Abstract
TRIM5α is an important host restriction factor that could potently block retrovirus infection. The SPRY domain of TRIM5α mediates post-entry restriction by recognition of and binding to the retroviral capsid. Human TRIM5α also functions as an innate immune sensor to activate AP-1 and NF-κB signaling, which subsequently restrict virus replication. Previous studies have shown that the AP-1 and NF-κB signaling activation relies on the RING motif of TRIM5α. In this study, we have demonstrated that the SPRY domain is essential for rhesus macaque TRIM5α to activate AP-1 but not NF-κB signaling. The AP-1 activation mainly depends on all of the β-sheet barrel on SPRY structure of TRIM5α. Furthermore, the SPRY-mediated auto-ubiquitination of TRIM5α is required for AP-1 activation. This study reports that rhesus macaque TRIM5α mainly undergoes Lys27-linked and Met1-linked auto-polyubiquitination. Finally, we found that the TRIM5α signaling function was positively correlated with its retroviral restriction activity. This study discovered an important role of the SPRY domain in immune signaling and antiviral activity and further expanded our knowledge of the antiviral mechanism of TRIM5α.
Keywords: antiviral response, auto-ubiquitination, TAB2, TAK1, TRIM5alpha
Introduction
During the time that they have been exposed to retroviruses, mammalian cells have evolved many intracellular proteins that function as innate defenses against retroviral pathogens (1–6). Among these proteins, the tripartite motif containing 5α (TRIM5α) is particularly significant, as it expresses multifunctional antiviral activity. First, rhesus macaque TRIM5α (RhTRIM5α)2 inhibits retrovirus infection at the post-entry step by recognizing the retroviral capsid and then accelerating its premature uncoating, resulting in proteasomal degradation of the viral reverse transcription complex (4, 7). Second, RhTRIM5α decreases HIV-1 production through degradation of HIV-1 Gag proteins (8). Finally, human TRIM5α (huTRIM5α) has been identified as a novel pattern recognition receptor, sensing the retrovirus capsid lattice, and contributes to innate immune signaling (9).
TRIM5α is a member of the tripartite motif (TRIM) protein family, members of which are encoded by over 100 genes in humans (10). TRIM proteins have a common structure named RBCC that features three major motifs: N-terminal RING, B-box, and coiled-coil domains. Over half of the TRIMs contain a SPRY/B30.2 domain at the C terminus that mediates protein interactions. In most cases, the RING domain has E3 ligase activity (11, 12) and activates Ubc13-ubiquitin (Ub) conjugate through its dimerization (13). The B-box and coiled-coil domains promote its oligomerization, which is required for the TRIM protein to form cytoplasmic or nuclear bodies (14–18). Furthermore, the RBCC motif was found to contribute to TRIM assembly, which is critical for the TRIM protein's ubiquitination activity (19). The SPRY domain of TRIM5α mediates post-entry restriction by recognizing and binding to the retroviral capsid (20–22). However, the SPRY domain has recently been considered to serve some unknown functions as well as binding to the capsid (23, 24). For instance, four putative SUMO-interacting motifs were reported in the SPRY domain, and SUMO-interacting motifs are responsible for the antiviral activity of TRIM5α (25–27). The SPRY domain may have additional functions that require further investigation.
HuTRIM5α regulates immune signaling mainly by interacting with mitogen-activated protein kinase kinase kinase 7 (TAK1) and then activates downstream pathways, including activator protein-1 (AP-1) and nuclear factor-κB (NF-κB) signaling (6, 9, 28). TAK1 is an important MAP3K activated by unanchored polyubiquitin chains (29). Quite a few studies indicate that TAK1 polyubiquitination is involved in signaling pathways (30–32). HuTRIM5α recruits E2 Ub-conjugating enzymes UBC13-UEV1A by the RING domain to generate free Lys63-linked polyubiquitination, resulting in TAK1 activation (9). However, the roles of the C-terminal function of TRIM5α in signaling activation and the mechanisms involved are still to be investigated.
This study was planned to evaluate (i) whether and how the SPRY domain of RhTRIM5α contributes to innate immune signaling and (ii) whether this function is correlated to its antiviral activity. In this study, it was demonstrated for the first time that the SPRY domain of RhTRIM5α was vital for activating the AP-1 signal, but not NF-κB. The molecular basis of SPRY for the activation of AP-1 by RhTRIM5α was mapped to the β-sheet barrel on the SPRY structure. Moreover, AP-1 activation of RhTRIM5α was found to be positively correlated with its auto-ubiquitination. A significant finding was that RhTRIM5α mainly undergoes Lys27- and Met1-linked auto-ubiquitination.
Results
SPRY is indispensable for TRIM5α activating AP-1 signaling
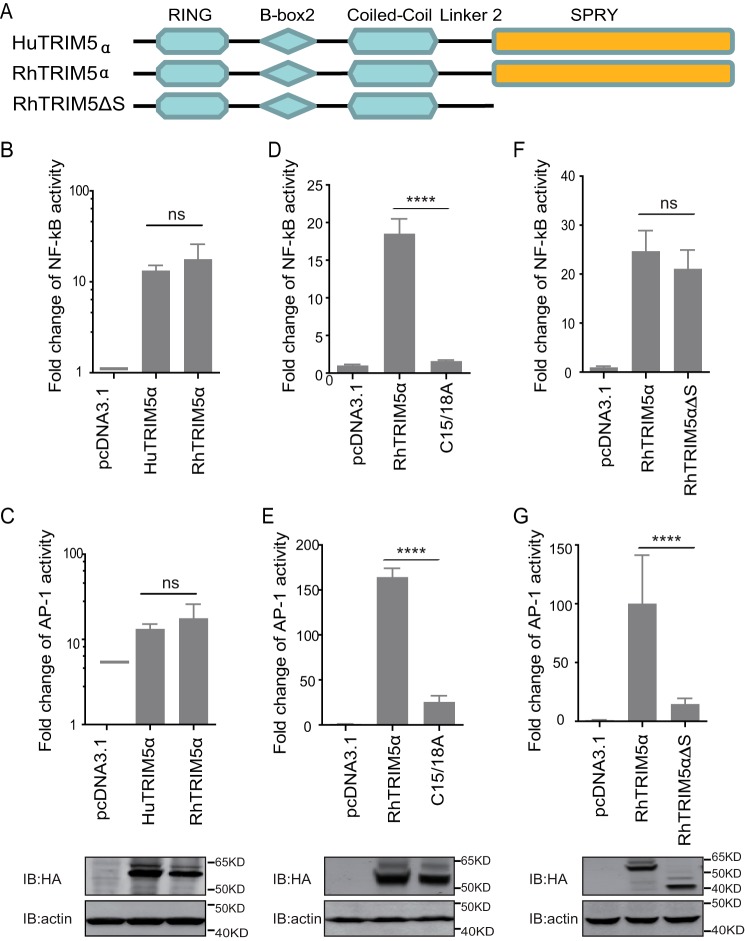
HuTRIM5α has been identified to activate AP-1 and NF-κB. RhTRIM5α and huTRIM5α were compared for their NF-κB and AP-1 signaling activation capacity. We found that RhTRIM5α stimulated both NF-κB and AP-1 transcriptional reporters with magnitudes similar to those of human TRIM5α (Fig. 1, A–C). We then tested RhTRIM5αC15/18A (C15/18A), a RING E3 Ub-ligase domain mutation construct (24). It was found that C15/18A had a reduced capacity to activate NF-κB and AP-1 of around 5–6-fold (Fig. 1, D and E), which is similar to the findings of a previous study that used HuTRIM5α (9). C terminus–truncated RhTRIM5 with a deletion of the SPRY domain (RhTRIM5ΔS) was constructed to test and determine the functionality of the SPRY domain of TRIM5α in signal transduction, (Fig. 1A). Surprisingly, RhTRIM5ΔS did not alter the capacity to activate NF-κB, indicating that the SPRY domain may not be necessary for NF-κB signal activation (Fig. 1F). However, RhTRIM5ΔS dramatically lost the capacity to activate AP-1 (Fig. 1G), suggesting that SPRY plays a critical role in AP-1 signaling activation.
Figure 1.
SPRY is indispensable for TRIM5α-mediated activation of AP-1 signaling. A, schematic diagram of the indicated TRIM5 proteins and truncations with deletion (Δ) of the C-terminal domain. The colored region indicates the RING, B-box 2, coiled-coil, and C-terminal SPRY domain. B–G, HEK293T cells were transfected with the indicated pcDNA3.1-based expression plasmids and luciferase reporters for NF-κB (B, D, and F) or AP-1 (C, E, and G), followed by luciferase assay after 24 h. Bars, mean luciferase activity levels ± S.D. (error bars). All of these data were acquired from at least three independent experiments. Shown is immunoblot analysis (bottom) of HEK293T cells transfected with the indicated TRIM proteins and deletion mutants (top). Relative luciferase activity was measured and statistically analyzed by unpaired t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, no significant). p values of <0.05 were considered statistically significant.
