Abstract
Lipid metabolism plays a critical role in female reproduction. During oogenesis, maturing oocytes accumulate high levels of neutral lipids that are essential for both energy production and the synthesis of other lipid molecules. Metabolic pathways within the ovary are partially regulated by protein kinases that link metabolic status to oocyte development. Although the functions of several kinases in this process are well established, the roles that many other kinases play in coordinating metabolic state with female germ cell development are unknown. Here, we demonstrate that the catalytic activity of casein kinase 2 (CK2) is essential for Drosophila oogenesis. Using an unbiased biochemical screen that leveraged an unusual catalytic property of the kinase, we identified a novel CK2 substrate in the Drosophila ovary, the lipid droplet–associated protein Jabba. We show that Jabba is essential for modulating ovarian lipid metabolism and for regulating female fertility in the fly. Our findings shed light on a CK2-dependent signaling pathway governing lipid metabolism in the ovary and provide insight into the long-recognized but poorly understood association between energy metabolism and female reproduction.
Keywords: Drosophila, lipid droplet, lipid metabolism, protein kinase, reproduction, signal transduction, CK2, Jabba, casein kinase 2, oogenesis
Introduction
In most female animals, metabolism regulates reproductive potential, and emerging evidence indicates a clear correlation between fatty acid metabolism and oogenesis (1, 2). During oogenesis, developing oocytes accumulate large amounts of sterols and triglycerides that can be oxidized for ATP production or used for the synthesis of other essential lipid species (3, 4). Consequently, perturbations in lipid metabolism such as metabolic syndrome and obesity are associated with infertility and several disorders of human reproduction, most notably polycystic ovary syndrome (5, 6). However, despite the requirement for proper lipid homeostasis in the female reproductive system, relatively little is known regarding the molecular mechanisms that regulate lipid metabolism in the ovary.
The model organism Drosophila melanogaster has proven to be a powerful system for dissecting the fundamental role of lipid metabolism in disease states and in normal developmental processes such as gametogenesis (7–9). Oogenesis in flies occurs in 14 continuous but morphologically distinct stages. At the anterior end of the ovary, germ line stem cells reside within a structure known as the germarium. These cells give rise to an oocyte that develops within an egg chamber (or follicle) composed of 15 germ line–derived nurse cells enclosed by an epithelium of hundreds of somatic follicle cells (10). Neutral lipid synthesis within the nurse cells is dramatically enhanced during mid-oogenesis (11), and these lipids, primarily in the form of lipid droplets, are transferred during development from the nurse cells to the oocyte through actin-based ring canals (12). The importance of lipid metabolism in oogenesis is highlighted by mutation of the midway gene that encodes Drosophila diacylglycerol acyltransferase: in midway mutants, oogenesis fails halfway through the process with a concomitant reduction in the level of neutral lipids within the ovary (13). Likewise, mutations in the gene encoding Cct1, a phospholipid biosynthetic enzyme, result in loss of germ line stem cell maintenance and inappropriate positioning of the oocyte within the Drosophila egg chamber (14). Several other studies in both flies and mammals have also demonstrated that lipids accumulate during specific stages of oocyte development and are required for oogenesis (3, 11, 15, 16), further underscoring the importance of lipid metabolism in the ovary. As key mediators of signaling cascades, protein and lipid kinases are crucial for regulating oogenesis and early embryonic development (17–19). During oogenesis and embryogenesis, specific kinases transduce developmental cues that establish axes of polarity and gradients of morphogens and coordinate the activities of somatic follicle cells with germ line cells for the formation of a mature oocyte (20–22). Although the functions of several kinases in the metabolic control of oogenesis are well described (23–25), the identification and characterization of the complete complement of kinases whose activities couple intermediary metabolism and female reproduction remain far from complete. Here, we describe a novel role for casein kinase 2 (CK2)2 during Drosophila oogenesis. We report that CK2 phosphorylates the lipid droplet–associated protein Jabba and that Jabba is essential for mediating lipid metabolism in the ovary. These findings expand the functional repertoire of this pleiotropic kinase and highlight a novel function for CK2 in the metabolic regulation of female reproduction.
Results
CK2 kinase activity is essential for oogenesis
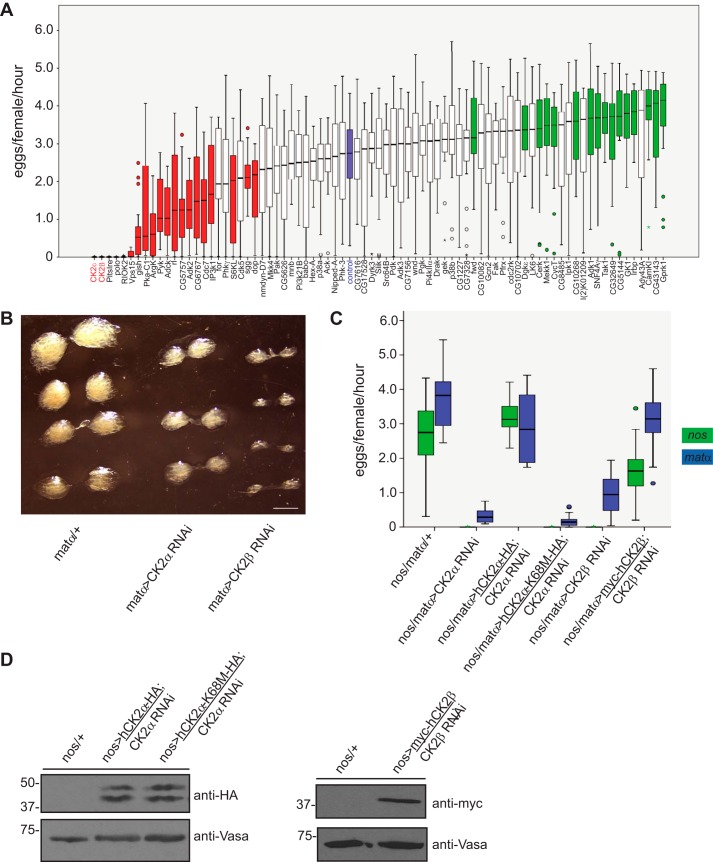
To identify kinases with previously uncharacterized roles in oogenesis, we individually reduced expression of a subset of the Drosophila kinome by shRNA in the female germ line using the Gal4-UAS system. Included in this RNAi screen were several kinases with known roles in Drosophila oogenesis such as polo (26) and protein kinase A (PKA) (27) that served as positive controls. Although knockdown of most kinases caused no change in oogenesis, reduced expression of several kinase-encoding genes resulted in either an increase or decrease in the number of eggs laid (Fig. 1A). Among the genes in this latter category that most significantly affected oogenesis were those encoding the two subunits of casein kinase 2 (CK2α and CK2β). Concomitant with decreased egg production, reduction of CK2 expression resulted in a significant decrease in overall ovarian size (Fig. 1B).
Figure 1.
CK2 kinase activity is essential for Drosophila oogenesis. A, identification of novel kinase regulators of Drosophila oogenesis. The indicated 78 kinases (on the x axis) were knocked down in the female germ line using nanos-Gal4:VP16-driven shRNA. Shown are box plots depicting egg laying rates of these flies (n = 20). Bars indicate the first to third quartiles, horizontal black lines denote the median, and circles denote outliers. Red bars indicate kinases whose reduced expression resulted in decreased numbers of eggs laid, whereas green bars indicate kinases whose reduced expression caused an increase in the number of eggs laid. The blue bar denotes the egg laying rate of flies expressing a control shRNA. Whiskers represent the upper and lower limits of the range. Statistical significance was determined using a Mann-Whitney U test. B, reduced ovarian size upon reduction of CK2 expression. Ovaries from flies of the indicated genotypes were dissected and imaged by light microscopy. Scale bar, 1 mm. C, egg laying rates from flies of the indicated genotypes. Expression of shRNA and/or transgenes was driven using either nanos-Gal4 (green) or matα-Gal4 (blue). Data are presented as in A. D, transgenic expression of epitope-tagged human CK2α and CK2β in the Drosophila female germ line. Ovary lysates were prepared, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies. Note that CK2α-HA migrates as a doublet.
CK2 is an evolutionarily conserved, ubiquitous serine/threonine kinase whose activity has been implicated in multiple physiological and pathological processes (28). The enzyme is highly promiscuous with a growing list of hundreds of putative substrates and is estimated to be responsible for phosphorylation of ∼10–15% of the eukaryotic proteome (29). CK2 forms heterotetramers composed of two catalytic subunits (CK2α or the highly homologous CK2α′) and two regulatory subunits (CK2β) (30). CK2 is also an unusual kinase; it is considered constitutively active (31), and its precise mechanism of regulation has yet to be fully defined.
