Abstract
Experimental models of neuroendocrine tumour disease are scarce, and no comprehensive characterisation of existing gastroenteropancreatic neuroendocrine tumour (GEPNET) cell lines has been reported. In this study, we aimed to define the molecular characteristics and therapeutic sensitivity of these cell lines. We therefore performed immunophenotyping, copy number profiling, whole-exome sequencing and a large-scale inhibitor screening of seven GEPNET cell lines. Four cell lines, GOT1, P-STS, BON-1 and QGP-1, displayed a neuroendocrine phenotype while three others, KRJ-I, L-STS and H-STS, did not. Instead, these three cell lines were identified as lymphoblastoid. Characterisation of remaining authentic GEPNET cell lines by copy number profiling showed that GOT1, among other chromosomal alterations, harboured losses on chromosome 18 encompassing the SMAD4 gene, while P-STS had a loss on 11q. BON-1 had a homozygous loss of CDKN2A and CDKN2B, and QGP-1 harboured amplifications of MDM2 and HMGA2. Whole-exome sequencing revealed both disease-characteristic mutations (e.g. ATRX mutation in QGP-1) and, for patient tumours, rare genetic events (e.g. TP53 mutation in P-STS, BON-1 and QGP-1). A large-scale inhibitor screening showed that cell lines from pancreatic NETs to a greater extent, when compared to small intestinal NETs, were sensitive to inhibitors of MEK. Similarly, neuroendocrine NET cells originating from the small intestine were considerably more sensitive to a group of HDAC inhibitors. Taken together, our results provide a comprehensive characterisation of GEPNET cell lines, demonstrate their relevance as neuroendocrine tumour models and explore their therapeutic sensitivity to a broad range of inhibitors.
Keywords: neuroendocrine tumour, GEPNET, immunophenotyping, copy number alterations, exome sequencing, inhibitor screening, cell lines, vorinostat, trametinib
Introduction
Patient tumour-derived cell lines, as models of tumour disease, have been widely used for studying the molecular mechanisms of tumours and their response to therapy. However, cell lines do not perfectly reflect their tumour of origin and in terms of genomic alterations, protein expression and therapeutic sensitivity, they can differ substantially (Stein et al. 2004, Sandberg & Ernberg 2005, Ertel et al. 2006, Gillet et al. 2011, Domcke et al. 2013).
The establishment of cell lines from human neuroendocrine tumours (NETs) of the gastrointestinal tract and pancreas (GEPNETs) has proved to be difficult. This has been attributed, in part, to the low proliferation rate of GEPNETs (Modlin et al. 2008). Despite these challenges, several cell lines have been established from small intestinal NETs (SINETs). The first cell line derived from a human SINET was the KRJ-I cell line, reported in 1996 (Pfragner et al. 1996). KRJ-I was established from a primary tumour and has been used to characterise the molecular mechanisms and therapeutic sensitivity of SINET disease (Modlin et al. 2006, Kidd et al. 2007, 2008). The GOT1 cell line was established in our laboratory in 2001 from a patient with metastatic SINET disease (Kölby et al. 2001). GOT1 cells were derived from a liver metastasis and expressed neuroendocrine markers, including somatostatin receptor type 2 (SSTR2). This cell line has been used as a model for optimisation of SSTR2-mediated peptide receptor radionuclide therapy (PRRT) (Kölby et al. 2005, Bernhardt et al. 2007, Forssell-Aronsson et al. 2013, Dalmo et al. 2017, Spetz et al. 2017). More recently, in 2009, three cell lines were established from a patient with metastatic SINET disease (Pfragner et al. 2009). The P-STS cell line was derived from the patient’s primary tumour, L-STS from a lymph node metastasis and H-STS from a liver metastasis. A basic characterisation of these cell lines has been reported, and P-STS has been used to study the secretory response of SINETs (Rinner et al. 2012, Pfanzagl et al. 2017).
The generation of cell lines from human pancreatic NETs (PanNETs) has also been proven difficult, and only few cell lines are available. Among the most frequently used PanNET cell lines are QGP-1 and BON-1. QGP-1 was established in 1980 from a somatostatin-producing islet cell carcinoma (Kaku et al. 1980, Iguchi et al. 1990) and it has been used to study the molecular mechanisms that regulate tumour cell proliferation (Doihara et al. 2009, Valentino et al. 2014). BON-1 was established in 1991 from a lymph node metastasis of a PanNET patient (Evers et al. 1991) and has been used as a model for studying the molecular mechanisms of PanNETs and their sensitivity to therapy. Albeit so far lesser used, the establishment of additional GEPNET cell lines has been reported, e.g. CM, LCC-18 and CNDT2.5 (Gueli et al. 1987, Lundqvist et al. 1991, Van Buren et al. 2007). However, the authenticity of CNDT2.5 has been questioned (Ellis et al. 2010). There are also GEPNET cell lines derived from transgenic murine rodents, e.g. STC-1 (intestinal NET from SV40-expressing transgenic mouse), βTC and MIN6 (insulinomas from SV40-expressing transgenic mouse), RIN and INS-1 (X-ray induced insulinomas from NEDH rat) (Gazdar et al. 1980, Rindi et al. 1990, Efrat et al. 1991, Asfari et al. 1992, Ishihara et al. 1993).
The current status can be summarised as follows. Authentic GEPNET cell lines are rare, their genomic and mutational characteristics largely unknown and comprehensive information regarding their therapeutic sensitivity is lacking. Recently, efforts have been made to characterise PanNET-derived cell lines BON-1 and QGP-1 by exome sequencing and genome-wide copy number analysis. These studies have raised questions regarding their relevance as models due to the absence of PanNET-associated mutations (Boora et al. 2015, Vandamme et al. 2015). There have been no corresponding efforts to characterise NET cell lines derived from SINETs. The aim of the present study was to re-evaluate the authenticity of GEPNET cell lines and to define their genomic profile and their therapeutic sensitivity. In this paper, we report the immunophenotyping, genome-wide copy number profiling, whole-exome sequencing and comprehensive inhibitor screening of seven GEPNET cell lines. We confirmed the neuroendocrine phenotype of GOT1, P-STS, BON-1 and QGP-1 and investigated their genomic profile. The inhibitor screening established the therapeutic sensitivity profiles of the cell lines and predicted the cell tumour type-specific efficacy of MEKi and HDACi.
Materials and methods
Cell lines, primary cell cultures and cell culturing
The GOT1 cell line was established from a liver metastasis of a midgut carcinoid from a female patient (Kölby et al. 2001). GOT1 was cultured in RPMI-1640 supplemented with 10% foetal bovine serum (FBS), 5 μg/mL insulin and 5 μg/mL transferrin. KRJ-I, P-STS, L-STS and H-STS were all gifts from Prof. R Pfragner and cultured in M199:Ham’s F12 (1:1) supplemented with 10% FBS. KRJ-I was established from a male patient with a multifocal carcinoid of the small intestine. P-STS (primary tumour), L-STS (lymph node metastasis) and H-STS (liver metastasis) were all established from the same male patient with metastatic carcinoid of the terminal ileum. The PanNET cell line BON-1, a gift from Prof. B Wiedenmann, was established from the lymph node of a carcinoid tumour of the pancreas in a male patient and cultured in DMEM:Ham’s F12 (1:1) with 10% FBS. QGP-1, which has been isolated from a pancreatic carcinoma of a male patient, was acquired from Japanese Collection of Research Bioresources Cell Bank (JCRB) and maintained in RPMI-1640 supplemented with 10% FBS. MCF10A, BJ and HUV-EC-C were acquired from ATCC. MCF10A (human mammary gland epithelium) was maintained in DMEM:Ham’s F12 (1:1) supplemented with 5% horse serum, 10 µg/mL insulin, 20 ng/mL epidermal growth factor (EGF), 0.5 µg/mL hydrocortisone and 0.1 µg/mL cholera toxin. BJ (human foreskin fibroblasts) was maintained in Eagle’s minimum essential medium supplemented with 10% FBS. HUV-EC-C (human vascular endothelium) was maintained in F12K medium supplemented with 10% FBS, 100 µg/mL heparin and 50 µg/mL endothelial cell growth supplement. The medium of all cell lines also contained 200 IU/mL penicillin and 200 μg/mL streptomycin. For doubling time experiments, cells were seeded onto 25 cm2 tissue cell culture flasks. At 24, 48, 72, 96 and 120 h, flasks (n = 2) were trypsinised and counted using Countess cell counter (Invitrogen).
Primary cell cultures of small intestinal and pancreatic NETs were generated from biopsies collected at the time of surgery, as previously described (Arvidsson et al. 2010). Both PanNET cultures (PanNET patient 1 and 2) were derived from primary tumours, ‘SINET patient 1’ from a lymph node metastasis and ‘SINET patient 2’ from a liver metastasis. The cells were kept in RPMI-1640 supplemented with 4% FBS, 200 IU/mL penicillin and 200 μg/mL streptomycin. For the characterisation and inhibitor sensitivity experiments, only first-passage primary cell cultures were used. All cell lines and primary cell cultures were incubated at 37°C in a humidified incubator with an atmosphere of 5% CO2.
