ABSTRACT
Oncolytic virotherapy is an emerging treatment modality that uses replication-competent viruses to destroy cancer cells. M1 is a naturally occurring alphavirus (Togaviridae) which shows potent oncolytic activities against many cancers. Accumulation of unfolded proteins during virus replication leads to a transcriptional/translational response known as the unfolded protein response (UPR), which might counteract the antitumor effect of the oncolytic virus. In this report, we show that either pharmacological or biological inhibition of IRE1α or PERK, but not ATF6, substantially increases the oncolytic effects of the M1 virus. Moreover, inhibition of IRE1α blocks M1 virus-induced autophagy, which restricts the antitumor effects of the M1 virus through degradation of viral protein, in glioma cells. In addition, IRE1α suppression significantly increases the oncolytic effect of M1 virus in an orthotopic glioma model. From a molecular pathology study, we found that IRE1α is expressed at lower levels in higher-grade gliomas, suggesting greater antitumor efficacy of the oncolytic virus M1. Taken together, these findings illustrate a defensive mechanism of glioma cells against the oncolytic virus M1 and identify possible approaches to enhance the oncolytic viral protein accumulation and the subsequent lysis of tumor cells.
IMPORTANCE Although oncolytic virotherapy is showing great promise in clinical applications, not all patients are benefiting. Identifying inhibitory signals in refractory cancer cells for each oncolytic virus would provide a good chance to increase the therapeutic effect. Here we describe that infection with the oncolytic virus M1 triggers the unfolded protein response (UPR) and subsequent autophagy, while blocking the UPR-autophagy axis significantly potentiates the antitumor efficacy of M1 in vitro and in vivo. A survey of cancer tissue banks revealed that IRE1α, a key element in the UPR pathway, is commonly downregulated in higher-grade human gliomas, suggesting favorable prospects for the application of M1. Our work provides a potential predictor and target for enhancement of the therapeutic effectiveness of the M1 virus. We predict that the mechanism-based combination therapy will promote cancer virotherapy in the future.
KEYWORDS: IRE1α, autophagy, glioblastoma, oncolytic viruses, unfolded protein response
INTRODUCTION
Oncolytic viruses (OVs) are multimodal antitumor agents advancing deeply into clinical trials, and talimogene laherparepvec (T-VEC) has been approved by the U.S. Food and Drug Administration (FDA) (1, 2). OVs are designed to exploit unique attributes of the tumor microenvironment because many of the pathways subverted by cancer cells are involved in the ability of cancer cells to combat virus infection (3). However, after systemic or intratumoral injection of an oncolytic virus, many processes are critical for efficient elimination and clearance of the neoplasm, such as viral entry, induction of inflammatory factors, viral replication, viral protein accumulation, cell lysis, and release of progeny virions to surrounding tumor cells (4). These processes against cancer cells can be impeded by several host factors that limit efficient tumor elimination (5–10).
One cellular process, the unfolded protein response (UPR), may lead to restricted oncolytic viral protein translation and accumulation. The UPR is an evolutionarily conserved cellular process that is used to cope with protein folding stress (11). Accumulation of misfolded proteins in the endoplasmic reticulum (ER) lumen activates three major pathways: the IRE1α-XBP1 pathway, the protein kinase-like endoplasmic reticulum kinase (PERK)-eIF2α pathway, and the ATF6 pathway. The IRE1 pathway is the most ancient branch of the UPR, being conserved from yeast to mammals (12). The ER stress response to misfolded proteins slows protein translation, increases ER chaperone production, and enhances ER-associated protein degradation (ERAD), which together serve to restore cellular homeostasis (13). PERK phosphorylates the translation initiation factor eIF2α, leading to attenuation of global protein synthesis. Finally, ATF6α belongs to a family of transcriptional factors that translocate to the nucleus and activate the ERAD pathway in response to unfolded proteins. When ER stress levels become insurmountable after activation of the UPR, downstream pathways are initiated, leading to cell apoptosis.
Autophagy is an evolutionarily conserved process involving the formation of double-membrane vesicles called autophagosomes (14). Autophagy plays key roles in maintaining cellular homeostasis by eliminating unwanted proteins and damaged organelles through self-digestion in cell lysosomes, thereby promoting cell survival. Moreover, autophagy activation protects cells against certain pathogens (15). Recent reports have revealed that the ER serves as a subcellular platform for autophagy initiation (16–18). Several lines of evidence have suggested that the UPR pathway regulates autophagy through the PERK-eIF2α pathway and the IRE1α-XBP1 pathway (14, 19–22).
As a group, the genus Alphavirus has a very wide geographic distribution (23–25). Many alphaviruses show potential anticancer activities. Semliki Forest virus has been reported as a novel candidate against unresectable osteosarcoma and prolongs survival in experimental lung cancer. Sindbis virus also shows an oncolytic effect in various cancers (26–29). The M1 virus, isolated in the 1960s from Hainan Province in China, belongs to the Togavirus family of viruses, which are arthropod borne and have mosquitoes as the usual vector. The genome of M1 virus is 11,690 nucleotides (nt) in length and contains two open reading frames encoding four nonstructural proteins (nsP1, nsP2, nsP3, and nsP4) and five structural proteins (C, E3, E2, 6K, and E1) (9, 24). For a group of patients with fever of unknown cause in Hainan Province, complement fixation tests showed that 26% of serum specimens from those patients had complement fixation antibody to M1 virus (30). However, there has been no further report about the pathology of M1 virus as far as we know. In addition, the M1 virus is very similar to Getah virus, which is a mosquito-borne pathogen that can cause a mild, self-limited illness in horses and reproductive losses in pigs but is not pathogenic in humans. We have previously identified that the alphavirus M1 is a potent potential oncolytic virus targeting many cancers (31–33) but not normal cells. Nevertheless, the oncolytic effect of M1 on glioma is not definite, and the mechanism of the antitumor effect is not fully understood.
In this study, we sought to investigate the oncolytic efficacy of M1 in glioma and uncover the host anti-M1 mechanisms, aiming to identify predictors and targets for personalized and intensified oncolytic virotherapy.
RESULTS
Oncolytic virus M1 inhibits glioma in vitro and in vivo.
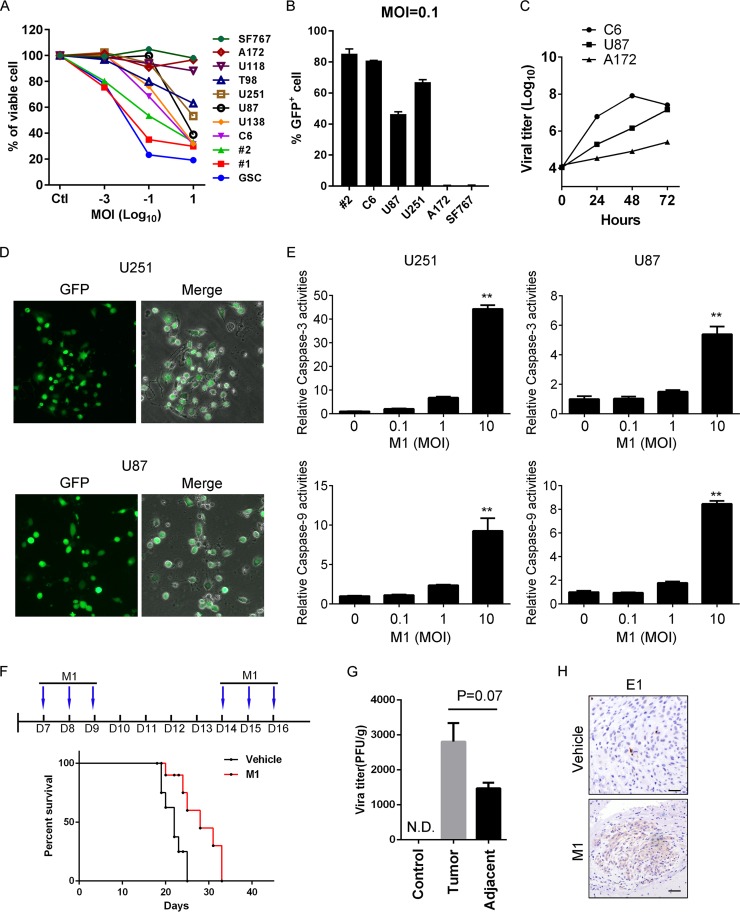
We previously have found that the alphavirus M1 selectively kills a diverse range of cancer cells, such as hepatocellular carcinoma, colorectal cancer, and bladder cancer cells, while the oncolytic effects of M1 on glioma are not clearly identified (33). To investigate the sensitivity of glioma cells to the oncolytic virus M1, we screened eight glioma cell lines, two primary glioma cell lines, and one glioma stem cell (GSC) line. We found that the M1 virus efficiently inhibited the viability of most of these cancer cells (Fig. 1A). Next, we used the M1-GFP virus, which can express green fluorescent protein (GFP) during replication in host cells, to infect representatives from each class (very resistant, with average resistance, or very sensitive to M1) and detected the susceptibilities of these cancer cells. The data in Fig. 1B show that the M1 virus can infect a higher percentage of sensitive cell lines, whereas it can barely infect very resistant cells. Similar results are observed in viral replication curves for three glioma cell lines with different responsiveness (Fig. 1C). We speculated that glioma cell lines with average responsiveness and permissiveness may possess greater chances for potentiation than very resistant or sensitive ones, and we thus focused on investigating this class of cells. Figure 1D shows representative virus infections in the U87 and U251 cell lines at a multiplicity of infection (MOI) of 10. Caspase-3/7 and caspase-9 are critical innate apoptotic pathway executors. In caspase activity assays, the data show that M1 virus infection activated caspase-3 and caspase-9 in the relatively sensitive cell lines U87 and U251 (Fig. 1E). These data suggest that infection by the M1 virus induces apoptosis in glioma cells in vitro.