The primary sequence identity between Drosophila and human CK2α and Drosophila and human CK2β is 85 and 88%, respectively (32, 33). We leveraged this high degree of homology to validate the results of our screen. Specifically, we tested whether female fertility could be restored in flies with reduced expression of endogenous CK2α or CK2β by transgenic expression of the respective epitope-tagged human CK2α or CK2β subunits (Fig. 1C). For these rescue experiments, we used two different germ line–specific Gal4 drivers to distinguish between germ line stem cell-autonomous versus non-autonomous functions of CK2. In particular, nanos-Gal4 drives expression throughout all stages of oogenesis including within the germ line stem cells of the germarium (34), whereas matα-Gal4 drives expression slightly later in the process beginning in stage 2 egg chambers outside of the germ line stem cell compartment (35). Transgenic expression of both human CK2α and CK2β was able to functionally compensate for reduced expression of the respective endogenous Drosophila CK2 subunits (Fig. 1, C and D). Moreover, the use of both Gal4 drivers produced similar results (Fig. 1C), indicating that CK2 has important functions outside of the stem cell compartment (although from these data we cannot entirely exclude a distinct germ line stem cell–specific role for CK2). We then addressed whether the kinase activity of CK2 is essential for female fertility in the fly by expressing a catalytically dead point mutant of human CK2α (CK2α-K68M) (34) in the context of Drosophila CK2α RNAi. Western blotting of ovary lysates demonstrated equal expression of both wildtype and kinase-dead human CK2α transgenes (Fig. 1D). Results from an egg laying using these flies revealed that expression of kinase-dead CK2α-K68M could not rescue the reduced fertility induced by knockdown of endogenous Drosophila CK2α (Fig. 1C). Collectively, these results indicate that the kinase activity of CK2 is critical for Drosophila oogenesis.
A biochemical screen identifies CK2 substrates in the Drosophila ovary
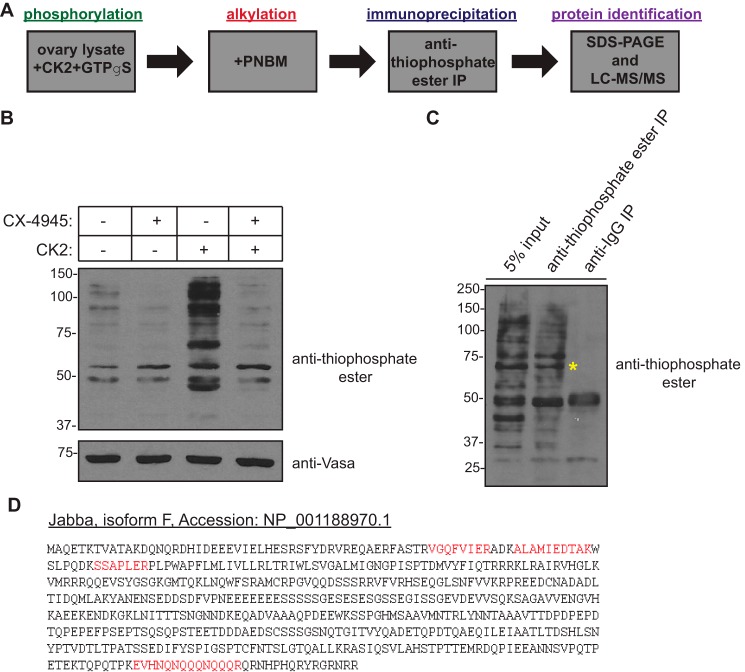
To gain additional insight into the role of CK2 kinase activity in oogenesis, we designed a novel biochemical strategy to identify CK2-specific substrates in the Drosophila ovary (Fig. 2A). Here, we took advantage of another unusual property of CK2, namely its dual cosubstrate specificity, or its ability to use either ATP or GTP as a phosphoryl donor (36). Ovary lysate from wildtype flies was incubated in the presence of excess recombinant CK2 holoenzyme and GTPγS, allowing for thiophosphorylation of CK2-specific substrates. The alkylating agent p-nitrobenzyl mesylate (PNBM) was then added to the reaction, generating a thiophosphate ester moiety on these CK2 substrates. This modification could be detected with an anti-thiophosphate ester antibody and could also be used to immunoprecipitate these thiophosphorylated proteins from the reaction for subsequent identification by mass spectrometry. We first confirmed the validity of this approach by incubation of ovary lysate with the CK2 kinase inhibitor CX-4945 (silmitasertib) in both the presence and absence of excess CK2 holoenzyme. Western blotting of these lysates using an anti-thiophosphate ester antibody revealed reduced total levels of thiophosphorylated proteins in the presence of CX-4945 (Fig. 2B), confirming that the observed signal was dependent upon CK2 activity. This antibody was then used to immunoprecipitate the thiophosphorylated proteins, and these proteins were resolved by SDS-PAGE and subjected to anti-thiophosphate ester Western blotting. A unique band of ∼60 kDa was present upon immunoprecipitation with the anti-thiophosphate ester antibody but not with the control antibody (Fig. 2C). This band was excised from a corresponding Coomassie Blue–stained gel and submitted for protein identification by liquid chromatography/tandem mass spectrometry (LC-MS/MS).
Figure 2.
A biochemical screen identifies substrates of CK2 in the Drosophila ovary. A, schematic of the strategy used to identify novel ovarian substrates of CK2. B, experimental validation of the approach outlined in A. Ovary lysate was incubated for 30 min at 30 °C with or without recombinant human CK2 holoenzyme and in the presence or absence of CX-4945 (100 μm) as indicated. GTPγS (2.5 mm) was then added, and the reactions were incubated further for 1 min at 30 °C followed by treatment with PNBM for 60 min at room temperature. Kinase reactions were subsequently resolved by SDS-PAGE and immunoblotted with the indicated antibodies. C, immunoprecipitation (IP) of putative CK2 substrates from Drosophila ovary lysate. An anti-thiophosphate ester antibody was used to immunoprecipitate thiophosphorylated proteins. Immunoprecipitated proteins were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies. The yellow asterisk indicates the position of the band that was excised from a corresponding Coomassie Blue–stained gel for protein identification by LC-MS/MS. D, primary amino acid sequence of Jabba (isoform F). Highlighted in red are unique peptides, representing 7% coverage of the protein, identified by mass spectrometry.
Jabba is a novel CK2 substrate and is essential for oogenesis
One of the proteins identified within this band was a specific isoform (isoform F) of a recently discovered insect-specific lipid droplet–associated protein named Jabba (Fig. 2D). Long considered inert structures for the storage of neutral fats, lipid droplets have emerged as dynamic organelles that function as hubs of lipid and energy metabolism and as platforms for signaling cascades (37, 38). Lipid droplets have well characterized functions in mediating lipid storage and mobilization (37) and are composed of a protein-studded phospholipid monolayer surrounding a hydrophobic core of neutral lipids (primarily triglycerides and sterol esters). The proteins associated with the lipid droplet membrane are intimately involved in the regulation of lipid metabolism and lipid droplet biogenesis, although no such function for Jabba has thus far been ascribed. Interestingly, Jabba physically interacts with histones and sequesters them on lipid droplets for use during early Drosophila embryogenesis (39, 40). Apart from this particular function, however, little is known regarding the physiological role of Jabba in the ovary. The Jabba locus is predicted to encode eight isoforms of the protein (isoforms B–I) of various molecular weights that result from alternative splicing (39). The F isoform of Jabba identified here as a putative CK2 substrate is one of two larger isoforms and, like other Jabba isoforms, does not contain any known functional domains.
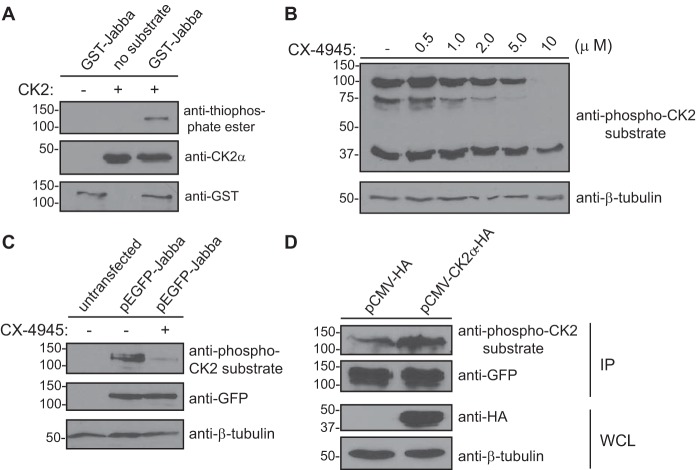
To validate the results obtained by mass spectrometry and confirm that Jabba is indeed a substrate of CK2, we expressed and purified GST-Jabba from bacteria. This fusion protein was used as a substrate in an in vitro kinase assay with recombinant CK2 holoenzyme. CK2 phosphorylated GST-Jabba (Fig. 3A), demonstrating that Jabba is a bona fide substrate of this kinase in vitro.
Figure 3.
Jabba is a novel CK2 substrate. A, in vitro kinase assay using GST-Jabba and CK2. GST-Jabba (or no substrate) was incubated in the presence or absence of recombinant CK2 holoenzyme and ATPγS followed by treatment with PNBM. Kinase reactions were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies. Western blots shown are representative of three independent experiments. B, inhibition of CK2 kinase activity using the small-molecule inhibitor CX-4945. HEK293 cells were treated (or not) with the indicated concentrations of CX-4945 for 24 h. Cell lysates (40 μg of total protein) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. C, HEK293 cells were transfected (or not) with a plasmid encoding GFP-Jabba. Cell lysates (5 μg of total protein) were resolved by SDS-PAGE and immunoblotted with the indicated antibodies. D, HEK293 cells were cotransfected with plasmids encoding GFP-Jabba and pCMV-HA (empty vector) or pCMV-CK2α-HA. Cells were lysed followed by immunoprecipitation of GFP-Jabba and immunoblotting of the precipitated protein with the indicated antibodies. Western blots shown are representative of three independent experiments. IP, immunoprecipitate; WCL, whole cell lysate.
