Abstract
Cardioviruses cause diseases in many animals including, in rare cases, humans. Although they share common features with all picornaviruses, cardioviruses have unique properties that distinguish them from other family members, including enteroviruses. One feature shared by all picornaviruses is the covalent attachment of VPg to the 5′ end of genomic RNA via a phosphotyrosyl linkage. For enteroviruses, this linkage is cleaved by a host cell protein, TDP2. Since TDP2 is divergently required during enterovirus infections, we determined if TDP2 is necessary during infection by the prototype cardiovirus, EMCV. We found that EMCV yields are reduced in the absence of TDP2. We observed a decrease in viral protein accumulation and viral RNA replication in the absence of TDP2. In contrast to enterovirus infections, we found that TDP2 is modified at peak times of EMCV infection. This finding suggests a unique mechanism for cardioviruses to regulate TDP2 activity during infection.
Keywords: picornavirus, cardiovirus, encephalomyocarditis virus, EMCV, 5′ tyrosyl-DNA phosphodiesterase 2, TDP2, virus-host interactions
1. INTRODUCTION
The cardiovirus genus of the Picornaviridae family is divided into three species (A, B, C) and includes encephalomyocarditis virus (EMCV), Theiler’s murine encephalomyelitis virus (TMEV), and the emerging human virus, Saffold virus (SAFV) (Ghildyal et al., 2009) [http://www.picornaviridae.com/cardiovirus/cardiovirus.htm]. EMCV is primarily known for diseases it causes in both wild and domestic animals worldwide, including mice, swine, and non-human primates; however, infections in humans have also been reported [reviewed in (Carocci and Bakkali-Kassimi, 2012)]. The wide range of animals susceptible to infection makes EMCV a potential zoonotic agent. Additionally, cultured human cells that express the EMCV receptor, sialoglycoprotein vascular cell adhesion molecule 1 (VCAM-1), are susceptible to EMCV infection (Huber, 1994). EMCV infections in swine and rodents can lead to myocarditis, encephalitis, paralysis, type I diabetes, or even mortality [reviewed in (Carocci and Bakkali-Kassimi, 2012)]. No vaccine or antiviral therapeutic against EMCV infection is currently available. Since cardioviruses have a wide host range and the ability to cause multiple diseases, it is important to further understand the viral mechanisms employed by cardioviruses for development of antiviral therapeutics.
EMCV, like members of the enterovirus genus of picornaviruses, has a small, positive-sense RNA genome (~7.8 kb). Although members of the picornavirus family share many similarities in their genomic RNAs, the cardiovirus genome has distinct features that lead to differences in viral replication and host modulation mechanisms. Although it is covalently linked to the small viral protein, VPg (viral protein genome-linked), via a phosphotyrosyl linkage, the 5′ noncoding region (NCR) of EMCV differs from that of enteroviruses (i.e., coxsackievirus, poliovirus, human rhinovirus) in that it possesses a poly(C) tract, followed by pseudoknots with unknown functions, and a type II internal ribosome entry site (IRES) [reviewed in (Wimmer et al., 1993)]. The approximately 430-nucleotide type II IRES is more highly structured than the type I IRES that enteroviruses possess and is subdivided into five domains known as H-L. The IRES mediates initiation of cap-independent translation of the viral polyprotein, which includes a protein not expressed by enteroviruses. This EMCV-specific protein is the leader (L) protein, which is phosphorylated during infection and contains an N-terminal zinc finger domain (Cornilescu et al., 2008; Dvorak et al., 2001). The L protein, despite having no enzymatic activity, disrupts nucleo-cytoplasmic trafficking by indirectly phosphorylating the nucleoporin proteins and binding to Ran GTPase (Porter et al., 2006; Porter and Palmenberg, 2009). This is different from enterovirus-mediated disruption of nucleo-cytoplasmic trafficking, which is carried out by their viral proteinases 2A (Castello et al., 2011; Park et al., 2010) and 3C/3CD (Ghildyal et al., 2009); for review, see (Flather and Semler, 2015). Another major difference between EMCV and the enteroviruses is that unlike enterovirus 2A proteinases, the EMCV 2A protein has no proteolytic activity. In contrast to enterovirus 2A cleavage of eIF4G to shut off host cell translation, EMCV 2A expression triggers hypo-phosphorylation of the translation initiation factor 4E-BP1, leading to inhibition of cap-dependent translation, albeit at a much slower rate than during poliovirus infection (Etchison et al., 1982; Jen et al., 1980; Svitkin et al., 1998). EMCV 2A was also shown to inhibit apoptosis (Carocci et al., 2011). Despite these mechanistic differences, EMCV, like enteroviruses, effectively alters the cellular environment to promote viral replication.
EMCV uses an additional strategy to enhance its replication cycle by inhibiting the host antiviral response. Previous studies showed that EMCV L protein interferes with IRF-3 dimerization, a step necessary for transcription of the interferon-alpha/beta genes (Hato et al., 2007). Additionally, EMCV, like poliovirus, targets the viral RNA sensor, RIG-I (Barral et al., 2009; Papon et al., 2009). It is important to note that not all host proteins function similarly during enterovirus and cardiovirus infections. For example, the cellular mRNA decay factor AUF1 acts as a restriction factor during coxsackievirus, poliovirus, or human rhinovirus infection, but during EMCV infection, levels of virus restriction appear to be host-cell dependent (Cathcart et al., 2013; Cathcart and Semler, 2014; Rozovics et al., 2012; Wong et al., 2013). In addition, host protein poly(rC) binding protein 2 (PCBP2) is required as an IRES trans-acting factor for enterovirus translation but not for EMCV translation (Walter et al., 1999). These previous findings illustrate some of the different pathways that are used by cardioviruses (compared to enteroviruses) to carry out viral gene expression and alter the cellular environment during infection.
In this report, we focused on the role of host protein 5′ tyrosyl-DNA phosphodiesterase 2 (TDP2) during EMCV infection. TDP2 possesses the VPg unlinkase activity that hydrolyzes the covalent phosphotyrosyl bond between VPg and the 5′ end of enterovirus RNAs (Virgen-Slane et al., 2012). TDP2 was also shown to be necessary for efficient enterovirus replication in murine cells (Maciejewski et al., 2015). Although it has not been confirmed that TDP2 functions as VPg unlinkase during cardiovirus infections, previous studies demonstrated that a cellular activity from mouse ascites Krebs II cells can hydrolyze the VPg-RNA linkage found in both poliovirus and EMCV RNA (Drygin et al., 1988; Drygin and Siianova, 1986). This activity was predicted to be a phosphodiesterase involved in repair of RNA and topoisomerase complexes (Gulevich et al., 2001, 2002). These characteristics resemble those of TDP2, which primarily removes tyrosine adducts resulting from topoisomerase II-mediated double-stranded DNA breaks (Cortes Ledesma et al., 2009). In this study, we sought to determine if TDP2 plays a role during EMCV infection and if its activity is modified by viral functions. Our previous work found that TDP2 was divergently required for enterovirus infections, with coxsackievirus B3 replication showing the highest levels of dependence on TDP2 (Maciejewski et al., 2015). Based on these findings, we hypothesize that picornaviruses use TDP2/VPg unlinkase activity to mark viral RNAs for use in viral translation, RNA replication, or encapsidation. Results obtained from the current study showed that TDP2 re-localizes from the nucleus to the cytoplasm of EMCV infected cells. We found that EMCV yields are significantly reduced during infection of mouse embryonic fibroblasts genetically ablated for TDP2. We also observed a reduction of EMCV protein accumulation in the absence of TDP2. Finally, in contrast to what is observed during enterovirus infections, we found that TDP2 appears to be proteolytically cleaved at peak times of EMCV infection, providing yet another example of the differential modification of host proteins during cardiovirus infections compared to enterovirus infections.
