Abstract
Advances in sequencing have facilitated nucleotide-resolution genome-wide transcriptomic profiles across multiple mouse eye tissues. However, these RNA sequencing (RNA-seq) based eye developmental transcriptomes are not organized for easy public access, making any further analysis challenging. Here, we present a new database “Express” (http://www.iupui.edu/~sysbio/express/) that unifies various mouse lens and retina RNA-seq data and provides user-friendly visualization of the transcriptome to facilitate gene discovery in the eye. We obtained RNA-seq data encompassing 7 developmental stages of lens in addition to that on isolated lens epithelial and fibers, as well as on 11 developmental stages of retina/isolated retinal rod photoreceptor cells from publicly available wild-type mouse datasets. These datasets were pre-processed, aligned, quantified and normalized for expression levels of known and novel transcripts using a unified expression quantification framework. Express provides heatmap and browser view allowing easy navigation of the genomic organization of transcripts or gene loci. Further, it allows users to search candidate genes and export both the visualizations and the embedded data to facilitate downstream analysis. We identified total of >81,000 transcripts in the lens and >178,000 transcripts in the retina across all the included developmental stages. This analysis revealed that a significant number of the retina-expressed transcripts are novel. Expression of several transcripts in the lens and retina across multiple developmental stages was independently validated by RT-qPCR for established genes such as Pax6 and Lhx2 as well as for new candidates such as Elavl4, Rbm5, Pabpc1, Tia1 and Tubb2b. Thus, Express serves as an effective portal for analyzing pruned RNA-seq expression datasets presently collected for the lens and retina. It will allow a wild-type context for the detailed analysis of targeted gene-knockout mouse ocular defect models and facilitate the prioritization of candidate genes from Exome-seq data of eye disease patients.
Keywords: mouse, transcriptome, expression levels, RNA-seq, development stages, eye tissues
Introduction
The eye is a complex sensory organ that consists of an anterior segment that comprises of the cornea, iris, lens, ciliary body and anterior sclera, and a posterior segment that comprises of the retina, choroid and the optic nerve. Eye development is coordinated by a complex regulatory program that involves a myriad of signaling, transcriptional and post-transcriptional events (Lachke and Maas, 2010; Cvekl and Ashery-Padan, 2014; Zagozewski et al., 2014; Dash et al., 2016). With the advancement of sequencing technologies (such as Next Generation Sequencing (NGS)) and its broad application on a genome wide scale (Consortium, 2004; Chin et al., 2011; Consortium, 2015), it is possible to explore the mechanisms governing the developmental “oculome” (Lachke and Maas, 2010). Indeed, over the past decade, several studies have reported on the transcriptome of specific eye tissues at various development stages (Lachke et al., 2012; Khan et al., 2015; Tian et al., 2015; Chaitankar et al., 2016; Anand and Lachke, 2017; Kakrana et al., 2017).
RNA sequencing (RNA-seq) provides a high-resolution comprehensive platform to define cell or tissue-specific transcriptomes and monitor changes therein (Kang et al., 2015). Further, it has advanced our knowledge and understanding of the structure and composition of protein-coding and non-coding transcript isoforms in higher eukaryotes, in turn facilitating the downstream functional and comparative analysis (Chen et al., 2014; Calahorro et al., 2015; Fernandez-Valverde et al., 2015; Zimmermann et al., 2015). Transcriptome studies reported for various developmental stages on the lens (Hoang et al., 2014; Khan et al., 2015; Khan et al., 2016) and retina (Busskamp et al., 2014; Roger et al., 2014; Sundermeier et al., 2014; Uren et al., 2014; Andzelm et al., 2015; Ruzycki et al., 2015; Zhang et al., 2015) were mostly limited to comparative gene expression analysis, by restricting to known or annotated genes. However, the complete transcriptome and various isoforms in the context of developmental stages in tissues of eye are not fully characterized. In this study, we present a comprehensive and user-friendly platform termed “Express”, which enables the investigation of the transcriptomic profiles in mouse lens and retina tissues across various development stages. Express provides a one-stop portal for investigating gene expression at the resolution of individual transcripts encoded by not just the annotated coding and non-coding genes, but importantly also many novel gene loci in the mouse genome. Express facilitates this by allowing users to view the transcript level expression profiles of a gene across multiple developmental stages as heatmaps and simultaneously enables the visualization of the genomic location of the transcripts in an embedded genome browser. Users can view and download the various visualizations as well as the underlying data to facilitate rational design of experiments to study transcript structure, expression and splicing alterations across different developmental stages.
Materials and Methods
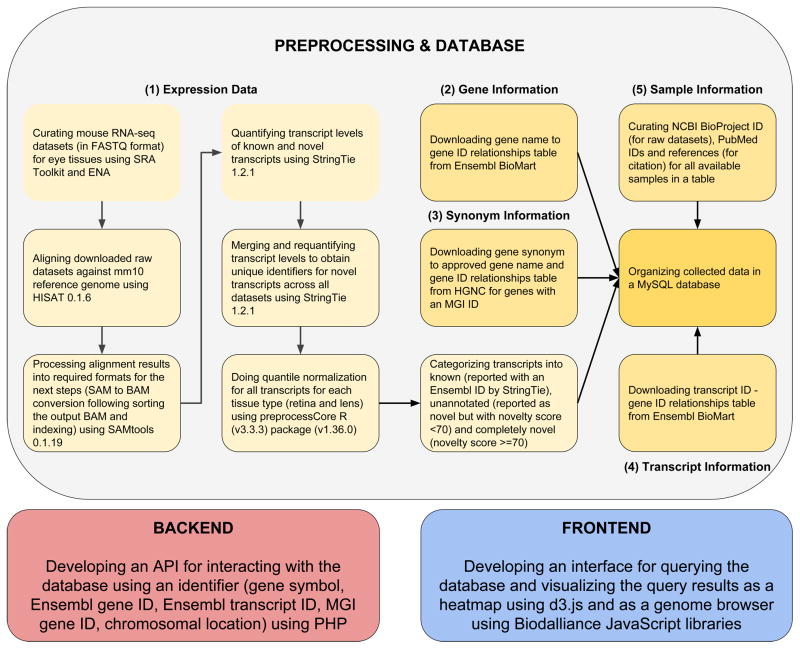
To obtain a comprehensive understanding of the transcriptome during development in lens and retinal tissues in mouse eye, we collected multiple publicly available RNA-seq datasets corresponding to the raw RNA sequence reads of mouse eye subcomponents from different developmental stages (Table 1 and Table 2). Briefly, these datasets were aligned to the mouse reference genome, quantified for expression levels of known and novel transcripts followed by the normalization of the expression levels across samples. Resulting raw and normalized expression levels were then organized into an open-source Relational DataBase Management System (RDBMS) My Structured Query Language (MySQL) database as illustrated in the workflow (Fig. 1). PHP: Hypertext Preprocessor (PHP) backend Application Program Interface (API) helps query the database and a user-friendly frontend enables the visualization of the query results as heatmap and browser views across development stages. Each of the major steps employed in processing and analysis were described in further detail in the following sections.
Table 1.
RNA-seq samples for mouse lens. The table shows SRA ID (Sequence Read Archive) for the sample, PMID (PubMed) for the study, developmental stage, read type, read length, read count, base count, and overall alignment rate using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts).
| # | SRA ID | PMID | Development Stage | Read Type | Read Length | Read Count | Base Count | Overall Alignment Rate (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | SRR2039769 | 26225632 | E15 | PE | 100 | 13772390 | 2754478000 | 94 |
| 2 | SRR2039770 | 26225632 | E15 | PE | 100 | 13542500 | 2708500000 | 95 |
| 3 | SRR953395 | 24161570 | E15.5 | SE | 52 | 48552190 | 2524713880 | 94 |
| 4 | SRR953394 | 24161570 | E15.5 | SE | 52 | 47574424 | 2473870048 | 94 |
| 5 | SRR953393 | 24161570 | E15.5 | SE | 52 | 42525381 | 2211319812 | 94 |
| 6 | SRR2039771 | 26225632 | E18 | PE | 100 | 17810970 | 3562194000 | 93 |
| 7 | SRR2039772 | 26225632 | E18 | PE | 100 | 18019388 | 3603877600 | 93 |
| 8 | SRR1222595 | 25489224 | P0 | SE | 51 | 33174286 | 1691888586 | 88 |
| 9 | SRR1222596 | 25489224 | P0 | SE | 51 | 29919226 | 1525880526 | 87 |
| 10 | SRR1222672 | 25489224 | P0 | SE | 51 | 29965660 | 1528248660 | 89 |
| 11 | SRR1222673 | 25489224 | P0 | SE | 51 | 28652759 | 1461290709 | 86 |
| 12 | SRR1222674 | 25489224 | P0 | SE | 51 | 30661663 | 1563744813 | 89 |
| 13 | SRR1222675 | 25489224 | P0 | SE | 51 | 24833352 | 1266500952 | 89 |
| 14 | SRR2039773 | 26225632 | P0 | PE | 100 | 17766309 | 3553261800 | 93 |
| 15 | SRR2039774 | 26225632 | P0 | PE | 100 | 14533000 | 2906600000 | 93 |
| 16 | SRR2039775 | 26225632 | P3 | PE | 100 | 15495833 | 3099166600 | 93 |
| 17 | SRR2039776 | 26225632 | P3 | PE | 100 | 13072393 | 2614478600 | 93 |
| 18 | SRR2039777 | 26225632 | P6 | PE | 100 | 16965754 | 3393150800 | 93 |
| 19 | SRR2039778 | 26225632 | P6 | PE | 100 | 17658286 | 3531657200 | 93 |
| 20 | SRR2039779 | 26225632 | P9 | PE | 100 | 18874309 | 3774861800 | 93 |
| 21 | SRR2039780 | 26225632 | P9 | PE | 100 | 13563853 | 2712770600 | 93 |
Table 2.
