Abstract
Chondrocytes at different maturation states in the growth plate produce matrix vesicles (MVs), membrane organelles found in the extracellular matrix, with a wide range of contents, such as matrix processing enzymes and receptors for hormones. We have shown that MVs harvested from growth zone (GC) chondrocyte cultures contain abundant small RNAs, including miRNAs. Here, we determined whether RNA also exists in MVs produced by less mature resting zone (RC) chondrocytes and, if so, whether it differs from the RNA in MVs produced by GC cells. Our results showed that RNA, small RNA specifically, was present in RC-MVs, and it was well-protected from RNase by the phospholipid membrane. A group of miRNAs was enriched in RC-MVs compared RC-cells, suggesting that miRNAs are selectively packaged into MVs. High throughput array and RNA sequencing showed that ~39% miRNAs were differentially expressed between RC-MVs and GC-MVs. Individual RT-qPCR also confirmed that miR-122-5p and miR-150-5p were expressed at significantly higher levels in RC-MVs compared to GC-MVs. This study showed that growth plate chondrocytes at different differentiation stages produce different MVs with different miRNA contents, further supporting extracellular vesicle miRNAs play a role as “matrisomes” that mediate the cell–cell communication in cartilage and bone development.
Introduction
Endochondral bone formation consists of a developmental cascade of chondrocyte maturation that is well-regulated in temporal and spatial dimensions1. This is reflected in a zonal change in cell morphology and function from resting to proliferative, prehypertrophic and hypertrophic chondrocytes followed by calcification of the extracellular matrix, vasculogenesis, and bone formation on the calcified cartilage scaffold. Central to the process of growth plate development is the role of small membrane-bound extracellular microvesicles, called matrix vesicles (MV)2. MVs are enriched in alkaline phosphatase specific activity compared to the plasma membrane and the first calcium phosphate crystals are observed on the inner leaflet of the MV membrane. Depending on the state of chondrocyte maturation within the growth plate, MVs have a different phospholipid composition3 and are selectively packed with different matrix metalloproteinases4–6, alkaline phosphatase activity3,7–9, growth factors10, and receptors for hormones11–13, suggesting they function as regulators of the endochondral bone formation environment14–16.
We have established a cell culture model to study the phenotypic transition of chondrocytes from the resting zone (RC) to the prehypertrophic/upper hypertrophic zone (GC), in which these two cell types are cultured separately after discarding the intervening proliferative cell zone3. Characterization of these two distinct cell populations has shown that they are different in morphology3, proliferation rate3, extracellular matrix components3, basal cation flux and membrane fluidity17,18, membrane phospholipid composition3,19, basal production of prostaglandin E220 and production of vitamin D metabolites 1,25-dihydroxyvitamin D3 [1,25(OH)2D3] and 24,25-dihydroxyvitamin D3 [24,25(OH)2D3]21.
In addition, RC and GC chondrocytes exhibit marked differences in their response to vitamin D metabolites and growth factors5,11,22. Whereas RC cells respond primarily to 24 R,25(OH)2D3, GC chondrocytes respond primarily to 1α,25(OH)2D3. Testosterone stimulates alkaline phosphatase specific activity in GC cells, with no effects on RC cells23; TGF-β1 induces a dose-dependent increase in proliferation of RC cells, but this effect was not seen in GC cells24. Interestingly, even though RC chondrocytes will form nodules in long-term culture, these cells do not mineralize their matrix. However, bone morphogenetic protein 2 (BMP2) or 24 R,25(OH)2D3 treatment of RC cells induces a phenotypic shift into GC chondrocytes25, which are able to calcify their extracellular matrix.
Both types of chondrocytes produce MVs, but with distinctly different properties. MVs produced by GC cells possess greater specific activities of alkaline phosphatase (the MV marker enzyme)3 and phospholipase A2 than do MVs produced by RC cells19. RC MVs contain neutral metalloproteinases, whereas MVs produced by GC chondrocytes contain acidic matrix metalloproteinases4,5,20,26. RC MVs also have different phospholipid profiles, membrane fluidities and membrane receptors compared to MVs produced by GC cells3,11,12,19. Protein kinase C (PKC) activity in RC-MVs is regulated by 24 R,25(OH)2D3; however, it is stimulated by 1α,25(OH)2D3 in GC-MVs27,28. RC-MVs are usually not able to calcify, however, mineralization occurs in GC-MVs29,30. Taken together, these findings suggest that chondrocytes at different maturation status in the growth plate produce different MVs with different contents, which modulate their different local environment and satisfy different metabolic needs.
Recent studies show that microRNAs (miRNA), which are short (20–22-nucleotide), endogenous, single-stranded, non-coding RNA molecules that regulate gene expression at the post-transcriptional level31, are involved in endochondral bone formation32. miR-145 was downregulated during TGFβ3-induced chondrogenic differentiation of mesenchymal stem cells (MSCs), and targeted Sox9 mRNA transcripts33. miR-140 was expressed in cartilage during embryonic development, contributes to craniofacial development, and regulated histone deacetylase 4 (HDAC4), but there was no evidence to indicate its involvement in postnatal tissue34. miR-1 was shown to be specifically expressed in the hypertrophic zone in postnatal growth plate, and repressed HDAC435. These studies substantiate the hypothesis that specific miRNA regulate growth plate cartilage.
miRNA is packaged in membrane bound vesicles termed endosomes, which are found in biofluids such as blood, plasma, urine and saliva, as well as culture medium36. Emerging as novel mediators in cell-cell communications, extracellular miRNA is involved in various physiological and pathological processes37. Recently, our lab showed that small RNA, and miRNA in particular, is highly enriched in matrix vesicles produced by GC chondrocytes38. Moreover, our results indicate that a group of miRNAs is selectively packed into these MVs, suggesting that MVs function as “matrisomes,” providing a mechanism for information transfer in the non-vascularized growth plate. However, it is not known whether small RNA also exists in MVs produced by RC chondrocytes and, if so, whether there is difference in their miRNA composition. The purpose of this study was to determine the presence of RNA in RC-MVs, and then use PCR array and next generation sequencing to identify their miRNA profile. We also compared the RC-MV miRNAs to those present in GC-MVs.
Results
Characterization of RC-MVs
Nanosight analysis showed that MVs isolated from RC chondrocyte cultures were approximately 100 nm in diameter (Fig. 1A). These vesicles demonstrated a more than 2-fold increase in alkaline phosphatase specific activity compared to the RC plasma membrane, which is a marker for MVs (Table 1). In addition, activity in RC-MVs was lower than in MVs from GC chondrocyte cultures (Fig. 1B), which exhibited a 6-fold enrichment over plasma membrane activity (Fig. 1C), being consistent with our previous observations3.
Figure 1.
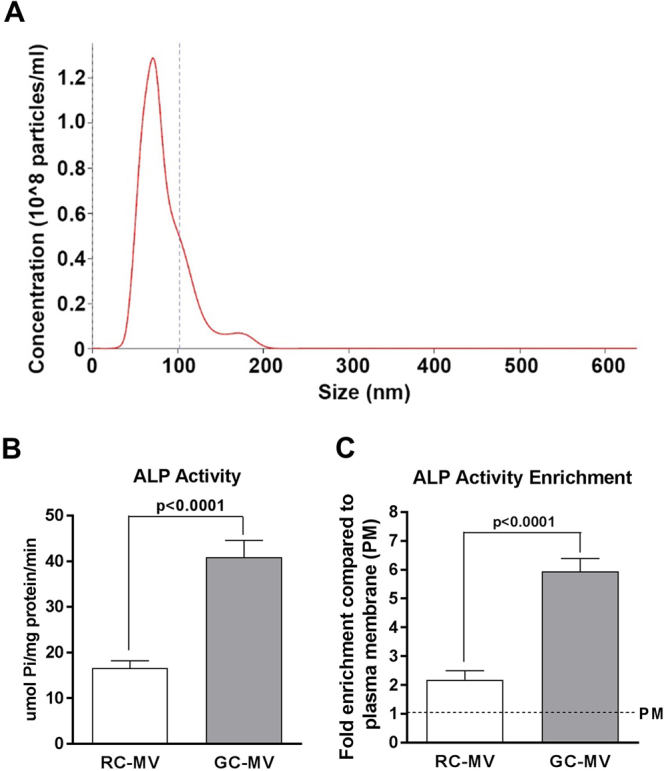
Characterization of RC-MVs. (A) Nanosight was used to measure the size of RC-MVs, which showed an average diameter around 80 nm. (B) Alkaline phosphatase (ALP) specific activity was compared between RC-MVs and GC-MVs. GC cells produced MVs with higher ALP specific activity than RC cells (n = 5 or 6). (C) In RC cell cultures, less ALP activity enrichment was seen in MVs compared to that in GC cell cultures (n = 5 or 6). PM=plasma membrane.
