ABSTRACT
Members of the apolipoprotein B mRNA-editing enzyme catalytic polypeptide-like (APOBEC3 [A3]) family of DNA cytidine deaminases are intrinsic restriction factors against retroviruses. In felids such as the domestic cat (Felis catus), the A3 genes encode the A3Z2, A3Z3, and A3Z2Z3 antiviral cytidine deaminases. Only A3Z3 and A3Z2Z3 inhibit viral infectivity factor (Vif)-deficient feline immunodeficiency virus (FIV). The FIV Vif protein interacts with Cullin (CUL), Elongin B (ELOB), and Elongin C (ELOC) to form an E3 ubiquitination complex to induce the degradation of feline A3s. However, the functional domains in FIV Vif for the interaction with Cullin are poorly understood. Here, we found that the expression of dominant negative CUL5 prevented the degradation of feline A3s by FIV Vif, while dominant negative CUL2 had no influence on the degradation of A3. In coimmunoprecipitation assays, FIV Vif bound to CUL5 but not CUL2. To identify the CUL5 interaction site in FIV Vif, the conserved amino acids from positions 47 to 160 of FIV Vif were mutated, but these mutations did not impair the binding of Vif to CUL5. By focusing on a potential zinc-binding motif (K175-C161-C184-C187) of FIV Vif, we found a conserved hydrophobic region (174IR175) that is important for the CUL5 interaction. Mutation of this region also impaired the FIV Vif-induced degradation of feline A3s. Based on a structural model of the FIV Vif-CUL5 interaction, the 52LW53 region in CUL5 was identified as mediating binding to FIV Vif. By comparing our results to the human immunodeficiency virus type 1 (HIV-1) Vif-CUL5 interaction surface (120IR121, a hydrophobic region that is localized in the zinc-binding motif), we suggest that the CUL5 interaction surface in the diverse HIV-1 and FIV Vifs is evolutionarily conserved, indicating a strong structural constraint. However, the FIV Vif-CUL5 interaction is zinc independent, which contrasts with the zinc dependence of HIV-1 Vif.
IMPORTANCE Feline immunodeficiency virus (FIV), which is similar to human immunodeficiency virus type 1 (HIV-1), replicates in its natural host in T cells and macrophages that express the antiviral restriction factor APOBEC3 (A3). To escape A3s, FIV and HIV induce the degradation of these proteins by building a ubiquitin ligase complex using the viral protein Vif to connect to cellular proteins, including Cullin 5. Here, we identified the protein residues that regulate this interaction in FIV Vif and Cullin 5. While our structural model suggests that the diverse FIV and HIV-1 Vifs use conserved residues for Cullin 5 binding, FIV Vif binds Cullin 5 independently of zinc, in contrast to HIV-1 Vif.
KEYWORDS: APOBEC3, Cullin 5, Vif, feline immunodeficiency virus, human immunodeficiency virus
INTRODUCTION
The antiretroviral APOBEC3 (A3) restriction factors are found in clade-specific numbers in placental mammals (1). Interestingly, only this group of animals is a host of lentiviruses, while other retroviruses are also found in animals outside placental mammals, e.g., in fish, reptiles, and birds. Humans carry seven A3 genes (A3A to A3D and A3F to A3H), and cats have four genes (A3Z2a to A3Z2c and A3Z3) (2–4). In addition, cats express A3Z2-Z3s by readthrough transcription and alternative splicing (3, 4). A3 proteins are packaged into nascent lentiviral particles during virus assembly and release, if the virus does not counteract A3 encapsidation. During the next round of infection, viral core-incorporated A3s deaminate cytidines in the single-stranded negative-strand DNA generated during reverse transcription, thus causing G-to-A hypermutations in the coding strand (5, 6). The best-characterized mechanism of lentiviruses to prevent this strong restriction is based on a viral protein called viral infectivity factor (Vif). In virus-producing cells, human immunodeficiency virus type 1 (HIV-1) Vif targets, for example, APOBEC3G (A3G) for degradation by forming an SCF (SKP1–Cullin/RBX–F-box protein)-like E3 ubiquitin ligase containing Cullin 5 (CUL5) as well as Elongin B (ELOB) and Elongin C (ELOC) through a novel SOCS (suppressor of cytokine signaling) box that binds ELOC (7, 8). The CUL5-SCF E3 ligase is also required for Vif activity against A3s in the related simian immunodeficiency virus of macaques (SIVmac) (8). In these complexes, Vif works as a substrate receptor for A3. In most cases, the interaction of Vif with A3s is species specific, and the intricate interfaces are still unresolved and a matter of ongoing investigation (9–13).
Feline immunodeficiency virus (FIV) also carries a vif gene, and Vif is essential for the virus to replicate in cats and cells that express feline A3s (3, 4, 9, 14–17). Similar to primate lentiviruses, FIV Vif interacts with CUL5, ELOB, and ELOC to form an E3 complex to induce the degradation of feline A3s (18). However, HIV-1 and SIV Vifs also need core-binding factor subunit beta (CBF-β) to stabilize and form the E3 ligase complex, whereas FIV and other nonprimate lentiviruses (e.g., maedi-visna virus [MVV], caprine arthritis encephalitis virus [CAEV], and bovine immunodeficiency virus [BIV]) do not require CBF-β to induce A3 degradation (19–24). Instead, MVV Vif hijacks cellular cyclophilin A (CYPA) as a cofactor to reconstitute the E3 ligase, while BIV Vif appears to operate independently of any cofactors (19). Whether FIV Vif recruits any additional protein to its E3 A3 complex is unclear.
To form an E3 ubiquitin ligase complex, HIV-1 Vif utilizes the BC box (SLQ motif) and the CUL5 box (HCCH motif) to interact with Elongin B/C and Cullin 5, respectively (7, 8, 25–28). CUL5 also binds to ELOC, so destabilizing the binding of ELOC to Vif by mutating the SLQ motif also removes CUL5 from the E3 complex (7, 25, 27). HIV-1 Vif utilizes the HCCH motif to bind zinc, which in turn is important for HIV-1 Vif-induced A3G degradation and binding to CUL5 (27–32). In the HIV-1 Vif-CUL5 costructure, the HCCH motif stabilizes Vif helix 3, which contains a hydrophobic interface that is involved in the direct CUL5 interaction (33). However, the interface between FIV Vif and CUL5 is unknown (18). In this study, we first demonstrate that FIV Vif interacts with CUL5, and not CUL2, to induce the degradation of feline A3s and then identify specific residues in Vif and CUL5 that are important for this binding. Our data support a conserved surface for the interaction of HIV-1 and FIV Vifs with CUL5.
RESULTS
CUL5 and not CUL2 is required for FIV Vif degradation of feline A3s.
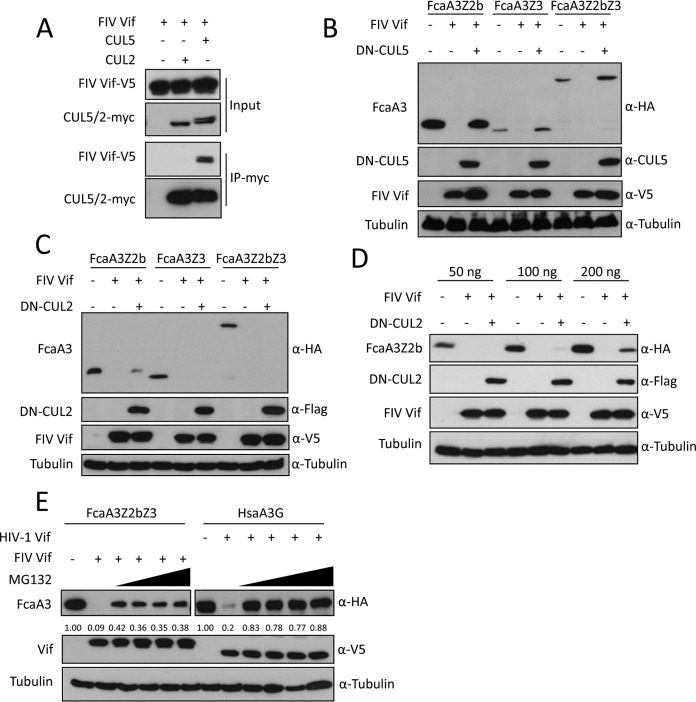
A previous study reported that FIV Vif interacts with CUL5 to form an E3 ubiquitin ligase complex that can induce the degradation of feline A3 proteins by the proteasome (18). However, the mechanism by which FIV Vif binds to CUL5 remains unclear, and whether FIV Vif binds to other Cullin family proteins is not known. Thus, we first investigated the interaction of FIV Vif with CUL5 and CUL2. The FIV Vif protein used in this study was from FIV clone 34TF10, which is derived from domestic cats (Felis catus), here referred to as FIV. To test the Vif-CUL interaction, we coimmunoprecipitated Vif and CUL using lysates of human embryonic kidney HEK293T cells transfected with expression plasmids for FIV Vif-V5, CUL2-myc, or CUL5-myc. We detected FIV Vif in the CUL5-immunoprecipitated complex, while no FIV Vif was observed in the CUL2 immunoprecipitation (Fig. 1A). CUL2 and CUL5 interact with Rbx by using their C terminus that is critical for E3 ubiquitin ligase activity (for a recent review, see reference 34). Dominant negative CUL5 (DN-CUL5) or DN-CUL2 has a C-terminal deletion, which can inhibit SOCS box protein-induced substrate degradation (35). Thus, we expressed the indicated feline A3s together with FIV Vif and DN-CUL5 or DN-CUL2. Immunoblots of protein extracts from transfected HEK293T cells were used as readouts for the degradation of the respective A3 proteins. The results showed that FIV Vif efficiently degraded all three tested feline A3s (Felis catus A3Z2b [FcaA3Z2b], FcaA3Z3, and FcaA3Z2bZ3) in the absence of DN-CUL5 (Fig. 1B), which is consistent with data from previous studies (4, 11, 14). The presence of DN-CUL5 enhanced the cellular protein level of Vif and abolished the degradation of the A3s (Fig. 1B). In contrast, DN-CUL2 did not affect the expression of Vif or Vif-dependent FcaA3Z3 and FcaA3Z2bZ3 degradation (Fig. 1C). However, in the presence of high levels of FcaA3Z2b, the expression of DN-CUL2 slightly impaired its degradation by FIV Vif, possibly due to a DN-CUL2-dependent exhaustion of endogenous Elongin B/C (Fig. 1D). Additionally, we investigated the impact of the proteasome inhibitor MG132 on Vif-mediated A3 degradation. We found that the degradation of FcaA3Z2bZ3 by FIV Vif was sensitive to MG132 treatment but comparably less sensitive than the HIV-1 Vif-induced degradation of human A3G (Fig. 1E). The reasons for these different responses to MG132 are unclear and might indicate different kinetics of degradation.
