ABSTRACT
Sterile alpha motif and HD domain-containing protein 1 (SAMHD1) regulates intracellular deoxynucleoside triphosphate (dNTP) levels and functions as a retroviral restriction factor through its dNTP triphosphohydrolase (dNTPase) activity. Human SAMHD1 interacts with cell cycle regulatory proteins cyclin A2, cyclin-dependent kinase 1 (CDK1), and CDK2. This interaction mediates phosphorylation of SAMHD1 at threonine 592 (T592), which negatively regulates HIV-1 restriction. We previously reported that the interaction is mediated, at least in part, through a cyclin-binding motif (RXL, amino acids [aa] 451 to 453). To understand the role of the RXL motif in regulating SAMHD1 activity, we performed structural and functional analyses of RXL mutants and the effect on HIV-1 restriction. We found that the RXL mutation (R451A and L453A, termed RL/AA) disrupted SAMHD1 tetramer formation and abolished its dNTPase activity in vitro and in cells. Compared to wild-type (WT) SAMHD1, the RL/AA mutant failed to restrict HIV-1 infection and had reduced binding to cyclin A2. WT SAMHD1 and RL/AA mutant proteins were degraded by Vpx from HIV-2 but were not spontaneously ubiquitinated in the absence of Vpx. Analysis of proteasomal and autophagy degradation revealed that WT and RL/AA SAMHD1 protein levels were enhanced only when both pathways of degradation were simultaneously inhibited. Our results demonstrate that the RXL motif of human SAMHD1 is required for its HIV-1 restriction, tetramer formation, dNTPase activity, and efficient phosphorylation at T592. These findings identify a new functional domain of SAMHD1 important for its structural integrity, enzyme activity, phosphorylation, and HIV-1 restriction.
IMPORTANCE SAMHD1 is the first mammalian dNTPase identified as a restriction factor that inhibits HIV-1 replication by decreasing the intracellular dNTP pool in nondividing cells, although the critical mechanisms regulating SAMHD1 function remain unclear. We previously reported that mutations of a cyclin-binding RXL motif in human SAMHD1 significantly affect protein expression levels, half-life, nuclear localization, and phosphorylation, suggesting an important role of this motif in modulating SAMHD1 functions in cells. To further understand the significance and mechanisms of the RXL motif in regulating SAMHD1 activity, we performed structural and functional analyses of the RXL motif mutation and its effect on HIV-1 restriction. Our results indicate that the RXL motif is critical for tetramer formation, dNTPase activity, and HIV-1 restriction. These findings help us understand SAMHD1 interactions with other host proteins and the mechanisms regulating SAMHD1 structure and functions in cells.
KEYWORDS: HIV-1, SAMHD1, cyclin binding, dNTPase, infection, restriction, structure
INTRODUCTION
Sterile alpha motif and HD domain-containing protein 1 (SAMHD1) is a mammalian deoxynucleoside triphosphate (dNTP) triphosphohydrolase (dNTPase) that is well characterized as a protein that restricts retroviruses and DNA viruses (1–5) by limiting the intracellular dNTP pool in nondividing cells (6, 7). There are multiple mechanisms regulating SAMHD1 activity, allowing dNTPase activity to be switched on or off when required. This regulation is critical for SAMHD1 to maintain cellular dNTP homeostasis (8). Many transformed, proliferating cell lines express low levels of SAMHD1, which has partly been attributed to epigenetic regulation, including methylation of the SAMHD1 promoter (9, 10) and increased microRNA 181 to downregulate SAMHD1 expression (11, 12). Overexpression of SAMHD1 in dividing HEK293T or HeLa cells leads to decreased intracellular dNTP levels, although the effect is not strong enough to restrict immunodeficiency virus type 1 (HIV-1) infection (13). In contrast, knockout of SAMHD1 in monocytic THP-1 cells renders the cells more permissive to HIV-1 infection and affects distribution of cell cycle and apoptosis (14). Furthermore, exogenous expression of SAMHD1 inhibits proliferation and induces apoptosis in T-cell lymphoma-derived HuT78 cells (15). These studies suggest that SAMHD1 is also functional in dividing cells (14, 16).
Regulation of SAMHD1 function is correlated with the cell cycle. SAMHD1 interacts with cell cycle-related proteins that are highly expressed in dividing cells and phosphorylate SAMHD1 at threonine 592 (T592), abolishing HIV-1 restriction (17–23) and destabilizing the tetramer (23, 24). The homotetramer is accepted to be the biologically active form of SAMHD1 and is required for HIV-1 restriction (25). However, conflicting reports indicate that SAMHD1 protein levels remain unchanged or vary with different stages of the cell cycle, and it is not clear if there is a mechanism to regulate total SAMHD1 protein levels in cells (26–28). Cell cycle regulation is closely tied to intracellular dNTP levels, which are elevated during S-phase and thus require a decrease in SAMHD1 expression or activity at the appropriate phases of the cell cycle (22).
The molecular mechanism of SAMHD1 hydrolysis of dNTPs has been characterized structurally and biochemically (23, 25, 29, 30). Allosteric sites in SAMHD1 bind activating dNTPs, which induces a conformation change and tetramerization of the protein into the active tetramer (29–35). Recent work has identified oxidation of cysteine residues in SAMHD1 as an additional mechanism of its functional regulation (36).
SAMHD1 restriction is circumvented by the Vpx protein from either HIV-2 or most simian immunodeficiency viruses (SIV) or the Vpr protein from certain SIV lineages (37), which target SAMHD1 for proteasomal degradation (4, 5, 38, 39). Aside from SAMHD1-targeted proteasomal degradation orchestrated by “helper” proteins, such as Vpx (40) or cyclin L2 in macrophages (41), there is little knowledge regarding the natural cellular regulation of SAMHD1 protein levels. Our previous work demonstrated that endogenous SAMHD1 has a half-life of 6.5 h in cycloheximide-treated cycling THP-1 cells (42). However, the fate of the protein and the cellular mechanisms of spontaneous SAMHD1 degradation remain unknown. We previously reported that a mutant of human SAMHD1 in the cyclin-binding motif (RXL) (R451A and L453A, termed RL/AA) that affects its cyclin A/CDK interaction (42). The RL/AA mutant was expressed poorly in cells, had reduced T592 phosphorylation, exhibited a reduced half-life and reduced ability to form tetramers compared to those of the wild-type (WT) protein, and was mislocalized to intranuclear aggregations (42). However, the impact of the RL/AA mutant on SAMHD1 function, HIV-1 restriction, and characterization of SAMHD1 spontaneous cellular degradation is unknown. Further investigation of these issues could allow clearer understanding of how SAMHD1 function is regulated within cells.
In this study, we provide evidence that the RXL motif is critical for the structural integrity of functional SAMHD1 tetramers, dNTPase activity, and thereby HIV-1 restriction. We also compared the mechanisms by which WT SAMHD1 and the tetramerization-deficient mutant (RL/AA) were degraded in cells. Interestingly, our results suggest that spontaneous degradation of SAMHD1 protein in cells can occur through simultaneous mechanisms involving proteasomal and autophagy/lysosome pathways. Understanding how SAMHD1 is regulated in cells is important for gaining clear mechanistic insights into SAMHD1 and its roles in cell cycle regulation, nucleotide homeostasis, viral restriction, and innate immune responses and potential avenues to manipulate SAMHD1 regulation in cells.
RESULTS
The RXL motif in SAMHD1 is critical for HIV-1 restriction and dNTPase activity in cells.
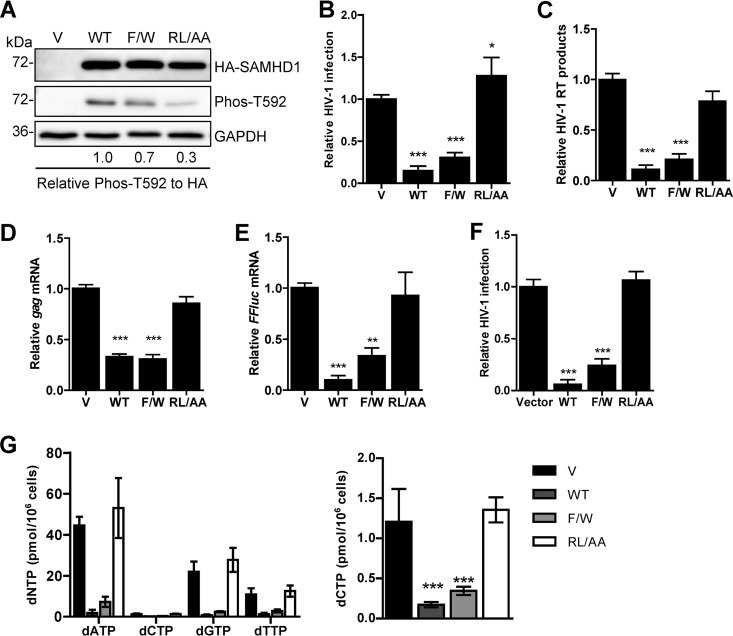
We previously demonstrated that mutation of the RXL motif in SAMHD1 resulted in low protein expression and loss of multimerization when overexpressed in HEK293T cells (42). However, the effect of the RL/AA mutant on SAMHD1-mediated HIV-1 restriction in a nondividing cell model remains unknown. To address this, we generated stable U937 cells that express WT full-length SAMHD1 or the RL/AA mutant. Monocytic U937 cells do not express endogenous SAMHD1 and have been used as a model to evaluate SAMHD1-mediated HIV-1 restriction (5, 18). We also included an additional SAMHD1 mutant, the F454W mutant (referred to as F/W), which we previously reported to have T592 phosphorylation, localization, and tetramer formation in cells comparable to those of WT SAMHD1 (42). We therefore predicted that the F/W mutant should restrict HIV-1 infection.
