ABSTRACT
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants, and an effective vaccine is not yet available. We previously generated an RSV live-attenuated vaccine (LAV) candidate, DB1, which was attenuated by a low-fusion subgroup B F protein (BAF) and codon-deoptimized nonstructural protein genes. DB1 was immunogenic and protective in cotton rats but lacked thermostability and stability of the prefusion conformation of F compared to strains with the line19F gene. We hypothesized that substitution of unique residues from the thermostable A2-line19F strain could thermostabilize DB1 and boost its immunogenicity. We therefore substituted 4 unique line19F residues into the BAF protein of DB1 by site-directed mutagenesis and rescued the recombinant virus, DB1-QUAD. Compared to DB1, DB1-QUAD had improved thermostability at 4°C and higher levels of prefusion F as measured by enzyme-linked immunosorbent assays (ELISAs). DB1-QUAD was attenuated in normal human bronchial epithelial cells, in BALB/c mice, and in cotton rats but grew to wild-type titers in Vero cells. In mice, DB1-QUAD was highly immunogenic and generated significantly higher neutralizing antibody titers to a panel of RSV A and B strains than did DB1. DB1-QUAD was also efficacious against wild-type RSV challenge in mice and cotton rats. Thus, substitution of unique line19F residues into RSV LAV DB1 enhanced vaccine thermostability, incorporation of prefusion F, and immunogenicity and generated a promising vaccine candidate that merits further investigation.
IMPORTANCE We boosted the thermostability and immunogenicity of an RSV live-attenuated vaccine candidate by substituting 4 unique residues from the RSV line19F protein into the F protein of the heterologous vaccine strain DB1. The resultant vaccine candidate, DB1-QUAD, was thermostable, attenuated in vivo, highly immunogenic, and protective against RSV challenge in mice and cotton rats.
KEYWORDS: respiratory syncytial virus, vaccines
INTRODUCTION
Respiratory syncytial virus (RSV) is the leading cause of lower respiratory tract infections in infants. It is estimated that RSV causes up to 3.2 million annual hospitalizations and 118,000 deaths in children <5 years of age worldwide (1). Multiple strategies to prevent and treat RSV are under investigation, including small-molecule antivirals (2, 3), prophylactic antibodies (4, 5), maternal immunization strategies (6, 7), and both live (8, 9) and nonreplicating (10, 11) vaccines. However, to date, an effective therapeutic drug has yet to come to market, and the only prophylaxis commercially available is the monoclonal antibody palivizumab (12, 13), which is costly and recommended only for certain high-risk groups (14). Thus, a safe and effective RSV vaccine remains a high public health priority and need.
However, the history of RSV vaccine development has been marred by the initial attempt to administer a formalin-inactivated virus to infants and young children in the 1960s. Not only did this vaccine fail to protect infants from subsequent RSV infection, but it also tragically primed seronegative vaccine recipients for enhanced RSV disease upon natural infection. A high proportion of vaccinated children required hospitalization, and 2 infants died from complications of RSV (15). This phenomenon of enhanced disease has since been replicated in animal models (16, 17) and has been attributed to altered immunologic priming with subsequent generation of poorly neutralizing antibodies (18), immune complex deposition (19), and a Th2-biased cellular immune response (20). The same phenomenon has been observed after administration of some other protein-based nonreplicating vaccines (21), which has precluded their use in seronegative infants to date. In contrast, enhanced disease has not been observed following administration of live-attenuated vaccines (LAVs) (22). Thus, from a safety and regulatory standpoint, LAVs represent a more acceptable strategy for the vaccination of the target population of seronegative infants.
RSV live-attenuated vaccines have several advantages, including the potential needle-free intranasal (i.n.) administration of a vaccine virus that broadly stimulates humoral and cellular immunity, in addition to mucosal immunity (23). However, LAVs present their own challenges, including the potential lack of genetic and thermal stability. Perhaps the greatest challenge in the development of an RSV LAV has been to achieve an effective balance of vaccine attenuation and immunogenicity. Traditional approaches to cold adaptation have generated highly attenuated viruses, many of which have lacked sufficient immunogenicity (24). Conversely, more recent reverse genetics approaches have generated immunogenic vaccine strains that have been insufficiently attenuated in clinical trials (25). Thus, there is a need to boost the intrinsic immunogenicity of RSV without compromising attenuation in vivo.
We previously described a vaccine strain, DB1, with the genotype RSV-A2-dNS-ΔSH-BAF. DB1 was attenuated by codon deoptimization of nonstructural protein (NS1 and NS2) genes, deletion of the short hydrophobic protein (SH) gene, and incorporation of a low-fusion F protein (BAF) consensus sequence gene from the Buenos Aires clade (26). We demonstrated that DB1 was attenuated in cotton rats (Sigmodon hispidus) and in primary human airway cells differentiated at the air-liquid interface (ALI) but maintained immunogenicity and efficacy in cotton rats. However, we subsequently found that DB1 also lacked thermostability and expressed relatively small amounts of the key RSV antigen, prefusion F (27). We identified a chimeric RSV strain, A2-line19F, with high thermostability, pre-F expression, and immunogenicity compared to its parental strain, A2 (27). Substitution of 2 unique line19F residues into the BAF protein of DB1 partially restored DB1 thermostability to A2-line19F levels (27). We therefore hypothesized that substitution of additional line19F residues into DB1 could confer enhanced thermostability, pre-F expression, and immunogenicity.
In this study, we substituted 4 unique line19F residues into the BAF of DB1 by sequential site-directed mutagenesis to generate the vaccine strain DB1-QUAD. We subsequently measured DB1-QUAD thermostability at 4°C and incorporation of prefusion F by MPE-8 and motavizumab enzyme-linked immunosorbent assays (ELISAs). We then analyzed DB1-QUAD attenuation, immunogenicity, and efficacy at protecting against RSV challenge in both BALB/c mice and cotton rats.
RESULTS
Design of RSV live-attenuated vaccine DB1-QUAD.
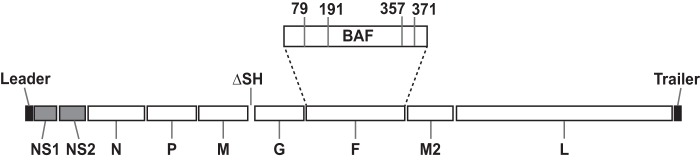
We previously described the generation of an RSV LAV candidate, DB1, with the genotype RSV-A2-dNS-ΔSH-BAF (26) and demonstrated that the vaccine expressed the intact viral repertoire of genetically modified proteins. The rationale for DB1's design was to combine two attenuating genetic modifications known to preserve immunogenicity (dNS and ΔSH) with a low-fusion F protein (BAF) to further attenuate the vaccine candidate. We showed that DB1 was attenuated in primary human epithelial cells and in cotton rats yet was broadly immunogenic and protective against RSV challenge. However, we subsequently found that DB1 lacked thermostability and incorporation of prefusion F, a key RSV immunogen (27). Substitution of 2 unique residues (K for T at amino acid 357 [K357T] and N371Y) from the thermostable A2-line19F strain into the BAF protein of DB1 partially recapitulated the thermostability phenotype. In this study, therefore, we hypothesized that substitution of additional line19F residues could fully thermostabilize DB1 to A2-line19F levels and further boost its immunogenicity by stabilizing prefusion F. We thus substituted 4 unique line19F residues, I79M, K191R, T357K, and N371Y, into the BAF protein by sequential site-directed mutagenesis in the pcDNA3.1+ vector. We cloned the modified BAF gene, BAF-I79M/K191R/T357K/N371Y, into the antigenomic bacterial artificial chromosome (BAC) pSynk-DB1 and rescued the recombinant virus denoted DB1-QUAD (Fig. 1).
FIG 1.
Schematic of DB1-QUAD live-attenuated RSV vaccine genome. DB1-QUAD includes codon deoptimization (gray shading) of NS1 and NS2 genes, deletion of the SH gene, replacement of the A2 F gene with a consensus sequence of the Buenos Aires (BA) F gene, and substitution of stabilizing line19F residues into the BAF sequence at positions 79, 191, 357, and 371.
Viral growth kinetics of DB1-QUAD in immortalized and primary cell cultures.
We measured DB1-QUAD growth kinetics in both immortalized cell lines and primary normal human bronchial epithelial (NHBE) cells differentiated at the ALI. In Vero cells, a cell line approved for vaccine production, we found that DB1-QUAD grew to titers similar to those of DB1 and A2-line19F (Fig. 2A). We hypothesized that this was attributable to the interferon deficiency of Vero cells, which limited the attenuating effects of codon-deoptimized nonstructural protein genes. In BEAS-2B cells, a human bronchial epithelial cell line, DB1-QUAD again grew to titers similar to those of DB1 and A2-line19F; however, the titers were statistically significantly lower at 96 h postinfection (p.i.) (Fig. 2B). Finally, we measured DB1-QUAD growth kinetics in NHBE cells at the ALI, a primary cell culture system that closely approximates viral attenuation in RSV-seronegative infants (28). Compared to A2, both DB1 and DB1-QUAD were significantly attenuated in NHBE ALI cells on days 4 and 5 p.i. (Fig. 2C), which was likely attributable to both the codon-deoptimized nonstructural protein genes and the poorly fusogenic F protein.
FIG 2.