2. RESULTS
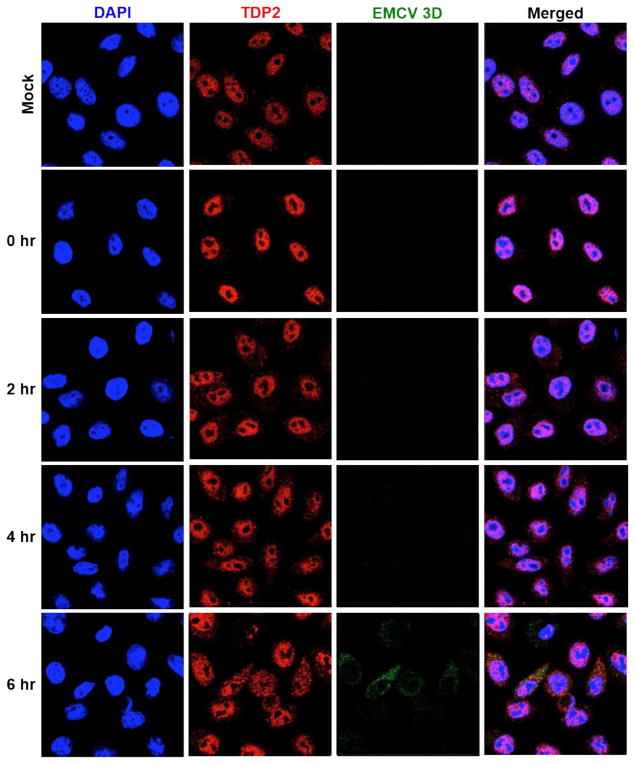
2.1. TDP2 is re-localized from the nucleus to the cytoplasm during EMCV infection
TDP2 is predominantly a nuclear protein that is present at lower concentrations in the cytoplasm (Li et al., 2011). It dramatically re-localizes from the nucleus to the cytoplasm by 4 hours after poliovirus infection (Virgen-Slane et al., 2012). TDP2 was also shown to localize to the cell periphery in sites distinct from putative viral replication and encapsidation at peak times of poliovirus infection (Virgen-Slane et al., 2012). These findings suggested that TDP2 may be sequestered from viral RNA replication and encapsidation sites as a mechanism to modulate the VPg unlinking activity of TDP2 during infection. To determine if this re-localization pattern occurs for picornaviruses other than poliovirus, we examined TDP2 re-localization during EMCV infection using confocal microscopy. HeLa cells were either mock- or EMCV-infected and fluorescently labeled using antibodies against TDP2 or EMCV 3D polymerase. As shown in Fig. 1, TDP2 is primarily localized to the nucleus in mock-infected cells and at 0 hr post-infection. At 2 and 4 hr post-infection, TDP2 shows increasing re-localization from the nucleus to the cytoplasm. At peak times of infection (6 hr), TDP2 is dispersed throughout the cell cytoplasm and nucleus. Staining of the RNA-dependent RNA polymerase 3D was used as a marker for viral replication in EMCV-infected cells and is detected at peak times of infection (6 hr). TDP2 and EMCV 3D are closely localized in the cell cytoplasm. These results are in contrast to the distinctive pattern observed for TDP2 during poliovirus infection, suggesting an alternative mechanism for modulating TDP2 catalytic activity throughout EMCV infection.
Fig. 1. TDP2 is re-localized from the nucleus to the cytoplasm during EMCV infection.
HeLa cells were seeded on coverslips and either mock- or EMCV-infected at an MOI of 20. Cells were then fixed with formaldehyde at 0, 2, 4, or 6 hr post-infection. Proteins were visualized by confocal microscopy using antibodies against human TDP2 (red) or EMCV 3D (green). Nuclei were counterstained with DAPI (blue).
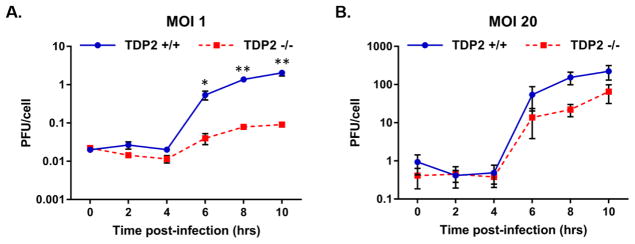
2.2. The absence of TDP2 during EMCV infection causes a decrease in viral yields
Since enteroviruses require TDP2 for efficient replication, we investigated the impact of TDP2 on cardiovirus infection. We infected TDP2 wild type (TDP2+/+) and knockout (TDP2−/−) mouse embryonic fibroblasts (MEFs) with EMCV at an MOI of 1 or 20 and quantified viral yields by plaque assay every 2 hr post-infection for 10 hr (Fig. 2). In the absence of TDP2, viral yields were reduced by 6 hr post-infection at either MOI. At lower MOI, EMCV replication in TDP2−/− cells was reduced more than 10-fold compared to TDP2+/+ MEFs (Fig. 2A). At higher MOI, the effect of TDP2 knockout on EMCV replication was slightly diminished, and less than a 10-fold reduction of virus yield was observed in TDP2−/− cells (Fig. 2B). The diminished effect of TDP2 knockout on EMCV replication at higher MOI indicates that the inhibitory effect of TDP2 knockout on virus production can be partially overcome by increased virus input. These results suggest that TDP2 is required for efficient replication of EMCV in MEFs, similar to the observed effect of TDP2 on replication of the enteroviruses poliovirus, human rhinovirus, and coxsackievirus B3 (Maciejewski et al., 2015).
Fig. 2. EMCV requires TDP2 for efficient viral replication.
