ABSTRACT
Hepatitis E virus (HEV) is a clinically important positive-sense RNA virus. The ORF1 of HEV encodes a nonstructural polyprotein of 1,693 amino acids. It is not clear whether the ORF1 polyprotein (pORF1) is processed into distinct enzymatic domains. Many researchers have attempted to understand the mechanisms of pORF1 processing. However, these studies gave various results and could never convincingly establish the mechanism of pORF1 processing. In this study, we demonstrated the possible role of thrombin and factor Xa in pORF1 processing. We observed that the HEV pORF1 polyprotein bears conserved cleavage sites of thrombin and factor Xa. Using a reverse genetics approach, we demonstrated that an HEV replicon having mutations in the cleavage sites of either thrombin or factor Xa could not replicate efficiently in cell culture. Further, we demonstrated in vitro processing when we incubated recombinant pORF1 fragments with thrombin, and we observed the processing of pORF1 polyprotein. The treatment of a liver cell line with a serine protease inhibitor as well as small interfering RNA (siRNA) knockdown of thrombin and factor Xa resulted in significant reduction in the replication of HEV. Thrombin and factor Xa have been well studied for their roles in blood clotting. Both of these proteins are believed to be present in the active form in the blood plasma. Interestingly, in this report, we demonstrated the presence of biologically active thrombin and factor Xa in a liver cell line. The results suggest that factor Xa and thrombin are essential for the replication of HEV and may be involved in pORF1 polyprotein processing of HEV.
IMPORTANCE Hepatitis E virus (HEV) causes a liver disorder called hepatitis in humans, which is mostly an acute and self-limiting infection in adults. A high mortality rate of about 30% is observed in HEV-infected pregnant women in developing countries. There is no convincing opinion about HEV ORF1 polyprotein processing owing to the variability of study results obtained so far. HEV pORF1 has cleavage sites for two host cellular serine proteases, thrombin and factor Xa, that are conserved among HEV genotypes. For the first time, this study demonstrated that thrombin and factor Xa cleavage sites on HEV pORF1 are obligatory for HEV replication. Intracellular biochemical activities of the said serine proteases are also essential for efficient HEV replication in cell culture and must be involved in pORF1 processing. This study sheds light on the presence and roles of clotting factors with respect to virus replication in the cells.
KEYWORDS: positive-stranded RNA virus, hepatitis E virus, polyprotein processing, thrombin, factor Xa
INTRODUCTION
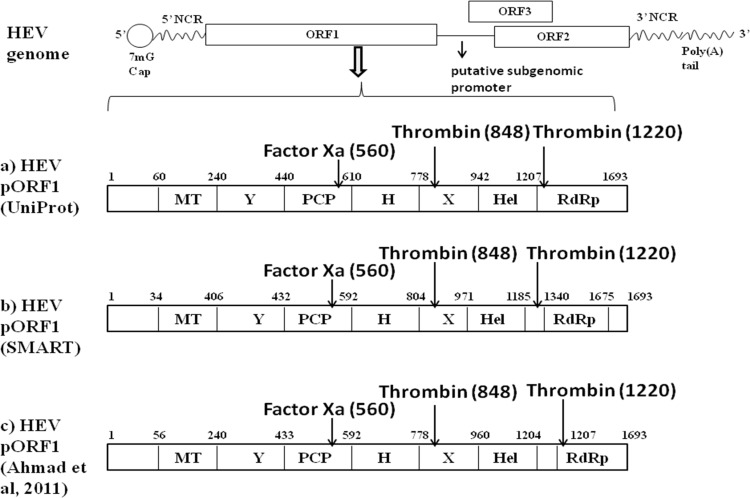
Hepatitis E virus (HEV) is a nonenveloped, single-stranded RNA virus, which is a member of the genus Hepevirus of the family Hepeviridae (1). It causes an enterically transmitted liver disorder called hepatitis in humans, which is mostly an acute and self-limiting infection in adults; however, chronic cases of HEV infection have also been reported (2). In human patients, extrahepatic manifestations such as neurological disorders and kidney injury have been observed (3). In young adults, the HEV case fatality rate is 0.5 to 3%; however, a higher mortality rate of up to 30% was observed in infected pregnant women. HEV infects several domestic animals, which also act as reservoirs for zoonosis (4). In spite of being a medically important pathogen, HEV remains understudied due to the lack of an efficient cell culture system and small-animal model. Infectious replicon and permissive cell lines have been widely used in the field, resulting in a better understanding of the molecular mechanisms of HEV. HEV replication is established by transfection of in vitro-transcribed capped RNA of an HEV replicon into permissive cell lines such as HepG2, Huh7, and PLC/PRF5 (1, 2). These cell culture systems may not be a faithful model for some pathophysiological or immunological studies; however, these systems are suitable for studying viral transcription, translation, and virus-host interactions. The genome of HEV is a positive sense, single-stranded RNA. It is 7.2 kb long and has a 7-methylguanosine cap at the 5′ end and a poly(A) tail at the 3′ end (1, 5, 6). The genome is schematically represented in Fig. 1. It has three open reading frames (ORFs), namely, ORF1, ORF2, and ORF3 (7). An additional open reading frame, ORF4, has also been observed to be expressed in genotype 1 under endoplasmic reticulum stress conditions (8).
FIG 1.
Schematic representation of hepatitis E virus (HEV) genome. The HEV genome consists of positive-sense single-stranded RNA which encodes three ORFs. ORF1 codes for nonstructural proteins, ORF2 codes for a structural protein, and ORF3 codes for a multifunctional phosphoprotein. Numbers denote the predicted boundaries of the different regions of the Sar55 strain of genotype 1 hepatitis E virus (GenBank accession number AF444002.1). NCR, noncoding regions. (a) HEV pORF1 domains predicted by Uniprot (GenBank accession number P33424). (b) HEV pORF1 domains predicted using SMART (Simple Modular Architecture Research Tool). (c) HEV pORF1 domains according to Ahmad et al. (5). Putative cleavage sites for thrombin and factor Xa (positions of arginine) are indicated by arrows on the HEV pORF1.
ORF1 encodes nonstructural proteins (nsPs). It is the largest ORF and has 5,079 bases. ORF2 is 1,980 nucleotides long and codes for a major structural protein. ORF3 is the smallest ORF, consisting of only 345 nucleotides, and is believed to code for a multifunctional phosphoprotein (5, 9–12). ORF1 is directly translated from positive-sense genomic RNA while ORF2 and ORF3 are translated through a subgenomic RNA intermediate (13–15). ORF1 of the HEV genome codes for a 186-kDa polyprotein with several functional domains (5). Computer-assisted alignment of the domains in ORF1 showed that there are the following putative functional domains: (i) methyl transferase (MT), (ii) a Y domain, (iii) papain-like cysteine protease (PCP), (iv) a proline-rich hinge domain, (v) an X domain, (vii) RNA helicase, and (vii) RNA-dependent RNA polymerase (RdRp) (16). The functional activities of MT (17), PCP (18, 19), helicase (Hel) (20, 21), and RdRp (22) have been biochemically demonstrated.
In most of the positive-sense RNA viruses, polyprotein processing is carried out by virus-encoded proteases. In these viruses, polyproteins expressed in Escherichia coli, mammalian cells, or in vitro translation systems show cis and trans proteolytic processing. Processing of the HEV ORF1 polyprotein remains an unsolved question in the field of HEV biology. Many research groups attempted to understand mechanisms of ORF1 polyprotein processing using different expression systems (23–28). However, these studies gave various results, and the mechanism of HEV polyprotein processing has remained elusive.
HEV is closely related to rubella virus and Beet necrotic yellow vein virus (BNYVV), which is a plant virus (16). HEV also belongs to the alphavirus-like superfamily. Rubella virus protease has been shown to cleave the polyprotein encoding nonstructural proteins both in cis and in trans (29). For Sindbis virus, a classical alphavirus, a virally encoded papain-like protease has been reported to function in cis and in trans for the processing of polyprotein (30). Expression of HEV pORF1 in a baculoviral expression system demonstrated processing in several smaller fragments; however, this study has not correlated polyprotein processing and HEV replication (25). Attempts to study HEV ORF1 polyprotein processing using an HEV replicon system have been carried out. One of the studies observed processing only in the context of replication; in contrast, another group reported the lack of processing in this scenario (27, 28). These studies, however, lack confirmation by biochemical assay or a reverse genetics approach. Precise cleavage sites on the HEV pORF1 and the viral or cellular protease taking part in the processing could also not be concluded.