FIG 1.
Oncolytic effects of the M1 virus in vitro and in vivo. (A) A panel of glioma cell lines and primary glioma cells from patients (1 and 2) were infected with the M1 virus at 10, 0.1, 0.001, and 0 PFU/cell. Cell viabilities were determined at 48 h postinfection. MOI, multiplicity of infection. (B) Cells were infected with the M1-GFP virus at 0.1 PFU/cell, and GFP positive cells were analyzed by flow cytometry at 48 h postinfection. (C) M1 virus replication in different glioma cell lines. Cells were infected with the M1 virus (0.1 PFU/cell), and supernatants were collected to determine the viral replication. (D) Representative photographs of U87 and U251 cells infected with the M1-GFP virus (10 PFU/cell) at 72 h postinfection. (E) U87 and U251 cells were infected with the M1 virus for 24 h, and caspase-3/7/9 activities were determined. (F) Timeline of treatment for in vivo experiment and survival analysis of glioma-bearing mice. Mice were orthotopically inoculated with 3 × 105 U87 cells. After 1 week, the M1 virus was injected through the tail vein. (G and H) Virus titer and expression of E1 viral protein from tissues derived from U87 orthotopic glioma model mice. N.D., not detectable. *, P < 0.05; **, P < 0.01.
To confirm the oncolytic effect on glioma cells in vivo, we next sought to determine whether the M1 virus also kills glioma cells in an orthotropic xenograft model. We used the human malignant glioma cell line U87 and injected the cells into the brains of nude mice. To determine whether this translated into a significant anticancer effect, the M1 virus was intravenously injected 7 days after cancer implantation for a total of 6 times. With a survival analysis, we found that the M1 virus slightly but significantly prolonged the survival of the glioma-bearing mice (Fig. 1F). We also detected the virus titers and viral proteins in the same orthotropic model and found that the M1 virus is enriched in tumor sites (Fig. 1G and H). The sum of these results suggested that although the M1 virus prolongs the survival of glioma-bearing mice, the antitumor effects remain unsatisfying. Thus, we next studied the mechanism following infection with the M1 virus and sought to improve the antitumor effects.
The UPR pathway is activated in M1-infected glioma cells.
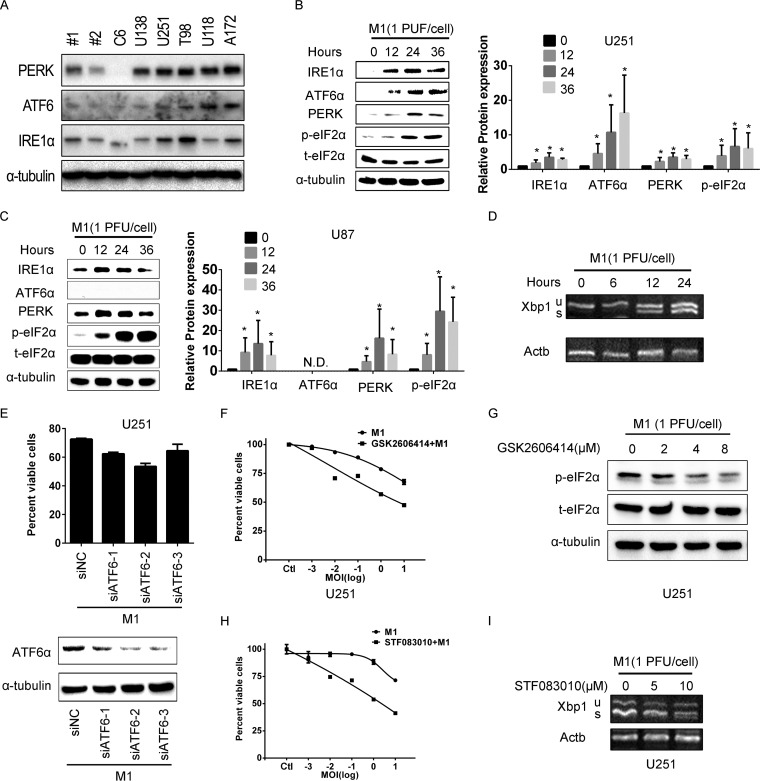
During the study of the antitumor effect of M1 virus in various cancers, such as hepatocellular carcinoma, colorectal cancer, and bladder cancer cells, we found that M1 virus induced ER stress-mediated apoptosis in very sensitive cancer cell lines (33). In this study, we found that the UPR pathway effectors were defective in M1-sensitive glioma cells (Fig. 2A). Therefore, we hypothesized that the UPR pathway-mediated protein degradation can maintain cellular homeostasis and protect cells from viral protein overload, which results in irreversible ER stress-mediated apoptosis. We detected the UPR effectors during M1 virus infection in glioma cells and found that UPR pathways were significantly activated by the M1 virus in a time-dependent manner in both the U87 and U251 cell lines (Fig. 2B and C). IRE1α is an endoribonuclease that initiates an unconventional splicing of the XBP1 mRNA, excising a 26-nt sequence, and shifts the reading frame to produce a functional activating isoform, XBP1(s), from XBP1(u). By detecting the spliced XBP1 variant, we observed that M1 virus infection activates IRE1α in U251 glioma cells (Fig. 2D).
FIG 2.
M1 virus infection induces the unfolded protein response in glioma cells. (A) UPR pathway protein expression levels in M1-sensitive and M1-refractory glioma cell lines. M1-sensitive and M1-refractory cancer cells were collected, and the protein expression levels were determined by Western blotting. (B and C) UPR pathway protein expression during M1 virus infection in malignant glioma cell lines. U251 (B) and U87 (C) cells were infected with M1 (1 PFU/cell), and protein expression levels were determined by Western blotting. The right panels show the gray scale analysis result. (D) XBP1(s) expression is increased during M1 virus infection. Actin was used as a loading control. (E) Knockdown of ATF6 does not affect the oncolytic effects of the M1 virus. Cell viabilities were determined using MTT assays (upper panel), and protein expression was determined by Western blotting (lower panel). (F and G) U251 cells were treated with PERK inhibitor GSK2606414 (4 μM) and then infected with the M1 virus at the indicated MOI. Cell viabilities were determined at 48 h postinfection (F), and protein expression was determined at 24 h postinfection (G). (H and I) Inhibition of IRE1α endonuclease activity increases the oncolytic effect of the M1 virus. Cells were pretreated with STF083010 (10 μM) for 1 h and then infected with the M1 virus. Cell viabilities were determined at 48 h postinfection (H), and XBP1(s) expression was determined at 24 h postinfection (I).
