Abstract
Hyperopic refractive error is detected by retinal neurons, which generate GO signals through a direct emmetropization signaling cascade: retinal pigment epithelium (RPE) into choroid and then into sclera, thereby increasing axial elongation. To examine signaling early in this cascade, we measured gene expression in the retina and RPE after short exposure to hyperopia produced by minus-lens wear. Gene expression in each tissue was compared with gene expression in combined retina+RPE. Starting 24 days after normal eye opening, three groups of juvenile tree shrews (n = 7 each) wore a monocular −5 D lens. The untreated fellow eye served as a control. The “6h” group wore the lens for 6 hours (h); the “24h” group wore the lens for 24 h; each group provided separate retina and RPE tissues. Group “24hC” wore the lens for 24 h and provided combined retina+RPE tissue. Quantitative PCR was used to measure the relative differences (treated eye vs. control eye) in mRNA levels for 66 candidate genes. In the retina after 6 h, mRNA levels for seven genes were significantly regulated: EGR1 and FOS (early intermediate genes) were down-regulated in the treated eyes. Genes with secreted protein products, BMP2 and CTGF, were down-regulated, whilst FGF10, IL18, and SST were up-regulated. After 24 h the pattern changed; only one of the seven genes still showed differential expression; BMP2 was still down-regulated. Two new genes with secreted protein products, IGF2 and VIP, were up-regulated. In the RPE, consistent with its role in receiving, processing, and transmitting GO signaling, differential expression was found for genes whose protein products are at the cell surface, intracellular, in the nucleus, and are secreted. After 6 h, mRNA levels for 17 genes were down-regulated in the treated eyes, whilst four genes (GJA1, IGF2R, LRP2, and IL18) were up-regulated. After 24 h the pattern was similar; mRNA levels for 14 of the same genes were still down-regulated; only LRP2 remained up-regulated. mRNA levels for six genes no longer showed differential expression, whilst nine genes, not differentially expressed at 6 h, now showed differential expression. In the combined retina+RPE after 24 h, mRNA levels for only seven genes were differentially regulated despite the differential expression of many genes in the RPE. Four genes showed the same expression in combined tissue as in retina alone, including up-regulation of VIP despite significant VIP down-regulation in RPE. Thus, hyperopia-induced GO signaling, as measured by differential gene expression, differs in the retina and the RPE. Retinal gene expression changed between 6 h and 24 h of treatment, suggesting evolution of the retinal response. Gene expression in the RPE was similar at both time points, suggesting sustained signaling. The combined retina+RPE does not accurately represent gene expression in either retina or, especially, RPE. When gene expression signatures were compared with those in choroid and sclera, GO signaling, as encoded by differential gene expression, differs in each compartment of the direct emmetropization signaling cascade.
Keywords: myopia, animal models, refractive error, axial elongation, gene expression, direct emmetropization signaling cascade
1. INTRODUCTION
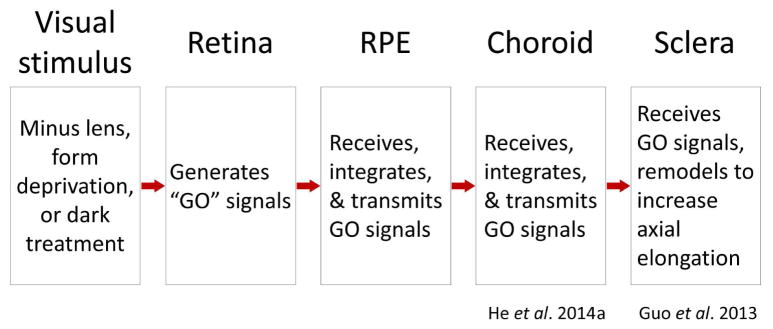
During normal postnatal eye development, an emmetropization feedback mechanism plays a critical role in modulating axial elongation so as to produce eyes that are in good focus. It uses refractive error to adjust the axial elongation rate so that the photoreceptors come to lie very near the focal plane (emmetropia) (Norton, 1999; Wallman and Winawer, 2004; Wildsoet, 1997). In many species, the target refraction appears to be mild hyperopia that is easily cleared with accommodation. Refractive error is detected by retinal neurons that generate signals which travel in a “direct” (within the eye) signaling pathway into the retinal pigment epithelium (RPE), then into choroid, and finally into the sclera (Figure 1) where remodeling occurs that modulates the axial elongation rate. This direct emmetropization cascade is able to adjust the axial elongation rate even when central communication is interrupted (Norton et al., 1994; Schaeffel et al., 1990; Troilo, 1990; Wildsoet, 2003). However, the emmetropization mechanism functions less well without communication to central structures (Wildsoet, 2003; Wildsoet and McFadden, 2010), suggesting that important signals also leave the retina through the optic nerve and reach central brain structures that control accommodation and send efferent connections to ocular structures (Dillingham et al., 2013; Gamlin, 1999; Gamlin et al., 1998). This study focuses on signaling in the direct pathway.
Figure 1.
Compartments of the direct emmetropization pathway signaling cascade. Refractive hyperopia causes retinal neurons to generate GO signals that pass into, and are transformed in, each compartment. In the sclera, in mammals, the result is remodeling that increases the axial elongation rate of the eye.
Imposing refractive error with lenses, held in front of an eye, stimulates a response from the emmetropization mechanism. A minus-power lens produces a hyperopic shift in the eye’s refractive state. In response, the retina produces what have been described as “GO” signals (Guo et al., 2014; He et al., 2014a; Rohrer and Stell, 1994; Schaeffel and Howland, 1988). The precise nature of the retinal signals remains unknown but appears to involve amacrine (and bipolar) cells and the retinal dopaminergic pathways (Fischer et al., 1997; Iuvone et al., 1991; Stell et al., 2003; Stell et al., 2004; Stone and Khurana, 2010; Stone et al., 1990; Young et al., 1994). In addition, amacrine cells are the earliest in the retinal circuit with sufficient receptive field complexity to potentially detect defocus and its direction (Zhong et al., 2004). As illustrated in Figure 1, the signals then pass into a separate compartment, the RPE, which receives, integrates, and then transmits the GO signals to the choroid. The nature of the signaling in the RPE is still unknown, as is how retinal GO signals move from the retina to the RPE, and then into the choroid, though there are numerous possibilities (Rymer and Wildsoet, 2005; Zhang and Wildsoet, 2015). The choroid, in turn, receives, integrates, and then transmits the GO signals to the final compartment, the sclera (He et al., 2014a, b; Nickla and Wallman, 2010). The sclera serves as an effector that responds to the GO signals by remodeling the extracellular matrix (Christian et al., 2013; Frost et al., 2012; Gao et al., 2011; Grytz and Siegwart, 2015; Guo et al., 2013; Moring et al., 2007; Norton and Rada, 1995; Seko et al., 1994; Summers Rada et al., 2006). In tree shrews, small mammals closely related to primates, the sclera is a fibrous tissue comprised largely of type 1 collagen. GO signals increase the viscoelasticity of the sclera and appears to allow normal intraocular pressure to expand the vitreous chamber, increasing the axial length (Phillips et al., 2000; Siegwart and Norton, 1999). Over time, as the eye elongates, the lens-induced hyperopia diminishes. When the elongation of the globe returns the eye to its age-normal refractive state, scleral remodeling returns toward normal, axial elongation slows, and the eye stabilizes at “emmetropia” (the same slight hyperopia as in normal eyes) while wearing the lens (He et al., 2014b; Norton et al., 2010). If refractive state is measured with the lens removed, the elongated eye is myopic. Thus, understanding the functioning of the emmetropization mechanism, and the direct signaling cascade, is an important step in learning how myopia develops and, perhaps, how myopia progression may be controlled.
This lab has examined, in tree shrews, alterations in gene expression produced in the choroid and in the sclera in response to induced hyperopia and during recovery from lens-induced myopia (Gao et al., 2011; Guo et al., 2013; Guo et al., 2014; He et al., 2014a, b; Siegwart and Norton, 2005). Minus-lens wear produces a GO pattern (“signature”) of change in the expression of many genes in both of these compartments of the direct emmetropization pathway, and the expression signatures are different in both the choroid and the sclera. In the present study, we examined mRNA changes in response to minus-lens wear in tree shrews in the first two compartments of the signaling cascade: retina and RPE.
Because of difficulties involved in separating the tissues while also preserving RNA integrity, most previous studies of emmetropization-related gene expression have not examined the RPE as a separate tissue. Rather, RPE typically has been examined either together with the retina (He et al., 2011; McGlinn et al., 2007; Stone et al., 2011) or with the choroid (Shelton et al., 2008). In either combination, one might suspect that detecting mRNA changes specific to the RPE might be difficult because it is a monolayer while these other tissues contain many more cells with a variety of functions that may mask changes that occur only in RPE. More recent studies in chick have examined expression of several genes in RPE alone (Zhang et al., 2013; Zhang et al., 2012b). The present study examined alterations in mRNA expression in tree shrews produced by minus-lens wear in the RPE, in the retina, each studied separately, and in the two tissues combined.
One consideration was the timing: how long after the onset of minus-lens wear should the tissues be sampled? The GO signals produced by minus-lens wear travel rapidly, but not instantaneously, through the compartments of the direct emmetropization pathway. In the tree shrew sclera (at the end of the direct pathway), few changes in gene expression have been found after one day of minus-lens wear, but many genes are affected after two days (Gao et al., 2011; Guo et al., 2013). In the choroid, the expression of many genes is altered after two days of minus-lens wear or form deprivation (He et al., 2014b); a much weaker response occurred after one day of minus-lens treatment (He, unpublished data). In tree shrew retina, altered expression of early intermediate genes has been found after 1 – 3 h of recovery from form-deprivation (Stell et al., 2004). In chick retina, minus lens-induced gene expression changes have been found after 1 h (Ashby et al., 2010), 2 h, and 6 h (Stone et al., 2011). In chick RPE, gene expression changes have been found as early as after 2 h of lens treatment (Zhang et al., 2013; Zhang et al., 2012b). We elected to examine gene expression after 6 h, a moderate delay after the onset of lens wear but one that we expected to produce altered gene expression in both the retina and RPE. We also examined these tissues after 24 h of minus-lens wear; a time before substantial refractive changes occur but a time at which GO signals should be well-developed in both retina and RPE. Our hypothesis was that we would find changes in gene expression in both tissues at both time points, and that the expression of different genes would be affected in each tissue. We also suspected that differential gene expression restricted to the RPE would be difficult to detect in the combined retina+RPE tissue.