Single cycle growth analyses were carried out in TDP2+/+ and TDP2−/− MEFs following EMCV infection at an MOI of 1 (A) or 20 (B). Cells and supernatant were collected every 2 hr up to 10 hr post-infection. Virus yields were quantified by plaque assay on HeLa cells and represented as plaque forming units per cell (PFU/cell). Viral yields are plotted on a logarithmic scale and values represent the mean of triplicate experiments ± standard error of the mean (SEM). *, P < 0.05; **, P < 0.01 (Student’s t test)
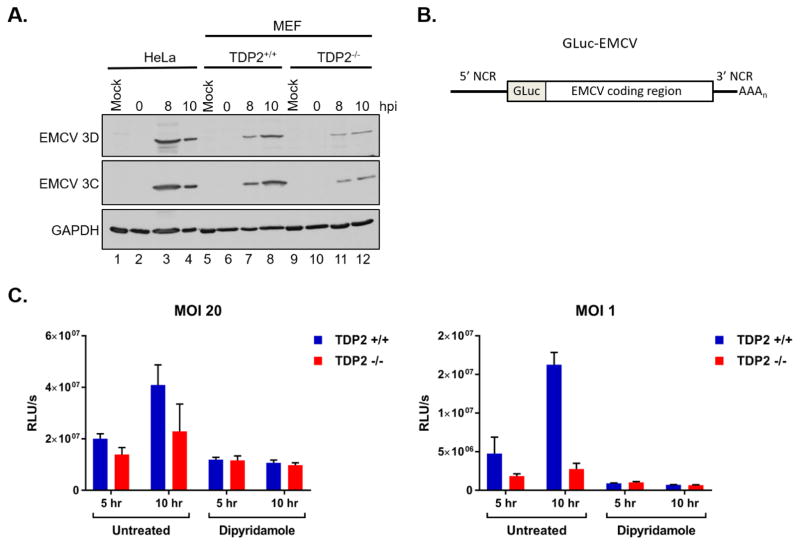
2.3. TDP2 is necessary for efficient EMCV replication
We next investigated the role of TDP2 during EMCV translation and RNA synthesis. Viral protein accumulation during EMCV infection of TDP2+/+ and TDP2−/− MEFs was analyzed by Western blot using antibodies against the EMCV nonstructural proteins 3C and 3D. We found that 3C and 3D expression was clearly detected at 8 and 10 hr post-infection in EMCV-infected HeLa cells and TDP2+/+ MEFs (Fig. 3A, lanes 3–4 and lanes 7–8). In agreement with the single-cycle growth analyses, EMCV 3C and 3D expression was reduced in the TDP2−/− MEFs (Fig. 3A, lanes 11 and 12). To separate the effect of TDP2 on viral translation from that on viral RNA synthesis, a recombinant Gaussia luciferase-expressing EMCV (GLuc-EMCV) was employed in combination with the viral RNA synthesis inhibitor, dipyridamole (Fata-Hartley and Palmenberg, 2005; Tonew et al., 1977) (Fig. 3B). TDP2+/+ and TDP2−/− MEFs were infected with GLuc-EMCV at an MOI of 1 or 20 and cells were left untreated or treated with 150 μM dipyridamole both during and after virus adsorption. Luciferase activity was measured 5 and 10 hr post-infection. In untreated cells, where viral translation and RNA synthesis were allowed to occur normally, there was a reduction in luciferase activity in TDP2−/− MEFs compared to TDP2+/+ at 5 and 10 hr post-infection at both MOIs (Fig. 3C). Similar to the single-cycle growth analyses, the effect of TDP2 knockout on luciferase activity was more significant at lower MOI, although the trend can still be observed at high MOI. Since these results do not distinguish between the effect of TDP2 on viral translation or RNA synthesis, dipyridamole treatment was used to measure the effect of TDP2 on viral translation only. Interestingly, there was no difference in luciferase activity measured from TDP2+/+ or TDP2−/− cells following inhibition of viral RNA synthesis (Fig. 3C). These results suggest that TDP2 is not required for translation of input EMCV RNA, but is required for subsequent rounds of viral translation and RNA synthesis.
Fig. 3. EMCV viral protein accumulation is reduced in the absence of TDP2.
(A) HeLa, TDP2+/+ MEFs, and TDP2−/− MEFs were mock- or EMCV-infected at an MOI of 20. Cells and supernatant were collected at 0, 8, or 10 hr post-infection (hpi) and used to generate protein lysates. Lysates were subjected to SDS-PAGE and Western blot analysis using an anti-Mengo 3D, anti-EMCV 3C, or anti-GAPDH antibody to detect protein accumulation. (B) Schematic of the pdnGLuc-VFETQG-m16.1 construct used to generate the GLuc-EMCV virus. GLuc-EMCV encodes Gaussia luciferase immediately following the 5′ noncoding region (5′ NCR) and upstream of the complete EMCV coding region, 3′ NCR, and poly(A) tract. (C) TDP2+/+ and TDP2−/− MEFs were infected with GLuc-EMCV at an MOI of 1 or 20 and luciferase activity was measured at 5 and 10 hr post-infection. To inhibit viral RNA synthesis, cells were treated with 150 μM dipyridamole during and after virus adsorption. Luminescence is represented as relative light units per second (RLU/s) normalized to cell count. Data represent the means of triplicate experiments ± standard error of the mean (SEM). *, P < 0.05; ***, P < 0.001 (Student’s t test)
2.4. TDP2 is modified during EMCV infection
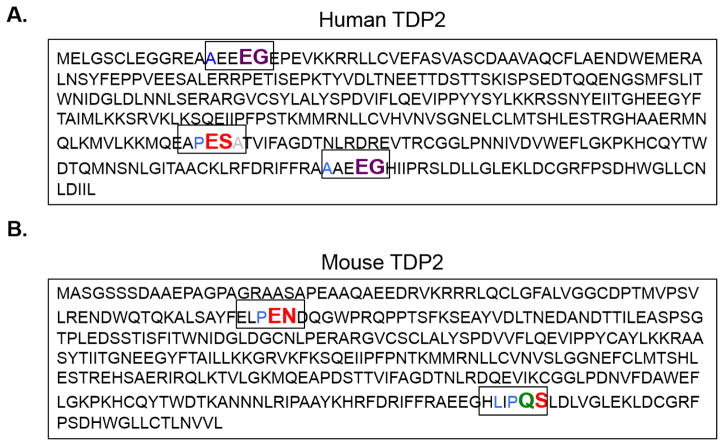
Numerous studies have revealed that picornaviruses modulate host proteins via their virus-encoded proteinases to aid in carrying out their replication cycle; for review, see (Chase and Semler, 2012). Modification of cellular proteins by viral proteinases is primarily accomplished through cleavage of proteins at specific amino acid residues. Enterovirus proteinase 3C and its proteolytically active precursor 3CD recognize the residues QG, QA, QN, and QS with an amino acid with an aliphatic side chain in the P4 position as putative cleavage sites (Blair and Semler, 1991). Additionally, rhinovirus 3C and 3CD recognize EG as putative cleavage sites. In contrast, cardiovirus 3C proteinase recognizes QG, QA, QS, EN, and ES with a proline in the P2 or P2′ position as putative 3C cleavage sites (Palmenberg, 1990). To determine if TDP2 might be cleaved during picornavirus infections, we analyzed both the human and mouse TDP2 sequence for putative 3C or 3CD cleavage sites since EMCV does not encode a proteinase 2A equivalent (Hellen et al., 1992; Palmenberg, 1990). We found that human TDP2 encodes two putative rhinovirus 3C/3CD cleavage sites and one cardiovirus 3C cleavage site (Fig. 4A). We next used the online ExPasy server (http://web.expasy.org/compute_pi/) (Artimo et al., 2012; Gasteiger et al., 2003) to calculate the predicted molecular masses of the human TDP2 fragments that would result from these putative cleavage sites (Table 1). The first rhinovirus EG cleavage site would result in 1.8 and 39.1 kDa TDP2 protein fragments. The second rhinovirus EG cleavage site would result in 36.9 and 4.0 kDa TDP2 protein fragments. The cardiovirus ES cleavage site would result in 28.6 and 12.3 kDa TDP2 protein fragments. Mouse TDP2 encodes two putative cardiovirus 3C cleavage sites and one enterovirus 3C/3CD cleavage site (Fig. 4B). We also used ExPasy to calculate predicted molecular masses of the mouse TDP2 fragments that would result from cleavage at these sites (Table 1). The first cardiovirus EN cleavage site would result in 8.1 and 32.9 kDa TDP2 protein fragments. The second cardiovirus and only enterovirus QS cleavage site would result in 37.7 and 3.3 kDa TDP2 protein fragments. We then mapped these putative cleavage sites to the published crystal structure of the human and mouse TDP2 catalytic domain on the Protein Data Bank using the Visual Molecular Dynamics software (data not shown) (Humphrey et al., 1996; Schellenberg et al., 2012; Schellenberg et al., 2016). We found that these cleavage sites in the catalytic domain were exposed and predicted to be accessible to 3C/3CD cleavage.