To obtain similar levels of WT or mutant SAMHD1 expression, we adjusted the volume of lentiviral vectors used when transducing U937 cells. Immunoblotting of cell lysates confirmed comparable levels of WT and mutant SAMHD1 expression between cell lines and demonstrated that the RL/AA mutant had reduced T592 phosphorylation relative to WT SAMHD1 (Fig. 1A), using an antibody previously confirmed for specificity to T592 phosphorylation of SAMHD1 (19). Differentiated U937 cells were infected with single-cycle, luciferase reporter HIV-1 to determine viral restriction by SAMHD1. Compared to the vector control cells, WT SAMHD1 and the F/W mutant restricted HIV-1 infection by 95% and 67%, respectively, whereas the RL/AA mutant did not restrict HIV-1 infection (Fig. 1B). We observed a 1.3-fold enhancement in single-cycle HIV-1 infection of the RL/AA mutant expressing a stable cell line compared to vector control cells (Fig. 1B). However, the effect of the increased HIV-1 infection by the RL/AA mutant was not evident at viral reverse transcription (RT) (Fig. 1C) or mRNA (Fig. 1D and E) levels, implying that a potential posttranscriptional regulatory mechanism might be involved. To determine if the increased HIV-1 infection by the RL/AA mutant expression was specific to a reporter gene, we performed single-cycle HIV-1 infection assays using a reporter virus expressing enhanced green fluorescent protein (GFP) in the HIV-1 envelope open reading frame (43). GFP reporter HIV-1 infection in RL/AA mutant-expressing cells was comparable to that in vector control cells (Fig. 1F). Furthermore, we infected these cells with an HIV-1LAI-based single-cycle virus and likewise found no enhancement of Gag expression in the RL/AA-expressing cells compared to the vector (data not shown). These results suggest that the slightly increased HIV-1 infection by the RL/AA mutant expression is unique to luciferase reporter virus.
FIG 1.
The RXL motif in SAMHD1 is required for HIV-1 restriction and dNTPase activity. (A) U937 cells stably expressing vector (V) or WT or mutant (F/W or RL/AA) SAMHD1 were differentiated with PMA for 24 h and then cultured for a further 24 h prior to detection of protein expression. GAPDH was used as a loading control. Relative Phos-T592 was calculated by normalizing densitometry values for total SAMHD1 (HA) signals to GAPDH. The relative ratio of Phos-T592 to total SAMHD1 is shown; the value for the WT was set as 1. (B) PMA-differentiated U937 cells were infected with single-cycle HIV-1-Luc/VSV-G (multiplicity of infection of 0.5). Lysates were collected 48 h postinfection, and infection was quantified by luciferase assay. All samples were normalized by protein content. Data were calculated relative to the value for the vector cells, which was set as 1. Data represent results from one experiment from four independent repeats. (C) HIV-1 late reverse transcription (RT) products from infected cells were quantified via qPCR of total genomic DNA. GAPDH levels were used as a normalization control. Data were calculated relative to the value for vector cells, which was set as 1. Data represent results from one experiment with four biological replicates. (D and E) The mRNA levels of HIV-1 gag and the reporter firefly luciferase gene (FFluc) were measured in infected cells by RT-qPCR. GAPDH mRNA levels were used as a control. Data were calculated relative to the value for the vector cells, which was set as 1 using the 2−ΔΔCT method. Data represent the averages of 3 independent experiments. (F) PMA-differentiated U937 cells were infected with single-cycle HIV-1-ΔEnv-GFP/VSV-G as described for panel B. Viral infection was quantified as GFP-positive cells using flow cytometry. Data were calculated relative to the value for the vector cells, which was set as 1. Data represent the averages of two independent experiments, each with triplicate samples. Error bars represent SDs. (G) dNTP analysis of PMA-differentiated U937 cell lines. Data represent the averages of two experiments. To better compare lower dCTP levels among the samples, the dCTP levels are also presented in a different scale (right graph). The data are means ± SDs. Statistical analysis was performed by one-way ANOVA with Dunnett's multiple-comparison test. ***, P < 0.001; **, P < 0.01.
To characterize at which stage of HIV-1 replication the RL/AA mutant lost restriction activity, we measured HIV-1 late reverse transcription products in the luciferase reporter virus-infected cells. We found that the RL/AA mutant was unable to reduce viral cDNA synthesis, whereas both WT SAMHD1 and the F/W mutant blocked HIV-1 reverse transcription effectively, by 90% and 79%, respectively (Fig. 1C). These results suggest that the RL/AA mutant was defective in reducing HIV-1 late reverse transcription, likely due to the lack of dNTPase activity. We next measured the levels of mRNA transcription to determine whether the RL/AA mutant had any effects on later stages of viral replication. Compared to vector control cells, levels of luciferase and gag mRNA driven by the HIV-1 promoter were consistent with late RT levels and decreased significantly in WT SAMHD1 or F/W mutant-expressing cells, but not for the RL/AA mutant (Fig. 1D and E). To examine whether the RL/AA mutant had dNTPase activity, we measured intracellular dNTP levels in U937 cells expressing WT or mutant SAMHD1. We observed that only the RL/AA mutant was defective for dNTP hydrolysis (Fig. 1G). Thus, the RXL motif in human SAMHD1 is critical for its HIV-1 restriction and dNTPase activity in cells.
The RXL mutant disrupts SAMHD1 tetramer formation and abolishes dNTPase activity in vitro.
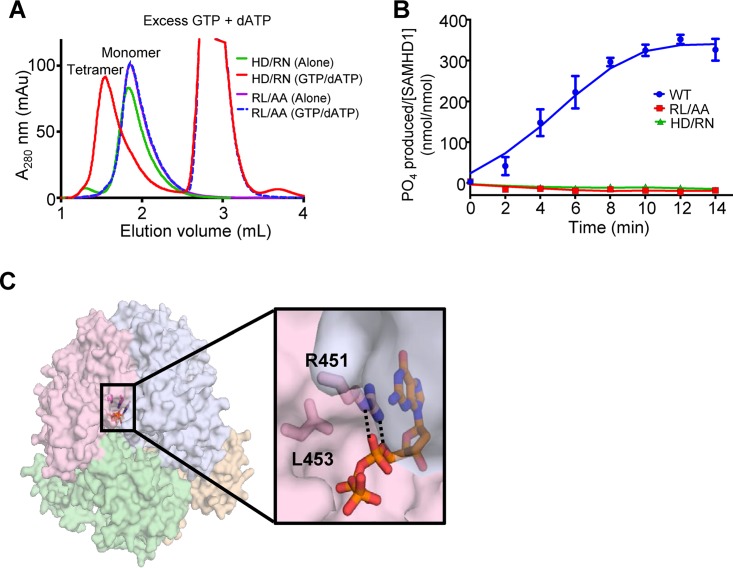
To determine why the RXL motif was required for HIV-1 restriction and dNTPase activity, we assessed the effect of the RL/AA mutant on SAMHD1 structure and dNTP hydrolysis. We utilized in vitro assays to assess tetramerization of SAMHD1 in the presence or absence of dATP/GTP. SAMHD1 tetramers are induced by binding of GTP or dGTP at a primary allosteric site. Further binding of any dNTP at a second allosteric site then occurs and the enzyme is activated (30). We found that the RL/AA mutant remained as a monomer, irrespective of the presence of dATP/GTP, whereas an H206R/D207N mutant (referred to as HD/RN) that abrogates the catalytic activity of SAMHD1 without affecting tetramerization (29) clearly formed tetramers in the presence of dATP/GTP (Fig. 2A). An in vitro dGTP hydrolysis assay showed that compared to WT SAMHD1, both the RL/AA and HD/RN mutants were catalytically inactive (Fig. 2B). These data suggest that the inability of the RL/AA mutant to tetramerize prevents the dNTPase activity and consequently HIV-1 restriction. Analysis of the published X-ray crystal structure (PDB ID 4BZB [29]) shows that the R451 and L453 residues are part of the allosteric nucleotide-binding pocket and that the RL/AA mutation disrupts the hydrogen bonds with an allosteric dNTP, likely destabilizing the tetramer interface (Fig. 2C). Together, these data indicate that the RL/AA mutant is ineffective at blocking HIV-1 restriction because it causes defects in binding to allosteric dNTPs, leading to destabilization of SAMHD1 tetramers, and therefore abolishes its dNTPase activity.
FIG 2.
The RXL mutant disrupts SAMHD1 tetramer formation and abolishes dNTPase activity. (A) Size exclusion chromatography of SAMHD1 mutants in the presence or absence of dNTPs. The H206R/D207N (HD/RN) mutant SAMHD1 is a monomer (green trace) in the absence of allosteric nucleotides and forms tetramers in the presence of GTP and dATP (red trace). In contrast, the R451A/L453A (RL/AA) mutant in either the presence (blue dashes) or absence (purple) of nucleotides elutes as a monomer. (B) WT SAMHD1 (blue) hydrolyzes dGTP to produce inorganic phosphates, which are measurable by the malachite green assay. Similar to the HD/RN mutant (green) that has previously been shown to be inactive; the RL/AA mutant (red) also lacks the dNTPase activity. (C) The RL/AA mutation disrupts the activity of SAMHD1 by destabilizing the tetramer interface. As shown in the published crystal structure (PDB ID 4BZB) (29), the SAMHD1 tetramer is represented. Each SAMHD1 monomer subunit is depicted by a different color. Residues R451 and L453 are positioned at the interface between two subunits of the SAMHD1 tetramer where R451 directly interacts (hydrogen bonding [dashed lines] and stacking) with dGTP bound to allosteric site 1. Mutating these residues disrupts the interaction with allosteric dNTPs and thus inhibits SAMHD1 tetramerization.
RXL mutation of SAMHD1 reduces T592 phosphorylation and cyclin A2 binding.
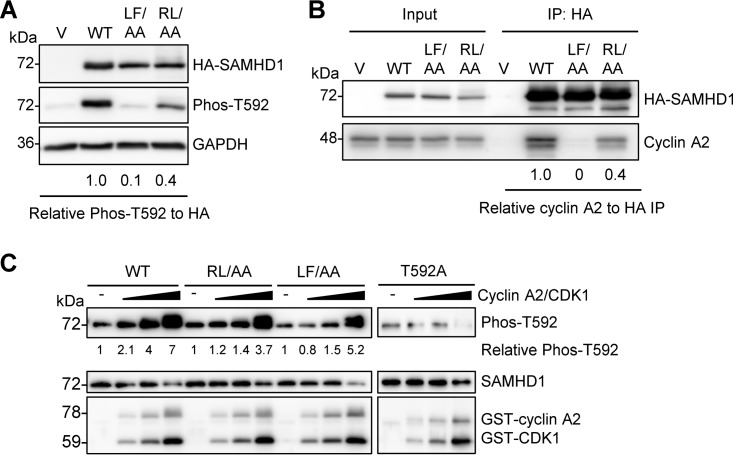
A previous study also identified a cyclin-binding motif (L620/F621) in the C-terminal region of human SAMHD1 critical for its interaction with cyclin A2 and showed that alanine substitution for the dihydrophobic residues leucine and phenylalanine (L620A/F621A, referred to as LF/AA) was sufficient to disrupt interaction with cyclin A2 (28). To better understand the significance of these two cyclin-binding motifs in cells, we directly compared the RL/AA and LF/AA mutants with WT SAMHD1. First, we assessed the levels of T592 phosphorylation of SAMHD1 overexpressed in HEK293T cells. Compared to WT SAMHD1, both mutants were defective for phosphorylation, with the relative level of T592 phosphorylation reduced by 90% and 60% for the LF/AA and RL/AA mutants, respectively (Fig. 3A). Next, we compared cyclin A2 interaction with WT SAMHD1 and the mutants in HEK293T cells using coimmunoprecipitation (co-IP) assays. We observed that compared to WT SAMHD1, both mutants were impaired in their interactions with cyclin A2, with the LF/AA mutant displaying almost undetectable interaction while the RL/AA mutant was decreased by 60% (Fig. 3B), consistent with the effects on phosphorylation that we observed (Fig. 3A). These data suggest that while the cyclin-binding motif (L620/F621) of human SAMHD1 is required for its interaction with cyclin A2, the RXL cyclin-binding motif partially contributes to the interaction, though possibly through an indirect mechanism.