RNA-seq samples for mouse retina. The table shows SRA ID (Sequence Read Archive) for the sample, PMID (PubMed) for the study, developmental stage, read type, read length, read count, base count, and overall alignment rate using HISAT (Hierarchical Indexing for Spliced Alignment of Transcripts).
| No | SRA ID | PMID | Development Stage | Read Type | Read Length | Read Count | Base Count | Overall Alignment Rate (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | SRR1023063 | 24382353 | P2 | SE | 76 | 29939891 | 2275431716 | 93 |
| 2 | SRR1023064 | 24382353 | P2 | SE | 76 | 36280541 | 2757321116 | 94 |
| 3 | SRR1784052 | 26324254 | P10 | SE | 50 | 27028041 | 1351402050 | 94 |
| 4 | SRR1784053 | 26324254 | P10 | SE | 50 | 24267068 | 1213353400 | 94 |
| 5 | SRR1784054 | 26324254 | P10 | SE | 50 | 28777255 | 1438862750 | 94 |
| 6 | SRR1574329 | 25801704 | P11 | PE | 90 | 108113500 | 19460430000 | 96 |
| 7 | SRR1574330 | 25801704 | P11 | PE | 90 | 106003809 | 19080685620 | 97 |
| 8 | SRR1574333 | 25801704 | P11 | SE | 90 | 176162120 | 8631943880 | 87 |
| 9 | SRR1574334 | 25801704 | P11 | SE | 90 | 176206610 | 8634123890 | 87 |
| 10 | SRR1023073 | 24382353 | P21 | SE | 76 | 39574659 | 3007674084 | 94 |
| 11 | SRR1023074 | 24382353 | P21 | SE | 76 | 38855951 | 2953052276 | 94 |
| 12 | SRR1784070 | 26324254 | P21 | SE | 50 | 34792825 | 1739641250 | 95 |
| 13 | SRR1784071 | 26324254 | P21 | SE | 50 | 31461693 | 1573084650 | 95 |
| 14 | SRR1784072 | 26324254 | P21 | SE | 50 | 37264811 | 1863240550 | 95 |
| 15 | SRR1176996 | 24812086 | P28 | SE | 50 | 45056397 | 2252819850 | 92 |
| 16 | SRR1176997 | 24812086 | P28 | SE | 50 | 51508183 | 2575409150 | 91 |
| 17 | SRR1176998 | 24812086 | P28 | SE | 50 | 52339450 | 2616972500 | 92 |
| 18 | SRR1687694 | 25712131 | P30 | PE | 100 | 16507045 | 3334423090 | 84 |
| 19 | SRR1687695 | 25712131 | P30 | PE | 100 | 16384187 | 3309605774 | 84 |
| 20 | SRR1687696 | 25712131 | P30 | PE | 100 | 14348324 | 2898361448 | 81 |
| 21 | SRR1687697 | 25712131 | P30 | PE | 100 | 14251738 | 2878851076 | 81 |
| 22 | SRR1687698 | 25712131 | P30 | PE | 100 | 25674252 | 5186198904 | 83 |
| 23 | SRR1427139 | 25002228 | P30 | SE | 51 | 44636149 | 2276443599 | 68 |
| 24 | SRR1427140 | 25002228 | P30 | SE | 51 | 40396217 | 2060207067 | 66 |
| 25 | SRR1427141 | 25002228 | P40 | SE | 51 | 52430453 | 2673953103 | 74 |
| 26 | SRR1427142 | 25002228 | P40 | SE | 51 | 46344921 | 2363590971 | 75 |
| 27 | SRR1213798 | 25489233 | P48 | PE | 90 | 6779886 | 1220379480 | 96 |
| 28 | SRR1213799 | 25489233 | P48 | PE | 90 | 6803332 | 1224599760 | 97 |
| 29 | SRR1213800 | 25489233 | P48 | PE | 90 | 6826280 | 1228730400 | 96 |
| 30 | SRR1427143 | 25002228 | P50 | SE | 51 | 42904725 | 2188140975 | 70 |
| 31 | SRR1427144 | 25002228 | P50 | SE | 51 | 41243569 | 2103422019 | 68 |
| 32 | SRR1427145 | 25002228 | P60 | SE | 51 | 56910039 | 2902411989 | 48 |
| 33 | SRR1427146 | 25002228 | P60 | SE | 51 | 46737505 | 2383612755 | 56 |
| 34 | SRR1427147 | 25002228 | P90 | SE | 51 | 40366735 | 2058703485 | 76 |
| 35 | SRR1427148 | 25002228 | P90 | SE | 51 | 45096977 | 2299945827 | 71 |
Fig. 1.
Overview of the transcriptome profiling and database construction for Express. Transcriptomes of mouse lens and retina spanning several development stages (with biological replicates) were collected from published sources listed in Tables 1 and 2. Curated RNA sequence data was quality filtered using FASTX Toolkit. High quality raw sequence reads were processed and aligned to mouse reference genome mm10 using HISAT and outputs were collected as Sequence Alignment Map (SAM) files. Post-processing (i.e. conversion of SAM to sorted Binary Alignment Map (BAM)) of aligned reads was accomplished using SAMTools. Aligned and post-processed RNA-seq BAM files associated with each developmental stage were utilized for identifying and quantifying the expression levels of known and novel transcripts across respective development stages of tissue subtypes using StringTie. Quantile normalization was performed for samples per tissue type using preprocess R package. The novel transcripts reported by StringTie were categorized into unannotated (novelty score < 70) and completely novel transcripts (novelty score >= 70). These normalized expression levels of known, unannotated and completely novel transcripts were organized into a table. Gene information mapping gene names to gene IDs was downloaded from Ensembl BioMart. Synonym information mapping gene synonyms to approved gene names and gene IDs was downloaded from Hugo Gene Nomenclature Committee (HGNC) for the genes with an MGI ID. Sample information was manually curated for samples and NCBI BioProject ID, PubMed ID and study reference were obtained per sample. These collected data were then organized into a My Structured Query Language (MySQL) database. Following abbreviations and web resources have been employed in this study: SRA - Sequence Read Archive (https://www.ncbi.nlm.nih.gov/sra), ENA - European Nucleotide Archive (https://www.ebi.ac.uk/ena), HISAT - https://ccb.jhu.edu/software/hisat/, SAM - Sequence Alignment Map, BAM - Binary Alignment Map, StringTie - https://ccb.jhu.edu/software/stringtie/, R - https://www.r-project.org/about.html, Ensembl Biomart - https://www.ensembl.org/biomart, HGNC - HUGO Gene Nomenclature Committee, MGI - Mouse Genome Informatics (www.informatics.jax.org), MySQL - My Structured Query Language, API - Application Programming Interface, PHP - Hypertext Preprocessor.
Data collection and processing
We collected the raw RNA-seq reads of multiple development stages (each with its biological replicate) of mouse eye from Gene Expression Omnibus (GEO) (Barrett et al., 2013) and European Nucleotide Archive (ENA) (Gibson et al., 2016). Table 1 and 2 show the relevant source of the RNA-seq datasets along with several metrics for lens and retina respectively, resulting from the alignment of the reads to the mouse reference genome (mm10). We downloaded the single end datasets in FASTQ format (A text-based format for storing both the nucleotide sequence and its corresponding quality scores) using the Sequence Read Archive (SRA) Toolkit (fastq-dump command), and the paired end datasets were directly downloaded from ENA (European Nucleotide Archive). We ensured the quality of the aligned sequence reads was a minimum of Phred quality score 20 for each sample using FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/).
We employed Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT, version 0.1.6) for aligning short reads from RNA-seq experiments onto reference genome (Kim et al., 2015). HISAT (with default parameters) can rapidly align the quality filtered reads collected from different sources (Table 1 and 2) against the mouse reference genome mm10. SAM (Sequence Alignment/Map) files obtained as outputs from HISAT were post-processed using SAMtools (version 0.1.19) (Wheeler et al., 2008; Li et al., 2009) for converting SAM to BAM (Binary Alignment/Map) followed by sorting the output BAM files, and finally these BAM files were indexed using SAMtools. The sorted BAM files obtained after post-processing were used to quantify the expression levels of known and novel transcripts across development stages.