FIG 1.
CUL5 is required for FIV Vif-induced degradation of feline APOBEC3s. (A) FIV Vif interacts with CUL5 but not CUL2. myc-CUL5 or myc-CUL2 expression plasmids were cotransfected with the FIV Vif-V5 expression plasmid. Cell lysates were immunoprecipitated with anti-myc beads and then analyzed by immunoblotting with anti-V5 antibody for FIV Vif and anti-myc antibody for CUL2 and CUL5. (B and C) Dominant negative CUL5 (DN-CUL5), but not DN-CUL2, disrupts the degradation of feline A3s induced by FIV Vif. HEK293T cells were cotransfected with 300 ng expression plasmids for FcaA3Z2b-HA, FcaA3Z3-HA, or FcaA3Z2bZ3-HA and 700 ng of DN-CUL5–FLAG or DN-CUL2–FLAG with 30 ng of FIV Vif-V5. pcDNA3.1 was used as a control plasmid to replace the FIV Vif or dominant negative CUL5/2 expression plasmids. Cells were analyzed by immunoblotting using anti-HA, anti-V5, anti-CUL5, anti-Flag, and antitubulin antibodies. (D) HEK293T cells were cotransfected with 50, 100, or 200 ng expression plasmids for FcaA3Z2b-HA and 700 ng of DN–CUL2-FLAG with 30 ng of FIV Vif-V5. Immunoblotting was performed as described above for panel B. (E) FIV Vif induces the degradation of FcaA3s in a proteasome-dependent manner. HEK293T cells were transfected with HsaA3G-HA or FcaZ2bZ3-HA and HIV-1 Vif-V5 or FIV Vif-V5 expression plasmids; pcDNA3.1 was used as an empty plasmid control. The transfected cells were treated with the proteasome inhibitor MG132 (2.5, 5, 7.5, or 10 μM) or DMSO as a control at 36 h posttransfection. Cells were harvested 12 h later (48 h after transfection) and then analyzed by immunoblotting with anti-HA, anti-V5, and antitubulin antibodies. The percentages of FcaA3 and HsaA3G were calculated relative to those in the absence of FIV Vif or HIV-1 Vif (set as 1.00).
FIV Vif N-terminal residues are not essential for CUL5 binding.
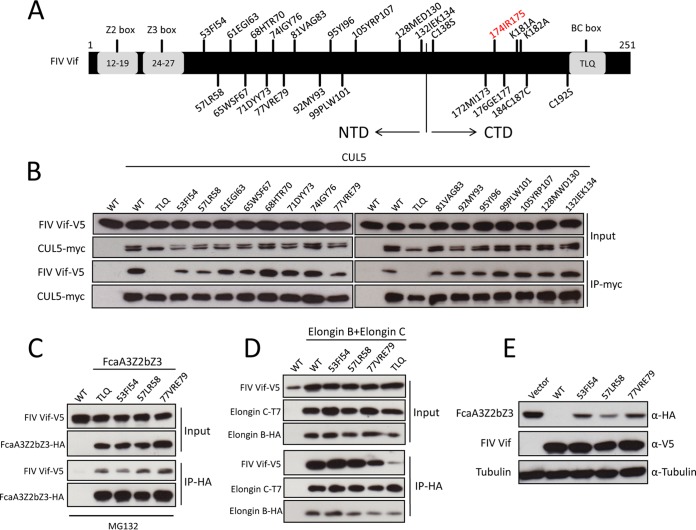
A previous study suggested that FIV Vif may utilize a novel mechanism for binding CUL5 (18). To investigate which domain of FIV Vif is involved in the CUL5 interaction, we analyzed FIV Vif sequences from different FIV strains. We found several conserved residues at the N-terminal protein region (residues 53 to 132). Thus, we replaced the N-terminal conserved residues (53FI54, 57LR58, 61EGI63, 65WSF67, 68HTR70, 71DYY73, 74IGY76, 77VRE79, 81VAG83, 92MY93, 95YI96, 99PLW101, 105YRP107, 128MED130, and 132IEK134) of FIV Vif with alanines (Fig. 2A). All FIV Vif mutants were coexpressed with CUL5-myc in HEK293T cells. FIV Vif with a TLQ-AAA mutation served as a control because destroying ELOC binding destabilizes the CUL5 interaction (18). Coimmunoprecipitation (co-IP) assays followed by immunoblot analyses were used to evaluate the binding of FIV Vif mutants to CUL5. The results showed that FIV Vif with mutations in 53FI54, 57LR58, and 77VRE79 bound CUL5, but the binding affinity appeared to be weaker than that of wild-type FIV Vif (Fig. 2B). These three FIV Vif mutants had similar binding affinities for FcaA3Z2bZ3 (Fig. 2C) and ELOB/C (Fig. 2D). Consistent with a reduced CUL5 interaction, these Vif mutants partially lost the function to induce FcaA3Z2bZ3 degradation (Fig. 2E). All other Vif mutants showed no change in CUL5 binding (Fig. 2B). Overall, it appears that the residues in the N terminus (residues 53 to 132) of FIV Vif are not important for the CUL5 interaction.
FIG 2.
Relevance of FIV Vif N-terminal residues for interaction with CUL5. (A) Schematic structure of FIV Vif. The numbers indicate the positions of amino acids mutated to alanines. The relative positions of the feline A3Z2 and A3Z3 interaction sites (Z2 box and Z3 box) and the Elongin B/C interaction site (BC box) are represented. NTD, N-terminal domain. (B) Coimmunoprecipitation of wild-type (WT) FIV Vif and mutants with CUL5. myc-CUL5 plasmids or the empty pcDNA3.1 plasmid was cotransfected with expression plasmids for wild-type FIV Vif-V5 or FIV Vif mutants. Immunoprecipitated (IP) complexes were analyzed by immunoblotting with anti-V5 for FIV Vif and anti-myc for CUL5. (C and D) HEK293T cells were transfected with expression plasmids for FcaZ2bZ3-HA, wild-type FIV Vif-V5, or the indicated FIV Vif mutants or the pcDNA3.1 empty plasmid (C) or with FIV Vif-V5, the indicated FIV Vif mutants, T7-ELOC, or HA-ELOB and the pcDNA3.1 empty plasmid (D). Cells were harvested 48 h after transfection, and proteins of cell lysates (input) and immunoprecipitated complexes were analyzed by using immunoblots stained with anti-V5 antibody for FIV Vif, anti-HA antibody for FcaZ2bZ3-HA and HA-Elongin B, and anti-T7 antibody for T7-Elongin C. (E) HEK293T cells were transfected with expression plasmids for FcaA3Z2bZ3-HA and wild-type FIV Vif-V5 or the indicated FIV Vif mutants or with the empty pcDNA3.1 plasmid. Cells were harvested and analyzed by immunoblotting with anti-HA, anti-V5, and antitubulin antibodies, respectively.
Identification of determinants in the C terminus of FIV Vif that regulate binding to CUL5.
