Abstract
Activating transcription factor 5 (ATF5) is a member of the ATF/cAMP response element–binding protein family of transcription factors. ATF5 regulates stress responses and cell survival, proliferation, and differentiation and also plays a role in viral infections, cancer, diabetes, schizophrenia, and the olfactory system. Moreover, it was found to also have a critical cell cycle–dependent structural function at the centrosome. However, the mechanism that controls the localization of ATF5 at the centrosome is unclear. Here we report that ATF5 is small ubiquitin-like modifier (SUMO) 2/3–modified at a conserved SUMO-targeting consensus site in various types of mammalian cells. We found that SUMOylation of ATF5 is elevated in the G1 phase of the cell cycle and diminished in the G2/M phase. ATF5 SUMOylation disrupted the interaction of ATF5 with several centrosomal proteins and dislodged ATF5 from the centrosome at the end of the M phase. Of note, blockade of ATF5 SUMOylation deregulated the centrosome cycle, impeded ATF5 translocation from the centrosome, and caused genomic instability and G2/M arrest in HeLa cells. Our results indicate that ATF5 SUMOylation is an essential mechanism that regulates ATF5 localization and function at the centrosome.
Keywords: SUMOylation, genomic instability, cell cycle, centrosome, transcription factor, ATF5, GCP-2, PCNT, cell cycle arrest
Introduction
ATF55 is a member of the ATF/cAMP response element–binding protein family of transcription factors that is implicated in diverse cellular functions and diseases. It regulates the cell stress response and cell survival, proliferation, and differentiation. It also plays a role in the process of viral infection and in the development of cancer, diabetes, schizophrenia, and the olfactory system (1, 2). ATF5 displays a dual nuclear and centrosomal localization with roles that are consistent with its biological function. In the nucleus, ATF5 acts as a transcription factor that can either stimulate or repress the transcription of genes, such as stimulating Bcl-2 (3–5), Mcl-1 (6, 7), Egr-1 (8, 9), and IE86 (10) and repressing cAMP response element–binding protein (11). In the centrosome, ATF5 serves as a structural protein that controls the cell cycle– and centriole age–dependent centriole–PCM interaction and maintains centrosome integrity (12).
Posttranslational modification of proteins by SUMO regulates essential cellular processes, including transcription and the cell stress response, and plays important roles in cancer, neurological diseases, and aging (13–17). SUMOylation regulates these distinct processes via modulation of specific protein–protein interactions (18). In vertebrates, three SUMO isoforms are expressed. SUMO1 shares 43% identity with SUMO2 and SUMO3, whereas the latter two are closely related (sharing 97% identity) and are often collectively referred to as SUMO2/3. Paralog-specific modification has been reported for various substrates (19). SUMO proteins are covalently attached to their targets via an isopeptide bond with the ϵ amino group of a lysine residue. The conjugation process involves an enzymatic cascade comprising the E1-activating enzyme Aos1 (also known as Uba2 and Sae1), the E2-conjugating enzyme Ubc9, and additional E3 ligases, including protein inhibitor of activated STAT (PIAS) family members, Ran-binding protein 2 (RanBP2), and human polycomb protein 2 (hPC2, also known as PC2 and CBX4) (20). De-SUMOylation is mediated by the action of SUMO-specific isopeptidases that belong to the family of ubiquitin-like proteases (Ulps, also known as sentrin-specific proteases (SENPs)). De-SUMOylation often targets specific proteins and takes place at specific locations in cells (21, 22). Notably, in most cases, only a small fraction of a given target protein is SUMOylated, making it difficult to identify novel SUMO substrates and elucidate the functional consequences of the modification (18).
In this study, we show that ATF5 is a substrate of SUMOylation in various mammalian cell types and that it is preferentially modified by SUMO2/3. We found that ATF5 SUMOylation, which appears to take place in the centrosome and at the end of the M phase of the cell cycle, disrupts ATF5 interaction with a number of centrosomal proteins and causes its down-regulation at the centrosome. Interruption of ATF5 SUMOylation results in centrosomal defects and genomic instability, impeding cell cycle progression.
Results
ATF5 is SUMO2/3-modified in various types of mammalian cells
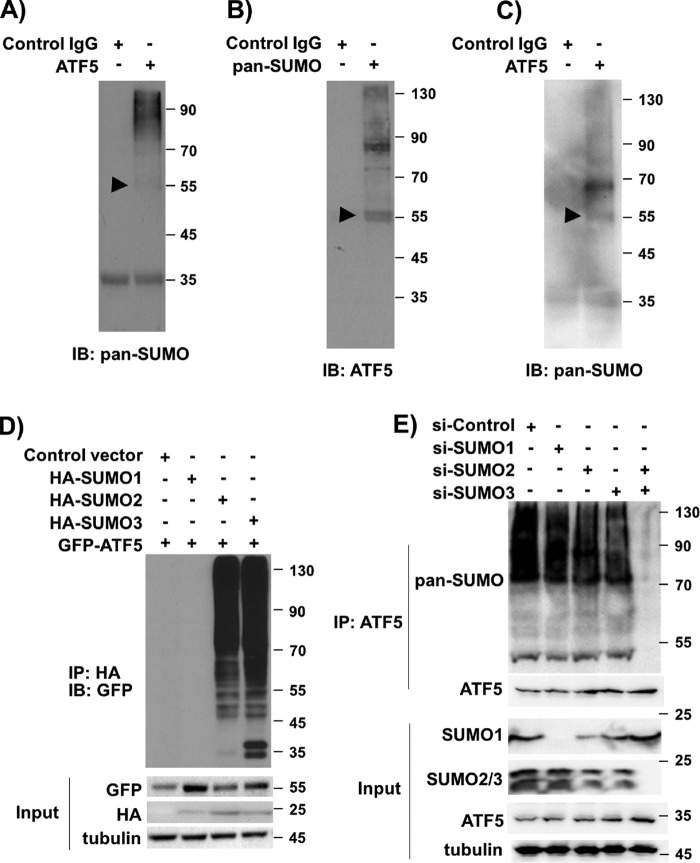
MS analysis of tandem affinity–purified FLAG-HA-ATF5 immunoprecipitates prepared from HEK293 and C6 glioma cells (12) revealed that fragments of ATF5 are associated with SUMOs (data not shown), raising the possibility that ATF5 is SUMOylated in those cells. To determine whether ATF5 is SUMOylated in HeLa cells, we prepared ATF5 immunoprecipitates and performed immunoblot analysis using a pan-SUMO antibody. Multiple protein bands, including one that migrated at 55 kDa and corresponded to mono-SUMOylated ATF5, were observed on the Western blot, indicating that ATF5 is indeed SUMOylated in HeLa cells (Fig. 1A). In reverse immunoprecipitation analysis, immunoblotting of SUMO immunoprecipitates with an anti-ATF5 antibody also showed ATF5 species of high molecular weight corresponding to mono- and poly-SUMOylated ATF5 (Fig. 1B). Similar immunoprecipitation and Western blot analyses using additional cell lines, including the breast cancer cell line MCF-7, the hepatocellular carcinoma cell line HepG2, the human lung fibroblast line IMR-90, and monkey kidney fibroblast-like COS-7 cells, all indicated the presence of SUMOylated ATF5 in the cells (Fig. 1C and data not shown). Thus, we conclude that ATF5 is SUMOylated in a variety of mammalian cells.
