ABSTRACT
Feline immunodeficiency virus (FIV) infection in domestic cats is the smallest usable natural model for lentiviral infection studies. FLA-E*01801 was applied to FIV AIDS vaccine research. We determined the crystal structure of FLA-E*01801 complexed with a peptide derived from FIV (gag positions 40 to 48; RMANVSTGR [RMA9]). The A pocket of the FLA-E*01801 complex plays a valuable restrictive role in peptide binding. Mutation experiments and circular-dichroism (CD) spectroscopy revealed that peptides with Asp at the first position (P1) could not bind to FLA-E*01801. The crystal structure and in vitro refolding of the mutant FLA-E*01801 complex demonstrated that Glu63 and Trp167 in the A pocket play important roles in restricting P1D. The B pocket of the FLA-E*01801 complex accommodates M/T/A/V/I/L/S residues, whereas the negatively charged F pocket prefers R/K residues. Based on the peptide binding motif, 125 FLA-E*01801-restricted FIV nonapeptides (San Diego isolate) were identified. Our results provide the structural basis for peptide presentation by the FLA-E*01801 molecule, especially A pocket restriction on peptide binding, and identify the potential cytotoxic T lymphocyte (CTL) epitope peptides of FIV presented by FLA-E*01801. These results will benefit both the reasonable design of FLA-E*01801-restricted CTL epitopes and the further development of the AIDS vaccine.
IMPORTANCE Feline immunodeficiency virus (FIV) is a viral pathogen in cats, and this infection is the smallest usable natural model for lentivirus infection studies. To examine how FLA I presents FIV epitope peptides, we crystallized and solved the first classic feline major histocompatibility complex class I (MHC-I) molecular structure. Surprisingly, pocket A restricts peptide binding. Trp167 blocks the left side of pocket A, causing P1D to conflict with Glu63. We also identified the FLA-E*01801 binding motif X (except D)-(M/T/A/V/I/L/S)-X-X-X-X-X-X-(R/K) based on structural and biochemical experiments. We identified 125 FLA-E*01801-restricted nonapeptides from FIV. These results are valuable for developing peptide-based FIV and human immunodeficiency virus (HIV) vaccines and for studying how MHC-I molecules present peptides.
KEYWORDS: crystal structure, CTL immunology, domestic cat, MHC-I, feline immunodeficiency virus (FIV)
INTRODUCTION
Feline immunodeficiency virus (FIV), which induces a lethal immunodeficiency in domestic cats that parallels AIDS in humans (1), is a typical lentivirus that shares a similar genomic structure, immunopathogenesis, and host cell dependency factor with human immunodeficiency virus (HIV) (2–5). Similar to humans infected with HIV, felines infected with FIV exhibit a typically protracted asymptomatic phase that is sustained for several years prior to developing the terminal immunodeficiency syndrome. Previous studies have shown that despite a lack of clinical symptoms, a long-term decline in CD4+ T cells, a concomitant increase in CD8+ cells and hypergammaglobulinemia occur during the asymptomatic phase (6). Thus, the FIV/cat model has been an invaluable tool for lentiviral infection research and vaccine evaluation (7, 8).
Studies have shown that major histocompatibility complex class I (MHC-I) molecules are critical for activating cytotoxic T lymphocytes (CTLs) to control HIV infection (9–12). CD8+ T-cell proliferation responses control FIV infection, and MHC-I molecule expression on microglial cells is transiently upregulated at the onset of FIV reproduction (13–17). Elevated MHC expression can also be observed in HIV and simian immunodeficiency virus (SIV) (18, 19). The CTL immune response is stimulated after MHC-I presents viral peptides to specific T-cell receptors (TCRs) (20–22). These results show that the domestic cat MHC-I may act against FIV similarly to the macaque MHC-I against SIV and to the human MHC-I against HIV.
The epitope peptides are fixed in the peptide binding groove (PBG) of the MHC-I heavy chain by six pockets (A to F) (23). MHC-I polymorphisms allow these pockets to be highly variable. One MHC-I molecule can bind only CTL epitopes with specific motifs based on compatibility with the PBG pockets (24, 25). Therefore, clarifying the pocket's peptide binding mechanism is the foundation for studying MHC-I-induced antiviral CTL responses and for developing polypeptide vaccines. Interactions between the bound peptide and the pockets have been explored using crystal structures of the MHC-I heavy chain, epitope peptides, and β2-microglobulin (β2m) complexes in species ranging from mammals to chickens (15, 26–31). The N- and C-terminal residues of the peptide insert into the pockets at the N and C termini of the PBG, and the middle of the PBG peptide arch contacts the TCR. The B and F pockets can accommodate the side chains at position 2 (P2) and the C-terminal (PC) residues of the peptides, respectively, to fix the peptide in place (32). The D pocket can accommodate the side chains of the P3 residue and is vital for peptide binding (29). The C or E pocket usually interacts with the side chain of the residue at the lowest point in the middle of the peptide (29, 32). These five pockets interact with the peptide residue side chains to determine the peptide binding motif of the MHC-I molecule, particularly the B, D, and F pockets, as they consist of highly variable residues that have different properties and strong interactions with their preferred residues. Compared to the B to F pockets, the A pocket is highly conserved and primarily interacts with the main chain of the P1 residue. The side chain of the P1 residue stretches out of pocket A; therefore, pocket A is not a factor in determining the peptide binding motif of the MHC-I molecule (29, 33). However, the domestic cat MHC-I peptide binding motifs and the FIV epitopes have not yet been characterized.
Felis catus MHC-I, also termed feline lymphocyte antigen class I (FLA I), was first cloned in 1988 (34), and its genomic region was sequenced in 2008 (35). Three loci (FLA I-E, -H, and -K) are believed to encode the classic FLA I molecules that induce CTL responses in F. catus (35). Despite the prominent roles of FIV and FLA I in modeling studies of the anti-HIV CTL response, little is known about how FLA I presents FIV peptides.
FLA-E*01801 (previously known as FLA-B*n06) has been used to study the immune effect of the FIV vaccine (36). To illustrate the structural basis of FIV peptide presentation by FLA I, we solved the crystal structure of FLA-E*01801 in complex with a nonameric peptide derived from an FIV gag protein (RMA9 [RMANVSTGR]). Surprisingly, the A pocket of the FLA-E*01801 complex restricted peptide binding and blocked peptide binding via interactions with the P1D residue. According to mutation analysis and in vitro refolding analyses, Trp167 in the A pocket is directly related to this unusual peptide binding method. The crystal structure of the mutated FLA-E*01801 complex with Ser167 indicated that the confined space of the A pocket caused by the large bulk of Trp167 might explain why the A pocket cannot accommodate the P1D residue. We also confirmed the peptide binding motifs of the B and F pockets and identified the FIV nonamer peptides that may be presented by FLA-E*01801. Our study reveals the structural basis of FLA I for use in the development of FIV and HIV vaccines and improves our understanding of how MHC-I molecules present viral peptides.
RESULTS
Overall structure of the FLA-E*01801/peptide complexes.
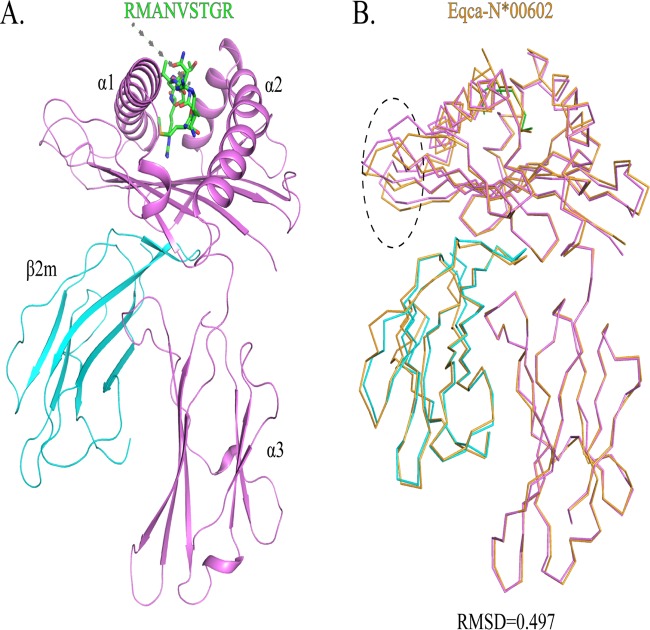
Nine peptides from FIV, feline circovirus (FCV), and feline panleukopenia virus (FPV) were predicted in silico (http://www.cbs.dtu.dk/services/NetMHCpan/) to be capable of binding to FLA-E*01801, and in an in vitro refolding experiment, two peptides formed stable complexes with FLA-E*01801 and fβ2m (see Table S1 in the supplemental material). One peptide, RMA9, was crystallized with FLA-E*01801 and feline β2m (fβ2m) in the P212121 space group with a resolution of 2.1 Å (Table 1). Only one FLA-E*01801 complex was contained within one asymmetric unit. The FLA-E*01801 complex displayed a canonical MHC-I complex structure, consisting of α1, α2, and α3 heavy-chain domains and the light-chain domain fβ2m. The peptide is located in the PBG formed by the α1 and α2 domains (Fig. 1A). The FLA-E*01801 complex structure is more similar to that of the horse MHC-I (Eqca-N*00602) than to other known MHC-I structures according to the Dali server (http://ekhidna.biocenter.helsinki.fi/dali_server). The root mean square difference (RMSD) of these structures is only 0.497 Å, indicating an identical structural conformation (Fig. 1B). The most noteworthy difference lies in the flexible loops in the α1 domain.