To identify pathways in the UPR that might render glioma cells refractory to oncolytic virus infection, we employed small interfering RNAs (siRNAs) or inhibitors to block the UPR pathway. Cells were transfected with siRNA targeting ATF6 for 24 h and were then infected with the M1 virus. As shown in Fig. 2E, knockdown of ATF6 did not affect the viability of glioma cells during M1 virus infection. However, we found that pharmacological inhibition of PERK with GSK2606414 inhibited the phosphorylation of eIF2α induced by the M1 virus and significantly enhanced the oncolytic effect of M1 (Fig. 2F and G). The third branch of the UPR pathway, the IRE1α-XBP1 pathway, is also an important effector during virus infection. The data show that STF083010, an IRE1α nuclease inhibitor, significantly increased the oncolytic effects of the M1 virus in cancer cells (Fig. 2H). The inhibition of IRE1α endonuclease activity was determined by detecting spliced XBP1 (Fig. 2I). These data demonstrate that direct inhibition of IRE1α shows greater potential for sensitizing glioma cells to the oncolytic virus M1.
Inhibition of IRE1α increases the oncolytic effects of the M1 virus.
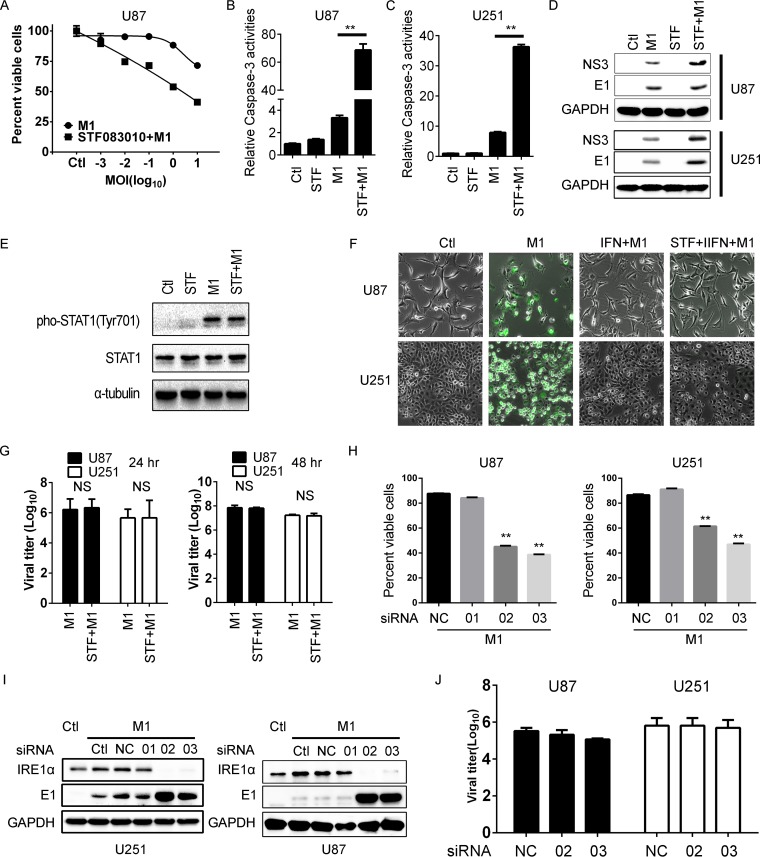
We next chose another glioma cell line, U87, to validate the combined antitumor effect of IRE1α inhibition and M1 virus. Consistent with the above findings, we observed that pretreatment with an IRE1α inhibitor significantly increased the antitumor effects of M1 virus (Fig. 3A). With caspase activity detection, we found that IRE1α inhibition increased oncolytic virus-induced apoptosis (1 PFU/cell) (Fig. 3B and C). To study the functional role of IRE1α, we first detected the viral protein expression and found that inhibition of IRE1α increased viral protein accumulation (Fig. 3D). To determine whether IRE1α inhibition affects type I interferon (IFN) signaling, we first tested the STAT1 phosphorylation and found that inhibition of IRE1α cannot abrogate the STAT1 phosphorylation induced by M1 virus (Fig. 3E). We further studied whether IRE1α inhibition can counteract the effects of type I IFN signaling. We found that IFN-α can significantly suppress the virus replication in U251 and U87 glioma cell lines, but IRE1α inhibition did not overcome the antiviral effects of IFN-α (Fig. 3F). Consistently, with titer determination, we found that IRE1α inhibition did not affect M1 viral replication (Fig. 3G). Based on these observations, we concluded that IRE1α inhibition did not affect the innate antiviral response and thus that IRE1α inhibition likely does not compromise the antitumor immunity induced by the virus.
FIG 3.
Inhibition of IRE1α increases the oncolytic effects of the M1 virus. (A) Determination of the antitumor effect of the M1 virus during IRE1α inhibition. U87 malignant glioma cells were pretreated with STF083010 (10 μM) for 1 h and were then infected with the M1 virus. Cell viabilities were determined at 48 h postinfection. (B and C) Caspase activities were determined in U87 (B) and U251 (C) cells treated with a combination of the M1 virus and STF083010. (D) Viral protein expression levels after STF083010 treatment. U87 and U251 cells were pretreated with STF083010 (10 μM) for 1 h and then infected with the M1 virus. Protein expression levels were determined 4 h later. STF, STF083010. (E) Phosphorylation of STAT1 during M1 virus infection (1 PFU/cell) with or without STF083010 treatment (10 μM) at 48 h postinfection. (F) Viral replication (10 PFU/cell) with or without IFN-α (1000 U/ml) treatment during STF083010 treatment (10 μM) at 48 h postinfection. (G) Viral titer determination in glioma cells after STF083010 treatment. Supernatants were collected at 24 or 48 h postinfection. (H) Cell viability assay after knockdown of IRE1α. Cells were transfected with scramble RNA or IRE1α siRNAs for 24 h and were then infected with the M1 virus (1 PFU/cell); cell viabilities were determined at 48 h postinfection. NC, negative control. (I) Knockdown efficiency determination and viral protein detection. Cells were transfected with scramble RNA or IRE1α siRNAs for 48 h and then infected with the M1 virus (1 PFU/cell) for 8 h. Protein expression levels were determined using Western blots. (J) Viral titer determination after knockdown of IRE1α. Cells were treated as described for panel H, and supernatants were collected at 48 h postinfection to determine viral titer. **, P < 0.01.
To confirm the specificity of the IRE1α inhibitor, we used siRNAs to knock down IRE1α expression. Consistent with the above results, we found that knockdown of IRE1α also increased the sensitivity to the oncolytic virus M1 compared with transfection with nontargeting RNA or low-efficiency siRNA (1) (Fig. 3H). The knockdown efficiency and viral protein expression are shown in Fig. 3I. Additionally, with titer determination, we found that knockdown of IRE1α did not affect viral replication in glioma cells (Fig. 3J). Taken together, these results suggested that activation of IRE1α can inhibit the viral protein load and subsequent oncolysis in glioma cells with average sensitivity. Inhibition of IRE1α increases the oncolytic effects of the M1 virus by overcoming this limitation.
IRE1α mediates M1 virus-induced autophagy.