Figure 1.
ATF5 is SUMOylated in human and mouse cells. A–C, immunoblotting (IB) of immunoprecipitates with the indicated antibodies detected SUMOylated ATF5 in various mammalian cells. A and B, HeLa cells; C, IMR-90 human lung fibroblasts. D, immunoblotting with an HA antibody of GFP immunoprecipitates (IP) from HeLa cells transfected with the indicated DNA constructs. The expression levels of the transfected genes are shown as input. E, immunoblotting with a pan-SUMO and an ATF5 antibody of ATF5 immunoprecipitates from HeLa cells transfected with the indicated siRNAs. The arrowheads in A–C indicate the position of the expected mono-SUMOylated ATF5 at 55 kDa.
To determine which SUMO species target ATF5, we co-transfected into HeLa cells GFP-ATF5 with DNA vectors empty or expressing HA-SUMO1, HA-SUMO2, and HA-SUMO3, respectively, and performed immunoblot analysis on GFP immunoprecipitates using an HA antibody. As shown in Fig. 1D, SUMO1 is only marginally detectable in GFP immunoprecipitates, whereas SUMO2 and 3 are readily detected in the GFP immunoprecipitates. These results suggest that ATF5 is preferentially modified by SUMO2/3. In support of this conclusion, knockdown of SUMO2/3 in HeLa cells blocked ATF5 SUMOylation (Fig. 1E).
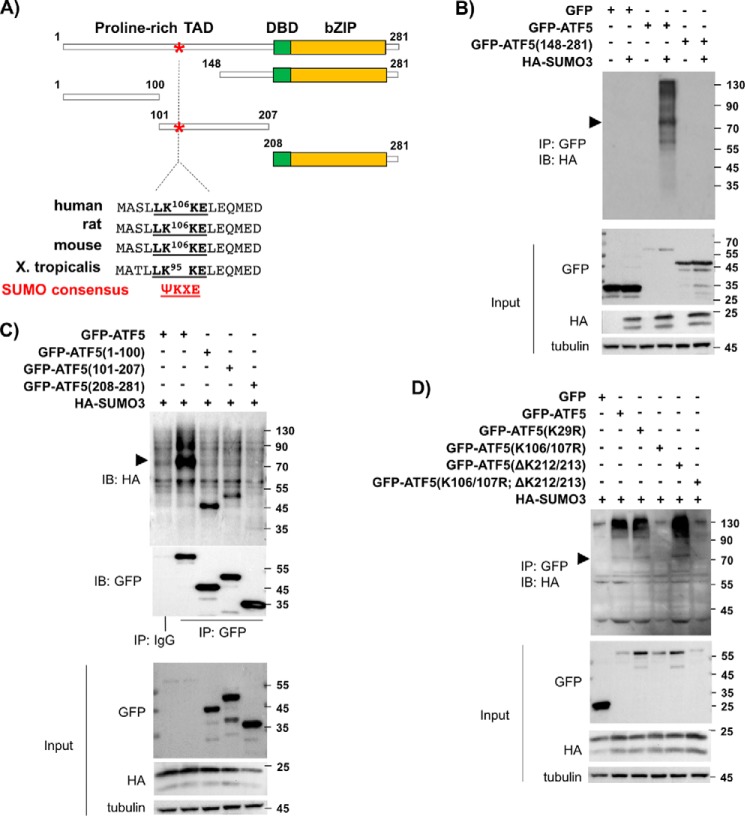
Lys106/Lys107 is the specific SUMO acceptor lysine on ATF5
Analysis of the protein amino acid sequences of ATF5 from human, rat, mouse, and frog identified a conserved sequence (LK106KE in human, rat, and mouse ATF5 and LK95KE in frog; Fig. 2A) that matches the SUMOylation consensus motif ΨKXE (23). To determine whether the LKKE site is involved in ATF5 SUMOylation, we first tested the ability of several ATF5 truncation mutants (12, 24), including ATF5(148–281), ATF5(1–100), ATF5(101–207), and ATF5(208–281), to be SUMOylated by SUMO3. As shown in Fig. 2, B and C, unlike the robust SUMOylation of the full-length ATF5, none of the ATF5 fragments was able to be SUMOylated effectively by SUMO3. As ATF5(100–207) is diffusely distributed in the cell and excluded from the centrosome (data not shown), and ATF5(148–281) is centrosome-bound as the full-length ATF5 (12) but does not contain the SUMOylation consensus site LKXE, our results seem to suggest that both the SUMOylation consensus site on ATF5 and the localization of ATF5 at the centrosome are required for ATF5 SUMOylation. Indeed, SUMOylation analysis using a number of ATF5 lysine mutants, including K29R, K106R/K107R, ΔK212/213, and K106R/K107R;ΔK212/213, confirmed that Lys-to-Arg mutation at 106/107 effectively abolished the overall SUMOylation level of the full-length ATF5 (Fig. 2D). Taken together, our data demonstrate that Lys106/Lys107 on ATF5 is the primary SUMOylation acceptor site for SUMO2/3.
