Abstract
The stress-induced unfolded protein response (UPR) in the endoplasmic reticulum (ER) involves various signaling cross-talks and controls cell fate. B-cell receptor (BCR) signaling, which can trigger UPR, induces gammaherpesvirus lytic replication and serves as a physiological mechanism for gammaherpesvirus reactivation in vivo. However, how the UPR regulates BCR-mediated gammaherpesvirus infection is unknown. Here, we demonstrate that the ER stressors tunicamycin and thapsigargin inhibit BCR-mediated murine gammaherpesvirus 68 (MHV68) lytic replication by inducing expression of the UPR mediator Bip and blocking activation of Akt, ERK, and JNK. Both Bip and the downstream transcription factor ATF4 inhibited BCR-mediated MHV68 lytic gene expression, whereas UPR-induced C/EBP homologous protein (CHOP) was required for and promoted BCR-mediated MHV68 lytic replication by suppressing upstream Bip and ATF4 expression. Bip knockout was sufficient to rescue BCR-mediated MHV68 lytic gene expression in CHOP knockout cells, and this rescue was blocked by ectopic ATF4 expression. Furthermore, ATF4 directly inhibited promoter activity of the MHV68 lytic switch transactivator RTA. Altogether, we show that ER stress–induced CHOP inhibits Bip and ATF4 expression and that ATF4, in turn, plays a critical role in CHOP-mediated regulation of BCR-controlled MHV68 lytic replication. We conclude that ER stress–mediated UPR and BCR signaling pathways are interconnected and form a complex network to regulate the gammaherpesvirus infection cycle.
Keywords: cell signaling, endoplasmic reticulum stress (ER stress), gene regulation, lymphocyte, microbiology, viral transcription, virology
Introduction
Gammaherpesviruses are lymphotropic viruses that are associated with the development of lymphoproliferative diseases and other nonlymphoid cancers, which contain two genera, lymphocryptoviruses and rhadinoviruses (1). Human gammaherpesviruses include Epstein-Barr virus (EBV),2 which belongs to the lymphocryptoviruses, and Kaposi sarcoma–associated herpesvirus (KSHV), which is a member of the rhadinoviruses (2–5). Because of the narrow host tropism of human gammaherpesviruses, murine gammaherpesvirus 68 (MHV68), isolated from bank voles and yellow-necked field mice, provides a valuable mouse model to define gammaherpesviral pathogenesis (6, 7). MHV68 infection of laboratory mice mimics EBV infection in human. During chronic infection, MHV68 drives the infected B cells to undergo proliferation and differentiation into memory B cells as latency reservoir and plasma cells as reactivation reservoir (8–10), sharing the common feature of gammaherpesviruses that establish latency in B lymphocytes.
Although various stimuli that activate B cells could induce the lytic cycle of gammaherpesvirus from latency, anti-immunoglobulin (anti-Ig) cross-linking by binding to surface Ig mimics the effect of antigen binding to Ig molecules of antigen-specific B cells and subsequently induces the B-cell receptor (BCR) signaling pathway, which serves as another physiologically relevant activator of gammaherpesvirus lytic reactivation (11, 12). Anti-Ig–mediated BCR signaling via the activation of cellular kinases can trigger the viral reactivation of EBV, KSHV, or MHV68 latently infected B cells (13–17). The induction of the gammaherpesvirus lytic cycle is initiated by the activation of a conserved lytic switch gene, which is encoded by highly conserved immediately-early genes BRLF1 in EBV and ORF50 in KSHV and MHV68. ORF50 is also commonly referred to as replication and transcription activator (RTA), and its expression is sufficient to disrupt EBV, KSHV, and MHV68 latency in some cell lines (11, 18–22). In the case of EBV, there is an additional immediate-early transcriptional activator, Zta, encoded by the BZLF1 gene that is essential and sufficient to trigger the EBV lytic cycle (11, 22).
Antigen binding to surface Ig induces aggregation of the BCR and leads to phosphorylation of the immunoreceptor tyrosine–based activation motif (ITAM) tyrosines by the SRC family kinases Lyn, Fyn, and Blk. The tyrosine kinase SYK is subsequently recruited to phosphorylated ITAM and forms a signalosome with the SRC family kinases and other adaptors, activating downstream Akt, phosphatidylinositol 3-kinase (PI3K), c-Jun N-terminal kinases (JNK), MAPK/extracellular signal–regulated kinases (ERK), NF-κB, and other signaling pathways (23). The activation of BCR signaling pathways like PI3K and downstream ERK and p38 is important for BCR-mediated EBV activation (24).
The unfolded protein response (UPR) is an endoplasmic reticulum (ER)-to-nucleus signaling pathway initiated by the protein-folding demand overwhelming the folding capacity of the ER, which is an ER stress response pathway that controls cell fate (25–27). UPR is initiated and mediated by three ER transmembrane stress sensors, protein kinase RNA-like ER kinase (PERK), inositol-requiring protein 1α (IRE1α), and activating transcription factor 6 (ATF6) (28, 29). In the resting state, these sensors are associated with binding immunoglobulin protein (Bip). ER stress accumulates unfolded proteins, activates the three ER stress sensors by dissociating Bip, and induces PERK-, IRE1α-, and ATF6-mediated UPR response pathways, leading to UPR-related gene expression such as ATF4 and C/EBP homologous protein (CHOP) (29). ER stress differentially regulates gammaherpesvirus lytic replication, such as the ER stress inducer thapsigargin (TG), which inhibits ER Ca2+-ATPase from recovering luminal ER calcium stores (30), triggers EBV lytic replication in lymphoblastoid cell lines (31), whereas the induction of ER stress by 2-deoxy-d-glucose inhibits KSHV and MHV68 lytic gene expression (32).
It has been demonstrated that BCR signaling is a physiologic UPR trigger that induces an adaptive UPR characterized by up-regulation of Bip and CHOP (33). Surface immunoglobulin M-mediated BCR signaling induces a UPR that is dependent on BCR signaling molecule BTK and SYK in chronic lymphocytic leukemia cells, and the activation level of UPR correlates with disease progression (34). As both BCR and UPR signaling mediate gammaherpesvirus lytic replication, in line with the induction of UPR by BCR signaling, we investigated the role of UPR in BCR-mediated gammaherpesvirus lytic replication. Here, we show that ER stress caused by TG and tunicamycin (TM) inhibited BCR-mediated MHV68 viral DNA replication and lytic gene expression in MHV68-immortalized SL-1 lymphoma B cells concomitantly with the inhibition of constitutive Akt, ERK, and JNK activation after prolonged TG or TM treatment preceded by Bip and CHOP induction. Ectopic CHOP expression promoted BCR-mediated MHV68 lytic gene expression but did not activate the transcription of the MHV68 RTA promoter, whereas CHOP knockout abolished BCR-mediated MHV68 lytic replication without influencing BCR signaling, which can be fully rescued by Bip knockout. Importantly, CHOP inhibited Bip and downstream transcription factor ATF4 expression. ATF4 directly inhibited RTA promoter activity, suppressed BCR-mediated MHV68 lytic gene expression, and correspondingly contributed to the regulatory role of CHOP in BCR-mediated MHV68 lytic replication.