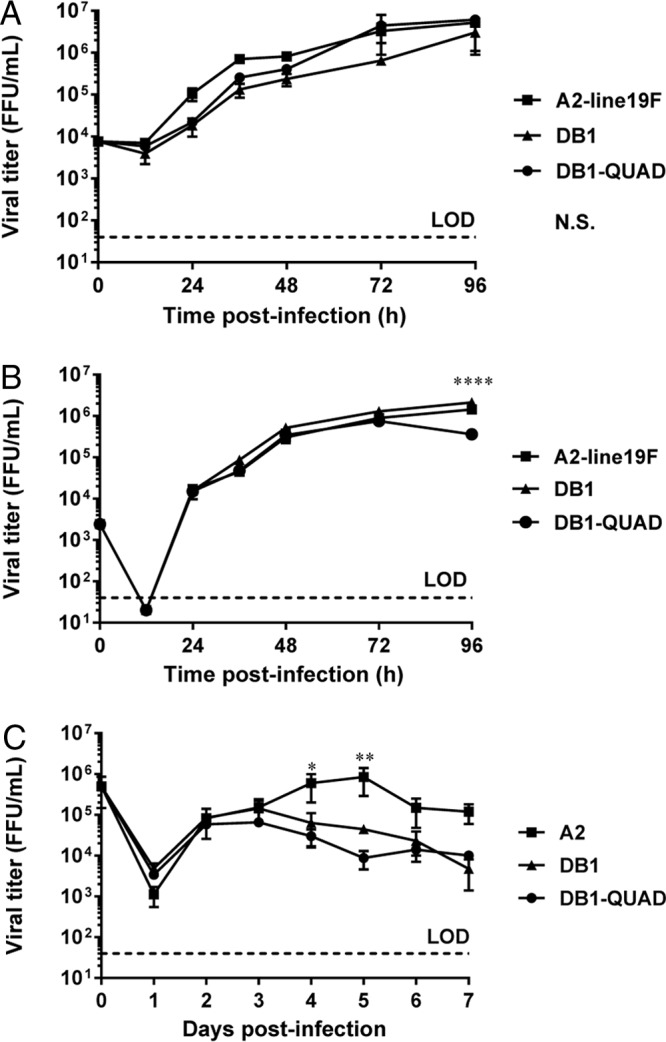
Viral growth kinetics of DB1-QUAD in immortalized and primary cell cultures. (A and B) Vero (A) and BEAS-2B (B) cells were infected with mKate2-labeled A2-line19F, DB1, and DB1-QUAD at an MOI of 0.01. Cells were collected at the indicated time points and titrated by FFU assay. The data represent the means ± SEM of two or three experimental replicates titrated in duplicate. (C) NHBE cells from a single donor were infected at an MOI of 2 and titrated by FFU assay. The data represent the means ± SEM of two experimental replicates titrated in duplicate. *, P < 0.05; **, P < 0.005; ****, P < 0.00005; N.S., not significant by two-way ANOVA and Tukey's multiple-comparison test. LOD, limit of detection.
DB1-QUAD thermostability, incorporation of prefusion F, and F fusion activity.
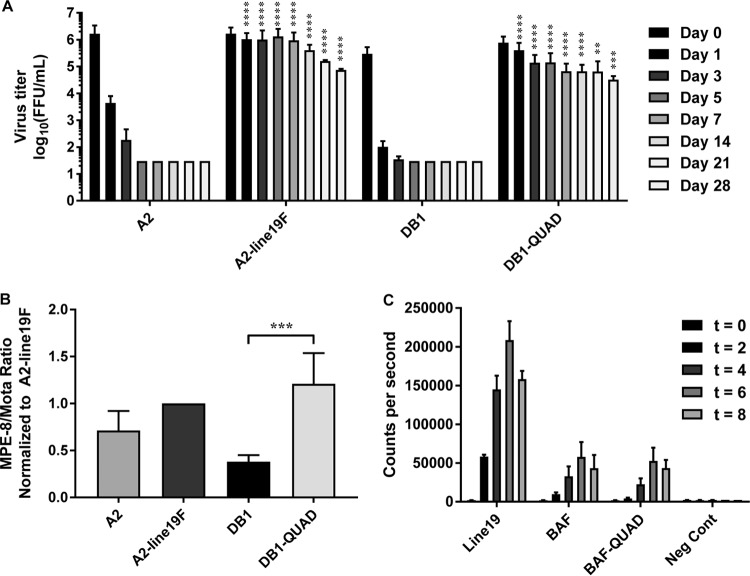
We next measured DB1-QUAD thermostability by incubating viral stocks at 4°C and measuring the decline in viral titers on days 0 through 28 of incubation. This temperature was chosen due to its relevance for cold-chain production and vaccine storage and distribution. Whereas A2 and DB1 titers declined precipitously and became undetectable after 5 days at 4°C, A2-line19F and DB1-QUAD (containing the 4 unique line19F residues) titers remained detectable after 28 days (Fig. 3A). DB1-QUAD decreased by only 1.38 log10 focus-forming units (FFU)/ml over this time course. Thus, substitution of line19F residues into DB1 to generate DB1-QUAD conferred enhanced thermostability at 4°C.
FIG 3.
DB1-QUAD thermostability, incorporation of prefusion F protein, and F fusion activity. (A) Thermostability was analyzed at 4°C by serially titrating vials of mKate2-labeled A2, A2-line19F, DB1, and DB1-QUAD at time points ranging from 0 to 28 days. The data represent means and SD of log-transformed titers of four experimental replicates for time points 0 to 14 days and two replicates for time points 21 to 28 days, all titrated in duplicate. The asterisks denote statistical differences between A2 and A2-line19F and between DB1 and DB1-QUAD at the respective time points. There were no statistical differences between A2-line19F and DB1-QUAD titers. (B) The ratio of prefusion F to total F protein expression was measured using MPE-8 (pre-F) and motavizumab (total F) ELISAs. The data represent means and SD of at least three experimental replicates for each virus. (C) The fusion activities of line19F, BAF, and BAF-79-191-357-371 were measured using a DSP assay. 293T cells were transfected with F expression plasmids in the presence of BMS-433771 fusion inhibitor, and cell-cell fusion activity was quantified by luciferase activity at time points 0, 2, 4, 6, and 8 h post-cell mixing. Three experimental replicates were performed, and representative data from one experiment are shown. **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005 by two-way ANOVA and Tukey's multiple-comparison test (A) or by Student's t test (B).
We then measured the incorporation of the prefusion conformation of the F protein into DB1-QUAD compared to DB1 and A2-line19F. Prefusion F is the predominant immunogen of RSV, and the majority of neutralizing antibodies generated during human infection are directed against the antigen (29). To quantify the relative amounts of pre- and postfusion F incorporation, we performed ELISAs on viral stocks using either MPE-8 (pre-F-specific) or motavizumab (pre- and post-F-specific) primary antibodies, as previously described (27). We found that substitution of the line19F residues into DB1 to generate DB1-QUAD significantly increased the relative proportion of pre-F to total F expression (Fig. 3B).
We then measured the fusion activity of the modified BAF-I79M/K191R/T357K/N371Y protein compared to those of BAF and line19F to determine if substitution of the line19F residues altered the fusion activity of the glycoprotein. To do this, we obtained codon-optimized sequences of BAF-I79M/K191R/T357K/N371Y, BAF, and line19F and cloned them into pcDNA3.1+ expression vectors. We then transfected these expression vectors into 293T cells and performed dual-split-protein (DSP) cell-to-cell fusion activity assays, as previously described (26, 30). We found that substitution of the line19F residues into BAF did not significantly affect fusion activity (Fig. 3C). As a quality control, we also measured F surface expression of transfected cells using flow cytometry and confirmed that expression levels did not differ significantly for the respective F proteins (data not shown).
DB1-QUAD attenuation, efficacy, immunogenicity, and pulmonary cytokines in BALB/c mice.
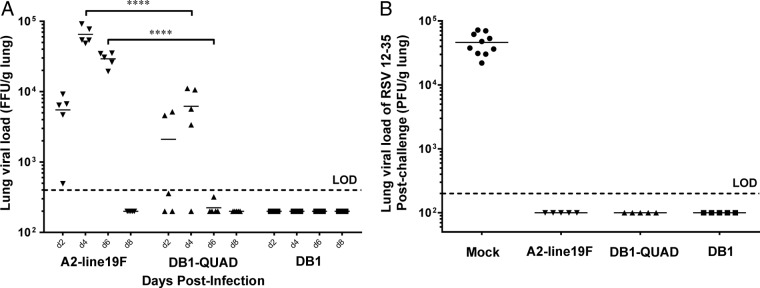
To measure vaccine attenuation in vivo, we then infected groups of 5 BALB/c mice i.n. with 106 FFU of monomeric Katushka (mKate2)-labeled A2-line19F, DB1-QUAD, and DB1. We harvested the mouse left lungs on days 2, 4, 6, and 8 p.i. and measured lung viral titers. We found that DB1-QUAD was significantly attenuated on days 4 and 6 p.i. compared to A2-line19F (Fig. 4A), whereas DB1 was completely undetectable. Thus, substitution of line19F residues into DB1 was moderately deattenuating in the BALB/c mouse lung model.
FIG 4.
DB1-QUAD attenuation and efficacy in BALB/c mice. (A) To measure viral attenuation, groups of 5 mice were inoculated intranasally with 106 FFU of mKate2-labeled A2-line19F, DB1, or DB1-QUAD, and lung viral loads were measured by FFU assay on days 2, 4, 6, and 8 postinfection. (B) To measure vaccine efficacy, groups of 5 mice were similarly inoculated with virus or mock infected. On day 96 p.i., the mice were challenged with 106 PFU of RSV 12-35, and lung viral loads were measured by standard plaque assays 4 days later. Two experimental replicates of 5 mice per group were performed. Each point represents a single animal, and the horizontal lines represent means. ****, P < 0.00005 by two-way ANOVA with Tukey's multiple-comparison test.
