ABSTRACT
Hepatitis C virus (HCV) is the leading cause of chronic hepatitis in humans. Several host molecules participate in HCV cell entry, but this process remains unclear. The complete unraveling of the HCV entry process is important to further understand viral pathogenesis and develop therapeutics. Human hepatitis A virus (HAV) cellular receptor 1 (HAVCR1), CD365, also known as TIM-1, functions as a phospholipid receptor involved in cell entry of several enveloped viruses. Here, we studied the role of HAVCR1 in HCV infection. HAVCR1 antibody inhibited entry in a dose-dependent manner. HAVCR1 soluble constructs neutralized HCV, which did not require the HAVCR1 mucinlike region and was abrogated by a mutation of N to A at position 94 (N94A) in the Ig variable (IgV) domain phospholipid-binding pocket, indicating a direct interaction of the HAVCR1 IgV domain with HCV virions. However, knockout of HAVCR1 in Huh7 cells reduced but did not prevent HCV growth. Interestingly, the mouse HAVCR1 ortholog, also a phospholipid receptor, did not enhance infection and a soluble form failed to neutralize HCV, although replacement of the mouse IgV domain with the human HAVCR1 IgV domain restored the enhancement of HCV infection. Mutations in the cytoplasmic tail revealed that direct HAVCR1 signaling is not required to enhance HCV infection. Our data show that the phospholipid-binding function and other determinant(s) in the IgV domain of human HAVCR1 enhance HCV infection. Although the exact mechanism is not known, it is possible that HAVCR1 facilitates entry by stabilizing or enhancing attachment, leading to direct interactions with specific receptors, such as CD81.
IMPORTANCE Hepatitis C virus (HCV) enters cells through a multifaceted process. We identified the human hepatitis A virus cellular receptor 1 (HAVCR1), CD365, also known as TIM-1, as a facilitator of HCV entry. Antibody blocking and silencing or knockout of HAVCR1 in hepatoma cells reduced HCV entry. Our findings that the interaction of HAVCR1 with HCV early during infection enhances entry but is not required for infection support the hypothesis that HAVCR1 facilitates entry by stabilizing or enhancing virus binding to the cell surface membrane and allowing the correct virus-receptor positioning for interaction with the main HCV receptors. Furthermore, our data show that in addition to the phospholipid-binding function of HAVCR1, the enhancement of HCV infection involves other determinants in the IgV domain of HAVCR1. These findings expand the repertoire of molecules that HCV uses for cell entry, adding to the already complex mechanism of HCV infection and pathogenesis.
KEYWORDS: hepatitis C virus, viral receptors, viral entry
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped, positive-sense, single-stranded RNA virus that is classified within the Flaviviridae. If left untreated, HCV infection can lead to chronic liver disease, which can progress to fibrosis, cirrhosis, or hepatocellular carcinoma (1). HCV is associated with the majority of liver transplantations in the United States. A protective vaccine against HCV infection is still not available, but there have been tremendous advances in antiviral drug treatment for HCV over recent years. Current HCV therapies can clear infections in the vast majority of people treated (2). However, the cost of therapy, the potential for reinfection, and the development of resistant viruses can limit the treatment success (2). Further understanding of the process of HCV entry would contribute to the development of additional strategies to prevent or treat infections.
HCV entry into cells is a complex and multifaceted process (reviewed in reference 3) involving initial nonspecific attachment via low-density-lipoprotein receptors and glycosaminoglycans followed by specific interaction with several host cell proteins, including the tetraspanin CD81 (4), scavenger receptor B1 (SRB1) (5), claudin 1 (CLDN1) (6), and occludin (OCLN) (7). Following receptor binding, virions are internalized via clathrin-mediated endocytosis (8), where the lower pH triggers fusion of viral and endosomal membranes, delivering the nucleocapsid into the cytoplasm (9). More recently, additional cofactors, including the receptor tyrosine kinases (10), Niemann-Pick C1-like 1 (11), transferrin receptor (12), E-cadherin (13), and CD36 (14), have been shown to facilitate particle entry.
Hepatitis A virus (HAV) cellular receptor 1 (HAVCR1), also known as TIM-1, was first identified as an HAV cellular receptor (15). HAV is a small, nonenveloped, positive-strand RNA virus that interacts with the immunoglobulin variable (IgV) domain of HAVCR1 (TIM-1) (16). Six cysteine residues in the IgV domain of HAVCR1 define a phospholipid-binding pocket, the function of which is abrogated by an N94A mutation that prevents coordination of the phosphate group with a metal ion present in the pocket (17). HAVCR1 functions as a pattern recognition receptor (PRR) of damage-associated molecular patterns (DAMPS) by recognizing phosphatidylserine exposed at the cell surface of apoptotic cells (18). HAVCR1 is expressed in T, B, and dendritic cells and is involved in the modulation of allergic, asthmatic, and autoimmune responses (reviewed in references 19 and 20). Furthermore, HAVCR1 is expressed on regulatory T cells (21) and B cells (22), where it functions as a PRR modulating tolerance.
HAVCR1 (TIM-1) has been implicated in cell entry of enveloped viruses by apoptotic mimicry via interaction with envelope phospholipids (23). Because HAVCR1 polymorphisms have been associated with the outcome of HCV infection (24, 25) and HAVCR1 (TIM-1) enhances HCV infection in cell culture (26), we further examined the interaction of HCV with HAVCR1 to understand the mechanism(s) involved in the infection enhancement. To do so, we studied entry into human hepatoma cells using cell culture-derived (HCVcc) and HCV envelope-pseudotyped (HCVpp) viruses. We evaluated the effects of anti-HAVCR1 antibodies and soluble HAVCR1 on HCV cell entry and infection and confirmed that HAVCR1 knockdown (KD) by small hairpin RNA (shRNA) and knockout (KO) by clustered regularly interspaced short palindromic repeat–CRISPR-associated protein 9 (CRISPR-Cas9) technology reduced HCV infection. Our data indicated that determinants in the IgV domain of HAVCR1 other than the phospholipid receptor function are required for the enhancement of HCV infection.
RESULTS
Antibody to HAVCR1 blocks HCV entry and is synergistic with anti-CD81 antibody.
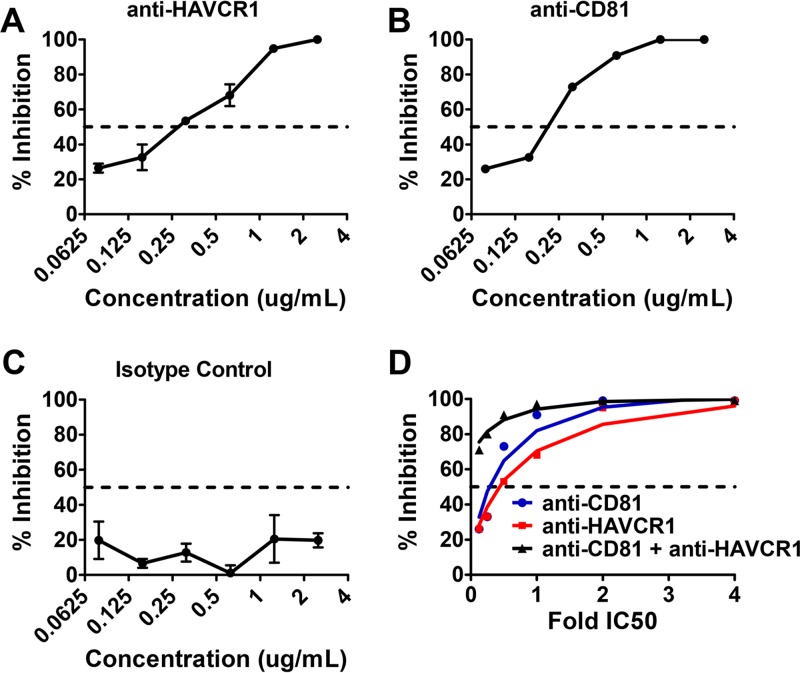
To determine if HAVCR1 is involved in HCV infection, we tested the ability of anti-HAVCR1 monoclonal antibody (MAb) 1D12 to block the entry of cell-culture-derived J6/JFH1 HCV particles (HCVcc) into human hepatoma Huh7.5 cells. We compared the inhibitory effect to that of a MAb targeting the known HCV entry factor, CD81. The anti-HAVCR1 MAb inhibited HCVcc infection of Huh7.5 cells in a dose-dependent manner (Fig. 1A). Similarly, antibody to CD81, a key determinant for HCV entry (27) that interacts with the HCV envelope E2 protein at a number of binding sites (28, 29), also inhibited HCVcc infection (Fig. 1B). An isotype control had no effect on HCV entry (Fig. 1C). The anti-CD81 MAb was slightly more potent than the anti-HAVCR1 MAb, with a calculated 50% inhibitory concentration (IC50) of 210 ng/ml versus 277 ng/ml. We also observed a synergistic effect of the anti-CD81 and 1D12 MAbs in blocking HCV infection by using a constant ratio combination of the two antibodies (Fig. 1D). This synergy was further evaluated by calculating a combination index (CI) (30) that can vary between synergistic (CI < 0.85), additive (CI = 0.85 to 1.1), and antagonistic (nonadditive) (CI > 1.1) effects. This value (mean CI = 0.7) confirmed that anti-CD81 and anti-HAVCR1 antibodies synergistically inhibited HCVcc infection. These data suggest that HAVCR1 plays a role in facilitating HCV entry into cells.
