ABSTRACT
The glycosylated Gag protein (gPr80) of murine leukemia viruses (MLVs) has been shown to exhibit multiple roles in facilitating retrovirus release, infection, and resistance to host-encoded retroviral restriction factors, such as APOBEC3, SERINC3, and SERINC5. One way in which gPr80 helps MLVs to escape host innate immune restriction is by increasing capsid stability, a feature that protects viral replication intermediates from being detected by cytosolic DNA sensors. gPr80 also increases the resistance of MLVs to deamination and restriction by mouse APOBEC3 (mA3). How the gPr80 accessory protein, with its three N-linked glycosylation sites, contributes to these resistance mechanisms is still not fully understood. Here we further characterized the function of gPr80 and, more specifically, revealed that the asparagines targeted for glycosylation in gPr80 also contribute to capsid stability through their parallel involvement in the Pr65 Gag structural polyprotein. In fact, we demonstrate that sensitivity to deamination by the mA3 and human A3 proteins is directly linked to capsid stability. We also show that full-length gPr80 is detected in purified viruses. However, our results suggest that gPr80 is inserted in the NexoCcyto orientation of a type I integral membrane protein. Additionally, our experiments have revealed the existence of a large population of Env-deficient virus-like particles (VLPs) harboring gPr80 inserted in the opposite (NcytoCexo) polarity, which is typical of type II integral membrane proteins. Overall this study provides new insight into the complex nature of the MLV gPr80 accessory protein.
IMPORTANCE Viruses have evolved numerous strategies to infect, spread in, and persist in their hosts. Here we analyze the details of how the MLV-encoded glycosylated Gag (gPr80) protein protects the virus from being restricted by host innate immune defenses. gPr80 is a variant of the structural Pr65 Gag protein with an 88-amino-acid extended leader sequence that directs the protein for translation and glycosylation in the endoplasmic reticulum. This study dissects the specific contributions of gPr80 glycans and capsid stability in helping the virus to infect cells, spread, and counteract the effects of the host intrinsic restriction factor APOBEC3. Overall this study provides further insight into the elusive role of the gPr80 protein.
KEYWORDS: APOBEC3, MLV, SERINC3, SERINC5, glyco-Gag
INTRODUCTION
Infection, spread, and persistence of retroviruses in an animal population are directly related to the ability of these viruses to evolve under the selective pressures of host immune defenses. The success of HIV-1 as a pathogen is credited, among other strategies, to its accessory protein Vif, which induces the proteasomal degradation of APOBEC3 (A3) proteins that are potent host-intrinsic retroviral restriction factors (reviewed in reference 1). In contrast, murine gammaretroviruses, such as Moloney murine leukemia virus (M-MLV), do not express a Vif-like protein but rather have a glycosylated version of the structural Gag protein, termed glyco-Gag (gPr80), to defend against the deleterious effects of mouse A3 (mA3) (2–4).
gPr80 originates from translation of the structural Gag polyprotein (Pr65) beginning at an alternate (CUG) start codon upstream of the canonical AUG initiation codon, a process that adds 88 amino acids to the original sequence (Fig. 1A) (5–8). This N-terminal leader segment of gPr80 includes a signal peptide that directs the completion of polyprotein synthesis to the endoplasmic reticulum (ER), where it undergoes N-linked glycosylation. It also contains a signal/anchor site upstream of the junction of the leader and matrix (MA) Gag domains that directs its insertion into the membrane (9, 10). The mature protein can be further proteolytically processed in the infected cell to yield a membrane-associated, ∼55-kDa N-terminal product and a secreted, ∼40-kDa C-terminal product (9, 11). While the N-terminal protein segment associates with the cell membrane as a type II integral membrane protein (10), it has also been detected in released retroviral particles, while the C-terminal fragment has not (10, 11). Three sites have been identified experimentally in the gPr80 protein of M-MLV that account for all detectable N-linked glycosylation: N113, located in the sequence of the viral matrix (MA), and N480 and N505, located in the capsid (CA) (Fig. 1A) (4).
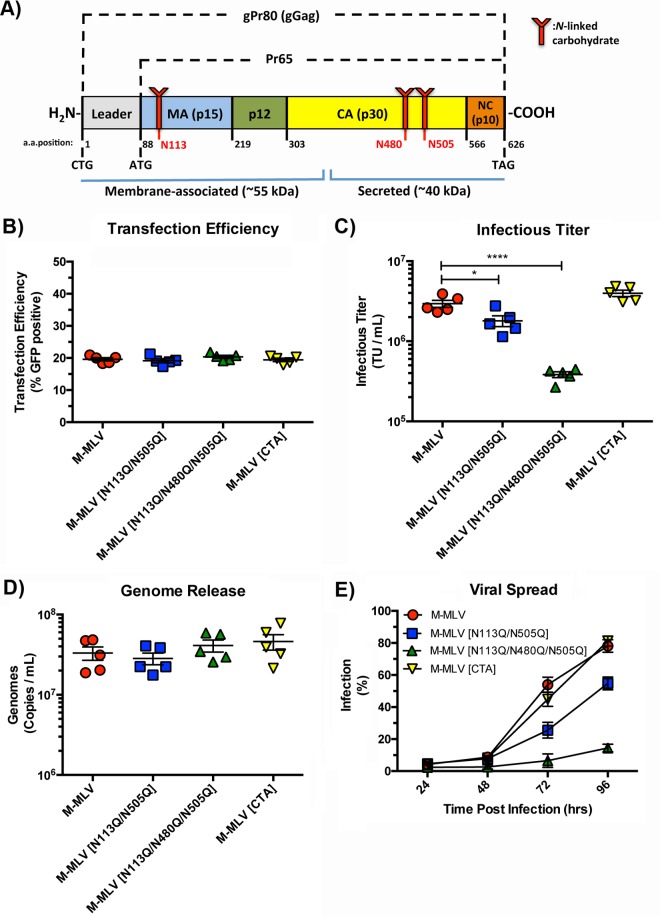
FIG 1.
Replicative fitness of gPr80-deficient and mutant viruses. (A) Schematic representation of the gPr80 and Pr65 proteins. The locations of the three N-linked glycosylation sites are indicated in red. The approximate regions encompassing the N-terminal membrane-associated domain and the C-terminal secreted domain are indicated with blue lines; the leader region contains the ER localization signal peptide. (B) Transfection efficiencies of virus expression constructs in transfected 293T cells. (C) Infectious titers of viruses released from transfected 293T cells. Viruses were harvested after 48 h and filtered through 0.45-μm filters. Equal volumes of the different viruses were then titrated on NIH 3T3 cells, and infection was measured by flow cytometry 24 h after infection. (D) Viral genomes released into the supernatants of the same transfected 293T cells as those described above were measured by digital droplet PCR (ddPCR). Samples were treated with DNase I prior to RNA extraction. (E) Replicative fitness of M-MLV and its glycosylation mutants. Input virus was normalized via the number of TU per milliliter. Infection was monitored by measuring EGFP expression at each time point following infection by flow cytometry. Data are the compilation of five independent transfection experiments done in triplicate. P values were calculated by Student's t test. *, P ≤ 0.05; ****, P ≤ 0.0001. P values for the data sets in panels B and D were not significant for comparisons to the M-MLV control (ns, P > 0.05).
Multiple functions that help the virus to infect its host have now been attributed to gPr80. Although expression of the protein has little effect on viral infection under most in vitro laboratory conditions (3, 12, 13), it enhances a late step of virus assembly and release through lipid rafts (14, 15) and is also involved in promoting virus replication, pathogenesis, and neurovirulence in vivo (12–18), especially when mA3 is expressed (2–4, 19). As first reported by Kolokithas et al., gPr80 is required to support the replication of the neurovirulent CasFrKP MLV in vivo when mA3 is expressed (2). Murine A3 is a cytidine deaminase that inhibits the infection of most MLVs by interfering with the early stages of reverse transcription, mainly through enzyme-independent strategies (3, 20–24). In contrast, human A3 proteins acting on MLVs and mA3 acting on retroviruses from species other than mice primarily restrict and inactivate retroviruses through intense deamination of cytosines in single-stranded DNA replication intermediates produced during reverse transcription (25, 26). Generation of large numbers of C-to-T mutations in the targeted minus-strand proviral DNA is a process called hypermutation. While most MLVs are only modestly susceptible to mA3 restriction in vitro (22, 23, 27), some MLVs, such as AKV, are more sensitive to mA3, and this has been associated with fewer N-linked glycosylation sites present on the gPr80 protein of AKV (4, 23). We previously showed, through site-directed mutagenesis, that larger numbers of N-linked glycosylation sites on gPr80 appear to correlate with increased resistance to mA3 deamination and restriction in vitro and ex vivo in murine splenocytes (4).
Another important feature of gPr80, revealed in two studies by Stavrou et al., showed that gPr80 increases MLV capsid stability (3, 19). Capsid integrity is important for creating a shielded environment that protects viral replication intermediates from immune cytosolic DNA sensors that can induce an interferon response, and also from mA3 expressed in the cytosol of the recipient cells (3, 19). The importance of gPr80 in antagonizing mA3 was further showcased by the demonstration that gPr80-deficient viruses with a stop codon in the N-terminal leader peptide reverted back to gPr80-proficient viruses when they were passaged in mice expressing mA3 but not in mA3 knockout mice (3, 19). These studies therefore suggest that one of gPr80's primary roles might in fact be to counteract the antiviral effects of mA3.
In light of a previous report by our group showing that the state of N-linked glycosylation is involved in modulating the sensitivity of MLVs to restriction and deamination of mA3 (4), here we further investigate the specific roles of gPr80 carbohydrates in capsid stability and resistance to editing by mouse and human A3 proteins. For our study, we used a replication-competent M-MLV reporter virus that has a CTG-to-CTA mutation to abolish gPr80 translation initiation and M-MLV mutant viruses in which the asparagines targeted for glycosylation have individually been converted to glutamine. We reveal that gPr80 contributes to increased capsid stability regardless of the presence of glycosylation; however, these asparagines are also essential for maintaining the structural integrity of the capsid. Furthermore, use of glycosylation inhibitors and cells unable to carry out complex N-linked glycosylation showed that diminished capsid stability is required for mA3 to deaminate viral DNA. Finally, we also provide evidence that full-length gPr80 may provide structural support to virions as a type I integral membrane protein, implying that the glycosylated C terminus of the protein may help to improve their stability. Our study provides additional insight into the mechanisms by which the elusive gPr80 protein antagonizes host innate immune detection and restriction.
RESULTS
gPr80-deficient MLV exhibits a replicative fitness similar to that of the wild-type virus.
We previously demonstrated that replacement of N-linked glycosylated amino acids (N → Q) of the gPr80 protein of gammaretroviruses increases the sensitivity of these viruses to deamination by virion-packaged mA3 and human A3G (4). While one glycosylation site maps to position N113, located in the viral matrix and the membrane-associated segment of gPr80, the two other sites (N480 and N505) map to the viral capsid and are within the secreted segment of gPr80 (Fig. 1A). Additionally, it has also been reported that gPr80 expression contributes to capsid stability, thereby increasing protection of replication intermediates against cytosolic sensors and mA3 (3, 19). However, it is unclear how these glycans inhibit editing by A3 proteins or whether they also partake in increasing capsid structural integrity.