We then addressed whether CK2 phosphorylates Jabba in a cellular context by inhibiting CK2 kinase activity in mammalian cells with CX-4945. We first determined an effective dose of the inhibitor by treating HEK293 cells with increasing concentrations of CX-4945. Cell lysates were prepared the following day and immunoblotted with an anti-phospho-CK2 substrate antibody (Fig. 3B). The results indicated significant inhibition of CK2 kinase activity at a concentration of 10 μm. HEK293 cells were then transfected or not with a construct encoding GFP-Jabba and either treated or not with 10 μm CX-4945. Cell lysates were subsequently prepared and immunoblotted with anti-phospho-CK2 substrate, anti-GFP, and anti-β-tubulin antibodies (Fig. 3C). The results demonstrate a reduction in the phospho-CK2 substrate signal at the molecular weight at which Jabba migrates by SDS-PAGE, suggesting that CK2 phosphorylates Jabba in cells. It should be noted here that GFP-Jabba runs at a molecular weight that is higher than expected; however, the identity of this band as Jabba was confirmed by mass spectrometry, and the aberrant migration of this protein is described in more detail below (Fig. 4D). To further substantiate this finding, we determined whether overexpression of CK2 would result in increased phosphorylation of Jabba. HEK293 cells were cotransfected with a plasmid encoding GFP-Jabba and a vector encoding CK2α-HA or empty vector as a control. Cells were subsequently lysed, and Western blotting of whole cell lysates confirmed overexpression of CK2α and equal expression of Jabba in both samples. Jabba was subsequently immunoprecipitated with an anti-GFP antibody, and immunoprecipitates were resolved by SDS-PAGE followed by immunoblotting with anti-GFP and anti-phospho-CK2 substrate antibodies (Fig. 3D). The results demonstrate increased phosphorylation of Jabba in cells overexpressing CK2α, providing further evidence that CK2 phosphorylates Jabba not only in vitro but also in cells.
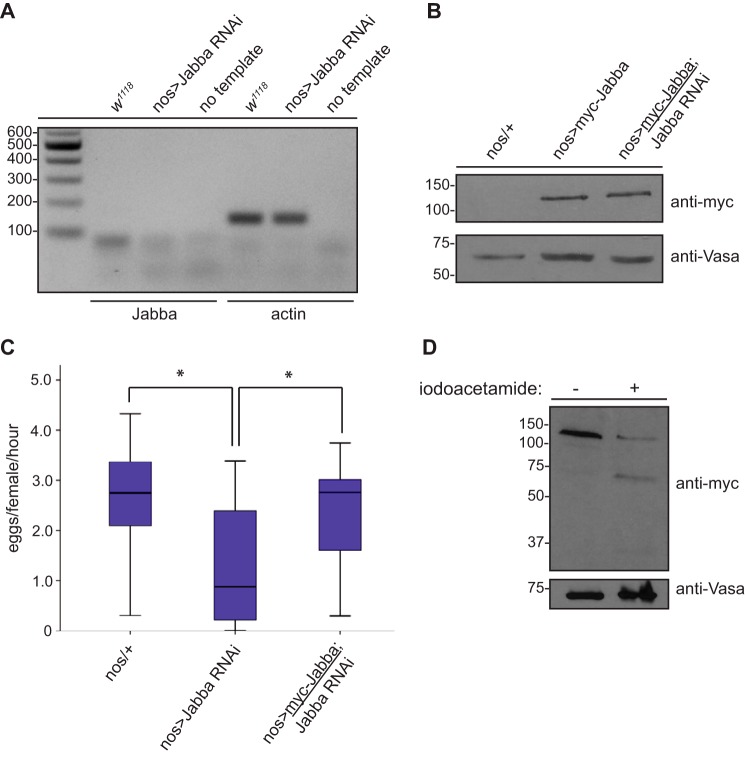
Figure 4.
Jabba is essential for Drosophila oogenesis and forms dimers in vivo. A, reduced Jabba mRNA levels in Jabba-RNAi ovaries by RT-PCR analysis. Total RNA was isolated from dissected ovaries from flies of the indicated genotypes. Resultant cDNA (or no template) was used in a polymerase chain reaction with primers to amplify specific regions of the genes encoding Jabba (expected product size, 93 bp) or actin (expected product size,137 bp) as a control. PCR products were run on a 2% agarose gel, stained with ethidium bromide, and imaged. B, expression of myc-Jabba and shRNA-resistant myc-Jabba in the Drosophila female germ line. Ovary lysates were prepared, resolved by SDS-PAGE, and immunoblotted with the indicated antibodies. C, egg laying rates from flies of the indicated genotypes. Data are presented as in Fig. 1A. n = 20; * denotes a p value <0.05 as determined by a Student's t test. D, Jabba (isoform F) forms dimers in vivo. Ovarian lysates from flies expressing myc-Jabba in the female germ line were treated (or not treated) with iodoacetamide (83.3 mm) at room temperature for 30 min. Lysates were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies.
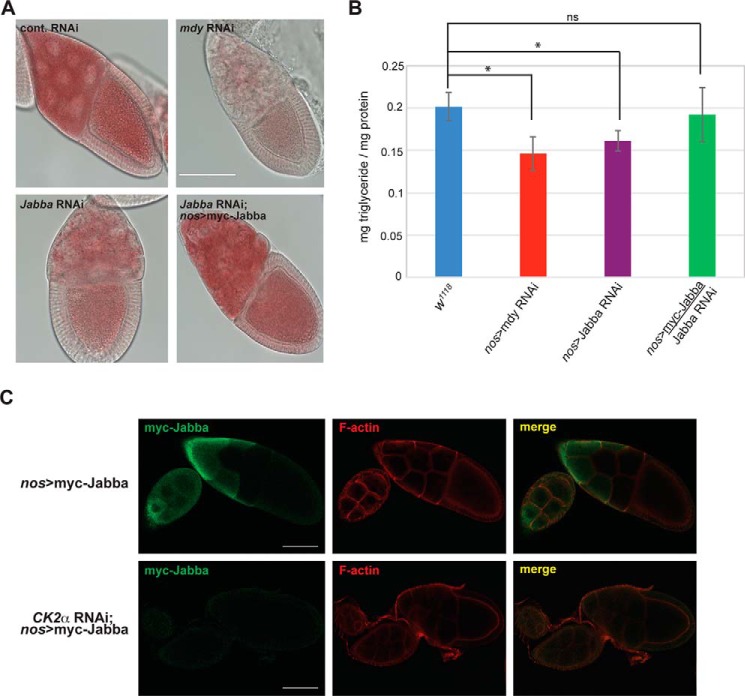
Jabba mutant embryos display reduced hatching at elevated temperatures, an altered histone H2Av/H2A ratio, and synthetic lethality upon reduced expression of histones (39, 40). Although these data clearly demonstrate that Jabba is important during Drosophila embryogenesis, the physiological role(s) that Jabba plays in earlier stages of development (i.e. during oogenesis) where its expression is first detected (39) is not known. We therefore tested whether isoform F of Jabba is required for oogenesis by reducing its expression in the female germ line. We first determined by RT-PCR analysis that the shRNA targeting Jabba is effective at reducing Jabba mRNA in the Drosophila ovary. Ovaries from Jabba RNAi flies displayed reduced Jabba (isoform F) transcript levels compared with ovaries from wildtype flies (Fig. 4A). Moreover, knockdown of this specific Jabba isoform by shRNA resulted in reduced egg production as assessed by an egg laying assay (Fig. 4C). We confirmed that this phenotype was indeed due to reduced expression of Jabba by transgenic expression of an epitope-tagged shRNA-resistant form of the protein in the Jabba RNAi genetic background. Importantly, Western blotting of ovary lysates from these flies demonstrated levels of Jabba that were similar to expression of the transgene in the absence of the targeting shRNA (Fig. 4B), and an egg laying assay confirmed that oogenesis was, in fact, restored in these flies (Fig. 4C). Collectively, these data demonstrate that isoform F of Jabba is essential for Drosophila oogenesis, phenocopying the effect of reduced CK2 expression (Fig. 1C).
An intriguing observation made during the course of this experiment was that Jabba migrated at approximately twice its predicted molecular weight on SDS-PAGE (Fig. 4B), suggesting that this protein may form covalent dimers. To determine whether this was indeed the case, Drosophila ovary lysate expressing myc-tagged Jabba was treated with the alkylating agent iodoacetamide or DMSO as a control (Fig. 4D). Treatment with iodoacetamide resulted in the appearance of a lower molecular weight band of the predicted size of Jabba, indicative of the formation of dimers in vivo and consistent with data from a recent study (41). However, this explanation cannot completely account for the aberrant migration of this protein by SDS-PAGE because other tagged forms of Jabba run at molecular weights that are inconsistent with dimer formation (Fig. 3). This is perhaps suggestive of additional, as yet unidentified, post-translational modifications of Jabba such as lipidation.