Macaca fascicularis TRIM5α is deficient in AP-1 activation
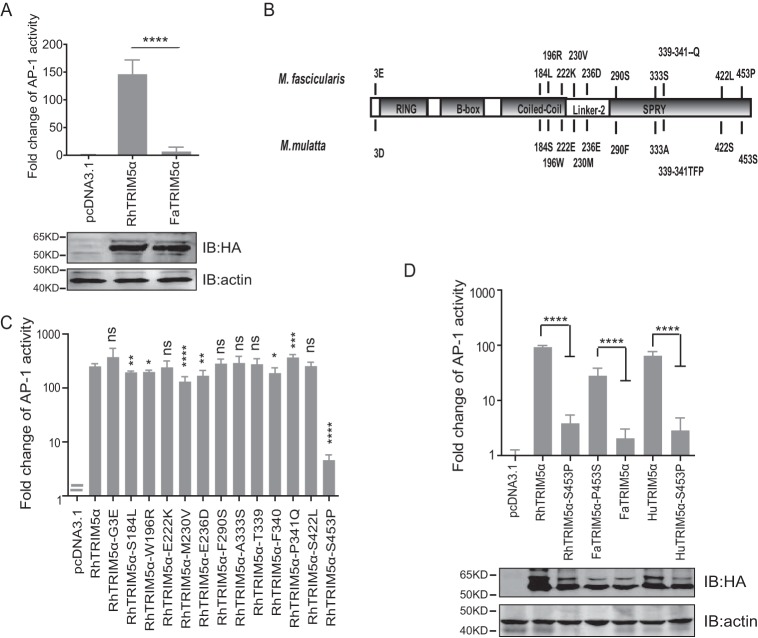
M. fascicularis, a closely related species of rhesus macaque, also encodes TRIM5α loci. Interestingly, it was found that M. fascicularis TRIM5α (FaTRIM5α) was deficient in activating AP-1 (Fig. 2A). By comparing the sequences of FaTRIM5α and RhTRIM5α, 13 amino acid differences were identified, including one located in the N terminus, three in the coiled-coil domain, two in Linker-2, and the other seven in SPRY (Fig. 2B). We further explored whether a single amino acid substitution could alter the signaling activity of RhTRIM5α. The results revealed that only the mutation of proline to serine at the 453 site on the SPRY (RhTRIM5α-S453P) aborted its ability to activate AP-1 (Fig. 2C). To confirm this result, HuTRIM5α-S453P and FaTRIM5α-P453S were constructed. As expected, HuTRIM5α-S453P lost the capacity to activate AP-1, whereas FaTRIM5α-P453S acquired this function (Fig. 2D). These results demonstrated that the mutation of serine at 453 of TRIM5α significantly influences its AP-1 activation.
Figure 2.
FaTRIM5α failed to activate AP-1. A, HEK293T cells were transfected with the individual TRIM expression plasmids and AP-1 luciferase reporter and TK. Reporter assays were performed at 24 h after transfection. Bars, mean luciferase activity levels ± S.D. (error bars) (n = 4). IB analysis (bottom) of HEK293T cells transfected with the indicated TRIM5α proteins (top). B, schematic diagram of the different amino acids between RhTRIM5α and FaTRIM5α. C, reporter assays of a panel of RhTRIM5α mutations, for which amino acids were replaced by corresponding ones in FaTRIM5α, were performed similarly as in A. D, HEK293T cells were transfected with the AP-1 promoter reporter plasmid and with the mutants of RhTRIM5α, FaTRIM5α, and HuTRIM5α (expression levels were detected by IB) and were then subjected to a Dual-Luciferase assay. Bars, mean luciferase activity levels ± S.D. (error bars). All of these data were acquired from at least three independent experiments. Relative luciferase activity was measured and statistically analyzed by unpaired t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant). p values of <0.05 were considered statistically significant.
Whole β-sheet barrel of SPRY was significant for TRIM5α-mediated AP-1 activation
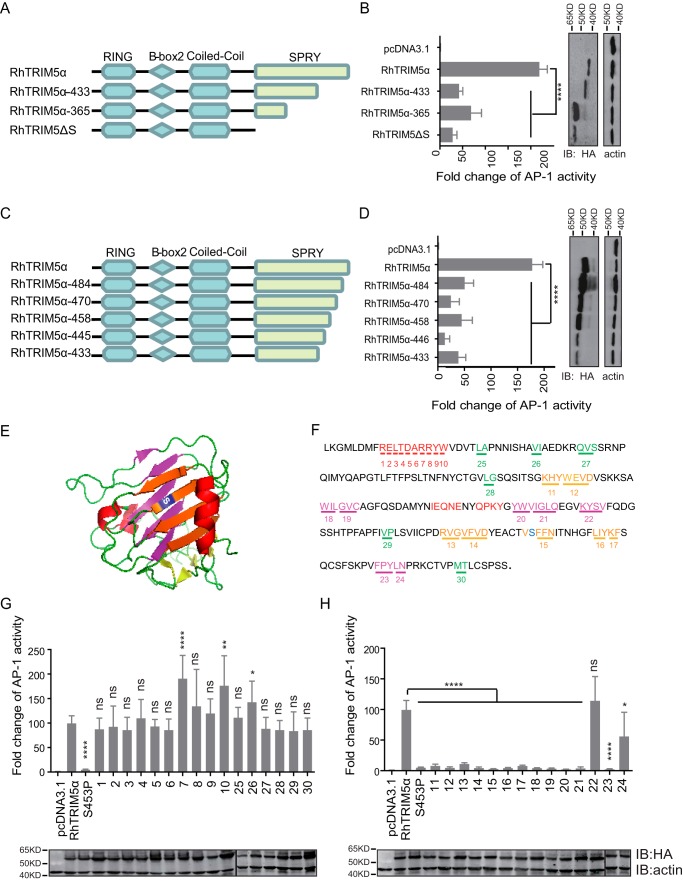
To identify the critical motif of RhTRIM5α for AP-1 activation, we constructed and tested its truncated variants, RhTRIM5α-365 and RhTRIM5α-433 (Fig. 3A). We found that both of the molecules showed a dramatic loss in their capacity to activate AP-1 (Fig. 3B). To confirm the key motif of RhTRIM5α, other truncated variants were tested (Fig. 3C). RhTRIM5α-484, RhTRIM5α-458, RhTRIM5α-433, RhTRIM5α-470, and RhTRIM5α-445 had 70–90% reductions in AP-1 activation compared with wild-type RhTRIM5α (Fig. 3D). These results imply that intact SPRY is required for RhTRIM5α to activate AP-1.
Figure 3.
The entire β-sheet barrel of SPRY was significant for TRIM5α-mediated AP-1 activation. A and C, schematic diagram of the indicated WT RhTRIM5α and truncations. The amino acid numbers are shown. B and D, the indicated plasmids were cotransfected with AP-1 reporter and TK into HEK293T cells, followed by a luciferase assay after 24 h. HEK293T cells were used for IB analysis with anti-HA and anti-actin antibodies. E, schematic representation of the RhTRIM5α SPRY domain crystal structure (Protein Data Bank code 2LM3) (38). The critical amino acid residue Ser453 is shown in blue and located on a larger sheet of SPRY. The larger sheets of the same side of Ser453 are shown in orange, β-sheets on the opposite side of Ser453 are colored purple, the biggest α-helix is colored red, and marginal small β-sheets are colored green. F, amino acid sequence of the RhTRIM5α SPRY domain. The colors indicate the same structures as in A. The numbers indicate the mutants of RhTRIM5α where residues were substituted by Ala. G and H, HEK293T cells were transfected with the individual TRIM expression plasmids and AP-1 luciferase reporter and TK. Reporter assays were performed at 24 h after transfection. Bars, mean luciferase activity levels ± S.D. (error bars). All of these data were acquired from at least three independent experiments. Relative luciferase activity was measured and statistically analyzed by unpaired t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001; ****, p < 0.0001; ns, not significant). p values of <0.05 were considered statistically significant. Shown is IB analysis (bottom) of HEK293T cells transfected with the indicated TRIM5α (top).
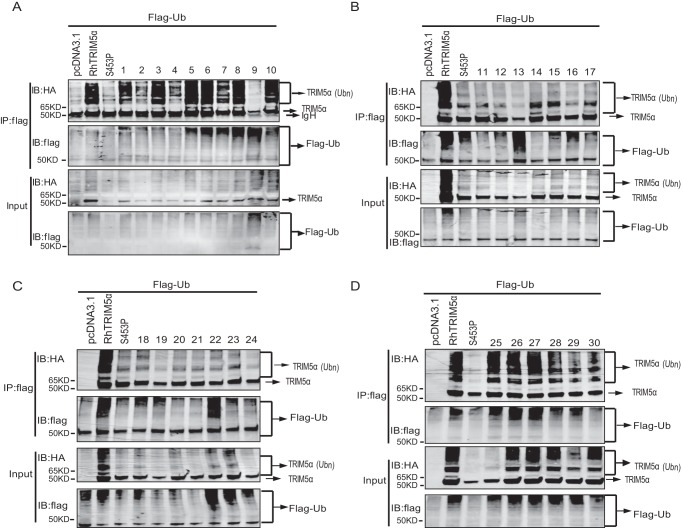
SPRY has a typical β-sheet barrel structure. To determine the key amino acids of TRIM5α for AP-1 activation, the SPRY structure was artificially marked into four parts: the biggest α-helix (red), the β-sheets on the same side of Ser453 (orange), the β-sheets on the opposite side of Ser453 (purple), and marginal small β-sheets (green) (Fig. 3E). Further, serial point mutations were constructed, named 1–30 (Fig. 3F), where the marked amino acids were mutated into arginine. Sites on the α-helix (marked in red) and the small β-sheets (marked in green) did not affect AP-1 activation (Fig. 3G). However, most variants on the β-sheet barrel lost this function, except for the number 22 mutant (Fig. 3H), which is a smaller β-sheet. These results indicated that the whole β-sheet barrel of SPRY was vital for TRIM5α-mediated AP-1 activation.