We first examined the effect of N-linked glycans on various parameters linked to replicative fitness, notably particle release, infectious titer, and spread. Our interest focused on a gPr80-deficient virus with the gPr80 CTG (leucine) initiation codon mutated to a CTA (leucine) to prevent translation of gPr80 (M-MLV[CTA]), and also on mutant M-MLVs containing a double (N113Q/N505Q) or triple (N113Q/N480Q/N505Q) substitution to prevent glycosylation to different degrees (Fig. 1A). We previously showed that a virus with the N480Q mutation has severely compromised infectivity; this is why we chose to use only the triple mutant, for simplicity and convenience (4). We first investigated how N-linked glycans affect viral genome release and infectious titers. Virus expression plasmids were transfected into 293T cells, and 48 h later, virus was harvested and passed through a 0.45-μm filter. Transfection efficiencies were similar for all constructs, as measured by expression of the viral envelope-enhanced green fluorescent protein (Env-EGFP) fusion protein (Fig. 1B). Equal volumes of virus-containing supernatant were used to infect NIH 3T3 cells to evaluate the viral titer (Fig. 1C). Titers of wild-type M-MLV and the CTA mutant were nearly identical. A slightly reduced titer was observed for the double mutant, and the triple mutant had a nearly 10-fold lower titer. To ensure that differences in titer are not a reflection of compromised particle release, we measured viral genomes in the transfected cell supernatants, which confirmed that particle release levels were nearly identical (Fig. 1D). Finally, we measured the replicative fitness of the viruses. NIH 3T3 cells were infected with equal transducing units (TU) of virus, and viral replication was monitored for 96 h. The replicative fitness levels of M-MLV and M-MLV[CTA] were nearly identical, while both glycosylation mutants (M-MLV[N113Q/N505Q] and M-MLV[N113Q/N480Q/N505Q]) had reduced propagation efficiencies (Fig. 1E). The similar spread curves for M-MLV and M-MLV[CTA] are not surprising, as these data confirm previously published observations (28, 29). These results are also consistent with our previous report showing that an N-to-Q substitution at capsid residue 480 severely impairs the infectivity of viruses harboring that mutation (4).
Mutations at sites of N-linked glycosylation reduce capsid stability.
We next investigated the effects of the asparagine mutations and the absence of gPr80 protein expression on capsid stability. We first harvested virus-containing cell supernatants from infected cells and treated them with 10% Triton X-100 to remove the viral envelope. The naked capsids were then spun through a sucrose cushion containing either 2% Triton X-100 or 0.2% SDS and resolved further through a 20% sucrose cushion (Fig. 2A). These concentrations were shown in our preliminary optimization assays to be at the threshold for destabilizing naked wild-type M-MLV capsids (data not shown). Virus pellets were then resuspended in RIPA lysis buffer and analyzed by Western blotting using anti-EGFP to stain the viral envelope and monoclonal anti-p30 capsid antibodies. In agreement with previous findings (3), the capsid of the gPr80-deficient M-MLV[CTA] strain was completely dissolved by the detergents, indicating less robust capsid stability than that of the wild-type virus (Fig. 2B). We also observed reduced recovery of p30 with the glycosylation-null mutant M-MLV[N113Q/N480Q/N505Q] and the single point mutant M-MLV[N480Q] (Fig. 2B). This assay did not show an important difference in capsid stability between the glycosylation mutant M-MLV[N113Q/N505Q] and the wild-type virus.
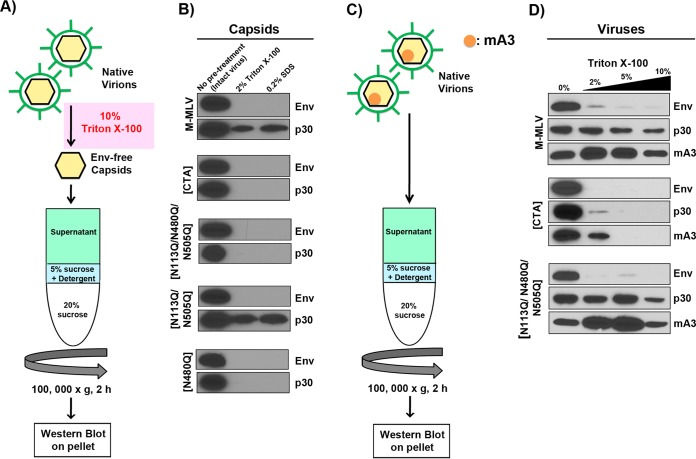
FIG 2.
Glycosylation mutants have diminished capsid stability. (A) Schematic representation of the procedure used for assessment of the stability of envelope-free viral cores in the presence of mild detergent. Native enveloped viruses were first treated with 10% Triton X-100 to strip the viral envelope. Envelope-free viral cores were then submitted to velocity sedimentation through a 5% sucrose step containing either 2% Triton X-100 or 0.2% SDS and then through a 20% sucrose step. (B) Western blots were carried out on intact core pellets by using monoclonal anti-p30 (R187) and anti-EGFP antibodies. EGFP was expressed as a fusion protein with the viral envelope glycoprotein (Env). (C) Schematic representation of the methodology used to assess packaging of mA3 inside viral cores. Native, enveloped virions were submitted to velocity sedimentation through a 5% sucrose step containing 0%, 2%, 5%, or 10% Triton X-100 and then through a 20% sucrose step. (D) Western blots were carried out on intact viral cores as described above; mA3 was detected using an anti-FLAG monoclonal antibody.
gPr80 does not affect the level of mA3 incorporation into the virion.
Our next question was whether mA3 was packaged similarly within the viral cores in all viruses tested or was excluded because of the gPr80 glycans. Previous studies clearly showed that detergent-treated MLVs harbored mA3 proteins within the viral core (22, 27). To investigate this issue, we produced virus as detailed above, but in the presence of mA3. We then layered the supernatant containing enveloped virus directly onto sucrose cushions containing increasing concentrations of detergent (Fig. 2C). This approach exposes virions to detergent for only a short time, as they pass from 5% sucrose to 20% sucrose, gently and gradually stripping away the envelope, matrix, and proteins located outside the viral capsid core as the detergent concentration increases. Four samples were analyzed individually, each with either 0%, 2%, 5%, or 10% Triton X-100 in the 5% sucrose layer. Our results show that the capsid of M-MLV[CTA] was easily dissolved using the lowest concentration of detergent, while the capsids of the triple mutant and wild-type viruses remained intact under all conditions, as judged by their relative abundances in the blots (Fig. 2D). These results highlight that the capsid of the gPr80-deficient virus is less stable than that of the virus with triple N-to-Q substitutions. We can also conclude from this experiment that the levels of virion-packaged mA3 were comparable for all viruses and that mA3 continued to be associated with p30 even when no viral envelope glycoprotein could be detected.
M-MLV asparagine mutants have compromised capsid stability.
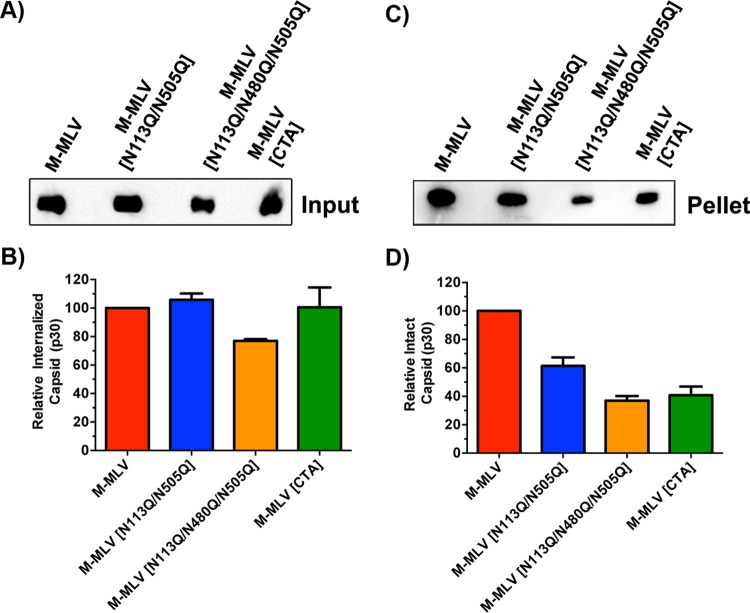
As an alternative approach to evaluate the structural integrity of the mutated viruses, we conducted a fate-of-capsid assay (3, 30–33). M-MLV, MLV[N113Q/N505Q], M-MLV[N113Q/N480Q/N505Q], and M-MLV[CTA] preparations were normalized for p30 levels prior to infection of NIH 3T3 cells. At 3 h postinfection, the cells were treated with pronase, washed, lysed, processed with a Dounce homogenizer, and pelleted through a 40% sucrose cushion to isolate intact capsid. Aliquots of the input samples pelleted through the dense sucrose were taken to normalize the pelleted fractions. As illustrated in Fig. 3A and B, M-MLV, M-MLV[N113Q/N505Q], and M-MLV[CTA] all had similar levels of capsid that successfully entered the cell. However, the M-MLV[N113Q/N480Q/N505Q] mutant had an approximately 20% lower capsid level at this early stage of infection, implying that there may be additional underlying defects with this virus. As expected, and in accordance with previous findings (3), M-MLV capsids were noticeably more stable than those of M-MLV[CTA] (Fig. 3C and D). In addition, M-MLV[N113Q/N505Q] and M-MLV[N113Q/N480Q/N505Q] were approximately 40% and 60% less stable, respectively, than the wild-type virus (Fig. 3C and D). These data are directly reflective of the results of our previous assay (Fig. 2B). Taken with the measure of viral fitness (Fig. 1), these data suggest that mutagenesis at key glycosylated residues of gPr80 affects the structural integrity of Pr65 products, the matrix protein (N113), and the capsid protein (N480/N505). However, the severely reduced capsid stability of M-MLV[CTA], coupled with its unaltered replicative fitness, suggests that these parameters are not related, at least for this virus.
FIG 3.
Intracellular fate-of-capsid assay. (A) Western blot showing levels of CA p30 input protein. (B) Graphical representation of densitometry measurements for the samples from panel A. (C) Western blot revealing cytosol-derived capsids recovered following pelleting through a 40% sucrose cushion. (D) Graphical representation of densitometry measurements for the samples from panel C. Viruses from this assay were produced from NIH 3T3 cells chronically infected for 96 h, filtered through 0.45-μm filters, and further purified through a 20% sucrose cushion by ultracentrifugation. Capsid levels were normalized by p30 ELISA prior to infection.
M-MLV capsid stability and sensitivity to mA3 deamination are independent of N-linked glycosylation of gPr80.
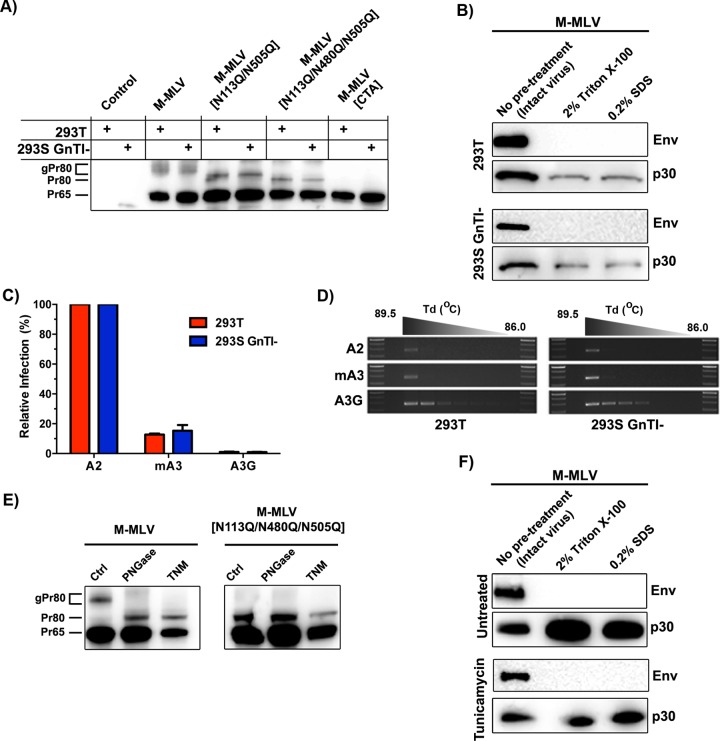
A limitation in studying glycosylation mutants is that both the gPr80 and Pr65 polyproteins are affected by alterations of the glycosylation residues. To further investigate the isolated impact of posttranslational glycosylation, we used two approaches. First, we employed the use of 293S GnTI− cells. Given that these cells do not have N-acetylglucosaminyltransferase I (GnTI) activity, and thus are unable to conduct complex N-linked glycosylation, they allowed us to uncouple the impact of diminished glycosylation complexity from gPr80 function. Surprisingly, loss of complex glycosylation was barely noticeable by SDS-PAGE performed on gPr80 in cell lysates (Fig. 4A). M-MLV was produced and isolated from transfected 293S GnTI− and 293T cells and evaluated for p30 capsid stability as in Fig. 2B. No difference was observed between these viruses (Fig. 4B). However, this does not exclude the possibility that simple glycans may still impair A3 activity. Infections were carried out with viruses produced from both cell types, and genomic DNAs from the infected cells were analyzed by differential DNA denaturation PCR (3D-PCR) for hypermutation. There was no important difference in terms of A3 restriction (Fig. 4C) or deamination intensity (Fig. 4D). This suggests that glycosylation complexity has no or little influence on the antagonistic effect of gPr80 on A3 function, at least in viruses with unaltered capsid stability.