CK2 phosphorylates multiple regions of Jabba
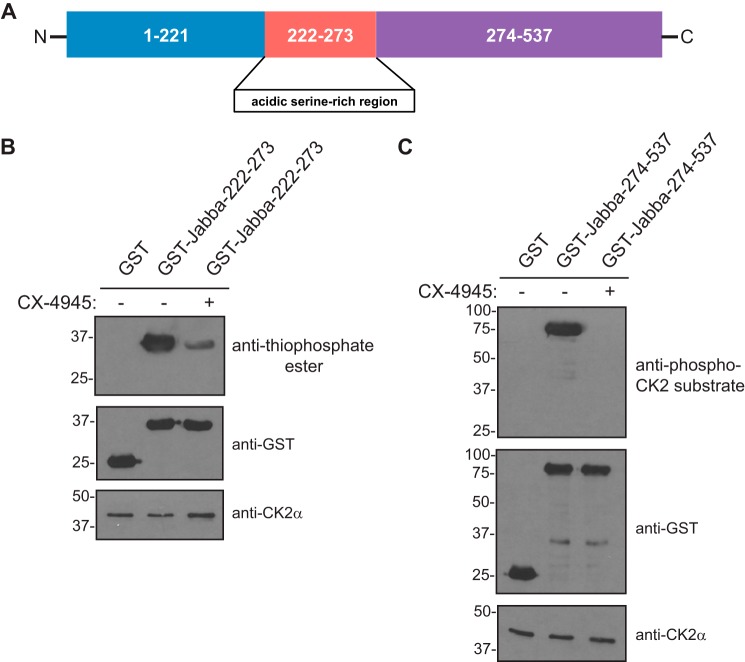
CK2 is an acidophilic kinase and preferentially phosphorylates serine or threonine residues in the context of surrounding glutamates or aspartates (29). Given this defining property, we assessed which regions of Jabba may be phosphorylated by CK2. Analysis of the primary sequence of Jabba revealed two major regions of the protein containing serine or threonine residues flanked by highly acidic stretches of amino acids that appeared to be within ideal consensus sites for CK2-dependent phosphorylation. One such region was an acidic serine-rich stretch consisting of residues 222–273, whereas the other encompassed the C-terminal portion of the protein comprising amino acids 274–537 (Figs. 2D and 5A). To test whether either or both of these regions of Jabba are phosphorylated by CK2, we expressed and purified GST-tagged truncation fragments of these regions from bacteria for use in in vitro kinase assays with CK2. The results revealed that both regions of Jabba are in fact phosphorylated by CK2 in vitro (Fig. 5, B and C), suggesting that CK2-mediated Jabba phosphorylation likely occurs at multiple regions and multiple sites in vivo.
Figure 5.
CK2 phosphorylates multiple regions of Jabba. A, schematic diagram of Jabba (isoform F) depicting various truncation fragments generated. B, in vitro kinase assay using GST-Jabba(222–273) and CK2. GST or GST-Jabba(222–273) was incubated with recombinant CK2 holoenzyme and ATPγS in the presence or absence of CX-4945 (10 μm) followed by treatment with PNBM. Kinase reactions were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies. Western blots shown are representative of three independent experiments. C, in vitro kinase assay using GST-Jabba(274–537) and CK2. GST or GST-Jabba(274–537) was incubated with recombinant CK2 holoenzyme and ATP in the presence or absence of CX-4945 (10 μm). Kinase reactions were resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies. Western blots shown are representative of three independent experiments.
Jabba localizes to lipid droplets in a CK2-independent manner
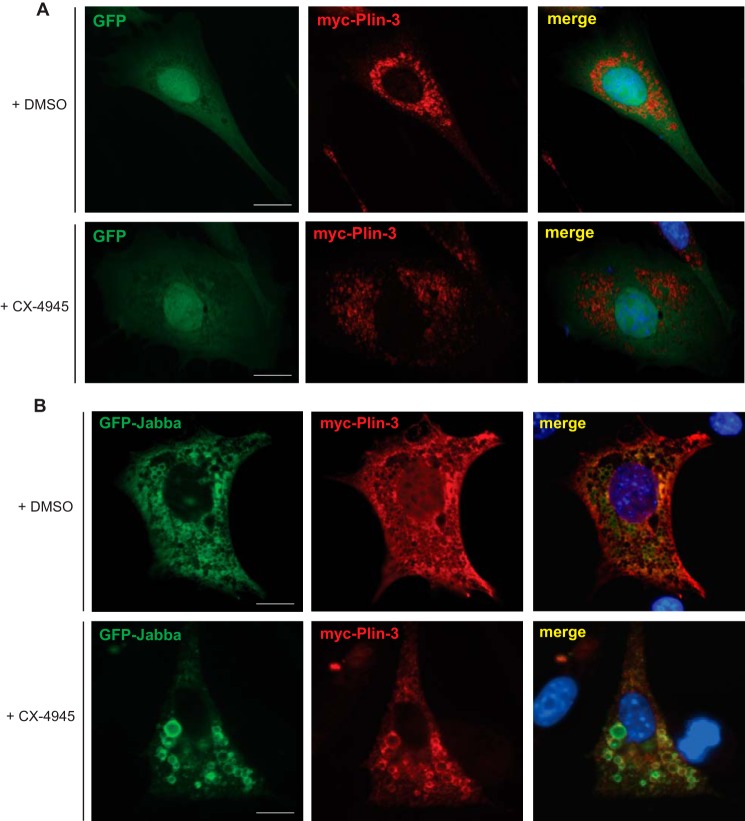
The molecular mechanisms by which proteins are targeted to lipid droplets are diverse and incompletely understood. Documented modes of localization include the embedding of amphipathic helices or hydrophobic hairpins of lipid droplet–associated proteins within the phospholipid monolayer or neutral lipid core of the organelle (42). Alternative targeting mechanisms include post-translational modifications such as lipidation (43–45) or binding to other lipid droplet–associated proteins that are often phosphorylated (46, 47). We therefore sought to determine whether CK2 kinase activity was required for localization of Jabba to lipid droplets. To address this question in a tractable system, we tested whether Jabba localized to lipid droplets in mammalian cells. 3T3-L1 murine preadipocytes were cotransfected with plasmids encoding GFP or GFP-tagged Jabba and myc-tagged perilipin-3, a member of the perilipin family of proteins that localizes to cytoplasmic lipid droplet membranes (48). These cells were subsequently treated with oleic acid to induce lipid droplet formation and stained with an anti-myc antibody to visualize lipid droplets. Although GFP displayed cytoplasmic and nuclear localization as expected (Fig. 6A), Drosophila Jabba strikingly localized to mammalian lipid droplet membranes as determined by its colocalization with perilipin-3 (Fig. 6B, top panels). To investigate whether this localization was dependent on CK2 kinase activity, cells expressing GFP or GFP-Jabba were treated with CX-4945 and imaged by fluorescence microscopy. These results indicate that Jabba still localizes to lipid droplets in the presence of the inhibitor (Fig. 6B, bottom panels), suggesting that the mechanism by which Jabba is targeted to this organelle is not dependent on CK2-mediated phosphorylation. These results are also consistent with a recent report indicating that a hydrophobic segment at the N terminus of Jabba is both necessary and sufficient for lipid droplet localization (41).
Figure 6.
Jabba localizes to lipid droplets in a CK2-independent manner. A, 3T3-L1 preadipocytes were cotransfected with plasmids encoding myc-perilipin-3 (Plin-3) and GFP. Cells were treated with oleic acid. Cells were then treated with DMSO (top panels) or CX-4945 (10 μm) (bottom panels), fixed, and processed for immunofluorescence microscopy with anti-myc antibodies. Cells were counterstained with DAPI to label nuclei (blue). Scale bars, 10 μm. B, 3T3-L1 preadipocytes were cotransfected with plasmids encoding myc-perlipin-3 (Plin-3) and GFP-Jabba and incubated with oleic acid. Cells were then treated with DMSO (top panels) or CX-4945 (10 μm) (bottom panels), fixed, and processed for immunofluorescence microscopy with anti-myc antibody. Cells were counterstained with DAPI to label nuclei (blue). Scale bars, 10 μm.
Jabba and CK2 regulate ovarian lipid metabolism
In comparison with wildtype embryos, lipid droplets in Jabba mutant embryos are misshapen and unevenly distributed; however, total embryonic triglyceride levels remain unchanged (39), suggesting that Jabba is not critical for lipid metabolism, at least in embryos. Nonetheless, to gain insight into the molecular basis for the reduced fertility noted upon Jabba knockdown (Fig. 4C), we qualitatively assessed levels of neutral lipids in ovaries from flies with reduced expression of Jabba by staining with Oil Red O (Fig. 7A). As a control for this experiment, we used flies in which expression of Midway was decreased by RNAi, a perturbation that results in a marked reduction in neutral lipids within the germ line (13). Although knockdown of Midway resulted in dramatically reduced levels of ovarian triglycerides and lipid droplet density, knockdown of Jabba caused a similar decrease as determined by Oil Red O staining (Fig. 7A). Importantly, this apparent reduction in lipid droplets could be rescued by expression of shRNA-resistant Jabba in the Jabba RNAi genetic background (Fig. 7A). To corroborate these results, we used a colorimetric assay to quantify total ovarian triglyceride levels from flies in which expression of Midway or Jabba was reduced by RNAi (Fig. 7B). Individual knockdown of both genes in the female germ line resulted in reduced levels of ovarian triglycerides compared with ovaries from wildtype (w1118) flies, and this reduction was again rescued by expression of shRNA-resistant Jabba in the Jabba RNAi genetic background (Fig. 7B). Taken together, these data indicate that the CK2 substrate Jabba is essential for maintaining neutral lipid levels and for regulating lipid homeostasis within the Drosophila ovary.