Table 1.
Alkaline phosphatase (ALP) specific activity of MVs from resting zone chondrocytes.
| ALP activity (µmol Pi/mg protein/min) (mean ± s.e.m.) | Fold enrichment | |
|---|---|---|
| Total cell lysate | 4.04 ± 0.45c,d | 1 |
| Mitochondria | 4.67 ± 2.58d | 1.16 |
| Plasma membrane | 8.00 ± 1.00d,e | 1.98 |
| MVs | 16.65 ± 3.79a,b,c,e | 4.12 |
| Pellet from culture medium | 2.33 ± 0.43c,d | 0.57 |
Note: ap < 0.05 compared with total cell lysate; bp < 0.05 compared with mitochondria; cp < 0.05 compared with plasma membrane; dp < 0.05 compared with MVs; ep < 0.05 compared with pellet from culture medium; n = 5 or 6. All the comparisons were done within the same cell type. (a–eBased on one way ANOVA). The enrichment folds compared with total cell lysate are presented on the right.
Small RNA is selectively packaged in matrix vesicles produced by resting zone chondrocytes
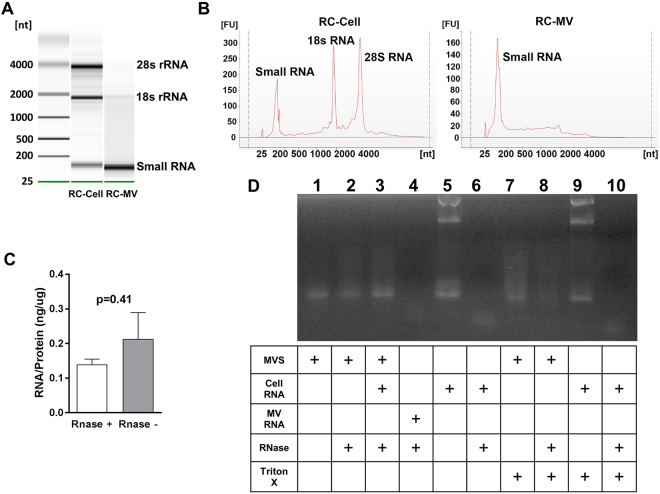
RC cells had both large ribosomal RNA (28 s and 18 s) and small RNA, while MVs produced by these cells only exhibited small RNA that was less than 200 nt (Fig. 2A). This was confirmed by Bioanalyzer (Fig. 2B). These RNAs were packed inside MVs and were protected from enzymatic digestion by RNase. RNase treatment did not decrease the yield of RNA extracted from MVs (Fig. 2C). If the membrane was intact, RNase did not digest the MV RNA (Fig. 2D). However, when Triton X-100 was added to disrupt the membrane, RNase completely digested the RNA in the MV suspension. These results suggested that the RNA in the MVs was protected by the phospholipid bilayer membrane.
Figure 2.
RNA exists in RC-MVs. (A) MV RNA was compared to cell RNA. In a 2% agarose gel, RC-MV RNA was heterogeneous in size but contained little or no large ribosomal RNA species (18S- and 28S-rRNA) compared to parent cells. Enriched small RNAs were observed in MVs. (B) A similar pattern was observed in the Bioanalyzer. (C) RC-MVs were treated by RNase A before RNA extraction by TRIzol. No significant differences in the RNA:protein ratios of MVs with or without RNase pretreatment (n = 6) were observed. (D) RNase was not able to digest the RNA component in MVs. However, it was able to digest the total cell RNA. When the MVs were pre-treated with the membrane detergent Triton X-100, RNA degradation was observed, suggesting that RNA in RC-MVs is protected by the intact lipid membrane.
Selective miRNAs are enriched in RC-MVs
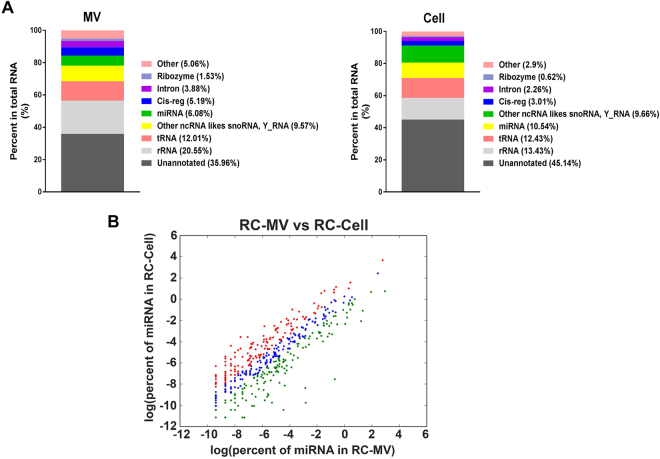
We used a PCR array to determine the miRNA profile of RC-MVs, and compared that to the total cell miRNA profile. A distinct pattern was seen in RC-MVs with a selective group of miRNAs that were highly enriched (Table 2). In this data set, 13 miRNAs were only detected in RC-MVs but not RC-cells; 10 other miRNAs were found to have at least more than 2-fold enrichment in RC-MVs, which included miR-150-5p, miR-126a-3p, and miR-129-2-3p. Conversely, miRNA expression levels within the cells that were elevated compared to MVs included miR-29b-3p, miR-503-5p, and miR-27a-3p. We also obtained a global view of RC-MV small RNA using high throughput, unbiased RNA-Seq and compared the results to parent chondrocytes (Fig. 3A). The annotated small RNAs in RC-MVs included rRNA (20.55%), transfer RNA (12.01%), miRNAs (6.08%), and non-coding RNA like snoRNA, Y_RNA, etc (9.57%). In contrast, chondrocytes had significantly more miRNAs (10.54%).
Table 2.
miRNAs with different expression levels between RC-MVs and RC-cells in miRNA PCR array.