Transcript quantification and discovery from the aligned RNA-seq datasets was accomplished using StringTie (version 1.2.1) (Pertea et al., 2015). StringTie is a novel network flow algorithm based on a fast and highly efficient assembler to quantify the transcripts of each genomic locus considering all possible multiple splice events. In addition to annotated transcripts, it can also provide the information of possible novel transcripts in each sample. The transcript level expression data for each sample quantified using StringTie were stored as GTFs (Gene Transfer Format files) providing expression levels for both known and novel transcripts. All the GTFs obtained for each sample were grouped and provided as an input for StringTie “merge” mode along with mouse reference genome (mm10) to obtain a reference annotation file (in GTF) including novel transcripts. Next, the reference merged GTF was used in re-running StringTie with the sorted BAM files for the corresponding samples, to obtain GTFs per sample having the same transcript identifier for a given novel transcript across all the samples. The known transcripts were defined as the transcripts that were annotated as reference mouse transcripts in the Ensembl database (Yates et al., 2016). In contrast, novel transcripts were defined as the transcripts that were exclusively predicted by StringTie with little or no overlap with existing mouse transcript annotations in mm10. We examined the length of the discovered transcript onto annotated reference transcript coordinates and calculated a novelty score for each novel transcript by using the below formula,
The novel transcripts having a novelty score (NS) ≥70 were considered as completely novel and the novel transcripts having novelty score <70 were considered as unannotated transcripts. Since sequencing or processing artifacts at various steps of the transcript quantification analysis could potentially contribute to high number of transcript isoforms, we have classified transcripts into three categories namely a) known transcripts which are annotated in Ensembl database (https://www.ensembl.org/Mus_musculus/Info/Index) b) completely novel transcripts i.e., transcripts which exhibit a novelty score of at least 70 and c) the remaining transcripts were classified as unannotated transcripts and excluded from all the downstream analysis. A quantification matrix was generated for both the lens and retinal transcriptomes with respect to different development stages by extracting the TPM (Transcripts Per Million reads sequenced) values from StringTie outputs. These matrices were utilized for downstream analysis such as normalization of expression levels and the corresponding datasets (raw and normalized expression levels) were employed to show the transcript levels across stages in lens and retina in Express.
Normalization of transcript expression levels across samples in a tissue
Although RNA-seq samples originating from the same laboratory are unlikely to have significant technical variation among the replicates and developmental stages, there could still be variations arising due to factors like tissue preparation, RNA extraction and sequencing depth differences. In our analyzed datasets for both lens and retina, RNA sequencing datasets originating from multiple labs and protocols were analyzed. Hence, in addition to providing the default option of raw expression levels of a transcript across developmental stages, we have performed a widely adopted quantile normalization of the samples using the preprocessCore package (Bolstad, 2001) in R and used the resulting normalized expression data for showing the expression heatmaps in Express. Quantile normalization is a global adjustment method that assumes the statistical distribution of each sample under study is the same (Bolstad et al., 2003). Normalization is achieved by forcing the observed distributions to be the same and the average distribution, obtained by taking the average of each quantile across samples, is used as the reference. Its application on both microarray and RNA sequencing data has consistently shown its superior performance compared to other competing methods (Bolstad et al., 2003; Dillies et al., 2013). Hence, we used this normalization on our RNA-seq expression profile matrices across developmental stages in lens and retina respectively. Raw or normalized expression levels of replicates of a developmental stage were averaged for display purposes on Express. In addition to the quantile normalization, to exhibit only high quality relevant transcripts, the end user has the option to select only highly expressed transcripts for visualization. This is facilitated by including a selection filter which allows the visualization of the expression levels for only those transcripts of a gene which have at least a certain level of expression observed in at least one of the developmental stages shown.
Database construction and implementation
In order to build the Express database of transcriptome profiles encompassing known and novel transcripts across multiple development stages in eye tissues in mouse, we employed several steps. These steps are illustrated in Fig. 1. Briefly, as described in the above sections, the aligned, quantified and then normalized datasets organized as final matrices for each tissue type were stored into an SQL table. Express stores both the raw as well as the quantile normalized expression levels of transcripts in Transcripts Per Million Reads (TPM) units. Moreover, the sample metadata information is manually curated with NCBI BioProject ID, PubMed ID and a reference for citing the corresponding dataset. Also, the table containing Ensembl gene ID, MGI (Mouse Genome Informatics) gene ID and chromosomal location for all genes in mouse genome was downloaded from Ensembl BioMart and the table containing gene synonym, approved gene name and Ensembl gene ID was downloaded from HGNC (HUGO Gene Nomenclature Committee) for genes that are linked to an MGI gene ID. Similarly, we obtained transcript ID - gene ID relationships table from Ensembl BioMart for linking gene information to the expression data. We then organized these tables into a MySQL database whose schema is shown in Fig. S1.
User interface and access
Backend
We developed a PHP: Hypertext Preprocessor (PHP) Application Programming Interface (API) for interacting with the database using a query (e.g. gene symbol, Ensembl gene ID, MGI gene ID, Ensembl transcript ID or chromosomal location) for the user given TPM cutoff and tissue type. Upon sending the query, TPM cutoff and tissue type to the API, the query type is identified, and the corresponding quantile normalized transcript level expression data is retrieved from the database. Next, the expression values are normalized between 0 and 1 per transcript and the final data is returned in JSON (JavaScript Object Notation) format to be visualized by the frontend. The backend PHP API can also be used programmatically to obtain expression data, which is documented on documentation page of Express (http://www.iupui.edu/~sysbio/express/docs.html).
Frontend
The frontend interacts with the user to accept input (a tissue type, a TPM cutoff, value type and a query) and a visualization of the retrieved data from the MySQL database is provided. We show the structure of queried transcripts in a genome browser developed using Biodalliance JavaScript library (http://www.biodalliance.org). We obtained mouse transcript structures from GENCODE version M7 (GRCm38.p4) in BigBed format (A binary file format that is created by conversion from a Browser Extensible Data format file) available on http://www.gencodegenes.org/mouse_biodalliance.html. To this BigBed file, we added the structures of novel transcripts discovered by StringTie from our analysis. We renamed the default identifiers obtained from StringTie to include corresponding tissue type in the identifier for easy understanding in the genome browser. To modify the GENCODE transcript annotation, we first converted the BigBed file into BED file, added the structures of novel transcripts and then converted back to BigBed format using UCSC utilities (Kent et al., 2010). Also, the expression data per transcript across multiple developmental stages is shown as a heatmap developed by using d3.js JavaScript library (https://d3js.org). The heatmap is sorted by transcript groups (as introduced in the section “Data collection and preprocessing”) as known transcripts, completely novel transcripts and unannotated transcripts. The transcripts in each group are also sorted by the averaged expression value for all developmental stages for keeping highly expressed transcripts at the top. The front end provides two select boxes for choosing an available TPM cutoff (0, 1, 2, and 5), and the tissue type and a textbox for entering the query. The frontend interface allows the user to choose a minimum expression cutoff for a transcript, which enables the display of only those transcripts resulting from search exhibiting this minimum expression level cutoff in at least one developmental stage. The default cutoff is set to 5 TPM. The value type select box can also be used to query for raw expression values or quantile normalized expression values. After search is performed, the results are shown as a heatmap along with a genome browser to view the transcript structure. The heatmap and browser view can be toggled using the button on the right-hand side of the navigation bar. Also, using the Export dropdown menu, it is possible to export heatmap view and browser view in SVG (Scalable Vector Graphics) format and heatmap data in TSV (Tab Separated Values).
Experimental validation of the RNA-seq identified transcripts for lens and retinal expressed genes
The University of Delaware animal facility hosted all the mice used in these experiments, which were performed following the guidelines defined in the Association for Research in Vision and Ophthalmology (ARVO) statement for the use of animals in ophthalmic and vision research. C57Bl/6 mouse lenses were microdissected at three stages, namely, embryonic day (E) 15.5, post-natal day (P) 0 and P10. Retina was dissected from four stages, namely P10, P20, P30 and P48. The day of detection of vaginal plug was defined as E0.5. Each of three biological replicates at E15.5 comprised of six lenses, and at P0 and P10 comprised of two lenses. Each of the biological replicates for retinal expression comprised of 2 retinas from P10, P20, P30 and P48. Total RNA was extracted from lenses using RNeasy Mini kit (Qiagen Inc, Valencia, CA) and cDNA was synthesized using Bio-Rad iScript™ cDNA Synthesis Kit (Bio-Rad Laboratories, Hercules, CA), for use as a template in quantitative PCR (RT-qPCR) analysis. Forward and reverse primers were designed on the longest isoform of the transcript on exonic sequence flanking an intronic region such that the product sizes were < 300 bp (Table 3). RT-qPCR was performed using Power SYBR Green PCR Master Mix (Invitrogen life technology, Grand Island, NY). Several house-keeping genes namely, Actb, B2m (Beta 2-microglobulin), and Hprt were used for normalization (Wigle et al., 1999; Shaham et al., 2013; Cavalheiro et al., 2014; Mamuya et al., 2014; He et al., 2016). Fold-change differences between target gene expression compared to specific housekeeping gene expression was estimated using the ΔΔCt method. The first comparison of gene expression in the ΔΔCt method was performed independently with several housekeeping genes. The second comparison was calculated based on expression at E15.5 (lens samples), and at P10 (retina samples). Statistical significance was calculated using two-way ANOVA as described (Bookout and Mangelsdorf, 2003).
Table 3.