Results
ER stress inhibits BCR-mediated MHV68 lytic replication
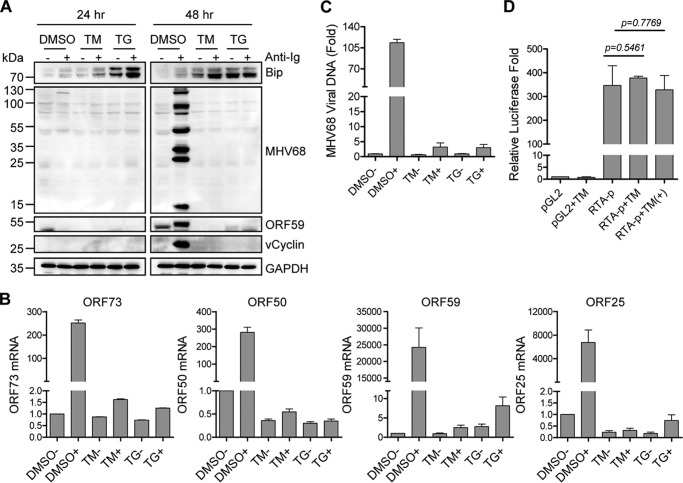
Anti-Ig cross-linking not only efficiently induces EBV lytic cycle (15, 16) but also effectively triggers MHV68 lytic replication in B cells expressing surface Ig (14, 35, 36). Furthermore, ER stress inducers TM and TG can also trigger EBV lytic activation in Burkitt's lymphoma cells and lymphoblastoid cell lines (31, 37). Based on the link between BCR-mediated signaling and UPR (33, 38), we questioned whether UPR interferes with BCR-mediated gammaherpesvirus lytic replication. To test this possibility, we used TM, which blocks N-linked protein glycosylation, and TG, which disrupts ER calcium homeostasis. MHV68-immortalized SL-1 cells were treated with 5 μg/ml TM or 5 μm TG in the presence or absence of 5 μg/ml F(ab′)2 anti-mouse IgG for 24 and 48 h. Immunoblot analyses were performed using specific antibodies against Bip, MHV68 vCyclin, ORF59, and lytic antigens as well as loading control GAPDH. As expected, Bip expression was induced at 24 and 48 h after TM or TG stimulation (Fig. 1A). Consistent with a previous report that BCR signaling induces UPR (33), anti-Ig treatment up-regulated Bip expression regardless of TG or TM treatment (Fig. 1A). However, both TM and TG stimulation completely blocked protein expression of MHV68 vCyclin, ORF59, and lytic antigens mediated by surface Ig cross-linking (Fig. 1A). Likewise, quantitative RT-PCR analyses showed that TM and TG significantly decreased mRNA levels of the MHV68 latency-associated gene ORF73, immediate-early gene ORF50, early gene ORF59, and late gene ORF25 induced by surface Ig cross-linking (Fig. 1B). Although ORF73 is a latency-associated gene, its expression can be induced upon MHV68 lytic reactivation (35).
Figure 1.
ER stress inhibits BCR-mediated MHV68 lytic replication. A, MHV68 lytic protein expression after induction of ER stress and anti-Ig stimulation. SL-1 cells were treated with TM (5 μg/ml), TG (5 μm), or DMSO together with (+) or without (−) anti-mouse IgG (5 μg/ml) for 24 and 48 h. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot was marked as indicated (kDa). B, mRNA expression of MHV68 viral genes after 48 h of TM or TG treatment. SL-1 cells were cultured in the presence (+) or absence (−) of anti-mouse IgG (5 μg/ml). Total RNA isolated from treated cells was subjected to quantitative RT-PCR analyses with specific primers corresponding to MHV68 ORF73, ORF50, ORF59, and ORF25, respectively. Viral gene mRNA was normalized to GAPDH mRNA. Relative fold was calculated by comparison with DMSO treatment without anti-Ig. Each sample was done in triplicate. C, MHV68 viral DNA replication after 48 h of TM or TG treatment. SL-1 cells were cultured in the presence (+) or absence (−) of anti-mouse IgG (5 μg/ml). Genomic DNA was isolated from treated cells and subjected to quantitative PCR analyses with primers corresponding to the ORF50 coding region. MHV68 viral DNA level was normalized to GAPDH. Relative fold was calculated by comparison with DMSO treatment without anti-Ig. Each sample was done in triplicate. D, effect of TM on RTA promoter activity. M12 cells were co-transfected with a Renilla reporter and RTA luciferase promoter (RTA-p) or vector pGL2 with or without TM (5 μg/ml) and anti-mouse IgG (5 μg/ml) treatment (+). Luciferase activity was normalized to Renilla activity. Histograms represent the mean ± S.D. of triplicate samples (two experiments). A p value of < 0.05 was considered significant.
Next, we determined the effects of TM and TG on MHV68 viral DNA replication. SL-1 cells were exposed to TM or TG together with anti-Ig stimulation for 48 h. To detect the MHV68 viral genome, we performed quantitative PCR with specific primers corresponding to the ORF50 (RTA) coding region. Both TM and TG dramatically reduced MHV68 viral DNA induced by surface Ig cross-linking, whereas DMSO had little effect (Fig. 1C), indicating that TM and TG inhibit BCR-mediated MHV68 viral DNA replication. We then tested whether TM or TG had any effect on the activation of the MHV68 lytic switch protein RTA promoter. M12 murine B cells were transfected with an RTA luciferase promoter construct in the presence or absence of TM treatment with or without anti-Ig stimulation. TM treatment had no direct, significant effect on RTA promoter activity regardless of anti-Ig treatment (Fig. 1D). Altogether, these data illustrate that UPR by TM or TG inhibits BCR-mediated MHV68 lytic replication.
Prolonged ER stress inhibits Akt, ERK, and JNK activation
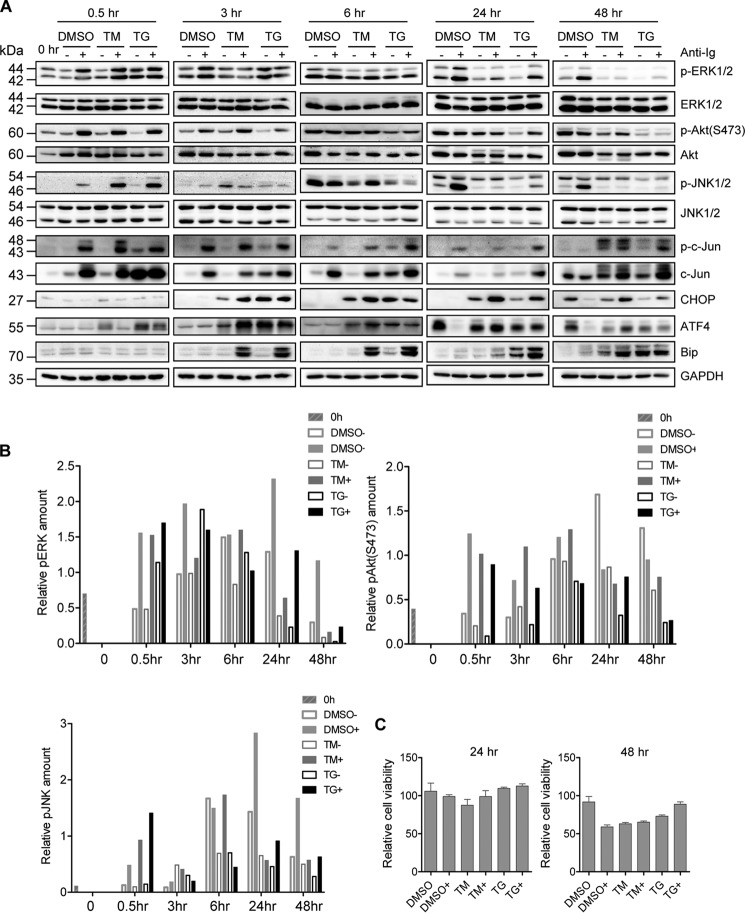
The BCR-mediated signaling pathways, including the PI3K pathway, are important for BCR-mediated EBV and KSHV lytic reactivation (16, 17, 24). As such, we wondered whether TM and TG block MHV68 lytic replication by inhibiting the BCR signaling pathways. To test this possibility, we performed a time-course experiment in SL-1 cells treated with TM or TG in the presence or absence of anti-Ig. We examined three main BCR signal transduction pathways by detecting the level of phosphorylated ERK, Akt, and JNK. Immunoblot analyses showed that neither TM or TG reduced the activation of ERK, Akt, and JNK at 0.5 h post-treatment; instead, they increased the magnitude of JNK activation upon anti-Ig treatment (Fig. 2, A and B), which is in agreement with a previous report that ER stress activates JNK (39). At 3 and 6 h post-stimulation by TM or TG, we started to observe a significant increase in the UPR-induced transcription factors ATF4 and CHOP regardless of surface Ig cross-linking, and Bip up-regulation primarily in the presence of surface Ig cross-linking. The induction of Bip, ATF4, and CHOP maintained over the course of the experiment, although the magnitude of induction varied at the later time points (24 and 48 h) (Fig. 2A). ATF4 induction was prior to CHOP induction and showed a pattern similar to CHOP, supporting the idea that ATF4 is an upstream transcription factor of CHOP upon ER stress. Concomitantly, the activation of ERK, Akt, and JNK was dramatically blocked at 24 and 48 h TM or TG post-stimulation (Fig. 2, A and B). We also detected JNK downstream effector c-Jun activation and observed that both TM and TG did not inhibit c-Jun phosphorylation as compared with control DMSO treatment (Fig. 2A). To rule out the effect of ER stress-related apoptosis on BCR activation and MHV68 lytic replication, we measured the cell viability for SL-1 cells treated with TM or TG in the presence or absence of anti-mouse IgG for 24 and 48 h. We observed a slight increase instead of the decrease in cell viability upon TM or TG treatment as compared with DMSO treatment in the presence of anti-mouse IgG (Fig. 2C), suggesting that apoptosis does not play a role in the attenuation of BCR activation and inhibition of MHV68 lytic replication by TM or TG upon anti-Ig stimulation. Altogether, these data illustrate that TM and TG have no effect on the initial activation of BCR signal transduction but block the BCR-mediated constitutive activation of ERK, Akt, and JNK at the later time point after 24 h of prolonged ER stress treatment, which might contribute to the inhibition of BCR-mediated MHV68 lytic replication at 48 h post–anti-Ig stimulation.