Analysis of STR, Epstein-Barr virus and Mycoplasma
DNA was isolated from cell lines at the same passage as used for the inhibitor screening using the DNeasy Blood & Tissue Kit (Qiagen). DNA from cell lines, and from GOT1 patient tumour tissue, was subjected to STR analysis at a DANAK/ILAC DS/EN ISO 15189:2008 accredited laboratory (IdentiCell, Department of Molecular Medicine (MOMA) at Aarhus University Hospital, Denmark) (Supplementary Fig. 1, see section on supplementary data given at the end of this article). Cell lines were regularly tested for Mycoplasma species by PCR as described in the study by van Kuppeveld et al. (1994) at a Swedac SS-EN ISO 15189 accredited laboratory (Bacteriological laboratory, Sahlgrenska University Hospital, Gothenburg, Sweden). DNA from GEPNETs and lymphoblastoid cell lines was subjected to EBV DNA quantification using artus EBV PCR Kit (Qiagen). The PCR was performed using a 7500 Fast-Real-time PCR system (Applied Biosystems).
Cell blocks and immunohistochemistry
Cell lines and primary cell cultures in exponential growth phase were detached and fixed in 4% buffered formaldehyde for 1 h followed by methanol fixation. The paraffin blocks were created using a Cellient automated cell block system (HOLOGIC). Sectioning and staining were carried out as previously described (Andersson et al. 2016). The following primary antibodies were used: anti-chromogranin A (PHE5; Chemicon; diluted 1:1000), anti-synaptophysin (SY38; Dako; diluted 1:200), anti-NCAM (ERIC1; Santa Cruz; diluted 1:10), anti-NSE (BBS/NC/VI-H14; Dako; ready-to-use), anti-PGP9.5 (Z5116; Dako; diluted 1:400), anti-CD57 (HNK-1; BD Biosciences; diluted 1:40), anti-VMAT1 (C-19; Santa Cruz; diluted 1:1000), anti-5HT (H209; Dako; diluted 1:10), anti-cytokeratin 8/18 (5D3; Leica Biosystems; diluted 1:2000), anti-pan-cytokeratin (AE1/AE3; Leica; ready-to-use), anti-SSTR2A (UMB-1; Abcam; diluted 1:100), anti-Ki67 (MIB-1; Dako; ready-to-use), anti-CD45 (2B11+PD7/26; Dako; diluted 1:200) and anti-CD20 (L26; Dako; ready-to-use). Each staining was scored by a board-certified pathologist (O N) into four categories; ‘0’ (<1% weakly stained cells), ‘+’ (<50% weakly stained cells), ‘++’ (moderate staining in >50% of cells) and ‘+++’ (strong staining in >50% of cells). A biopsy of a SINET served as a positive control (‘+++’). For Ki67, manual counting was performed on printed images and the percentage of labelled tumour cell nuclei was calculated (Reid et al. 2015).
Confocal laser scanning microscopy
Adherent cells (GOT1, P-STS, QGP-1 and BON-1) were grown on µ-slide 8-well chamber slides (ibidi, Martinsried, Germany) for 3 days and fixed in 4% buffered formaldehyde in PBS. Cells growing in suspensions (KRJ-I, L-STS, and H-STS) were spun onto microscope slides with a Cytospin 2 cytocentrifuge (Shandon, UK) before fixation. The slides were incubated with primary antibodies for 1 h at room temperature in 1% BSA and 0.2% Triton X-100 in PBS and for 1 h with secondary antibodies conjugated to Alexa Fluor 594 (rabbit anti-mouse, cat. no. A11062 or goat anti-rabbit, cat. no. A11037; ThermoFisher) or to FITC (rabbit anti-goat, cat. no. F0250; Dako). All experiments included negative controls wherein the primary antibody was omitted. The fluorescent cells were analysed using a Zeiss LSM 700 confocal microscope and Zen black software was supplied by the manufacturer. Images were captured using a Zeiss LSM 700 inverted microscope with a 63× oil immersion objective.
Analysis of copy number alterations
DNA from cell lines was extracted as mentioned earlier, and arrayCGH analysis was performed using 4x180K Human SurePrint G3 ISCA CGH + SNP microarrays (Agilent Technologies) as recommended by the manufacturer and as previously described (Barrett et al. 2004, Persson et al. 2008). Data analysis was carried out using Nexus Copy Number software, v.8.0 (BioDiscovery Inc., El Segundo, CA, USA). The FASST2 segmentation algorithm was used to define non-random regions of copy number alterations (CNAs) across the genome with a significance threshold ranging from P = 10−8 to P = 10−18, where the higher stringency threshold was used for arrays with QC >0.2. The log2 ratio thresholds for aberration calls of gain and loss were set to 0.2 and −0.2 for arrays with a significance threshold P≥10−8, and to 0.3 and −0.3 for arrays with a significance threshold P<10−8. Thresholds for amplification and homozygous loss were set to 1.5 and −1.5 in all cases. Sex chromosomes and regions partially or completely covered by a previously reported copy number variation (Database of Genomic Variants; http://dgvbeta.tcag.ca/dgv/app/news?ref=NCBI37/hg19) were excluded from the analysis (Iafrate et al. 2004). Each aberration was checked manually to confirm the accuracy of the call.
Whole-exome sequencing
DNA from cell lines was extracted as described earlier and subjected to whole-exome sequencing at GATC Biotech (Cologne, Germany). FastQC (version 0.11.2) was used to assess the quality of the data. Paired end reads were aligned to the human reference genome (hg19) using a Burrows-Wheeler Aligner (mem version BWA_0.7.13) (Li & Durbin 2009). Samtools (version 1.3.1) was used to sort, index and assess mapping statistics. Picard (version 2.2.4) was used to remove duplicates. The Genome Analysis ToolKit (GATK, version 3.1-1) (McKenna et al. 2010) was used for realignment and variant calling. Variant calling was performed following GATK best practices with the tool HaplotypeCaller. In HaplotypeCaller, the following hard filters were applied for quality filtering; for SNPs: QD <2.0, MQ <40.0, FS >60.0, ReadPosRankSum <−8.0, MQRankSum <−12.5 and for indels: QD <2.0, FS >200.0, ReadPosRankSum <−20.0. Called variants that passed the GATK quality filtering were further filtered against 1000 Genomes (Auton et al. 2015) to remove variants with a MAF >0.01. Remaining variants were annotated with the knownGene database using the tool ANNOVAR (Wang et al. 2010).
Screening library and inhibitor sensitivity
The screening library consisted of 1224 compounds (Inhibitor library, no. L1100; Selleckchem). The library was prepared by two-step dilution of the compounds from 10 mM to 100 µM in DMSO and batched in 96-well plates. Inhibitors were subjected to a maximum of five freeze-thaw cycles. From frozen stocks, cells were expanded 2–5 passages before being used in screening experiments. Seeding density was adjusted for each cell line so that control cells were approximately 70‒80% confluent at treatment endpoint in 100 µL cell medium/well in black solid-bottom 96-well plates. The plates were incubated for 72 h at 37°C to allow for cell attachment. Each treatment plate included DMSO control wells (n = 8), and each experiment included an additional plate with DMSO control wells (n = 96), as well as one cell-free control plate. The screening library was diluted 1:50 in 100 µL medium and added to the treatment plates (1:2; end concentration 1 µM). Cell viability was estimated using a fluorescence-based assay to measure the reducing capacity of metabolically active cells (alamarBlue, DAL1100; Life Technologies). All assay plates were incubated for 72 h at 37°C, followed by addition of 1:100 alamarBlue reagent. The plates were further incubated for 6 h at 37°C and were then analysed using a 96-well fluorescence plate reader (Victor3 multilabel reader, ex. 560 nm/em. 640 nm). For vorinostat and trametinib dose–response curves, first-passage primary cultures were thawed and seeded onto 96-well plates, allowed to settle for 48 h before treatment with vorinostat or trametinib for 72 h. Each concentration was added in triplicate and plates were analysed as described earlier. Fitting of the curves was done in GraphPad Prism software, v7.02 using inhibitor vs. response nonlinear fit with variable slope.
Statistical analyses
For each cell line, inhibitor fluorescence intensities generated by the cell viability assay were log-transformed and normalized by subtracting the average and dividing by the standard deviation generating a Z-score. Average Z-scores were calculated for the SINET and PanNET cell lines respectively. The difference in inhibitor intensity between SINET and PanNET was then calculated and a corresponding p-score was derived from a fitted normal distribution. Testing for groups of inhibitors was done by comparing their differences to the differences of all other inhibitors using a Wilcoxon–Mann–Whitney test. False discovery rates were estimated using the Benjamini–Hochberg algorithm. Differences in the effect of MEKi and HDACi to SINET, PanNET and non-tumourigenic cell lines were analysed using Wilcoxons signed-rank test. Differences in the response of primary cell cultures to different doses of MEKi and HDACi was analysed using two-sided unpaired Student’s t-test for each dose and corrected for multiple testing with Benjamini–Hochberg algorithm.
