Abstract
Phospholipase D (PLD) plays a key role in both cell membrane lipid reorganization and architecture, as well as a cell signaling protein via the product of its enzymatic reaction, phosphatidic acid (PA). PLD is involved in promoting breast cancer cell growth, proliferation, and metastasis and both gene and protein expression are upregulated in breast carcinoma human samples. In spite of all this, the ultimate reason as to why PLD expression is high in cancer cells vs. their normal counterparts remains largely unknown. Until we understand this and the associated signaling pathways, it will be difficult to establish PLD as a bona fide target to explore new potential cancer therapeutic approaches. Recently, our lab has identified several molecular mechanisms by which PLD expression is high in breast cancer cells. First, PA, a mitogen, functions as a protein and mRNA stabilizer that counteracts natural decay and degradation. Second, there is a repertoire of microRNAs (miRs) that keep PLD mRNA translation at low levels in normal cells, but their effects change with starvation and during endothelial-to-mesenchymal transition (EMT) in cancer cells. Third, there is a novel way of post-transcriptional regulation of PLD involving 3'-exonucleases, specifically the deadenylase, Poly(A)-specific Ribonuclease (PARN) which tags the mRNA for degradation. This enables PLD accumulation and ultimately breast cancer cell growth, proliferation, and metastasis. We review in depth the emerging field of post-transcriptional regulation of PLD, which is only recently beginning to be understood. Since, surprisingly, so little is known about post-transcriptional regulation of PLD and related phospholipases (PLC or PLA), this new knowledge could help our understanding of how post-transcriptional deregulation of a lipid enzyme expression impacts tumor growth.
Keywords: Signal transduction, phospholipases, post-transcriptional control, RNA
Introduction: Post-transcriptional regulation of messenger RNA
Transcriptional control is the predominant form of regulation for most genes [1]. However, after RNA polymerase has bound to the gene's promoter and RNA synthesis has started, post-transcriptional regulation can still control the amount of gene that is ultimately expressed and for many genes, post-transcriptional control is essential. Post-transcriptional control of gene expression is important for cellular functions across biological contexts. It comprises a complex regulatory network that contributes to cell-type and organism specific gene expression patterns.
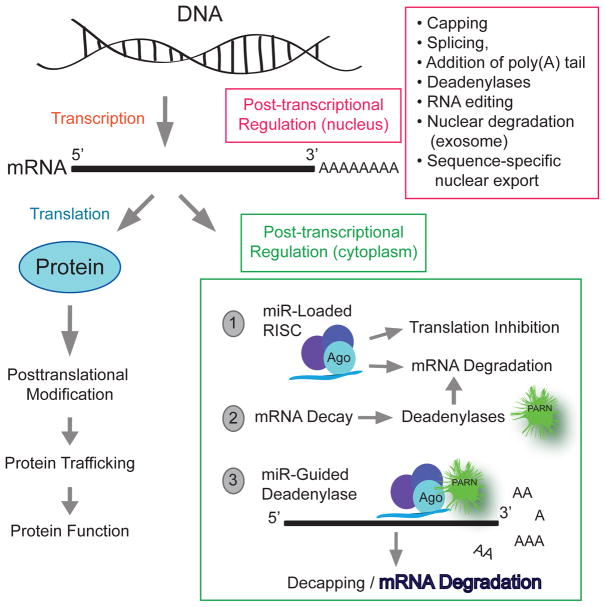
Pre-mRNA synthesized in the nucleus undergoes a series of modifications that include capping, splicing, addition of poly(A) tail, RNA editing, nuclear degradation (exosome), sequence-specific nuclear export mRNA, all of which occur in the cell nucleus. More post-transcriptional control mechanisms occur in the cytoplasm, such as stability and lifetime in the cytosol and small regulatory RNAs, specifically microRNA (miRs),that can be considered translation regulators (Figure 1).
Figure 1. Post-transcriptional control of gene expression.
It comprises a complex regulatory network that contributes to cell-type and organism specific gene expression patterns. The pre-mRNA has to go through some modifications to become a mature mRNA molecule that can leave the nucleus and be translated. Those modifications include capping, splicing, addition of poly(A) tail, RNA editing, nuclear degradation (exosome), sequence-specific nuclear export mRNA, stability and lifetime in the cytosol and small regulatory RNAs, specifically microRNA (miRs). This review will focus on the three events (labeled “1”, “2” and “3”) occurring in the cytoplasm.
Microarray analysis has indicated that close to 50% of the changes in inducible gene expression occur at the level of mRNA stability [2] highlighting the exquisite level of post-transcriptional regulation. All of these post-transcriptional regulations that occur in both the nucleus and the cytoplasm determine the level of gene expression and how much of the transcripts are ultimately translated into proteins [3]. For the purpose of this review we will concentrate on the mRNA stability and miR post-transcriptional events.
PLD as an example to apply new post-transcriptional control mechanisms
Extensive studies exist on the enzymatic regulation of phospholipid phospholipases A, C and D. However, little is known about their transcriptional and especially post-transcriptional regulation. This is surprising considering the central role these phospholipases play in lipid metabolism and cell signaling. The discrete number of articles found in the scientific literature, some of which are cited below, offer a glimpse of how valuable post-transcriptional control could be for lipid enzyme signaling. For example, regarding PLA2, insulin-like growth factor-I (IGF-I) destabilizes mRNA of the type II sPLA2. Conversely, IL-1beta stimulates the transcription rate and gives rise to a very stable mRNA [4]. PLA, regulation of lipolytic activities by PLA2 depends on the transcriptional regulators LetA/S and RpoS, inducing the expression of virulence traits, and on post-transcriptional activators like the zinc metalloprotease ProA [5]. Inhibition of endogenous miR-338 with anti-miR-338 increased the mRNA and protein expression of PLA2G4B in decidual cells with a proposed role in human pregnancy and parturition [6].
Downstream phospholipid catabolism by PLA2 produces arachidonic acid that can be used in prostaglandin synthesis by PGEs. A group of selected miRs regulate mRNA expression M-type phospholipase A2 receptor (PLA2R1) in normal human mammary epithelial cells and cancer cell lines [7]. There are several RNA sequence elements within the 3'UTRs of the genes involved in the PGE(2) pathway, that are predicted to be binding sites for miRNAs and RNA-binding proteins, both of which appear to be central regulators of PGE(2) synthesis and function [8]. Regarding another phospholipase, PLC, a particular group of miRs (miR-200b, miR-200c, and miR-429) target PLC (PLCG1) during regulation of PG function [8]. Inhibition of miR-214 in C2C12 cells enhances protein expression of PLCβ1 and promotes C2C12 BMP-2-induced osteogenesis through PLCβ1 [9]. Thus, numerous microRNAs have recently emerged as post-transcriptional gene repressors for phospholipases (like PLA2 and PLCβ) or phosphatases. However, very little is known about deadenylases post-transcriptionally regulating phospholipases.
PLD background
The conversion of PC to PA is catalyzed by the enzyme PLD [10] and is, in general, dependent on the presence of the co-factor phosphatidylinositol 4,5-bisphosphate (PIP2) (an anionic lipid localized primarily to the plasma membrane) [11, 12]. PLD has been found in a variety of cells and tissues. Its activity has been reported predominantly in the plasma membrane, as well as in cytoplasmic locations, the mitochondrial membrane, the Golgi endoplasmic reticulum (ER), the nucleus, the nuclear membrane and subcellular compartments [13, 14]. There is also an interplay between PLD and Diacylglycerol Kinase (DGK). PLD generates phosphatidic acid by catalyzing the hydrolysis of phosphatidylcholine (PC), which could be de-phosphorylated to generate DAG. Inversely, DGK catalyzes the phosphorylation of DAG to synthesize PA. Thus, both enzymes regulate the levels of DAG and PA, increasing the later and decreasing the former. This is important for intracellular vesicle trafficking cycling as both PA and DAG are both needed for exocytosis [15]. Many of these functions are negated or diminished during PLD loss of function [16, 17]. In elegant studies, Ryu’s group [17] and Frohman’s group [18] have provided direct evidence of the function of PLD2 in several pathological conditions such as cancer, vascular disease, immunological disease, and neurological disease.
