Abstract
“The Atlas of Mouse Development” by Kaufman is a classic paper atlas that is the de facto standard for the definition of mouse embryo anatomy in the context of standard histological images. We have redigitized the original haematoxylin and eosin–stained tissue sections used for the book at high resolution and transferred the hand-drawn annotations to digital form. We have augmented the annotations with standard ontological assignments (EMAPA anatomy) and made the data freely available via an online viewer (eHistology) and from the University of Edinburgh DataShare archive. The dataset captures and preserves the definitive anatomical knowledge of the original atlas, provides a core image set for deeper community annotation and teaching, and delivers a unique high-quality set of high-resolution histological images through mammalian development for manual and automated analysis.
Keywords: embryo, mouse, section, atlas, anatomy, imaging
Data Description
“The Atlas of Mouse Development” [1] is a book detailing the anatomy of mouse embryo development and stands as the definitive work in the field. The atlas is based on a lifetime of work by Kaufman, who established a unique set of histological sections of about 450 mouse embryos, many of which are full serial section-series, from which he selected carefully staged samples for the histological images within the book. The combination of the histological section series and the printed book represent a unique resource and captures the current understanding of classical mouse anatomy. In taxonomic terms, these physical sections are the reference specimens for the definition of mouse embryo anatomy, and the digitized images with the associated annotations are a digital holotype for the definition of anatomical terms and the progression of mouse embryo development. In addition, the paper atlas has given rise to the Mouse Atlas programme in Edinburgh [2] and to the EMAPA mouse anatomy ontology [3,4]. The original index for the book was used to develop the primary list of anatomical terms in the ontology, and EMAPA is now recognized as the standard mouse embryo ontology used to annotate mouse embryo data including embryo phenotype data [5,6].
In generating the eHistology Atlas, new images of the histological sections were acquired at high resolution, and the annotations have been transferred to a database. These images and annotations are now freely available from the eMouseAtlas web resource as eHistology (Fig. 1) and have been described by Graham et al. [7]. The new high-resolution images and the associated image coordinates for each annotation are fully freely available under a Creative Commons CC BY 4.0 licence. In addition, we have an agreement with Elsevier to present the images in a form similar to the original atlas plate layout (the web resource), and Elsevier is able to use the new images for their own version of an online version if they want to. Here we describe the dataset of the 937 high-resolution histology images with anatomy annotations and how they have been made available for further study and analysis.
Figure 1:
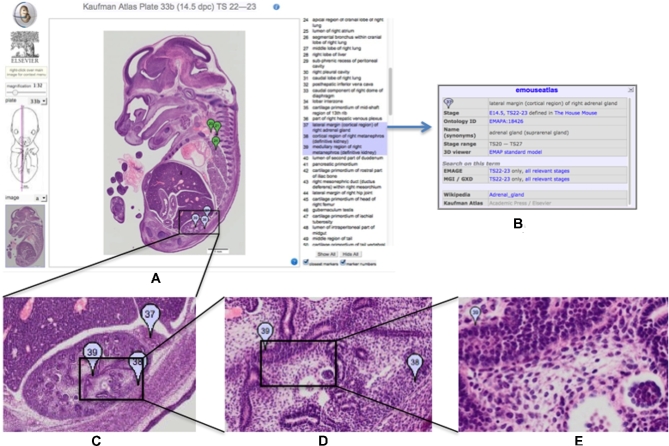
Kaufman Atlas eHistology Viewer. A, Screen shot showing the user interface with annotation list. The 3 selected terms showing as a blue “flags” numbers 37, 38, 39 are over the developing kidney. B, The pop-up dialog with extra information on selecting the “37.” C–E, Progressively higher-resolution images corresponding to zooming-in on the image. At full resolution, the pixel spacing is 0.34 × 0.34 microns and reveals cellular architecture and arrangements.