Results
Immunophenotyping confirmed expression of neuroendocrine tumour markers and SSTR2 in four GEPNET cell lines
NETs are defined by their expression of neuroendocrine markers, notably chromogranin A and synaptophysin. The degree of neuroendocrine differentiation of NET cell lines was evaluated by performing immunohistochemical staining on cell blocks. Each staining was scored into four categories, using biopsies from SINETs as positive controls.
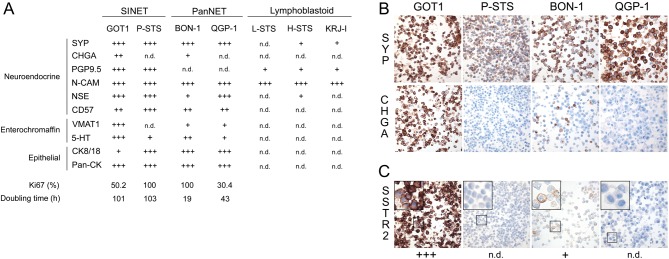
We found strong staining for synaptophysin in GOT1, P-STS, QGP-1 and BON-1, while staining for chromogranin A was moderate in GOT1, weak in BON-1 and absent in P-STS and QGP-1 (Fig. 1A and B). In addition, PGP9.5 staining was strong in SINET cell lines and negative in PanNET cell lines. NCAM staining was strong in all investigated cell lines. Neuron-specific enolase (NSE) showed strong staining in GOT1, P-STS and QGP-1, but was weak in BON-1. P-STS had strong staining for CD57, while GOT1, BON-1 and QGP-1 had moderate levels.
Figure 1.
Expression of neuroendocrine markers and somatostatin receptor 2 in GEPNET cell lines. (A) Scoring of protein expression based on immunohistochemical staining of cell blocks. 5-HT was evaluated from confocal laser scanning microscopy. (B and C) Immunohistochemical staining of cell blocks, illustrating expression of the neuroendocrine markers synaptophysin and chromogranin A (B), and clinically relevant SSTR2 (C) in GEPNET cell lines. CHGA, chromogranin A; SYP, synaptophysin; PGP9.5, ubiquitin carboxyl-terminal hydrolase isozyme L1; N-CAM, neural cell adhesion molecule 1; NSE, gamma-enolase; 5-HT, serotonin; VMAT1, chromaffin granule amine transporter 1; cytokeratin 8/18, keratin type II cytoskeletal 8/type I cytoskeletal 18; pan-CK, pan-cytokeratin; SSTR2, somatostatin receptor type 2; n.d., not detected.
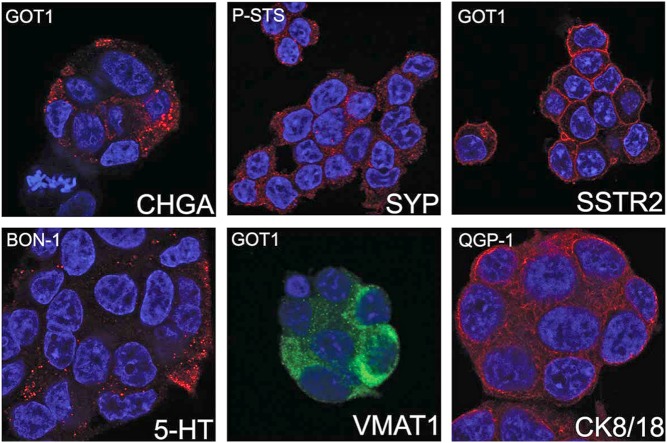
Linage markers of SINETs (VMAT1 and 5-HT) were also investigated. VMAT1 was strongly stained in GOT1, weakly stained in BON-1 and QGP-1, and not stained in P-STS, while 5-HT was strong in GOT1, moderate in BON-1 and weak in P-STS and QGP-1. The localisation of synaptophysin, chromogranin A, VMAT1 and 5-HT to secretory granules was confirmed by confocal laser scanning microscopy (Fig. 2).
Figure 2.
The subcellular localisation of neuroendocrine markers in GEPNET cell lines. The staining pattern of chromogranin A (CHGA), synaptophysin (SYP), chromaffin granule amine transporter 1 (VMAT1) and serotonin (5-HT) was consistent with localisation to secretory granules. The staining of somatostatin receptor type 2 (SSTR2) and cytokeratin 8/18 (CK8/18) was confirmed to be membranous and cytoskeletal, respectively.
GEPNETs are derived from progenitor cells in the intestinal crypts, or pancreatic ducts, which express epithelial markers such as cytokeratins. To confirm the epithelial phenotype of NET cell lines, we stained for high-molecular-weight cytokeratins (AE1/AE3) and cytokeratin 8/18. The staining for high-molecular-weight cytokeratins was strong in all cell lines, while staining for cytokeratin 8/18 was weak to moderate in GOT1, P-STS, BON-1 and QGP-1 (Fig. 1).
Neuroendocrine marker, linage-specific marker and epithelial marker expression for GOT1, P-STS, BON-1 and QGP-1 overall well reflected that of clinical samples of GEPNETs. However, the Ki67 index of the cell lines was not representative of typical GEPNETs. GOT1 originated from a SINET liver metastasis with positivity for SYP, CgA, 5-HT, CK8/18 and with a Ki67 index of 1.3% (Grade I; data not shown). Furthermore, the Ki67 index of the cell lines correlated poorly to their doubling time (Fig. 1).
For the three putative SINET cell lines KRJ-I, L-STS and H-STS, we however, with the exception of NCAM, found only weak or no staining of neuroendocrine markers and could not detect any linage-specific markers or cytokeratins. These observations, in addition to the fact that these three cell lines grow non-adherently and form aggregates in cell culture (in contrast to the other GEPNET cell lines), made us question their authenticity. We hypothesised that KRJ-I, L-STS and H-STS were EBV-driven lymphoblastoid cell lines and thus performed immunohistochemical staining for lymphoid (CD45) and B cell markers (CD20). Both markers were highly expressed in KRJ-I, L-STS and H-STS, and the cell lines also contained high levels of EBV DNA, while the GOT1, P-STS, BON-1 and QGP-1 were negative (Supplementary Fig. 2). We therefore conclude that these cell lines are lymphoblastoid, and do not represent true GEPNET cell lines.
Somatostatin receptors are frequently over-expressed in GEPNETs and are used as a target for imaging and therapy. We performed staining for the somatostatin receptor subtype 2 (SSTR2) and found strong staining in GOT1 cells, weak staining in BON-1 and no staining in P-STS and QGP-1 (Fig. 1C). Confocal laser scanning microscopy showed strong membranous localisation of the SSTR2 protein in GOT1 cells (Fig. 2).
GEPNET cell lines harbour multiple copy number alterations
Chromosomal changes have been linked to tumourigenesis and disease progression in patients with GEPNETs. Recurrent copy number alterations (CNAs) differ between SINETs and PanNETs, suggesting different mechanisms for tumour initiation and progression. To study CNAs in GEPNET cell lines, we performed whole-genome copy number profiling and found CNAs in all GEPNET cell lines (range 7‒44) (Supplementary Table 1).