TABLE 1.
Data collection and refinement statistics for FLA-E*01801 and FLA-E*01801-167W/S
| Parameter | Value(s)a for: |
|
|---|---|---|
| FLA-E*01801 | FLA-E*01801-167W/S | |
| Data collection | ||
| Space group | P212121 | P212121 |
| Cell dimensions | ||
| a, b, c (Å) | 47.421, 83.379, 121.636 | 47.12, 82.64, 121.13 |
| α, β, γ (°) | 90, 90, 90 | 90, 90, 90 |
| Resolution (Å) | 50.0–2.1 (2.103–2.157) | 50.00–2.90 (3.06–2.90) |
| No. of reflections | ||
| Total | 953,393 | 18,631 |
| Unique | 27,016 | 18,625 |
| Rsym or Rmergeb | 0.063 (0.542) | 0.092 (0.568) |
| I/σI | 30.971 (4.2) | 7.3 (2.8) |
| Completeness (%) | 99.55 (98.95) | 83.8 (68.5) |
| Redundancy | 4.5 (4.7) | 4.1 (4.1) |
| Refinement | ||
| Resolution (Å) | 50.00–2.10 | 50.00–2.90 |
| No. of reflections | 27,016 | 8,745 |
| Rwork/Rfree (%)c | 21.33/25.52 | 22.08/25.00 |
| RMSDs | ||
| Bond lengths (Å) | 0.005 | 0.007 |
| Bond angles (°) | 0.919 | 1.077 |
| Average B factor | 35.4 | 25.6 |
| Ramachandran plot quality | ||
| Most favored region (%) | 97.35 | 94.6 |
| Allowed region (%) | 2.39 | 2.92 |
| Disallowed (%) | 0.27 | 0.00 |
Values in parentheses are for the highest-resolution shell.
Rmerge = Σhkl Σi |Ii(hkl) − <I(hkl)> |/Σhkl Σi Ii(hkl), where Ii(hkl) is the observed intensity and <I(hkl)> is the average intensity from multiple measurements.
R = Σhkl || Fobs | − k | Fcalc | |Σhkl | Fobs |, where Rfree is calculated for a randomly chosen 5% of reflections and Rwork is calculated for the remaining 95% of reflections used for structure refinement.
FIG 1.
Structure of the domestic cat MHC class I molecule FLA-E*01801. (A) Stereo view of the FLA-E*01801 complex. The α1, α2, and α3 domains form the FLA-E*01801 complex heavy chain, which is illustrated in purple; fβ2m is cyan; and the pol peptide (RMA9 [RMANVSTGR]) is a stick model colored by atom type (C, green; N, blue; and O, red). (B) Superposition of the structure of the FLA-E*01801 molecule with that of equine MHC-I Eqca-N*00602. The RMSD is 0.479, and the area with the greatest difference is marked with an oval in the α1 domain.
Species-specific features of the FLA I sequence.
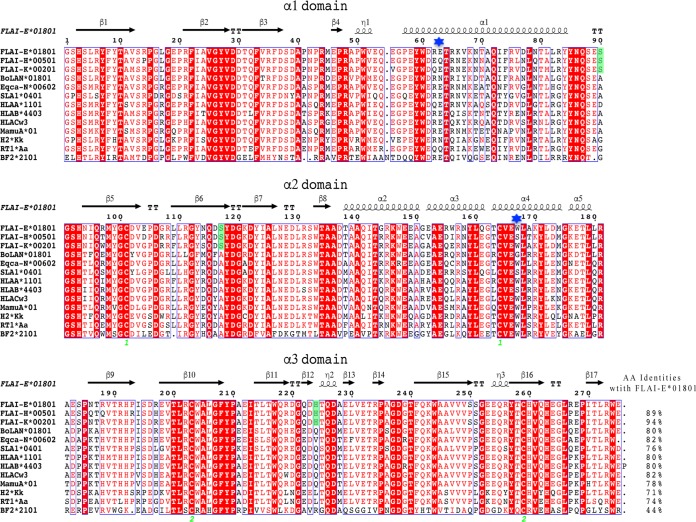
To analyze FLA I diversity, FLA-E, FLA-H and FLA-K sequences were aligned with typical MHC-I molecules from cattle, horse, swine, human, monkey, mouse, rat, and chicken (Fig. 2). All FLA I alleles coincide with the prospective model of polymorphic and conserved sites in the α1/α2 domains (36). Three hypervariable regions (HVRs) were identified, namely, positions 62 to 81 in the α-helix of the α1 domain, positions 94 to 116 in the α-helix, and positions 94 to 116 in the β-strand of the α2 domain. Similarly, those three segments correspond to the same residue stretches in other vertebrates. The homology of FLA I in mammals is higher than that in nonmammals. FLA-E*01801 is 81.82% identical to HLA-Cw3 molecules, which are most similar to MHC-I molecules. NCBI BLAST database searches found 3 amino acid differences among the FLA I alleles and other crystallized vertebrate class I molecules. In FLA I alleles, Ser90, Ser117, and His224 are highly conserved, and Ser117 is located in the F pocket, which may influence peptide binding.
FIG 2.
Structure-based amino acid sequence alignment of FLA-E*01801 and other representative crystallized MHC-I molecules. Black arrows above the alignment show β-strands; cylinders indicate α-helices. Green numbers indicate residues that form disulfide bonds. Conserved residues are in red. Residues in green are species-specific amino acids that differ between cats and other animals according to NCBI BLAST database searches. The residues at position 63 and 167 are marked by blue stars. The total amino acid (AA) identities between FLA-E*01801 and the listed MHC-I molecules are shown at the end of each sequence.
Viral peptide presentation and pocket characteristics of the FLA-E*01801 complex.
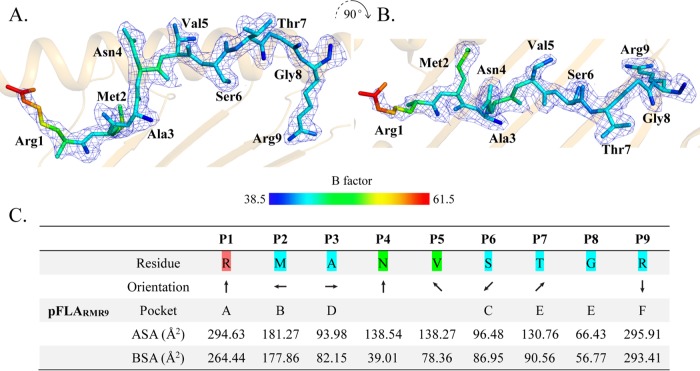
The RMA9 peptide adopts an overall “M” conformation (Fig. 3A and B), and the electron density map indicates that the conformation is stable. The B factor of the RMA9 N terminus is relatively high compared to that of the other regions. Information on the exposed accessible surface area (ASA) and buried surface area (BSA) of each RMA9 residue is shown in Fig. 3C.
FIG 3.
Structure of RMA9 presented by FLA-E*01801. (A and B) The peptide is presented with electron density in two vertical directions, and the peptide coloration is based on the isotropic B factors. (C) Side chain orientation of RMA9 in the FLA-E*01801 structure in the side view from the peptide N terminus to the C terminus. An arrow pointing up indicates that an amino acid residue is oriented toward the TCR, an arrow pointing down indicates that an amino acid residue is oriented toward the bottom of the PBG, an arrow pointing left indicates orientation toward the α1 helix domain; and an arrow pointing right indicates orientation toward the α2 helix domain. The residues of the accommodating pockets are listed under the relevant anchors in the PBG. ASA and BSA represent the exposed area and the buried area of each RMA9 peptide residue, respectively.