The motivation for the eHistology resource was to capture the anatomical knowledge in a permanently accessible open and digital form delivered with a viewer providing a view of the underlying histology data not possible in the printed atlas. The high-resolution images provide a rich resource of carefully staged mouse histology, which could be used for deeper analysis of tissue development and as a teaching resource. See Fig. 1 for an illustration of the resolution now available for these images. Embryogenesis is a highly dynamic process, and in Fig. 2 we highlight some of the advantages of capturing images at cellular resolution, e.g., the ability to zoom in and morphologically identify mitotically dividing cells and apoptotic cells undergoing programmed cell death. This is simply not possible in the print version of the atlas and represents a significant contribution to the community.
Figure 2:
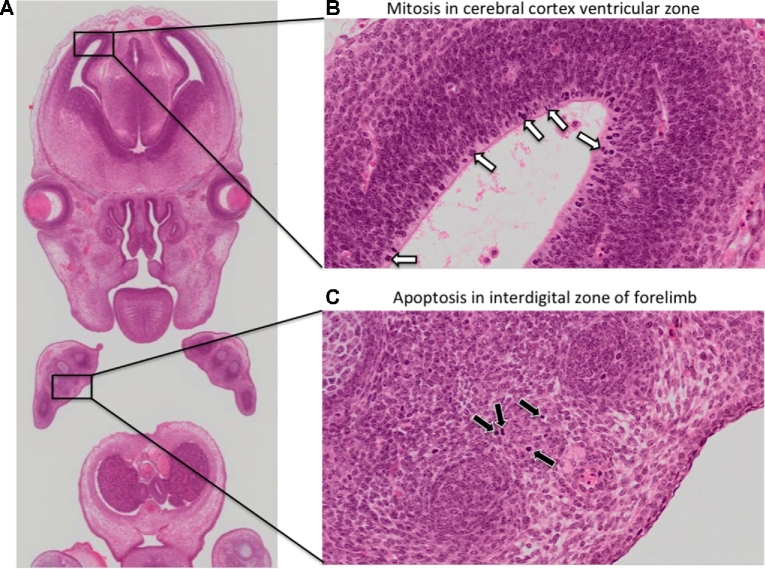
Observing mitosis and apoptosis in cellular resolution eHistology atlas images. A key advantage of capturing histology images at high resolution is the ability to morphologically identify mitotically dividing cells and apoptotic cells in embryo atlas images. A, Zoomed-out view of a coronal image of an E14.5 embryo. B, On the zoomed-in view, neuroblasts in the ventricular zone of the cerebral cortex show intense haematoxylin staining (white arrows), a morphological feature associated with chromosome condensation in mitotically dividing cells. C, On the zoomed-in view, scattered cells in the interdigital zone of the forelimb are pyknotic (black arrows), a morphological feature associated with nuclear condensation. The pyknotic cells additionally show signs of cell shrinkage. These are morphological hallmarks of apoptosis.
All the data are available under a creative commons licence (CC BY 4.0). In the future, we envisage the annotations being extended on a tissue-by-tissue basis through community curation. The eHistology viewer is open source and is available from the Mouse Atlas technical GitHub repository (github.com/ma-tech) (eHistology; RRID:SCR_015887).
Providing secure and long-term accessibility for research data is a difficult problem. A recent study of the longevity of 375 biomedical resources/databases [8] available on the web in 1997 found that 62.3% had ceased to be available, 14.4% were static, and only 23.3% were available as an active resource. The authors concluded that survival depended primarily on institutional interest and that a strategy dependent on external funding will very likely fail. To ensure long-term preservation of the image data and supporting annotations, we have therefore registered this dataset with the University of Edinburgh DataShare [9] repository [10], with policies registered in OpenDOAR (Directory of Open Access Repositories) [11]. Specifically, the preservation policy includes indefinite preservation of the original data with format migration to ensure continued readability and accessibility. In addition, and for convenience, these data are also hosted in the GigaDB repository [12].