FIG 1.
Inhibition of HCVcc entry into Huh7.5 cells by anti-HAVCR1 antibody. Huh7.5 cells were treated with decreasing concentrations of the following antibodies following 2-fold dilutions from 4 μg/ml to 0.0625 μg/ml: anti-HAVCR1 MAb (1D12) (A), anti-CD81 MAb (JS-81) (B), and isotype control (C). Cells were treated with antibodies 1 h prior to infection with J6/JFH1. After 72 h, HCVcc infection was evaluated by automatic counting of stained foci and the percentage of inhibition calculated by comparing focus counts in treated and mock-treated cells. (D) Huh7.5 cells were treated with increasing concentrations of anti-CD81 and anti-HAVCR1 MAbs individually or in combination. The percentage of inhibition and dose effect plot was created using CalcuSyn 2.0 software. Testing for all assays was performed in duplicate. Error bars represent standard errors of the means. Data are representative of three independent experiments.
HAVCR1 functions early during HCV infection.
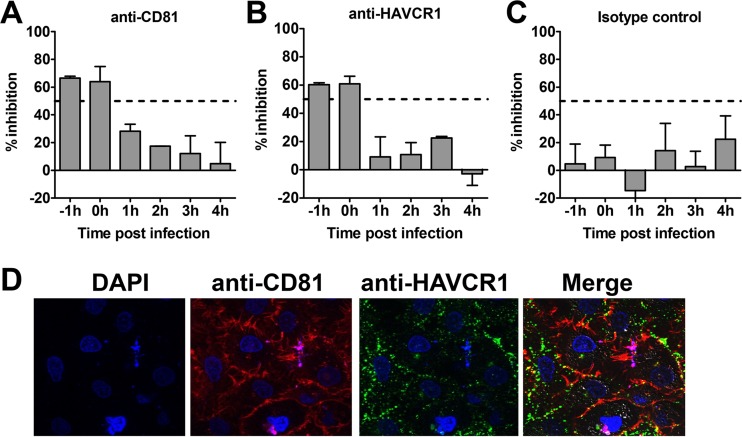
Our data using antibodies further confirmed that HAVCR1 is involved in the HCV entry process. To understand the mechanism of action and assess whether this molecule is involved in attachment and/or at postbinding steps, we assessed the inhibitory effects of anti-HAVCR1 MAb treatment at different times before and after virus binding. Similar studies have been used to analyze the time of action of other HCV receptors (12, 14, 27). Huh7.5 cells were infected with JFH1 HCVcc carrying the Nanoluciferase reporter gene (JFH1-Nanoluc) at 4°C for 1 h to allow virus binding to cells but not entry, washed three times, and incubated at 37°C to allow virus cell entry to proceed. Antibodies to CD81, HAVCR1, or an isotype control were added before and after infection at hourly intervals, and luciferase activities were compared to the activity in mock-infected cells to assess infectivity (Fig. 2). The amount of antibody added corresponded to the IC50 determined in prior experiments. Consistent with previous studies (9), anti-CD81 antibody inhibited infection by 50% when added during virus binding and directly after transfer to 37°C but had no significant effect at 1 to 4 h postinfection (Fig. 2A). Similarly, anti-HAVCR1 MAb 1D12 inhibited approximately 50% of HCV infection when added at −1 and 0 h but had no effect at 1 to 4 h postinfection (Fig. 2B). As expected, the isotype control had no significant effect on HCVcc infectivity (Fig. 2C). These data indicate that HAVCR1 functions early during HCVcc infection and suggest that this receptor may act at the initial binding stage of entry. To analyze whether there is an association between HAVCR1 and CD81 at the cell surface, we used confocal microscopy. This revealed that despite strong signals on the surface of cells for both molecules, there was no evidence of colocalization between CD81 and HAVCR1 at the surface of uninfected cells (Fig. 2D).
FIG 2.
Comparison of inhibitory activities and cell membrane localization of CD81 and HAVCR1. JFH1-Nanoluc was added to cells at 4°C for 1 h before moving the cells to 37°C to allow entry and infection to proceed. Antibody against CD81 (JS-81) (A), HAVCR1 (1D12) (B), or mouse isotype control (C) was added hourly to cells before and after HCVcc binding. The amount of antibody used was the concentration that was previously found to result in 50% inhibition of infection or the highest equivalent concentration in the case of the isotype control. The levels of luciferase expression at different time points were measured, and the percentages of inhibition were calculated by comparing the signals in mock-treated and infected cells at each time point. Error bars represent standard errors of the means. (D) Confocal analysis of Huh7.5 cells surface stained with 4′,6-diamidino-2-phenylindole (DAPI), anti-CD81 MAb (JS-81), and anti-HAVCR1 MAb (HAVCR1-1 MAb).
Soluble HAVCR1 inhibits HCV entry in a dose-dependent manner.
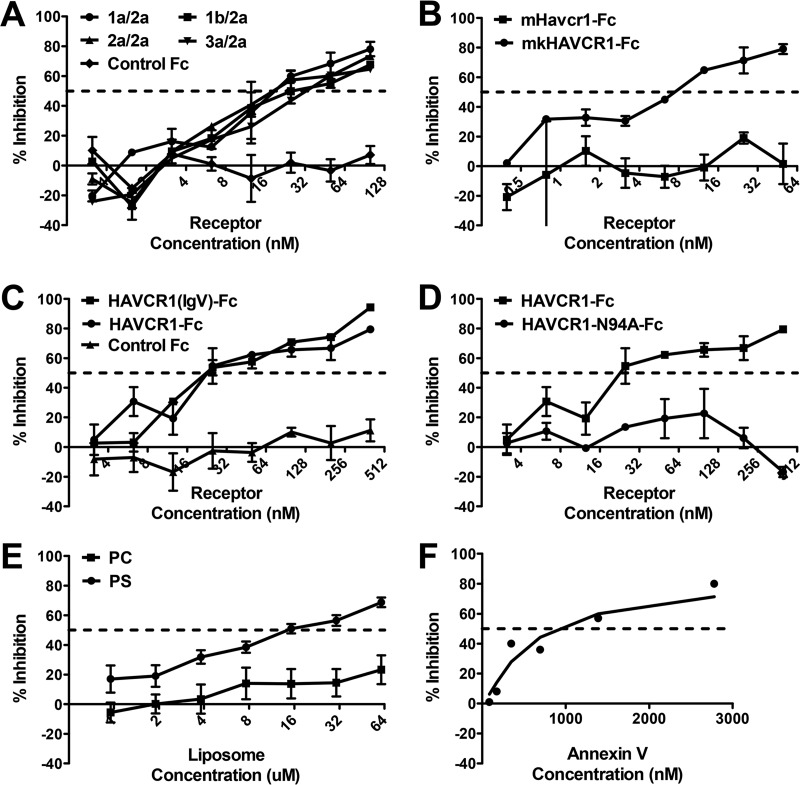
If HCVcc entry into cells is mediated through binding of the virus particles to HAVCR1, we hypothesized that soluble HAVCR1 would compete for cellular binding to the viral particles and inhibit entry. To determine whether HAVCR1 can directly interact with HCVcc, we treated virus with a soluble HAVCR1 in the form of an Fc fusion protein (HAVCR1-Fc) prior to infection of Huh7.5 cells. HAVCR-Fc inhibited the infection of four HCVcc chimeric viruses containing E1/E2 glycoproteins from different HCV genotypes similarly, in a dose-dependent manner (Fig. 3A), whereas no inhibition was seen using the negative control Fc (Fig. 3A). From these studies, we calculated the IC50 values of HAVCR1 for the 1a/2a, 1b/2a, 2a/2a, and 3a/2a HCVcc as 18.8 nM, 25.6 nM, 18.3 nM, and 32.6 nM, respectively. Interestingly, we found that an Fc fusion of the monkey homolog of HAVCR1, mkHAVCR1-Fc, also inhibited HCV entry in a dose-dependent manner (Fig. 3B) (IC50 = 7.46 nM), similarly to HAVCR1-Fc. However, an Fc fusion of the mouse ortholog of HAVCR1, Havcr1-Fc, showed no inhibitory activity against HCVcc even at high concentrations (Fig. 3B). Because mouse Havcr1 is also a phospholipid receptor, our data indicated that the presence of a phospholipid-binding pocket alone is not sufficient for the HAVCR1-HCV interaction.
FIG 3.
Dose-dependent inhibition of HCVcc entry with soluble HAVCR1 receptors, phosphatidyl serine, or annexin V. HCVcc was incubated 1 h prior to infection of Huh7.5 cells with increasing amounts of soluble HAVCR1 receptor containing the IgV- and mucinlike domains (HAVCR1-Fc) (A), soluble monkey HAVCR1 (mkHAVCR1-Fc) or mouse Havcr1 ortholog (mHavcr1-Fc) (B), HAVCR1-Fc or soluble HAVCR1 containing the IgV-like domain [HAVCR1(IgV)-Fc] (C), HAVCR1-Fc or the same receptor with an N94A mutation in the phosphatidylserine binding domain (HAVCR1-N94A-Fc) (D), phosphatidylserine (PS) or phosphatidylcholine (PC) liposomes (E), or annexin V (F). Testing for all assays was performed in duplicate. Error bars represent standard errors of the means. Data are representative of three independent experiments. All experiments were performed with HCVcc J6/JFH1 except for that shown in panel A, which used HCVcc chimeric viruses 1a/2a, 1b/2a, 2a/2a (J6/JFH1), and 3a/2a.
