Abstract
Intrinsically disordered proteins (IDPs) play important roles in many physiological processes such as signal transduction and transcriptional regulation. Computer simulations that are based on empirical force fields have been increasingly used to understand the biophysics of disordered proteins. In this review, we focus on recent improvement of protein force fields, including polarizable force fields, concerning their accuracy in modeling intrinsically disordered proteins. Some recent benchmarks and applications of these force fields are also overviewed.
Introduction
The abundance of intrinsically disordered proteins (IDPs), which include proteins with disordered regions, in the human proteome has recently been recognized.[1,2] IDPs are characterized by the lacking of any well-defined three-dimensional tertiary structures in contrast to the common paradigm that a protein functions by folding into a single native structure. Instead, an IDP exists as an ensemble of flexible conformations that interconvert with each other, which often involves transient forming and breaking of secondary structure elements. The primary sequences of IDPs feature an enrichment of polar and charged amino acids, with decreased amounts of non-polar residues that normally drive hydrophobic core formation. The conformational flexibility of IDPs not only allows them to serve as flexible linkers between functional domains, but more importantly allows them to play essential roles in protein-protein interaction network as IDPs can adopt different conformations when binding to different partners.
The central role of IDPs in eukaryotic protein interaction networks makes them involved in many pathological conditions, especially in cancers and neurodegenerative diseases.[3] The advantage of IDPs’ structural plasticity for their regulatory roles, such as signal transduction and transcriptional regulation, also makes them occur at a high frequency among tumor-related proteins such as p53 and PTEN.[3] Since IDPs can sample a large variety of conformational states, they are prone to aggregate under certain environments. The assembly and aggregation of IDPs leads to the generation of fibrils, hallmarks of many neurodegenerative diseases. Examples include α-synuclein in Parkinson’s disease, the β-amyloid (Aβ) peptide and tau protein in Alzheimer’s disease, and polyglutamine (polyQ) in Hungtington’s disease. Although the importance of conformational dynamics has been appreciated in the computer-aided drug design (CADD), IDPs represent a very challenging case for therapeutic targeting. Instead of binding to a particular IDP conformation, a ligand needs to modulate the IDP’s conformational dynamics and its interactions with binding partners.
Experimental tools to investigate IDP conformational ensembles include small-angle X-ray scattering (SAXS), nuclear magnetic resonance (NMR) and Förster resonance energy transfer (FRET) spectroscopy. However, the observables from these experiments are ensemble averaged over the interconverting conformational states of IDPs. [4,5] Even with single molecule experiments, the number of degrees of freedom for an IDP conformational ensemble still far exceeds the number of available experimental observables. To address such an underdetermined problem, theoretical models need to be introduced to extract detailed structural information from these experiments. These methods can be based on polymer physics such as the Gaussian chain model or more detailed atomistic models such as the computer simulations based on molecular mechanics force fields (FFs).
Protein force fields are empirically developed potential energy functions for polypeptides. Combined with proper sampling methods such as molecular dynamics (MD) or Monte Carlo (MC) simulations, they can be used to generate structural ensembles for any IDP without a posteriori knowledge. The atomistic details obtained from force field-based simulations can be used to help interpret experimental results, or sometime resolve the conflicts between different experimental measurements.[6] It’s also possible to derive IDP ensembles based on mutual information of force fields and experiments. Possibilities include driving MD simulations with the guide of experimental data,[7,8] or post processing force-field generated ensembles to match experimental data in a Bayesian fashion.[9–13] These atomistic models of IDP conformations serve as the starting point for structure-based drug design.
The quality of IDP ensembles, either generated completely in silico or determined jointly by combining computations and experiments, depends critically on the accuracy of underlying computational models. To this end, IDPs represent important benchmark systems for protein FFs, which were originally developed for folded proteins and are continuously under further development. In this article we will review some of the recent progress in force field development and simulations for IDPs.
Improvement of Protein Force Fields for IDP Simulations
Protein force fields are by definition the potential energy functions and corresponding parameters to describe the bonded and non-bonded interactions between the particles, typically atoms that define the amino acids, as well as the interactions between polypeptides and water. The ability for a protein force field to model these interactions is in principle transferable between folded protein and IDPs, so that any general improvement of protein FFs, though usually not directly targeting IDPs, often leads to more accurate representation of IDPs. Two types of protein FF improvements are particularly relevant for IDP simulations. The first one is to balance the propensity of the sampling of secondary structures, as the conformational dynamics of IDPs may contain frequent formation and breaking of α-helices and β-sheets. This often involves the refinement of the backbone ϕ, ψ dihedral parameters targeting short peptides that fold into α-helices such as the (AAQAA)3 peptide[14] or β-hairpins (for example the GB1 hairpin[15] and chigolin[16]) as model systems. The second one is to improve the modeling of the balance of the protein-water and protein-protein interactions, which often results in the introduction of atom pair-specific Lenard-Jones (L-J) parameters (e.g. NBFIX in CHARMM nomenclature) in protein FFs. Useful target data include quantum mechanical data on water-model compound and model compound-model compound interactions and experimental hydration free energies[17] and, more recently, the osmotic pressures of model compounds, where the model compounds are backbone or side-chain analogs.[18,19] The balance between protein-protein and protein-water interactions is particularly important for IDPs, in which no stable hydrophobic cores are formed to bury non-polar residues. This also highlights the importance of using the correct combination of protein FF and water model in IDP simulations, as it has been shown that the equilibrium between folded and unfolded states can be modified with even a subtle change in the water model used in the simulations.[20,21] In the remaining part of this section, we will overview recent general improvements in major protein FFs, including the Amber, CHARMM, and OPLS FFs.