Methods
Histology
Details of the mouse strains used, histological sectioning, and staining are provided by Kaufman (1994) [1]. Briefly, the embryos were “isolated from spontaneously cycling (C57BL X CBA) F1 hybrid females that had been previously mated to genetically similar F1 hybrid males.” The embryos were fixed, dehydrated, embedded in paraffin wax, and sectioned at 7-micron thickness. The mounted sections were then stained with haematoxylin and eosin.
Slide digitization
Digitization of the original histology slides was accomplished using the Olympus DotSlide slide scanner system. Using a × 20 objective lens, this generated full-colour images with a pixel resolution of 0.34 microns. Calibration was accomplished as part of the digitization process, allowing the inclusion of scale bars and the option to measure the distance between 2 points. In 2 instances (Plate 5 and Plate 14), the original sections could not be sourced and were presumed lost. In these instances, the original photographic negatives were used in place of the original slides to generate cellular-resolution grey-scale images.
Annotation and linking to the EMAPA ontology
Annotation was accomplished using a manual procedure whereby “flags” were positioned on points corresponding to the matching points as used in each plate in the book. The flags were placed using an editor’s version of the eHistology interface [7]. Each flag was linked to the anatomical term or phrase used in the book and also an EMAPA ontology term and an associated Wikipedia link. There were more than 10 000 flag labels used to annotate the eHistology sections, and linking them to EMAPA IDs was achieved through a combination of string matching and manual assignment of terms [13]. Linking to Wikipedia was accomplished using a manual process that utilized parent terms in the partonomic ontology tree to find the closest match for a given anatomical term or tissue.
eHistology viewer
Each eHistology image is described in an Edinburgh DataShare Digital Object Identifier (DOI), and this description includes the URL link to the eHistology viewer for that image. In this way, we provide a persistent means of accessing the zoom viewer for that image. An example DOI for a single high-resolution image is dx.doi.org/10.7488/ds/1232. This link resolves to a specific page at the Edinburgh Datashare web resource[14], [15], which in turn provides a link through to the current URL for the eHistology viewer.
By starting with a fully persistent DOI, the user will always be able to locate the data and is protected from any change to the hosting domain and URL of the eHistology viewer [16]. For convenience, we also provide an interactive index to the new images based on the plate and image designations of the original atlas.
Code availability
The data are provided in open-standard tif or jpeg image formats. All metadata are in plain txt format, and the Supplementary Data are in the Microsoft Excel open xml format xlsx. The code used for the online histology viewer is provided at the ma-tech GitHub archive, and specifically we use the WlzIIPSrv tiled image server and the eAtlasViewer javascript application.
Data records
Each record has an assigned DOI that resolves to a set of data files comprising a jpeg or tiff encoded image, Dublin core and other metadata files, and the set of annotations associated with the image. The image data volumes range up to 2 Gb, with a total volume of 118 GB for the full series in compressed “zip” format. Table 1 lists the files with each dataset. Each University of Edinburgh DataShare submission requires a subset of the Dublin Core (dublincore.org) data elements to be completed and allows a further set of optional elements; these are detailed in Table 2. Table 3 provides a partial listing of the datasets available as an example of the data content. The full listing of all 937 images is provided in the Supplementary Excel formatted data file SciDataKaufmanTable3.xlsx and corresponds to all of the histology section images of the original atlas for Plate numbers 2–41.