Figure 2.
ER stress inhibits constitutive activation of BCR-signaling Akt, ERK, and JNK pathways. SL-1 cells were treated with TM (5 μg/ml), TG (5 μm), or DMSO, together with (+) or without (−) anti-mouse IgG (5 μg/ml) for 0.5, 3, 6, 24, and 48 h. A, immunoblot analyses were performed using specific antibodies as indicated. The molecular mass of each protein is marked as indicated (kDa). The samples for 24 and 48 h were the same as those in Fig. 1A. The same Bip detection at 24 and 48 h shown in Fig. 1A was also used in this panel. B, quantitation of phosphorylated ERK, Akt, and JNK by normalizing to GAPDH for the immunoblot analyses described in A. C, cell viability of SL-1 cells stimulated with DMSO, TM (5 μg/ml), or TG (5 μm) in the presence (+) or absence of anti-mouse IgG (5 μg/ml) for 24 and 48 h. The result for each treatment was normalized to that of the untreated control. Histograms represent the mean ± S.D. of triplicate samples (two experiments).
To determine the link between inhibition of ERK, Akt, and JNK activation and the inhibition of BCR-mediated MHV68 lytic replication, SL-1 cells were treated with Akt inhibitor VIII, ERK inhibitor PD98059, and JNK-specific inhibitor SP600125, respectively, together with surface Ig cross-linking. All of these inhibitors completely blocked MHV68 lytic antigen expression induced by anti-Ig stimulation (Fig. 3A), indicating that ERK, Akt, and JNK activation of BCR signaling is required for BCR-mediated MHV68 lytic gene expression. Next, we examined the cell viability for SL-1 cells treated with inhibitors in the presence of anti-mouse IgG for 48 h and observed that only 100 μm Akt inhibitor reduced cell viability significantly as compared with DMSO (Fig. 3B). Thus, cell cytotoxicity induced by inhibitors does not play an important role in MHV68 lytic gene expression, because the lower concentration of inhibitor treatment still showed the inhibition of MHV68 lytic gene expression without eliciting cytotoxicity. Therefore, TM and TG possibly block BCR-mediated MHV68 lytic replication through inhibiting the constitutive BCR signaling pathways. Given that TM and TG-induced CHOP and Bip expression preceded the inhibition of BCR-mediated ERK, Akt, and JNK activation (Fig. 2), it is also likely that ER stress–induced CHOP, ATF4, or Bip is the main regulator for UPR-mediated inhibition of BCR-mediated MHV68 lytic replication.
Figure 3.

Activation of Akt, ERK, and JNK is required for BCR-mediated MHV68 lytic gene expression. A, SL-1 cells were treated with different concentration of Akt1/2-specific inhibitor (AktI), ERK inhibitor PD98059 (ERKI), or JNK inhibitor SP600125 (JNKI), respectively, together with (+) or without (−) anti-mouse IgG (5 μg/ml) for 48 h. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot is marked as indicated (kDa). B, cell viability of SL-1 cells treated with DMSO, Akt1/2-specific inhibitor, ERK inhibitor PD98059, or JNK inhibitor SP600125 in the presence (+) of anti-mouse IgG (5 μg/ml) for 48 h. The result for each treatment normalized to that of the untreated control. Histograms represent mean ± S.D. of triplicate samples (two experiments). A p value of < 0.05 was considered significant.
CHOP is required for BCR-mediated MHV68 lytic replication
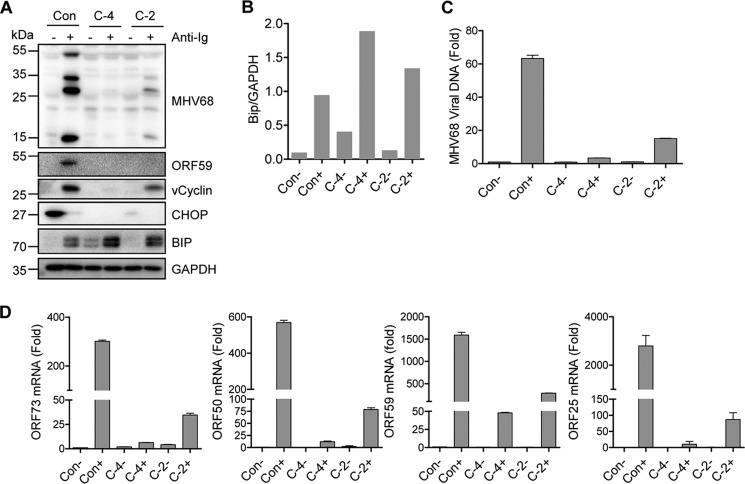
We analyzed the possibility of the ER stress–induced downstream transcription factor CHOP being involved in BCR-mediated MHV68 lytic replication. To delineate the role of CHOP, we used the CRISP/Cas9 system with puromycin selection to generate CHOP knockout SL-1 cells and performed a limiting dilution culture to select individual clones in which the CHOP gene was efficiently knocked out and confirmed by sequencing analyses. We chose the clone C-4 in which the CHOP genomic sequences between 161939 and 162032 were deleted and CHOP expression was completely knocked out and clone C-2 in which the CHOP genomic sequences between 161990 and 162062 were deleted and CHOP expression was partially knocked out for further analyses. Considering the potential effect of puromycin on cell physiology, SL-1 cells transfected with CRISP/Cas9 vector alone and selected in parallel with puromycin were used as control cells. Surprisingly, CHOP knockout completely inhibited the expression of MHV68 vCyclin, ORF59, and lytic antigens induced by surface Ig cross-linking in C-4 cells, whereas MHV68 lytic gene expression was partially blocked in C-2 cells, correlated with a partial knockout of the CHOP gene (Fig. 4A). It is worth noting that Bip expression was increased in CHOP knockout C-4 and C-2 cells regardless of anti-Ig treatment, and the increased level of Bip expression corresponded to the knockout level of CHOP (Fig. 4, A and B), indicating that CHOP inhibits Bip expression.
Figure 4.
CHOP is required for BCR-mediated MHV68 lytic replication. CHOP knockout individual SL-1 clones C-4 and C-2 as well as control cells (Con) were treated with anti-mouse IgG (5 μg/ml) for 48 h and subjected to further analyses. A, immunoblot analyses detected by indicated antibodies. The molecular mass for each blot was marked as indicated (kDa). B, quantitation of Bip by normalizing to GAPDH for the immunoblot analyses in A. C, MHV68 viral DNA detected by quantitative PCR with specific primers corresponding to ORF50. Viral DNA level was normalized to GAPDH. Relative fold was calculated by comparison with DMSO treatment without anti-Ig. Each sample was done in triplicate. D, mRNA expression of the MHV68 viral genes detected by quantitative RT-PCR with specific primers against ORF73, ORF50, ORF59, and ORF25. Viral gene mRNA was normalized to GAPDH mRNA. Relative fold was calculated by comparison with DMSO treatment without anti-Ig. Each sample was done in triplicate.
Next, we examined the effect of CHOP on MHV68 viral DNA replication and temporal gene expression during lytic reactivation. Clones C-4 and C-2 and control cells were treated with anti-Ig for 48 h followed by quantitative PCR analyses to detect MHV68 viral genome and quantitative RT-PCR analyses to detect MHV68 gene expression. As compared with control cells, MHV68 viral DNA replication was significantly blocked in C-4 cells and partially blocked in C-2 cells in response to anti-Ig treatment (Fig. 4C). Similarly, mRNA expression of MHV68 ORF73, ORF50, ORF59, and ORF25 was also dramatically inhibited in C-4 and C-2 cells (Fig. 4D). Altogether, these data demonstrate that CHOP is required for BCR-mediated MHV68 viral DNA replication and lytic gene expression.
