ABSTRACT
In previous studies, we showed that the cellular RNA-binding protein AUF1 supports the replication process of the flavivirus West Nile virus. Here we demonstrate that the protein also enables effective proliferation of dengue virus and Zika virus, indicating that AUF1 is a general flavivirus host factor. Further studies demonstrated that the AUF1 isoform p45 significantly stimulates the initiation of viral RNA replication and that the protein's RNA chaperone activity enhances the interactions of the viral 5′UAR and 3′UAR genome cyclization sequences. Most interestingly, we observed that AUF1 p45 destabilizes not only the 3′-terminal stem-loop (3′SL) but also 5′-terminal stem-loop B (SLB) of the viral genome. RNA structure analyses revealed that AUF1 p45 increases the accessibility of defined nucleotides within the 3′SL and SLB and, in this way, exposes both UAR cyclization elements. Conversely, AUF1 p45 does not modulate the fold of stem-loop A (SLA) at the immediate genomic 5′ end, which is proposed to function as a promoter of the viral RNA-dependent RNA polymerase (RdRp). These findings suggest that AUF1 p45, by destabilizing specific stem-loop structures within the 5′ and 3′ ends of the flaviviral genome, assists genome cyclization and concurrently enables the RdRp to initiate RNA synthesis. Our study thus highlights the role of a cellular RNA-binding protein inducing a flaviviral RNA switch that is crucial for viral replication.
IMPORTANCE The genus Flavivirus within the Flaviviridae family includes important human pathogens, such as dengue, West Nile, and Zika viruses. The initiation of replication of the flaviviral RNA genome requires a transformation from a linear to a cyclized form. This involves considerable structural reorganization of several RNA motifs at the genomic 5′ and 3′ ends. Specifically, it needs a melting of stem structures to expose complementary 5′ and 3′ cyclization elements to enable their annealing during cyclization. Here we show that a cellular RNA chaperone, AUF1 p45, which supports the replication of all three aforementioned flaviviruses, specifically rearranges stem structures at both ends of the viral genome and in this way permits 5′-3′ interactions of cyclization elements. Thus, AUF1 p45 triggers the RNA switch in the flaviviral genome that is crucial for viral replication. These findings represent an important example of how cellular (host) factors promote the propagation of RNA viruses.
KEYWORDS: AUF1, dengue virus, flavivirus, RNA chaperone, RNA cyclization, RNA-protein interaction, RNA replication, West Nile virus, Zika virus, host factor
INTRODUCTION
Dengue virus (DENV) is one of the most important viral human pathogens, annually infecting about 400 million people (1). Clinical manifestations of DENV infections range from asymptomatic or cold-like disease to severe dengue hemorrhagic fever and shock syndrome. Along with other human pathogens, such as West Nile virus (WNV), Japanese encephalitis virus (JEV), and tick-borne encephalitis virus (TBEV), DENV belongs to the genus Flavivirus within the Flaviviridae family. Another Flavivirus member, Zika virus (ZIKV), has gained recent global attention due to the linkage of ZIKV infections to severe neurological disorders, with several outbreaks in Micronesia, the South Pacific, and the Americas (2).
The Flavivirus genome is an approximately 11-kb-long, single-stranded RNA of positive polarity. Following entry and uncoating, the viral RNA is directly translated in the cytoplasm of the infected host cell, generating three structural (capsid [C], precursor of membrane [prM], and envelope [E]) and seven nonstructural (NS1, NS2A, NS2B, NS3, NS4A, NS4B, and NS5) proteins. Viral RNA replication occurs in distinct virus-induced organelle-like membrane structures (viral replication factories) in the cytoplasm. In the first step, negative-strand intermediates are synthesized, which then serve as templates for the generation of progeny positive-strand genomes (3–5). Replication initiates by specific binding of the viral RNA-dependent RNA polymerase (RdRp) NS5 to a promoter element, stem-loop A (SLA), which is formed by the 5′ end of the flaviviral genome (see Fig. 2B and 4). Thus, an essential prerequisite to enable the RdRp to start negative-strand RNA synthesis at the genomic 3′ end is a 5′-3′ cyclization of the viral RNA (6). Genome cyclization involves interactions of complementary cyclization sequences, termed CS, UAR, and DAR sequences, as well as considerable structural rearrangements of the 5′ and 3′ termini of the viral RNA (see Fig. 2B and 7) (reviewed in references 7 to 10).
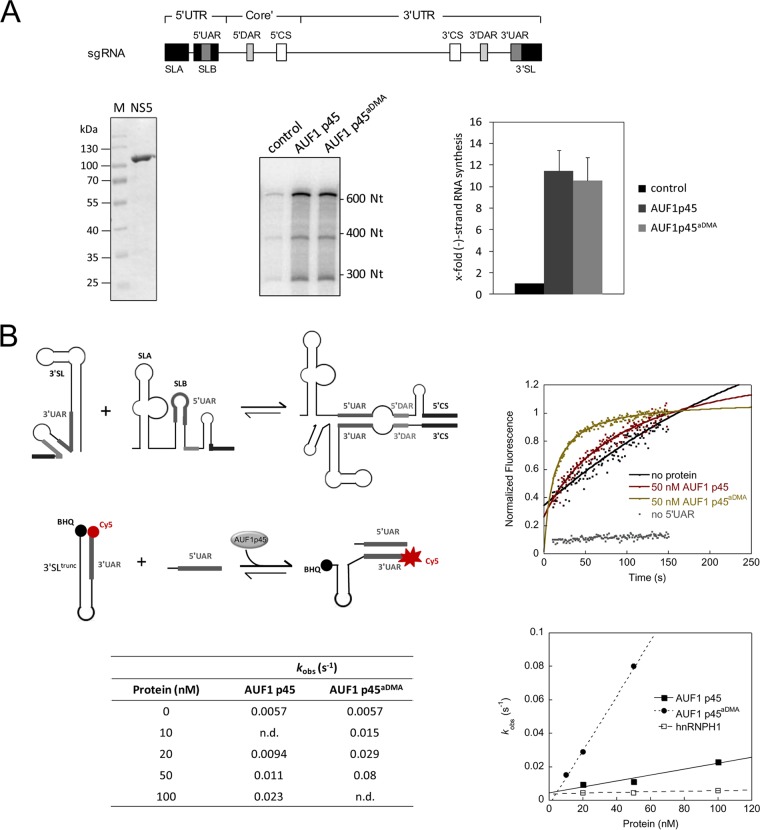
FIG 2.
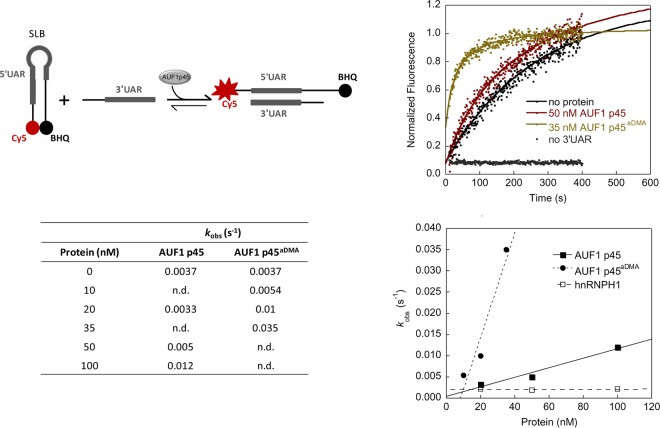
DENV RNA synthesis and 5′-3′ RNA-RNA interaction are stimulated by AUF1 p45. (A) (Top) Schematic representation of organization of the DENV sgRNA (635 nt) used in the assay. The RNA consists of the 5′UTR, 3′UTR, and a part of the core coding sequence. SLA, stem-loop A; SLB, stem-loop B; 3′SL, 3′stem-loop; CS, conserved sequence; UAR, upstream AUG region; DAR, downstream AUG region. (Bottom left) Degree of purity of E. coli-purified DENV NS5 used in the assay. Aliquots of 3 μg were analyzed in a Coomassie-stained SDS gel in parallel with a molecular marker (M). (Bottom middle) In vitro replicase assay with DENV sgRNA and NS5 protein. The assay was performed in the absence (control) or presence of 100 nM AUF1 p45 or AUF1 p45aDMA. The radiolabeled RNA products were analyzed by denaturing 5% PAGE. (Bottom right) Quantitative analysis of de novo RNA products of the replicase assay. The control reaction value was set to 1. Error bars reflect standard deviations (n = 3). (B) (Top left) Scheme of the structural rearrangement of the 5′ and 3′ termini, specifically of the 5′UAR and 3′UAR elements, during cyclization of the DENV RNA genome. (Middle left) Scheme of the fluorescence-based 3′SLtrunc-5′UAR interaction assay to detect AUF1 p45-mediated conformational rearrangements of DENV RNA by dequenching of Cy5. (Top right) The assay was performed with labeled 3′SLtrunc incubated with the indicated concentrations of recombinant AUF1 p45 or AUF1 p45aDMA. Following the addition of 5′UAR RNA, the fluorescence signals were measured, plotted as a function of time, and fitted according to a first-order reaction (no protein; equation 1) or second-order reaction (in the presence of protein; equation 2). When we performed the assay in the presence of 50 nM AUF1 p45 but without the complementary 5′UAR RNA, an increase of fluorescence was not detected. (Bottom left) Observed rate constants (kobs) for the DENV 3=SLtrunc-5=UAR interaction in the absence and presence of different concentrations of AUF1 p45 and AUF1 p45aDMA. (Bottom right) The observed rate constants (kobs) that were measured for the RNA-RNA interaction in the presence of the indicated concentrations of recombinant AUF1 p45, AUF1 p45aDMA, or a control protein, hnRNPH1, were plotted as a function of protein concentration.