Figure 7.
Jabba and CK2 regulate lipid metabolism during Drosophila oogenesis. A, ovaries from flies of the indicated genotypes were dissected and stained with Oil Red O for visualization of neutral lipids. Shown are representative stage 10 egg chambers. Scale bar, 100 μm. B, quantitation of total triglyceride levels from ovaries of flies of the indicated genotypes. Data are presented as the mean (n = 4) with error bars denoting S.D. * indicates a p value <0.05. ns, not significant. C, confocal immunofluorescence microscopy of egg chambers from flies of the indicated genotypes. Dissected egg chambers were fixed and stained with anti-myc antibody to detect Jabba-labeled lipid droplets (green) and counterstained with rhodamine phalloidin to label F-actin (red). Shown are single optical confocal sections. Scale bars, 50 μm.
To investigate a role for CK2 in lipid metabolism during oogenesis, we used confocal immunofluorescence microscopy to analyze Jabba-labeled lipid droplets in the Drosophila ovary upon reduced expression of the kinase. In wildtype egg chambers, dense clusters of Jabba-associated lipid droplets that localized primarily to the subcortical regions of nurse cells were observed (Fig. 7C). In contrast, upon reduction of CK2 expression by RNAi, Jabba-labeled lipid droplets were markedly reduced (Fig. 7C), indicating an overall decrease in the number of ovarian lipid droplets in flies of this genotype. These results suggest that CK2 plays an integral role in regulating lipid metabolism in the Drosophila ovary, although the precise functions of this kinase and its substrate Jabba have yet to be determined.
Discussion
In the present study, we have identified the lipid droplet–associated protein Jabba as a novel CK2 substrate and defined a new role for CK2 in regulating lipid metabolism during Drosophila oogenesis. In the context of development, CK2 has been shown to be essential for early mammalian embryogenesis: mice lacking the CK2α subunit die in midgestation due to neural tube and cardiovascular defects (49). Interestingly, mice that are homozygous null for the highly homologous CK2α′ subunit are viable, but males are sterile and display abnormal sperm morphology and degeneration of germ cells at all stages of spermatogenesis (50, 51). Although these findings implicate CK2 in male germ cell development, our results now demonstrate a role for this kinase in regulating female germ cell development, expanding the functions of this already multifaceted kinase.
Kinase-mediated phosphorylation is an established mechanism for regulating the activity of metabolic enzymes via covalent modification. Among protein kinases that control lipid metabolism and lipid droplet biology, PKA is undoubtedly the best characterized, regulating a key step in the mobilization of triglycerides stored in adipose tissue. PKA phosphorylates hormone-sensitive lipase and perilipin, promoting their interaction and initiating the degradation of triacylglycerols and the subsequent release of fatty acids from lipid droplets. Phosphorylation of the lipid droplet–associated protein CGI-58/ABHD5 by PKA results in its dissociation from the lipid droplet membrane and enhances its capacity to coactivate adipose triglyceride lipase (52). In addition, phosphorylation of perilipin A by PKA facilitates lipolysis by causing lipid droplet fragmentation and cytoplasmic dispersion (53). Strikingly, deletion of the two genes in yeast that encode the β subunit of CK2 (CKB1 and CKB2) also affects lipid droplet morphology. However, deletion of CK2β results in the opposite phenotype and the accumulation of “supersized” lipid droplets (54). Notably, we observed a similar phenotype (i.e. the presence of larger lipid droplets) when CK2 kinase activity was inhibited in mammalian cells, although intriguingly, this phenotype seems to be more prominent for lipid droplets that are associated with Jabba (Fig. 6, A and B). Collectively, these results suggest that CK2 regulates lipid droplet morphology in various ways in multiple species and, as a consequence, may modulate lipid metabolism by regulating access of cytosolic lipases to lipid droplets.
Previous work has demonstrated that Jabba is essential for Drosophila development only under certain conditions such as histone depletion (40) and that mutation of Jabba does not affect overall triglyceride levels in embryos (39). Our findings indicate that Jabba may play a more prominent role earlier in the process by regulating lipid metabolism prior to embryogenesis during female germ cell development. Indeed, expression of Jabba is first detected within the nurse cells and oocyte of the Drosophila egg chamber (39) where its function has thus far been unknown. Although we have not identified the specific amino acids that are phosphorylated by CK2, we speculate that Jabba is heavily phosphorylated by this kinase on multiple serine/threonine residues. Moreover, we predict that other isoforms of Jabba are likely phosphorylated by this kinase as they all contain the highly acidic serine-rich middle region that is phosphorylated by CK2 (Fig. 5B). Taken together, these findings further suggest that there may be isoform-specific functions of Jabba, perhaps mediated by post-translational modification of their divergent C termini.
Accumulating evidence from several model organisms indicates that CK2 plays key roles in the regulation of lipid metabolism. In budding yeast, CK2 phosphorylates both Opi1p, the transcription factor that regulates phospholipid synthesis (55), and Pah1, the phosphatase that catalyzes the penultimate step in the synthesis of triacylglycerol (56). In mammalian cells, CK2 is essential for adipocyte differentiation (57, 58), and in Drosophila embryos, data suggest that endogenous CK2 partially localizes to lipid droplets (59), all lending support to the notion that CK2 may function as a master regulator of lipid metabolism. Our results bolster these findings by demonstrating a functional role for CK2 in modulating lipid metabolism in the Drosophila ovary. Whether CK2 regulates lipid metabolism in the human ovary has yet to be determined, but the possibility is tantalizing and warrants further investigation.
Experimental procedures
Antibodies and reagents
Anti-CK2α (C-18), anti-CK2β (FL-215), anti-GFP (E6), anti-c-myc (9E10), anti-GST (Z-5), anti-β-tubulin (H-235), anti-GAPDH (6C5), and anti-Vasa (d-260) antibodies were from Santa Cruz Biotechnology. Anti-HA and anti-phospho-CK2 substrate ((pS/pT)DXE where pS is phosphoserine and pT is phosphothreonine) antibodies were from Cell Signaling Technology. Anti-thiophosphate ester antibody (51-8) and PNBM were from Abcam. ATP, ATPγS, GTPγS, DAPI, oleic acid, Oil Red O, and iodoacetamide were purchased from Sigma-Aldrich. Rhodamine phalloidin was purchased from Invitrogen. The CK2 inhibitor CX-4945 was purchased from Selleckchem.
Cell culture
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% FBS and 1% penicillin/streptomycin. 3T3-L1 cells were purchased from the American Type Culture Collection (ATCC) and grown in DMEM supplemented with 10% bovine calf serum and 1% penicillin/streptomycin. Cells were grown in a humidified atmosphere with 5% CO2 at 37 °C. Lipofectamine 2000 (Invitrogen) was used for transfection according to the manufacturer's instructions. To induce lipid droplet formation, 3T3-L1 cells were stimulated overnight with 0.4 mm oleic acid.
Plasmid construction
Drosophila Jabba (isoform F) was synthesized as a gene block fragment (Integrated DNA Technologies) flanked with BglII and SalI restriction sites and ligated into the corresponding sites of pEGFP-C1 (Clontech) and pGEX-6P-1 (GE Healthcare). shRNA-resistant Jabba was also synthesized as a gene block fragment (Integrated DNA Technologies) and contains nine silent mutations in the region targeted by the shRNA. Constructs encoding human CK2α-ΗΑ (pZW6) and myc-CK2β (pZW12) for mammalian cell expression were obtained from Addgene (plasmids deposited by David Litchfield). Murine perilipin-3 was PCR-amplified from a complementary DNA (cDNA) clone (OriGene) as an EcoRI-BglII fragment and cloned into the corresponding sites of pCMV-myc (Clontech). For bacterial expression of GST-tagged Jabba fusion proteins, residues 1–537, 222–273, and 274–537 were PCR-amplified as BglII-EcoRI fragments and cloned into the BamHI-EcoRI sites of pGEX-6P-1. Oligonucleotides were purchased from Integrated DNA Technologies. Primer sequences are available upon request. All constructs were fully sequenced.