Ectopic CHOP expression promotes BCR-mediated MHV68 lytic gene expression without activating the RTA promoter
To further define the role of CHOP in BCR-mediated MHV68 lytic replication, C-4 and control cells were transfected with a CHOP-expressing plasmid with an AU1 tag or vector. At 24 h post-transfection, the cells were treated with anti-Ig for 48 h. CHOP overexpression not only augmented the expression of MHV68 ORF59, vCyclin, and lytic antigens mediated by surface Ig cross-linking in control cells but also rescued MHV68 lytic antigen expression induced by anti-Ig cross-linking in C-4 cells (Fig. 5A). Both vCyclin and ORF59 expression were also slightly induced by CHOP overexpression in C-4 cells (Fig. 5A). To further confirm this finding, we transfected C-4 cells with the CHOP-expressing plasmid followed by anti-Ig treatment and immunofluorescent staining analyses. Notably, CHOP-expressing cells showed the expression of MHV68 lytic antigen (Fig. 5B, left panel). CHOP expression significantly increased the frequency of MHV68 lytic antigen–positive cells (Fig. 5B, right panel), indicating that CHOP expression could rescue MHV68 lytic gene expression in C-4 cells upon anti-Ig treatment.
Figure 5.
CHOP overexpression promotes BCR-mediated MHV68 lytic gene expression. A, CHOP knockout C-4 cells and control cells (Con) were transfected with vector or CHOP-expressing plasmid with AU1 tag followed by anti-mouse IgG (5 μg/ml) treatment for 48 h at 24 h post-transfection. Immunoblot analyses were performed with the indicated antibodies. The AU1 antibody was used to detect ectopic CHOP expression. Actin was used as a loading control. The molecular mass was marked as indicated (kDa). B, immunofluorescent imaging of MHV68 lytic gene expression in CHOP-expressing C-4 cells. CHOP knockout C-4 cells were transfected with CHOP-IRES-tdTomato–expressing plasmids followed by anti-mouse Ig treatment for 48 h and indirect immunofluorescent staining with specific antibodies against MHV68 lytic antigen (green), CHOP-IRES-tdtomato is shown in red. DNA is stained blue with DAPI. The percentage of MHV68 lytic antigen–positive cells in CHOP-expressing cells and CHOP-negative cells was quantified as shown in the right panel. C, CHOP regulation of RTA promoter activity. M12 cells were co-transfected with Renilla reporter and RTA luciferase promoter (RTA-p) or vector pGL2, together with the CHOP-expressing plasmid with AU1 epitope tag or vector, in the presence (+) or absence (−) of anti-mouse IgG (5 μg/ml) treatment. CHOP expression was detected by AU1 antibody. Luciferase activity was normalized to Renilla activity. Histograms represent mean ± S.D. of triplicate samples (two experiments). A p value of < 0.05 was considered significant.
The essential role of CHOP in BCR-mediated MHV68 lytic replication prompted us to investigate whether CHOP could activate the promoter of the MHV68 lytic switch gene RTA directly. To test this possibility, murine M12 B cells were transfected with the MHV68 RTA luciferase reporter plasmid together with a CHOP-expressing plasmid or vector. Instead of activating the RTA promoter, ectopic CHOP expression inhibited RTA promoter activity in the absence of anti-Ig stimulation and had no significant effect on the RTA promoter in the presence of anti-Ig treatment (Fig. 5C). Thus, we concluded that CHOP not only is required but also promotes BCR-mediated MHV68 lytic replication independently of CHOP regulation of RTA transcription.
CHOP is not essential for BCR signaling activation but is required for constitutive JNK activation
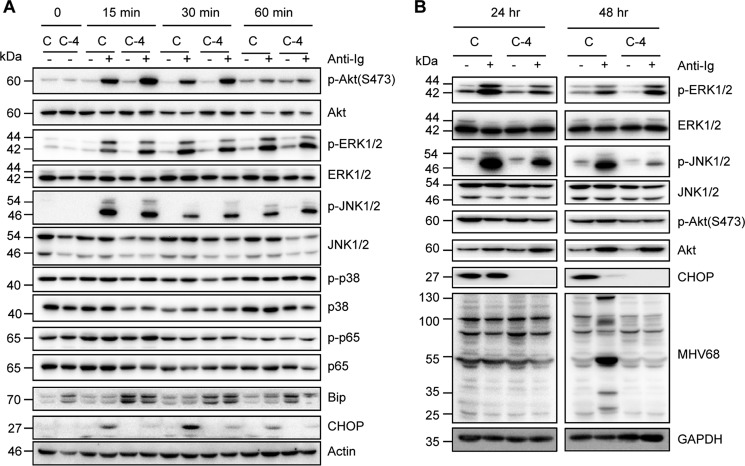
Based on the requirement of both CHOP and BCR signal transduction for BCR-mediated MHV68 lytic replication, we examined whether CHOP interfered with BCR signaling pathways. To test this possibility, a time-course experiment was performed in CHOP knockout C-4 and control cells treated with anti-Ig. We did not observe any reduced activity for Akt, ERK, JNK, p38, or p65 in C-4 cells as compared with control cells at 15, 30, and 60 min post–anti-Ig treatment, instead, Akt and JNK activity was slightly increased in C-4 cells at 15, 30, and 60 min post–anti-Ig stimulation (Fig. 6A), suggesting that CHOP is not required for the initiation of BCR signaling activation. However, JNK activity was significantly blocked in C-4 cells at 24 and 48 h post–anti-Ig treatment, whereas ERK activity was only inhibited at 24 h but not at 48 h post–anti-Ig treatment in C-4 cells (Fig. 6B), indicating that CHOP is required for constitutive JNK activation, which is required for MHV68 lytic replication. Overall, our data demonstrate that CHOP is not necessary for the initiation of BCR signal transduction but is required for constitutive JNK activation, which might be due to the indirect effect of CHOP and ultimately contribute to the role of CHOP in facilitating BCR-mediated MHV68 lytic replication.
Figure 6.
CHOP has no effect on the initiation of B cell signaling but indirectly inhibits constitutive JNK activation. CHOP knockout C-4 cells and control cells (C) were treated with anti-mouse IgG (5 μg/ml) followed by immunoblot analyses with the indicated antibodies: A, 0, 15, 30, and 60 min; B, 24 and 48 h. The molecular mass for each blot is marked as indicated (kDa).
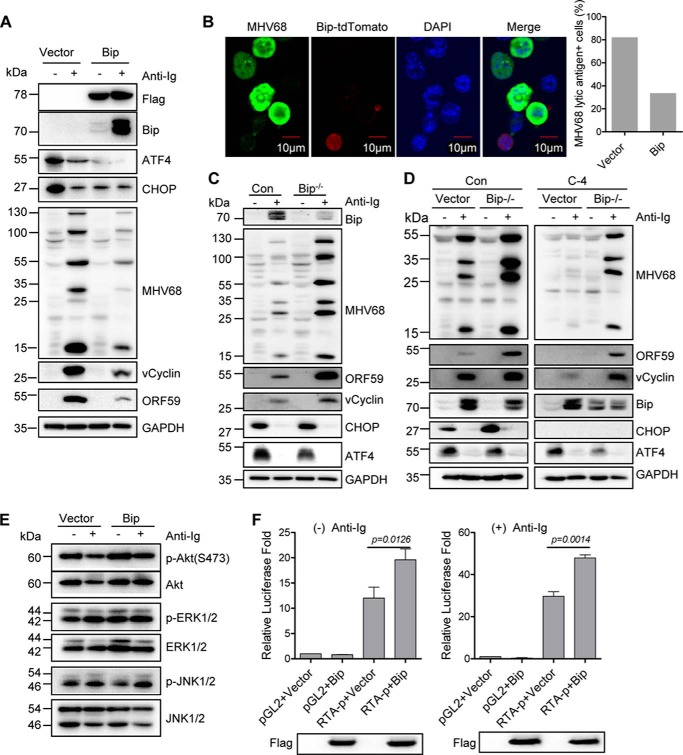
Bip inhibits BCR-mediated MHV68 lytic gene expression and mediates CHOP regulation of MHV68 lytic gene expression
As mentioned above, we consistently observed the up-regulation of Bip expression in CHOP knockout cells, irrespective of anti-Ig stimulation (Figs. 4A, 5A, and 6A), which prompted us to examine whether Bip regulates BCR-mediated MHV68 lytic replication and mediates CHOP function. To test this possibility, SL-1 cells were transfected with a Bip-expressing plasmid with a FLAG epitope tag or vector followed by anti-Ig treatment for 48 h. Ectopic Bip expression significantly inhibited BCR-mediated expression of MHV68 ORF59, vCyclin, and lytic antigens (Fig. 7A). It is worth noting that Bip overexpression led to the decrease of ATF4 and CHOP expression regardless of anti-Ig treatment, consistent with the previous report that ectopic Bip expression does not lead to ER stress and is able to attenuate ER stress signal (40). Immunofluorescent staining also showed that MHV68 lytic antigen was detected only in cells without Bip expression but not in the cells expressing Bip upon anti-Ig stimulation (Fig. 7B, left panel). Bip-expressing cells showed a significant percentage of reduction of MHV68 lytic antigen–positive cells as compared with the cells transfected with vector alone (Fig. 7B, right panel), supporting the role of Bip in inhibiting BCR-mediated MHV68 lytic gene expression. Consistent with this finding, Bip knockout increased the expression of the MHV68 lytic antigen, ORF59, and vCyclin in SL-1 cells upon anti-Ig stimulation (Fig. 7C).
