluorescent patches within a background of diffuse red fluorescence [Fig. 1(b)]. The yellowish-green features in the overlay of these two fluorescence images indicate colocalization between some Kv2.1 clusters in the plasma membrane with the endoplasmic reticulum tubules near the cell surface [Fig. 1(c)]. Kv2.1 clusters were also detected on HEK cells that had been transfected so they expressed green fluorescent protein-labeled Kv2.1 (GFP-Kv2.1), but not DsRed2-ER, and labeled with ER-Tracker. Figure 1(d) shows a confocal fluorescence microscopy image of a transfected cell that has large (700 nm to 2 μm) GFP-Kv2.1 clusters in the flat portion of the plasma membrane that was adjacent to the glass substrate. GFP-Kv2.1 clusters were also present on the top surface of the cell, which is the region analyzed using SIMS, but fluorescence visualization is more favorable on the flat cell bottom where GFP-Kv2.1 clusters preferentially form.36 The fluorescence from the ER-Tracker highlights the endoplasmic reticulum in the confocal fluorescence microscopy image shown in Fig. 1(e). The overlay of the green and red fluorescence images in the corresponding bright-field optical image [Fig. 1(f)] highlights the partial colocalization of the GFP-Kv2.1 clusters and endoplasmic reticulum, which is consistent with previous reports.30 This colocalized red and green fluorescence, which appears yellowish-green in Fig. 1(f), denotes regions where the endoplasmic reticulum and GFP-Kv2.1 clusters are separated in the z-direction by not more than the depth of the confocal imaging plane. Because Kv2.1 clusters in the plasma membrane are bound to the endoplasmic reticulum, this colocalization indicates that the endoplasmic reticulum and the plasma membrane are 10–15 nm apart at these sites.34
Fig. 1.
[(a)–(c)] TIRF microscopy images of biotinylated Kv2.1, detected by fluorescent (CF640) streptavidin binding to the biotin moiety attached to an extracellular domain on the Kv2.1 channel, and DsRed2-ER in the endoplasmic reticulum. Arrows specify the regions where the Kv2.1 clusters on the cell surface overlap with the DsRed2-ER near the plasma membrane. (a) Clusters of Kv2.1 were present on the bottom surface of the cell adjacent to the glass substrate. (b) Bright red patches denote the endoplasmic reticulum tubules within the TIR illumination field. (c) Overlay of both fluorescence signals. [(d)–(f)] Confocal fluorescence microscopy images of HEK cells that were transfected to express GFP-Kv2.1 and labeled with ER-Tracker. Arrows indicate the regions where the green and red fluorescence overlaps. (d) On the bottom surface of the representative cell, the GFP-labeled Kv2.1 ion channels formed clusters with sizes that ranged from 700 nm to 2 μm. These clusters associate with junctions between the plasma membrane and endoplasmic reticulum. (e) Fluorescence microscopy images of the ER-Tracker that locate the endoplasmic reticulum. (f) Merging the two fluorescence images and visualizing them on top of the bright-field optical image produce yellowish-green features that indicate partial colocalization between GFP-Kv2.1 clusters and the endoplasmic reticulum, as indicated by the yellow arrows. All scale bars are 10 μm.