FIG 3.
RXL mutation of SAMHD1 reduces T592 phosphorylation and cyclin A2 binding. (A) HEK293T cells expressing vector or WT, LF/AA, or RL/AA SAMHD1 protein were analyzed by immunoblotting for Phos-T592, HA-SAMHD1, or GAPDH. Relative Phos-T592 was calculated by normalizing densitometry values for Phos-T592 and total SAMHD1 (HA) signals to GAPDH. The relative ratio of Phos-T592 to total SAMHD1 is shown; the value for the WT was set as 1. Immunoblot data are representative of two independent experiments; for densitometry, the average of two experiments is depicted. (B) Co-IP of cyclin A2 from HEK293T cells overexpressing vector or WT, LF/AA, or RL/AA SAMHD1 protein. Transfected cells were lysed and HA-tagged SAMHD1 was bound to HA-conjugated agarose. SAMHD1-interacting proteins were eluted and analyzed by immunoblotting for HA or cyclin A. Data are representative of results from two independent experiments. Cyclin A2 IP densitometry was normalized to HA-SAMHD1 IP products. Relative amounts were calculated according to the value for WT SAMHD1, which was set as 1. (C) Recombinant WT, RL/AA, LF/AA, or T592A SAMHD1 protein was used for in vitro kinase assays. SAMHD1 proteins were incubated with increasing amounts of cyclin A2/CDK1 complex in the presence of ATP. Kinase-free samples were used as a negative control. Reactions were analyzed by SDS-PAGE and immunoblotting for SAMHD1, Phos-T592, and GST. Data are representative of results from three independent experiments. The densitometry value of the T592 phosphorylated species of SAMHD1 was calculated, with the value for the kinase-free samples set as 1 and each protein phosphorylation calculated relative to the kinase-free control.
To independently determine the direct effect of the RL/AA mutation on T592 phosphorylation of SAMHD1, we performed in vitro kinase assays with purified recombinant SAMHD1 proteins. As expected, we observed dose-dependent T592 phosphorylation of WT SAMHD1 protein (up to 7-fold enhancement), but not a phospho-ablative SAMHD1 mutant (T592A) (18), in incubations with increasing amounts of glutathione S-transferase (GST)-tagged cyclin A2 and CDK1 proteins (Fig. 3C). Interestingly, we observed slightly decreased levels of T592 phosphorylation for the RL/AA (3.7-fold) and LF/AA (5.2-fold) mutants compared to that for WT SAMHD1 (Fig. 3C), suggesting that these two mutants are phosphorylated less efficiently than WT SAMHD1. Because the CDK motif (TPQK; residues 592 to 595) of human SAMHD1 (21) is intact and outside the mutated region of RL/AA and LF/AA, this could explain our observation of in vitro T592 phosphorylation of WT and mutant SAMHD1. Lower levels of T592 phosphorylation signal in the controls without cyclin A2/CDK1, and the T592A samples with or without cyclin A2/CDK1, were likely due to background antibody cross-reactivity with total SAMHD1 protein (Fig. 3C).
SAMHD1 protein levels are not significantly affected by inhibiting the proteasome or lysosome degradation in cells.
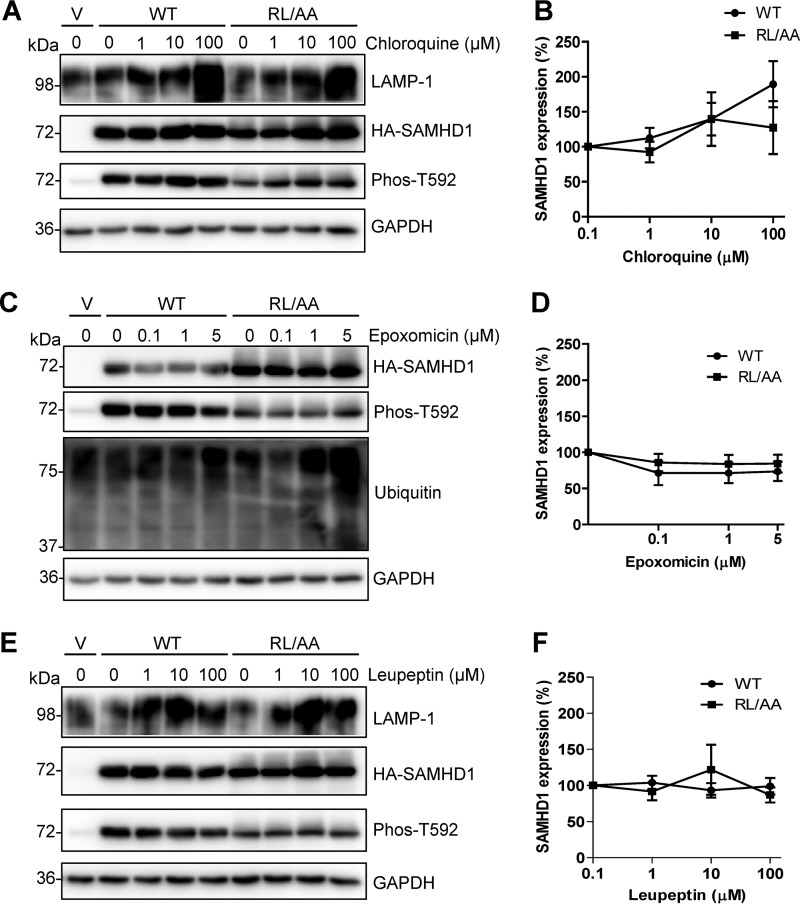
Our previous study showed that the RL/AA mutant was expressed poorly in cells (over 10-fold lower than WT) and has a 6-fold-shorter half-life (t1/2) than that of WT SAMHD1 (42). We sought to determine whether the RL/AA mutant experienced enhanced cellular degradation and whether it was degraded via the same mechanism as WT SAMHD1. We treated HEK293T cells overexpressing WT or RL/AA SAMHD1 with different inhibitors. Epoxomicin is a selective proteasome inhibitor (44); chloroquine is a lysosomotropic agent that inhibits lysosomal hydrolases and prevents autophagosomal fusion and degradation, while leupeptin is an acid protease inhibitor and causes accumulation of autolysosomes (45). SAMHD1 protein levels were analyzed by immunoblotting and quantified by densitometry after 16 h of treatment with a range of inhibitor concentrations.
Chloroquine-treated cells showed that WT and RL/AA SAMHD1 protein levels had a slight enhancement at 10 and 100 μM (Fig. 4A); however, the trend across six independent experiments was not significant (Fig. 4B). Epoxomicin caused a slight decrease in protein levels, possibly due to cytotoxicity at higher concentrations (Fig. 4C and D). Leupeptin also did not show a significant effect on either WT or RL/AA SAMHD1 protein level (Fig. 4E and F). Enhanced LAMP-1 protein expression was used as a positive control for lysosomal inhibition (Fig. 4A and E) (46), and ubiquitin was used as a control for epoxomicin (Fig. 4C). Changes in the levels of phosphorylated T592 correlated with changes in the total levels of SAMHD1 (Fig. 4A, C, and E), as expected, as the majority of SAMHD1 in cycling cells is phosphorylated (21). Furthermore, we examined whether the inhibitor treatments altered SAMHD1 localization using immunofluorescence microscopy. However, analysis of WT SAMHD1 or RL/AA mutant expression after treatment with each inhibitor showed no significant change in localization, or association with lysosomal-associated membrane protein 1 (LAMP-1) (data not shown). Overall, these data suggest that overexpressed SAMHD1 in HEK293T cells is not spontaneously degraded through lysosomes or the proteasome.
FIG 4.
SAMHD1 protein levels are not affected by inhibiting the proteasome or lysosome degradation in cells. HEK293T cells expressing WT or RL/AA SAMHD1 protein were treated with chloroquine (A), epoxomicin (C), or leupeptin (E) at the indicated concentrations for 16 h. Vector plasmid DNA-transfected cells were used as a negative control. Lysates were harvested and analyzed by immunoblotting using antibodies specific to LAMP-1 or ubiquitin, HA-tagged SAMHD1 (HA), and Phos-T592 SAMHD1. GAPDH was used as a loading control. (B, D, and F) Graphs depicting SAMHD1 were generated from densitometry analysis of WT or RL/AA SAMHD1 (HA) immunoblots in HEK293T cells treated as described for panels A, C, and E. All densitometry was normalized to the value for GAPDH. Negative controls with no inhibitor treatment were set to 100% for each WT or RL/AA experiment, and all densitometry values were calculated as a percentage of this. The graphs represent a summary of 6 independent transfection experiments.