2. MATERIALS AND METHODS
2.1 Experimental groups
The juvenile tree shrews (Tupaia glis belangeri) used in this study were produced in our breeding colony and raised by their mothers on a 14-h light/10-h dark cycle. Tree shrew pups open their eyes about three weeks after birth (Norton and McBrien, 1992). The first day both eyes are open is designated as day 1 of visual experience (DVE). All procedures complied with the ARVO Statement for the Use of Animals in Ophthalmic and Visual Research and were approved by the Institutional Animal Care and Use Committee of the University of Alabama at Birmingham. Experimental groups were balanced to include both males and females, and avoided pups from the same parents wherever possible.
Three groups of animals (n = 7 per group) were used in this study. Starting at 24 ± 1 DVE, the “6h” group wore a monocular −5 D (spherical power) lens for 6 h and provided separate retina and RPE tissues; the “24h” group wore the monocular −5 D lens for 24 h, also providing separate retina and RPE tissues. The “24hC” group also wore a −5 D lens for 24 h and provided combined retina+RPE tissues.
2.2 Lens treatment
At 21 ± 1 DVE (10 DVE for one animal in the 24hC group), animals were anesthetized (17.5 mg ketamine, 1.2 mg xylazine; supplemented with 0.5 – 2.0% isoflurane as needed) and received a dental acrylic pedestal (Siegwart and Norton, 1994). After pedestal installation, all animals were placed in individual cages with standard colony fluorescent lighting, 100 – 300 lux on the floor of the cage. At 24 ± 1 DVE, a goggle frame holding a −5 D lens (12 mm diameter PMMA contact lens; Conforma Contact Lenses, Norfolk, VA) was clipped to the pedestal at approximately 9:30 AM, firmly holding the lens in front of the randomly selected treated eye. The untreated fellow eye served as a control and had unrestricted vision through an open goggle frame. For the 6h group, the animals wore the lens continuously in their home cage and were prevented from entering their dark nest tubes to ensure that they received ample visual stimulation. Animals in the 24h and 24hC groups were allowed to enter and exit their nest tubes at will. For these groups, the goggles were briefly (<3 min) removed in the late afternoon to clean the lens under dim illumination. During goggle cleaning, animals were kept in their darkened nest tube to minimize exposure to visual stimuli.
2.3 Refractive and axial measures
For the 6h group, non-cycloplegic refractive measures were made, in awake animals, at the start of treatment with a Nidek ARK-700A infrared autorefractor (Marco Ophthalmic, Jacksonville, FL). Because refractive changes and axial elongation were not expected in such a short treatment period, post-treatment refractive measures were omitted. For the 24h and 24hC groups, measures were made at the start and end of the treatment period. Cycloplegic refractive measures were omitted to prevent any interference by atropine on retino-scleral signaling. However, previous studies have shown that non-cycloplegic measures provide a valid estimate of the refractive state, and of induced myopia, in tree shrews (Norton et al., 2000; Norton et al., 2003). All refractive values were corrected for the small-eye artifact (Glickstein and Millodot, 1970), previously shown to be approximately +4 D in tree shrews (Norton et al., 2003).
At the time the pedestal was attached, ocular component dimensions were measured. In the 24hC group, A-scan ultrasound was used whilst the animals were anesthetized to receive the pedestal. In the 6h and 24h groups, ocular components were measured in awake animals with a Lenstar LS-900 optical biometer (Haag-Streit USA, Mason, OH). These measures were made before the start of minus-lens treatment to ensure that the two eyes did not differ significantly in axial length before treatment began.
2.4 Tissue dissection
On completion of the minus-lens treatment, animals were euthanized (17.5 mg ketamine and 1.2 mg xylazine, followed by 50 mg xylazine); both eyes were enucleated and placed into RNAlater solution (Life Technologies, Carlsbad, CA). Extraocular muscles, conjunctiva, and orbital fat were trimmed from the exterior surface of the eye and the cornea dissected away just behind the corneoscleral junction. While viewing through a surgical microscope, the lens and vitreous humor were removed; the retina and RPE, which were tightly bound to each other, were then lifted from the eyecup. For the 24hC group, the combined retina+RPE tissue was simply collected and frozen in liquid nitrogen. For the 6h and 24h groups, the combined retina+RPE tissue was not frozen but transferred into ice-cold sterile PBS to rinse away the residual RNAlater (~10 seconds). The combined tissue was then carefully transferred to a fresh aliquot of PBS and left on ice for ~20 minutes, with occasional gentle agitation, to separate the RPE from the retina. The retina was carefully transferred to a further PBS aliquot to rinse away any residual RPE; the ‘clean’ retina was then collected and frozen in liquid nitrogen. The suspended RPE fragments were gently centrifuged and the supernatant removed; RPE samples were processed immediately to isolate RNA.
2.5 Gene expression analysis
The frozen combined retina+RPE, frozen separate retina, and the freshly-collected RPE were each homogenized with a disposable pellet pestle (Fisher Scientific, Pittsburgh, PA) from which total RNA was isolated using a RiboPure kit (Life Technologies) per the manufacturer’s instructions, with the addition of an on-filter DNase treatment. The purified RNA was quantified (NanoDrop Technologies, Wilmington, DE), with an average yield (mean ± SD) of 38.2 ± 3.5 μg for combined retina+RPE, 18.5 ± 2.4 μg for separate retina, and 2.7 ± 0.7 μg for separate RPE. RNA quality was confirmed by denaturing gel electrophoresis (RNA FlashGel; Lonza, Rockland, ME). cDNA was synthesized from 1 μg of total RNA in a final reaction volume of 20 μl using a Superscript III RT kit (Life Technologies) with minor modifications (2.5 μM anchored oligo (dT)20 primers and DTT omitted). The resultant cDNA was diluted 5-fold before use.
Tree shrew-specific quantitative PCR (qPCR) primers were designed for 66 genes of interest (Table 1) and the reference gene RNA polymerase II (POLR2A) using Beacon Designer v7.7 (Premier Biosoft International, Palo Alto, CA). Most of the candidate genes were selected because they were located at nodes in an Ingenuity Pathway Analysis interaction network seeded from previous literature (Hammond and Wildsoet, 2012; Stone et al., 2011; Zhang et al., 2012a; Zhang et al., 2013; Zhang et al., 2012b; Zhang et al., 2010); others were chosen based on previous data from this lab (He et al., 2011; He et al., 2014b). The candidate genes included ones whose protein products represented four major groupings: cell surface interaction (abbreviated in tables and text as “cell surface”), intracellular processing (“intracellular”), transcriptional regulation (“transcription”), and secreted proteins (“secreted”). Altered gene expression in the cell surface and intracellular categories may indicate responses involved in receiving GO signals; differential expression in the transcription and secreted categories may be associated with the generation of GO signaling that is transmitted to the next compartment of the direct emmetropization signaling cascade.
Table 1.
Genes examined by cellular location/functional category, with its UniProt ID
| Gene symbol | Protein name | UniProt ID |
|---|---|---|
|
|
|
|
| Cell surface | ||
| AQP4 | Aquaporin 4 | P55087 |
| BMPR1B | Bone morphogenetic protein receptor 1B | O00238 |
| BMPR2 | Bone morphogenetic protein receptor 2 | Q13873 |
| DRD1 | Dopamine receptor D1 | P21728 |
| DRD2 | Dopamine receptor D2 | P14416 |
| FGFR2 | Fibroblast growth factor receptor 2 | P21802 |
| GJA1 | Gap junction α1 | P17302 |
| GRM5 | Metabotropic glutamate receptor 5 | P41594 |
| IGF2R | Insulin-like growth factor 2 receptor | P11717 |
| KCNJ2 | Inward rectifier potassium channel 2 | P63252 |
| KDR | Vascular endothelial growth factor receptor 2 | P35968 |
| LRP2 | Low-density lipoprotein receptor-related protein 2 | P98164 |
| OPN4 | Melanopsin | Q9UHM6 |
| P2RY1 | P2Y purinoceptor 1 | P47900 |
| SLC1A6 | Excitatory amino acid transporter 4 | P48664 |
| SSTR2 | Somatostatin receptor 2 | P30874 |
| STX2 | Syntaxin 2 | P32856 |
| VIPR1 | Vasoactive intestinal polypeptide receptor 1 | P32241 |
| Intracellular | ||
| CHAT | Choline O-acetyltransferase | P28329 |
| CRABP1 | Cellular retinoic acid-binding protein 1 | P29762 |
| DBH | Dopamine β hydroxylase | P09172 |
| NOS1 | Nitric oxide synthase 1 | P29475 |
| PCSK1 | Neuroendocrine convertase 1 | P29120 |
| RASGRF1 | Ras-specific guanine nucleotide-releasing factor 1 | Q13972 |
| RLBP1 | Retinaldehyde binding protein 1 | P12271 |
| RPE65 | Retinoid isomerohydrolase | Q16518 |
| SLC18A2 | Synaptic vesicular amine transporter | Q05940 |
| TH | Tyrosine 3-hydroxylase | P07101 |
| TYR | Tyrosinase | P14679 |
| TYRP1 | Tyrosinase-related protein 1 | P17643 |
| Transcription | ||
| EGR1 | Early growth response protein 1 | P18146 |
| FOS | Proto-oncogene c-Fos | P01100 |
| HIF1A | Hypoxia-inducible factor 1α | Q16665 |
| PER2 | Period circadian clock 2 | O15055 |
| RARB | Retinoic acid receptor β | P10826 |
| RXRB | Retinoid X receptor β | P28702 |
| Secreted | ||
| ACHE | Acetylcholinesterase | P22303 |
| APOA1 | Apolipoprotein A1 | P02647 |
| APOE | Apolipoprotein E | P02649 |
| BMP2 | Bone morphogenetic protein 2 | P12643 |
| BMP4 | Bone morphogenetic protein 4 | P12644 |
| CHRDL1 | Chordin-like protein 1 | Q9BU40 |
| CTGF | Connective tissue growth factor | P29279 |
| FGF1 | Fibroblast growth factor 1 | P05230 |
| FGF10 | Fibroblast growth factor 10 | O15520 |
| FIGF | Vascular endothelial growth factor D | O43915 |
| GCG | Glucagon | P01275 |
| IGF1 | Insulin-like growth factor 1 | P05019 |
| IGF2 | Insulin-like growth factor 2 | P01344 |
| IL1B | Interleukin 1β | P01584 |
| IL18 | Interleukin 18 | Q14116 |
| LIPG | Endothelial lipase | Q9Y5X9 |
| MMP2 | Matrix metallopeptidase 2 | P08253 |
| NOV | Protein NOV homolog | P48745 |
| NPY | Pro-neuropeptide Y | P01303 |
| NRG1 | Neuregulin 1 | Q02297 |
| NTS | Neurotensin | P30990 |
| NYX | Nyctalopin | Q9GZU5 |
| PENK | Proenkephalin A | P01210 |
| SERPINF1 | Pigment epithelium-derived factor | P36955 |
| SOSTDC1 | Sclerostin domain-containing protein 1 | Q6X4U4 |
| SST | Somatostatin | P61278 |
| TAC1 | Protachykinin 1 | P20366 |
| TGFB1 | Transforming growth factor β1 | P01137 |
| TGFB2 | Transforming growth factor β2 | P61812 |
| VIP | Vasoactive intestinal peptide | P01282 |
None of the treatment conditions affected the expression of the reference gene. Primer sequences, amplicon size, and efficiencies are listed in Supplementary Table S1. All primers were designed to work under the same cycling conditions. All amplicons were located within the coding region and most spanned at least one intron; amplicon identity was verified by gel electrophoresis and sequencing.