Efforts from Best and Hummer to balance the secondary structure propensity for the Amber series of protein force fields led to Amber ff99SB* and ff03*,[22] which corrected the bias of underestimating and overestimating the helical content in ff99SB[23] and ff03[24], respectively. Both ff99SB* and ff03* were developed to be used together with the TIP3P water model.[25] A subsequent refinement of ff03* yielded the ff03w FF to be used with the four-site TIP4P/2005 water model,[26] which has been used in a variety of IDP simulations.[6,27–30] Other Amber protein FF development included ff14SB,[31] an improvement over ff99SB with new side chain dihedral parameters and empirical adjustment to the backbone ϕ energy profile. Cerutti et al derived ff14ipq,[32] which contains a completely new charge set using the implicitly polarized charge (IPoIQ) model.[33] The bond, angle and L-J parameters in ff14ipq were taken from ff99SB, while torsional parameters were fitted using gas phase quantum mechanics (QM) calculations at the MP2/cc-pVTZ level. A further reparametrization of bonded and non-bonded parameters based on the IPoIQ method yielded ff15ipq.[34] The ff14ipq and ff15ipq FFs were developed to be used with the TIP4P-Ew[35] and the SPC/Eb[36] water models, respectively. The AMBER-FB15 force field[37] was also developed based on ff99SB, with a focus on the optimizing the FF parameters for bonded interactions using QM RI-MP2/aug-cc-pVTZ calculations and the ForceBalance procedure.[38]
The CHARMM36 (C36) protein FF[39] was published in 2012 and contains multiple improvements over its predecessor, the CHARMM22/CMAP FF (also known as CHARMM27),[40,41] including refinement of the backbone CMAP potentials and new side-chain dihedral parameters. CMAP is a 2-dimensional (2D) ϕ,ψ grid-based energy correction map[42] first introduced in 2002 to improve the treatment of the protein backbone conformations. C36 is able to reproduce a variety of NMR observables for folded proteins[43], shows enhanced cooperativity of helix and hairpin formation,[44] and yields high accuracy in protein structure refinement.[45] However, application of the C36 protein FF to simulate several IDPs revealed a potential deficiency that conformational states containing left handed helicies were overly populated.[30] A further refinement of the CMAP potentials was performed to address this issue, which together with the introduction of a NBFIX term for improved modeling of salt-bridge interactions yielded the CHARMM36m (C36m) FF.[21] C36m was developed as a transferable FF between folded and disordered proteins, and has been validated using a wide range of IDPs. The C36m protein FF, as other CHARMM additive FFs, has to be used with the CHARMM modified TIP3P water model.[46] Recently, Chen and co-workers optimized the GBMV2 implicit solvent model based on the C36 protein FF with particular emphasis on IDP simulations.[47] Other implicit solvent models useful in studying IDPs include the FACTS model[48] and the ABSINTH model,[49] which have been used to investigate the conformational states of Aβ40 and Aβ42,[50] the homodimzerization of polyQ,[51] and the aggregation of phenylalanine at pathological concentration in phenylketonuria.[52]
Two refinements of the OPLS protein force fields have been published recently. Robertson et al reported the OPLS-AA/M force field[53] and Harder et al presented the OPLS3 force field.[54] Both revisions involved the reparametrization of peptide backbone ϕ, ψ and side chain χ1, χ2 dihedrals by fitting to QM torsional energy scans for blocked dipeptides. The OPLS3 protein FF, together with its improvement in small molecule FF parameters, leads to enhanced accuracy in predicting protein-ligand binding affinities.[54]
While most additive protein FF development has focused on refitting torsional parameters, another possibility being explored in recent years is to expand the parameter space by using residue-specific backbone dihedral parameters. This means that protein backbone ϕ, ψ parameters will depend on the amino acid type, thus introducing an explicit coupling between the backbone and the sidechain parameters and allows more flexibility in parametrization. Based on this idea, Wu and coworkers developed RSFF1[55] based on the OPLS-AA/L[56] force field and RSFF2[57] based on Amber ff99SB, using rotamer distribution from protein coil library as the major target data. In a similar way, Chen and coworkers proposed to add residue-specific CMAP corrections in Amber FFs to better model IDPs. This was done initially for eight disorder-promoting amino acids (A, G, P, R, Q, S, E, and K), and then extended to all amino acids, yielding the ff99IDPs[58,59] and the ff14IDPSFF force fields[60], respectively.
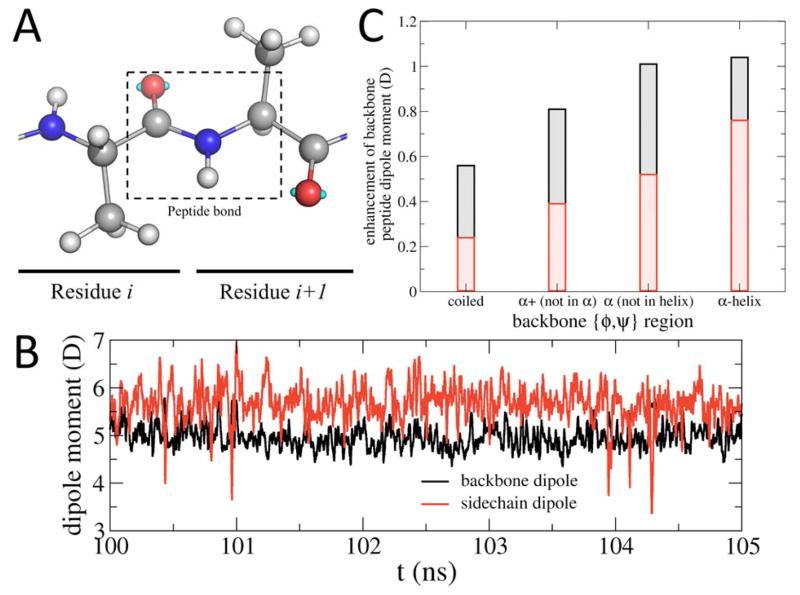
We also note the significant progress in polarizable FFs as two parameter sets for peptide and protein simulations have been published, including the AMOEBA-2013 FF using the distributed multipole and induced dipole formalism[61] and the Drude-2013 force field based on the Drude oscillator formalism.[62] A recent study demonstrates the equivalency between the two formalisms by mapping the Drude FF into an AMOEBA-like model termed MPID.[63] Application of the Drude-2013 FF showed it to correctly treat the coopertivity of folding of the helical (AAQAA)3 peptide[64] and showed cooperative unfolding of a β-amyloid fragment,[65] both associated with variations in the dipole moment of the peptide backbone that cannot occur in additive protein FFs (Figure 1). With improved algorithms and implementation of polarizable force fields in a wider-range of software packages,[66] we expect greater utilization of polarizable protein FFs in modeling and simulating IDPs.
Figure 1.