Mass spectrometry
Mass spectrometry was performed by MS Bioworks, LLC (Ann Arbor, MI). Briefly, in-gel digestion was performed on the submitted gel bands (an endogenous band of ∼60 kDa from Drosophila ovarian lysate and GFP-tagged Jabba immunoprecipitated from mammalian cell lysate) using a robot (ProGest, DigiLab). The sample was washed with 25 mm ammonium bicarbonate followed by acetonitrile and then reduced at 60 °C followed by alkylation with 50 mm iodoacetamide at room temperature. The sample was then digested with trypsin (Promega) at 37 °C for 4 h and quenched with formic acid. The supernatant was analyzed directly without further processing. Half of the digested sample was analyzed by nano-LC-MS/MS with a Thermo Fisher nLC-1000 HPLC system interfaced to a Thermo Fisher Q Exactive mass spectrometer. Peptides were loaded onto a trapping column and eluted over a 75-μm analytical column at 350 nl; both columns were packed with Acclaim PepMap (Thermo Fisher). The mass spectrometer was operated in data-dependent mode with MS and MS/MS performed in the Orbitrap at a resolution of 70,000 and 17,500 full width at half-maximum, respectively. The 15 most abundant ions were selected for MS/MS. MS/MS samples were analyzed using Mascot (version 2.5.1, Matrix Science, London, UK). Mascot was used to search the UniProt D. melanogaster database assuming strict digestion with trypsin. Scaffold software (version 4.6.2, Proteome Software Inc., Portland, OR) was used to validate MS/MS-based peptide and protein identifications. Peptide identifications were accepted if they could be established at a greater than 50.0% probability by the Scaffold local false discovery rate algorithm. Protein identifications were accepted if they could be established at greater than 90.0% probability and contained at least two identified peptides. Protein probabilities were assigned by the Protein Prophet algorithm (60). Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
Fly stocks and transgenic fly lines
Stocks were maintained and all crosses were performed at 25 °C on molasses-based food. All RNAi fly stocks were generated by the Transgenic RNAi Project (Harvard University Medical School) and obtained through the Bloomington Drosophila Stock Center (Indiana University). Human CK2α, CK2α-K68M, and CK2β were PCR-amplified from vectors pZW6, pGV15, and pZW12 (Addgene; deposited by David Litchfield), respectively, using attB-modified primers and cloned into pDONR-Zeo (Invitrogen). The cloned inserts were transferred into the relevant Drosophila destination vectors using Gateway cloning technology to create N- or C-terminal epitope-tagged constructs, as indicated, for Drosophila embryo microinjection. Transgenic flies were generated by BestGene, Inc. All transgenes map to the second chromosome.
RNAi screen and rescue experiments
Virgin nos-Gal4:VP16/TM3 Sb females were crossed with UAS-RNAi males. Four to six young (<7-day-old) nos-Gal4:VP16/UAS-RNAi females were then crossed with w1118 males, and eggs were collected overnight (16–18 h) on grape juice agar plates. The eggs were then counted using a dissecting microscope, and the number of eggs laid per female per hour was calculated. After 20 egg collections, the egg laying rate of nos-Gal4:VP16/UAS-RNAi females was compared with that of nos-Gal4:VP16/attP2 females using the Mann-Whitney U test. For the CK2 rescue experiments, virgin nos-Gal4:VP16/TM3 Sb females were crossed with UAS-CK2α-HA; UAS-CKIIα RNAi/TM6b or UAS-myc-CK2β/CyO; UAS-CKIIβ RNAi/TM6b males. Four to six young (<7-day-old) UAS-CK2α-HA/+; nos-Gal4:VP16/UAS-CKIIα RNAi or UAS-myc-CK2β/+; nos-Gal4:VP16/UAS-CKIIβ RNAi females were crossed with w1118 males, and their eggs were collected overnight (16–18 h) on grape juice agar plates. The number of eggs laid per female per hour was calculated. After 20 egg collections, the egg laying rate of rescue females was compared with that of nos-Gal4:VP16/UAS-CKIIα RNAi or nos-Gal4:VP16/UAS-CKIIβ RNAi females using the Mann-Whitney U test. All experiments were performed at 25 °C.
Oil Red O staining
This protocol was performed essentially as described (61). Briefly, ovaries were dissected and fixed in 4% formaldehyde for 30 min, washed twice in PBS, and washed twice in 100% propylene glycol. Fixed ovaries were then stained in a solution of 0.5% Oil Red O dissolved in propylene glycol previously filtered through Whatman Number 1 filter paper and preheated to 60 °C. Samples were incubated for 1 h at 60 °C and then washed twice with 85% propylene glycol and twice with PBS at room temperature. Stained specimens were mounted on microscope slides in glycerol and imaged using an EVOS FL microscope (Life Technologies) with a 40× objective.
Colorimetric triglyceride assay
This protocol was performed essentially as described (61). Virgin female flies were raised on medium with yeast and males for 3 days at 25 °C. Ovaries were dissected in cold PBS, washed, transferred to cold PBS + 0.05% Tween 20, homogenized on ice using a motorized pestle, and heated at 70 °C for 10 min. Triglycerides were measured from each sample in triplicate by incubating with Triglyceride Reagent (Sigma, T2449) at 37 °C for 45 min. Reactions were pelleted, and the supernatant was incubated with Free Glycerol Reagent (Sigma, F6428) at 37 °C for 5 min. Absorbance was measured at 540 nm using a Spark® multimode microplate reader (Tecan Life Sciences). Concentrations were determined by comparing readings with a glycerol standard curve of Glycerol Standard Solution (Sigma, G7793) subjected to the same reactions. Protein concentration was determined using a Bradford assay (Bio-Rad Protein Assay Dye Reagent Concentrate) and by measuring absorbance at 595 nm. Triglyceride levels were then compared with total protein content.
Recombinant protein expression and purification
BL21(DE3) cells (Novagen) were transformed with plasmids encoding GST, GST-Jabba, GST-Jabba(222–273), or GST-Jabba(274–537). Protein expression was induced at 16 °C for 24 h with 0.5 mm isopropyl 1-thio-β-d-galactopyranoside. Following centrifugation, the pellet was resuspended in 15 ml of lysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm DTT, 1 mm EDTA with protease inhibitors (Roche Applied Science)). The lysate was sonicated on ice and centrifuged at 20,000 × g for 20 min at 4 °C. The supernatant was added to 0.5 ml (packed volume) of glutathione-agarose beads (Gold Biotechnology) and incubated at 4 °C for 2 h with end-over-end tumbling. The slurry was transferred to a column and washed extensively with wash buffer. GST fusion proteins were eluted with excess glutathione in lysis buffer (pH 7.4), and appropriate fractions were pooled. The protein was dialyzed overnight at 4 °C into dialysis buffer (20 mm Tris, pH 7.4, 150 mm NaCl, 1 mm DTT). Glycerol was added to a final concentration of 10%, and the proteins were snap frozen and stored at −80 °C.
In vitro kinase assays
Recombinant CK2 holoenzyme (New England Biolabs) was incubated with 25 ng of recombinant purified protein in kinase buffer (50 mm HEPES, pH 7.5, 0.65 mm MgCl2, 0.65 mm MnCl2, 12.5 mm NaCl) with 500 μm ATP or ATPγS as indicated. Kinase reactions were incubated at 30 °C for 30 min. When appropriate, PNBM (dissolved in DMSO) was added to a final concentration of 2.5 mm, and reactions were incubated at room temperature for 1 h followed by the addition of 6× boiling sample buffer to stop the reaction. Reactions were then resolved by SDS-PAGE followed by immunoblotting with the indicated antibodies.
SDS-PAGE and Western blotting
To prepare lysates for immunoblotting or immunoprecipitation, dissected Drosophila ovaries or mammalian cells were lysed in ice-cold lysis buffer (20 mm Tris, pH 8.0, 200 mm NaCl, 1% Triton X-100 with protease inhibitors and phosphatase inhibitors (1 mm sodium orthovanadate, 10 mm sodium fluoride, and 1 mm β-glycerophosphate)). Proteins were resolved on 10 or 12.5% polyacrylamide gels and transferred to nitrocellulose membranes (GE Healthcare) using the Pierce G2 Fast Blotter (Thermo Scientific). Membranes were blocked in 5% nonfat milk in TBS followed by overnight incubation with the indicated primary antibody in the same buffer. Membranes were washed three times the following day in TBS + 0.5% Tween (TBS-T), incubated for 1 h with the appropriate HRP-conjugated secondary antibody (Cell Signaling Technology), washed three times again in TBS-T, and processed for signal detection using enhanced chemiluminescence (Santa Cruz Biotechnology). For detection of proteins from the same reaction/lysate that migrate at similar molecular weights (i.e. phosphorylated and non-phosphorylated forms of the same protein), samples were split and run on separate gels.
Immunofluorescence microscopy
3T3-L1 cells were seeded on glass coverslips and transfected the following day with the indicated constructs for 6 h. The medium was then replaced with fresh medium containing 400 μm oleic acid for overnight incubation. Cells were fixed the following day in 4% formaldehyde and processed for immunofluorescence microscopy. Coverslips were mounted on glass slides in Vectashield mounting medium (Vector Laboratories), and cells were imaged using an EVOS FL Auto microscope with a 40× objective. For confocal microscopy, dissected ovaries were fixed and immunostained as described (25) and imaged using an Olympus FluoView® FV3000 confocal laser-scanning microscope equipped with a 30× silicone immersion oil objective (1.05 numerical aperture). Images were acquired using FluoView software (Olympus) and processed using Photoshop CS6 (Adobe).
RT-PCR
Ovaries were dissected from flies of the indicated genotypes, and total RNA was extracted using the RNeasy Mini kit (Qiagen). cDNA was synthesized using SuperScript IV First-Strand Synthesis System (Invitrogen) and used as a template in PCR with gene-specific primers. The following primer pairs were used: Jabba-For, 5′-GCC ACC TCC TCC GAA GAC ATA TTC-3′; Jabba-Rev, 5′-CCT GGG TTC CTA GGC TAG TAT TGA A-3′; Actin42A-For, 5′-AAG AGG TTG CAG CTT TAG TGG-3′; Actin42A-Rev, 5′-TCC CAT TCC TAC CAT TAC GCC-3′. PCR products were run on a 2% agarose gel and stained with ethidium bromide.