Based on available studies on HEV ORF1 polyprotein processing, it is suggested that polyprotein processing occurs only in the context of virus replication (28). This may be due to interactions of viral and cellular factors, which are required for HEV polyprotein processing. HEV protease may not be biologically active when it is part of the entire polyprotein, and it may require activation or processing by some cellular factors. Along with viral proteases, host proteases have also been proven to play crucial roles in the processing of viral proteins. The hepatitis C virus genome is translated into a single entire polyprotein consisting of structural and nonstructural protein domains. A host signal peptide protease (SPP) is involved in the separation of nonstructural and structural proteins by proteolytic cleavage (31, 32). SPP is an aspartic protease capable of carrying out intramembrane cleavage of proteins usually involved in cleaving the signal peptide from proteins entering the endoplasmic reticulum (31). For hepatitis A virus (HAV), a host protease called factor Xa has been shown to process procapsid protein to the mature capsid of the virus particle (33). Caspase-like proteases and Ca2+-dependent calpain proteases have been demonstrated to process and activate HAV NS5A protein (34). Proteolytic processing of the spike protein of severe acute respiratory syndrome (SARS) coronavirus by factor Xa has also been associated with viral infectivity (35). Initially, we attempted to search putative polyprotein processing cleavage sites on HEV pORF1 using the ExPASy peptide cutter tool (ExPASy Bioinformatics Resource Portal). We attempted to search potential polyprotein cleavage sites with following criteria: (i) the site should be conserved in HEV genotypes; (ii) the protease should be present in hepatocytes, which are the primary site of HEV replication; and (iii) the putative polyprotein cleavage site should be near the boundaries of predicted functional domains of the HEV polyprotein. Surprisingly, we found a putative factor Xa cleavage site and two thrombin cleavage sites on HEV pORF1. Putative cleavage sites are schematically shown in Fig. 1. Also, these cleavage sites are highly conserved throughout HEV genotypes. Multiple-sequence alignment for amino acid sequences of a total of 290 strains of HEV genotypes has been performed by using the ClustalW program in Bioedit software (see Table S1 in the supplemental material), and the thrombin and factor Xa cleavage site positions on pORF1 of representative sequences have been listed in Table 1.
TABLE 1.
Thrombin and factor Xa cleavage sites on pORF1 are conserved in HEV genotypes
| HEV genotype | Strain or isolate | NCBI accession no. | Thrombin cleavage site position (aa)a |
Factor Xa cleavage site position (aa)a | |
|---|---|---|---|---|---|
| 1st site | 2nd site | ||||
| I | Sar55 | AF444002.1 | 848 | 1220 | 560 |
| Burma | M73218.1 | 848 | 1220 | 560 | |
| India | X98292.1 | 848 | 1220 | 560 | |
| China | D11092.1 | 848 | 1220 | 560 | |
| TK15/92 (Nepal) | AF051830.1 | 848 | 1220 | 560 | |
| US1 | AF060668 | 853 | 1225 | — | |
| II | Mexican | M74506.1 | 846 | 1218 | 526 |
| III | JE03-1760F | AB425830.1 | 858 | 1230 | — |
| Meng | AF082843.1 | 863 | 1235 | — | |
| pSHEV-3 | AY575859.1 | 863 | 1235 | — | |
| IV | HE-JA30 | LC022745.1 | 862 | 1234 | 560 |
| T1 | AJ272108.1 | 862 | — | 560 | |
The dash denotes the absence of the site in the genome.
We therefore hypothesized that HEV ORF1 polyprotein processing is carried out by cellular factor Xa and thrombin. Thrombin and factor Xa are serine proteases, which are best known for their role in the blood coagulation cascade (36). Thrombin and factor Xa are primarily synthesized in the liver as precursor proenzymes known as prothrombin and factor X, respectively. Prothrombin (molecular mass [MM] of ∼69 kDa) is cleaved into active thrombin (MM of ∼32 kDa) by factor Xa in the plasma (37). As a result of vascular insult, factor X is activated to factor Xa by an intrinsic or extrinsic pathway by a complex of proteases involving factors IX, VIII, and VII along with tissue factor (38). Activated factor Xa triggers thrombin activation and release, which further converts soluble fibrinogen to insoluble fibrin, forming a clot at the site of injury (36). Thrombin also has proinflammatory effects mediated by the activation of protease-activating receptors on the surface of monocytes, lymphocytes, endothelium, and dendritic cells (36). Thrombin is a multifunctional protein that has several roles, namely, activation of platelets through receptor binding, activation of other coagulation factors, and signaling through chemotactic properties (39). Thrombin has a role in neuronal cell physiology through proteolysis of tau proteins, the failure of which has been associated with neurodegenerative diseases (40). Both factor Xa and thrombin belong to the family of vitamin K-dependent coagulation factors characterized by the NH2-terminal domain of G-carboxyglutamic acid (Gla domain) (41). They hydrolyzed a peptide bond on the carboxyl side of a basic amino acid, mostly arginine (42). Prothrombin and fibrinogen are the most common substrates of factor Xa and thrombin, respectively (38, 43).
In this study, we have shown that thrombin and factor Xa proteases play a crucial role in HEV replication, probably through ORF1 polyprotein processing. Thrombin and factor Xa cleavage sites are conserved throughout all HEV genotypes and are obligatory for the replication of HEV in cell culture. Though prothrombin and factor X are primarily synthesized in hepatocytes, biologically active thrombin and factor Xa are predominantly found in blood plasma. In this study, we also demonstrated that biologically active thrombin and factor Xa are associated with hepatocytes and are essential for HEV replication in cell culture. In total, this study provides new insights into replication and ORF1 polyprotein processing in the biology of HEV.
RESULTS
Thrombin and factor Xa cleavage sites in pORF1 are essential for HEV replication in cell culture.
To explore the possible role of host proteases in HEV replication and ORF1 polyprotein processing, we analyzed proteases having cleavage sites on pORF1 of HEV. The ExPASy Peptide Cutter tool was used for prediction of potential peptide cleavage sites on the HEV ORF1 sequence. We could find factor Xa and thrombin cleavage sites in the pORF1 sequence. Both thrombin and factor Xa cleavages sites are conserved across HEV genotypes (Table 1). Thrombin cleavage at the arginine residue at position 1220 releases HEV polymerase from pORF1 while cleavage at the arginine at amino acid position 848 separates the predicted helicase domain from the rest of the pORF1. The factor Xa cleavage site is located at the C terminus of the HEV putative Papain-like cysteine protease.
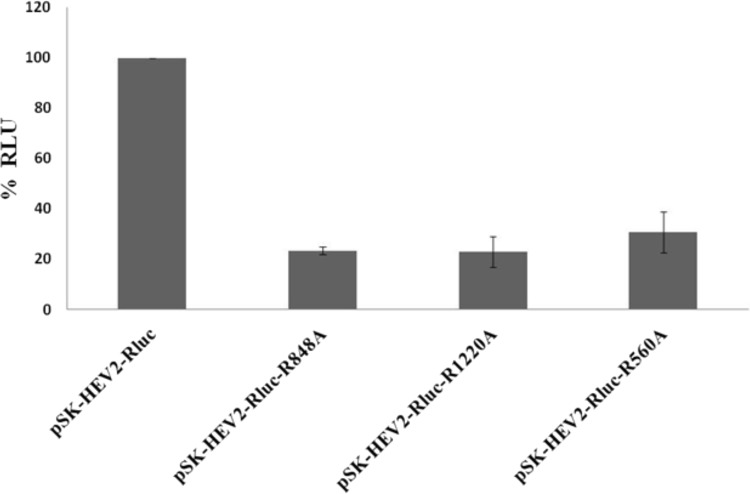
We further utilized a reverse genetics approach to confirm the role of thrombin and factor Xa cleavage sites in HEV replication in cell culture. pSK-HEV-2-Rluc subgenomic replicon mutants lacking either one of the thrombin cleavage sites or the factor Xa cleavage site were generated using site-directed mutagenesis. In vitro-transcribed RNAs from wild-type and mutant replicons were transfected into the Huh7-S10-3 cell line. At 3 days posttransfection, replication of the subgenomic HEV replicon inside the cells was assessed by a dual-luciferase assay. We observed a reduction in HEV replication in terms of percent Renilla luciferase units for mutant replicons compared to that of a wild-type replicon in Huh7-S10-3 cells. These results indicate that cleavage site sequences for thrombin and factor Xa, which are present on pORF1, are essential for HEV replication in cell culture (Fig. 2). This may be due to the inability of thrombin and factor Xa to cleave and activate pORF1 functional domains in mutant replicons.
FIG 2.
Thrombin and factor Xa cleavage sites are required for HEV replication. Huh7-S10-3 cells were transfected with capped RNA transcripts of a pSK-HEV2-Rluc wild-type or thrombin/factor Xa cleavage site mutant subgenomic replicon (pSK-HEV2-Rluc-R848A, pSK-HEV2-Rluc-R1220A, or pSK-HEV2-Rluc-R560A). A dual-luciferase assay was carried out to detect replication in terms of Renilla luciferase activity at 3 days posttransfection. Renilla luciferase values were normalized to firefly luciferase counts as an internal control. The experiment was done in triplicate, and the graph represents average percent Renilla luciferase units (RLU) indicating HEV replication in cell culture.
Thrombin cleaves at the predicted cleavage sites on HEV pORF1 in vitro.