We then measured vaccine efficacy by again infecting groups of 5 mice intranasally with 106 FFU of mKate2-labeled A2-line19F, DB1-QUAD, DB1, or minimum essential medium (MEM) (mock infection). We collected sera on days 0, 28, 56, and 100 p.i. and challenged the mice with 106 PFU of RSV subgroup A clinical strain 12-35 on day 96 p.i. On day 100 p.i., the mice were sacrificed and the left lungs were harvested, homogenized, and titrated by standard immunoplaque assay. The results showed that DB1-QUAD completely protected the mice against wild-type RSV challenge (Fig. 4B). Similarly, DB1 completely protected the mice against wild-type challenge despite undetectable lung viral loads, as was previously described, likely due to boosted intrinsic immunogenicity attributable to the incorporated genetic modifications (26).
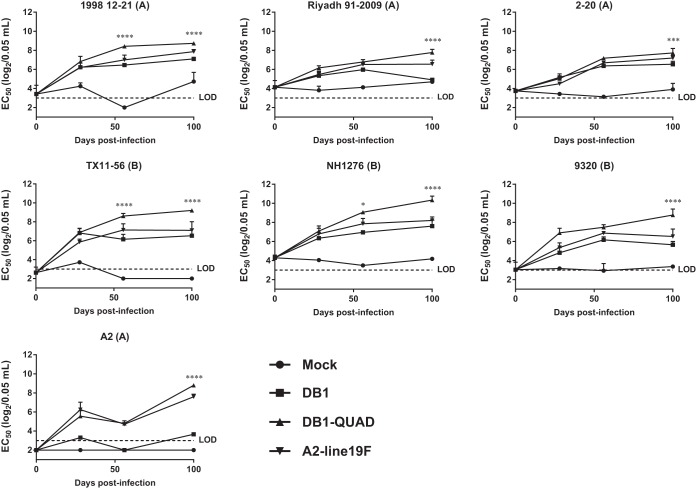
We then measured DB1-QUAD immunogenicity in mice by performing microneutralization assays against a panel of mKate2-labeled chimeric RSV strains using pooled serum specimens collected on days 0, 28, 56, and 100 p.i. The generation of this panel was previously described; it consists of chimeric RSV strains with mKate2 in the first gene position and clinical F and G genes within an A2 backbone (26). We found that DB1-QUAD generated significantly higher neutralizing antibody (nAb) titers than DB1 or A2-line19F against all 7 panel strains on day 100 p.i. (Fig. 5). Thus, incorporation of line19F residues into DB1 significantly boosted its immunogenicity in BALB/c mice.
FIG 5.
Neutralizing antibody responses generated by DB1-QUAD to a panel of recombinant RSVs. To assess immunogenicity, groups of 5 BALB/c mice were inoculated i.n. with either MEM (mock infection) or 106 FFU of mKate2-labeled A2-line19F, DB1, or DB1-QUAD. Sera were collected and pooled on days 0, 28, 56, and 100 p.i., and neutralizing antibody responses to a panel of recombinant mKate2-labeled RSVs were measured by FFU reduction assays. The data represent mean 50% effective concentrations (EC50) plus SD of two experimental replicates of pooled sera of 5 mice per group. *, P < 0.05; ***, P < 0.0005; ****, P < 0.00005 by two-way ANOVA with Tukey's multiple-comparison test.
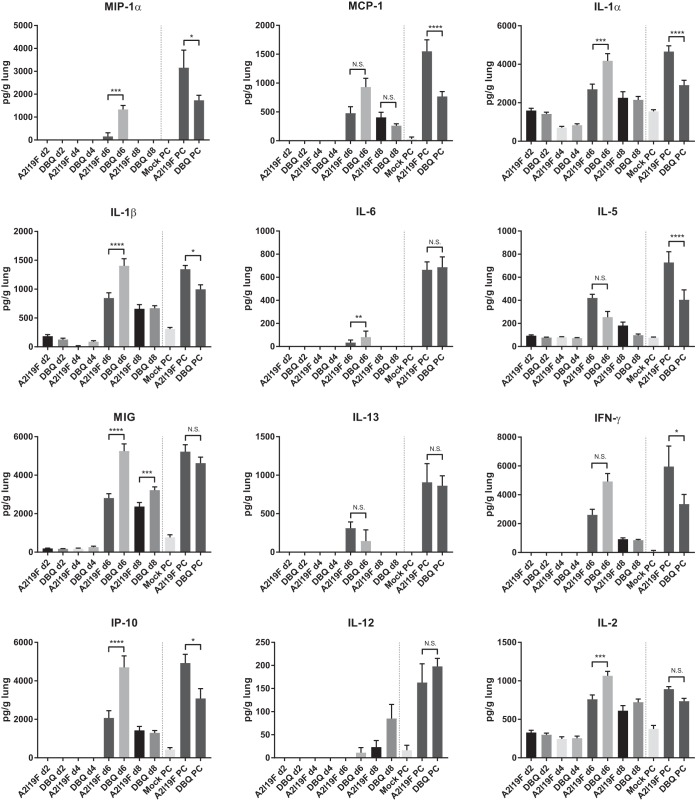
We subsequently analyzed pulmonary cytokines generated by DB1-QUAD in mouse lung homogenates postvaccination and post-RSV 12-35 challenge (Fig. 6). Compared to A2-line19F, DB1-QUAD generated an overall higher Th1-type and proinflammatory response in mouse lungs, with significant differences noted on day 6 p.i. for cytokines MIG, IP-10 (Th1 type), MIP-1α, interleukin 1α (IL-1α), and IL-1β (proinflammatory). In contrast, Th2-type cytokines, which are associated with enhanced mucus production and airway hyperreactivity, were not increased in DB1-QUAD-vaccinated mice compared to A2-line19F-vaccinated mice. These results suggest that DB1-QUAD generated a more robust, Th1-biased antiviral immune response than A2-line19F.
FIG 6.
Pulmonary cytokine analysis after vaccination and postchallenge. The cell-free supernatants of centrifuged lung homogenates obtained from mice infected with 106 FFU kDB1-QUAD or kA2-line19F on days 2, 4, 6, and 8 p.i. were analyzed using a mouse cytokine magnetic 20-plex panel kit (Invitrogen, Frederick, MD) and the Luminex 100/200 system. Similarly, lung homogenates from mice infected with MEM (mock infection), 106 FFU kDB1-QUAD, or 106 FFU kA2-line19F and subsequently challenged with 106 FFU RSV 12-35 on day 96 p.i. were obtained and analyzed. The concentration of each analyte was determined by comparison to a standard curve according to the manufacturer's instructions. IL-4 levels were undetectable for all the specimens. Concentrations (picograms per milliliter) were normalized to the weight of homogenized lung (range, 70.6 to 125.6 μg). The data show means and SD. *, P < 0.05; **, P < 0.005; ***, P < 0.0005; ****, P < 0.00005 by two-way ANOVA with Tukey's multiple-comparison test.
Following RSV 12-35 challenge on day 96 p.i., both A2-line19F- and DB1-QUAD-vaccinated mice demonstrated significantly higher levels of lung cytokines on day 4 postchallenge than unvaccinated mice, consistent with a secondary immune response (boost). Interestingly, DB1-QUAD-vaccinated mice experienced lower levels of pulmonary cytokines postchallenge than A2-line19F-vaccinated mice. This may have been because DB1-QUAD vaccination protected the mice against subsequent pulmonary inflammation postchallenge. Alternatively, the heterologous challenge strain (RSV 12-35) may not have boosted the immune response to DB1-QUAD to the same extent as it did to a homologous (A2-line19F) prime.
DB1-QUAD attenuation, efficacy, and immunogenicity in cotton rats.
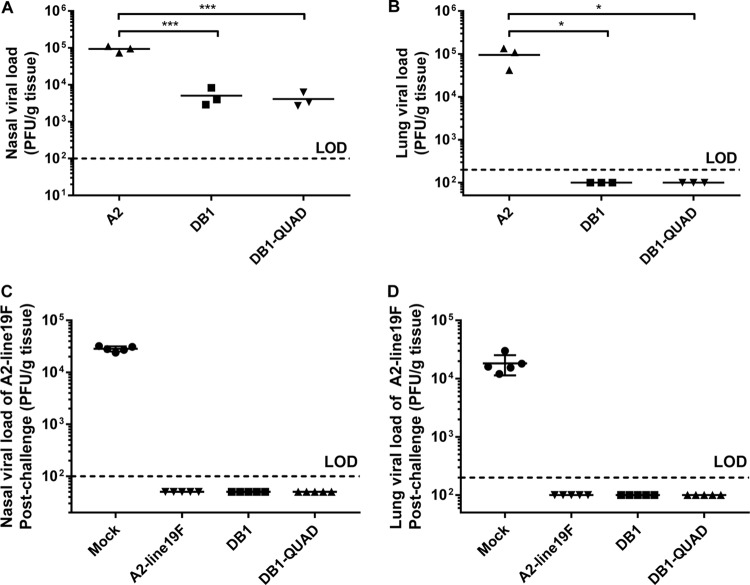
We then measured viral attenuation in S. hispidus cotton rats, an animal model that is more permissive to RSV infection than BALB/c mice. To do this, we infected groups of 5 cotton rats i.n. with 106 FFU of mKate2-labeled A2-line19F, DB1-QUAD, or DB1. On day 4 p.i., we sacrificed the animals and harvested the nasal turbinates and left lungs for titration. We found that DB1-QUAD was attenuated by 1.35 log10 PFU in the nasal homogenates (Fig. 7A) and was undetectable in cotton rat lungs (Fig. 7B). Thus, incorporation of line19F residues did not significantly affect vaccine attenuation levels in the cotton rat model.
FIG 7.