Examining the orientation of the RMA9 side chains revealed that the side chains of residues P2, P3, P6, and P9 are inserted into the corresponding pockets and the side chains of residues P1, P4, P5, and P7 extend out of the PBG. Previous MHC-I structural studies showed the residues P2 and P9 to be the primary anchor residues and that P3 and P6 also affect peptide binding. The P4, P5, and P7 residues at the top of the arch formed by RMA9 are most likely recognized by specific TCRs.
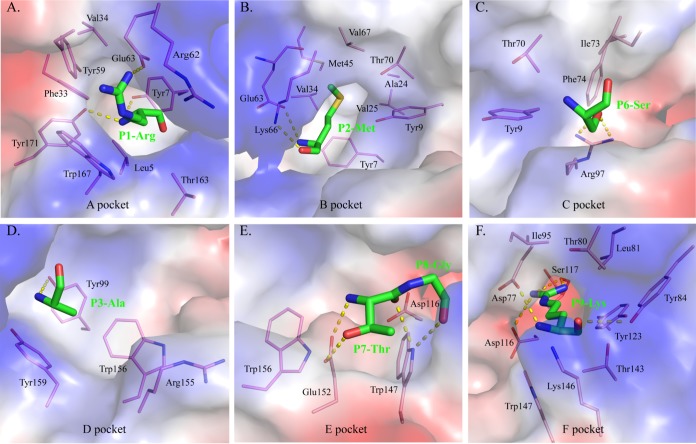
The A pocket of FLA-E*01801, which consists of Leu5, Tyr7, Phe33, Val34, Tyr59, Arg62, Glu63, Thr163, Trp167, and Tyr171, fixes the position of P1R via hydrogen bonding and strong van der Waals forces (VDWs) (Fig. 4A; Table 2). In general, the A pocket is nonrestrictive in the PBG because it binds the main chain of the P1 residue in a common mode among MHC-I structures, and the side chain of the P1 residue extends upward out of the A pocket (33).
FIG 4.
Composition of the FLA-E*01801 pockets. The pockets are shown as surfaces colored by their vacuum electrostatics. Red represents negatively charged residues, blue represents positively charged residues, and gray indicates noncharged residues. The residues composing these pockets are shown as labeled sticks. Residues bound by these pockets are shown as green sticks. The hydrogen bonds between the residues and pockets are shown as yellow dashed lines. (A) Pocket A with the P1 residue R; (B) pocket B with the P2 residue M; (C) pocket C with the P6 residue S; (D) pocket D with the P3 residue A; (E) pocket E with the P7 and P8 residues; (F) pocket F with the P9 residue K.
TABLE 2.
Hydrogen bonds and van der Waals interactions between RMA9 peptide and FLA-E*n01801
| Complex | Hydrogen bonds and salt bridges |
van der Waals contact residuesa | |||
|---|---|---|---|---|---|
| Peptide |
Heavy chain |
||||
| Residue | Atom | Residue | Atom | ||
| P1-Arg | N | Tyr171 | OH | Leu5, Tyr7, Tyr59, Arg62, Glu63, Lys66, Tyr159, Thr163, Trp167, Tyr171 (93) | |
| Tyr7 | |||||
| NH2 | Glu63 | OE2 (S) | |||
| O | Tyr159 | OH | |||
| P2-Met | N | Glu63 | OE1 | Tyr7, Tyr9, Met45, Glu63, Lys66, Val67, Tyr99, Tyr159 (70) | |
| O | Lys66 | NZ | |||
| P3-Ala | N | Tyr99 | OH | Tyr9, Lys66, Tyr99, Trpt156, Tyr159 (31) | |
| P4-Asn | Lys66 (10) | ||||
| P5-Val | Asn69, Thr70, Ile73 (17) | ||||
| FLA-E*n01801 | P6-Ser | Og | Arg97 | NH1 | Ile73, Arg97, Glu152 (13) |
| Arg97 | NH2 | ||||
| P7-Thr | N | Glu152 | OE2 | Ile73, Lys146, Trp147, Ala150, Glu152 (26) | |
| Glu152 | OE1 | ||||
| Og1 | Arg155 | NH1 | |||
| O | Trp147 | NE1 | |||
| P8-Gly | O | Lys146 | NZ | Ile73, Asp77, Lys146, Trp147 (21) | |
| Trp147 | NE1 | ||||
| P9-Arg | N | Asp77 | OD1 | Asp77, Thr80, Leu81, Tyr84, Ile95, Gln96, Asp116, Ser117, Tyr118, Tyr123, Thr143, Lys146, Trp147 (94) | |
| NH1 | Ser117 | O | |||
| NH2 | Ser117 | O | |||
| Asp116 | OD2 (S) | ||||
| O | Tyr84 | OH | |||
Numbers in parentheses are the amounts of van der Waals force.
The B and F pockets are the primary anchor sites at the N and C termini of the PBG and restrict peptide binding. The side chains of the P2M and P9R residues are inserted into the B and F pockets of FLA-E*01801, respectively. The compositions of these two pockets and their interactions with P2M and P9R are shown in Table 2 and Fig. 4B and F. The environment of the B pocket is hydrophobic, whereas that of the F pocket is negatively charged. P2M and P9R fit perfectly into the B and F pockets, respectively, and determine the binding stability of RMA9 and FLA-E*01801.
The C, D, and E pockets usually accommodate the residue side chains in the middle of the peptide and affect peptide binding. The amino acid compositions of these three FLA-E*01801 pockets are shown in Fig. 4C to E. The C and D pockets interact with the side chains of the P3 and P6 residues, respectively. The E pocket interacts with the main chain of the P7 and P8 residues because the P7 side chain is oriented upward, and P8G has no side chain (Table 2).
Pocket A restricts FLA-E*01801 peptide binding.
The F pocket of FLA-E*01801 prefers positively charged residues due to its negatively charged environment. Three of the tested peptides (RMA9, KMV9 [KMVSIFMEK], and DTV9 [DTVTNTIGK]) fit this requirement, and two can bind FLA-E*01801 efficiently (with the exception of DTV9) (Fig. 5A). As the primary anchor residue, P2T of DTV9 was expected to be the main reason for the poor refolding if this peptide is not suitable for the B pocket. However, we mutated P2T of DTV9 to P2M, and the refolding results showed that P2M did not restore the refolding of DMV9 and FLA-E*01801 (Fig. 5A). We also mutated P2M to P2T, as found in KMV9 and RMA9, and found that these new mutated peptides formed a complex with FLA-E*01801 similar to that of the wild-type peptide (Fig. 5A). After excluding the primary anchor residue at P2, the secondary anchor residues at P3 and P6 became the most likely factors responsible for the poor refolding of DTV9. The P6 residues of DTV9 and RMA9 were mutated to Phe to remain consistent with KMV9, and the mutated peptides were tested by in vitro refolding. However, the results showed that the P6 residue was likewise not the cause (Fig. 5A). A single anchor residue could not alter the DTV9 refolding, but a combination of anchor residues might cause the poor refolding. We mutated the P2 and P6 residues of DTV9 such that all the anchor residues matched those of KMV9. However, this mutated peptide did not improve the refolding results (Fig. 5A). Together, these mutations demonstrated that none of the canonical anchor residues caused the poor refolding of DTV9.
FIG 5.
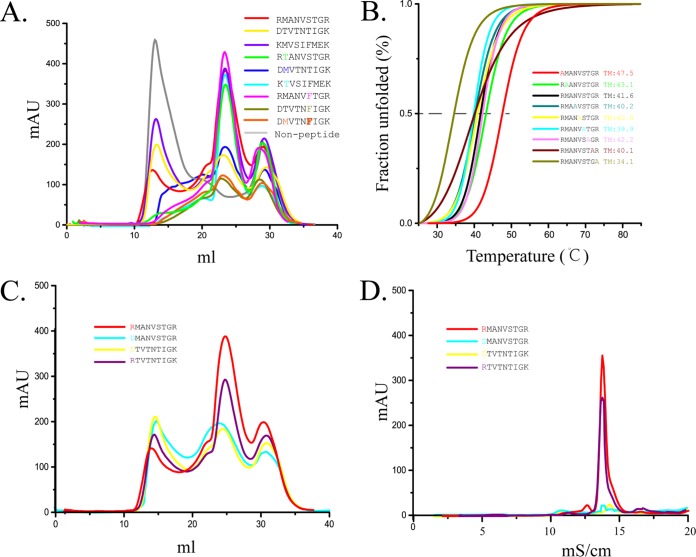
Peptide binding with FLA-E*01801 indicated by in vitro refolding. Anchor residues of the FLA-E*01801 complex were tested by alanine scanning, CD spectroscopy, and peptide mutation. (A) FLA-E*01801 with three different wild-type peptides (RMA9, KMV9, and DTV9) and six mutant peptides (RMA9-P2T, KMV9-P2T, DTV9-P2M, RMA9-P6F, DTV9-P6F, and DTV9-P2MP6F) refolded in vitro and analyzed by chromatography on a Superdex 200 16/60 column. FLA-E*01801 without a peptide was included as a negative control. Peaks 1, 2, and 3 represent the heavy-chain polymer, the correctly refolded FLA-E*01801 complex, and the excess fβ2m, respectively. The relevant concentration ratios and altitudes of peak 2 formed by complexes with different peptides represent the refolding efficiencies. (B) Thermal stabilities of the FLA-E*01801 complex. The thermal stabilities of FLA-E*01801 with nine peptides (RMA9, RMA9-P1A, RMA9-P2A, RMA9-P4A, RMA9-P5A, RMA9-P6A, RMA9-P7A, RMA9-P8A, and RMA9-P9A) were tested by CD spectroscopy. (C) Gel filtration chromatograms of FLA-E*01801 complex with RMA9, RMA9-P1D, DTV9, and DTV9-P1R. (D) Anion-exchange chromatography of FLA-E*01801 complexes with RMA9, RMA9-P1D, DTV9, and DTV9-P1R. The complexes with RMA9 and DTV9-P1R were eluted at NaCl concentrations of 12% to 15%. The refolded complex proteins with RMA9-P1D and DTV9 disaggregated at NaCl concentrations of 12% to 15%, indicating instability.