| miRNA unique in RC-MVs | miRNAs shared in RC-MVs and RC-cells | miRNA unique in RC-MVs | |
|---|---|---|---|
| miRNA | miRNAs | Fold_change | miRNA |
| rno-miR-451-5p | rno-miR-150-5p | 10.23 | rno-miR-27a-3p |
| rno-miR-223-3p | rno-miR-126a-3p | 6.73 | rno-miR-219-5p |
| rno-miR-142-3p | rno-miR-129-2-3p | 4.61 | rno-miR-190a-5p |
| rno-miR-122-5p | rno-miR-10a-5p | 3.45 | rno-miR-147 |
| rno-miR-139-5p | rno-miR-125a-3p | 2.75 | rno-miR-325-3p |
| rno-miR-339-5p | rno-miR-490-3p | 2.16 | rno-miR-499-5p |
| rno-miR-142-5p | rno-miR-186-5p | 2.14 | rno-miR-598-3p |
| rno-miR-144-3p | rno-miR-196b-5p | 2.12 | rno-miR-196a-5p |
| rno-miR-211-5p | rno-miR-34b-3p | 2.11 | rno-miR-298-5p |
| rno-miR-133a-3p | rno-miR-132-3p | 2.04 | rno-miR-181d-5p |
| rno-miR-133b-3p | rno-miR-136-5p | −2.07 | rno-miR-540-5p |
| rno-miR-200c-3p | rno-miR-218a-5p | −2.15 | rno-miR-203a-3p |
| rno-miR-330-5p | rno-miR-324-5p | −2.15 | rno-miR-130b-3p |
| rno-miR-193-3p | −2.18 | rno-miR-384-5p | |
| rno-let-7i-5p | −2.38 | rno-miR-384-3p | |
| rno-miR-29c-3p | −2.40 | rno-miR-296-5p | |
| rno-miR-301a-3p | −2.51 | rno-miR-135a-5p | |
| rno-miR-210-3p | −2.52 | rno-miR-187-3p | |
| rno-miR-376a-3p | −2.54 | rno-miR-291a-5p | |
| rno-miR-195-5p | −2.58 | rno-miR-181c-5p | |
| rno-miR-34c-5p | −2.71 | rno-miR-342-5p | |
| rno-miR-32-5p | −4.21 | ||
| rno-miR-503-5p | −4.59 | ||
| rno-miR-29b-3p | −5.65 | ||
| rno-miR-33-5p | −5.80 | ||
| rno-miR-34b-5p | −6.35 | ||
Note: miRNAs that were only detected in MVs from resting zone chondrocytes were shown in the left column. miRNAs that were only detected in the resting zone chondrocytes were shown in the right column. miRNAs that were detected both in resting zone MVs and cells, but with at least a 2-fold change and p < 0.05 were shown in the middle column. miRNAs that are in Italic area: highly enriched in RC MVs. miRNAs that in underline area: mostly remained in RC cells. miRNAs highlighted in bold italic were selected for further validation by RT-PCR.
Figure 3.
The profiles of small RNAs in RC-MVs and parent cells. (A) Complex populations of coding and non-coding small RNAs were found in RC-MVs in proportions that were distinct from those in the RC cells. (B) miRNAs in RC-MVs and RC-cells identified by RNA-seq were plotted. Red dots represent miRNAs with at least a 2-fold expression level increase in MVs than those in cells. Green dots represent miRNAs with more than a 2-fold expression level increase in cells than that in MVs. Blue indicates the overlap between these two groups. A large number of miRNAs were differentially expressed between RC-MVs and RC-cells.
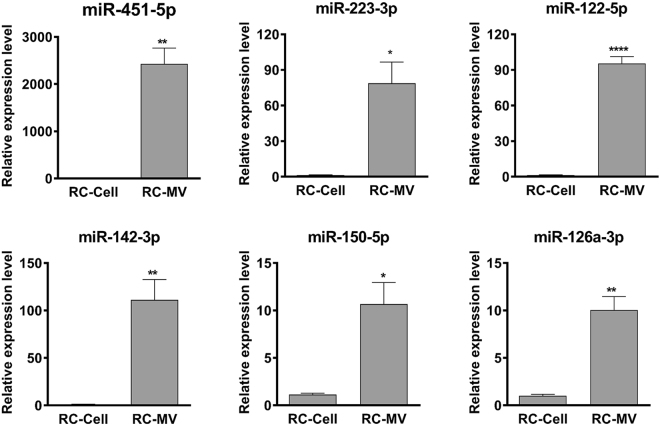
When the RC-MV miRNA pool was examined more closely, we determined that it was dominated by a small number of miRNAs. The most abundant miRNA in RC-MVs was miR-22-3p, which accounted for 20.27% of the total miRNA reads, followed by miR-21-5p (17.21%) and miR-143-3p (11.77%). The top 3 most abundant miRNAs occupied over 50% of the miRNA pool (Table 3), which was similar to that of the parent cell miRNA pool (Table 4). However, we also found distinct differences in the miRNA profile between RC-MVs and RC-cells (Fig. 3B). 139 (32.1%) miRNAs were found to be enriched in MVs by at least two-fold. The top 20 most RC-MV enriched miRNAs (compared to RC-cells) identified from RNA sequencing are listed in Table 5. We validated these findings using real time PCR. 6 RC-MV miRNAs (miR-451-5p, miR-223-3p, miR-122-5p, miR-142-3p, miR-150-5p, miR-126a-3p) were selected because they were only detected or were highly enriched in RC-MV based on the PCR-array (Table 2, highlighted in red). We confirmed that these miRNAs were expressed at significantly higher levels in RC-MV compared to RC-Cell (Fig. 4), suggesting that the packaging of miRNAs into resting zone chondrocyte matrix vesicles is a selective process.
Table 3.
Top 20 most abundant miRNAs in RC-MV.
| miRNA | % in RC-MVs |
|---|---|
| rno-miR-22-3p | 20.27 |
| rno-miR-21-5p | 17.21 |
| rno-miR-143-3p | 11.77 |
| rno-miR-127-3p | 6.84 |
| rno-miR-320-3p | 3.50 |
| rno-miR-134-5p | 3.43 |
| rno-miR-541-5p | 1.93 |
| rno-let-7c-5p | 1.88 |
| rno-miR-152-3p | 1.77 |
| rno-let-7b-5p | 1.62 |
| rno-let-7i-5p | 1.54 |
| rno-miR-381-3p | 1.50 |
| rno-miR-100-5p | 1.49 |
| rno-miR-99b-5p | 1.36 |
| rno-miR-370-3p | 1.27 |
| rno-miR-181a-5p | 1.26 |
| rno-miR-341 | 1.22 |
| rno-miR-148a-5p | 1.12 |
| rno-miR-125b-5p | 1.03 |
| rno-miR-140-3p | 0.95 |
Table 4.
Top 20 most abundant miRNAs in RC-cells.
| miRNA | % in RC-Cells |
|---|---|
| rno-miR-21-5p | 42.70 |
| rno-miR-143-3p | 11.21 |
| rno-let-7i-5p | 5.00 |
| rno-miR-27b-3p | 3.11 |
| rno-miR-148a-3p | 2.67 |
| rno-let-7f-5p | 2.35 |
| rno-miR-22-3p | 2.23 |
| rno-miR-199a-5p | 2.07 |
| rno-miR-127-3p | 1.73 |
| rno-miR-199a-3p | 1.46 |
| rno-miR-125b-5p | 1.34 |
| rno-let-7c-5p | 1.34 |
| rno-miR-24-3p | 0.96 |
| rno-miR-100-5p | 0.91 |
| rno-miR-26a-5p | 0.91 |
| rno-miR-30a-5p | 0.79 |
| rno-miR-379-5p | 0.77 |
| rno-miR-152-3p | 0.75 |
| rno-let-7g-5p | 0.73 |
| rno-miR-99b-5p | 0.67 |
Table 5.
Top 20 selectively packaged miRNAs in MVs in RNA sequencing.
| miRNA | % in RC-MVs | % in RC-Cells | Fold enrichment in MVs |
|---|---|---|---|
| rno-miR-122-5p | 4.01E − 01 | 4.91E − 04 | 816.82 |
| rno-miR-451-5p | 6.14E − 02 | 7.67E − 05 | 800.54 |
| rno-miR-144-3p | 1.30E − 02 | 3.07E − 05 | 423.20 |
| rno-miR-23a-5p | 6.40E − 03 | 1.23E − 04 | 52.16 |
| rno-miR-320-3p | 3.50E + 00 | 1.29E − 01 | 27.10 |
| rno-miR-490-3p | 9.02E − 03 | 3.84E − 04 | 23.51 |
| rno-miR-615 | 1.57E − 02 | 1.23E − 03 | 12.78 |
| rno-miR-296-3p | 1.75E − 02 | 1.41E − 03 | 12.39 |
| rno-miR-365-5p | 1.33E − 02 | 1.12E − 03 | 11.92 |
| rno-miR-134-5p | 3.43E + 00 | 2.88E − 01 | 11.90 |
| rno-miR-664-2-5p | 1.37E − 02 | 1.18E − 03 | 11.60 |
| rno-miR-770-3p | 8.78E − 02 | 8.15E − 03 | 10.78 |
| rno-miR-148a-5p | 1.12E + 00 | 1.04E − 01 | 10.76 |
| rno-miR-129-5p | 1.91E − 01 | 1.97E − 02 | 9.72 |
| rno-miR-22-3p | 2.03E + 01 | 2.23E + 00 | 9.10 |
| rno-miR-666-5p | 1.14E − 02 | 1.30E − 03 | 8.71 |
| rno-miR-219a-1-3p | 7.85E − 03 | 9.82E − 04 | 7.99 |
| rno-miR-423-5p | 3.24E − 01 | 4.09E − 02 | 7.94 |
| rno-miR-760-3p | 2.24E − 02 | 2.98E − 03 | 7.51 |
| rno-miR-127-5p | 5.82E − 01 | 8.26E − 02 | 7.05 |
Figure 4.