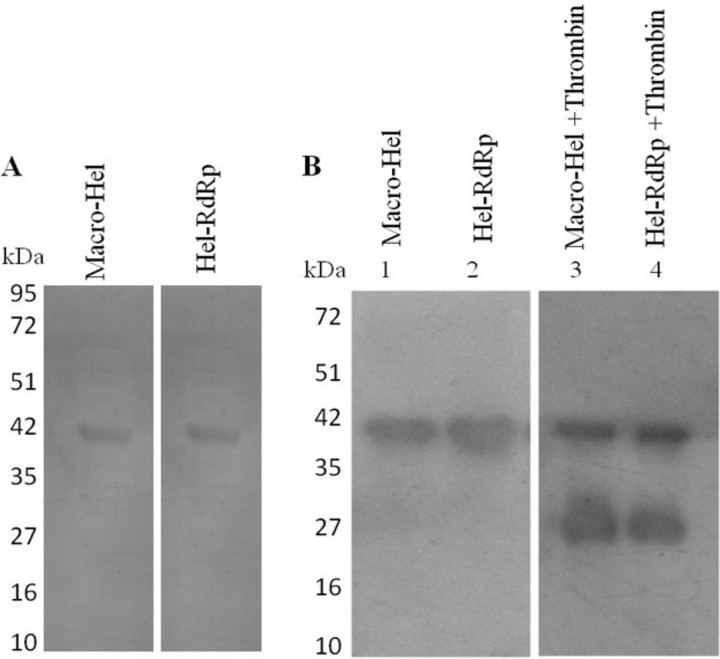
To test whether thrombin can cleave HEV pORF1, an assay was designed in which truncated pORF1 fragments were digested in vitro with thrombin. Truncated regions of the macro domain-helicase (macro-Hel; amino acids [aa] 622 to 984 of ORF1) and helicase-RdRp (Hel-RdRp; aa 994 to 1356 of ORF1) proteins were thus expressed as N-terminal His-tagged recombinant proteins of ∼40 kDa in bacteria. The proteins macro-Hel and Hel-RdRp bear thrombin cleavage sites at amino acid positions 848 and 1220, respectively. The proteins were purified by Ni-nitrilotriacetic acid (NTA) affinity column chromatography under denaturing conditions, followed by on-column renaturation. Initially, the proteins were purified by Ni-NTA affinity chromatography under native conditions; however, we could not obtain the proteins in soluble form. Expression and purification of proteins were detected using 12% SDS-PAGE followed by Coomassie brilliant blue staining, which showed the expected band corresponding to both proteins at ∼40 kDa (Fig. 3A). The purified N-terminal 6×His-tagged proteins were incubated with human thrombin at room temperature for 4 h as described in Materials and Methods. Thrombin treatment resulted in cleaving of the macro-Hel and Hel-RdRp proteins. An N-terminal His-tagged digestion product of ∼25 kDa was detected by Western blotting using anti-His antibody (Fig. 3B). The results of this in vitro assay indicate that thrombin is capable of cleaving HEV pORF1.
FIG 3.
In vitro digestion of pORF1 by thrombin. (A) The HEV pORF1 regions encompassing truncated Hel-RdRp and macro-Hel domains were expressed in bacteria as N-terminal 6×His-tagged proteins of ∼40 kDa. Proteins were purified using Ni-NTA affinity chromatography, and the purified proteins were subjected to 12% SDS-PAGE followed by Coomassie brilliant blue staining. (B) The expressed fusion proteins were incubated for 4 h at room temperature with bovine thrombin in thrombin cleavage buffer. Digestion of proteins in the presence of thrombin was checked with Western blotting using anti-His antibody produced in mouse. Digestion with thrombin yielded a ∼25-kDa band in the respective lanes.
Biologically active thrombin and factor Xa are present in Huh7-S10-3 cells.
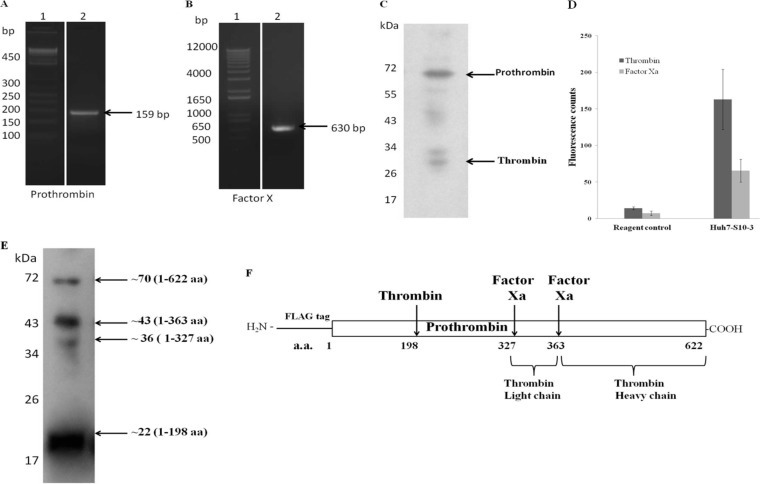
Thrombin and factor Xa have been well studied for their roles in the blood clotting pathway. Both of these proteins are believed to be present in the active form in the blood plasma or on the surface of platelets in the blood. The intracellular biological activity of these proteins and their function beyond that in the blood clotting pathway have not been much explored. Though most of the blood clotting factors are synthesized in the liver, they are in the inactive zymogen form. Thus, we wanted to study whether biologically active thrombin and factor Xa are present in the active form in Huh7-S10-3 cells. At the transcript level, we could detect the presence of both prothrombin and factor X mRNAs in cells using reverse transcriptase PCR (RT-PCR) (Fig. 4A and B). Also, the presence of prothrombin and thrombin in Huh7-S10-3 cell lysate was detected by Western blotting. Bands for full-length prothrombin (69 kDa) and thrombin (32 kDa) were observed. It can be concluded from these results that prothrombin is processed inside the cells, and therefore active thrombin as well as active factor Xa must be associated with Huh7-S10-3 cells (Fig. 4C).
FIG 4.
Biologically active thrombin and factor Xa present in Huh7-S10-3 cells. Total RNA was isolated form Huh7-S10-3 cells, and RT-PCR was carried out using specific primers. (A) Prothrombin cDNA amplification of 159 bp was observed on a 2% agarose gel. Lane 1, 50-bp DNA ladder. (B) Factor X cDNA amplification of 630 bp was observed on a 2% agarose gel. Lane 1, 50-bp DNA ladder. (C) Presence of prothrombin (∼69 kDa) and thrombin (∼32 kDa) was detected in Huh7-S10-3 cell lysate by Western blotting using anti-thrombin antibody. (D) Fluorogenic assay for measuring intracellular thrombin and factor Xa activity. Huh7-S10-3 cell lysate was incubated with thrombin or factor Xa fluorogenic substrate in cleavage buffer for 1 h at 37°C. The reagent control reaction mixture excludes the Huh7-S10-3 cell lysate. Values in the graph represent mean fluorescence counts of three independent experiments. (E) The human prothrombin gene was cloned into pCDNA3.1, and the protein was overexpressed in Huh7-S10-3 cells. Western blotting was carried out to detect processing of exogenously expressed FLAG-prothrombin in Huh7-S10-3 cells with anti-FLAG antibody. (F) Schematic representation of N-terminal FLAG-tagged prothrombin molecule. The arrows indicate the cleavage sites for factor Xa and thrombin while numbers denote the positions of corresponding amino acids of the cleavage sites.
For detection of cell-associated activity of thrombin and factor Xa in Huh7-S10-3 cells, lysis was done in 100 μl of passive lysis buffer, and 20 μl of cell lysate was added as a test sample in a fluorogenic assay for thrombin and factor Xa activity. The assay included specific substrates for thrombin and factor Xa bearing the respective cleavage sites. The cleavage product emits fluorescence, which is an indicator of the protease activity. This is a highly specific and sensitive assay. We observed significant activity of thrombin and factor Xa associated with Huh7-S10-3 cells (Fig. 4D). In another set of experiments, Huh7-S10-3 cells were transfected with in vitro-transcribed RNA of a pSK-HEV2-Rluc replicon in 24-well plates. At 3 days posttransfection, mock-transfected cells and RNA transfected cells were harvested. A dual-luciferase assay was done to confirm HEV replication in transfected cells. However, we did not observe a significant alteration in the activity of thrombin and factor Xa in Huh7-S10-3 cells after HEV RNA transfection (data not shown).
In order to further confirm our findings, we studied the processing of exogenously added prothrombin in Huh7 S10 cells. We cloned an N-terminal FLAG-tagged human prothrombin gene in the pCDNA3.1 vector. Plasmid pCDNA3.1-prothrombin was transfected in Huh7-S10-3 cells. Transient expression and processing of prothrombin were confirmed by immunoblotting using anti-FLAG antibody (Fig. 4E). Immunoblotting showed bands corresponding to full-length prothrombin (∼70 kDa) along with smaller fragments of ∼43 kDa, ∼36 kDa, and ∼22 kDa, suggesting intracellular processing of prothrombin (Fig. 4F). Overall, our results indicate that biologically active factor Xa and thrombin are associated with Huh7-S10-3 cells.
Ben-HCl, a serine protease inhibitor, represses HEV replication in cell culture.