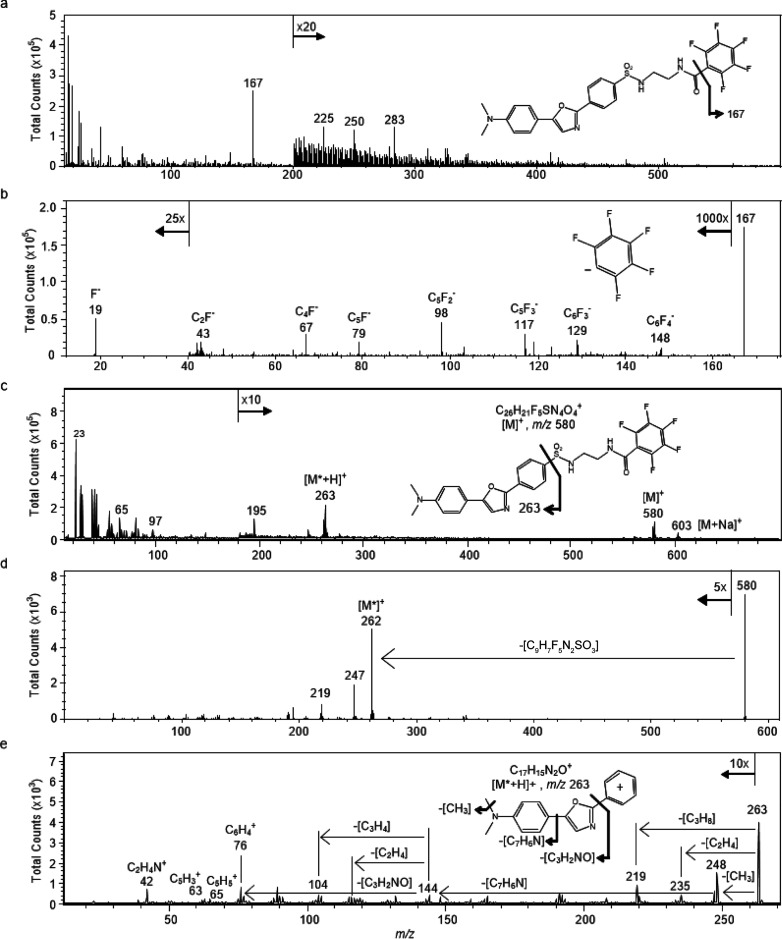
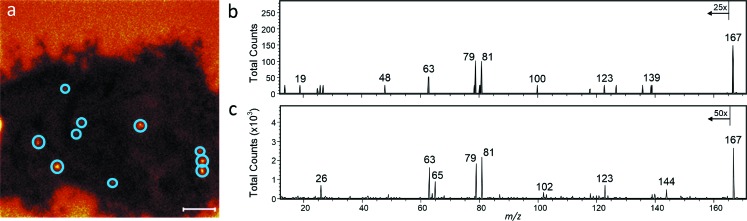
To determine whether the ER-Tracker molecule [Fig. 2(a), inset] produces characteristic fragment ions that may be used for imaging by TOF-SIMS, the pure compound was dissolved in DMSO, dried on a silicon substrate, and analyzed via TOF-SIMS tandem MS imaging in both the positive and negative ion modes. In the negative ion mode, a prominent peak at m/z 167 was present in the MS1 spectrum [Fig. 2(a)]. This ion was fragmented for tandem MS identification. The product ion spectrum of the m/z 167 precursor produced an intense fragment ion at m/z 19, which corresponds to a fluoride ion [Fig. 2(b)]. Less intense peaks from several fluorocarbon fragment ions, including C6F4−, C5F3−, C5F2−, C4F−, and C2F−, were also detected. Based on the fragmentation pattern, exact mass, and structure of the parent ER-Tracker molecule, the m/z 167 ion corresponds to a pentafluorophenyl anion [Fig. 2(b), inset]. The full positive ion spectrum of the ER-Tracker stain [Fig. 2(c)] shows peaks at m/z 580 and 603 which correspond to the molecular ion (C26H21F5N4O4S) and the sodiated molecular ion, respectively, as well as several lower mass peaks, such as m/z 263, 195, 65, and 23. The molecular ion (C26H21F5N4O4S) at m/z 580 and the peak at m/z 263 were fragmented for tandem MS identification. The product ion spectrum of the m/z 580 precursor contains prominent peaks at m/z 262, 247, and 219 [Fig. 2(d)]. The prominent peak at m/z 263 was fragmented for tandem MS identification. The product ion spectrum of the m/z 263 precursor, ostensibly the 3-(5-(4-(dimethylamino)phenyl)oxazol-2-yl)benzene-1-ylium (Dapoxyl dye) [C17H15N2O]+ cation [Fig. 2(e), inset], contained multiple fragment ions. This includes a fragment at m/z 235 from the loss of CO, which has been reported for substituted oxazoles.9 The peaks at m/z 248 and 219 result from the elimination of CH3 and CH3NHCH2, respectively, from the N,N-dimethylaniline moiety, and a peak at m/z 144 reflecting the loss of the entire N,N-dimethylaniline moiety [Fig. 2(e), inset]. The product ion spectrum of the m/z 263 precursor also contains peaks at m/z 89, 76, and 65 which correspond to [C7H5]+, [C6H4]+, and [C5H5]+, respectively, from fragmentation of the aromatic rings. This fragmentation pattern indicates that the precursor ion at m/z 263 corresponds to the Dapoxyl cation.
Fig. 2.