To identify the vital restriction pockets for peptide binding, the peptide RMV9 was mutated by alanine scanning, and circular-dichroism (CD) spectroscopy was used to assess the stability of the FLA-E*01801 complexes with these mutant peptides (Fig. 5B). The midpoint transition temperature (Tm) value of the wild-type RMV9 peptide, which was used as the control, was 41.6°C. The Tm value of the P9A mutant peptide was significantly lower (34.1°C), indicating that the P9 residue was the primary anchor residue. Mutating the remaining anchor residues (P2, P3, and P6) did not show significant changes with respect to the wild-type RMA9 peptide. These CD results were consistent with the mutation experiments described above. The P1A mutant peptide had the highest Tm value (47.5°C) among the tested peptides. This result suggested that the difference in the P1 residue might be the reason for the unexpected poor refolding in DTV9.
Consequently, the P1 residues of RMA9 and DTV9 were interconverted and tested by in vitro refolding. The results revealed that the P1 residue was the key factor. The refolding of RMA9-P1D was similar to that of DTV9, and the refolding of DTV9-P1R was similar to that of RMA9 (Fig. 5C). Because P1R and P1D are charged residues, we also used anion-exchange chromatography to test their electronic stabilities. Both complexes with the P1R peptides were stable in a strongly ionic environment, whereas the complexes with the P1D peptides were dissociated (Fig. 5D).
As shown in previous studies, the A pocket does not restrict MHC-I peptide binding because pocket A binds primarily with the main chain of the P1 residue via conserved residues, such as Tyr7 and Tyr171, and the side chain of the P1 residue stretches out of this pocket (Fig. 4A). However, our experimental data demonstrated that the P1 residue is the vital anchor residue and that the A pocket can restrict the binding of peptides to FLA-E*01801.
Glu63 and Trp167 play important roles in restricting pocket A of FLA-E*01801.
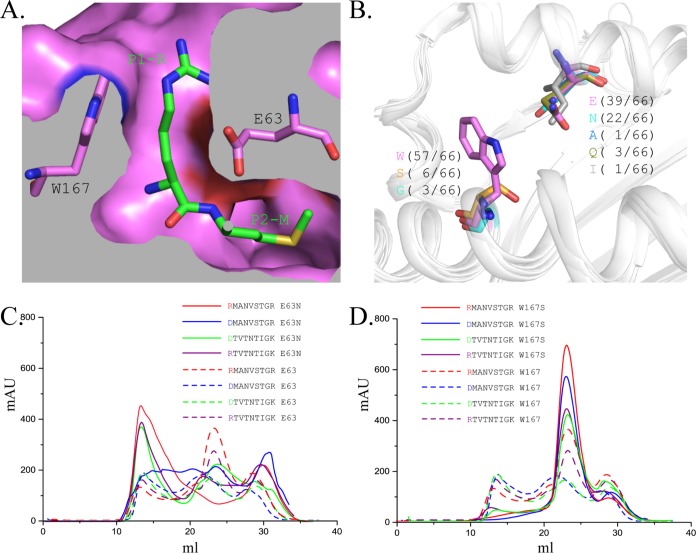
The filtration of the P1 residues of FLA-E*01801 must be determined by the residues of pocket A, which interact with the side chain of residue P1. Glu63 and Trp167 were the most likely candidates because they fixed the side chain of P1R via hydrogen bonds, a salt bridge, and VDWs (Fig. 6A). The residues at those two positions are not conserved in other solved MHC-I structures, and Asn63 and Ser167 appear with the second highest frequencies (Fig. 6B). To test the relative binding abilities, Glu63 and Trp167 were mutated into Asn63 and Ser167 (named FLA-E*01801-63E/N and FLA-E*01801-63E/N), respectively.
FIG 6.
Pocket A restriction was confirmed by in vitro refolding with the mutant FLA-E*01801-167W/S and mutant peptides. (A) The relative positions of Glu63, Trp167, and P1-R in the three-dimensional structure are shown. (B) Statistical analysis of the residue types at positions 63 and 167 in the solved MHC-I structures. The residues at positions 63 and 167 are shown as sticks, and the other parts of the α1 and α2 domains are shown as cartoons. (C) Gel filtration chromatograms of the wild-type FLA-E*01801 (dashed lines) and mutant FLA-E*01801-63E/N (solid lines) with the peptides RMA9, DTV9, RMA9-P1D, and DTV9-P1R. (D) Gel filtration chromatograms of the wild-type FLA-E*01801 (dashed lines) and the FLA-E*01801-167W/S (solid lines) with peptides RMA9, DTV9, RMA9-P1D, and DTV9-P1R. The refolding efficiencies of FLA-E*01801-167W/S are higher than those of the wild-type FLA-E*01801.
Peptides with P1R or P1D were refolded with the two mutated FLA-E*01801 heavy chains and fβ2m. The refolding results for FLA-E*01801-63E/N showed that no stable complexes were formed, even with the crystallized RMA9 (Fig. 6C). One reason may be that the residue at position 63 is part of both the A and B pockets. The change from Glu63 to Asn63 might alter the FLA-E*01801 B pocket, making P2M and P2T unsuitable anchor residues. The refolding results for FLA-E*01801-167W/S showed that all four peptides formed stable complexes (Fig. 6D). Moreover, the refolding efficiencies of FLA-E*01801-167W/S were much higher than the refolding efficiency of the wild-type FLA-E*01801. Therefore, Trp167 is likely one of the critical residues that determines the A pocket restriction.
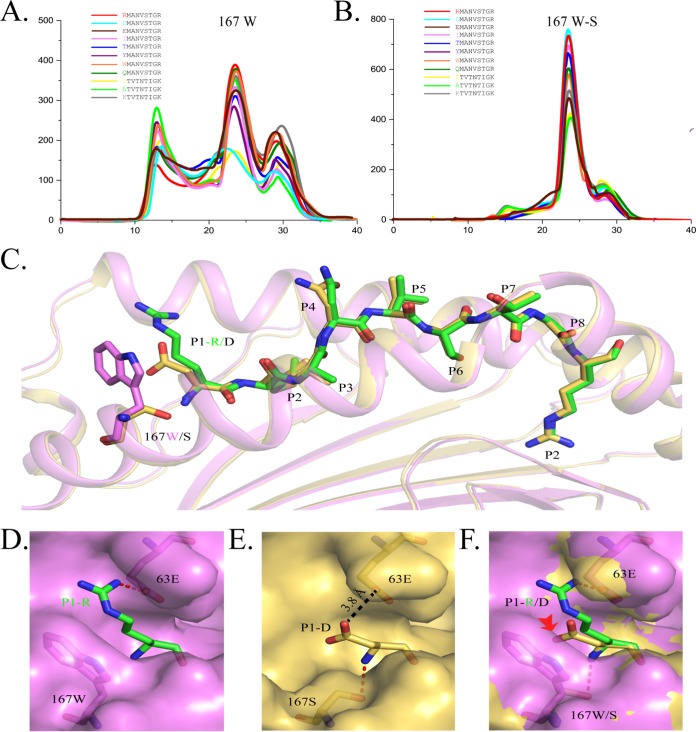
Next, we investigated which peptides could be accommodated by the A pocket of FLA-E*01801. Amino acids can be classified into several different types based on their side chain properties, such as size, polarity, and charge characteristics. The P1 residue of RMA9 or DTV9 was mutated to various residues representing the different amino acid types and tested in the in vitro refolding assay. With the exception of Asp, all residues were able to bind to the A pocket, regardless of whether they were small (Ala) or large (Tyr and Trp), hydrophilic (Thr and Asn) or hydrophobic (Ile), or positively (Arg and Lys) or negatively (Glu) charged (Fig. 7A). For example, when considering Asn and Glu, one has a similar size to Asp, and the other has the same electronic characteristics as Asp. Only the combination of the size and electronic characteristics of P1D might be sufficient for rejection by the A pocket. All peptides could form stable complexes with the Ser167 mutated heavy chain and had higher refolding efficiencies (Fig. 7B).