Validation of the miRNA enriched in RC-MVs compared with parent cells. Six miRNAs (miR-451-5p, miR-223-3p, miR-122-5p, miR-142-3p, miR-150-5p, miR-126a-3p) were selected for RT-qPCR assays. Consistent with previous array results, the expression of all these miRNAs were significantly higher in MVs compared to cells. These results suggest that certain miRNAs are selectively exported into MVs. *p < 0.05 in t-test. **p < 0.01 in t-test. ****p < 0.0001 in t-test (n = 3).
Functional annotation of RC-MV enriched miRNAs
miRNAs function by binding to target mRNAs and inhibiting their translation or promoting degradation. We identified the target genes of the top most enriched miRNA in RC-MVs and analyze the signaling pathways that are potentially involved. Based on results from KEGG signaling pathway analysis, key functions of target genes included roles in stem cell regulation, Hippo signaling, Wnt signaling, Rap1 signaling, ErbB signaling, focal adhesion, and cell cycle, many of which have been shown to be involved in chondrocyte proliferation and differentiation (Table 6).
Table 6.
KEGG pathways targeted by the top 10 RC-MV enriched miRNAs.
| KEGG pathway | P value |
|---|---|
| Axon guidance | 2.30E − 13 |
| Signaling pathways regulating pluripotency of stem cells | 6.70E − 13 |
| Hippo signaling pathway | 2.60E − 12 |
| Wnt signaling pathway | 3.20E − 09 |
| Focal adhesion | 6.80E − 09 |
| MAPK signaling pathway | 1.30E − 08 |
| Thyroid hormone signaling pathway | 3.50E − 08 |
| PI3K-Akt signaling pathway | 1.40E − 07 |
| Rap1 signaling pathway | 1.60E − 07 |
| TGF-beta signaling pathway | 3.00E − 07 |
| cAMP signaling pathway | 7.40E − 07 |
| ErbB signaling pathway | 8.20E − 07 |
| cGMP-PKG signaling pathway | 2.30E − 06 |
| Calcium signaling pathway | 2.50E − 06 |
| Endocytosis | 3.50E − 06 |
| Oxytocin signaling pathway | 3.60E − 06 |
| Neurotrophin signaling pathway | 4.30E − 06 |
| Ras signaling pathway | 5.90E − 06 |
| Circadian entrainment | 7.80E − 06 |
| Insulin resistance | 1.40E − 05 |
| Dorso-ventral axis formation | 1.70E − 05 |
MV miRNA component is dependent on the cell maturation status
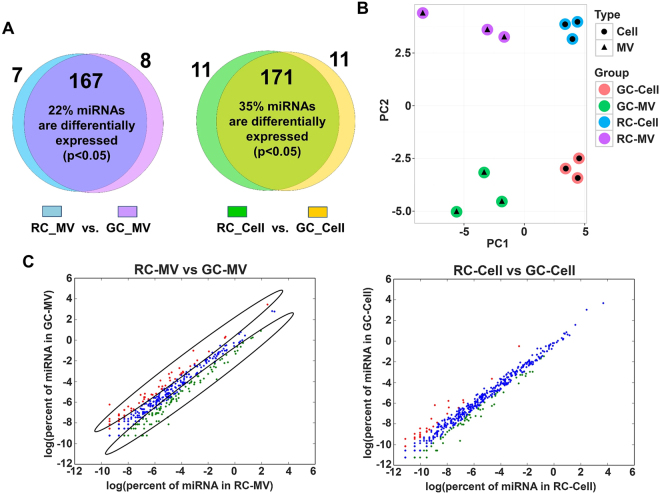
We have previously shown distinct differences in MVs that can be ascribed to the level of chondrocyte maturation. We recently reported that GC-MVs were packed with selective miRNAs. Based on these studies, we hypothesized that miRNAs were selectively packed into RC and GC MVs. To investigate unique maturation-dependent differences, we compared the PCR array results of RC-MVs to those of GC-MVs. In the 182 miRNAs detected, 10 miRNAs were identified in GC-MVs and 7 were found in RC-MVs (Fig. 5A). 35 miRNAs were differently expressed between RC-MVs and GC-MVs with at least a more than 2-fold difference (increase or decrease) (Table 7). We also identified a group of miRNAs that showed different expression levels between RC cells and GC cells (Table 8). Using principle component analysis, we observed differences between RC-MVs (purple dots, top left) and GC-MVs (green dots, bottom left) (Fig. 5B). While 39% of miRNAs showed differential expression level with more than 2-fold difference between RC-MVs (green dots, circled) and GC-MVs (red dots, circled), we observed a unique separation between these miRNA (Fig. 5C), further supporting our hypothesis. Therefore, the selective packaging of miRNAs into MVs exhibits spatial and maturation dependence. Interestingly, less miRNAs were found differentially expressed between RC-cells and GC-cells than between RC-MVs and GC-MVs (Fig. 5C), suggesting more heterogeneous miRNA profiles exist in the extracellular vesicles than that in the cytoplasm. It is worth mentioning that the difference between RC-MVs and GC-MVs was less dramatic than between RC-MVs and RC-cells or between GC-MVs and GC-cell, since the distance at the x-axis represents the most prominent difference followed by the distance at the y-axis. This indicates that the difference between MVs and the cells that produce them is more significant than that between two cell maturation statuses (RC vs. GC) (Fig. 5C).
Figure 5.
Distinct differences in the miRNAs between RC-MVs and GC-MVs. (A) Different miRNAs patterns were identified from PCR-array between two maturation status of chondrocytes, both in MVs and the parent cells. (B) Principal component analysis (PCA) of the miRNA PCR-array data for all the four different types of samples: RC-MVs, RC-cells, GC-MVs and GC-cells. The differences among these samples were clear, as larger difference showed on the x-axis direction compared to that on the y-axis. This suggests that the differences between MVs and cells were larger than between two different cell maturation stages. (C) Left panel: miRNAs in RC-MVs and GC-MVs identified by RNA-seq were plotted. Red dots represent miRNAs with at least a 2-fold expression level increase in GC-MVs than in RC-MVs. Green dots represent miRNAs with a more than 2-fold expression level increase in RC-MVs than in GC-MVs. Blue indicates the overlap between these two groups. A large number of miRNAs were differentially expressed between RC-MVs and GC-MVs. Right panel: similar comparison was done between RC-cells and GC-cells. Interestingly, the difference between RC-cells and GC-cells was not as large as that was seen between RC-MVs and GC-MVs.