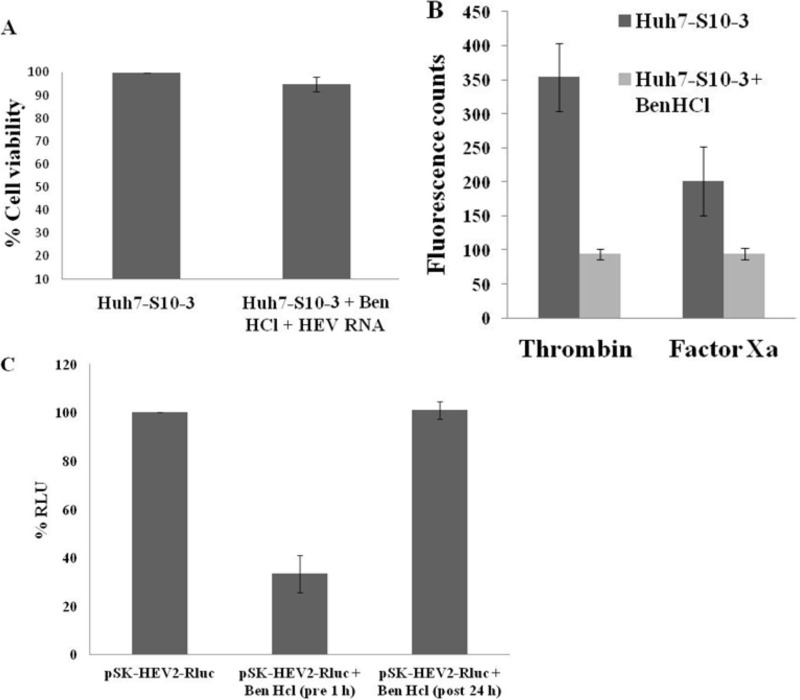
Benzamidine hydrochloride (Ben-HCl) is a cell-permeable serine protease inhibitor. Ben-HCl is known to inhibit the activity of thrombin and factor Xa (35). The toxicity of the inhibitor was tested by alamarBlue assay, and we showed that the inhibitor is not cytotoxic when a 4 mM concentration is used (Fig. 5A). Further, to check the effect of Ben-HCl on cell-associated activities of thrombin and factor Xa, Ben-HCl treatment was given up to 24 h, and the cells were harvested at 48 h posttreatment. Fluorometric assays showed decreased intracellular activities of thrombin and factor Xa in inhibitor-treated cells compared to levels in untreated cells (Fig. 5B).
FIG 5.
Effect of serine protease inhibitor, Ben-HCl, on HEV replication in cell culture. (A) Cells were treated with 4 mM Ben-HCl for 1 h followed by HEV RNA transfection at 37°C. Untransfected cells which were not treated with Ben-HCl were kept as controls. A cell viability assay was performed after 24 h of RNA transfection in a 96-well plate using alamarBlue reagent by measuring absorbance at 570 nm. Values plotted are means of three independent experiments. (B) Fluorogenic assay to check the effect of Ben-HCl treatment on intracellular thrombin and factor Xa activity in Huh7-S10-3 cells. Cells were treated with Ben-HCl (4 mM) for 24 h. Cells were harvested at 48 h posttreatment, and lysate from treated or untreated cells was incubated with thrombin or factor Xa fluorogenic substrate in cleavage buffer for 1 h at 37°C. Values plotted are means of three experiments. (C) Capped RNA transcript of a pSK-HEV2-Rluc subgenomic replicon was transfected into Huh7-S10-3 cells treated with Ben-HCl (4 mM) before 1 h or after 24 h of transfection. Cells were harvested at 3 days posttransfection, and a dual-luciferase assay was performed. The graph represents percent Renilla luciferase units indicating HEV replication in cells with or without Ben-HCl treatment. RLU, Renilla luciferase units.
To test the effect of Ben-HCl on HEV replication, Huh7-S10-3 cells were preincubated in 4 mM Ben-HCl (Sigma) for 1 h before transfection at 37°C. Cells were transfected with the capped transcript of the pSK-HEV-2-Rluc replicon. Cells were harvested at 3 days posttransfection, and a dual-luciferase assay was performed to check HEV replication. We observed more than a 50% reduction in Renilla luciferase units in cells treated with Ben-HCl, suggesting the importance of a serine protease in the replication of HEV in cell culture. An alamarBlue assay was carried out to check the viability of cells at 24 h after HEV RNA transfection for both Ben-HCl-treated and untreated cells (Fig. 5A). More than 95% cell viability was observed in inhibitor-treated cells. ORF1 polyprotein processing is one of the earliest events to occur during the HEV replication cycle. To investigate whether thrombin and factor Xa play a role in the initial stages of HEV replication, Ben-HCl was added at 24 h posttransfection of HEV RNA to Huh cells. At 3 days posttransfection, a dual-luciferase assay was performed. In contrast to cells pretreated with Ben-HCl, where a stark decrease in HEV replication could be observed, cells treated with Ben-HCl at 24 h post-RNA transfection showed HEV replication comparable to that of untreated cells (Fig. 5C). This observation implies the possibility of serine proteases playing an important role in the early events of the HEV life cycle in cell culture.
The activity of thrombin and factor Xa is essential for HEV replication in cultured cells.
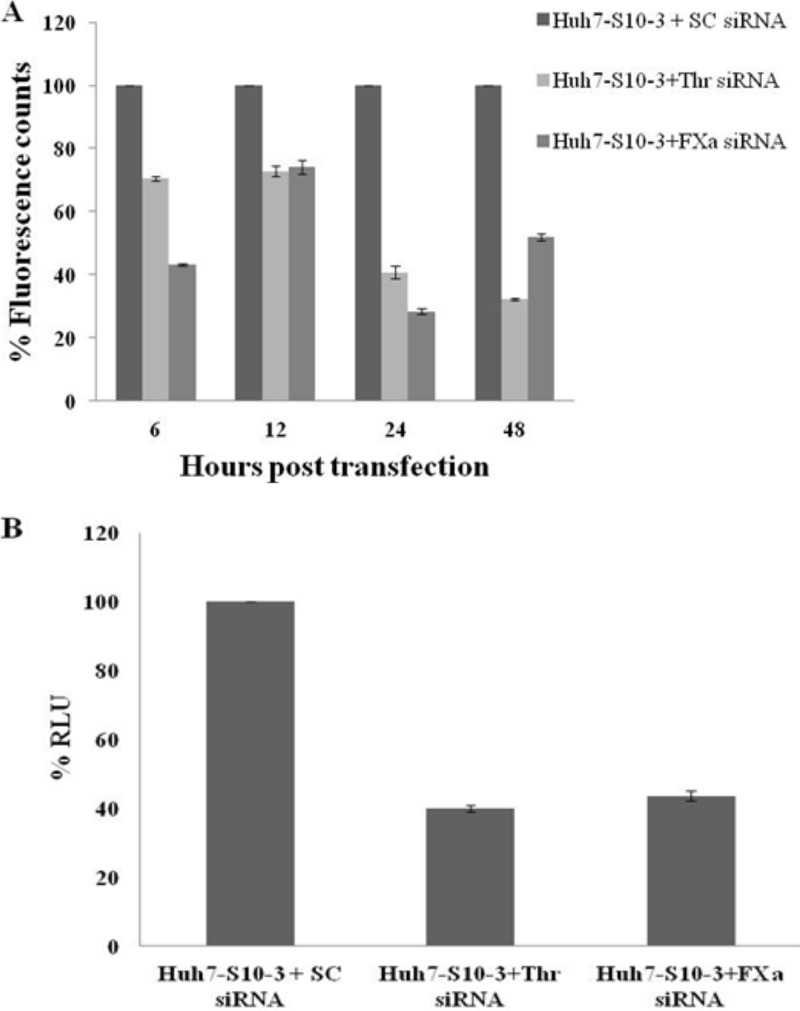
To validate the role of thrombin and factor Xa in HEV replication, small interfering (siRNA)-mediated knockdown of thrombin and factor Xa was performed. For optimization of siRNA concentration and treatment duration, Huh7-S10-3 cells were transfected with 10 pM or 20 pM of either thrombin- or factor Xa-specific siRNAs. Cells were harvested at 6 h, 12 h, 24 h, and 48 h posttransfection of siRNA. Thrombin and factor Xa activities in cell lysates of either control cells, thrombin siRNA-treated cells, or factor Xa siRNA-treated cells were measured using fluorometric assays as explained earlier. There was no significant difference in the effects of 10 pM and 20 pM siRNAs; hence, a 10 pM concentration was used in all further siRNA experiments (data not shown). As evident from Fig. 6A, for both thrombin and factor Xa siRNAs, the maximum reduction in the intracellular activity of the target protein was found at 24 h of posttransfection. To check the effect of thrombin and factor Xa knockdown on HEV replication, capped transcript of pSK-HEV2-Rluc was transfected 24 h post-siRNA transfection in Huh7-S10-3 cells. For all siRNA knockdown experiments, scrambled siRNA (10 pM) transfected cells were used as a control. Cells were harvested at 3 days post-HEV RNA transfection, and replication was monitored by a dual-luciferase assay as described previously. We observed a significant reduction in the levels of HEV replication in thrombin or factor Xa siRNA transfected cells compared to levels in control cells (Fig. 6B). The results suggest that the activities of cellular thrombin and factor Xa are essential for HEV replication.