Relative gene expression was measured by qPCR on a StepOnePlus Real-Time PCR System using Power SYBR Green PCR Master Mix (both, Life Technologies). Reactions were performed in triplicate in a 15 μl volume containing 300 nM each primer and 0.4 μl cDNA template. Cycling parameters were the same for all assays: initial denaturation at 95°C for 10 minutes, followed by 40 cycles of 95°C for 15 seconds, 62°C for 60 seconds. Single gene products were obtained for all reactions as assessed by melt curve analysis. Relative gene expression was calculated using the ΔΔCt method (Livak and Schmittgen, 2001) to first normalize the expression level of the target gene to that of the reference gene, and then to compare the relative expression of the target gene for treated vs. control eyes. The geometric group mean (for the 7 biological replicates) of these expression ratios was used to calculate the fold change in gene expression for each of the target genes.
For both refractive and gene expression data, paired t-tests were used to assess treated-eye vs. control-eye differences. In all cases, p<0.05 was considered significant and no adjustment for possible false discovery rate was applied. Linear regressions between expression differences were made in SigmaPlot (Systat Software, San Jose, CA).
3. RESULTS
3.1 Refraction
When treated with a monocular minus lens, groups of tree shrews at this age typically do not develop a significant myopia in the treated eye until after two days of minus-lens wear (Norton et al., 2010). In the 6h group, pre-treatment measures showed that the refractions of the to-be-treated eyes (1.4 ± 0.5 D; mean ± SEM) were not significantly different from those of the control eyes (0.9 ± 0.4 D). These eyes were not re-measured at the end of the 6-h treatment period because no change was expected after this short exposure. In the 24h group, the post-treatment refractive difference (treated eyes - control eyes) showed the treated eyes to have developed a small, significant myopia of −0.4 ± 0.2 D. In the 24hC group, the difference (0.1 ± 0.1 D) was not significant.
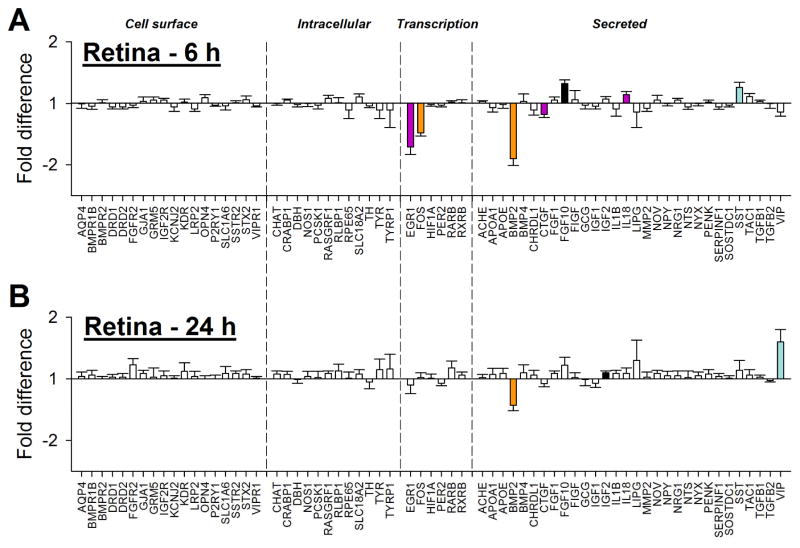
3.2 Gene expression – Retina
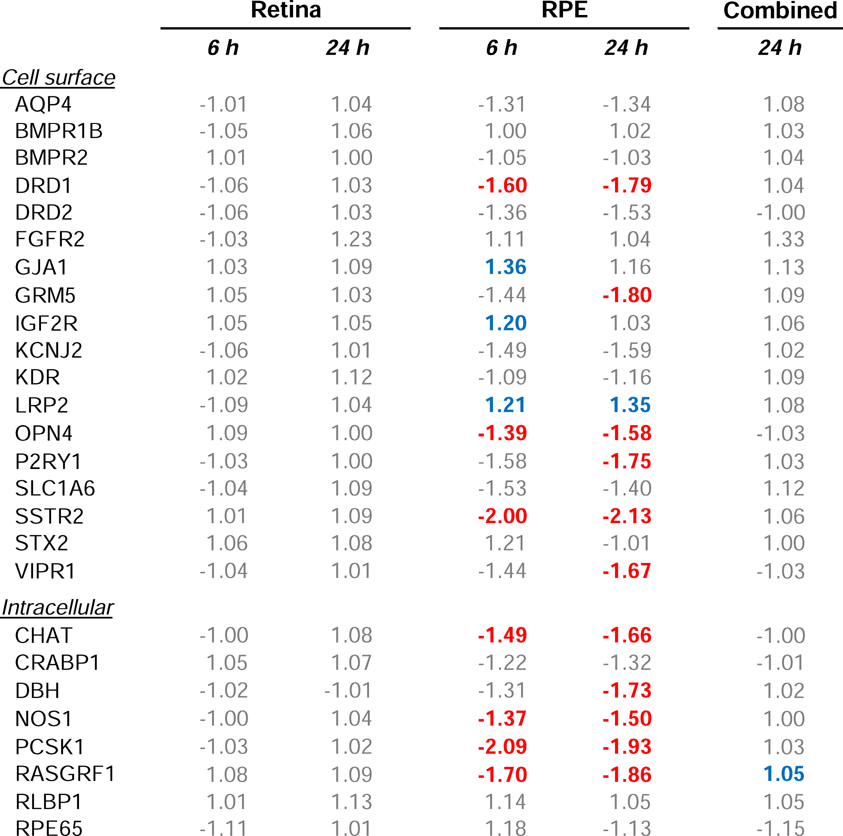
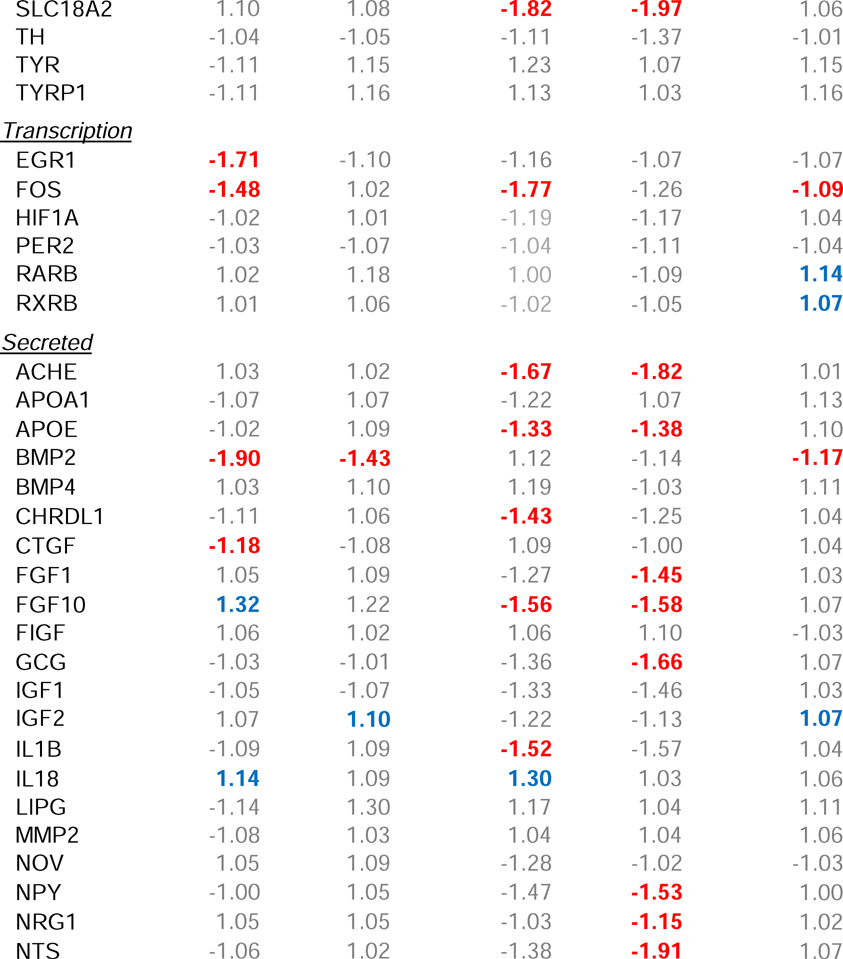
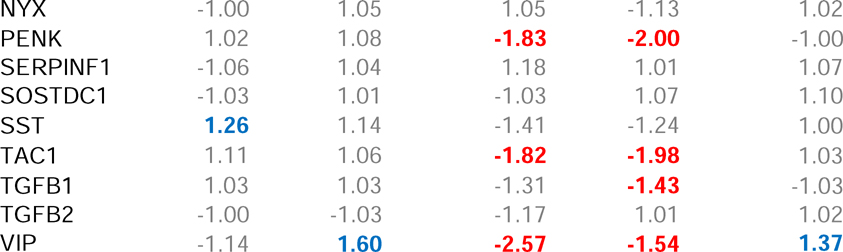
Minus-lens wear produced significant alteration in gene expression in the treated eye for genes whose protein products are involved in two of the four major groupings, transcriptional regulation (“transcription”) and those that are secreted (“secreted”) (Table 2), but not in genes whose protein products are involved in cell surface interactions (“cell surface”) or intracellular processing (“intracellular”). Six h of minus-lens wear produced significant differences in mRNA expression levels in the treated vs. control eyes for seven of the 66 genes examined (Figure 2A). The mRNA expression values are listed in Table 2. mRNA levels for the early intermediate genes EGR1 and FOS (transcription) were down-regulated in the treated eyes, as were BMP2 and CTGF (secreted); FGF10, IL18, and SST (secreted) were up-regulated. After 24 h, fewer genes were differentially expressed (Figure 2B, Table 2); BMP2 was still down-regulated, but mRNA expression of the other six genes was not significantly different. mRNA levels for two other genes, IGF2 and VIP (secreted), were up-regulated in the treated eyes.