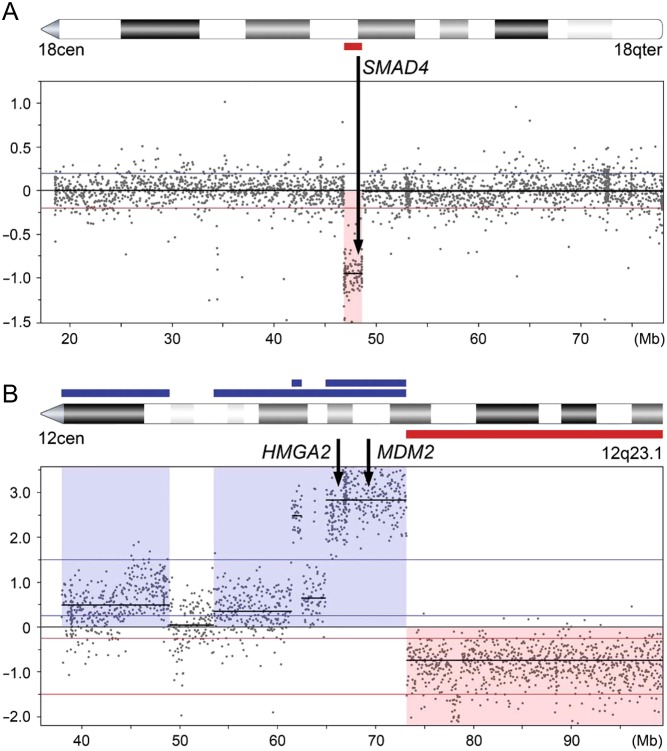
The SINET cell lines GOT1 and P-STS had a lower frequency of alterations and a predominance of chromosomal losses. Loss of parts or whole chromosome 18 is the most common genomic event in SINETs (found in 60‒70% of the tumours). The GOT1 cell line showed loss of a 1.8 Mb segment of 18q, including the tumour suppressor SMAD4 (Fig. 3A). The GOT1 cell line originated from a tumour that had loss of whole chromosome 18 and like GOT1, with predominance of losses and without gains of whole chromosomes (data not shown). The P-STS cell line had no losses on chromosome 18, but instead showed losses involving 11q, which is also a frequent alteration in SINETs (Kulke et al. 2008, Andersson et al. 2009). The PanNET cell lines BON-1 and QGP-1 had a higher frequency of CNAs. The BON-1 cell line had a predominance of gains, including whole chromosomes 2, 5, 7, 10 and 12, which are also frequent gains in PanNETs (Hu et al. 2010, Gebauer et al. 2014). BON-1 also harboured a homozygous loss of the CDKN2A/B tumour suppressor genes. The QGP-1 cell line had the highest number of CNAs and was the only cell line with gene amplifications. There were three amplicons on chromosome 12, one in 12p12.1, including SOX5, one in 12q14.1 and one in 12q12.2–q21.1 including MDM2 and HMGA2 (Fig. 3B). The lymphoblastoid cell lines L-STS and H-STS had no alterations, while KRJ-I harboured three small CNAs.
Figure 3.
Copy number alterations detected in four GEPNET cell lines. (A) GOT1 harboured a loss of a 1.8 Mb segment on chromosome 18q, encompassing the SMAD4 gene. (B) Of the three amplicons on chromosome 12 that QGP-1 harboured, one spanned 12q12.2–q21.1 including the HMGA2 and MDM2 genes.
GEPNET cell lines harbour mutations in several tumour suppressor genes, including TP53
Mutations in GEPNETs are rare, and the only identified recurrent gene mutation in SINETs is of CDKN1B, which occurs in less than a tenth of all tumours (Francis et al. 2013). Recurrent mutations in PanNETs have been found in several genes, including ATRX, DAXX, as well as genes involved in the Akt/mTOR pathway, DNA repair and chromatin remodelling (Jiao et al. 2011, Scarpa et al. 2017). In order to evaluate the mutational profiles of the GEPNET cell lines, we performed whole-exome sequencing and searched for tumour disease-associated genes.
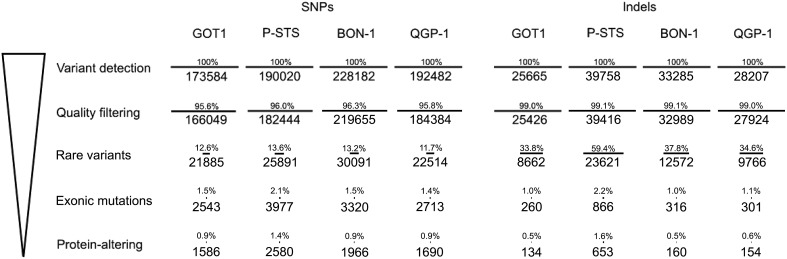
We detected an average of 196,067 single-nucleotide polymorphisms (SNPs) per cell line (range 173,584‒228,182). After quality filtering according to GATK best practices with the tool HaplotypeCaller, the number of reads was narrowed down to 188,133 on average (range 166,049‒219,655). In order to remove frequently occurring SNPs, all variants were further filtered against the 1000 Genomes Project database (Auton et al. 2015), where only SNPs with a frequency less than 0.01 were included. This generated an average of 25,095 rare SNPs per cell line. On average 1956 SNPs (range 1586‒2580) were located within exons and predicted to cause an alteration in the translated protein (Fig. 4 and Supplementary Table 2). Using the same filtering process, the number of indels was narrowed down from an average of 31,729 per cell line (range 25,665‒39,758) to an average of 275 (range 134‒653) protein-modifying events (Supplementary Table 3). Among the four GEPNET cell lines, P-STS had the highest number of protein-altering SNPs and indels and also displayed a higher degree of transitions compared to the other cell lines (Supplementary Fig. 3).
Figure 4.
Whole-exome sequencing of GEPNET cell lines. Successive filtering with the remaining number of SNPs or indels in each step is shown. Percentage is relative to the number of variants detected in the detection step.
We first investigated the cell lines for mutations in genes linked to hereditary endocrine tumour syndromes, i.e. MEN1, VHL, NF1 and TSC2 syndromes (Capelli et al. 2009, Corbo et al. 2010). Among these genes, only heterozygous TSC2 mutations in P-STS and BON-1 were found (Fig. 5). None of the cell lines harboured mutations in the MEN1, VHL or NF1 genes. Next, we searched for mutations in genes previously reported to be recurrently mutated in SINETs (Francis et al. 2013) and PanNETs (Jiao et al. 2011, Scarpa et al. 2017). CDKN1B, which is recurrently mutated in SINETs, was not affected in any of the cell lines. ATRX, MTOR, SETD2 and DIS3L2, all of which have been found recurrently mutated in PanNETs, had bi-allelic inactivations in one or several cell lines. QGP-1 harboured homozygous mutations in ATRX, while GOT1 had a heterozygous mutation. QGP-1 had only one MTOR gene copy, which was mutated. SETD2, a gene coding for a histone modifier protein, had only one gene copy in GOT1, P-STS and QGP-1. This gene copy was mutated in P-STS and QGP-1 causing bi-allelic inactivation. DNA repair gene CHEK2 showed a homozygous mutation in BON-1. Finally, we studied other cancer-associated genes, by analysing the 127 mutated genes identified in the Tumor Cancer Genome Atlas (TCGA) Pan-Cancer effort. (Kandoth et al. 2013) Out of these cancer-associated genes, 46 genes had heterozygous or homozygous mutations in one or more cell lines (Supplementary Table 4). Among affected genes, we found the key tumour suppressor TP53 mutated in three out of four cell lines. TP53 is seldom inactivated in GEPNETs, but here found mutated in P-STS, BON-1 and QGP-1. GOT1 was the only GEPNET cell line with wild-type TP53. The cell-cycle regulators CDKN2A and CDKN2B were inactivated by homozygous loss in BON-1. The SMAD4 gene, involved in cell growth inhibition signalling, was lost in GOT1, had a heterozygous mutation in P-STS and a homozygous mutation in BON-1.
Figure 5.
Genomic events involving genes linked to hereditary endocrine tumour syndromes, genes recurrently mutated in GEPNETs, and cancer-associated genes. Four genes have been hereditary linked to GEPNETs, none of which had bi-allelic inactivation in the cell lines. Out of previously identified recurrently mutated genes in GEPNETS, four had bi-allelic inactivations: ATRX (QGP-1), MTOR (QGP-1), SETD2 (P-STS and QGP-1), and CHEK2 (BON-1). Out of the 127 genes identified by the Tumor Cancer Genome Atlas, 49 had one or more protein-altering mutations in the cell lines; these genes included key tumour suppressors TP53, CDKN2A, CDKN2B and SMAD4.
PanNET and SINET cells are sensitive to inhibitors of MEK and HDAC
GEPNET cell lines have been used to study selected inhibitors for their efficacy and mechanism of action (Grozinsky-Glasberg et al. 2012). No comprehensive screening for effective inhibitors of GEPNETs has however been reported. One of this study’s aims was to define a therapeutic sensitivity profile for GEP-NETs. To this end, we performed an inhibitor screening with 1224 inhibitors, on the GEPNET cell lines GOT1, P-STS, BON-1 and QGP-1 using the non-tumourigenic cell lines MCF10A, BJ and HUV-EC-C as controls.