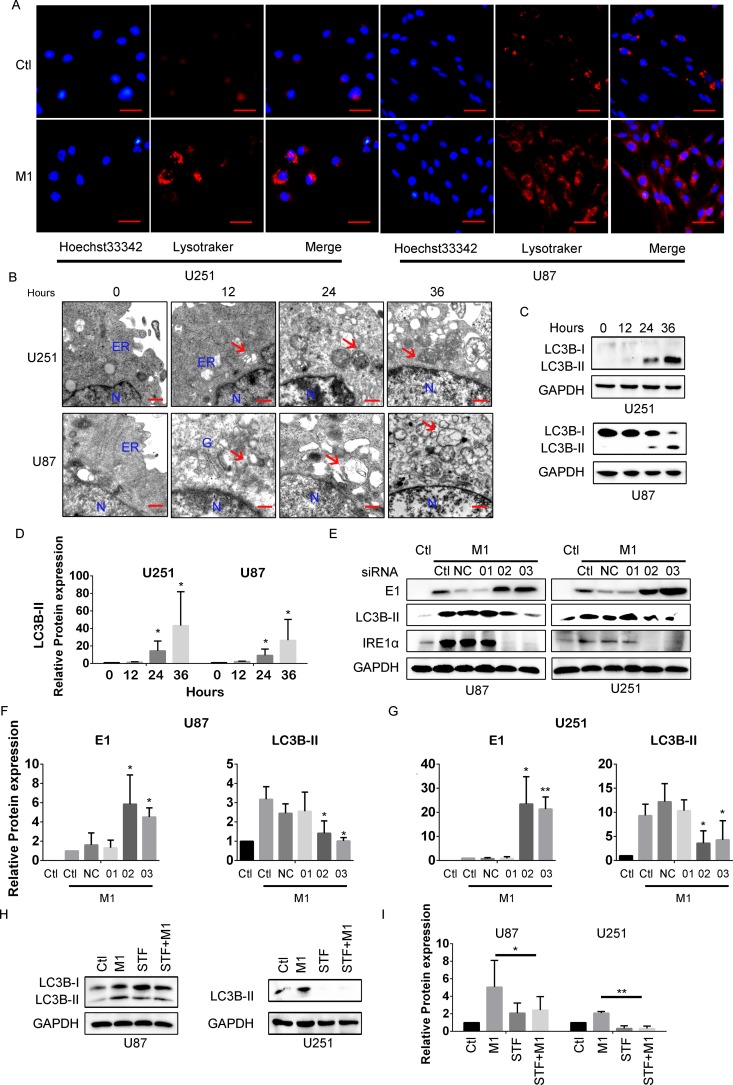
Autophagy is a self-digestion process, whose activation protects cells against certain pathogens through direct phagocytosis. Relationships between the UPR and autophagy have been extensively studied (34). Thus, we sought to determine if M1 virus infection induces autophagy in glioma cell lines. With LysoTracker staining to specifically indicate late-phase autophagosomes, we observed that M1 virus infection induced punctum formation in glioma cell lines (Fig. 4A). To validate this result, we used transmission electron microscopy to observe glioma cells after the M1 virus infection (Fig. 4B). Furthermore, M1 virus infection induced LC3B-II expression, which is commonly used as an autophagy marker, in glioma cancer cell lines (Fig. 4C and D).
FIG 4.
M1 virus infection induces autophagy through IRE1α. (A) LysoTracker staining was used to visualize intracellular later-phase autophagosomes. Cells were infected with the M1 virus (1 PFU/cell) for 24 h, and LysoTracker staining was performed according to the manufacturer's procedure. Hoechst 33342 staining was performed 10 min before capture of photographs. Scale bars, 0.25 μm. (B) Ultrastructural observation of cancer cells after infection with the M1 virus. U87 and U251 malignant glioma cells were infected with the M1 virus (1 PFU/cell) and observed with a transmission electron microscope. ER, endoplasmic reticulum. N, Nucleus. The red arrows indicate autophagosomes. Scale bars, 500 nm. (C) Expression of the autophagy marker LC3B using Western blotting. (D) Quantification of the data from panel C. (E) LC3B detection after knockdown of IRE1α. U87 and U251 malignant glioma cells were transfected with scramble RNA or IRE1α siRNAs (50 nM) for 48 h. The indicated protein expression levels were determined 8 h after viral infection. (F and G) Quantification of the data from panel E. (H and I) LC3B expression after STF083010 treatment for 24 h. Whole-cell lysates were collected, and Western blotting was performed. *, P < 0.05; **, P < 0.01.
The IRE1α-XBP1 pathway has been reported to induce autophagy under various conditions. Next, we sought to determine if induction of autophagy by the M1 virus is mediated by IRE1α (15). In accordance with the above results, M1 infection induced LC3B-II expression in glioma cell lines. However, with knockdown of IRE1α expression, the M1 virus-induced autophagy was also inhibited (Fig. 4E to G). To confirm this result, glioma cells were pretreated with the IRE1α inhibitor STF083010, and this abrogated the M1 virus infection-induced autophagy (Fig. 4H and I). These results strongly suggest that the IRE1α-autophagy axis plays critical roles during M1 virus infection.
IRE1α-induced autophagy restricts oncolytic effects of M1.
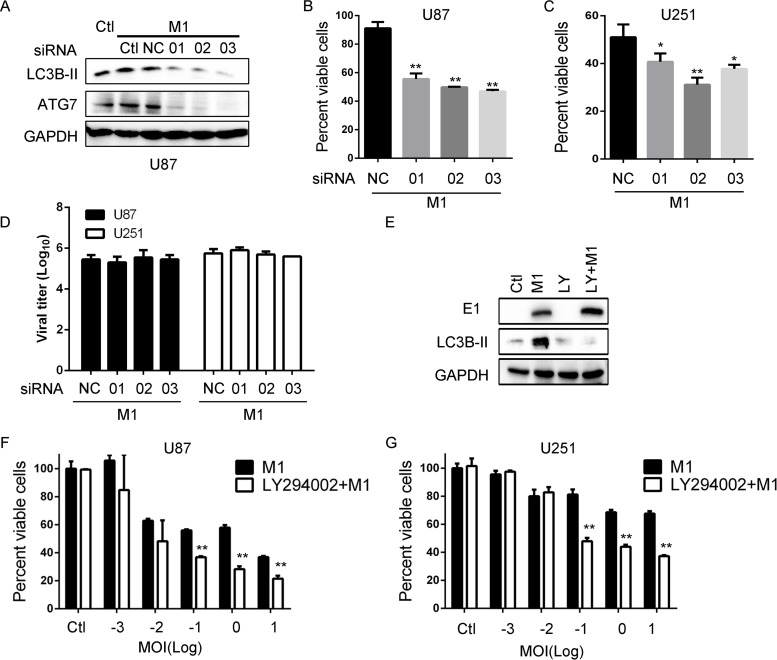
Given that IRE1α inhibition substantially increased the oncolytic effects and viral protein load of M1 virus, we sought to find out whether autophagy mediates the inhibition of oncolysis. ATG7 has been reported as a critical autophagy initiation factor (35). To clarify the role of autophagy, we used siRNAs targeting ATG7 and found that knockdown of ATG7 inhibits autophagy induction during M1 virus infection (Fig. 5A). Moreover, like IRE1α inhibition, we found with a cell viability assay that ATG7 knockdown significantly increased the oncolytic effects of M1 in glioma cell lines (Fig. 5B and C). Also, in accordance with IRE1α inhibition, knockdown of ATG7 does not affect the viral titer (Fig. 5D). To further validate the results, we used the autophagy inhibitor LY294002 to block the induction of autophagy (Fig. 5E). We found that inhibition of autophagy with LY294002 increases viral protein expression and cell viability inhibition (Fig. 5E to G). Taken together, these results support our previous observation that inhibition of autophagic degradation of viral protein increases the oncolytic effect of the M1 virus, and IRE1α coordinately regulates the autophagy in response to viral infection.
FIG 5.
IRE1α-mediated autophagy restricts the oncolytic effects of the M1 virus. (A) LC3B detection after knockdown of ATG7. Cells were transfected with scramble RNA or siATG7 (50 nM) for 48 h. The indicated protein expression levels were determined at 8 h postinfection. (B and C) Cell viability determination after knockdown of ATG7. U87 and U251 cells were transfected with siRNA for 24 h. Cell viabilities were determined at 48 h postinfection. (D) Viral titer assay after ATG7 knockdown. Cells were treated as described for panel B, and supernatants were collected at 48 h postinfection. (E) Viral protein detection after treatment with the autophagy inhibitor LY294002. U87 cells were pretreated with LY294002 (20 μM) for 1 h and then infected with the M1 virus (1 PFU/cell). E1 and LC3B protein expression levels were determined with a Western blot. (F and G) Cell viability assay of the oncolytic effect of the M1 virus after LY294002 treatment. Cells were pretreated with LY294002 for 1 h and infected with the M1 virus for 48 h. *, P < 0.05; **, P < 0.01.