DB1-QUAD attenuation and efficacy in cotton rats. (A and B) To measure viral attenuation, groups of 3 cotton rats were inoculated i.n. with 106 FFU of mKate2-labeled A2, DB1, or DB1-QUAD, and viral loads in nasal (A) and lung (B) homogenates were measured by standard plaque assays. (C and D) To measure vaccine efficacy, groups of 5 cotton rats were similarly inoculated with either MEM (mock infection) or mKate2-labeled A2-line19F, DB1, or DB1-QUAD. The animals were challenged with A2-line19F on day 42 p.i., and viral loads in nasal (C) and lung (D) homogenates were quantified by plaque assays 4 days later. Each point represents a single animal, and the horizontal lines represent means. *, P < 0.05; ***, P < 0.0005 by two-way ANOVA with Tukey's multiple-comparison test.
We next assessed vaccine efficacy in cotton rats by measuring protection against RSV challenge. To do this, we again infected groups of 5 cotton rats i.n. with 106 FFU of mKate2-labeled A2-line19F, DB1-QUAD, DB1, or MEM (mock infection). We collected serum samples on days 0, 21, and 42 p.i. and challenged the animals with 106 FFU of A2-line19F on day 42 p.i. Four days later, we sacrificed the animals and harvested the nasal turbinates and left lungs for titration as described above. We found that A2-line19F, DB1, and DB1-QUAD vaccination completely protected cotton rats against A2-line19F challenge in both the upper and lower airways (Fig. 7C and D).
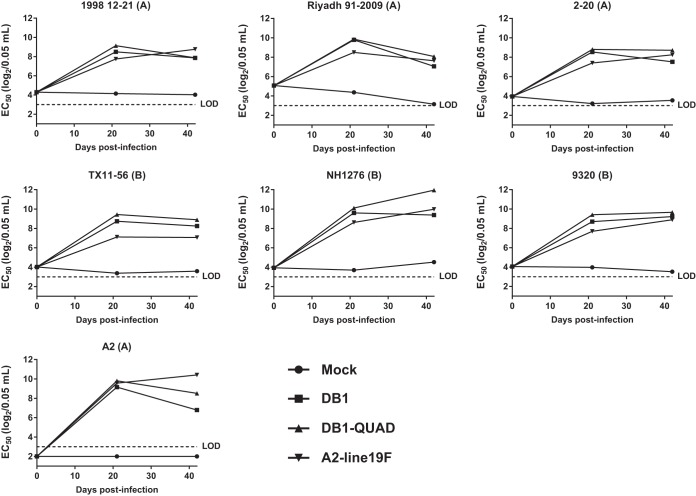
We subsequently pooled the serum samples for each group of animals at each time point and measured the breadth of immunogenicity by performing microneutralization assays for the RSV panel of 7 viruses as described above. Because there was only one replicate of pooled sera for each group of cotton rats at each time point, statistical comparisons could not be performed. However, we found that DB1-QUAD generated the highest nAb titers against the RSV B strains on the panel on days 21 and 42 p.i. (Fig. 8).
FIG 8.
Immunogenicity of DB1-QUAD in cotton rats. To assess immunogenicity, groups of 5 cotton rats were inoculated i.n. with either MEM (mock infection) or 106 FFU of mKate2-labeled A2-line19F, DB1, or DB1-QUAD. Sera were collected and pooled on days 0, 21, and 42 p.i., and neutralizing antibody responses to a panel of recombinant mKate2-labeled RSVs were measured by FFU reduction assays. The data represent mean EC50s of a single replicate of pooled sera from 5 cotton rats per group.
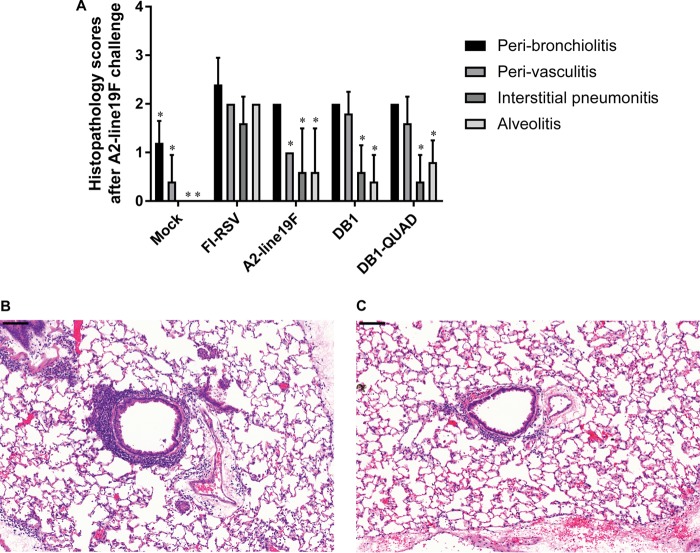
To analyze for vaccine-attributable enhanced disease following RSV challenge in cotton rats, we inoculated groups of 5 cotton rats i.n. with either MEM (mock infection), DB1, DB1-QUAD, or A2-line19F or intramuscularly (i.m.) with formalin-inactivated RSV (FI-RSV). Animals vaccinated with FI-RSV received an identical boost vaccination at day 21 p.i. We then challenged the animals with A2-line19F and harvested the lungs 6 days later for histopathologic scoring. Although our FI-RSV-vaccinated positive controls did not demonstrate as robust pulmonary histopathology as has been reported in previous analyses, we found that DB1 and DB1-QUAD had significantly lower histopathologic scores for interstitial pneumonitis and alveolitis than FI-RSV-vaccinated animals, with mean scores of less than 2 for all measured parameters (Fig. 9).
FIG 9.
Histopathology in cotton rats. To evaluate for vaccine-attributable enhanced disease postchallenge, groups of 5 cotton rats were inoculated i.n. with either MEM (mock infection), A2-line19F, DB1, or DB1-QUAD or i.m. with FI-RSV (lot 100; 1:100). Animals vaccinated with FI-RSV received an identical boost on day 21 postvaccination. (A) The animals were challenged with A2-line19F on day 42 p.i., lungs were harvested 4 days later, and histopathology scores were assigned. The data represent means plus SD. *, P < 0.05. (B and C) Representative hematoxylin and eosin stains for rats vaccinated with FI-RSV (B) and DB1-QUAD (C). Scale bars, 100 μm.
DISCUSSION
We boosted the thermostability and immunogenicity of an RSV live-attenuated vaccine candidate by substituting 4 unique line19F residues into the BAF protein of DB1. The resultant vaccine candidate, DB1-QUAD, had increased thermostability at 4°C and had a significantly higher proportion of prefusion to total F expression than DB1. BALB/c mice vaccinated with DB1-QUAD also had significantly higher neutralizing antibody titers to a panel of RSV A and B strains on days 56 and 100 p.i. than DB1-vaccinated mice. Although DB1-QUAD was moderately deattenuated in BALB/c mouse lungs compared to DB1, it remained fully attenuated in cotton rats and in normal human bronchial epithelial cells, which better model attenuation levels in seronegative infants. Thus, incorporation of unique line19F residues into the vaccine strain DB1 conferred enhanced thermostability, incorporation of prefusion F, and immunogenicity without compromising attenuation in cotton rats or primary cells.
Achieving a balance of attenuation and immunogenicity in an RSV LAV has proven challenging. However, DB1-QUAD was attenuated, broadly immunogenic, and efficacious in two animal models. In BALB/c mice, DB1-QUAD was attenuated by 1.02 log10 and 2.12 log10 FFU in lung homogenates on days 4 and 6 p.i., respectively, compared to unattenuated A2-line19F. However, DB1-QUAD elicited significantly higher neutralizing antibody titers to 7 chimeric RSV strains on day 100 p.i. than DB1 and A2-line19F. DB1-QUAD was also completely efficacious in protecting mice against the RSV subgroup A challenge strain 12-35. In cotton rats, DB1-QUAD was 1.35 log10 PFU attenuated in the upper airways, compared to unattenuated A2, and was completely undetectable in the lungs. Nevertheless, the vaccine generated broadly neutralizing antibodies to the panel of RSV A and B strains, with titers similar in magnitude to those generated by A2-line19F. DB1-QUAD completely protected cotton rats from A2-line19F challenge in both the upper and lower airways. Thus, DB1-QUAD achieved a balance of attenuation and immunogenicity in two animal models and was highly efficacious at protecting against RSV challenge.
The limitations of this study include its reliance upon the mouse and cotton rat models, which are only semipermissive to RSV infection and may not adequately predict human responses to infection. In terms of viral attenuation, for example, DB1-QUAD was 1.02 log10 and 2.12 log10 FFU attenuated in mouse lungs on days 4 and 6 p.i., respectively. In contrast, the vaccine was 1.35 log10 PFU attenuated in cotton rat nares and was completely undetectable in cotton rat lungs. The vaccine was also approximately 1 log10 FFU attenuated in NHBE cells, which better predict attenuation levels in seronegative infants. However, the base vaccine candidate, DB1, appeared to be more attenuated in NHBE cells in previously published results, which may have been a function of donor variability (26). Nevertheless, it remains unclear if DB1-QUAD would be sufficiently attenuated in the target population of young infants. Indeed, previous studies showed that LAV candidates with high attenuation levels in animal models were insufficiently attenuated in human clinical trials (9, 31). Similarly, although DB1-QUAD is highly immunogenic in the two animal models, future studies are needed to characterize the human immune responses to the vaccine.
Other remaining questions about DB1-QUAD include whether the thermostabilizing effects of the line19F mutations are sufficient for cold-chain production, distribution, and storage in a real-world setting. Although DB1-QUAD decreased in titer by only 1.38 log10 units over a period of 28 days at 4°C, a more thermostable construct could certainly be advantageous. The relative contributions of viral deattenuation versus stabilization of pre-F in terms of the boost in vaccine immunogenicity observed in BALB/c mice are also unclear. In cotton rats, the line19F residues did not deattenuate the vaccine strain, and the boost in immunogenicity was not as pronounced as in mice. However, only one replicate of pooled sera from each group of 5 cotton rats was available for neutralizing antibody assays, so statistical comparisons could not be performed for the animals.