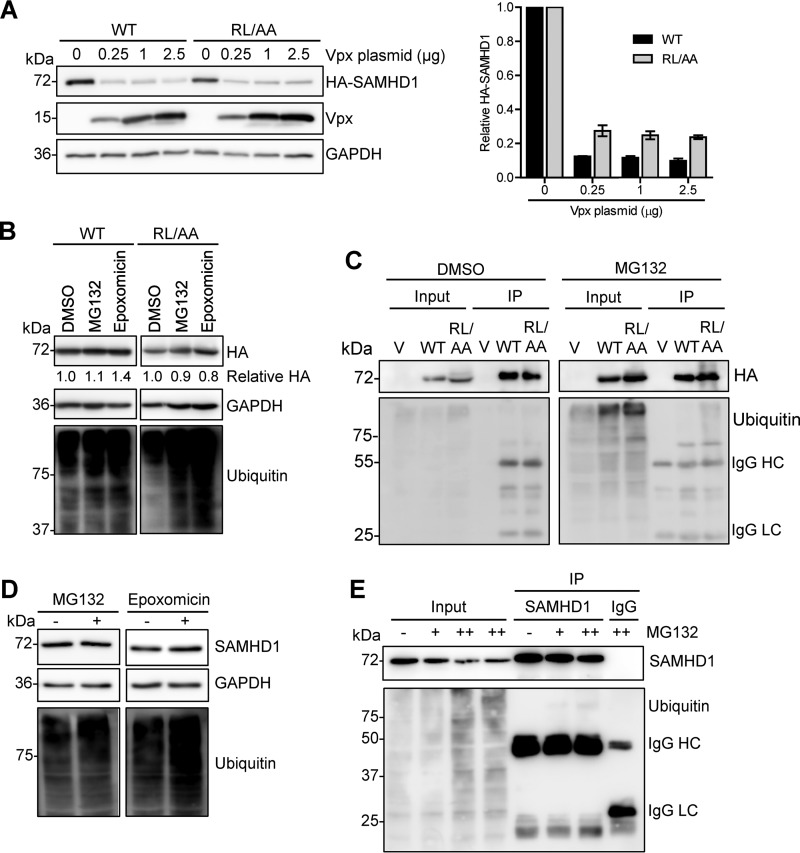
WT SAMHD1 and RL/AA mutant proteins are degraded by HIV-2 Vpx but are not spontaneously ubiquitinated.
Several lentiviruses, including HIV-2 and certain SIV, encode the Vpx protein, which targets SAMHD1 for proteasomal degradation (4, 5). As we were unable to alter SAMHD1 protein levels using epoxomicin or lysosomal inhibitors, we first verified whether Vpx could target the RL/AA mutant for proteasomal degradation. We cotransfected HEK293T cells with plasmids expressing Vpx from the HIV-2ROD strain (13) and WT SAMHD1 or the RL/AA mutant. Analysis of SAMHD1 protein levels confirmed that both WT SAMHD1 and the RL/AA mutant could be efficiently degraded by coexpression of Vpx (Fig. 5A). Degradation of the RL/AA mutant was approximately 2-fold less efficient than that of WT SAMHD1 in spite of increasing amounts of Vpx (Fig. 5A and B). The RL/AA mutant is not in the vicinity of the C-terminal Vpx-interacting domain in SAMHD1 (47); hence, it should not interfere with the ability of this mutant to be targeted to the proteasome. These results suggest that a portion of the mislocalized RL/AA mutant (42) may be partially resistant or inaccessible to Vpx-mediated degradation in cells.
FIG 5.
WT SAMHD1 and RL/AA mutant proteins are degraded by Vpx from HIV-2 but are not spontaneously ubiquitinated. (A) HEK293T cells were cotransfected with plasmids expressing WT (0.1 μg) or RL/AA (1 μg) SAMHD1 along with a plasmid expressing HIV-2ROD Vpx. Empty vectors (pLenti-puro and pCG-myc) were used to normalize all DNA transfection amounts. Lysates were harvested and HA-tagged SAMHD1 and Vpx expression was confirmed by immunoblotting using anti-HA and Vpx-specific antibodies, respectively. GAPDH was used as a loading control. Relative SAMHD1 protein levels were calculated by densitometry and normalized to the value for GAPDH. The value for cells not treated with Vpx was set as 1. The graph depicting densitometry shows averaged data from two independent experiments. (B) HEK293T cells transfected with plasmid DNA expressing WT or RL/AA SAMHD1 were pretreated with MG132 (1 μM) or epoxomicin (1 μM) for 3 h. Cells were washed and fresh medium was added without inhibitors. Lysates were harvested 6 h after the removal of MG132. Lysates were immunoblotted for expression of HA and ubiquitin, and GAPDH was used as a loading control. Relative SAMHD1 was calculated as described for panel A. The value for DMSO controls was set as 1. (C) HEK293T cells expressing WT or RL/AA SAMHD1 were treated with MG132 (1 μM) and cells were lysed in lysis buffer containing protease inhibitor MG132 and N-ethylmaleimide. HA-IP was performed and IP products were analyzed by immunoblotting. Cells transfected with plasmid expressing empty vector were used as a negative control. IgG heavy chain (HC) and light chain (LC) contaminating bands are indicated in panels C and E. (D) THP-1 cells were pretreated with MG132 (0.5 μM) or epoxomicin (0.1 μM) for 3 h. Cells were washed and fresh medium without inhibitors was added. Cell lysates were harvested 6 h after the removal of MG132. (E) THP-1 cells were treated with MG132 for 12 h (0.1 or 1 μM, indicated by “+” and “++,” respectively). Cells were lysed in cell lysis buffer as described for panel C. SAMHD1 IP was performed with the cell lysates. Nonspecific mouse IgG was used as a negative control. IP products were analyzed by immunoblotting with antibodies specific to SAMHD1 and ubiquitin.
Based on our data in Fig. 4 indicating that 16 h of treatment with epoxomicin did not rescue SAMHD1 levels, we tested an additional proteasome inhibitor (MG132) (48) and treated cells for a shorter time (3 h, with harvesting of cells 6 h posttreatment), based on the half-life of SAMHD1 (42). We used a lower concentration of each inhibitor (1 μM), as we observed decreased protein levels overall with higher concentrations or longer treatment times. HEK293T cells overexpressing WT SAMHD1 or the RL/AA mutant were treated with either MG132 or epoxomicin, and SAMHD1 protein levels were assessed. WT or RL/AA SAMHD1 protein levels were not significantly affected by each inhibitor at 6 h posttreatment, while the levels of ubiquitin control were elevated compared to those of dimethyl sulfoxide (DMSO) controls (Fig. 5B). Next, we asked whether SAMHD1 could be ubiquitinated in dividing cells in the absence of Vpx. We treated cells with MG132 prior to IP of SAMHD1. IP products were then assessed for the presence of ubiquitinated species. While the ubiquitin signal increased in MG132-treated cell lysates, we could not detect strong ubiquitinated species in the WT or RL/AA SAMHD1 IP samples (Fig. 5C), suggesting that overexpressed SAMHD1 is not polyubiquitinated in HEK293T cells.
To test whether these results were recapitulated in THP-1 cells expressing endogenous SAMHD1, we repeated experiments; consistent with the results for HEK293T cells, the endogenous SAMHD1 protein level was not significantly altered 6 h after treatment with either inhibitor (Fig. 5D). IP experiments with endogenous SAMHD1 confirmed the data for HEK293T cells, and there was not strong evidence indicating that SAMHD1 is polyubiquitinated in these cells (Fig. 5E). Upon darker exposure, a weak band between 100 and 150 kDa was detectable with the ubiquitin antibody; however, it was not detected in HEK293T experiments. Thus, SAMHD1 is not actively polyubiquitinated and targeted to the proteasome for degradation in cells in the absence of Vpx.
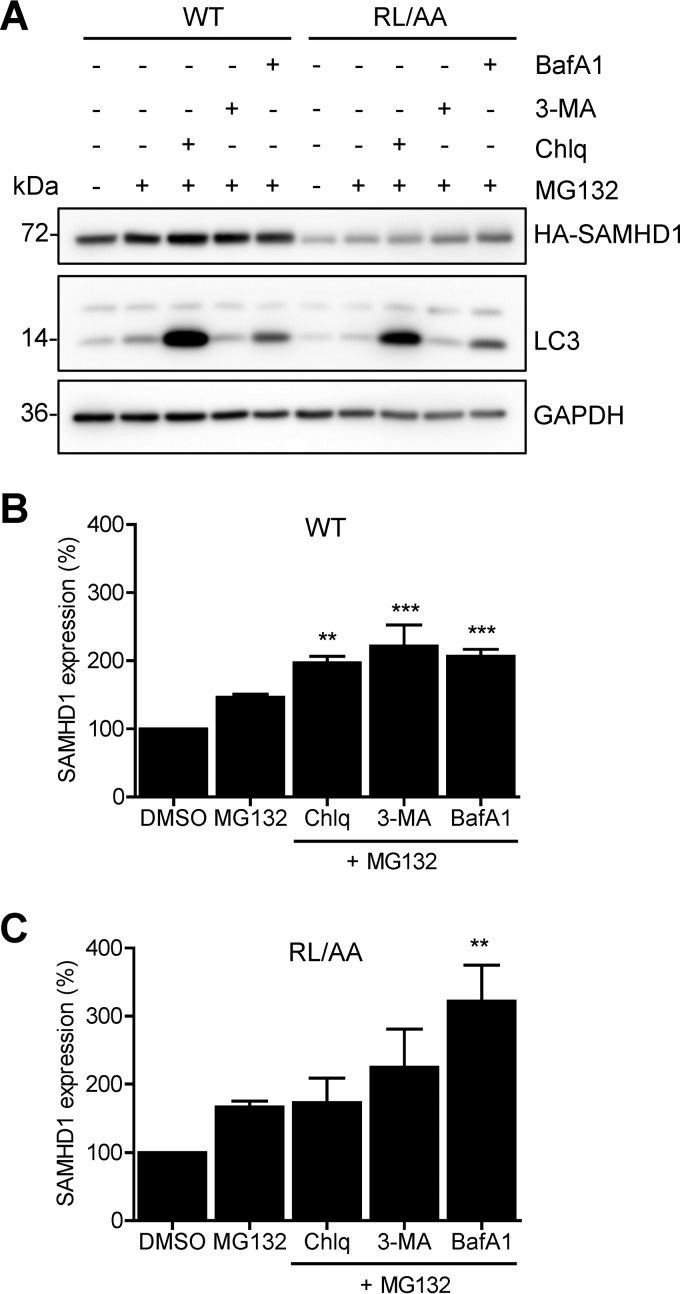
Blocking both autophagy/lysosome and proteasomal degradation enhanced WT and RL/AA SAMHD1 protein expression.
Given that we did not observe ubiquitination of SAMHD1 in cells, and protein levels were not rescued by treatment with proteasome or lysosome inhibitors (Fig. 5B to E), we questioned whether activation of autophagy would induce SAMHD1 protein turnover. Autophagy is a dynamic process of protein degradation that can be upregulated in response to certain stimuli, such as nutrient starvation (49). Autophagy can be inhibited by bafilomycin A1 (BafA1), which inhibits autophagosome-lysosome fusion, or by 3-methyladenine (3-MA), which blocks autophagosome formation via inhibition of class III phosphoinositide 3-kinase (50).