Little is known about PLD post-transcriptional regulation. However, recent publications from our lab are indicating a previously unsuspected level of post-transcriptional regulation of PLD mRNA that can affect how much and when PLD is expressed. As such we will discuss post-transcriptional regulation of PLD as a new paradigm in this review as a novel and interesting alternative perspective on PLD. Understanding control of PLD expression is important because PLD has been documented as having a direct involvement in promoting breast cancer cell growth, proliferation, and metastasis [19–21].
The three post-transcriptional control mechanisms considered in this review
We posit that normally operating mechanisms in non-cancerous cells keeping PLD at a desired level are deregulated in cancer cells. Thus, we will concentrate on the understudied field of PLD post-transcriptional regulation and will review the following three mechanisms: (a) miRs, (b) deadenylases, and (c) a combination of the two (Figure 1).
In (a), microRNAs are a well-studied means of post-transcriptional regulation of protein levels within the cell. The association of miR-loaded RISC on targeted mRNA functions to inhibit translation of the mRNA by either inhibiting ribosomal function or by inducing the degradation of the mRNA [22].
For the second mechanism of post-transcriptional control considered here (b), the involvement of deadenylases, specifically, 3' exonucleatic cleavage of mRNA poly-A tails by deadenylases, we will concentrate on PARN as it was downregulated in patient invasive breast carcinoma samples compared to adjacent normal control tissue and at the same time Phospholipase D (PLD) was upregulated in these same breast carcinoma samples.
For the third mechanism (c), mRNA deadenylation is under tight control of cis-acting regulatory elements, both of which are located in the 3’ UTR of eukaryotic mRNAs. In a unique and still somewhat controversial mechanism, miRs promote deadenylase-induced mRNA decay.
PLD in cancer
The mammalian phospholipase D (PLD) family members are important signaling molecules that hydrolyze membrane lipids [18, 24, 25]. The classical PLD isoforms (PLD1 and PLD2) are ubiquitously expressed and are the best characterized. PLD1 localizes predominately to cytoplasmic membranes while PLD2 localizes to the plasma membrane [18, 24, 25]. Their enzymatic function is to hydrolyze phosphatidylcholine (PC) to free choline and phosphatidic acid (PA) [18, 24, 25]. PA is a critical secondary messenger signal within the cell, regulating pathways leading to cell growth and proliferation, vesicle trafficking, and cell migration [18, 24, 25].
Several authors have reported an increase in PLD gene expression, protein expression, and enzymatic activity in multiple cancer types including breast [19, 26–30], gastric [31, 32], colorectal [33], renal [34], thyroid [35], and brain [36]. Regarding the enzymatic activity, the product PA is highly mitogenic and has been shown to be involved in regulation of tumorigenesis, cell proliferation, cell invasion, and cell movement including metastasis [37–39]. PA mediates induction of HIF-1α [40]. Regarding expression of PLD protein, it has been shown that PLD1 is elevated in breast cancer xenotransplants and Frohman’s group studied PLD1 in the tumor microenvironment (TME) and angiogenesis [28]. Our laboratory has recently shown [26, 27, 41] that increased PLD1 and PLD2 protein expression in breast cancer cells correlates with increases in PLD lipase activity [27]. Fite et al. demonstrated that several miRNAs regulate PLD2 in non-cancerous human breast cells, but upon epithelial-mesenchymal transition (EMT), these miRNA were downregulated, allowing for the increase in PLD2 protein mass observed in an invasive breast cancer cell line [27].
It is to be expected that keeping mRNA transcripts for PLD at manageable levels is desirable in normal cells, but in cancer, PLD1 and PLD2 mRNA transcripts are maintained at more elevated levels and subsequently translated into protein more efficiently.
Regulating protein expression post-transcriptionally by non-coding RNAs
Between 70–90% of the human genome is transcribed into RNA but only approximately 2% of the genome encodes protein [42], meaning the majority of RNA are non-coding RNA molecules. Non-coding RNAs are classified as either long (lncRNAs) (greater than 200 nucleotides) or short (less than 200 nucleotides) [43]. Types of long non-coding RNAs include: sense lncRNAs, antisense lncRNAs, bidirectional lncRNAs, intronic lncRNAs, and long intergenic ncRNAs (lincRNAs). Sense and antisense lncRNAs overlap exons of a protein-coding gene while bidirectional lncRNAs are transcribed opposite to another transcript and intronic lncRNAs are transcribed from within introns. LincRNA transcripts derive from regions between two protein-coding genes [44]. Long non-coding RNAs function as competitor RNA, or “sponges”, for miRNA targeting or function as mediators of epigenetic silencing [44]. Their expression is cell and tissue specific and increasing evidence supports lncRNA involvement in cancer progression [43, 45, 46]. The other category, small non-coding RNAs, include tRNAs and rRNAs, in addition to miRNAs, siRNAs, snoRNAs, snRNAs, and piRNAs [44]. MicroRNAs (miRNAs; miRs) and endogenous siRNA are means of posttranscriptional regulation within the cell and function to inhibit protein translation of target mRNA. Small nucleolar RNAs (snoRNA) are involved in post-transcriptional regulation of rRNA [47]. Small nuclear RNA (snRNA) associate with proteins of the spliceosome and therefore function in intron removal in mRNA processing [48]. PIWI-interacting RNA (piRNA) serve to silence transposable elements thereby maintaining genome integrity in germ cells [49]. We focus herein on discussing miRNA and cancer (Figure 2).
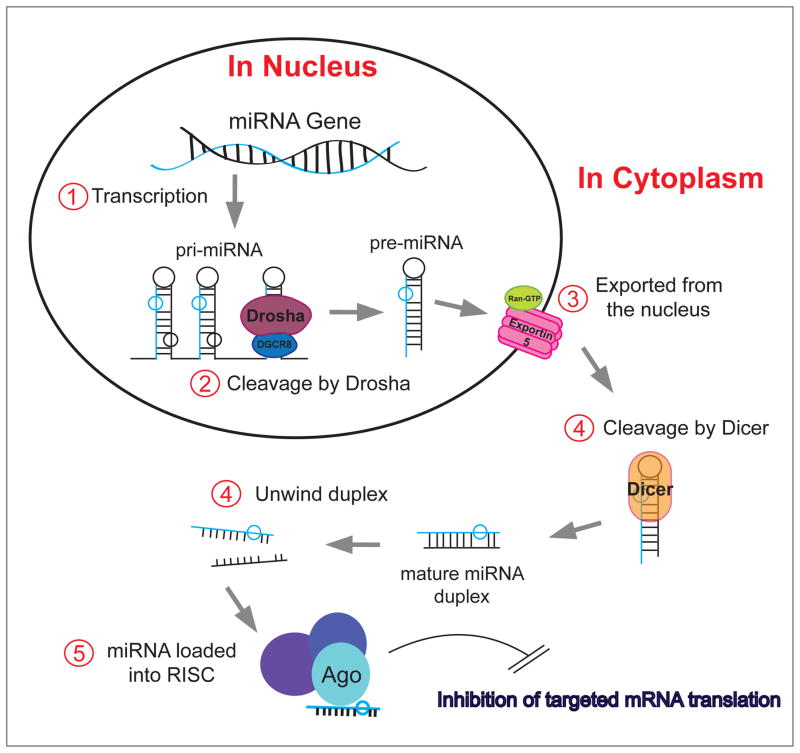
Figure 2. MicroRNA (miRNA, miR) and cancer.
miRs are short (~22 nucleotide) single-stranded RNA molecules. After transcription (1), the miR precursors undergo several processing steps in the nucleus and cytoplasm. In the nucleus a key step is their cleavage by endonucleases, such as Drosha (2) and the resulting pre-miR is exported from the nucleus (3). In the cytoplasm, Dicer (4) produces a mature miR duplex that, after unwinding (4), becomes associated with Ago2 in the RISC (RNA-induced silencing complex) (5). Once the miR is loaded onto the RISC, the entire complex targets mRNA based on sequence complementarity with the seed sequence (first 8 amino acids) of the miR. The association of miR-loaded RISC on mRNA functions to inhibit translation of the mRNA by either inhibiting ribosomal function or by inducing the degradation of the mRNA. This process is deregulated in several pathologies including cancer.
miRs in cancer
MicroRNA (miRNA, miR) are short (~22 nucleotide) single-stranded RNA molecules [50–52]. After transcription, the miR precursors undergo several processing steps in the nucleus and cytoplasm, in which they are cleaved by endonucleases, Drosha and Dicer. Cleavage by Dicer produces a mature miR that becomes associated with Ago2 in the RISC (RNA-induced silencing complex) [53–56]. Once the miR is loaded onto the RISC, the entire complex targets mRNA based on sequence complementarity with the seed sequence (first 8 amino acids) of the miR. The association of miR-loaded RISC on mRNA functions to inhibit translation of the mRNA by either inhibiting ribosomal function or by inducing the degradation of the mRNA [22]. MicroRNA are one of the main factors in post-transcriptional regulation to regulate protein levels within the cell. It is predicted that miRs regulate the majority of the human transcriptome and are therefore involved in virtually every signaling pathway. Furthermore, deregulation of miRs contributes to numerous pathologies, including cancer [22, 57].