The optimal immunization strategy and target population for an RSV vaccine also remain matters of debate. Although the majority of RSV mortality occurs in infants aged 0 to 3 months, active vaccination of these infants may be problematic due to the immaturity of the immune system, the persistence of maternal antibodies, and the potential for enhanced disease following administration of nonreplicating vaccines. The target population for an RSV LAV such as DB1-QUAD would likely be older seronegative infants (>3 months of age). An efficacious LAV administered to this age group could directly prevent a substantial proportion of RSV morbidity and mortality with lower risk of enhanced disease. Perhaps more importantly, if herd immunity could be achieved (32), such a vaccine could also protect the youngest infants from RSV exposure and subsequent morbidity and mortality. Indeed, epidemiologic studies have demonstrated that the most common sources of RSV exposure in young infants are household contacts with older infants and young children (33). Recent mathematical modeling studies have demonstrated that prevention of RSV disease in these older infants could confer vast public health benefits in terms of herd protection and reduction of hospitalizations among young infants (32), and because children are the most common sources of RSV in the elderly, this herd protection could likely confer benefits on older adults, as has been observed with other pediatric vaccines (34, 35). Thus, an efficacious RSV LAV could provide vast public health benefits both from the direct prevention of RSV in vaccine recipients and from indirect herd protection of other vulnerable populations.
In conclusion, substituting line19F residues into the RSV LAV DB1 conferred enhanced thermostability, increased incorporation of prefusion F, and increased immunogenicity in BALB/c mice without altering fusion activity in vitro. The resultant vaccine candidate, DB1-QUAD, was attenuated in mice and cotton rats but was also broadly immunogenic and protective against RSV challenge. DB1-QUAD is a promising RSV vaccine candidate that merits further evaluation. The development and administration of a safe and efficacious RSV LAV to seronegative infants could provide an important public health benefit.
MATERIALS AND METHODS
Cell lines and primary cell cultures.
HEp-2 and Vero cells were obtained from the ATCC and maintained in MEM with Earle's salts and l-glutamine (Gibco) supplemented with 10% fetal bovine serum (FBS) (HyClone) and 1 μg/ml of penicillin, streptomycin sulfate, and amphotericin B solution (PSA) (Life Technologies, Grand Island, NY). BEAS-2B bronchial epithelial cells (a gift from Pierre Massion, Nashville, TN) were maintained in RPMI 1640 (Cellgro), and 293T cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) (Gibco), both supplemented with 10% FBS and 1 μg/ml PSA. BSR T7/5 cells (a gift from Ursula Buchholz, National Institutes of Health, Bethesda, MD) were maintained in Glasgow's minimum essential medium (GMEM) (Gibco) supplemented with 10% FBS and 1 μg/ml PSA and, every other passage, with Geneticin at 1 mg/ml as previously described (36).
Freshly isolated passage 0 (P0) NHBE cells from otherwise healthy donors were isolated by the Experimental Models Support Core at Emory University using ex vivo tissues from deceased individuals donated for transplant and research purposes. Donor NHBE cells were isolated within 36 h of tissue donation via protease digestion and seeded onto human type IV placental-collagen-coated 6.5-mm Costar Transwells (Corning; 3470) at a density of 150,000 cells/filter in bilateral Emory-ALI medium. Emory-ALI differentiation medium consists of an equal mixture of low-glucose DMEM and LHC basal medium supplemented with pyruvate, l-glutamine, insulin, human epidermal growth factor, hydrocortisone, bovine pituitary extract, triiodothyronine, human holotransferrin, epinephrine, O-phosphorylethanolamine, ethanolamine, retinoic acid, human serum albumin, heparin, calcium, trace elements, voriconazole (an antifungal), and Primocin (a broad-spectrum antibiotic mixture). After 48 h of attachment and growth in Emory-ALI differentiation medium, unattached cells and spent medium were aspirated and replaced with fresh Emory-ALI differentiation medium for an additional 48 h. Subsequently, ALI differentiation was initiated via the removal of the apical medium, while only the basolateral medium was replaced with fresh Emory-ALI differentiation medium. The cells were fed Emory-ALI medium every other day (Monday, Wednesday, and Friday) for at least 2 weeks at the ALI to allow differentiation of the monolayer into a pseudostratified mucociliary culture, as determined by a rise in transepithelial resistance (TER) via the formation of tight junctions, exclusion of bulk liquid from the apical space, and the presence of ciliated and Muc5ac-producing cells by immunofluorescence imaging. Experimentation was performed between weeks 2 and 4 of ALI culture.
Cloning and rescue of DB1-QUAD.
The cloning and rescue of RSV recombinant viruses kRSV-A2 (where k denotes the far-red-fluorescent protein mKate2 in the first gene position), kRSV-A2-line19F, and kRSV-DB1 (kRSV-dNS-ΔSH-BAF) and the panel of mKate2-labeled RSV strains containing clinical F and G genes from viruses RSVA/human/USA/A1998-12-21 (JX069802), A/2001/2-20 (JF279545 and JF279544), Riyadh 91/2009 (JF714706 and JF714710), NH1276 (JQ680988 and JQ736678), 9320 (AY353550), and TX11-56 (JQ680989 and JQ736679) within an A2 backbone have all been previously described (26). Codon deoptimization of nonstructural protein genes in kRSV-DB1 was performed according to human codon usage bias. Codon usage bias in mice (Mus musculus) is similar to that in humans (37), and the least used codons for each amino acid are identical between the two species (38).
To generate the live-attenuated vaccine strain DB1-QUAD, we first performed sequential site-directed mutagenesis to substitute residues I79M, K191R, T357K, and N371Y from the thermostable line19F strain into the BAF protein encoded by a cDNA expression vector (pcDNA3.1+; Life Technologies, Carlsbad, CA). We cloned this modified BAF gene, denoted BAF-I79M/K191R/T357K/N371Y, into the antigenomic BAC vector pSynk-DB1 (pSynkRSV-A2-dNS-ΔSH-BAF) to generate the quadruple mutant pSynkRSV-A2-dNS-ΔSH-BAF-I79M/K191R/T357K/N371Y, denoted pSynk-DB1-QUAD. To rescue this virus, we cotransfected the antigenomic BAC with four human codon-optimized helper plasmids that expressed RSV N, P, M2-1, or L protein into BSR T7/5 cells as previously described (39). Master and working stocks of kRSV-DB1-QUAD were propagated and harvested in Vero cells (40).
In vitro growth analyses.
Subconfluent Vero and BEAS-2B cells were infected in duplicate or triplicate at a multiplicity of infection (MOI) of 0.01 FFU/cell with mKate2-labeled A2-line19F, DB1, or DB1-QUAD in 6-well plates using 500 μl inoculum per well and rocking for 1 h at room temperature. Full medium was then added to each well, and the plates were incubated at 37°C in 5% CO2. At time points 0, 12, 24, 36, 48, 72, and 96 h p.i., the wells were harvested by scraping the cells into the medium, and samples were snap-frozen in liquid nitrogen until titration by FFU assay as described previously (26).
Differentiated NHBE cells from a single donor were infected in duplicate at an MOI of 2 FFU/cell with mKate2-labeled viruses A2, DB1, and DB1-QUAD by applying 150 μl inoculum to each insert and incubating at 37°C for 2 h following a phosphate-buffered saline (PBS) wash. The inoculum was then removed with a single apical PBS wash. To collect virus at specified time points, 150 μl full differentiation medium was added to each apical chamber and incubated for 10 min at 37°C. This step was repeated once for each well (300 μl total), and the apical supernatant was snap-frozen in liquid nitrogen and stored at −80°C until subsequent analysis was performed as described previously (41).
Thermostability assays.
To analyze thermostability, working stocks of mKate2-labeled A2, A2-line19F, DB1, and DB1-QUAD were thawed for 15 min to room temperature, pooled in 15-ml conical tubes, and diluted in MEM to yield starting titers of 106 FFU/ml. Samples were then incubated in water baths at 4°C. At specified time points, 180 μl was removed from each tube, snap-frozen in liquid nitrogen, and stored at −80°C until subsequent analysis was performed. Samples were then thawed for 15 min to room temperature and titrated in duplicate by FFU assay as described previously (27). Four experimental replicates were performed for time points 0 to 14 days. Two experimental replicates were continued until 28 days.
Pre-F antigen ELISAs.
Pre-F antigen and motavizumab ELISAs were performed as previously described (27). Briefly, virus stocks of mKate2-labeled A2-line19F, DB1, and DB1-QUAD were diluted in MEM, applied to 96-well Costar assay plates, high binding (Corning), and incubated at room temperature overnight for coating. The next day, the plates were washed with 150 μl per well of PBS-Tween (PBST) (0.05% Tween 20 in PBS) and blocked with 150 μl per well of 5% bovine serum albumin (BSA) in PBST at room temperature for 2 h. Primary antibodies MPE-8 (pre-F specific) and motavizumab (pre- and post-F specific) were then serially diluted in 1% BSA in PBST to yield dilutions ranging from 1:10,000 to 1:320,000. Following blocking and PBST washing, dilutions of the primary antibodies were applied to plates and incubated at room temperature for 2 h. The plates were washed three times with PBST, and the secondary antibody, anti-human-horseradish peroxidase (HRP) diluted 1:10,000 in 1% BSA, was then applied and incubated at room temperature for 2 h. The plates were again washed three times with PBST, and 100 μl of a premixed reactive substrate reagent mixture (R&D Systems) was applied to catalyze a colorimetric reaction. The reaction was quenched with 0.2 N sulfuric acid, and the plates were read at 450 nm on an ELISA plate reader. Background absorbance was subtracted from the sample absorbance readings and plotted to a curve. The ratio of the area under the curve for MPE-8 to the area under the curve for motavizumab was calculated to determine the ratio of prefusion to total F protein expression.