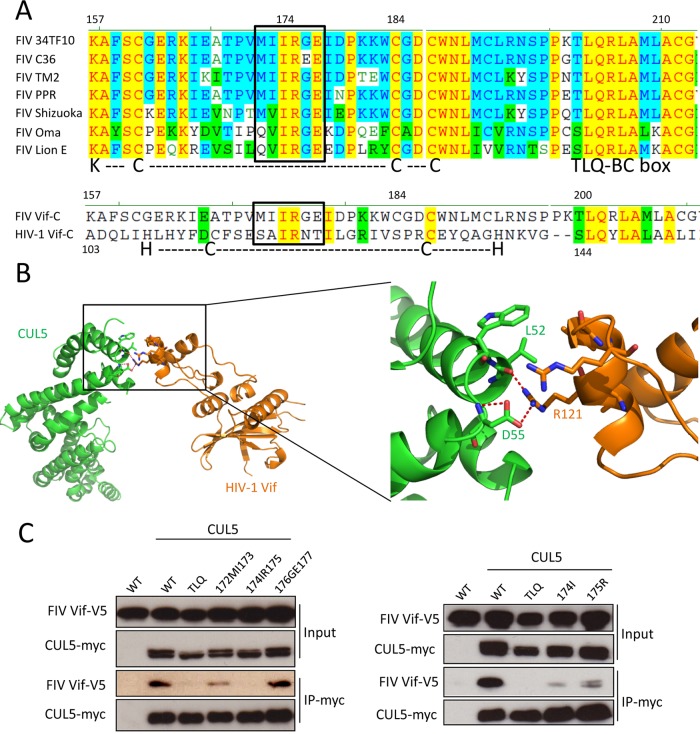
It has been shown that in HIV-1 Vif, CUL5 interacts with the C-terminal HCCH box (27, 28, 36). Thus, we analyzed the C-terminal region of Vifs from different FIV strains. As in HIV-1, all FIV Vifs contained a TLQ-BC box that is essential for ELOB/C binding (Fig. 3A). In addition, a KCCC motif that is similar to the HCCH motif of HIV-1 Vif is found in FIV Vif. In the KCCC motif, we identified a conserved hydrophobic domain (172MIIRGE177) (Fig. 3A). Despite the low sequence identity of only 13% in this region between FIV Vif and HIV-1 Vif, this hydrophobic domain matched when the C-terminal regions of both Vifs were aligned (Fig. 3A). In the complex structure of HIV-1 Vif-CUL5 (33), the 120IR121 region of HIV-1 Vif is involved in the direct interaction with CUL5 (Fig. 3B). To investigate whether the equivalent hydrophobic domain, 172MIIRGE177, of FIV Vif is also involved in CUL5 binding, we replaced 172MI173, 174IR175, or 176GE177 with alanines (Fig. 3C). The binding activities of these FIV Vif mutants with CUL5 were evaluated by co-IP assays. The results showed that the FIV Vif 176177GE-AA (mutating both 176G and 177E to alanines) mutant bound CUL5 similarly to wild-type FIV Vif and that the 172173MI-AA mutant slightly impaired the FIV Vif-CUL5 interaction, while the 174175IR-AA mutant impaired binding (Fig. 3C). Additionally, single point mutations were introduced at 174IR175, and the I174A and R175A mutations of FIV Vif also decreased the CUL5 interaction (Fig. 3C). Taken together, we conclude that the conserved 174IR175 motif of FIV Vif is the main CUL5-binding site.
FIG 3.
Identification of determinants in the C terminus of FIV Vif that regulate binding to CUL5. (A, top) Sequence alignment of FIV Vifs from different FIV strains, including 34TF10, C36, TM2, PRR, Shizuoka, Oma (Pallas's cats), and lion subtype E. (Bottom) Sequence alignment of C-terminal residues of FIV Vif (clone 34TF10) and HIV-1 Vif (clone NL4-3). KCCC is a motif of FIV Vif that is similar to the HIV-1 Vif zinc interaction motif HCCH. The boxed region indicates a conserved hydrophobic motif. Nonsimilar residues are indicated in black with no background, conservative residues are indicated in blue with a cyan background, blocks of similar residues are indicated in black with a green background, identical residues are indicated in red with a yellow background, and weakly similar residues are indicated in green with no background. (B) Structure of the HIV-1 Vif-CUL5 complex (orange, Vif; green, CUL5) (PDB accession number 4N9F). A closeup view of the HIV-1 Vif-CUL5 interface is shown. The residues that are involved in the HIV-1 Vif-CUL5 interaction are indicated. Red dashed lines represent hydrogen bonds. (C) The FIV Vif 174IR175 region is important for the interaction of FIV Vif with CUL5. myc-CUL5 expression plasmids or the empty pcDNA3.1 plasmid was cotransfected with expression plasmids for wild-type FIV Vif-V5 or the indicated FIV Vif mutants. Immunoprecipitated complexes were analyzed by immunoblotting with anti-V5 for FIV Vif and with anti-myc for CUL5.
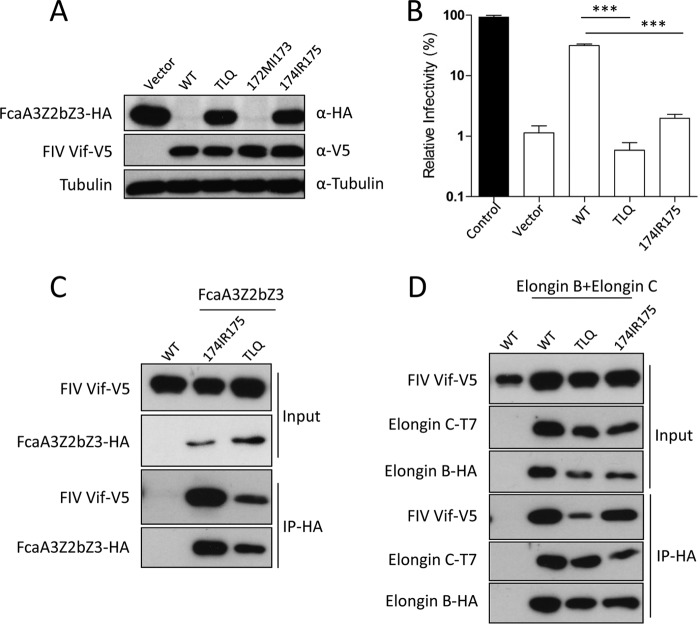
Next, we tested the degradation activity of the FIV Vif 174175IR-AA mutant toward feline A3. The results showed that the FIV Vif 174175IR-AA mutant did not induce the degradation of coexpressed FcaA3Z2bZ3 (Fig. 4A) and lost the ability to inhibit the anti-FIV activity of FcaA3Z2bZ3 (Fig. 4B). These data support the model that CUL5 binding of FIV Vif is essential for its antagonism of feline A3s. Furthermore, we asked whether impairment of the CUL5-binding sites of FIV Vif affects its interaction with feline A3 and ELOB/C. To address this question, HEK293T cells were cotransfected with expression plasmids for wild-type FIV Vif or alanine mutations of TLQ or 174IR175 together with expression plasmids for FcaA3Z2bZ3 or ELOB and ELOC. The immunoprecipitation results indicate that alanine mutations of FIV Vif TLQ and 174IR175 variants did not lead to a loss of binding to FcaA3Z2bZ3 (Fig. 4C). However, the FIV Vif TLQ-AAA variant lost its interaction with ELOB/C, as shown previously (18). The Vif 174175IR-AA variant bound to ELOB/C similarly to wild-type FIV Vif (Fig. 4D). Together, these results support that the 174IR175 region of FIV Vif is required for the interaction with CUL5.
FIG 4.
Mutation of the 174IR175 region in FIV Vif does not impair interaction with FcaA3s, ELOB, and ELOC. (A) The FIV Vif 174175IR-AA mutant has lost degradation activity against FcaA3Z2bZ3. Cells were transfected with expression plasmids for FcaA3Z2bZ3-HA and wild-type FIV Vif-V5 or the indicated FIV Vif mutants or with the empty pcDNA3.1 plasmid. Cells were harvested and analyzed by immunoblotting with anti-HA, anti-V5, and antitubulin antibodies, respectively. (B) The FIV Vif 174175IR-AA mutant does not antagonize FcaA3Z2bZ3 antiviral activity. Single-round FIVΔvif luciferase reporter virions were produced in the presence of feline A3 expression plasmids (FcaA3Z2bZ3) with wild-type FIV Vif or Vif mutants; pcDNA3.1(+) was added as a control for feline A3 (control) and FIV Vif (vector). The infectivity of reporter vectors was determined by quantification of luciferase activity in HEK293T cells transduced with normalized amounts of viral vector particles. (C and D) The FIV Vif 174175IR-AA mutant still has the capability to bind to FcaA3s, ELOB, and ELOC. HEK293T cells were transfected with expression plasmids for FcaZ2bZ3-HA, wild-type FIV Vif-V5, or the indicated FIV Vif mutants or with the empty pcDNA3.1 plasmid (C) or with wild-type FIV Vif-V5, the indicated FIV Vif mutants, T7-ELOC, or HA-ELOB and the empty pcDNA3.1 plasmid (D). Cells were harvested 48 h after transfection, and proteins of cell lysates (input) and immunoprecipitated complexes were analyzed by using Western blots stained with anti-V5 antibody for FIV Vif, anti-HA antibody for FcaZ2bZ3-HA and HA-Elongin B, and anti-T7 antibody for T7-Elongin C. ***, P value of <0.001.
Modeling the FIV Vif-CUL5 complex structure.