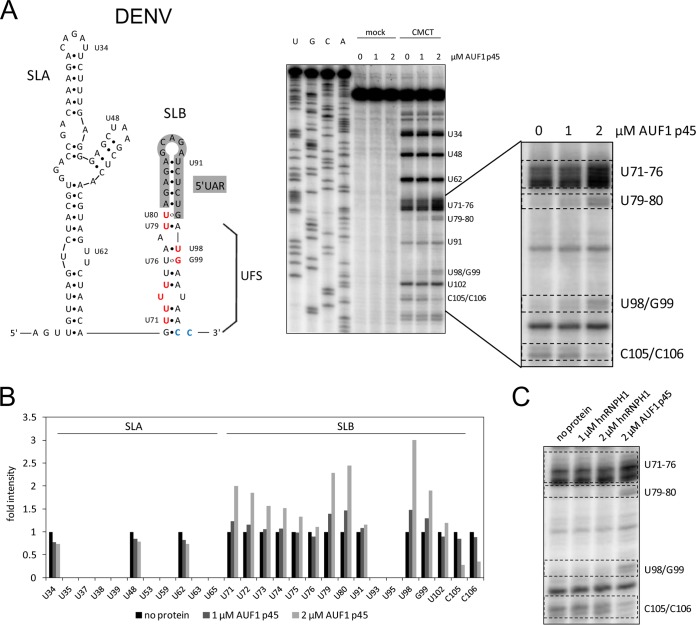
FIG 4.
AUF1 p45 specifically destabilizes SLB within the DENV 5′UTR. (A) (Left) Secondary structures of 5′-terminal stem-loop A (SLA) and stem-loop B (SLB) of the DENV genome. Nucleotide position numbering starts at the 5′ end of the RNA. The 5′UAR-flanking stem (UFS) is indicated. Nucleotides that are exposed by AUF1 p45 are highlighted in red. Nucleotides that are less well modified by CMCT (modifies U and G residues in single-stranded RNA) in the presence of AUF1 p45 are highlighted in blue. (Right) RNA structure probing of the DENV 5′ end by CMCT in the presence of the indicated concentrations of AUF1 p45 (lanes 8 to 10). Control reaction mixtures in CMCT buffer were included (lanes 5 to 7). Products of the primer extension reaction, carried out with a radiolabeled primer, were analyzed by 8% denaturing PAGE along with a sequencing ladder (lanes 1 to 4). Note that due to the stop of the reverse transcriptase upon encountering alkylated Watson-Crick positions, the resulting product is one base shorter. One region of the gel is enlarged on the right. (B) Quantification of the primer extension analysis shown in panel A. (C) RNA structure probing of the DENV 5′ end with CMCT in the presence of the indicated concentrations of AUF1 p45 and hnRNPH1. One region of the gel where we observed a change of the secondary structure in the presence of AUF1 p45 is shown (see panel A).
FIG 7.
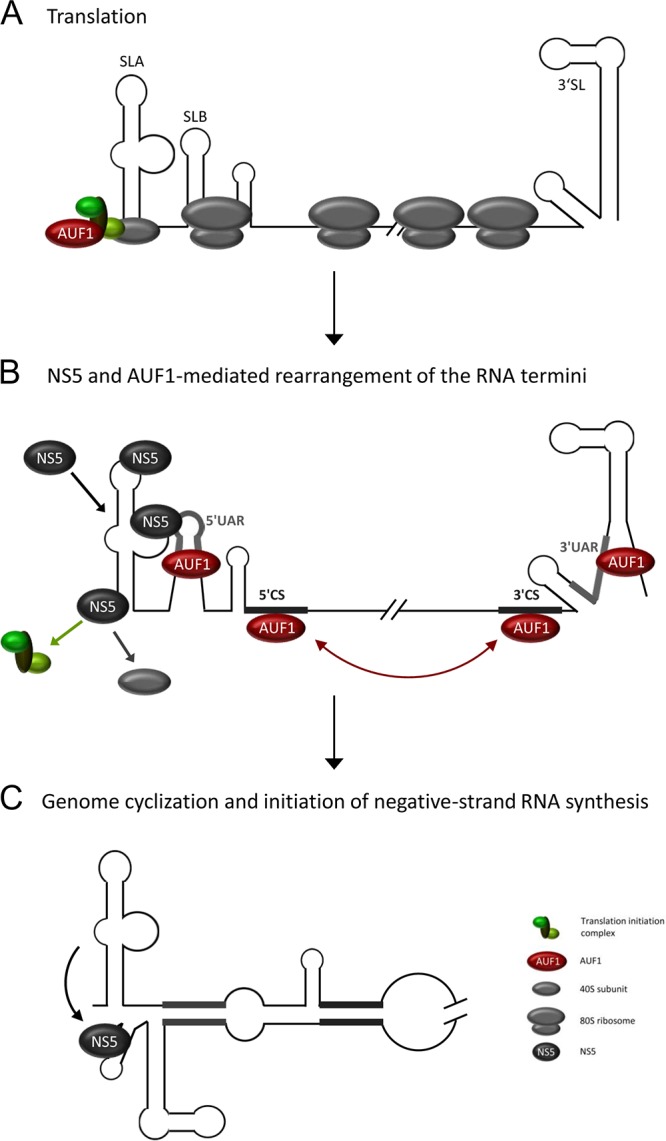
Model of AUF1's role during flavivirus genome cyclization. (A) Ribosomes (depicted in gray) engage in active translation of the flavivirus genome following the release of the viral RNA into the cytoplasm of the host cell. AUF1 (depicted in red) interacts with proteins of the translation initiation apparatus (depicted in different shades of green), e.g., eIF4G. (B) As a result of translation, the local concentration of the viral protein NS5 increases (depicted in blue), and this protein displaces the translation initiation factors at the 5′ end of the RNA. NS5 and AUF1 trigger a transition from translating mRNA to a replication template for negative-strand RNA synthesis. AUF1 binds 5′CS and 3′CS cyclization sequences and promotes RNA hybridization. AUF1 destabilizes SLB and 3′SL and exposes the 5′UAR and 3′UAR cyclization sequences. (C) The RNA switch from linear to circular, including the rearrangement of SLB, decreases the affinity of NS5 for SLA, and NS5 initiates negative-strand RNA synthesis at the 3′ end of the viral RNA.
The genomes of most RNA viruses encode only a small number of proteins; accordingly, viral replication often requires the additional activity of cellular factors. Several RNA-binding proteins (RBPs) have been implicated in interacting with flaviviral genomes and acting as host factors during RNA replication (reviewed in reference 11). Among them is AU-rich element-binding protein 1 (AUF1; also known as hnRNPD), whose canonical function involves the regulation of the stability and/or translation of mRNA targets, based on the recognition of AU-rich sequences within the mRNA's 3′ untranslated regions (3′UTR) (12, 13). Four AUF1 isoforms (p37, p40, p42, and p45) are generated by alternative splicing from a common pre-mRNA, which results in isoform-specific sequences encoded by exons 2 and 7 (14). While for most RBPs the mechanisms for how these proteins support flaviviral RNA replication are unknown, we provided an initial functional model for the host factor activity of the largest AUF1 isoform, AUF1 p45, based on studies with WNV. On the one hand, we could show that AUF1 p45 has an RNA chaperone activity that is suggested to assist the protein to destabilize the WNV 3′ stem-loop structure (3′SL) and in this way to promote structural rearrangements of the WNV genome (15). On the other hand, AUF1 p45 has an RNA annealing activity which enables the protein to hybridize complementary single-stranded RNA molecules. Thus, AUF1 p45 was proposed to accelerate interactions of the WNV 5′UAR-3′UAR and 5′CS-3′CS cyclization sequences (16). In mammalian cells, AUF1 p45 is frequently modified by methylation at five arginine residues within an arginine- and glycine-rich region (RGG/RG motif). The methylated protein variant, termed AUF1 p45aDMA, shows a considerably increased RNA chaperone activity, while its RNA annealing activity is unchanged in comparison to that of the nonmethylated protein (16).