FIG 4.
Influence of gPr80 glycosylation on capsid stability and resistance to mA3 deamination. (A) Lysates from control (untransfected) or transfected 293T and 293S GnTI− cells were analyzed by SDS-PAGE. Protein expression was detected using the polyclonal anti-p30 antibody. (B) Viruses were processed as described in the legend to Fig. 2B. Western blots were carried out using monoclonal anti-p30 and anti-EGFP antibodies. EGFP was expressed as a fusion protein with the viral envelope glycoprotein (Env). (C) 293T and 293S GnTI− cells were cotransfected with M-MLV and APOBEC expression plasmids, and supernatants were harvested 48 h later. Infection in NIH 3T3 cells was measured by EGFP expression 48 h later. (D) Deamination intensity analysis by 3D-PCR. The analysis was performed on genomic DNAs extracted from the infected NIH 3T3 cells used for panel C. The results show a 279-bp product of the EGFP gene that was amplified using a decreasing denaturing temperature gradient (Td) from 89.5°C to 86.0°C. Larger numbers of bands indicate more intensely mutated products. (E) Western blot analysis of gPr80 expression in tunicamycin-treated cells. Untreated cells (left lanes) and PNGase F-treated lysates (middle lanes) were compared to tunicamycin-treated (TNM) cells (right lanes) for both M-MLV and M-MLV[N113Q/N480Q/N505Q]. (F) Capsid stability of viruses produced in tunicamycin-treated cells, determined as described for panel B.
The inhibitor tunicamycin was next used to evaluate the effect of completely preventing N-linked glycosylation. Figure 4E clearly shows that there was a nearly complete abolition of glycosylation on gPr80. Capsid stability was next evaluated for viruses produced from tunicamycin-treated infected NIH 3T3 cells. Once again, there was no apparent difference in capsid stability between the wild-type and glycan-deficient viruses (Fig. 4F). This led us to propose that the glycans alone are not responsible for capsid stability. Unfortunately, 293S GnTI− cells produce greatly diminished levels of virus, which are not conducive to carrying out quantitative fate-of-capsid assays. Furthermore, tunicamycin-treated cells yielded virus that was noninfectious, likely due to collateral effects on the viral envelope glycoprotein.
No detectable interaction between gPr80 and A3 proteins in the cytosol.
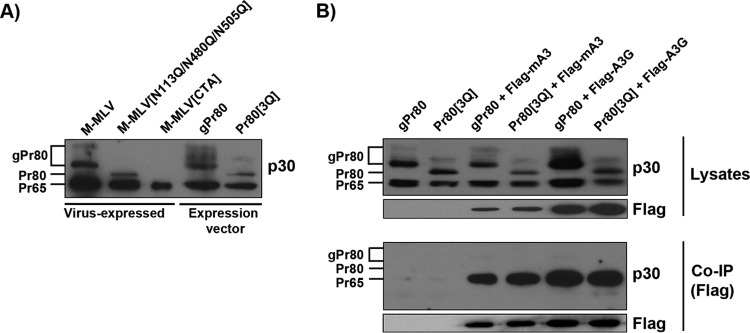
To gain further insight into how gPr80 antagonizes mA3 restriction and deamination, we next wanted to determine if the two proteins interact. It is well established that capsid inclusion of mA3 and human A3G in HIV-1 virions occurs through binding to the nucleocapsid (NC) region of the Gag polyprotein (34–39). Mouse A3 is also efficiently packaged into MLV particles (22, 23, 27); however, the 88-amino-acid leader sequence of gPr80 does not appear to be involved, and the exact epitopes of NC to which mA3 binds have not yet been precisely mapped (2, 39). We asked whether mA3 binds differently to gPr80 and the glycosylation-null protein Pr80[N113Q/N480Q/N505Q], henceforth called Pr80[3Q] for simplicity. The gPr80 and Pr80[3Q] expression vectors have an ATG at the site of gPr80 protein translation instead of a CTG to increase protein translation efficiency. For our analysis, we prepared cytosolic lysates of 293T cells transfected with viral vectors (first 3 lanes) or with gPr80 and Pr80[3Q] expression vectors (last 2 lanes) (Fig. 5A). Western blot analyses showed that bands for Pr65, Pr80, and gPr80 could clearly be visualized where expected for our constructs.
FIG 5.
Binding of gPr80 to mA3 and A3G in the cytosol. (A) Virus-expressed and plasmid-expressed gPr80 and Pr65 Gag proteins display similar profiles in transfected 293T cells. gPr80 and Pr80[3Q] were expressed from a CMV promoter. The gPr80 initiation codon was changed from CTG to ATG to improve expression. The Pr80[3Q] coding sequence was modified to replace the three glycosylated asparagines (N), at positions 113, 480, and 505, with glutamines (Q) (33). (B) Coimmunoprecipitation of FLAG-mA3 and FLAG-A3G with gPr80 and Pr80[3Q] in the soluble fraction of transfected 293T cell lysates. Protein interactions were revealed by Western blotting using anti-FLAG monoclonal and anti-p30 polyclonal antibodies.
Coimmunoprecipitation (co-IP) analyses were next performed by capturing either FLAG-mA3 or FLAG-A3G from extracts of 293T cells cotransfected with gPr80 or Pr80[3Q]. The beads were recovered after overnight incubation, and equal sample volumes were loaded onto the gels for all co-IP conditions. The blots were then stained with a p30 antibody. Our co-IP results clearly show that mA3 and A3G bound only to Pr65 (Fig. 5B). Despite repeated attempts, we were unable to detect a signal for gPr80 or Pr80[3Q] under our experimental conditions. However, these results cannot exclude the possibility that our lysate conditions were not conducive to detecting an interaction between mA3 and Pr80. Additionally, mA3 interactions with gPr80 inside virions at this stage are unknown.
N-linked glycans do not inhibit A3 deaminase activity.
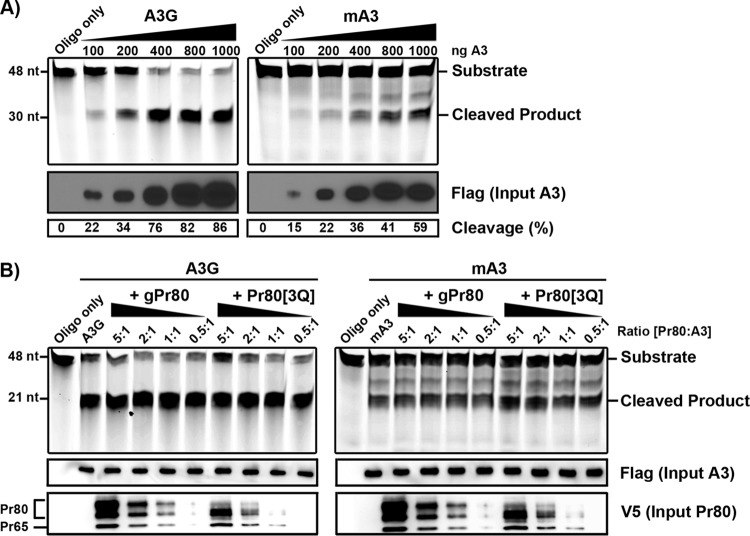
MLVs that express an unglycosylated Pr80 protein are more sensitive to hypermutation by mA3 (4). Although it was unlikely given the results shown in Fig. 4, we wanted to confirm through a different approach whether N-linked glycans or the gPr80 protein itself acts as a catalytic inhibitor of deamination. In order to address this issue, we purified FLAG-tagged mA3 and A3G and V5-tagged gPr80 and Pr80[3Q] from transfected 293T cells. We then performed an oligodeoxynucleotide deamination assay using oligonucleotides containing central 5′-CCC and 5′-TCC sequences for use in conjunction with A3G and mA3, respectively, as previously described (21, 40). The activities of both A3 proteins on their respective target oligonucleotides were first tested and showed cleavage efficiency as a function of the amount of input deaminase (Fig. 6A). The assay was then repeated using 800 ng of A3G or mA3 and by adding increasing amounts of purified gPr80 or Pr80[3Q]. We did not find any evidence that gPr80 acted as a catalytic inhibitor of A3 deaminase activity under our experimental conditions (Fig. 6B).
FIG 6.
Oligonucleotide deamination assays. (A) Oligonucleotide deamination was performed using purified mA3 and A3G. Lysates of 293T cells expressing FLAG-tagged mA3 or A3G were precipitated with anti-FLAG-conjugated agarose beads. Proteins were eluted from the beads and then used in oligonucleotide cleavage assays with 0.1 pmol/μl of FAM-labeled oligonucleotides containing TTC or CCC target sites. Deaminated oligonucleotides were treated with uracil DNA glycosylase (UDG), boiled, and resolved in an acrylamide gel. Input A3 protein and the efficiency of oligonucleotide cleavage are shown below the input gels. The percentage of cleaved oligonucleotide in each sample was determined using ImageJ software. (B) Oligonucleotide deamination assays in the presence of purified gPr80-V5 or Pr80[3Q]-V5 and 800 ng of mA3 or A3G were carried out as described for panel A. Molar ratios of gPr80/Pr80[3Q] to A3 are indicated above the gels. Input gPr80/Pr80[3Q] was assessed using a polyclonal anti-p30 antibody. The average cleavage efficiency was 84% for A3G and 46% for mA3 under all conditions in two independent experiments.
Capsid stability affects susceptibility to deamination by all seven human A3 proteins.
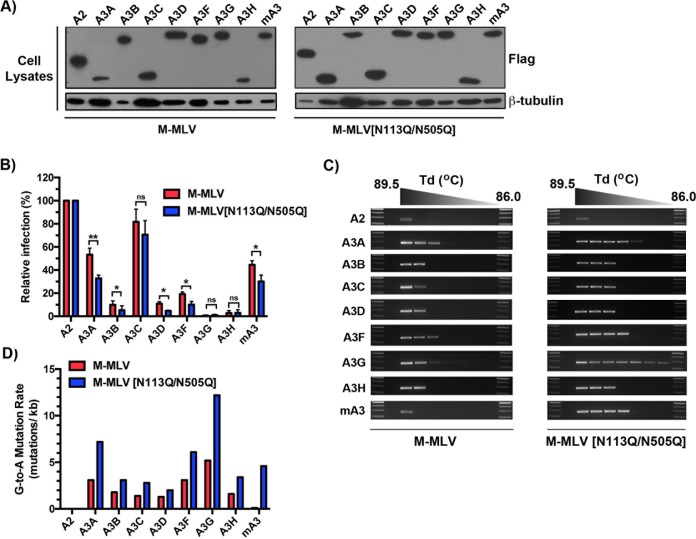
In a previous report, we showed that an absence of gPr80 or reduced glycosylation increases susceptibility to deamination by mA3 (4). Here we tested the sensitivities of M-MLV and M-MLV[N113Q/N505Q] to mA3 and each of the seven human A3 proteins. M-MLV[N113Q/N505Q], with two of the three possible sites mutated, was used because it displays only slightly reduced infectivity compared to that of the wild-type virus, along with a minor reduction in capsid stability, as opposed to the triple mutant, which has heavily compromised infectivity and capsid stability (Fig. 1E, 2B, and 3) (4). Viruses were coproduced in 293T cells along with the various APOBEC proteins (Fig. 7A), harvested from the supernatant, and normalized for p30 content by enzyme-linked immunosorbent assay (ELISA). Target NIH 3T3 cells were then infected at a multiplicity of infection (MOI) of 1 for the A2 cotransfection, while the others were infected with the corresponding amount of p30. Infection was assessed by measuring EGFP expression after 48 h (Fig. 7B). A3 proteins were expressed at comparable levels under all conditions (Fig. 7A).