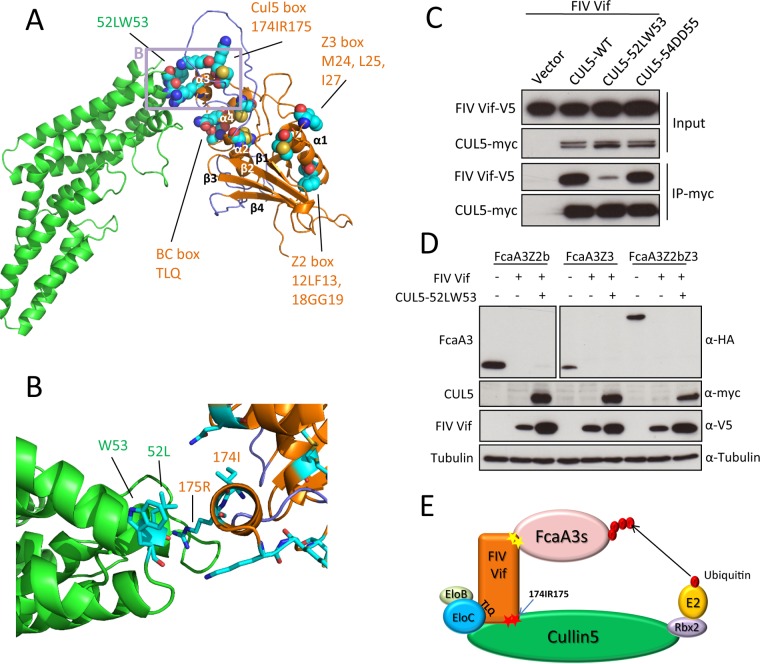
To identify further interacting residues in the FIV Vif-CUL5 complex, we built a homology model of the complex to guide mutational analyses. As described in Materials and Methods, we incorporated information on the predicted secondary structure of FIV Vif and our results on the importance of the 174IR175 region and the TLQ-BC box for CUL5 binding when generating the sequence alignment of FIV Vif to HIV-1 Vif. We did so to ameliorate the fact that the sequence identity between FIV and HIV-1 Vifs is only 5.2%. In this way, residues corresponding to the FIV 174IR175 and TLQ-BC box motifs were identified in HIV-1. The homology model of the FIV Vif-CUL5 complex showed a contact of the TLQ-BC box with CUL5 and revealed a close proximity of the 174IR175 region of Vif to the 52LWDD55 region of CUL5 (Fig. 5A and B). Thus, we mutated 52LW53 and 54DD55 of CUL5 to alanines and tested these CUL5 variants for interactions with FIV Vif. The results showed that the CUL5 52LW53-AA variant no longer bound to FIV Vif; however, the CUL5 54DD55-AA variant still bound Vif similarly to wild-type CUL5 (Fig. 5C). Additionally, we tested whether the CUL5 52LW53-AA mutant affects FIV Vif-induced feline A3 degradation. We expressed the indicated feline A3s together with FIV Vif and the CUL552LW53-AA mutant in HEK293T cells. Immunoblots of protein extracts from transfected cells were used as readouts for the degradation of the respective A3 proteins. The results showed that FIV Vif degraded feline A3s with an unchanged efficiency in the presence of this CUL5 mutant (Fig. 5D). A model representing the FIV Vif E3 complex is shown in Fig. 5E.
FIG 5.
FIV Vif-CUL5 three-dimensional structure model. (A) Homology model of the FIV Vif (orange)-CUL5 (green) complex in a cartoon representation. Helices α3 and α4 of Vif are interacting with CUL5. The model contains two unstructured loops (navy) before and after helix α2, as no structural template is available for these regions. These loops might be important for binding other parts of the complex. Residues that were subjected to mutational analysis are shown in a sphere representation. The region of the closeup shown in panel B is indicated by a light plum rectangle. (B) Closeup view of the homology model of the FIV Vif (orange)-CUL5 (green) complex in a cartoon representation, with interacting residues shown in a stick representation. (C) Expression plasmids for wild-type myc-CUL5 or the indicated mutants were cotransfected with the expression plasmid for wild-type FIV Vif-V5 into HEK293T cells. Cell lysates were immunoprecipitated with anti-myc beads and then analyzed by immunoblotting with anti-V5 antibody for FIV Vif and anti-myc antibody for CUL5. (D) HEK293T cells were cotransfected with expression plasmids for FcaA3Z2b-HA, FcaA3Z3-HA, or FcaA3Z2bZ3-HA and the CUL5 52LW53-AA mutant with FIV Vif-V5. pcDNA3.1 was used as a control plasmid to replace the FIV Vif or CUL5 expression plasmids. Cells were analyzed by immunoblotting using anti-HA, anti-V5, anti-CUL5, anti-Flag, and antitubulin antibodies. (E) Model of FIV Vif with the E3 ligase complex. The CUL5 interaction sites of FIV Vif (174IR175) are shown as red stars.
The FIV Vif-CUL5 interaction is zinc independent.
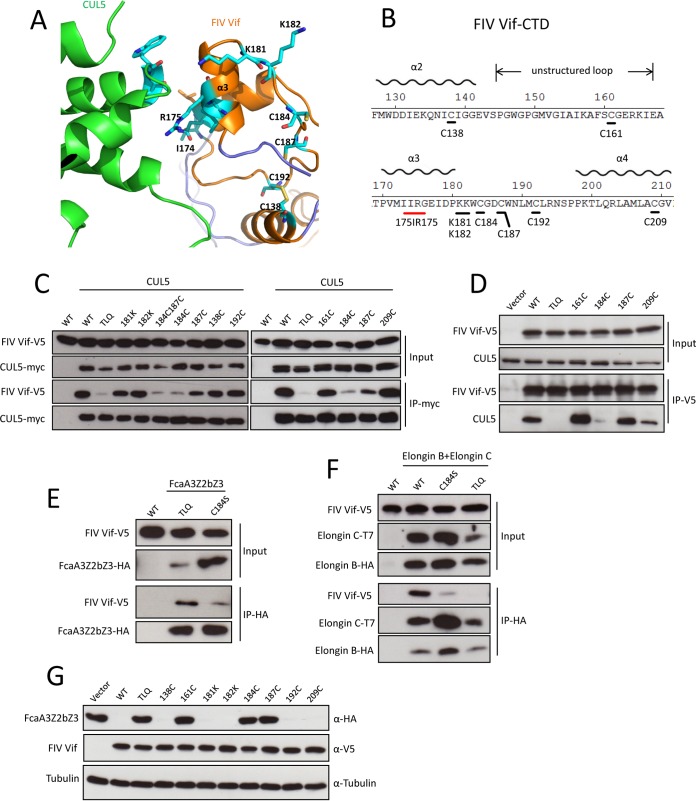
Our FIV Vif-CUL5 complex model suggests that helix 3 of the FIV Vif C-terminal domain (CTD) regulates CUL5 binding (Fig. 5 and 6A). There are two positively charged lysines (K181 and K182) and several potential zinc-binding cysteines (C138, C161, C184, C187, C192, and C209) at the FIV Vif CTD (Fig. 6B). To further address the FIV Vif-CUL5 interaction properties, we mutated K181 and K182 to alanines and several potential zinc-binding residues to serine (C138S, C161S, C184S, C187S, C192S, and C209S) (Fig. 6B). The immunoprecipitation results revealed that the C184S and C184C187-SS variants decreased the CUL5 interaction, but Vif mutations K181A, K182A, C187S, C138S, C161S, C192S, and C209S did not affect CUL5 binding (Fig. 6C). Additionally, we tested the interaction of several FIV C-to-S mutants (C161S, C184S, C187S, and C209S) with endogenous CUL5. The results showed that only the C184S mutant lost CUL5-binding activity (Fig. 6D). We further investigated the binding property of the FIV Vif C184S mutant with feline A3 and ELOB/C. The immunoprecipitation results indicate that the FIV Vif C184S mutant has a weaker binding affinity for FcaA3Z2bZ3 (Fig. 6E) than does the FIV Vif TLQ-AAA mutant, and both the FIV Vif TLQ-AAA and C184S mutants lost the interaction with ELOB/C (Fig. 6F). We further found that FIV Vif C161S, C184S, and C187S mutants completely lost the FcaA3Z2bZ3 degradation function, but the other cysteine mutants (C138S, C192S, and C209S) efficiently degraded FcaA3Z2bZ3 (Fig. 6G). Taken together, these data indicate that C184 of FIV Vif is not specific for the CUL5 interaction, and we speculate that C184 may regulate the integral structure of FIV Vif.
FIG 6.
C184 of FIV Vif is essential for Vif-CUL5, Vif-FcaA3, and Vif-ELOB/C interactions. (A) Closeup view of the homology model of the FIV Vif (orange)-CUL5 (green) complex in a cartoon representation, with residues that underwent mutational analysis shown in a stick representation. These residues are K181, K182, C184, C187, and C192 in the loop following helix 3 of FIV Vif; C138 that forms a disulfide bond with C192 is also shown. (B) Sequence representation of the FIV Vif C-terminal domain (CTD). (C) myc-CUL5 or the empty pcDNA3.1 plasmid was cotransfected with expression plasmids for wild-type FIV Vif-V5 or the indicated FIV Vif mutants. Immunoprecipitated complexes were analyzed by immunoblotting with anti-V5 for FIV Vif and with anti-myc for CUL5. (D) HEK293T cells were transfected with expression plasmids for wild-type FIV Vif-V5 or the indicated FIV Vif mutants. Immunoprecipitated complexes were analyzed by immunoblotting with anti-V5 for FIV Vif and with anti-CUL5 for CUL5. (E and F) The FIV Vif C184S mutant has lost the capability to bind to FcaA3s, ELOB, and ELOC. HEK293T cells were transfected with expression plasmids for FcaZ2bZ3-HA, wild-type FIV Vif-V5, or the indicated FIV Vif mutants or with the empty pcDNA3.1 plasmid (E) or with wild-type FIV Vif-V5, the indicated FIV Vif mutants, T7-ELOC, or HA-ELOB and the empty pcDNA3.1 plasmid (F). Cells were harvested 48 h after transfection, and proteins of cell lysates (input) and immunoprecipitated complexes were analyzed by using Western blots stained with anti-V5 antibody for FIV Vif, anti-HA antibody for FcaZ2bZ3-HA and HA-Elongin B, and anti-T7 antibody for T7-Elongin C. (G) HEK293T cells were transfected with expression plasmids for FcaA3Z2bZ3-HA and wild-type FIV Vif-V5 or the indicated FIV Vif mutants or with the empty pcDNA3.1 plasmid. Cells were harvested and analyzed by immunoblotting with anti-HA, anti-V5, and antitubulin antibodies, respectively.