Statistical analysis
Unless otherwise described, data are presented as the mean ± S.D. Statistical significance (p < 0.05) was determined by an unpaired Student's t test or a Mann-Whitney U test as indicated.
Author contributions
T. I. S. conceived and coordinated the study and wrote the paper. E. A. M. designed and conducted experiments and analyzed the experimental data in Figs. 1–4 and 7. S. M. L. performed experiments and analyzed the data in Fig. 2. M. D. S. and S. B. performed experiments and analyzed the data in Fig. 3. B. L. and T. I. S. performed experiments and analyzed the data in Fig. 6. N. K. S. performed experiments and analyzed the data in Fig. 5. All authors reviewed the results and approved the final version of the manuscript.
This work was supported by a Commonwealth Universal Research Enhancement grant from the Pennsylvania Department of Health (to T. I. S.) and by start-up funds from Drexel University College of Medicine (to T. I. S.). The authors declare that they have no conflicts of interest with the contents of this article.
- CK2
- casein kinase 2
- GTPγS
- guanosine 5′-O-(thiotriphosphate)
- PNBM
- p-nitrobenzyl mesylate
- ATPγS
- adenosine 5′-O-(thiotriphosphate).
References
- 1. Della Torre S., Benedusi V., Fontana R., and Maggi A. (2014) Energy metabolism and fertility: a balance preserved for female health. Nat. Rev. Endocrinol. 10, 13–23 10.1038/nrendo.2013.203 [DOI] [PubMed] [Google Scholar]
- 2. Fontana R., and Della Torre S. (2016) The deep correlation between energy metabolism and reproduction: a view on the effects of nutrition for women fertility. Nutrients 8, 87 10.3390/nu8020087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Dunning K. R., Anastasi M. R., Zhang V. J., Russell D. L., and Robker R. L. (2014) Regulation of fatty acid oxidation in mouse cumulus-oocyte complexes during maturation and modulation by PPAR agonists. PLoS One 9, e87327 10.1371/journal.pone.0087327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Dunning K. R., Russell D. L., and Robker R. L. (2014) Lipids and oocyte developmental competence: the role of fatty acids and β-oxidation. Reproduction 148, R15–27 10.1530/REP-13-0251 [DOI] [PubMed] [Google Scholar]
- 5. Lai H., Jia X., Yu Q., Zhang C., Qiao J., Guan Y., and Kang J. (2014) High-fat diet induces significant metabolic disorders in a mouse model of polycystic ovary syndrome. Biol. Reprod. 91, 127. 10.1095/biolreprod.114.120063 [DOI] [PubMed] [Google Scholar]
- 6. Essah P. A., and Nestler J. E. (2006) The metabolic syndrome in polycystic ovary syndrome. J. Endocrinol. Invest. 29, 270–280 10.1007/BF03345554 [DOI] [PubMed] [Google Scholar]
- 7. Teixeira L., Rabouille C., Rørth P., Ephrussi A., and Vanzo N. F. (2003) Drosophila Perilipin/ADRP homologue Lsd2 regulates lipid metabolism. Mech. Dev. 120, 1071–1081 10.1016/S0925-4773(03)00158-8 [DOI] [PubMed] [Google Scholar]
- 8. Grönke S., Beller M., Fellert S., Ramakrishnan H., Jäckle H., and Kühnlein R. P. (2003) Control of fat storage by a Drosophila PAT domain protein. Curr. Biol. 13, 603–606 10.1016/S0960-9822(03)00175-1 [DOI] [PubMed] [Google Scholar]
- 9. Baker K. D., and Thummel C. S. (2007) Diabetic larvae and obese flies-emerging studies of metabolism in Drosophila. Cell Metab. 6, 257–266 10.1016/j.cmet.2007.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. King R. C. (1970) Ovarian Development in Drosophila melanogaster, Academic Press, New York [Google Scholar]
- 11. Sieber M. H., and Spradling A. C. (2015) Steroid signaling establishes a female metabolic state and regulates SREBP to control oocyte lipid accumulation. Curr. Biol. 25, 993–1004 10.1016/j.cub.2015.02.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Mahajan-Miklos S., and Cooley L. (1994) Intercellular cytoplasm transport during Drosophila oogenesis. Dev. Biol. 165, 336–351 10.1006/dbio.1994.1257 [DOI] [PubMed] [Google Scholar]
- 13. Buszczak M., Lu X., Segraves W. A., Chang T. Y., and Cooley L. (2002) Mutations in the midway gene disrupt a Drosophila acyl coenzyme A: diacylglycerol acyltransferase. Genetics 160, 1511–1518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gupta T., and Schüpbach T. (2003) Cct1, a phosphatidylcholine biosynthesis enzyme, is required for Drosophila oogenesis and ovarian morphogenesis. Development 130, 6075–6087 10.1242/dev.00817 [DOI] [PubMed] [Google Scholar]
- 15. Wood B. R., Chernenko T., Matthäus C., Diem M., Chong C., Bernhard U., Jene C., Brandli A. A., McNaughton D., Tobin M. J., Trounson A., and Lacham-Kaplan O. (2008) Shedding new light on the molecular architecture of oocytes using a combination of synchrotron Fourier transform-infrared and Raman spectroscopic mapping. Anal. Chem. 80, 9065–9072 10.1021/ac8015483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Parra-Peralbo E., and Culi J. (2011) Drosophila lipophorin receptors mediate the uptake of neutral lipids in oocytes and imaginal disc cells by an endocytosis-independent mechanism. PLoS Genet. 7, e1001297 10.1371/journal.pgen.1001297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schneider L. E., and Spradling A. C. (1997) The Drosophila G-protein-coupled receptor kinase homologue Gprk2 is required for egg morphogenesis. Development 124, 2591–2602 [DOI] [PubMed] [Google Scholar]
- 18. Sopko R., Foos M., Vinayagam A., Zhai B., Binari R., Hu Y., Randklev S., Perkins L. A., Gygi S. P., and Perrimon N. (2014) Combining genetic perturbations and proteomics to examine kinase-phosphatase networks in Drosophila embryos. Dev Cell 31, 114–127 10.1016/j.devcel.2014.07.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhn H., Sopko R., Coughlin M., Perrimon N., and Mitchison T. (2015) The Atg1-Tor pathway regulates yolk catabolism in Drosophila embryos. Development 142, 3869–3878 10.1242/dev.125419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Felix M., Chayengia M., Ghosh R., Sharma A., and Prasad M. (2015) Pak3 regulates apical-basal polarity in migrating border cells during Drosophila oogenesis. Development 142, 3692–3703 10.1242/dev.125682 [DOI] [PubMed] [Google Scholar]
- 21. Tan J., Oh K., Burgess J., Hipfner D. R., and Brill J. A. (2014) PI4KIIIα is required for cortical integrity and cell polarity during Drosophila oogenesis. J. Cell Sci. 127, 954–966 10.1242/jcs.129031 [DOI] [PubMed] [Google Scholar]
- 22. Castanieto A., Johnston M. J., and Nystul T. G. (2014) EGFR signaling promotes self-renewal through the establishment of cell polarity in Drosophila follicle stem cells. Elife 3, e04437 10.7554/eLife.04437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Herr D. R., Fyrst H., Creason M. B., Phan V. H., Saba J. D., and Harris G. L. (2004) Characterization of the Drosophila sphingosine kinases and requirement for Sk2 in normal reproductive function. J. Biol. Chem. 279, 12685–12694 10.1074/jbc.M310647200 [DOI] [PubMed] [Google Scholar]
- 24. Sieber M. H., Thomsen M. B., and Spradling A. C. (2016) Electron transport chain remodeling by GSK3 during oogenesis connects nutrient state to reproduction. Cell 164, 420–432 10.1016/j.cell.2015.12.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Strochlic T. I., Stavrides K. P., Thomas S. V., Nicolas E., O'Reilly A. M., and Peterson J. R. (2014) Ack kinase regulates CTP synthase filaments during Drosophila oogenesis. EMBO Rep. 15, 1184–1191 10.15252/embr.201438688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Pek J. W., Ng B. F., and Kai T. (2012) Polo-mediated phosphorylation of Maelstrom regulates oocyte determination during oogenesis in Drosophila. Development 139, 4505–4513 10.1242/dev.082867 [DOI] [PubMed] [Google Scholar]
- 27. Lane M. E., and Kalderon D. (1995) Localization and functions of protein kinase A during Drosophila oogenesis. Mech. Dev. 49, 191–200 10.1016/0925-4773(94)00317-G [DOI] [PubMed] [Google Scholar]
- 28. Pinna L. A., and Allende J. E. (2009) Protein kinase CK2 in health and disease: protein kinase CK2: an ugly duckling in the kinome pond. Cell. Mol. Life Sci. 66, 1795–1799 10.1007/s00018-009-9148-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Meggio F., and Pinna L. A. (2003) One-thousand-and-one substrates of protein kinase CK2? FASEB J. 17, 349–368 10.1096/fj.02-0473rev [DOI] [PubMed] [Google Scholar]