FIG 7.
Impacts of glycosylation site mutations on deamination and restriction by all 7 members of the human A3 family. (A) FLAG-tagged APOBEC expression plasmids were cotransfected with M-MLV or M-MLV[N113Q/N505Q] into 293T cells. Two days following transfection, cells were harvested and assessed for FLAG expression by Western blot analysis. The expression of β-tubulin was used as a loading control, and viral capsid protein expression was assessed using a monoclonal anti-p30 antibody. (B) NIH 3T3 cells were infected with p30-normalized amounts of the viruses produced for panel A. Infection was monitored as a function of EGFP reporter protein expression 48 h later by flow cytometry. Infection assays were performed for two independent transfections, with triplicate infection values for each. Results are presented as normalized mean infection values ± standard deviations (SD) relative to that for the A2 control. (C) Deamination intensity analysis by 3D-PCR. The analysis was performed on genomic DNAs extracted from infected NIH 3T3 cells at 48 h postinfection as described in the legend to Fig. 4D. (D) Genomic DNAs from infected target cells were amplified by PCR and cloned, and the EGFP gene was sequenced. Mutation rates reflect the averages for 8 independently mutated clones. P values were calculated by Student's t test. *, P ≤ 0.05; **, P ≤ 0.01; ns, P > 0.05.
Hypermutation analysis of integrated proviral sequences by 3D-PCR revealed an overall increase in mutation intensity with the M-MLV[N113Q/N505Q] mutant for all A3 proteins except for deaminase-inactive A2 (Fig. 7C). The increase in mutation intensity was most noticeable for A3A, A3G, and mA3 in the double mutant virus. DNA editing was confirmed by sequencing of individual clones of amplified EGFP sequences from integrated proviruses (Fig. 7D). G-to-A mutation rates were calculated for 8 mutated clones and support the 3D-PCR data, whereby the most noticeable increases were seen with A3A, A3G, and mA3 packaged into the MLV double mutant. Despite increases in mutation intensities, the effect on viral infection in single-round assays was relatively weak for mA3 and some other A3 proteins (Fig. 7B). Nevertheless, mutated viruses may be damaged in ways not detectable by this flow cytometry assay. Overall, these results imply that capsid stability has a primary role in influencing the susceptibility of M-MLV to A3-mediated deamination across multiple animal species.
Detection of gPr80 in cell supernatant and virus preparations.
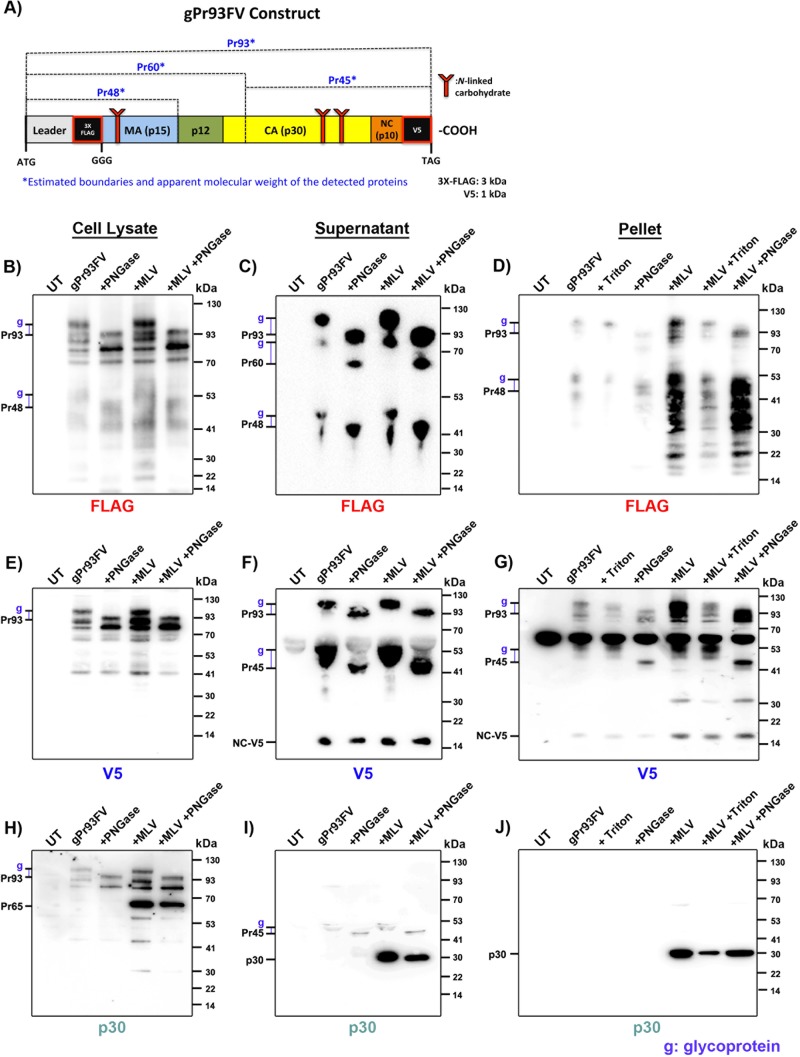
We next investigated how gPr80 glycosylated peptides interact with virions. It has been shown that the N-terminal cleavage product of gPr80 becomes associated with the cellular membrane and buds off with virions, while the C terminus (containing mostly CA and NC) is secreted by the infected cell (9, 11). The N-terminal leader sequence alone, with a functional ER signal and most of its transmembrane domain intact, is insufficient to increase capsid stability (3). This implies that the C terminus is likely responsible for capsid stability, either alone or in conjunction with the N-terminal peptide. However, according to these concepts, it is extremely challenging to explain how the glycosylated secreted C-terminal peptide has an effect on inhibiting deamination or increasing capsid stability if it is not actually present in viruses. Equally, it is unclear how the N-terminal peptide of gPr80 influences capsid stability if it contains mostly the leader, MA, and p12 constituents and is directed toward the luminal side of the viral envelope, as expected for type II integral membrane proteins (Fig. 1A). We generated a new gPr80 expression construct (gPr93FV), in which we changed the Pr65 AUG initiation codon to GGG to prevent its translation by leaky scanning and inserted a sequence coding for a 3× FLAG epitope tag immediately downstream of the leader sequence (Fig. 8A) (9). We also added a V5 epitope tag at the C terminus of the protein. The apparent molecular mass of the unglycosylated form of the protein in our gels is 93 kDa.
FIG 8.
Association of gPr80-derived proteins with viral particles. (A) Schematic representation of the dually tagged FLAG-V5-gPr80 protein (gPr93FV). The estimated positions of the detected peptides are indicated. Western blots were performed on cellular lysates of gPr93FV-transfected 293T cells (B, E, and H), 0.45-μm-filtered supernatants (C, F, and I), and pellets of supernatants ultracentrifuged through a 20% sucrose cushion (D, G, and J). Cotransfections with virus were performed with M-MLV[CTA] (+MLV). UT, untransfected. Samples prepared from cell lysates and supernatants were treated with PNGase F as indicated. Pellet samples were treated with 2% Triton X-100 for 30 min prior to ultracentrifugation as indicated. Tick marks to the left of each gel indicate glycoproteins of interest (g) along with their related unglycosylated forms used to estimate molecular masses. Samples were resolved by SDS-PAGE in triplicate and probed with anti-FLAG (B to D), anti-V5 (E to G), and monoclonal anti-p30 (H to J) antibodies. Data are representative of one experiment for at least three independent transfections.
The gPr93FV expression vector was transfected in the presence of M-MLV[CTA] to complement the absence of virus-encoded gPr80. Cell lysates, supernatants, and supernatants ultracentrifuged through a sucrose cushion (pellet) were analyzed by SDS-PAGE for gPr93 expression products by use of an anti-FLAG (Fig. 8B to D) or anti-V5 (Fig. 8E to G) antibody. Pr65 was detected only in cell lysates containing the viral vector (Fig. 8H). The mature p30 CA protein was detected in both the supernatants and supernatant pellets, also only when the virus was present (Fig. 8I and J). The unglycosylated form of gPr93 appeared as a 93-kDa band in our gels (Pr93) due to the presence of the various tags. Detection of products in the supernatants and pellets when gPr93FV was transfected alone is indicative of virus-like particles (VLPs) and/or extracellular vesicles (EVs) packaging Pr93 or its cleavage products.
A number of glycosylated FLAG-labeled proteins were detected (indicated by “g”), notably Pr93, Pr60, and Pr48, as evidenced by peptide-N-glycosidase F (PNGase F) treatment (Fig. 8A to 8D). Pr48 appears to be an N-terminal peptide that ends near or at the MA p15-p12 junction. Pr60 appears to be the equivalent of the ∼55-kDa membrane-associated peptide of gPr80 (Fig. 1A) (9, 11). These three glycoproteins were clearly detectable in cell supernatants (Fig. 8C), but only Pr93 was clearly visible in virus pellets (Fig. 8D). Triton X-100 was used prior to pelleting of the samples, where indicated, to dissolve EVs and strip the viral envelope. VLPs and virus cores are resistant to Triton X-100 treatment. This procedure ensures that the proteins detected are firmly associated with viral cores. Despite several attempts, multiple FLAG-containing cleavage products prevented clear identification of Pr60 and Pr48 in the virus pellets (Fig. 8D). There were many fewer detected bands containing the V5 tag in the pellets (Fig. 8G).
Full-length glycosylated Pr93 was also detected in cell lysates, supernatants, and pellets stained with anti-V5 (Fig. 8E to G). A large amount of this protein appeared to be associated with the viral envelope (and, to a much lesser extent, EVs and VLPs) in the pellets, as Triton treatment greatly diminished but did not abolish the signal (Fig. 8G). An additional glycosylated protein, of approximately 45 kDa (Pr45), was also detected in supernatants and pellets only (Fig. 8F and G). We believe this protein to be equivalent to the ∼40-kDa secreted cleavage product of gPr80 that has also been reported previously (Fig. 1A) (9, 11). While Pr45 could easily be detected in all supernatants, only a very faint signal could be detected in pellets, thereby indicating that most of this peptide is secreted in soluble form. However, in pelleted samples, the Pr45 peptide did appear to form detergent-resistant complexes, indicating that it may be cargo or a structural part of the capsid (Fig. 8G). Surprisingly, our p30 monoclonal antibody was extremely inefficient at detecting the full-length glycosylated Pr93 protein when the plasmid was transfected alone (Fig. 8H and J).
Glycosylated Gag has an unexpected orientation in the viral envelope.