Polyubiquitination of TAK1 was not sufficient for TRIM5-mediated AP-1 signaling
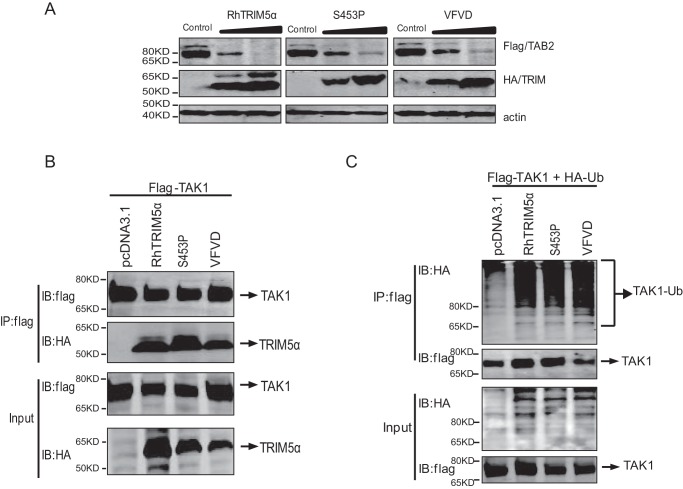
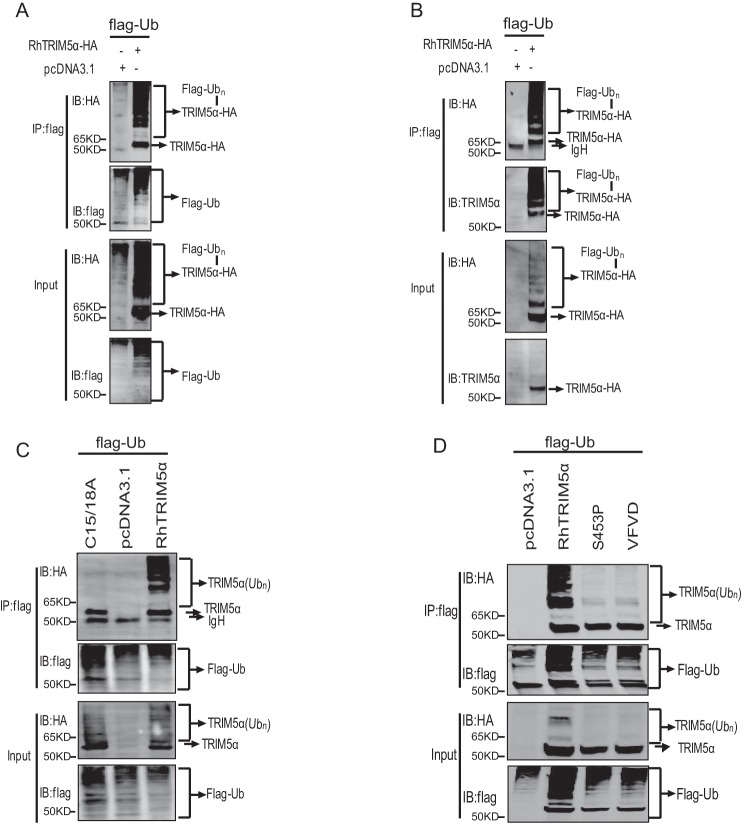
TAK1-binding protein 2 (TAB2) is involved in the direct interaction between the TAK1 complex and TRIM5α for AP-1 signaling activation. To explore the molecular mechanism of SPRY-mediated AP-1 activation, RhTRIM5α was compared with two selected inactive mutations in two nearby β-sheets of SPRY: RhTRIM5αS453P and RhTRIM5αVFVD (number 14 mutant). We found that RhTRIM5αS453P and RhTRIM5αVFVD degraded TAB2 to the same level as RhTRIM5α (Fig. 4A), indicating that TAB2 degradation is independent of TRIM5α-mediated AP-1 activation.
Figure 4.
Polyubiquitination of TAK1 was not sufficient for TRIM5-mediated AP-1 activation. A, HEK293T cells were cotransfected with 4 μg of TAB2-FLAG expression plasmid and increasing doses (0, 2, and 4 μg) of plasmids expressing RhTRIM5a (left), RhTRIM5αS453P (middle), or RhTRIM5αVFVD (right). Cells were lysed at 36 h post-transfection and examined by Western blotting using the indicated antibodies. B, co-immunoprecipitation (Co-IP) and IB analysis of HEK293T cells cotransfected with RhTRIM5α, RhTRIM5αS453P, or RhTRIM5αVFVD and FLAG-TAK1. C, HEK293T cells were cotransfected with 2.5 μg of FLAG-TAK1, 3 μg of Ub-HA, and 2.5 μg of RhTRIM5a, RhTRIM5αS453P, or RhTRIM5αVFVD. Thirty hours after transfection, cell lysates were immunoprecipitated with anti-FLAG beads followed by IB with anti-HA or anti-FLAG, as indicated.
HuTRIM5α can recruit E2 Ub-conjugating enzymes to generate free Lys63-linked polyubiquitin chains, which subsequently activate TAK1 (9). We next tested whether RhTRIM5αS453P and RhTRIM5αVFVD would bind to TAK1 and affect its ubiquitination. Both of the variants interacted well with TAK1 (Fig. 4B). The ubiquitination assay showed that RhTRIM5αS453P and RhTRIM5αVFVD could also catalyze TAK1 polyubiquitination (Fig. 4C). These data indicate that polyubiquitination of TAK1 was not sufficient for TRIM5-mediated AP-1 activation, suggesting that other unknown mechanisms may exist.
Auto-ubiquitination of TRIM5α mediated by SPRY is correlated with AP-1 signal activation
Auto-regulated polyubiquitination of many members of the TRIM family has been reported to be involved in regulating innate immune signaling (33, 34). In our study, the heavily polyubiquitinated TRIM5α was also detected by both HA antibody (Fig. 5A) and TRIM5α-specific polyclonal antibody (Fig. 5B), as per previous studies (11, 35–37), whereas the enzymatically inactive mutation, RhTRIM5αC15/18A, dramatically lost its polyubiquitination (Fig. 5C). This result indicates that TRIM5α ubiquitination is auto-regulated. It was further investigated whether RhTRIM5αS453P and RhTRIM5αVFVD also lost their auto-ubiquitination activities. As shown in Fig. 5D, RhTRIM5α was well auto-ubiquitinated, contrary to RhTRIM5αS453P and RhTRIM5αVFVD. This result indicates that RhTRIM5α auto-ubiquitination may be responsible for AP-1 activation. To confirm the role of TRIM5α auto-ubiquitination in AP-1 signaling, 29 variants of SPRY were tested for auto-regulating polyubiquitination. We found that mutations that activated AP-1 could auto-ubiquitinate (with the single exception of variant 9), whereas mutations that did not activate AP-1 failed to auto-ubiquitinate sufficiently (Fig. 6, A–D). These findings suggested that TRIM5α auto-ubiquitination was positively correlated with TRIM5α-mediated AP-1 activation.
Figure 5.
Auto-ubiquitination of RhTRIM5α was responsible for AP-1 activation. A and B, HEK293T cells were transfected with 3 μg of Ub-FLAG and 3 μg of HA-RhTRIM5α or pcDNA3.1 (negative control). Cell lysates were immunoprecipitated with anti-FLAG beads and were then used for IB analysis using the indicated antibodies. C and D, HEK293T cells were cotransfected with Ub-FLAG and RhTRIM5α, RhTRIM5αS453P, RhTRIM5αVFVD, or RhTRIM5αC15/18A using the same methods as in A. All results shown are representative of three independent experiments.
Figure 6.
The β-sheet barrel of SPRY was significant for TRIM5α auto-ubiquitination. HEK293T cells were cotransfected with Ub-FLAG and the 30 HA-RhTRIM5α variants or pcDNA as a negative control, indicated above the lanes. Thirty hours after transfection, cell lysates were immunoprecipitated (IP) with anti-FLAG beads, followed by immunoblots with anti-HA or anti-FLAG, as indicated. Variants labeled 1–10 in A showed mutations in the biggest α-helix. Variants labeled 11–24 in B and C showed mutations in the whole β-sheet barrel of the SPRY domain. Variants labeled 25–30 in D showed mutations in marginal small β-sheets. All experiments were performed three times, and a representative result is shown.