Variation of dipole moments in (AAQAA)3 during a MD simulation with the Drude polarizable FF. (A) Definition of the peptide bond used to compute backbone dipole moment. (B) Time series of the backbone dipole moment and the sidechain dipole moment of the central Gln residue in (AAQAA)3. (C) The average enhancement of peptide backbone dipole moments in different conformational regions as defined using the backbone ϕ,ψ dihedral angles. Shown are the total dipole moment enhancement, the intra-peptide enhancement (red) and the solvent enhancement (gray). Stronger enhancement of the backbone dipole moment, in particular the intra-peptide enhancement, is observed when a residue folds into an α-helix. Such enhancement provides additional free energy incentive for helix elongation, leading to the reproduction of the experimentally observed folding cooperativity. The enhancement is computed as the difference of the peptide bond dipole moment with and without their environments (water and the remaining part of the peptide). Results are averaged over all peptide bonds in the protein. See Ref. 59 for more details.
Sampling more extended states of IDPs with additive force fields
The size of an IDP, averaged over its heterogeneous conformational states, is a basic property that can be inferred from experiments such as SAXS and FRET measurement. Comparison between experimental and computational chain dimensions shows that most current additive protein FFs underestimate the radius of gyration (Rg) for IDPs. The problem that computationally generated ensembles are often overly compact is not limited to IDPs, but also observed for denatured proteins and random coils.
An effective way to computationally sample more extended states of proteins is to increase the strength of protein-water interactions while maintaining the protein-protein and water-water interactions. Different approaches haven been proposed to modify the protein-water interactions.[17,21,67,68] Best et al suggested to directly scale up the dispersion interactions between protein and water by a common factor.[67] In practice, this can be achieved by multiplying the VdW potential depth ε of water atoms by a scaling factor, and resetting the water-water and water-ion interactions with atom pair-specific LJ (e.g. NBFIX) terms. Based on the comparison between the computational and experimental mean FRET efficiencies of a 34-residue cold-shock protein fragment (Csp M34), a scaling factor of 1.10 was determined for the ff03w FF with TIP4P/2005 water model. The resulting ff03ws FF has been validated for the chain dimension of ACTR,[67] and used in several IDP studies.[29,30,69]
Piana et al adopted another approach by parametrizing a new water model, TIP4P-D,[68] with the oxygen ε value constrained to a larger value than in typical water models. Combining TIP4P-D with the Amber or CHARMM protein FFs generated conformational ensembles with significantly larger Rg for a set of IDPs including HIV-1 integrase domain (IN), protein L immunoglobulin-binding domain, cold-shock protein from Thermotoga maritima (CspTm), α-synuclein and Prothymosin α.[68] We note that the C6 dispersion coefficient is set to 900 kcal/mol/Å6 in the TIP4P-D water model, which compared with the value of 736 kcal/mol/Å6 in TIP4P/2005 is equivalent to increase protein-water dispersion interactions by a factor of 1.11 according to the Lorentz–Berthelot mixing rules. Accordingly, simulations with ff03w/TIP4P-D and with ff03ws have similar effects in enhancing protein-water dispersion interactions, although with TIP4P-D the electrostatics interaction between protein and water is also increased due to the stronger water dipole moment associated with TIP4P-D.
Recently we proposed another way to sample more extended states with the CHARMM protein force field, taking advantage of the fact that in the CHARMM modified TIP3P water model both oxygen and hydrogen atoms participate in L-J interactions.[46] We suggested keeping the VdW parameters of water oxygen atoms, and simply increasing the ε value on the water hydrogen atom. This is because the L-J potential contains both the repulsion (r −12) and the dispersion (r −6) parts such that changing ε of the water oxygen atom impacts both the L-J dispersive interactions and the repulsive wall. This will have a general impact on water-protein hydrogen bond interactions thereby affecting the structure of solvation shells. In contrast, the water hydrogen atom has a very small L-J radius so that it is effectively buried within the repulsive wall of its bonded oxygen atom and only contributes dispersion interactions with the environment. Thus, by modifying the hydrogen VdW parameters only, the Hamiltonian is disturbed in a minimal way, i.e. only the dispersion part of the VdW interaction with water in the simulation systems is changed. Using this approach increasing εH from −0.046 kcal/mol in the CHARMM TIP3P water to a more favorable value of −0.1 kcal/mol has been shown to reproduce the chain dimension and mean FRET efficiency of CspTm.[21]
While all these approaches have been shown to generate IDP conformational ensembles with larger polypeptide chain dimensions, for different IDPs the extent of the increase differs and the correlations with experimental observables can become better or worse.[21,29,30,70] This implies that without further development on the protein FF side, it might be difficult to find a universal VdW scaling factor, or oxygen C6 parameter, or modified TIP3P εH parameter that gives the correct chain dimensions for all types of IDPs. Nevertheless, having systematic ways to generate computational IDP ensembles with larger size is of importance itself, as they can be used as better inputs (e.g. broader priori distribution) for Bayesian interface. We would also like to point out that purely reproducing the experimentally derived Rg is not sufficient to claim that a computational model is accurate, as multiple ensembles of different qualities can all have the same Rg value. To this end it’s an underdetermined problem with few available experimental data. In addition, how the fine tuning of protein-water interactions impacts the ensemble properties of folded protein and the energy landscape of IDPs is not fully understood, requiring further investigations.
Recent FF benchmarks
In this section we review some recent benchmarks that compare IDP conformational ensembles generated with a set of different force fields to a variety of experimental data from SAXS, FRET and NMR measurements (Table 1). The SAXS data contain detailed information on the shape of proteins, and the intensity profiles computed from MD ensembles can be compared with experimental data directly. In FRET experiments, the FRET efficiency (quantum yield of the energy transfer) can be computed as (1+(r/R0)6)−1, where R0 is the Förster distance determined by the nature of donor/acceptor pair, and r is the distance between donor and acceptor, which is equivalent to the end-to-end distance if chromophores are inserted at the two chain termini. The ensemble averaged FRET efficiency can be directly compared between calculations and experiments, and in the case of single molecule FRET experiments, the distribution of efficiency can be compared. NMR chemical shift and scalar coupling data provide secondary structure propensities for individual amino acids, while NMR NOE data can contain useful information on long-range contact between residues. NMR techniques can also be used to extract chain dimension information of polypeptides, for example pulsed field gradient NMR can be used to measure the hydrodynamic radius.
Table 1.