The HAVCR1 phospholipid-binding pocket is involved in the interaction with HCV.
HAVCR1 is a class I integral membrane glycoprotein that contains an N-terminal IgV domain extended from the plasma membrane by a mucinlike domain, which is anchored to cellular membranes by a canonical transmembrane domain followed by a cytoplasmic tail (15). The HAVCR1 monkey homolog mkHAVCR1 and the mouse ortholog Havcr1 share the same organization (20) but differ in sequence identity. There is 53% sequence identity between mkHAVCR1 and Havcr1 and 79% identity between mkHAVCR1 and HAVCR1 (17). The IgV domain has been shown to contain the binding site for HAV (16) and a phospholipid-binding pocket (17) that binds phosphatidylserine (PS) but not phosphatidylcholine (PC) (18). PS is a phospholipid contained in the inner leaflet of the plasma membrane that is exposed at the outer leaflet in apoptotic cells and is also a component of viral envelopes (18). We found that the HAVCR1 mucinlike region is not required for the HAVCR1-HCV interaction because HAVCR1(IgV)-Fc, a construct containing only the IgV-like domain and not the mucinlike domain, had an inhibitory effect on HCVcc infection similar to that of HAVCR1-Fc, a construct containing both extracellular domains (Fig. 3C). HAVCR1(IgV)-Fc and HAVCR1-Fc had similar IC50 values, 22.7 nM and 22.36 nM, respectively. In contrast, an Fc fusion of HAVCR1 containing an N94A mutation in the phospholipid-binding pocket of the IgV domain (HAVCR1-N94A-Fc), which prevents binding of phospholipids, showed no inhibition of HCVcc infection (Fig. 3D). Taken together, these data indicate that the integrity of the phospholipid-binding pocket in HAVCR1 is necessary for the interaction with HCVcc particles.
To further analyze the role of PS in HCVcc entry via HAVCR1, we tested the inhibitory effects of PS-containing liposomes. We found that high concentrations of PS-liposomes, in the 10 μM range, only partially inhibited HCVcc infection, while the control liposomes containing PC showed no inhibitory effect (Fig. 3E). This requirement for relatively high concentrations of PS-liposomes to achieve inhibition of HCVcc entry is consistent with previously published data on the role of HAVCR-1 (TIM-1) in HCV entry (26). Similarly, treatment of viral particles with a micromolar concentration of annexin V, a cellular protein that binds to PS, partially inhibited HCVcc entry (Fig. 3F). Since a low nanomolar concentration of HAVCR-Fc results in similar levels of inhibition (Fig. 3A to D), our findings further suggest that the HAVCR1-HCV interaction is not mediated solely by phospholipids present on the viral membrane.
Knockdown of HAVCR1 expression reduces HCV entry.
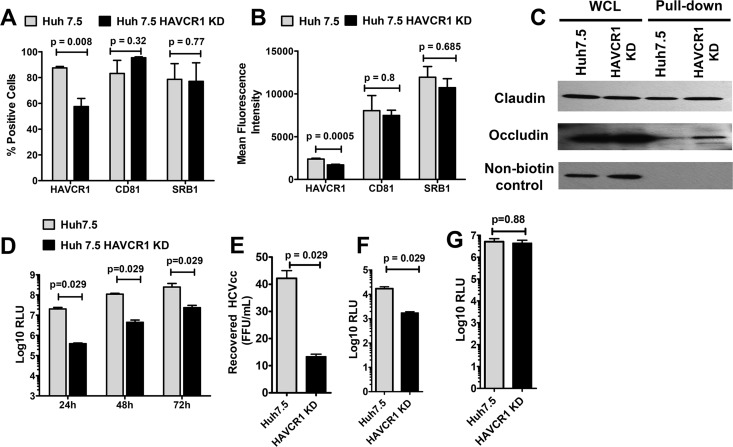
To further understand the HAVCR1-HCVcc interaction, we knocked down the expression of HAVCR1 in Huh7.5 cells by transfection of plasmids expressing HAVCR1-specific shRNA fragments (HAVCR1 KD cells). Fluorescence-activated cell sorting (FACS) analysis revealed that the proportion of cells expressing HAVCR1 (Fig. 4A) and the level of expression on a per-cell basis (mean fluorescence intensity) (Fig. 4B) were significantly reduced in the HAVCR1 KD cells compared to the results for the parental Huh7.5 cells, whereas the expression of CD81 and SRB1 (Fig. 4A and B) were not affected. We also verified the surface expression of claudin and occludin using biotinylation pulldown studies and found no difference between the Huh7.5 and the HAVCR1 KD cells (Fig. 4C). Infection with JFH1-Nanoluc showed significantly lower levels (P = 0.029) of luciferase in HAVCR1 KD cells than in parental cells for up to 72 h postinfection (Fig. 4D). Furthermore, the levels of HCVcc recovered from the supernatants of infected HAVCR1 KD cells were significantly lower (P = 0.029) than the levels recovered from parental cells (Fig. 4E). We also observed significantly lower levels (P = 0.029) of luciferase expression in HAVCR1 KD cells than in parent cells following infection with HCVpp (Fig. 4F). At the same time, we did not find any differences in the levels of luciferase expression following infection with control vesicular stomatitis virus envelope glycoprotein (VSV-G) pseudoparticles (Fig. 4G). These results indicate that knockdown of HAVCR1 decreased but did not prevent HCV infection of Huh7 cells.
FIG 4.
Reduced HAVCR1 expression on Huh7.5 cells results in reduced levels of HCVcc and HCVpp entry. Huh7.5 HAVCR1-1 knockdown (KD) cells were generated by transfection of parent Huh7.5 cells with a plasmid expressing HAVCR1 shRNA. (A) Percentages of parent and KD cells positive for HAVCR1, CD81, and SRB1 surface expression. Bars represent the mean values from 5 independent experiments for HAVCR1 and CD81 and 4 independent experiments for SRB1. (B) Mean fluorescence signals of HAVCR1, CD81, and SRB1 surface expression on parent and KD cells. Bars represent the mean values from 5 independent experiments for HAVCR1 and CD81 and 4 independent experiments for SRB1. (C) Western blot detection of claudin and occludin after cell surface biotinylation of Huh7.5 and HAVCR1 KD cells. Whole-cell lysate (WCL) is cell lysate prior to pulldown treatment. Nonbiotinylated control represents cells treated exactly as for the biotinylated cells but without the addition of EZ-Link sulfo-NHS-LCLC-biotin for cell surface biotinylation. Anti-claudin antibody was used for detection and shows the specificity of the NeutrAvidin beads for precipitation. (D) Luciferase activities at 24, 48, and 72 h postinfection in lysates of parent and KD cells infected with JFH1-Nanoluc. Bars represent the mean relative light units (RLU) calculated from 4 independent experiments using 9 replicates per experiment. (E) Titers of virus recovered from the supernatants of parent and KD cells at 72 h postinfection with JFH1-Nanoluc. Bars represent the mean titers from 4 independent experiments. (F) Luciferase activities in parent and KD cells 48 h after infection with HCVpp. (G) Luciferase activities in parent and KD cells 48 h after infection with VSV-G. Bars represent the mean values from 4 independent experiments. Error bars in all graphs represent standard errors of the means. Statistical analyses were performed using the Mann-Whitney test.
Knockout of HAVCR1 decreases but does not prevent HCV infection.