DSP assays.
A dual split protein (DSP) cell-to-cell fusion activity assay was performed to quantify the fusion activity of RSV F proteins as previously described (26, 30, 36, 42). First, mammalian codon-optimized sequences of line19F, BAF, and BAF-I79M/K191R/T357K/N371Y were obtained from GeneArt and cloned into pcDNA3.1+. Subsequently, 293T cells at 90% confluence in 6-well plates were transfected with 500 fmol of DSP8–11 or cotransfected with DSP1–7 and an F expression plasmid using 10 μl Lipofectamine 2000 (ThermoFisher Scientific, Waltham, MA) in 500 μl Opti-MEM (ThermoFisher Scientific, Waltham, MA). The plates were rocked at room temperature for 1 h, followed by the addition of 500 μl of Opti-MEM and BMS-433771 fusion inhibitor (provided by Jin Hong, Alios Biopharma, San Francisco, CA) at a concentration of 600 nM (43). The plates were incubated at 37°C overnight. Sixteen hours posttransfection, cells were harvested in 1 ml medium supplemented with a 1:1,000 dilution of EnduRen live-cell luciferase substrate (Promega, Madison, WI). Target and effector cells were mixed at a 1:1 ratio and seeded into 96-well opaque plates at a density of 4 × 104 cells per 100 μl per well. The cells were then maintained at 37°C, and luciferase activity was measured at time points 0, 2, 4, 6, and 8 h postmixing using a TopCount NXT microplate scintillation and luminescence counter (PerkinElmer, Waltham, MA).
To ensure that F protein surface expression levels were consistent among transfected wells, cells were transfected as described above and harvested at 16 h posttransfection into PBS, transferred to fluorescence-activated cell sorter (FACS) tubes, and washed in PBS containing 2% FBS and 0.1% sodium azide. The cells were stained using the RSV F-specific monoclonal antibody palivizumab and a phycoerythrin (PE)-conjugated anti-human secondary antibody (Southern Biotech, Birmingham, AL). Flow cytometric analysis was performed using a Becton Dickinson LSRII flow cytometer, and data were analyzed using FlowJo software (Tree Star Inc., Ashland, OR) as previously described (30).
Animals.
Six- to 8-week-old female BALB/c mice obtained from the Jackson Laboratory (Bar Harbor, ME) were housed under pathogen-free conditions until the time of use. Experiments were performed according to the rules and regulations set forth by the Emory University Institutional Animal Care and Use Committee (IACUC).
Six- to 8-week-old female S. hispidus cotton rats were obtained from the inbred colony maintained at Sigmovir Biosystems, Inc. (SBI) (Rockville, MD). All experiments with cotton rats were performed using protocols that followed federal guidelines and were in accordance with the guidance of the SBI IACUC. Cotton rats were individually housed in polycarbonate cages and provided with standard rodent chow and tap water ad libitum.
Viral attenuation, immunogenicity, efficacy, and pulmonary cytokines in BALB/c mice.
To quantify viral attenuation, BALB/c mice (5 per group) were anesthetized intraperitoneally with ketamine-xylazine and infected intranasally with 106 FFU of either kA2-line19F, kDB1, or kDB1-QUAD in 100 μl serum-free MEM. On days 2, 4, 6, and 8 p.i., groups of 5 mice were weighed and sacrificed, and the left lungs were harvested and homogenized for FFU titration. Titers below the assay's limit of detection were assigned values equal to half the limit of detection, as previously described (26, 27, 36, 40, 41), because their true values were unknown. No significant changes in weight were observed in any of the groups compared to preinfection weight.
To measure immunogenicity, mice (5 per group) were anesthetized intraperitoneally with ketamine-xylazine and infected intranasally with either MEM or 106 FFU of kA2-line19F, kDB1, or kDB1-QUAD in 100 μl of serum-free MEM. On days 0, 28, 56, and 100 p.i., sera were collected by submandibular bleeding. Serum samples were pooled for each group of mice and heat inactivated for 30 min at 56°C. Microneutralization assays against a panel of chimeric RSV strains with clinical F and G proteins in an A2 backbone with mKate2 in the first gene position were performed by measuring FFU reduction as previously described (26, 41). Two experimental replicates consisting of 5 mice per group were performed.
To determine vaccine efficacy, the same mice were challenged on day 96 p.i. with RSV subgroup A clinical strain 12-35 (40, 41). This delayed time point for challenge was chosen to assess the durability of protection conferred by vaccination, as previously described (26, 27, 41). Four days later, the mice were sacrificed and the left lungs were harvested and homogenized for viral titration by standard immunodetection plaque assays on HEp-2 cells (40). Homogenized lung specimens were stored at −80°C until they were used for pulmonary cytokine analysis.
To measure pulmonary cytokines postvaccination, lung homogenates from mice infected with 106 FFU kDB1-QUAD or kA2-line19F on days 2, 4, 6, and 8 p.i. were obtained as described above. Similarly, to measure pulmonary cytokines postchallenge, lung homogenates from mice infected with MEM (mock infection), 106 FFU kDB1-QUAD, or 106 FFU kA2-line19F and subsequently challenged with 106 FFU RSV 12-35 on day 96 p.i. were obtained as described above 4 days postchallenge. The cell-free supernatants of centrifuged lung homogenates were analyzed using a mouse cytokine magnetic 20-plex panel kit (Invitrogen, Frederick, MD) and the Luminex 100/200 system. The concentration of each analyte was determined by comparison to a standard curve according to the manufacturer's instructions. Concentrations (picograms per milliliter) were normalized to the weight of homogenized lung (range, 70.6 to 125.6 μg).
Viral attenuation, immunogenicity, efficacy, and histopathology in cotton rats.
To measure viral attenuation in cotton rats, animals (5 per group) were lightly anesthetized with isoflurane and inoculated intranasally with 106 FFU of either kA2-line19F, kDB1, or kDB1-QUAD in 100 μl serum-free MEM. The cotton rats were euthanized using CO2 on day 4 p.i., and the nasal turbinates and left lungs were harvested for homogenization in 3 ml of Hanks' balanced salt solution (HBSS) supplemented with 10% sucrose-phosphate-glutamate (SPG). Lung and nasal homogenates were clarified by centrifugation and diluted in Eagle's minimal essential medium (EMEM). For titration, confluent HEp-2 monolayers were infected in duplicate with diluted homogenates in 24-well plates. The plates were incubated at 37°C for 1 h and overlaid with 0.75% methylcellulose. Four days later, the overlay was removed and the cells were fixed in 0.1% crystal violet stain for 1 h, rinsed, and air dried. Plaques were counted, and the virus titer was expressed as PFU per gram of tissue (26, 44). Titers below the limit of detection were assigned values equal to half the limit of detection.
To assess immunogenicity, cotton rats (5 per group) were lightly anesthetized with isoflurane and infected intranasally with either MEM, or 106 FFU of kA2-line19F, kDB1, or kDB1-QUAD. Sera were collected by retroorbital bleeding for animals on days 0, 21, and 42 p.i. and pooled for each group. Microneutralization assays against the panel of chimeric RSV strains were performed as described above. To assess vaccine efficacy, the same cotton rats were challenged on day 42 p.i. with kA2-line19F as previously described (26, 27). Four days later, the animals were euthanized, and the nasal turbinates and left lungs were harvested, homogenized, and titrated by plaque assay as described above.
We evaluated for enhanced lung histopathology attributable to vaccination following RSV challenge in cotton rats as previously described (26, 45). Briefly, we vaccinated groups of 5 cotton rats intranasally with 105 FFU of either A2-line19F, DB1, DB1-QUAD, or MEM (mock infection) or we intramuscularly injected 100 μl FI-RSV (lot 100; 1:100). On day 21 postvaccination, we administered an identical boost to the FI-RSV-vaccinated animals. On day 42 postvaccination, we challenged the animals with 106 FFU of RSV strain A2-line19F, and we harvested the lungs 4 days later. The lungs were inflated intratracheally with 10% neutral buffered formalin, embedded in paraffin, cut into 4-μm sections, and stained with hematoxylin and eosin. Blinded histopathologic scoring was performed for four parameters (peribronchiolitis, perivasculitis, interstitial pneumonitis, and alveolitis), with a maximum value of 4 and a minimum of 0.
Statistical analyses.
Statistical analyses were performed using GraphPad (San Diego, CA) Prism software, version 6.0 or 7.0. The data represent means ± standard deviations (SD) or standard errors of the mean (SEM), as indicated. Statistical comparisons were performed using Student's t test, one-way analysis of variance (ANOVA), or two-way ANOVA with Tukey's post hoc multiple-comparison tests as indicated. P values of ≤0.05 were considered significant.
ACKNOWLEDGMENTS
We thank Michael C. Currier for technical assistance, Ursula Bucholz and Karl-Klaus Conzelmann for BSR-T7/5 cells, Nancy Ulbrandt for motavizumab, Jin Hong and Jerome Deval (Alios) for the fusion inhibitor BMS-433771, and the Emory+Children's Pediatric Research Center Flow Cytometry Core. NHBE cells were provided by the CF@LANTA RDP Experimental Models Support Core (CF@LANTA/Cystic Fibrosis Foundation Research and Development Program Center grant MCCART15R0) as part of the Emory+Children's Center for Cystic Fibrosis and Airways Disease Research.
We declare the following competing financial interests. J.C.G.B. is an employee of Sigmovir Biosystems, Inc., a contract research organization that uses the cotton rat model of infectious diseases. M.L.M. cofounded Meissa Vaccines, Inc., and serves as the chief executive officer for the company. M.L.M. and C.A.R. are coinventors of the RSV vaccine evaluated in this study.