Fig. 4. Putative picornavirus 3C and 3CD proteinase cleavage sites in the TDP2 sequence.
The picornavirus 3C proteinase and its precursor 3CD recognize specific residues at putative cleavage sites. Enterovirus 3C/3CD recognize QG, QA, QN, and QS residues with an amino acid with an aliphatic side chain in the P4 position (cleavage site colored in green). Rhinovirus 3C/3CD recognizes the same cleavage sites as the enterovirus cleavage sites as well as an additional site: EG, also with an aliphatic side chain in the P4 position (cleavage site colored in purple). Cardiovirus 3C proteinases recognize QG, QA, QS, EN and ES residues with a proline preferred in the P2 or P2′ position (cleavage sites colored in red). Putative cleavage sites are highlighted in their denoted color in both the (A) human and (B) mouse TDP2 sequences.
Table 1. Predicted TDP2 molecular weights of cleavage products generated from different 3C/3CD proteinase recognition sites.
Fragment TDP2 molecular weights due to cleavage at putative scissile bonds were predicted using ExPasy online software (Artimo et al., 2012; Gasteiger et al., 2003). The predicted cleavage sites in the human or mouse TDP2 sequence are listed.
| Predicted cleavage site | Picornavirus 3C/3CD | TDP2 host | N-terminus molecular weight | C-terminus molecular weight |
|---|---|---|---|---|
| EG | rhinovirus | human | 1.8 kDa | 39.1 kDa |
| EG | rhinovirus | human | 36.9 kDa | 4.0 kDa |
| ES | cardiovirus | human | 28.6 kDa | 12.3 kDa |
| EN | cardiovirus | mouse | 8.1 kDa | 32.9 kDa |
| QS | cardiovirus/enterovirus | mouse | 37.7 kDa | 3.3 kDa |
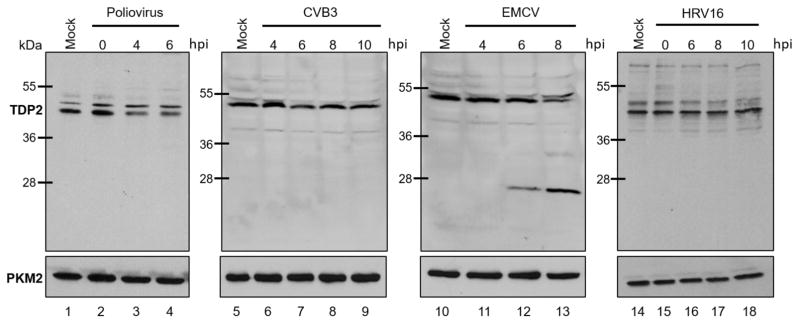
To determine if TDP2 is cleaved during infection, we performed a Western blot analysis using a polyclonal antibody that recognizes epitopes located at the N-terminus of human TDP2 on lysates generated from virus-infected HeLa cells at 0, 4, and 6 hr post-infection (for poliovirus), 4, 6, 8, and 10 hr post-infection (for CVB3), or 4, 6, and 8 hr post-infection (for EMCV) (Fig. 5). The different time points used for Western blot analysis for each these viruses were chosen to include peak times of intracellular replication for each individual virus during an infection of cultured HeLa cells. We found that TDP2 was cleaved beginning at 6 hr post-EMCV infection (Fig. 5, lane 12), resulting in a fragment similar in molecular mass to the N-terminal fragment (~28 kDa) predicted from the putative ES cleavage site in Table 1. The abundance of this putative cleavage fragment increased at 8 hr post-infection, with a concomitant decrease in intact TDP2 levels migrating slightly faster than the 55 kDa molecular mass marker protein (Fig. 5, lane 13). TDP2 was not cleaved or detectably modified during poliovirus or CVB3 infection (Fig. 5, lanes 1–9). Additionally, we performed a Western blot analysis using an antibody against human TDP2 using lysates from human rhinovirus 16 (HRV16)-infected HeLa cells from 0, 6, 8, and 10 hr post-infection (Fig. 5, lanes 14–18) and lysates from human rhinovirus 1A (HRV1A)-infected HeLa cells from 0 and 13 hr post-infection (data not shown). HRV16 belongs to the major receptor rhinovirus group, while HRV1A belongs to the minor receptor rhinovirus group (Gern et al., 1997). We found that TDP2 is not cleaved during rhinovirus infection despite encoding putative rhinovirus 3C/3CD cleavage sites. It was confirmed that TDP2 was not cleaved as late as 13 hr post-HRV16 infection (data not shown). Together these results show that TDP2 is cleaved during peak times of EMCV infection in HeLa cells but not during enterovirus infections, suggesting proteolytic cleavage as an alternative mechanism for regulating TDP2 activity during cardiovirus infection.
Fig. 5. TDP2 is cleaved during EMCV infection.
HeLa cells were mock-, poliovirus-, CVB3-, EMCV-, or HRV16-infected at an MOI of 20. Cells and supernatant were collected at specified times after infection (hours post-infection, hpi) and used to generate protein lysates. Lysates were subjected to SDS-PAGE and Western blot analysis using an anti-TDP2 or anti-PKM2 antibody (loading control) to detect proteins.