- 30. Litchfield D. W. (2003) Protein kinase CK2: structure, regulation and role in cellular decisions of life and death. Biochem. J. 369, 1–15 10.1042/bj20021469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sarno S., Ghisellini P., and Pinna L. A. (2002) Unique activation mechanism of protein kinase CK2. The N-terminal segment is essential for constitutive activity of the catalytic subunit but not of the holoenzyme. J. Biol. Chem. 277, 22509–22514 10.1074/jbc.M200486200 [DOI] [PubMed] [Google Scholar]
- 32. Saxena A., Padmanabha R., and Glover C. V. (1987) Isolation and sequencing of cDNA clones encoding α and β subunits of Drosophila melanogaster casein kinase II. Mol. Cell. Biol. 7, 3409–3417 10.1128/MCB.7.10.3409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lin W. J., Jakobi R., and Traugh J. A. (1994) Reconstitution of heterologous and chimeric casein kinase II with recombinant subunits from human and Drosophila: identification of species-specific differences in the β subunit. J. Protein Chem. 13, 217–225 10.1007/BF01891979 [DOI] [PubMed] [Google Scholar]
- 34. Turowec J. P., Duncan J. S., French A. C., Gyenis L., St Denis N. A., Vilk G., and Litchfield D. W. (2010) Protein kinase CK2 is a constitutively active enzyme that promotes cell survival: strategies to identify CK2 substrates and manipulate its activity in mammalian cells. Methods Enzymol. 484, 471–493 10.1016/B978-0-12-381298-8.00023-X [DOI] [PubMed] [Google Scholar]
- 35. Hudson A. M., and Cooley L. (2014) Methods for studying oogenesis. Methods 68, 207–217 10.1016/j.ymeth.2014.01.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Niefind K., Pütter M., Guerra B., Issinger O. G., and Schomburg D. (1999) GTP plus water mimic ATP in the active site of protein kinase CK2. Nat. Struct. Biol. 6, 1100–1103 10.1038/70033 [DOI] [PubMed] [Google Scholar]
- 37. Walther T. C., and Farese R. V. Jr. (2012) Lipid droplets and cellular lipid metabolism. Annu. Rev. Biochem. 81, 687–714 10.1146/annurev-biochem-061009-102430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Welte M. A. (2015) Expanding roles for lipid droplets. Curr. Biol. 25, R470–R481 10.1016/j.cub.2015.04.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li Z., Thiel K., Thul P. J., Beller M., Kühnlein R. P., and Welte M. A. (2012) Lipid droplets control the maternal histone supply of Drosophila embryos. Curr. Biol. 22, 2104–2113 10.1016/j.cub.2012.09.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Li Z., Johnson M. R., Ke Z., Chen L., and Welte M. A. (2014) Drosophila lipid droplets buffer the H2Av supply to protect early embryonic development. Curr. Biol. 24, 1485–1491 10.1016/j.cub.2014.05.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kolkhof P., Werthebach M., van de Venn A., Poschmann G., Chen L., Welte M., Stühler K., and Beller M. (2017) A luciferase-fragment complementation assay to detect lipid droplet-associated protein-protein interactions. Mol. Cell. Proteomics 16, 329–345 10.1074/mcp.M116.061499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kory N., Farese R. V. Jr, and Walther T. C. (2016) Targeting fat: mechanisms of protein localization to lipid droplets. Trends Cell Biol. 26, 535–546 10.1016/j.tcb.2016.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Leung K. F., Baron R., Ali B. R., Magee A. I., and Seabra M. C. (2007) Rab GTPases containing a CAAX motif are processed post-geranylgeranylation by proteolysis and methylation. J. Biol. Chem. 282, 1487–1497 10.1074/jbc.M605557200 [DOI] [PubMed] [Google Scholar]
- 44. Kitamura T., Takagi S., Naganuma T., and Kihara A. (2015) Mouse aldehyde dehydrogenase ALDH3B2 is localized to lipid droplets via two C-terminal tryptophan residues and lipid modification. Biochem. J. 465, 79–87 10.1042/BJ20140624 [DOI] [PubMed] [Google Scholar]
- 45. Suzuki M., Murakami T., Cheng J., Kano H., Fukata M., and Fujimoto T. (2015) ELMOD2 is anchored to lipid droplets by palmitoylation and regulates adipocyte triglyceride lipase recruitment. Mol. Biol. Cell 26, 2333–2342 10.1091/mbc.E14-11-1504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Sztalryd C., Xu G., Dorward H., Tansey J. T., Contreras J. A., Kimmel A. R., and Londos C. (2003) Perilipin A is essential for the translocation of hormone-sensitive lipase during lipolytic activation. J. Cell Biol. 161, 1093–1103 10.1083/jcb.200210169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Granneman J. G., Moore H. P., Krishnamoorthy R., and Rathod M. (2009) Perilipin controls lipolysis by regulating the interactions of AB-hydrolase containing 5 (Abhd5) and adipose triglyceride lipase (Atgl). J. Biol. Chem. 284, 34538–34544 10.1074/jbc.M109.068478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Lee B., Zhu J., Wolins N. E., Cheng J. X., and Buhman K. K. (2009) Differential association of adipophilin and TIP47 proteins with cytoplasmic lipid droplets in mouse enterocytes during dietary fat absorption. Biochim. Biophys. Acta 1791, 1173–1180 10.1016/j.bbalip.2009.08.002 [DOI] [PubMed] [Google Scholar]
- 49. Lou D. Y., Dominguez I., Toselli P., Landesman-Bollag E., O'Brien C., and Seldin D. C. (2008) The α catalytic subunit of protein kinase CK2 is required for mouse embryonic development. Mol. Cell. Biol. 28, 131–139 10.1128/MCB.01119-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Xu X., Toselli P. A., Russell L. D., and Seldin D. C. (1999) Globozoospermia in mice lacking the casein kinase II α′ catalytic subunit. Nat. Genet. 23, 118–121 10.1038/12729 [DOI] [PubMed] [Google Scholar]
- 51. Escalier D., Silvius D., and Xu X. (2003) Spermatogenesis of mice lacking CK2α′: failure of germ cell survival and characteristic modifications of the spermatid nucleus. Mol. Reprod. Dev. 66, 190–201 10.1002/mrd.10346 [DOI] [PubMed] [Google Scholar]
- 52. Sahu-Osen A., Montero-Moran G., Schittmayer M., Fritz K., Dinh A., Chang Y. F., McMahon D., Boeszoermenyi A., Cornaciu I., Russell D., Oberer M., Carman G. M., Birner-Gruenberger R., and Brasaemle D. L. (2015) CGI-58/ABHD5 is phosphorylated on Ser239 by protein kinase A: control of subcellular localization. J. Lipid Res. 56, 109–121 10.1194/jlr.M055004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Marcinkiewicz A., Gauthier D., Garcia A., and Brasaemle D. L. (2006) The phosphorylation of serine 492 of perilipin a directs lipid droplet fragmentation and dispersion. J. Biol. Chem. 281, 11901–11909 10.1074/jbc.M600171200 [DOI] [PubMed] [Google Scholar]
- 54. Fei W., Shui G., Zhang Y., Krahmer N., Ferguson C., Kapterian T. S., Lin R. C., Dawes I. W., Brown A. J., Li P., Huang X., Parton R. G., Wenk M. R., Walther T. C., and Yang H. (2011) A role for phosphatidic acid in the formation of “supersized” lipid droplets. PLoS Genet. 7, e1002201 10.1371/journal.pgen.1002201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chang Y. F., and Carman G. M. (2006) Casein kinase II phosphorylation of the yeast phospholipid synthesis transcription factor Opi1p. J. Biol. Chem. 281, 4754–4761 10.1074/jbc.M513064200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hsieh L. S., Su W. M., Han G. S., and Carman G. M. (2016) Phosphorylation of yeast Pah1 phosphatidate phosphatase by casein kinase II regulates its function in lipid metabolism. J. Biol. Chem. 291, 9974–9990 10.1074/jbc.M116.726588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Wilhelm N., Kostelnik K., Götz C., and Montenarh M. (2012) Protein kinase CK2 is implicated in early steps of the differentiation of pre-adipocytes into adipocytes. Mol. Cell. Biochem. 365, 37–45 10.1007/s11010-012-1241-y [DOI] [PubMed] [Google Scholar]
- 58. Schwind L., Schetting S., and Montenarh M. (2017) Inhibition of protein kinase CK2 prevents adipogenic differentiation of mesenchymal stem cells like C3H/10T1/2 cells. Pharmaceuticals 10, E22 10.3390/ph10010022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Cermelli S., Guo Y., Gross S. P., and Welte M. A. (2006) The lipid-droplet proteome reveals that droplets are a protein-storage depot. Curr. Biol. 16, 1783–1795 10.1016/j.cub.2006.07.062 [DOI] [PubMed] [Google Scholar]
- 60. Nesvizhskii A. I., Keller A., Kolker E., and Aebersold R. (2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 75, 4646–4658 10.1021/ac0341261 [DOI] [PubMed] [Google Scholar]
- 61. Tennessen J. M., Barry W. E., Cox J., and Thummel C. S. (2014) Methods for studying metabolism in Drosophila. Methods 68, 105–115 10.1016/j.ymeth.2014.02.034 [DOI] [PMC free article] [PubMed] [Google Scholar]