In the present study, we show that AUF1 p45 also propagates DENV and ZIKV replication, indicating that this RBP is a general host factor for mosquito-borne flaviviruses. Moreover, we demonstrate that AUF1 p45 induces specific restructuring events at both ends of the viral RNA that expose the UAR cyclization elements and in this way triggers the flaviviral RNA switch during genome cyclization.
RESULTS
AUF1 supports effective propagation of DENV and ZIKV in human cells.
Strong hints indicating a host factor function for AUF1 in the WNV replication process were obtained when we performed a small interfering RNA (siRNA)-mediated depletion (knockdown) of AUF1 and observed that viral propagation was markedly inhibited in the depleted cells (15). Accordingly, to test whether AUF1 has a similar role in DENV and ZIKV infections, we applied the established knockdown protocol (15). Mock-depleted and AUF1-depleted Huh7 cells were infected with DENV type 2 or ZIKV strain PF13, and the progress of infection was followed by immunofluorescence (IF) staining of the viral protein NS5 and/or by measuring the amounts of viral RNA in the culture supernatants. In the case of DENV infection, IF staining of NS5 revealed that in comparison to those of the control cells, which showed continuous viral spread between 72 and 120 h postinfection (p.i.), the AUF1-depleted cells displayed significantly lower infection rates (Fig. 1A). These data were confirmed when we measured the viral titers in the supernatants of the infected cells at different time points. For example, at 96 h p.i., the DENV RNA titer was about 10 times lower in the culture fluids of AUF1-depleted cells than in those of the controls (Fig. 1A). A similar picture was obtained when we analyzed the ZIKV infection experiments. In this case, at 48 h p.i., the amount of ZIKV RNA molecules was about 10 times smaller in the supernatants of AUF1-depleted cells than in those of mock-depleted cells (Fig. 1B). These experiments suggested that AUF1 is a host factor that strongly enhances the replication of different flaviviruses.
FIG 1.
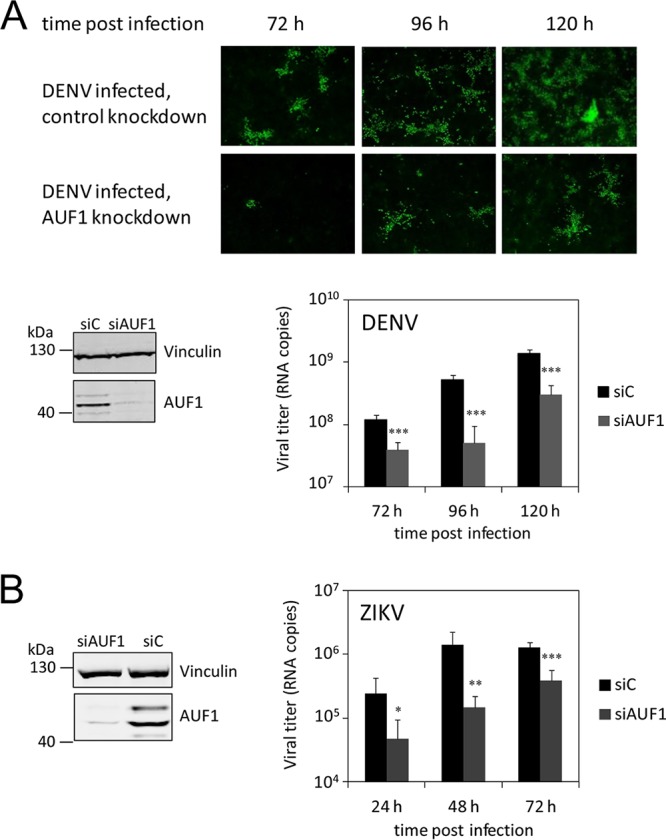
RNA interference (RNAi)-mediated depletion of AUF1 inhibits DENV and ZIKV propagation. (A) Huh7 cells were treated with a control siRNA (siC) and a siRNA targeting AUF1 mRNA (siAUF1). After 72 h, the cytoplasmic depletion of AUF1 was confirmed by Western blotting (bottom left), and the cells were infected with DENV. At the indicated time points, the expression of NS5 was analyzed by indirect immunofluorescence analysis (top), and culture fluids were assayed for virus titers by qRT-PCR (bottom right). Average results and standard deviations (n = 4 [two independent experiments performed in duplicate]) are shown. ***, P ≤ 0.001. (B) Same as panel A, but cells were infected with ZIKV. Culture fluids were assayed for virus titers by qRT-PCR. Average results and standard deviations (n = 6 [two independent experiments performed in triplicate]) are shown. *, P ≤ 0.05; **, P ≤ 0.01; ***, P ≤ 0.001.
AUF1 p45 supports DENV negative-strand RNA synthesis by promoting structural rearrangements of the viral RNA.
To determine whether AUF1 p45, in analogy to the previous observations with WNV, affected the first step of the DENV replication process, we performed a replicase assay that applied the purified viral polymerase NS5 and a subgenomic RNA (sgRNA) as a substrate. The DENV sgRNA included the 5′-terminal 184 nucleotides (nt) as well as the 3′-terminal 451 nt and thus contained all RNA elements known to be essential for initiation of de novo synthesis of negative-strand RNA by the viral replicase (Fig. 2A) (15, 17, 18). As also shown in Fig. 2A, these experiments yielded a dominant, newly synthesized RNA product in the applied gel system; since it migrated close to the 600-nt marker, we concluded that it corresponded to the full-length negative-strand copy of the DENV sgRNA. Additional, faster-migrating RNA species were assumed to represent either incompletely processed negative-strand molecules or hairpin by-products that were generated by priming and subsequent “copyback” of the replicase at the template's 3′ end (17). Note that during previous work we could show that RNA synthesis was undetectable when we performed the assay in the presence of only one nucleotide or with a DNA template (15). Importantly, when we supplemented the reaction mixture with AUF1 p45 purified from Escherichia coli, the NS5-mediated DENV RNA synthesis was markedly increased; this was also the case when we applied the methylated variant of the protein, AUF1 p45aDMA (Fig. 2A). These findings demonstrate that AUF1 p45 significantly promotes DENV negative-strand RNA synthesis.
In previously performed experiments with unrelated and WNV-derived RNA substrates, we could show that AUF1 p45 does not directly stimulate the viral RdRp but specifically modifies the structure of the viral RNA to improve the efficiency of negative-strand RNA synthesis (15). Accordingly, we next wanted to know whether and how AUF1 p45 is capable of promoting 5′-3′ interactions of the DENV genome. As an example, we investigated the interaction of the 5′UAR and 3′UAR cyclization sequences. This interaction requires considerable restructuring of the viral genome, as in the linear form the 5′UAR sequence is part of stem-loop B (SLB), while the 3′UAR sequence is part of the stem structure of the 3′SL (Fig. 2B; also see Fig. 7). Accordingly, to enable 5′UAR-3′UAR hybridization, both stem-loop motifs have to be destabilized or even denatured. To measure such an AUF1 p45-promoted restructuring of the 3′SL, we took advantage of a fluorescence-based assay (15). This assay applied an RNA oligonucleotide (3′SLtrunc) which mainly corresponded to the double-stranded, lower portion of the DENV 3′SL, which includes the 3′UAR sequence. The 3′SLtrunc-RNA was 5′ labeled with Cy5, while the 3′ end was associated with the black hole quencher (BHQ) dye. Thus, if the 3′SLtrunc-RNA formed a stem, the Cy5 fluorescence would be quenched due to the close proximity of BHQ (Fig. 2B). The second RNA used in the assay contained a portion of the DENV 5′UTR comprising the 5′UAR sequence and was unlabeled. (RNA sequences are summarized in Table S3 in the supplemental material.) Hybridization of both RNAs via the UAR elements requires a rearrangement of the 3′SLtrunc-RNA's stem structure, which dissociates the Cy5 fluorophore from BHQ, and the increasing fluorescence can be followed over time.