3. DISCUSSION
Enteroviruses are divergently dependent on TDP2 during their viral replication cycles (Maciejewski et al., 2015). TDP2, an enzyme primarily known for its DNA repair function in the uninfected cell, was identified as the cellular activity responsible for hydrolyzing the highly conserved phosphotyrosyl bond between the third amino acid (tyrosine) of VPg and the 5′ end of the viral RNA (Virgen-Slane et al., 2012). In the absence of this cellular protein, enterovirus yields are significantly reduced (Maciejewski et al., 2015). These decreases in viral yields are corroborated by an overall decrease in viral protein accumulation and viral RNA replication, with the most significant decrease observed during coxsackievirus B3 infection (Maciejewski et al., 2015). To determine if TDP2 plays a positive role during infection by a picornavirus belonging to a different genus, we chose to characterize the role of TDP2 during cardiovirus infection, specifically EMCV. Previous work confirmed that a cellular phosphodiesterase activity could hydrolyze the phosphotyrosyl bond between VPg and EMCV viral RNA (Gulevich et al., 2001). Although it has not been confirmed that TDP2 functions as VPg unlinkase during EMCV infection, we wanted to determine if the absence of TDP2 had an impact on EMCV infection.
A previous study carried out in our laboratory found that the primarily nuclear protein TDP2 was re-localized from the nucleus to the cytoplasm of poliovirus-infected HeLa cells as early as 2 hr post-infection (Virgen-Slane et al., 2012). At peak times of poliovirus infection (4 hr post-infection), TDP2 is localized to the cell periphery in sites that do not co-localize with viral replication and encapsidation markers (3A and capsid, respectively) (Virgen-Slane et al., 2012). The authors suggested a regulatory role for TDP2 and its catalytic activity during the course of poliovirus infection. To determine if TDP2 was re-localized during cardiovirus infection, we infected HeLa cells with EMCV, followed by fluorescent labeling of TDP2 and the EMCV viral replication marker 3D polymerase. Similar to poliovirus infection, TDP2 relocalized from the nucleus to the cell cytoplasm as early as 4 hr post-EMCV infection. TDP2 cytoplasmic dispersion increased by 4 and 6 hr post-EMCV infection. In contrast to peak times of poliovirus-infection, we did not observe re-localization of TDP2 to the cell periphery during peak times of EMCV infection. These results suggest that TDP2 may provide a function during early times in EMCV infection but is not excluded from putative viral replication sites at peak and later times during EMCV infection.
To explore the dependence of EMCV on TDP2 during infection, we used murine cells ablated for TDP2 and found that EMCV yields were reduced in the absence of TDP2 at high MOI (20). Since peak titers are not achieved in the TDP2+/+ cells until 10 hr post-infection, the decrease in viral titers in the absence of TDP2 is less pronounced in comparison to what we observed during enterovirus infections at a similar MOI. However, at low MOI (1) EMCV yields were more dramatically reduced (> 10-fold) between 6 and 10 hr post-infection in the absence of TDP2. For both MOIs used in our study, the single-cycle growth analysis revealed that EMCV replication is delayed in the absence of TDP2 and that TDP2 plays a pro-viral role during the cardiovirus replication cycle.
To further dissect the role of TDP2 during EMCV infection, we analyzed viral protein accumulation in the TDP2+/+ and TDP2−/− MEFs by Western blot. We observed a reduction in viral protein accumulation in the absence of TDP2. This latter phenotype is also consistent with our single-cycle growth analysis of EMCV infection in the TDP2−/− cells. To further characterize the role of TDP2 during EMCV infection, we took advantage of an infectious EMCV reporter construct (GLuc-EMCV) to generate recombinant virus. The GLuc-EMCV virus was used to measure viral translation (in the presence of the viral RNA synthesis inhibitor dipyridamole) and RNA replication in the presence and absence of TDP2 in MEFs. We found that translation of the input viral RNA was not decreased in the absence of TDP2 when viral RNA synthesis was inhibited. However, we found that viral translation and RNA replication were significantly reduced in the absence of TDP2 when the infection was allowed to proceed normally. These results are consistent with our single-cycle growth and Western blot analyses, indicating that TDP2 promotes steps following translation of input RNA.
After confirming a dependence on TDP2 during EMCV infection, we explored possible ways in which EMCV might regulate the activity of TDP2 during infection. Since our confocal microscopy imaging did not reveal the distinct re-localization pattern of TDP2 to the cell periphery away from potential viral replication sites, as seen during poliovirus infection, we investigated the possibility that EMCV infection results in modification of TDP2. Picornavirus proteinases are known to play major roles during infection by modifying host proteins to enhance their viral replication cycles (Chase and Semler, 2012). First, we analyzed both the human and mouse TDP2 sequences to identify putative viral proteinase cleavage sites. We calculated the predicted molecular masses of TDP2 that would result from such cleavage events and mapped the location of the putative cleavage sites on the three-dimensional structure of TDP2. We found these putative cleavage sites to be surface-exposed on the TDP2 catalytic domain, raising the possibility of cleavage of TDP2 during picornavirus infections. Our Western blot analysis confirmed that human TDP2 is cleaved/modified during EMCV infection. The observed molecular mass of the TDP2 fragment was similar to the predicted molecular mass of the N-terminal fragment that would result following 3C proteinase cleavage as determined by the putative cleavage sites. This N-terminal fragment is also predicted to be recognized by the polyclonal antibody used for these studies. However, results from ongoing transfection and biochemical experiments will be required to demonstrate that EMCV proteinase 3C/3CD is, in fact, responsible for generating the TDP2 cleavage product observed in Fig. 5 and whether this is a direct or indirect cleavage event occurring during EMCV infection.
TDP2 cleavage was not observed during enterovirus infections in HeLa cells, despite identifying two putative rhinovirus cleavage sites. It is possible that during rhinovirus infection, these putative cleavage sites are masked by TDP2 interactions with either viral or host proteins or that TDP2 is cleaved later than 13 hr post-infection. TDP2 cleavage was observed at 6 and 8 hr post-EMCV infection in HeLa cells (refer to Fig. 5), which is the peak kinetic window for EMCV RNA synthesis in this cell line (Martin and Palmenberg, 1996; Prather and Taylor, 1975). These results have parallels with those seen for PCBP2, a host protein necessary for enterovirus translation and RNA synthesis (Chase et al., 2014; Gamarnik and Andino, 1997; Parsley et al., 1997; Walter et al., 1999; Walter et al., 2002). PCBP2 is differentially cleaved among picornavirus infections in WisL human airway cells. PCBP2 is not cleaved during human rhinovirus infection of WisL cells but is cleaved during poliovirus infection of WisL cells (Chase and Semler, 2014). Since TDP2 is closely associated with the EMCV 3D polymerase at 6 hr post-infection, as shown by confocal microscopy, it is possible that EMCV cleaves TDP2 as a mechanism to regulate its catalytic activity during later times in the viral replication cycle. This would represent a divergent pathway utilized by EMCV to reduce the levels of active TDP2/VPg unlinkase associated with viral RNA replication complexes and/or sites of encapsidation during the later stages of infection when the accumulation of intact, VPg-linked RNAs is required for progeny virion assembly. Overall, these results support our hypothesis that TDP2 functions as a regulator of the viral replication cycle via its VPg unlinkase activity and marks viral RNA via the presence or absence of VPg for viral translation, RNA synthesis, or encapsidation. The presence or absence of VPg on viral RNA is not absolutely required for these steps to occur; however, our data suggest that VPg cleavage from viral RNA via TDP2 activity drives efficient cardiovirus replication.