FIG 7.
Determination of the suitable peptide binding motif for the A pocket by in vitro refolding with mutant peptides and the structure of RMA9-P1D presented by FLA-E*01801-167W/S. (A) Gel filtration chromatograms of the wild-type peptide RMA9 and its P1 mutants (P1D, P1I, P1T, P1Y, P1W, and P1Q) and of DTV9 and its P1 mutants (P1A and P1K). (B) Gel filtration chromatograms of the wild-type RMA9 and its P1 mutants (P1D, P1I, P1T, P1Y, P1W, and P1Q) and of DTV9 and its P1 mutants (P1A and P1K) with FLA-E*01801-167W/S. (D) Superposed structures of FLA-E*01801-167W/S and FLA-E*01801. The peptides RMA9-P1D (yellow) and RMA9 (green) are shown as stick models, and the residues in position 167 (Trp167 is magenta and Ser167 is yellow) are shown as stick models. (E) The surface of the A pocket (magenta) in FLA-E*01801 with P1R. P1R (green), Glu63 (magenta), and Trp167 (magenta) are shown as stick models. Hydrogen bonds can be formed by P1R and Glu63 (red dotted line). (F) The surface of the A pocket (yellow) in FLA-E*01801-167W/S with P1D. P1D, Glu63, and Ser167 are shown as stick models in yellow. Hydrogen bonds can be formed by P1D and Ser167 (red dotted line). The distance between the carboxyl groups of P1D and Glu63 in FLA-E*01801-167W/S is 3.8 Å (black dotted line). (D) Comparison of the A pockets of FLA-E*01801-167W/S (yellow) and FLA-E*01801 (magenta).
To determine why Trp167 restricts the A pocket while Ser167 does not, the crystal structure of the mutated FLA-E*01801-167W/S with RMA9-P1D was solved. The peptide presentations of the mutated and wild-type FLA-E*01801 are shown in Fig. 7C. Except for the mutated sites, the two are almost identical. Trp167 is large and closes the left side of the A pocket (Fig. 7D). In contrast, the A pocket containing the smaller Ser167, as in HLA-B*4402, is open, and the P1D residue side chain stretches through it at a sufficient distance to avoid repulsion by Glu63 (Fig. 7E). This superposition reveals that Trp167 blocks the P1 residue side chain and causes Asp to be repelled by Glu63 in the limited space. This hindrance should be reason for the rejection of P1D by the A pocket of FLA-E*01801.
Peptide binding motif of FLA-E*01801 and the FIV peptide binding map.
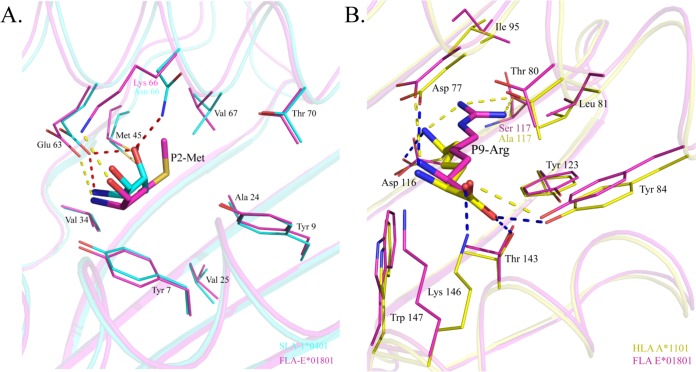
To determine the peptide binding motif of FLA-E*01801, the residues that can be accommodated by the B and F pockets were analyzed. The sequence and structural comparison revealed that the B pocket of FLA-E*01801 is most similar to the B pocket of SLA-1*0401 (Fig. 8A). Only one residue differs between the B pockets of these two MHC-I structures (66K/FLA-E*01801 and 66N/SLA-1*0401). Examination of SLA-1*0401 indicated that the B pocket can accommodate multiple residues, including M/T/A/V/I/L/S. We mutated P2M of RMA9 into other residues to test the residue binding scope of the FLA-E*01801 B pocket and found that it can bind to M/T/A/V/I/L/S (Table S1), which was consistent with the results of the previous study (29).
FIG 8.
Comparison of the B and F pockets of FLA-E*01801 with those of other solved MHC-I structures. The residues forming the pockets and the peptides accommodated by the pockets are shown as stick models. Dashed lines represent hydrogen bonds between the peptides and the pockets. (A) B pocket alignment between FLA-E*01801 and SLA-1*0401 (PDB code 3QQ3). (B) F pocket alignment between FLA-E*01801 and HLA-A*1101 (PDB code 1X7Q).
The F pocket of FLA-E*01801 is negatively charged and forms a strong salt bridge with the positively charged Arg. It is most similar to the F pocket of HLA-A*1101, and only one residue, S117 of FLA-E*01801 and A117 of HLA-A*1101, differs between them (Fig. 8B). Our refolding data confirmed that the F pocket of FLA-E*01801 prefers the positively charged residues Arg and Lys, as reported for HLA-A*1101 in previous studies (37) (Fig. 4A; see also Table S2).
Based on our structural and refolding data, we summarized the primary peptide FLA-E*01801 binding motif as follows: X (except D)-(M/T/A/V/I/L/S)-X-X-X-X-X-X-X-X-(R/K). The FIV proteins were screened, and 125 peptides fitting this motif were identified (Table S3). Among those peptides, NTP9 (NTPVFAIKK) and NAG9 (NAGKFRRAR) are reported in previous studies (38, 39). Their binding to FLA-E*01801 was tested by in vitro refolding (Table S1). Our peptides predicted by structure-based analysis to bind FLA-E*01801 were compared with the peptides predicted by NetMHCpan. Peptides fitting either only the NetMHCpan prediction or only our summarized motif were synthesized, and the in vitro refolding results showed that our structure-based prediction performed better than that of NetMHCpan. These finding are presented in Table S1.
DISCUSSION
A new peptide presentation mode was recently reported in which HLA-B*57:01 presents N-terminally extended peptides (40). Previously, the A pocket was thought not to restrict the P1 residue but to accommodate any residue for two reasons: the A pocket forms strong hydrogen bonds with the invariable P1 residue main chain through residues that are conserved among different alleles, such as Tyr7 and Tyr171 (Fig. 4A), and the variable side chain of the P1 residue stretches up and out of this pocket. Thus, the current methods used to predict and analyze the peptide binding motifs of certain MHC-I molecules do not account for the influence of the A pocket. In this study, we found that FLA-E*01801 is not able to bind peptides with P1D, indicating that the A pocket also restricts certain P1 residues. However, this result led to more questions requiring further study. First, is the A pocket restriction to P1 residues universal in other MHC-I alleles, or does it occur only in alleles with specific sequences? Based on the refolding and crystal structures of the wild-type and mutated FLA-E*01801, we believe that the A pocket does not accommodate P1D because its positively charged carboxyl group conflicts with the positively charged Glu63 carboxyl group in the limited space resulting from the large heterocycle of Trp167. We screened other published MHC-I structures that also contain Glu63 and Trp167, including HLA-A02/A11/B15/B27/B41/B52/B57/C08/CW3, H2-Db/Dd/Kb, and BF2-04/21; however, no bound peptide with P1D was found (Table S2). Searching the peptide repertoire of some extensively studied alleles in databases, e.g., HLA-A02 and H-2Kb in IEDB (http://www.iedb.org), showed that Asp was disfavored at the P1 position but was not absent. Two reasons are possible. The first is the variations of the A and B pocket in FLA-E*01801, such as Met5 and Gly62 in HLA-A*02:01 and Tyr45 in H-2Kb. These variations may alter the steric freedom of Trp167 and Glu63 and influence the accommodation of P1D. The second is the role of the remaining anchor residues of the bound peptides. Some strong anchor residues at other positions, such as P2 or P9, may compensate for the loss of binding affinity caused by P1D. This compensation suggests that P1D rejection may not be absolutely universal in MHC-I alleles containing Glu63 and Trp167. Second, to what extent does the A pocket restrict P1 residues? Typical restricting pockets, such as B or F, accommodate residues with similar properties, such as charge, polarity, and size. According to a comprehensive analysis of the P1-mutated peptides and other MHC-I structures with Glu63 and Trp167, the FLA-E*01801 A pocket appears to accommodate all residues except Asp, regardless of size, polarity, and charge (Fig. 6A; see also Table S1). Are alleles with different A pocket compositions also restricted to specific P1 residues? The possibility of other critical A pocket residues and their influence on peptide binding require further study.