Figure 2.
ATF5 is SUMOylated at a conserved SUMOylation consensus site, Lys106/Lys107. A, schematic of ATF5, its truncation mutants, and its SUMOylation consensus site. TAD, transcription activation domain; DBD, DNA binding domain; bZIP, basic zipper region. B, immunoblotting (IB) with an HA antibody of GFP immunoprecipitates (IP) from HeLa cells transfected with DNA empty vector (−) or expressing HA-SUMO3 (+) and other indicated DNA constructs. C and D, immunoblotting and immunoprecipitation were performed as in B, except the indicated DNA constructs were used. Pre-immune IgG (IgG) was used as a control. The arrowheads in B–D indicate the position of the expected mono-SUMOylated GFP-ATF5 at 81 kDa.
ATF5 SUMOylation is cell cycle–dependent
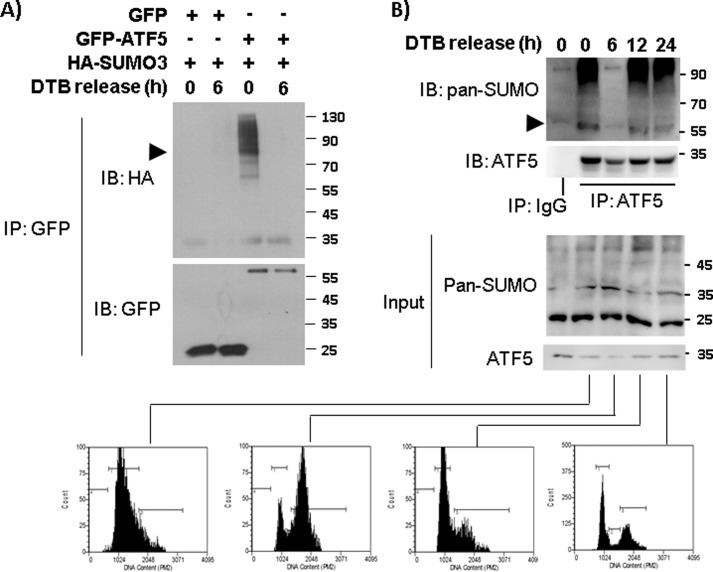
Because ATF5 functions in a cell cycle–dependent manner (12), we wished to determine whether ATF5 SUMOylation takes place during specific phase(s) of the cell cycle. HeLa cells co-transfected with HA-SUMO3 and vectors expressing GFP or GFP-ATF5 were synchronized at defined cell cycle phases by release from double thymidine blockade (DTB), and the level of ATF5 SUMOylation was assessed by immunoblotting GFP immunoprecipitates with an antibody against HA. This experiment showed that ATF5 SUMOylation is much elevated at G1 phase and almost undetectable at the G2/M phase (Fig. 3A). Immunoblotting with a pan-SUMO antibody of ATF5 immunoprecipitates obtained from DTB-released HeLa cells also showed that the endogenous ATF5 is conjugated with SUMO in G1 and S phases but not in G2/M phase (Fig. 3B). These data indicate that ATF5 is subject to cell cycle–dependent SUMOylation, with the highest and lowest levels of SUMOylation at G1 and M phase, respectively.
Figure 3.
ATF5 SUMOylation is cell cycle–dependent. A, SUMOylation of transfected ATF5 was monitored in synchronized HeLa cells. HeLa cells transfected with the indicated DNA constructs were synchronized by DTB and release for the indicated hours. Immunoblotting (IB) and immunoprecipitation (IP) with the indicated antibodies were performed as in Fig. 1. The arrowhead points to mono-SUMOylated GFP-ATF5 at 81 kDa. B, immunoblotting and immunoprecipitation were performed as in A, except that the indicated antibodies for endogenous ATF5 and SUMO were tested. A portion of the DTB-released cells from each time points (0, 6, 12, and 24 h) was stained with propidium iodine and analyzed by FACS to confirm cell cycle positions (bottom panel). The arrowhead points to mono-SUMOylated ATF5 at 55 kDa. Preimmune IgG (IgG) was used as control.
SUMOylation of ATF5 blocks ATF5 interaction with centrosomal proteins
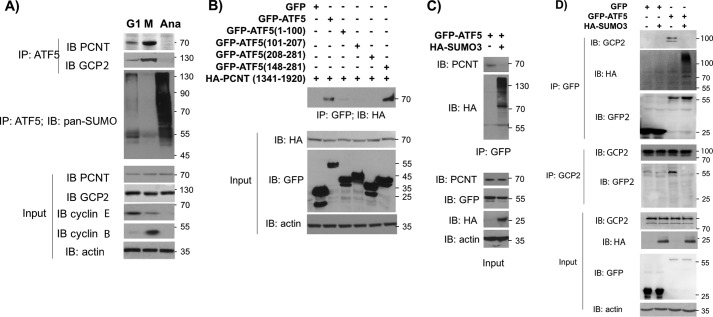
We have shown previously that ATF5 interacts with several centrosomal proteins, including pericentrin (PCNT) and GCP-2 and is present in the centrosome in a cell cycle–dependent manner (12). Consistent with this, we found that increasing the amount of PCNT and GCP2 was associated with ATF5 in HeLa cells from G1 to early M phase, and the association became undetectable at anaphase, when ATF5 SUMOylation suddenly increases (Fig. 4A). Furthermore, we found that both WT ATF5 and ATF5(148–281) associated with a PCNT truncation, PCNT(1341–1920) (12), in co-transfection assays, indicating that the ATF5 C-terminal region interacts with PCNT (Fig. 4B). To determine whether ATF5 SUMOylation affects ATF5 interaction with PCNT, we co-transfected GFP-ATF5 with an empty vector or a vector expressing HA-SUMO3 into HeLa cells and assessed ATF5 interaction with PCNT by immunoprecipitation–immunoblot analysis. As shown in Fig. 4C, immunoblotting of GFP immunoprecipitate with an anti-PCNT antibody revealed that ATF5 SUMOylation blocked ATF5 interaction with PCNT (Fig. 4C). Similarly, immunoblotting of GFP immunoprecipitate with a GCP-2 antibody or immunoblotting of GCP-2 immunoprecipitate with a GFP antibody both showed that SUMO3 overexpression interrupted ATF5 interaction with GCP-2 (Fig. 4D). Additional protein–protein association analyses demonstrated that SUMO3 overexpression reduced or blocked ATF5 interaction with GCP-4, EG5, and α-tubulin (data not shown). We therefore conclude that ATF5 SUMOylation interrupts ATF5 interaction with centrosomal proteins.