4. MATERIALS AND METHODS
4.1. Cell culture and virus stocks
Wild type (TDP2+/+) and TDP2 knockout (TDP2−/−) mouse embryonic fibroblast (MEF) cell lines were generated by Caldecott and colleagues (Zeng et al., 2012) and cultured in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% fetal bovine serum (FBS). HeLa cells were cultured in DMEM supplemented with 8% newborn calf serum (NCS). EMCV stocks were generated from the pEC9 infectious cDNA construct kindly provided by Dr. Ann C. Palmenberg at the University of Wisconsin, Madison.
4.2. Virus infections and single cycle growth analysis
Single cycle growth analyses of EMCV were performed on TDP2+/+ and TDP2−/− MEFs. Cells were infected with EMCV at a multiplicity of infection (MOI) of 1 or 20 in DMEM. Following a 30 min virus adsorption, cells were washed with phosphate-buffered saline (PBS), overlaid with DMEM supplemented with 10% FBS, and incubated at 37° C, 5% CO2. HeLa cells were infected with EMCV, poliovirus, CVB3, or HRV16 at an MOI of 20 as described above. Cells and supernatant were collected at the indicated times post-infection and subjected to 5 freeze-thaw cycles prior to titration by plaque assay on HeLa cells. Viral yields are reported as plaque forming units per cell (PFU/cell). Statistical significance was calculated by Student’s t test.
4.3. Immunofluorescence
HeLa cells were cultured on glass coverslips and infected with EMCV at an MOI of 20 in DMEM. Following a 30 min virus adsorption, cells were washed 3 times with PBS, overlaid with DMEM supplemented with 8% NCS, and incubated at 37° C, 5% CO2. Cells were washed with PBS and fixed with 3.7% formaldehyde in PBS for 20 minutes at 0, 2, 4, or 6 hr post-infection. Fixed cells were washed with PBS 3 times, permeabilized with 0.5% NP-40, and blocked with 1% goat serum in PBS. Cells were immunolabelled with anti-Mengo 3D mouse monoclonal antibody (1:200; Santa Cruz), which cross-reacts with EMCV 3D, and anti-TDP2 rabbit polyclonal antibody (1:200; Bethyl), in 1% bovine serum albumin (BSA) for 1 hr at room temperature. Following primary antibody incubation, the cells were incubated with Alexa Fluor 488 goat anti-mouse IgG and Alexa Fluor 594 goat anti-rabbit IgG (1:250; Molecular Probes) in 1% BSA for 1 hr at room temperature. Coverslips were washed with 1% BSA in PBS 3 times following each antibody incubation. Nuclei were stained with 4 μg/ml DAPI for 10 min at room temperature and coverslips were mounted on slides using Fluoro-Gel (Electron Microscopy Sciences). Cells were imaged using a LSM700 laser scanning confocal microscope and ZEN software (Zeiss).
4.4. Western blot analysis
Infections with EMCV, poliovirus, CVB3, and HRV16 were carried out as described above. Cells and their supernatants were harvested and pelleted at various time points post-infection. Samples were washed with PBS 3 times and pelleted. The pellet was re-suspended in NP-40 lysis buffer (50 mM Tris-HCl, pH 7.5, 5 mM EDTA, 150 mM NaCl, 1% NP-40) and incubated on ice for 30 minutes. Cell debris was pelleted and the supernatant collected. Protein concentration was measured using Bio-Rad Protein Assay Dye Reagent. For Western blot analysis, 50 to 60 μg of cell lysates were subjected to electrophoresis on SDS-containing 12.5% polyacrylamide gels, then transferred to a PVDF membrane. Membranes were incubated with mouse monoclonal anti-Mengo 3D (1:1,000; Santa Cruz) or rabbit polyclonal anti-EMCV 3C (1:7,500) to detect viral protein accumulation. Dr. T. Glen Lawson at Bates College kindly provided the rabbit polyclonal antibody to EMCV 3C. Rabbit polyclonal anti-TDP2 antibody (1:2,000; Bethyl) was used to detect TDP2 during enterovirus and cardiovirus infections. GAPDH was detected using rabbit monoclonal antibody anti-GAPDH (1:10,000; Abcam) and PKM2 was detected using rabbit polyclonal anti-PKM2 (1:2,000; Bethyl). Following primary antibody incubation, membranes were incubated with goat anti-rabbit or goat anti-mouse IgG (H+L) horseradish peroxidase (HRP)-conjugated antibodies (1:7,500; Bethyl). Proteins were detected using ECL Western Blotting Substrate (Life Technologies).
4.5. Luciferase assay
Gaussia luciferase (GLuc)-expressing EMCV was generated from the infectious GLuc-Mengovirus clone, pdnGLuc-VFETQG-m16.1, generously provided by Dr. Frank van Kuppeveld at the University of Utrecht, The Netherlands. GLuc-EMCV stocks were generated from transcripts of pdnGLuc-VFETQG-m16.1 transfected and subsequently passaged in HeLa cells. MEFs were infected with GLuc-EMCV at an MOI of 1 or 20 as described above. Either vehicle or 150 μM dipyridamole (Sigma) was added both during and after virus adsorption. Luciferase activity was measured 5 and 10 hr post-infection using Renilla Luciferase Assay Lysis System (Promega). Cells were lysed for 15 min in Renilla Luciferase Assay Lysis Buffer and luminescence was measured using a SIRIUS luminometer (Berthold Detection System) and represented as relative light units per second (RLU/s) normalized to cell count. Statistical significance was calculated by Student’s t test.
4.6. Visual Molecular Dynamics analysis of TDP2 catalytic domain and cleavage sites
The human and mouse TDP2 catalytic domain structure was downloaded from the protein data bank (PDB 5INO and PDB 5INM) and visualized using the Visual Molecular Dynamics software (http://www.ks.uiuc.edu/Research/vmd/) as a ribbon model (Humphrey et al., 1996; Schellenberg et al., 2012; Schellenberg et al., 2016). The conserved catalytic residues E152, N264, D262, and H351 for human TDP2 and E162, N274, D272, and His359 were selected and represented as a stick model. The putative picornavirus 3C/3CD cleavage sites were represented as a stick model. Catalytic residues and putative cleavage sites were labeled.
Highlights.
Based on previously published studies, we hypothesized that picornaviruses use TDP2/VPg unlinkase activity to mark viral RNAs for specific uses during picornavirus replication.
Data from the current study showed that TDP2 re-localizes from the nucleus to the cytoplasm of encephalomyocarditis virus (EMCV) infected cells.
EMCV yields are significantly reduced during infection of mouse embryonic fibroblasts genetically ablated for TDP2.
TDP2 appears to be proteolytically cleaved at peak times of EMCV infection, providing a new example of modification of host proteins during picornavirus infections.