C.A.R., S.O.T., S.A.M., S.L., and J.C.G.B. performed the experiments. C.A.R., C.C.S., S.L., and M.L.M. designed the studies and analyzed the data. C.A.R. wrote the paper.
M.L.M., C.A.R., and S.L. were funded by HHS, NIH, National Institute of Allergy and Infectious Diseases (NIAID) (1R01AI087798 and 1U19AI095227). C.A.R. was funded by HHS, NIH, National Institute of Child Health and Human Development (NICHD) (5K12HD072245).
REFERENCES
- 1.Shi T, McAllister DA, O'Brien KL, Simoes EAF, Madhi SA, Gessner BD, Polack FP, Balsells E, Acacio S, Aguayo C, Alassani I, Ali A, Antonio M, Awasthi S, Awori JO, Azziz-Baumgartner E, Baggett HC, Baillie VL, Balmaseda A, Barahona A, Basnet S, Bassat Q, Basualdo W, Bigogo G, Bont L, Breiman RF, Brooks WA, Broor S, Bruce N, Bruden D, Buchy P, Campbell S, Carosone-Link P, Chadha M, Chipeta J, Chou M, Clara W, Cohen C, de Cuellar E, Dang DA, Dash-Yandag B, Deloria-Knoll M, Dherani M, Eap T, Ebruke BE, Echavarria M, de Freitas Lazaro Emediato CC, Fasce RA, Feikin DR, Feng L, Gentile A, Gordon A, Goswami D, Goyet S, Groome M, Halasa N, Hirve S, Homaira N, Howie SRC, Jara J, Jroundi I, Kartasasmita CB, Khuri-Bulos N, Kotloff KL, Krishnan A, Libster R, Lopez O, Lucero MG, Lucion F, Lupisan SP, Marcone DN, McCracken JP, Mejia M, Moisi JC, Montgomery JM, Moore DP, Moraleda C, Moyes J, Munywoki P, Mutyara K, Nicol MP, Nokes DJ, Nymadawa P, da Costa Oliveira MT, Oshitani H, Pandey N, Paranhos-Baccalà G, Phillips LN, Picot VS, Rahman M, Rakoto-Andrianarivelo M, Rasmussen ZA, Rath BA, Robinson A, Romero C, Russomando G, Salimi V, Sawatwong P, Scheltema N, Schweiger B, Scott JAG, Seidenberg P, Shen K, Singleton R, Sotomayor V, Strand TA, Sutanto A, Sylla M, Tapia MD, Thamthitiwat S, Thomas ED, Tokarz R, Turner C, Venter M, Waicharoen S, Wang J, Watthanaworawit W, Yoshida LM, Yu H, Zar HJ, Campbell H, Nair H, RSV Global Epidemiology Network. 2017. Global, regional, and national disease burden estimates of acute lower respiratory infections due to respiratory syncytial virus in young children in 2015: a systematic review and modelling study. Lancet 390:946–958. doi: 10.1016/S0140-6736(17)30938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeVincenzo JP, Whitley RJ, Mackman RL, Scaglioni-Weinlich C, Harrison L, Farrell E, McBride S, Lambkin-Williams R, Jordan R, Xin Y, Ramanathan S, O'Riordan T, Lewis SA, Li X, Toback SL, Lin SL, Chien JW. 2014. Oral GS-5806 activity in a respiratory syncytial virus challenge study. N Engl J Med 371:711–722. doi: 10.1056/NEJMoa1401184. [DOI] [PubMed] [Google Scholar]
- 3.DeVincenzo JP, McClure MW, Symons JA, Fathi H, Westland C, Chanda S, Lambkin-Williams R, Smith P, Zhang Q, Beigelman L, Blatt LM, Fry J. 2015. Activity of oral ALS-008176 in a respiratory syncytial virus challenge study. N Engl J Med 373:2048–2058. doi: 10.1056/NEJMoa1413275. [DOI] [PubMed] [Google Scholar]
- 4.Zhu Q, McLellan JS, Kallewaard NL, Ulbrandt ND, Palaszynski S, Zhang J, Moldt B, Khan A, Svabek C, McAuliffe JM, Wrapp D, Patel NK, Cook KE, Richter BWM, Ryan PC, Yuan AQ, Suzich JA. 2017. A highly potent extended half-life antibody as a potential RSV vaccine surrogate for all infants. Sci Transl Med 9:eaaj1928. doi: 10.1126/scitranslmed.aaj1928. [DOI] [PubMed] [Google Scholar]
- 5.Carbonell-Estrany X, Simoes EA, Dagan R, Hall CB, Harris B, Hultquist M, Connor EM, Losonsky GA, Motavizumab Study Group. 2010. Motavizumab for prophylaxis of respiratory syncytial virus in high-risk children: a noninferiority trial. Pediatrics 125:e35–e51. doi: 10.1542/peds.2008-1036. [DOI] [PubMed] [Google Scholar]
- 6.Glenn GM, Fries LF, Thomas DN, Smith G, Kpamegan E, Lu H, Flyer D, Jani D, Hickman SP, Piedra PA. 2016. A randomized, blinded, controlled, dose-ranging study of a respiratory syncytial virus recombinant fusion (F) nanoparticle vaccine in healthy women of childbearing age. J Infect Dis 213:411–422. doi: 10.1093/infdis/jiv406. [DOI] [PubMed] [Google Scholar]
- 7.Munoz FM, Piedra PA, Glezen WP. 2003. Safety and immunogenicity of respiratory syncytial virus purified fusion protein-2 vaccine in pregnant women. Vaccine 21:3465–3467. doi: 10.1016/S0264-410X(03)00352-9. [DOI] [PubMed] [Google Scholar]
- 8.Bernstein DI, Malkin E, Abughali N, Falloon J, Yi T, Dubovsky F, MI-CP149 Investigators. 2012. Phase 1 study of the safety and immunogenicity of a live, attenuated respiratory syncytial virus and parainfluenza virus type 3 vaccine in seronegative children. Pediatr Infect Dis J 31:109–114. doi: 10.1097/INF.0b013e31823386f1. [DOI] [PubMed] [Google Scholar]
- 9.Karron RA, Luongo C, Thumar B, Loehr KM, Englund JA, Collins PL, Buchholz UJ. 2015. A gene deletion that up-regulates viral gene expression yields an attenuated RSV vaccine with improved antibody responses in children. Sci Transl Med 7:312ra175. doi: 10.1126/scitranslmed.aac8463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fries L, Shinde V, Stoddard JJ, Thomas DN, Kpamegan E, Lu H, Smith G, Hickman SP, Piedra P, Glenn GM. 2017. Immunogenicity and safety of a respiratory syncytial virus fusion protein (RSV F) nanoparticle vaccine in older adults. Immun Ageing 14:8. doi: 10.1186/s12979-017-0090-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simoes EA, Tan DH, Ohlsson A, Sales V, Wang EE. 2001. Respiratory syncytial virus vaccine: a systematic overview with emphasis on respiratory syncytial virus subunit vaccines. Vaccine 20:954–960. doi: 10.1016/S0264-410X(01)00388-7. [DOI] [PubMed] [Google Scholar]
- 12.Anonymous. 1998. Palivizumab, a humanized respiratory syncytial virus monoclonal antibody, reduces hospitalization from respiratory syncytial virus infection in high-risk infants. The IMpact-RSV Study Group. Pediatrics 102:531–537. [PubMed] [Google Scholar]
- 13.Feltes TF, Cabalka AK, Meissner HC, Piazza FM, Carlin DA, Top FH Jr, Connor EM, Sondheimer HM, Cardiac Synagis Study Group. 2003. Palivizumab prophylaxis reduces hospitalization due to respiratory syncytial virus in young children with hemodynamically significant congenital heart disease. J Pediatr 143:532–540. doi: 10.1067/S0022-3476(03)00454-2. [DOI] [PubMed] [Google Scholar]
- 14.American Academy of Pediatrics Committee on Infectious Disease, American Academy of Pediatrics Bronchiolitis Guidelines Committee. 2014. Updated guidance for palivizumab prophylaxis among infants and young children at increased risk of hospitalization for respiratory syncytial virus infection. Pediatrics 134:415–420. doi: 10.1542/peds.2014-1665. [DOI] [PubMed] [Google Scholar]
- 15.Kim HW, Canchola JG, Brandt CD, Pyles G, Chanock RM, Jensen K, Parrott RH. 1969. Respiratory syncytial virus disease in infants despite prior administration of antigenic inactivated vaccine. Am J Epidemiol 89:422–434. doi: 10.1093/oxfordjournals.aje.a120955. [DOI] [PubMed] [Google Scholar]
- 16.Connors M, Kulkarni AB, Firestone CY, Holmes KL, Morse HC III, Sotnikov AV, Murphy BR. 1992. Pulmonary histopathology induced by respiratory syncytial virus (RSV) challenge of formalin-inactivated RSV-immunized BALB/c mice is abrogated by depletion of CD4+ T cells. J Virol 66:7444–7451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kakuk TJ, Soike K, Brideau RJ, Zaya RM, Cole SL, Zhang JY, Roberts ED, Wells PA, Wathen MW. 1993. A human respiratory syncytial virus (RSV) primate model of enhanced pulmonary pathology induced with a formalin-inactivated RSV vaccine but not a recombinant FG subunit vaccine. J Infect Dis 167:553–561. doi: 10.1093/infdis/167.3.553. [DOI] [PubMed] [Google Scholar]
- 18.Murphy BR, Prince GA, Walsh EE, Kim HW, Parrott RH, Hemming VG, Rodriguez WJ, Chanock RM. 1986. Dissociation between serum neutralizing and glycoprotein antibody responses of infants and children who received inactivated respiratory syncytial virus vaccine. J Clin Microbiol 24:197–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Polack FP, Teng MN, Collins PL, Prince GA, Exner M, Regele H, Lirman DD, Rabold R, Hoffman SJ, Karp CL, Kleeberger SR, Wills-Karp M, Karron RA. 2002. A role for immune complexes in enhanced respiratory syncytial virus disease. J Exp Med 196:859–865. doi: 10.1084/jem.20020781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Graham BS, Henderson GS, Tang YW, Lu X, Neuzil KM, Colley DG. 1993. Priming immunization determines T helper cytokine mRNA expression patterns in lungs of mice challenged with respiratory syncytial virus. J Immunol 151:2032–2040. [PubMed] [Google Scholar]