Table 1:
Listing of the data files available with each dataset
| File | Description |
|---|---|
| license.txt | Licence agreement for the data deposited at DataShare providing permissions for distribution and migration as needed—CC BY 4.0. |
| README.txt | Short description of the data and data files. |
| citation.txt | How to cite use of this particular image. |
| details.txt | The text describing the embryo taken from the matching page of the printed atlas and provided in tab-delimited form for reading into a spreadsheet programme. |
| image.tif/image.jpg | Full resolution tiff or jpeg formatted image of the histological section. |
| Image.txt | Image pixel dimensions and pixel size in microns. |
| terms.txt | A tab-delimited table of the annotations for this image providing the Kaufman annotations, location in the image, annotation number, EMAPA ID, and EMAPA term, with synonyms in brackets. |
| url.txt | Text providing the URL for the image on the emousetlas.org web resource. |
Table 2:
DataShare Dublin Core elements used for the Kaufman datasets
| Element | Qualifier | DCMI | Label | Input type | Mandatory |
|---|---|---|---|---|---|
| Contributor | contributor | Depositor | name | true | |
| Contributor | other | contributor | Funder | name | true |
| Creator | creator | Data Creator | name | false | |
| Date | accessioned | Date | Date Accessioned | date | true |
| date | available | Date | Date Available | date | true |
| Identifier | citation | identifier | Citation | citation | false |
| Identifier | uri | identifier | Persistent Identifier | DOI/handle | true |
| Description | abstract | decription | Data Description (abstract) | text | false |
| Description | tableofcontents | decription | Data Description (TOC) | text | false |
| Publisher | publisher | Publisher | text | true | |
| Relation | isversionof | isVersionOf | Relation (Is Version Of) | text | false |
| Relation | isreferencedby | isReferencedBy | Relation (Is Referenced By) | text | false |
| Subject | subject | Subject Keywords | text | false | |
| Subject | classification | subject | Subject Classification | Controlled text | false |
| Title | Title | Title | text | true | |
| Title | alternative | Title | Alternative Title | text | false |
| Type | Type | Type | Controlled text | true |
The DCMI column provides the official Dublin Core term for the element, and Label is the heading for these data on the DataShare metadata listing.
Table 3:
Partial list of data records for the Kaufman Atlas image set
| Kaufman image | Age | Stage | Orientation | Position | Stain | DOI |
|---|---|---|---|---|---|---|
| Plate 02 image a | E5.5 | 7–8 | Sagittal | H&E | http://dx.doi.org/10.7488/ds/393 | |
| Plate 02 image b | E5.5 | 7–8 | Sagittal | H&E | http://dx.doi.org/10.7488/ds/394 | |
| Plate 02 image c | E5.5 | 7–8 | Sagittal | H&E | http://dx.doi.org/10.7488/ds/395 | |
| Plate 02 image d | E5.5 | 7–8 | Sagittal | H&E | http://dx.doi.org/10.7488/ds/396 | |
| Plate 03 image a | E6.5,7 | 9,10 | Transverse | 0.210 | H&E | http://dx.doi.org/10.7488/ds/397 |
| Plate 03 image b | E6.5,7 | 9,10 | Transverse | 0.370 | H&E | http://dx.doi.org/10.7488/ds/398 |
| Plate 03 image c | E6.5,7 | 9,10 | Transverse | 0.590 | H&E | http://dx.doi.org/10.7488/ds/399 |
| . | . | . | . | . | . | . |
| . | . | . | . | . | . | . |
| Full table in Supplementary Excel file SciDataKaufmanTable3.xlsx | ||||||
| . | . | . | . | . | . | . |
| . | . | . | . | . | . | . |
| . | . | . | . | . | . | . |
| Plate 40l image b | E17.5 | 26 | Transverse | 0.888 | H&E | http://dx.doi.org/10.7488/ds/1301 |
| Plate 40l image c | E17.5 | 26 | Transverse | 0.903 | H&E | http://dx.doi.org/10.7488/ds/1302 |
| Plate 40l image d | E17.5 | 26 | Transverse | 0.921 | H&E | http://dx.doi.org/10.7488/ds/1303 |
| Plate 40l image e | E17.5 | 26 | Transverse | 0.948 | H&E | http://dx.doi.org/10.7488/ds/1304 |
| Plate 41 image 41 | E17.5 | 26 | Sagittal | 0.496 | H&E | http://dx.doi.org/10.7488/ds/1305 |
The DOI resolves to a dataset of image data, metadata, and annotations, which can be downloaded individually or combined. Image volumes range up to about 2 Gb, with a total volume for the full set of compressed zip files of 118 Gb. The Position column gives an estimate of the relative distance through the embryo of the individual histology section. The values are between 0 and 1, corresponding to the proportionate distance left-right (sagittal sections), cranial-caudal (transverse sections), and dorsal-ventral (coronal sections).