Ionization and fragmentation of ER-Tracker standard were performed to identify characteristic fragments that might be used to image its distribution in cells using SIMS. (a) Full negative ion spectrum of ER-Tracker standard with the structure of the intact molecule (inset). (b) Product ion spectrum of the precursor ion m/z 167 contained fluoride ions at m/z 19 and several less intense fluorocarbon fragment ions, which confirms that the m/z 167 precursor ion was the pentafluorophenyl anion. (c) Full positive ion spectrum of the ER-Tracker standard. The inset shows the structure of the intact ER-Tracker molecule, where the fragment that corresponds to the fragment ion with m/z 263 is indicated. (d) Product ion spectrum of the molecular ion (C26H21F5N4O4S) at m/z 580. (e) The peaks in the product ion spectrum of the precursor ion m/z 263 result from the loss of CO, fragments of the N,N-dimethyl moiety, N,N-dimethylaniline, and aromatic ring fragments. This fragmentation pattern confirms that the precursor ion at m/z 263 was the Dapoxyl dye cation. The inset illustrates its fragmentation.
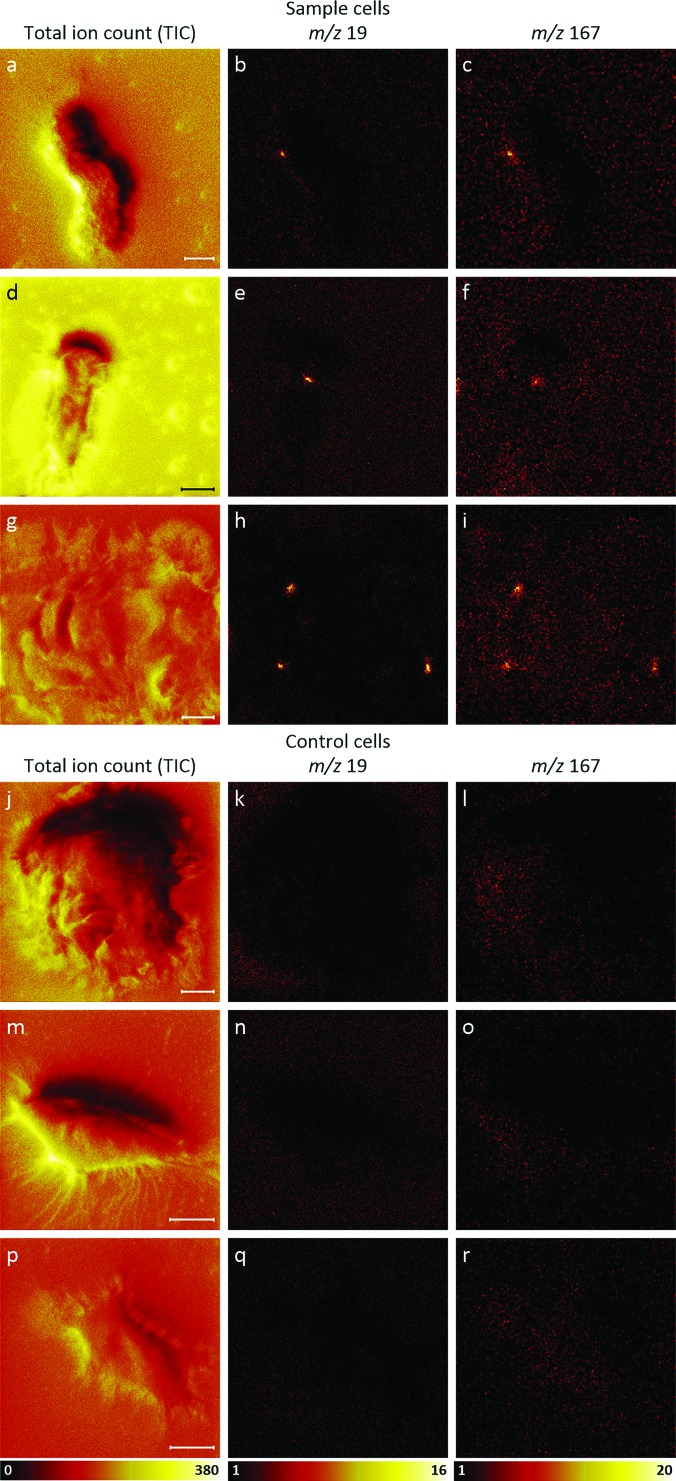
Transfected and ER-Tracker-labeled HEK cells were analyzed using TOF-SIMS tandem MS imaging in the negative ion mode to assess whether m/z 19 and 167 ions produced by the ER-Tracker stain could be detected on the surfaces of the cells, presumably at ER-PM junctions. Control HEK cells that were neither transfected nor labeled with ER-Tracker stain were analyzed for comparison. Images of the TICs and the intensities of the m/z 19 and 167 ions detected on three representative cells are shown in Fig. 3. These images reveal local elevations in the m/z 19 ions [Figs. 3(b), 3(e), and 3(h)] and m/z 167 ions [Figs. 3(c), 3(f), and 3(i)] at the same locations on these three transfected HEK cells. Comparison of these ion images with the TIC images acquired from each cell [Figs. 3(a), 3(d), and 3(g)] shows that the m/z 19 and 167 ion hotspots cannot be attributed to local elevations in all ion species that may result from topography or matrix effects. Figures 3(j)–3(r) show three control cells that were neither transfected nor labeled with ER-Tracker stain. No regional elevations in either the m/z 19 ions [Figs. 3(k), 3(n), and 3(q)] or the m/z 167 ions [Figs. 3(l), 3(o), and 3(r)] were detected on these cells. These findings suggest that the m/z 19 and 167 hotspots detected on the surfaces of the transfected cells corresponded to fluoride and pentafluorophenyl ion fragments, respectively, from the ER-Tracker stain, and they likely signify ER-PM contact sites. The measurement of the m/z 19 and 167 hotspots in the cell images revealed that the diameters of these features were approximately 500 nm to 2 μm in diameter, which agrees well with that expected for ER-PM junctions. Noteworthily, the detection of the ER-Tracker stain in the plasma membrane may seem unexpected because the endoplasmic reticulum is reportedly positioned 10–15 nm below the plasma membrane,34 which should be beyond the sampling depth of TOF-SIMS imaging. The m/z 255 image that was collected in parallel with the SIMS images shown in Figs. 3(g)–3(i) reveals that elevated m/z 255 signals were detected on the cell (Fig. S1).47 Because this ion is frequently used to visualize palmitic acid fragments from lipids,22,31,39,45 this suggests that the plasma membrane was present and the ER-Tracker stain had transferred to the plasma membrane.