Figure 7.
Bip inhibits BCR-mediated MHV68 lytic gene expression and mediates CHOP function on MHV68 lytic gene expression. A, SL-1 cells were transfected with a Bip-expressing plasmid with a FLAG tag or vector followed by anti-mouse IgG (5 μg/ml) treatment for 48 h at 24 h post-transfection. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot is marked as indicated (kDa). B, immunofluorescent imaging of MHV68 lytic antigen expression. SL-1 cells were transfected with a Bip-IRES-tdTomato–expressing plasmid and treated with anti-mouse IgG for 48 h followed by immunofluorescent staining with specific antibodies against MHV68 lytic antigen (green); CHOP-IRES-tdTomato is shown in red. DNA is stained blue with DAPI. The percentage of MHV68 lytic antigen–positive cells in Bip-expressing cells was quantified as shown in the right panel. C, Bip knockout SL-1 cells and control cells (Con) were treated with anti-mouse IgG (5 μg/ml) for 48 h and subjected to immunoblot analyses with the indicated antibodies. The molecular mass for each blot is marked as indicated. D, CHOP knockout C-4 cells and control cells (Con) were transfected with empty vector or the CRISP/Cas9 plasmid expressing Bip sgRNA, and single-cell clones were selected and treated with anti-mouse IgG (5 μg/ml) for 48 h. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot is marked as indicated. E, the same sample from A was detected with specific phosphorylated antibodies as indicated. The molecular mass for each blot is marked as indicated. F, M12 cells were co-transfected with Renilla reporter and RTA luciferase promoter (RTA-p) or vector pGL2 together with Bip-expressing plasmid with a FLAG epitope tag or vector. Bip expression was detected with FLAG antibody. Luciferase activity was normalized to Renilla activity. Histograms represent mean ± S.D. of triplicate samples (two experiments). A p value of < 0.05 was considered significant.
In line with the role of CHOP in inhibiting Bip expression and the effect of Bip on blocking BCR-mediated MHV68 lytic gene expression, it is likely that CHOP promotes BCR-mediated MHV68 lytic replication through negatively regulating Bip expression. To test this possibility, we generated Bip knockout in CHOP knockout C-4 and control cells by transfecting Bip single-guide RNA (sgRNA)–expressing CRISP/Cas9 plasmid or vector alone. The selected clone was confirmed by sequencing analyses and showed the deletion of Bip genomic sequences between 63760 and 63852 in Bip knockout C-4 cells. The selected individual clones were treated with anti-Ig stimulation for 48 h and subsequently subjected to immunoblot analyses. Strikingly, Bip knockout substantially rescued MHV68 lytic antigen, vCyclin, and ORF59 expression in CHOP knockout C-4 cells upon anti-Ig stimulation (Fig. 7D), suggesting that Bip is negatively regulated by CHOP and is responsible for the role of CHOP in promoting BCR-mediated MHV68 lytic gene expression. In Bip knockout cells, we consistently observed the reduction of ATF4 expression in the absence of anti-Ig (Fig. 7, C and D), suggesting that ATF4 is a downstream transcription factor mediated by Bip. However, Bip knockout had no significant effect on CHOP expression when Bip was knocked out in parental SL-1 cells (Fig. 7C), but it increased the level of CHOP when Bip was knocked out in the selected control cells (Fig. 7D), indicating that the second hit of nucleofection and puromycin selection might have some effect on cell physiology.
Next, we examined the effect of ectopic Bip expression on Akt, ERK, and JNK activation. Bip overexpression did not inhibit constitutive phosphorylation of Akt, ERK, and JNK (Fig. 7E), which rules out the possibility that Bip up-regulation contributes to the inhibition of Akt, ERK, and JNK activity by TM and TG at the later time points (24 and 48 h), as shown in Fig. 2. Furthermore, ectopic Bip expression activated rather than inhibited MHV68 RTA promoter transcription, irrespective of anti-Ig stimulation (Fig. 7F), suggesting that Bip inhibition of MHV68 lytic gene expression is not through direct regulation of the RTA promoter. However, Bip regulation of the CHOP function in BCR-mediated MHV68 lytic gene expression is most likely indirect.
ATF4 inhibits RTA promoter activity and plays a key role in CHOP regulation of MHV68 lytic replication
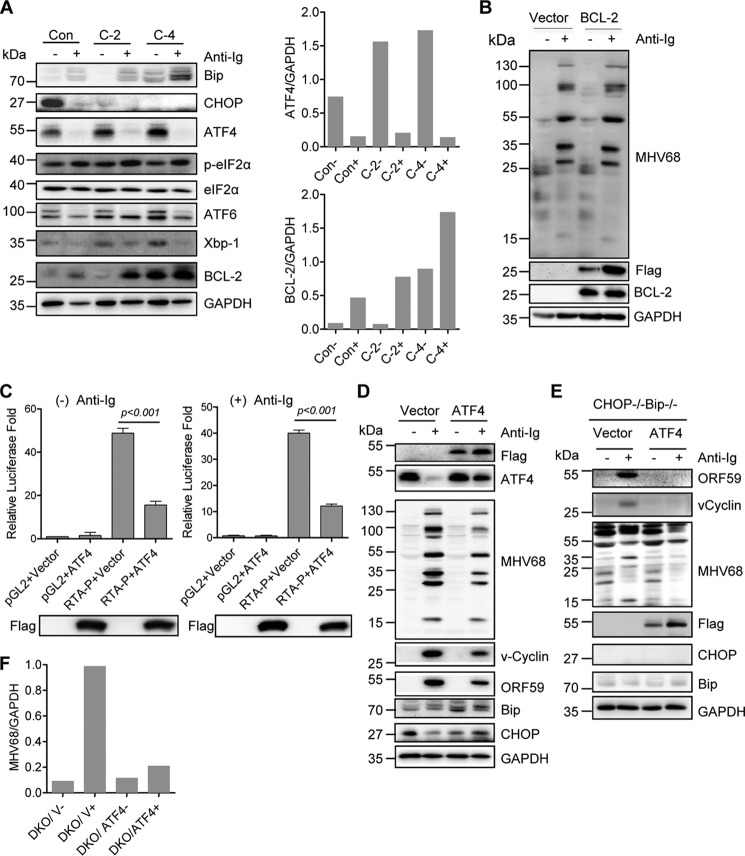
ER stress induces multiple cellular responses mediated by Bip-associated stress sensors IRE1α, ATF6, and PERK. PERK activation initiates the phosphorylation of eIF2α and induces the translation of transcription factor ATF4 and subsequent downstream CHOP expression, whereas active IRE1α processes transcription factor XBP1 to produce an active spliced form, XBP1s (28). Given that CHOP inhibited Bip expression and that Bip did not play a direct role in CHOP regulation of MHV68 lytic replication, we examined Bip downstream molecules in CHOP knockout C-2 and C-4 cells. Correspondingly, the expression of Bip downstream transcription factors ATF4, ATF6, and XBP1 was up-regulated in CHOP knockout cells (Fig. 8A), indicating that CHOP also inhibits the expression of Bip downstream transcription factors. CHOP has been shown to mediate ER stress–induced apoptosis and down-regulate BCL-2 expression (28). We also observed that BCL-2 expression was up-regulated in CHOP knockout C-2 and C-4 cells (Fig. 8A). Next, to test whether BCL-2 up-regulation had any effect on MHV68 lytic gene expression, SL-1 cells were transfected with a BCL-2–expressing plasmid with a FLAG tag or vector alone. BCL-2 overexpression had no effect on MHV68 lytic antigen expression upon anti-Ig stimulation (Fig. 8B). Considering XBP1s activation of gammaherpesviral lytic replication and ATF6 ER-Golgi translocation upon activation (28, 41, 42), we postulated that ATF4 would be more likely to play an important role. To test this possibility, M12 cells were first co-transfected with an ATF4-expressing plasmid and RTA luciferase promoter construct. Strikingly, ATF4 overexpression significantly blocked MHV68 RTA promoter activation in M12 cells regardless of anti-Ig treatment (Fig. 8C). Next, SL-1 cells were transfected with an ATF4-expressing plasmid with FLAG epitope or vector alone, together with or without anti-Ig stimulation. Consistently, ectopic ATF4 expression blocked the expression of MHV68 lytic antigen, ORF59, and vCyclin induced by anti-Ig (Fig. 8D). Furthermore, ectopic ATF4 expression alone slightly induced Bip expression but not CHOP expression in the absence of ER stress (Fig. 8D).