FIG 6.
The activity of thrombin and factor Xa is required for HEV replication. (A) For optimization of siRNA treatment, 10 pM of either factor Xa siRNA or thrombin siRNA was transfected into Huh7-S10-3 cells. Cells were harvested at 6, 12, 24, and 48 h posttransfection, and lysates were incubated with thrombin or factor Xa fluorogenic substrate in cleavage buffer for 1 h at 37°C. Fluorescence measured has been plotted in terms of percent fluorescence counts. Data were normalized using the total protein concentration in the respective cell lysate. (B) Capped RNA transcript of a pSK-HEV2-Rluc subgenomic replicon was transfected into Huh7-S10-3 cells transfected with either scrambled (SC) siRNA, factor Xa siRNA, or thrombin siRNA. HEV RNA transfection was done 24 h post-siRNA transfection. Cells were harvested at 3 days posttransfection, and a dual-luciferase assay was performed. The mean numbers of Renilla luciferase units from three independent experiments were converted to percentages, with values for control cells considered 100%. Cells transfected with 10 pM scrambled siRNA were used as controls.
DISCUSSION
Translation of viral proteins is the earliest event in the life cycle of positive-stranded RNA viruses. Since positive-sense RNA viruses have compact genomes, they encode a limited number of ORFs, which in most cases is restricted to one or two. These ORFs code for polyproteins, and regulation of the life cycle occurs primarily at polyprotein translational levels. In all positive-sense RNA viruses, nonstructural proteins are expressed as a polyprotein. These nonstructural polyproteins are processed by limited/restricted proteolysis by viral or cellular proteases. This tightly regulated posttranslational modification by a viral or cellular protease is responsible for temporal and spatial regulation of the biochemical activity of nonstructural proteins (44). All positive-sense RNA animal viruses exhibit nonstructural polyproteins processed to individual subunits of functional domains, such as capping enzyme, helicase, protease, and RNA-dependent RNA polymerase. The presence of RNA-dependent RNA polymerase (RdRp) is the hallmark of positive-sense RNA viruses. RdRp is a key enzyme responsible for the synthesis of the genomic strand and anti-genomic strand and transcription of the ORF present at the 3′ end. Activities of RdRp are regulated by temporal polyprotein processing and interactions with viral or host proteins. Polyprotein processing has been extensively studied in alphaviruses and flaviviruses (45, 46).
In alphaviruses, the processing of the polyprotein to generate precursor and end product nonstructural proteins (nsPs) is believed to regulate the synthesis of viral RNAs. Processing of the nonstructural polyprotein occurs through the action of a virus-encoded protease located within nsP2. The translation of the structural polyprotein, however, proceeds through a subgenomic RNA intermediate initiated at the C terminus of the nsP coding region. The structural polyprotein is proteolytically cleaved by an autocatalytic activity within the capsid protein, cellular signalase proteins, and a furin-like protease (46).
HEV is most closely related to members of the Alphavirus and Rubivirus genera of the family Togaviridae (30, 47). For the viruses belonging to both of these genera, viral polyprotein processing has been proved to be of significance for the regulation of RNA synthesis, which regulates the viral life cycle (48, 49). Many studies have attempted to understand the mechanisms of HEV pORF1 polyprotein processing using different expression systems. However, these studies gave varied results and could never convincingly establish the role of HEV protease in proteolytic processing. Overall, it is suggested that HEV pORF1 polyprotein processing occurs only in the context of virus replication or only to a minimum degree and is very tightly regulated. Thus, we hypothesized the existence of certain cellular factors required to trigger HEV polyprotein processing. To test this hypothesis, we searched conserved protease sites on the pORF1 polyprotein and observed the presence of one factor Xa and two thrombin cleavage sites. By employing a reverse genetics approach, we have shown that both the factor Xa and thrombin cleavage sites present on HEV pORF1 are necessary for HEV replication. In this study, we have altered thrombin and factor Xa cleavage sites at amino acid positions 560, 848, and 1220 by site-directed mutagenesis (change of R to A). The putative active sites of families of viral nonstructural proteins like PCP, helicase, and RdRp have been reported in the literature (17–22), and an R-to-A change carried out in this study does not disrupt any canonical active site. Also, HEV pORF1 domains such as PCP, macro, helicase, and RdRp have diverse biochemical activities. Thus, it is unlikely that all of these proteins require arginine for biochemical activities. We therefore believe that the mutations in the thrombin and factor Xa cleavage sites affect the processing and results in reduced replication rather than disruption of functional domains of pORF1. We further demonstrated that the cellular activities of factor Xa and thrombin are essential for the replication of HEV. HEV replicon mutants lacking either of the two thrombin cleavage sites or the factor Xa cleavage site were unable to replicate efficiently in a liver cell line. One of the mechanisms underlying the observed effect could be a result of polyprotein processing of HEV pORF1 via thrombin and factor Xa. We could observe the cleavage of pORF1 in vitro by thrombin. However, demonstrating such processing of pORF1 in replicon-mediated infection in cells is difficult owing to the unavailability of specific antibodies and low yield of HEV nonstructural proteins in cell culture.
It may also be possible that cleavage at the thrombin and factor Xa cleavage sites occurs in a chronological manner, regulating the release of pORF1 functional domains and thereby controlling the synthesis of positive and negative strands. This is evident in replication of alphaviruses, where nonstructural proteins are synthesized as a polyprotein, called P1234, which is cleaved into nsP1, nsP2, nsP3, and nsP4 by the viral protease domain of nsP2. The cleavage intermediate P123-nsP4 is required to synthesize the minus-strand RNA, whereas the completely processed complex is required for plus-strand synthesis (46).
The boundaries of functional domains of HEV pORF1 have still not been experimentally mapped. Based on sequence similarity with proteins from related viruses, the domains have been predicted. Based on the replication model for alphaviruses, we hypothesized that, after translation of pORF1, cleavage at the helicase-RdRp junction would separate RdRp from the rest of the polyprotein. Studies with alphavirus suggested that separation of RdRp is crucial for regulation of early stages of the viral life cycle (46). Also, the significance of viral polyprotein processing may not be limited to merely the separation of functional domains but also is crucial for regulation of replication. The sequential processing of nonstructural polyprotein may be essential for the regulation of activities of nonstructural proteins, and temporal processing may lead to loss of activity of particular nonstructural proteins during the life cycle of viruses.
HEV replicates primarily in liver cells, and clotting factors like thrombin and factor Xa are also synthesized in liver cells as zymogens. Active thrombin and factor Xa were believed to be present in plasma or on the surface of platelets; however, there are some reports suggesting the presence of intracellular thrombin. The presence of thrombin has been shown in brain cells using histochemical techniques (40). Binding and internalization of thrombin have also been observed in chick fibroblast cells (50). Thrombin has been reported to be associated with, and internalized in, cultured human endothelial cells (51). Despite the age-old discovery of these coagulation factors, their roles apart from those in blood clotting have remained underexplored. HEV ORF3 protein is known to interact with plasma proteins like fibrinogen and serine protease inhibitors, leading to disruption of the blood coagulation process (52). This suggests that HEV interacts with blood clotting pathway components which may play important roles in HEV replication and pathogenesis.
Transcripts corresponding to prothrombin and factor X were detected in Huh-S10-3 cells. We could also detect activities of thrombin and factor Xa in Huh-S10-3 hepatoma cells by using specific fluorogenic substrates. However, the activities of factor Xa and thrombin in the context of HEV replication in Huh7-S10-3 cells were found to be unaltered. A serine protease inhibitor called Ben-HCl showed a significant reduction in HEV replication compared to the level in control cells. This effect was evident only when cells were treated with Ben-HCl during the initial stages of the HEV life cycle, prior to RNA transfection. Ben-HCl treatment at 24 h post-HEV RNA transfection did not exhibit any effect on HEV replication. This suggests that thrombin and factor Xa are required only in the initial stages of HEV replication. A reduction in the activities of intracellular thrombin and factor Xa that was also observed in Huh-S10-3 cells treated with Ben-HCl confirms the efficiency of the inhibitor. Owing to the broad-spectrum activity of Ben-HCl, siRNAs were used to specifically inhibit factor Xa and thrombin activity. siRNA-mediated knockdown resulted in reduced intracellular activity of thrombin and factor Xa. Therefore, it can be concluded that prothrombin and factor X are processed inside the cells to active thrombin and factor Xa, respectively. In other words, genome-encoded prothrombin and factor X directly contribute to the intracellular activity of thrombin and factor Xa rather than uptake of active proteins by cells from plasma. In liver cell lines where thrombin or factor Xa was knocked down, HEV replication was significantly reduced. It is thus clear that the activities of thrombin and factor Xa are required for HEV replication in cell culture. In summary, this study shows that thrombin and factor Xa are crucial for HEV replication in cell culture. The results further suggest that the activities of the said host proteases are probably involved in HEV pORF1 processing.