Inhibition of IRE1α facilitates the oncolytic effects of M1 virus in vivo.
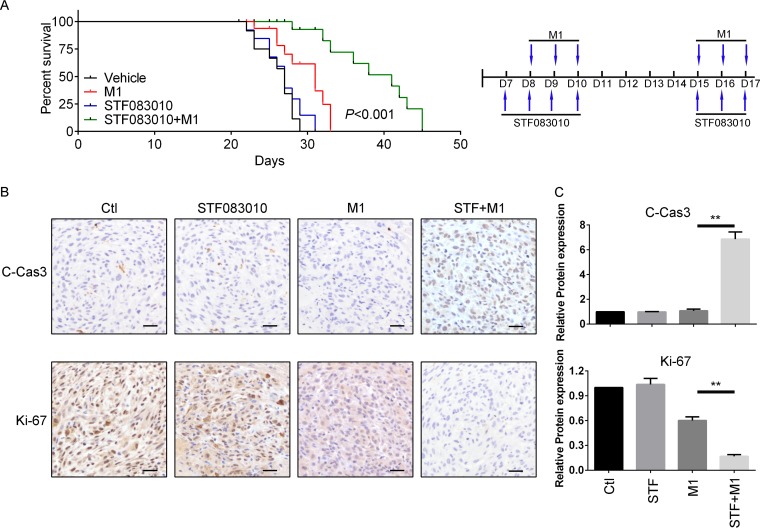
Since IRE1α activation renders tumor cells refractory to M1 virus infection via induction of autophagy signaling, we predicted that blocking IRE1α-autophagy signaling would increase the viral protein load and the oncolytic effect. We established an orthotopic glioma model with U87 cells in nude mice. Following implantation of U87 tumors into mice, we administered STF083010 or dimethyl sulfoxide (DMSO) and 24 h later injected the mice with the M1 virus 6 times. The data show that 50% survival improved from 27 days (no treatment) to 31 days (M1 alone), to 41 days (M1 plus STF083010) (Fig. 6A). To validate the antitumor effect, we detected cleaved caspase-3 and Ki67 expression in the tumor mass with immunohistochemistry staining. The data show that the M1 virus, together with the IRE1α inhibitor, significantly increased the level of cleaved caspase-3 and downregulated Ki67 expression (Fig. 6B). Quantification of the protein expression is shown in Fig. 6C. Taken together, our data demonstrate the fundamental importance of the IRE1α-autophagy axis in modulating the responsiveness of glioma cells to the oncolytic virus M1.
FIG 6.
IRE1α inhibition increases the oncolytic effects of the M1 virus in vivo. (A) Survival of tumor-bearing mice after treatment with STF083010, the M1 virus, or both. A U87 orthotopic glioma model was established, and mice were pretreated or not with STF083010 (20 mg/kg/day) 7 times through intraperitoneal injection 1 week after tumor inoculation. Mice were also treated or not with the M1 virus (2 × 107 PFU) 6 times through tail vein injections 1 week after tumor inoculation. (B) Expression of cleaved caspase-3 and Ki67 in orthotopic glioma tissue. Tissues derived from U87 orthotopic glioma model mice after treatment for 3 days are shown. (C) Quantification of data from panel B. **, P < 0.01.
IRE1α is downregulated during malignant progression of glioma.
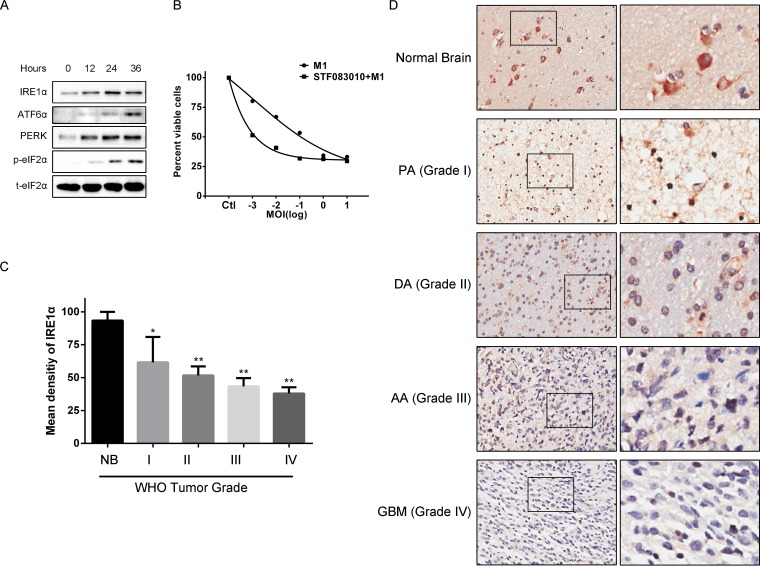
To determine the clinical relevance of IRE1α with respect to the M1 virus, we performed primary glioma cell culture and found that M1 virus infection significantly induced the UPR in primary malignant glioma cells (Fig. 7A). By using cell viability assays, we found that M1 infection alone inhibited primary glioma cell viability in a dose-dependent manner, and inhibition of IRE1α increased the oncolytic effect (Fig. 7B).
FIG 7.
IRE1α expression in clinical glioma samples. (A) Expression of UPR pathway proteins in primary glioma cancer cells after infection with the M1 virus. Primary glioma cells were isolated and cultured from clinical specimens. Cells were infected with the M1 virus (0.1 PFU/cell), and protein expression levels were determined with Western blots. (B) Cell viability assay of primary glioma cells. Primary glioma cells were pretreated with STF083010 for 1 h and then infected with the M1 virus for 48 h. (C) IRE1α expression in various stages of glioma and in normal brain. (D) Representative images of IRE1α expression in various stages of glioma and in normal brain. *, P < 0.05; **, P < 0.01.
Next, we sought to explore the IRE1α expression levels in glioma specimens. To identify if IRE1α is correlated with malignant progression of glioma in patients, we analyzed the IRE1α expression profiles of glioma tissues and normal brain tissues resected from 60 glioma patients. As shown in Fig. 7C and D, we observed that the IRE1α expression levels were reduced during malignant progression of glioma, suggesting that IRE1α is negatively correlated with glioma progression. These data strongly suggest that M1 virus may be applicable for treatment of high-grade malignant glioma.
DISCUSSION
Glioblastoma has the highest incidence and mortality rate among primary brain cancer patients. Despite current therapies, the survival benefits are measured in months (36). Based on previous reports, oncolytic virotherapy possesses the potential to repress glioblastoma (37–39). Here we found that the oncolytic virus M1 kills a diverse range of glioblastoma cell lines and exhibits therapeutic potential in vivo. However, although the antitumor effect shows significance in vivo, the results remain unsatisfying with M1 virus treatment alone, and the mechanism needs to be further investigated.
Many viruses have specific tissue tropisms which can be exploited to kill cancer cells while replicating and spreading within disease foci (40). For instance, recombinant measles virus particles that express a retargeted hemagglutinin can be reliably generated and efficiently enter CD46-negative rodent cells expressing the human epidermal growth factor (EGF) or the IGF1 receptor, respectively (41). The principles of tumor targeting strategies vary a lot in different oncolytic viruses (40, 42). It is of interest to identify the interaction of M1 virus with malignant glioma cells. Here, we identified that low expression of IRE1α in malignant glioma cells may lead to increased oncolytic activity of the M1 virus. The major UPR pathways are the IRE1α, ATF6, and PERK pathways. IRE1α is a transmembrane protein that senses the accumulation of proteins in the ER and induces various pathways to address misfolded proteins. Here, we determined that IRE1α is downregulated during glioma tumorigenesis, which may open a gate for M1 virus infection. It is reported that IRE1α inhibition promotes oncolytic virus-induced cell death without increasing the accumulation of viral protein in vitro in a caspase-2-dependent manner (8), while we here identified a different mechanism downstream of IRE1α. Moreover, we determined the expression of IRE1α in clinical glioma specimens and found that it is negatively correlated with malignant glioma progression. However, the relationships and underlying mechanisms between IRE1α and malignant progression of glioma remain to be further explored.