Figure 8.
ATF4 expression blocks MHV68 RTA transcription and suppresses MHV68 lytic gene expression rescued by Bip knockout in CHOP knockout cells. A, control cells (Con) and CHOP knockout clones C-2 and C-4 were stimulated with anti-mouse IgG (5 μg/ml) for 48 h and subjected to immunoblot analyses with the indicated antibodies. The molecular mass for each blot is marked as indicated (kDa). Quantitation of ATF4 and BCL-2 amount, respectively, by normalizing it to GAPDH is shown. B, SL-1 cells were transfected with a BCL-2-expressing plasmid with a FLAG tag or vector followed by anti-mouse IgG (5 μg/ml) treatment for 48 h at 24 h post-transfection. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot is marked as indicated. C, ATF4 inhibits RTA promoter activity. M12 cells were co-transfected with Renilla reporter and RTA luciferase promoter (RTA-p) or vector pGL2, together with an ATF4-expressing plasmid with FLAG tag or vector, in the presence (+) or absence (−) of anti-mouse IgG (5 μg/ml) treatment. ATF4 expression was detected with FLAG antibody. Luciferase activity was normalized to Renilla activity. Histograms represent mean ± S.D. of triplicate samples (two experiments). A p value of < 0.05 was considered significant. D, SL-1 cells were transfected with an ATF4-expressing plasmid with a FLAG tag or vector followed by anti-mouse IgG (5 μg/ml) treatment for 48 h at 24 h post-transfection. Immunoblot analyses were performed with specific antibodies as indicated. The molecular mass for each blot is marked as indicated. E, CHOP and Bip double knockout cells were transfected with vector or an ATF4-expressing plasmid followed by stimulation with anti-mouse IgG (5 μg/ml) for 48 h and subsequent immunoblot analyses with the indicated antibodies. The molecular mass for each blot is marked as indicated. F, quantitation of MHV68 lytic antigen by normalizing it to GAPDH for the immunoblot analyses in E. DKO, CHOP−/−Bip−/− cells; V, vector; + or −, presence or absence of anti-Ig.
As demonstrated above, Bip knockout rescues MHV68 lytic gene expression in CHOP knockout C-4 cells (Fig. 7D). However, Bip has no direct effect on MHV68 lytic gene expression. Thus, we wondered whether Bip knockout-derived rescue would result from a downstream transcription factor, ATF4, which could directly inhibit MHV68 RTA activation and lytic gene expression. To this end, an ATF4-expressing plasmid with a FLAG epitope tag was expressed in CHOP and Bip double knockout cells in the presence or absence of anti-Ig stimulation. Dramatically, ectopic ATF4 expression blocked MHV68 lytic gene expression rescued by Bip knockout in CHOP knockout cells upon anti-Ig–induced viral reactivation (Fig. 8, E and F), suggesting that the CHOP knockout phenotype can be directly restored by ATF4 expression when Bip is knocked out. Altogether, these results illustrate that ATF4 inhibits MHV68 lytic gene expression through directly blocking RTA promoter activity; ATF4 expression is negatively regulated by CHOP and, in turn, plays a key role in the CHOP requirement for MHV68 lytic gene expression.
Discussion
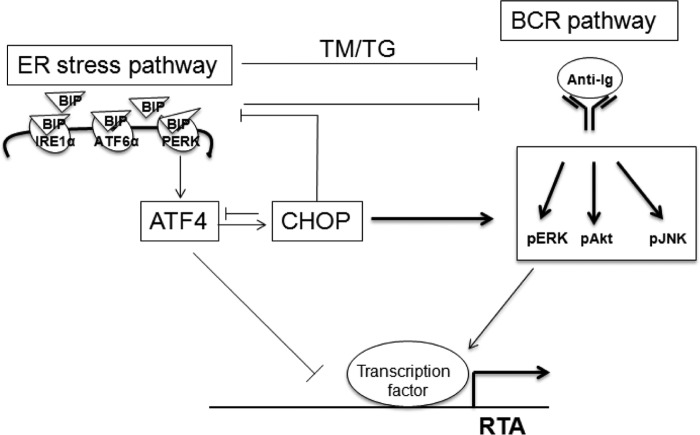
ER stress buffered by the activation of UPR is involved in the pathogenesis of many human diseases. The UPR signaling network regulates various processes such as metabolism, immunity, and cell differentiation (28, 29). BCR signaling and plasma cell differentiation activate UPR (33, 43, 44). UPR, BCR signaling, and plasma cell differentiation are interconnected and form a complex cross-talk network. BCR signaling mediated by surface Ig cross-linking is a common way to induce gammaherpesvirus lytic cycle in vitro and is viewed as the main physiological stimuli that govern EBV and MHV68 lytic reactivation from latency in vivo as latently infected memory B cells differentiate toward plasma cells (11, 12, 45); however, UPR also mediates EBV lytic replication in vitro (31, 37). Here, we have demonstrated that ER stress induced by TM and TG inhibits BCR-mediated MHV68 viral DNA replication and lytic gene expression. The UPR mediator Bip and the downstream transcription factor ATF4 inhibit BCR-mediated MHV68 lytic gene expression, whereas UPR-induced CHOP is essential and promotes BCR-mediated MHV68 lytic replication by negatively regulating Bip and ATF4 expression. ATF4 directly inhibit MHV68 RTA promoter activity. These results illustrate the complex cross-talk among the UPR, BCR, and gammaherpesvirus infection cycle (Fig. 9).
Figure 9.
Regulation model of BCR-mediated MHV68 lytic replication by ER stress. ER stress inhibits BCR-mediated MHV68 lytic replication. ER stress mediator Bip and downstream transcription factor ATF4 inhibit BCR-mediated MHV68 lytic gene expression, whereas ER stress–induced gene CHOP is essential and promotes BCR-mediated MHV68 lytic gene expression by negative regulation of Bip and ATF4 expression. ATF4 directly blocks MHV68 RTA promoter activity, which ultimately leads to the inhibition of MHV68 lytic gene expression and contributes to the essential function of CHOP on BCR-mediated MHV68 lytic replication.
Consistent with the previous report that BCR signaling is a physiologic UPR trigger (33, 34), we also observed up-regulation of CHOP and Bip expression upon anti-Ig treatment (Figs. 1A, 2A, and 4A). Furthermore, BCR signaling activation is very rapid and precedes the induction of CHOP and Bip by TM or TG (Fig. 2A). We consistently observed higher CHOP expression in SL-1 cells without anti-Ig stimulation as compared with the cells with anti-Ig stimulation at 48 h post-culture but not at 24 h post-culture (Figs. 2 and 6B). This might be due to certain signals derived from prolonged cell proliferation in the absence of anti-Ig, leading to CHOP induction, whereas in the presence of anti-Ig, SL-1 cells undergo a reactivation process and proliferation arrest. Furthermore, we consistently observed that the selected knockout clone following puromycin selection was more vulnerable than parental cells upon nucleofection and more susceptible to cell death. We chose the D17 nucleofection program, which showed a higher survival rate and lower transfection efficiency, to perform a CHOP overexpression rescue experiment in C-4 cells instead of using the T20 program, which showed a higher transfection efficiency and lower survival rate and had been used in parental SL-1 cells.