MATERIALS AND METHODS
Virus replicon and cells.
A subclone of the human hepatoma cell line Huh7-S10-3, which is permissive for the replication of HEV infectious clone or replicons, was obtained from Suzanne U. Emerson, NIH, Bethesda, MD. Cells were maintained in Dulbecco's modified Eagle's medium (DMEM) with GlutaMAX (Invitrogen) supplemented with 10% fetal bovine serum (FBS) (Invitrogen) and 100 U/ml penicillin and 100 mg/ml streptomycin (Sigma). HEV subgenomic replicon carrying a Renilla luciferase reporter gene (pSK-HEV2-Rluc) was obtained from Xiang Jin Meng, Virginia Tech, Blacksburg, VA.
Construction of recombinant vectors.
Regions of pORF1 containing the first thrombin cleavage site (at R848) spanning amino acids 622 to 984 of pORF1 (encompassing the macro domain and helicase domains) and a second cleavage site (at R1220) spanning amino acids 994 to 1356 of pORF1 (encompassing the helicase and RdRp domains) were cloned in the bacterial expression vector pET-28a (Novagen). For the construction of recombinant vectors, primers were designed with the restriction enzymes indicated in Table 2. PCR was performed with the appropriate primers and plasmid pSK-HEV2-Rluc as a template. The PCR product was directionally cloned in the vector pET-28a. Clones were confirmed by DNA sequencing. These clones are designated macro-Hel and Hel-RdRp.
TABLE 2.
Primer sequences used in the study
| Primer name | Sequence | Amplicon |
|---|---|---|
| His_Thr848_NcoI_F | CGCCCCATGGGACATCATCATCATCATCATGTCGCCGCCGGGCTTGAC | Macro-Hel |
| Thrombin_PCP_848_Hel_HindIII_R | GCCGCCAAGCTTTCAAGAGCGGGACTTGCCGGATC | |
| His_Thr1220_Nco_F | CGCCCCATGGGACATCATCATCATCATCATGTGGTCCCGACCCGGGAG | Hel-RdRp |
| Thrombin_Hel_1220_RdRp_R | GCCGCCAAGCTTTCAGACCATGGCCTCCACTA | |
| Thrombin_848HEV_F | GCCTACACATTAACCCCCGCGCCAATAATTCATGCC | Thr 848 site mutant |
| Thrombin_848HEV_R | GGCATGAATTATTGGCGCGGGGGTTAATGTGTAGGC | |
| Thrombin_1220HEV_F | CCATCTGTTATCCCTGCCGGCAATCCTGACGCC | Thr 1220 site mutant |
| Thrombin_1220HEV_R | GGCGTCAGGATTGCCGGCAGGGATAACAGATGG | |
| FXa_560HEV_F | CAGGTCGACGGGGCGATCGATTGTGAGACCCTT | Factor Xa 560 site mutant |
| FXa_560HEV_R | AAGGGTCTCACAATCGATCGCCCCGTCGACCTG | |
| ProTh_EcoRI_FLAG_F | CACTGGAATTCATGGACTACAAAGACGATGACGACAAGATGGCGCACGTCCGAGGCTTGC | Human prothrombin |
| ProTh_KpnI_HA_R | CACTGGGTACCCTAAGCGTAATCTGGAACATCGTATGGGTACTCTCCAAACTGATCAATGAC | |
| Fxa_RT_F | AGATTCAAGGTGAGGGTAGGGGACC | Factor Xa transcript detection |
| FXa_RT_R | GAGTGGGATCTCACTTTAATGGAGA | |
| Thr_RT_F | CTATTGTGAGGAGGCCGTGGAGG | Thr transcript detection |
| Thr_RT_R | AACAGAGGTCGCAGCCCACAGTC |
To test processing of prothrombin in Huh7-S10-3 cells, the prothrombin (F2) gene was cloned into pCDNA3.1/myc-His(-) (Invitrogen) to express an N-terminal FLAG-tagged prothrombin in Huh7-S10-3 cells. To amplify the prothrombin gene, RNA was isolated from Huh7-S10-3 cells using a RiboPure kit (Ambion). Isolated RNA was used for cDNA synthesis using SuperScript III reverse transcriptase (Invitrogen). From cDNA, the prothrombin gene was amplified and directionally cloned into vector pCDNA3.1. This clone is designated pCDNA3.1-FLAG-prothrombin.
Site-directed mutagenesis.
Site-directed mutagenesis was performed to mutate the factor Xa and thrombin cleavage sites on pORF1. For site-directed mutagenesis overlapping primers were designed containing the desired mutation (Table 2). Plasmid pSK-HEV2-Rluc was used as the backbone for site-directed mutagenesis. PCR was performed using Q5 high-fidelity DNA polymerase (NEB), pSK-HEV2-Rluc as a template, and mutagenic primers. The PCR product was digested with the enzyme DpnI (NEB) to remove the wild-type pSK-HEV2-Rluc template plasmid. Five microliters of DpnI-digested PCR mix was transformed into NEB-5α chemically competent cells (NEB). Clones with the desired mutation were confirmed by DNA sequencing. These clones are designated pSK-HEV2-Rluc-R848A and pSK-HEV2-Rluc-R1220A for thrombin cleavage site mutants and pSK-HEV2-Rluc-R560A for the factor Xa cleavage site mutant.
In vitro transcription and transfection.
pSK-HEV2-Rluc wild-type and mutant plasmids were linearized by BglII restriction enzyme. The capped transcript was synthesized from a linearized template by using a T7 riboprobe transcription RNA system (Promega). A 50-μl reaction mixture contained 5 μg of linearized pSK-HEV2-Rluc template, 10× transcription buffer, 5 μl of 100 mM dithiothreitol (DTT), 2 μl of RNasin (40 U/μl), 5 μl of ribonucleotide triphosphate (rNTP) mix (5 mM each) with low GTP (0.5 mM), 5 μl of 5 mM anti-reverse cap analog (ARCA; Invitrogen), and 2 μl of T7 RNA polymerase (20 U/μl). The reaction mixture was incubated at 37°C for 2 h. Equal amounts of the capped transcript (either wild-type or mutant transcript) of HEV subgenomic replicons and 10 ng of pGL4.10 (Promega) (a plasmid expressing firefly luciferase) were cotransfected into Huh7-S10-3 cells in a 48-well plate using DMRIE-C (1,2-dimyristyloxypropyl-3-dimethyl-hydroxy ethyl ammonium bromide and cholesterol) transfection reagent (Invitrogen) as described previously (14). Cells were maintained at 37°C and harvested at 3 days posttransfection. A dual-luciferase assay was carried out to measure HEV replication as described previously (14). Firefly luciferase was used as an internal control for monitoring HEV replication.
RT-PCR.
Total cellular RNA was isolated from cells by using a RiboPure RNA purification kit (Ambion). Reverse transcriptase PCR (RT-PCR) was carried out using a SuperScript III one-step RT-PCR system with platinum Taq DNA polymerase. Primers used for the amplification of prothrombin and factor X mRNA are listed in Table 2 (35, 40).
Thrombin and factor Xa activity assay.
To study levels of biologically active thrombin and factor Xa in Huh-S10-3 cells, thrombin and factor Xa assays were performed. The fluorogenic substrates Boc-Val-Pro-Arg-7-amido-4-methylcoumarin hydrochloride (Sigma) and Boc-Ile-Glu-Gly-Arg-7-amido-4-methylcoumarin acetate salt (Sigma) were used to study the activity of thrombin and factor Xa, respectively. Thrombin and factor Xa assays were done separately in 100 μl of a reaction mixture containing 50 μl of either cell lysate or a positive control (commercial bovine thrombin or factor Xa) in cleavage buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM CaCl2, pH 8) and 50 μl of a fluorogenic substrate (0.1 mM). The reaction mixture was incubated at 37°C for 1 h, and fluorescence was measured at excitation/emission wavelengths of 360/460 nm. Commercially available thrombin (Sigma) and factor Xa (Sigma) were used as positive controls and for obtaining standard curves.
In vitro thrombin digestion assay of pORF1 was performed by incubating 20 μl of purified macro-Hel and Hel-RdRp proteins with 1 μl of human thrombin (Sigma) in cleavage buffer at room temperature for 4 h. Proteins without thrombin addition were treated as undigested controls.
Ben-HCl treatment and cell viability assay.
Huh7-S10-3 cells were treated with 4 mM 3,3′-diaminobenzidine tetrahydrochloride (Ben-HCl) (Sigma) for 1 h before HEV RNA transfection to 24 h posttransfection at 37°C. A cell viability assay was performed in 96-well plates using alamarBlue (Invitrogen) reagent by measuring absorbance at 570 nm, normalized to 600-nm values. The viability of cells treated with 4 mM Ben-HCl was compared with that of untreated cells.
siRNA-mediated knockdown of thrombin and factor Xa in Huh7-S10-3 cells.