Technical Validation
The Images and associated data are all validated against the published atlas, which provides the detail of the genotype, defines the histological protocols, and establishes the correct staging of each embryo against the Theiler criteria. The section images used in the book are from specific tissue sections identified on the sets of microscope slides stored at the MRC Human Genetics Unit at the IGMM, University of Edinburgh. Each section was scanned digitally, then checked by a second curator to ensure validity. The annotations were originally captured using optical character recognition, and the text and spelling were checked by a second curator. All the end-point locations for the annotation terms have been double-checked, and a series of quality control steps have meant that inspection of the whole dataset has not revealed any errors.
Usage Notes
There are no constraints on the use of the images and associated data. The Supplementary Data file lists all samples and assays—1 for each section image—and also a “source,” which is the embryonic mouse specimen used by Kaufman in producing the histological sections. The “source” can be used to identify the set of physical glass slides, archived with the Centre for Research Collections of the University of Edinburgh, on which each histological section can be found. In principle, it is possible to obtain further images of the same or other sections in the series. “Age” is defined in embryonic days post-coitum, and “stage” refers to Theiler stage, a morphological staging system used to further define mouse embryo development. “Position” describes the relative position of the section in the embryo, with 0 representing, e.g., the most cranial section in a transverse series and 1 denoting the most caudal section. We additionally include details of the pixel resolution of each image, enabling accurate measurements to be made on each high-resolution embryo atlas image.
Availability of source code and requirements
Project name: eHistology
Project home page: http://www.emouseatlas.org/emap/eHistology/ [RRID:SCR_015887]; https://github.com/ma-tech/eHistologyWebapp
Operating system(s): platform independent
Programming language: The IIP Image server is a Fast CGI module written in C++.
Other requirements: The IIP Image server can be embedded within a host web server such as Apache, Lighttpd, MyServer, or Nginx.
License: software—GNU General Public License v2.0; Image Data—CC BY 4.0
Availability of supporting data
The image datasets described in this article are available at “Edinburgh DataShare” repository as Plates 02–15 (early gestation; E5.5-E8.5), Plates 16–29 (mid-gestation; E8.5-E13.0), Plates 30–41 (late gestation; E13.5-E17.5), and Plates S1-S6 (coronal supplement; E11.5-E15.5) [17]. Each image dataset is provided with all the information needed to generate the annotated views shown on the eHistology website. The data are provided as a series of files, including details.txt—a tab-delimited file with the embryo text description that appears at the top of the printed atlas page; image.jpg—full resolution jpeg image of the histological section (in some cases, this is a tif image); terms.txt—a tab-delimited file with the annotations for this image, including location of point annotation (x, y coordinates); and EMAPA ID and EMAPA terms associated with that point. Edinburgh DataShare has provided data DOIs for each of these image datasets, and links to these data DOIs are additionally hosted in GigaDB. An archival copy of the GitHub repository with software tools for the eHistology application is additionally available in GigaDB [12].
Abbreviations
DOI: Digital Object Identifier; H&E: haematoxylin and eosin; MRC: Medical Research Council; EMAPA: Edinburgh Mouse Atlas Project Anatomy.
Competing interests
The authors declare they have no competing interests in the publication of this data and manuscript. The new high-resolution images and the associated image coordinates for each annotation are fully freely available under a Creative Commons CC BY 4.0 licence. We have an agreement with Elsevier to present the images in a form similar to the original atlas plate layout on the eHistology web resource. However, this does not impact reuse of the image data and annotation described in this manuscript.