RhTRIM5α is modified with Met1-linked and Lys27-linked poly-Ub chains in HEK293T cells
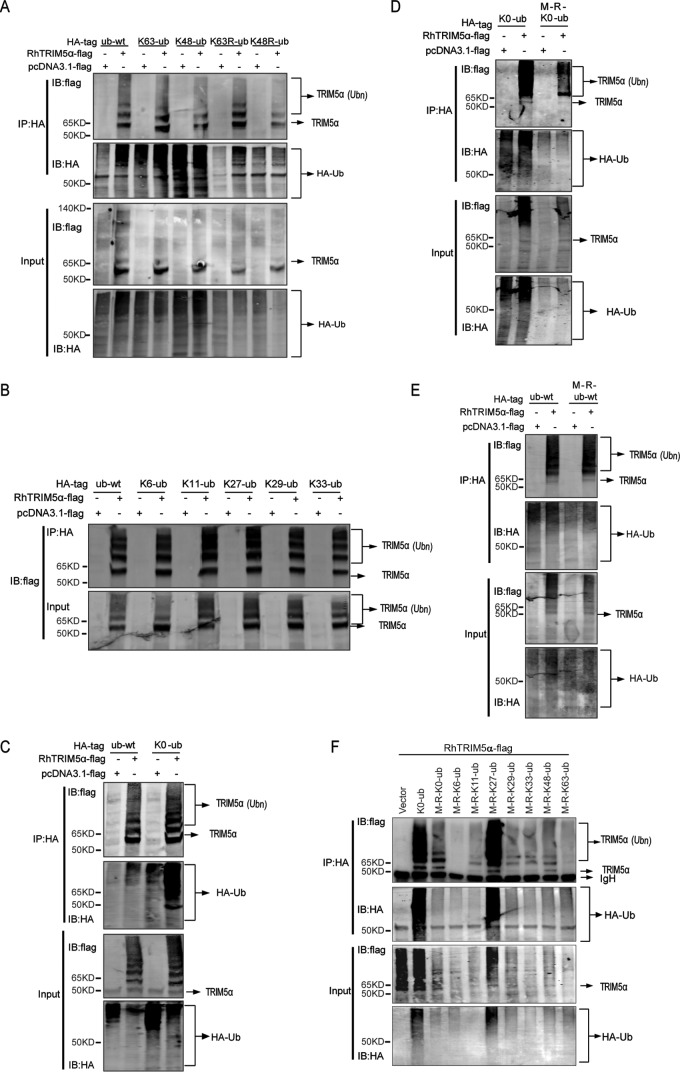
It is reported that HuTRIM5α is modified with Lys63-linked poly-Ub by Ube2W and Ube2N/Ube2V2. To analyze the type of ubiquitin modification of RhTRIM5α under conditions of its overexpression, we utilized K63-Ub (Lys63 only), K48-Ub, K63R (all residues except Arg63), and K48R mutant ubiquitin plasmids and found that RhTRIM5α was modified by both Lys63- and Lys48-linked ubiquitin chains (Fig. 7A). Ubiquitin has five lysine residues, which can be ubiquitinated to give rise to isopeptide-linked polyubiquitin chains with the exception of Lys48 and Lys63. To further identify other lysines' contribution to RhTRIM5α polyubiquitination, K6-, K11-, K27-, K29-, and K33-Ub plasmids were used to characterize the polyubiquitin linkages within the Ubn-RhTIM5α. It was clear that RhTRIM5α could be heavily polyubiquitinated in the presence of all of these Ub plasmids (Fig. 7B). In addition, Fig. 7C indicated that polyubiquitination of RhTIM5α was normal after expression of K0-Ub (seven Lys residues mutated to Arg, Lys-less). The results above indicated that RhTRIM5α undergoes “atypical” Met1-linked polyubiquitination.
Figure 7.
RhTRIM5α undergoes Met1-linked and Lys27-linked polyubiquitination in HEK293T cells. A–F, HEK293T cells were transfected with 2 μg of a series of Ub-HA and 4 μg of FLAG-RhTRIM5α or pcDNA3.1 (negative control). Cell lysates were immunoprecipitated with anti-HA beads and were then used for IB analysis. All results shown are representative of three independent experiments.
Intriguingly, formation of Ubn-RhTIM5α was efficiently blocked by expression of M-R-K0-Ub (seven Lys residues and Met1 mutated to Arg) (Fig. 7D). These results revealed that the polyubiquitin linkages within Ubn-RhTIM5α contained Met1-linked ubiquitin. Fig. 7E indicates that RhTIM5α undergoes other Lys-linked ubiquitination. We designed the experiments with a set of Ub plasmids containing only one Lys and no N-terminal Met1, because the mutant ubiquitin plasmids used in Fig. 7 (A and B) contained the N-terminal Met1 (the ORF of Ub), which could also be ubiquitinated. Fig. 7F shows that RhTIM5α undergoes Lys27-linked ubiquitination. Together, these results indicate that RhTRIM5α is modified with “atypical” Met1-linked and Lys27-linked poly-Ub chains in HEK293T cells.
AP-1 activity of RhTRIM5α was correlated with its antiviral activity
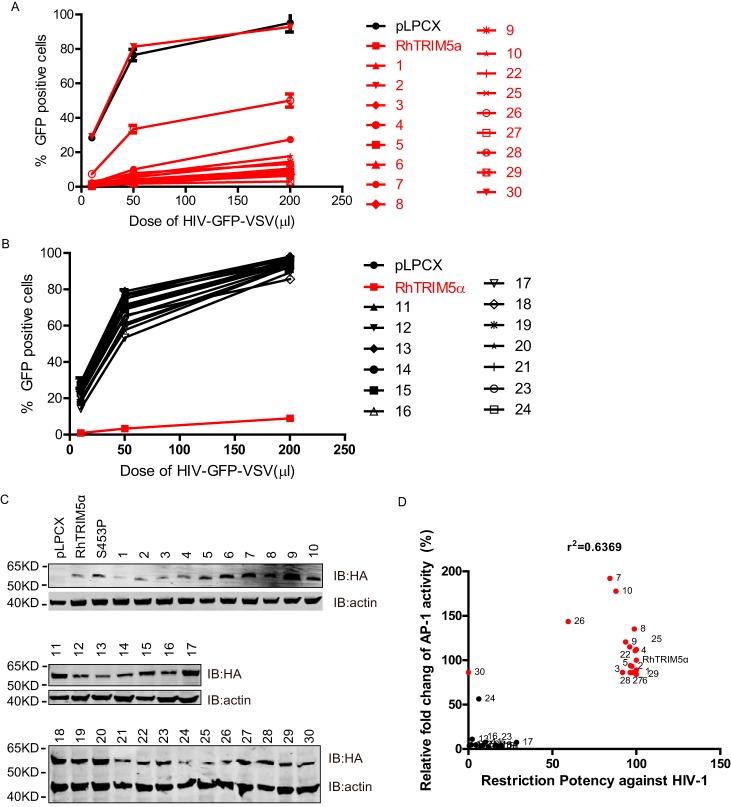
TRIM5α proteins have species-specific activities to inhibit retroviruses. RhTRIM5α does not inhibit the simian immunodeficiency virus of macaque strains, but it strongly inhibits HIV-1, feline immunodeficiency virus, and equine infectious anemia virus (39–41). To identify whether RhTRIM5α signal transduction activity was correlated with its antiviral activity, we performed a post-entry inhibition assay against HIV-1 using HeLa cells, which stably express all of the RhTRIM5α variants. The results indicated that RhTRIM5α could significantly inhibit HIV-1, whereas (with the exception of variant 30) the corresponding variants that failed to activate AP-1 also lost their antiviral activities (Fig. 8, A–C). To further understand the correlation between the capability of AP-1 activation and restriction of HIV-1, we plotted -fold change of AP-1 activities as a function of the restriction abilities of different RhTRIM5α variants. As shown in Fig. 8D, RhTRIM5α-mediated AP-1 activation and its anti-HIV-1 activity were positively correlated, R2 = 0.6369. These results demonstrated that the antiviral activities of RhTRIM5α variants have a significant positive correlation with their capacities to activate AP-1 when stably expressed in HeLa cells. This finding is consistent with the fact that AP-1–dependent gene up-regulation can result in antiviral activity.
Figure 8.
Correlation of RhTRIM5α-mediated AP-1 activity and its antiviral activity. A and B, HeLa cells stably expressing the RhTRIM5 gene of interest or empty vector were infected with an increasing dose of HIV-GFP-VSVG pseudotyped virus. The percentages of GFP-positive cells (infected cells) were enumerated at 48 h post-transfection by FACS. Lines in red represent RhTRIM5α variants that intensely activate AP-1 signaling. All experiments were performed three times, and a representative result is shown. C, HeLa cells stably expressing all RhTRIM5α variants were lysed and used for IB analysis with the indicated antibodies. D, the relative -fold change of AP-1 activity of different RhTRIM5α SPRY domain variants was calculated as described in the legend to Fig. 3. Similarly, the capability of the different variants against HIV-1 was calculated as described in A and B. Dots in red represent RhTRIM5α variants that intensely activate AP-1 signaling. Correlation analyses indicated that RhTRIM5α-mediated AP-1 activation and its anti-HIV-1 activity were positively correlated (R2 = 0.6369).
Discussion
Accumulating evidence suggests that the E3-ubiquitin ligase activity of the TRIM family of proteins plays multiple roles in innate antiviral immunity (42–44). Many TRIM proteins are reported to affect the ubiquitination levels of several signaling adaptors in the NF-κB and AP-1 signaling pathways that are dependent on their RING domains (43, 45). For example, TRIM21 negatively regulates DNA sensor signaling by enhancing Lys48-linked DDX41 ubiquitination (46), whereas TRIM4 promotes type I IFN production against virus infection by targeting RIG-I for Lys63-linked ubiquitination (47). Both regulations are dependent on their RING domains. TRIM5α has been reported to activate AP-1 and NF-κB signaling pathways dependent on the RING domain (9). In this study, we demonstrated that AP-1, but not NF-κB, signal activation by TRIM5α is dependent on the SPRY domain in addition to the RING domain.