Table 7.
miRNAs with different expression levels between RC-MVs and GC-MVs in miRNA PCR array.
| miRNA unique in RC-MVs | miRNAs shared in RC-MVs and GC-MVs | miRNA unique in GC-MVs | |
|---|---|---|---|
| miRNA | miRNAs | Fold_change | miRNA |
| rno-miR-380-5p | rno-miR-503-5p | 8.73 | rno-miR-325-3p |
| rno-miR-200c-3p | rno-miR-672-5p | 4.31 | rno-miR-326-3p |
| rno-miR-874-3p | rno-miR-450a-5p | 3.85 | rno-miR-219-5p |
| rno-miR-339-3p | rno-miR-204-5p | 3.10 | rno-miR-298-5p |
| rno-miR-340-3p | rno-miR-322-5p | 2.78 | rno-miR-130b-3p |
| rno-miR-186-5p | rno-miR-668 | 2.78 | rno-miR-760-3p |
| rno-miR-212-3p | rno-miR-362-5p | 2.37 | rno-miR-598-3p |
| rno-miR-125a-3p | 2.37 | rno-miR-181c-5p | |
| rno-miR-329-3p | 2.29 | rno-miR-592 | |
| rno-miR-181b-5p | 2.14 | rno-miR-499-5p | |
| rno-miR-130a-3p | −2.01 | ||
| rno-miR-129-5p | −2.05 | ||
| rno-miR-20a-5p | −2.11 | ||
| rno-miR-133a-3p | −2.17 | ||
| rno-miR-365-3p | −2.29 | ||
| rno-miR-18a-5p | −2.30 | ||
| rno-miR-143-3p | −2.39 | ||
| rno-miR-150-5p | −2.40 | ||
| rno-miR-195-5p | −2.48 | ||
| rno-miR-451-5p | −2.55 | ||
| rno-miR-497-5p | −2.57 | ||
| rno-miR-223-3p | −2.57 | ||
| rno-miR-582-5p | −2.60 | ||
| rno-miR-132-3p | −2.63 | ||
| rno-miR-133b-3p | −2.65 | ||
| rno-miR-29b-3p | −2.74 | ||
| rno-miR-192-5p | −2.76 | ||
| rno-miR-126a-3p | −2.81 | ||
| rno-miR-210-3p | −2.83 | ||
| rno-miR-122-5p | −2.88 | ||
| rno-miR-142-5p | −3.07 | ||
| rno-miR-142-3p | −3.57 | ||
| rno-miR-139-5p | −3.69 | ||
| rno-miR-144-3p | −4.78 | ||
| rno-miR-10a-5p | −5.93 | ||
Note: miRNAs that were only detected in MVs from RC were shown in the left column. miRNAs that were only detected in MVs from GC were shown in the right column. miRNAs that were detected in MVs from both RC and GC cells, but with at least a 2-fold change and p < 0.05 were shown in the middle column. miRNAs that are bold area: highly enriched in RC-MVs. miRNAs that in underline area: mostly enriched in GC-MVs.
Table 8.
miRNAs with different expression levels between RC-cells and GC-cells in miRNA PCR array.
| miRNA unique in RC-cells | miRNAs shared in RC-cells and GC-cells | miRNA unique in GC-cells | |
|---|---|---|---|
| miRNA | miRNAs | Fold_change | miRNA |
| rno-miR-203a-3p | rno-miR-503-5p | 4.62 | rno-miR-339-5p |
| rno-miR-384-5p | rno-miR-204-5p | 4.31 | rno-miR-139-5p |
| rno-miR-135a-5p | rno-miR-380-5p | 3.64 | rno-miR-211-5p |
| rno-miR-335 | rno-miR-450a-5p | 3.60 | rno-miR-760-3p |
| rno-miR-384-3p | rno-miR-322-5p | 3.07 | rno-miR-592 |
| rno-miR-291a-5p | rno-miR-542-5p | 2.69 | rno-miR-142-3p |
| rno-miR-27a-3p | rno-miR-376c-3p | 2.02 | rno-miR-330-5p |
| rno-miR-147 | rno-miR-143-3p | −2.21 | rno-miR-182 |
| rno-miR-339-3p | rno-miR-449a-5p | −2.27 | rno-miR-326-3p |
| rno-miR-484 | rno-miR-17-5p | −2.27 | rno-miR-141-3p |
| rno-miR-497-5p | −2.38 | rno-miR-206-3p | |
| rno-miR-132-3p | −2.54 | ||
| rno-miR-296-5p | −2.69 | ||
| rno-miR-196b-5p | −2.73 | ||
| rno-miR-582-5p | −4.06 | ||
| rno-miR-298-5p | −4.85 | ||
| rno-miR-10a-5p | −10.69 | ||
Note: miRNAs that were only detected in RC cells were shown in the left column. miRNAs that were only detected in GC cells were shown in the right column. miRNAs that were detected in cells from both RC and GC, but with at least a 2-fold change and p < 0.05 were shown in the middle column. miRNAs that are in bold area: highly enriched in RC-cells. miRNAs that in underline area: mostly enriched in GC-cells.
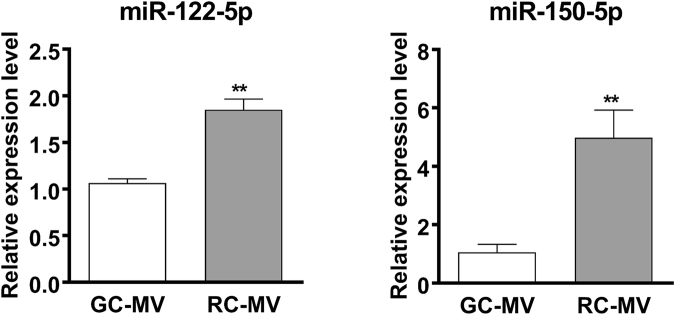
To further validate our observations in miRNA PCR-array and RNA-Seq, we selected a set of miRNAs that are highly expressed in RC-MVs and performed RT-qPCR to compare their expression levels between RC-MVs and GC-MVs. Although we did not observe significant differences in some of the selected miRNAs such as miR126a-3p and miR-451-5p, higher expression levels of miR-122-5p and miR-150-5p were consistently seen in RC-MVs compared with GC-MVs (Fig. 6). This further suggests that cells at different maturation status produced different matrix vesicles with different miRNA contents.
Figure 6.
RT-qPCR was used to further validate the differences in miRNA expression between RC-MVs and GC-MVs. miR-122-5p and miR-150-5p had significantly higher expression levels in RC-MVs than GC-MVs, suggesting the miRNAs contents in MVs are associated with the cell maturation status. **p < 0.01 in t-test. (n = 6).
Discussion
In this study, we demonstrate that RNA exists in MVs produced by costochondral growth plate resting zone cartilage cells. This RNA is packaged by the cells within the extracellular matrisomes and is protected by the MV membrane. RNase treatment of intact RC MVs didn’t digest their RNA component, however significant degradation occurred after the membrane was disrupted by Triton X-100. Moreover, a set of miRNAs was specifically enriched in the MVs, further supporting the hypothesis that their inclusion involves a regulated process.
A variety of RNA species were found in RC-MVs, similar to RNA species found in MVs produced by GC chondrocytes38. The majority of the RNA pool in RC-MVs was rRNA and tRNA. It is still not clear the biological meaning of these RNAs in the regulation of extracellular matrix and cell behavior. Recently, it was reported that tRNAs can be cleaved into small fragments that can also function as miRNAs39. Despite these similarities, however, PCR array and RNA-Seq showed that a large group of miRNAs was differentially expressed in MVs from RC and GC cells. Individual RT-PCR confirmed that there were significant differences in some miRNAs, such as miR-122-5p and miR-150-5p.
Chondrocytes at different maturation states in the growth plate not only produce zone-specific extracellular matrix3,40–42, but as noted above, their matrix vesicles differ in phospholipid profile3, enzyme content3–5, and response to hormones11,20,28. Our results add to this by demonstrating maturation-dependent differences in miRNA composition. This suggests that MV miRNAs may be involved in the regulation of zone-specific phenotypic behavior of the cells. Our previous observation that release of matrix vesicle contents is mediated by regulated secretion of 1α,25(OH)2D3 and 24R,25(OH)2D3 and their interaction with the MV membrane5,43–45, provides a mechanism for modulating the bioavailability of the miRNAs during endochondral development.
The relative differences between miRNA in RC and GC cells and the differences between RC-cells and RC-MVs or GC-cells and GC-MVs were greater than the differences between RC-MVs and GC-MVs. Chondrocytes in the resting zone of the growth plate will eventually express a growth zone phenotype, so it is likely that the phenotypic shift involves only a subset of components. MV species are heterogeneous46, so it is also possible that not all MVs are uniformly modulated during growth plate development.