Funding
The MRC Human Genetics Unit Mouse Atlas Programme was core-funded by the Medical Research Council (Awardee R.A. Baldock).
Authors’ contributions
R.A.B. leads the Mouse Atlas programme that generated these datasets, designed the dataset submissions to the DataShare system, and wrote the scripts that provided the upload formats needed for batch ingest to DataShare. He also wrote the first draft of the manuscript.
C.A. is the senior editor for the Mouse Atlas databases, cowrote the manuscript, and has performed much of the quality control on the datasets.
Supplementary Material
03 May 2017 Reviewed
09 May 2017 Reviewed
Acknowledgements
The authors would like to acknowledge the critical contribution to the collection, collation, and curation of these data by Liz Graham and Julie Moss and to the development of the data visualization aspects by Nick Burton. All these are co-authors on the key citation for this work [7]. The authors would also like to thank the support of the Edina DataShare team, in particular Pauline Ward and Robin Rice. Finally, this work has been funded as part of the MRC core-funded Mouse Atlas Programme at the MRC Human Genetics Unit, Edinburgh.
References
- 1. Kaufman MH. The Atlas of Mouse Development. Amsterdam, the Netherlands: Elsevier Academic Press; 1994. [Google Scholar]
- 2. Richardson L, Venkataraman S, Stevenson P et al. EMAGE mouse embryo spatial gene expression database: 2014 update. Nucl Acids Res 2014;42(D1):D835–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Hayamizu TF, Wicks MN, Davidson DR et al. EMAP/EMAPA ontology of mouse developmental anatomy: 2013 update. J Biomed Sem 2013;4(1):15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Hayamizu TF, Baldock RA, Ringwald M. Mouse anatomy ontologies: enhancements and tools for exploring and integrating biomedical data. Mamm Genome 2015;26(9–10):422–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brown SD, Moore MW. Towards an encyclopaedia of mammalian gene function: the International Mouse Phenotyping Consortium. Dis Model Mech 2012;5(3):289–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mohun T, Adams DJ, Baldock R et al. Deciphering the Mechanisms of Developmental Disorders (DMDD): a new programme for phenotyping embryonic lethal mice. Dis Model Mech 2013;6(3):562–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Graham E, Moss J, Burton N et al. The atlas of mouse development eHistology resource. Development 2015;14211:1909–11. [DOI] [PubMed] [Google Scholar]
- 8. Attwood TK, Agit B, Ellis ABM. Longevity of biological databases. EMBnet J 2015;21(0):e803 http://dx.doi.org/10.14806/ej.21.0.803. [Google Scholar]
- 9. http://datashare.is.ed.ac.uk/.
- 10. Rice R. Edinburgh datashare - reflections from a data repository manager. Bul Am Soc Info Sci Tech 2014;40(2):39–40. [Google Scholar]
- 11. http://www.opendoar.org/find.php?rID=1176&format=full.
- 12. Baldock RA, Armit C. Supporting data for “eHistology image and annotation data from the Kaufman Atlas of Mouse Development.” GigaScience Database 2017. http://dx.doi.org/10.5524/100365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Richardson L, Moss J, Graham E et al. Developing the eHistology Atlas. Database 2015; doi: 10.1093/database/bav105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. datashare.is.ed.ac.uk/handle/10283/1807.
- 15. Graham E, Moss J, Burton N et al. eHistology Kaufman Atlas Plate 38b image b. University of Edinburgh, College of Medicine and Veterinary Medicine; 2015. http://dx.doi.org/10.7488/ds/1232. [Google Scholar]
- 16. www.emouseatlas.org/eAtlasViewer_ema/application/ema/kaufman/plate_38b.php?image=b.
- 17. eHistology Atlas with Kaufman Annotations. https://datashare.is.ed.ac.uk/handle/10283/822.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
03 May 2017 Reviewed
09 May 2017 Reviewed