PERK phosphorylates eIF2α at Ser51 and reduces global protein translation to restrict invading viruses. GSK2606414 is a recently reported PERK-selective inhibitor (43) that can partially inhibit M1 virus-induced phosphorylation of eIF2α. However, the increase in the oncolytic effect of the M1 virus with GSK2606414 is limited. We speculate that there are two possibilities. First, IRE1α might play central roles in restricting oncolytic virus M1 infection. Second, there are three other eIF2α kinases that might compensate for the function of PERK (44, 45). ATF6 regulates the expression of diverse genes during ER stress, whereas to our knowledge, there is still a lack of efficient and selective inhibitors to target ATF6.
Autophagy is a vesicular process that results in the degradation of the sequestered component, which can then be recycled by the cell (15). Here we identify that IRE1α mediates viral protein degradation through induction of autophagy, which implies that autophagic inhibitors can also enhance the oncolytic effects of M1 virus. From autophagy induction and maturation, the processes are complex, and several autophagy-related genes are induced. Hence, the detailed relationships between autophagy and the oncolytic effect of M1 virus are also interesting subjects.
Above all, we found the mechanism by which malignant progression of glioma may result in IRE1α loss and in turn render glioma cells permissive and sensitive to the oncolytic virus M1. On the other hand, the oncolytic virus M1 synergizes with IRE1α inhibitor to kill glioma cells. As inhibition of IRE1α does not affect the IFN antiviral signaling and the viral replication, this combination strategy is a relatively safe approach. Thus, our findings may bring new insights into the relationships between UPR and oncolytic viruses, and the combination of UPR pathway inhibitors with the M1 virus may be a promising strategy for glioblastoma therapeutics.
MATERIALS AND METHODS
The alphavirus M1 used in this study was described previously (33). Briefly, M1 viruses were propagated in Vero cells (OptiPRO SFM, 12309-019; Thermo Fisher, Waltham, MA) for 60 to 72 h after virus infection at an MOI of 0.001. The supernatant was collected and centrifuged to remove the cell debris. Virus titers were determined by 50% tissue culture infective dose (TCID50) in the BHK-21 cell line. Cell lines were maintained at 37°C with 5% CO2 in Dulbecco's modified Eagle's medium (DMEM), supplemented with 10% fetal bovine serum (FBS) and 100 μg/ml penicillin-streptomycin. Cell lines were purchased from the American Type Culture Collection or the Shanghai Institute of Cell Biology. Primary glioma stem cells (GSCs) were cultured in nonadhesive flasks containing DMEM-F12 supplemented with B27 (17504044; Thermo Fisher), 20 μg/ml of both human EGF and fibroblast growth factor 2 (FGF-2) (PHG0311 and 13256029; Thermo Fisher), and 100 μg/ml penicillin-streptomycin. The reagents used in this study were STF083010 (10 mM in DMSO) (S7771; Selleck), LY294002 (10 mM in DMSO) (S1105; Selleck), and GSK2606414 (10 mM in DMSO) (S7303; Selleck).
Cell viability assay.
Cells were seeded in 96-well plates at 3,000 cells per well. Cells were treated with 20 μM STF083010 or 10 μM LY294002 and then infected with the M1 virus as described in the relevant figure legends.
Cells were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) at a concentration of 1 mg/ml, and plates were further cultured at 37°C for another 3 h. The medium was removed, and the precipitate was dissolved in 100 μl of DMSO. The optical absorbance was determined at 570 nm using an iMark microplate reader (Bio-Rad).
RNA interference experiments.
Specific siRNAs and nontargeting siRNA were synthesized by and obtained from RiboBio, China. Cells were placed in DMEM with 10% FBS but without penicillin-streptomycin and transfected with siRNAs using Lipofectamine RNAiMAX (Life Technologies) with Opti-MEM (Life Technologies).
Transmission electron microscopy.
U87 and U251 cells were infected with the M1 virus at 1 PFU/cell for 0, 12, 24, and 36 h. Briefly, cells were scraped from the culture flask and pelleted at 400 × g for 5 min at room temperature. They were then resuspended and washed with phosphate-buffered saline (PBS) once, pelleted at 600 × g for 5 min, and fixed on ice for 4 h in 0.1 M PBS (pH 7.4) containing 2.5% glutaraldehyde and 2% paraformaldehyde (PFA). Samples were then submitted to the Zhongshan School of Medicine (Sun Yat-sen University) Electron Microscopy Facility for standard transmission electron microscopy ultrastructural analysis.
Antibodies and Western blots.
Cells were lysed, and SDS-PAGE was performed. Antibodies were used against the following proteins: IRE1α (ab96481; Abcam), PERK (5683; Cell Signaling Technology), ATF6α (65880; Cell Signaling Technology), eIF-2α (5324; Cell Signaling Technology), phosphorylated eIF-2α (3398; Cell Signaling Technology), GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (AP0060; Bioworld), β-actin (AP0063; Bioworld), α-tubulin (2144; Cell Signaling Technology), phosphorylated JNK (9255; Cell Signaling Technology), LC3B (3868; Cell Signaling Technology), ATG7 (2631; Cell Signaling Technology), M1 E1 and NS3 (produced by Beijing Protein Innovation), Ki-67 (9449s; Cell Signaling Technology), and cleaved caspase-3 (9664s; Cell Signaling Technology).
Caspase activity detection.
The apoptosis cascade caspase-3/7 and caspase-9 activities were determined using a Caspase-Glo assay system (Promega) according to the manufacturer's protocols. The results were normalized to cell viability (MTT assay).
Animal models.
This study was approved by the Animal Ethical and Welfare Committee of Sun Yat-sen University. U87 cells were inoculated orthotopically into 4-week-old female BALB/c-nu/nu mice after intraperitoneal injection of chloral hydrate. After 5 to 7 days, mice were randomly divided into groups. These groups were injected with vehicle, M1 (by tail vein), STF083010 (by intraperitoneal injection), or STF083010 plus M1.
Immunohistochemistry staining.
The cleaved caspase-3 and Ki-67 protein expression was characterized by immunohistochemistry using specific antibodies. Briefly, tumor sections (4 μm) were dewaxed in xylene, dehydrated in descending concentrations of ethanol, immersed in 0.3% H2O2–methanol for 30 min, washed with PBS, and probed with monoclonal antibodies (1:100) or an isotype control at 4°C overnight. After washing, the sections were incubated with biotinylated goat anti-rabbit or anti-mouse IgG at room temperature for 2 h. Immunostaining was visualized with streptavidin/peroxidase complex and diaminobenzidine, and sections were then counterstained with hematoxylin. Photographs were captured in a double-blinded fashion. We then quantified the relative protein expression with Image-Pro Plus 6.0 software (Media Cybernetics) for at least three samples. The data were subjected to Student's t test for comparison with the indicated group.
Statistical analysis.
All statistical analyses were performed using SPSS 13.0 software. Most of the data were analyzed with Student's t test or one-way analysis of variance (ANOVA) followed by Dunnett's multiple post hoc tests. Unless otherwise indicated, the error bars indicate standard deviation (SD). Significance was defined as a P value of <0.05.
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (81702749, 81503088 and 81603127), the Science and Technology Planning Project of Guangdong Province, China (20160909), the Science and Technology Planning Project of Guangdong Province, China (2017A030313706), the Research and Development Project of Applied Science and Technology of Guangdong Province, China (2016B020237004), the Science and Technology Planning Project of Guangdong Province, China (2015B020211003), and the Medical Scientific Research Foundation of Guangdong Province, China (A2016435).