Measurements of the 5′UAR-3′UAR interactions revealed that these occurred only slowly in the absence of protein (Fig. 2B). Yet when we added nonmethylated or methylated AUF1 p45 to the reaction mixture, we observed a significant acceleration of the RNA-RNA hybridization, which was most apparent with AUF1 p45aDMA (Fig. 2B). That is, in the presence of 50 nM AUF1 p45, the rate constant was increased ca. 2-fold compared to that of the RNA-only reaction; in the presence of 50 nM AUF1 p45aDMA, we measured a ca. 14-fold increase (Fig. 2B). An acceleration of the 3′SLtrunc-5′UAR reaction was not detected when we performed the assay in the presence of the RBP hnRNPH1, which implicated a specific modulation of the viral cyclization elements by AUF1 p45 (Fig. 2B). The results demonstrated that the RNA chaperone activity of AUF1 p45 leads to a destabilization of the 3′SL of the DENV RNA and, in this manner, enhances 5′-3′ RNA-RNA interactions. In analogy to our previous findings, the destabilizing activity of AUF1 p45 on the DENV RNA was shown to be enhanced by arginine methylation of the protein.
AUF1 destabilizes the 3′SL as well as 5′SLB of DENV RNA.
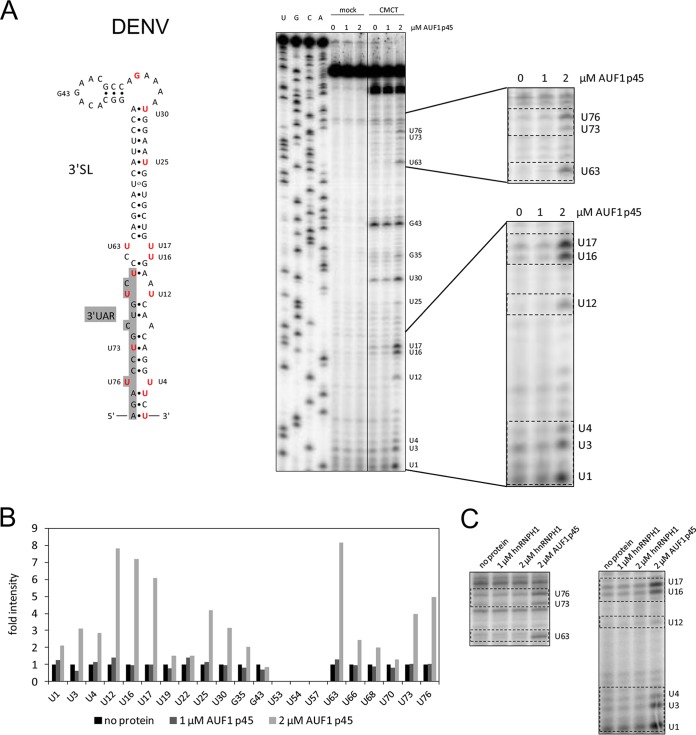
To characterize the destabilizing activity of AUF1 p45 with the DENV 3′SL in further detail, we performed chemical modification experiments. For this purpose, a transcript comprising the 3′SL of the DENV RNA was treated with the methylating agent 1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate (CMCT). CMCT specifically modifies unpaired U residues at N-3 and, to a lesser extent, G residues at N-1 (19). The sites of CMCT modification were determined by primer extension in parallel with a DNA sequencing reaction. As shown in Fig. 3A, the obtained U/G modification pattern was in close correspondence with a previously experimentally verified DENV 3′SL structure (20). However, the bulges in the lower part of the 3′SL, which include several unpaired U residues (U4/U76 and U12/U68), were observed to be modified only poorly by CMCT (Fig. 3A). Interestingly, this situation was very different when we performed structure probing in the presence of AUF1 p45. That is, several U residues in the bottom part of the 3′SL which localize within or close to the 3′UAR sequence and the complementary strand (U76, U73, U63, U17, U16, U12, U4, U3, and U1) (Fig. 3A, highlighted in red) turned out to be more accessible to chemical modification in the presence of high concentrations of AUF1 p45 (Fig. 3B). Experiments that were done with another RBP, hnRNPH1, revealed no changes of the RNA structure (Fig. 3C). Accordingly, these data directly visualized the RNA chaperone activity of AUF1 p45 on the 3′SL substrate: in particular, by destabilizing the bottom portion of the 3′SL, AUF1 p45 apparently increases the accessibility of the 3′UAR sequence for base pairing with the 5′UAR sequence.
FIG 3.
AUF1 p45 increases the accessibility of U residues predominantly in the bottom part of the DENV 3′SL. (A) (Left) Secondary structure of the 3′-terminal stem-loop (3′SL) of the DENV genome. Nucleotide position numbering starts at the 3′ end of the RNA. Nucleotides that were exposed in the presence of AUF1 p45 are highlighted in red. (Right) RNA structure probing of the DENV 3′SL with CMCT (modifies U and G residues in single-stranded RNA) in the presence of the indicated concentrations of AUF1 p45 (lanes 8 to 10). Control reaction mixtures in CMCT buffer were included (lanes 5 to 7). Products of the primer extension reaction, carried out with a radiolabeled primer, were analyzed by 8% denaturing PAGE along with a sequencing ladder (lanes 1 to 4). Note that due to the stop of the reverse transcriptase upon encountering alkylated Watson-Crick positions, the resulting product is one base shorter. Two regions of the gel are enlarged on the right. (B) Quantification of cDNA products of the primer extension analysis from panel A. (C) RNA structure probing of the DENV 3′SL with CMCT in the presence of the indicated concentrations of AUF1 p45 and hnRNPH1. Two regions of the gel where we observed changes of the secondary structure in the presence of AUF1 p45 are shown (see panel A).
A further important question was whether AUF1 p45 is also able to destabilize stem-loop B (SLB), which is folded by the 5′ end of the linear form of the flaviviral RNA. As outlined above, SLB encloses the 5′UAR cyclization sequence and needs to be restructured to enable 5′UAR-3′UAR hybridization during genome cyclization (Fig. 2B and 4A). To address this, we performed CMCT structure probing of a transcript that contained the DENV RNA's 5′ end, including the SLA as well as SLB sequences. Again, the CMCT modification data on the 5′ end of the DENV RNA in the absence of protein were in strong agreement with previous reports (21–23). Most interestingly, and comparable to the situation with the 3′SL, we observed the SLB RNA structure to be noticeably changed in the presence of AUF1 p45. That is, several U and G residues which are located within the bottom part of SLB (U71 to U74 and U98/G99), but also within and close to the 5′UAR sequence (U79-U80) (Fig. 4A, highlighted in red), were significantly more susceptible to CMCT modification (Fig. 4B). Besides this, two C residues, at positions 105 and 106, showed a markedly reduced accessibility to CMCT when AUF1 p45 was present (Fig. 4A [highlighted in blue] and B). In contrast, the presence of hnRNPH1 did not change the accessibility of the RNA to CMCT modification (Fig. 4C).
The observed protection of C105 and C106 suggested a potential binding site for AUF1 p45 operating on the DENV SLB. Another interesting observation showed that in contrast to that of SLB, the conformation of the neighboring NS5 promoter element SLA was not rearranged by AUF1 p45 (Fig. 4). Hence, these data indicated an evident preference of the RNA chaperone activity of AUF1 p45 to target flaviviral RNA motifs which are involved in the cyclization process (see Discussion).