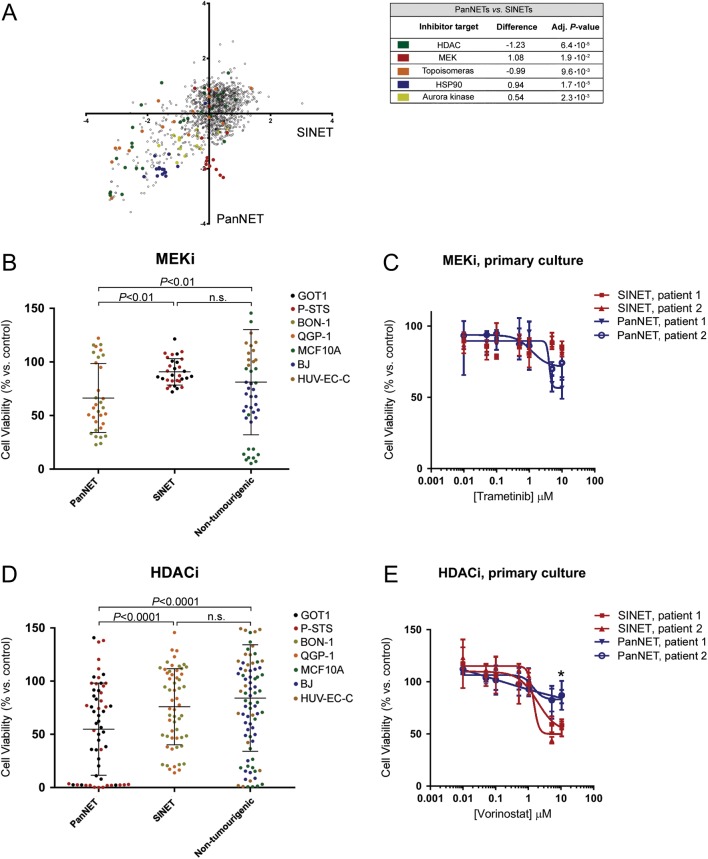
This resulted in data regarding the efficacy of a large number of inhibitors and protein targets (Supplementary Table 5). In the data, we observed an overall high degree of similarity in inhibitor sensitivity between cell lines derived from SINETs and PanNETs. We could however identify several groups of inhibitors that were significantly more selective for PanNET cells or SINET cells (Fig. 6A). Inhibitors of HDAC and topoisomerase were more efficient against SINETs, and inhibitors of MEK, HSP90 and Aurora kinase were more efficient against PanNETs. Looking at the anti-tumour effect of all MEKi in the screening library, SINET cells were largely unaffected, while most of the MEKi were efficient against PanNET cell lines and in a lesser degree against non-tumourigenic cells (Fig. 6B). To confirm these findings, we used first-passage primary tumour cells, prepared from patient SINETs and PanNETs collected at surgery. The authenticity of tumours was verified using immunohistochemistry (Supplementary Fig. 4 and Supplementary Table 6). FDA-approved MEK inhibitor trametinib was used to treat these cells (Fig. 6C). While many HDACi had no or limited effect, this was mostly true for PanNET cells and non-tumourigenic cells. A large number of HDACi had, in fact, a potent effect against SINET cells (Fig. 6D). In primary cells, FDA-approved HDACi vorinostat was seemingly more efficient against SINETs than PanNETs at the highest tested concentration (Fig. 6E).
Figure 6.
Therapeutic sensitivity of GEPNET cell lines and primary cell cultures. (A) Average Z-score representing effect on cell viability of individual inhibitors to SINETs (GOT1/P-STS) and PanNETs (BON-1/QGP-1), plotted against each other. Groups of inhibitors that are significantly more potent against SINETs or PanNETs are marked by colour. (B) The effect of all MEKi against SINET cells, PanNET cells and non-tumourigenic cells. MEKi are more potent against PanNET cells, compared to SINET and non-tumourigenic cells. (C) Comparing the sensitivity of PanNET and SINET first-passage primary cells to MEKi trametinib. (D) SINET cell lines are more sensitive to HDACi, compared to PanNET cells and non-tumourigenic cells. (E) First-passage primary SINET cells are seemingly more sensitive than primary PanNET cells to the HDACi vorinostat. (B and D) Bars indicate mean effect, error bars s.d. and P values generated from Wilcoxon signed-rank test. (C and E) Dose–response curves represent a mean of n = 3 and the error bars denote standard deviation (s.d.).
Discussion
We here report the first comprehensive characterisation of seven GEPNET-derived cell lines, including evaluation of their neuroendocrine phenotype, genomic alterations and therapeutic sensitivity profiles. We found a preserved neuroendocrine phenotype in four cell lines and that these cell lines have genomic alterations characteristic of GEPNETs, albeit while also harbouring additional genomic alterations. A large-scale inhibitor screening was performed, providing a basis for future therapeutic developments and identified HDAC and MEK as promising protein targets for therapy.
NETs are defined by their expression of neuroendocrine markers, e.g. synaptophysin and chromogranin A, which are components of small synaptic-like vesicles, and large dense-core vesicles, respectively (Rindi et al. 1986, Wiedenmann et al. 1986). Furthermore, GEPNETs are derived from progenitor cells in the intestinal crypts or pancreatic ducts, and consequently express epithelial markers, e.g. cytokeratins. We showed that four GEPNET cell lines, GOT1, P-STS, BON-1 and QGP-1, all expressed synaptophysin and NCAM strongly, while expression of chromogranin A, PGP9.5, NSE, CD57 and lineage-specific markers VMAT1 and 5-HT varied between the cell lines. In GOT1 and BON-1, we also found expression of SSTR2, a major target for symptomatic treatment of hormonal symptoms, tumour imaging and peptide receptor radiotherapy in GEPNET patients. These findings demonstrate the neuroendocrine phenotype of these cell lines, and their usefulness as models for studying somatostatin receptor-targeted radiotherapy in neuroendocrine tumours (Reubi et al. 2000). All four cell lines also stained positively for cytokeratins. We failed, however, to find any neuroendocrine differentiation or expression of cytokeratins in the KRJ-I, L-STS and H-STS cell lines. These cell lines grew as suspension cultures, forming floating aggregates. Based on this, we hypothesised that they might be lymphoblastoid cell lines. We went on to show that these cell lines expressed lymphoid markers CD45 and CD20 and contained high levels of EBV DNA, confirming our hypothesis. The lack of NET markers and cytokeratins, and the presence of lymphoid markers plus EBV DNA, has also been found in early passages of these cell lines at the laboratory in which they were established (R Pfragner, unpublished observations). While KRJ-I in the initial characterisation displayed a positivity for anti-chromogranin A and HISL19 staining, it was also shown that the culture consisted of clones with both high and low growth rates, and with varying anchorage dependency (Pfragner et al. 1996). The established cell line later consisted only of a suspension of single cells and multicellular spheroids. P-STS was shown to express neuroendocrine markers in all cells, while the staining frequency and intensity of neuroendocrine markers in L-STS and H-STS was variable (Pfragner et al. 2009). Based on these early observations, combined with the present investigation, has lead us to believe that the loss of a neuroendocrine phenotype in KRJ-I, L-STS and H-STS was because the cultures initially consisted of both neuroendocrine and lymphoblastoid cells. The neuroendocrine cells were then were completely taken over by the lymphoblastoid cells. We also characterised primary cell cultures, which as expected showed expression of all general neuroendocrine and epithelial markers while lymphoid markers could not be detected.
SINETs are often subcategorised into at least two groups based on their CNAs. The largest group of tumours is characterised by chromosomal losses, including loss of chromosome 18, and is associated with longer patient survival. A smaller group of tumours is characterised by multiple chromosomal gains and is associated with shorter patient survival (Kulke 2008, Andersson et al. 2009, Karpathakis et al. 2016). PanNETs harbour recurrent CNAs characterised by a predominance of gains (Gebauer et al. 2014). To study the genomic profile of the GEPNET cell lines, we performed arrayCGH and found that they had very different CNA profiles. The SINET cell lines GOT1 and P-STS were characterised by a predominance of chromosomal losses. The PanNET cell line BON-1, on the other hand, was characterised by a predominance of gains, including gain of several whole chromosomes. Among the cell lines investigated, QGP-1 had the highest frequency of CNAs, with an equal number of losses and gains. GOT1 had partial losses on 18q encompassing the tumour suppressor SMAD4, which is believed to be haploinsufficient (Howe et al. 1998, Xu et al. 2000, Alberici et al. 2006). BON-1 had homozygous loss of the tumour suppressor genes CDKN2A and CDKN2B, an event that has previously been reported in this cell line (Vandamme et al. 2015). The QGP-1 cell line had amplifications involving three segments on chromosome 12, including the genes MDM2 and HMGA2. Interestingly, amplification and overexpression of MDM2 is a well-known mechanism of TP53 inactivation occurring in 22% of all PanNETs, while overexpression of HMGA2 is associated with a malignant phenotype (Abe et al. 2003, Hu et al. 2010).
PanNETs frequently have somatic mutations in MEN1, ATRX or DAXX. ATRX, located on the X-chromosome, was mutated in QGP-1 (male) and had a heterozygous mutation in GOT1 (female), possibly leading to a bi-allelic inactivation of the gene. No mutations in DAXX or MEN1 were found in any of the cell lines. While expression studies have indicated that mTOR signalling is upregulated in most PanNET tumours (Perren et al. 2000, Missiaglia et al. 2010), whole-exome and whole-genome sequencing studies have found mutations in negative regulators of the signalling pathway in only 12–15% of patient tumours (Jiao et al. 2011, Scarpa et al. 2017). None of these genes were mutated in any of the GEPNET cell lines, but MTOR was altered in QGP-1. CDKN1B is the only gene so far reported to be recurrently mutated in a subset of SINETs (Francis 2013). We did not find any mutations affecting CDKN1B in any of the GEPNET cell lines. Due to selection pressure and stress during in vitro culturing, tumour cell lines often acquire novel mutations that may not be found in the tumour from which they were derived. Acquired mutations may affect the growth properties of cell lines, and influence their therapeutic sensitivity. TP53, encoding a tumour suppressor and key regulator of DNA repair and apoptosis, is usually unaffected in GEPNETs, but was found to be bi-allelically inactivated in P-STS, BON-1 and QGP1. We found a homozygous stop-loss mutation in BON-1 and a frameshift deletion in QGP-1, in accordance with a previous report (Vandamme et al. 2015).