UPR mediated by the ER stressors TM and TG triggers EBV lytic replication in some virus-positive lymphoma cell lines (31, 37). However, we showed that TM and TG stimulation alone failed to induce MHV68 lytic gene expression in SL-1 lymphoma cells as they do in EBV-positive lymphoma cells. The possible cause might be the MHV68 tight latency, which can only be reactivated by surface Ig cross-linking for 48 h in SL-1 cells (36), whereas the EBV latency–lytic cycle switch occurs more rapidly and efficiently by surface Ig cross-linking and other stimuli (15, 17). Additionally, TM and TG block BCR-mediated JNK, Akt, and ERK constitutive activation, which is an indirect effect, as suggested by our results. Other signaling pathways involved will require further investigation.
Bip inhibits BCR-mediated MHV68 lytic gene expression, which is different from the observation that Bip positively regulates tetradecanoylphorbol acetate–induced KSHV lytic cycle progression in primary effusion lymphoma cells (46). The discrepancy might be attributed to the distinct signaling pathways regulated by tetradecanoylphorbol acetate and BCR. As all known KSHV-positive lymphoma B cells do not naturally express surface Ig, we reasoned that UPR regulation of BCR-mediated lytic replication would possibly be conserved among gammaherpesviruses.
The UPR-induced gene CHOP mainly mediates apoptosis and cytokine production (47, 48). Here, we have demonstrated for the first time that CHOP is required for and promotes BCR-mediated MHV68 lytic replication by inhibiting Bip and downstream ATF4 expression. ER stress–induced UPR initiates IRE1α, PERK, and ATF6, three main signaling pathways, by dissociation with Bip, whereas CHOP is induced mainly by PERK-mediated signaling (28). Although we did not analyze the effect of IRE1α- and ATF6-mediated UPR signaling pathways on BCR-mediated MHV68 lytic replication, we do not exclude the possibility of either of those signaling pathways playing a role in BCR-mediated gammaherpesvirus lytic replication, even though our data appear not to support this hypothesis, because the activation of IRE1α downstream molecules JNK and XBP1s positively regulates MHV68 lytic replication, as shown in Fig. 3 and as demonstrated previously (49). Additionally, ATF6, representing a group of ER stress transducers, translocates to the Golgi under ER stress conditions and releases its cytosolic domain fragment, ATF6f, to control other gene expressions such as XBP1 (28). It is less likely that ATF6 would inhibit RTA transcription directly. The PERK-mediated ER stress–signaling branch induces translation of transcription factor ATF4, which subsequently regulates CHOP expression. Previous reports have shown that ATF4 plays a critical role in promoting murine cytomegalovirus DNA replication and late gene expression in a multiplicity of infection-dependent ways (50), whereas EBV LMP1 activates PERK and up-regulates ATF4, which in turn transactivates the LMP1 promoter (51). Therefore, it is reasonable to propose that ATF4 might be involved in the regulation of viral gene expression. Our data show that ATF4 inhibits MHV68 RTA transcription and lytic gene expression upon viral reactivation, in contrast to the observation that ATF4 promotes late gene expression during murine cytomegalovirus infection. This discrepancy might result from the distinct scenario between viral reactivation and acute infection. Further experiments are needed to investigate the role of ATF4 in gammaherpesvirus de novo infection to reveal whether ATF4 exhibits different functions between de novo infection and viral reactivation or between β-herpesviruses like cytomegalovirus and gammaherpesviruses in the future.
In summary, our study reveals a complex and interconnected regulatory network between UPR and BCR-mediated gammaherpesvirus lytic reactivation. This network cross-talk might shed light on a complex physiological scenario for the regulation of the gammaherpesvirus latency and lytic cycle in vivo. It would be interesting to characterize this network cross-talk in other gammaherpesviruses and MHV68 infection mouse models; this could be critical to extending our understanding of gammaherpesvirus pathogenesis.
Experimental procedures
Cell lines
MHV68-transformed SL-1 cells were cultured in RPMI 1640 supplemented with 5% heat-inactivated fetal bovine serum (FBS) and 1% penicillin-streptomycin as described previously (36). Virus-free murine B-cell line M12 was cultured in RPMI 1640 containing 10% FBS and 1% penicillin-streptomycin.
Plasmids
The MHV68 Rta-Luc plasmid containing the 410-bp proximal promoter region was a gift from Samuel Speck (52). The Bip-expressing plasmid with a C-terminal FLAG epitope tag was generated by insertion of the PCR-amplified Bip coding fragments into the pLVX-tdtomato or pLVX-puro vector (Clontech) with specific primers: forward primer with NotI restriction enzyme site, 5′-ATGCGGCCGCATGATGAAGTTCACTGTGGTGGCGG-3′; and reverse primer with BamHI restriction enzyme site and FLAG epitope tag, 5′-GCGGATCCCTACTTGTCATCGTCGTCCTTGTAGTCCAACTCATCTTTTTCTA-3′. The CHOP-expressing plasmid was similarly generated with specific primers: forward primer with NotI site, 5′-ATGCGGCCGCATGGCAGCTGAGTCCCTGCCTTTCA-3′; and reverse primer with BamHI site and AU1 epitope tag, 5′-GCGGATCCTCATATATAGCGATAGGTATCTGCTTGGTGCAGGCTGACCATG-3′. BCL-2- and ATF4-expressing plasmids with C-terminal FLAG tag were generated similarly with specific primers: ATF4, 5′-ATGCGGCCGCATGACCGAGATGAGCTTCCT-3′ and 5′-GCGGATCCTTACTTGTCATCGTCGTCCTTGTAGTCCGGAACTCTCTTCTTC-3′; BCL-2, 5′-GTGAATTCATGGCGCAAGCCGGGAGAACA-3′ and 5′-AGACTAGTTCACTTGTCATCGTCGTCCTTGTAGTCCTTGTGGCCCAGGTATGCACCCAGA-3′.
CHOP and Bip knockout plasmids were generated with lentiviral CRISPR/Cas9 system. Two pairs of sgRNA sequences targeting CHOP and Bip were designed according to the website http://crispr.mit.edu/3 and cloned into a lentiCRISPRv2 vector (Addgene) following the protocols provided by Addgene. The two sgRNA sequences targeting the CHOP gene were: 5′-CACCGGGCACCTATATCTCATCCCC-3′, and 5′-AAACGGGGATGAGATATAGGTGCCC-3′. The two sgRNA sequences targeting the Bip gene were: 5′-CACCGATGATGAAGTTCACTGTGGT-3′, and 5′-AAACACCACAGTGAACTTCATCATC-3′.
Generation of knockout cell lines
We used the CRISPR-Cas9 genome-editing system to generate CHOP knockout SL-1 cells. 2 × 106 SL-1 cells were transfected with 4 μg of lentiCRISPRv2-vector, lentiCRISPRv2-CHOP-1, or lentiCRISPRv2-CHOP-2, respectively, using the Amaxa Nucleofector II system (Lonza) according to the manufacturer's instruction. At 48 h post-nucleofection, cells were selected with puromycin at a concentration of 2 μg/ml for 2 weeks. Cells were subsequently cloned by limiting dilution culture. Individual clones were subjected to immunoblot and sequencing analyses to confirm the depletion of the target protein CHOP. LentiCRISPRv2 vector-nucleofected cells were selected in parallel and used as a control cell line. Bip knockout cells were similarly generated with Bip sgRNA-expressing plasmids. The primers used to sequence genomic CHOP and Bip DNA were: CHOP forward, 5′-TGCCCTTACCTATCGTGCAA-3′, and CHOP reverse, and 5′-CAGTGCAGGGTCACATGCTT-3′; Bip forward, 5′-TCGATACTGGCCGAGACAAC-3′, and Bip reverse, and 5′-AGTGGCAACCCCTAAATACTCC-3′.
Western blot, antibodies, and reagents
Western blot was performed following standard procedures. The antibodies used were as follows. GAPDH (sc-32233), Akt1 (sc-5298) and actin (sc-1616) were from Santa Cruz Biotechnology. phospho-Akt (Ser-473) (193H12), phospho-Akt (Thr-308) (244F9), PhosphoPlus p44/42 mitogen-activated protein kinase (MAPK) (Erk1/2) antibody kit (9100), PhosphoPlus p38 MAP kinase antibody kit (9210), PhosphoPlus SAPK/JNK antibody kit (9250), c-Jun (9165), and p-c-Jun (3270) were from Cell Signaling Technology. Goat F(ab′)2 anti-mouse IgG was obtained from Jackson ImmunoResearch. Polyclonal MHV68 antiserum to detect lytic antigen was obtained from C57BL/6 mice 6 weeks post-infection (53). vCyclin and ORF59 antibodies were described previously (35). Anti-FLAG antibody (A8592), secondary antibodies, anti-rabbit IgG (R2004) and anti-mouse IgG (M8642), were from Sigma-Aldrich. The AU1 epitope tag antibody was from Novus Biologicals. All Alexa Fluor-conjugated secondary antibodies were from Invitrogen. Tunicamycin and thapsigargin were from Cell Signaling Technology. Akt1/2 kinase inhibitor (A6730) was from Sigma. ERK inhibitor PD98059 (HY-12028) was from MedChem Express, and JNK inhibitor SP600125 was from InvivoGen.