Table 2.
Gene expression fold-differences comparing treated vs. control eyes. Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant.
Figure 2.
Retina gene expression fold-differences (treated vs. control eyes) in the (A) 6h and (B) 24h groups. Negative values represent down-regulation in the treated eyes. Headings separated by vertical dashed lines indicate cellular location/functional category of the gene’s protein product. Filled bars represent statistically significant differences (p<0.05); bar color is arbitrary and intended to help in comparing the same gene in the different conditions; error bars = SEM.
In Figure 3, the differential gene expression in the retina after 6 h is compared with the expression after 24 h for the 66 genes examined. BMP2 was the only gene significantly differentially expressed at both time points (down-regulated at both). The substantial down-regulation of EGR1 and FOS after 6 h of treatment was absent after 24 h. VIP, which was not differentially affected after 6 h, was up-regulated after 24 h. It thus appears that the pattern of gene expression, even amongst the small number of affected genes, was changing between these two time points.
Figure 3.

Comparison of the gene expression differences (treated vs. control eye) in retina after 6 h (Figure 2A) and 24 h (Figure 2B) of minus-lens wear. Values near the dashed line indicate genes that responded similarly at the two time points. Stars = significant fold-differences for both the 6h and 24h groups; triangles = significant fold-differences only for the 6h group; squares = significant fold-differences only for the 24h group; open circles = expression not significantly different at either time point.
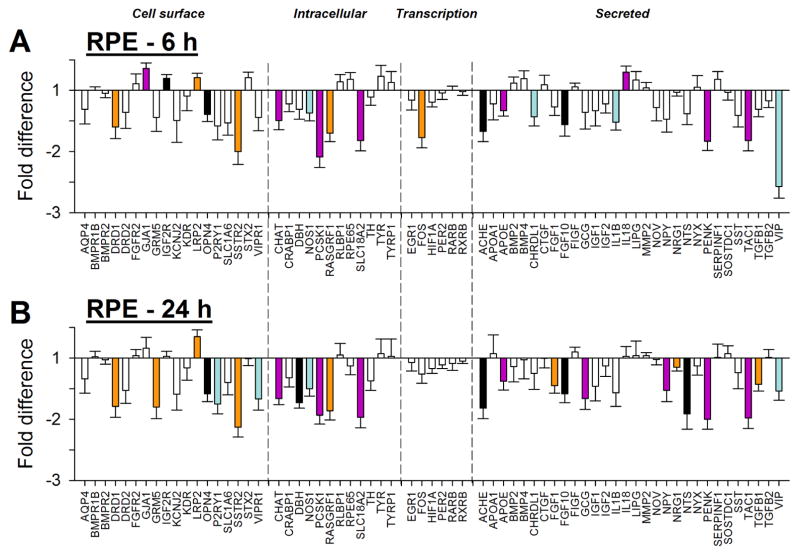
3.3 Gene expression – RPE
Minus-lens wear produced significant differences in gene expression for genes in all four of the major groupings: cell surface interaction, intracellular processing, transcriptional regulation, and secreted proteins. Six h of minus-lens wear produced significant differences in mRNA expression levels in the separate RPE tissue for 21 genes in the treated eyes relative to control eyes (Figure 4A, Table 2). The mRNA expression levels of 17 genes were down-regulated: DRD1, OPN4, and SSTR2 (cell surface); CHAT, NOS1, PCSK1, RASGRF1, and SLC18A2 (intracellular); FOS (transcription); ACHE, APOE, CDRDL1, FGF10, IL1B, PENK, TAC1, and VIP (secreted). Four genes were up-regulated: GJA1, IGF2R, and LRP2 (cell surface); IL18 (secreted).
Figure 4.
RPE gene expression fold-differences (treated vs. control eyes) in the (A) 6h and (B) 24h groups. Negative values represent down-regulation in the treated eyes. Headings separated by vertical dashed lines indicate cellular location/functional category of the gene’s protein product. Filled bars represent statistically significant differences (p<0.05); bar color is arbitrary and intended to help in comparing the same gene in the different conditions; error bars = SEM.
After 24 h of minus-lens treatment, a generally similar pattern of differential gene expression was found in the RPE, in terms of the number of genes that showed differential expression (25), which genes were, and were not, affected, and the direction of the differential gene expression (Table 2, Figure 4B). Fourteen genes remained significantly down-regulated and LRP2 remained up-regulated. mRNA for six genes no longer showed differential expression, while ten genes, not differentially expressed at 6 h, now were down-regulated in the treated eyes. The additional down-regulated genes were: GRM5, P2RY1, and VIPR1 (cell surface); DBH (intracellular); FGF1, GCG, NPY, NRG1, NTS, and TGFB1 (secreted).
In Figure 5, the differential gene expression in the RPE after 6 h is compared with the expression after 24 h for all of the 66 genes examined, including ones that did not show significant differential expression. At both time points, the same genes generally showed similar differences in mRNA levels, as indicated by the proximity of most genes to the 1:1 line in Figure 5. This was the case even for the cluster of genes in Figure 5 (blue square symbols) that were not significantly differentially expressed at 6 h but were significantly expressed at 24 h. The non-significant fold-differences at 6 h (Table 2) were only slightly smaller than the significant fold-differences at 24 h, suggesting that the altered expression at 24 h may have already started at 6 h. An exception to the similar pattern at 6 h and 24 h was mRNA expression for VIP, which was significantly differentially expressed at both time points, but was more strongly down-regulated after 6 h than after 24 h.
Figure 5.
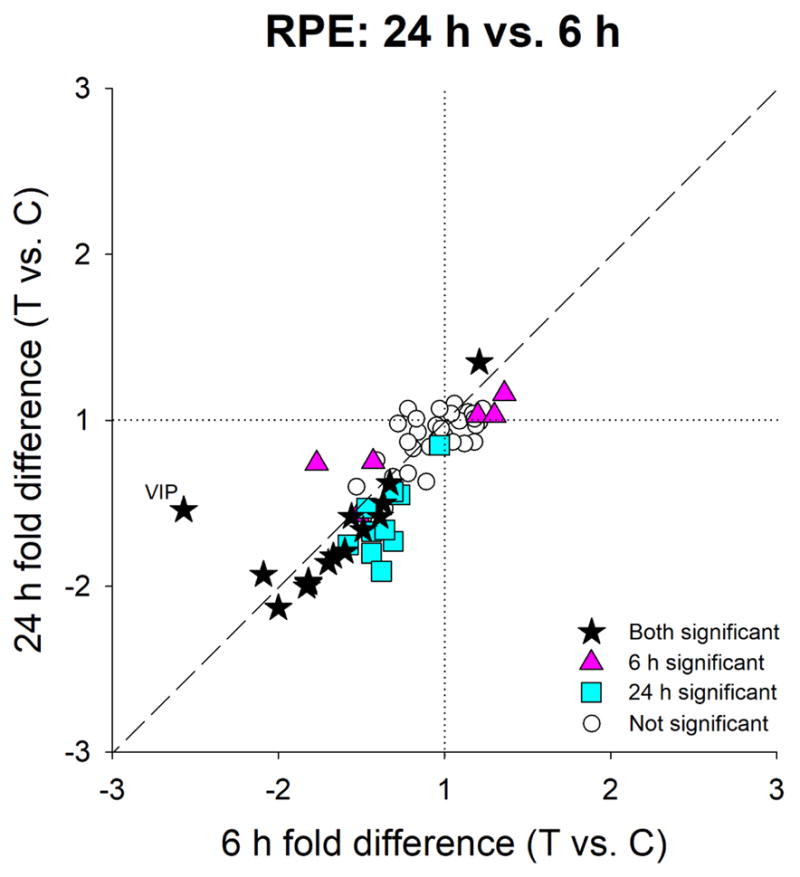
Comparison of the gene expression differences (treated vs. control eye) in RPE after 6 h (Figure 4A) and 24 h (Figure 4B) of minus-lens wear. Values near the dashed line indicate genes that responded similarly at the two time points. Stars = significant fold-differences for both 6h and 24h groups; triangles = significant fold-differences only for 6h; squares = significant fold-differences only for 24h; open circles = expression not significantly different at either time point.