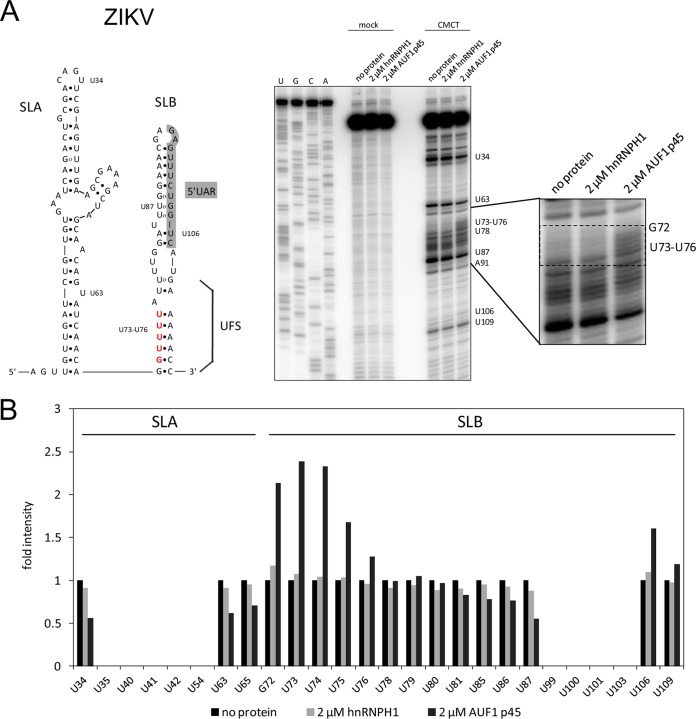
The final experiments of this study addressed two issues. On the one hand, we wanted to determine whether these observations with the DENV SLB were also applicable to slightly different flaviviral SLBs, namely, those of ZIKV and WNV. On the other hand, we were interested in measuring the kinetics of the destabilizing activity of AUF1 p45 on SLB. First, we performed CMCT structure probing by using an RNA that comprised the ZIKV SLA and SLB structures (Fig. 5A). Interestingly, in analogy to the situation with the DENV SLB, we observed the ZIKV SLB RNA structure to be noticeably changed in the presence of AUF1 p45. That is, several U and G residues which are located within the bottom part of SLB (G72 and U73 to U76) (Fig. 5A, highlighted in red) were significantly more susceptible to CMCT modification. This was not the case when the reaction mixture was analogously complemented with hnRNPH1 (Fig. 5A and B). Second, we adapted the fluorescence-based RNA-RNA interaction assay described above to the WNV SLB by using a Cy5- and BHQ-labeled WNV SLB and an unlabeled WNV 3′UAR sequence (Fig. 6). Again, in the absence of protein, the 5′UAR-3′UAR interactions occurred rather slowly. However, when we complemented the reaction mixture with nonmethylated or methylated AUF1 p45, we observed a significant acceleration of the RNA-RNA interactions. As in the former experiments, AUF1 p45aDMA showed a significantly higher activity than that of the nonmethylated protein (Fig. 6). For example, with 100 nM AUF1 p45 the observed rate constant (kobs) was increased 3-fold compared to that for the RNA-only reaction, while in the presence of 35 nM AUF1 p45aDMA it was increased 9-fold (Fig. 6). Experiments that were performed in the presence of the RBP hnRNPH1 showed no acceleration of the WNV SLB-3′UAR interactions (Fig. 6). Taken together, these results demonstrated that AUF1 p45, in particular the methylated form of the protein, has the capability of inducing conformational changes and destabilizing stem-loop structures in the 5′ as well as 3′ end of the flaviviral RNA. This enhances 5′-3′ interactions of the viral RNA and supports the initiation of negative-strand RNA synthesis.
FIG 5.
AUF1 p45 specifically destabilizes SLB within the ZIKV 5′UTR. (A) (Left) Secondary structures of 5′-terminal stem-loop A (SLA) and stem-loop B (SLB) of the ZIKV genome. Nucleotide position numbering starts at the 5′ end of the RNA. The 5′UAR-flanking stem (UFS) is indicated. Nucleotides that are exposed by AUF1 p45 are highlighted in red. (Right) RNA structure probing of the ZIKV 5′ end by CMCT in the presence of the indicated concentrations of AUF1 p45 or hnRNPH1 (lanes 8 to 10). Control reaction mixtures in CMCT buffer were included (lanes 5 to 7). Products of the primer extension reaction, carried out with a radiolabeled primer, were analyzed by 8% denaturing PAGE along with a sequencing ladder (lanes 1 to 4). Note that due to the stop of the reverse transcriptase upon encountering alkylated Watson-Crick positions, the resulting product is one base shorter. One region of the gel is enlarged on the right. (B) Quantification of the primer extension analysis shown in panel A.
FIG 6.
Stem-loop B (SLB) of WNV is destabilized by AUF1 p45. (Top left) Scheme of fluorescence-based assay to detect AUF1 p45-mediated conformational rearrangement of WNV SLB by dequenching of Cy5. (Top right) The assay was performed with Cy5- and BHQ-labeled SLB incubated with the indicated concentrations of recombinant AUF1 p45 or AUF1 p45aDMA. Following the addition of 3′UAR RNA, the fluorescence signals were measured, plotted as a function of time, and fitted according to a first-order reaction (no protein; equation 1) or second-order reaction (in the presence of protein; equation 2). When we performed the assay in the presence of 50 nM AUF1 p45 but without the complementary 3′UAR RNA, an increase of fluorescence was not detected. (Bottom left) Observed rate constants (kobs) for the WNV 5′SLB-3′UAR interaction in the absence and presence of different concentrations of AUF1 p45 and AUF1 p45aDMA. (Bottom right) The observed rate constants (kobs) that were measured for the RNA-RNA interaction in the presence of the indicated concentrations of recombinant AUF1 p45, AUF1 p45aDMA, or a control protein, hnRNPH1, were plotted as a function of protein concentration.
DISCUSSION
Cellular as well as viral RNA-binding proteins (RBPs) play crucial roles in the propagation of RNA viruses. However, a better understanding of the specific activities of these proteins is just beginning to emerge. An important aspect in this regard is that some RBPs not only bind to the RNA target but also function as RNA chaperones that promote the folding of RNAs and/or facilitate RNA-RNA interaction in an ATP-independent manner (24, 25).
The switch that the positive-strand viral genomic RNA undergoes from a translatable mRNA to a template for negative-strand RNA synthesis during replication is a decisive step in the flaviviral life cycle. While the mechanism of this RNA switch is not yet fully understood, it is well established that genome cyclization via complementary sequences within the 5′ and 3′ ends of the viral RNA represents a key event (6, 26–29). The capsid protein and the NS3 helicase of the virus were suggested to function as RNA chaperones which promote the transformation of the linear to the circular conformation of the viral RNA genome (30, 31). In addition, AUF1 p45, a cellular RNA chaperone, was shown to support WNV replication by adopting RNA chaperone and RNA annealing activities, both of which contribute considerably to the genome cyclization process (15, 16).
Here we show that cellular depletion of AUF1 inhibits DENV and ZIKV replication, suggesting that the protein acts as a universal flavivirus host factor. Our data are in agreement with the finding that AUF1 is one of the DENV RNA-interacting proteins identified by quantitative mass spectrometry-based proteomic analysis (qTUX-MS) during live infection of human cells (S. Thompson, personal communication). We obtained a series of data with DENV, WNV, and ZIKV, which substantially improved our understanding of the host factor's RNA chaperone activity. Thus, we can now explain that AUF1 p45 stimulates negative-strand RNA synthesis by destabilizing two conserved RNA structures, located within the viral genomic 5′ and 3′ ends. On the one hand, the RNA chaperone activity of AUF1 p45 was demonstrated to induce defined structural changes within the 3′SL motif, which exposes the 3′UAR cyclization sequence. On the other hand, we found that AUF1 p45 destabilizes the SLB motif within the 5′UTR and in that way exposes the 5′UAR cyclization sequence. Accordingly, the rearrangement of the 3′SL and 5′SLB by AUF1 p45 enables base pairing of the complementary 5′UAR and 3′UAR sequences as a prerequisite for flaviviral genome cyclization. Notably, the protein has no effect on the structure of the SLA motif, suggesting that AUF1 p45 targets specifically those flaviviral RNA motifs that are involved in the cyclization process.
Our observations are consistent with recent findings on DENV4, by Liu et al., showing that an increase of the stability of the AU-rich stem within the bottom part of SLB, named the 5′UAR-flanking stem (UFS) (Fig. 4A), negatively affects viral RNA replication (23). Intriguingly, this report further revealed that the UFS element is also crucial for the recruitment of NS5 to the SLA promoter and that unwinding of SLB, including the UFS, decreases the affinity of NS5 for the viral genome's 5′ end (23). Accordingly, SLB not only is involved in the cyclization process but also participates in the recruitment of the viral RdRp to the genome's 5′ end and in translocation of the RdRp to the 3′ end. Considering the high affinity of AUF1 for AU-rich and U/GU-rich sequences (32), we hypothesize that while it interacts with this region, AUF1 may trigger the RNA switch that regulates the translocation of the RdRp to the genome's 3′ end. Filomatori et al. previously showed that binding of NS5 to the SLA element induces conformational changes downstream of the promoter, i.e., within the same region that we found here to be exposed upon AUF1 p45 binding. As these structural changes were proposed to render the SLA-NS5 complex competent for RNA synthesis (33), this supports the idea that NS5 and AUF1 p45 act in concert to induce multiple structural changes within the flaviviral RNA that promote genome cyclization and subsequent initiation of negative-strand RNA synthesis.