IDP systems used in force field benchmark studies.
| Sequence | Exp. data | Force fields tested | |
|---|---|---|---|
| RS peptide | GAMGPSYG(RS)8 | SAXS, NMR | ff99SB*-ILDNa, ff03wc, ff03wsc, C22*b, d, C36a, b, C36mb |
| Histatin 5 | DSHAKRHHGYKRKFHEKHHSHRGY | SAXS, NMR | ff99SB-ILDNa, d, ff99SBNMR-ILDNa, G53a6f, G54a7f, ff03wsb |
| Polygutamine | (Q)30 | FRET | ff99a, ff99SBa, ff99SB*a, ff03a, ff03*a, ff03wc, C22/CMAPb, C22*b, C36a, G53a6f, G54a7f, OPLS-AA/Le, C36mb |
| β-amyloid | DAEFRHDSGYEVHHQKLVFFAEDVGSNKGAIIGLMVGGVVIA | NMR | ff99SBg, ff99SB*-ILDNg, ff99SBNMR-ILDNg, C22*g, b, G53a6f, OPLS-AAa, ff99SB-ILDNa, ff14SBa |
| hIAAP | KCNTATCATQRLANFLVHSSNNFGAILSSTNVGSNTY | NMR | ff99SB*-ILDNa, e, ff03wc, e, C22/CMAPb, C22*b, e, G53a6f, OPLS-AA/Le |
with the TIP3P water model;
CHARMM modified TIP3P water model;
TIP4P-2005 water model;
TIP4P-D water model;
TIP4P water model;
SPC water model;
TIP4P-Ew water model.
The RS peptide is a highly charged IDP with a repeating arginine and serine sequence. Rauscher et al carried out temperature replica exchange simulations to sample the conformational states of the RS peptide using a wide range of Amber and CHARMM force fields (see Table 1).[30] The CHARMM22* (C22*)[71] and C36m FFs, both simulated with the CHARMM modified TIP3P water model, were found to best reproduce SAXS and NMR observables, such as radius of gyration and hydrodynamics radius. Different approaches that adjust the protein-water dispersion interactions were also tested, and all of them (ff03ws,[67] C22* with TIP4P-D,[68] and C36m with alternative εH=−0.1 kcal/mol TIP3P water model[21]) were found to generate more expanded ensembles as expected. However, the agreement with experimental chain dimensions became worse as the conformational ensembles were overly expanded.
Histatin 5 is another 24 residue IDP that has been used to compare force fields. Two Amber FFs (ff99SB-ILDN and ff99SBNMR-ILDN) and two GROMOS FFs (G53a6 and G54a7) were found to generate too collapsed ensembles that are not consistent with experimental measurement. Approaches with adjusted protein-water dispersion interactions, both ff03ws and ff99SB-ILDN/TIP4P-D, generated more expanded conformational ensembles with satisfactory reproduction of experimental SAXS profiles.[29,70]
PolyQ is implicated in a series of expanded CAG repeat neurodegenerative diseases. Fluitt and de Pablo carried out replica exchange simulations of (Q)30 using 12 different FFs, and found out that most were able to capture the lack of secondary structure of polyQ peptides.[72] We noted that simulations of (Q)30 with the newly developed C36m FF predicts a mean FRET efficiency of 0.20 ± 0.01, very close to the experimentally extrapolated value of 0.22.[73] Force field comparisons have also been carried out for other IDPs including Aβ monomer[74] and dimer,[75] and the human islet amyloid polypeptide (hIAAP)[76], although experimental data to benchmark the FFs are relatively limited, mostly involving localized secondary structural information inferred from NMR measurements.
Challenges in simulating IDP-related biophysical processes
To understand the atomistic details of how IDPs function, computer simulations have to go beyond characterizing the conformational landscape of single IDPs, and to model IDP-related biophysical processes, such as post-translational modifications (PTMs), coupled binding and folding, and IDP aggregations. PTMs such as phosphorylation, acetylation, and glycosylation are common in IDPs and can be crucial to their function.[77] The force field parameters for PTMs are readily available; for example, the CHARMM36 force field, supplemented with the CHARMM General force field[78] and carbohydrate parameters[79,80] provides good coverage for PTMs, and Forcefield_PTM[81] contains parameters consistent with the Amber ff03 FF. A recent computational study of two IDPs found no significant difference in their conformational ensembles with and without glycosylation.[82] To some extent this is consistent with a recent experimental study suggesting that the global conformational properties of a single IDP are conserved when it undergoes multisite phosphorylation.[83] These studies imply that the effect of PTMs might only be revealed when an IDP interacts with its binding partners.
The recognition and interaction between two IDPs typically involves the process of coupled binding and folding, where both IDPs acquire folded tertiary structures when they encounter each other and form a complex. This process is poorly understood, and different mechanisms have been proposed to answer questions such as whether binding or folding happens first. In the conformational selection scheme a pre-formed conformation is selected by IDP’s binding partner such that the folding happens first, while in the induced fit scheme an IDP first binds to its partner and subsequently folds into its bound conformation. These two schemes should be considered limiting ideal cases, with the actual process probably lying in between, as shown recently in an explicit solvent atomistic simulations of the coupled binding and folding between the transcription factor c-myb and the cotranscription factor CREB binding protein.[84] A related process is IDP aggregation, where IDP monomers assemble into oligomers as well as fibril structures. We noted that the investigation of these processes raises the bar on the accuracy of protein force fields, especially in their abilities in modeling protein-protein interactions. Matthes et al compared three FFs (ff99SB*-ILDN, C36, and GROMOS96 43a1) in modeling the self-assembly of amyloidogenic peptide fragments,[85] and more such benchmark studies are needed to validate and to further improve protein FFs.
Conclusion
In this review we overview recent advance in empirical force field development and atomistic simulations of intrinsically disordered proteins. Readers interested in other computational models and theoretical methods for IDPs are referred two recent reviews.[86,87] In general, IDPs represent difficult challenges for atomistic force fields, but also represent big opportunities as FF-based simulations remains the most powerful way to unravel the atomistic details on IDPs’ conformational dynamics. We expect that the accuracy and transferability of protein force fields will continue being improved, both with respect to better parametrization and more advanced potential energy functions, most notable being models the include the explicit treatment of electronic polarizability. We found that the radius of gyration for the unfolded states of (AAQAA)3 is 10% larger with the polarizable Drude FF than with the additive C36 FF.[64] These improvements, coupled with better conformational sampling techniques and hardware developments, will lead to a better understanding of how IDPs function.
Highlights.