To date, GEPNET cell lines have only been used to study the potency and mechanisms of action of a limited number of inhibitors. In the present study, we performed a comprehensive inhibitor screening to obtain a sensitivity profile representing a wide range of therapeutic principles. The screening identified inhibitors that are selectively efficient to SINET or PanNET cell lines. To study these findings in another model, we used first-passage primary cells. The primary cell cultures expressed neuroendocrine markers similar to the investigated GEPNET cell lines. Experiments showed that vorinostat killed SINET primary cells more efficiently than PanNET primary cells and that trametinib was more efficient against PanNET primary cells. This effect was however only prominent at high doses, suggesting that observed effects might be due to off-target activity. HDAC inhibition has also previously been suggested as a treatment option for GEPNET patients (Baradari et al. 2006, Arvidsson et al. 2016, Sun et al. 2016).
From these findings combined, we conclude that the four cell lines GOT1, P-STS, BON-1 and QGP-1 are indeed authentic neuroendocrine tumour cell lines, harbouring genomic events characteristic of GEPNETs, but also cancer-associated mutations that most likely have occurred in cell culture. The identification of lymphoblastoid cell lines among putative GEPNETs emphasises the need to thoroughly characterise GEPNET cell lines used as models for neuroendocrine tumour disease. Our observations also emphasises the need to be cautious when drawing conclusions from studies performed on GEPNET cell lines. One major concern is the origin of GEPNET cell lines and whether they were derived from well-differentiated neuroendocrine tumours or poorly differentiated neuroendocrine carcinomas. The difficulties in establishing GEPNET cell lines are usually attributed to the low proliferation rate of these tumours. One may argue that available GEPNET cell lines were established from more aggressive tumours that should be classified as neuroendocrine carcinomas. The occurrence of TP53 mutations in three of the investigated cell lines (P-STS, BON-1 and QGP-1) further strengthens this concern. The original publications on GEPNET cell lines do not contain sufficient data to determine the grade of the tumours from which cell lines were derived. We have therefore reinvestigated the origin of the GOT1 cell line and found it to be a liver metastasis from a well-differentiated, serotonin-producing (enterochromaffin cell type) ileal NET. The liver metastasis was of grade 1 (WHO 2010) and harboured a loss of chromosome 18. We conclude that it remains to be proven to what extent GEPNET cell lines are representative for the tumour disease from which they were derived. Experimental data obtained from GEPNET cell lines should therefore be carefully validated using patient tumours.
Supplementary Material
Declaration of interest
The authors declare that there are no conflicts of interests that could be perceived as prejudicing the impartiality of the research reported.
Funding
This study was supported by grants from the Swedish Cancer Society, the Assar Gabrielsson Research Foundation, the Wilhelm and Martina Lundgren Foundation for Scientific Research, the Sahlgrenska University Hospital Funds, the BioCARE National Strategic Research Program and by ALF grants from the Sahlgrenska University Hospital.
Acknowledgements
The authors thank the Centre for Cellular Imaging and Bioinformatics Core Facility at the Sahlgrenska Academy, University of Gothenburg and the National Microscopy Infrastructure (NMI; VR-RFI 2016-00978), for the use of imaging equipment and for support from the staff. The expert technical assistance of Gülay Altiparmak, Monica Dohsé and Akif Demir is greatly appreciated. We also thank Pierre Åman for valuable advice.
References
- Abe N, Watanabe T, Suzuki Y, Matsumoto N, Masaki T, Mori T, Sugiyama M, Chiappetta G, Fusco A, Atomi Y. 2003. An increased high-mobility group A2 expression level is associated with malignant phenotype in pancreatic exocrine tissue. British Journal of Cancer 89 2104–2109. ( 10.1038/sj.bjc.6601391) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alberici P, Jagmohan-Changur S, De Pater E, Van Der Valk M, Smits R, Hohenstein P, Fodde R. 2006. Smad4 haploinsufficiency in mouse models for intestinal cancer. Oncogene 25 1841–1851. ( 10.1038/sj.onc.1209226) [DOI] [PubMed] [Google Scholar]
- Andersson E, Swärd C, Stenman G, Ahlman H, Nilsson O. 2009. High-resolution genomic profiling reveals gain of chromosome 14 as a predictor of poor outcome in ileal carcinoids. Endocrine-Related Cancer 16 953–966. ( 10.1677/ERC-09-0052) [DOI] [PubMed] [Google Scholar]
- Andersson E, Arvidsson Y, Swärd C, Hofving T, Wängberg B, Kristiansson E, Nilsson O. 2016. Expression profiling of small intestinal neuroendocrine tumors identifies subgroups with clinical relevance, prognostic markers and therapeutic targets. Modern Pathology 29 616–629. ( 10.1038/modpathol.2016.48) [DOI] [PubMed] [Google Scholar]
- Arvidsson Y, Bergström A, Arvidsson L, Kristiansson E, Ahlman H, Nilsson O. 2010. Hypoxia stimulates CXCR4 signalling in ileal carcinoids. Endocrine-Related Cancer 17 303–316. ( 10.1677/ERC-09-0085) [DOI] [PubMed] [Google Scholar]
- Arvidsson Y, Johanson V, Pfragner R, Wängberg B, Nilsson O. 2016. Cytotoxic effects of valproic acid on neuroendocrine tumour cells. Neuroendocrinology 103 578–591. ( 10.1159/000441849) [DOI] [PubMed] [Google Scholar]
- Asfari M, Janjic D, Meda P, Li G, Halban PA, Wollheim CB. 1992. Establishment of 2-mercaptoethanol-dependent differentiated insulin-secreting cell lines. Endocrinology 130 167–178. ( 10.1210/endo.130.1.1370150) [DOI] [PubMed] [Google Scholar]
- Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR. 2015. A global reference for human genetic variation. Nature 526 68–74. ( 10.1038/nature15393) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baradari V, Huether A, Hopfner M, Schuppan D, Scherubl H. 2006. Antiproliferative and proapoptotic effects of histone deacetylase inhibitors on gastrointestinal neuroendocrine tumor cells. Endocrine-Related Cancer 13 1237–1250. ( 10.1677/erc.1.01249) [DOI] [PubMed] [Google Scholar]
- Barrett MT, Scheffer A, Ben-Dor A, Sampas N, Lipson D, Kincaid R, Tsang P, Curry B, Baird K, Meltzer PS, et al 2004. Comparative genomic hybridization using oligonucleotide microarrays and total genomic DNA. PNAS 101 17765–17770. ( 10.1073/pnas.0407979101) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernhardt P, Oddstig J, Kölby L, Nilsson O, Ahlman H, Forssell-Aronsson E. 2007. Effects of treatment with Lu-177-DOTA-Tyr(3)-octreotate on uptake of subsequent injection in carcinoid-bearing nude mice. Cancer Biotherapy and Radiopharmaceuticals 22 644–653. ( 10.1089/cbr.2007.333) [DOI] [PubMed] [Google Scholar]
- Boora GK, Kanwar R, Kulkarni AA, Pleticha J, Ames M, Schroth G, Beutler AS, Banck MS. 2015. Exome-level comparison of primary well-differentiated neuroendocrine tumors and their cell lines. Cancer Genetics 208 374–381. ( 10.1016/j.cancergen.2015.04.002) [DOI] [PubMed] [Google Scholar]
- Capelli P, Martignoni G, Pedica F, Falconi M, Antonello D, Malpeli G, Scarpa A. 2009. Endocrine neoplasms of the pancreas: pathologic and genetic features. Archives of Pathology and Laboratory Medicine 133 350–364. ( 10.1043/1543-2165-133.3.350) [DOI] [PubMed] [Google Scholar]
- Corbo V, Dalai I, Scardoni M, Barbi S, Beghelli S, Bersani S, Albarello L, Doglioni C, Schott C, Capelli P, et al 2010. MEN1 in pancreatic endocrine tumors: analysis of gene and protein status in 169 sporadic neoplasms reveals alterations in the vast majority of cases. Endocrine-Related Cancer 17 771–783. ( 10.1677/ERC-10-0028) [DOI] [PubMed] [Google Scholar]