Many investigators are pursuing the potential of microRNAs as biomarkers in various pathologies including cancer [58–61]. MicroRNAs make attractive candidates for their stability in body fluid samples such as blood and urine, which are less invasive and less painful diagnostic samples than tissue biopsies. Additionally, microRNAs are being investigated as biomarkers for response to therapy [62, 63]. Several miRNA-based interventions are currently in clinical trials. The first was Miravirsen, which is currently in Phase II clinical trial. This drug targets the liver-specific miR-122 in the context of Hepatitis C. Hepatitis C virus uses the liver miR-122 in stabilizing its genome and downregulation of miR-122 would reduce viral production [64, 65]. MiR-122 was an ideal target because it is only expressed at meaningful levels in the liver and targeting it would therefore not have abundant off-target effects in many other cell types [65, 66]. MRX34 was a miR-34 mimic intended to treat melanoma patients. However, MRX34 was pulled from Phase I clinical trial in 2016 due to severe adverse effects [67]. The delivery system of MRX34 was intravenous injection of miR-34-containing liposomes. This study highlights the challenge in designing miRNA-based interventions. Since one miRNA can have numerous targets and the targets can vary between cell types, administering a systemic miRNA-based intervention could impact many tissues.
Another recent trial took a targeted approach with TargomiRs and a Phase I clinical trial was completed in early 2017. These specific TargomiRs are miR-16 mimics designed for lung cancers including mesothelioma and non-small cell lung cancer (NSCLC) [68]. The miR mimics are packaged into nanoparticles with anti-EGFR expressed on the nanoparticle surface with the intention to target the nanoparticles to EGFR-expressing cells. Both mesothelioma and NSCLC are known for their high expression of EGFR and as such, if successful, the anti-EGFR coated nanoparticles should specifically target these cancer cells and release the TargomiRs selectively.
miRs, PLD: an interlocked positive and negative feedback mechanism
Two recent studies in our lab serve to clear the way to understand how miRs regulate PLD expression. In the first, our laboratory identified a set of microRNAs predicted to have “high score” binding abilities to PLD 3’UTRs [27]. Matrigel-based cell invasion assays and breast cancer cell lines showed cell invasion was reduced in the presence of these miRs due to a decrease in PLD protein. Additionally, expression of these miRs likely decrease as a result of the epithelial-mesenchymal transition (EMT). EMT induces genetic and epigenetic changes in the cell allowing cancer cells to adopt a more mesenchymal cell phenotype with increased ability to migrate. E-cadherin triggers expression of miRs in pre-EMT breast cancer cells, thereby keeping PLD levels low [27]. Exogenous addition of these miRs negatively affects PLD protein levels in the post-EMT MDA-MB-231 cells leading to a reduction in cell invasion. The invasive properties of MDA-MB-231 cells through Matrigel matrix were increased with PLD overexpression and then negated following miR overexpression.
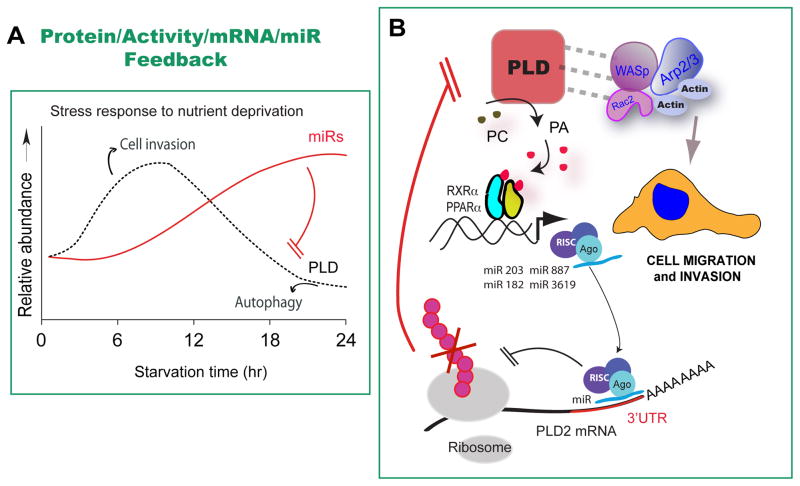
In a second study from our lab [26], we asked the question of what happens to PLD and miR expression in tumor-like conditions including nutrient starvation, hypoxia, or culture cell density? PLD expression is upregulated in nutrient-starved cells. Expression of certain microRNA increases after prolonged (> 12 hours) starvation. In fact, the earlier identified repertoire of microRNAs directly target and regulate PLD itself during cancer cell starvation. In conditions of cellular stress, such as hypoxia and starvation, cancer cells increase PLD expression and activity as well as cell invasion. Prolonged starvation of cultured cells reverses that phenotype. The mechanism for this biphasic process is: Initially, PLD mediates cell invasion. PLD associates with motility proteins and PA promotes the formation of “positive” membrane curvature needed for the formation of lamellipodia and motility cellular structures in the cell membrane. With prolonged starvation, autophagy and survival become paramount to cell migration. As PLD rises, increased PA levels promote transcription of several miR genes. Although no specific transcription factor has been demonstrated, Mahankali et al [69] have demonstrated that PPAR binds to PA and regulates the expression of both PLD and EGFR. Several miRs target the 3’UTR of PLD transcripts downregulating PLD translation, so the PLD-led process comes to an end in prolonged starvation. In other words, PLD protein, activity, mRNA, and miR operate in a feedback loop in cancer cells (Figure 3).
Figure 3. PLD protein, activity, mRNA, and miR operate in a feedback loop in cancer cells.
(A) There is a causal relation between certain miRs and PLD expression. (B) During starvation, PLD (a “stress protein”) rises and mediates cell invasion. This could help cells at the core of a primary solid tumor escape hypoxia and low nutrient conditions. However, with prolonged starvation, PA levels (possibly to association to trasncription factors of the PPAR family) promote expression of several miR genes, which un turn target the 3’UTR of PLD transcripts, consequently downregulating PLD translation. In this way, the PLD-led process comes to an end in prolonged starvation.
Another way of regulating protein expression post-transcriptionally: mRNA decay regulated by deadenylases
For mRNA decay, the rate-limiting step is the shortening of mRNA poly(A) tails. This is accomplished through the action of a group of 3’ to 5’ exonucleases and is one of the most potent methods for preventing mRNA translation and induce mRNA transcript turnover [70–73]. The 3’ to 5’ exonucleases involved in this poly(A) tail shortening are known as deadenylases, of which there are many. The two major eukaryotic deadenylase complexes are known as Ccr4-Not and Pan2/Pan3 [73, 74]. A third deadenylase, poly(A)-specific ribonuclease (PARN), has also been identified [75–77].
Deadenylases (Ccr4-Not, Pan2/Pan3, and PARN)
Ccr4-Not
The Ccr4-Not complex consists of 9 components and the human orthologs are termed CNOT1-CNOT10 [78, 79]. While many studies have been in the yeast system, interest is growing in understanding the role of the human CNOT complex in physiology and pathology. The Ccr4-Not yeast complex regulates transcription by associating with transcription factors, however, this mechanism of transcriptional control is not well understood [74]. The better characterized mechanism of the complex is its deadenylation of mRNA [74], which is conserved in humans [80]. In yeast, Ccr4 contains the deadenylase activity and is a global regulator of mRNA decay in yeast. Notably, it regulates the deadenylation of mRNA encoding for ribosomal proteins and is therefore important in the control of ribosomal protein levels within the cell [81, 82].