General improvement of protein force fields, even not directly targeting IDPs, could lead to more accurate representation of IDPs.
Increasing protein-water dispersion interactions allows additive force fields to effectively sample more extended states of IDPs, and different approaches have been proposed and tested.
Further improvement of force field transferability between folded and disordered proteins will involve the explicit inclusion of electronic polarizability in potential energy functions.
Acknowledgments
Financial support from the NIH to ADM (GM051501 and GM072558) is acknowledged.
Footnotes
Reference Annotations
Papers of outstanding interest:
Ref. [6]: The authors studied the effect of chemical denaturation (urea and guanidinium chloride) on the conformational ensemble of an IDP, and used simulations to explain the inconsistency between SAXS and FRET experimental results.
Ref. [21]: The authors presented CHARMM36m, a refinement of the C36 protein force field with improved accuracy in generating IDP conformational ensembles. Also proposed was an alternative water model to sample more extended IDP conformations.
Ref. [67]: The authors showed that scaling the water-protein VdW interactions led to more extended conformations for disordered proteins and more realistic protein-protein affinities.
Ref. [68]: The authors developed a new water model, TIP4P-D, with a larger C6 coefficient on the oxygen atom. The TIP4P-D water model, combined with different protein FFs, led to better agreement with experimental IDP chain dimensions for a variety of IDPs.
Papers of special interest:
Ref. [30]: The authors presented a benchmark of protein force fields in modeling the RS peptide.
Ref. [70]: The authors compared the accuracy of protein force fields in the simulations of a model IDP, and discussed the representativeness of protein disorder models.
Ref. [85]: The authors compared different protein force field in their quality to model the self-assembly of amyloidogenic peptide fragments.
References
- 1.Csizmok V, Follis AV, Kriwacki RW, Forman-Kay JD. Dynamic Protein Interaction Networks and New Structural Paradigms in Signaling. Chemical Reviews. 2016;116:6424–6462. doi: 10.1021/acs.chemrev.5b00548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wright PE, Dyson HJ. Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol. 2015;16:18–29. doi: 10.1038/nrm3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Uversky VN, Davé V, Iakoucheva LM, Malaney P, Metallo SJ, Pathak RR, Joerger AC. Pathological Unfoldomics of Uncontrolled Chaos: Intrinsically Disordered Proteins and Human Diseases. Chemical Reviews. 2014;114:6844–6879. doi: 10.1021/cr400713r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brucale M, Schuler B, Samorì B. Single-Molecule Studies of Intrinsically Disordered Proteins. Chemical Reviews. 2014;114:3281–3317. doi: 10.1021/cr400297g. [DOI] [PubMed] [Google Scholar]
- 5.Jensen MR, Zweckstetter M, Huang, Blackledge M. Exploring Free-Energy Landscapes of Intrinsically Disordered Proteins at Atomic Resolution Using NMR Spectroscopy. Chemical Reviews. 2014;114:6632–6660. doi: 10.1021/cr400688u. [DOI] [PubMed] [Google Scholar]
- 6.Zheng W, Borgia A, Buholzer K, Grishaev A, Schuler B, Best RB. Probing the Action of Chemical Denaturant on an Intrinsically Disordered Protein by Simulation and Experiment. Journal of the American Chemical Society. 2016;138:11702–11713. doi: 10.1021/jacs.6b05443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavalli A, Camilloni C, Vendruscolo M. Molecular dynamics simulations with replica-averaged structural restraints generate structural ensembles according to the maximum entropy principle. The Journal of Chemical Physics. 2013;138:094112. doi: 10.1063/1.4793625. [DOI] [PubMed] [Google Scholar]
- 8.Roux B, Weare J. On the statistical equivalence of restrained-ensemble simulations with the maximum entropy method. The Journal of Chemical Physics. 2013;138:084107. doi: 10.1063/1.4792208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fisher CK, Huang A, Stultz CM. Modeling Intrinsically Disordered Proteins with Bayesian Statistics. Journal of the American Chemical Society. 2010;132:14919–14927. doi: 10.1021/ja105832g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hummer G, Köfinger J. Bayesian ensemble refinement by replica simulations and reweighting. The Journal of Chemical Physics. 2015;143:243150. doi: 10.1063/1.4937786. [DOI] [PubMed] [Google Scholar]
- 11.Brookes DH, Head-Gordon T. Experimental Inferential Structure Determination of Ensembles for Intrinsically Disordered Proteins. Journal of the American Chemical Society. 2016;138:4530–4538. doi: 10.1021/jacs.6b00351. [DOI] [PubMed] [Google Scholar]
- 12.Boomsma W, Ferkinghoff-Borg J, Lindorff-Larsen K. Combining Experiments and Simulations Using the Maximum Entropy Principle. PLOS Computational Biology. 2014;10:e1003406. doi: 10.1371/journal.pcbi.1003406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Löhr T, Jussupow A, Camilloni C. Metadynamic metainference: Convergence towards force field independent structural ensembles of a disordered peptide. The Journal of Chemical Physics. 2017;146:165102. doi: 10.1063/1.4981211. [DOI] [PubMed] [Google Scholar]
- 14.Shalongo W, Dugad L, Stellwagen E. Distribution of Helicity within the Model Peptide Acetyl(AAQAA)3amide. Journal of the American Chemical Society. 1994;116:8288–8293. [Google Scholar]
- 15.Fesinmeyer RM, Hudson FM, Andersen NH. Enhanced Hairpin Stability through Loop Design: The Case of the Protein G B1 Domain Hairpin. Journal of the American Chemical Society. 2004;126:7238–7243. doi: 10.1021/ja0379520. [DOI] [PubMed] [Google Scholar]
- 16.Honda S, Yamasaki K, Sawada Y, Morii H. 10 Residue Folded Peptide Designed by Segment Statistics. Structure. 2004;12:1507–1518. doi: 10.1016/j.str.2004.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Nerenberg PS, Jo B, So C, Tripathy A, Head-Gordon T. Optimizing Solute–Water van der Waals Interactions To Reproduce Solvation Free Energies. The Journal of Physical Chemistry B. 2012;116:4524–4534. doi: 10.1021/jp2118373. [DOI] [PubMed] [Google Scholar]