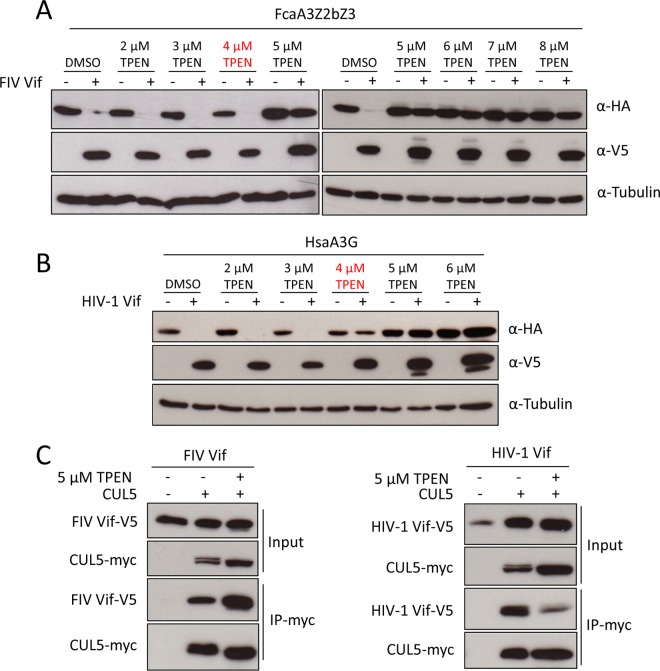
To test the zinc dependency of the FIV Vif-CUL5 interaction, the cell-permeable zinc chelator TPEN [N,N,N′,N′-tetrakis(2-pyridylmethyl)ethane-1,2-diamine] was applied in the following experiments. We first tested the effect of TPEN on the ability of FIV Vif to induce the degradation of feline A3s. HEK293T cells were cotransfected with FIV Vif or HIV-1 Vif and FcaA3Z2bZ3 or Homo sapiens A3G (HsaA3G) and treated with increasing amounts of TPEN. The immunoblotting results obtained from lysates of these cells indicated that FIV Vif efficiently degraded FcaA3Z2bZ3 in the presence of low concentrations of TPEN (2 to 4 μM), while higher concentrations of TPEN (5 to 8 μM) blocked FIV Vif-induced FcaA3Z2bZ3 degradation (Fig. 7A). A similar observation was made in the HIV-1 Vif-HsaA3G degradation assay; however, 4 μM TPEN blocked HIV-1 Vif-induced HsaA3G degradation, while this concentration of TPEN had no influence on FcaAZ2bZ3 degradation by FIV Vif (Fig. 7A and B). Because higher concentrations of TPEN may nonspecifically influence A3 degradation, we tested the interaction of HIV-1 Vif or FIV Vif with CUL5 by co-IP assays using lysates of cells treated with 5 μM TPEN. The results showed that 5 μM TPEN repressed the HIV-1 Vif-CUL5 interaction, thus supporting the previous model that the HIV-1 Vif-CUL5 interaction is regulated by zinc (Fig. 7C) (27–29, 36). In stark contrast, treatment with 5 μM TPEN did not impair FIV Vif-CUL5 binding. These data suggest that the FIV Vif-CUL5 interaction is zinc independent.
FIG 7.
FIV Vif binding to CUL5 is zinc independent. (A and B) HEK293T cells were transfected with expression plasmids for FcaZ2bZ3-HA (A) or HsaA3G-HA (B), and HIV-1 Vif-V5, or FIV Vif-V5. pcDNA3.1 was used as the empty plasmid control. Transfected cells were treated with the zinc chelator TPEN (2, 3, 4, 5, 6, 7, or 8 μM) or DMSO as a control at 36 h posttransfection. Cells were harvested 12 h later (48 h after transfection) and then analyzed by immunoblotting with anti-HA, anti-V5, and antitubulin antibodies. (C) The myc-CUL5 expression plasmid was cotransfected with FIV Vif-V5 or HIV-1 Vif-V5 expression plasmids into HEK293T cells. The transfected cells were treated with 5 μM TPEN or DMSO at 36 h posttransfection. Cell lysates were immunoprecipitated with anti-myc beads and then analyzed by immunoblotting with anti-V5 antibody for FIV Vif and HIV-1 Vif and with anti-myc antibody for CUL5.
DISCUSSION
FIV Vif interacts with CUL5, ELOB, and ELOC to form an E3 complex to induce the degradation of feline A3s (18). The properties of the interaction of FIV Vif with feline A3s and ELOB/C were previously identified (9, 11, 18). However, the interface between FIV Vif and CUL5 was not characterized.
Feline CUL5 and human CUL5 differ in only one amino acid, and this mutation does not affect its interaction with FIV Vif (18). It was also shown that FIV Vif can induce the degradation of feline A3s in both feline kidney CRFK and HEK293T cells, thus supporting that FIV Vif can interact with human and feline CUL5 proteins (11, 18). In this study, we first identified that three FIV Vif N-terminal mutants (53FI54, 57LR58, and 77VRE79) partially lost CUL5 binding, compared to wild-type FIV Vif (Fig. 2B). These Vif mutants showed the capacity to induce significant amounts of feline A3 but clearly less efficiently than wild-type Vif. According to our FIV Vif-CUL5 structural model, the N terminus of FIV Vif does not directly bind CUL5 (Fig. 5A). Thus, we speculate that these residues rather interact with an unknown factor that regulates FIV Vif-CUL5 binding. Indeed, a previous study suggested that FIV Vif requires an unknown factor to stabilize the FIV Vif-CUL5-ELOB/C complex (19).
CUL5-type ubiquitin ligases have a variety of adaptors that induce the degradation of different cellular substrates (for a recent review, see reference 34). The adaptors of CUL5 share one common domain, i.e., the SOCS box, which consists of a BC box and a CUL5 box (34). The CUL5 box has a common sequence, -LPθP-θ-YL, in which θ represents a hydrophobic residue, and the CUL5 box is localized downstream of the BC box (34). Such a CUL5-like box is also found in HIV-1 Vif (PPLP motif), but this region does not interact with CUL5 (26, 37). In fact, HIV-1 Vif uses a hydrophobic region of helix 3 to interact with CUL5 (28, 33, 36). In FIV Vif, a typical CUL5 box is also missing (18). However, we found that FIV Vif, similarly to HIV Vif, uses a hydrophobic region of helix 3 upstream of the BC box, at positions 174 and 175, to interact with CUL5 (Fig. 3 and 5). Vif proteins are not unique in applying unusual CUL5 boxes. For example, the adenovirus serotype 5 (Ad5) protein E4orf6 does not have a typical CUL5 box either, but it still interacts with CUL5 and forms an E3 ligase complex to degrade p53 (38).
Three HIV/SIV accessory proteins (Vpr, Vpx, and Vif) bind zinc, which is essential for the assembly of their E3 ligase complexes (39–41). Zinc is also required for BIV Vif-CUL2 binding, while the MVV Vif-CUL5 interaction does not need zinc (42). In this study, we used the cell-permeable zinc chelator TPEN to investigate whether this chelator impairs the function of FIV Vif or HIV-1 Vif in A3 degradation. We found that TPEN inhibited both FcaA3 and HsaA3 degradation induced by FIV Vif and HIV-1 Vif, respectively (Fig. 7A and B). It is important to point out that high concentrations of TPEN will repress cellular pathways, such as those of the cellular lysosome and autophagy (43). Thus, it is possible that high concentrations of TPEN may impact many cellular degradation pathways. However, we found that 4 μM TPEN inhibited HIV-1 Vif-induced HsaA3G degradation, while this concentration of TPEN had no influence on FcaAZ2bZ3 degradation by FIV Vif (Fig. 7A and B). In addition, the presence of 5 μM TPEN did not allow the isolation of HIV-1 Vif-CUL5 complexes, whereas FIV Vif-CUL5 complexes could still be detected (Fig. 7). Thus, our data support that zinc is not important for the FIV Vif-CUL5 interaction, as discussed previously (18). Other studies have demonstrated that TPEN treatment blocks the function of HIV-1 Vif and Vpr and SIVmac Vpx (39). These findings indicate that in the group of lentiviruses, there are zinc-dependent (e.g., HIV-1 and BIV) and zinc-independent (e.g., FIV and MVV) Vif proteins. The level of inter-Vif diversity is high, and HIV-1 Vif shares only around 16% and 5.2% identical residues with BIV and FIV Vifs, respectively. Thus, the requirement and structural consequences of zinc binding in Vifs are currently unclear. However, whether other metals (Mg2+ or Ca2+) bind FIV or MVV Vif needs more investigation. The SOCS3-CUL5 and E4orf6-CUL5 interactions are also zinc independent (38, 44), which together indicate that zinc is not necessary for the binding of CUL5.