Pan2/Pan3
Pan2/Pan3 and Ccr4-Not complexes are responsible for the majority of mRNA deadenylation activity in eukaryotes [73, 83]. Pan stands for Poly(A) Nuclease and, similar to Ccr4-Not, is most highly studied in yeast [73, 84]. The Pan complex has 3’ to 5’ exonuclease activity and can only degrade the 3’ poly(A) tail with no exonuclease activity on the rest of the mRNA [73]. Unlike the Ccr4-Not complex, the Pan complex consists of only two proteins, Pan2 and Pan3 [73], with Pan2 having exonuclease activity [85, 86]. Pan2/Pan3 predominately localizes to P-bodies in the cytoplasm and it remains unclear if the complex functions in the nucleus [73, 87].
PARN
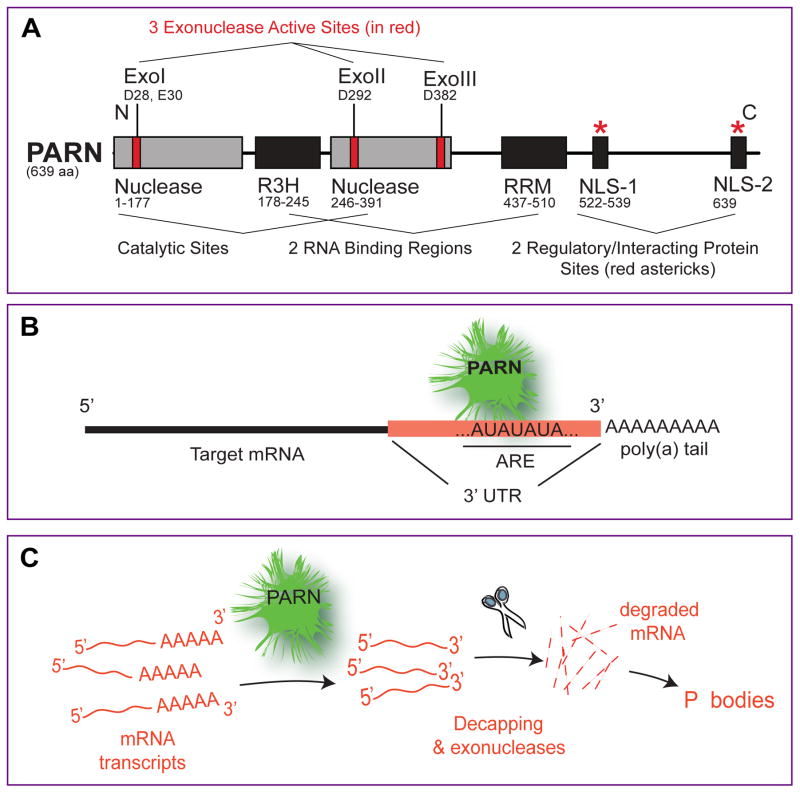
PARN is a Mg2+ ion-dependent, poly(A)-specific 3’ to 5’ exonuclease [88]. PARN protein contains a large nuclease domain as well as two distinct RNA-binding domains. PARN has 5’ GTP cap-binding capabilities due to the RNA recognition motif (RRM) RNA-binding domain. This enhances PARN processivity. A second RNA binding domain, the R3H, is essential for PARN dimer stability [89]. Importantly, PARN shows a preference for the targeting of mRNAs containing AU-rich elements in their 3’ untranslated regions (UTRs) [90–93] (Figure 4).
Figure 4. The 3’-mRNA exonuclease PARN.
(A) Modular scheme of the PARN protein. The PARN protein consists of three distinct exonuclease (deadenylase) sites (Exo I, Exo II, and Exo III) contained in two nuclease regions also with two RNA binding regions (R3H and RRM). (B) PARN targets mRNA 3' UTR containing long stretches of AU combinations called, AU- rich elements (AREs). Such elements are present in the 3' UTR of pLD1 mRNA. (C) PARN contributes to mRNA degradation through the cleavage of mRNA poly(A) tails (deadenylase function) followed by decapping and by exonuclease activity; cleaved mRNAs accumulate in “P bodies” in the cell that can be observed under the microscope.
PARN has been shown to participate in non-sense mediated decay and also in the process of human telomerase RNA component (TERC) maturation. In the nucleus, nuclear cap binding protein complex (CBC) inhibits PARN deadenylation activity, which potentially interferes with PARN's function in nonsense-mediated decay and TERC maturation [94]. PARN counteracts this oligoadenylation by cleaving off the oligo-A tails on the TERC, allowing it to serve its crucial role in telomerase [95–97]. In the cytoplasm, the integration of mRNA decay and translation initiation has been proposed where PARN plays an important role [98].
PARN deficiencies have been shown to be a cause in the development of a severe form of dyskeratosis congenita, a telomere disease, due to the progressive shortening of telomeres [96, 99–102]. Previous studies have also begun to characterize PARN’s mRNA targets. PARN can target transcripts involved in cell migration, adhesion, p53 signaling, BRCA1 DNA damage response, and oncogenes such as c-myc, c-fos, and c-jun to keep these transcripts levels controlled under normal conditions [103–110].
Deadenylases in PLD-associated breast cancer
After PARN has removed the poly-A tail the remaining mRNA is “marked” for further or total degradation. PARN preferentially targets mRNA 3' UTR containing long stretches of AU combinations called, AU- rich elements (AREs). As described herein earlier, PLD is a cell-signaling molecule well known for promoting breast cancer cell growth, proliferation, and metastasis, and was found to be upregulated in human breast carcinoma samples compared to the normal adjacent breast tissues.
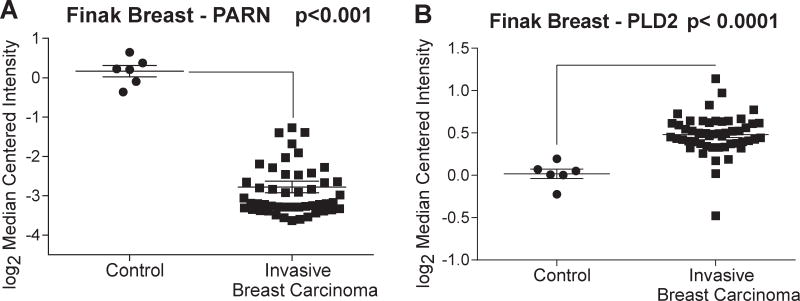
It is known that PARN regulates the transcript levels of several proteins involved in cancer development and progression, can be activated by the tumor-suppressor BARD1, and when overexpressed in SCC patients, they survive 7.0 months longer than in patients that underexpress it [103]. Interestingly, PARN expression levels in human breast carcinoma samples were decreased compared to the normal control tissues, the opposite results of PLD (Fig. 5), and would be an interesting area of further study.
Figure 5. An inverse relation exists between PARN and PLD expression in breast cancer.
Microarray data from the Finak Breast dataset were downloaded from the Oncomine database [115]. The data compares PARN levels of deadenylase (PARN) (A) and PLD2 (B).
miR and deanylation regulation of protein translation
While still somewhat controversial in the field, there is evidence that all three major cellular deadenylases, Ccr4-Not, Pan2/Pan3, and PARN can exert their function through miR-dependent mechanisms. Recently, PARN has been identified as capable of using a similar miR-dependent mechanism. PARN protein was shown physically bound to Ago2 in the RNA-Induced Silencing Complex (RISC). Once the RISC was loaded with miR-504 or miR-125b, the RISC, along with PARN, were brought to the TP53 mRNA 3’ UTR for which these miRs targeted. This resulted in subsequent TP53 transcript degradation under non-stress conditions. Upon UV-induced DNA damage, this degradation of TP53 mRNA was abolished, allowing TP53 translation into functional p53 protein to then exert DNA damage response [23, 106].
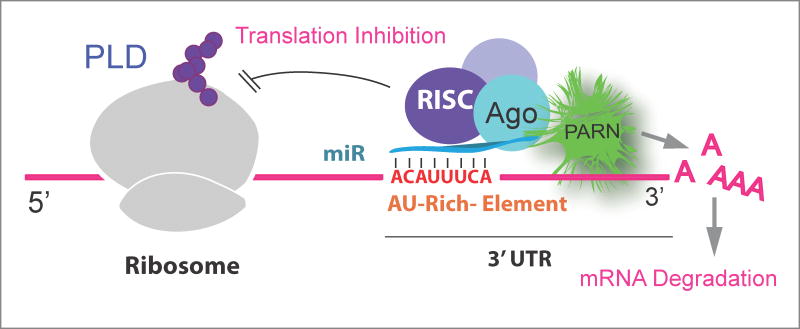
It is also possible that a combination of PARN deadenylase activity and a putative presence of a miR that would synergize in destabilizing PLD transcripts impeding protein translation (Fig. 6).