Fig. 3.
TOF-SIMS tandem MS negative ion images of three transfected, ER-Tracker stained cells and three unstained (control) cells that were not transfected. Images of the [(a), (d), and (g)] TIC, [(b), (e), and (h)] m/z 19 signals, and [(c), (f), and (i)] m/z 167 signals acquired from each sample cell reveal colocalized hotspots of fluoride and pentafluorophenyl ions, respectively, from the ER-Tracker stain on each cell. Comparison between the ion images and the corresponding TIC images shows that these hotspots are not regions where all the negative ion signals were elevated. Images of the [(j), (m), and (p)] TIC, [(k), (n), and (q)] m/z 19 signals, and [(l), (o), and (r)] m/z 167 signals acquired on each of the three control cells show no local elevations in these two ER-Tracker-associated ions on the cells. The m/z 19 ion images in this figure were binned to 256 × 256 pixels, and the m/z 167 ion images in this figure were binned to 128 × 128 pixels for better visualization. All scale bars are 10 μm.
To visualize the morphology of the endoplasmic reticulum tubules within the transfected cell shown in Figs. 3(g)–3(i), a depth profile analysis was performed, producing a stack of images at increasing depth, up to approximately 40 nm from the cell surface. The first m/z 19 ion image in the stack [Fig. 4(a)] reveals three fluoride ion hotspots on the cell's surface. Additional hotspots are visible in the last m/z 19 ion image in the stack [Fig. 4(b)], which was acquired approximately 40 nm beneath the cell's surface. Noteworthily, the elevated m/z 19 signals surrounding the cell in Fig. 4(b) indicate that the silicon chip contains fluorine that could be detected after sputtering away the poly-l-lysine coating. 3D-renderings of the m/z 19 ion images were created by scaling them to the same ion intensity range, correcting for potential instrument drift by manually aligning them so distinctive features (i.e., the cell/substrate interface) were in registration, and rotating the resulting stack to facilitate visualizations [Figs. 4(c) and 4(d)]. 3D-renderings that show a smaller region within the cell were also created to emphasize the m/z 19 signals from the endoplasmic reticulum tubules [Figs. 4(e) and 4(f)]. These 3D image stacks show that the elevated m/z 19 signals detected within the cell have cylindrical structures that appear at different distances below the cell surface. For example, the three localized elevations in the m/z 19 signals positioned near the right side of the image stacks shown in Figs. 4(c) and 4(e) each become visible at different distances from the plasma membrane. The largest of these three fluorine-rich structures was detected at the plasma membrane and extends throughout the entire image stack. The two adjacent fluorine-rich cylinders were detected further below the cell's surface. The bottom-side views [Figs. 4(d) and 4(f)] also highlight the difference in the depth that these fluoride-rich cylindrical features appear below the cell's surface. The geometries of the fluorine-rich features shown in the 3D image stacks are consistent with endoplasmic reticulum tubules that either formed junctions with the plasma membrane or were located just below the membrane.