Figure 4.
SUMOylation of ATF5 interrupts ATF5 interaction with the centrosomal proteins PCNT and GCP2. A, immunoblotting (IB) and immunoprecipitation (IP) were performed using the indicated antibodies. Cell extracts were prepared from HeLa cells synchronized by DTB and release for 9, 12, and 20 h to obtain cells at G2/M (M), Anaphase (Ana), and G1 phase cells. Cyclin E and cyclin B were used as G1 and M phase markers, respectively. B–D, immunoblotting and immunoprecipitation were performed as in A, except the indicated antibodies were used. Cell extracts were prepared from HeLa cells transfected with the indicated DNA constructs.
ATF5 SUMOylation removes ATF5 from the centrosome
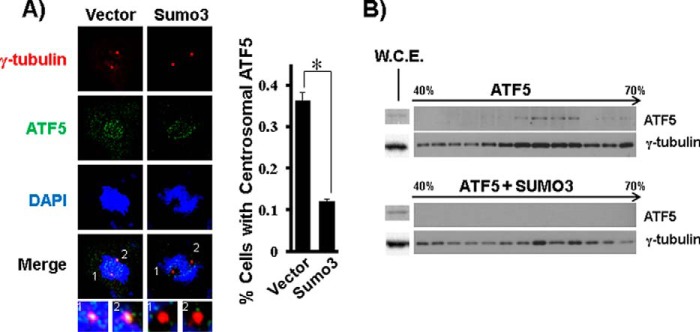
We next examined whether ATF5 SUMOylation affects ATF5 localization to the centrosome. COS-7 (Fig. 5A) and HeLa (data not shown) cells were transfected with a DNA empty vector (control) or a vector expressing HA-SUMO3, and ATF5 localization at the centrosome was assessed by co-staining of ATF5 and γ-tubulin, a centrosome marker, in transfected cells. About 37% of the vector-transfected cells displayed ATF5 localization at the centrosome, which is consistent with our previous observation (12), whereas only 11% of the SUMO3-transfected cells showed ATF5 colocalization with γ-tubulin (Fig. 5A). ATF5 association with the centrosome under conditions of SUMOylation was further assessed by centrosome sedimentation analysis. Immunoblotting of samples from sucrose gradient sedimentation showed that both overexpressed and endogenous ATF5 co-sedimented with γ-tubulin in large protein complexes corresponding to the centrosome fractions (12). Co-expression of SUMO3 caused ATF5 to disappear from the centrosome even though the overall expression of ATF5 is consistently elevated (Fig. 5B and data not shown), possibly because of a reduction in ubiquitination-dependent protein degradation. Together, these data show that ATF5 SUMOylation results in ATF5 dissociation from the centrosome.
Figure 5.
ATF5 SUMOylation drives ATF5 out of the centrosome. A, COS-7 cells transfected with a DNA empty vector or expressing HA-SUMO3 were co-stained with γ-tubulin (red) and ATF5 (green) and counterstained with DAPI (blue). Representative images of fluorescence microscopy are shown. Magnified images (∼×20) of the centrosomes in the merged pictures are shown at the bottom. Percentages of cells in which ATF5 is present at the mitotic centrosome, as indicated by co-staining with γ-tubulin, are tallied at the right. At least 50 cells were examined for each experimental condition. *, p < 0.01. B, centrosomes were purified by sucrose gradient centrifugation from COS-7 cells transfected as in A, and the fractions were examined by immunoblotting for the presence of γ-tubulin and ATF5. Whole-cell extracts (W.C.E.) were used to determine the expression levels of γ-tubulin and ATF5 in the cells (left).
SUMOylation-defective ATF5 mutants are resistant to SUMOylation-driven ATF5 dissociation with PCNT and the centrosome
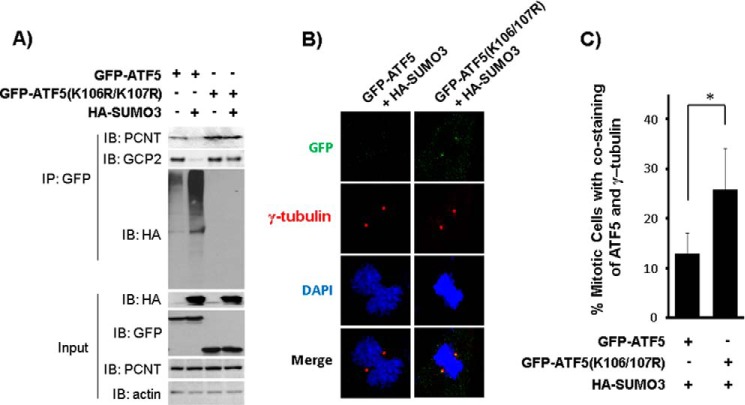
Our data showing that ATF5 SUMOylation disrupts ATF5 interaction with centrosomal proteins (Fig. 4) and drives ATF5 out of the centrosome (Fig. 5) raised the possibility that SUMOylation of ATF5 regulates ATF5 association with the centrosome. Based on our observation that ATF5 SUMOylation is undetectable during G2/M phase and reaches its peak level at G1 phase of the cell cycle (Fig. 3), we hypothesized that ATF5 SUMOylation is required for completing the centrosome cycle by removing ATF5 from the centrosome at the end of G2/M phase. To test this hypothesis, we first determined the properties of the SUMOylation-defective ATF5 mutants ATF5(K106R/K107R) and ATF5(148–281) with regard to their ability to interact with PCNT and their localization at the centrosome. HeLa cells were co-transfected with a vector expressing HA-SUMO3 and a vector expressing (at a low level) GFP-ATF5 or GFP-ATF5(K106R/K107). ATF5 interaction with PCNT and ATF5 localization at the centrosome were determined by immunoprecipitation and immunofluorescence microscopy, respectively. Immunoblotting of GFP immunoprecipitates showed that ATF5 interaction with PCNT and GCP-2 was disrupted when ATF5 was SUMOylated by overexpressed SUMO3 (Fig. 6A, compare the first lane with the second lane), whereas the SUMOylation-defective ATF5(K106R/K107R) interacted with PCNT and GCP-2 steadily irrespective of SUMO3 expression (Fig. 6A, compare the third lane with the fourth lane). ATF5 localization in the centrosome was determined by staining the transfected cells with GFP, γ-tubulin, and DAPI. As shown in Fig. 6B, C (also compare with Fig. 5A), SUMOylation-induced reduction in GFP-ATF5 localization at the M-phase centrosomes was significantly alleviated for the SUMOylation-defective GFP-ATF5(K106R/K107R). Similar results were obtained when ATF5(148–281), which is centrosome-bound (12) and associated with PCNT (Fig. 4B), and is also defective for SUMOylation (Fig. 2), was tested (data not shown). These results show that blockade of ATF5 SUMOylation prevents ATF5 from SUMOylation-induced dissociation with PCNT and GCP-2 and from SUMOylation-driven ATF5 down-regulation at the centrosome.