As the sequence of FIV Vif is 59 amino acids longer than that of HIV-1 Vif and no other structural template is available, an unstructured loop is found in our model between helices 2 and 3 (Fig. 5A). This loop could be involved in the binding of CUL5 or other cofactors; however, without a structural template, its function remains unknown. Whether this loop is specific for FIV Vif requires further investigation. Despite the low sequence identity between FIV and HIV-1 Vifs and their difference in sequence lengths, our homology model of the FIV Vif-CUL5 complex is apparently accurate enough to predict interacting residues between these proteins. Our data from the CUL5 alanine variant of 52LW53 (Fig. 5C) and our homology model suggest that the interaction between Vif and CUL5 is also mediated by a hydrophobic contact. This is similar to the importance of the hydrophobic motif in the CUL5 BC box. In our homology model, helix 3 of Vif, which interacts with CUL5, is followed by C184. This cysteine was shown to be important for the CUL5 interaction and also for the feline A3 or ELOB/C interaction, which differs from the results of a previous study (18). As C184 is neither located in the Vif-CUL5 interface according to our homology model nor involved in zinc binding, we speculate that it might contribute to stabilizing the integral structure of FIV Vif. Overall, while our model certainly cannot be expected to be a perfect structural representation of the FIV Vif-CUL5 complex given the low sequence identity between FIV and HIV-1 Vifs, we were able to successfully predict interacting residues between the two proteins in those regions with higher sequence similarity.
Our data suggest that the Vif-CUL5 interaction in FIV structurally mirrors the one in HIV-1, even if the zinc dependencies of both interactions are different. After cross-species transmissions of lentiviruses, the lentiviruses adapt to rather variable A3 proteins, which are under positive selection. Positive selection is not seen in mammalian CUL5, and thus, it makes sense that the mechanism by which Vif interacts with CUL5 shows evolutionarily preserved interfaces.
MATERIALS AND METHODS
Plasmids.
Domestic cat A3s expressing a carboxy-terminal hemagglutinin (HA) tag were described previously (3, 45). The FIV 34TF10 (codon-optimized) Vif gene was inserted into pcWPRE containing a C-terminal V5 tag (4, 9). All the FIV Vif mutants were produced by fusion PCR and inserted into pcWPRE by using EcoRI and NotI restriction sites (11). The HIV-1 Vif expression plasmid with a C-terminal V5 tag was described previously (4). Human A3G with a C-terminal HA tag, a gift of Nathanial Landau, was previously described (46). pcDNA3-DN-hCUL5-FLAG (catalog number 15823) (35), pcDNA3-myc-CUL5 (catalog number 19895) (47), pcDNA3-DN-hCUL2-FLAG (catalog number 15819) (35), pcDNA3-myc3-CUL2 (catalog number 19892) (47), T7-Elongin C-pcDNA3 (catalog number 19998) (48), and HA-Elongin B-pcDNA3.1(+)-Zeo (catalog number 20000) (49) were obtained from Addgene (Cambridge, MA, USA). The CUL5 mutations were produced by fusion PCR and cloned into pcDNA3-myc-CUL5 by using BamHI and XbaI to replace wild-type CUL5. The replication-deficient packaging construct pFP93, a gift of Eric Poeschla (50); the FIV luciferase vector pLinSin (4); and a vesicular stomatitis virus G (VSV-G) expression plasmid, pMD.G, were used to produce FIV single-cycle luciferase viruses (FIV-Luc), which were described previously (4).
Cell cultures and transfections.
HEK293T cells (ATCC CRL-3216) were maintained in Dulbecco's high-glucose modified Eagle's medium (DMEM; Biochrom, Berlin, Germany) supplemented with 10% fetal bovine serum (FBS), 2 mM l-glutamine, penicillin (100 U/ml), and streptomycin (100 μg/ml) at 37°C with 5% CO2. The FcaA3Z2bZ3 degradation experiments were performed in 12-well plates, and 2 × 105 HEK293T cells were transfected with 300 ng feline A3Z2b or feline A3Z3 or A3Z2bZ3 expression plasmids together with 60 ng of a codon-optimized FIV Vif expression plasmid, or pcDNA3.1(+) as a control, and 700 ng pcDNA3-DN-hCUL5-FLAG or pcDNA3-DN-hCUL2-FLAG expression plasmids, or pcDNA3.1(+)as a control. To produce FIV-luciferase viruses, HEK293T cells were cotransfected with 300 ng of an FIV packaging construct, 300 ng of an FIV-luciferase vector, 300 ng of an A3 expression plasmid, 100 ng of a VSV-G expression plasmid, and 30 ng of an FIV Vif expression plasmid in 12-well plates; in some experiments, pcDNA3.1(+) was used instead of Vif or A3 expression plasmids. To test the interaction between CUL5 and FIV Vif, HEK293T cells were cotransfected with 1,000 ng pcDNA3-myc-CUL5 and 1,000 ng FIV Vif-V5 or FIV Vif mutants in 6-well plates; pcDNA3.1(+) was used as a control. For the interaction between ELOB, ELOC, and FIV Vif, HEK293T cells were cotransfected with 700 ng T7-Elongin C-pcDNA3, 700 ng HA-Elongin B-pcDNA3.1(+)-Zeo, and 700 ng FIV Vif-V5 or FIV Vif mutants in 6-well plates; pcDNA3.1(+) was used as a control. All transfections were performed by using Lipofectamine LTX (Thermo Fisher Scientific, Schwerte, Germany) according to the manufacturer's instructions. At 48 h posttransfection, cells and supernatants were collected.
For transfections in the presence of MG132 (catalog number 474790; Calbiochem) and TPEN (catalog number 16858-02-9; Calbiochem), the culture medium was replaced with fresh DMEM containing different concentrations of MG132 or TPEN or dimethyl sulfoxide (DMSO). After the cells were treated for 16 h, the cell lysates were used for immunoprecipitation or immunoblotting as described below.
Antibodies.
The following antibodies were used for the present study: mouse anti-HA antibody (1:7,500 dilution) (catalog number MMS-101P; Covance, Münster, Germany), mouse anti-V5 antibody (1:4,500 dilution) (catalog number MCA1360; ABDserotec, Düsseldorf, Germany), mouse anti-α-tubulin antibody (1:4,000 dilution) (clone B5-1-2; Sigma-Aldrich, Taufkirchen, Germany), mouse anti-T7 tag monoclonal antibody (1:1,000 dilution) (catalog number 69522, mouse monoclonal IgG2b; Merck, Germany), mouse anti-CUL5 monoclonal antibody (1:1,000 dilution) (catalog number sc-373822; Santa Cruz, USA), mouse anti-myc monoclonal antibody (1:100 dilution) (catalog number MCA2200; MbD Serotec, Canada), and mouse anti-Flag M2 monoclonal antibody (1:1,000 dilution) (catalog number F1804; Sigma, USA). The secondary antibody was horseradish peroxidase (HRP)-conjugated rabbit anti-mouse antibody (1:10,000) (anti-mouse IgG-HRP; GE Healthcare, Munich, Germany).
Immunoprecipitation.
HEK293T cells were transfected with FIV Vif-V5 together with pcDNA3-myc-CUL5 or T7-Elongin C-pcDNA3, HA-Elongin B-pcDNA3.1(+)-Zeo, or FcaA3Z2bZ3-HA expression plasmids. At 48 h posttransfection, the cells were harvested and lysed in IP lysis buffer (50 mM Tris-HCl [pH 8], 10% glycerol, 0.8% NP-40, 150 mM NaCl) with protease inhibitor cocktail set III (Calbiochem, Darmstadt, Germany) on ice for 20 min. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. The supernatants were incubated with 15 μl rabbit anti-c-myc agarose affinity gel antibody beads (catalog number A7470; Sigma, USA) or 15 μl rat anti-HA affinity matrix beads (catalog number 16598600; Roche, USA). After 2 h of incubation at 4°C with end-over-end rotation, the samples were washed 4 times with lysate buffer on ice. Bound proteins were eluted by boiling the beads for 5 min at 95°C in sodium dodecyl sulfate (SDS) loading buffer. The eluted materials were subsequently analyzed by immunoblotting. To detect the interaction of FIV Vif with endogenous CUL5, 5 × 105 HEK293T cells in 6-well plates were transfected with wild-type FIV Vif or the indicated mutant expression plasmids. The cells were harvested as described above, and 500 μl cell lysis buffer was incubated with 2 μg mouse anti-V5 antibody (catalog number MCA1360; ABDserotec, Düsseldorf, Germany) and 20 μl protein A/G plus agarose (Santa Cruz, Heidelberg, Germany) for 4 h at 4°C with end-over-end rotation. The samples were washed 4 times after 4 h of incubation, and bound proteins were analyzed by immunoblotting.
Immunoblotting.
Transfected HEK293T cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (25 mM Tris-HCl [pH 7.6], 150 mM NaCl, 1% NP-40, 1% sodium deoxycholate, 0.1% SDS, protease inhibitor cocktail set III [Calbiochem, Darmstadt, Germany]) on ice for 20 min. Cell lysates were clarified by centrifugation at 10,000 × g for 30 min at 4°C. Test proteins were boiled for 5 min at 95°C in SDS loading buffer and resolved on an SDS-PAGE gel. The proteins were then transferred onto an Immobilon polyvinylidene difluoride (PVDF) transfer membrane (catalog number IPVH 00010; Merck Millipore, Germany) by semidry transfer (Bio-Rad, Hercules, CA) at 25 V for 50 min. After blocking in 5% nonfat milk (catalog number A0830; PanReac AppliChem, Germany), the membranes were detected with various primary antibodies against proteins of interest followed by secondary antibodies and developed with ECL chemiluminescence reagents (GE Healthcare).
Homology modeling.