Noteworthily, the detection of m/z 167 ions on the surfaces of both the transfected and control cells (Fig. 3) suggests that fragments of cellular components contributed to the m/z 167 signal. This possibility was investigated by analyzing tandem MS data of the m/z 167 ions detected in regions of interest (ROIs) from TOF-SIMS cell images. Fragmentation of the m/z 167 ions collected from the fluoride ion hotspots in the transfected cell, indicated by circles in Fig. 5(a), yielded a fragment ion at m/z 19 [Fig. 5(b)]. The presence of the m/z 19 peak in the product ion spectrum collected at the fluoride ion hotspots suggested that the 1 Da precursor selection window at m/z 167 contained the pentafluorophenyl ion from the ER-Tracker molecule. However, the product ion spectrum also contained multiple peaks that were not present in the spectrum of the pure ER-Tracker molecule [Fig. 2(b)], which indicates the presence of a chemical interference at m/z 167. The product ion spectrum of the m/z 167 precursor ions detected on a control cell [Fig. 5(c)] looked similar to that acquired from the hotspots on the transfected cell, except the MS2 spectrum from the control cell did not contain a peak at m/z 19. These findings indicate the pentafluorophenyl ion from the ER-Tracker stain, as well as fragments of cellular components, contributed to the m/z 167 peak detected in the fluorine-rich regions on the transfected cell. Thus, the fluoride ions at m/z 19 were more specific for detecting the ER-Tracker stain and thus the endoplasmic reticulum.
Fig. 5.
Parallel imaging MS/MS allows for MS1 imaging simultaneously with MS2 imaging. (a) Tandem MS was performed on the m/z 167 precursor detected in the ROIs (blue circles) corresponding to the fluoride hotspots on the transfected and ER-Tracker-labeled cell. No pixel binning was applied to this image. The scale bar is 10 μm. (b) The resulting product ion spectrum contained a fluoride ion at m/z 19, which is consistent with the product ion spectra of the m/z 167 ion produced by the ER-Tracker standard. (c) In control cells, fragmentation of the m/z 167 precursor yielded product ion spectra that lacked the fluoride ion peak at m/z 19.
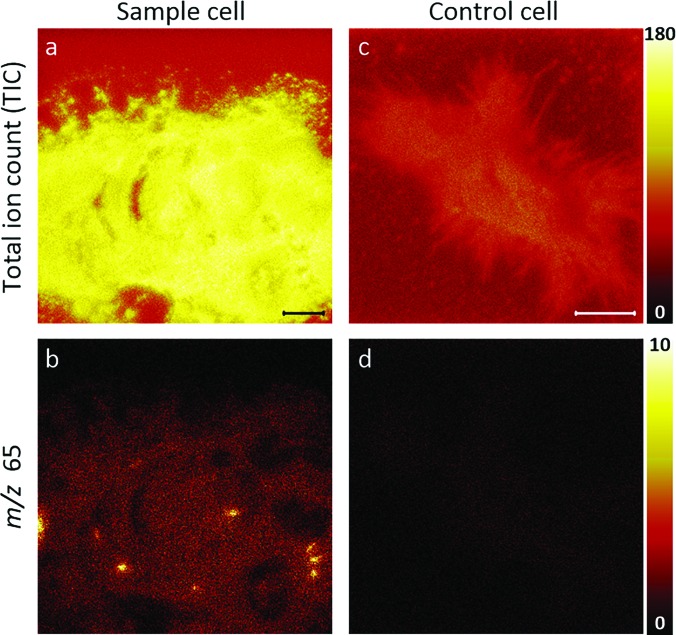
The same location on this transfected cell was also imaged in the positive ion mode to assess whether ions from the positive ion spectra of the ER-Tracker standard could be detected. The resulting TIC and m/z 65 ion images are shown in Figs. 6(a) and 6(b), respectively. The ER-Tracker-associated m/z 65 ion was selected for visualization because it had a high signal intensity, and it does not overlap with reported lipid fragment ions.46 The m/z 65 ion image reveals hotspots [Fig. 6(b)] at the same locations as the m/z 19 ion hotspots that had been detected by negative ion TOF-SIMS imaging [Fig. 4(b)]. For comparison, the TIC and m/z 65 ion images acquired from representative nontransfected and unlabeled (control) cells are shown in Figs. 6(c) and 6(d), respectively. The absence of regional elevations in m/z 65 on the control cell [Fig. 6(d)] and the localization of these ions to the same site where the endoplasmic reticulum tubules were detected on the transfected and ER-Tracker-labeled cell suggest that the m/z 65 signal may be [C5H5]+ ion fragments from ER-Tracker. However, we cannot rule out the possibility that this ion resulted from some other component that is found in endoplasmic reticulum tubules.