Figure 6.
A SUMOylation-defective ATF5 mutant is recalcitrant to SUMOylation-driven dissociation of ATF5 from PCNT and GCP-2 and from the centrosome. A, interaction of PCNT and GCP-2 with wildtype ATF5 was disrupted upon SUMO3 overexpression, whereas that with SUMOylation-defective GFP-ATF5(K106R/K107R) was unaffected. Immunoblotting (IB) and immunoprecipitation (IP) with the indicated antibodies were performed as in Fig. 4, except that the indicated DNA constructs were used in transfection of COS-7 cells. B and C, immunofluorescence staining and statistical analysis were performed as in Fig. 5A, except the indicated DNA constructs were transfected. *, p < 0.02.
Interference with ATF5 SUMOylation deregulates the centrosome cycle and causes genomic instability and G2/M arrest in HeLa cells
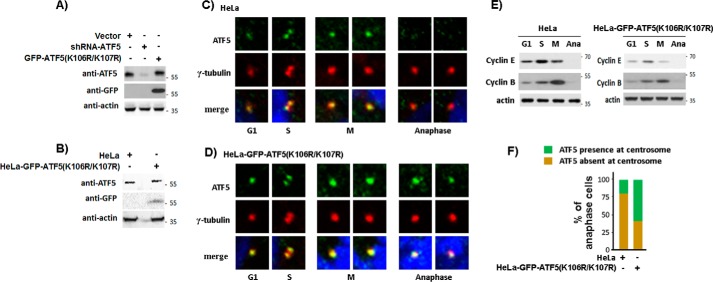
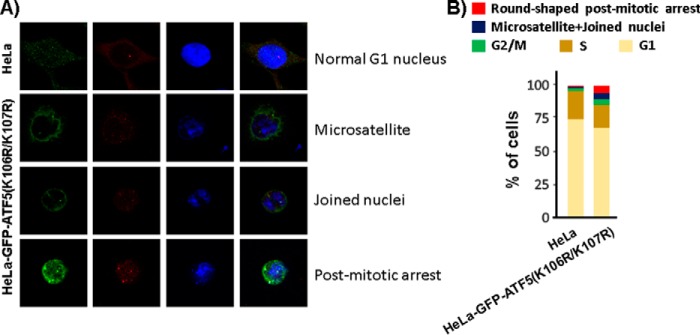
To assess whether defect in ATF5 SUMOylation affects the centrosome cycle, we created a HeLa cell line in which endogenous ATF5 was replaced by GFP-ATF5(K106R/K107R). In this cell line, an shRNA targeting the endogenous ATF5 at the K106/K107 region was co-expressed with SUMOylation-defective GFP-ATF5(K106R/K107R) that is resistant to the shRNA (Fig. 7A, B). The parental (HeLa) and SUMOylation-defective ATF5 cell line, HeLa-GFP-ATF5(K106R/K107R), were growing in growth media and immunostained with antibodies against ATF5 and γ-tubulin, and counterstained with DAPI. As shown in Fig. 7, C and E, and consistent with our previous report (12), we observed abrupt down-regulation of centrosomal ATF5 at the anaphase of the cell cycle following a steady accumulation from the G1 to M phase in HeLa cells. However, this anaphase down-regulation of centrosomal ATF5 was undetectable in the majority of the HeLa-GFP-ATF5(K106R/K107R) cells (Fig. 7, D–F). Thus, defective ATF5 SUMOylation specifically affects the timely ATF5 down-regulation in the centrosome that normally takes place at the end of M phase (12). Consistent with the abnormalities known to be associated with a defective centrosome or the mitotic spindle, we found that, in comparison with the parental HeLa cells, the HeLa-GFP-ATF5(K106R/K107R) cells exhibited a 3-fold increase of the G2/M cell population, suggesting a retarded G2/M phase. More strikingly, we observed a 7-fold increase in genomic instability, as manifested by joined nuclei and microsatellites, and an 11-fold increase in post-mitotic arrested cells, which are smaller, round, mononuclear cells that appeared to be no longer growing after mitosis (Fig. 8, A and B). Together, these results show that interference with ATF5 SUMOylation deregulates the centrosome cycle and causes genomic instability and G2/M arrest.
Figure 7.
Blockade of ATF5 SUMOylation prevents ATF5 down-regulation at the anaphase centrosome. A and B, characterization of the HeLa cell line in which ATF5 was replaced by GFP-ATF5(K106R/K107R). A, Western blot analysis monitoring ATF5 expression in transiently transfected HeLa cells. B, protein levels in parental (HeLa) and HeLa-GFP-ATF5(K106R/K107R) cells were determined. C and D, parental (HeLa) or engineered HeLa-GFP-ATF5(K106R/K107R) cells were co-stained with γ-tubulin (red) and ATF5 (green) and counterstained with DAPI (blue) as in Fig. 5A. Presence of ATF5 at the anaphase (Ana) centrosome showed the most difference. E, cells at specific cell cycle time points in C and D were confirmed by Western blot analysis monitoring cyclin E and cyclin B expression. F, percentages of anaphase cells in which ATF5 is present or absent at the centrosome in C and D were tallied. At least 50 cells were examined for each experimental condition.