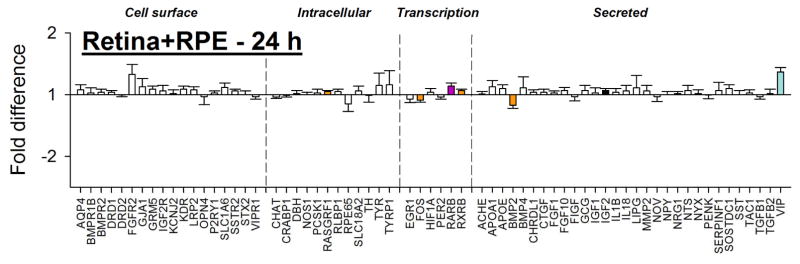
3.4 Gene expression – Combined retina+RPE
In the combined retina+RPE tissue (Figure 6 and Table 2), 24 h of minus-lens wear produced significant differences in mRNA expression levels for only seven genes. As was the case for differential expression in the separate retinal tissue, the affected genes were in the transcription and secreted categories, but not in the cell surface and intracellular groups (Table 2). Four genes (FOS and BMP2, down-regulated; IGF2 and VIP, up-regulated) showed the same expression in combined tissue as in retina alone at either 6 h or 24 h. mRNA for VIP was up-regulated in the combined tissue, as in retina alone, despite being significantly down-regulated in RPE. mRNA for three genes (RASGRF1, RARB, and RXRB) showed significant up-regulation in the combined retina+RPE tissue, but were not up-regulated in either separate retina or RPE. Indeed, RASGRF1 was down-regulated in RPE at 24 h. Overall, as may be seen in Table 2 and by comparing combined tissue (Figure 6) with separate retina (Figure 2B) and separate RPE (Figure 4B), it is evident that the gene expression pattern of the combined retina+RPE primarily represented gene expression in the retina. The significant expression differences in many genes that occurred in the RPE were not detected in the combined retina+RPE tissue.
Figure 6.
Combined retina+RPE gene expression fold-differences (treated vs. control eyes) in the 24hC group. Negative values represent down-regulation in the treated eyes. Headings separated by vertical dashed lines indicate cellular location/functional category of the gene’s protein product. Filled bars represent statistically significant differences (p<0.05); error bars = SEM.
4. DISCUSSION
This is the first report in tree shrews (mammals closely related to primates) that examined GO signaling with the same panel of genes in the retina and in the RPE, the first two compartments of the direct emmetropization signaling cascade. The pattern of differential gene expression in the retina changed between 6 h and 24 h, suggesting that the retinal GO signal has an initial and later component. The pattern of differential gene expression in the RPE was different than in the retina, and was similar at both time points, suggesting longer-term involvement in signals modulating axial elongation. We also found that the many differentially-expressed genes in RPE were not detected in combined retina+RPE. Comparison of the present results with previously-examined GO signaling in tree shrew choroid and sclera showed that GO signaling is represented by different gene expression “signatures” in each compartment of the direct emmetropization pathway.
4.1 Retinal gene expression
The identity of the retinal neurons that generate signals sent though the direct emmetropization pathway is not well understood, but likely includes populations of amacrine cells and bipolar cells (Fischer et al., 1999; Pickett-Seltner and Stell, 1995; Rohrer et al., 1995; Stell et al., 2003; Stell et al., 2004; Stone et al., 1988; Zhong et al., 2004) that are small in number in comparison to the many photoreceptors that convert light into neural activity along with the bipolar, horizontal, and ganglion cells that process visual signals. This larger population of neurons is involved in extracting information from the visual scene that is sent through central visual pathways to produce visual perception, guide accommodation, and produce eye movements. Still other neurons, including intrinsically-photosensitive retinal ganglion cells, are involved with pupillary responses and circadian rhythms. Thus, GO signaling produced by minus-lens wear in the small retinal neuronal population is difficult to detect against a much larger background of vision-related activity produced by minus-lens wear.
An exception may be emmetropization-related neurons that secrete unique neuromodulators/neurotransmitters, such as the sparse populations of amacrine cells (and perhaps bipolar and/or ganglion cells) that use dopamine (Rohrer et al., 1995), vasoactive intestinal polypeptide (VIP) (Stone et al., 1988), glucagon (in chicks) (Vessey et al., 2005), and possibly other secreted peptides (somatostatin, bone morphogenic protein 2) (Zhang et al., 2013) as signaling messengers. Alterations in mRNA expression in these neurons may be detectable, even though the amount of altered mRNA involved is small in comparison with total retinal mRNA, because it is not masked by the use of the same molecules in other retinal activities. Thus, it is not surprising that we found increased mRNA levels in the retina of the treated eyes after 24 h of minus-lens wear for VIP, which is produced only by a small population of amacrine cells (Casini and Brecha, 1991), even though mRNA for VIP is less abundant than is mRNA for most of the other genes in our sample (based on relative Ct values). A similar up-regulation of VIP was found in form-deprived monkeys (Stone et al., 1988; Tkatchenko et al., 2006). We also found differential expression of other genes with secreted protein products: BMP2, SST, and IGF2. BMP2 is broadly distributed in the retina, but seems to be concentrated in the outer segment region of the photoreceptors and in the RPE (Li et al., 2016; Zhang et al., 2012b) where it may play a regulatory role in retinal differentiation and ocular growth (Belecky-Adams and Adler, 2001; McGlinn et al., 2007; Sakuta et al., 2006; Stone et al., 2011; Zhang et al., 2012b). BMP2 was down-regulated in the tree shrew retina sample; it was the only gene in the retinal sample that was differentially expressed at both time points, suggesting that it may participate in a continuing signal, while the expression of other genes may participate early or slightly later in creating and maintaining retinal GO signaling. SST mediates nitric oxide production by activating the somatostatin 2 receptor in rat retina (Vasilaki et al., 2002), and nitric oxide is considered to be a potential regulator of ocular growth (Fujikado et al., 1997; Nickla et al., 2009).
We also found changes in mRNA expression for the early-response genes EGR1 and FOS (transcription), down-regulated at 6 h but not at 24 h. The EGR1 down-regulation in GO is in agreement with studies of retina in minus lens-treated chick (Ashby and Feldkaemper, 2010) and form-deprived mouse (Brand et al., 2007), although Ashby et al. (2010) found that down-regulation of ZENK (the avian analog of EGR1) continued when minus-lens treatment lasted for 10 days. The protein products of EGR1 and FOS affect many downstream pathways and may play a role in the early phases of retinal GO signaling. The reduction we found in EGR1 and FOS also is consistent with the up-regulation during STOP signaling of EGR1 and FOS proteins in tree shrew retinal amacrine cells, 1 – 3 h after removal of form deprivation or a minus lens (Stell et al., 2003; Stell et al., 2004).
Our finding that retinal signaling changed between 6 h and 24 h is similar to the result of many previous studies that examined altered gene expression in GO signaling (typically in retina+ RPE) at multiple time points in chicks (McGlinn et al., 2007; Stone and Khurana, 2010), mice (Brand et al., 2007), and monkeys (Zhong et al., 2004). This suggests that the strong, early (within hours of altered visual input) down-regulation of EGR1, FOS, and BMP2 are related to the onset of the GO signaling. After 24 h, the only differential gene expression in tree shrew retina was for genes in the secreted category, including VIP. These may be involved in output of GO signaling from the retina to the RPE.
4.2 RPE gene expression
The RPE is a polarized monolayer of cells located adjacent to the photoreceptors. In addition to its roles in light-cycle trafficking, in photoreceptor disk phagocytosis, and in establishing and maintaining the blood-retina barrier (Bito, 1977), it has become evident that emmetropization-related signals enter the RPE, are processed there, and then pass on to the choroid.
In the RPE, we detected emmetropization-related altered expression after 6 h for 21 genes; overall, 31 of the 66 candidate genes in our sample showed differential expression. The signature of differentially-expressed genes was similar at both time points. Fifteen of the genes that showed differential expression did so both after 6 h and 24 h of lens wear, including VIP, which was strongly down-regulated at 6 h. Of the 17 genes whose mRNA levels were down-regulated after 6 h, 14 still showed significant down-regulation after 24 h. The additional 10 genes whose mRNA was significantly down-regulated after 24 h also showed negative, but non-significant, fold-differences after 6 h (Figure 5, Table 2). These genes may have started to change their mRNA expression after 6 h but the size of the effect was too small to be statistically significant. Thus, the RPE GO signaling involves a relatively constant group of genes whose altered expression increases over time, at least within the first day of minus-lens wear.
Although the RPE shares a common embryological origin with the retina, it is a distinct monolayer of cells, with different functions than the retina, in which one might expect signaling produced by minus-lens wear must be transformed from retinal neural signals to another, potentially non-neural, signal. The differential gene expression signature in the RPE was consistent with a role in receiving, transforming, and transmitting GO signals produced by minus-lens wear. The differential mRNA expression for genes whose protein products are located at the cell surface and involved in intracellular signaling, not found in the retinal sample (Table 2), reinforces the suggestion that retinally-generated signals are received by RPE cells. DRD1 (a dopamine receptor) was down-regulated, as were NOS1 (involved in nitric oxide production) and VIPR1 (encodes a VIP receptor). The down-regulation, both at 6 h and 24 h, of mRNA for SSTR2, which encodes a somatostatin receptor, suggests a possible role for somatostatin in communicating emmetropization-related GO signaling from the retina to the RPE, though how retinally-generated somatostatin might reach the RPE is uncertain.
The differential expression of mRNA levels for many genes in RPE whose protein products are involved in intracellular signaling and transcription (Table 2) is consistent with the generation of a GO signal in RPE that is different from that in the retina. The differential expression of genes whose protein products are secreted is consistent with a role in transmitting emmetropization-related signals to the next compartment of the direct emmetropization pathway, the choroid.
It was interesting to find, in tree shrew RPE, that expression of mRNA for neither BMP2 nor BMP4 was significantly altered after either 6 h or 24 h. In contrast, down-regulation of mRNA for BMP2 has been found in chick RPE after 2 h of minus-lens wear (Zhang et al., 2012b). However, expression of BMP2 was not significantly reduced in chick after 48 h. It is unclear if the differing results are due to the different durations of minus-lens wear in the two studies, or to a species difference.
4.3 Gene expression in combined retina+RPE
The gene expression pattern of the 66 target genes in the combined retina+RPE after 24 h of minus-lens wear was different from the expression pattern in either the retina alone or the RPE alone (Table 2). It more closely resembled the pattern in the retinal sample, as might be expected from the larger amount of retinal mRNA compared with mRNA from the RPE. In the combined retina+RPE, seven genes showed significant regulation. mRNA for three genes (BMP2, down-regulated; IGF2 and VIP, up-regulated) showed the same differential expression in the retina+RPE as in the retina. However, four genes that were not differentially expressed in the retina showed differential expression in the combined tissue.