- Dalmo J, Spetz J, Montelius M, Langen B, Arvidsson Y, Johansson H, Parris TZ, Helou K, Wängberg B, Nilsson O, et al 2017. Priming increases the anti-tumor effect and therapeutic window of 177Lu-octreotate in nude mice bearing human small intestine neuroendocrine tumor GOT1. EJNMMI Research 7 6 ( 10.1186/s13550-016-0247-y) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doihara H, Nozawa K, Kojima R, Kawabata-Shoda E, Yokoyama T, Ito H. 2009. QGP-1 cells release 5-HT via TRPA1 activation; a model of human enterochromaffin cells. Molecular and Cellular Biochemistry 331 239 ( 10.1007/s11010-009-0165-7) [DOI] [PubMed] [Google Scholar]
- Domcke S, Sinha R, Levine DA, Sander C, Schultz N. 2013. Evaluating cell lines as tumour models by comparison of genomic profiles. Nature Communication 4 2126 ( 10.1038/ncomms3126) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efrat S, Surana M, Fleischer N. 1991. Glucose induces insulin gene transcription in a murine pancreatic beta-cell line. Journal of Biological Chemistry 266 11141–11143. [PubMed] [Google Scholar]
- Ellis LM, Samuel S, Sceusi E. 2010. Varying opinions on the authenticity of a human midgut carcinoid cell line – letter. Clinical Cancer Research 16 5365–5366. ( 10.1158/1078-0432.CCR-10-2550) [DOI] [PubMed] [Google Scholar]
- Ertel A, Verghese A, Byers SW, Ochs M, Tozeren A. 2006. Pathway-specific differences between tumor cell lines and normal and tumor tissue cells. Molecular Cancer 5 55 ( 10.1186/1476-4598-5-55) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evers BM, Townsend CM, Jr, Upp JR, Allen E, Hurlbut SC, Kim SW, Rajaraman S, Singh P, Reubi JC, Thompson JC. 1991. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology 101 303–311. ( 10.1016/0016-5085(91)90004-5) [DOI] [PubMed] [Google Scholar]
- Forssell-Aronsson E, Spetz J, Ahlman H. 2013. Radionuclide therapy via SSTR: future aspects from experimental animal studies. Neuroendocrinology 97 86–98. ( 10.1159/000336086) [DOI] [PubMed] [Google Scholar]
- Francis JM, Kiezun A, Ramos AH, Serra S, Pedamallu CS, Qian ZR, Banck MS, Kanwar R, Kulkarni AA, Karpathakis A, et al 2013. Somatic mutation of CDKN1B in small intestine neuroendocrine tumors. Nature Genetics 45 1483–1486. ( 10.1038/ng.2821) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gazdar AF, Chick WL, Oie HK, Sims HL, King DL, Weir GC, Lauris V. 1980. Continuous, clonal, insulin- and somatostatin-secreting cell lines established from a transplantable rat islet cell tumor. PNAS 77 3519–3523. ( 10.1073/pnas.77.6.3519) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebauer N, Schmidt-Werthern C, Bernard V, Feller AC, Keck T, Begum N, Rades D, Lehnert H, Brabant G, Thorns C. 2014. Genomic landscape of pancreatic neuroendocrine tumors. World Journal of Gastroenterology 20 17498–17506. ( 10.3748/wjg.v20.i46.17498) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillet JP, Calcagno AM, Varma S, Marino M, Green LJ, Vora MI, Patel C, Orina JN, Eliseeva TA, Singal V, et al 2011. Redefining the relevance of established cancer cell lines to the study of mechanisms of clinical anti-cancer drug resistance. PNAS 108 18708–18713. ( 10.1073/pnas.1111840108) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grozinsky-Glasberg S, Shimon I, Rubinfeld H. 2012. The role of cell lines in the study of neuroendocrine tumors. Neuroendocrinology 96 173–187. ( 10.1159/000338793) [DOI] [PubMed] [Google Scholar]
- Gueli N, Toto A, Palmieri G, Carmenini G, Delpino A, Ferrini U. 1987. In vitro growth of a cell line originated from a human insulinoma. Journal of Experimental and Clinical Cancer Research 6 281–285. [Google Scholar]
- Howe JR, Roth S, Ringold JC, Summers RW, Jarvinen HJ, Sistonen P, Tomlinson IP, Houlston RS, Bevan S, Mitros FA, et al 1998. Mutations in the SMAD4/DPC4 gene in juvenile polyposis. Science 280 1086–1088. ( 10.1126/science.280.5366.1086) [DOI] [PubMed] [Google Scholar]
- Hu W, Feng Z, Modica I, Klimstra DS, Song L, Allen PJ, Brennan MF, Levine AJ, Tang LH. 2010. Gene amplifications in well-differentiated pancreatic neuroendocrine tumors inactivate the p53 pathway. Genes and Cancer 1 360–368. ( 10.1177/1947601910371979) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, Qi Y, Scherer SW, Lee C. 2004. Detection of large-scale variation in the human genome. Nature Genetics 36 949–951. ( 10.1038/ng1416) [DOI] [PubMed] [Google Scholar]
- Iguchi H, Hayashi I, Kono A. 1990. A somatostatin-secreting cell line established from a human pancreatic islet cell carcinoma (somatostatinoma): release experiment and immunohistochemical study. Cancer Research 50 3691. [PubMed] [Google Scholar]
- Ishihara H, Asano T, Tsukuda K, Katagiri H, Inukai K, Anai M, Kikuchi M, Yazaki Y, Miyazaki JI, Oka Y. 1993. Pancreatic beta cell line MIN6 exhibits characteristics of glucose metabolism and glucose-stimulated insulin secretion similar to those of normal islets. Diabetologia 36 1139–1145. ( 10.1007/BF00401058) [DOI] [PubMed] [Google Scholar]
- Jiao Y, Shi C, Edil BH, de Wilde RF, Klimstra DS, Maitra A, Schulick RD, Tang LH, Wolfgang CL, Choti MA, et al 2011. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science 331 1199–1203. ( 10.1126/science.1200609) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaku M, Nishiyama T, Yagawa K, Abe M. 1980. Establishment of a carcinoembryonic antigen-producing cell line from human pancreatic carcinoma. Gann 71 596–601. [PubMed] [Google Scholar]
- Kandoth C, McLellan MD, Vandin F, Ye K, Niu B, Lu C, Xie M, Zhang Q, McMichael JF, Wyczalkowski MA, et al 2013. Mutational landscape and significance across 12 major cancer types. Nature 502 333–339. ( 10.1038/nature12634) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karpathakis A, Dibra H, Pipinikas C, Feber A, Morris T, Francis J, Oukrif D, Mandair D, Pericleous M, Mohmaduvesh M, et al. 2016. Prognostic impact of novel molecular subtypes of small intestinal neuroendocrine tumor. Clinical Cancer Research 22 250–258. ( 10.1158/1078-0432.CCR-15-0373) [DOI] [PubMed] [Google Scholar]
- Kidd M, Modlin IM, Pfragner R, Eick GN, Champaneria MC, Chan AK, Camp RL, Mane SM. 2007. Small bowel carcinoid (enterochromaffin cell) neoplasia exhibits transforming growth factor-beta1-mediated regulatory abnormalities including up-regulation of C-Myc and MTA1. Cancer 109 2420–2431. ( 10.1002/cncr.22725) [DOI] [PubMed] [Google Scholar]
- Kidd M, Modlin IM, Gustafsson BI, Drozdov I, Hauso O, Pfragner R. 2008. Luminal regulation of normal and neoplastic human EC cell serotonin release is mediated by bile salts, amines, tastants, and olfactants. American Journal of Physiology: Gastrointestinal and Liver Physiology 295 G260–G272. ( 10.1152/ajpgi.00056.2008) [DOI] [PubMed] [Google Scholar]
- Kulke MH, Freed E, Chiang DY, Philips J, Zahrieh D, Glickman JN, Shivdasani RA. 2008. High-resolution analysis of genetic alterations in small bowel carcinoid tumors reveals areas of recurrent amplification and loss. Genes, Chromosomes and Cancer 47 591–603. ( 10.1002/gcc.20561) [DOI] [PubMed] [Google Scholar]