Figure 8.
Defective ATF5 SUMOylation causes genomic instability and G2/M arrest in HeLa cells. A, representative images of cells with genomic instability, including microsatellite and joined nuclei, and of cells that were post-mitotically arrested. B, cells as shown in A were scored. A total of 1200 cells were counted for both HeLa and HeLa-GFP-ATF5(K106R/K107R) cells.
Discussion
Protein SUMOylation is known to regulate protein–protein interaction and their subcellular localization, modulating essential cellular processes, including genomic instability and cell cycle progression. We have shown previously that ATF5 is a PCM protein that forms the critical middle in the PGT–ATF5–PCNT tripartite between the PCM and the mother centriole (12). We demonstrated here that ATF5 is SUMOylated at the end of mitosis and that ATF5 SUMOylation interrupts ATF5–PCNT interaction and promotes ATF5 disengagement from the centrosome at anaphase (Figs. 3–5). Although timely SUMOylation at anaphase is required for the completion of the centrosome cycle, a failure in ATF5 SUMOylation disrupts the dynamic cell cycle–dependent ATF5 cycling at the centrosome, causing excessive genomic instability and post-mitotic cell cycle arrest (Figs. 6–8).
Our data showed directly that ATF5 SUMOylation disrupts ATF5 interaction with PCNT and GCP2 and drives ATF5 away from the centrosome (Figs. 4 and 5). In addition, several lines of evidence seem to suggest that non-SUMOylated ATF5 is as specific and active in participation in cell cycle–dependent ATF5 regulation and that ATF5 SUMOylation, which occurs at the end of M phase, is the driver for such events. ATF5 accumulation at the centrosome starts from G1 and reaches its peak at G2/M phase (Fig. 7C and (12)), which coincides with a pattern of ATF5 SUMOylation in the cell that is high at G1 and undetectable at G2/M phase (Fig. 3). SUMOylated ATF5 does not interact with centrosomal proteins and cannot be incorporated into the centrosome (Figs. 4 and 5). SUMOylation-defective ATF5 mutants, ATF5(148–281) (12) and ATF5(K106R/K107R), interact with PCNT similarly as wildtype ATF5, if not stronger (Figs. 4B and 6A), and both are accumulated at the centrosome as the wildtype ATF5 (ATF5(148–281) in Ref. 12 and ATF5(K106R/K107R) in Fig. 7). Wildtype ATF5 is sensitive, whereas SUMOylation-defective ATF5 mutants are resistant, to SUMOylation-promoted ATF5 loss from the centrosome (Figs. 5 and 6B). Replacement of wildtype ATF5 with a SUMOylation-defective ATF5 mutant in HeLa cells blocks anaphase down-regulation of ATF5 at the centrosome (Fig. 7). Not coincidentally, we observed that co-expression of ATF5 with de-SUMOylases, SENP2 and PIAS1, reverses most of the phenotypes caused by ATF5 SUMOylation (data not shown). Based on these analyses, we propose that ATF5 SUMOylation is a key regulatory mechanism that controls the centrosome cycle. Thus, the ATF5 centrosome cycle can be viewed from a novel perspective with regard to ATF5 SUMOylation status. Non-SUMOylated ATF5 accumulates at the centrosome from G1 to M phase, which forms the linchpin between the mother centriole and the PCM. ATF5 SUMOylation accelerates suddenly at the end of M phase, resulting in disruption of the interaction between ATF5 and PCNT and the precipitous loss of ATF5 at the centrosome. Therefore, both the timely ATF5 SUMOylation and de-SUMOylation are critical for the proper regulation of ATF5 at the centrosome.
The conserved SUMOylation site in ATF5 from human, rat, mouse, and frog (Fig. 2A) suggests that ATF5 SUMOylation has an essential function. Although we currently do not know whether ATF5 SUMOylation also modulates its other functions, such as transcription potential, we did observe that SUMOylation affects ATF5 association with the mitotic chromosomes and enrichment in the G1 nucleus (data not shown). With the centrosome-related defects in genomic instability and postmitotic cell cycle arrest reported here, it is anticipated that further investigation of ATF5 SUMOylation may lead to additional novel findings involving roles of ATF5 in several human diseases, such as cancer and various neurodisorders.
Experimental procedures
DNA constructs and site-specific mutagenesis
The DNA constructs expressing ATF5 full-length, ATF5(1–100), ATF5(101–207), ATF5(148–281), ATF5(208–281), and ATF5(K29R) have been described previously (8, 12, 24, 25). ATF5(K106R/K107R), ATF5(ΔK212/213), and ATF5(K106R/K107R;ΔK212/213) were created using GFP-ATF5 as template and a QuikChange site-directed mutagenesis kit (Stratagene) as described previously (9). Primers and reaction conditions for the generation of these mutants are available upon request. pcDNA3-HA-SUMO1, pcDNA3-HA-SUMO2, and pcDNA3-HA-SUMO3 were generous gifts from Dr. J. A. Iniguez-Lluhi (26). All PCR-generated genes were verified by DNA sequencing.
Cells and transfection
HeLa, MCF-7, HepG2, COS-7, and IMR-90 were purchased from the AATC. HeLa-GFP-ATF5(K106R/K107R) was created by stable transfection of HeLa cells with a DNA construct expressing shRNA that targets endogenous ATF5 at the Lys106/Lys107 region and a DNA construct expressing SUMOylation-defective GFP-ATF5(K106R/K107R), which is resistant to the siRNA. All cells were maintained in Dulbecco's modified Eagle's medium plus 10% fetal bovine serum (Atlanta Biologicals). All culturing media contained 100 μg/ml streptomycin and 100 international units/ml penicillin. Cells were transfected using Lipofectamine 2000 (Invitrogen) according to the instructions of the manufacturer or using polyethyleneimine (1 μg/ml) with a ratio of 1:3 to DNA and a similar procedure. Transfection of control siRNA (si-37007) and siRNAs against SUMO1 (sc-29498), 2 (sc-41081), 3 (41083), and 2/3 (sc-37167) were made using siRNA transfection reagent (sc-29528) and siRNA transfection medium (sc-36868), all from Santa Cruz Biotechnology.