Similarly, and more dramatically, the gene expression in the combined retina+RPE did not provide an accurate picture of RPE gene expression. Most of the genes that were differentially expressed in RPE did not show differential expression in the combined retina+RPE. The inability to detect differentially-expressed RPE genes in the combined tissue likely is because the monolayer of RPE cells contributed only a small fraction (about 1/7th) of the total RNA in the retina+RPE sample. Apparently, the large amount of mRNA in the combined tissue, from genes in retina that were not differentially expressed, made it impossible to detect the differential expression in the RPE. In instances, such as VIP, where there was significant, but opposite, differential gene expression in the two tissues, the combined tissue (up-regulation) reflected the expression in the retina; this over-rode the significant down-regulation of VIP in the RPE. These results clearly show that to study expression changes in RPE, it must be examined as a separate tissue.
Although the combined retina+RPE does not provide the same result as retina or RPE alone, it is still useful to compare our results with other studies that have examined the combined tissues. In our sample of 66 candidate genes in the combined retina+RPE, mRNA levels were significantly regulated in only seven genes after 24 h. In contrast, Stone et al. (2011), who also used minus-lens wear as the GO stimulus, examined combined retina+RPE with an array of over 28,000 genes and found differential expression of over 2,600 gene transcripts in chicks, measured after 6 h or 3 days. Nine of the genes in our study were found by Stone et al. to change in chick at the 6-h or 3-day time points. Only two of these genes were found to change significantly in our tree shrew combined retina+RPE: BMP2 was down-regulated in both; VIP was down-regulated in chick retina+RPE and up-regulated in tree shrew (as in monkey) (Stone et al., 2011; Tkatchenko et al., 2006). The other seven differentially-expressed genes in chick (six of them down-regulated) were not differentially expressed in tree shrew combined retina+RPE. The other five genes that were differentially regulated in tree shrew combined tissue were not affected in chick. How many of these differences were due to species differences, or other factors, is unclear.
4.4 Strengths and limitations
A strength of this study is that it examined gene expression in tree shrews, mammals with excellent vision and ocular structures similar to humans, such as an all-fibrous sclera. To the extent that results in tree shrew are similar to those in other species, it suggests that similar mechanisms are involved in the direct emmetropization signaling cascade. Differing results may reflect species differences, but also may be due to experimental details.
Another strength is that the data from each gene in each eye of each animal was examined, in triplicate; mRNA was not pooled. This allowed us to detect the relatively low fold-differences that we, and others, have found in previous studies. That signaling in the direct emmetropization signaling cascade involves small fold-differences is reassuring; it suggests that the minus-lens wear used to generate GO signaling causes only physiological-level perturbations of the ongoing gene expression.
A limitation is the number of candidate genes and time points that could be examined. For each gene, tree shrew-specific primers were developed and validated, a time-consuming process needed to ensure consistent and accurate results. Because many genes might participate in GO signaling, and it is not known whether any particular gene is critically important, we considered with some care which candidate genes to include. Also, we considered that absence of differential expression was as important as a positive response. However, it is virtually certain there are other genes, that we did not examine, whose expression may be regulated in retina and/or in RPE, that may be important parts of GO signaling in the direct emmetropization cascade. In addition, expression may be altered by circadian rhythms. Despite these limitations, the sample was sufficient to clearly show that gene expression signatures are very different in the retina and the RPE, and to show the importance of examining each tissue separately.
Detecting differential expression of mRNA levels is not necessarily an indication of alterations in the amount of the protein products. In complex biological systems, it has been found that alterations in mRNA abundance are often poorly correlated with changes in protein levels (Csárdi et al., 2015; Gygi et al., 1999; Kumar et al., 2016; Liu et al., 2016; Maier et al., 2009). This may be due, in part, to the fact that up- or down-regulation of mRNA may be necessary simply to maintain a certain level of protein in tissues where many interacting factors and feedback loops are at play. Rather, altered mRNA abundance serves as an indicator of a response by the cells in the target tissue.
Although the signature of differential gene expression involves altered expression in numerous genes, even in our limited sample, this approach is not able to determine if there is a single molecule (or a few key molecules) that is primarily responsible for GO signaling in each compartment. Systems biology analyses provide useful information about protein interactions but do not show which of the secreted proteins whose mRNA levels change may be “key” signaling elements. The large number of genes that showed differential expression suggests that altered abundance of several, or even many, proteins may constitute the GO signal.
4.5 Signaling in the direct emmetropization pathway
A contribution of the present study, taken together with our previous studies of gene expression in the choroid (He et al., 2014a, b) and sclera (Guo et al., 2013; Guo et al., 2014) is the conclusion that GO signaling, as assessed with altered gene expression, differs in each compartment of the pathway. In the retina, RPE, choroid, and the sclera, different categories of genes are up- or down-regulated in each compartment, as are the specific genes that show differential expression and whether they are up- or down-regulated.
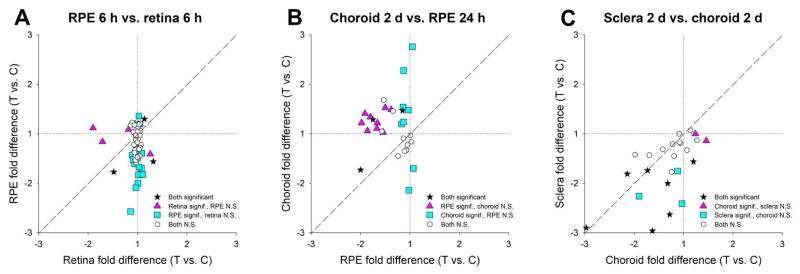
In the retina, altered gene expression was consistent with the origination of GO signaling (detection of the imposed hyperopia) that may be transmitted by secreted molecules to cell-surface receptors in the RPE. In the RPE, the GO signaling also involved altered gene expression for numerous genes whose protein products are secreted, a potential mechanism for transmitting GO signaling to the choroid. Both the larger number of genes and the specific genes that showed differential expression provided evidence that the GO signaling in RPE is distinct from that in the retina. As shown in Fig. 7A, after 6 h of minus-lens wear, only three genes were differentially expressed in both the retina and the RPE: FOS (down-regulated in both), IL18 (up-regulated in both), and FGF10 (up-regulated in retina, down-regulated in RPE). After 24 h, VIP was up-regulated in retina, whilst it was down-regulated in RPE after both 6 h and 24 h of lens wear. None of the other five genes that showed differential expression in retina were significantly regulated in RPE, and none of the other 27 genes found to be differentially expressed in RPE were differentially expressed in retina. Thus, the gene expression signatures were very different in the first two compartments of the direct emmetropization signaling cascade.
Figure 7.
Comparison of the GO gene expression across compartments of the direct emmetropization signaling cascade. (A) Differential expression in retina and RPE after 6 h of minus-lens wear. (B) Differential expression in RPE after 24 h and choroid after 2 days of minus-lens wear. (C) Differential expression in choroid and sclera after 2 days of minus-lens wear. Values near the dashed line indicate genes that responded similarly in both compartments. Stars = significant fold-differences for both compartments; triangles = significant fold-differences only for the compartment plotted on the x-axis; squares = significant fold-differences only for the compartment plotted on the y-axis; open circles = expression not significantly different in either compartment. Choroid data from He et al. 2014a. Sclera data from Guo et al. 2013.
As shown by comparing differential gene expression in RPE and choroid, GO signaling in the choroid is very different from GO signaling in the RPE. In a previous paper, we examined differential gene expression in choroid for 30 of the same genes sampled in the RPE (He et al., 2014a), including representatives from all four categories in Table 2 (values are provided in supplementary Table S2). As shown in Figure 7B, most of the differentially-expressed genes in RPE were down-regulated after 24 h, whereas many of the differentially-expressed genes in choroid after 2 days of minus-lens wear were up-regulated. Most of the differentially-expressed genes in RPE were not differentially expressed in choroid and vice versa. Three genes were differentially expressed in both RPE and choroid. Of these, two were down-regulated in RPE and up-regulated in choroid; one was down-regulated in both. The very different gene expression signature in these two compartments, albeit at different times after the start of minus-lens wear and with a limited sample of genes, suggests that GO signaling is encoded very differently in these compartments of the direct emmetropization signaling cascade.
GO signaling in the sclera also was different from that in the choroid, but the difference was not as striking as between the RPE and choroid. Twenty-four genes were examined in both compartments after two days of minus-lens wear (Guo et al., 2013; He et al., 2014a), including representatives of all four categories in Table 2. As shown in Figure 7C, the general pattern was down-regulation (values are provided in Table S3). All of the differentially-expressed genes in sclera were down-regulated, including three that were not differentially expressed in choroid. Seven genes were down-regulated in both, but with different fold-differences in choroid and sclera. One gene was up-regulated in choroid and down-regulated in sclera. Two were up-regulated in choroid but not differentially expressed in sclera.
Thus, viewed across all four compartments of the direct emmetropization signaling cascade, the gene expression signatures in response to the same GO stimulus are different in each compartment. In each there is differential gene expression that may be part of a signal for increased axial elongation rate, but the GO signal, as assessed with altered mRNA levels, is tissue specific; GO signaling is transformed as it is passes through the compartments of the direct pathway.
It also may be important that most of the genes that were differentially expressed were down-regulated in each compartment. One might wonder if this consistent down-regulation may imply that GO signaling in this pathway involves lowered signal activity. Perhaps, a certain flow of secreted molecules is needed to control the axial elongation rate in juvenile tree shrews that have achieved a near-to-emmetropia low hyperopia. Reducing the flow of these molecules may, surprisingly, constitute GO signaling.
A topic not considered in the present study is whether the GO signaling in each compartment of the direct pathway differs depending on the visual stimulus. The present study only used minus-lens wear. In the choroid and the sclera, it has been found that nearly identical differential gene expression signatures are produced by form deprivation and by dark treatments (Guo et al., 2013; He et al., 2014b). It would be expected that these stimuli would produce different patterns of differential gene expression in the retina, but it may be of interest for future studies to learn if the GO signaling in the RPE differs with the visual stimulus, or if the signaling is simplified such that any retinal GO signal produces the same response in the rest of the emmetropization signaling cascade.
Supplementary Material
Primers used: Sequences, amplicon sizes, and efficiencies
Gene expression fold-differences comparing treated vs. control eyes. Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant.
Gene expression fold-differences comparing treated vs. control eyes. Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant.