Figure 6. Hypothetical post-transcriptional regulation of PLD.
This is a hypothetical model showing a combination of PARN deadenylase activity and a putative presence of a miR that would synergize in destabilizing PLD transcripts hampering translation.
Future directions
In the study of PLD post-transcriptional regulation [26, 27, 41] new questions that are arising are: if PARN can regulate PLD, what controls this process and what role does PARN have in the patho-physiology and development of breast cancer? It has been shown that phosphorylation is important in PARN regulation. Under UV to induce DNA damage, PARN is phosphorylated at S557 by MK2 and no longer binds and regulates Gadd45α mRNA [111]. Under serum starvation conditions, PARN can be phosphorylated which increases its 5’ GTP cap binding affinity to out compete eIF4E for access to the cap [112]. Poly(A) binding proteins (PABPs) can inhibit PARN action by binding RNA and excluding PARN from its target [113]. Cap-binding proteins (CBPs) can also repress PARN [94, 113]. CUG-binding protein anchors PARN to mRNA targets thus increasing PARN processing [110, 114]. The role PARN has in the patho-physiology and development of breast cancer also needs further investigation.
Acknowledgments
The following grants to Dr. Cambronero (J.G-C.) have supported this work: HL056653-14 from the National Institutes of Health (NIH) and 13GRNT17230097 from the American Heart Association.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Alberts B. Molecular biology of the cell. 4. Garland Science; New York: 2002. [Google Scholar]
- 2.Weng JK, Tanurdzic M, Chapple C. Functional analysis and comparative genomics of expressed sequence tags from the lycophyte Selaginella moellendorffii. BMC genomics. 2005;6:85. doi: 10.1186/1471-2164-6-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Garneau NL, Wilusz J, Wilusz CJ. The highways and byways of mRNA decay. Nature reviews. Molecular cell biology. 2007;8:113–126. doi: 10.1038/nrm2104. [DOI] [PubMed] [Google Scholar]
- 4.Jacques C, Bereziat G, Humbert L, Olivier JL, Corvol MT, Masliah J, Berenbaum F. Posttranscriptional effect of insulin-like growth factor-I on interleukin-1beta-induced type II-secreted phospholipase A2 gene expression in rabbit articular chondrocytes. The Journal of clinical investigation. 1997;99:1864–1872. doi: 10.1172/JCI119353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerji S, Aurass P, Flieger A. The manifold phospholipases A of Legionella pneumophila - identification, export, regulation, and their link to bacterial virulence. International journal of medical microbiology : IJMM. 2008;298:169–181. doi: 10.1016/j.ijmm.2007.11.004. [DOI] [PubMed] [Google Scholar]
- 6.Montenegro D, Romero R, Kim SS, Tarca AL, Draghici S, Kusanovic JP, Kim JS, Lee DC, Erez O, Gotsch F, Hassan SS, Kim CJ. Expression patterns of microRNAs in the chorioamniotic membranes: a role for microRNAs in human pregnancy and parturition. The Journal of pathology. 2009;217:113–121. doi: 10.1002/path.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Menschikowski M, Hagelgans A, Nacke B, Jandeck C, Sukocheva O, Siegert G. Epigenetic control of phospholipase A2 receptor expression in mammary cancer cells. BMC cancer. 2015;15:971. doi: 10.1186/s12885-015-1937-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore AE, Young LE, Dixon DA. MicroRNA and AU-rich element regulation of prostaglandin synthesis. Cancer metastasis reviews. 2011;30:419–435. doi: 10.1007/s10555-011-9300-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ramazzotti G, Bavelloni A, Blalock W, Piazzi M, Cocco L, Faenza I. BMP-2 Induced Expression of PLCbeta1 That is a Positive Regulator of Osteoblast Differentiation. Journal of cellular physiology. 2016;231:623–629. doi: 10.1002/jcp.25107. [DOI] [PubMed] [Google Scholar]
- 10.Frohman MA, Sung TC, Morris AJ. Mammalian phospholipase D structure and regulation. Biochimica et biophysica acta. 1999;1439:175–186. doi: 10.1016/s1388-1981(99)00093-1. [DOI] [PubMed] [Google Scholar]
- 11.Exton JH. Phospholipase D. Annals of the New York Academy of Sciences. 2000;905:61–68. doi: 10.1111/j.1749-6632.2000.tb06538.x. [DOI] [PubMed] [Google Scholar]
- 12.Powner DJ, Wakelam MJ. The regulation of phospholipase D by inositol phospholipids and small GTPases. FEBS letters. 2002;531:62–64. doi: 10.1016/s0014-5793(02)03410-5. [DOI] [PubMed] [Google Scholar]
- 13.Foster DA, Xu L. Phospholipase D in cell proliferation and cancer. Molecular cancer research : MCR. 2003;1:789–800. [PubMed] [Google Scholar]
- 14.Chi X, Wang S, Huang Y, Stamnes M, Chen JL. Roles of rho GTPases in intracellular transport and cellular transformation. International journal of molecular sciences. 2013;14:7089–7108. doi: 10.3390/ijms14047089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tu-Sekine B, Goldschmidt H, Raben DM. Diacylglycerol, phosphatidic acid, and their metabolic enzymes in synaptic vesicle recycling. Advances in biological regulation. 2015;57:147–152. doi: 10.1016/j.jbior.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gomez-Cambronero J. Phosphatidic acid, phospholipase D and tumorigenesis. Advances in biological regulation. 2014;54:197–206. doi: 10.1016/j.jbior.2013.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghim J, Chelakkot C, Bae YS, Suh PG, Ryu SH. Accumulating insights into the role of phospholipase D2 in human diseases. Advances in biological regulation. 2016;61:42–46. doi: 10.1016/j.jbior.2015.11.010. [DOI] [PubMed] [Google Scholar]
- 18.Nelson RK, Frohman MA. Physiological and pathophysiological roles for phospholipase D. Journal of lipid research. 2015;56:2229–2237. doi: 10.1194/jlr.R059220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henkels KM, Boivin GP, Dudley ES, Berberich SJ, Gomez-Cambronero J. Phospholipase D (PLD) drives cell invasion, tumor growth and metastasis in a human breast cancer xenograph model. Oncogene. 2013;32:5551–5562. doi: 10.1038/onc.2013.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gomez-Cambronero J. Phospholipase D in Cell Signaling: From a Myriad of Cell Functions to Cancer Growth and Metastasis. Journal of Biological Chemistry. 2014;289:22557–22566. doi: 10.1074/jbc.R114.574152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frohman MA. The phospholipase D superfamily as therapeutic targets. Trends in Pharmacological Sciences. 2015;36:137–144. doi: 10.1016/j.tips.2015.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oliveto S, Mancino M, Manfrini N, Biffo S. Role of microRNAs in translation regulation and cancer. World journal of biological chemistry. 2017;8:45–56. doi: 10.4331/wjbc.v8.i1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang X, Devany E, Murphy MR, Glazman G, Persaud M, Kleiman FE. PARN deadenylase is involved in miRNA-dependent degradation of TP53 mRNA in mammalian cells. Nucleic acids research. 2015;43:10925–10938. doi: 10.1093/nar/gkv959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bruntz RC, Lindsley CW, Brown HA. Phospholipase D signaling pathways and phosphatidic acid as therapeutic targets in cancer. Pharmacological reviews. 2014;66:1033–1079. doi: 10.1124/pr.114.009217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gomez-Cambronero J. Phospholipase D in cell signaling: from a myriad of cell functions to cancer growth and metastasis. The Journal of biological chemistry. 2014;289:22557–22566. doi: 10.1074/jbc.R114.574152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fite K, Elkhadragy L, Gomez-Cambronero J. A Repertoire of MicroRNAs Regulates Cancer Cell Starvation by Targeting Phospholipase D in a Feedback Loop That Operates Maximally in Cancer Cells. Molecular and cellular biology. 2016;36:1078–1089. doi: 10.1128/MCB.00711-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fite K, Gomez-Cambronero J. Down-regulation of MicroRNAs (MiRs) 203, 887, 3619 and 182 Prevents Vimentin-triggered, Phospholipase D (PLD)-mediated Cancer Cell Invasion. The Journal of biological chemistry. 2016;291:719–730. doi: 10.1074/jbc.M115.686006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen Q, Hongu T, Sato T, Zhang Y, Ali W, Cavallo JA, van der Velden A, Tian H, Di Paolo G, Nieswandt B, Kanaho Y, Frohman MA. Key roles for the lipid signaling enzyme phospholipase d1 in the tumor microenvironment during tumor angiogenesis and metastasis. Science signaling. 2012;5:ra79. doi: 10.1126/scisignal.2003257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Uchida N, Okamura S, Nagamachi Y, Yamashita S. Increased phospholipase D activity in human breast cancer. Journal of cancer research and clinical oncology. 1997;123:280–285. doi: 10.1007/BF01208639. [DOI] [PubMed] [Google Scholar]