We modeled the complex of FIV Vif and CUL5 based on the X-ray crystal structure of the complex of HIV-1 Vif and CUL5 as a template (33). Owing to the low sequence identity between FIV and HIV-1 Vifs, we performed modeling in an iterative fashion. We used ClustalW2 (33) to align the sequences, using the (T/S)LQ-BC box and the conserved 174IR175 motif as anchor points to guide the alignment. Next, we modeled the complex by employing Modeler v9.10 (51) and subsequently manually curated the sequence alignment based on the resulting models. Accounting for a possible zinc dependency of FIV Vif, cysteines in the FIV Vif sequence, which in the models were structurally close to zinc-binding cysteine and histidine residues in HIV-1 Vif, were aligned to these zinc binding residues in the HIV-1 sequence (C113, C114, H108, and H139). Furthermore, we used information on the secondary structure of FIV Vif, predicted by PSIPRED (52), to guide the manual curation of the sequence alignment. The final model was then used to predict interacting residues. The model is accessible at the Protein Model Data Base (PMDB) (https://bioinformatics.cineca.it/PMDB/main.php) with accession number PM0081296.
Nucleotide sequence accession numbers.
The FIV Vif sequences were obtained from GenBank, and the accession numbers are AY600517.1 for FIV C36, M25381.1 for FIV 34TF10, M36968.1 for FIV PRR, M59418.1 for FIV TM-2, LC079040.1 for FIV Shizuoka, AY713445 for FIV Oma, EU117991 for FIV lion subtype B, EU117992 for FIV lion subtype E, U03982 for FIV puma subtype A, and DQ192583 for FIV puma subtype B. The feline APOBEC3 and human APOBEC3G GenBank accession numbers are AY971954 for FcaA3Z2b, EU109281 for FcaA3Z3, EU109281 for FcaA3Z2bZ3, and NM_021822 for HsaA3G.
Statistical analysis.
Data are represented as the means with standard deviations (SD) in all bar diagrams. Statistically significant differences between two groups were analyzed by using unpaired Student's t test with GraphPad Prism version 5 (GraphPad Software, San Diego, CA, USA). A P value of ≤0.05 was considered statistically significant; a P value of <0.001 was considered extremely significant, a P value of 0.001 to 0.01 was considered very significant, a P value of 0.01 to 0.05 was considered significant, and a P value of >0.05 was considered not significant.
ACKNOWLEDGMENTS
We thank Wioletta Hörschken for excellent technical assistance. We thank Eric Poeschla for plasmid pFP93. We are grateful for computational support by the Zentrum für Informations und Medientechnologie at the Heinrich Heine University Düsseldorf.
Q.G. and Z.Z. are supported by the China Scholarship Council. C.M. is supported by the Heinz-Ansmann Foundation for AIDS Research.
REFERENCES
- 1.Münk C, Willemsen A, Bravo IG. 2012. An ancient history of gene duplications, fusions and losses in the evolution of APOBEC3 mutators in mammals. BMC Evol Biol 12:71. doi: 10.1186/1471-2148-12-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.LaRue RS, Andresdottir V, Blanchard Y, Conticello SG, Derse D, Emerman M, Greene WC, Jonsson SR, Landau NR, Löchelt M, Malik HS, Malim MH, Münk C, O'Brien SJ, Pathak VK, Strebel K, Wain-Hobson S, Yu XF, Yuhki N, Harris RS. 2009. Guidelines for naming nonprimate APOBEC3 genes and proteins. J Virol 83:494–497. doi: 10.1128/JVI.01976-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Münk C, Beck T, Zielonka J, Hotz-Wagenblatt A, Chareza S, Battenberg M, Thielebein J, Cichutek K, Bravo IG, O'Brien SJ, Löchelt M, Yuhki N. 2008. Functions, structure, and read-through alternative splicing of feline APOBEC3 genes. Genome Biol 9:R48. doi: 10.1186/gb-2008-9-3-r48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zielonka J, Marino D, Hofmann H, Yuhki N, Löchelt M, Münk C. 2010. Vif of feline immunodeficiency virus from domestic cats protects against APOBEC3 restriction factors from many felids. J Virol 84:7312–7324. doi: 10.1128/JVI.00209-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang H, Yang B, Pomerantz RJ, Zhang C, Arunachalam SC, Gao L. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98. doi: 10.1038/nature01707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mangeat B, Turelli P, Caron G, Friedli M, Perrin L, Trono D. 2003. Broad antiretroviral defence by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103. doi: 10.1038/nature01709. [DOI] [PubMed] [Google Scholar]
- 7.Mehle A, Goncalves J, Santa-Marta M, McPike M, Gabuzda D. 2004. Phosphorylation of a novel SOCS-box regulates assembly of the HIV-1 Vif-Cul5 complex that promotes APOBEC3G degradation. Genes Dev 18:2861–2866. doi: 10.1101/gad.1249904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yu X, Yu Y, Liu B, Luo K, Kong W, Mao P, Yu XF. 2003. Induction of APOBEC3G ubiquitination and degradation by an HIV-1 Vif-Cul5-SCF complex. Science 302:1056–1060. doi: 10.1126/science.1089591. [DOI] [PubMed] [Google Scholar]
- 9.Zhang Z, Gu Q, Jaguva Vasudevan AA, Hain A, Kloke BP, Hasheminasab S, Mulnaes D, Sato K, Cichutek K, Häussinger D, Bravo IG, Smits SH, Gohlke H, Münk C. 2016. Determinants of FIV and HIV Vif sensitivity of feline APOBEC3 restriction factors. Retrovirology 13:46. doi: 10.1186/s12977-016-0274-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang Z, Gu Q, Jaguva Vasudevan AA, Jeyaraj M, Schmidt S, Zielonka J, Perkovic M, Heckel JO, Cichutek K, Häussinger D, Smits SH, Münk C. 2016. Vif proteins from diverse human immunodeficiency virus/simian immunodeficiency virus lineages have distinct binding sites in A3C. J Virol 90:10193–10208. doi: 10.1128/JVI.01497-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu Q, Zhang Z, Cano Ortiz L, Franco AC, Häussinger D, Münk C. 2016. Feline immunodeficiency virus Vif N-terminal residues selectively counteract feline APOBEC3s. J Virol 90:10545–10557. doi: 10.1128/JVI.01593-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Harris RS, Anderson BD. 2016. Evolutionary paradigms from ancient and ongoing conflicts between the lentiviral Vif protein and mammalian APOBEC3 enzymes. PLoS Pathog 12:e1005958. doi: 10.1371/journal.ppat.1005958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Simon V, Bloch N, Landau NR. 2015. Intrinsic host restrictions to HIV-1 and mechanisms of viral escape. Nat Immunol 16:546–553. doi: 10.1038/ni.3156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stern MA, Hu C, Saenz DT, Fadel HJ, Sims O, Peretz M, Poeschla EM. 2010. Productive replication of Vif-chimeric HIV-1 in feline cells. J Virol 84:7378–7395. doi: 10.1128/JVI.00584-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shen X, Leutenegger CM, Stefano Cole K, Pedersen NC, Sparger EE. 2007. A feline immunodeficiency virus vif-deletion mutant remains attenuated upon infection of newborn kittens. J Gen Virol 88:2793–2799. doi: 10.1099/vir.0.83268-0. [DOI] [PubMed] [Google Scholar]
- 16.Yoshikawa R, Takeuchi JS, Yamada E, Nakano Y, Misawa N, Kimura Y, Ren F, Miyazawa T, Koyanagi Y, Sato K. 2017. Feline immunodeficiency virus evolutionarily acquires two proteins, Vif and protease, capable of antagonizing feline APOBEC3. J Virol 91:e00250-17. doi: 10.1128/JVI.00250-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Troyer RM, Thompson J, Elder JH, VandeWoude S. 2013. Accessory genes confer a high replication rate to virulent feline immunodeficiency virus. J Virol 87:7940–7951. doi: 10.1128/JVI.00752-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang J, Zhang W, Lv M, Zuo T, Kong W, Yu X. 2011. Identification of a Cullin5-ElonginB-ElonginC E3 complex in degradation of feline immunodeficiency virus Vif-mediated feline APOBEC3 proteins. J Virol 85:12482–12491. doi: 10.1128/JVI.05218-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kane JR, Stanley DJ, Hultquist JF, Johnson JR, Mietrach N, Binning JM, Jonsson SR, Barelier S, Newton BW, Johnson TL, Franks-Skiba KE, Li M, Brown WL, Gunnarsson HI, Adalbjornsdottir A, Fraser JS, Harris RS, Andresdottir V, Gross JD, Krogan NJ. 2015. Lineage-specific viral hijacking of non-canonical E3 ubiquitin ligase cofactors in the evolution of Vif anti-APOBEC3 activity. Cell Rep 11:1236–1250. doi: 10.1016/j.celrep.2015.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ai Y, Zhu D, Wang C, Su C, Ma J, Ma J, Wang X. 2014. Core-binding factor subunit beta is not required for non-primate lentiviral Vif-mediated APOBEC3 degradation. J Virol 88:12112–12122. doi: 10.1128/JVI.01924-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Han X, Liang W, Hua D, Zhou X, Du J, Evans SL, Gao Q, Wang H, Viqueira R, Wei W, Zhang W, Yu XF. 2014. Evolutionarily conserved requirement for core binding factor beta in the assembly of the human immunodeficiency virus/simian immunodeficiency virus Vif-cullin 5-RING E3 ubiquitin ligase. J Virol 88:3320–3328. doi: 10.1128/JVI.03833-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yoshikawa R, Takeuchi JS, Yamada E, Nakano Y, Ren F, Tanaka H, Munk C, Harris RS, Miyazawa T, Koyanagi Y, Sato K. 2015. Vif determines the requirement for CBF-beta in APOBEC3 degradation. J Gen Virol 96:887–892. doi: 10.1099/jgv.0.000027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jager S, Kim DY, Hultquist JF, Shindo K, LaRue RS, Kwon E, Li M, Anderson BD, Yen L, Stanley D, Mahon C, Kane J, Franks-Skiba K, Cimermancic P, Burlingame A, Sali A, Craik CS, Harris RS, Gross JD, Krogan NJ. 2012. Vif hijacks CBF-beta to degrade APOBEC3G and promote HIV-1 infection. Nature 481:371–375. doi: 10.1038/nature10693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang W, Du J, Evans SL, Yu Y, Yu XF. 2012. T-cell differentiation factor CBF-beta regulates HIV-1 Vif-mediated evasion of host restriction. Nature 481:376–379. doi: 10.1038/nature10718. [DOI] [PubMed] [Google Scholar]