Small interfering RNAs (siRNAs) targeted against the F2 (thrombin) and F10 (factor Xa) genes were procured from Invitrogen and Sigma, respectively. Huh7-S10-3 cells were transfected with 10 pM siRNA using DMRIE-C reagent (Invitrogen) in a 24-well plate. Knockdown efficiencies of siRNAs were studied by harvesting cells at 6, 12, 24, and 48 h post-siRNA transfection by measuring thrombin and factor Xa intracellular activity using a fluorometric assay. Total protein concentration of control and siRNA-treated cell lysate was estimated by bicinchoninic acid (BCA) assay (Sigma). Activity in siRNA-treated cells was compared to that in cells transfected with a scrambled siRNA. Further, thrombin or factor Xa knockdown cells were transfected with capped transcript of pSK-HEV2-Rluc 24 h after siRNA transfection.
Protein expression and purification.
For the production of recombinant proteins, plasmids pET-28a-Hel-RdRp and pET-28a-Macro-Hel were transformed independently into Rosetta (DE3)pLysS cells (Novagen). In all of these plasmids the PCR product was cloned in frame with an N-terminal 6-histidine tag. Recombinant protein induction was done with 1.0 mM isopropyl-β-d-thiogalactopyranoside (IPTG) in Luria-Bertani (LB) medium. For optimization of protein expression, induction was carried out at different temperatures (25°C to 37°C) and time intervals (15 min to overnight). Recombinant proteins were purified from bacterial culture pellets by using a ProBond nickel-chelating resin column (Invitrogen).
For purification of recombinant proteins under native conditions, a 50-ml culture of induced cells was lysed on ice using 4 mg of lysozyme (Sigma) and 5 μl of Triton X-100 (Sigma) in 4 ml of native purification buffer (NPB) (50 mM NaH2PO4 and 0.5 M NaCl, pH 8) and incubated on ice for 1 h. Cell lysate was spun at 10,000 × g for 30 min at 4°C. The clear supernatant was loaded on a ProBond resin column preequilibrated with NPB. The cell lysate was incubated with resin in the column on ice for 1 h. The column was washed two times with 4 ml of native wash buffer 1 (10 mM imidazole in NPB) and four times with 4 ml of native wash buffer II (20 mM imidazole in NPB). Bound proteins were eluted in native elution buffer (250 mM imidazole in NPB).
For purification of recombinant proteins under hybrid conditions, 50-ml culture of induced cells was lysed using 4 ml of guanidinium lysis buffer (6 M guanidine hydrochloride, 20 mM sodium phosphate, pH 7.8, and 500 mM NaCl). Cell lysate was spun at 10,000 × g for 30 min at 25°C. The clear supernatant was loaded on a ProBond resin column preequilibrated with denaturing binding buffer (8 M urea, 20 mM sodium phosphate, pH 7.8, and 500 mM NaCl). The cell lysate was incubated with resin at room temperature for 1 h. The column was washed two times with 4 ml of denaturing binding buffer and four times with 4 ml of denaturing wash buffer (8 M urea, 20 mM sodium phosphate, pH 6.0, and 500 mM NaCl) at room temperature. To elute proteins under hybrid purification conditions, the column was washed four times with 8 ml of prechilled native wash buffer (20 mM imidazole in NPB). Bound proteins were eluted in native elution buffer (250 mM imidazole in NPB). Ten fractions of 1 ml each were collected, and Amicon Ultra centrifugal filters (Millipore) were used for buffer exchange and protein concentration.
Immunoblot analysis.
Protein samples were mixed with Laemmli's buffer, subjected to SDS-PAGE analysis, and transferred to polyvinylidene difluoride (PVDF) membranes (Sigma). The membranes were incubated overnight for blocking in Western blocker solution (Sigma) at 4°C. The membranes were washed with phosphate-buffered saline with 0.1% Tween 20 (PBST) and then incubated with primary antibody. For all recombinant proteins expressed in E. coli, Western blot analysis was done using anti-His monoclonal antibodies (Sigma) (dilution of 1:3,000). For detection of cell-associated thrombin from Huh7-S10-3 cells, anti-thrombin antibody (Invitrogen) (1:1,000) was used. For detection of recombinant FLAG-tagged prothrombin in Huh7 cells, anti-FLAG antibody (Sigma) (1:1,000) was used. After incubation with primary antibody, membranes were washed with PBST and probed with the appropriate secondary antibody. For recombinant proteins expressed in bacteria, membranes were developed using 3,3′-diaminobenzidine tetrahydrochloride (Sigma) as a colorimetric substrate. For detection of cell-associated thrombin and recombinant prothrombin in Huh7-S10-3 cells, enhanced chemiluminescence (ECL) substrate (Sigma) was used, and blot images were captured on X-ray films.
Supplementary Material
ACKNOWLEDGMENTS
We are thankful to K. M. Paknikar, Director, MACS-ARI, Pune, for all the support. We are grateful to X. J. Meng and S. U. Emerson for providing the HEV subgenomic replicon and human hepatoma cell line Huh7-S10-3, respectively.
This work is supported by financial assistance from a DST-INSPIRE grant and intramural grants from the Agharkar Research Institute, Pune. K.D.P. acknowledges the Council of Scientific and Industrial Research (CSIR), Government of India, for a junior research fellowship.
We declare that we have no conflicts of interest.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01853-17.
REFERENCES
- 1.Cao D, Meng XJ. 2012. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1:e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vasickova P, Psikal I, Kralik P, Widen F, Hubalek Z, Pavlik I. 2007. Hepatitis E virus: a review. Vet Med (Praha) 52:365–384. [Google Scholar]
- 3.Feng Z. 2016. Causation by HEV of extrahepatic manifestations remains unproven. Liver Int 36:477–479. doi: 10.1111/liv.13085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Meng XJ. 2010. Hepatitis E virus: animal reservoirs and zoonotic risk. Vet Microbiol 140:256. doi: 10.1016/j.vetmic.2009.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ahmad IR, Holla P, Jameel S. 2011. Molecular virology of hepatitis E Virus. Virus Res 161:47–58. doi: 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aggarwal R. 2011. Hepatitis E: historical, contemporary and future perspectives. J Gastroenterol Hepatol 26:72–82. doi: 10.1111/j.1440-1746.2010.06540.x. [DOI] [PubMed] [Google Scholar]
- 7.Tam AW, Smith MM, Guerra ME, Huang CC, Bradley DW, Fry KE, Reyes GR. 1991. Hepatitis E virus (HEV): molecular cloning and sequencing of the full-length viral genome. Virology 185:120–131. doi: 10.1016/0042-6822(91)90760-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nair VP, Anang S, Subramani C, Madhvi A, Bakshi K, Srivastava A, Shalimar Nayak B, Ranjith Kumar CT, Surjit M. 2016. Endoplasmic reticulum stress induced synthesis of a novel viral factor mediates efficient replication of genotype-1 hepatitis E virus. PLoS Pathog 12:e1005521. doi: 10.1371/journal.ppat.1005521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nan Y, Ma Z, Wang R, Yu Y, Kannan H, Fredericksen B, Zhang YJ. 2014. Enhancement of interferon induction by ORF3 product of hepatitis E virus. J Virol 88:8696–8705. doi: 10.1128/JVI.01228-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korkaya H, Jameel S, Gupta D, Tyagi S, Kumar R, Zafrullah M. 2001. The ORF3 protein of hepatitis E virus binds to Src homology 3 domains and activates MAPK. J Biol Chem 276:42389–42400. doi: 10.1074/jbc.M101546200. [DOI] [PubMed] [Google Scholar]
- 11.Kannan H, Fan S, Patel D, Bossis I, Zhang YJ. 2009. The hepatitis E virus open reading frame 3 product interacts with microtubules and interferes with their dynamics. J Virol 83:6375–6382. doi: 10.1128/JVI.02571-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zafrullah M, Ozdener MH, Panda SK, Jameel S. 1997. The ORF3 protein of hepatitis E virus is a phosphoprotein that associates with the cytoskeleton. J Virol 71:9045–9053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Graff J, Torian U, Nguyen H, Emerson SU. 2006. A bicistronic subgenomic mRNA encodes both the ORF2 and ORF3 proteins of hepatitis E virus. J Virol 80:5919–5926. doi: 10.1128/JVI.00046-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cao D, Huang YW, Meng XJ. 2010. The nucleotides on the stem-loop RNA structure in the junction region of the hepatitis E virus genome are critical for virus replication. J Virol 84:13040–13044. doi: 10.1128/JVI.01475-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang YW, Opriessnig T, Halbur PG, Meng XJ. 2007. Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J Virol 81:3018–3026. doi: 10.1128/JVI.02259-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Koonin EV, Gorbalenyat AE, Purdy MA, Rozanov MN, Reyes GR, Bradley DW. 1992. Computer-assisted assignment of functional domains in the nonstructural polyprotein of hepatitis E virus: delineation of an additional group of positive-strand RNA plant and animal viruses. Proc Natl Acad Sci U S A 89:8259–8263. doi: 10.1073/pnas.89.17.8259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Magden J, Takeda N, Li T, Auvinen P, Ahola T, Miyamura T. 2001. Virus-specific mRNA capping enzyme encoded by hepatitis E virus. J Virol 75:6249–6255. doi: 10.1128/JVI.75.14.6249-6255.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paliwal D, Panda SK, Kapur N, Varma SP, Durgapal H. 2014. Hepatitis E virus (HEV) protease: a chymotrypsin-like enzyme that processes both non-structural (pORF1) and capsid (pORF2) protein. J Gen Virol 95:1689–1700. doi: 10.1099/vir.0.066142-0. [DOI] [PubMed] [Google Scholar]