It has been well documented that the SPRY domain is used by RhTRIM5α to recognize and interact with HIV-1 CA. Previous studies showed that the SPRY domain serves unknown functions uncoupled from binding retroviral CA (23, 24). In this study, we have demonstrated that the SPRY domain functions to regulate RhTRIM5α-mediated AP-1 signaling. Moreover, we mapped the key amino acids of SPRY for AP-1 signal and illustrated that the whole β-sheet barrel of the SPRY domain was required for this function. We conclude that the β-sheet barrel of the SPRY domain influences the AP-1 pathway by regulating the auto-ubiquitination of TRIM5α.
TRIM5α has been reported to activate the AP-1 and NF-κB signaling pathways by recruiting the E2 Ub-conjugating enzymes UBC13-UEV1A to synthesize free Lys63-linked polyubiquitin chains, which could activate TAK1 in the presence of TAB2 and TAB3. TAB2 and TAB3 are required for TAK1 activation as redundant receptors to preferentially bind to Lys63-linked polyubiquitin chains (48, 49). Many signal transduction molecules have been reported to negatively regulate TAB2-dependent NF-κB and AP-1 signaling via degradation of TAB2, including TRIM22 (50), mouse TRIM30α (51), and TRIM38. TRIM38 inhibits IL-1β- and TNFα-triggered signaling by mediating the degradation of TAB2/3, which depends on the SPRY domain but not the RING domain (52). However, there were two controversial findings for RhTRIM5α in 2011 (24, 53). One study indicated the potential of RhTRIM5α to promote TAB2 degradation, resulting in the repression of HIV-1 LTR promoter activity by the negative regulation of NF-κB activation (53); another study demonstrated that the capacity of TRIM5α to negatively regulate TAB2 levels is present in human and mouse TRIM5α but not in rhesus TRIM5α (24). Our data supported the former finding that RhTRIM5α intensely degrades TAB2 (Fig. 4A). It also identified that this function is independent of the SPRY domain when RhTRIM5α is overexpressed (data not shown). Interestingly, and similar to TRIM38, RhTRIM5α overexpression activates NF-κB and AP-1 in reporter assays regardless of the fact that it intensely degrades TAB2 (44, 52). Therefore, NF-κB and AP-1 activation by TRIM5α will increase when TAB2 is compensated (44, 52). TRIM5α may also exhibit an unknown function in other pathways, as well as the pathway sensing the retrovirus CA lattice. This result is similar to the action of TRIM21, which has been reported to show differential regulation in different cells or with different stimuli (54–58). The role of TRIM5α in other signaling pathways needs to be further investigated.
Several reports have suggested that TAK1 polyubiquitination is involved in TAK1-mediated activation of the NF-κB and AP-1 signaling pathways (29, 32). Many molecules have been reported to regulate innate immunity by ubiquitinating TAK1. For instance, TRIM8 enhances TNFα- and IL-1β-triggered NF-κB activation by targeting TAK1 for Lys63-linked polyubiquitination (31). In addition, TRAF6-catalyzed Lys63-linked polyubiquitination sufficiently activates NF-κB and AP-1 by activating TAK1 (32). Likewise, Pertel et al. (9) provided evidence for a model in which TRIM5α interacts with TAK1 to activate NF-κB and AP-1 signaling by catalyzing unanchored Lys63-linked polyubiquitination. In this study, we also found that RhTRIM5α interacts with TAK1 for polyubiquitination. However, the inactive mutants RhTRIM5αS453P and RhTRIM5αVFVD reserved their capacities to bind and polyubiquitinate TAK1. These results revealed that TAK1 polyubiquitination is not sufficient for TRIM5α-mediated AP-1 activation.
TRIM5α auto-ubiquitination has been reported previously. There are many E2 ubiquitin ligases involved in TRIM5α auto-ubiquitination, including UbcH5B, Ubc2W, Ube2N/Ube2V2, and Ube2D3 (11, 35, 58). The RING and B-box 2 domains have been reported to contribute to auto-ubiquitination of TRIM5α (36, 59). Furthermore, previous studies have concluded that auto-ubiquitination is a critical event in the TRIM5α restriction mechanism (11, 36). Coincidentally, in our studies, we revealed that auto-ubiquitination also plays an important role in AP-1 activation. It was also found that AP-1 activity of RhTRIM5α was correlated with its antiviral activity, which is consistent with the work of the Luban laboratory on six murine Trim5 homologues restricting retroviruses (60). Therefore, at least a portion of the antiviral activity of RhTRIM5α is mediated through AP-1 activation. We further detected that RhTRIM5α auto-ubiquitination and AP-1 activation are both affected by the SPRY domain and especially by the β-sheet barrel of the SPRY structure. These results suggested that the SPRY domain is vital for RhTRIM5α auto-polyubiquitination to further activate AP-1.
It has been reported that HuTRIM5α employed Ube2W and Ube2N/Ube2V2 to anchor and elongate the Lys63-linked poly-Ub chains, respectively, in a process of HuTRIM5α auto-polyubiquitination in cells and in vitro (38). Our studies, however, have revealed that RhTRIM5α was not modified with Lys63-linked poly-Ub chains when overexpressed in 293T cells. A surprising find was that RhTRIM5α undergoes “atypical” Met1-linked and Lys27-linked auto-ubiquitination in HEK293T cells. Although the reason for this difference has not been confirmed, it is now clear that the Lys63-linked and Met1-linked ubiquitin chains can mediate signal transduction by non-degradative mechanisms (61). Several key signal transduction molecules have so far been identified to be Met1-Ub substrates, including NEMO (62), RIPK1 (63), RIPK2 (64) and others. Our study is the first to state that RhTRIM5α is a new target in cells for Met1-Ub linkage, although we have no in vitro experiments for polyubiquitination of RhTRIM5α. However, few substrates are known for chains linked through Lys27, and this area is poorly understood and little investigated. Several studies suggest that Lys27-linked chains may play roles in the DNA response and recruitment of proteins (65–67), but to date, the role of Lys27-linked chains of RhTRIM5α is unidentified.
It has been demonstrated that the auto-polyubiquitination of TRIM protein functions as a platform that facilitates protein-protein interactions in signal transduction (68, 69). TRIM14 recruits the NF-κB essential modulator (NEMO) to the MAVS signalosome via Lys63-linked polyubiquitin chains (70); Lys63-linked auto-polyubiquitination of TRIM9S (short splice variant) serves as a platform between GSK3β and TBK1, leading to the activation of IRF3 signal, and auto-ubiquitination of TRIM11 is required for its binding of p62, resulting in the suppression of AIM2 inflammasome. However, the molecular details about the role of anto-ubiquitination of RhTRIM5α in the innate immunity require further study.
Overall, our results have revealed that the SPRY domain of RhTRIM5α is critical for AP-1 signaling, and this function may correlate with auto-ubiquitination of RhTRIM5α. We conclude that RhTRIM5α was modified with Met1-linked and Lys27-linked polyubiquitination in 293T cells. In addition, we discovered that the RhTRIM5α-mediated signal activity was positively correlated with its antiviral activity.
Experimental procedures
Cell culture and transfection
HEK293T and HeLa cell lines were maintained in high-glucose DMEM (Hyclone) supplemented with l-glutamine (2 mm), penicillin (100 units/ml), streptomycin (100 μg/ml; Gibco), and FBS (10%; Gibco). Cells were plated 16 h before transfection on 6-well plates (Corning, Inc.) at a concentration of 2 × 106/5 × 105 cells per 2 ml of tissue culture medium per well. Cells were transfected using poly Jet (SignaGen Laboratories)/calcium phosphate according to the manufacturer's instructions.
Protein extraction and Western blotting
After the cells were transfected for 24–36 h, they were washed once with ice-cold PBS and subsequently lysed in ice-cold radioimmune precipitation assay (RIPA) lysis buffer (CST, 9086) containing a protease inhibitor mixture (Sigma, P8340). The lysates were centrifuged at 12,000 × g for 10 min at 4 °C to remove the cell debris. The proteins were separated on a 12% Tris/glycine gel or 4–12% BisTris gel (Genscript) and blotted onto PVDF membranes (Millipore). The filters were blocked in 5% BSA in Tris-buffered saline (TBS) and were then probed with the indicated primary antibodies followed by DyLight 680 or 800–labeled secondary antibody (KPL). Signals were visualized using a LI-COR Odyssey imaging system with both 700 and 800 channels.