- 25.Yu Y, Xiao Z, Ehrlich ES, Yu X, Yu XF. 2004. Selective assembly of HIV-1 Vif-Cul5-ElonginB-ElonginC E3 ubiquitin ligase complex through a novel SOCS box and upstream cysteines. Genes Dev 18:2867–2872. doi: 10.1101/gad.1250204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bergeron JR, Huthoff H, Veselkov DA, Beavil RL, Simpson PJ, Matthews SJ, Malim MH, Sanderson MR. 2010. The SOCS-box of HIV-1 Vif interacts with ElonginBC by induced-folding to recruit its Cul5-containing ubiquitin ligase complex. PLoS Pathog 6:e1000925. doi: 10.1371/journal.ppat.1000925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Luo K, Xiao Z, Ehrlich E, Yu Y, Liu B, Zheng S, Yu XF. 2005. Primate lentiviral virion infectivity factors are substrate receptors that assemble with cullin 5-E3 ligase through a HCCH motif to suppress APOBEC3G. Proc Natl Acad Sci U S A 102:11444–11449. doi: 10.1073/pnas.0502440102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xiao Z, Xiong Y, Zhang W, Tan L, Ehrlich E, Guo D, Yu XF. 2007. Characterization of a novel Cullin5 binding domain in HIV-1 Vif. J Mol Biol 373:541–550. doi: 10.1016/j.jmb.2007.07.029. [DOI] [PubMed] [Google Scholar]
- 29.Mehle A, Thomas ER, Rajendran KS, Gabuzda D. 2006. A zinc-binding region in Vif binds Cul5 and determines cullin selection. J Biol Chem 281:17259–17265. doi: 10.1074/jbc.M602413200. [DOI] [PubMed] [Google Scholar]
- 30.Xiao Z, Ehrlich E, Luo K, Xiong Y, Yu XF. 2007. Zinc chelation inhibits HIV Vif activity and liberates antiviral function of the cytidine deaminase APOBEC3G. FASEB J 21:217–222. doi: 10.1096/fj.06-6773com. [DOI] [PubMed] [Google Scholar]
- 31.Paul I, Cui J, Maynard EL. 2006. Zinc binding to the HCCH motif of HIV-1 virion infectivity factor induces a conformational change that mediates protein-protein interactions. Proc Natl Acad Sci U S A 103:18475–18480. doi: 10.1073/pnas.0604150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Giri K, Scott RA, Maynard EL. 2009. Molecular structure and biochemical properties of the HCCH-Zn2+ site in HIV-1 Vif. Biochemistry 48:7969–7978. doi: 10.1021/bi900677w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo Y, Dong L, Qiu X, Wang Y, Zhang B, Liu H, Yu Y, Zang Y, Yang M, Huang Z. 2014. Structural basis for hijacking CBF-beta and CUL5 E3 ligase complex by HIV-1 Vif. Nature 505:229–233. doi: 10.1038/nature12884. [DOI] [PubMed] [Google Scholar]
- 34.Okumura F, Joo-Okumura A, Nakatsukasa K, Kamura T. 2016. The role of cullin 5-containing ubiquitin ligases. Cell Div 11:1. doi: 10.1186/s13008-016-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin J, Ang XL, Shirogane T, Harper JW. 2005. Identification of substrates for F-box proteins. Methods Enzymol 399:287–309. doi: 10.1016/S0076-6879(05)99020-4. [DOI] [PubMed] [Google Scholar]
- 36.Xiao Z, Ehrlich E, Yu Y, Luo K, Wang T, Tian C, Yu XF. 2006. Assembly of HIV-1 Vif-Cul5 E3 ubiquitin ligase through a novel zinc-binding domain-stabilized hydrophobic interface in Vif. Virology 349:290–299. doi: 10.1016/j.virol.2006.02.002. [DOI] [PubMed] [Google Scholar]
- 37.Wolfe LS, Stanley BJ, Liu C, Eliason WK, Xiong Y. 2010. Dissection of the HIV Vif interaction with human E3 ubiquitin ligase. J Virol 84:7135–7139. doi: 10.1128/JVI.00031-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Luo K, Ehrlich E, Xiao Z, Zhang W, Ketner G, Yu XF. 2007. Adenovirus E4orf6 assembles with Cullin5-ElonginB-ElonginC E3 ubiquitin ligase through an HIV/SIV Vif-like BC-box to regulate p53. FASEB J 21:1742–1750. doi: 10.1096/fj.06-7241com. [DOI] [PubMed] [Google Scholar]
- 39.Wang H, Guo H, Su J, Rui Y, Zheng W, Gao W, Zhang W, Li Z, Liu G, Markham RB, Wei W, Yu XF. 2017. Inhibition of Vpx-mediated SAMHD1 and Vpr-mediated host helicase transcription factor degradation by selective disruption of viral CRL4 (DCAF1) E3 ubiquitin ligase assembly. J Virol 91:e00225-17. doi: 10.1128/JVI.00225-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Schwefel D, Groom HC, Boucherit VC, Christodoulou E, Walker PA, Stoye JP, Bishop KN, Taylor IA. 2014. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505:234–238. doi: 10.1038/nature12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu Y, Zhou X, Barnes CO, DeLucia M, Cohen AE, Gronenborn AM, Ahn J, Calero G. 2016. The DDB1-DCAF1-Vpr-UNG2 crystal structure reveals how HIV-1 Vpr steers human UNG2 toward destruction. Nat Struct Mol Biol 23:933–940. doi: 10.1038/nsmb.3284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang J, Wu J, Wang W, Wu H, Yu B, Wang J, Lv M, Wang X, Zhang H, Kong W, Yu X. 2014. Role of cullin-elonginB-elonginC E3 complex in bovine immunodeficiency virus and maedi-visna virus Vif-mediated degradation of host A3Z2-Z3 proteins. Retrovirology 11:77. doi: 10.1186/s12977-014-0077-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee SJ, Koh JY. 2010. Roles of zinc and metallothionein-3 in oxidative stress-induced lysosomal dysfunction, cell death, and autophagy in neurons and astrocytes. Mol Brain 3:30. doi: 10.1186/1756-6606-3-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kim YK, Kwak MJ, Ku B, Suh HY, Joo K, Lee J, Jung JU, Oh BH. 2013. Structural basis of intersubunit recognition in elongin BC-cullin 5-SOCS box ubiquitin-protein ligase complexes. Acta Crystallogr D Biol Crystallogr 69:1587–1597. doi: 10.1107/S0907444913011220. [DOI] [PubMed] [Google Scholar]
- 45.Münk C, Zielonka J, Constabel H, Kloke BP, Rengstl B, Battenberg M, Bonci F, Pistello M, Löchelt M, Cichutek K. 2007. Multiple restrictions of human immunodeficiency virus type 1 in feline cells. J Virol 81:7048–7060. doi: 10.1128/JVI.02714-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mariani R, Chen D, Schröfelbauer B, Navarro F, König R, Bollman B, Münk C, Nymark-McMahon H, Landau NR. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31. doi: 10.1016/S0092-8674(03)00515-4. [DOI] [PubMed] [Google Scholar]
- 47.Ohta T, Michel JJ, Schottelius AJ, Xiong Y. 1999. ROC1, a homolog of APC11, represents a family of cullin partners with an associated ubiquitin ligase activity. Mol Cell 3:535–541. doi: 10.1016/S1097-2765(00)80482-7. [DOI] [PubMed] [Google Scholar]
- 48.Lonergan KM, Iliopoulos O, Ohh M, Kamura T, Conaway RC, Conaway JW, Kaelin WG Jr. 1998. Regulation of hypoxia-inducible mRNAs by the von Hippel-Lindau tumor suppressor protein requires binding to complexes containing elongins B/C and Cul2. Mol Cell Biol 18:732–741. doi: 10.1128/MCB.18.2.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ohh M, Park CW, Ivan M, Hoffman MA, Kim TY, Huang LE, Pavletich N, Chau V, Kaelin WG. 2000. Ubiquitination of hypoxia-inducible factor requires direct binding to the beta-domain of the von Hippel-Lindau protein. Nat Cell Biol 2:423–427. doi: 10.1038/35017054. [DOI] [PubMed] [Google Scholar]
- 50.Loewen N, Barraza R, Whitwam T, Saenz DT, Kemler I, Poeschla EM. 2003. FIV vectors. Methods Mol Biol 229:251–271. [DOI] [PubMed] [Google Scholar]
- 51.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5:Unit 5.6. doi: 10.1002/0471250953.bi0506s15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jones DT. 1999. Protein secondary structure prediction based on position-specific scoring matrices. J Mol Biol 292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]