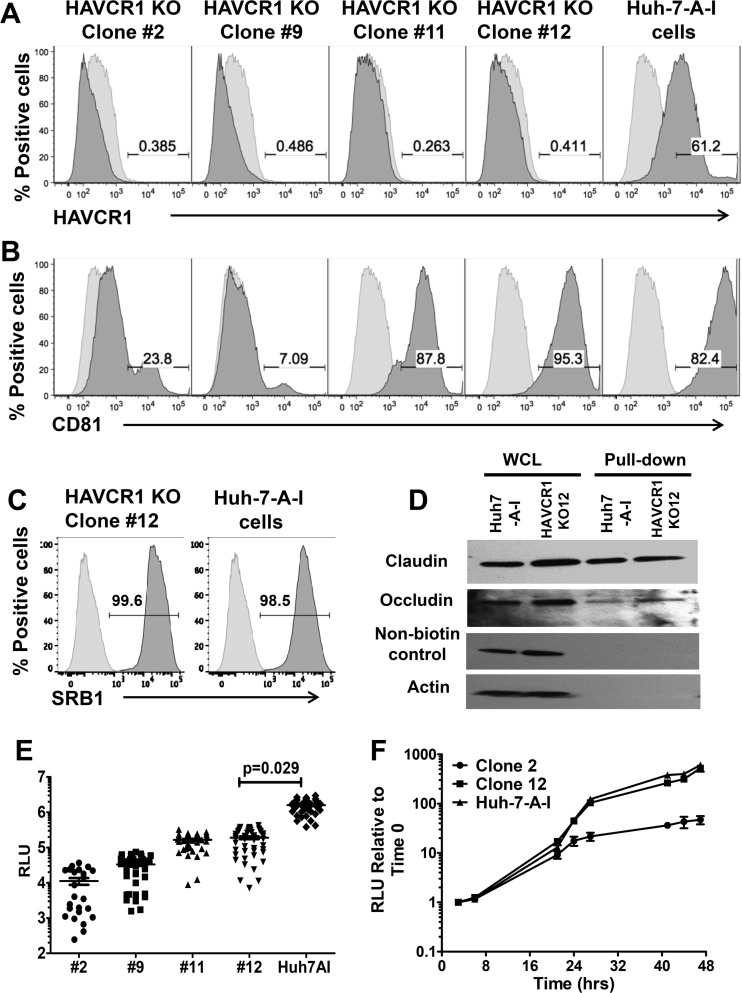
Because HAVCR1 knockdown affected HCV infection, we further studied HCV infection in knockout cells. To do so, we used CRISPR-Cas9 technology to knock out the HAVCR1 gene in Huh7-A-I cells (N. Amharref, M. I. Costafreda, M. Manangeeswarwn, J. Jacques, K. Konduru, A. C. Shurtleff, J. M. Casasnovas, and G. Kaplan, submitted for publication), a cell clone derived from Huh7 cells. Flow cytometry analysis confirmed that four KO cell clones (clones 2, 9, 11, and 12) did not express HAVCR1 (Fig. 5A). However, two of the HAVCR1 KO cell clones, clones 11 and 12, expressed normal levels of CD81, whereas clones 2 and 9 expressed significantly lower levels of CD81 at the cell surface than did Huh7-A-I cells (Fig. 5B). Infection of HAVCR1 KO cells with JFH1-Nanoluc showed a significant decrease in luciferase signal at 72 h postinfection in all clones compared to the signal in the parent Huh7-A-I cells (P = 0.029) (Fig. 5E). The greatest reductions were seen for clones 2 and 9, presumably due to the additional reductions in CD81 expression. However, clones 11 and 12, which had levels of CD81 expression on the surface comparable to the level on Huh7-A-I cells, also showed significantly reduced levels of HCVcc infection (P = 0.029). Clone 12 was confirmed to express comparable levels of SRB1 through FACS staining (Fig. 5C) and of claudin and occludin using biotinylation of surface proteins followed by pulldown with streptavidin beads (Fig. 5D). This observed decrease in the infectivity of clone 12 was due to reduced entry of HCVcc and not RNA replication, as similar luciferase levels were observed in parental cells and clone 12 HAVCR1 KO cells transfected with RNA transcribed from the JFH1-Nanoluc plasmid in vitro (Fig. 5F). The transfection of RNA into the cells bypasses the cell entry process by delivering RNA directly into the cytoplasm. In contrast, RNA replication was reduced in clone 2 at 24 to 47 h posttransfection compared to the RNA replication in clone 12 and Huh7-A-I. Clone 2 was found to have minimal levels of CD81 expression (Fig. 5B), and these data are consistent with previous results indicating that reduced CD81 expression levels have a negative impact on HCV RNA replication (31).
FIG 5.
Complete silencing of HAVCR1 expression in Huh7.5 cells significantly reduces HCVcc infection. Surface expression levels of receptors on Huh7.5 cells, Huh7-A-I cells, and different clones of Huh7-A-I HAVCR1 knockout (KO) cells were assessed by FACS using MAbs specific to HAVCR1 (REA383) (A), CD81 (JS-81) (B), and SRB1 (m1B9) (C). Data for positive cells are shown in dark gray, and data for mouse PE-isotype control are shown in light gray. Data are representative of three independent experiments. (D) Western blot detection of claudin and occludin after cell surface biotinylation of Huh7-A-I and clone 12 KO cells. Whole-cell lysate (WCL) is cell lysate prior to pulldown treatment. Nonbiotinylated control represents cells treated exactly as for the biotinylated cells but without the addition of EZ-Link sulfo-NHS-LCLC-biotin for cell surface biotinylation. Anticlaudin antibody was used for detection and shows the specificity of the NeutrAvidin beads for precipitation. Antiactin antibody was used for detection of actin, a cytoplasmic protein that is not expressed on the cell surface, demonstrating biotinylation of surface proteins only. (E) Luciferase activities in lysates from Huh7-A-I HAVCR1 KO clones and parent cell line at 72 h postinfection with JFH1-Nanoluc. Data were obtained from 3 independent experiments, and each experiment was carried out using 12 replicates for each cell clone. Each symbol represents an individual well from the 3 independent experiments. Horizontal bars represent the mean luciferase activities. P values were calculated using the mean values from each of the 3 independent experiments. (F) Time course of luciferase expression relative to the expression at 3 h posttransfection in Huh7-A-I HAVCR1 KO clones 2 and 12 and parent cells following transfection with JFH1-Nanoluc RNA transcribed in vitro. Data are the mean values from 6 replicates at each time point. Error bars represent standard errors of the means. Data are representative of two independent experiments. RLU, relative light units.
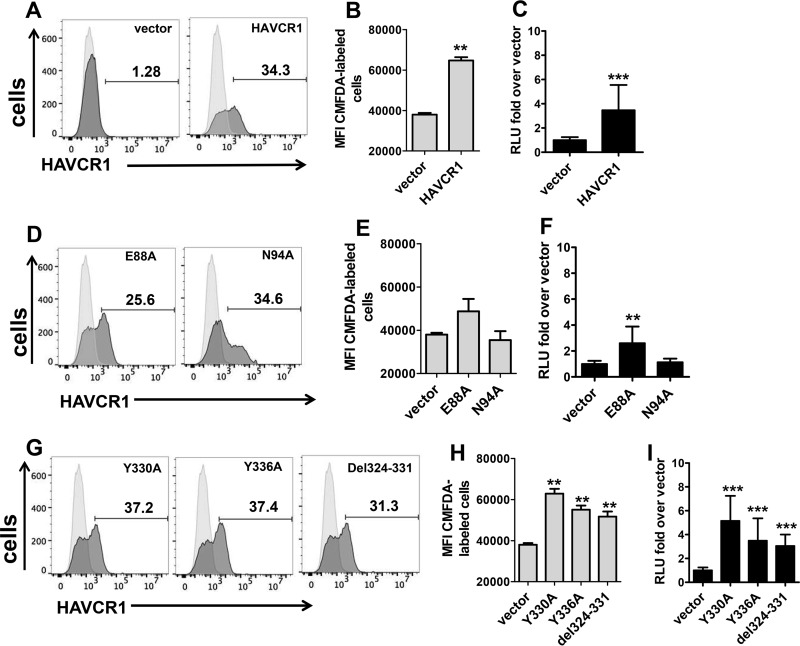
To further verify that the reduced infectivity of HCV in the HAVCR1 KO cells was due to the lack of HAVCR1 expression, we transfected HAVCR1 KO clone 12 cells with a plasmid coding for HAVCR1. Transient transfection of HAVCR1 resulted in the expression of the receptor at the cell surface (Fig. 6A), which was determined to be active because these cells bound significantly higher levels of apoptotic cells than vector-transfected cells (Fig. 6B). Furthermore, transfection of HAVCR1 restored the enhancement of HCV infection (Fig. 6C), which confirmed that HAVCR1 expression increases but it is not required for HCV infection.
FIG 6.
Expression of HAVCR1 in KO clone 12 enhances HCV infection. HAVCR1 KO clone 12 cells were transfected with plasmids expressing HAVCR1, mutated forms of HAVCR1 (E88A, N94A, Y330A, Y336A, and del324–331 mutations), or vector. (A) Expression of HAVCR1 on the surface of clone 12 cells transfected with vector or plasmid expressing HAVCR1 48 h posttransfection was assessed using anti-HAVCR1-1 MAb. (B) Apoptotic binding activities of clone 12 cells transfected with vector or plasmid expressing HAVCR1. (C) Luciferase expression in cell lysates of clone 12 cells transfected with HAVCR1 expression plasmid and infected with JFH1-Nanoluc. Data at 24 h postinfection are shown. Four independent experiments were performed; each time point included 3 replicates. (D) Expression of E88A or N94A mutant on the surface of transfected clone 12 cells 48 h posttransfection assessed using anti-HAVCR1-1 MAb. (E) Apoptotic binding of clone 12 cells transfected with vector or plasmids expressing E88A and N94A mutant forms of HAVCR1. (F) Luciferase expression in cell lysates of clone 12 cells transfected with plasmids expressing E88A or N94A mutants and infected with JFH1-Nanoluc. Data are from 4 independent experiments, each including 3 replicates, at 24 h postinfection. (G) Expression of Y330A, Y336A, or del324–331 mutant on the surface of transfected clone 12 cells 48 h posttransfection was assessed using anti-HAVCR1-1 MAb. (H) Apoptotic binding of clone 12 cells transfected with vector or plasmids expressing Y330A, Y336A, and del324–331 mutant forms of HAVCR1. (I) Luciferase expression in cell lysates of clone 12 cells transfected with Y330A, Y336A, or del324–331 mutant plasmids and infected with JFH1-Nanoluc. Data are from 4 independent experiments, each including 3 replicates, at 24 h postinfection. Horizontal bars represent the mean values. Asterisks represent significance values obtained using the Mann-Whitney test to compare the mean values for vector-transfected cells with the mean values for each of the other transfectants. NS, not significant; **, P < 0.001; ***, P < 0.0001. For FACS histograms, HAVCR1 expression on transfected cells is shown as filled dark-gray areas, and data for untransfected cells stained with anti-HAVCR1-1 MAb are shown as filled light-gray areas. RLU, relative light units; MFI, mean fluorescence intensity.
The integrity of the HAVCR1 phospholipid-binding pocket is required to enhance HCV infection.