Fig. 6.
Unbinned TOF-SIMS positive ion images of a transfected, ER-Tracker stained cell [(a) and (b)] and an unstained (control) cell that was not transfected [(c) and (d)]. Images of the (a) TIC and (b) m/z 65 signals acquired from the transfected cell. The local elevations in m/z 65 signals shown in (b) correspond to the same locations where the m/z 19 hotspots were detected on this cell [Fig. 4(b)]. This colocalization suggests that the m/z 65 signals might be cyclopentadienyl ions [C5H5]+ from the ER-Tracker stain or some other component that is found in endoplasmic reticulum tubules. Consistent with these possibilities, images of the (c) TIC and (d) m/z 65 signals acquired on the control cell show no local elevations in the m/z 65 signals. Scale bars are 10 μm.
IV. SUMMARY AND CONCLUSIONS
The recently developed TOF-SIMS Parallel Imaging MS/MS was used to image the endoplasmic reticulum within GFP-Kv2.1-expressing HEK cells. We found that the commercial ER-Tracker fluorescent dye yields characteristic secondary ions, namely, fluoride, which allow for its detection and imaging by TOF-SIMS. Depth profiling analysis and 3D visualization of the ER-Tracker-associated peaks allowed for imaging the morphologies of the endoplasmic reticulum tubules that were near the plasma membrane along their z-direction. Future work will focus on imaging the endoplasmic reticulum within the entire cell volume and assessing the lipid composition within this organelle. The use of the ER-Tracker to label the endoplasmic reticulum is also expected to permit probing the lipid composition at different locations within the endoplasmic reticulum, including ER-PM junctions. Such efforts will greatly benefit from the ability to confirm MS1 peak identities through tandem mass spectrometry, while simultaneously imaging using TOF-SIMS Parallel Imaging MS/MS.
ACKNOWLEDGMENTS
This material is based upon the work partially supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award No. R01GM109888 and by the National Science Foundation under Grant No. CHE-1508662. Any opinions, findings, and conclusions or recommendations expressed in this material are those of the authors and do not necessarily reflect the views of the National Science Foundation or the National Institutes of Health. The authors also acknowledge the Core Facilities at the Carl R. Woese Institute for Genomic Biology, which were used for confocal microscopy imaging. This paper is dedicated to the memory of John S. Hammond who passed away on 9 September 2017. John leaves a strong legacy at Physical Electronics (PHI) where his work spanned from 1981 until February 2017. He will be missed at PHI and within the worldwide scientific community where his love for science, learning, and new technologies never wavered. In his last few years at PHI, John was a coinventor of the TOF-SIMS tandem MS imaging technology and he was a champion for molecular imaging and identification of subcellular structures and organelles. This paper, a step toward the eventuality of that vision, is dedicated to John. He was a colleague, a friend, and an inspiration—G. L. Fisher.
Contributor Information
Gregory L. Fisher, Email: .
Mary L. Kraft, Email: .