- Kölby L, Bernhardt P, Ahlman H, Wängberg B, Johanson V, Wigander A, Forssell-Aronsson E, Karlsson S, Ahrén B, Stenman G, et al 2001. A transplantable human carcinoid as model for somatostatin receptor-mediated and amine transporter-mediated radionuclide uptake. American Journal of Pathology 158 745–755. ( 10.1016/S0002-9440(10)64017-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kölby L, Bernhardt P, Johanson V, Schmitt A, Ahlman H, Forssell-Aronsson E, Macke H, Nilsson O. 2005. Successful receptor-mediated radiation therapy of xenografted human midgut carcinoid tumour. British Journal of Cancer 93 1144–1151. ( 10.1038/sj.bjc.6602845) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25 1754–1760. ( 10.1093/bioinformatics/btp324) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundqvist M, Mark J, Funa K, Heldin NE, Morstyn G, Wedell B, Layton J, Oberg K. 1991. Characterization of a cell-line (LCC-18) from a cultured human neuroendocrine-differentiated colonic-carcinoma. European Journal of Cancer 27 1663–1668. ( 10.1016/0277-5379(91)90441-F) [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al 2010. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Research 20 1297–1303. ( 10.1101/gr.107524.110) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Missiaglia E, Dalai I, Barbi S, Beghelli S, Falconi M, della Peruta M, Piemonti L, Capurso G, Di Florio A, delle Fave G, et al 2010. Pancreatic endocrine tumors: expression profiling evidences a role for AKT-mTOR pathway. Journal of Clinical Oncology 28 245–255. ( 10.1200/JCO.2008.21.5988) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modlin IM, Kidd M, Pfragner R, Eick GN, Champaneria MC. 2006. The functional characterization of normal and neoplastic human enterochromaffin cells. Journal of Clinical Endocrinology and Metabolism 91 2340–2348. ( 10.1210/jc.2006-0110) [DOI] [PubMed] [Google Scholar]
- Modlin IM, Moss SF, Chung DC, Jensen RT, Snyderwine E. 2008. Priorities for improving the management of gastroenteropancreatic neuroendocrine tumors. Journal of the National Cancer Institute 100 1282–1289. ( 10.1093/jnci/djn275) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perren A, Komminoth P, Saremaslani P, Matter C, Feurer S, Lees JA, Heitz PU, Eng C. 2000. Mutation and expression analyses reveal differential subcellular compartmentalization of PTEN in endocrine pancreatic tumors compared to normal islet cells. American Journal of Pathology 157 1097–1103. ( 10.1016/S0002-9440(10)64624-X) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Persson F, Winnes M, Andrén Y, Wedell B, Dahlenfors R, Asp J, Mark J, Enlund F, Stenman G. 2008. High-resolution array CGH analysis of salivary gland tumors reveals fusion and amplification of the FGFR1 and PLAG1 genes in ring chromosomes. Oncogene 27 3072–3080. ( 10.1038/sj.onc.1210961) [DOI] [PubMed] [Google Scholar]
- Pfanzagl B, Mechtcheriakova D, Meshcheryakova A, Aberle SW, Pfragner R, Jensen-Jarolim E. 2017. Activation of the ileal neuroendocrine tumor cell line P-STS by acetylcholine is amplified by histamine: role of H3R and H4R. Scientific Reports 7 1313 ( 10.1038/s41598-017-01453-5) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfragner R, Wirnsberger G, Niederle B, Behmel A, Rinner I, Mandl A, Wawrina F, Luo J, Adamiker D, Hoger H, et al 1996. Establishment of a continuous cell line from a human carcinoid of the small intestine (KRJ-I). International Journal of Oncology 8 513–520. ( 10.3892/ijo.8.3.513) [DOI] [PubMed] [Google Scholar]
- Pfragner R, Behmel A, Hoger H, Beham A, Ingolic E, Stelzer I, Svejda B, Moser VA, Obenauf AC, Siegl V, et al 2009. Establishment and characterization of three novel cell lines – P-STS, L-STS, H-STS – derived from a human metastatic midgut carcinoid. Anticancer Research 29 1951–1961. [PubMed] [Google Scholar]
- Reid MD, Bagci P, Ohike N, Saka B, Erbarut Seven I, Dursun N, Balci S, Gucer H, Jang KT, Tajiri T, et al 2015. Calculation of the Ki67 index in pancreatic neuroendocrine tumors: a comparative analysis of four counting methodologies. Modern Pathology 28 686–694. ( 10.1038/modpathol.2014.156) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubi JC, Schar JC, Waser B, Wenger S, Heppeler A, Schmitt JS, Macke HR. 2000. Affinity profiles for human somatostatin receptor subtypes SST1-SST5 of somatostatin radiotracers selected for scintigraphic and radiotherapeutic use. European Journal of Nuclear Medicine and Molecular 27 273–282. ( 10.1007/s002590050034) [DOI] [PubMed] [Google Scholar]
- Rindi G, Buffa R, Sessa F, Tortora O, Solcia E. 1986. Chromogranin A, B and C immunoreactivities of mammalian endocrine cells. Distribution, distinction from costored hormones/prohormones and relationship with the argyrophil component of secretory granules. Histochemistry 85 19–28. ( 10.1007/BF00508649) [DOI] [PubMed] [Google Scholar]
- Rindi G, Grant SG, Yiangou Y, Ghatei MA, Bloom SR, Bautch VL, Solcia E, Polak JM. 1990. Development of neuroendocrine tumors in the gastrointestinal tract of transgenic mice. Heterogeneity of hormone expression. American Journal of Pathology 136 1349–1363. [PMC free article] [PubMed] [Google Scholar]
- Rinner B, Galle B, Trajanoski S, Fischer C, Hatz M, Maierhofer T, Michelitsch G, Moinfar F, Stelzer I, Pfragner R, et al 2012. Molecular evidence for the bi-clonal origin of neuroendocrine tumor derived metastases. BMC Genomics 13 594 ( 10.1186/1471-2164-13-594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandberg R, Ernberg I. 2005. Assessment of tumor characteristic gene expression in cell lines using a tissue similarity index (TSI). PNAS 102 2052–2057. ( 10.1073/pnas.0408105102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarpa A, Chang DK, Nones K, Corbo V, Patch AM, Bailey P, Lawlor RT, Johns AL, Miller DK, Mafficini A, et al 2017. Whole-genome landscape of pancreatic neuroendocrine tumours. Nature 543 65–71. ( 10.1038/nature21063) [DOI] [PubMed] [Google Scholar]
- Spetz J, Langen B, Rudqvist N, Parris TZ, Helou K, Nilsson O, Forssell-Aronsson E. 2017. Hedgehog inhibitor sonidegib potentiates 177Lu-octreotate therapy of GOT1 human small intestine neuroendocrine tumors in nude mice. BMC Cancer 17 528 ( 10.1186/s12885-017-3524-x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein WD, Litman T, Fojo T, Bates SE. 2004. A Serial Analysis of Gene Expression (SAGE) database analysis of chemosensitivity: comparing solid tumors with cell lines and comparing solid tumors from different tissue origins. Cancer Research 64 2805–2816. ( 10.1158/0008-5472.CAN-03-3383) [DOI] [PubMed] [Google Scholar]
- Sun L, Qian Q, Sun G, Mackey LV, Fuselier JA, Coy DH, Yu C-Y. 2016. Valproic acid induces NET cell growth arrest and enhances tumor suppression of the receptor-targeted peptide–drug conjugate via activating somatostatin receptor type II. Journal of Drug Targeting 24 169–177. ( 10.3109/1061186X.2015.1066794) [DOI] [PubMed] [Google Scholar]
- Valentino JD, Li J, Zaytseva YY, Mustain WC, Elliott VA, Kim JT, Harris JW, Campbell K, Weiss H, Wang C. et al 2014. Cotargeting the PI3K and RAS pathways for the treatment of neuroendocrine tumors. Clinical Cancer Research 20 1212–1222. ( 10.1158/1078-0432.CCR-13-1897) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Buren G, 2nd, Rashid A, Yang AD, Abdalla EK, Gray MJ, Liu W, Somcio R, Fan F, Camp ER, Yao JC, et al 2007. The development and characterization of a human midgut carcinoid cell line. Clinical Cancer Research 13 4704–4712. ( 10.1158/1078-0432.CCR-06-2723) [DOI] [PubMed] [Google Scholar]
- Vandamme T, Peeters M, Dogan F, Pauwels P, Van Assche E, Beyens M, Mortier G, Vandeweyer G, de Herder W, Van Camp G, et al 2015. Whole-exome characterization of pancreatic neuroendocrine tumor cell lines BON-1 and QGP-1. Journal of Molecular Endocrinology 54 137–147. ( 10.1530/JME-14-0304) [DOI] [PubMed] [Google Scholar]
- van Kuppeveld FJM, Johansson KE, Galama JMD, Kissing J, Bolske G, Vanderlogt JTM, Melchers WJG. 1994. Detection of mycoplasma contamination in cell-cultures by a mycoplasma group-specific PCR. Applied and Environmental Microbiology 60 149–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Research 38 e164 ( 10.1093/nar/gkq603) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiedenmann B, Franke WW, Kuhn C, Moll R, Gould VE. 1986. Synaptophysin: a marker protein for neuroendocrine cells and neoplasms. PNAS 83 3500–3504. ( 10.1073/pnas.83.10.3500) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu XL, Brodie SG, Yang X, Im YH, Parks WT, Chen L, Zhou YX, Weinstein M, Kim SJ, Deng CX. 2000. Haploid loss of the tumor suppressor Smad4/Dpc4 initiates gastric polyposis and cancer in mice. Oncogene 19 1868–1874. ( 10.1038/sj.onc.1203504) [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.



 This work is licensed under a
This work is licensed under a