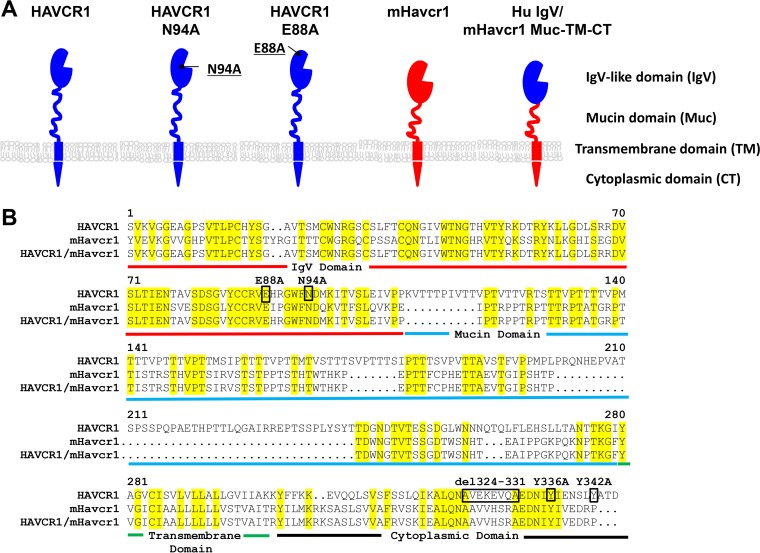
Our neutralization data using soluble forms of HAVCR1 indicated that the integrity of the phospholipid-binding pocket in the IgV domain is required for the HAVCR1-HCV interaction (Fig. 3D). To determine whether the phospholipid-binding pocket is also required to enhance HCV infectivity, we transfected HAVCR1 KO clone 12 cells with an HAVCR1 expression plasmid containing an N94A mutation in the phospholipid-binding pocket or a control E88A mutation outside the pocket. A schematic representation of HAVCR1 with the different domains and mutations studied is presented in Fig. 7A, and the exact positions of the mutations are shown in Fig. 7B. The transfectants expressed HAVCR1 N94A or E88A at the cell surface (Fig. 6D). As expected, the N94A but not the E88A mutation blocked the phospholipid-binding function of HAVCR1 (Fig. 6E). More importantly, HCV infection was not enhanced in HAVCR1 N94A-transfected cells, whereas HAVCR1 E88A significantly increased infectivity compared to the results for vector-transfected cells (Fig. 6F). Our results indicated that the integrity of the phospholipid-binding pocket is needed for the direct HAVCR1-HCV interaction and is also required for the enhancement of HCV infection.
FIG 7.
Schematic representation of the HAVCR1, mHavcr1, and chimeric proteins used in transient-transfection experiments. (A) Receptor domains and mutations in the human and mouse proteins. (B) Sequence alignment of HAVCR1, mouse Havcr1 (mHavcr1), and chimeric protein. Yellow highlighting denotes residues conserved between the molecules. The different receptor regions are defined by colored underlining as follows: red, IgV domain; blue, mucin domain; green, transmembrane domain; black, cytoplasmic domain. Mutated or deleted residues in HAVCR1 are shown in black boxes.
Direct signaling via HAVCR1 is not required to enhance HCV infection.
The HAVCR1 cytoplasmic tail contains a conserved tyrosine residue (Y330) that is phosphorylated upon HAVCR1 cross-linking (32) and a nonconserved tyrosine residue (Y336) of unknown function. A binding motif for SH2 domains of Src family kinases that does not require phosphorylation of Y330 for kinase binding (33) is also present between amino acids 324 and 331 of the HAVCR1 cytoplasmic tail. To determine whether direct signaling via HAVCR1 is required for the enhancement of HCV infection, we made HAVCR1 mutants containing Y330A, Y336A, or the deletion of amino acid residues 324 to 331 (del324–331) (Fig. 7A and B). Transient transfection of HAVCR1 KO clone 12 cells with these constructs resulted in cell surface expression of the cytoplasmic tail mutants (Fig. 6G), and these mutants were confirmed as active PS receptors (Fig. 6H). Interestingly, all these cytoplasmic tail mutants significantly (P < 0.001) enhanced HCV infection (Fig. 6I) compared to the results for the vector control, indicating that direct signaling via HAVCR1 is not required to increase HCV cell entry.
The IgV domain of human HAVCR1 is responsible for facilitating HCV entry.
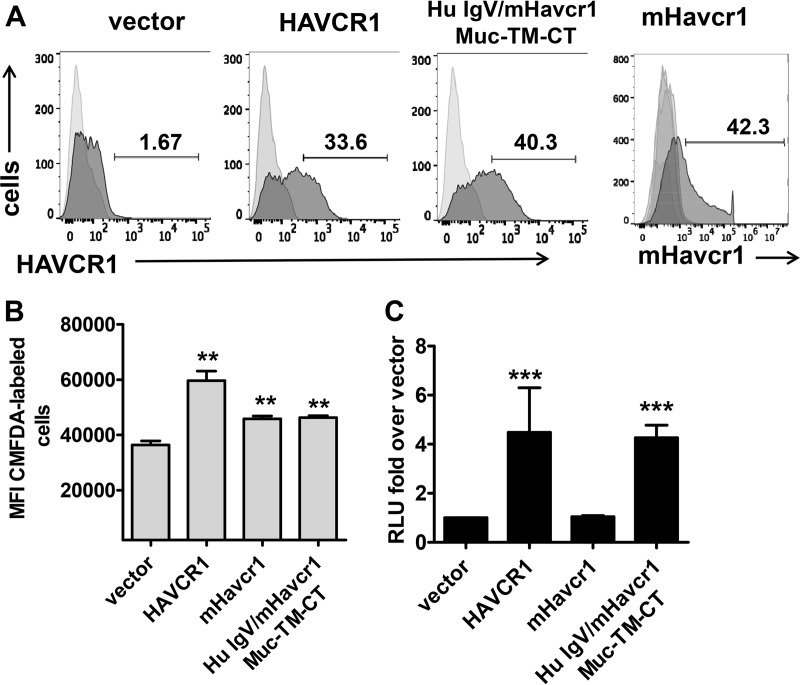
To determine whether the IgV domain of human HAVCR1 is sufficient to enhance HCV infection, we generated a human/mouse chimeric receptor by replacing the mouse IgV domain with that of human HAVCR1 (Hu IgV/mHavcr1) while retaining the mouse mucin domain (Muc), transmembrane domain (TM), and cytoplasmic tail (CT) (Fig. 7A and B). Transiently transfected HAVCR1 KO clone 12 cells expressed Hu IgV/mHavcr1 (Muc-TM-CT) at the cell surface (Fig. 8A), which was active as a PS receptor (Fig. 8B), and the cells showed significantly (P < 0.001) enhanced HCV infection compared to the results for mHavcr1- and vector-transfected cells (Fig. 8C), despite the expression of mHavcr1 on the surface of the cells (Fig. 8A). These data indicated that an active PS receptor such as mHavcr1 is not sufficient to increase HCV infection and that additional determinants present in human HAVCR1 are required for the enhancement.
FIG 8.
The IgV domain of human HAVCR1 is responsible for facilitating HCV entry. (A) Expression of HAVCR1 or mHavcr1 on the surface of transfected clone 12 cells 48 h posttransfection. Receptor expression is shown as filled dark-gray areas, and data for stained, untransfected cells are shown as filled light-gray areas. (B) Apoptotic binding of clone 12 cells transfected with vector or plasmids expressing HAVCR1, mHavcr1, or chimeric human/mouse receptor. MFI, mean fluorescence intensity. (C) Luciferase expression in cell lysates of clone 12 cells transfected with expression plasmids and infected with JFH1-Nanoluc. RLU, relative light units. Data shown are from 5 experiments with transient transfection of the control vector and HAVCR1 and 3 independent experiments for mHavcr1, including 3 replicates each, at 24 h postinfection. Asterisks represent significance values obtained using the Mann-Whitney test to compare the data for vector-transfected cells with the data for each of the other transfectants or the parent cell line. NS, not significant; **, P < 0.001; ***, P < 0.0001.
DISCUSSION
Viruses usurp the apoptotic cell clearance mechanism to gain entry into cells via a mechanism known as viral apoptotic mimicry, in which PS exposed at the virion surface binds to PS receptors on cells to facilitate entry (for a review, see reference 23). Apoptotic mimicry involving several direct PS receptors (HAVCR1 [also known as TIM-1], HAVCR2, CD300a, and CD300f) and indirect PS receptors like MFGE8 and GAS6 that bind to cell surface receptors has been reported for alphaviruses, flaviviruses, filoviruses, arenaviruses, poxviruses, rhabdoviruses, and baculoviruses (23). Here, we show through a series of studies using antibodies, soluble receptors, and knockdown (KD) and knockout (KO) cells that HAVCR1 (TIM-1) facilitates the entry of HCV.
Our initial experiments showed that a MAb against the IgV-like domain of HAVCR1 inhibited HCV entry in a dose-dependent manner (Fig. 1) and that this inhibition was synergistic when combined with antibody to CD81. Moreover, soluble HAVCR1 receptor blocked HCV infection, indicating a direct interaction between HAVCR1 and HCV particles, while the deletion of the mucinlike region of HAVCR1 did not affect neutralization. The N94A mutation in HAVCR1 that disrupts the PS-binding pocket blocked neutralization of HCVcc by HAVCR1-Fc and did not enhance the infection of HAVCR1 KO cell transfectants expressing HAVCR1.