References
- 1. van Meer G. and de Kroon A. I. P. M., J. Cell Sci. 124, 5 (2011). 10.1242/jcs.071233 [DOI] [PubMed] [Google Scholar]
- 2. van Meer G., Voelker D. R., and Feigenson G. W., Nat. Rev. Mol. Cell Biol. 9, 112 (2008). 10.1038/nrm2330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Di Paolo G. and De Camilli P., Nature 443, 651 (2006). 10.1038/nature05185 [DOI] [PubMed] [Google Scholar]
- 4. Okeke E., Dingsdale H., Parker T., Voronina S., and Tepikin A. V., J. Physiol. 594, 2837 (2016). 10.1113/JP271142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Chung J. et al. , Science 349, 428 (2015). 10.1126/science.aab1370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Gatta A. T., Wong L. H., Sere Y. Y., Calderón-Noreña D. M., Cockcroft S., Menon A. K., and Levine T. P., eLife 4, e07253 (2015). 10.7554/eLife.07253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ikonen E., Nat. Rev. Mol. Cell Biol. 9, 125 (2008). 10.1038/nrm2336 [DOI] [PubMed] [Google Scholar]
- 8. Brügger B., Annu. Rev. Biochem. 83, 79 (2014). 10.1146/annurev-biochem-060713-035324 [DOI] [PubMed] [Google Scholar]
- 9. Vance J. E., Traffic 16, 1 (2015). 10.1111/tra.12230 [DOI] [PubMed] [Google Scholar]
- 10. Kraft M. L. and Klitzing H. A., Biochim. Biophys. Acta 1841, 1108 (2014). 10.1016/j.bbalip.2014.03.003 [DOI] [PubMed] [Google Scholar]
- 11. Kurczy M. E., Piehowski P. D., Van Bell C. T., Heien M. L., Winograd N., and Ewing A. G., Proc. Natl. Acad. Sci. 107, 2751 (2010). 10.1073/pnas.0908101107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ostrowski S. G., Van Bell C. T., Winograd N., and Ewing A. G., Science 305, 71 (2004). 10.1126/science.1099791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Passarelli M. K. and Winograd N., Biochim. Biophys. Acta 1811, 976 (2011). 10.1016/j.bbalip.2011.05.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Wilson R. L., Frisz J. F., Klitzing H. A., Zimmerberg J., Weber P. K., and Kraft M. L., Biophys. J. 108, 1652 (2015). 10.1016/j.bpj.2015.02.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Frisz J. F., Klitzing H. A., Lou K. Y., Hutcheon I. D., Weber P. K., Zimmerberg J., and Kraft M. L., J. Biol. Chem. 288, 16855 (2013). 10.1074/jbc.M113.473207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Frisz J. F. et al. , Proc. Natl. Acad. Sci. U. S. A. 110, E613 (2013). 10.1073/pnas.1216585110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Klitzing H. A., Weber P. K., and Kraft M. L., Nanoimaging 950, 483 (2013). 10.1007/978-1-62703-137-0_26 [DOI] [PubMed] [Google Scholar]
- 18. Tian H., Fletcher J. S., Thuret R., Henderson A., Papalopulu N., Vickerman J. C., and Lockyer N. P., J. Lipid Res. 55, 1970 (2014). 10.1194/jlr.D048660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Breitenstein D., Rommel C. E., Möllers R., Wegener J., and Hagenhoff B., Angew. Chem. Int. Ed. 46, 5332 (2007). 10.1002/anie.200604468 [DOI] [PubMed] [Google Scholar]
- 20. Brison J., Benoit D. S. W., Muramoto S., Robinson M., Stayton P. S., and Castner D. G., Surf. Interface Anal. 43, 354 (2011). 10.1002/sia.3415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Brison J., Robinson M. A., Benoit D. S. W., Muramoto S., Stayton P. S., and Castner D. G., Anal. Chem. 85, 10869 (2013). 10.1021/ac402288d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fletcher J. S., Lockyer N. P., Vaidyanathan S., and Vickerman J. C., Anal. Chem. 79, 2199 (2007). 10.1021/ac061370u [DOI] [PubMed] [Google Scholar]
- 23. Fletcher J. S., Rabbani S., Henderson A., Blenkinsopp P., Thompson S. P., Lockyer N. P., and Vickerman J. C., Anal. Chem. 80, 9058 (2008). 10.1021/ac8015278 [DOI] [PubMed] [Google Scholar]
- 24. Fletcher J. S., Rabbani S., Henderson A., Lockyer N. P., and Vickerman J. C., Rapid Commun. Mass Spectrom. 25, 925 (2011). 10.1002/rcm.4944 [DOI] [PubMed] [Google Scholar]
- 25. Graham D. J., Wilson J. T., Lai J. J., Stayton P. S., and Castner D. G., Biointerphases 11, 02A304 (2016). 10.1116/1.4934795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Robinson M. A., Graham D. J., and Castner D. G., Anal. Chem. 84, 4880 (2012). 10.1021/ac300480g [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Van Nuffel S., Parmenter C., Scurr D. J., Russell N. A., and Zelzer M., Analyst 141, 90 (2016). 10.1039/C5AN01743B [DOI] [PubMed] [Google Scholar]