HEK293T cells overexpressing WT or RL/AA SAMHD1 were nutrient starved to activate autophagy and treated with inhibitors targeting early or late stages of autophagy. However, neither nutrient starvation alone nor treatment with BafA1 or 3-MA affected SAMHD1 protein levels (data not shown). These data suggest that SAMHD1 is not actively degraded in dividing cells. To address the possibility that SAMHD1 was basally degraded through both pathways simultaneously, we tested combinations of the inhibitors and their effects on SAMHD1 protein levels. HEK293T cells overexpressing WT SAMHD1 or RL/AA mutants were treated with either DMSO, MG132 alone, or MG132 in combination with chloroquine, 3-MA, or BafA1. Immunoblotting analysis revealed that WT SAMHD1 protein levels modestly, but statistically significantly, increased (up to 2-fold) with combination inhibitor treatment, compared to DMSO or MG132 controls (Fig. 6A and B). Likewise, the RL/AA mutant increased from approximately 2- to 3-fold with combination inhibitor treatment (Fig. 6A and C). Interestingly, the effect appeared stronger on the RL/AA mutant than on WT SAMHD1 (Fig. 6B and C), although we cannot rule out this is because of the lower expression levels of the RL/AA mutant. Overall, these results suggest that degradation of SAMHD1 protein in cells may occur through coordinated lysosomal/autophagosomal and proteasomal pathways.
FIG 6.
Blocking both autophagy/lysosome and proteasomal degradation enhances WT and RL/AA SAMHD1 protein expression. (A) HEK293T cells expressing HA-tagged WT or RL/AA SAMHD1 were treated with inhibitors as indicated for 24 h. The inhibitors were MG132 (1 μM), BafA1 (100 nM), 3-MA (5 mM), and chloroquine (Chlq; 100 μM); DMSO was used as a control. Lysates were analyzed by immunoblotting with antibodies to SAMHD1 (HA) and the autophagy marker LC3. GAPDH was used as a loading control. (B and C) For quantification purposes, the densitometry levels of WT or RL/AA SAMHD1 bands were normalized to that of GAPDH. The value for DMSO-treated cells was set at 100%, and all samples were calculated as a percentage of this. Graphs depict average densitometry results from four independent transfection experiments. Statistical analysis was performed by one-way ANOVA with Dunnett's multiple-comparison test. ***, P < 0.001; **, P < 0.01.
DISCUSSION
SAMHD1 dNTPase function is tightly linked to its ability to restrict HIV-1 (6, 7). Previously, an R451E mutant was predicted to abolish hydrogen bonding with dNTP molecules, affecting dNTPase activity and tetramerization in vitro (34). Here, we extend these findings in a cellular context and describe a double alanine mutation at residues R451 and L453 in the HD domain of SAMHD1 that is sufficient to abrogate SAMHD1-mediated HIV-1 restriction, through loss of tetramerization and rendering the protein dNTPase inactive. Interestingly, other studies reported differing results about the requirement for tetramerization for HIV-1 restriction (25, 51). Furthermore, anti-HIV activity of some SAMHD1 mutants in cells does not correlate with in vitro dNTPase activity or tetramerization, indicating that domains demonstrated to regulate tetramerization in vitro might not be crucial for cellular function. This suggests that other mechanisms play a role in regulating SAMHD1 function at a cellular level. Indeed, residues Y146 and Y154 contribute to stability of tetramers in vitro, but HIV-1 restriction is not affected by Y146S/Y154S mutations (52). Our data confirm that residues R451 and L453 are necessary for HIV-1 restriction, highlighting that binding of dNTPs is an absolute requirement for HIV-1 restriction in cells. In contrast to this, mouse SAMHD1 isoform 2 is catalytically active in the absence of GTP, which has been linked to more efficient tetramerization, mediated through the carboxyl-terminal domain (53). However, it is possible that different species of SAMHD1 experience different mechanisms of regulation in a cell-type-dependent manner.
We compared two cyclin-binding motifs in human SAMHD1 regarding their T592 phosphorylation in cells. Yan et al. demonstrated that residues L620 and F621 of SAMHD1 were crucial for T592 phosphorylation in cells and in vitro, and interaction with cyclin A2 (28), but not for tetramerization and HIV-1 restriction (25). However, our data demonstrate both the RL/AA and LF/AA mutants could be phosphorylated in vitro, with lower efficiencies than for WT SAMHD1. It is possible that proximity of the LF/AA mutation to the CDK recognition motif in the disordered C terminus of SAMHD1 may render the CDK motif inaccessible or unable to be phosphorylated by CDK2, used in their in vitro assay, but not CDK1, used in ours. Nevertheless, our in vitro kinase assays demonstrated phosphorylation of SAMHD1 in the absence of tetramer-activating GTP/dATP, indicating that SAMHD1 can be phosphorylated in a monomeric state.
Notably, several studies used a variety of cyclin-kinase complexes with contradictory effects on SAMHD1 T592 phosphorylation in vitro (17, 21, 22, 27, 28), indicating that these experiments do not always reproduce cellular conditions. Moreover, we did not observe direct interaction between purified recombinant SAMHD1 and GST-tagged cyclin A2/CDK1 complex (data not shown), which could be due to the GST tag obstructing interaction between the proteins. This observation raises the possibility that direct cyclin A2 binding may not be required for in vitro phosphorylation. Having a localized concentration of substrate and CDK in vitro could explain why the RL/AA and LF/AA mutants were phosphorylated in the absence of cyclin A2 binding. Furthermore, knockdown of cyclin A2 and CDK1/2 in cells is sufficient to reduce SAMHD1 phosphorylation in cells, indicating the requirement of these proteins to mediate phosphorylation of SAMHD1 in cells (18) and therefore demonstrating the importance of both mutants in mediating cyclin A2 interaction in cells, compared to in vitro data.
Cyclins can modulate CDK substrate recognition and activity in cells through spatial and cell cycle-dependent constraints (54, 55). Co-IP experiments performed with cells demonstrate that the RL/AA mutant is deficient in binding to cyclin A2, although the interaction is not completely abrogated compared to the LF/AA mutant. As both the RL and LF residues adhere to conserved canonical cyclin-binding motifs discussed previously (42), this suggests that SAMHD1 could contain multiple cyclin-binding motifs with differing functions and/or redundancies of function. Indeed, SAMHD1 contains four phosphorylation sites (S6, T21, S33, and T592), whose functions are not fully understood (20, 21). Our results suggest two possibilities for the diminished interaction between RL/AA SAMHD1 and cyclin A2. The reduced interaction could be attributed to mislocalization of the RL/AA mutant, limiting access to cyclin A2/CDK complexes in cells, or mutation of the RXL motif in SAMHD1 was sufficient to partially abrogate the interaction with the hydrophobic patch of cyclin A, which is a feature of cyclins binding to RXL motif-containing substrates (56–59), but was not sufficient to abolish interaction.
We investigated whether the RL/AA mutant was selectively targeted for spontaneous degradation during protein turnover, or when autophagy had been activated. Our data indicate that SAMHD1 is not spontaneously degraded through the major proteasomal, lysosomal, or autophagy pathways. Coupled with our observation that SAMHD1 is not clearly mono- or polyubiquitinated in the absence of Vpx, this provides further evidence that SAMHD1 is not targeted for proteasomal degradation in cycling cells. Furthermore, while we did not absolutely exclude SAMHD1 ubiquitination, the presence of a higher-molecular-weight ubiquitin-specific band (∼140 kDa) enriched in SAMHD1 IP from THP-1 cells could represent a SAMHD1 interacting protein. Or it could represent SAMHD1 multimonoubiquitination, which could serve as a molecular signal for other cellular responses, such as DNA damage or inflammation, but is not linked to proteasomal degradation (60). Contrary to our observations, a recent publication suggested that CD81 regulates SAMHD1 expression and that SAMHD1 protein expression can be enhanced 2- to 3-fold by MG132 treatment in HeLa cells (61). The small effects of MG132 treatment on SAMHD1 protein observed could be attributed to the low endogenous SAMHD1 expression levels in HeLa cells, whereas our system used overexpressed SAMHD1 in HEK293T cells or highly expressed endogenous SAMHD1 in THP-1 cells.
Interestingly, we observed rescue of SAMHD1 protein levels only in the presence of a combination of inhibitors targeting the proteasome and autophagy. Autophagy and the proteasome are closely linked systems (62), and inhibition of the proteasome has been shown to activate autophagy (63, 64). There are reports that some proteins are substrates of both pathways (65–67). Therefore, it is possible that SAMHD1 could experience some degradation through both the proteasome and autophagy as it is regulated during the cell cycle. This could explain why we were unable to rescue protein with single inhibitor treatments. In addition, experiments presented here were performed in cells actively undergoing de novo protein synthesis, unlike our half-life experiments using cycloheximide treatment of cells (42).
Of note, autophagy can be activated under oxidative stress (68), and a recent study showed that SAMHD1 is regulated through oxidation of cysteine residues and relocates to areas outside the nucleus in the presence of growth factors (36). This could suggest that only under oxidative stress, or cellular stress, could SAMHD1 degradation be initiated. Nuclear protein lamin B1 can be degraded by autophagy only when cells experience oncogenic insult (69). In addition, ubiquitin-independent mechanisms of proteasomal degradation do exist (70), including that of oxidized proteins (71). It is possible that our combination treatment to inhibit multiple degradation pathways caused cellular stress, explaining why we observed rescue of SAMHD1 protein levels.
In summary, our results demonstrate that the RXL motif of SAMHD1 is critical for formation and structural integrity of the SAMHD1 tetramer, dNTPase activity, and HIV-1 restriction. These findings highlight the necessity of SAMHD1 tetramerization initiated by allosteric binding of dNTPs in cells in order to maintain an HIV-1 restriction function. In addition, our data imply that the RXL motif is important to ensure efficient phosphorylation and regulation of SAMHD1 functions in cells.
MATERIALS AND METHODS
Plasmids.
pLenti-puro constructs expressing WT hemagglutinin (HA)-tagged human SAMHD1 and the empty vector control were provided by Nathaniel Landau (New York University). The pLenti-puro constructs expressing HA-tagged RL/AA, F/W, or LF/AA mutant SAMHD1 were generated using a QuikChange multisite-directed mutagenesis kit (Agilent Technologies) and described previously (18). The pCG-myc-Vpx plasmid expressing myc-tagged Vpx from HIV-1ROD (13), and pCG vector were provided by Jacek Skowronski (Case Western Reserve University).
In vitro size exclusion chromatography.
N-terminally 6×His-tagged WT SAMHD1 or mutant constructs were expressed in Escherichia coli and purified as previously described (29). Purified protein samples (2 mg/ml; 50 μl) with or without 500 μM GTP and 2 mM dATP were applied to a Superdex 200 Increase 5/150 GL column (GE Healthcare) preequilibrated in reaction buffer (50 mM Tris-HCl [pH 8.0], 150 mM NaCl, 5 mM MgCl2, and 0.5 mM Tris(2-carboxyethyl)phosphine [TCEP]). The UV absorbance at 280 nm was measured as the protein sample was eluted from the column.