Juvenile tree shrews wore a monocular −5 D lens for 6 hours or 24 hours
mRNA expression was measured for 66 genes in retina, RPE, and combined retina+RPE
In retina, differential expression changed between 6 h and 24 h
In RPE, the pattern of expression was similar at both time points
Abundant differential gene expression in RPE was not detected in retina+RPE
Acknowledgments
This study was supported by NIH grants EY005922 and EY003039 (P30). Li He was supported in part by a supplement to EY005922 and by funds from the Department of Vision Sciences. This work was performed in partial fulfillment of the requirements for the degree of Doctor of Philosophy at the University of Alabama at Birmingham (Li He). Preliminary results were presented in abstract form.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ashby R, Kozulin P, Megaw PL, Morgan IG. Alterations in ZENK and glucagon RNA transcript expression during increased ocular growth in chickens. Mol Vis. 2010;16:639–649. [PMC free article] [PubMed] [Google Scholar]
- Ashby RS, Feldkaemper MP. Gene expression within the amacrine cell layer of chicks after myopic and hyperopic defocus. Invest Ophthalmol Vis Sci. 2010;51:3726–3735. doi: 10.1167/iovs.09-4615. [DOI] [PubMed] [Google Scholar]
- Belecky-Adams T, Adler R. Developmental expression patterns of bone morphogenetic proteins, receptors, and binding proteins in the chick retina. J Comp Neurol. 2001;430:562–572. [PubMed] [Google Scholar]
- Bito LZ. The physiology and pathophysiology of intraocular fluids. Exp Eye Res. 1977;25:273–289. doi: 10.1016/s0014-4835(77)80024-9. [DOI] [PubMed] [Google Scholar]
- Brand C, Schaeffel F, Feldkaemper MP. A microarray analysis of retinal transcripts that are controlled by image contrast in mice. Mol Vis. 2007;13:920–932. [PMC free article] [PubMed] [Google Scholar]
- Casini G, Brecha NC. Vasoactive intestinal polypeptide-containing cells in the rabbit retina: Immunohistochemical localization and quantitative analysis. J Comp Neurol. 1991;305:313–327. doi: 10.1002/cne.903050212. [DOI] [PubMed] [Google Scholar]
- Christian PG, Harkin DG, Rayner C, Schmid KL. Comparative effects of posterior eye cup tissues from myopic and hyperopic chick eyes on cultured scleral fibroblasts. Exp Eye Res. 2013;107:11–20. doi: 10.1016/j.exer.2012.11.005. [DOI] [PubMed] [Google Scholar]
- Csárdi G, Franks A, Choi DS, Airoldi EM, Drummond DA. Accounting for experimental noise reveals that mRNA levels, amplified by post-transcriptional processes, largely determine steady-state protein levels in yeast. PLOS Genetics. 2015;11:e1005206. doi: 10.1371/journal.pgen.1005206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillingham CM, Guggenheim JA, Erichsen JT. Disruption of the centrifugal visual system inhibits early eye growth in chicks. Invest Ophthalmol Vis Sci. 2013;54:3632–3643. doi: 10.1167/iovs.12-11548. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and focus-dependent expression of the transcription factor ZENK in the chick retina. Nature Neurosci. 1999;2:706–712. doi: 10.1038/11167. [DOI] [PubMed] [Google Scholar]
- Fischer AJ, Seltner RLP, Stell WK. N-methyl- D-aspartate-induced excitotoxicity causes myopia in hatched chicks. Can J Ophthalmol. 1997;32:373–377. [PubMed] [Google Scholar]
- Frost MR, Guo L, Norton TT. Whole transcriptome analysis of tree shrew sclera during the development of lens-induced myopia. Invest Ophthalmol Vis Sci. 2012;53 doi: 10.1167/iovs.11-8354. ARVO E-abstract 3452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujikado T, Kawasaki Y, Suzuki A, Ohmi G, Tano Y. Retinal function with lens-induced myopia compared with form-deprivation myopia in chicks. Graefes Arch Clin Exp Ophthalmol. 1997;235:320–324. doi: 10.1007/BF01739642. [DOI] [PubMed] [Google Scholar]
- Gamlin PDR. Subcortical neural circuits for ocular accommodation and vergence in primates. Oph Phys Optics. 1999;19:81–89. doi: 10.1046/j.1475-1313.1999.00434.x. [DOI] [PubMed] [Google Scholar]
- Gamlin PDR, Zhang H, Harlow A, Barbur JL. Pupil responses to stimulus color, structure and light flux increments in the rhesus monkey. Vision Res. 1998;38:3353–3358. doi: 10.1016/s0042-6989(98)00096-0. [DOI] [PubMed] [Google Scholar]
- Gao H, Frost MR, Siegwart JT, Jr, Norton TT. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis. 2011;17:903–919. [PMC free article] [PubMed] [Google Scholar]
- Glickstein M, Millodot M. Retinoscopy and eye size. Science. 1970;168:605–606. doi: 10.1126/science.168.3931.605. [DOI] [PubMed] [Google Scholar]
- Grytz R, Siegwart JT., Jr Changing material properties of the tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol Vis Sci. 2015;56:2065–2078. doi: 10.1167/iovs.14-15352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Frost MR, He L, Siegwart JT, Norton TT. Gene expression signatures in tree shrew sclera in response to three myopiagenic conditions. Invest Ophthalmol Vis Sci. 2013;54:6806–6819. doi: 10.1167/iovs.13-12551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo L, Frost MR, Siegwart JT, Jr, Norton TT. Scleral gene expression during recovery from myopia compared with expression during myopia development in tree shrew. Mol Vis. 2014;20:1643–1659. [PMC free article] [PubMed] [Google Scholar]
- Gygi SP, Rochon Y, Franza BR, Aebersold R. Correlation between protein and mRNA abundance in yeast. Mol Cell Biol. 1999;19:1720–1730. doi: 10.1128/mcb.19.3.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond DS, Wildsoet CF. Lens defocus-induced changes In the ocular somatostatin signaling pathway of chicks. Invest Ophthalmol Vis Sci. 2012;53 ARVO E-abstract 3429. [Google Scholar]
- He L, Frost MR, Siegwart JT, Jr, Filios SR, Norton TT. Retinal gene expression signatures in tree shrew in response to three myopiagenic visual conditions: Minus lens, form deprivation, and darkness. Invest Ophthalmol Vis Sci. 2011;52 ARVO E-abstract 6301. [Google Scholar]
- He L, Frost MR, Siegwart JT, Jr, Norton TT. Gene expression signatures in tree shrew choroid during lens-induced myopia and recovery. Exp Eye Res. 2014a;123:56–71. doi: 10.1016/j.exer.2014.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Frost MR, Siegwart JT, Jr, Norton TT. Gene expression signatures in tree shrew choroid in response to three myopiagenic conditions. Vision Res. 2014b;102:52–63. doi: 10.1016/j.visres.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iuvone PM, Tigges M, Stone RA, Lambert S, Laties AM. Effects of apomorphine, a dopamine receptor agonist, on ocular refraction and axial elongation in a primate model of myopia. Invest Ophthalmol Vis Sci. 1991;32:1674–1677. [PubMed] [Google Scholar]
- Kumar D, Bansal G, Narang A, Basak T, Abbas T, Dash D. Integrating transcriptome and proteome profiling: Strategies and applications. Proteomics. 2016;16:2533–2544. doi: 10.1002/pmic.201600140. [DOI] [PubMed] [Google Scholar]
- Li H, Wu J, Cui D, Zeng J. Retinal and choroidal expression of BMP-2 in lens-induced myopia and recovery from myopia in guinea pigs. Mol Med Rep. 2016;13:2671–2676. doi: 10.3892/mmr.2016.4843. [DOI] [PubMed] [Google Scholar]
- Liu Y, Beyer A, Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maier T, Güell M, Serrano L. Correlation of mRNA and protein in complex biological samples. FEBS Letters. 2009;583:3966–3973. doi: 10.1016/j.febslet.2009.10.036. [DOI] [PubMed] [Google Scholar]
- McGlinn AM, Baldwin DA, Tobias JW, Budak MT, Khurana TS, Stone RA. Form-deprivation myopia in chick induces limited changes in retinal gene expression. Invest Ophthalmol Vis Sci. 2007;48:3430–3436. doi: 10.1167/iovs.06-1538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moring AG, Baker JR, Norton TT. Modulation of glycosaminoglycan levels in tree shrew sclera during lens-induced myopia development and recovery. Invest Ophthalmol Vis Sci. 2007;48:2947–2956. doi: 10.1167/iovs.06-0906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickla DL, Damyanova P, Lytle G. Inhibiting the neuronal isoform of nitric oxide synthase has similar effects on the compensatory choroidal and axial responses to myopic defocus in chicks as does the non-specific inhibitor l-NAME. Exp Eye Res. 2009;88:1092–1099. doi: 10.1016/j.exer.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickla DL, Wallman J. The multifunctional choroid. Prog Retin Eye Res. 2010;29:144–168. doi: 10.1016/j.preteyeres.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TT. Animal models of myopia: Learning how vision controls the size of the eye. ILAR Journal. 1999;40:59–77. doi: 10.1093/ilar.40.2.59. [DOI] [PubMed] [Google Scholar]
- Norton TT, Amedo AO, Siegwart JT., Jr The effect of age on compensation for a negative lens and recovery from lens-induced myopia in tree shrews (Tupaia glis belangeri) Vision Res. 2010;50:564–576. doi: 10.1016/j.visres.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton TT, Essinger JA, McBrien NA. Lid-suture myopia in tree shrews with retinal ganglion cell blockade. Vis Neurosci. 1994;11:143–153. doi: 10.1017/s0952523800011184. [DOI] [PubMed] [Google Scholar]
- Norton TT, McBrien NA. Normal development of refractive state and ocular component dimensions in the tree shrew (Tupaia belangeri) Vision Res. 1992;32:833–842. doi: 10.1016/0042-6989(92)90026-f. [DOI] [PubMed] [Google Scholar]