Based on a previous model of flavivirus replication (34), we propose the following role for AUF1 p45. One of the canonical functions of AUF1 is the control of mRNA translation via the recognition of AU-rich elements within the mRNA's 3′UTR (13). AUF1 has been shown to interact with proteins of the translation apparatus, such as the translation initiation factor eIF4G and poly(A)-binding protein (PABP) (35, 36). Therefore, it is conceivable that AUF1 contacts the flaviviral RNA during translation initiation (Fig. 7A). While translation is occurring, the local concentrations of viral proteins, such as NS5, increase (Fig. 7B). The high affinity of NS5 for SLA (33) then leads to the displacement of translation initiation factors, while AUF1 p45 triggers genome cyclization by its RNA remodeling activities. AUF1 thus destabilizes the 5′SLB and 3′SL, exposing the 5′UAR and 3′UAR cyclization sequences, respectively, through its RNA chaperone activity (Fig. 7B) (15; this study). Furthermore, AUF1 accelerates the hybridization of the conserved 5′CS and 3′CS cyclization sequences via its RNA annealing activity (Fig. 7B) (16). The RNA switch from a linear to a circular structure, including the rearrangement of SLB, decreases the affinity of NS5 for SLA, and NS5 initiates negative-strand RNA synthesis at the 3′ end of the viral RNA (Fig. 7C) (23).
Our study demonstrates how the RNA chaperone activity of the largest AUF1 isoform, AUF1 p45, contributes to the RNA switch during flavivirus genome cyclization, triggering the initiation of negative-strand RNA synthesis. While earlier complementation experiments with the WNV polymerase revealed primarily AUF1 p45 to stimulate the first step of RNA replication in vitro (15), a still insufficiently addressed question concerns whether other AUF1 isoforms in the cell may substitute for AUF1 p45 or perhaps act in a different way. Considering that the four AUF1 isoforms were indicated to show different RNA binding properties and to remodel bound RNA substrates into divergent structures (37), this will be an important aspect of future investigations.
A second vital aspect to be examined concerns earlier reported pro- or antiviral activities of AUF1 in the life cycles of other RNA viruses. For example, AUF1 was shown to promote the translation of the hepatitis C virus (HCV) genome by binding to the HCV internal ribosome entry site (IRES) (38). Conversely, AUF1 was observed to inhibit the translation of various picornavirus genomes, such as those of poliovirus, rhinovirus, and enterovirus, again by binding to specific RNA structures within the viral IRES (39; reviewed in reference 40). While the precise mechanisms are unknown, it is plausible that these activities of AUF1 are also associated with its RNA restructuring capabilities. A final intriguing question that remains is whether and how the RNA remodeling activities of AUF1 proteins contribute to their functions in the cellular RNA metabolism.
MATERIALS AND METHODS
Plasmid encoding DENV sgRNA.
All DENV constructs were derived from an infectious cDNA clone of DENV 2 strain 16681 (kindly provided by Richard Kinney). Primer sequences are supplied in Table S2 in the supplemental material. The plasmid pDENVsgRNA, which encodes the DENV sgRNA (encompassing the 5′-terminal 184 nt and the 3′-terminal 451 nt of the DENV genomic RNA), was generated by PCR amplification of a plasmid coding for a DENV2 replicon by use of oligonucleotide primers DENVsgRNAFor and DENVsgRNARev, followed by phosphorylation and religation.
Plasmid encoding ZIKV 5′UTR.
To obtain pUC18-ZIKV-5′UTR, the corresponding cDNA was generated by reverse transcription-PCR (RT-PCR) amplification of RNA isolated from the supernatant of ZIKV-infected Huh7 cells (strain PF13) by using the primers 5′UTRZIKV_RT, 5UTRZIKVFor, and 5UTRZIKVRev. This DNA fragment was inserted between the HindIII and SmaI restriction sites in pUC18.
Plasmids encoding SUMO fusion proteins.
The pETSUMO expression system was used for the expression of recombinant proteins in E. coli. For DENV NS5, the NS5 coding sequence was amplified by PCR from a plasmid coding for a DENV2 replicon by using primers NS5DENVFor and NS5DENVRev, and the resulting DNA fragment was inserted between the BsaI and BamHI restriction sites of the pETSUMO expression vector. The plasmids encoding AUF1 p45 and PRMT1-AUF1 p45 were described previously (15, 16).
RNA extraction and qRT-PCR.
For the quantification of virus titers after infection with DENV or ZIKV, the viral RNA was extracted from culture fluids by using the MagNA Pure system (Roche). DENV RNA was quantified by a two-step quantitative RT-PCR (qRT-PCR) protocol targeting the NS5-3′UTR region. Standard RT was performed using Superscript III reverse transcriptase (Invitrogen). PCR mixtures contained buffer (Invitrogen), 3 mM MgCl2, 100 ng/μl bovine serum albumin (BSA), primer FLA EMF1 for (10 pmol; Metabion), primer FLA VD8 rev (10 pmol; Metabion), deoxynucleoside triphosphates (dNTPs) (250 μM [each]; Qiagen), SYBR Green, Platinum Taq polymerase (2 U; Invitrogen), and cDNA. ZIKV RNA was quantified by a one-step qRT-PCR protocol using fluorescence resonance energy transfer (FRET) probes targeting the envelope coding region. RT-PCR mixtures contained QuantiFast multiplex RT-PCR master mix (Qiagen), QuantiFast RT mix (Qiagen), RNase inhibitor (Fermentas), primer ZIKV_for (10 pmol; Metabion), primer ZIKV_rev (10 pmol; Metabion), probe ZIKA_FL (4 pmol; TibMolBiol), probe ZIKA_LCRed640 (4 pmol; TibMolBiol), and RNA. See Table S2 in the supplemental material for details of the primers and probes. To quantify the RNA copy number, the data were related to a standard curve assessed by plasmid dilution. Further details of the RT and PCR conditions are available upon request.
Western blotting and antibodies.
Anti-AUF1 antibody was purchased from Millipore, and anti-vinculin antibody was purchased from Sigma. Secondary antibodies were purchased from Li-Cor. Western blots were performed by using standard protocols and following the manufacturers' instructions.
Replicase assay.
The template sgRNA was transcribed by use of T7 RNA polymerase (standard protocol) from a PCR product that was generated from pDENVsgRNA by using primers 5UTRDENVFor and 3UTRnativDENV (Table S2). The assay was performed in a total volume of 40 μl in buffer containing 50 mM HEPES-NaOH, 55 mM KCl, 5 mM MgCl2, 0.5 mM MnCl2, and 1 mM dithiothreitol, pH 8.0. It contained 500 μM (each) ATP, GTP, and UTP, 0.1 μM CTP, 10 μCi [α-32P]CTP, 10 nM template RNA, and 60 nM purified recombinant NS5. Supplementation was performed such that the proteins were preincubated under the assay conditions with the template RNA, and NS5 was added subsequently. When proteins were added, the buffer conditions of the control reaction mixture were adapted to keep the ionic strength constant. The reaction was carried out for 60 min at 22°C and stopped by phenol-chloroform extraction and ethanol precipitation of the RNA. Alternatively, the RNA products were purified using a High Pure RNA isolation kit (Roche). The RNA products were analyzed by electrophoresis on a 5% denaturing polyacrylamide gel (7 M urea), visualized by phosphorimaging, and quantified by use of ImageQuant software (GE).
Expression and purification of DENV NS5.
DENV NS5 was expressed as a SUMO fusion protein in Escherichia coli BL21-CodonPlus (DE3)-RP cells. The protein was purified from the soluble fraction by nickel-agarose affinity chromatography (HisTrap HP; GE Healthcare) and, after cleavage with SUMO-protease, by heparin-Sepharose affinity chromatography and gel filtration (HiLoad 16/60 Superdex 200; GE Healthcare). Purified NS5 was stored at −20°C in 50 mM Tris-HCl, 300 mM NaCl, 40% (vol/vol) glycerol, pH 7.6, until use. The protein concentration was determined by measuring the absorbance at 280 nm, using an ε value of 218,135 M−1 cm−1.
Expression and purification of nonmethylated and methylated AUF1 p45 from E. coli.
Nonmethylated and methylated AUF1 p45 (coexpressed with protein arginine methyltransferase 1) proteins were purified from the soluble fraction of Escherichia coli BL21-CodonPlus (DE3)-RP cells as previously described (16). UV-visible absorption spectra were measured using a Jasco V-550 spectrophotometer. The protein concentration was determined by measuring the absorbance at 280 nm, using an ε value of 58,915 M−1 cm−1. The proteins were stored at −80°C in 20 mM Tris-HCl, pH 7.6, 150 mM KCl, 1 mM Tris(2-carboxyethyl)phosphine (TCEP).