In vitro malachite green assay.
To compare the dNTPase activities of different SAMHD1 constructs, a colorimetric malachite green assay was performed, as previously described (72). All reactions were performed at 25°C in the reaction buffer described above. Each reaction, performed with 40 μl containing 10 μM pyrophosphatase, 0.5 μM SAMHD1, and 125 μM substrate or allosteric activator, was quenched with 40 μl of 20 mM EDTA after time points from 0 to 14 min. Then, 20 μl of malachite green reagent was added to the solution and developed for 15 min before the absorbance at 650 nm was measured.
In vitro kinase assay.
Kinase assays were performed for 2 h at 30°C as described previously (27). Recombinant full-length WT SAMHD1 and RL/AA, LF/AA, and T592A mutant proteins were purified as described previously (29, 30). Assays were performed using a titration of cyclin A2/CDK1 complex according to the manufacturer's instructions (Sigma-Aldrich; catalog number C0244). Purified recombinant SAMHD1 protein (25 ng) was used in the reaction. T592 phosphorylation of SAMHD1 was detected using the T592-phospho-specific antibody described previously (19). GST-cyclin A2 and GST-CDK1 were detected using a GST antibody (Santa Cruz; sc-459).
Cell culture.
THP-1 cells were cultured in RPMI 1640 medium as described previously (14). HEK293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin. Stable U937 cells were cultured in RPMI 1640 as described previously (7). U937 cells were differentiated using phorbol 12-myristate 13-acetate (PMA; 100 ng/ml) for 24 h, PMA was removed and cells were cultured for a further 24 h. All cell lines were maintained at 37°C and 5% CO2 and tested negative for mycoplasma contamination using a universal mycoplasma detection kit (ATCC; number 30-101-2K).
Generation of stable U937 cell lines.
Lentiviral stocks for overexpression of HA-tagged SAMHD1 mutants were generated by transfection of HEK293T cells with pLenti-puro vector or SAMHD1-expressing plasmids, pMDL packaging plasmid, pRSV-rev, and pVSV-G. Forty-eight hours posttransfection, lentiviral stocks were harvested, filtered, and concentrated through a sucrose cushion. Resuspended lentivirus was used to spinoculate U937 cells in the presence of Polybrene (8 μg/ml), after which cells were cultured in normal RPMI 1640 for 3 days prior to selection with 1 μg/ml of puromycin as described previously (7). Overexpression of SAMHD1 in PMA-differentiated cells was confirmed by SDS-PAGE and immunoblotting analysis.
Measurement of intracellular dNTP levels.
PMA-differentiated U937 stable cell lines were harvested and processed for dNTP measurements using the single-nucleotide incorporation assay as described previously (73).
Lysosome and proteasome inhibitor treatments.
Transfected HEK293T cells expressing WT or RL/AA SAMHD1 or THP-1 cells were treated with specific inhibitors as described in the figure legends. Briefly, cells were treated with proteasomal or lysosomal inhibitors for the times and at the concentrations indicated in the figures. The inhibitors included MG132 (Santa Cruz; sc-201270), epoxomicin (Enzo Life Sciences, Inc.; BML-PI127), 3-MA (tlrl-3ma) and bafilomicin A1 (tlrl-baf1) (Invivogen), and chloroquine (C6628) and leupeptin (L2023) (Sigma-Aldrich). All samples were made up to the same volume of dimethyl sulfoxide (DMSO), and DMSO alone was used as a negative control.
Protein quantification, immunoblotting, and antibodies.
Cells were collected, washed with phosphate-buffered saline (PBS), and lysed using 1× cell lysis buffer (Cell Signaling; number 9803) containing protease inhibitor cocktail (Sigma-Aldrich; P8340). Protein lysate was quantified via a Pierce bicinchoninic acid (BCA) protein assay kit (Pierce), followed by immunoblotting as described previously (42). Immunoblotting was performed using antibodies against the following proteins (catalog numbers are in parentheses): HA-11 (901501) from Biolegend and SAMHD1 (8007) and Phos-T592 (8005) from ProSci. Ubiquitin (3936), LC3A/B (12741), and LAMP-1 (9091) were from Cell Signaling Technology, and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (AHP1628) was from Bio-Rad. HIV-2 Vpx monoclonal antibody (clone 6D2.6) (74) was obtained from the NIH AIDS Reagent Program (catalog number 2710).
Densitometry quantification of immunoblotting.
Densitometry of immunoblots was performed on low-exposure unaltered images using ImageJ software. All densitometry values were normalized to their respective GAPDH values.
Co-IP.
HEK293T cells were transfected with an empty vector or plasmids encoding HA-tagged WT or RL/AA SAMHD1. At 16 h posttransfection, cells were treated with MG132, collected and lysed for HA-IP, and analyzed by immunoblotting as described previously (18). For THP-1 cells, cells were treated as described in the figure legend and lysates generated. Dynabeads (protein G; Thermo Fisher Scientific) were incubated with 5 μg of SAMHD1 antibody (Abcam; 67820) or mouse IgG control, according to the manufacturer's directions. After binding of antibody to the beads, cell lysates were incubated with the Dynabeads at 4°C for 2 h. Beads were then washed with 3 times with PBS with 0.1% Tween. Bound proteins were eluted from beads by boiling in 1× SDS sample buffer, and the supernatants were used for immunoblotting.
HIV-1 infection assay.
To quantitate HIV-1 infection in SAMHD1-expressing stable U937 cell lines, cells were differentiated with PMA and infected with single-cycle NL4-3ΔenvE-R+ or NL4-3-deltaE-EGFP (43). Cells were infected with vesicular stomatitis virus protein G (VSV-G)-pseudotyped HIV-1 at a multiplicity of infection of 0.5 for 2 h before removal of virus. Cells were then cultured for 48 h prior to collection of cell lysates for luciferase assay (Promega) or flow cytometry. All luciferase assays were normalized to protein content. For infected cells processed for downstream quantitative PCR (qPCR) analysis of late RT products and mRNA, virus stocks were DNase I treated according to the manufacturer's instructions as described previously (75).
Quantitative PCR.
To measure HIV-1 late reverse transcription products in infected cells, 16 h postinfection, total cellular DNA was isolated using a DNeasy blood and tissue kit (Qiagen). Cellular DNA (50 ng) was used for iTaq Universal SYBR green Supermix-based qPCR (Bio-Rad). HIV-1NLAD8 proviral DNA plasmid was used as a standard (106 to 102 copies). Unspliced GAPDH primers were used for normalization of samples as described previously (76). For quantification of Gag and luciferase mRNA in infected cells, total cellular RNA was extracted using the RNeasy minikit (Qiagen). Equal amounts of total RNA from each sample were used as a template for first-strand cDNA synthesis using the Superscript III first-strand synthesis system and oligo(dT) primers (Thermo Fisher Scientific). SYBR green PCR analysis was performed using HIV-1 gag or firefly luciferase gene-specific primers. Quantification of spliced GAPDH mRNA was used for normalization as described previously (76). Calculation of relative gene expression was performed using the threshold cycle (2−ΔΔCT) method as described previously (77).
Statistical analysis.
One-way ANOVA with Dunnett's multiple-comparison test was performed using GraphPad Prism 5.0. Statistical significance was defined at a P value of <0.05.
ACKNOWLEDGMENTS
This work was supported by a grant (AI104483) from the National Institutes of Health (NIH) to L.W. L.W. is also supported in part by NIH grants (AI120209 and GM128212). C.S. and B.K. are supported by NIH grants GM104198 and AI049781. O.B. was supported by NIH T32 grant GM007223 and a National Science Foundation Graduate Research Fellowship, and K.K. was supported by NIH T32 grant GM008283.
The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
We thank Nathaniel Landau and Jacek Skowronski for plasmids and the Wu lab members for valuable discussions. The following were obtained through the NIH AIDS Reagent Program, Division of AIDS, NIAID, NIH: HIV-2 Vpx monoclonal antibody (6D2.6) from John C. Kappes, and pNL4-3-deltaE-EGFP (catalog number 11100) from Haili Zhang, Yan Zhou, and Robert Siliciano.
L.W. conceived the study and designed experiments with C.S.G. C.S.G., S.H.K., V.V.M., O.B., K.K., and C.S. performed experiments and analyzed data with L.W. B.K. and Y.X. contributed to experimental design and data analysis. C.S.G. and L.W. wrote the manuscript.