Antibodies
The uses and sources of antibodies against ATF5, GFP, GCP-2, GCP-4, HA, and γ-tubulin have been described previously (12). Antibodies against pan-SUMO and SUMO2/3 were obtained from Abgent (AP1290a) and Santa Cruz Biotechnology (sc-32873), respectively. Antibodies against cyclin B and cyclin E were from Abcam (ab72) and Santa Cruz Biotechnology (sc-247), respectively.
Immunoblotting, immunoprecipitation, and immunofluorescence microscopy
These were performed as described previously (8, 12, 24, 25).
DTB
Exponentially growing cells were treated with thymidine (2 mm) for 16 h at 37 °C, washed three times with PBS, and refed with normal growth medium for 8 h at 37 °C. The cells were treated again with thymidine (2 mm) for 16 h, washed three times with PBS, and returned to normal growth medium at the indicated time points to obtain synchronized cells at G1 and G2/M phases.
FACS
HeLa cells were synchronized by DTB, released into Dulbecco's modified Eagle's medium, and harvested during the second S/G2/M cycle. A portion of cells was stained with propidium iodide and analyzed by FACS as described previously (27).
Centrosome sedimentation analysis
This was done according to Bornens et al. (28). Briefly, confluent cells were incubated at 37 °C for 2 h with 2 mg/ml final concentration of nocodazole (Sigma) and successively washed with ice-cold PBS, 8% sucrose in 0.1% PBS, and 8% sucrose in H20. Cells were then lysed with buffer A (50 mm Tris (pH 7.4), 80 mm NaCl, 25 mm EDTA (pH 8.0), and 1% Triton X-100) plus protease inhibitor for 10 min on ice with vigorous rotation. After incubation overnight at 4 °C, the lysate was sonicated for 10 strokes at low output with a microtip and subjected to two passages through a 23-gauge needle and one filtration through a 40-μm nylon mesh (BD Falcon). The filtrate was cleared by centrifugation at 2500 × g for 10 min, and the supernatant was laid on top of a 3.5-ml 60% sucrose cushion and subjected to centrifugation at 10,000 × g for 30 min. The 60% sucrose fraction containing centrosomes was recovered and diluted to 20% to 25% with distilled water (with 3 m NaCl to make a final NaCl concentration at 0.5 m for the preparation of stripped centrosomes), which was then added on top of a discontinuous sucrose gradient (from the bottom to the top, containing 1.7, 1, and 1 ml of 70%, 50%, and 40% sucrose solutions, respectively) in a 13-ml SW28 Beckman ultraclear tube. After centrifugation at 40,000 g for 1 h, 18 fractions (170 μl each) were collected, starting from the bottom of the gradient. The fractions were frozen in liquid nitrogen and then stored at −80 °C for future use.
Author contributions
Y. Y., K. G., E. K., E. L., M. H., K. L., D. Q., Y. X., H. K., and D. X. L. data curation; Y. Y., K. G., E. K., K. L., and D. X. L. formal analysis; Y. Y., K. G., and Y. X. validation; Y. Y., E. K., E. L., K. L., D. Q., and Y. X. methodology; M. H., K. L., D. Q., B. W., and D. X. L. conceptualization; M. H. and B. W. investigation; D. X. L. funding acquisition; D. X. L. writing-original draft; D. X. L. writing-review and editing.
Acknowledgment
We thank Dr. J. A. Iniguez-Lluhi for providing the pcDNA3-HA-SUMO1, pcDNA3-HA-SUMO2, and pcDNA3-HA-SUMO3 DNA constructs.
This work was supported in part by a startup package from Washington State University College of Pharmacy, a research scholar grant from the American Cancer Society, and an NCI, National Institutes of Health R15 award (to D. X. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- ATF
- activating transcription factor
- SUMO
- small ubiquitin-like modifier
- HA
- hemagglutinin
- DTB
- double thymidine blockade
- PCNT
- pericentrin
- DAPI
- 4′,6-diamidino-2-phenylindole
- shRNA
- short hairpin RNA
- PCM
- pericentriolar material.
References
- 1. Greene L. A., Lee H. Y., and Angelastro J. M. (2009) The transcription factor ATF5: role in neurodevelopment and neural tumors. J. Neurochem. 108, 11–22 10.1111/j.1471-4159.2008.05749.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Lengel K., Kim E., and Liu D. X. (2015) ATF5 is an essential protein in the centrosome. Cell Cycle 14, 3215–3216 10.1080/15384101.2015.1086202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dluzen D., Li G., Tacelosky D., Moreau M., and Liu D. X. (2011) BCL-2 is a downstream target of ATF5 that mediates the prosurvival function of ATF5 in a cell type-dependent manner. J. Biol. Chem. 286, 7705–7713 10.1074/jbc.M110.207639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chen A., Qian D., Wang B., Hu M., Lu J., Qi Y., and Liu D. X. (2012) ATF5 is overexpressed in epithelial ovarian carcinomas and interference with its function increases apoptosis through the downregulation of Bcl-2 in SKOV-3 cells. Int. J. Gynecol. Pathol. 31, 532–537 10.1097/PGP.0b013e31824df26b [DOI] [PubMed] [Google Scholar]
- 5. Hu M., Wang B., Qian D., Li L., Zhang L., Song X., and Liu D. X. (2012) Interference with ATF5 function enhances the sensitivity of human pancreatic cancer cells to paclitaxel-induced apoptosis. Anticancer Res. 32, 4385–4394 [PubMed] [Google Scholar]