Expression and purification of hnRNPH1 from E. coli.
Expression and purification of hnRNPH1 were performed as previously described (15).
Cell culture, transfection conditions, infection experiments, and IFA.
Human hepatoma cells (Huh7) were cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen) supplemented with 10% fetal calf serum (FCS) (Pan-Biotech), 1% penicillin-streptomycin (Invitrogen), 0.1% d-biotin, and 0.1% hypoxanthine (Sigma). Duplex siRNA targeting the mRNAs of AUF1 was purchased from Eurofins MWG Operon (Table S1). Transfection was performed with 70% confluent Huh7 cells and Lipofectamine RNAiMAX (Invitrogen), using the manufacturer's instructions. To test the effect of AUF1 depletion on the propagation of infectious DENV and ZIKV, approximately 5 × 105 Huh7 cells were infected with DENV strain 16681 or ZIKV strain PF13 (1 h at a 50% tissue culture infective dose [TCID50] of 1 × 104/ml). A rabbit polyclonal antiserum (kindly provided by Ralf Bartenschlager) was applied to detect the synthesis of NS5 by indirect immunofluorescence analysis (IFA).
Fluorescence-based RNA-RNA interaction assay.
The fluorescence-based RNA-RNA interaction assay was performed as described previously (15). Briefly, different concentrations of the AUF1 p45 wild-type (WT) protein or a variant thereof were added to 10 nM 5′-Cy5-labeled DENV 3SLtrunc-RNA or 5′-Cy5-labeled WNV SLB RNA (purchased from IBA, Göttingen, Germany) (Fig. 2 and 6; Table S3) in assay buffer (50 mM HEPES-NaOH, pH 8.0, 100 mM KCl, 5 mM MgCl2). Next, 100 nM nonlabeled DENV 5′UAR RNA or WNV 3′UAR RNA (Table S3) was added, and readings were taken for another 150 s. Changes in the fluorescence signals were monitored in a Fluoromax-4 spectrofluorometer (Jobin Yvon) at 22°C, with the following parameters: excitation at 643 nm, emission at 667 nm, and slit widths of 10 and 10 nm. Fluorescence intensities relative to the starting fluorescence were plotted against the time and fitted by KaleidaGraph (Synergy) to a first-order reaction (equation 1) when protein was omitted and a second-order reaction (equation 2) when protein was included, yielding the corresponding observed rate constants (kobs), as follows:
| (1) |
| (2) |
where ΔF is the total change of the relative fluorescence amplitude, Foffset is the fluorescence intensity at the start point of the reaction, Fmax is the maximum signal amplitude, kobs is the observed rate constant, and t is time.
Chemical probing of RNA secondary structure and primer extension analysis.
The DENV 3′-terminal 109 nt and 5′-terminal 151 nt (each fused to a binding site for primer extension) were transcribed by use of T7 RNA polymerase (standard protocol) from a PCR product that was generated from pDENVsgRNA by using oligonucleotide primer pairs 3CycDENVFor/3UTRDENVPolyCA and 5UTRDENVFor/5UTRDENVPolyCA. The ZIKV 5′-terminal 151 nt (fused to a binding site for primer extension) were transcribed by use of T7 RNA polymerase (standard protocol) from a PCR product that was generated from pUC18-ZIKV-5′UTR by using oligonucleotide primer pair 5UTRZIKVFor/5UTRZIKVPolyCA. Evaluation of the RNA secondary structure was done using the protocol published by Yu et al. (41), with minor modifications. The modifying agent used was CMCT [1-cyclohexyl-(2-morpholinoethyl)carbodiimide metho-p-toluene sulfonate]. First, 0.5 μg RNA (174 nM 5′RNA or 230 nM 3′RNA) was preincubated in the absence or presence of 1 or 2 μM AUF1 p45 or hnRNPH1 in 50 mM potassium borate, pH 8.0, 100 mM KCl, 5 mM MgCl2. CMCT was added to the reaction mixture, to a final concentration of 100 mM, followed by incubation for 10 min at 22°C. The reaction was stopped with 325 μl stop buffer [0.2 M piperazine-N,N′-bis(2-ethanesulfonic acid) (PIPES)–NaOH, pH 6.4, 0.3 M sodium acetate, 5 mM EDTA]. The modified RNA was extracted with phenol-chloroform and then precipitated with ethanol, with 10 μg tRNA as a carrier. Chemically modified RNA molecules were mixed with 1 pmol of 5′-end-labeled DNA primer (3UTRDENVPolyCA_RT). Primer extension was performed at 42°C for 60 min with 100 U of RevertAid reverse transcriptase (ThermoScientific) under the conditions recommended by the manufacturer. The locations of modification sites were determined by 8% denaturing polyacrylamide gel electrophoresis (with 8 M urea) of the primer extension products. In parallel, the corresponding DNA sequence was determined by dideoxynucleotide sequencing (DNA cycle sequencing kit; Jena Bioscience) with the same 5′-end-labeled primer. The cDNA products were visualized by phosphorimaging and quantified by use of ImageQuant software (GE).
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Richard Kinney for the dengue virus cDNA clone and to Ralf Bartenschlager for providing reagents. We thank Gary Sawers for critically reading the manuscript.
This work was supported by the Deutsche Forschungsgemeinschaft (grants BE1885/7-1/2 and BE1885/12-1 to S.F. and S.-E.B., Graduiertenkolleg 1026 to T.S. and R.P.G., and Graduiertenkolleg 1591 to S.E. and S.-E.B.).
We have no conflicts of interest to report.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JVI.01647-17.
REFERENCES
- 1.Bhatt S, Gething PW, Brady OJ, Messina JP, Farlow AW, Moyes CL, Drake JM, Brownstein JS, Hoen AG, Sankoh O, Myers MF, George DB, Jaenisch T, Wint GR, Simmons CP, Scott TW, Farrar JJ, Hay SI. 2013. The global distribution and burden of dengue. Nature 496:504–507. doi: 10.1038/nature12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Weaver SC, Costa F, Garcia-Blanco MA, Ko AI, Ribeiro GS, Saade G, Shi PY, Vasilakis N. 2016. Zika virus: history, emergence, biology, and prospects for control. Antiviral Res 130:69–80. doi: 10.1016/j.antiviral.2016.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Welsch S, Miller S, Romero-Brey I, Merz A, Bleck CK, Walther P, Fuller SD, Antony C, Krijnse-Locker J, Bartenschlager R. 2009. Composition and three-dimensional architecture of the dengue virus replication and assembly sites. Cell Host Microbe 5:365–375. doi: 10.1016/j.chom.2009.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Paul D, Bartenschlager R. 2015. Flaviviridae replication organelles: oh, what a tangled web we weave. Annu Rev Virol 2:289–310. doi: 10.1146/annurev-virology-100114-055007. [DOI] [PubMed] [Google Scholar]
- 5.Nagy PD, Strating JR, van Kuppeveld FJ. 2016. Building viral replication organelles: close encounters of the membrane types. PLoS Pathog 12:e1005912. doi: 10.1371/journal.ppat.1005912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filomatori CV, Lodeiro MF, Alvarez DE, Samsa MM, Pietrasanta L, Gamarnik AV. 2006. A 5′ RNA element promotes dengue virus RNA synthesis on a circular genome. Genes Dev 20:2238–2249. doi: 10.1101/gad.1444206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fernandez-Sanles A, Rios-Marco P, Romero-Lopez C, Berzal-Herranz A. 2017. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front Microbiol 8:546. doi: 10.3389/fmicb.2017.00546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brinton MA, Basu M. 2015. Functions of the 3′ and 5′ genome RNA regions of members of the genus Flavivirus. Virus Res 206:108–119. doi: 10.1016/j.virusres.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Selisko B, Wang C, Harris E, Canard B. 2014. Regulation of flavivirus RNA synthesis and replication. Curr Opin Virol 9:74–83. doi: 10.1016/j.coviro.2014.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gebhard LG, Filomatori CV, Gamarnik AV. 2011. Functional RNA elements in the dengue virus genome. Viruses 3:1739–1756. doi: 10.3390/v3091739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bidet K, Garcia-Blanco MA. 2014. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J 462:215–230. doi: 10.1042/BJ20140456. [DOI] [PubMed] [Google Scholar]