Primers for qPCR validation of transcripts.
| Gene | Forward primer sequence (5′ -> 3′) | Reverse primer sequence (5′ -> 3′) |
|---|---|---|
| Pax6 | AGTTCTTCGCAACCTGGCTA | ACTTGGACGGGAACTGACAC |
| Elavl4 | GGCAGAAGAAGCCATCAAAG | GCAAATTGTCCAGCCTGAAT |
| Rbm5 | GCACCACAGTGACTACCACCT | GCACGTAGGTCTCCTTCTCG |
| Lhx2 | CGCGCTTAGCTGTAACGAGAA | CGCTTTGTCTTTTGGCTGCT |
| Pabpc1 | GAGACCAGCTTCCTCACAGG | ACCTTGCACATGAACAGCAG |
| Tia1 | GGCTTGGTGGAAGACAAATC | TCACCCCTCCACAGTACACA |
| Tubb2b | ATCGGTGCCAAGATCGGT | CCTGAAGATCTGCCCAAATG |
Results and Discussion
Overview of Express database
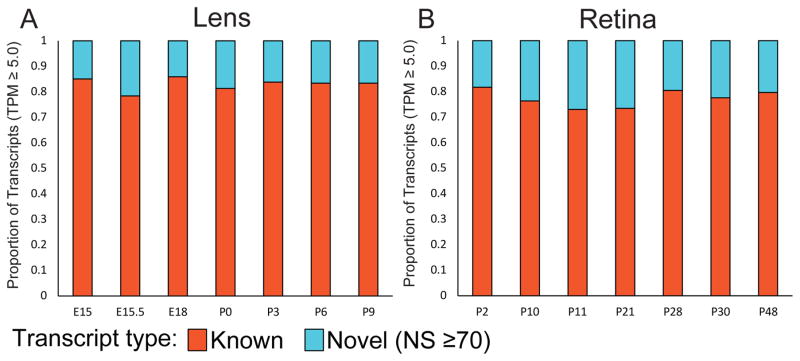
Express is a database of transcriptome profiles encompassing known and novel transcripts across multiple development stages in mouse eye tissues. Several steps involved in preprocessing, postprocessing, quantification and normalization of collected data followed by its organization in Express are illustrated in Fig. 1 (see Materials and Methods). Express contains 81779 distinct transcripts for mouse lens and 178367 distinct transcripts for mouse retinal samples. Novel transcripts are defined as those that are not annotated in the reference genome annotation (see Materials and Methods). The proportions of the known and completely novel transcripts for each developmental stage at 5 TPM threshold in lens and retina are shown in Figs. 2A and 2B, respectively (Fig. S2 shows the distributions for different TPM thresholds). In the following sections, we illustrate the composition of the datasets and functionality of the database as well as present validations of several genes in lens and retinal tissues, to demonstrate the utility of Express for studying eye development.
Fig. 2.
Histograms showing the proportion of known and completely novel transcripts documented in Express across developmental stages at 5 Transcripts Per Million (TPM) mapped reads threshold. (A) Proportion of transcripts for each stage available for lens samples from E15 to P9. (B) Proportion of transcripts for each stage available for retinal samples from P2 to P90. Multiple datasets associated with a given developmental stage are merged to facilitate the ease of comparison across stages.
Analysis of lens and retinal RNA-seq data for building Express
Express contains gene and transcript level expression data obtained from 21 lens and 35 retinal RNA-seq mouse samples. As shown in Table 1 for lens samples and in Table 2 for retinal samples, the information about the datasets used in this study is given per dataset as Sequence Read Archive (SRA) ID, PubMed (PM) ID, developmental stage, read type (single-end or paired-end), read length (in bp), read count, base count and overall alignment rate. The lens samples include developmental stages from E15 to P9 whose read types and read lengths vary with alignment rates ranging from 86% to 94%. The retinal samples include developmental stages from P2 to P90 whose read types and read lengths also vary. Majority of the retinal samples exhibited a high overall alignment rate varying from 80% to 97% (See Materials and Methods, Table 1 and 2). We downloaded the raw datasets for lens and retinal samples from Gene Expression Omnibus (GEO) (Barrett et al., 2013) and European Nucleotide Archive (ENA) (Gibson et al., 2016). We performed a quality control using FASTX-Toolkit. Reads with a Phred quality score lower than 20 were filtered out. Next, we aligned them to the mouse reference genome (mm10) using Hierarchical Indexing for Spliced Alignment of Transcripts (HISAT) and quantified the aligned datasets for transcript expression levels for known and novel transcripts identified by StringTie, which is an established method for identifying new transcripts (See Materials and Methods, Fig. 1). The gene and transcript information from mouse reference genome were also downloaded from Ensembl (Yates et al., 2016) and organized along with the expression data in a MySQL database (See Materials and Methods, Fig. 1). To control for technical variation in expression levels between samples, we performed a quantile normalization of all the samples in a given tissue type (See Materials and Methods). Both raw as well as normalized expression levels in Transcripts Per Million (TPM) reads sequenced units, are stored in the database and are available to download from the Express website. We also developed a PHP: Hypertext Preprocessor (PHP) backend to interact with the database and a frontend to interact with the user and to visualize the query results (See Materials and Methods, Fig. 1).
User guide for exploring Express database
To retrieve gene expression data from Express, we have added the following features to the web interface. Step-by-step instructions for using Express are also available as a User Manual (see Figure 3A and the web interface of Express following the webpage -http://www.iupui.edu/~sysbio/express/user-guide.html).
Fig. 3.
User guide for employing Express to investigate eye gene expression is highlighted in panel 3A. 1) User selects parameters, 2) Enters query gene or chromosomal region, 3) Selects view options, 3a. with heatmap view can visualize gene expression in various developmental stages, 3b. with Browser view can visualizes different transcripts, 4) Uses magnification slider to controls chromosomal range, 5) Can use the Export dropdown menu to download heatmap view, raw or normalized gene expression data or browser view. Heatmaps showing the expression profiles of selected (B) transcription factors (C) crystallin genes and (D) non-crystallin genes in multiple development stages of mouse lens. Expression data is normalized by the maximum expression level of a given transcript across stages and visualized as heatmap in Express.
The parameters to investigate the expression of a gene are (a) tissue type, namely lens, retina and lens cell subtype; (b) expression level, namely gene or transcript (splice isoform) level; (c) TPM (transcripts per million) cutoff of 0, 1, 2 and 5 and tpm values which could be raw or values after quantile normalization.
The query to investigate gene expression could be gene name, ENSEMBL ID or chromosome location.
-
The output can be viewed in (a) heatmap or (b) browser view using toggle buttons on the top right side of the web interface.
Heatmap view shows gene expression at different developmental stages. A heatmap is provided on the panel with gradations of blue color intensity. Higher intensity of blue indicates high gene expression compared to other developmental stages investigated in this study. The output is shown as gene name, ENSEMBL ID and chromosome location and the relative expression of query gene.
Browser view shows all the genes and transcripts expressed in lens and retina in the query chromosomal location. Unannotated genes are displayed as MSTRG.XXXX.XXXXX.X.
The chromosomal window on the browser view can be increased or decreased using the magnification slider provided on the top right of the browser view panel.
The heatmap view and the browser view can be downloaded as high-resolution images using a dropdown export menu provided on the top right-hand side of the web interface. Along with the views, the raw and quantile normalized values for each isoform of a gene can be downloaded using the heatmap data option on the export menu.
Further sources for the RNA seq-data used for the analysis in this study are provided at the bottom of the web interface.
As shown in Fig. 3A, for instance – on investigating the gene expression profile of Bfsp1 in the lens at the gene level, at a threshold of 5 TPM for raw expression level, an output is generated with both heatmap and browser view. In the browser view, all genes expressed in the lens and retina at the chromosomal location as Bfsp1 can be visualized. In the heatmap view, relative expression of Bfsp1 at various developmental stages is shown. We can compare the expression of splice forms of Bfsp1 at different developmental stages using the transcript level option i.e. while the expression of isoform ENSMUST00000099296 increases with development, the expression of ENSMUST00000028907 is highest at P0.
Express re-captures the general expression trends in developing lens tissue for various transcription factors as shown in Fig. 3B. For example, Pax6, Sox2, Mab21l1, Foxe3, Pitx3 and Mafg exhibit high expression in early (embryonic) lens development stages compared to late (postnatal) stages. In contrast, Maf, Prox1, and Sox1 expression is lower in early lens developmental stages and higher in later stages. Similarly, Express-based analysis of gene expression for Crystallin genes in lens development revealed an increased expression of Crystallin genes in postnatal stages compared to embryonic stages in developing lens tissue as shown in Fig. 3C. Express also shows varying expression profile of various non-crystallin genes that are linked to human congenital/pediatric cataract (Fig. 3D). For example, Chmp4b, Gemin4, Pxdn, Vim, Agk, Fyco1 and Wfs1 are expressed highly in early embryonic stages, while Epha2, Gja3, Gja8, Lim2, Mip, Bfsp1 and Bfsp2 exhibit high expression in late embryonic and postnatal stages.