Kai Li, Cheng Hu, Fan Xing, Mingshi Gao, Jiankai Liang, Xiao Xiao, Jing Cai, Yaqian Tan, Yuan Li, Wenli Chen, Bingzheng Lu, and Jialuo Mai performed the experiments. Jun Hu, Wenbo Zhu, Wei Yin, Pengxin Qiu, and Xingwen Su analyzed and interpreted the experimental results. Kai Li, Guangmei Yan, Haipeng Zhang, and Yuan Lin designed the study and wrote and reviewed the manuscript.
We declare no conflict of interest.
REFERENCES
- 1.Andtbacka RH, Kaufman HL, Collichio F, Amatruda T, Senzer N, Chesney J, Delman KA, Spitler LE, Puzanov I, Agarwala SS, Milhem M, Cranmer L, Curti B, Lewis K, Ross M, Guthrie T, Linette GP, Daniels GA, Harrington K, Middleton MR, Miller WH Jr, Zager JS, Ye Y, Yao B, Li A, Doleman S, VanderWalde A, Gansert J, Coffin RS. 2015. Talimogene laherparepvec improves durable response rate in patients with advanced melanoma. J Clin Oncol 33:2780–2788. doi: 10.1200/JCO.2014.58.3377. [DOI] [PubMed] [Google Scholar]
- 2.Kelly E, Russell SJ. 2007. History of oncolytic viruses: genesis to genetic engineering. Mol Ther 15:651–659. doi: 10.1038/sj.mt.6300108. [DOI] [PubMed] [Google Scholar]
- 3.Ilkow CS, Swift SL, Bell JC, Diallo JS. 2014. From scourge to cure: tumour-selective viral pathogenesis as a new strategy against cancer. PLoS Pathog 10:e1003836. doi: 10.1371/journal.ppat.1003836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Marchini A, Scott EM, Rommelaere J. 2016. Overcoming barriers in oncolytic virotherapy with HDAC inhibitors and immune checkpoint blockade. Viruses 8:9. doi: 10.3390/v8010009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stojdl DF, Lichty BD, ten Oever BR, Paterson JM, Power AT, Knowles S, Marius R, Reynard J, Poliquin L, Atkins H, Brown EG, Durbin RK, Durbin JE, Hiscott J, Bell JC. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275. doi: 10.1016/S1535-6108(03)00241-1. [DOI] [PubMed] [Google Scholar]
- 6.Hou W, Sampath P, Rojas JJ, Thorne SH. 2016. Oncolytic virus-mediated targeting of PGE2 in the tumor alters the immune status and sensitizes established and resistant tumors to immunotherapy. Cancer Cell 30:108–119. doi: 10.1016/j.ccell.2016.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.O'Shea CC, Johnson L, Bagus B, Choi S, Nicholas C, Shen A, Boyle L, Pandey K, Soria C, Kunich J, Shen Y, Habets G, Ginzinger D, McCormick F. 2004. Late viral RNA export, rather than p53 inactivation, determines ONYX-015 tumor selectivity. Cancer Cell 6:611–623. doi: 10.1016/j.ccr.2004.11.012. [DOI] [PubMed] [Google Scholar]
- 8.Mahoney DJ, Lefebvre C, Allan K, Brun J, Sanaei CA, Baird S, Pearce N, Gronberg S, Wilson B, Prakesh M, Aman A, Isaac M, Mamai A, Uehling D, Al-Awar R, Falls T, Alain T, Stojdl DF. 2011. Virus-tumor interactome screen reveals ER stress response can reprogram resistant cancers for oncolytic virus-triggered caspase-2 cell death. Cancer Cell 20:443–456. doi: 10.1016/j.ccr.2011.09.005. [DOI] [PubMed] [Google Scholar]
- 9.Li K, Zhang H, Qiu J, Lin Y, Liang J, Xiao X, Fu L, Wang F, Cai J, Tan Y, Zhu W, Yin W, Lu B, Xing F, Tang L, Yan M, Mai J, Li Y, Chen W, Qiu P, Su X, Gao G, Tai PW, Hu J, Yan G. 2016. Activation of cyclic adenosine monophosphate pathway increases the sensitivity of cancer cells to the oncolytic virus M1. Mol Ther 24:156–165. doi: 10.1038/mt.2015.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Machitani M, Sakurai F, Wakabayashi K, Tachibana M, Fujiwara T, Mizuguchi H. 2017. Enhanced oncolytic activities of the telomerase-specific replication-competent adenovirus expressing short-hairpin RNA against Dicer. Mol Cancer Ther 16:251–259. doi: 10.1158/1535-7163.MCT-16-0383. [DOI] [PubMed] [Google Scholar]
- 11.Hetz C, Chevet E, Oakes SA. 2015. Proteostasis control by the unfolded protein response. Nat Cell Biol 17:829–838. doi: 10.1038/ncb3184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hetz C, Martinon F, Rodriguez D, Glimcher LH. 2011. The unfolded protein response: integrating stress signals through the stress sensor IRE1alpha. Physiol Rev 91:1219–1243. doi: 10.1152/physrev.00001.2011. [DOI] [PubMed] [Google Scholar]
- 13.Schroder M, Kaufman RJ. 2005. The mammalian unfolded protein response. Annu Rev Biochem 74:739–789. doi: 10.1146/annurev.biochem.73.011303.074134. [DOI] [PubMed] [Google Scholar]
- 14.Rzymski T, Milani M, Pike L, Buffa F, Mellor HR, Winchester L, Pires I, Hammond E, Ragoussis I, Harris AL. 2010. Regulation of autophagy by ATF4 in response to severe hypoxia. Oncogene 29:4424–4435. doi: 10.1038/onc.2010.191. [DOI] [PubMed] [Google Scholar]
- 15.Jheng JR, Ho JY, Horng JT. 2014. ER stress, autophagy, and RNA viruses. Front Microbiol 5:388. doi: 10.3389/fmicb.2014.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. 2008. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J Cell Biol 182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ma XH, Piao SF, Dey S, McAfee Q, Karakousis G, Villanueva J, Hart LS, Levi S, Hu J, Zhang G, Lazova R, Klump V, Pawelek JM, Xu X, Xu W, Schuchter LM, Davies MA, Herlyn M, Winkler J, Koumenis C, Amaravadi RK. 2014. Targeting ER stress-induced autophagy overcomes BRAF inhibitor resistance in melanoma. J Clin Invest 124:1406–1417. doi: 10.1172/JCI70454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ciechomska IA, Gabrusiewicz K, Szczepankiewicz AA, Kaminska B. 2013. Endoplasmic reticulum stress triggers autophagy in malignant glioma cells undergoing cyclosporine a-induced cell death. Oncogene 32:1518–1529. doi: 10.1038/onc.2012.174. [DOI] [PubMed] [Google Scholar]
- 19.Vidal RL, Hetz C. 2013. Unspliced XBP1 controls autophagy through FoxO1. Cell Res 23:463–464. doi: 10.1038/cr.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Luan Q, Jin L, Jiang CC, Tay KH, Lai F, Liu XY, Liu YL, Guo ST, Li CY, Yan XG, Tseng HY, Zhang XD. 2015. RIPK1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy 11:975–994. doi: 10.1080/15548627.2015.1049800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Matus S, Nassif M, Glimcher LH, Hetz C. 2009. XBP-1 deficiency in the nervous system reveals a homeostatic switch to activate autophagy. Autophagy 5:1226–1228. doi: 10.4161/auto.5.8.10247. [DOI] [PubMed] [Google Scholar]
- 22.Park MA, Curiel DT, Koumenis C, Graf M, Chen CS, Fisher PB, Grant S, Dent P. 2008. PERK-dependent regulation of HSP70 expression and the regulation of autophagy. Autophagy 4:364–367. doi: 10.4161/auto.5593. [DOI] [PubMed] [Google Scholar]
- 23.Phillips DA, Murray JR, Aaskov JG, Wiemers MA. 1990. Clinical and subclinical Barmah Forest virus infection in Queensland. Med J Aust 152:463–466. [DOI] [PubMed] [Google Scholar]