Acknowledgments
We are grateful T. Glen Lawson for the EMCV 3C antibody and Frank van Kuppeveld for the GLuc-EMCV construct. We are indebted to the UC Irvine Optical Biology Core Facility for use of the Zeiss LSM700 laser scanning confocal microscope. We thank MyPhuong Tran and Joseph H. Nguyen for experimental assistance. We also thank Autumn Holmes, Eric Baggs, and Dylan Flather for their critical review of the manuscript. This research was supported by Public Health Service grant AI110782 from the National Institutes of Health to B.L.S. This material is based, in part, upon work supported by the National Science Foundation Graduate Research Fellowship Program under Grant No. DGE-1321846 to S.M. W.U. is a predoctoral trainee supported by U. S. Public Health Service training grant AI007319 from the National Institutes of Health. The UC Irvine Optical Biology Core facility is a Chao Family Comprehensive Cancer Center shared resource supported by the National Cancer Institute of the National Institutes of Health under award number P30CA062203.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Artimo P, Jonnalagedda M, Arnold K, Baratin D, Csardi G, de Castro E, Duvaud S, Flegel V, Fortier A, Gasteiger E, Grosdidier A, Hernandez C, Ioannidis V, Kuznetsov D, Liechti R, Moretti S, Mostaguir K, Redaschi N, Rossier G, Xenarios I, Stockinger H. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012;40:W597–603. doi: 10.1093/nar/gks400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barral PM, Sarkar D, Fisher PB, Racaniello VR. RIG-I is cleaved during picornavirus infection. Virology. 2009;391:171–176. doi: 10.1016/j.virol.2009.06.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blair WS, Semler BL. Role for the P4 amino acid residue in substrate utilization by the poliovirus 3CD proteinase. J Virol. 1991;65:6111–6123. doi: 10.1128/jvi.65.11.6111-6123.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carocci M, Bakkali-Kassimi L. The encephalomyocarditis virus. Virulence. 2012;3:351–367. doi: 10.4161/viru.20573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carocci M, Cordonnier N, Huet H, Romey A, Relmy A, Gorna K, Blaise-Boisseau S, Zientara S, Kassimi LB. Encephalomyocarditis virus 2A protein is required for viral pathogenesis and inhibition of apoptosis. J Virol. 2011;85:10741–10754. doi: 10.1128/JVI.00394-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castello A, Alvarez E, Carrasco L. The multifaceted poliovirus 2A protease: regulation of gene expression by picornavirus proteases. J Biomed Biotechnol. 2011;2011:369648. doi: 10.1155/2011/369648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart AL, Rozovics JM, Semler BL. Cellular mRNA decay protein AUF1 negatively regulates enterovirus and human rhinovirus infections. J Virol. 2013;87:10423–10434. doi: 10.1128/JVI.01049-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cathcart AL, Semler BL. Differential restriction patterns of mRNA decay factor AUF1 during picornavirus infections. J Gen Virol. 2014;95:1488–1492. doi: 10.1099/vir.0.064501-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AJ, Daijogo S, Semler BL. Inhibition of poliovirus-induced cleavage of cellular protein PCBP2 reduces the levels of viral RNA replication. Journal of virology. 2014;88:3192–3201. doi: 10.1128/JVI.02503-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AJ, Semler BL. Viral subversion of host functions for picornavirus translation and RNA replication. Future Virol. 2012;7:179–191. doi: 10.2217/fvl.12.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chase AJ, Semler BL. Differential cleavage of IRES trans-acting factors (ITAFs) in cells infected by human rhinovirus. Virology. 2014;449:35–44. doi: 10.1016/j.virol.2013.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornilescu CC, Porter FW, Zhao KQ, Palmenberg AC, Markley JL. NMR structure of the mengovirus Leader protein zinc-finger domain. FEBS Lett. 2008;582:896–900. doi: 10.1016/j.febslet.2008.02.023. [DOI] [PubMed] [Google Scholar]
- Cortes Ledesma F, El Khamisy SF, Zuma MC, Osborn K, Caldecott KW. A human 5′-tyrosyl DNA phosphodiesterase that repairs topoisomerase-mediated DNA damage. Nature. 2009;461:674–678. doi: 10.1038/nature08444. [DOI] [PubMed] [Google Scholar]
- Drygin YF, Shabanov AA, Bogdanov AA. An enzyme which specifically splits a covalent bond between picornaviral RNA and VPg. FEBS Lett. 1988;239:343–346. doi: 10.1016/0014-5793(88)80948-7. [DOI] [PubMed] [Google Scholar]
- Drygin YF, Siianova E. Characteristics of an enzyme hydrolyzing the covalent bond between RNA and protein VPg of the encephalomyocarditis virus. Biokhimiia. 1986;51:249–259. [PubMed] [Google Scholar]
- Dvorak CM, Hall DJ, Hill M, Riddle M, Pranter A, Dillman J, Deibel M, Palmenberg AC. Leader protein of encephalomyocarditis virus binds zinc, is phosphorylated during viral infection, and affects the efficiency of genome translation. Virology. 2001;290:261–271. doi: 10.1006/viro.2001.1193. [DOI] [PubMed] [Google Scholar]
- Etchison D, Milburn SC, Edery I, Sonenberg N, Hershey JW. Inhibition of HeLa cell protein synthesis following poliovirus infection correlates with the proteolysis of a 220,000-dalton polypeptide associated with eucaryotic initiation factor 3 and a cap binding protein complex. J Biol Chem. 1982;257:14806–14810. [PubMed] [Google Scholar]
- Fata-Hartley CL, Palmenberg AC. Dipyridamole reversibly inhibits mengovirus RNA replication. J Virol. 2005;79:11062–11070. doi: 10.1128/JVI.79.17.11062-11070.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flather D, Semler BL. Picornaviruses and nuclear functions: targeting a cellular compartment distinct from the replication site of a positive-strand RNA virus. Front Microbiol. 2015;6:594. doi: 10.3389/fmicb.2015.00594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamarnik AV, Andino R. Two functional complexes formed by KH domain containing proteins with the 5′ noncoding region of poliovirus RNA. RNA. 1997;3:882–892. [PMC free article] [PubMed] [Google Scholar]
- Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gern JE, Galagan DM, Jarjour NN, Dick EC, Busse WW. Detection of rhinovirus RNA in lower airway cells during experimentally induced infection. Am J Respir Crit Care Med. 1997;155:1159–1161. doi: 10.1164/ajrccm.155.3.9117003. [DOI] [PubMed] [Google Scholar]
- Ghildyal R, Jordan B, Li D, Dagher H, Bardin PG, Gern JE, Jans DA. Rhinovirus 3C protease can localize in the nucleus and alter active and passive nucleocytoplasmic transport. J Virol. 2009;83:7349–7352. doi: 10.1128/JVI.01748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gulevich AY, Yusupova RA, Drygin YF. A phosphodiesterase from ascites carcinoma Krebs II cells specifically cleaves the bond between VPg and RNA of encephalomyocarditis virus. Biochemistry (Mosc) 2001;66:345–349. doi: 10.1023/a:1010220401336. [DOI] [PubMed] [Google Scholar]