The domestic cat is an attractive model for studying viral pathology and immunology in humans, especially for researching AIDS. The interaction of FIV with its primary receptor changes during the disease development, which is parallel to the way HIV switches its receptor as AIDS progresses (41). FIV is the only lentivirus for which a commercial vaccine is available for prevention in either human or veterinary medicine. It is beneficial to research the protective mechanisms of lentivirus vaccines in humans and, at the same time, has comparative value in AIDS vaccine research. In this study, we determined the primary peptide binding motif of FLA-E*01801 and depicted its FIV peptide binding map. Some peptides, such as NTPVFAIKK, were predicted to be restricted by HLA A*0301, A*6601 and C*0102 (38). Interestingly, NTPVFAIKK is conserved in both HIV and FIV. Another peptide, NAGKFRRAR, is a part of a long peptide that can induce immunity (39), but most peptides were identified the first time. These potential FIV epitopes will benefit both the further study of FLA I-mediated anti-FIV responses and vaccine development. The alleles that include the same or similar HVRs likely have overlapped peptide binding preferences and present the same epitope peptides.
Studying MHC-I structures from species other than humans and mice, including swine, cattle, monkeys, horses, dogs, and chickens, can provide new perspectives and a better understanding of issues such as the influence of PBG flexibility and shape and the critical role of a single residue in the pocket. In this report, we provide the first crystal structure of FLA I with an FIV peptide to facilitate the study of FLA I-mediated anti-FIV responses. In addition, we found that the A pocket restricts the P1 residues, illustrating the impact of the A pocket on the peptide binding motif.
MATERIALS AND METHODS
Peptide synthesis.
Forty-seven peptides were used in these experiments (Table S1). Nine epitope peptides that potentially bound to FLA-E*01801 were predicted by the NetMHCpan4.0 server (http://www.cbs.dtu.dk/services/NetMHC/) based on the gag protein of FIV (GenBank accession no. GU055218), the VP2 protein of FCV (GenBank accession no. APB53908.1), and the VP1 protein of FPV (GenBank accession no. AAA47154.1). These peptides were predicted automatically using the closest homologue (BoLA-D18.4) and purified by reverse-phase high-performance liquid chromatography (HPLC) (SciLight Biotechnology) with >90% purity. The other 38 mutant peptides used to determine the peptide binding motif of FLA-E*01801 were synthesized and purified in the same manner.
Protein preparation.
The extracellular regions of the class I H-chain FLA-E*01801 (GenBank accession no. EU915360.1; residues 25 to 299 of the mature protein) and feline β2m (fβ2m) (GenBank accession no. AY829266.1; residues 21 to 118 of the mature protein) were synthesized with the amino acid preferences of Escherichia coli and cloned into the pMA-T vector (Suzhou; GENEWIZ). To obtain the desired protein, the target fragments were digested with NdeI and XhoI, inserted into the prokaryotic expression vector pET21a(+) (Novagen), and expressed in E. coli Rosetta (DE3). FLA-E*01801 and fβ2m were expressed as inclusion bodies and purified as previously described (42).
To confirm the relationship between Glu63 and Trp167 in FLA-E*01801 and the P1 residues, Glu63 was mutated to Asn63 and Trp167 to Ser167 by overlap PCR (the primers used for the mutation were 5′-TTTCGCCAGGCTTTCCACGCA-3′, 5′-TGCGTGGAAAGCCTGGCGAAA-3′, 5′-TTTACGGGTGTTACGATCCCA-3′, and 5′-TGGGATCGTAACACCCGTAAA-3′, where the underlined sequences altered codons encoding Asn and Ser, respectively). The mutated genes (FLA-E*01801-63E/N and FLA-E*01801-167W/S) were inserted into the pET21(+) vector and expressed in Rosetta (DE3) cells. Recombinant FLA-E*01801-63E/N and FLA-E*01801-167W/S were expressed in inclusion bodies and further purified as described above.
FLA-E*01801 complex and FLA-E*01801-167W/S complex assembly.
Gradual dilutions were used to form the FLA-E*01801 and FLA-E*01801-167W/S complexes (43). Briefly, the FLA-E*01801 and fβ2m inclusion bodies were individually added to a solution of 6 M guanidine HCl and 50 mM Tris-HCl (pH 8.0). The complex was refolded by gradual dilution of the FLA-E*01801–fβ2m–RMA9 peptide at a 1:1:3 molar ratio followed by incubation for 48 h at 277 K. The refolded complex was concentrated and purified with a Superdex 200 16/60 column, followed by Resource Q anion-exchange chromatography (GE Healthcare). The purified proteins were buffer exchanged three times (10 μM Tris-HCl and 10 μM NaCl, pH 8).
FLA-E*01801-167W/S was refolded with fβ2m and RMA9-P1D in the same manner. The complex was formed by refolding and then purified by gel filtration and anion-exchange chromatography as described above.
Crystallization and data collection.
The crystallization conditions for the FLA-E*01801 complex were screened using the hanging-drop vapor diffusion method and the PEG/Ion kit (Hampton Research, Riverside, CA) at 277 K. The FLA-E*01801 crystal was obtained with PEG/ion solution no. 23 (0.2 M ammonium formate and 20% [wt/vol] polyethylene glycol 3350). Prior to X-ray diffraction, the crystals were soaked for several seconds in reservoir solution containing 25% glycerol as a cryoprotectant and then flash-cooled in a stream of gaseous nitrogen at 100 K (44). Diffraction data were collected to a resolution of 2.1 Å at Beamline BL17U (wavelength, 0.97892 Å) of the Shanghai Synchrotron Radiation Facility (Shanghai, China) using an R-AXIS IV++ imaging plate detector. The data were autoindexed and integrated using the DENZO program (45) and subsequently scaled and merged using the HKL-2000 software package (HKL Research).
To determine why Asp did not fit into the A pocket, the FLA-E*01801-167W/S complex was crystallized as described above, and a crystal of the FLA-E*01801-167W/S complex was obtained with PEG/ion 2 solution no. 28 (0.2 M sodium formate at pH 7.0 and 20% [wt/vol] polyethylene glycol 3350) at 277 K. Diffraction data for FLA-E*01801-167W/S were collected at a resolution of 2.9 Å. The crystallographic statistics for the models of the two complexes are listed in Table 1.
Structural determination and analysis.
The structure of the FLA-E*01801 complex was determined by molecular replacement with the MOLREP program using BoLA-A11N*01801 (Protein Data Bank code 3PWV, with the peptide excluded) as a search model. The structure of the FLA-E*01801-167W/S complex was determined by molecular replacement using the final model of the FLA-E*01801 complex. The comprehensive model was built manually using COOT (46), and refinement was restrained with REFMACS5 (47). Refinement rounds were implemented using the phenix refine program in the PHENIX package with isotropic atomic displacement parameter (ADP) refinement and bulk solvent modeling (48). Finally, the PROCHECK program was used to assess the stereochemical quality of the final model (49). The structural illustrations and the electron density-related figures were drawn using PyMOL (https://www.pymol.org). Multiple-sequence alignment was performed with Clustal Omega (http://www.ebi.ac.uk/Tools/msa/clustalo/) (50) and ESPript 3.0 (http://espript.ibcp.fr/ESPript/ESPript/) (51). ASA and BSA were calculated with PDBePISA (http://www.ebi.ac.uk/msd-srv/prot_int/pistart.html), and the B factor was calculated with CCP4.
Thermal stabilities of the peptide–MHC-I molecules.
The thermal stabilities of the FLA-E*01801 complexes with the mutant RMA9 peptides were tested on a CD instrument (Chirascan; Applied Photophysic, Ltd.). CD spectra were measured at 20°C using a Jasco J-810 spectrometer equipped with a water-circulating cell holder. The protein concentration was 10 μM in 20 mM (pH 8.0) Tris-HCl and 50 mM NaCl. A 1-mm optical path length cell was used to monitor the CD value at 218 nm as the temperature was increased from 25°C to 85°C at a rate of 1°C/min. The temperature of the protein solution was detected using a thermistor. The ratio of unfolded protein to the mean residue ellipticity (θ) was calculated using a standard method. The unfolded fraction (expressed as a percentage) is shown as (θ − θN)/(θU − θN), where θN and θU are the mean residue ellipticity values in the fully folded and the fully unfolded states, respectively. The midpoint transition temperature (Tm) was confirmed by denaturation curve data in the Origin 9.1 program (OriginLab) (52).