REFERENCES
- 1.Baldauf HM, Pan X, Erikson E, Schmidt S, Daddacha W, Burggraf M, Schenkova K, Ambiel I, Wabnitz G, Gramberg T, Panitz S, Flory E, Landau NR, Sertel S, Rutsch F, Lasitschka F, Kim B, Konig R, Fackler OT, Keppler OT. 2012. SAMHD1 restricts HIV-1 infection in resting CD4(+) T cells. Nat Med 18:1682–1687. doi: 10.1038/nm.2964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gramberg T, Kahle T, Bloch N, Wittmann S, Mullers E, Daddacha W, Hofmann H, Kim B, Lindemann D, Landau NR. 2013. Restriction of diverse retroviruses by SAMHD1. Retrovirology 10:26. doi: 10.1186/1742-4690-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hollenbaugh JA, Gee P, Baker J, Daly MB, Amie SM, Tate J, Kasai N, Kanemura Y, Kim DH, Ward BM, Koyanagi Y, Kim B. 2013. Host factor SAMHD1 restricts DNA viruses in non-dividing myeloid cells. PLoS Pathog 9:e1003481. doi: 10.1371/journal.ppat.1003481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lahouassa H, Daddacha W, Hofmann H, Ayinde D, Logue EC, Dragin L, Bloch N, Maudet C, Bertrand M, Gramberg T, Pancino G, Priet S, Canard B, Laguette N, Benkirane M, Transy C, Landau NR, Kim B, Margottin-Goguet F. 2012. SAMHD1 restricts the replication of human immunodeficiency virus type 1 by depleting the intracellular pool of deoxynucleoside triphosphates. Nat Immunol 13:223–228. doi: 10.1038/ni.2236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Antonucci JM, St Gelais C, de Silva S, Yount JS, Tang C, Ji X, Shepard C, Xiong Y, Kim B, Wu L. 2016. SAMHD1-mediated HIV-1 restriction in cells does not involve ribonuclease activity. Nat Med 22:1072–1074. doi: 10.1038/nm.4163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kohnken R, Kodigepalli KM, Wu L. 2015. Regulation of deoxynucleotide metabolism in cancer: novel mechanisms and therapeutic implications. Mol Cancer 14:176. doi: 10.1186/s12943-015-0446-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Silva S, Hoy H, Hake TS, Wong HK, Porcu P, Wu L. 2013. Promoter methylation regulates SAMHD1 gene expression in human CD4+ T cells. J Biol Chem 288:9284–9292. doi: 10.1074/jbc.M112.447201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.de Silva S, Wang F, Hake TS, Porcu P, Wong HK, Wu L. 2014. Downregulation of SAMHD1 expression correlates with promoter DNA methylation in Sezary syndrome patients. J Invest Dermatol 134:562–565. doi: 10.1038/jid.2013.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jin C, Peng X, Liu F, Cheng L, Lu X, Yao H, Wu H, Wu N. 2014. MicroRNA-181 expression regulates specific post-transcriptional level of SAMHD1 expression in vitro. Biochem Biophys Res Commun 452:760–767. doi: 10.1016/j.bbrc.2014.08.151. [DOI] [PubMed] [Google Scholar]
- 12.Kohnken R, Kodigepalli KM, Mishra A, Porcu P, Wu L. 2017. MicroRNA-181 contributes to downregulation of SAMHD1 expression in CD4+ T-cells derived from Sezary syndrome patients. Leuk Res 52:58–66. doi: 10.1016/j.leukres.2016.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.St Gelais C, de Silva S, Amie SM, Coleman CM, Hoy H, Hollenbaugh JA, Kim B, Wu L. 2012. SAMHD1 restricts HIV-1 infection in dendritic cells (DCs) by dNTP depletion, but its expression in DCs and primary CD4+ T-lymphocytes cannot be upregulated by interferons. Retrovirology 9:105. doi: 10.1186/1742-4690-9-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bonifati S, Daly MB, St Gelais C, Kim SH, Hollenbaugh JA, Shepard C, Kennedy EM, Kim DH, Schinazi RF, Kim B, Wu L. 2016. SAMHD1 controls cell cycle status, apoptosis and HIV-1 infection in monocytic THP-1 cells. Virology 495:92–100. doi: 10.1016/j.virol.2016.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodigepalli KM, Li M, Liu SL, Wu L. 2017. Exogenous expression of SAMHD1 inhibits proliferation and induces apoptosis in cutaneous T-cell lymphoma-derived HuT78 cells. Cell Cycle 16:179–188. doi: 10.1080/15384101.2016.1261226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Badia R, Pujantell M, Torres-Torronteras J, Menendez-Arias L, Marti R, Ruzo A, Pauls E, Clotet B, Ballana E, Este JA, Riveira-Munoz E. 2017. SAMHD1 is active in cycling cells permissive to HIV-1 infection. Antiviral Res 142:123–135. doi: 10.1016/j.antiviral.2017.03.019. [DOI] [PubMed] [Google Scholar]
- 17.Cribier A, Descours B, Valadao AL, Laguette N, Benkirane M. 2013. Phosphorylation of SAMHD1 by cyclin A2/CDK1 regulates its restriction activity toward HIV-1. Cell Rep 3:1036–1043. doi: 10.1016/j.celrep.2013.03.017. [DOI] [PubMed] [Google Scholar]
- 18.St Gelais C, de Silva S, Hach JC, White TE, Diaz-Griffero F, Yount JS, Wu L. 2014. Identification of cellular proteins interacting with the retroviral restriction factor SAMHD1. J Virol 88:5834–5844. doi: 10.1128/JVI.00155-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang F, St Gelais C, de Silva S, Zhang H, Geng Y, Shepard C, Kim B, Yount JS, Wu L. 2016. Phosphorylation of mouse SAMHD1 regulates its restriction of human immunodeficiency virus type 1 infection, but not murine leukemia virus infection. Virology 487:273–284. doi: 10.1016/j.virol.2015.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Welbourn S, Dutta SM, Semmes OJ, Strebel K. 2013. Restriction of virus infection but not catalytic dNTPase activity is regulated by phosphorylation of SAMHD1. J Virol 87:11516–11524. doi: 10.1128/JVI.01642-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.White TE, Brandariz-Nunez A, Valle-Casuso JC, Amie S, Nguyen LA, Kim B, Tuzova M, Diaz-Griffero F. 2013. The retroviral restriction ability of SAMHD1, but not its deoxynucleotide triphosphohydrolase activity, is regulated by phosphorylation. Cell Host Microbe 13:441–451. doi: 10.1016/j.chom.2013.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pauls E, Ruiz A, Badia R, Permanyer M, Gubern A, Riveira-Munoz E, Torres-Torronteras J, Alvarez M, Mothe B, Brander C, Crespo M, Menendez-Arias L, Clotet B, Keppler OT, Marti R, Posas F, Ballana E, Este JA. 2014. Cell cycle control and HIV-1 susceptibility are linked by CDK6-dependent CDK2 phosphorylation of SAMHD1 in myeloid and lymphoid cells. J Immunol 193:1988–1997. doi: 10.4049/jimmunol.1400873. [DOI] [PubMed] [Google Scholar]
- 23.Arnold LH, Groom HC, Kunzelmann S, Schwefel D, Caswell SJ, Ordonez P, Mann MC, Rueschenbaum S, Goldstone DC, Pennell S, Howell SA, Stoye JP, Webb M, Taylor IA, Bishop KN. 2015. Phospho-dependent regulation of SAMHD1 oligomerisation couples catalysis and restriction. PLoS Pathog 11:e1005194. doi: 10.1371/journal.ppat.1005194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tang C, Ji X, Wu L, Xiong Y. 2015. Impaired dNTPase activity of SAMHD1 by phosphomimetic mutation of Thr-592. J Biol Chem 290:26352–26359. doi: 10.1074/jbc.M115.677435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yan J, Kaur S, DeLucia M, Hao C, Mehrens J, Wang C, Golczak M, Palczewski K, Gronenborn AM, Ahn J, Skowronski J. 2013. Tetramerization of SAMHD1 is required for biological activity and inhibition of HIV infection. J Biol Chem 288:10406–10417. doi: 10.1074/jbc.M112.443796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Franzolin E, Pontarin G, Rampazzo C, Miazzi C, Ferraro P, Palumbo E, Reichard P, Bianchi V. 2013. The deoxynucleotide triphosphohydrolase SAMHD1 is a major regulator of DNA precursor pools in mammalian cells. Proc Natl Acad Sci U S A 110:14272–14277. doi: 10.1073/pnas.1312033110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kretschmer S, Wolf C, Konig N, Staroske W, Guck J, Hausler M, Luksch H, Nguyen LA, Kim B, Alexopoulou D, Dahl A, Rapp A, Cardoso MC, Shevchenko A, Lee-Kirsch MA. 2015. SAMHD1 prevents autoimmunity by maintaining genome stability. Ann Rheum Dis 74:e17. doi: 10.1136/annrheumdis-2013-204845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yan J, Hao C, DeLucia M, Swanson S, Florens L, Washburn MP, Ahn J, Skowronski J. 2015. Cyclin A2-cyclin-dependent kinase regulates SAMHD1 protein phosphohydrolase domain. J Biol Chem 290:13279–13292. doi: 10.1074/jbc.M115.646588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ji X, Wu Y, Yan J, Mehrens J, Yang H, DeLucia M, Hao C, Gronenborn AM, Skowronski J, Ahn J, Xiong Y. 2013. Mechanism of allosteric activation of SAMHD1 by dGTP. Nat Struct Mol Biol 20:1304–1309. doi: 10.1038/nsmb.2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ji X, Tang C, Zhao Q, Wang W, Xiong Y. 2014. Structural basis of cellular dNTP regulation by SAMHD1. Proc Natl Acad Sci U S A 111:E4305–E4314. doi: 10.1073/pnas.1412289111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amie SM, Bambara RA, Kim B. 2013. GTP is the primary activator of the anti-HIV restriction factor SAMHD1. J Biol Chem 288:25001–25006. doi: 10.1074/jbc.C113.493619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hansen EC, Seamon KJ, Cravens SL, Stivers JT. 2014. GTP activator and dNTP substrates of HIV-1 restriction factor SAMHD1 generate a long-lived activated state. Proc Natl Acad Sci U S A 111:E1843–E1851. doi: 10.1073/pnas.1401706111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koharudin LM, Wu Y, DeLucia M, Mehrens J, Gronenborn AM, Ahn J. 2014. Structural basis of allosteric activation of sterile alpha motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J Biol Chem 289:32617–32627. doi: 10.1074/jbc.M114.591958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhu C, Gao W, Zhao K, Qin X, Zhang Y, Peng X, Zhang L, Dong Y, Zhang W, Li P, Wei W, Gong Y, Yu XF. 2013. Structural insight into dGTP-dependent activation of tetrameric SAMHD1 deoxynucleoside triphosphate triphosphohydrolase. Nat Commun 4:2722. doi: 10.1038/ncomms3722. [DOI] [PubMed] [Google Scholar]
- 35.Zhu CF, Wei W, Peng X, Dong YH, Gong Y, Yu XF. 2015. The mechanism of substrate-controlled allosteric regulation of SAMHD1 activated by GTP. Acta Crystallogr D Biol Crystallogr 71:516–524. doi: 10.1107/S1399004714027527. [DOI] [PubMed] [Google Scholar]