Development of Express as a user-friendly tool
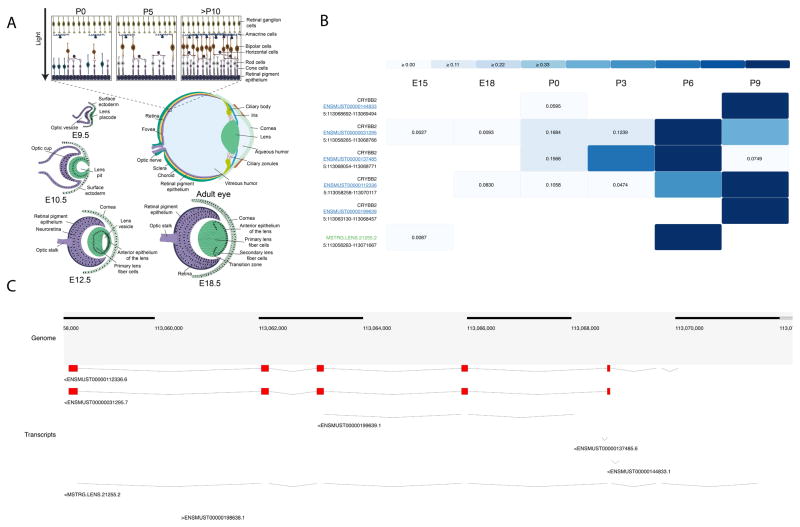
Express provides transcript level expression data for mouse lens and retina across different developmental stages for known and novel transcripts as identified by StringTie. The mouse developmental stages are expressed as embryonic (E) or post-natal (P) followed by a number that indicates the number of days after fertilization or birth, respectively (e.g. E18 corresponds to an embryo dissected 18 days after the vaginal plug was observed, while P0 corresponds to the day of birth). RNA-seq data is available for 7 developmental stages of the lens (E15, E15.5, E18, P0, P3, P6, and P9) and 11 development stages of the retina (P2, P10, P11, P21, P28, P30 P40, P48, P50, P60 and P90). In express, users can also search for cell-type specific expression profiles where available. For instance, a representation for lens dataset such as P0:E and P0:F stands for the epithelial and fiber compartments in lens. The fraction of transcripts for each developmental stage for lens and retinal samples is shown in Fig. 2. Although majority of the lens developmental stages exhibit ~17% of completely novel transcripts, the proportion of completely novel transcripts in retina were found to be significantly higher and varying with expression threshold. Observed fraction of completely novel transcripts was found to be <25% across majority of the retina stages when transcripts were filtered to include only those expressed greater than 5 TPM in retinal samples (see Fig. 2 and Supplementary Fig. S2). The number of retina-expressed transcripts that are identified to be novel in this study is comparable to that previously reported in the human retina (Farkas et al., 2013), and therefore supports the finding that retinal cells potentially express a large number of uncharacterized transcripts. In Express, users can filter to view only those transcripts resulting from a search that satisfy one of the four levels of confidence in expression levels – 1) transcripts exhibiting a non-zero expression level in TPM in at least one developmental stage, 2) transcripts with at least 1 TPM in at least one developmental stage 3) transcripts with at least 2 TPM in at least one developmental stage and 4) transcripts expressed with at least 5 TPM in at least one developmental stage (default threshold). A summary of eye developmental stages for ready comparison of ocular morphological changes with Express data stages is shown in Fig. 4A.
Fig. 4.
Overview of the mouse eye development and user interface. (A) In the initial stages of eye development, the optic vesicle interacts with the overlying non-neural surface ectoderm at embryonic day (E) 9.5 in mouse and induces its thickening to form the lens placode. Subsequently at E10.5 the optic vesicle and the lens placode interact to develop into the optic cup and the lens pit, respectively. The lens pit closes to the form the lens vesicle and the overlying ectoderm contributes to the corneal epithelium. The posterior cells of the lens vesicle differentiate to form the primary lens fiber cells while cells of the anterior epithelium of the lens divide to form new epithelial cells that migrate towards the transition zone. Cells at the transition zone exit the cell cycle and terminally differentiate to form the secondary fiber cells. Further, the fiber cells migrate towards the center of the lens, and as they terminally differentiate, undergo organelle degradation, resulting in an organelle free zone in the center of the lens by E18.5. Further development and differentiation events lead to the formation of the adult eye where the anterior region consists of the cornea, iris, cilliary body and cilliary zonules. The posterior of the lens consists of the retina, retinal pigment epithelium, choroid and sclera. A more detailed diagram of the retina shows that it is composed of several distinct cell types, including the retinal ganglion cells, amacrine cells, bipolar cells, horizontal cells and the rod and cone photoreceptors. Retinal ganglion cells and cone cells are differentiated and functional by E18.5 and by postnatal day (P) 5, amacrine cells, bipolar cells, horizontal cells and rod cells are formed. By P10, all the neuronal cells in the retina have completely connected synaptic junctions. Rod and cone cells synapse with horizontal cells for communicating with other photoreceptors and with bipolar cells, which further synapse with amacrine cells. The amacrine cells in turn synapse with the retinal ganglion cells. (B) Heatmap view from Express resulting from a query for the chromosomal location “5:113058250-113072250” when lens is selected as tissue with TPM cutoff = 5 using d3.js JavaScript package. The transcripts are sorted by novelty category (known transcripts colored in blue and linked to its Ensembl transcript page, completely novel transcripts colored in green and unannotated transcripts colored in black). The transcripts within each novelty category are also sorted by averaged normalized expression values for the row. In lens datasets some development stages are marked as “E” for epithelium and “F” for fiber cells and unmarked development stages represent whole tissue. Also in retina some development stages are marked as “C” for cone and “R” for rod cells and unmarked development stages represent whole tissue. The marked datasets are derived from the given cell type. (C) Browser view from Express from a query for the chromosomal location “5:113058250-113072250” using BioDalliance JavaScript package.
At 5 TPM cut-off, the lens samples were found to exhibit ~16% completely novel transcripts across stages. In contrast, the retinal samples were found to comprise of ~22% completely novel transcripts. When lower expression thresholds were used the fraction of completely novel transcripts significantly increased in retinal samples. It is likely that the high number of novel transcripts in retinal samples is likely due to the several distinct types of cells in the retina (Fig. 4A and Supplementary Fig. S2). Indeed, the total number of transcripts identified in mouse retina in this study are very similar to the numbers reported in human retinal samples (Farkas et al., 2013). Express allows a heatmap view for any specific query, as shown in the example query of the chromosomal location “5:113058250-113072250” for the lens (Fig. 4B). A legend is provided at the top to indicate the intensity of the color for normalized expression values across the heatmap (dark color corresponds to high expression), while the heatmap itself shows transcripts in rows and different development stages in columns that represent progressive stages. The row labels show the gene symbol, Ensembl transcript ID linked to its official page or novel transcript ID and the chromosomal location of the transcript. The heatmap view can be exported using the Export dropdown menu in the navigation bar in SVG format, which can provide high-quality vector image for use in publications. Fig. 4C shows the browser view for the same query (for the chromosomal location “5:113058250-113072250”). The mouse genome is provided in the “Genome” track and the “Transcripts” track provides the view showing the various transcript structures for known and novel transcripts encoded by this genomic location. The browser is interactive and one can drag and navigate through neighboring transcript structures in the browser. The browser view can also be exported using the Export dropdown menu in the navigation bar in SVG format, which can serve to provide high-quality vector image for use in publications. In addition, users can also download both the raw and normalized expression levels resulting from the search for a gene of interest as tab-delimited text files.
Validation of transcript-expression in lens and retina
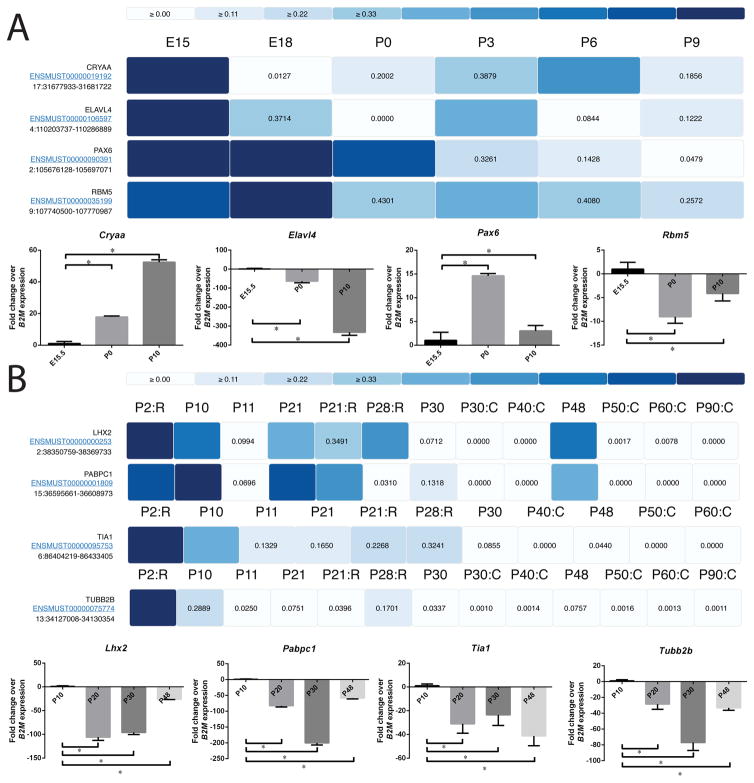
We identified several genes and their corresponding transcripts that were found to significantly altered across the developmental stages in lens and retina. We validated the expression pattern of these genes as well as other established genes as a representative set of very significantly altering transcripts across stages to evaluate expression levels reported in Express. In particular, we downloaded the expression profile of the selected transcripts (in the form of a heatmap) from Express for each tissue subtype and experimentally validated their levels for multiple development stages using RT-qPCR (see Materials and Methods). In the lens, we validated the expression of Pax6, Elavl4 and Rbm5 (Fig. 5A, Fig. S3A). Pax6 (Paired box 6) is a transcription factor essential for eye development in mice and humans. Mutations in Pax6 have been linked to congenital cataract, aniridia and anophthalmia in humans (Glaser et al., 1994) and haplo-insuffciency of Pax6 in mice results in small eyes (Sey) in mice (Hogan et al., 1986; Hill et al., 1991). RT-qPCR shows that Pax6 expression is elevated in early postnatal stages in agreement with Express (Fig. 5A). Elavl4 (ELAV (Embryonic Lethal, Abnormal Vision, Drosophila)-like 4 (Hu antigen D) belongs to ELAV protein family and is expressed in the mouse lens and frog retina (Amato et al., 2005; Bitel et al., 2010). Elevated expression of Elavl4 in the mouse lens increases the expression level of its targets (GAP43 and CamKIIα), which is a similar outcome to its overexpression in brain tissue (Bitel et al., 2010). We find that the expression of Elavl4 in mouse lens is high during embryonic stages and reduces gradually in postnatal stages (Fig. 5A), as predicted by the transcriptome datasets in Express. Rbm5 (RNA binding motif protein 5) belongs to the Rbm protein family and is associated with lung cancer (Shao et al., 2012; Li et al., 2014; Su et al., 2014; Yang et al., 2016). We find that Rbm5 is expressed highly at embryonic stages and its expression reduces during early postnatal stages (Fig. 5A). While this is the first report of Rbm5 expression in the lens, another member of the Rbm family, Rbm24 is expressed in the vertebrate eye and its deficiency in zebrafish causes microphthalmia (Lachke et al., 2012; Maragh et al., 2014).