Mutation experiments.
FLA-E*01801-63E/N and FLA-E*01801-167W/S were refolded with fβ2m and the peptides RMA9, RMA9-P1D, RMA9-P1I, RMA9-P1T, RMA9-P1Y, RMA9-P1W, RMA9-P1Q, DTV9, DTV9-P1A, and DTV9-P1K, separately, and purified by gel filtration chromatography as described above.
Eighteen RMA9 peptide P2 mutants were synthesized to determine the binding motif in the B pocket, and six peptide mutants (RMA9-P2T, RMA9-P6F, DTV9-P2M, DTV9-P6F, DTV9-P2MP6F, and KMV9-P2T) were synthesized to determine the primary anchor residues at P2 and P6. Their binding affinities with FLA-E*01801 were detected by in vitro refolding as described above. Eight peptide mutants (RMA9-P1D, RMA9-P1I, RMA9-P1T, RMA9-P1Y, RMA9-P1W, RMA9-P1Q, KTV9-P1D, and KTV9-P1A) were used to determine which peptides could fit into the A pocket of FLA-E*01801. Their binding affinities with FLA-E*01801 and FLA-E*01801-167W/S were detected by in vitro refolding as described above. Additionally, four peptides (ARM9, KQR9, NTP9, and NAG9) were used to compare the binding of NetMHCpan-predicted peptides and our structured-based predicted peptides to FLA-E*01801. Their binding affinities with FLA-E*01801 were detected by in vitro refolding as described above.
Accession number(s).
The FLA-E*01801 and FLA-E*01801-167W/S complex structures were deposited in the Protein Data Bank with the following accession numbers: FLA-E*01801, 5XMF, and FLA-E*01801-167W/S, 5XMM.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the 863 Project of the China Ministry of Science and Technology (grant no. 2013AA102503), the National Natural Science Foundation of China (grant no. 31201887), and the 973 Project of the China Ministry of Science and Technology (grant no. 2013CB835302).
We acknowledge the assistance of the staff at the Shanghai Synchrotron Radiation Facility of China (SSRF) for diffraction data collection.
We declare no financial or commercial conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01631-17.
REFERENCES
- 1.Hartmann K. 2011. Clinical aspects of feline immunodeficiency and feline leukemia virus infection. Vet Immunol Immunopathol 143:190–201. doi: 10.1016/j.vetimm.2011.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Llano M, Saenz DT, Meehan A, Wongthida P, Peretz M, Walker WH, Teo W, Poeschla EM. 2006. An essential role for LEDGF/p75 in HIV integration. Science 314:461–464. doi: 10.1126/science.1132319. [DOI] [PubMed] [Google Scholar]
- 3.Kanzaki LI, Looney DJ. 2004. Feline immunodeficiency virus: a concise review. Front Biosci 9:370–377. doi: 10.2741/1235. [DOI] [PubMed] [Google Scholar]
- 4.Talbott RL, Sparger EE, Lovelace KM, Fitch WM, Pedersen NC, Luciw PA, Elder JH. 1989. Nucleotide sequence and genomic organization of feline immunodeficiency virus. Proc Natl Acad Sci U S A 86:5743–5747. doi: 10.1073/pnas.86.15.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Olmsted RA, Barnes AK, Yamamoto JK, Hirsch VM, Purcell RH, Johnson PR. 1989. Molecular cloning of feline immunodeficiency virus. Proc Natl Acad Sci U S A 86:2448–2452. doi: 10.1073/pnas.86.7.2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ackley CD, Yamamoto JK, Levy N, Pedersen NC, Cooper MD. 1990. Immunologic abnormalities in pathogen-free cats experimentally infected with feline immunodeficiency virus. J Virol 64:5652–5655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elder JH, Lin YC, Fink E, Grant CK. 2010. Feline immunodeficiency virus (FIV) as a model for study of lentivirus infections: parallels with HIV. Curr HIV Res 8:73–80. doi: 10.2174/157016210790416389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abbott JR, Sanou MP, Coleman JK, Yamamoto JK. 2011. Evolutionarily conserved T-cell epitopes on FIV for designing an HIV/AIDS vaccine. Vet Immunol Immunopathol 143:246–254. doi: 10.1016/j.vetimm.2011.06.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen SS, Tingstedt JL, Larsen TK, Brandt L, Gerstoft J, Kronborg G, Pedersen C, Fomsgaard A, Karlsson I. 2016. HIV-specific CD8+ T cell-mediated viral suppression correlates with the expression of CD57. J Acquir Immune Defic Syndr 71:8–16. [DOI] [PubMed] [Google Scholar]
- 10.Lécuroux C, Saez-Cirion A, Girault I, Versmisse P, Boufassa F, Avettand-Fenoel V, Rouzioux C, Meyer L, Pancino G, Lambotte O, Sinet M, Venet A. 2014. Both HLA-B*57 and plasma HIV RNA levels contribute to the HIV-specific CD8+ T cell response in HIV controllers. J Virol 88:176–187. doi: 10.1128/JVI.02098-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Benito JM, Lopez M, Soriano V. 2004. The role of CD8+ T-cell response in HIV infection. AIDS Rev 6:79–88. [PubMed] [Google Scholar]
- 12.Buseyne F, Le Gall S, Boccaccio C, Abastado JP, Lifson JD, Arthur LO, Riviere Y, Heard JM, Schwartz O. 2001. MHC-I-restricted presentation of HIV-1 virion antigens without viral replication. Nat Med 7:344–349. doi: 10.1038/85493. [DOI] [PubMed] [Google Scholar]
- 13.Aranyos AM, Roff SR, Pu R, Owen JL, Coleman JK, Yamamoto JK. 2016. An initial examination of the potential role of T-cell immunity in protection against feline immunodeficiency virus (FIV) infection. Vaccine 34:1480–1488. doi: 10.1016/j.vaccine.2016.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sanou MP, Roff SR, Mennella A, Sleasman JW, Rathore MH, Yamamoto JK, Levy JA. 2013. Evolutionarily conserved epitopes on human immunodeficiency virus type 1 (HIV-1) and feline immunodeficiency virus reverse transcriptases detected by HIV-1-infected subjects. J Virol 87:10004–10015. doi: 10.1128/JVI.00359-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Omori M, Pu R, Tanabe T, Hou W, Coleman JK, Arai M, Yamamoto JK. 2004. Cellular immune responses to feline immunodeficiency virus (FIV) induced by dual-subtype FIV vaccine. Vaccine 23:386–398. doi: 10.1016/j.vaccine.2004.05.032. [DOI] [PubMed] [Google Scholar]
- 16.Pu R, Omori M, Okada S, Rine SL, Lewis BA, Lipton E, Yamamoto JK. 1999. MHC-restricted protection of cats against FIV infection by adoptive transfer of immune cells from FIV-vaccinated donors. Cell Immunol 198:30–43. doi: 10.1006/cimm.1999.1574. [DOI] [PubMed] [Google Scholar]
- 17.Hein A, Martin JP, Dorries R. 2005. Early pathological changes in the central nervous system of acutely feline-immunodeficiency-virus-infected cats. Virology 343:162–170. doi: 10.1016/j.virol.2005.08.014. [DOI] [PubMed] [Google Scholar]
- 18.Clements JE, Babas T, Mankowski JL, Suryanarayana K, Piatak M Jr, Tarwater PM, Lifson JD, Zink MC. 2002. The central nervous system as a reservoir for simian immunodeficiency virus (SIV): steady-state levels of SIV DNA in brain from acute through asymptomatic infection. J Infect Dis 186:905–913. doi: 10.1086/343768. [DOI] [PubMed] [Google Scholar]
- 19.Gray F, Chimelli L, Mohr M, Clavelou P, Scaravilli F, Poirier J. 1991. Fulminating multiple sclerosis-like leukoencephalopathy revealing human immunodeficiency virus infection. Neurology 41:105–109. doi: 10.1212/WNL.41.1.105. [DOI] [PubMed] [Google Scholar]
- 20.Piepenbrink KH, Blevins SJ, Scott DR, Baker BM. 2013. The basis for limited specificity and MHC restriction in a T cell receptor interface. Nat Commun 4:1948. doi: 10.1038/ncomms2948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Clayberger C, Parham P, Rothbard J, Ludwig DS, Schoolnik GK, Krensky AM. 1987. HLA-A2 peptides can regulate cytolysis by human allogeneic T lymphocytes. Nature 330:763–765. doi: 10.1038/330763a0. [DOI] [PubMed] [Google Scholar]