- 30.Noh DY, Ahn SJ, Lee RA, Park IA, Kim JH, Suh PG, Ryu SH, Lee KH, Han JS. Overexpression of phospholipase D1 in human breast cancer tissues. Cancer letters. 2000;161:207–214. doi: 10.1016/s0304-3835(00)00612-1. [DOI] [PubMed] [Google Scholar]
- 31.Zhang J, Bian Z, Zhou J, Song M, Liu Z, Feng Y, Zhe L, Zhang B, Yin Y, Huang Z. MicroRNA-638 inhibits cell proliferation by targeting phospholipase D1 in human gastric carcinoma. Protein & cell. 2015;6:680–688. doi: 10.1007/s13238-015-0187-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Uchida N, Okamura S, Kuwano H. Phospholipase D activity in human gastric carcinoma. Anticancer research. 1999;19:671–675. [PubMed] [Google Scholar]
- 33.Yoshida M, Okamura S, Kodaki T, Mori M, Yamashita S. Enhanced levels of oleate-dependent and Arf-dependent phospholipase D isoforms in experimental colon cancer. Oncology research. 1998;10:399–406. [PubMed] [Google Scholar]
- 34.Zhao Y, Ehara H, Akao Y, Shamoto M, Nakagawa Y, Banno Y, Deguchi T, Ohishi N, Yagi K, Nozawa Y. Increased activity and intranuclear expression of phospholipase D2 in human renal cancer. Biochemical and biophysical research communications. 2000;278:140–143. doi: 10.1006/bbrc.2000.3719. [DOI] [PubMed] [Google Scholar]
- 35.Kim YR, Byun HS, Won M, Park KA, Kim JM, Choi BL, Lee H, Hong JH, Park J, Seok JH, Kim DW, Shong M, Park SK, Hur GM. Modulatory role of phospholipase D in the activation of signal transducer and activator of transcription (STAT)-3 by thyroid oncogenic kinase RET/PTC. BMC cancer. 2008;8:144. doi: 10.1186/1471-2407-8-144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park MH, Ahn BH, Hong YK, Min do S. Overexpression of phospholipase D enhances matrix metalloproteinase-2 expression and glioma cell invasion via protein kinase C and protein kinase A/NF-kappaB/ Sp1-mediated signaling pathways. Carcinogenesis. 2009;30:356–365. doi: 10.1093/carcin/bgn287. [DOI] [PubMed] [Google Scholar]
- 37.Lee JG, Lee SH, Park DW, Bae YS, Yun SS, Kim JR, Baek SH. Phosphatidic acid as a regulator of matrix metalloproteinase-9 expression via the TNF-alpha signaling pathway. FEBS letters. 2007;581:787–793. doi: 10.1016/j.febslet.2007.01.048. [DOI] [PubMed] [Google Scholar]
- 38.Hao NB, Lu MH, Fan YH, Cao YL, Zhang ZR, Yang SM. Macrophages in tumor microenvironments and the progression of tumors. Clinical & developmental immunology. 2012;2012:948098. doi: 10.1155/2012/948098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lee YS, Lan Tran HT, Van Ta Q. Regulation of expression of matrix metalloproteinase-9 by JNK in Raw 264.7 cells: presence of inhibitory factor(s) suppressing MMP-9 induction in serum and conditioned media. Experimental & molecular medicine. 2009;41:259–268. doi: 10.3858/emm.2009.41.4.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han S, Huh J, Kim W, Jeong S, Min do S, Jung Y. Phospholipase D activates HIF-1-VEGF pathway via phosphatidic acid. Experimental & molecular medicine. 2014;46:e126. doi: 10.1038/emm.2014.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Miller TE, Gomez-Cambronero J. A feedback mechanism between PLD and deadenylase PARN for the shortening of eukaryotic poly(A) mRNA tails that is deregulated in cancer cells. Biology open. 2017;6:176–186. doi: 10.1242/bio.021261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kung JT, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193:651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, Bell I, Cheung E, Drenkow J, Dumais E, Patel S, Helt G, Ganesh M, Ghosh S, Piccolboni A, Sementchenko V, Tammana H, Gingeras TR. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 44.Heery R, Finn SP, Cuffe S, Gray SG. Long Non-Coding RNAs: Key Regulators of Epithelial-Mesenchymal Transition, Tumour Drug Resistance and Cancer Stem Cells. Cancers. 2017;9 doi: 10.3390/cancers9040038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ning S, Zhang J, Wang P, Zhi H, Wang J, Liu Y, Gao Y, Guo M, Yue M, Wang L, Li X. Lnc2Cancer: a manually curated database of experimentally supported lncRNAs associated with various human cancers. Nucleic acids research. 2016;44:D980–985. doi: 10.1093/nar/gkv1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stepanov GA, Filippova JA, Komissarov AB, Kuligina EV, Richter VA, Semenov DV. Regulatory role of small nucleolar RNAs in human diseases. BioMed research international. 2015;2015:206849. doi: 10.1155/2015/206849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Valadkhan S. snRNAs as the catalysts of pre-mRNA splicing. Current opinion in chemical biology. 2005;9:603–608. doi: 10.1016/j.cbpa.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 49.Hyun S. Small RNA Pathways That Protect the Somatic Genome. International journal of molecular sciences. 2017;18 doi: 10.3390/ijms18050912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allegra A, Alonci A, Campo S, Penna G, Petrungaro A, Gerace D, Musolino C. Circulating microRNAs: new biomarkers in diagnosis, prognosis and treatment of cancer (review) International journal of oncology. 2012;41:1897–1912. doi: 10.3892/ijo.2012.1647. [DOI] [PubMed] [Google Scholar]
- 51.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 52.Fabian MR, Sonenberg N. The mechanics of miRNA-mediated gene silencing: a look under the hood of miRISC. Nature structural & molecular biology. 2012;19:586–593. doi: 10.1038/nsmb.2296. [DOI] [PubMed] [Google Scholar]
- 53.Ohtsuka M, Ling H, Doki Y, Mori M, Calin GA. MicroRNA Processing and Human Cancer. Journal of clinical medicine. 2015;4:1651–1667. doi: 10.3390/jcm4081651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 55.Kim VN. MicroRNA biogenesis: coordinated cropping and dicing. Nature reviews. Molecular cell biology. 2005;6:376–385. doi: 10.1038/nrm1644. [DOI] [PubMed] [Google Scholar]