Fig. 5.
Heatmaps showing the expression profiles of selected transcripts in multiple development stages of mouse eye tissues. Expression data were normalized by the maximum expression level of a given transcript across stages and visualized as heatmap in Express. Expression profile of selected transcripts for (A) lens and (B) retina were downloaded from Express and shown in form of heatmap. In retina datasets some development stages are marked as “C” for cone and “R” for rod cells and unmarked development stages represent whole tissue. The marked datasets are derived from the given cell type. Their expression profile was also verified for multiple development stages using qPCR with B2M as housekeeping control and shown as additional panels.
Further, we also validated the expression of Express-predicted genes Lhx2, Pabpc1, Tia1 and Tubb2b in the retina (Fig. 5B, Fig. S3B). Lhx2 (LIM homeobox 2) encodes an eye field transcription factor that is expressed from the earliest stages of optic development. LHX2 mutations in human as well as its knockout in mice causes anophthalmia (Porter et al., 1997; Desmaison et al., 2010). As indicated by Express, RT-qPCR show that Lhx2 expression reduces in the retina in late postnatal stages (Fig. 5B). Pabpc1 (Poly A-binding protein, cytoplasmic 1) binds to the poly A tail of mRNA and modulates its susceptibility to cap-mediated mRNA decay (Walters et al., 2010). RT-qPCR shows that Pabpc1 is expressed highly at early postnatal stages and its expression reduces significantly at later developmental stages (Fig. 5B) until P30 when its expression increases again. Tia1 (T-Cell-Restricted Intracellular Antigen-1) promotes the recruitment of U1 snRNP to splice sites and is implicated in lymphoma and leukemia (Forch et al., 2002; Milne et al., 2009; Koreishi et al., 2010), which is also expressed in the mouse lens (Lachke et al., 2011). We find that Tia1 expression gradually decreases in the retina with age (Fig. 5B). Tubb2b (Tubulin, Beta 2B Class IIb) is a component of microtubules. Tubb2b mutations result in congenital fibrosis of extraocular muscles (CFOEM), which leads to ptosis (drooping eyelids) and restricted eye movements in humans (Cederquist et al., 2012). RT-qPCR confirms that Tubb2b expression is high at P10 and reduces sharply at P30 before increasing again at P48 (Fig. 5B) as predicted by Express. We also verified the levels of the control genes compared across time points for reference as shown in Fig. S3C.
We also investigated how Express compares with the established expression pattern for the gamma-Crystallin family of genes. A previous study describes the expression of different Cryg family transcripts at the mouse stages E16.5, P1, P10, P20, P30, P40, P80, P120, and P180 (Goring et al. 1992). We compared Cryg gene expression for the stages in Express that are closest in developmental time to the stages in the Goring et al. study. Specifically, we compared Cryg expression in Express for E15, P0 and P9 that are close to the stages E16.5, P1 and P10 in the Goring et al. study. Using raw expression and TPM cut-off of 5, we find that there is good agreement between the Express and previous findings for the general trends of the Cryg genes, namely Cryge, Crygf, Crygb, Crygc and Crygd (Fig. 6). Cryga showed a slight deviation from the Goring et al. study in that it did not exhibit a slight reduction at P0 prior to being high at P10 (although it exhibits general agreement with the previous study in that the expression of Cryga was higher at P9 compared to E15). Therefore, these findings offer further support that gene expression data in Express reflects the experimentally validated and established gene expression patterns in the lens.
Fig. 6.
Gene expression analysis of Cryg family of genes. Raw Expression profiles of Cryg genes in mouse lens were downloaded from the Express database to investigate if they are in agreement with previously described patterns (reference: Goring et al.,1992). Expression values for specific Cryg genes in the stages closest to the developmental time points in the previous study are plotted (E15 in Express in lieu of E16.5 in Goring et al. 1992, P0 in lieu of P1, P9 in lieu of P10). The general expression patterns for the Cryg genes correlate well between Express and the previous findings. Specifically, Crygb, Crygc, Crygd and Crygf expression increases as development progresses and is highest at P9, while Cryge expression elevates at P0 and beyond. The only minor deviation is exhibited by Cryga whose expression increases from E15.5 through P9 in Express, instead of the slight decrease at P1 prior to increasing again at P10 as previously described.
Conclusion
A number of studies in the past have focused on studying the expression landscape of genes using microarrays across developmental stages (Farjo et al., 2002; Blackshaw et al., 2004; Zhang et al., 2006) in mouse eye development and specialized databases (Lachke et al., 2012; King et al., 2015) have been built. However, our understanding of the transcript structure, expression and their splicing alterations is just beginning to emerge during lens development across model systems (Srivastava et al., 2017). Here, we present Express which to our knowledge is the first large-scale carefully pruned transcriptomic resource based on eye tissue RNA-seq data to provide a user-friendly portal for studying and visualizing the expression levels of both the known and novel transcript isoforms across developmental stages in mouse eye tissues. Further, we validate several transcripts using RT-qPCR across multiple developmental stages in mouse lens and retinal tissues to confirm that the Express-quantified levels of transcripts are in agreement with the detected expression levels from RNA-seq quantification pipeline employed in this study. We found several transcripts encoding RNA-binding proteins to be highly expressed in embryonic development that are significant down-regulated in post-natal stages suggesting that post-transcriptional control of gene expression may function in early eye development.
Our analysis suggests that retinal samples exhibit a significant number of novel transcripts comparable to a recent analysis of human retinal transcriptomes (Farkas et al., 2013). It can be speculated that these novel RNA transcripts may reflect cell type specific functions. Hence resources like Express can not only further our understanding of the tissue-specific developmental transcriptome but can also serve to improve gene annotations in mouse. Although several of these novel transcripts identified across developmental stages in our analysis could be non-coding, with some isoforms resulting from errors in transcript assembly process, we include them in Express since some of these transcripts could have regulatory roles that are yet to be discovered. However, since Express enables users to filter the gene and transcripts resulting from a search based on their expression levels, a user can choose to analyze and explore only the most promising candidates.
We anticipate that future versions of Express, which will continue to be manually curated and pruned to comprise of only high quality RNA-seq expression data across eye tissues, can be a useful resource for prioritization of candidate genes from exome sequencing analysis for patients with ocular defects as well as for providing a functional and developmental context to investigate the significance of differentially expressed genes in mouse mutants with eye defects.
Supplementary Material
Highlights.
A unified database to study developmental transcriptomes in eye tissues is presented
Transcriptome profiles encompassing multiple mouse lens and retinal RNA-sequencing datasets
User-friendly visualization of transcriptomes using heatmap and browser centric views
Both known and novel transcript isoforms can be navigated and visualized easily
Several transcripts were independently validated by qRT-PCR in multiple stages
Acknowledgments
Research reported in this publication was in part supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number R01GM123314 (SCJ) and the National Eye Institute of the National Institutes of Health under Award Number R01EY021505 (SAL). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health. SAL is a Pew Scholar in the Biomedical Sciences.
Footnotes
Authors’ contribution
GB, RS and SCJ designed the study, implemented the computational approaches and performed the data analysis. GB, RS, and SD interpreted the data. SD and SAL designed and validated the predicted targets experimentally. RS, GB, SD wrote the manuscript. All authors read and approved the final manuscript.
Competing interests
The authors declare no competing financial interests.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Amato MA, Boy S, Arnault E, Girard M, Della Puppa A, Sharif A, Perron M. Comparison of the expression patterns of five neural RNA binding proteins in the Xenopus retina. J Comp Neurol. 2005;481:331–339. doi: 10.1002/cne.20387. [DOI] [PubMed] [Google Scholar]
- Anand D, Lachke SA. Systems biology of lens development: A paradigm for disease gene discovery in the eye. Exp Eye Res. 2017;156:22–33. doi: 10.1016/j.exer.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andzelm MM, Cherry TJ, Harmin DA, Boeke AC, Lee C, Hemberg M, Pawlyk B, Malik AN, Flavell SW, Sandberg MA, Raviola E, Greenberg ME. MEF2D drives photoreceptor development through a genome-wide competition for tissue-specific enhancers. Neuron. 2015;86:247–263. doi: 10.1016/j.neuron.2015.02.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A. NCBI GEO: archive for functional genomics data sets--update. Nucleic Acids Res. 2013;41:D991–995. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitel CL, Perrone-Bizzozero NI, Frederikse PH. HuB/C/D, nPTB, REST4, and miR-124 regulators of neuronal cell identity are also utilized in the lens. Mol Vis. 2010;16:2301–2316. [PMC free article] [PubMed] [Google Scholar]
- Blackshaw S, Harpavat S, Trimarchi J, Cai L, Huang H, Kuo WP, Weber G, Lee K, Fraioli RE, Cho SH, Yung R, Asch E, Ohno-Machado L, Wong WH, Cepko CL. Genomic analysis of mouse retinal development. PLoS Biol. 2004;2:E247. doi: 10.1371/journal.pbio.0020247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolstad B. Probe Level Quantile Normalization of High Density Oligonucleotide Array Data. Unpublished manuscript. 2001 http://bmbolstad.com/stuff/qnorm.pdf.
- Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- Bookout AL, Mangelsdorf DJ. Quantitative real-time PCR protocol for analysis of nuclear receptor signaling pathways. Nucl Recept Signal. 2003;1:e012. doi: 10.1621/nrs.01012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busskamp V, Krol J, Nelidova D, Daum J, Szikra T, Tsuda B, Juttner J, Farrow K, Scherf BG, Alvarez CP, Genoud C, Sothilingam V, Tanimoto N, Stadler M, Seeliger M, Stoffel M, Filipowicz W, Roska B. miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron. 2014;83:586–600. doi: 10.1016/j.neuron.2014.06.020. [DOI] [PubMed] [Google Scholar]
- Calahorro F, Holden-Dye L, O’Connor V. Analysis of splice variants for the C. elegans orthologue of human neuroligin reveals a developmentally regulated transcript. Gene Expr Patterns. 2015;17:69–78. doi: 10.1016/j.gep.2015.02.002. [DOI] [PubMed] [Google Scholar]
- Cavalheiro GR, Matos-Rodrigues GE, Gomes AL, Rodrigues PM, Martins RA. c-Myc regulates cell proliferation during lens development. PLoS One. 2014;9:e87182. doi: 10.1371/journal.pone.0087182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cederquist GY, Luchniak A, Tischfield MA, Peeva M, Song Y, Menezes MP, Chan WM, Andrews C, Chew S, Jamieson RV, Gomes L, Flaherty M, Grant PE, Gupta ML, Jr, Engle EC. An inherited TUBB2B mutation alters a kinesin-binding site and causes polymicrogyria, CFEOM and axon dysinnervation. Hum Mol Genet. 2012;21:5484–5499. doi: 10.1093/hmg/dds393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaitankar V, Karakulah G, Ratnapriya R, Giuste FO, Brooks MJ, Swaroop A. Next generation sequencing technology and genomewide data analysis: Perspectives for retinal research. Prog Retin Eye Res. 2016 doi: 10.1016/j.preteyeres.2016.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Kostadima M, Martens JH, Canu G, Garcia SP, Turro E, Downes K, Macaulay IC, Bielczyk-Maczynska E, Coe S, Farrow S, Poudel P, Burden F, Jansen SB, Astle WJ, Attwood A, Bariana T, de Bono B, Breschi A, Chambers JC, Choudry FA, Clarke L, Coupland P, van der Ent M, Erber WN, Jansen JH, Favier R, Fenech ME, Foad N, Freson K, van Geet C, Gomez K, Guigo R, Hampshire D, Kelly AM, Kerstens HH, Kooner JS, Laffan M, Lentaigne C, Labalette C, Martin T, Meacham S, Mumford A, Nurnberg S, Palumbo E, van der Reijden BA, Richardson D, Sammut SJ, Slodkowicz G, Tamuri AU, Vasquez L, Voss K, Watt S, Westbury S, Flicek P, Loos R, Goldman N, Bertone P, Read RJ, Richardson S, Cvejic A, Soranzo N, Ouwehand WH, Stunnenberg HG, Frontini M, Rendon A Consortium B. Transcriptional diversity during lineage commitment of human blood progenitors. Science. 2014;345:1251033. doi: 10.1126/science.1251033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin L, Andersen JN, Futreal PA. Cancer genomics: from discovery science to personalized medicine. Nat Med. 2011;17:297–303. doi: 10.1038/nm.2323. [DOI] [PubMed] [Google Scholar]
- ENCODE Project Consortium. The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004;306:636–640. doi: 10.1126/science.1105136. [DOI] [PubMed] [Google Scholar]
- GTEx Consortium. Human genomics. The Genotype-Tissue Expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–660. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cvekl A, Ashery-Padan R. The cellular and molecular mechanisms of vertebrate lens development. Development. 2014;141:4432–4447. doi: 10.1242/dev.107953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dash S, Siddam AD, Barnum CE, Janga SC, Lachke SA. RNA-binding proteins in eye development and disease: implication of conserved RNA granule components. Wiley Interdiscip Rev RNA. 2016 doi: 10.1002/wrna.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desmaison A, Vigouroux A, Rieubland C, Peres C, Calvas P, Chassaing N. Mutations in the LHX2 gene are not a frequent cause of micro/anophthalmia. Mol Vis. 2010;16:2847–2849. [PMC free article] [PubMed] [Google Scholar]
- Dillies MA, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloe D, Le Gall C, Schaeffer B, Le Crom S, Guedj M, Jaffrezic F, French StatOmique C. A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform. 2013;14:671–683. doi: 10.1093/bib/bbs046. [DOI] [PubMed] [Google Scholar]
- Farjo R, Yu J, Othman MI, Yoshida S, Sheth S, Glaser T, Baehr W, Swaroop A. Mouse eye gene microarrays for investigating ocular development and disease. Vision Res. 2002;42:463–470. doi: 10.1016/s0042-6989(01)00219-x. [DOI] [PubMed] [Google Scholar]
- Farkas MH, Grant GR, White JA, Sousa ME, Consugar MB, Pierce EA. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14:486. doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez-Valverde SL, Calcino AD, Degnan BM. Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genomics. 2015;16:387. doi: 10.1186/s12864-015-1588-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forch P, Puig O, Martinez C, Seraphin B, Valcarcel J. The splicing regulator TIA-1 interacts with U1-C to promote U1 snRNP recruitment to 5′ splice sites. EMBO J. 2002;21:6882–6892. doi: 10.1093/emboj/cdf668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson R, Alako B, Amid C, Cerdeno-Tarraga A, Cleland I, Goodgame N, Ten Hoopen P, Jayathilaka S, Kay S, Leinonen R, Liu X, Pallreddy S, Pakseresht N, Rajan J, Rossello M, Silvester N, Smirnov D, Toribio AL, Vaughan D, Zalunin V, Cochrane G. Biocuration of functional annotation at the European nucleotide archive. Nucleic Acids Res. 2016;44:D58–66. doi: 10.1093/nar/gkv1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glaser T, Jepeal L, Edwards JG, Young SR, Favor J, Maas RL. PAX6 gene dosage effect in a family with congenital cataracts, aniridia, anophthalmia and central nervous system defects. Nat Genet. 1994;7:463–471. doi: 10.1038/ng0894-463. [DOI] [PubMed] [Google Scholar]
- He S, Limi S, McGreal RS, Xie Q, Brennan LA, Kantorow WL, Kokavec J, Majumdar R, Hou H, Jr, Edelmann W, Liu W, Ashery-Padan R, Zavadil J, Kantorow M, Skoultchi AI, Stopka T, Cvekl A. Chromatin remodeling enzyme Snf2h regulates embryonic lens differentiation and denucleation. Development. 2016;143:1937–1947. doi: 10.1242/dev.135285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill RE, Favor J, Hogan BL, Ton CC, Saunders GF, Hanson IM, Prosser J, Jordan T, Hastie ND, van Heyningen V. Mouse small eye results from mutations in a paired-like homeobox-containing gene. Nature. 1991;354:522–525. doi: 10.1038/354522a0. [DOI] [PubMed] [Google Scholar]
- Hoang TV, Kumar PK, Sutharzan S, Tsonis PA, Liang C, Robinson ML. Comparative transcriptome analysis of epithelial and fiber cells in newborn mouse lenses with RNA sequencing. Mol Vis. 2014;20:1491–1517. [PMC free article] [PubMed] [Google Scholar]
- Hogan BL, Horsburgh G, Cohen J, Hetherington CM, Fisher G, Lyon MF. Small eyes (Sey): a homozygous lethal mutation on chromosome 2 which affects the differentiation of both lens and nasal placodes in the mouse. J Embryol Exp Morphol. 1986;97:95–110. [PubMed] [Google Scholar]
- Kakrana A, Yang A, Anand D, Djordjevic D, Ramachandruni D, Singh A, Huang H, Ho JWK, Lachke SA. iSyTE 2.0: a database for expression-based gene discovery in the eye. Nucleic Acids Res. 2017 doi: 10.1093/nar/gkx837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang MG, Byun K, Kim JH, Park NH, Heinsen H, Ravid R, Steinbusch HW, Lee B, Park YM. Proteogenomics of the human hippocampus: The road ahead. Biochim Biophys Acta. 2015;1854:788–797. doi: 10.1016/j.bbapap.2015.02.010. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Lee MC, Pourmand N, Talbot CC, Jr, Riazuddin SA. Transcriptome Profiling of Developing Murine Lens Through RNA Sequencing. Invest Ophthalmol Vis Sci. 2015;56:4919–4926. doi: 10.1167/iovs.14-16253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan SY, Hackett SF, Riazuddin SA. Non-coding RNA profiling of the developing murine lens. Exp Eye Res. 2016;145:347–351. doi: 10.1016/j.exer.2016.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12:357–360. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King R, Lu L, Williams RW, Geisert EE. Transcriptome networks in the mouse retina: An exon level BXD RI database. Mol Vis. 2015;21:1235–1251. [PMC free article] [PubMed] [Google Scholar]