- 22.Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC. 1987. Structure of the human class I histocompatibility antigen, HLA-A2 Nature 329:506–512. [DOI] [PubMed] [Google Scholar]
- 23.Saper MA, Bjorkman PJ, Wiley DC. 1991. Refined structure of the human histocompatibility antigen HLA-A2 at 2.6 A resolution. J Mol Biol 219:277–319. doi: 10.1016/0022-2836(91)90567-P. [DOI] [PubMed] [Google Scholar]
- 24.Hirohashi Y, Torigoe T, Maeda A, Nabeta Y, Kamiguchi K, Sato T, Yoda J, Ikeda H, Hirata K, Yamanaka N, Sato N. 2002. An HLA-A24-restricted cytotoxic T lymphocyte epitope of a tumor-associated protein, survivin. Clin Cancer Res 8:1731–1739. [PubMed] [Google Scholar]
- 25.Batalia MA, Collins EJ. 1997. Peptide binding by class I and class II MHC molecules. Biopolymers 43:281–302. doi:. [DOI] [PubMed] [Google Scholar]
- 26.Yao S, Liu J, Qi J, Chen R, Zhang N, Liu Y, Wang J, Wu Y, Gao GF, Xia C. 2016. Structural illumination of equine MHC class I molecules highlights unconventional epitope presentation manner that is evolved in equine leukocyte antigen alleles. J Immunol 196:1943–1954. doi: 10.4049/jimmunol.1501352. [DOI] [PubMed] [Google Scholar]
- 27.Xiao J, Xiang W, Chai Y, Haywood J, Qi J, Ba L, Qi P, Wang M, Liu J, Gao GF. 2016. Diversified anchoring features the peptide presentation of DLA-88*50801: first structural insight into domestic dog MHC class I. J Immunol 197:2306–2315. doi: 10.4049/jimmunol.1600887. [DOI] [PubMed] [Google Scholar]
- 28.Zhang J, Chen Y, Qi J, Gao F, Liu Y, Liu J, Zhou X, Kaufman J, Xia C, Gao GF. 2012. Narrow groove and restricted anchors of MHC class I molecule BF2*0401 plus peptide transporter restriction can explain disease susceptibility of B4 chickens. J Immunol 189:4478–4487. doi: 10.4049/jimmunol.1200885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang N, Qi J, Feng S, Gao F, Liu J, Pan X, Chen R, Li Q, Chen Z, Li X, Xia C, Gao GF. 2011. Crystal structure of swine major histocompatibility complex class I SLA-1 0401 and identification of 2009 pandemic swine-origin influenza A H1N1 virus cytotoxic T lymphocyte epitope peptides. J Virol 85:11709–11724. doi: 10.1128/JVI.05040-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borbulevych OY, Piepenbrink KH, Gloor BE, Scott DR, Sommese RF, Cole DK, Sewell AK, Baker BM. 2009. T cell receptor cross-reactivity directed by antigen-dependent tuning of peptide-MHC molecular flexibility. Immunity 31:885–896. doi: 10.1016/j.immuni.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chu F, Lou Z, Chen YW, Liu Y, Gao B, Zong L, Khan AH, Bell JI, Rao Z, Gao GF. 2007. First glimpse of the peptide presentation by rhesus macaque MHC class I: crystal structures of Mamu-A*01 complexed with two immunogenic SIV epitopes and insights into CTL escape. J Immunol 178:944–952. doi: 10.4049/jimmunol.178.2.944. [DOI] [PubMed] [Google Scholar]
- 32.Fan S, Wu Y, Wang S, Wang Z, Jiang B, Liu Y, Liang R, Zhou W, Zhang N, Xia C. 2016. structural and biochemical analyses of swine major histocompatibility complex class I complexes and prediction of the epitope map of important influenza A virus strains. J Virol 90:6625–6641. doi: 10.1128/JVI.00119-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rammensee HG, Friede T, Stevanoviic S. 1995. MHC ligands and peptide motifs: first listing. Immunogenetics 41:178–228. doi: 10.1007/BF00172063. [DOI] [PubMed] [Google Scholar]
- 34.Pollack MS, Hayes A, Mooney S, Pedersen NC, Cook RG. 1988. The detection of conventional class I and class II I-E homologue major histocompatibility complex molecules on feline cells. Vet Immunol Immunopathol 19:79–91. doi: 10.1016/0165-2427(88)90048-7. [DOI] [PubMed] [Google Scholar]
- 35.Yuhki N, Mullikin JC, Beck T, Stephens R, O'Brien SJ. 2008. Sequences, annotation and single nucleotide polymorphism of the major histocompatibility complex in the domestic cat. PLoS One 3:e2674. doi: 10.1371/journal.pone.0002674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Holmes JC, Holmer SG, Ross P, Buntzman AS, Frelinger JA, Hess PR. 2013. Polymorphisms and tissue expression of the feline leukocyte antigen class I loci FLAI-E, FLAI-H, and FLAI-K. Immunogenetics 65:675–689. doi: 10.1007/s00251-013-0711-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li L, Bouvier M. 2004. Structures of HLA-A*1101 complexed with immunodominant nonamer and decamer HIV-1 epitopes clearly reveal the presence of a middle, secondary anchor residue. J Immunol 172:6175–6184. doi: 10.4049/jimmunol.172.10.6175. [DOI] [PubMed] [Google Scholar]
- 38.Reche PA, Keskin DB, Hussey RE, Ancuta P, Gabuzda D, Reinherz EL. 2006. Elicitation from virus-naive individuals of cytotoxic T lymphocytes directed against conserved HIV-1 epitopes. Med Immunol 5:1. doi: 10.1186/1476-9433-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Finerty S, Stokes CR, Gruffydd-Jones TJ, Hillman TJ, Barr FJ, Harbour DA. 2001. Targeted lymph node immunization can protect cats from a mucosal challenge with feline immunodeficiency virus. Vaccine 20:49–58. doi: 10.1016/S0264-410X(01)00323-1. [DOI] [PubMed] [Google Scholar]
- 40.Pymm P, Illing PT, Ramarathinam SH, O'Connor GM, Hughes VA, Hitchen C, Price DA, Ho BK, McVicar DW, Brooks AG, Purcell AW, Rossjohn J, Vivian JP. 2017. MHC-I peptides get out of the groove and enable a novel mechanism of HIV-1 escape. Nat Struct Mol Biol 24:387–394. doi: 10.1038/nsmb.3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hosie MJ, Techakriengkrai N, Beczkowski PM, Harris M, Logan N, Willett BJ. 2017. The comparative value of feline virology research: can findings from the feline lentiviral vaccine be translated to humans? Vet Sci 4:7. doi: 10.3390/vetsci4010007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen W, Gao F, Chu F, Zhang J, Gao GF, Xia C. 2010. Crystal structure of a bony fish beta2-microglobulin: insights into the evolutionary origin of immunoglobulin superfamily constant molecules. J Biol Chem 285:22505–22512. doi: 10.1074/jbc.M109.095000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou M, Xu Y, Lou Z, Cole DK, Li X, Liu Y, Tien P, Rao Z, Gao GF. 2004. Complex assembly, crystallization and preliminary X-ray crystallographic studies of MHC H-2Kd complexed with an HBV-core nonapeptide. Acta Crystallogr D Biol Crystallogr 60:1473–1475. doi: 10.1107/S0907444904013587. [DOI] [PubMed] [Google Scholar]
- 44.Pflugrath JW. 2004. Macromolecular cryocrystallography–methods for cooling and mounting protein crystals at cryogenic temperatures. Methods 34:415–423. doi: 10.1016/j.ymeth.2004.03.032. [DOI] [PubMed] [Google Scholar]
- 45.Rossmann MG, van Beek CG. 1999. Data processing. Acta Crystallogr D Biol Crystallogr 55:1631–1640. [DOI] [PubMed] [Google Scholar]
- 46.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 47.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystall D Biol Crystallogr 53:240–255. [DOI] [PubMed] [Google Scholar]
- 48.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 49.Laskowski RA, Moss DS, Thornton JM. 1993. Main-chain bond lengths and bond angles in protein structures. J Mol Biol 231:1049–1067. [DOI] [PubMed] [Google Scholar]
- 50.Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 25:4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Gouet P, Robert X, Courcelle E. 2003. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tobita T, Oda M, Morii H, Kuroda M, Yoshino A, Azuma T, Kozono H. 2003. A role for the P1 anchor residue in the thermal stability of MHC class II molecule I-Ab. Immunol Lett 85:47–52. doi: 10.1016/S0165-2478(02)00206-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.