- 21.Schneider-Ohrum K, Cayatte C, Bennett AS, Rajani GM, McTamney P, Nacel K, Hostetler L, Cheng L, Ren K, O'Day T, Prince GA, McCarthy MP. 2017. Immunization with low doses of recombinant postfusion or prefusion respiratory syncytial virus F primes for vaccine-enhanced disease in the cotton rat model independently of the presence of a Th1-biasing (GLA-SE) or Th2-biasing (alum) adjuvant. J Virol 91:e02180-. doi: 10.1128/JVI.02180-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wright PF, Karron RA, Belshe RB, Shi JR, Randolph VB, Collins PL, O'Shea AF, Gruber WC, Murphy BR. 2007. The absence of enhanced disease with wild type respiratory syncytial virus infection occurring after receipt of live, attenuated, respiratory syncytial virus vaccines. Vaccine 25:7372–7378. doi: 10.1016/j.vaccine.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karron RA, Buchholz UJ, Collins PL. 2013. Live-attenuated respiratory syncytial virus vaccines. Curr Top Microbiol Immunol 372:259–284. doi: 10.1007/978-3-642-38919-1_13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karron RA, Buonagurio DA, Georgiu AF, Whitehead SS, Adamus JE, Clements-Mann ML, Harris DO, Randolph VB, Udem SA, Murphy BR, Sidhu MS. 1997. Respiratory syncytial virus (RSV) SH and G proteins are not essential for viral replication in vitro: clinical evaluation and molecular characterization of a cold-passaged, attenuated RSV subgroup B mutant. Proc Natl Acad Sci U S A 94:13961–13966. doi: 10.1073/pnas.94.25.13961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Karron RA, Wright PF, Belshe RB, Thumar B, Casey R, Newman F, Polack FP, Randolph VB, Deatly A, Hackell J, Gruber W, Murphy BR, Collins PL. 2005. Identification of a recombinant live attenuated respiratory syncytial virus vaccine candidate that is highly attenuated in infants. J Infect Dis 191:1093–1104. doi: 10.1086/427813. [DOI] [PubMed] [Google Scholar]
- 26.Rostad CA, Stobart CC, Gilbert BE, Pickles RJ, Hotard AL, Meng J, Blanco JC, Moin SM, Graham BS, Piedra PA, Moore ML. 2016. A recombinant respiratory syncytial virus vaccine candidate attenuated by a low-fusion F protein is immunogenic and protective against challenge in cotton rats. J Virol 90:7508–7518. doi: 10.1128/JVI.00012-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stobart CC, Rostad CA, Ke Z, Dillard RS, Hampton CM, Strauss JD, Yi H, Hotard AL, Meng J, Pickles RJ, Sakamoto K, Lee S, Currier MG, Moin SM, Graham BS, Boukhvalova MS, Gilbert BE, Blanco JC, Piedra PA, Wright ER, Moore ML. 2016. A live RSV vaccine with engineered thermostability is immunogenic in cotton rats despite high attenuation. Nat Commun 7:13916. doi: 10.1038/ncomms13916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wright PF, Ikizler MR, Gonzales RA, Carroll KN, Johnson JE, Werkhaven JA. 2005. Growth of respiratory syncytial virus in primary epithelial cells from the human respiratory tract. J Virol 79:8651–8654. doi: 10.1128/JVI.79.13.8651-8654.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ngwuta JO, Chen M, Modjarrad K, Joyce MG, Kanekiyo M, Kumar A, Yassine HM, Moin SM, Killikelly AM, Chuang GY, Druz A, Georgiev IS, Rundlet EJ, Sastry M, Stewart-Jones GB, Yang Y, Zhang B, Nason MC, Capella C, Peeples ME, Ledgerwood JE, McLellan JS, Kwong PD, Graham BS. 2015. Prefusion F-specific antibodies determine the magnitude of RSV neutralizing activity in human sera. Sci Transl Med 7:309ra162. doi: 10.1126/scitranslmed.aac4241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hotard AL, Lee S, Currier MG, Crowe JE Jr, Sakamoto K, Newcomb DC, Peebles RS Jr, Plemper RK, Moore ML. 2015. Identification of residues in the human respiratory syncytial virus fusion protein that modulate fusion activity and pathogenesis. J Virol 89:512–522. doi: 10.1128/JVI.02472-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wright PF, Karron RA, Belshe RB, Thompson J, Crowe JE Jr, Boyce TG, Halburnt LL, Reed GW, Whitehead SS, Anderson EL, Wittek AE, Casey R, Eichelberger M, Thumar B, Randolph VB, Udem SA, Chanock RM, Murphy BR. 2000. Evaluation of a live, cold-passaged, temperature-sensitive, respiratory syncytial virus vaccine candidate in infancy. J Infect Dis 182:1331–1342. doi: 10.1086/315859. [DOI] [PubMed] [Google Scholar]
- 32.Kinyanjui TM, House TA, Kiti MC, Cane PA, Nokes DJ, Medley GF. 2015. Vaccine induced herd immunity for control of respiratory syncytial virus disease in a low-income country setting. PLoS One 10:e0138018. doi: 10.1371/journal.pone.0138018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Munywoki PK, Koech DC, Agoti CN, Kibirige N, Kipkoech J, Cane PA, Medley GF, Nokes DJ. 2015. Influence of age, severity of infection, and co-infection on the duration of respiratory syncytial virus (RSV) shedding. Epidemiol Infect 143:804–812. doi: 10.1017/S0950268814001393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson EJ, Shippee DB, Weinrobe MH, Davila MD, Katz BZ, Reddy S, Cuyugan MG, Lee SY, Simons YM, Yogev R, Noskin GA. 2013. Indirect protection of adults from rotavirus by pediatric rotavirus vaccination. Clin Infect Dis 56:755–760. doi: 10.1093/cid/cis1010. [DOI] [PubMed] [Google Scholar]
- 35.Centers for Disease Control and Prevention. 2005. Direct and indirect effects of routine vaccination of children with 7-valent pneumococcal conjugate vaccine on incidence of invasive pneumococcal disease—United States, 1998-2003. MMWR Morb Mortal Wkly Rep 54:893–897. [PubMed] [Google Scholar]
- 36.Stokes KL, Currier MG, Sakamoto K, Lee S, Collins PL, Plemper RK, Moore ML. 2013. The respiratory syncytial virus fusion protein and neutrophils mediate the airway mucin response to pathogenic respiratory syncytial virus infection. J Virol 87:10070–10082. doi: 10.1128/JVI.01347-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Castro-Chavez F. 2011. Most used codons per amino acid and per genome in the code of man compared to other organisms according to the rotating circular genetic code. Neuroquantology 9:500. doi: 10.14704/nq.2011.9.4.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Elzanowski A, Ostell J. 2016. The genetic codes. https://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi Accessed 6 December 2017.
- 39.Hotard AL, Shaikh FY, Lee S, Yan D, Teng MN, Plemper RK, Crowe JE Jr, Moore ML. 2012. A stabilized respiratory syncytial virus reverse genetics system amenable to recombination-mediated mutagenesis. Virology 434:129–136. doi: 10.1016/j.virol.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stokes KL, Chi MH, Sakamoto K, Newcomb DC, Currier MG, Huckabee MM, Lee S, Goleniewska K, Pretto C, Williams JV, Hotard A, Sherrill TP, Peebles RS Jr, Moore ML. 2011. Differential pathogenesis of respiratory syncytial virus clinical isolates in BALB/c mice. J Virol 85:5782–5793. doi: 10.1128/JVI.01693-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Meng J, Lee S, Hotard AL, Moore ML. 2014. Refining the balance of attenuation and immunogenicity of respiratory syncytial virus by targeted codon deoptimization of virulence genes. mBio 5:e01704–e01714. doi: 10.1128/mBio.01704-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ishikawa H, Meng F, Kondo N, Iwamoto A, Matsuda Z. 2012. Generation of a dual-functional split-reporter protein for monitoring membrane fusion using self-associating split GFP. Protein Eng Des Sel 25:813–820. doi: 10.1093/protein/gzs051. [DOI] [PubMed] [Google Scholar]
- 43.Cianci C, Langley DR, Dischino DD, Sun Y, Yu KL, Stanley A, Roach J, Li Z, Dalterio R, Colonno R, Meanwell NA, Krystal M. 2004. Targeting a binding pocket within the trimer-of-hairpins: small-molecule inhibition of viral fusion. Proc Natl Acad Sci U S A 101:15046–15051. doi: 10.1073/pnas.0406696101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Prince GA, Jenson AB, Horswood RL, Camargo E, Chanock RM. 1978. The pathogenesis of respiratory syncytial virus infection in cotton rats. Am J Pathol 93:771–791. [PMC free article] [PubMed] [Google Scholar]
- 45.Prince GA, Curtis SJ, Yim KC, Porter DD. 2001. Vaccine-enhanced respiratory syncytial virus disease in cotton rats following immunization with Lot 100 or a newly prepared reference vaccine. J Gen Virol 82:2881–2888. doi: 10.1099/0022-1317-82-12-2881. [DOI] [PubMed] [Google Scholar]