Our studies using HAVCR1 knockdown and knockout cells showed reduced levels of HCV infection, providing additional confirmation of a role for HAVCR1 in HCV entry. These studies indicated that the HAVCR1-mediated enhancement of HCV infection is at the cell entry level, since HAVCR1 KO and parental cells supported similar HCV growth upon transfection of viral RNA. Deletion of the HAVCR1 gene in the knockout cells, which was confirmed through sequence analysis (data not shown) and flow cytometry, did not completely prevent HCVcc infection, suggesting that HAVCR1 is not required but instead facilitates or enhances cell entry by playing a role of cell entry cofactor. The inhibition of HCV infection by anti-HAVCR1 MAb treatment is consistent with this hypothesis because it is only effective before infection or during absorption.
Further analyses of KO cells rescued with plasmids expressing mutant forms of HAVCR1 or the mouse orthologue (mHavcr1) provided additional insights into regions critical for interactions of the receptor with HCV particles. Mutations in the cytoplasmic tail of HAVCR1 that could mediate direct signal transduction did not affect the enhancement of HCV infection. Additionally, mHavcr1, the mouse ortholog of HAVCR1 that is also a PS receptor, did not neutralize HCVcc or enhance infection in KO cells. Exchange of the IgV domain of mHavcr1 with the IgV domain of human HAVCR1 restored the enhancement of HCV infection. Taken together, our results indicate that the IgV domain of HAVCR1 is required to enhance HCV infection. However, binding of PS to a PS receptor alone is not sufficient to mediate the HAVCR1-HCV interaction, since mHavcr1 did not enhance HCV infection, suggesting that the HAVCR1 IgV may also interact with other molecules at the virion surface, such as E1E2. Our results are consistent with previous studies showing that the human HAVCR1 (TIM-1) homologs HAVCR2 (also known as TIM-3) and TIM-4, which are also PS receptors, did not enhance HCV infection (26) and indicate that apoptotic mimicry may not be the only mechanism by which HAVCR1 enhances HCV infection.
A high concentration of PS-liposomes, in the 10 μM range, was required to partially block HCVcc infection in Huh7.5 cells, compared to the low, 0.1-to-1 μM range needed to block HAVCR1-mediated binding and phagocytosis of apoptotic cells (18). These results support our hypothesis that either the interaction of HCV with HAVCR1 is not entirely mediated through binding of PS to the PS-binding pocket or the PS present on the HCV virion surface does not interact efficiently with HAVCR1. Consistent with these findings, we also observed that a high concentration of annexin V, in the 1,000 μM range, is required to partially inhibit HCV infection, compared to the 0.03 μM range of HAVCR1-Fc needed to achieve similar inhibition, further suggesting that the interaction of HCV with the IgV of HAVCR1 may not be restricted to PS interactions with the PS-binding pocket. The infection of dengue virus (DV), another member of the Flaviviridae family, is enhanced by HAVCR1, TIM-4, and members of the TAM family (34), which are unrelated PS-binding receptors, suggesting that DV has less stringent requirements for enhancement of infection by PS receptors.
In humans, there is some evidence that HAVCR1 (TIM-1) may be expressed in different cell types, including hepatocytes, multiple epithelial cells, and immune system cells like B- and T cells, macrophages, and dendritic cells (35). It is possible that HCV may use HAVCR1 for initial binding to the surface membrane and correct positioning of the virus for interaction with the main HCV receptors. Alternatively, HAVCR1 may enhance HCV cell entry by an indirect mechanism that increases the activation of other receptors and coreceptors. HCV may also use HAVCR1 to bind cells circulating in the blood and facilitate transportation to the site of replication in the liver. Our findings provide mechanistic insights into the interaction of HCV with the HAVCR1 IgV domain, expand the repertoire of molecules that HCV uses for cell entry, and add to the already complex mechanism of infection for this virus. Further studies will be required to determine whether HAVCR1 (TIM-1) plays any role in the pathogenesis of HCV, as suggested by epidemiological studies (24, 25, 36), and whether it could be involved in cell-to-cell spread or used as a target in novel therapeutic strategies.
MATERIALS AND METHODS
HCVcc production and inhibition of infection.
We used four HCVcc chimeric viruses carrying the structural proteins from four different genotypes, 1a, 1b, 2a and 3a, on the JFH1 2a backbone (37). J6/JFH1 (2a/2a) was kindly provided by Charles Rice (Rockefeller University, NY). The 1a/2a, 1b/2a, and 3a/2a chimeric viruses were produced and their titers determined on human hepatoma Huh7.5 cells as described previously (38).
Inhibition assays were performed using decreasing concentrations of the following antibodies following 2-fold dilutions from 4 μg/ml to 0.0625 μg/ml: anti-HAVCR1 MAb (1D12) (BioLegend, San Diego, CA) and anti-human CD81 MAb (JS-81) (BD Pharmingen, San Jose, CA). Antibodies were added to Huh7.5 cells, and after 1 h at 37°C, the cells were washed and infected with HCVcc and the foci stained as previously described (39). Foci were counted using an automated counting system (Cellular Technology Limited, Cleveland, OH) and BioSpot 5.0 Professional software (Cellular Technology Limited, Cleveland, OH). A mouse IgG1 isotype control antibody (BioLegend, San Diego, CA) was included in all inhibition assays.
Inhibition assays using soluble receptors, PS, PC, or annexin V were performed in a similar fashion following 2-fold dilutions except that the molecules were incubated with 100 focus-forming units of HCVcc for 1 h at 37°C before infection. The soluble receptor-HCVcc mixture was then added to cells and incubated for an additional 3 h, after which cells were washed twice with phosphate-buffered saline (PBS) and fresh medium was added. After 72 h, we performed focus staining of Huh7.5 cells (39).
Infection with JFH1 luciferase-expressing HCVcc and HCVpp.
JFH1 HCVcc carrying the Renilla luciferase reporter gene (JFH1-luc) was kindly provided by Curt Hagedorn (University of Arkansas) (40). JFH1 expressing NanoLuc luciferase (JFH1-Nanoluc) was generated by cloning the NanoLuc reporter gene (Promega, Madison, WI) into the NS5A region of JFH1-luc using the MluI restriction site, thereby replacing the Renilla luciferase gene with the Nanoluciferase gene. HCV-pseudotyped particles (HCVpp) carrying the envelope protein of HCV GT1a/H77 and VSV-G-pseudotyped particles were generated as described previously (41). Cells in 96-well plates were infected with JFH1-Nanoluc or HCVpp stock. After 3 washes with PBS, fresh Dulbecco's modified Eagle's medium (DMEM) was added and the cells incubated for an additional 72 h. To determine luciferase activity, cells were lysed by freezing and thawing in reporter lysis buffer (Promega, Madison WI) and 20-μl amounts were used in a luciferase assay system (Promega, Madison WI). The signal was read using a MicroLumat plus LB96V luminometer (Berthold Technologies, Oak Ridge TN).
Generation and purification of soluble receptors.
Recombinant proteins containing the extracellular domain of human HAVCR1, the monkey homolog (mkHAVCR1), or the mouse ortholog (mHavcr1) fused to the human IgG1 Fc fragment (HAVCR1-Fc) were generated and purified using protein G as previously described (42).
Generation of Huh7.5 HAVCR1 knockdown (KD) cells.
To knock down the expression of HAVCR1, Huh7.5 cells were electroporated (Amaxa Nucleofector; Lonza, Allendale, NJ) with two plasmids expressing short hairpin RNAs (shRNAs) of HAVCR1 (AAGGAGGTTCAACAACTAAGT and CGACACTGCAGGGAGCAATAA) under the control of the U1 promoter and the hygromycin resistance gene (SureSilencing shRNA plasmids for human HAVCR1) (SABiosciences Corp., Frederick, MD). Transfectants were selected with 0.150 mg/ml of hygromycin, and reduced expression of HAVCR1 was verified by quantitative PCR and FACS analysis using anti-HAVCR1-1 MAb.
Analysis of HCV infection in HAVCR1 KO cells.
HAVCR1 in Huh7-A-I cells, a clone of Huh7 cells permissive for wild-type HAV infection (43), was knocked out using CRISPR-Cas9 technology (Amharref et al., submitted), and single-cell clones were isolated and termed Huh7-A-I HAVCR1 KO cells. The lack of expression of HAVCR1 in Huh7-A-I HAVCR1 knockout (KO) cells was verified by FACS analysis. Huh7-A-I and Huh7-A-I HAVCR1 KO cells were infected with HCVcc and HCVpp as described above.
Transfection of Huh7-A-I cells.
Huh7-A-I parental and HAVCR1 KO cells were transfected, using Lipofectamine 3000 (Thermo Fisher Scientific, Waltham, MA), with plasmids expressing human HAVCR1 (pEFHAVCR1) wild type (21), human HAVCR1 containing an N-to-A mutation at residue 94 in the phospholipid-binding pocket that prevents binding of apoptotic cells (17, 18) (pEFHAVCR1-N94A), or the mouse ortholog Havcr1 (pEFmHavcr1) (21) under the EF1 alpha promoter. Mutants with the following additional mutations were constructed using the same expression vector: an E-to-A mutation at residue 88, a Y-to-A mutation at residue 330, a Y-to-A mutation at residue 336, and a deletion of residues 324 to 331.
RNA transfections were performed using a Gene Pulser electroporation system (Bio-Rad, Hercules, CA) with settings as previously described (44). RNA was transcribed in vitro from a plasmid containing the JFH-Nanoluc genome linearized with XbaI (New England BioLabs, Ipswich, MA) using a T7 in vitro transcription system according to the manufacturer's instructions (Ambion, Austin, TX).