Further investigation is needed to determine whether there are general properties that define matrisomes or if they are tissue or cell specific. Others have reported the presence of RNA in the extracellular matrix of mineralizing tissues47, and miRNAs have been found in dermis, bladder and small intestinal submucosa, which are associated with extracellular matrix-bound nanovesicles48. Compared to other extracellular vesicles such as exosomes, MVs are unique in many ways. MVs have a strong affinity to collagen and stay in the extracellular matrix, while exosomes are released into the culture media3,38. The biogenesis mechanisms of MVs and exosomes are different as well. MVs are formed by a cell “budding” mechanism, but exosomes are formed through membrane invagination49. MVs released by resting zone and growth zone chondrocytes also express higher alkaline phosphatase activity than the plasma membrane and compared to membrane vesicles they secrete into the medium3. Furthermore, the heterogeneity of matrix vesicles produced by chondrocytes46, indicates that they may perform multiple functions in the extracellular matrix. A recent study also found that mesenchymal stem cells secret at least three different types of microvescicles with different RNA material50, raising the possibility that miRNAs may be differentially packaged in subsets of matrisomes and exosomes to modulate specific cell functions.
Extracellular miRNAs, exosomal miRNAs in particular, have been shown to play an important role in cell–cell communication in physiological and pathological processes37. Similarly, MV miRNAs may also function as a modulator during chondrogenesis by transferring to other cells in the local environment in an autocrine or paracrine manner, and thereby, regulating their gene expression and biological behavior. Functional analysis suggests that MV miRNAs are associated with stem cell regulation. miR-122-5p, which has been related to liver function and HCV infection51, was more highly expressed in RC-MVs than GC-MVS. Others have shown that miR-122-5p targets the IGF-1R signaling pathway, which is a major player in the musculoskeletal system52. Our preliminary data showed that overexpression of miR-122-5p enhanced chondrocyte proliferation and maintained the stemness of these cells (data not shown). miR-150-5p, which is also higher in RC-MVs was reported to target matrix metalloproteinases53,54. Therefore, RC-MV enriched miRNAs may help maintain the local environment needed for RC cells to maintain a stronger proliferative capability than GC cells.
In conclusion, we demonstrated that RNA is present in MVs derived from resting zone chondrocytes, and selective miRNAs are enriched. We also observed that miRNAs are differentially expressed between RC-MVs and GC-MVs, although these differences are not as prominent as those between RC-MVs and RC-cells. Our results further suggest the importance of extracellular matrix RNA during chondrogenesis and endochondral bone formation.
Methods
Chondrocyte cultures
Chondrocytes were isolated by enzymatic digestion of the costochondral cartilage of 125 g male Sprague Dawley rats and cultured as described by Boyan et al. previously3. All procedures conducted on animals followed a protocol approved by the Institutional Animal Care and Use Committee (IACUC) at Virginia Commonwealth University, and all experiments were performed in accordance with relevant guidelines and regulations. Rib cages were removed and placed in Dulbecco’s modified Eagle’s medium (DMEM, Life Technologies). The resting zone and growth zone cartilage was carefully dissected out, sliced, and incubated in 1% trypsin (Life Technologies) for 1 hour and 0.02% collagenase for 3 hours. After filtering by 40-micro nylon mesh, cells were collected by centrifugation, resuspended in DMEM, and plated at a density of 25,000 cells/cm2. Fetal bovine serum (FBS, Life Technologies) was centrifuged at 184,000 × g for 4 h to remove the bovine serum exosomes. Chondrocytes were incubated in DMEM containing 10% exosome-free FBS, 1% penicillin/streptomycin and 50 mg/ml ascorbic acid (Sigma-Aldrich). Cell culture medium was changed 24 hours after plating and then at 72-hour intervals. At confluence (5–7 days), cells were subcultured using the same plating densities and allowed to return to confluence. All experiments used confluent cells in their fourth passage.
MV isolation and characterization
MVs were isolated from chondrocyte cultures by differential centrifugation of the trypsin digested cell layer, as described in detail previously3. Briefly, after trypsinization (0.25% Trypsin-EDTA, Life Technologies) for 10 min, 1 × DPBS containing 10% exosome-free FBS was added to stop the reaction. The MVs were isolated by sequential centrifugation at 500 g for 5 min, 21,000 g for 20 min, and 184,000 g for 70 min. The MV pellets were washed with 0.9% NaCl and recentrifuged at 184,000 g for 70 min. The same centrifugation protocol was used to pellet the microvesicles in cell culture media. Alkaline phosphatase specific activity was measured and normalized to protein content. In addition, to validate that MVs were isolated but not plasma membrane debris, plasma membranes were isolated from the cell pellet as described previously and alkaline phosphatase activity determined. Alkaline phosphatase specific activity was enriched in MVs at least 2 folds compared to plasma membrane, which was consistent with our previous studies. The original MVs solution was diluted 200 times for nanoparticle tracking analysis (Nanosight, Malvern, UK).
RNA extraction and detection
RNA was isolated from MVs or cells using TRizol (Life Technologies). RNA was eluted with RNase-free water and quantified by NanoDrop spectrophotometer (Thermo Scientific). For agarose gels, 100 ng RNA was run and visualized on a 2.2% agarose gel FashGelTM System (Lonza). Bioanalyzer analyses were performed using 300 ng RNA with an RNA 600 Nano Kit (Agilent) and an Agilent 2100 Bioanalyzer (Agilent) according to manufacturer’s protocol. To confirm that RNA was inside and not outside the MVs, the vesicles were treated with RNase A (10 mg/ml, Qiagen) before RNA extraction as described previously38. MVs were also incubated with 0.05% Triton-X 100 in order to break down the membrane in some experiments.
miRNA PCR profiling in MVs
cDNA library preparation and miRNA profiling were performed in MV and cell RNA from RC and GC chondrocytes as described previously38. The microRNA PCR array (miRCURY LNA Universal RT microRNA PCR array) included 223 rat miRNAs. Details in data normalization and analysis were provided in supplementary materials.
Reverse transcription and quantitative real-time PCR (RT-qPCR)
50 ng RNA was reverse transcribed to cDNA with miScript II RT kit (Qiagen), followed by real-time qPCR using the StepOnePlus Real-time PCR System and miScript SYBR Green PCR Kit (Qiagen). Fluorescence values were quantified as starting quantities using known dilutions of standard control. The reverse primer was the universal primer in the kit. The sequences for the forward primers were synthesized by Eurofins MWG Operon: rno-miR-451-5p: AAACCGTTACCATTACTGAGTT; rno-miR-223-3p: TGTCAGTTTGTCAAATACCC; rno-miR-122-5p: TGGAGTGTGACAATGGTGTT; rno-miR-142-3p: TGTAGTGTTTCCTACTTTATGGA; rno-miR-150-5p: TCTCCCAACCCTTGTACCAGT; rno-miR-126a-3p: TCGTACCGTGAGTAATAATGCG; rno-miR-652-3p: AATGGCGCCACTAGGGTT.
RNA sequencing (RNA-Seq)
RNA was isolated from MVs or cells using TRizol. Next generation sequencing was performed in an Illumina HiSeq. 2500 (Illumina, San Diego, CA). Data normalization and analysis were conducted with the miARma-Seq tool (see supplemental materials). Briefly, sequence quality was assessed with FASTQC; sequence reads were aligned with Bowtie2 to the NCBI Rattus norvegicus annotation release 105 (Rnor_6.0). The resulting bam files were used by miARma-Seq for tabulating discovered miRNA. Reads were also aligned to Rfam 12.3 with Bowtie2 for categorization of RNA reads. To quantify the differential expression of miRNA between samples, the count of each miRNA was normalized as the percent of all discovered miRNAs in that sample. The fold-change was calculated as the percent expression of MV miRNA/Cell miRNA. KEGG Pathway enrichment analyses were performed based on the target genes of selected miRNAs.
Statistical analysis
Alkaline phosphatase specific activity is presented as mean ± standard error of the mean (SEM) for 6 independent MV samples per variable. RT-qPCR is presented as mean ± SEM for 3 independent samples per variable. Data were examined by ANOVA with post-hoc Bonferroni’s modification of Student’s t-test. A p value of less than 0.05 was considered statistically significant. Statistical analyses for miRNA array and RNA-Seq data were performed according to our previous protocol38.