- Norton TT, Rada JA. Reduced extracellular matrix in mammalian sclera with induced myopia. Vision Res. 1995;35:1271–1281. doi: 10.1016/0042-6989(94)00243-f. [DOI] [PubMed] [Google Scholar]
- Norton TT, Siegwart JT, German AJ, Jr, Robertson J, Wu WW. Comparison of cycloplegic streak retinoscopy with autorefractor measures in tree shrew eyes with, and without, myopia. Invest Ophthalmol Vis Sci. 2000;41 ARVO Abstract 2990. [Google Scholar]
- Norton TT, Wu WW, Siegwart JT., Jr Refractive state of tree shrew eyes measured with cortical visual evoked potentials. Optom Vis Sci. 2003;80:623–631. doi: 10.1097/00006324-200309000-00006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips JR, Khalaj M, McBrien NA. Induced myopia associated with increased scleral creep in chick and tree shrew eyes. Invest Ophthalmol Vis Sci. 2000;41:2028–2034. [PubMed] [Google Scholar]
- Pickett-Seltner RL, Stell WK. The effect of vasoactive intestinal peptide on development of form deprivation myopia in the chick: A pharmacological and immunocytochemical study. Vision Res. 1995;35:1265–1270. doi: 10.1016/0042-6989(94)00244-g. [DOI] [PubMed] [Google Scholar]
- Rohrer B, Iuvone PM, Stell WK. Stimulation of dopaminergic amacrine cells by stroboscopic illumination or fibroblast growth factor (bFGF, FGF-2) injections: possible roles in the prevention of form deprivation myopia in the chick. Brain Res. 1995;686:169–181. doi: 10.1016/0006-8993(95)00370-6. [DOI] [PubMed] [Google Scholar]
- Rohrer B, Stell WK. Basic fibroblast growth factor (bFGF) and transforming growth factor beta (TGF-β) act as stop and go signals to modulate postnatal ocular growth in the chick. Exp Eye Res. 1994;58:553–561. doi: 10.1006/exer.1994.1049. [DOI] [PubMed] [Google Scholar]
- Rymer J, Wildsoet CF. The role of the retinal pigment epithelium in eye growth regulation and myopia: A review. Vis Neurosci. 2005;22:251–261. doi: 10.1017/S0952523805223015. [DOI] [PubMed] [Google Scholar]
- Sakuta H, Takahashi H, Shintani T, Etani K, Aoshima A, Noda M. Role of bone morphogenic protein 2 in retinal patterning and retinotectal projection. J Neurosci. 2006;26:10868–10878. doi: 10.1523/JNEUROSCI.3027-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffel F, Howland HC. Mathematical model of emmetropization in the chicken. J Opt Soc Am A, Opt Image Sci Vis. 1988;5:2080–2086. doi: 10.1364/josaa.5.002080. [DOI] [PubMed] [Google Scholar]
- Schaeffel F, Troilo D, Wallman J, Howland HC. Developing eyes that lack accommodation grow to compensate for imposed defocus. Vis Neurosci. 1990;4:177–183. doi: 10.1017/s0952523800002327. [DOI] [PubMed] [Google Scholar]
- Seko Y, Tanaka Y, Tokoro T. Scleral cell-growth is influenced by retinal-pigment epithelium in vitro. Graefes Arch Clin Exp Ophthalmol. 1994;232:545–552. doi: 10.1007/BF00181998. [DOI] [PubMed] [Google Scholar]
- Shelton L, Troilo D, Lerner MR, Gusev Y, Brackett DJ, Summers Rada JA. Microarray analysis of choroid/RPE gene expression in marmoset eyes undergoing changes in ocular growth and refraction. Mol Vis. 2008;14:1465–1479. [PMC free article] [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Selective regulation of MMP and TIMP mRNA levels in tree shrew sclera during minus lens compensation and recovery. Invest Ophthalmol Vis Sci. 2005;46:3484–3492. doi: 10.1167/iovs.05-0194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Goggles for controlling the visual environment of small animals. Lab Anim Sci. 1994;44:292–294. [PubMed] [Google Scholar]
- Siegwart JT, Jr, Norton TT. Regulation of the mechanical properties of tree shrew sclera by the visual environment. Vision Res. 1999;39:387–407. doi: 10.1016/s0042-6989(98)00150-3. [DOI] [PubMed] [Google Scholar]
- Stell WK, Tao J, Karkhanis A, Siegwart JT, Jr, Norton TT. Intensity- and focus-dependent modulation of activity (expression of immediate-early gene products) in retinal interneurons of the tree shrew, Tupaia glis belangeri. J Vis. 2003;3:53. [Google Scholar]
- Stell WK, Tao J, Karkhanis A, Siegwart JT, Jr, Norton TT. Amacrine cells responsive to optical conditions regulating eye growth in the tree shrew, Tupaia glis belangeri. Invest Ophthalmol Vis Sci. 2004;45 ARVO E-abstract 1159. [Google Scholar]
- Stone RA, Khurana TS. Gene profiling in experimental models of eye growth: Clues to myopia pathogenesis. Vision Res. 2010;50:2322–2333. doi: 10.1016/j.visres.2010.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RA, Laties AM, Raviola E, Wiesel TN. Increase in retinal vasoactive intestinal polypeptide after eyelid fusion in monkeys. Proceedings of the National Academy of Science USA. 1988;85:257–260. doi: 10.1073/pnas.85.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone RA, Lin T, Iuvone PM, Laties AM. Postnatal control of ocular growth: Dopaminergic mechanisms. In: Bock G, Widdows K, editors. Myopia and the control of eye growth (Ciba Foundation Symposium) John Wiley & Sons; Chichester: 1990. pp. 45–57. discussion 57–62. [DOI] [PubMed] [Google Scholar]
- Stone RA, McGlinn AM, Baldwin DA, Tobias JW, Iuvone PM, Khurana TS. Image defocus and altered retinal gene expression in chick: Clues to the pathogenesis of ametropia. Invest Ophthalmol Vis Sci. 2011;52:5765–5777. doi: 10.1167/iovs.10-6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers Rada JA, Shelton S, Norton TT. The sclera and myopia. Exp Eye Res. 2006;82:185–200. doi: 10.1016/j.exer.2005.08.009. [DOI] [PubMed] [Google Scholar]
- Tkatchenko AV, Walsh PA, Tkatchenko TV, Gustincich S, Raviola E. Form deprivation modulates retinal neurogenesis in primate experimental myopia. Proc Natl Acad Sci USA. 2006;103:4681–4686. doi: 10.1073/pnas.0600589103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troilo D. Experimental studies of emmetropization in the chick. In: Bock G, Widdows K, editors. Myopia and the control of eye growth (Ciba Foundation Symposium) John Wiley & Sons; Chichester: 1990. pp. 89–102. discussion 102–114. [DOI] [PubMed] [Google Scholar]
- Vasilaki A, Mouratidou M, Schulz S, Thermos K. Somatostatin mediates nitric oxide production by activating sst2 receptors in the rat retina. Neuropharmacology. 2002;43:899–909. doi: 10.1016/s0028-3908(02)00181-8. [DOI] [PubMed] [Google Scholar]
- Vessey KA, Rushforth DA, Stell WK. Glucagon- and secretin-related peptides differentially alter ocular growth and the development of form-deprivation myopia in chicks. Invest Ophthalmol Vis Sci. 2005;46:3932–3942. doi: 10.1167/iovs.04-1027. [DOI] [PubMed] [Google Scholar]
- Wallman J, Winawer J. Homeostasis of eye growth and the question of myopia. Neuron. 2004;43:447–468. doi: 10.1016/j.neuron.2004.08.008. [DOI] [PubMed] [Google Scholar]
- Wildsoet CF. Active emmetropization - evidence for its existence and ramifications for clinical practice. Oph Phys Optics. 1997;17:279–290. [PubMed] [Google Scholar]
- Wildsoet CF. Neural pathways subserving negative lens-induced emmetropization in chicks – Insights from selective lesions of the optic nerve and ciliary nerve. Curr Eye Res. 2003;27:371–382. doi: 10.1076/ceyr.27.6.371.18188. [DOI] [PubMed] [Google Scholar]
- Wildsoet CF, McFadden SA. Optic nerve section does not prevent form deprivation-induced myopia or recovery from it in the mammalian eye. Invest Ophthalmol Vis Sci. 2010;51 ARVO E-abstract 1737. [Google Scholar]
- Young TL, Raviola E, Russell ME, Wiesel TN. Upregulation of vasoactive intestinal polypeptide (VIP) gene expression in the retina of myopic eyes following lid fusion in monkeys. Invest Ophthalmol Vis Sci. 1994;35 ARVO abstract 2069. [Google Scholar]
- Zhang Y, Liu Y, Ho C, Hammond D, Wildsoet CF. Differential expression of BMP7, TGF-β2, and noggin in chick RPE after imposed optical defocus. Invest Ophthalmol Vis Sci. 2012a;53 ARVO E-abstract 3458. [Google Scholar]
- Zhang Y, Liu Y, Ho C, Wildsoet CF. Effects of imposed defocus of opposite sign on temporal gene expression patterns of BMP4 and BMP7 in chick RPE. Exp Eye Res. 2013;109:98–106. doi: 10.1016/j.exer.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Wildsoet CF. Bidirectional, optical sign-dependent regulation of BMP2 gene expression in chick retinal pigment epithelium. Invest Ophthalmol Vis Sci. 2012b;53:6072–6080. doi: 10.1167/iovs.12-9917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu Y, Xu J, Nimri N, Wildsoet CF. Microarray analysis of RPE gene expression in chicks during long-term imposed myopic defocus. Invest Ophthalmol Vis Sci. 2010;51 ARVO E-abstract 3680. [Google Scholar]
- Zhang Y, Wildsoet CF. RPE and choroid mechanisms underlying ocular growth and myopia. In: Hejtmancik JF, Nickerson JM, editors. Progress in Molecular Biology and Translational Science. Academic Press; 2015. pp. 221–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong X, Ge J, Smith EL, III, Stell WK. Image defocus modulates activity of bipolar and amacrine cells in macaque retina. Invest Ophthalmol Vis Sci. 2004;45:2065–2074. doi: 10.1167/iovs.03-1046. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Primers used: Sequences, amplicon sizes, and efficiencies
Gene expression fold-differences comparing treated vs. control eyes. Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant.
Gene expression fold-differences comparing treated vs. control eyes. Red text = significant down-regulation, blue = significant up-regulation, grey = expression difference not statistically significant.