- 24.Wen JS, Zhao WZ, Liu JW, Zhou H, Tao JP, Yan HJ, Liang Y, Zhou JJ, Jiang LF. 2007. Genomic analysis of a Chinese isolate of Getah-like virus and its phylogenetic relationship with other alphaviruses. Virus Genes 35:597–603. doi: 10.1007/s11262-007-0110-3. [DOI] [PubMed] [Google Scholar]
- 25.Nagata LP, Hu WG, Parker M, Chau D, Rayner GA, Schmaltz FL, Wong JP. 2006. Infectivity variation and genetic diversity among strains of Western equine encephalitis virus. J Gen Virol 87:2353–2361. doi: 10.1099/vir.0.81815-0. [DOI] [PubMed] [Google Scholar]
- 26.Ketola A, Hinkkanen A, Yongabi F, Furu P, Maatta AM, Liimatainen T, Pirinen R, Bjorn M, Hakkarainen T, Makinen K, Wahlfors J, Pellinen R. 2008. Oncolytic Semliki forest virus vector as a novel candidate against unresectable osteosarcoma. Cancer Res 68:8342–8350. doi: 10.1158/0008-5472.CAN-08-0251. [DOI] [PubMed] [Google Scholar]
- 27.Saito K, Uzawa K, Kasamatsu A, Shinozuka K, Sakuma K, Yamatoji M, Shiiba M, Shino Y, Shirasawa H, Tanzawa H. 2009. Oncolytic activity of Sindbis virus in human oral squamous carcinoma cells. Br J Cancer 101:684–690. doi: 10.1038/sj.bjc.6605209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzme R, Tseng JC, Levin B, Ibrahim S, Meruelo D, Pellicer A. 2012. Sindbis viral vectors target hematopoietic malignant cells. Cancer Gene Ther 19:757–766. doi: 10.1038/cgt.2012.56. [DOI] [PubMed] [Google Scholar]
- 29.Maatta AM, Makinen K, Ketola A, Liimatainen T, Yongabi FN, Vaha-Koskela M, Pirinen R, Rautsi O, Pellinen R, Hinkkanen A, Wahlfors J. 2008. Replication competent Semliki Forest virus prolongs survival in experimental lung cancer. Int J Cancer 123:1704–1711. doi: 10.1002/ijc.23646. [DOI] [PubMed] [Google Scholar]
- 30.Li XD, Qiu FX, Yang H, Rao YN, Calisher CH. 1992. Isolation of Getah virus from mosquitos collected on Hainan Island, China, and results of a serosurvey. Southeast Asian J Trop Med Public Health 23:730–734. [PubMed] [Google Scholar]
- 31.Li K, Liang J, Lin Y, Zhang H, Xiao X, Tan Y, Cai J, Zhu W, Xing F, Hu J, Yan G. 2016. A classical PKA inhibitor increases the oncolytic effect of M1 virus via activation of exchange protein directly activated by cAMP 1. Oncotarget 7:48443–48455. doi: 10.18632/oncotarget.10305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang H, Lin Y, Li K, Liang J, Xiao X, Cai J, Tan Y, Xing F, Mai J, Li Y, Chen W, Sheng L, Gu J, Zhu W, Yin W, Qiu P, Su X, Lu B, Tian X, Liu J, Lu W, Dou Y, Huang Y, Hu B, Kang Z, Gao G, Mao Z, Cheng SY, Lu L, Bai XT, Gong S, Yan G, Hu J. 2016. Naturally existing oncolytic virus M1 is nonpathogenic for the nonhuman primates after multiple rounds of repeated intravenous injections. Hum Gene Ther 27:700–711. doi: 10.1089/hum.2016.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lin Y, Zhang H, Liang J, Li K, Zhu W, Fu L, Wang F, Zheng X, Shi H, Wu S, Xiao X, Chen L, Tang L, Yan M, Yang X, Tan Y, Qiu P, Huang Y, Yin W, Su X, Hu H, Hu J, Yan G. 2014. Identification and characterization of alphavirus M1 as a selective oncolytic virus targeting ZAP-defective human cancers. Proc Natl Acad Sci U S A 111:E4504-12. doi: 10.1073/pnas.1408759111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kruse KB, Brodsky JL, McCracken AA. 2006. Autophagy: an ER protein quality control process. Autophagy 2:135–137. doi: 10.4161/auto.2.2.2388. [DOI] [PubMed] [Google Scholar]
- 35.Piya S, Andreeff M, Borthakur G. 2016. Targeting autophagy to overcome chemoresistance in acute myleogenous leukemia. Autophagy. doi: 10.1080/15548627.2016.1245263:1-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shchors K, Massaras A, Hanahan D. 2015. Dual targeting of the autophagic regulatory circuitry in gliomas with repurposed drugs elicits cell-lethal autophagy and therapeutic benefit. Cancer Cell 28:456–471. doi: 10.1016/j.ccell.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 37.Kleijn A, Kloezeman J, Treffers-Westerlaken E, Fulci G, Leenstra S, Dirven C, Debets R, Lamfers M. 2014. The therapeutic efficacy of the oncolytic virus Delta24-RGD in a murine glioma model depends primarily on antitumor immunity. Oncoimmunology 3:e955697. doi: 10.4161/21624011.2014.955697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zemp FJ, McKenzie BA, Lun X, Reilly KM, McFadden G, Yong VW, Forsyth PA. 2014. Cellular factors promoting resistance to effective treatment of glioma with oncolytic myxoma virus. Cancer Res 74:7260–7273. doi: 10.1158/0008-5472.CAN-14-0876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wakimoto H, Kesari S, Farrell CJ, Curry WT Jr, Zaupa C, Aghi M, Kuroda T, Stemmer-Rachamimov A, Shah K, Liu TC, Jeyaretna DS, Debasitis J, Pruszak J, Martuza RL, Rabkin SD. 2009. Human glioblastoma-derived cancer stem cells: establishment of invasive glioma models and treatment with oncolytic herpes simplex virus vectors. Cancer Res 69:3472–3481. doi: 10.1158/0008-5472.CAN-08-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miest TS, Cattaneo R. 2014. New viruses for cancer therapy: meeting clinical needs. Nat Rev Microbiol 12:23–34. doi: 10.1038/nrmicro3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schneider U, Bullough F, Vongpunsawad S, Russell SJ, Cattaneo R. 2000. Recombinant measles viruses efficiently entering cells through targeted receptors. J Virol 74:9928–9936. doi: 10.1128/JVI.74.21.9928-9936.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.McFadden G. 2005. Poxvirus tropism. Nat Rev Microbiol 3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Axten JM, Medina JR, Feng Y, Shu A, Romeril SP, Grant SW, Li WH, Heerding DA, Minthorn E, Mencken T, Atkins C, Liu Q, Rabindran S, Kumar R, Hong X, Goetz A, Stanley T, Taylor JD, Sigethy SD, Tomberlin GH, Hassell AM, Kahler KM, Shewchuk LM, Gampe RT. 2012. Discovery of 7-methyl-5-(1-{[3-(trifluoromethyl)phenyl]acetyl}-2,3-dihydro-1H-indol-5-yl)-7H-p yrrolo[2,3-d]pyrimidin-4-amine (GSK2606414), a potent and selective first-in-class inhibitor of protein kinase R (PKR)-like endoplasmic reticulum kinase (PERK). J Med Chem 55:7193–7207. doi: 10.1021/jm300713s. [DOI] [PubMed] [Google Scholar]
- 44.Garcia MA, Meurs EF, Esteban M. 2007. The dsRNA protein kinase PKR: virus and cell control. Biochimie 89:799–811. doi: 10.1016/j.biochi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 45.Dey M, Cao C, Sicheri F, Dever TE. 2007. Conserved intermolecular salt bridge required for activation of protein kinases PKR, GCN2, and PERK. J Biol Chem 282:6653–6660. doi: 10.1074/jbc.M607897200. [DOI] [PubMed] [Google Scholar]