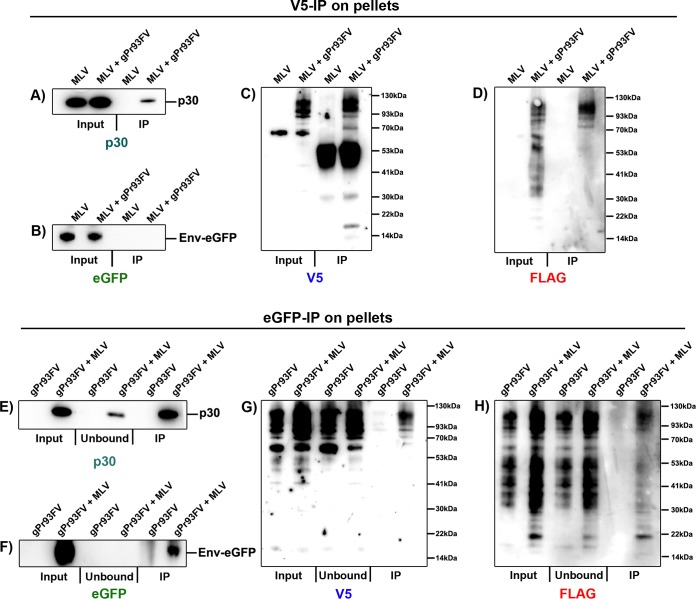
It was revealed in Fig. 8D that full-length glycosylated Pr93 is detectable in pellets. Because viral pellets contain a mixture of virus, EVs, and VLPs that copurify by ultracentrifugation, we sought to determine which Pr93 fragments are specifically virus associated. To achieve this, we produced M-MLV[CTA] viruses in the conditional presence of gPr93FV, as described in the previous section, purified them by velocity sedimentation through a sucrose cushion, and further isolated them by use of antibodies conjugated to magnetic beads directed against surface viral epitopes (V5 and EGFP). Full-length gPr80 was previously characterized as a type II integral membrane protein, and as such, our gPr93FV protein is expected to have its C-terminal V5 tag exposed to the external surface of the viral envelope (9). Additionally, our viruses have an envelope glycoprotein fused with EGFP (Env-EGFP) expressed on their surfaces (41). Recent work from our lab revealed that the M-MLV envelope glycoprotein is nearly undetectable on EVs and thereby constitutes a robust virus selection marker (42).
V5 immunoprecipitation (IP) revealed many of the same V5-tagged proteins and fragments in the pelleted fraction as those described in the previous section (Fig. 9A to D). However, staining of V5-immunoprecipitated particles with anti-FLAG revealed almost exclusively full-length glycosylated Pr93 products (Fig. 9D). Unconjugated magnetic beads had no detectable binding to Pr93 (data not shown). In terms of viral constituents, the p30 capsid was readily detectable, albeit at much lower levels than in the input (Fig. 9A). Most surprisingly, however, Env-EGFP levels in the immunoprecipitated samples were below the limit of detection of the assay (Fig. 9B). This implies that most particles that contain full-length glycosylated Pr93 with an antibody-accessible V5 tag are largely devoid of the viral envelope glycoprotein and therefore constitute VLPs, not infectious virus.
FIG 9.
Glycosylated Gag adopts a type I integral membrane protein conformation in the envelope of viral particles. 293T cells were transfected similarly to those in Fig. 8. Supernatants were pelleted through a 20% sucrose cushion by velocity sedimentation and resuspended in PBS. (A to D) The viral preparations were immunoprecipitated by use of anti-V5-conjugated magnetic beads and washed with PBS. Input fractions (lysates) and anti-V5 IP purified particles were analyzed by SDS-PAGE and probed with p30 (A), EGFP (B), V5 (C), and FLAG (D) antibodies. (E to H) Viral preparations were also immunoprecipitated using anti-EGFP-conjugated magnetic beads. Input (lysates), flowthrough (unbound), and anti-GFP (IP)-purified enveloped particles were analyzed by SDS-PAGE and probed with p30 (E), EGFP (F), V5 (G), and FLAG (H) antibodies. Data are representative of one experiment for three independent transfections.
In light of the data from the V5 IP isolation, we sought to analyze the population of particles that have accessible Env-EGFP on their surfaces (Fig. 9E to H). Confirming the accessibility of envelope glycoprotein and the efficacy of this method, the EGFP blot revealed high levels of Env-EGFP in both the input and IP fractions but none in the unbound fraction (Fig. 9F). This was regardless of the length of the exposure (data not shown). In agreement with the V5 IP results, readily detectable p30 capsid protein was present in the unbound fraction of this isolation (Fig. 9E). Otherwise, as expected, abundant p30 capsid protein was detected in the IP fraction. In the search for Pr93, the FLAG blot revealed low levels of virus-associated full-length glycosylated Pr93 and a range of smaller FLAG-containing cleavage products (Fig. 9H). However, large amounts of full-length Pr93 and FLAG cleavage products remained in the EGFP-unbound fraction. Similarly, the V5 blot also showed low levels of full-length Pr93 associated with the virus and large amounts of it in the unbound fraction (Fig. 9G).
Given that V5+ particles contained undetectable levels of Env-EGFP and that full-length V5 and FLAG products were detected in Env-EGFP+ particles, the data suggest that glycosylated Pr93 is oriented as a type I integral membrane protein (NexoCcyto) in a large population of potentially infectious viruses. The observation that glycosylated Gag can insert itself into the cellular membrane with a type I polarity was previously described by Fujisawa et al., but their study did not report that this phenomenon also occurs in egressed viruses (9). Finally, relatively large amounts of pelletable V5+ VLPs may also underline the discovery of a new mechanism whereby gPr80 induces the release of Env-deficient VLPs with exposed Gag sequences on the surface.
DISCUSSION
Early research on gPr80 dates back to the mid-1970s, when a glycosylated form of MLV Gag was discovered at the surfaces of AKR lymphomas (43, 44). It was soon established that there were in fact two almost identical Gag proteins that were distinguishable only by the presence of glycosylation and by an additional, short N-terminal leader sequence found on the glycosylated form (5, 6, 8). The first functional studies using gPr80-deficient viruses in cell culture systems were somewhat disappointing, as the mutant viruses were replication competent, had normal reverse transcriptase activity, and, most importantly, were nearly as infectious as their wild-type counterparts (12, 13). However, these early infection assays were performed in NIH 3T3 and Mus dunni cells, which are now known to express undetectable levels of the mA3 restriction factor. It is currently understood that one of the important roles of gPr80 is to antagonize restriction and deamination by mA3 (2–4). Looking back at early reports of infection assays in mice, it is clear that gPr80 has an important role in infection, spread, and disease pathogenesis in vivo, as mA3 is naturally expressed in cells targeted by murine retroviruses (14, 16, 17).
Several studies have clearly shown that mA3 restricts retroviruses in vivo and in primary mouse splenocytes (20, 23, 24). The strongest evidence that mA3 exerts selective pressure on retroviruses is the reversion of gPr80-deficient viruses to the wild type in mA3-expressing mice but not mA3 knockout mice (3, 17). However, the impact of deamination by mA3 may not be as prominent as that of deamination by human A3G, which restricts MLVs and HIV mainly through intense hypermutation (1). Most murine retroviruses, such as M-MLV, Friend MLV, and mouse mammary tumor virus (MMTV), generally show no or very few signs of hypermutation; AKV MLV, on the other hand, shows clear but very modest levels of mA3-induced mutations (20, 23, 24). Nevertheless, even a mild increase in MLV susceptibility to deamination by mA3 would likely contribute to extinguishing the virus infection over time, as MLVs do not encode an A3 antagonist such as HIV Vif. Although we found only a small decrease in infectivity as the mutation frequency increased by altering the asparagines at positions 113 and 505 in our assays (Fig. 7B), it is possible that the true effects of these mutations on viral replicative fitness may not have been observed under our experimental conditions. Single-round infection assays, such as those used in this part of our study, are unable to reveal if overall virus replicative fitness and viral spreading are compromised by A3 mutations. Mutations in other parts of the virus may be deleterious, for example, if critical residues of the catalytic centers of the integrase or reverse transcriptase are mutated. If this were the case, these in vitro assays would not immediately detect these perturbations.
In light of our previous observation that the number of N-linked glycans directly affects MLV sensitivity to mA3 deamination (4), we were curious to better understand the mechanism behind this phenomenon. It has now been ruled out that gPr80 negatively affects virion packaging or core localization of mA3 (Fig. 2) (2). We also asked whether gPr80 or its sugars could interfere with the catalytic activity of mA3. We found no evidence that catalytic activity was affected, regardless of the mA3-to-gPr80 ratio, in an oligonucleotide cleavage assay with purified proteins (Fig. 6). In fact, we found no evidence that mA3 and gPr80 naturally interact inside the cytosol (Fig. 5). mA3 is selectively packaged into MLV virions through an interaction with the MLV Gag protein. Lack of detectable protein interactions in the soluble fraction of cytosolic extracts may be indicative that gPr80 is maintained in an isolated cellular compartment up until the moment it reaches the cellular membrane, when it is recruited into assembling viral particles. In support of this, recent work has shown that gPr80 is produced in the rough endoplasmic reticulum and traffics through the Golgi apparatus to the cell surface (29). Given that neither N-linked glycans nor the gPr80 protein itself affects the catalytic activity of mA3 in a test tube assay and that mutations at glycosylation sites on both the N terminus and the C terminus of gPr80 influence deamination intensity (4), we proceeded to evaluate the role of capsid stability in more detail. The absence of gPr80 expression had an important impact on capsid stability, as did mutations at all three glycosylation residues (Fig. 2 and 3). Decreased capsid stability is the common feature of all MLVs displaying increased sensitivity to A3 deamination. What has not been resolved fully is the importance of N-linked glycosylation in preventing this mutator activity. Isolated disruption of glycosylation by use of inhibitors or GnTI-deficient cells appeared to have no impact on deamination intensity and/or capsid stability. If glycosylation does play a role in preventing A3 deamination in some manner, it does so in concert with decreased capsid stability.
Along the same lines, the improved capsid stability provided by gPr80 has been shown to prevent the premature decapsidation and cytosolic exposure of replication intermediates that could trigger innate immune responses (3, 19). Until now, there was only evidence that the ∼55-kDa N-terminal cleavage product of gPr80 could be detected in viral particles (2, 11). Furthermore, a previous study showed that expression of the leader sequence of gPr80 alone did not rescue diminished capsid stability (3). The mystery of how gPr80 increases capsid stability therefore remains. Our efforts to detect gPr80 peptides associated with viral particles have revealed several new pieces of information (Fig. 8 and 9). By expressing a dually tagged construct along with gPr80-deficient M-MLV, we were able to use high-affinity antibodies to detect interactions that have not been characterized before. We clearly showed that full-length gPr80 (i.e., gPr93) and several of its C- and N-terminal-domain peptides are associated with viruses, and more specifically with detergent-resistant viral cores. Additionally, full-length gPr80 was also detected in cell supernatants and in pellets by use of antibodies against both V5 and FLAG epitope tags (Fig. 8). However, most gPr80 appeared to be part of the envelopes of viruses, VLPs, and EVs, as the signal was greatly diminished upon detergent treatment. More in-depth dissection of this issue by use of antibodies to capture external epitopes revealed that nearly all full-length gPr80 is present as a type I integral membrane protein in the envelope of infectious virus and as a type II protein in Env-deficient VLPs (Fig. 9). However, this does not exclude the possibility that some infectious viruses harbor the previously observed ∼55-kDa N-terminal peptide of gPr80 as a type II membrane protein (9, 11). In fact, there is evidence that this fragment exists in our EGFP pulldowns stained with anti-FLAG; however, the abundance of FLAG-containing cleavage products makes it difficult to clearly resolve it (Fig. 9H). Importantly, however, a demonstration that complementation of M-MLV[CTA] with gPr93FV restores capsid stability and deamination resistance will be essential to lend support to the physiological relevance of the observations made using the current laboratory system. Additionally, a formal demonstration that gPr80 also adopts such an unusual topology on the surface of the replicative wild-type virus is essential.