Data availability statement
The datasets generated during the current study have been submitted to NCBI GEO database (GSE106348).
Electronic supplementary material
Acknowledgements
We thank Nicholas Rodriguez for his help in the bioinformatics analysis. This work was funded by NIDCR grant DE027146 and Virginia Commonwealth University Startup Support (ZL), Shanghai Jiao Tong University School of Medicine 085 Project (JZ) and Department of Defense W81XWH-11-1-0306 (BDB).
Author Contributions
Z.L., Z.S. and B.D.B. conceived and designed the experiments; Z.L., J.Z., M.J.M and A.N.R. performed the experiments, Z.L, J.Z., M.J.M., N.A., Z.S. and B.D.B. analyzed the data; S.L.H. contributed reagents/materials/analysis tools; Z.L., M.J.M., Z.S. and B.D.B. wrote the paper.
Competing Interests
The authors declare no competing interests.
Footnotes
Electronic supplementary material
Supplementary information accompanies this paper at 10.1038/s41598-018-21517-4.
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Kronenberg HM. Developmental regulation of the growth plate. Nature. 2003;423:332–336. doi: 10.1038/nature01657. [DOI] [PubMed] [Google Scholar]
- 2.Anderson HC. Vesicles associated with calcification in the matrix of epiphyseal cartilage. J. Cell Biol. 1969;41:59–72. doi: 10.1083/jcb.41.1.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boyan BD, Schwartz Z, Swain LD, Carnes DL, Jr, Zislis T. Differential expression of phenotype by resting zone and growth region costochondral chondrocytes in vitro. Bone. 1988;9:185–194. doi: 10.1016/8756-3282(88)90008-7. [DOI] [PubMed] [Google Scholar]
- 4.Dean DD, et al. Matrix vesicles are enriched in metalloproteinases that degrade proteoglycans. Calcif. Tissue Int. 1992;50:342–349. doi: 10.1007/BF00301632. [DOI] [PubMed] [Google Scholar]
- 5.Dean DD, Boyan BD, Muniz OE, Howell DS, Schwartz Z. Vitamin D metabolites regulate matrix vesicle metalloproteinase content in a cell maturation-dependent manner. Calcif. Tissue Int. 1996;59:109–116. doi: 10.1007/s002239900096. [DOI] [PubMed] [Google Scholar]
- 6.D’Angelo M, Billings PC, Pacifici M, Leboy PS, Kirsch T. Authentic matrix vesicles contain active metalloproteases (MMP). a role for matrix vesicle-associated MMP-13 in activation of transforming growth factor-beta. J. Biol. Chem. 2001;276:11347–11353. doi: 10.1074/jbc.M009725200. [DOI] [PubMed] [Google Scholar]
- 7.Wuthier RE, et al. Isolation and characterization of calcium-accumulating matrix vesicles from chondrocytes of chicken epiphyseal growth plate cartilage in primary culture. J. Biol. Chem. 1985;260:15972–15979. [PubMed] [Google Scholar]
- 8.Glaser JH, Conrad HE. Formation of matrix vesicles by cultured chick embryo chondrocytes. J. Biol. Chem. 1981;256:12607–12611. [PubMed] [Google Scholar]
- 9.Pawlowski A, Makower AM, Madsen K, Wroblewski J, Friberg U. Cell fractions from rat rib growth cartilage. Biochemical characterization of matrix molecules. Exp. Cell. Res. 1986;164:211–222. doi: 10.1016/0014-4827(86)90468-4. [DOI] [PubMed] [Google Scholar]
- 10.Nahar NN, Missana LR, Garimella R, Tague SE, Anderson HC. Matrix vesicles are carriers of bone morphogenetic proteins (BMPs), vascular endothelial growth factor (VEGF), and noncollagenous matrix proteins. J. Bone Miner. Metab. 2008;26:514–519. doi: 10.1007/s00774-008-0859-z. [DOI] [PubMed] [Google Scholar]
- 11.Schwartz Z, Knight G, Swain LD, Boyan BD. Localization of vitamin D3-responsive alkaline phosphatase in cultured chondrocytes. J. Biol. Chem. 1988;263:6023–6026. [PubMed] [Google Scholar]
- 12.Nemere I, et al. Identification of a membrane receptor for 1,25-dihydroxyvitamin D3 which mediates rapid activation of protein kinase C. J. Bone Miner. Res. 1998;13:1353–1359. doi: 10.1359/jbmr.1998.13.9.1353. [DOI] [PubMed] [Google Scholar]
- 13.Pedrozo HA, et al. Physiological importance of the 1,25(OH)2D3 membrane receptor and evidence for a membrane receptor specific for 24,25(OH)2D3. J. Bone Miner. Res. 1999;14:856–867. doi: 10.1359/jbmr.1999.14.6.856. [DOI] [PubMed] [Google Scholar]
- 14.Wuthier RE, Lipscomb GF. Matrix vesicles: structure, composition, formation and function in calcification. Front. Biosci. 2011;16:2812–2902. doi: 10.2741/3887. [DOI] [PubMed] [Google Scholar]
- 15.Shapiro IM, Landis WJ, Risbud MV. Matrix vesicles: Are they anchored exosomes? Bone. 2015;79:29–36. doi: 10.1016/j.bone.2015.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Golub EE. Role of matrix vesicles in biomineralization. Biochim. Biophys. Acta. 2009;1790:1592–1598. doi: 10.1016/j.bbagen.2009.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schwartz Z, Langston GG, Swain LD, Boyan BD. Inhibition of 1,25-(OH)2D3- and 24,25-(OH)2D3-dependent stimulation of alkaline phosphatase activity by A23187 suggests a role for calcium in the mechanism of vitamin D regulation of chondrocyte cultures. J. Bone Miner. Res. 1991;6:709–718. doi: 10.1002/jbmr.5650060708. [DOI] [PubMed] [Google Scholar]
- 18.Langston GG, et al. Effect of 1,25(OH)2D3 and 24,25(OH)2D3 on calcium ion fluxes in costochondral chondrocyte cultures. Calcif. Tissue Int. 1990;47:230–236. doi: 10.1007/BF02555924. [DOI] [PubMed] [Google Scholar]
- 19.Schwartz Z, Schlader DL, Swain LD, Boyan BD. Direct effects of 1,25-dihydroxyvitamin D3 and 24,25-dihydroxyvitamin D3 on growth zone and resting zone chondrocyte membrane alkaline phosphatase and phospholipase-A2 specific activities. Endocrinology. 1988;123:2878–2884. doi: 10.1210/endo-123-6-2878. [DOI] [PubMed] [Google Scholar]
- 20.Schwartz Z, Swain LD, Kelly DW, Brooks B, Boyan BD. Regulation of prostaglandin E2 production by vitamin D metabolites in growth zone and resting zone chondrocyte cultures is dependent on cell maturation. Bone. 1992;13:395–401. doi: 10.1016/8756-3282(92)90456-7. [DOI] [PubMed] [Google Scholar]
- 21.Schwartz Z, et al. Production of 1,25-dihydroxyvitamin D3 and 24,25-dihydroxyvitamin D3 by growth zone and resting zone chondrocytes is dependent on cell maturation and is regulated by hormones and growth factors. Endocrinology. 1992;130:2495–2504. doi: 10.1210/endo.130.5.1572278. [DOI] [PubMed] [Google Scholar]
- 22.Schwartz Z, et al. 1alpha,25(OH)2D3 regulates chondrocyte matrix vesicle protein kinase C (PKC) directly via G-protein-dependent mechanisms and indirectly via incorporation of PKC during matrix vesicle biogenesis. J. Biol. Chem. 2002;277:11828–11837. doi: 10.1074/jbc.M110398200. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz Z, et al. Gender-specific, maturation-dependent effects of testosterone on chondrocytes in culture. Endocrinology. 1994;134:1640–1647. doi: 10.1210/endo.134.4.8137726. [DOI] [PubMed] [Google Scholar]