- 19.Karpe YA, Lole KS. 2011. Deubiquitination activity associated with hepatitis E virus putative papain-like cysteine protease. J Gen Virol 92:2088–2092. doi: 10.1099/vir.0.033738-0. [DOI] [PubMed] [Google Scholar]
- 20.Karpe YK, Lole KS. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol 84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Karpe YK, Lole KS. 2010. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol 84:9637–9641. doi: 10.1128/JVI.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Agrawal S, Gupta D, Panda SK. 2001. The 3′ end of hepatitis E virus (HEV) genome binds specifically to the viral RNA-dependent RNA polymerase. Virology 282:87–101. doi: 10.1006/viro.2000.0819. [DOI] [PubMed] [Google Scholar]
- 23.Ansari IH, Nanda SK, Durgapal H, Agrawal S, Mohanty SK, Gupta D, Shahid J, Panda SK. 2000. Cloning, sequencing, and expression of the hepatitis E virus (HEV) nonstructural open reading frame 1 (ORF1). J Med Virol 60:275–283. doi:. [DOI] [PubMed] [Google Scholar]
- 24.Ropp SL, Tam AW, Beames B, Purdy M, Frey TK. 2000. Expression of the hepatitis E virus ORF1. Arch Virol 145:1321–1337. doi: 10.1007/s007050070093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sehgal D, Thomas S, Chakraborty M, Jameel S. 2006. Expression and processing of the hepatitis E virus ORF1 nonstructural polyprotein. Virology 3:38. doi: 10.1186/1743-422X-3-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perttila J, Spuul P, Ahola T. 2013. Early secretory pathway localization and lack of processing for hepatitis E virus replication protein pORF1. J Gen Virol 94:807–816. doi: 10.1099/vir.0.049577-0. [DOI] [PubMed] [Google Scholar]
- 27.Suppiah S, Zhou Y, Frey TK. 2011. Lack of processing of the expressed ORF1 gene product of hepatitis E virus. Virology 8:245. doi: 10.1186/1743-422X-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Panda SK, Ansari IH, Durgapal H, Agrawal S, Jameel S. 2000. The in vitro-synthesized RNA from a cDNA clone of hepatitis E virus is infectious. J Virol 74:2430–2437. doi: 10.1128/JVI.74.5.2430-2437.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Yang D, Chong P, Hwang D, Liang Y, Gillam S. 1998. Proteolytic processing of rubella virus nonstructural proteins. Virology 246:74–82. doi: 10.1006/viro.1998.9179. [DOI] [PubMed] [Google Scholar]
- 30.Collins PL, Fuller FJ, Marcus PS, Hightower LE, Ballt LA. 1982. Synthesis and processing of Sindbis virus nonstructural proteins in vitro. Virology 118:363–379. doi: 10.1016/0042-6822(82)90356-7. [DOI] [PubMed] [Google Scholar]
- 31.McLauchlan J, Lemberg MK, Hope G, Martoglio B. 2002. Intramembrane proteolysis promotes trafficking of hepatitis C virus core protein to lipid droplets. EMBO J 21:3980–3988. doi: 10.1093/emboj/cdf414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carrère-Kremer S, Montpellier C, Lorenzo L, Brulin B, Cocquerel L, Belouzard S, Penin F, Dubuisson J. 2004. Regulation of hepatitis C virus polyprotein processing by signal peptidase involves structural determinants at the p7 sequence junctions. J Biol Chem 279:41384–41392. doi: 10.1074/jbc.M406315200. [DOI] [PubMed] [Google Scholar]
- 33.Rachow A, Gauss-Müller V, Probst C. 2003. Homogenous hepatitis A virus particles proteolytic release of the assembly signal 2a from procapsids by factor Xa. J Biol Chem 278:29744–29751. doi: 10.1074/jbc.M300454200. [DOI] [PubMed] [Google Scholar]
- 34.Kalamvoki M, Mavromara P. 2004. Calcium-dependent calpain proteases are implicated in processing of the hepatitis C virus NS5A protein. J Virol 78:11865–11878. doi: 10.1128/JVI.78.21.11865-11878.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Du L, Kao RY, Zhou Y, He Y, Zhao G, Wong C, Jiang S, Yuen K, Jin DY, Zheng BJ. 2007. Cleavage of spike protein of SARS coronavirus by protease factor Xa is associated with viral infectivity. Biochem Biophys Res Commun 359:174–179. doi: 10.1016/j.bbrc.2007.05.092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Palta S, Saroa R, Palta A. 2014. Overview of the coagulation system. Indian J Anaesth 58:515–523. doi: 10.4103/0019-5049.144643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butkowski RJ, Elion J, Downing MR, Mann KG. 1977. Primary structure of human prethrombin 2 and alpha-thrombin. J Biol Chem 252:4942–4957. [PubMed] [Google Scholar]
- 38.Jackson CM, Nemerson Y. 1980. Blood coagulation. Annu Rev Biochem 49:765–811. doi: 10.1146/annurev.bi.49.070180.004001. [DOI] [PubMed] [Google Scholar]
- 39.Licari LG, Kovacic JP. 2009. Thrombin physiology and pathophysiology. J Vet Emerg Crit Care (San Antonio) 19:11–22. doi: 10.1111/j.1476-4431.2009.00383.x. [DOI] [PubMed] [Google Scholar]
- 40.Arai T, Miklossy J, Klegeris A, Guo JP, McGeer PL. 2006. Thrombin and prothrombin are expressed by neurons and glial cells and accumulate in neurofibrillary tangles in Alzheimer disease brain. J Neuropathol Exp Neurol 65:19–25. doi: 10.1097/01.jnen.0000196133.74087.cb. [DOI] [PubMed] [Google Scholar]
- 41.Crawley JT, Zanardelli S, Chion CK, Lane DA. 2007. The central role of thrombin in hemostasis. J Thromb Haemost 5(Suppl 1):95–101. doi: 10.1111/j.1538-7836.2007.02500.x. [DOI] [PubMed] [Google Scholar]
- 42.Jenny RJ, Mann KG, Lundblad RL. 2003. A critical review of the methods for cleavage of fusion proteins with thrombin and factor Xa. Protein Expr Purif 31:1–11. doi: 10.1016/S1046-5928(03)00168-2. [DOI] [PubMed] [Google Scholar]
- 43.Fenton JW, Fasco MJ, Stackrow AB. 1977. Human thrombins. Production, evaluation, and properties of alpha-thrombin. J Biol Chem 252:3587–3598. [PubMed] [Google Scholar]
- 44.Tautz N, Tews BA, Meyers G. 2015. The molecular biology of pestiviruses. Adv Virus Res 93:47–160. doi: 10.1016/bs.aivir.2015.03.002. [DOI] [PubMed] [Google Scholar]
- 45.Blitvich BJ, Firth AE. 2015. Insect-specific flaviviruses: a systematic review of their discovery, host range, mode of transmission, superinfection exclusion potential and genomic organization. Viruses 7:1927–1959. doi: 10.3390/v7041927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rupp JC, Sokoloski KJ, Gebhart NN, Hardy RW. 2015. Alphavirus RNA synthesis and non-structural protein functions. J Gen Virol 96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Marr LD, Wang CY, Frey TK. 1994. Expression of the rubella virus non-structural protein ORF and demonstration of proteolytic processing. Virology 198:586–592. doi: 10.1006/viro.1994.1070. [DOI] [PubMed] [Google Scholar]
- 48.Liang Y, Gillam S. 2000. Mutational analysis of the rubella virus nonstructural polyprotein and its cleavage products in virus replication and RNA synthesis. J Virol 74:5133–5141. doi: 10.1128/JVI.74.11.5133-5141.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shirako Y, Strauss JH. 1994. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol 68:1874–1885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zetter BR, Chen LB, Buchanan JM. 1977. Binding and internalization of thrombin by normal and transformed chick cells. Proc Natl Acad Sci U S A 74:596–600. doi: 10.1073/pnas.74.2.596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lollar P, Hoak JC, Owen WG. 1980. Binding of thrombin to cultured human endothelial cells. J Biol Chem 255:10279–10283. [PubMed] [Google Scholar]
- 52.Ratra R, Kar-Roy A, Lal SK. 2009. ORF3 protein of hepatitis E virus interacts with the Bb chain of fibrinogen resulting in decreased fibrinogen secretion from HuH-7 cells. J Gen Virol 90:1359–1370. doi: 10.1099/vir.0.009274-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.