Flow cytometry analysis of receptor expression on cells.
To characterize receptor expression on the surface of Huh7.5 cells, we performed staining with the following antibodies: anti-CD81 mouse MAb (JS-81; BD Pharmingen, San Jose, CA); anti-SRB1 antibody (m1B9; BioLegend, San Diego, CA), anti-HAVCR1 phycoerythrin (PE)-labeled mouse MAb (REA384; Miltenyi Biotec, Inc., Auburn, CA), or purified HAVCR1-1 MAb, an antibody directed against the IgV domain of HAVCR1 that blocks binding of phospholipids. One million cells were incubated with antibodies for 1 h at 4°C, washed with PBS–1% bovine serum albumin (BSA) and incubated (in the case of anti-CD81 and HAVCR1-1) with secondary anti-mouse antibodies labeled with PE (Thermo Fisher Scientific, Waltham, MA) for an additional 30 min at 4°C. Following washing, cells were analyzed using an LSR Fortessa instrument. (BD Biosciences, San Diego CA). Data were collected for 104 cells and analyzed using FlowJo 9.4 software.
Cell surface protein biotinylation.
For isolation and quantification of occludin and claudin on the surface of cells, we used a biotinylation pulldown approach (45). Briefly, 5 × 105 cells were seeded onto 100-mm dishes 24 h before staining. Cells were washed twice with PBS, and membrane proteins were biotinylated by incubating cells for 30 min on ice with 1 ml of EZ-Link sulfo-NHS-LC-LC-biotin (sulfosuccinimidyl-6-[biotinamido]-6-hexanamido hexanoate) (2.5 mg/ml) (Life Technologies; Thermo Fisher Scientific, Waltham, MA). Cells were washed twice in 5 ml of cold 100 mM glycine in Dulbecco's PBS (DPBS) and 3 times in 20 mM glycine in DPBS and lysed in 400 μl of lysis buffer (1% Triton X-100 in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, protease inhibitor cocktail [Sigma, St. Louis, MO]) for 1 h at 4°C, and the collected cell lysate was incubated overnight with rotation at 4°C with 40 μl of NeutrAvidin beads (Pierce; Thermo Fisher Scientific, Waltham, MA). Following incubation, the bead-lysate mixture was washed 4 times with 1 ml of lysis buffer, 2 times with 1 ml of buffer containing 0.1% Triton X-100 in Tris-HCl, pH 7.4, 350 mM NaCl, and 5 mM EDTA, and once in 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, and 1 mM EDTA. The beads with precipitated protein were resuspended in 40 μl of 2× gel loading buffer. Samples were heated at 70°C for 10 min and centrifuged, and supernatants analyzed by Western blotting. A portion of the whole-cell lysate was retained for Western blot analysis. The protein concentration was quantified using a standard protein assay, and equal amounts were loaded onto SDS-PAGE gels. The following antibodies were used for Western blot analysis: rabbit anti-claudin antibody (Thermo Fisher Scientific, Waltham, MA), mouse anti-occludin antibody (BD Bioscience, San Jose, CA), and mouse anti-actin MAb (Pierce; Thermo Fisher Scientific, Rockford, IL). The following peroxidase-labeled secondary antibodies were used: anti-rabbit IgG (GE Healthcare, UK) and anti-mouse IgG (H+L) (KPL, Gaithersburg, MD). Signals were detected with the SuperSignal west pico plus chemiluminescent substrate (Pierce; Thermo Fisher Scientific, Waltham, MA).
Binding of apoptotic cells.
Binding of apoptotic cells was assessed as previously described (21). In brief, apoptotic Jurkat cells were produced by treatment with 50 μM etoposide for 8 h and stained with 5 μM CMFDA (5-chloromethylfluorescein diacetate) (Jurkat-CMFDA cells). Subconfluent monolayers (50%) in 12-well plates of Huh7-A-I and Huh7-A-I HAVCR1 KO cells transiently transfected with the plasmids coding for HAVCR1 variants or empty vector were incubated with 106 apoptotic Jurkat-CMFDA cells in 500 μl of complete medium for 30 min at room temperature. Cell monolayers were washed 3 times, and the binding of Jurkat-CMFDA cells was quantitated in a Synergy HT fluorescence plate reader (485 nm/528 nm excitation/emission).
Confocal analysis of Huh7.5 cells.
Huh7.5 cells were seeded onto Labtek chambered coverglasses (Nunc, Rochester, NY) pretreated with 0.1 mg/ml poly-l-lysine (Sigma, St. Louis, MO) 24 h before use. Anti-CD81 MAb (JS-81) and anti-HAVCR1-1 MAb were labeled with Alexa Fluor 546 and Alexa Fluor 488, respectively, using an antibody labeling kit (Molecular Probes, Eugene, OR) according to the manufacturer's instructions. Live cells were incubated with labeled antibodies, washed, and then fixed with 2% paraformaldehyde for 20 min at room temperature. Cells were visualized using the Leica SP8 DMI6000 confocal microscope system (Mannheim, Germany) equipped with a 63× oil objective lens (numeric aperture [NA] 1.4). A white light laser was used for excitation at wavelengths of 488 and 546 nm.
Statistical analyses.
Statistical calculations were performed using GraphPad Prism software (version 5.0) (San Diego, CA) and Mann-Whitney testing. A P value of <0.05 was considered significant.
ACKNOWLEDGMENTS
Financial support for this work was provided by Food and Drug Administration intramural funds (program number Z01 BK 04010-11 LHV to M.E.M. and program number Z01 BP 06004-05 LEP to G.K.).
We thank Ruoxuan Xiang for statistical advice.
REFERENCES
- 1.Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. 2000. Pathogenesis, natural history, treatment, and prevention of hepatitis C. Ann Intern Med 132:296–305. doi: 10.7326/0003-4819-132-4-200002150-00008. [DOI] [PubMed] [Google Scholar]
- 2.Pawlotsky JM, Feld JJ, Zeuzem S, Hoofnagle JH. 2015. From non-A, non-B hepatitis to hepatitis C virus cure. J Hepatol 62:S87–S99. doi: 10.1016/j.jhep.2015.02.006. [DOI] [PubMed] [Google Scholar]
- 3.Lindenbach BD, Rice CM. 2013. The ins and outs of hepatitis C virus entry and assembly. Nat Rev Microbiol 11:688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. 1998. Binding of hepatitis C virus to CD81. Science 282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 5.Scarselli E, Ansuini H, Cerino R, Roccasecca RM, Acali S, Filocamo G, Traboni C, Nicosia A, Cortese R, Vitelli A. 2002. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J 21:5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans MJ, von Hahn T, Tscherne DM, Syder AJ, Panis M, Wolk B, Hatziioannou T, McKeating JA, Bieniasz PD, Rice CM. 2007. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature 446:801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 7.Ploss A, Evans MJ, Gaysinskaya VA, Panis M, You H, de Jong YP, Rice CM. 2009. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature 457:882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meertens L, Bertaux C, Dragic T. 2006. Hepatitis C virus entry requires a critical postinternalization step and delivery to early endosomes via clathrin-coated vesicles. J Virol 80:11571–11578. doi: 10.1128/JVI.01717-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Koutsoudakis G, Kaul A, Steinmann E, Kallis S, Lohmann V, Pietschmann T, Bartenschlager R. 2006. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J Virol 80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lupberger J, Zeisel MB, Xiao F, Thumann C, Fofana I, Zona L, Davis C, Mee CJ, Turek M, Gorke S, Royer C, Fischer B, Zahid MN, Lavillette D, Fresquet J, Cosset FL, Rothenberg SM, Pietschmann T, Patel AH, Pessaux P, Doffoel M, Raffelsberger W, Poch O, McKeating JA, Brino L, Baumert TF. 2011. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat Med 17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sainz B Jr, Barretto N, Martin DN, Hiraga N, Imamura M, Hussain S, Marsh KA, Yu X, Chayama K, Alrefai WA, Uprichard SL. 2012. Identification of the Niemann-Pick C1-like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat Med 18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Martin DN, Uprichard SL. 2013. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc Natl Acad Sci U S A 110:10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li Q, Sodroski C, Lowey B, Schweitzer CJ, Cha H, Zhang F, Liang TJ. 2016. Hepatitis C virus depends on E-cadherin as an entry factor and regulates its expression in epithelial-to-mesenchymal transition. Proc Natl Acad Sci U S A 113:7620–7625. doi: 10.1073/pnas.1602701113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng JJ, Li JR, Huang MH, Ma LL, Wu ZY, Jiang CC, Li WJ, Li YH, Han YX, Li H, Chen JH, Wang YX, Song DQ, Peng ZG, Jiang JD. 2016. CD36 is a co-receptor for hepatitis C virus E1 protein attachment. Sci Rep 6:21808. doi: 10.1038/srep21808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kaplan G, Totsuka A, Thompson P, Akatsuka T, Moritsugu Y, Feinstone SM. 1996. Identification of a surface glycoprotein on African green monkey kidney cells as a receptor for hepatitis A virus. EMBO J 15:4282–4296. [PMC free article] [PubMed] [Google Scholar]