Cloning and plasmids
RhTRIM5α and FaTRIM5α were cloned into pcDNA3.1(+) at the EcoRI and XhoI sites and fused to an N-terminal HA tag using PCR. All mutants, including the truncated variants, were introduced by site-directed mutagenesis, and the mutations were confirmed by sequencing. Human TRIM5α, AP-1-luc, pRL-TK (Renilla luciferase internal control reporter plasmid), and pCDNA-TAK1 were gifts from Dr. Jeremy Luban. NF-κB-luc plasmid was from Beyotime. FLAG-TAB2 was cloned into the pEF/V5-HisB vector from pCDNA-TAB2 (a gift from Dr. Jeremy Luban) using BamHI and XhoI sites, including the N-3×FLAG tag and the TAB2 ORF. Ub plasmids were constructed with an HA or FLAG tag at their N termini in-house. For creation of TRIM5-expressing HeLa cell lines, pLPCX, the MLV Gag polymerase expression vector pCGP, and pVSV-G were purchased from Clontech. pFUGW is an HIV-1-based transfer vector with enhanced green fluorescent protein expression, and psPAX2 and pMD2.G encode HIV-1-gag-pol and the vesicular stomatitis virus glycoprotein, respectively.
Antibodies
Rabbit anti-FLAG (F7425, Sigma; 1:1,000), mouse anti-FLAG (TA50011, Origene; 1:2,000), mouse anti-HA (H9658, Sigma; 1:10,000), rabbit anti-HA (H6908, Sigma; 1:1,000), mouse anti-β-actin (A5441, Sigma; 1:20,000), rabbit anti-TRIM5α (ab59000, Abcam; 1:1,000), DyLight 800-labeled antibody to mouse IgG (H+L) (072-07-18-06, KPL; 1:10,000), and DyLight 680-labeled antibody to rabbit IgG (H+L) (072-06-15-16, KPL; 1:5,000).
Luciferase assays
HEK293T cells were plated on 96-well culture plates (Corning) at a concentration of 5 × 104 cells/well 14 h before transfection. Cells were transfected with poly Jet (SignaGen Laboratories), using 0.3 μl of poly Jet/well, with 1 ng of the internal control reporter plasmid pRL-TK, 10 ng of firefly luciferase experimental reporter plasmid AP-1-luc or NF-κB-luc, and 90 ng of individual TRIM plasmids or empty pcDNA3.1 as a control, following the manufacturer's instructions. Each experimental condition was performed in quadruplicate. After the cells were transfected for 24 h, luciferase activity was measured with a Dual-Glo luciferase assay system (Promega) using an EIA/RIA plate (Corning, 3693). Firefly luciferase values were normalized to Renilla luciferase values, and the data are represented as -fold inductions compared with empty pcDNA3.1(+). Three independent transfection experiments were performed.
Immunoprecipitation
Thirty hours after transfection, cells were washed once with ice-cold PBS and subsequently lysed in 300 μl of ice-cold RIPA lysis buffer (CST, 9086) containing a protease inhibitor mixture (Sigma, P8340) and PMSF. Cell lysates were scraped off the surface of the plate and transferred to prechilled 2-ml microcentrifuge tubes. The lysates were centrifuged at 12,000 × g for 10 min to remove cell debris. Then 50 μl of the clarified lysate was diluted in 5× loading buffer, boiled at 100 °C for 5 min, and stored at −80 °C, and the remaining 250 μl of cell lysate was incubated with monoclonal anti-HA/anti-FLAG-agarose (A2095/A2220, Sigma) at 4 °C overnight. The beads were then washed four times with prechilled PBS and resuspended in 50 μl of 2× loading buffer. The immunoprecipitated proteins and input lysates were detected by Western blotting.
In vivo TAK1 polyubiquitination
HEK293T cells were co-transfected with the HA-tagged TRIM expression vector, FLAG-tagged TAK1, and HA-tagged Ub expression vector at a 1:1:1 ratio using a normal calcium phosphate method. Protein was extracted in 300 μl of ice-cold RIPA lysis buffer 30 h post-transfection. The TAK1-ubiquitin complexes were immunoprecipitated using anti-FLAG antibody M2-conjugated beads (A2220, Sigma) and immunoblotted with anti-HA antibody to detect ubiquitinated proteins.
In vivo TRIM5 auto-ubiquitination
HEK293T cells were co-transfected with plasmids encoding HA/FLAG-tagged WT or mutant TRIM5 together with a plasmid expressing FLAG/HA-tagged ubiquitin at a 1:1 ratio using a normal calcium phosphate method. Thirty-six hours later, cells were harvested and lysed in RIPA buffer. Extracts were immunoprecipitated using anti-HA-agarose or anti-FLAG-agarose (the tag of ubiquitin). Immunoprecipitated proteins were Western blotted and separately probed with an anti-FLAG or anti-HA antibody to detect polyubiquitinated TRIM5.
Infection with HIV-1 expressing GFP
Pseudotyped HIV-1 expressing GFP was prepared as shown before (71). For infections, HeLa cells stably transduced with TRIM5 genes of interest or empty vector were seeded at a density of 1.5 × 105 cells/well in a 24-well plate, incubated with this pseudotyped virus (described above) for 48 h. The cells were collected and then analyzed using flow cytometry (Beckman, FC500MCL/MPL).
Author contributions
L. N. and X. W. conceptualization; L. N., Y.-D. T., and C. W. data curation; L. N., Y.-D. T., and C. W. formal analysis; L. N. and X. W. supervision; L. N. and X. W. funding acquisition; L. N., C. W., and C. L. methodology; L. N. writing-original draft; L. N. and X. W. project administration; C. W. validation.
Acknowledgments
We thank Dr. Jeremy Luban for the expression plasmids encoding TAK1, TAB2, HuTRIM5α, AP-1-luc, and TK and Changjiang Weng for discussions.
This work was supported by National Natural Science Foundation of China Grants 31602034 (to L. N.) and 31222054 and 81561128010 (to X. W.). The authors declare that they have no conflicts of interest with the contents of this article.
- huTRIM5α
- FaTRIM5α, and RhTRIM5α, human, M. fascicularis, and rhesus macaque TRIM5α, respectively
- SUMO
- small ubiquitin-like modifier
- TRIM
- tripartite motif
- Ub
- ubiquitin
- RIPA
- radioimmune precipitation assay
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- TK
- thymidine kinase
- IB
- immunoblotting
- IP
- immunoprecipitation.
References
- 1. Best S., Le Tissier P., Towers G., and Stoye J. P. (1996) Positional cloning of the mouse retrovirus restriction gene Fv1. Nature 382, 826–829 10.1038/382826a0 [DOI] [PubMed] [Google Scholar]
- 2. Goff S. P. (2004) Genetic control of retrovirus susceptibility in mammalian cells. Annu. Rev. Genet. 38, 61–85 10.1146/annurev.genet.38.072902.094136 [DOI] [PubMed] [Google Scholar]
- 3. Bieniasz P. D. (2004) Intrinsic immunity: a front-line defense against viral attack. Nat. Immunol. 5, 1109–1115 10.1038/ni1125 [DOI] [PubMed] [Google Scholar]
- 4. Stremlau M., Owens C. M., Perron M. J., Kiessling M., Autissier P., and Sodroski J. (2004) The cytoplasmic body component TRIM5α restricts HIV-1 infection in Old World monkeys. Nature 427, 848–853 10.1038/nature02343 [DOI] [PubMed] [Google Scholar]
- 5. Zhang J., Hu M. M., Wang Y. Y., and Shu H. B. (2012) TRIM32 protein modulates type I interferon induction and cellular antiviral response by targeting MITA/STING protein for K63-linked ubiquitination. J. Biol. Chem. 287, 28646–28655 10.1074/jbc.M112.362608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kuang Y. Q., Liu H. L., and Zheng Y. T. (2015) The innate immune roles of host factors TRIM5α and Cyclophilin A on HIV-1 replication. Med. Microbiol. Immunol. 204, 557–565 10.1007/s00430-015-0417-y [DOI] [PubMed] [Google Scholar]
- 7. Stremlau M., Perron M., Lee M., Li Y., Song B., Javanbakht H., Diaz-Griffero F., Anderson D. J., Sundquist W. I., and Sodroski J. (2006) Specific recognition and accelerated uncoating of retroviral capsids by the TRIM5α restriction factor. Proc. Natl. Acad. Sci. U.S.A. 103, 5514–5519 10.1073/pnas.0509996103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sakuma R., Noser J. A., Ohmine S., and Ikeda Y. (2007) Rhesus monkey TRIM5α restricts HIV-1 production through rapid degradation of viral Gag polyproteins. Nat. Med. 13, 631–635 10.1038/nm1562 [DOI] [PubMed] [Google Scholar]
- 9. Pertel T., Hausmann S., Morger D., Züger S., Guerra J., Lascano J., Reinhard C., Santoni F. A., Uchil P. D., Chatel L., Bisiaux A., Albert M. L., Strambio-De-Castillia C., Mothes W., Pizzato M., et al. (2011) TRIM5 is an innate immune sensor for the retrovirus capsid lattice. Nature 472, 361–365 10.1038/nature09976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Han K., Lou D. I., and Sawyer S. L. (2011) Identification of a genomic reservoir for new TRIM genes in primate genomes. PLoS Genet. 7, e1002388 10.1371/journal.pgen.1002388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Fletcher A. J., Christensen D. E., Nelson C., Tan C. P., Schaller T., Lehner P. J., Sundquist W. I., and Towers G. J. (2015) TRIM5α requires Ube2W to anchor Lys63-linked ubiquitin chains and restrict reverse transcription. EMBO J. 34, 2078–2095 10.15252/embj.201490361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Napolitano L. M., and Meroni G. (2012) TRIM family: pleiotropy and diversification through homomultimer and heteromultimer formation. IUBMB Life 64, 64–71 10.1002/iub.580 [DOI] [PubMed] [Google Scholar]