- Gulevich AY, Yusupova RA, Drygin YF. VPg unlinkase, the phosphodiesterase that hydrolyzes the bond between VPg and picornavirus RNA: a minimal nucleic moiety of the substrate. Biochemistry (Mosc) 2002;67:615–621. doi: 10.1023/a:1016124202274. [DOI] [PubMed] [Google Scholar]
- Hato SV, Ricour C, Schulte BM, Lanke KH, de Bruijni M, Zoll J, Melchers WJ, Michiels T, van Kuppeveld FJ. The mengovirus leader protein blocks interferon-alpha/beta gene transcription and inhibits activation of interferon regulatory factor 3. Cell Microbiol. 2007;9:2921–2930. doi: 10.1111/j.1462-5822.2007.01006.x. [DOI] [PubMed] [Google Scholar]
- Hellen CU, Lee CK, Wimmer E. Determinants of substrate recognition by poliovirus 2A proteinase. J Virol. 1992;66:3330–3338. doi: 10.1128/jvi.66.6.3330-3338.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber SA. VCAM-1 is a receptor for encephalomyocarditis virus on murine vascular endothelial cells. J Virol. 1994;68:3453–3458. doi: 10.1128/jvi.68.6.3453-3458.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- Jen G, Detjen BM, Thach RE. Shutoff of HeLa cell protein synthesis by encephalomyocarditis virus and poliovirus: a comparative study. J Virol. 1980;35:150–156. doi: 10.1128/jvi.35.1.150-156.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Sun SY, Khuri FR, Li R. Pleiotropic functions of EAPII/TTRAP/TDP2: cancer development, chemoresistance and beyond. Cell Cycle. 2011;10:3274–3283. doi: 10.4161/cc.10.19.17763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maciejewski S, Nguyen JH, Gomez-Herreros F, Cortes-Ledesma F, Caldecott KW, Semler BL. Divergent requirement for a DNA repair enzyme during enterovirus infections. mBio. 2015;7:e01931–01915. doi: 10.1128/mBio.01931-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin LR, Palmenberg AC. Tandem mengovirus 5′ pseudoknots are linked to viral RNA synthesis, not poly(C)-mediated virulence. J Virol. 1996;70:8182–8186. doi: 10.1128/jvi.70.11.8182-8186.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmenberg AC. Proteolytic processing of picornaviral polyprotein. Annu Rev Microbiol. 1990;44:603–623. doi: 10.1146/annurev.mi.44.100190.003131. [DOI] [PubMed] [Google Scholar]
- Papon L, Oteiza A, Imaizumi T, Kato H, Brocchi E, Lawson TG, Akira S, Mechti N. The viral RNA recognition sensor RIG-I is degraded during encephalomyocarditis virus (EMCV) infection. Virology. 2009;393:311–318. doi: 10.1016/j.virol.2009.08.009. [DOI] [PubMed] [Google Scholar]
- Park N, Skern T, Gustin KE. Specific cleavage of the nuclear pore complex protein Nup62 by a viral protease. J Biol Chem. 2010;285:28796–28805. doi: 10.1074/jbc.M110.143404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsley TB, Towner JS, Blyn LB, Ehrenfeld E, Semler BL. Poly (rC) binding protein 2 forms a ternary complex with the 5′-terminal sequences of poliovirus RNA and the viral 3CD proteinase. RNA. 1997;3:1124–1134. [PMC free article] [PubMed] [Google Scholar]
- Porter FW, Bochkov YA, Albee AJ, Wiese C, Palmenberg AC. A picornavirus protein interacts with Ran-GTPase and disrupts nucleocytoplasmic transport. Proc Natl Acad Sci U S A. 2006;103:12417–12422. doi: 10.1073/pnas.0605375103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FW, Palmenberg AC. Leader-induced phosphorylation of nucleoporins correlates with nuclear trafficking inhibition by cardioviruses. J Virol. 2009;83:1941–1951. doi: 10.1128/JVI.01752-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prather SO, Taylor MW. Host-dependent restriction of mengovirus replication. II Effect of host restriction on late viral RNA synthesis and viral maturation. J Virol. 1975;15:872–881. doi: 10.1128/jvi.15.4.872-881.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozovics JM, Chase AJ, Cathcart AL, Chou W, Gershon PD, Palusa S, Wilusz J, Semler BL. Picornavirus modification of a host mRNA decay protein. mBio. 2012;3:e00431–00412. doi: 10.1128/mBio.00431-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Appel CD, Adhikari S, Robertson PD, Ramsden DA, Williams RS. Mechanism of repair of 5′-topoisomerase II-DNA adducts by mammalian tyrosyl-DNA phosphodiesterase 2. Nat Struct Mol Biol. 2012;19:1363–1371. doi: 10.1038/nsmb.2418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schellenberg MJ, Perera L, Strom CN, Waters CA, Monian B, Appel CD, Vilas CK, Williams JG, Ramsden DA, Williams RS. Reversal of DNA damage induced Topoisomerase 2 DNA-protein crosslinks by Tdp2. Nucleic Acids Res. 2016;44:3829–3844. doi: 10.1093/nar/gkw228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svitkin YV, Hahn H, Gingras AC, Palmenberg AC, Sonenberg N. Rapamycin and wortmannin enhance replication of a defective encephalomyocarditis virus. J Virol. 1998;72:5811–5819. doi: 10.1128/jvi.72.7.5811-5819.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tonew M, Tonew E, Mentel R. The antiviral activity of dipyridamole. Acta Virol. 1977;21:146–150. [PubMed] [Google Scholar]
- Virgen-Slane R, Rozovics JM, Fitzgerald KD, Ngo T, Chou W, van der Heden van Noort GJ, Filippov DV, Gershon PD, Semler BL. An RNA virus hijacks an incognito function of a DNA repair enzyme. Proc Natl Acad Sci USA. 2012;109:14634–14639. doi: 10.1073/pnas.1208096109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Nguyen JH, Ehrenfeld E, Semler BL. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA. 1999;5:1570–1585. doi: 10.1017/s1355838299991483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter BL, Parsley TB, Ehrenfeld E, Semler BL. Distinct poly(rC) binding protein KH domain determinants for poliovirus translation initiation and viral RNA replication. J Virol. 2002;76:12008–12022. doi: 10.1128/JVI.76.23.12008-12022.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wimmer E, Hellen CU, Cao X. Genetics of poliovirus. Annu Rev Genet. 1993;27:353–436. doi: 10.1146/annurev.ge.27.120193.002033. [DOI] [PubMed] [Google Scholar]
- Wong J, Si X, Angeles A, Zhang J, Shi J, Fung G, Jagdeo J, Wang T, Zhong Z, Jan E, Luo H. Cytoplasmic redistribution and cleavage of AUF1 during coxsackievirus infection enhance the stability of its viral genome. FASEB J. 2013;27:2777–2787. doi: 10.1096/fj.12-226498. [DOI] [PubMed] [Google Scholar]
- Zeng Z, Sharma A, Ju L, Murai J, Umans L, Vermeire L, Pommier Y, Takeda S, Huylebroeck D, Caldecott KW, El-Khamisy SF. TDP2 promotes repair of topoisomerase I-mediated DNA damage in the absence of TDP1. Nucleic Acids Res. 2012;40:8371–8380. doi: 10.1093/nar/gks622. [DOI] [PMC free article] [PubMed] [Google Scholar]