- Koreishi AF, Saenz AJ, Persky DO, Cui H, Moskowitz A, Moskowitz CH, Teruya-Feldstein J. The role of cytotoxic and regulatory T cells in relapsed/refractory Hodgkin lymphoma. Appl Immunohistochem Mol Morphol. 2010;18:206–211. doi: 10.1097/PAI.0b013e3181c7138b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Maas RL. Building the developmental oculome: systems biology in vertebrate eye development and disease. Wiley Interdiscip Rev Syst Biol Med. 2010;2:305–323. doi: 10.1002/wsbm.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Alkuraya FS, Kneeland SC, Ohn T, Aboukhalil A, Howell GR, Saadi I, Cavallesco R, Yue Y, Tsai AC, Nair KS, Cosma MI, Smith RS, Hodges E, Alfadhli SM, Al-Hajeri A, Shamseldin HE, Behbehani A, Hannon GJ, Bulyk ML, Drack AV, Anderson PJ, John SW, Maas RL. Mutations in the RNA granule component TDRD7 cause cataract and glaucoma. Science. 2011;331:1571–1576. doi: 10.1126/science.1195970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachke SA, Ho JW, Kryukov GV, O’Connell DJ, Aboukhalil A, Bulyk ML, Park PJ, Maas RL. iSyTE: integrated Systems Tool for Eye gene discovery. Invest Ophthalmol Vis Sci. 2012;53:1617–1627. doi: 10.1167/iovs.11-8839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li G, Yi S, Yang F, Zhou Y, Ji Q, Cai J, Mei Y. Identification of mutant genes with high-frequency, high-risk, and high-expression in lung adenocarcinoma. Thorac Cancer. 2014;5:211–218. doi: 10.1111/1759-7714.12080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R Genome Project Data Processing S. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mamuya FA, Wang Y, Roop VH, Scheiblin DA, Zajac JC, Duncan MK. The roles of alphaV integrins in lens EMT and posterior capsular opacification. J Cell Mol Med. 2014;18:656–670. doi: 10.1111/jcmm.12213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maragh S, Miller RA, Bessling SL, Wang G, Hook PW, McCallion AS. Rbm24a and Rbm24b are required for normal somitogenesis. PLoS One. 2014;9:e105460. doi: 10.1371/journal.pone.0105460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milne K, Kobel M, Kalloger SE, Barnes RO, Gao D, Gilks CB, Watson PH, Nelson BH. Systematic analysis of immune infiltrates in high-grade serous ovarian cancer reveals CD20, FoxP3 and TIA-1 as positive prognostic factors. PLoS One. 2009;4:e6412. doi: 10.1371/journal.pone.0006412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33:290–295. doi: 10.1038/nbt.3122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Porter FD, Drago J, Xu Y, Cheema SS, Wassif C, Huang SP, Lee E, Grinberg A, Massalas JS, Bodine D, Alt F, Westphal H. Lhx2, a LIM homeobox gene, is required for eye, forebrain, and definitive erythrocyte development. Development. 1997;124:2935–2944. doi: 10.1242/dev.124.15.2935. [DOI] [PubMed] [Google Scholar]
- Roger JE, Hiriyanna A, Gotoh N, Hao H, Cheng DF, Ratnapriya R, Kautzmann MA, Chang B, Swaroop A. OTX2 loss causes rod differentiation defect in CRX-associated congenital blindness. J Clin Invest. 2014;124:631–643. doi: 10.1172/JCI72722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruzycki PA, Tran NM, Kefalov VJ, Kolesnikov AV, Chen S. Graded gene expression changes determine phenotype severity in mouse models of CRX-associated retinopathies. Genome Biol. 2015;16:171. doi: 10.1186/s13059-015-0732-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaham O, Gueta K, Mor E, Oren-Giladi P, Grinberg D, Xie Q, Cvekl A, Shomron N, Davis N, Keydar-Prizant M, Raviv S, Pasmanik-Chor M, Bell RE, Levy C, Avellino R, Banfi S, Conte I, Ashery-Padan R. Pax6 regulates gene expression in the vertebrate lens through miR-204. PLoS Genet. 2013;9:e1003357. doi: 10.1371/journal.pgen.1003357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao C, Zhao L, Wang K, Xu W, Zhang J, Yang B. The tumor suppressor gene RBM5 inhibits lung adenocarcinoma cell growth and induces apoptosis. World J Surg Oncol. 2012;10:160. doi: 10.1186/1477-7819-10-160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srivastava R, Budak G, Dash S, Lachke SA, Janga SC. Transcriptome analysis of developing lens reveals abundance of novel transcripts and extensive splicing alterations. Sci Rep. 2017;7:11572. doi: 10.1038/s41598-017-10615-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Z, Yin J, Zhao L, Li R, Liang H, Zhang J, Wang K. Lentiviral vector-mediated RBM5 overexpression downregulates EGFR expression in human non-small cell lung cancer cells. World J Surg Oncol. 2014;12:367. doi: 10.1186/1477-7819-12-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundermeier TR, Zhang N, Vinberg F, Mustafi D, Kohno H, Golczak M, Bai X, Maeda A, Kefalov VJ, Palczewski K. DICER1 is essential for survival of postmitotic rod photoreceptor cells in mice. FASEB J. 2014;28:3780–3791. doi: 10.1096/fj.14-254292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian L, Kazmierkiewicz KL, Bowman AS, Li M, Curcio CA, Stambolian DE. Transcriptome of the human retina, retinal pigmented epithelium and choroid. Genomics. 2015;105:253–264. doi: 10.1016/j.ygeno.2015.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren PJ, Lee JT, Doroudchi MM, Smith AD, Horsager A. A profile of transcriptomic changes in the rd10 mouse model of retinitis pigmentosa. Mol Vis. 2014;20:1612–1628. [PMC free article] [PubMed] [Google Scholar]
- Walters RW, Bradrick SS, Gromeier M. Poly(A)-binding protein modulates mRNA susceptibility to cap-dependent miRNA-mediated repression. RNA. 2010;16:239–250. doi: 10.1261/rna.1795410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, Feolo M, Geer LY, Helmberg W, Kapustin Y, Khovayko O, Landsman D, Lipman DJ, Madden TL, Maglott DR, Miller V, Ostell J, Pruitt KD, Schuler GD, Shumway M, Sequeira E, Sherry ST, Sirotkin K, Souvorov A, Starchenko G, Tatusov RL, Tatusova TA, Wagner L, Yaschenko E. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigle JT, Chowdhury K, Gruss P, Oliver G. Prox1 function is crucial for mouse lens-fibre elongation. Nat Genet. 1999;21:318–322. doi: 10.1038/6844. [DOI] [PubMed] [Google Scholar]
- Yang C, Sun C, Liang X, Xie S, Huang J, Li D. Integrative analysis of microRNA and mRNA expression profiles in non-small-cell lung cancer. Cancer Gene Ther. 2016;23:90–97. doi: 10.1038/cgt.2016.5. [DOI] [PubMed] [Google Scholar]
- Yates A, Akanni W, Amode MR, Barrell D, Billis K, Carvalho-Silva D, Cummins C, Clapham P, Fitzgerald S, Gil L, Giron CG, Gordon L, Hourlier T, Hunt SE, Janacek SH, Johnson N, Juettemann T, Keenan S, Lavidas I, Martin FJ, Maurel T, McLaren W, Murphy DN, Nag R, Nuhn M, Parker A, Patricio M, Pignatelli M, Rahtz M, Riat HS, Sheppard D, Taylor K, Thormann A, Vullo A, Wilder SP, Zadissa A, Birney E, Harrow J, Muffato M, Perry E, Ruffier M, Spudich G, Trevanion SJ, Cunningham F, Aken BL, Zerbino DR, Flicek P. Ensembl 2016. Nucleic Acids Res. 2016;44:D710–716. doi: 10.1093/nar/gkv1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zagozewski JL, Zhang Q, Eisenstat DD. Genetic regulation of vertebrate eye development. Clin Genet. 2014;86:453–460. doi: 10.1111/cge.12493. [DOI] [PubMed] [Google Scholar]
- Zhang N, Tsybovsky Y, Kolesnikov AV, Rozanowska M, Swider M, Schwartz SB, Stone EM, Palczewska G, Maeda A, Kefalov VJ, Jacobson SG, Cideciyan AV, Palczewski K. Protein misfolding and the pathogenesis of ABCA4-associated retinal degenerations. Hum Mol Genet. 2015;24:3220–3237. doi: 10.1093/hmg/ddv073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang SS, Xu X, Liu MG, Zhao H, Soares MB, Barnstable CJ, Fu XY. A biphasic pattern of gene expression during mouse retina development. BMC Dev Biol. 2006;6:48. doi: 10.1186/1471-213X-6-48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimmermann C, Stevant I, Borel C, Conne B, Pitetti JL, Calvel P, Kaessmann H, Jegou B, Chalmel F, Nef S. Research resource: the dynamic transcriptional profile of sertoli cells during the progression of spermatogenesis. Mol Endocrinol. 2015;29:627–642. doi: 10.1210/me.2014-1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.