- 13. Yudina Z., Roa A., Johnson R., Biris N., de Souza Aranha Vieira D. A., Tsiperson V., Reszka N., Taylor A. B., Hart P. J., Demeler B., Diaz-Griffero F., and Ivanov D. N. (2015) RING dimerization links higher-order assembly of TRIM5α to synthesis of K63-linked polyubiquitin. Cell Rep. 12, 788–797 10.1016/j.celrep.2015.06.072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kar A. K., Mao Y., Bird G., Walensky L., and Sodroski J. (2011) Characterization of a core fragment of the rhesus monkey TRIM5α protein. BMC Biochem. 12, 1 10.1186/1471-2091-12-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Diaz-Griffero F., Qin X. R., Hayashi F., Kigawa T., Finzi A., Sarnak Z., Lienlaf M., Yokoyama S., and Sodroski J. (2009) A B-box 2 surface patch important for TRIM5α self-association, capsid binding avidity, and retrovirus restriction. J. Virol. 83, 10737–10751 10.1128/JVI.01307-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Langelier C. R., Sandrin V., Eckert D. M., Christensen D. E., Chandrasekaran V., Alam S. L., Aiken C., Olsen J. C., Kar A. K., Sodroski J. G., and Sundquist W. I. (2008) Biochemical characterization of a recombinant TRIM5α protein that restricts human immunodeficiency virus type 1 replication. J. Virol. 82, 11682–11694 10.1128/JVI.01562-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Javanbakht H., Yuan W., Yeung D. F., Song B., Diaz-Griffero F., Li Y., Li X., Stremlau M., and Sodroski J. (2006) Characterization of TRIM5α trimerization and its contribution to human immunodeficiency virus capsid binding. Virology 353, 234–246 10.1016/j.virol.2006.05.017 [DOI] [PubMed] [Google Scholar]
- 18. Reymond A., Meroni G., Fantozzi A., Merla G., Cairo S., Luzi L., Riganelli D., Zanaria E., Messali S., Cainarca S., Guffanti A., Minucci S., Pelicci P. G., and Ballabio A. (2001) The tripartite motif family identifies cell compartments. EMBO J. 20, 2140–2151 10.1093/emboj/20.9.2140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Keown J. R., Yang J. X., Douglas J., and Goldstone D. C. (2016) Characterisation of assembly and ubiquitylation by the RBCC motif of Trim5α. Sci. Rep. 6, 26837 10.1038/srep26837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Nakayama E. E., Miyoshi H., Nagai Y., and Shioda T. (2005) A specific region of 37 amino acid residues in the SPRY (B30.2) domain of African green monkey TRIM5α determines species-specific restriction of simian immunodeficiency virus SIVmac infection. J. Virol. 79, 8870–8877 10.1128/JVI.79.14.8870-8877.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohkura S., Yap M. W., Sheldon T., and Stoye J. P. (2006) All three variable regions of the TRIM5α B30.2 domain can contribute to the specificity of retrovirus restriction. J. Virol. 80, 8554–8565 10.1128/JVI.00688-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Stremlau M., Perron M., Welikala S., and Sodroski J. (2005) Species-specific variation in the B30.2(SPRY) domain of TRIM5α determines the potency of human immunodeficiency virus restriction. J. Virol. 79, 3139–3145 10.1128/JVI.79.5.3139-3145.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Yang Y., Brandariz-Nuñez A., Fricke T., Ivanov D. N., Sarnak Z., and Diaz-Griffero F. (2014) Binding of the rhesus TRIM5α PRYSPRY domain to capsid is necessary but not sufficient for HIV-1 restriction. Virology 448, 217–228 10.1016/j.virol.2013.10.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tareen S. U., and Emerman M. (2011) Human Trim5α has additional activities that are uncoupled from retroviral capsid recognition. Virology 409, 113–120 10.1016/j.virol.2010.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Dutrieux J., Portilho D. M., Arhel N. J., Hazan U., and Nisole S. (2015) TRIM5α is a SUMO substrate. Retrovirology 12, 28 10.1186/s12977-015-0155-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Nepveu-Traversy M. E., Demogines A., Fricke T., Plourde M. B., Riopel K., Veillette M., Diaz-Griffero F., Sawyer S. L., and Berthoux L. (2016) A putative SUMO interacting motif in the B30.2/SPRY domain of rhesus macaque TRIM5α important for NF-κB/AP-1 signaling and HIV-1 restriction. Heliyon 2, e00056 10.1016/j.heliyon.2015.e00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Arriagada G., Muntean L. N., and Goff S. P. (2011) SUMO-interacting motifs of human TRIM5alpha are important for antiviral activity. PLoS Pathog. 7, e1002019 10.1371/journal.ppat.1002019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Tareen S. U., and Emerman M. (2011) Trim5 TAKes on pattern recognition. CelL Host Microbe 9, 349–350 10.1016/j.chom.2011.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Xia Z. P., Sun L., Chen X., Pineda G., Jiang X., Adhikari A., Zeng W., and Chen Z. J. (2009) Direct activation of protein kinases by unanchored polyubiquitin chains. Nature 461, 114–119 10.1038/nature08247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lamb A., Yang X. D., Tsang Y. H., Li J. D., Higashi H., Hatakeyama M., Peek R. M., Blanke S. R., and Chen L. F. (2009) Helicobacter pylori CagA activates NF-κB by targeting TAK1 for TRAF6-mediated Lys 63 ubiquitination. EMBO Rep. 10, 1242–1249 10.1038/embor.2009.210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Li Q., Yan J., Mao A. P., Li C., Ran Y., Shu H. B., and Wang Y. Y. (2011) Tripartite motif 8 (TRIM8) modulates TNF - and IL-1-triggered NF-B activation by targeting TAK1 for K63-linked polyubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 19341–19346 10.1073/pnas.1110946108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sorrentino A., Thakur N., Grimsby S., Marcusson A., von Bulow V., Schuster N., Zhang S., Heldin C. H., and Landström M. (2008) The type I TGF-β receptor engages TRAF6 to activate TAK1 in a receptor kinase-independent manner. Nat. Cell Biol. 10, 1199–1207 10.1038/ncb1780 [DOI] [PubMed] [Google Scholar]
- 33. Qin Y., Liu Q., Tian S., Xie W., Cui J., and Wang R. F. (2016) TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 26, 613–628 10.1038/cr.2016.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Liu T., Tang Q., Liu K., Xie W., Liu X., Wang H., Wang R. F., and Cui J. (2016) TRIM11 suppresses AIM2 inflammasome by degrading AIM2 via p62-dependent selective autophagy. Cell Rep. 16, 1988–2002 10.1016/j.celrep.2016.07.019 [DOI] [PubMed] [Google Scholar]
- 35. Yamauchi K., Wada K., Tanji K., Tanaka M., and Kamitani T. (2008) Ubiquitination of E3 ubiquitin ligase TRIM5α and its potential role. FEBS J. 275, 1540–1555 10.1111/j.1742-4658.2008.06313.x [DOI] [PubMed] [Google Scholar]
- 36. Lienlaf M., Hayashi F., Di Nunzio F., Tochio N., Kigawa T., Yokoyama S., and Diaz-Griffero F. (2011) Contribution of E3-ubiquitin ligase activity to HIV-1 restriction by TRIM5 rh: structure of the RING domain of TRIM5. J. Virol. 85, 8725–8737 10.1128/JVI.00497-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kim J., Tipper C., and Sodroski J. (2011) Role of TRIM5α RING domain E3 ubiquitin ligase activity in capsid disassembly, reverse transcription blockade, and restriction of simian immunodeficiency virus. J. Virol. 85, 8116–8132 10.1128/JVI.00341-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Biris N., Yang Y., Taylor A. B., Tomashevski A., Guo M., Hart P. J., Diaz-Griffero F., and Ivanov D. N. (2012) Structure of the rhesus monkey TRIM5α PRYSPRY domain, the HIV capsid recognition module. Proc. Natl. Acad. Sci. U.S.A. 109, 13278–13283 10.1073/pnas.1203536109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hatziioannou T., Perez-Caballero D., Yang A., Cowan S., and Bieniasz P. D. (2004) Retrovirus resistance factors Ref1 and Lv1 are species-specific variants of TRIM5α. Proc. Natl. Acad. Sci. U.S.A. 101, 10774–10779 10.1073/pnas.0402361101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Keckesova Z., Ylinen L. M., and Towers G. J. (2004) The human and African green monkey TRIM5α genes encode Ref1 and Lv1 retroviral restriction factor activities. Proc. Natl. Acad. Sci. U.S.A. 101, 10780–10785 10.1073/pnas.0402474101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Saenz D. T., Teo W., Olsen J. C., and Poeschla E. M. (2005) Restriction of feline immunodeficiency virus by Ref1, Lv1, and primate TRIM5α proteins. J. Virol. 79, 15175–15188 10.1128/JVI.79.24.15175-15188.