While the absence of the envelope glycoprotein on VLPs implies that these particles are noninfectious, it is entirely possible that they are still endocytosed, given the conserved YXXL motif present in the leader sequence of gPr80 (45). However, unlike Env-EGFP-containing particles, they will not be able to escape the endosome. The presence of these VLPs stuck in endosomes may explain the results observed with our fate-of-capsid assay. While it normalized to the same level of capsid input, gPr80-deficient M-MLV had noticeably less endocytosed intact capsid than that in the wild-type virus (Fig. 3). Prior to this finding, the equal infectivities of these two viruses (Fig. 1C and E) were somewhat perplexing. Now it is clear that an endocytosed intact virion or VLP may not be indicative of a productive infection when gPr80 is present. While this applies to our ecotropic MLV, it may vary with other amphotropic or xenotropic strains (28, 29). This begs the question as to whether VLPs (and EVs) that express gPr80 as a type II membrane protein influence adaptive and humoral immune responses in mice.
Given the very low levels of envelope glycoprotein on these VLPs, the role of gPr80 in antagonizing SERINC3 and SERINC5 incorporation into virions comes to mind. Recently, in a fashion similar to that for HIV-1 Nef, gPr80 was shown to antagonize the effects of newly identified restriction factors, known as SERINC3 and SERINC5, by preventing their incorporation into virions (29, 46–48). In fact, a Nef-deficient HIV-1 strain has its infectivity fully restored when complemented with gPr80 if SERINC3 or -5 is present (29, 46, 47). However, it is clear that the roles of gPr80 and Nef do not fully overlap, as Nef complementation does not rescue infectivity of gPr80-deficient MLV under some conditions (28). But given that the Nef-like effect of gPr80 is mediated by its cytoplasmic N-terminal domain, dependent on the presence of the YXXL motif (45) and the type of viral envelope glycoprotein present (29, 49), it is tempting to speculate that VLPs may serve as decoys for the incorporation of SERINC proteins, sequestering them away from infectious virus. In all regards, further work should explore this new and intriguing avenue.
Overall our findings provide added insight into the mechanism behind gPr80 protection of MLV from host restriction. However, our understanding of gPr80 is not yet complete. In vivo or ex vivo evidence of the gPr80 membrane orientation and VLP release must be obtained. Additionally, if gPr80 is associated with EVs and VLPs alike, what are their impacts on host immune responses? Moreover, the relationships between Env, Nef, gPr80, and SERINC3 and -5 are still largely unresolved.
MATERIALS AND METHODS
Cells.
Human embryonic kidney epithelium cells (293T), 293S GnTI− cells (ATCC CRL-3022), and mouse embryonic fibroblasts (NIH 3T3) were cultured in Dulbecco's modified Eagle's medium (DMEM)/high-glucose medium (Wisent) supplemented with 10% fetal bovine serum (FBS) (Gibco), 100 U/ml penicillin, and 100 μg/ml streptomycin (Multicell; Wisent Inc., Canada) and propagated at 37°C in a 5% CO2 incubator. 293S GnTI− cells lack N-acetylglucosaminyltransferase I (GnTI) activity and therefore lack complex N-glycans.
Expression plasmids and viruses.
The pMOV-eGFP expression vector encoding replicative M-MLV has been described before (23, 41). Expression vectors for the FLAG-tagged C57BL/6 allele of mouse APOBEC3 delta exon 5 (referred to here as FLAG-mA3), human APOBEC2 (A2), human APOBEC3C (A3C), human APOBEC3F (A3F), and human APOBEC3G (A3G) have been described before (23, 50). The DNA coding sequences of human APOBEC3A (A3A), human APOBEC3B (A3B), and human APOBEC3H (A3H) were amplified from mRNAs of human peripheral blood mononuclear cells (PBMCs). Products were then cloned downstream of a FLAG epitope tag in the pcDNA3.1 expression vector. The cDNA for human APOBEC3D (A3D) was provided by the NIH AIDS Research and Reference Reagent Program in a pcDNA3.1 backbone. A FLAG epitope tag was inserted upstream of the A3D coding sequence.
M-MLV[N480Q], M-MLV[N113Q/N505Q], M-MLV[N113Q/N480Q/N505Q], and M-MLV[CTA] were described in a previous report by our group (4). For cloning of recombinant gPr80 (wild type) and Pr80[N113Q/N480Q/N505Q], the cDNA sequences were amplified by PCR from the corresponding M-MLV expression vectors. The forward primer contained a Kozak sequence and the ATG initiation codon instead of the nonconventional CTG initiation codon. The reverse primer contained the cDNA for the V5 epitope tag inserted before the stop codon at the 3′ end of the coding sequence. The sequences of the primers were as follows: MoMLVPr80-EcoRI-ATG-FWD, 5′-ACAAGAATTCGCCACCATGGGAGACGTCC-3′; and MoMLVpr80-V5-BamHI-REV, 5′-GTGTGGATCCCTACGTAGAATCGAGACCGAGGAGAGGGTTAGGGATAGGCTTACCGTCATCTAGGGTC-3′. The PCR amplicon was then directionally cloned using EcoRI and BamHI restriction sites into the pcDNA3.1 expression vector (Invitrogen). Constructs were sequenced for accuracy.
To assess the incorporation of gPr80 into M-MLV[CTA] and distinguish the N-terminal and C-terminal fragments, we developed a new construct. As outlined for the primers below, a 3× FLAG tag was added just prior to the traditional ATG codon for the Pr65 protein, which was replaced with GGG. Briefly, the gPr80 leader sequence was amplified with a primer within the CMV promoter and cloned using the NheI site in the 5′ multiple-cloning site (MCS). This region already contained a Kozak sequence and ATG, as described above. The reverse primer bound just before the Pr65 start codon and added SacII and AgeI sites after the leader sequence. A 3× FLAG tag was developed by filling in with the two primers described below. These primers had SacII and AgeI sites at the 5′ and 3′ ends, respectively. Finally, the Pr65 coding sequence was amplified using a forward primer with an AgeI site at the 5′ end and a GGG codon instead of ATG, while the reverse primer sequence was virtually unchanged from that of the parental V5-tagged gPr80 vector. The leader sequence was cloned into the pcDNA3.1 backbone first, and then the 3× FLAG tag was inserted by using the SacII and AgeI sites. The Pr65 sequence was inserted into a separate pcDNA3.1 backbone, and the leader sequence with the FLAG tag was inserted into the 5′ end of this vector by using the NheI and AgeI sites. The primers used were as follows: leader sequence SacII AgeI REV, CATACCGGTCCGCGGATTTTCAGACAAATACAGAAACACAGTCAGAC; SacII 3× FLAG FWD, ATTCCGCGGGATTACAAGGACCATGACGGAGACTACAAAGATCACGACA; 3× FLAG AgeI REV, CATACCGGTGCCCTTGTCATCGTCGTCCTTGTAGTCGATGTCGTGATCT; AgeI Pr65 FWD, GGGACCGGTGGGGGCCAGACTGTTACCACTCCC; and V5 SacII REV, CTAGGTACCCTACGTAGAATCGAGACCGAGGAGAGG. The vector sequence was confirmed by Sanger sequencing.
Western blotting and antibodies.
Western blot analyses were conducted following the procedures detailed by Gerpe et al. (4). Samples presented in Fig. 8 and 9 were run in 4 to 12% Bis-Tris NuPAGE gels with morpholinepropanesulfonic acid (MOPS)-SDS running buffer (Thermo Fisher). The following antibodies were used for this study: horseradish peroxidase (HRP)-conjugated anti-FLAG (Sigma), anti-EGFP (Clontech), HRP-conjugated anti-mouse IgG (Cell Signaling), HRP-conjugated anti-rabbit IgG (Abcam), HRP-conjugated anti-rat IgG (Sigma), polyclonal anti-V5 (EMD Millipore), monoclonal anti-V5, and HRP-conjugated anti-β-tubulin (Abcam). The polyclonal anti-p30 antibody was kindly provided by Hung Fan (University of California, Irvine). The rat anti-p30 monoclonal antibody R187 was purified from a B cell hybridoma (CRL-1912; ATCC).
Transfections, infections, and p30 ELISA.
Transfection and viral infectivity assays were performed as previously described (4). Briefly, 293T or 293S GnTI− cells were seeded at 3.0 × 105 cells per well in a 6-well plate or at 1.0 × 106 cells per 10-cm dish 24 h prior to transfection. Cotransfections in 6-well plates were performed with 800 ng of viral expression vector and various amounts (50 ng to 250 ng) of A3 expression plasmids by use of GeneJuice transfection reagent (Novagen, EMD Millipore) according to the manufacturer's instructions. For work in 10-cm dishes, 10 μg of viral expression vector and various amounts of A3 expression plasmids were cotransfected using CaCl2. Cells were then cultured for 2 days before viruses were harvested.
One day prior to infection, NIH 3T3 target cells were seeded at 1.0 × 105 cells per well in 12-well plates. At the time of infection, the medium was replaced with fresh medium containing Polybrene at a final concentration of 8 μg/ml, and virus-containing supernatants from 293T producer cells were harvested, cleared by centrifugation, and passed through 0.45-μm filters. Depending on the assay, target cells were infected with equal volumes of virus-containing supernatant, or the amount of p30 capsid protein in each sample was determined by p30 ELISA (QuickTiter ELISA kit; Cell Biolabs Inc.) and normalized viral supernatants used to infect NIH 3T3 target cells by spinoculation. At 48 h postinfection, the infected cells were partitioned for flow cytometry analysis of EGFP reporter gene expression and for genomic DNA (gDNA) extraction for subsequent mutation analysis.
To calculate the viral titer in transducing units (TU) per milliliter, equal volumes of virus-containing 293T cell supernatant were titrated on 1.0 × 105 NIH 3T3 cells per well in a 12-well plate. At 24 h postinfection, these cells were analyzed by flow cytometry. Infections ranging from 2 to 30% were assumed to have one productive integration per cell and used to calculated the number of TU per milliliter with the following formula: TU/ml = number of infected cells/volume of viral supernatant.
Genome measurements.
Viral genomes were extracted from supernatants by use of a QIAamp Viral RNA minikit (Qiagen) following the manufacturer's guidelines, with one exception. Just prior to washing, the dried silica column was incubated with 50 μl of solution containing 100 U of DNase I (RNase free) and 1× reaction buffer (New England BioLabs). This was allowed to sit for 30 min at room temperature prior to washing of the column to completely remove the transfected plasmid DNA. The RNA eluate was then reverse transcribed and analyzed by droplet digital PCR, using a QX200 system (Bio-Rad), for the presence of the GFP coding sequence. The primers used for this analysis were R279-FWD and R279-REV, as previously described (51). Data were analyzed using QuantaSoft and extrapolated based on the dilutions used for the assay.
Viral core stability assay.
Chronically infected NIH 3T3 cells were seeded in 10-cm plates and used to produce M-MLV, M-MLV[N480Q], M-MLV[N113Q/N505Q], M-MLV[N113Q/N480Q/N505Q], or M-MLV[CTA] over a period of 96 h (4). Supernatants were then collected and filtered through 0.45-μm cartridge filters prior to treatment for 20 min at room temperature with 10% Triton X-100 to strip viral envelopes. Samples were then layered on top of a sucrose step gradient. The step gradient consisted of 5 ml of 20% (wt/vol) sucrose–phosphate-buffered saline (PBS) overlaid with 2 ml of 5% (wt/vol) sucrose-PBS containing 2% Triton X-100 or 0.2% SDS. Gradients were centrifuged for 2 h at 100,000 × g in a 70Ti rotor, and viral cores were resuspended in 100 μl of RIPA lysis buffer (150 mM NaCl, 1% NP-40, 0.2% SDS, 1 mM EDTA, 0.5% sodium deoxycholate, 50 mM Tris-HCl, pH 8.0) supplemented with Complete EDTA-free protease inhibitor cocktail (Roche) for 20 min on ice. The samples were then mixed with Laemmli loading buffer, boiled, and analyzed by Western blotting for the presence of envelope and p30 capsid proteins, using anti-EGFP and monoclonal anti-p30 antibodies, respectively. For determination of the impact of glycosylation on stability, the same was done with chronically infected NIH 3T3 cells for M-MLV in the presence of tunicamycin (Sigma) at a concentration of 0.2 μg/ml. However, in this experiment, 3 × 106 cells were seeded in 10-cm dishes, incubated for 6 h, and washed 3 times with PBS, and then the medium was refreshed with the conditional presence of tunicamycin. The tunicamycin concentration was chosen to allow for virus production for approximately 36 h, as increased exposure or longer incubations had severe impacts on cellular viability. Alternatively, 293T or 293S GnTI− cells were seeded in 10-cm plates and transfected with M-MLV, and the supernatants were treated as described above.
mA3 core packaging assay.