- 6. Sheng Z., Li L., Zhu L. J., Smith T. W., Demers A., Ross A. H., Moser R. P., and Green M. R. (2010) A genome-wide RNA interference screen reveals an essential CREB3L2-ATF5-MCL1 survival pathway in malignant glioma with therapeutic implications. Nat. Med. 16, 671–677 10.1038/nm.2158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Izumi S., Saito A., Kanemoto S., Kawasaki N., Asada R., Iwamoto H., Oki M., Miyagi H., Ochi M., and Imaizumi K. (2012) The endoplasmic reticulum stress transducer BBF2H7 suppresses apoptosis by activating the ATF5-MCL1 pathway in growth plate cartilage. J. Biol. Chem. 287, 36190–36200 10.1074/jbc.M112.373746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Liu D. X., Qian D., Wang B., Yang J. M., and Lu Z. (2011) p300-Dependent ATF5 acetylation is essential for Egr-1 gene activation and cell proliferation and survival. Mol. Cell Biol. 31, 3906–3916 10.1128/MCB.05887-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Li G., Li W., Angelastro J. M., Greene L. A., and Liu D. X. (2009) Identification of a novel DNA binding site and a transcriptional target for activating transcription factor 5 in c6 glioma and mcf-7 breast cancer cells. Mol. Cancer Res. 7, 933–943 10.1158/1541-7786.MCR-08-0365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hu M., Wang B., Qian D., Wang M., Huang R., Wei L., Li L., Zhang L., and Liu D. X. (2017) Human cytomegalovirus immediate-early protein promotes survival of glioma cells through interacting and acetylating ATF5. Oncotarget 8, 32157–32170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Angelastro J. M., Ignatova T. N., Kukekov V. G., Steindler D. A., Stengren G. B., Mendelsohn C., and Greene L. A. (2003) Regulated expression of ATF5 is required for the progression of neural progenitor cells to neurons. J. Neurosci. 23, 4590–4600 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Madarampalli B., Yuan Y., Liu D., Lengel K., Xu Y., Li G., Yang J., Liu X., Lu Z., and Liu D. X. (2015) ATF5 connects the pericentriolar materials to the proximal end of the mother centriole. Cell 162, 580–592 10.1016/j.cell.2015.06.055 [DOI] [PubMed] [Google Scholar]
- 13. Gong L., Qi R., and Li D. W. (2017) Sumoylation pathway as potential therapeutic targets in cancer. Curr. Mol. Med. 16, 900–905 10.2174/1566524016666161223105201 [DOI] [PubMed] [Google Scholar]
- 14. Gong L., Sun Q., and Li D. W. (2017) Sumoylation in cellular senescence and aging. Curr. Mol. Med. 16, 871–876 10.2174/1566524016666161223104915 [DOI] [PubMed] [Google Scholar]
- 15. Seeler J. S., and Dejean A. (2017) SUMO and the robustness of cancer. Nat. Rev. Cancer 17, 184–197 10.1038/nrc.2016.143 [DOI] [PubMed] [Google Scholar]
- 16. Liu F. Y., Liu Y. F., Yang Y., Luo Z. W., Xiang J. W., Chen Z. G., Qi R. L., Yang T. H., Xiao Y., Qing W. J., and Li D. W. (2017) SUMOylation in neurological diseases. Curr. Mol. Med. 16, 893–899 10.2174/1566524017666170109125256 [DOI] [PubMed] [Google Scholar]
- 17. Flotho A., and Melchior F. (2013) Sumoylation: a regulatory protein modification in health and disease. Annu. Rev. Biochem. 82, 357–385 10.1146/annurev-biochem-061909-093311 [DOI] [PubMed] [Google Scholar]
- 18. Geiss-Friedlander R., and Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 10.1038/nrm2293 [DOI] [PubMed] [Google Scholar]
- 19. Vertegaal A. C., Andersen J. S., Ogg S. C., Hay R. T., Mann M., and Lamond A. I. (2006) Distinct and overlapping sets of SUMO-1 and SUMO-2 target proteins revealed by quantitative proteomics. Mol. Cell. Proteomics 5, 2298–2310 10.1074/mcp.M600212-MCP200 [DOI] [PubMed] [Google Scholar]
- 20. Anckar J., and Sistonen L. (2007) SUMO: getting it on. Biochem. Soc. Transact. 35, 1409–1413 10.1042/BST0351409 [DOI] [PubMed] [Google Scholar]
- 21. Liang J., Singh N., Carlson C. R., Albuquerque C. P., Corbett K. D., and Zhou H. (2017) Recruitment of a SUMO isopeptidase to rDNA stabilizes silencing complexes by opposing SUMO targeted ubiquitin ligase activity. Genes Dev. 31, 802–815 10.1101/gad.296145.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hay R. T. (2007) SUMO-specific proteases: a twist in the tail. Trends Cell Biol. 17, 370–376 10.1016/j.tcb.2007.08.002 [DOI] [PubMed] [Google Scholar]
- 23. Johnson E. S. (2004) Protein modification by SUMO. Annu. Rev. Biochem. 73, 355–382 10.1146/annurev.biochem.73.011303.074118 [DOI] [PubMed] [Google Scholar]
- 24. Li G., Xu Y., Guan D., Liu Z., and Liu D. X. (2011) HSP70 protein promotes survival of C6 and U87 glioma cells by inhibition of ATF5 degradation. J. Biol. Chem. 286, 20251–20259 10.1074/jbc.M110.211771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Liu X., Liu D., Qian D., Dai J., An Y., Jiang S., Stanley B., Yang J., Wang B., Liu X., and Liu D. X. (2012) Nucleophosmin (NPM1/B23) interacts with activating transcription factor 5 (ATF5) protein and promotes proteasome- and caspase-dependent ATF5 degradation in hepatocellular carcinoma cells. J. Biol. Chem. 287, 19599–19609 10.1074/jbc.M112.363622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mukherjee S., Cruz-Rodríguez O., Bolton E., and Iñiguez-Lluhi J. A. (2012) The in vivo role of androgen receptor SUMOylation as revealed by androgen insensitivity syndrome and prostate cancer mutations targeting the proline/glycine residues of synergy control motifs. J. Biol. Chem. 287, 31195–31206 10.1074/jbc.M112.395210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Liu X., Yang J. M., Zhang S. S., Liu X. Y., and Liu D. X. (2010) Induction of cell cycle arrest at G1 and S phases and cAMP-dependent differentiation in C6 glioma by low concentration of cycloheximide. BMC Cancer 10, 684 10.1186/1471-2407-10-684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bornens M., Paintrand M., Berges J., Marty M. C., and Karsenti E. (1987) Structural and chemical characterization of isolated centrosomes. Cell Motil. Cytoskeleton 8, 238–249 10.1002/cm.970080305 [DOI] [PubMed] [Google Scholar]