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rajsbaum R., García-Sastre A., and Versteeg G. A. (2014) TRIMmunity: the roles of the TRIM E3-ubiquitin ligase family in innate antiviral immunity. J. Mol. Biol. 426, 1265–1284 10.1016/j.jmb.2013.12.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Versteeg G. A., Benke S., García-Sastre A., and Rajsbaum R. (2014) InTRIMsic immunity: positive and negative regulation of immune signaling by tripartite motif proteins. Cytokine Growth Factor Rev. 25, 563–576 10.1016/j.cytogfr.2014.08.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Uchil P. D., Hinz A., Siegel S., Coenen-Stass A., Pertel T., Luban J., and Mothes W. (2013) TRIM protein-mediated regulation of inflammatory and innate immune signaling and its association with antiretroviral activity. J. Virol. 87, 257–272 10.1128/JVI.01804-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Versteeg G. A., Rajsbaum R., Sánchez-Aparicio M. T., Maestre A. M., Valdiviezo J., Shi M., Inn K. S., Fernandez-Sesma A., Jung J., and García-Sastre A. (2013) The E3-ligase TRIM family of proteins regulates signaling pathways triggered by innate immune pattern-recognition receptors. Immunity 38, 384–398 10.1016/j.immuni.2012.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Z., Bao M., Lu N., Weng L., Yuan B., and Liu Y. J. (2013) The E3 ubiquitin ligase TRIM21 negatively regulates the innate immune response to intracellular double-stranded DNA. Nat. Immunol. 14, 172–178 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Yan J., Li Q., Mao A. P., Hu M. M., and Shu H. B. (2014) TRIM4 modulates type I interferon induction and cellular antiviral response by targeting RIG-I for K63-linked ubiquitination. J. Mol. Cell Biol. 6, 154–163 10.1093/jmcb/mju005 [DOI] [PubMed] [Google Scholar]
- 48. Kanayama A., Seth R. B., Sun L., Ea C. K., Hong M., Shaito A., Chiu Y. H., Deng L., and Chen Z. J. (2004) TAB2 and TAB3 activate the NF-κB pathway through binding to polyubiquitin chains. Mol. Cell. 15, 535–548 10.1016/j.molcel.2004.08.008 [DOI] [PubMed] [Google Scholar]
- 49. Besse A., Lamothe B., Campos A. D., Webster W. K., Maddineni U., Lin S. C., Wu H., and Darnay B. G. (2007) TAK1-dependent signaling requires functional interaction with TAB2/TAB3. J. Biol. Chem. 282, 3918–3928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qiu H., Huang F., Xiao H., Sun B., and Yang R. (2013) TRIM22 inhibits the TRAF6-stimulated NF-κB pathway by targeting TAB2 for degradation. Virol. Sin. 28, 209–215 10.1007/s12250-013-3343-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Shi M., Deng W., Bi E., Mao K., Ji Y., Lin G., Wu X., Tao Z., Li Z., Cai X., Sun S., Xiang C., and Sun B. (2008) TRIM30 α negatively regulates TLR-mediated NF-κB activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9, 369–377 10.1038/ni1577 [DOI] [PubMed] [Google Scholar]
- 52. Hu M. M., Yang Q., Zhang J., Liu S. M., Zhang Y., Lin H., Huang Z. F., Wang Y. Y., Zhang X. D., Zhong B., and Shu H. B. (2014) TRIM38 inhibits TNFα- and IL-1β-triggered NF-κB activation by mediating lysosome-dependent degradation of TAB2/3. Proc. Natl. Acad. Sci. U.S.A. 111, 1509–1514 10.1073/pnas.1318227111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gong J., Shen X. H., Qiu H., Chen C., and Yang R. G. (2011) Rhesus monkey TRIM5α represses HIV-1 LTR promoter activity by negatively regulating TAK1/TAB1/TAB2/TAB3-complex-mediated NF-κB activation. Arch. Virol. 156, 1997–2006 10.1007/s00705-011-1097-6 [DOI] [PubMed] [Google Scholar]
- 54. Yang K., Shi H. X., Liu X. Y., Shan Y. F., Wei B., Chen S., and Wang C. (2009) TRIM21 is essential to sustain IFN regulatory factor 3 activation during antiviral response. J. Immunol. 182, 3782–3792 10.4049/jimmunol.0803126 [DOI] [PubMed] [Google Scholar]
- 55. Espinosa A., Dardalhon V., Brauner S., Ambrosi A., Higgs R., Quintana F. J., Sjöstrand M., Eloranta M. L., Ní G. J., Winqvist O., Sundelin B., Jefferies C. A., Rozell B., Kuchroo V. K., and Wahren-Herlenius M. (2009) Loss of the lupus autoantigen Ro52/Trim21 induces tissue inflammation and systemic autoimmunity by disregulating the IL-23-Th17 pathway. J. Exp. Med. 206, 1661–1671 10.1084/jem.20090585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ozato K., Yoshimi R., Chang T. H., Wang H., Atsumi T., and Morse H. R. (2009) Comment on “Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts”. J. Immunol. 183, 7619, 720–721 10.4049/jimmunol.0990103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Yoshimi R., Chang T. H., Wang H., Atsumi T., Morse H. C. 3rd, and Ozato K. (2009) Gene disruption study reveals a nonredundant role for TRIM21/Ro52 in NF-κB-dependent cytokine expression in fibroblasts. J. Immunol. 182, 7527–7538 10.4049/jimmunol.0804121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Higgs R., Ní G. J., Ben Larbi N., Breen E. P., Fitzgerald K. A., and Jefferies C. A. (2008) The E3 ubiquitin ligase Ro52 negatively regulates IFN-β production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181, 1780–1786 10.4049/jimmunol.181.3.1780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Diaz-Griffero F., Li X., Javanbakht H., Song B., Welikala S., Stremlau M., and Sodroski J. (2006) Rapid turnover and polyubiquitylation of the retroviral restriction factor TRIM5. Virology 349, 300–315 10.1016/j.virol.2005.12.040 [DOI] [PubMed] [Google Scholar]
- 60. Lascano J., Uchil P. D., Mothes W., and Luban J. (2015) TRIM5 retroviral restriction activity correlates with the ability to induce innate immune signaling. J. Virol. 90, 308–316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Baud V., and Karin M. (2009) Is NF-κB a good target for cancer therapy? Hopes and pitfalls. Nat. Rev. Drug Discov. 8, 33–40 10.1038/nrd2781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Tokunaga F., Sakata S., Saeki Y., Satomi Y., Kirisako T., Kamei K., Nakagawa T., Kato M., Murata S., Yamaoka S., Yamamoto M., Akira S., Takao T., Tanaka K., and Iwai K. (2009) Involvement of linear polyubiquitylation of NEMO in NF-κB activation. Nat. Cell Biol. 11, 123–132 10.1038/ncb1821 [DOI] [PubMed] [Google Scholar]
- 63. Fiil B. K., Damgaard R. B., Wagner S. A., Keusekotten K., Fritsch M., Bekker-Jensen S., Mailand N., Choudhary C., Komander D., and Gyrd-Hansen M. (2013) OTULIN restricts Met1-linked ubiquitination to control innate immune signaling. Mol. Cell 50, 818–830 10.1016/j.molcel.2013.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mevissen T. E., Hospenthal M. K., Geurink P. P., Elliott P. R., Akutsu M., Arnaudo N., Ekkebus R., Kulathu Y., Wauer T., El Oualid F., Freund S. M., Ovaa H., and Komander D. (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154, 169–184 10.1016/j.cell.2013.05.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Gatti M., Pinato S., Maiolica A., Rocchio F., Prato M. G., Aebersold R., and Penengo L. (2015) RNF168 promotes noncanonical K27 ubiquitination to signal DNA damage. Cell Rep. 10, 226–238 10.1016/j.celrep.2014.12.021 [DOI] [PubMed] [Google Scholar]
- 66. Liu Z., Chen P., Gao H., Gu Y., Yang J., Peng H., Xu X., Wang H., Yang M., Liu X., Fan L., Chen S., Zhou J., Sun Y., Ruan K., et al. (2014) Ubiquitylation of autophagy receptor Optineurin by HACE1 activates selective autophagy for tumor suppression. Cancer Cell 26, 106–120 10.1016/j.ccr.2014.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Palicharla V. R., and Maddika S. (2015) HACE1 mediated K27 ubiquitin linkage leads to YB-1 protein secretion. Cell Signal. 27, 2355–2362 10.1016/j.cellsig.2015.09.001 [DOI] [PubMed] [Google Scholar]
- 68. Swatek K. N., and Komander D. (2016) Ubiquitin modifications. Cell Res. 26, 399–422 10.1038/cr.2016.39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hu H., and Sun S. C. (2016) Ubiquitin signaling in immune responses. Cell Res. 26, 457–483 10.1038/cr.2016.40 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zhou Z., Jia X., Xue Q., Dou Z., Ma Y., Zhao Z., Jiang Z., He B., Jin Q., and Wang J. (2014) TRIM14 is a mitochondrial adaptor that facilitates retinoic acid-inducible gene-I-like receptor-mediated innate immune response. Proc. Natl. Acad. Sci. U.S.A. 111, E245–E254 10.1073/pnas.1316941111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Na L., Tang Y. D., Liu J. D., Yu C. Q., Sun L. K., Lin Y. Z., Wang X. F., Wang X., and Zhou J. H. (2014) TRIMe7-CypA, an alternative splicing isoform of TRIMCyp in rhesus macaque, negatively modulates TRIM5α activity. Biochem. Biophys. Res. Commun. 446, 470–474 10.1016/j.bbrc.2014.02.132 [DOI] [PubMed] [Google Scholar]