M-MLV or mutant virus plasmids (10 μg) and 1 μg of mA3 expression plasmid were cotransfected into 293T cells. Three days after transfection, virus-containing supernatants were filtered through 0.45-μm cartridge filters and layered on top of a sucrose step gradient. The step gradient consisted of 5 ml of 20% (wt/vol) sucrose-PBS overlaid with 2 ml of 5% (wt/vol) sucrose-PBS with 0, 2, 5, or 10% Triton X-100. Gradients were centrifuged and processed as described above.
A3-gPr80 interaction assay.
293T cells in 10-cm dishes were cotransfected with 5 μg of FLAG-tagged A3G or mA3 and 20 μg of gPr80-V5 or Pr80[3Q]-V5 (Pr80[N113Q/N480Q/N505Q]-V5) and cultured for 72 h. Three days after transfection, complexes were purified using anti-FLAG-conjugated agarose beads. Following incubation, bound complexes were spun down at 8,200 × g for 30 s at 4°C, followed by four washes with 200 μl of wash buffer 1 (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 1% Igepal CA-630, 0.5% sodium deoxycholate, 0.1% SDS). A last wash in 100 μl of wash buffer 2 (20 mM Tris-HCl [pH 7.4]) was then performed, and the samples were resuspended in 40 μl of 5× Laemmli loading buffer. Protein-protein interactions were then determined by Western blotting using polyclonal anti-p30 and anti-FLAG antibodies.
Purification of FLAG-tagged A3 proteins.
293T cells transfected with 1 μg of FLAG-tagged mA3 or A3G expression plasmid were lysed with M2 (Sigma) affinity gel lysis buffer for 30 min on ice. Following lysis, samples were cleared by centrifugation at 12,000 × g for 10 min at 4°C and mixed with 40 μl of anti-FLAG-conjugated agarose beads (Sigma). Mixtures were rotated for 3 h at 4°C, followed by two washes with cold 1× PBS prior to the elution step. Elution was carried out by incubating the beads twice with 50 μl of 0.1 M glycine, pH 3.5, for 5 min, and eluates were collected in fresh tubes containing 3 μl of 1 M Tris-HCl, pH 8.0. The protein concentrations in immunoprecipitated samples were determined by Bradford assay (Sigma).
Purification of gPr80-V5 and Pr80[3Q]-V5.
Two million 293T cells were seeded in a 10-cm dish and cultured in decomplemented medium the day prior to transfection with 20 μg of gPr80-V5-pcDNA or Pr80[3Q]-V5-pcDNA plasmid. The cells were harvested 96 h later and washed four times with 1× PBS. The cells were then resuspended in 0.8 ml of M2 affinity gel lysis buffer containing 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1 mM EDTA, and 1% Triton X-100. Lysates were then homogenized by enforced passage through a 19-gauge needle and rotated at 4°C for 30 min. Lysates were cleared by centrifugation at 12,000 × g for 10 min prior to addition of anti-V5-conjugated agarose beads (ab1229; Abcam, Cambridge, United Kingdom). Stock anti-V5 beads were first spun down at 3,000 × g for 2 min to remove glycerol and then washed with 1× TBS (50 mM Tris-HCl with 150 mM NaCl, pH 7.4). The anti-V5 beads (20 μl) were then added to the lysates and rotated overnight at 4°C. The samples were then spun down at 3,000 × g for 2 min and washed with 1× TBS before proceeding to the elution step. Elution was carried out three times with 30 μl 0.1 M triethylamine HCl, pH 11.5, for 10 min each. Samples containing the eluted proteins were spun at 3,000 × g for 2 min after each 10 min of incubation. Eluates were pooled and added to a tube containing 6 μl of 1 M MOPS-HCl, pH 3.0. Before loading into the gel, 30 μl of each sample was resuspended in 5× Laemmli buffer and boiled for 5 min.
Oligonucleotide cleavage.
Various amounts of purified FLAG-A3 proteins, with or without purified gPr80-V5 or Pr80[3Q]-V5, were incubated with 0.1 pmol/μl of a 6-carboxyfluorescein (FAM)-labeled oligonucleotide with TTC or CCC target sites in the presence of 50 μg/ml of RNase A in a final volume of 100 μl for 5 h at 37°C, followed by incubation for 1 h at 37°C in the presence of 5 U of uracil DNA glycosylase (UDG). Both oligonucleotides used as substrates for mA3 and A3G have been described previously (21, 40). Reactions were terminated by the addition of 60 μl of formamide loading dye (95% deionized formamide, 0.025% bromophenol blue, 0.025% xylene cyanol FF, 5 mM EDTA) containing 0.2 M NaOH and heating at 95°C for 20 min. Samples were then resolved in 15% Tris-borate-EDTA (TBE)–urea polyacrylamide gels and visualized using a Typhoon imaging system (GE Healthcare). Quantification analyses were performed using ImageJ software.
3D-PCR analysis and DNA sequencing.
3D-PCR analysis was conducted as detailed by Gerpe et al. (4). In brief, first-round PCR was performed on 10 ng of genomic DNA from infected NIH 3T3 target cells in a two-step protocol using PrimeSTAR high-fidelity polymerase (TaKaRa) and primers eGFP-FWD (5′-CGAGGAGCTGTTCACA-3′) and eGFP-REV (5′-CAGCTCGTCCATGCCGAGAGTGAT-3′). PCR conditions were 98°C for 1 min followed by 32 cycles of denaturation at 98°C for 10 s, annealing at 58°C for 5 s, and extension at 72°C for 1 min, with a final extension at 72°C for 2 min. Two microliters of a 1:1,000 dilution of the first-round PCR products was used per reaction mixture to perform a second-round gradient PCR using previously described primers (R279-FWD and R279-REV) (51). The PCR denaturation gradient was from 89.5°C to 86.0°C. Products were resolved in 2% agarose gels. Proviral DNA integrated into NIH 3T3 cells was isolated, amplified, and cloned as described elsewhere (52). The region amplified corresponds to the 717-bp coding sequence of the EGFP reporter gene. Cloned DNA was sequenced.
Fate-of-capsid assay.
NIH 3T3 producer cells produced virus as described earlier for 96 h. Supernatants were collected, filtered through 0.45-μm filters, and centrifuged through a 20% sucrose cushion to clear free protein. Virus was resuspended in fresh complete medium, and the capsid level was normalized by ELISA. This assay was performed similarly to the methods of previous groups (3, 30–33), with some slight modifications. Briefly, 2.5 × 105 uninfected NIH 3T3 cells were seeded in 6-well plates 1 day prior to infection. One plate each was dedicated to M-MLV, M-MLV[N113Q/N480Q], M-MLV[N113Q/N480Q/N505Q], and M-MLV[CTA]. Approximately 1 × 108 TU of M-MLV was used to infect each well in the presence of 8 μg/ml Polybrene; equivalent levels of p30 were used for all viruses. The plates were subjected to spinoculation at 1,600 × g for 30 min in a precooled centrifuge at 4°C. The cells were then incubated at 37°C in the presence of 5% CO2 for 3 h. Following incubation, the cells were washed 3 times with ice-cold PBS, followed by treatment with 1 ml pronase (7 mg/ml) for 5 min at room temperature. The cells from each plate were pooled and then washed 3 times with ice-cold PBS by centrifugation. The pellets were lysed with 2.5 ml hypotonic lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM KCl, and 1 mM EDTA) supplemented with protease inhibitors and incubated on ice for 15 min. A Dounce homogenizer (7 ml) was used with the B pestle to continue lysis for 15 strokes. Cellular debris was cleared by centrifugation at 1,600 × g at 4°C. A 100-μl aliquot of cleared lysate was used for “input” analysis by Western blotting. The remainder was diluted in cold PBS and centrifuged at 125,000 × g for 2 h at 4°C through a 40% sucrose cushion in a 70Ti rotor. The pellet was then analyzed by Western blotting. For the pellet, the signal was enhanced through the use of a sensitive detection reagent (Thermo Fisher).
Intact virion immunoprecipitations.
For anti-V5 IPs, M-270 epoxy Dynabeads (Thermo Fisher) were covalently conjugated to a monoclonal (Thermo Fisher) or polyclonal (Millipore) V5 antibody according to the manufacturer's instructions. For anti-EGFP IPs, a μMACS GFP isolation kit (Miltenyi) was used according to the manufacturer's instructions. 293T cells were seeded and transfected as described above and in accordance with the parameters outlined in Fig. 9. The supernatants from these cells were filtered through a 0.45-μm syringe filter and ultracentrifuged through a 20% sucrose cushion prior to initiation of immunoprecipitation. Antibody-conjugated beads were then incubated with virions for 3 h at 4°C with constant rotation. The magnetic beads were then washed with 5 ml of PBS before analysis by SDS-PAGE. As with the gels shown in Fig. 8, these IPs were run in NuPAGE 4 to 12% gradient gels.
Sequencing.
All the DNA sequencing in this study was performed at the McGill University and Génome Québec Innovation Centre, Montréal, Canada.
ACKNOWLEDGMENTS
M.-A.L. holds a Canada Research Chair in Molecular Virology and Intrinsic Immunity.
This research was supported by a grant from the Canadian Institutes of Health Research (grant 89774) and an Early Researcher Award from the Ontario Ministry of Research and Innovation to M.-A.L. T.M.R. holds a QEII Graduate Scholarship of Ontario.