- 18.Yoo J, Aksimentiev A. Improved Parameterization of Amine–Carboxylate and Amine–Phosphate Interactions for Molecular Dynamics Simulations Using the CHARMM and AMBER Force Fields. Journal of Chemical Theory and Computation. 2016;12:430–443. doi: 10.1021/acs.jctc.5b00967. [DOI] [PubMed] [Google Scholar]
- 19.Miller MS, Lay WK, Li S, Hacker WC, An J, Ren J, Elcock AH. Reparametrization of Protein Force Field Nonbonded Interactions Guided by Osmotic Coefficient Measurements from Molecular Dynamics Simulations. Journal of Chemical Theory and Computation. 2017;13:1812–1826. doi: 10.1021/acs.jctc.6b01059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Boonstra S, Onck PR, van der Giessen E. CHARMM TIP3P Water Model Suppresses Peptide Folding by Solvating the Unfolded State. The Journal of Physical Chemistry B. 2016 doi: 10.1021/acs.jpcb.6b01316. [DOI] [PubMed] [Google Scholar]
- 21.Huang J, Rauscher S, Nawrocki G, Ran T, Feig M, de Groot BL, Grubmuller H, MacKerell AD., Jr CHARMM36m: an improved force field for folded and intrinsically disordered proteins. Nat Meth. 2017;14:71–73. doi: 10.1038/nmeth.4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Best RB, Hummer G. Optimized Molecular Dynamics Force Fields Applied to the Helix–Coil Transition of Polypeptides. The Journal of Physical Chemistry B. 2009;113:9004–9015. doi: 10.1021/jp901540t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Comparison of multiple Amber force fields and development of improved protein backbone parameters. Proteins. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Duan Y, Wu C, Chowdhury S, Lee MC, Xiong G, Zhang W, Yang R, Cieplak P, Luo R, Lee T, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. Journal of Computational Chemistry. 2003;24:1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 25.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79:926–926. [Google Scholar]
- 26.Abascal JLF, Vega C. A general purpose model for the condensed phases of water: TIP4P/2005. The Journal of Chemical Physics. 2005;123:234505. doi: 10.1063/1.2121687. [DOI] [PubMed] [Google Scholar]
- 27.Knott M, Best RB. A Preformed Binding Interface in the Unbound Ensemble of an Intrinsically Disordered Protein: Evidence from Molecular Simulations. PLOS Computational Biology. 2012;8:e1002605. doi: 10.1371/journal.pcbi.1002605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mittal J, Yoo TH, Georgiou G, Truskett TM. Structural Ensemble of an Intrinsically Disordered Polypeptide. The Journal of Physical Chemistry B. 2013;117:118–124. doi: 10.1021/jp308984e. [DOI] [PubMed] [Google Scholar]
- 29.Henriques J, Cragnell C, Skepö M. Molecular Dynamics Simulations of Intrinsically Disordered Proteins: Force Field Evaluation and Comparison with Experiment. Journal of Chemical Theory and Computation. 2015;11:3420–3431. doi: 10.1021/ct501178z. [DOI] [PubMed] [Google Scholar]
- 30.Rauscher S, Gapsys V, Gajda MJ, Zweckstetter M, de Groot BL, Grubmüller H. Structural Ensembles of Intrinsically Disordered Proteins Depend Strongly on Force Field: A Comparison to Experiment. Journal of Chemical Theory and Computation. 2015;11:5513–5524. doi: 10.1021/acs.jctc.5b00736. [DOI] [PubMed] [Google Scholar]
- 31.Maier JA, Martinez C, Kasavajhala K, Wickstrom L, Hauser KE, Simmerling C. ff14SB: Improving the Accuracy of Protein Side Chain and Backbone Parameters from ff99SB. Journal of Chemical Theory and Computation. 2015;11:3696–3713. doi: 10.1021/acs.jctc.5b00255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cerutti DS, Swope WC, Rice JE, Case DA. ff14ipq: A Self-Consistent Force Field for Condensed-Phase Simulations of Proteins. Journal of Chemical Theory and Computation. 2014;10:4515–4534. doi: 10.1021/ct500643c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cerutti DS, Rice JE, Swope WC, Case DA. Derivation of Fixed Partial Charges for Amino Acids Accommodating a Specific Water Model and Implicit Polarization. The Journal of Physical Chemistry B. 2013;117:2328–2338. doi: 10.1021/jp311851r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Debiec KT, Cerutti DS, Baker LR, Gronenborn AM, Case DA, Chong LT. Further along the Road Less Traveled: AMBER ff15ipq, an Original Protein Force Field Built on a Self-Consistent Physical Model. Journal of Chemical Theory and Computation. 2016;12:3926–3947. doi: 10.1021/acs.jctc.6b00567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Horn HW, Swope WC, Pitera JW, Madura JD, Dick TJ, Hura GL, Head-Gordon T. Development of an improved four-site water model for biomolecular simulations: TIP4P-Ew. The Journal of Chemical Physics. 2004;120:9665–9678. doi: 10.1063/1.1683075. [DOI] [PubMed] [Google Scholar]
- 36.Takemura K, Kitao A. Water Model Tuning for Improved Reproduction of Rotational Diffusion and NMR Spectral Density. The Journal of Physical Chemistry B. 2012;116:6279–6287. doi: 10.1021/jp301100g. [DOI] [PubMed] [Google Scholar]
- 37.Wang L-P, McKiernan KA, Gomes J, Beauchamp KA, Head-Gordon T, Rice JE, Swope WC, Martínez TJ, Pande VS. Building a More Predictive Protein Force Field: A Systematic and Reproducible Route to AMBER-FB15. The Journal of Physical Chemistry B. 2017;121:4023–4039. doi: 10.1021/acs.jpcb.7b02320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang L-P, Martinez TJ, Pande VS. Building Force Fields: An Automatic, Systematic, and Reproducible Approach. The Journal of Physical Chemistry Letters. 2014;5:1885–1891. doi: 10.1021/jz500737m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Best RB, Zhu X, Shim J, Lopes P, Mittal J, Feig M, MacKerell AD. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi1 and chi2 dihedral angles. J Chem Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.MacKerell AD, Jr, Bashford D, Dunbrack Bellott, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, et al. All-Atom Empirical Potential for Molecular Modeling and Dynamics Studies of Proteins?? J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 41.MacKerell AD, Jr, Feig M, Brooks CL. Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 42.MacKerell AD, Feig M, Brooks CL. Improved Treatment of the Protein Backbone in Empirical Force Fields. Journal of the American Chemical Society. 2003;126:698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 43.Huang J, MacKerell AD. CHARMM36 all-atom additive protein force field: Validation based on comparison to NMR data. Journal of Computational Chemistry. 2013;34:2135–2145. doi: 10.1002/jcc.23354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Best RB, Mittal J, Feig M, MacKerell AD. Inclusion of Many-Body Effects in the Additive CHARMM Protein CMAP Potential Results in Enhanced Cooperativity of alpha-Helix and beta-Hairpin Formation. Biophysical journal. 2012;103:1045–1051. doi: 10.1016/j.bpj.2012.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Feig M, Mirjalili V. Protein structure refinement via molecular-dynamics simulations: What works and what does not? Proteins: Structure, Function, and Bioinformatics. 2016;84:282–292. doi: 10.1002/prot.24871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Durell SR, Brooks BR, Ben-Naim A. Solvent-Induced Forces between Two Hydrophilic Groups. The Journal of Physical Chemistry. 1994;98:2198–2202. [Google Scholar]
- 47.Lee KH, Chen J. Optimization of the GBMV2 implicit solvent force field for accurate simulation of protein conformational equilibria. Journal of Computational Chemistry. 2017;38:1332–1341. doi: 10.1002/jcc.24734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Haberthür U, Caflisch A. FACTS: Fast analytical continuum treatment of solvation. Journal of Computational Chemistry. 2008;29:701–715. doi: 10.1002/jcc.20832. [DOI] [PubMed] [Google Scholar]
- 49.Vitalis A, Pappu RV. ABSINTH: A new continuum solvation model for simulations of polypeptides in aqueous solutions. Journal of Computational Chemistry. 2009;30:673–699. doi: 10.1002/jcc.21005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitalis A, Caflisch A. Micelle-Like Architecture of the Monomer Ensemble of Alzheimer’s Amyloid-β Peptide in Aqueous Solution and Its Implications for Aβ Aggregation. Journal of Molecular Biology. 2010;403:148–165. doi: 10.1016/j.jmb.2010.08.003. [DOI] [PubMed] [Google Scholar]
- 51.Vitalis A, Wang X, Pappu RV. Atomistic Simulations of the Effects of Polyglutamine Chain Length and Solvent Quality on Conformational Equilibria and Spontaneous Homodimerization. Journal of Molecular Biology. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Adler-Abramovich L, Vaks L, Carny O, Trudler D, Magno A, Caflisch A, Frenkel D, Gazit E. Phenylalanine assembly into toxic fibrils suggests amyloid etiology in phenylketonuria. Nat Chem Biol. 2012;8:701–706. doi: 10.1038/nchembio.1002. [DOI] [PubMed] [Google Scholar]
- 53.Robertson MJ, Tirado-Rives J, Jorgensen WL. Improved Peptide and Protein Torsional Energetics with the OPLS-AA Force Field. Journal of Chemical Theory and Computation. 2015;11:3499–3509. doi: 10.1021/acs.jctc.5b00356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Harder E, Damm W, Maple J, Wu C, Reboul M, Xiang JY, Wang L, Lupyan D, Dahlgren MK, Knight JL, et al. OPLS3: A Force Field Providing Broad Coverage of Drug-like Small Molecules and Proteins. Journal of Chemical Theory and Computation. 2016;12:281–296. doi: 10.1021/acs.jctc.5b00864. [DOI] [PubMed] [Google Scholar]
- 55.Jiang F, Zhou C-Y, Wu Y-D Residue-Specific Force Field Based on the Protein Coil Library. RSFF1: Modification of OPLS-AA/L. The Journal of Physical Chemistry B. 2014;118:6983–6998. doi: 10.1021/jp5017449. [DOI] [PubMed] [Google Scholar]
- 56.Kaminski GA, Friesner RA, Tirado-Rives J, Jorgensen WL. Evaluation and Reparametrization of the OPLS-AA Force Field for Proteins via Comparison with Accurate Quantum Chemical Calculations on Peptides†. J Phys Chem B. 2001;105:6474–6487. [Google Scholar]
- 57.Zhou C-Y, Jiang F, Wu Y-D Residue-Specific Force Field Based on Protein Coil Library. RSFF2: Modification of AMBER ff99SB. The Journal of Physical Chemistry B. 2015;119:1035–1047. doi: 10.1021/jp5064676. [DOI] [PubMed] [Google Scholar]
- 58.Wang W, Ye W, Jiang C, Luo R, Chen H-F. New Force Field on Modeling Intrinsically Disordered Proteins. Chemical Biology & Drug Design. 2014;84:253–269. doi: 10.1111/cbdd.12314. [DOI] [PubMed] [Google Scholar]
- 59.Ye W, Ji D, Wang W, Luo R, Chen H-F. Test and Evaluation of ff99IDPs Force Field for Intrinsically Disordered Proteins. Journal of Chemical Information and Modeling. 2015;55:1021–1029. doi: 10.1021/acs.jcim.5b00043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Song D, Luo R, Chen H-F. The IDP-Specific Force Field ff14IDPSFF Improves the Conformer Sampling of Intrinsically Disordered Proteins. Journal of Chemical Information and Modeling. 2017;57:1166–1178. doi: 10.1021/acs.jcim.7b00135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Shi Y, Xia Z, Zhang J, Best R, Wu C, Ponder JW, Ren P. Polarizable Atomic Multipole-Based AMOEBA Force Field for Proteins. Journal of Chemical Theory and Computation. 2013;9:4046–4063. doi: 10.1021/ct4003702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lopes PEM, Huang J, Shim J, Luo Y, Li H, Roux B, MacKerell AD. Polarizable Force Field for Peptides and Proteins Based on the Classical Drude Oscillator. Journal of Chemical Theory and Computation. 2013;9:5430–5449. doi: 10.1021/ct400781b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Huang J, Simmonett AC, Pickard FC, MacKerell AD, Brooks BR. Mapping the Drude polarizable force field onto a multipole and induced dipole model. The Journal of Chemical Physics. 2017;147:161702. doi: 10.1063/1.4984113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huang J, MacKerell Alexander D., Jr Induction of Peptide Bond Dipoles Drives Cooperative Helix Formation in the (AAQAA)3 Peptide. Biophysical Journal. 2014;107:991–997. doi: 10.1016/j.bpj.2014.