- 28. Vreja I. C., Kabatas S., Saka S. K., Kröhnert K., Höschen C., Opazo F., Diederichsen U., and Rizzoli S. O., Angew. Chem. Int. Ed. 54, 5784 (2015). 10.1002/anie.201411692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lau K. H., Christlieb M., Schroder M., Sheldon H., Harris A. L., and Grovenor C. R. M., J. Microsc. 240, 21 (2010). 10.1111/j.1365-2818.2010.03380.x [DOI] [PubMed] [Google Scholar]
- 30. Wilson R. L., Frisz J. F., Hanafin W. P., Carpenter K. J., Hutcheon I. D., Weber P. K., and Kraft M. L., Bioconjugate Chem. 23, 450 (2012). 10.1021/bc200482z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Chen L.-J., Shah S. S., Silangcruz J., Eller M. J., Verkhoturov S. V., Revzin A., and Schweikert E. A., Int. J. Mass Spectrom. 303, 97 (2011). 10.1016/j.ijms.2011.01.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Saka S. K., Vogts A., Kröhnert K., Hillion F., Rizzoli S. O., and Wessels J. T., Nat. Commun. 5, 3664 (2014). 10.1038/ncomms4664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Shibata Y., Voeltz G. K., and Rapoport T. A., Cell 126, 435 (2006). 10.1016/j.cell.2006.07.019 [DOI] [PubMed] [Google Scholar]
- 34. Fox P. D., Haberkorn C. J., Akin E. J., Seel P. J., Krapf D., and Tamkun M. M., J. Cell Sci. 128, 2096 (2015). 10.1242/jcs.166009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Misonou H., Mohapatra D. P., Menegola M., and Trimmer J. S., J. Neurosci. 25, 11184 (2005). 10.1523/JNEUROSCI.3370-05.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. O'Connell K. M. S. and Tamkun M. M., J. Cell Sci. 118, 2155 (2005). 10.1242/jcs.02348 [DOI] [PubMed] [Google Scholar]
- 37. Tamkun M. M., O'Connell K. M. S., and Rolig A. S., J. Cell Sci. 120, 2413 (2007). 10.1242/jcs.007351 [DOI] [PubMed] [Google Scholar]
- 38. Mohapatra D. P. and Trimmer J. S., J. Neurosci. 26, 685 (2006). 10.1523/JNEUROSCI.4620-05.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Fisher G. L., Bruinen A. L., Potocnik N. O., Hammond J. S., Bryan S. R., Larson P. E., and Heeren R. M. A., Anal. Chem. 88, 6433 (2016). 10.1021/acs.analchem.6b01022 [DOI] [PubMed] [Google Scholar]
- 40. Fisher G. L., Hammond J. S., Bryan S. R., Larson P. E., and Heeren R. M. A., Microsc. Microanal. 23, 843 (2017). 10.1017/S1431927617000654 [DOI] [PubMed] [Google Scholar]
- 41. O'Connell K. M. S., Loftus R., and Tamkun M. M., Proc. Natl. Acad. Sci. U. S. A. 107, 12351 (2010). 10.1073/pnas.1003028107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. O'Connell K. M. S., Rolig A. S., Whitesell J. D., and Tamkun M. M., J. Neurosci. 26, 9609 (2006). 10.1523/JNEUROSCI.1825-06.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Akin E. J., Solé L., Johnson B., Beheiry M. e., Masson J.-B., Krapf D., and Tamkun M. M., Biophys. J. 111, 1235 (2016). 10.1016/j.bpj.2016.08.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fox P. D., Haberkorn C. J., Weigel A. V., Higgins J. L., Akin E. J., Kennedy M. J., Krapf D., and Tamkun M. M., Mol. Biol. Cell 24, 2703 (2013). 10.1091/mbc.E12-12-0895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dowlatshahi Pour M., Jennische E., Lange S., Ewing A. G., and Malmberg P., Sci. Rep. 6, 32797 (2016). 10.1038/srep32797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Anderton C. R., Vaezian B., Lou K., Frisz J. F., and Kraft M. L., Surf. Interface Anal. 44, 322 (2012). 10.1002/sia.3806 [DOI] [Google Scholar]
- 47.See supplementary material at https://doi.org/10.1116/1.5019736E-BJIOBN-13-310803 for negative ion SIIMS images showing m/z 19 and 255 signals on the surface of a cell.
- 48. Yeager A. N., Weber P. K., and Kraft M. L., Biointerphases 11, 02A309 (2016). 10.1116/1.4939681 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- See supplementary material at https://doi.org/10.1116/1.5019736E-BJIOBN-13-310803 for negative ion SIIMS images showing m/z 19 and 255 signals on the surface of a cell.