- 24.Schwartz Z, Bonewald LF, Caulfield K, Brooks B, Boyan BD. Direct effects of transforming growth factor-beta on chondrocytes are modulated by vitamin D metabolites in a cell maturation-specific manner. Endocrinology. 1993;132:1544–1552. doi: 10.1210/endo.132.4.8462452. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz Z, Sylvia VL, Liu Y, Dean DD, Boyan BD. Treatment of resting zone chondrocytes with bone morphogenetic protein-2 induces maturation into a phenotype characteristic of growth zone chondrocytes by downregulating responsiveness to 24,25(OH)2D3 and upregulating responsiveness to 1,25-(OH)2D3. Endocrine. 1998;9:273–280. doi: 10.1385/ENDO:9:3:273. [DOI] [PubMed] [Google Scholar]
- 26.Dean DD, et al. Vitamin D regulation of metalloproteinase activity in matrix vesicles. Connect. Tissue Res. 1996;35:331–336. doi: 10.3109/03008209609029208. [DOI] [PubMed] [Google Scholar]
- 27.Schwartz Z, et al. 24R, 25-(OH)(2)D(3) mediates its membrane receptor-dependent effects on protein kinase C and alkaline phosphatase via phospholipase A(2) and cyclooxygenase-1 but not cyclooxygenase-2 in growth plate chondrocytes. J. Cell. Physio. 2000;182:390–401. doi: 10.1002/(SICI)1097-4652(200003)182:3<390::AID-JCP10>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- 28.Sylvia VL, et al. Maturation-dependent regulation of protein kinase C activity by vitamin D3 metabolites in chondrocyte cultures. J. Cell. Physio. 1993;157:271–278. doi: 10.1002/jcp.1041570209. [DOI] [PubMed] [Google Scholar]
- 29.Boskey AL, et al. Studies of matrix vesicle-induced mineralization in a gelatin gel. Bone Miner. 1992;17:257–262. doi: 10.1016/0169-6009(92)90747-2. [DOI] [PubMed] [Google Scholar]
- 30.Boskey AL, Boyan BD, Schwartz Z. Matrix vesicles promote mineralization in a gelatin gel. Calcif. Tissue Int. 1997;60:309–315. doi: 10.1007/s002239900234. [DOI] [PubMed] [Google Scholar]
- 31.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 32.Lian JB, et al. MicroRNA control of bone formation and homeostasis. Nat. Rev. Endocrinol. 2012;8:212–227. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang B, et al. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PloS one. 2011;6:e21679. doi: 10.1371/journal.pone.0021679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Papaioannou G, et al. MicroRNA-140 Provides Robustness to the Regulation of Hypertrophic Chondrocyte Differentiation by the PTHrP-HDAC4 Pathway. J. Bone Miner. Res. 2015;30:1044–1052. doi: 10.1002/jbmr.2438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Li P, et al. MicroRNA-1 regulates chondrocyte phenotype by repressing histone deacetylase 4 during growth plate development. FASEB J. 2014;28:3930–3941. doi: 10.1096/fj.13-249318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Creemers EE, Tijsen AJ, Pinto YM. Circulating microRNAs: novel biomarkers and extracellular communicators in cardiovascular disease? Circ. Res. 2012;110:483–495. doi: 10.1161/CIRCRESAHA.111.247452. [DOI] [PubMed] [Google Scholar]
- 37.Tkach M, Thery C. Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. Cell. 2016;164:1226–1232. doi: 10.1016/j.cell.2016.01.043. [DOI] [PubMed] [Google Scholar]
- 38.Lin Z, et al. Selective enrichment of microRNAs in extracellular matrix vesicles produced by growth plate chondrocytes. Bone. 2016;88:47–55. doi: 10.1016/j.bone.2016.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sharma U, et al. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aydelotte MB, Greenhill RR, Kuettner KE. Differences between sub-populations of cultured bovine articular chondrocytes. II. Proteoglycan metabolism. Connect. Tissue Res. 1988;18:223–234. doi: 10.3109/03008208809016809. [DOI] [PubMed] [Google Scholar]
- 41.Aydelotte MB, Kuettner KE. Differences between sub-populations of cultured bovine articular chondrocytes. I. Morphology and cartilage matrix production. Connect. Tissue Res. 1988;18:205–222. doi: 10.3109/03008208809016808. [DOI] [PubMed] [Google Scholar]
- 42.Radhakrishnan P, Lewis NT, Mao JJ. Zone-specific micromechanical properties of the extracellular matrices of growth plate cartilage. Ann Biomed Eng. 2004;32:284–291. doi: 10.1023/B:ABME.0000012748.41851.b4. [DOI] [PubMed] [Google Scholar]
- 43.Boyan BD, et al. Latent transforming growth factor-beta is produced by chondrocytes and activated by extracellular matrix vesicles upon exposure to 1,25-(OH)2D3. J. Biol. Chem. 1994;269:28374–28381. [PubMed] [Google Scholar]
- 44.Boyan BD, Sylvia VL, McKinney N, Schwartz Z. Membrane actions of vitamin D metabolites 1alpha,25(OH)2D3 and 24R,25(OH)2D3 are retained in growth plate cartilage cells from vitamin D receptor knockout mice. J. Biol. Chem. 2003;90:1207–1223. doi: 10.1002/jcb.10716. [DOI] [PubMed] [Google Scholar]
- 45.Boyan BD, Wong KL, Fang M, Schwartz Z. 1alpha,25(OH)2D3 is an autocrine regulator of extracellular matrix turnover and growth factor release via ERp60 activated matrix vesicle metalloproteinases. J. Steroid Biochem. Mol. Biol. 2007;103:467–472. doi: 10.1016/j.jsbmb.2006.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kakuta S, Malamud D, Golub EE, Shapiro IM. Isolation of matrix vesicles by isoelectric focusing in Pevikon-Sephadex. Bone. 1985;6:187–191. doi: 10.1016/8756-3282(85)90052-3. [DOI] [PubMed] [Google Scholar]
- 47.Slavkin HC, Bringas P, Bavetta LA. Ribonucleic acid within the extracellular matrix during embryonic tooth formation. J. Cell. Physio. 1969;73:179–190. doi: 10.1002/jcp.1040730304. [DOI] [PubMed] [Google Scholar]
- 48.Huleihel L, et al. Matrix-bound nanovesicles within ECM bioscaffolds. Sci. Adv. 2016;2:e1600502. doi: 10.1126/sciadv.1600502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Leach RJ, et al. Osteosarcoma hybrids can preferentially target alkaline phosphatase activity to matrix vesicles: evidence for independent membrane biogenesis. J. Bone Miner. Res. 1995;10:1614–1624. doi: 10.1002/jbmr.5650101103. [DOI] [PubMed] [Google Scholar]
- 50.Lai RC, et al. MSC secretes at least 3 EV types each with a unique permutation of membrane lipid, protein and RNA. J. Extracell. Vesicles. 2016;5:29828. doi: 10.3402/jev.v5.29828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thakral S, Ghoshal K. miR-122 is a unique molecule with great potential in diagnosis, prognosis of liver disease, and therapy both as miRNA mimic and antimir. Curr. Gene Ther. 2015;15:142–150. doi: 10.2174/1566523214666141224095610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang B, Wang H, Yang Z. MiR-122 inhibits cell proliferation and tumorigenesis of breast cancer by targeting IGF1R. PloS one. 2012;7:e47053. doi: 10.1371/journal.pone.0047053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Li T, et al. miR-150-5p inhibits hepatoma cell migration and invasion by targeting MMP14. PloS one. 2014;9:e115577. doi: 10.1371/journal.pone.0115577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sakr M, et al. miR-150-5p and miR-133a suppress glioma cell proliferation and migration through targeting membrane-type-1 matrix metalloproteinase. Gene. 2016;587:155–162. doi: 10.1016/j.gene.2016.04.058. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated during the current study have been submitted to NCBI GEO database (GSE106348).