- 56.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes & development. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Iorio MV, Croce CM. Causes and consequences of microRNA dysregulation. Cancer J. 2012;18:215–222. doi: 10.1097/PPO.0b013e318250c001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Imani S, Zhang X, Hosseinifard H, Fu S, Fu J. The diagnostic role of microRNA-34a in breast cancer: a systematic review and meta-analysis. Oncotarget. 2017;8:23177–23187. doi: 10.18632/oncotarget.15520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chang JC, Kundranda M. Novel Diagnostic and Predictive Biomarkers in Pancreatic Adenocarcinoma. International journal of molecular sciences. 2017;18 doi: 10.3390/ijms18030667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Smith B, Agarwal P, Bhowmick NA. MicroRNA applications for prostate, ovarian and breast cancer in the era of precision medicine. Endocrine-related cancer. 2017;24:R157–R172. doi: 10.1530/ERC-16-0525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bradshaw G, Sutherland HG, Haupt LM, Griffiths LR. Dysregulated MicroRNA Expression Profiles and Potential Cellular, Circulating and Polymorphic Biomarkers in Non-Hodgkin Lymphoma. Genes. 2016;7 doi: 10.3390/genes7120130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ahmad P, Sana J, Slavik M, Slampa P, Smilek P, Slaby O. MicroRNAs Involvement in Radioresistance of Head and Neck Cancer. Disease markers. 2017;2017:8245345. doi: 10.1155/2017/8245345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shen S, Lin Y, Yuan X, Shen L, Chen J, Chen L, Qin L, Shen B. Biomarker MicroRNAs for Diagnosis, Prognosis and Treatment of Hepatocellular Carcinoma: A Functional Survey and Comparison. Scientific reports. 2016;6:38311. doi: 10.1038/srep38311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ottosen S, Parsley TB, Yang L, Zeh K, van Doorn LJ, van der Veer E, Raney AK, Hodges MR, Patick AK. In vitro antiviral activity and preclinical and clinical resistance profile of miravirsen, a novel antihepatitis C virus therapeutic targeting the human factor miR-122. Antimicrobial agents and chemotherapy. 2015;59:599–608. doi: 10.1128/AAC.04220-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gebert LF, Rebhan MA, Crivelli SE, Denzler R, Stoffel M, Hall J. Miravirsen (SPC3649) can inhibit the biogenesis of miR-122. Nucleic acids research. 2014;42:609–621. doi: 10.1093/nar/gkt852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shrivastava S, Mukherjee A, Ray RB. Hepatitis C virus infection, microRNA and liver disease progression. World journal of hepatology. 2013;5:479–486. doi: 10.4254/wjh.v5.i9.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Adams BD, Parsons C, Slack FJ. The tumor-suppressive and potential therapeutic functions of miR-34a in epithelial carcinomas. Expert opinion on therapeutic targets. 2016;20:737–753. doi: 10.1517/14728222.2016.1114102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reid G, Kao SC, Pavlakis N, Brahmbhatt H, MacDiarmid J, Clarke S, Boyer M, van Zandwijk N. Clinical development of TargomiRs, a miRNA mimic-based treatment for patients with recurrent thoracic cancer. Epigenomics. 2016;8:1079–1085. doi: 10.2217/epi-2016-0035. [DOI] [PubMed] [Google Scholar]
- 69.Mahankali M, Farkaly T, Bedi S, Hostetler HA, Gomez-Cambronero J. Phosphatidic Acid (PA) can Displace PPARalpha/LXRalpha Binding to The EGFR Promoter Causing its Transrepression in Luminal Cancer Cells. Scientific reports. 2015;5:15379. doi: 10.1038/srep15379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Funakoshi Y, Doi Y, Hosoda N, Uchida N, Osawa M, Shimada I, Tsujimoto M, Suzuki T, Katada T, Hoshino S. Mechanism of mRNA deadenylation: evidence for a molecular interplay between translation termination factor eRF3 and mRNA deadenylases. Genes & development. 2007;21:3135–3148. doi: 10.1101/gad.1597707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mitchell P, Tollervey D. mRNA stability in eukaryotes. Current opinion in genetics & development. 2000;10:193–198. doi: 10.1016/s0959-437x(00)00063-0. [DOI] [PubMed] [Google Scholar]
- 72.Nousch M, Techritz N, Hampel D, Millonigg S, Eckmann CR. The Ccr4-Not deadenylase complex constitutes the main poly(A) removal activity in C. elegans. Journal of cell science. 2013;126:4274–4285. doi: 10.1242/jcs.132936. [DOI] [PubMed] [Google Scholar]
- 73.Wolf J, Passmore LA. mRNA deadenylation by Pan2-Pan3. Biochemical Society transactions. 2014;42:184–187. doi: 10.1042/BST20130211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Collart MA, Panasenko OO. The Ccr4--not complex. Gene. 2012;492:42–53. doi: 10.1016/j.gene.2011.09.033. [DOI] [PubMed] [Google Scholar]
- 75.Korner CG, Wormington M, Muckenthaler M, Schneider S, Dehlin E, Wahle E. The deadenylating nuclease (DAN) is involved in poly(A) tail removal during the meiotic maturation of Xenopus oocytes. The EMBO journal. 1998;17:5427–5437. doi: 10.1093/emboj/17.18.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Mian IS. Comparative sequence analysis of ribonucleases HII, III, II PH and D. Nucleic acids research. 1997;25:3187–3195. doi: 10.1093/nar/25.16.3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Moser M, Ruschoff J, Buettner R. Comparative analysis of AP-2 alpha and AP-2 beta gene expression during murine embryogenesis. Developmental dynamics : an official publication of the American Association of Anatomists. 1997;208:115–124. doi: 10.1002/(SICI)1097-0177(199701)208:1<115::AID-AJA11>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 78.Gavin AC, Bosche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Hofert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. [DOI] [PubMed] [Google Scholar]
- 79.Morel AP, Sentis S, Bianchin C, Le Romancer M, Jonard L, Rostan MC, Rimokh R, Corbo L. BTG2 antiproliferative protein interacts with the human CCR4 complex existing in vivo in three cell-cycle-regulated forms. Journal of cell science. 2003;116:2929–2936. doi: 10.1242/jcs.00480. [DOI] [PubMed] [Google Scholar]
- 80.Yamashita A, Chang TC, Yamashita Y, Zhu W, Zhong Z, Chen CY, Shyu AB. Concerted action of poly(A) nucleases and decapping enzyme in mammalian mRNA turnover. Nature structural & molecular biology. 2005;12:1054–1063. doi: 10.1038/nsmb1016. [DOI] [PubMed] [Google Scholar]
- 81.Azzouz N, Panasenko OO, Deluen C, Hsieh J, Theiler G, Collart MA. Specific roles for the Ccr4-Not complex subunits in expression of the genome. Rna. 2009;15:377–383. doi: 10.1261/rna.1348209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Grigull J, Mnaimneh S, Pootoolal J, Robinson MD, Hughes TR. Genome-wide analysis of mRNA stability using transcription inhibitors and microarrays reveals posttranscriptional control of ribosome biogenesis factors. Molecular and cellular biology. 2004;24:5534–5547. doi: 10.1128/MCB.24.12.5534-5547.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Wiederhold K, Passmore LA. Cytoplasmic deadenylation: regulation of mRNA fate. Biochemical Society transactions. 2010;38:1531–1536. doi: 10.1042/BST0381531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Sachs AB, Deardorff JA. Translation initiation requires the PAB-dependent poly(A) ribonuclease in yeast. Cell. 1992;70:961–973. doi: 10.1016/0092-8674(92)90246-9. [DOI] [PubMed] [Google Scholar]
- 85.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic acids research. 1997;25:5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zuo Y, Deutscher MP. Exoribonuclease superfamilies: structural analysis and phylogenetic distribution. Nucleic acids research. 2001;29:1017–1026. doi: 10.1093/nar/29.5.1017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng D, Ezzeddine N, Chen CY, Zhu W, He X, Shyu AB. Deadenylation is prerequisite for P-body formation and mRNA decay in mammalian cells. The Journal of cell biology. 2008;182:89–101. doi: 10.1083/jcb.200801196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez J, Ren YG, Thuresson AC, Hellman U, Astrom J, Virtanen A. A 54-kDa fragment of the Poly(A)-specific ribonuclease is an oligomeric, processive, and cap-interacting Poly(A)-specific 3' exonuclease. The Journal of biological chemistry. 2000;275:24222–24230. doi: 10.1074/jbc.M001705200. [DOI] [PubMed] [Google Scholar]
- 89.He GJ, Yan YB. Self-association of poly(A)-specific ribonuclease (PARN) triggered by the R3H domain. Biochimica et biophysica acta. 2014;1844:2077–2085. doi: 10.1016/j.bbapap.2014.09.010. [DOI] [PubMed] [Google Scholar]