- 16.Thompson P, Lu J, Kaplan GG. 1998. The Cys-rich region of hepatitis A virus cellular receptor 1 is required for binding of hepatitis A virus and protective monoclonal antibody 190/4. J Virol 72:3751–3761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santiago C, Ballesteros A, Martinez-Munoz L, Mellado M, Kaplan GG, Freeman GJ, Casasnovas JM. 2007. Structures of T cell immunoglobulin mucin protein 4 show a metal-ion-dependent ligand binding site where phosphatidylserine binds. Immunity 27:941–951. doi: 10.1016/j.immuni.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kobayashi N, Karisola P, Pena-Cruz V, Dorfman DM, Jinushi M, Umetsu SE, Butte MJ, Nagumo H, Chernova I, Zhu B, Sharpe AH, Ito S, Dranoff G, Kaplan GG, Casasnovas JM, Umetsu DT, Dekruyff RH, Freeman GJ. 2007. TIM-1 and TIM-4 glycoproteins bind phosphatidylserine and mediate uptake of apoptotic cells. Immunity 27:927–940. doi: 10.1016/j.immuni.2007.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Z, Ju Z, Frieri M. 2013. The T-cell immunoglobulin and mucin domain (Tim) gene family in asthma, allergy, and autoimmunity. Allergy Asthma Proc 34:e21–e26. doi: 10.2500/aap.2013.34.3646. [DOI] [PubMed] [Google Scholar]
- 20.Freeman GJ, Casasnovas JM, Umetsu DT, DeKruyff RH. 2010. TIM genes: a family of cell surface phosphatidylserine receptors that regulate innate and adaptive immunity. Immunol Rev 235:172–189. doi: 10.1111/j.0105-2896.2010.00903.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manangeeswaran M, Jacques J, Tami C, Konduru K, Amharref N, Perrella O, Casasnovas JM, Umetsu DT, DeKruyff RH, Freeman GJ, Perrella A, Kaplan GG. 2012. Binding of hepatitis A virus to its cellular receptor 1 inhibits T-regulatory cell functions in humans. Gastroenterology 142:1516–1525. doi: 10.1053/j.gastro.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ding Q, Yeung M, Camirand G, Zeng Q, Akiba H, Yagita H, Chalasani G, Sayegh MH, Najafian N, Rothstein DM. 2011. Regulatory B cells are identified by expression of TIM-1 and can be induced through TIM-1 ligation to promote tolerance in mice. J Clin Invest 121:3645–3656. doi: 10.1172/JCI46274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Amara A, Mercer J. 2015. Viral apoptotic mimicry. Nat Rev Microbiol 13:461–469. doi: 10.1038/nrmicro3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Abad-Molina C, Garcia-Lozano JR, Montes-Cano MA, Torres-Cornejo A, Torrecillas F, Aguilar-Reina J, Romero-Gomez M, Lopez-Cortes LF, Nunez-Roldan A, Gonzalez-Escribano MF. 2012. HAVCR1 gene haplotypes and infection by different viral hepatitis C virus genotypes. Clin Vaccine Immunol 19:223–227. doi: 10.1128/CVI.05305-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wojcik G, Latanich R, Mosbruger T, Astemborski J, Kirk GD, Mehta SH, Goedert JJ, Kim AY, Seaberg EC, Busch M, Thomas DL, Duggal P, Thio CL. 2014. Variants in HAVCR1 gene region contribute to hepatitis C persistence in African Americans. J Infect Dis 209:355–359. doi: 10.1093/infdis/jit444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang J, Qiao L, Hou Z, Luo G. 2017. TIM-1 promotes hepatitis C virus cell attachment and infection. J Virol 91:e01583-. doi: 10.1128/JVI.01583-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koutsoudakis G, Herrmann E, Kallis S, Bartenschlager R, Pietschmann T. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J Virol 81:588–598. doi: 10.1128/JVI.01534-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Owsianka AM, Timms JM, Tarr AW, Brown RJ, Hickling TP, Szwejk A, Bienkowska-Szewczyk K, Thomson BJ, Patel AH, Ball JK. 2006. Identification of conserved residues in the E2 envelope glycoprotein of the hepatitis C virus that are critical for CD81 binding. J Virol 80:8695–8704. doi: 10.1128/JVI.00271-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Drummer HE, Boo I, Maerz AL, Poumbourios P. 2006. A conserved Gly436-Trp-Leu-Ala-Gly-Leu-Phe-Tyr motif in hepatitis C virus glycoprotein E2 is a determinant of CD81 binding and viral entry. J Virol 80:7844–7853. doi: 10.1128/JVI.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chou TC, Talalay P. 1984. Quantitative analysis of dose-effect relationships: the combined effects of multiple drugs or enzyme inhibitors. Adv Enzyme Regul 22:27–55. doi: 10.1016/0065-2571(84)90007-4. [DOI] [PubMed] [Google Scholar]
- 31.Zhang YY, Zhang BH, Ishii K, Liang TJ. 2010. Novel function of CD81 in controlling hepatitis C virus replication. J Virol 84:3396–3407. doi: 10.1128/JVI.02391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Binne LL, Scott ML, Rennert PD. 2007. Human TIM-1 associates with the TCR complex and up-regulates T cell activation signals. J Immunol 178:4342–4350. doi: 10.4049/jimmunol.178.7.4342. [DOI] [PubMed] [Google Scholar]
- 33.Curtiss ML, Hostager BS, Stepniak E, Singh M, Manhica N, Knisz J, Traver G, Rennert PD, Colgan JD, Rothman PB. 2011. Fyn binds to and phosphorylates T cell immunoglobulin and mucin domain-1 (Tim-1). Mol Immunol 48:1424–1431. doi: 10.1016/j.molimm.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Meertens L, Carnec X, Lecoin MP, Ramdasi R, Guivel-Benhassine F, Lew E, Lemke G, Schwartz O, Amara A. 2012. The TIM and TAM families of phosphatidylserine receptors mediate dengue virus entry. Cell Host Microbe 12:544–557. doi: 10.1016/j.chom.2012.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Su EW, Lin JY, Kane LP. 2008. TIM-1 and TIM-3 proteins in immune regulation. Cytokine 44:9–13. doi: 10.1016/j.cyto.2008.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mosbruger TL, Duggal P, Goedert JJ, Kirk GD, Hoots WK, Tobler LH, Busch M, Peters MG, Rosen HR, Thomas DL, Thio CL. 2010. Large-scale candidate gene analysis of spontaneous clearance of hepatitis C virus. J Infect Dis 201:1371–1380. doi: 10.1086/651606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wakita T, Pietschmann T, Kato T, Date T, Miyamoto M, Zhao Z, Murthy K, Habermann A, Krausslich HG, Mizokami M, Bartenschlager R, Liang TJ. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat Med 11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kachko A, Loesgen S, Shahzad-Ul-Hussan S, Tan W, Zubkova I, Takeda K, Wells F, Rubin S, Bewley CA, Major ME. 2013. Inhibition of hepatitis C virus by the cyanobacterial protein Microcystis viridis lectin: mechanistic differences between the high-mannose specific lectins MVL, CV-N, and GNA. Mol Pharm 10:4590–4602. doi: 10.1021/mp400399b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kachko A, Kochneva G, Sivolobova G, Grazhdantseva A, Lupan T, Zubkova I, Wells F, Merchlinsky M, Williams O, Watanabe H, Ivanova A, Shvalov A, Loktev V, Netesov S, Major ME. 2011. New neutralizing antibody epitopes in hepatitis C virus envelope glycoproteins are revealed by dissecting peptide recognition profiles. Vaccine 30:69–77. doi: 10.1016/j.vaccine.2011.10.045. [DOI] [PubMed] [Google Scholar]
- 40.Liu S, Nelson CA, Xiao L, Lu L, Seth PP, Davis DR, Hagedorn CH. 2011. Measuring antiviral activity of benzimidazole molecules that alter IRES RNA structure with an infectious hepatitis C virus chimera expressing Renilla luciferase. Antiviral Res 89:54–63. doi: 10.1016/j.antiviral.2010.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kachko A, Frey SE, Sirota L, Ray R, Wells F, Zubkova I, Zhang P, Major ME. 2015. Antibodies to an interfering epitope in hepatitis C virus E2 can mask vaccine-induced neutralizing activity. Hepatology 62:1670–1682. doi: 10.1002/hep.28108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tami C, Silberstein E, Manangeeswaran M, Freeman GJ, Umetsu SE, DeKruyff RH, Umetsu DT, Kaplan GG. 2007. Immunoglobulin A (IgA) is a natural ligand of hepatitis A virus cellular receptor 1 (HAVCR1), and the association of IgA with HAVCR1 enhances virus-receptor interactions. J Virol 81:3437–3446. doi: 10.1128/JVI.01585-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Konduru K, Kaplan GG. 2006. Stable growth of wild-type hepatitis A virus in cell culture. J Virol 80:1352–1360. doi: 10.1128/JVI.80.3.1352-1360.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Blight KJ, McKeating JA, Rice CM. 2002. Highly permissive cell lines for subgenomic and genomic hepatitis C virus RNA replication. J Virol 76:13001–13014. doi: 10.1128/JVI.76.24.13001-13014.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tarradas A, Selga E, Beltran-Alvarez P, Perez-Serra A, Riuro H, Pico F, Iglesias A, Campuzano O, Castro-Urda V, Fernandez-Lozano I, Perez GJ, Scornik FS, Brugada R. 2013. A novel missense mutation, I890T, in the pore region of cardiac sodium channel causes Brugada syndrome. PLoS One 8:e53220. doi: 10.1371/journal.pone.0053220. [DOI] [PMC free article] [PubMed] [Google Scholar]