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lemkul JA, Huang J, MacKerell AD. Induced Dipole–Dipole Interactions Influence the Unfolding Pathways of Wild-Type and Mutant Amyloid β-Peptides. The Journal of Physical Chemistry B. 2015;119:15574–15582. doi: 10.1021/acs.jpcb.5b09978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lemkul JA, Huang J, Roux B, MacKerell AD. An Empirical Polarizable Force Field Based on the Classical Drude Oscillator Model: Development History and Recent Applications. Chemical Reviews. 2016;116:4983–5013. doi: 10.1021/acs.chemrev.5b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Best RB, Zheng W, Mittal J. Balanced Protein–Water Interactions Improve Properties of Disordered Proteins and Non-Specific Protein Association. Journal of Chemical Theory and Computation. 2014;10:5113–5124. doi: 10.1021/ct500569b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Piana S, Donchev AG, Robustelli P, Shaw DE. Water Dispersion Interactions Strongly Influence Simulated Structural Properties of Disordered Protein States. The Journal of Physical Chemistry B. 2015;119:5113–5123. doi: 10.1021/jp508971m. [DOI] [PubMed] [Google Scholar]
- 69.Best Robert B, Hofmann H, Nettels D, Schuler B. Quantitative Interpretation of FRET Experiments via Molecular Simulation: Force Field and Validation. Biophysical Journal. 2015;108:2721–2731. doi: 10.1016/j.bpj.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Henriques J, Skepö M. Molecular Dynamics Simulations of Intrinsically Disordered Proteins: On the Accuracy of the TIP4P-D Water Model and the Representativeness of Protein Disorder Models. Journal of Chemical Theory and Computation. 2016;12:3407–3415. doi: 10.1021/acs.jctc.6b00429. [DOI] [PubMed] [Google Scholar]
- 71.Piana S, Lindorff-Larsen K, Shaw DE. How robust are protein folding simulations with respect to force field parameterization? Biophys J. 2011;100:L47-49–L47-49. doi: 10.1016/j.bpj.2011.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Fluitt Aaron M, de Pablo Juan J. An Analysis of Biomolecular Force Fields for Simulations of Polyglutamine in Solution. Biophysical Journal. 2015;109:1009–1018. doi: 10.1016/j.bpj.2015.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Walters RH, Murphy RM. Examining Polyglutamine Peptide Length: A Connection between Collapsed Conformations and Increased Aggregation. Journal of Molecular Biology. 2009;393:978–992. doi: 10.1016/j.jmb.2009.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Carballo-Pacheco M, Strodel B. Comparison of force fields for Alzheimer’s A β42: A case study for intrinsically disordered proteins. Protein Science. 2017;26:174–185. doi: 10.1002/pro.3064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Man VH, Nguyen PH, Derreumaux P. High-Resolution Structures of the Amyloid-β 1–42 Dimers from the Comparison of Four Atomistic Force Fields. The Journal of Physical Chemistry B. 2017;121:5977–5987. doi: 10.1021/acs.jpcb.7b04689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hoffmann KQ, McGovern M, Chiu C-c, de Pablo JJ. Secondary Structure of Rat and Human Amylin across Force Fields. PLOS ONE. 2015;10:e0134091. doi: 10.1371/journal.pone.0134091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bah A, Vernon RM, Siddiqui Z, Krzeminski M, Muhandiram R, Zhao C, Sonenberg N, Kay LE, Forman-Kay JD. Folding of an intrinsically disordered protein by phosphorylation as a regulatory switch. Nature. 2015;519:106–109. doi: 10.1038/nature13999. [DOI] [PubMed] [Google Scholar]
- 78.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Guvench O, Hatcher E, Venable RM, Pastor RW, MacKerell AD. CHARMM Additive All-Atom Force Field for Glycosidic Linkages between Hexopyranoses. Journal of Chemical Theory and Computation. 2009;5:2353–2370. doi: 10.1021/ct900242e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Guvench O, Mallajosyula SS, Raman EP, Hatcher E, Vanommeslaeghe K, Foster TJ, Jamison FW, MacKerell AD. CHARMM Additive All-Atom Force Field for Carbohydrate Derivatives and Its Utility in Polysaccharide and Carbohydrate–Protein Modeling. Journal of Chemical Theory and Computation. 2011;7:3162–3180. doi: 10.1021/ct200328p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Khoury GA, Thompson JP, Smadbeck J, Kieslich CA, Floudas CA. Forcefield_PTM: Ab Initio Charge and AMBER Forcefield Parameters for Frequently Occurring Post-Translational Modifications. Journal of Chemical Theory and Computation. 2013;9:5653–5674. doi: 10.1021/ct400556v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zerze GH, Mittal J. Effect of O-Linked Glycosylation on the Equilibrium Structural Ensemble of Intrinsically Disordered Polypeptides. The Journal of Physical Chemistry B. 2015;119:15583–15592. doi: 10.1021/acs.jpcb.5b10022. [DOI] [PubMed] [Google Scholar]
- 83.Martin EW, Holehouse AS, Grace CR, Hughes A, Pappu RV, Mittag T. Sequence Determinants of the Conformational Properties of an Intrinsically Disordered Protein Prior to and upon Multisite Phosphorylation. Journal of the American Chemical Society. 2016;138:15323–15335. doi: 10.1021/jacs.6b10272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ithuralde RE, Roitberg AE, Turjanski AG. Structured and Unstructured Binding of an Intrinsically Disordered Protein as Revealed by Atomistic Simulations. Journal of the American Chemical Society. 2016;138:8742–8751. doi: 10.1021/jacs.6b02016. [DOI] [PubMed] [Google Scholar]
- 85.Matthes D, Gapsys V, Brennecke JT, de Groot BL. An Atomistic View of Amyloidogenic Self-assembly: Structure and Dynamics of Heterogeneous Conformational States in the Pre-nucleation Phase. Scientific Reports. 2016;6:33156. doi: 10.1038/srep33156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Best RB. Computational and theoretical advances in studies of intrinsically disordered proteins. Current Opinion in Structural Biology. 2017;42:147–154. doi: 10.1016/j.sbi.2017.01.006. [DOI] [PubMed] [Google Scholar]
- 87.Levine ZA, Shea J-E. Simulations of disordered proteins and systems with conformational heterogeneity. Current Opinion in Structural Biology. 2017;43:95–103. doi: 10.1016/j.sbi.2016.11.006. [DOI] [PubMed] [Google Scholar]