- 90.Helfer S, Schott J, Stoecklin G, Forstemann K. AU-rich element-mediated mRNA decay can occur independently of the miRNA machinery in mouse embryonic fibroblasts and Drosophila S2-cells. PloS one. 2012;7:e28907. doi: 10.1371/journal.pone.0028907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Korner CG, Wahle E. Poly(A) tail shortening by a mammalian poly(A)-specific 3'-exoribonuclease. The Journal of biological chemistry. 1997;272:10448–10456. doi: 10.1074/jbc.272.16.10448. [DOI] [PubMed] [Google Scholar]
- 92.Lai WS, Kennington EA, Blackshear PJ. Tristetraprolin and its family members can promote the cell-free deadenylation of AU-rich element-containing mRNAs by poly(A) ribonuclease. Molecular and cellular biology. 2003;23:3798–3812. doi: 10.1128/MCB.23.11.3798-3812.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lin WJ, Duffy A, Chen CY. Localization of AU-rich element-containing mRNA in cytoplasmic granules containing exosome subunits. The Journal of biological chemistry. 2007;282:19958–19968. doi: 10.1074/jbc.M702281200. [DOI] [PubMed] [Google Scholar]
- 94.Balatsos NA, Nilsson P, Mazza C, Cusack S, Virtanen A. Inhibition of mRNA deadenylation by the nuclear cap binding complex (CBC) The Journal of biological chemistry. 2006;281:4517–4522. doi: 10.1074/jbc.M508590200. [DOI] [PubMed] [Google Scholar]
- 95.Berndt H, Harnisch C, Rammelt C, Stohr N, Zirkel A, Dohm JC, Himmelbauer H, Tavanez JP, Huttelmaier S, Wahle E. Maturation of mammalian H/ACA box snoRNAs: PAPD5-dependent adenylation and PARN-dependent trimming. Rna. 2012;18:958–972. doi: 10.1261/rna.032292.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Shukla S, Schmidt JC, Goldfarb KC, Cech TR, Parker R. Inhibition of telomerase RNA decay rescues telomerase deficiency caused by dyskerin or PARN defects. Nature structural & molecular biology. 2016;23:286–292. doi: 10.1038/nsmb.3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Boyraz B, Moon DH, Segal M, Muosieyiri MZ, Aykanat A, Tai AK, Cahan P, Agarwal S. Posttranscriptional manipulation of TERC reverses molecular hallmarks of telomere disease. The Journal of clinical investigation. 2016;126:3377–3382. doi: 10.1172/JCI87547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Martinez J, Ren YG, Nilsson P, Ehrenberg M, Virtanen A. The mRNA cap structure stimulates rate of poly(A) removal and amplifies processivity of degradation. The Journal of biological chemistry. 2001;276:27923–27929. doi: 10.1074/jbc.M102270200. [DOI] [PubMed] [Google Scholar]
- 99.Mason PJ, Bessler M. mRNA deadenylation and telomere disease. The Journal of clinical investigation. 2015;125:3304. doi: 10.1172/JCI82903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stuart BD, Choi J, Zaidi S, Xing C, Holohan B, Chen R, Choi M, Dharwadkar P, Torres F, Girod CE, Weissler J, Fitzgerald J, Kershaw C, Klesney-Tait J, Mageto Y, Shay JW, Ji W, Bilguvar K, Mane S, Lifton RP, Garcia CK. Exome sequencing links mutations in PARN and RTEL1 with familial pulmonary fibrosis and telomere shortening. Nature genetics. 2015;47:512–517. doi: 10.1038/ng.3278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tummala H, Walne A, Collopy L, Cardoso S, de la Fuente J, Lawson S, Powell J, Cooper N, Foster A, Mohammed S, Plagnol V, Vulliamy T, Dokal I. Poly(A)-specific ribonuclease deficiency impacts telomere biology and causes dyskeratosis congenita. The Journal of clinical investigation. 2015;125:2151–2160. doi: 10.1172/JCI78963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Moon DH, Segal M, Boyraz B, Guinan E, Hofmann I, Cahan P, Tai AK, Agarwal S. Poly(A)-specific ribonuclease (PARN) mediates 3'-end maturation of the telomerase RNA component. Nature genetics. 2015;47:1482–1488. doi: 10.1038/ng.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Maragozidis P, Papanastasi E, Scutelnic D, Totomi A, Kokkori I, Zarogiannis SG, Kerenidi T, Gourgoulianis KI, Balatsos NA. Poly(A)-specific ribonuclease and Nocturnin in squamous cell lung cancer: prognostic value and impact on gene expression. Molecular cancer. 2015;14:187. doi: 10.1186/s12943-015-0457-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee JE, Lee JY, Trembly J, Wilusz J, Tian B, Wilusz CJ. The PARN deadenylase targets a discrete set of mRNAs for decay and regulates cell motility in mouse myoblasts. PLoS genetics. 2012;8:e1002901. doi: 10.1371/journal.pgen.1002901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Shyu AB, Belasco JG, Greenberg ME. Two distinct destabilizing elements in the c-fos message trigger deadenylation as a first step in rapid mRNA decay. Genes & development. 1991;5:221–231. doi: 10.1101/gad.5.2.221. [DOI] [PubMed] [Google Scholar]
- 106.Devany E, Zhang X, Park JY, Tian B, Kleiman FE. Positive and negative feedback loops in the p53 and mRNA 3' processing pathways. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3351–3356. doi: 10.1073/pnas.1212533110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Boele J, Persson H, Shin JW, Ishizu Y, Newie IS, Sokilde R, Hawkins SM, Coarfa C, Ikeda K, Takayama K, Horie-Inoue K, Ando Y, Burroughs AM, Sasaki C, Suzuki C, Sakai M, Aoki S, Ogawa A, Hasegawa A, Lizio M, Kaida K, Teusink B, Carninci P, Suzuki H, Inoue S, Gunaratne PH, Rovira C, Hayashizaki Y, de Hoon MJ. PAPD5-mediated 3' adenylation and subsequent degradation of miR-21 is disrupted in proliferative disease. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11467–11472. doi: 10.1073/pnas.1317751111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Buiting K, Korner C, Ulrich B, Wahle E, Horsthemke B. The human gene for the poly(A)-specific ribonuclease (PARN) maps to 16p13 and has a truncated copy in the Prader-Willi/Angelman syndrome region on 15q11-->q13. Cytogenetics and cell genetics. 1999;87:125–131. doi: 10.1159/000015378. [DOI] [PubMed] [Google Scholar]
- 109.Cevher MA, Zhang X, Fernandez S, Kim S, Baquero J, Nilsson P, Lee S, Virtanen A, Kleiman FE. Nuclear deadenylation/polyadenylation factors regulate 3' processing in response to DNA damage. The EMBO journal. 2010;29:1674–1687. doi: 10.1038/emboj.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Moraes KC, Wilusz CJ, Wilusz J. CUG-BP binds to RNA substrates and recruits PARN deadenylase. Rna. 2006;12:1084–1091. doi: 10.1261/rna.59606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Reinhardt HC, Hasskamp P, Schmedding I, Morandell S, van Vugt MA, Wang X, Linding R, Ong SE, Weaver D, Carr SA, Yaffe MB. DNA damage activates a spatially distinct late cytoplasmic cell-cycle checkpoint network controlled by MK2-mediated RNA stabilization. Molecular cell. 2010;40:34–49. doi: 10.1016/j.molcel.2010.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Seal R, Temperley R, Wilusz J, Lightowlers RN, Chrzanowska-Lightowlers ZM. Serum-deprivation stimulates cap-binding by PARN at the expense of eIF4E, consistent with the observed decrease in mRNA stability. Nucleic acids research. 2005;33:376–387. doi: 10.1093/nar/gki169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Gao M, Fritz DT, Ford LP, Wilusz J. Interaction between a poly(A)-specific ribonuclease and the 5' cap influences mRNA deadenylation rates in vitro. Molecular cell. 2000;5:479–488. doi: 10.1016/s1097-2765(00)80442-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lee JE, Lee JY, Wilusz J, Tian B, Wilusz CJ. Systematic analysis of cis-elements in unstable mRNAs demonstrates that CUGBP1 is a key regulator of mRNA decay in muscle cells. PloS one. 2010;5:e11201. doi: 10.1371/journal.pone.0011201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Finak G, Bertos N, Pepin F, Sadekova S, Souleimanova M, Zhao H, Chen H, Omeroglu G, Meterissian S, Omeroglu A, Hallett M, Park M. Stromal gene expression predicts clinical outcome in breast cancer. Nature medicine. 2008;14:518–527. doi: 10.1038/nm1764. [DOI] [PubMed] [Google Scholar]