- 36.Mauney CH, Rogers LC, Harris RS, Daniel LW, Devarie-Baez NO, Wu H, Furdui CM, Poole LB, Perrino FW, Hollis T. 18 April 2017. The SAMHD1 dNTP triphosphohydrolase is controlled by a redox switch. Antioxid Redox Signal doi: 10.1089/ars.2016.6888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fregoso OI, Ahn J, Wang C, Mehrens J, Skowronski J, Emerman M. 2013. Evolutionary toggling of Vpx/Vpr specificity results in divergent recognition of the restriction factor SAMHD1. PLoS Pathog 9:e1003496. doi: 10.1371/journal.ppat.1003496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schwefel D, Groom HC, Boucherit VC, Christodoulou E, Walker PA, Stoye JP, Bishop KN, Taylor IA. 2014. Structural basis of lentiviral subversion of a cellular protein degradation pathway. Nature 505:234–238. doi: 10.1038/nature12815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schwefel D, Boucherit VC, Christodoulou E, Walker PA, Stoye JP, Bishop KN, Taylor IA. 2015. Molecular determinants for recognition of divergent SAMHD1 proteins by the lentiviral accessory protein Vpx. Cell Host Microbe 17:489–499. doi: 10.1016/j.chom.2015.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ahn J, Hao C, Yan J, DeLucia M, Mehrens J, Wang C, Gronenborn AM, Skowronski J. 2012. HIV/simian immunodeficiency virus (SIV) accessory virulence factor Vpx loads the host cell restriction factor SAMHD1 onto the E3 ubiquitin ligase complex CRL4DCAF1. J Biol Chem 287:12550–12558. doi: 10.1074/jbc.M112.340711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kyei GB, Cheng X, Ramani R, Ratner L. 2015. Cyclin L2 is a critical HIV dependency factor in macrophages that controls SAMHD1 abundance. Cell Host Microbe 17:98–106. doi: 10.1016/j.chom.2014.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.St Gelais C, Kim SH, Ding L, Yount JS, Ivanov D, Spearman P, Wu L. 2016. A putative cyclin-binding motif in human SAMHD1 contributes to protein phosphorylation, localization, and stability. J Biol Chem 291:26332–26342. doi: 10.1074/jbc.M116.753947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H, Zhou Y, Alcock C, Kiefer T, Monie D, Siliciano J, Li Q, Pham P, Cofrancesco J, Persaud D, Siliciano RF. 2004. Novel single-cell-level phenotypic assay for residual drug susceptibility and reduced replication capacity of drug-resistant human immunodeficiency virus type 1. J Virol 78:1718–1729. doi: 10.1128/JVI.78.4.1718-1729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Meng L, Mohan R, Kwok BH, Elofsson M, Sin N, Crews CM. 1999. Epoxomicin, a potent and selective proteasome inhibitor, exhibits in vivo antiinflammatory activity. Proc Natl Acad Sci U S A 96:10403–10408. doi: 10.1073/pnas.96.18.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chesarino NM, McMichael TM, Yount JS. 2015. E3 ubiquitin ligase NEDD4 promotes influenza virus infection by decreasing levels of the antiviral protein IFITM3. PLoS Pathog 11:e1005095. doi: 10.1371/journal.ppat.1005095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen PM, Gombart ZJ, Chen JW. 2011. Chloroquine treatment of ARPE-19 cells leads to lysosome dilation and intracellular lipid accumulation: possible implications of lysosomal dysfunction in macular degeneration. Cell Biosci 1:10. doi: 10.1186/2045-3701-1-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Wu L. 2013. Cellular and biochemical mechanisms of the retroviral restriction factor SAMHD1. ISRN Biochem 2013:728392. doi: 10.1155/2013/728392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen M, Cortay JC, Logan IR, Sapountzi V, Robson CN, Gerlier D. 2005. Inhibition of ubiquitination and stabilization of human ubiquitin E3 ligase PIRH2 by measles virus phosphoprotein. J Virol 79:11824–11836. doi: 10.1128/JVI.79.18.11824-11836.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mizushima N, Yoshimori T, Levine B. 2010. Methods in mammalian autophagy research. Cell 140:313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu YT, Tan HL, Shui G, Bauvy C, Huang Q, Wenk MR, Ong CN, Codogno P, Shen HM. 2010. Dual role of 3-methyladenine in modulation of autophagy via different temporal patterns of inhibition on class I and III phosphoinositide 3-kinase. J Biol Chem 285:10850–10861. doi: 10.1074/jbc.M109.080796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Brandariz-Nuñez A, Valle-Casuso JC, White TE, Nguyen L, Bhattacharya A, Wang Z, Demeler B, Amie S, Knowlton C, Kim B, Ivanov DN, Diaz-Griffero F. 2013. Contribution of oligomerization to the anti-HIV-1 properties of SAMHD1. Retrovirology 10:131. doi: 10.1186/1742-4690-10-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bhattacharya A, Wang Z, White T, Buffone C, Nguyen LA, Shepard CN, Kim B, Demeler B, Diaz-Griffero F, Ivanov DN. 2016. Effects of T592 phosphomimetic mutations on tetramer stability and dNTPase activity of SAMHD1 can not explain the retroviral restriction defect. Sci Rep 6:31353. doi: 10.1038/srep31353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bloch N, Glasker S, Sitaram P, Hofmann H, Shepard CN, Schultz ML, Kim B, Landau NR. 2017. A highly active isoform of lentivirus restriction factor SAMHD1 in mouse. J Biol Chem 292:1068–1080. doi: 10.1074/jbc.M116.743740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Huet X, Rech J, Plet A, Vie A, Blanchard JM. 1996. Cyclin A expression is under negative transcriptional control during the cell cycle. Mol Cell Biol 16:3789–3798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maridor G, Gallant P, Golsteyn R, Nigg EA. 1993. Nuclear localization of vertebrate cyclin A correlates with its ability to form complexes with cdk catalytic subunits. J Cell Sci 106(Part 2):535–544. [DOI] [PubMed] [Google Scholar]
- 56.Adams PD, Sellers WR, Sharma SK, Wu AD, Nalin CM, Kaelin WG Jr. 1996. Identification of a cyclin-cdk2 recognition motif present in substrates and p21-like cyclin-dependent kinase inhibitors. Mol Cell Biol 16:6623–6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Adams PD, Li X, Sellers WR, Baker KB, Leng X, Harper JW, Taya Y, Kaelin WG Jr. 1999. Retinoblastoma protein contains a C-terminal motif that targets it for phosphorylation by cyclin-cdk complexes. Mol Cell Biol 19:1068–1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Takeda DY, Wohlschlegel JA, Dutta A. 2001. A bipartite substrate recognition motif for cyclin-dependent kinases. J Biol Chem 276:1993–1997. doi: 10.1074/jbc.M005719200. [DOI] [PubMed] [Google Scholar]
- 59.Stevenson-Lindert LM, Fowler P, Lew J. 2003. Substrate specificity of CDK2-cyclin A. What is optimal? J Biol Chem 278:50956–50960. doi: 10.1074/jbc.M306546200. [DOI] [PubMed] [Google Scholar]
- 60.Pickart CM, Fushman D. 2004. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol 8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 61.Rocha-Perugini V, Suarez H, Alvarez S, Lopez-Martin S, Lenzi GM, Vences-Catalan F, Levy S, Kim B, Munoz-Fernandez MA, Sanchez-Madrid F, Yanez-Mo M. 4 September 2017. CD81 association with SAMHD1 enhances HIV-1 reverse transcription by increasing dNTP levels. Nat Microbiol doi: 10.1038/s41564-017-0019-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lilienbaum A. 2013. Relationship between the proteasomal system and autophagy. Int J Biochem Mol Biol 4:1–26. [PMC free article] [PubMed] [Google Scholar]
- 63.Ding WX, Ni HM, Gao W, Yoshimori T, Stolz DB, Ron D, Yin XM. 2007. Linking of autophagy to ubiquitin-proteasome system is important for the regulation of endoplasmic reticulum stress and cell viability. Am J Pathol 171:513–524. doi: 10.2353/ajpath.2007.070188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Pandey UB, Nie Z, Batlevi Y, McCray BA, Ritson GP, Nedelsky NB, Schwartz SL, DiProspero NA, Knight MA, Schuldiner O, Padmanabhan R, Hild M, Berry DL, Garza D, Hubbert CC, Yao TP, Baehrecke EH, Taylor JP. 2007. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature 447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 65.Ding WX, Yin XM. 2008. Sorting, recognition and activation of the misfolded protein degradation pathways through macroautophagy and the proteasome. Autophagy 4:141–150. doi: 10.4161/auto.5190. [DOI] [PubMed] [Google Scholar]
- 66.Webb JL, Ravikumar B, Atkins J, Skepper JN, Rubinsztein DC. 2003. Alpha-synuclein is degraded by both autophagy and the proteasome. J Biol Chem 278:25009–25013. doi: 10.1074/jbc.M300227200. [DOI] [PubMed] [Google Scholar]
- 67.Cuervo AM, Stefanis L, Fredenburg R, Lansbury PT, Sulzer D. 2004. Impaired degradation of mutant alpha-synuclein by chaperone-mediated autophagy. Science 305:1292–1295. doi: 10.1126/science.1101738. [DOI] [PubMed] [Google Scholar]
- 68.Kiffin R, Christian C, Knecht E, Cuervo AM. 2004. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell 15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dou Z, Xu C, Donahue G, Shimi T, Pan JA, Zhu J, Ivanov A, Capell BC, Drake AM, Shah PP, Catanzaro JM, Ricketts MD, Lamark T, Adam SA, Marmorstein R, Zong WX, Johansen T, Goldman RD, Adams PD, Berger SL. 2015. Autophagy mediates degradation of nuclear lamina. Nature 527:105–109. doi: 10.1038/nature15548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Murakami Y, Matsufuji S, Kameji T, Hayashi S, Igarashi K, Tamura T, Tanaka K, Ichihara A. 1992. Ornithine decarboxylase is degraded by the 26S proteasome without ubiquitination. Nature 360:597–599. [DOI] [PubMed] [Google Scholar]
- 71.Grune T, Reinheckel T, Davies KJ. 1997. Degradation of oxidized proteins in mammalian cells. FASEB J 11:526–534. [PubMed] [Google Scholar]
- 72.Seamon KJ, Stivers JT. 2015. A high-throughput enzyme-coupled assay for SAMHD1 dNTPase. J Biomol Screen 20:801–809. doi: 10.1177/1087057115575150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Diamond TL, Roshal M, Jamburuthugoda VK, Reynolds HM, Merriam AR, Lee KY, Balakrishnan M, Bambara RA, Planelles V, Dewhurst S, Kim B. 2004. Macrophage tropism of HIV-1 depends on efficient cellular dNTP utilization by reverse transcriptase. J Biol Chem 279:51545–51553. doi: 10.1074/jbc.M408573200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kappes JC, Parkin JS, Conway JA, Kim J, Brouillette CG, Shaw GM, Hahn BH. 1993. Intracellular transport and virion incorporation of vpx requires interaction with other virus type-specific components. Virology 193:222–233. [DOI] [PubMed] [Google Scholar]
- 75.St Gelais C, Roger J, Wu L. 2015. Non-POU domain-containing octamer-binding protein negatively regulates HIV-1 infection in CD4(+) T cells. AIDS Res Hum Retroviruses 31:806–816. doi: 10.1089/aid.2014.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dong C, Janas AM, Wang JH, Olson WJ, Wu L. 2007. Characterization of human immunodeficiency virus type 1 replication in immature and mature dendritic cells reveals dissociable cis- and trans-infection. J Virol 81:11352–11362. doi: 10.1128/JVI.01081-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Livak KJ, Schmittgen TD. 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) method. Methods 25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]