- 12.Moore AE, Chenette DM, Larkin LC, Schneider RJ. 2014. Physiological networks and disease functions of RNA-binding protein AUF1. Wiley Interdiscip Rev RNA 5:549–564. doi: 10.1002/wrna.1230. [DOI] [PubMed] [Google Scholar]
- 13.White EJ, Matsangos AE, Wilson GM. 2017. AUF1 regulation of coding and noncoding RNA. Wiley Interdiscip Rev RNA 8:e1393. doi: 10.1002/wrna.1393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wagner BJ, DeMaria CT, Sun Y, Wilson GM, Brewer G. 1998. Structure and genomic organization of the human AUF1 gene: alternative pre-mRNA splicing generates four protein isoforms. Genomics 48:195–202. doi: 10.1006/geno.1997.5142. [DOI] [PubMed] [Google Scholar]
- 15.Friedrich S, Schmidt T, Geissler R, Lilie H, Chabierski S, Ulbert S, Liebert UG, Golbik RP, Behrens SE. 2014. AUF1 p45 promotes West Nile virus replication by an RNA chaperone activity that supports cyclization of the viral genome. J Virol 88:11586–11599. doi: 10.1128/JVI.01283-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Friedrich S, Schmidt T, Schierhorn A, Lilie H, Szczepankiewicz G, Bergs S, Liebert UG, Golbik RP, Behrens SE. 2016. Arginine methylation enhances the RNA chaperone activity of the West Nile virus host factor AUF1 p45. RNA 22:1574–1591. doi: 10.1261/rna.055269.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.You S, Padmanabhan R. 1999. A novel in vitro replication system for dengue virus. Initiation of RNA synthesis at the 3′-end of exogenous viral RNA templates requires 5′- and 3′-terminal complementary sequence motifs of the viral RNA. J Biol Chem 274:33714–33722. [DOI] [PubMed] [Google Scholar]
- 18.Nomaguchi M, Teramoto T, Yu L, Markoff L, Padmanabhan R. 2004. Requirements for West Nile virus (−)- and (+)-strand subgenomic RNA synthesis in vitro by the viral RNA-dependent RNA polymerase expressed in Escherichia coli. J Biol Chem 279:12141–12151. doi: 10.1074/jbc.M310839200. [DOI] [PubMed] [Google Scholar]
- 19.Waldsich C. (ed). 2014. RNA folding. Springer, New York, NY. [Google Scholar]
- 20.Chapman EG, Moon SL, Wilusz J, Kieft JS. 2014. RNA structures that resist degradation by Xrn1 produce a pathogenic dengue virus RNA. Elife 3:e01892. doi: 10.7554/eLife.01892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Polacek C, Foley JE, Harris E. 2009. Conformational changes in the solution structure of the dengue virus 5′ end in the presence and absence of the 3′ untranslated region. J Virol 83:1161–1166. doi: 10.1128/JVI.01362-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.de Borba L, Villordo SM, Iglesias NG, Filomatori CV, Gebhard LG, Gamarnik AV. 2015. Overlapping local and long-range RNA-RNA interactions modulate dengue virus genome cyclization and replication. J Virol 89:3430–3437. doi: 10.1128/JVI.02677-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu ZY, Li XF, Jiang T, Deng YQ, Ye Q, Zhao H, Yu JY, Qin CF. 2016. Viral RNA switch mediates the dynamic control of flavivirus replicase recruitment by genome cyclization. Elife 5:e17636. doi: 10.7554/eLife.17636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rajkowitsch L, Chen D, Stampfl S, Semrad K, Waldsich C, Mayer O, Jantsch MF, Konrat R, Blasi U, Schroeder R. 2007. RNA chaperones, RNA annealers and RNA helicases. RNA Biol 4:118–130. doi: 10.4161/rna.4.3.5445. [DOI] [PubMed] [Google Scholar]
- 25.Grohman JK, Gorelick RJ, Lickwar CR, Lieb JD, Bower BD, Znosko BM, Weeks KM. 2013. A guanosine-centric mechanism for RNA chaperone function. Science 340:190–195. doi: 10.1126/science.1230715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Khromykh AA, Meka H, Guyatt KJ, Westaway EG. 2001. Essential role of cyclization sequences in flavivirus RNA replication. J Virol 75:6719–6728. doi: 10.1128/JVI.75.14.6719-6728.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo MK, Tilgner M, Bernard KA, Shi PY. 2003. Functional analysis of mosquito-borne flavivirus conserved sequence elements within 3′ untranslated region of West Nile virus by use of a reporting replicon that differentiates between viral translation and RNA replication. J Virol 77:10004–10014. doi: 10.1128/JVI.77.18.10004-10014.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Alvarez DE, Lodeiro MF, Luduena SJ, Pietrasanta LI, Gamarnik AV. 2005. Long-range RNA-RNA interactions circularize the dengue virus genome. J Virol 79:6631–6643. doi: 10.1128/JVI.79.11.6631-6643.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dong H, Zhang B, Shi PY. 2008. Terminal structures of West Nile virus genomic RNA and their interactions with viral NS5 protein. Virology 381:123–135. doi: 10.1016/j.virol.2008.07.040. [DOI] [PubMed] [Google Scholar]
- 30.Ivanyi-Nagy R, Darlix JL. 2012. Core protein-mediated 5′-3′ annealing of the West Nile virus genomic RNA in vitro. Virus Res 167:226–235. doi: 10.1016/j.virusres.2012.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gebhard LG, Kaufman SB, Gamarnik AV. 2012. Novel ATP-independent RNA annealing activity of the dengue virus NS3 helicase. PLoS One 7:e36244. doi: 10.1371/journal.pone.0036244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yoon JH, De S, Srikantan S, Abdelmohsen K, Grammatikakis I, Kim J, Kim KM, Noh JH, White EJ, Martindale JL, Yang X, Kang MJ, Wood WH III, Noren Hooten N, Evans MK, Becker KG, Tripathi V, Prasanth KV, Wilson GM, Tuschl T, Ingolia NT, Hafner M, Gorospe M. 2014. PAR-CLIP analysis uncovers AUF1 impact on target RNA fate and genome integrity. Nat Commun 5:5248. doi: 10.1038/ncomms6248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Filomatori CV, Iglesias NG, Villordo SM, Alvarez DE, Gamarnik AV. 2011. RNA sequences and structures required for the recruitment and activity of the dengue virus polymerase. J Biol Chem 286:6929–6939. doi: 10.1074/jbc.M110.162289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Garcia-Blanco MA, Vasudevan SG, Bradrick SS, Nicchitta C. 2016. Flavivirus RNA transactions from viral entry to genome replication. Antiviral Res 134:244–249. doi: 10.1016/j.antiviral.2016.09.010. [DOI] [PubMed] [Google Scholar]
- 35.Laroia G, Cuesta R, Brewer G, Schneider RJ. 1999. Control of mRNA decay by heat shock-ubiquitin-proteasome pathway. Science 284:499–502. doi: 10.1126/science.284.5413.499. [DOI] [PubMed] [Google Scholar]
- 36.Lu JY, Bergman N, Sadri N, Schneider RJ. 2006. Assembly of AUF1 with eIF4G-poly(A) binding protein complex suggests a translation function in AU-rich mRNA decay. RNA 12:883–893. doi: 10.1261/rna.2308106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zucconi BE, Ballin JD, Brewer BY, Ross CR, Huang J, Toth EA, Wilson GM. 2010. Alternatively expressed domains of AU-rich element RNA-binding protein 1 (AUF1) regulate RNA-binding affinity, RNA-induced protein oligomerization, and the local conformation of bound RNA ligands. J Biol Chem 285:39127–39139. doi: 10.1074/jbc.M110.180182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paek KY, Kim CS, Park SM, Kim JH, Jang SK. 2008. RNA-binding protein hnRNP D modulates internal ribosome entry site-dependent translation of hepatitis C virus RNA. J Virol 82:12082–12093. doi: 10.1128/JVI.01405-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lin JY, Li ML, Brewer G. 2014. mRNA decay factor AUF1 binds the internal ribosomal entry site of enterovirus 71 and inhibits virus replication. PLoS One 9:e103827. doi: 10.1371/journal.pone.0103827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ullmer W, Semler BL. 2016. Diverse strategies used by picornaviruses to escape host RNA decay pathways. Viruses 8:335. doi: 10.3390/v8120335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu H, Grassmann CW, Behrens SE. 1999. Sequence and structural elements at the 3′ terminus of bovine viral diarrhea virus genomic RNA: functional role during RNA replication. J Virol 73:3638–3648. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.