REFERENCES
- 1.Harris RS, Dudley JP. 2015. APOBECs and virus restriction. Virology 479–480:131–145. doi: 10.1016/j.virol.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kolokithas A, Rosenke K, Malik F, Hendrick D, Swanson L, Santiago ML, Portis JL, Hasenkrug KJ, Evans LH. 2010. The glycosylated Gag protein of a murine leukemia virus inhibits the antiretroviral function of APOBEC3. J Virol 84:10933–10936. doi: 10.1128/JVI.01023-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stavrou S, Nitta T, Kotla S, Ha D, Nagashima K, Rein AR, Fan H, Ross SR. 2013. Murine leukemia virus glycosylated Gag blocks apolipoprotein B editing complex 3 and cytosolic sensor access to the reverse transcription complex. Proc Natl Acad Sci U S A 110:9078–9083. doi: 10.1073/pnas.1217399110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerpe MCR, Renner TM, Belanger K, Lam C, Aydin H, Langlois MA. 2015. N-linked glycosylation protects gammaretroviruses against deamination by APOBEC3 proteins. J Virol 89:2342–2357. doi: 10.1128/JVI.03330-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edwards SA, Fan H. 1979. Gag-related polyproteins of Moloney murine leukemia virus: evidence for independent synthesis of glycosylated and unglycosylated forms. J Virol 30:551–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Evans LH, Dresler S, Kabat D. 1977. Synthesis and glycosylation of polyprotein precursors to the internal core proteins of Friend murine leukemia virus. J Virol 24:865–874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prats AC, De Billy G, Wang P, Darlix JL. 1989. CUG initiation codon used for the synthesis of a cell surface antigen coded by the murine leukemia virus. J Mol Biol 205:363–372. doi: 10.1016/0022-2836(89)90347-1. [DOI] [PubMed] [Google Scholar]
- 8.Schultz AM, Rabin EH, Oroszlan S. 1979. Post-translational modification of Rauscher leukemia virus precursor polyproteins encoded by the gag gene. J Virol 30:255–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fujisawa R, McAtee FJ, Zirbel JH, Portis JL. 1997. Characterization of glycosylated Gag expressed by a neurovirulent murine leukemia virus: identification of differences in processing in vitro and in vivo. J Virol 71:5355–5360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pillemer EA, Kooistra DA, Witte ON, Weissman IL. 1986. Monoclonal antibody to the amino-terminal L sequence of murine leukemia virus glycosylated Gag polyproteins demonstrates their unusual orientation in the cell membrane. J Virol 57:413–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fujisawa R, McAtee FJ, Favara C, Hayes SF, Portis JL. 2001. N-terminal cleavage fragment of glycosylated Gag is incorporated into murine oncornavirus particles. J Virol 75:11239–11243. doi: 10.1128/JVI.75.22.11239-11243.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schwartzberg P, Colicelli J, Goff SP. 1983. Deletion mutants of Moloney murine leukemia virus which lack glycosylated Gag protein are replication competent. J Virol 46:538–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan H, Chute H, Chao E, Feuerman M. 1983. Construction and characterization of Moloney murine leukemia virus mutants unable to synthesize glycosylated gag polyprotein. Proc Natl Acad Sci U S A 80:5965–5969. doi: 10.1073/pnas.80.19.5965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Low A, Datta S, Kuznetsov Y, Jahid S, Kothari N, McPherson A, Fan H. 2007. Mutation in the glycosylated Gag protein of murine leukemia virus results in reduced in vivo infectivity and a novel defect in viral budding or release. J Virol 81:3685–3692. doi: 10.1128/JVI.01538-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nitta T, Tam R, Kim JW, Fan H. 2011. The cellular protein La functions in enhancement of virus release through lipid rafts facilitated by murine leukemia virus glycosylated Gag. mBio 2:e00341-. doi: 10.1128/mBio.00341-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corbin A, Prats AC, Darlix JL, Sitbon M. 1994. A nonstructural gag-encoded glycoprotein precursor is necessary for efficient spreading and pathogenesis of murine leukemia viruses. J Virol 68:3857–3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chun R, Fan H. 1994. Recovery of glycosylated gag virus from mice infected with a glycosylated gag-negative mutant of Moloney murine leukemia virus. J Biomed Sci 1:218–223. [DOI] [PubMed] [Google Scholar]
- 18.Portis JL, Fujisawa R, McAtee FJ. 1996. The glycosylated gag protein of MuLV is a determinant of neuroinvasiveness: analysis of second site revertants of a mutant MuLV lacking expression of this protein. Virology 226:384–392. doi: 10.1006/viro.1996.0666. [DOI] [PubMed] [Google Scholar]
- 19.Stavrou S, Blouch K, Kotla S, Bass A, Ross SR. 2015. Nucleic acid recognition orchestrates the anti-viral response to retroviruses. Cell Host Microbe 17:478–488. doi: 10.1016/j.chom.2015.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. 2007. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature 445:927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 21.Nair S, Sanchez-Martinez S, Ji X, Rein A. 2014. Biochemical and biological studies of mouse APOBEC3. J Virol 88:3850–3860. doi: 10.1128/JVI.03456-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Browne EP, Littman DR. 2008. Species-specific restriction of APOBEC3-mediated hypermutation. J Virol 82:1305–1313. doi: 10.1128/JVI.01371-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langlois M-A, Kemmerich K, Rada C, Neuberger MS. 2009. The AKV murine leukemia virus is restricted and hypermutated by mouse APOBEC3. J Virol 83:11550–11559. doi: 10.1128/JVI.01430-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Takeda E, Tsuji-Kawahara S, Sakamoto M, Langlois MA, Neuberger MS, Rada C, Miyazawa M. 2008. Mouse APOBEC3 restricts Friend leukemia virus infection and pathogenesis in vivo. J Virol 82:10998–11008. doi: 10.1128/JVI.01311-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, Watt IN, Neuberger MS, Malim MH. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809. doi: 10.1016/S0092-8674(03)00423-9. [DOI] [PubMed] [Google Scholar]
- 26.Mariani R, Chen D, Schrofelbauer B, Navarro F, Konig R, Bollman B, Munk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 27.Rulli SJ Jr, Mirro J, Hill SA, Lloyd P, Gorelick RJ, Coffin JM, Derse D, Rein A. 2008. Interactions of murine APOBEC3 and human APOBEC3G with murine leukemia viruses. J Virol 82:6566–6575. doi: 10.1128/JVI.01357-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pizzato M. 2010. MLV glycosylated-Gag is an infectivity factor that rescues Nef-deficient HIV-1. Proc Natl Acad Sci U S A 107:9364–9369. doi: 10.1073/pnas.1001554107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ahi YS, Zhang S, Thappeta Y, Denman A, Feizpour A, Gummuluru S, Reinhard B, Muriaux D, Fivash MJ, Rein A. 2016. Functional interplay between murine leukemia virus glycogag, Serinc5, and surface glycoprotein governs virus entry, with opposite effects on gammaretroviral and Ebolavirus glycoproteins. mBio 7:e01985-. doi: 10.1128/mBio.01985-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang Y, Luban J, Diaz-Griffero F. 2014. The fate of HIV-1 capsid: a biochemical assay for HIV-1 uncoating. Methods Mol Biol 1087:29–36. doi: 10.1007/978-1-62703-670-2_3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Perron MJ, Stremlau M, Lee M, Javanbakht H, Song B, Sodroski J. 2007. The human TRIM5alpha restriction factor mediates accelerated uncoating of the N-tropic murine leukemia virus capsid. J Virol 81:2138–2148. doi: 10.1128/JVI.02318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wight DJ, Boucherit VC, Wanaguru M, Elis E, Hirst EM, Li W, Ehrlich M, Bacharach E, Bishop KN. 2014. The N-terminus of murine leukaemia virus p12 protein is required for mature core stability. PLoS Pathog 10:e1004474. doi: 10.1371/journal.ppat.1004474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stremlau M, Perron M, Lee M, Li Y, Song B, Javanbakht H, Diaz-Griffero F, Anderson DJ, Sundquist WI, Sodroski J. 2006. Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5alpha restriction factor. Proc Natl Acad Sci U S A 103:5514–5519. doi: 10.1073/pnas.0509996103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alce TM, Popik W. 2004. APOBEC3G is incorporated into virus-like particles by a direct interaction with HIV-1 Gag nucleocapsid protein. J Biol Chem 279:34083–34086. doi: 10.1074/jbc.C400235200. [DOI] [PubMed] [Google Scholar]
- 35.Cen S, Guo F, Niu M, Saadatmand J, Deflassieux J, Kleiman L. 2004. The interaction between HIV-1 Gag and APOBEC3G. J Biol Chem 279:33177–33184. doi: 10.1074/jbc.M402062200. [DOI] [PubMed] [Google Scholar]
- 36.Luo K, Liu B, Xiao Z, Yu Y, Yu X, Gorelick R, Yu XF. 2004. Amino-terminal region of the human immunodeficiency virus type 1 nucleocapsid is required for human APOBEC3G packaging. J Virol 78:11841–11852. doi: 10.1128/JVI.78.21.11841-11852.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schafer A, Bogerd HP, Cullen BR. 2004. Specific packaging of APOBEC3G into HIV-1 virions is mediated by the nucleocapsid domain of the gag polyprotein precursor. Virology 328:163–168. doi: 10.1016/j.virol.2004.08.006. [DOI] [PubMed] [Google Scholar]
- 38.Zennou V, Perez-Caballero D, Gottlinger H, Bieniasz PD. 2004. APOBEC3G incorporation into human immunodeficiency virus type 1 particles. J Virol 78:12058–12061. doi: 10.1128/JVI.78.21.12058-12061.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang L, Li X, Ma J, Yu L, Jiang J, Cen S. 2008. The incorporation of APOBEC3 proteins into murine leukemia viruses. Virology 378:69–78. doi: 10.1016/j.virol.2008.05.006. [DOI] [PubMed] [Google Scholar]
- 40.Rausch JW, Chelico L, Goodman MF, Le Grice SF. 2009. Dissecting APOBEC3G substrate specificity by nucleoside analog interference. J Biol Chem 284:7047–7058. doi: 10.1074/jbc.M807258200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sliva K, Erlwein O, Bittner A, Schnierle BS. 2004. Murine leukemia virus (MLV) replication monitored with fluorescent proteins. Virol J 1:14. doi: 10.1186/1743-422X-1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tang VA, Renner TM, Fritzsche AK, Burger D, Langlois MA. 2017. Single-particle discrimination of retroviruses from extracellular vesicles by nanoscale flow cytometry. Sci Rep 7:17769. doi: 10.1038/s41598-017-18227-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ledbetter J, Nowinski RC, Emery S. 1977. Viral proteins expressed on the surface of murine leukemia cells. J Virol 22:65–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tung JS, Yoshiki T, Fleissner E. 1976. A core polyprotein of murine leukemia virus on the surface of mouse leukemia cells. Cell 9:573–578. doi: 10.1016/0092-8674(76)90039-8. [DOI] [PubMed] [Google Scholar]
- 45.Usami Y, Popov S, Gottlinger HG. 2014. The Nef-like effect of murine leukemia virus glycosylated Gag on HIV-1 infectivity is mediated by its cytoplasmic domain and depends on the AP-2 adaptor complex. J Virol 88:3443–3454. doi: 10.1128/JVI.01933-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosa A, Chande A, Ziglio S, De Sanctis V, Bertorelli R, Goh SL, McCauley SM, Nowosielska A, Antonarakis SE, Luban J, Santoni FA, Pizzato M. 2015. HIV-1 Nef promotes infection by excluding SERINC5 from virion incorporation. Nature 526:212–217. doi: 10.1038/nature15399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Usami Y, Wu Y, Gottlinger HG. 2015. SERINC3 and SERINC5 restrict HIV-1 infectivity and are counteracted by Nef. Nature 526:218–223. doi: 10.1038/nature15400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sood C, Marin M, Chande A, Pizzato M, Melikyan GB. 2017. SERINC5 protein inhibits HIV-1 fusion pore formation by promoting functional inactivation of envelope glycoproteins. J Biol Chem 292:6014–6026. doi: 10.1074/jbc.M117.777714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Beitari S, Ding S, Pan Q, Finzi A, Liang C. 2017. Effect of HIV-1 Env on SERINC5 antagonism. J Virol 91:e02214-16. doi: 10.1128/JVI.02214-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Langlois M, Beale R, Conticello S, Neuberger M. 2005. Mutational comparison of the single-domained APOBEC3C and double-domained APOBEC3F/G anti-retroviral cytidine deaminases provides insight into their DNA target site specificities. Nucleic Acids Res 33:1913–1923. doi: 10.1093/nar/gki343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Belanger K, Savoie M, Aydin H, Renner TM, Montazeri Z, Langlois MA. 2014. Deamination intensity profiling of human APOBEC3 protein activity along the near full-length genomes of HIV-1 and MoMLV by HyperHRM analysis. Virology 448:168–175. doi: 10.1016/j.virol.2013.10.008. [DOI] [PubMed] [Google Scholar]
- 52.Belanger K, Savoie M, Gerpe MCR, Couture JF, Langlois MA. 2013. Binding of RNA by APOBEC3G controls deamination-independent restriction of retroviruses. Nucleic Acids Res 41:7438–7452. doi: 10.1093/nar/gkt527. [DOI] [PMC free article] [PubMed] [Google Scholar]