Nucleofection and luciferase assay
All SL-1 and M12 cells were transfected by nucleofection. Cells were washed with PBS, resuspended in 100 μl of transfection buffer using an Ingenio® kit (MIR 50118, Mirus), and nucleofected with plasmids using the Amaxa Nucleofector II system (Lonza) according to the manufacturer's instructions. The program T20 was used for SL-1, M12, and other selected cells, whereas the program D17 was used for CHOP knockout C-4 cells in the CHOP overexpression rescue experiments. The MHV68 RTA promoter luciferase assay was performed as described previously (54).
Luciferase assays were carried out in triplicate. Luciferase activity was measured using the Dual-Luciferase reporter gene assay kit (Beyotime Biotechnology) according to the manufacturer's instructions and normalized to Renilla activity.
Quantitative RT-PCR and quantitative PCR
Total RNA was extracted using TRIzol reagent (Life Technologies) and reverse-transcribed with the FastQuant RT kit (TIANGEN) according to the manufacturer's instruction. Quantitative PCR was performed with a SuperReal PreMix Plus (SYBR Green) kit (TIANGEN) on the 7900HT sequence detection system (Life Technologies) using the following primers: ORF50 forward, 5′-GGCCGCAGACATTTAATGAC-3′, and ORF50 reverse, 5′-GCCTCAACTTCTCTGGATATGCC-3′; MHV68-ORF73 forward, 5′-GGGAACTCAAACGCCAAACC-3′, and MHV68-ORF73 reverse, 5′-GGTGTCTTCGCATTCCCTGA-3′; MHV68-ORF59 forward, 5′-GACACGGGGTGGGAATAAGG-3′, and MHV68-ORF59 reverse, 5′-GGGGCCCCATCTACCTCTAA-3′; MHV68-ORF25 forward, 5′-CAGCGGCGTCTTTGAAACAA-3′, and MHV68-ORF25 reverse, 5′-GTAGCCGCAGGTATTGTGGT-3′; and GAPDH forward, 5′-AACGACCCCTTCATTGACCT-3′ and GAPDH reverse, 5′-ATGTTAGTGGGGTCTCG CTC-3′. The GAPDH housekeeping gene was used as an internal control for normalization. For viral DNA quantification, genomic DNA was isolated with a Tianamp genomic DNA kit (DP304-02, TianGen) and used for quantitative PCR. Real-time DNA PCR for the g50 gene was used to measure the MHV68 genome. Relative genome copy numbers were calculated based on normalization with the GAPDH housekeeping gene.
Immunofluorescence
Transfected cells were fixed with 4% paraformaldehyde for 30 min and washed three times with PBS. After penetration with methanol:acetone (1:1) for 20 min, cells were washed with PBS three times and subjected to blocking with 5% BSA followed by incubation with primary antibodies and detection with secondary antibodies (Alexa-488 goat anti-mouse IgG for MHV68 lytic antigen, Alexa-568 donkey anti-rabbit IgG for AU1, and Alexa-568 donkey anti-mouse IgG for FLAG). Images were acquired using a confocal microscope (Olympus FV1200). Quantification was calculated by counting three different images.
Cytotoxicity and cell viability assays
Cytotoxic activity and cell viability were determined using a colorimetric Cell Counting Kit-8 (Beyotime, C0037) according to the manufacturer's instructions. Briefly, The SL-1 cells were seeded in 96-well plates at a density of 5000 cells/well and stimulated with individual stimuli. Cell viability was determined by measuring the absorbance at A450 using a microplate reader. Individual treatment was assayed in triplicate, and at least two independent experiments were performed for each stimulus.
Statistical analyses
Statistical analysis was performed using Prism (GraphPad Software). The data are reported as means ± S.D. Differences between groups of research subjects were analyzed for statistical significance with two-tailed Student's t tests. A p value of 0.05 was considered significant.
Author contributions
X. Z. performed the experiments, analyzed the data, and drafted the article. S. D., Z. L., S. L., and C. Z. performed the experiments and analyzed the data. X. L. conceived and designed the experiments and drafted and edited the manuscript.
Acknowledgment
We thank Dr. Samuel Speck (Emory University) for providing us with MHV68 Rta-Luc plasmids, vCyclin, and ORF59 antibodies.
This work was supported by Grant 2016YFA0502100 from the National Key R&D Program of China, Grant 81371825 from the Natural Science Foundation of China, and Grant 2060299 from the Chinese Academy of Sciences “100 Talents” Program. The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party-hosted site.
- EBV
- Epstein-Barr virus
- KSHV
- Kaposi sarcoma–associated herpesvirus
- MHV68
- murine gammaherpesvirus 68
- BCR
- B-cell receptor
- UPR
- unfolded protein response
- ER
- endoplasmic reticulum
- PERK
- protein kinase RNA-like ER kinase
- IRE1α
- inositol-requiring protein 1α
- ATF6
- activating transcription factor 6
- Bip
- binding immunoglobulin protein
- CHOP
- C/EBP homologous protein
- TG
- thapsigargin
- TM
- tunicamycin
- RTA
- replication and transcription activator
- sgRNA
- single-guide RNA.
References
- 1. Barton E., Mandal P., and Speck S. H. (2011) Pathogenesis and host control of gammaherpesviruses: Lessons from the mouse. Annu. Rev. Immunol. 29, 351–397 10.1146/annurev-immunol-072710-081639 [DOI] [PubMed] [Google Scholar]
- 2. Cesarman E. (2014) Gammaherpesviruses and lymphoproliferative disorders. Annu. Rev. Pathol. 9, 349–372 10.1146/annurev-pathol-012513-104656 [DOI] [PubMed] [Google Scholar]
- 3. Thorley-Lawson D. A., Schooley R. T., Bhan A. K., and Nadler L. M. (1982) Epstein-Barr virus superinduces a new human B cell differentiation antigen (B-LAST 1) expressed on transformed lymphoblasts. Cell 30, 415–425 10.1016/0092-8674(82)90239-2 [DOI] [PubMed] [Google Scholar]
- 4. Schulz T. F., and Chang Y. (2007) KSHV gene expression and regulation, in Human Herpesviruses: Biology, Therapy, and Immunoprophylaxis, Chapter 28, (Arvin A., Campadelli-Fiume G., Mocarski E., Moore P. S., Roizman B., Whitley R., and Yamanishi K., eds) Cambridge, Cambridge University Press; [PubMed] [Google Scholar]
- 5. Arvanitakis L., Geras-Raaka E., Varma A., Gershengorn M. C., and Cesarman E. (1997) Human herpesvirus KSHV encodes a constitutively active G-protein-coupled receptor linked to cell proliferation. Nature 385, 347–350 10.1038/385347a0 [DOI] [PubMed] [Google Scholar]
- 6. Bussey K. A., Reimer E., Todt H., Denker B., Gallo A., Konrad A., Ottinger M., Adler H., Stürzl M., Brune W., and Brinkmann M. M. (2014) The gammaherpesviruses Kaposi's sarcoma-associated herpesvirus and murine gammaherpesvirus 68 modulate the Toll-like receptor-induced proinflammatory cytokine response. J. Virol. 88, 9245–9259 10.1128/JVI.00841-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chastel C., Beaucournu J. P., Chastel O., Legrand M. C., and Le Goff F. (1994) A herpesvirus from an European shrew (Crocidura russula). Acta Virol. 38, 309 [PubMed] [Google Scholar]
- 8. Liang X., Collins C. M., Mendel J. B., Iwakoshi N. N., and Speck S. H. (2009) Gammaherpesvirus-driven plasma cell differentiation regulates virus reactivation from latently infected B lymphocytes. PLoS Pathog. 5, e1000677 10.1371/journal.ppat.1000677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Willer D. O., and Speck S. H. (2003) Long-term latent murine Gammaherpesvirus 68 infection is preferentially found within the surface immunoglobulin D-negative subset of splenic B cells in vivo. J. Virol. 77, 8310–8321 10.1128/JVI.77.15.8310-8321.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Weck K. E., Kim S. S., Virgin H. W. IV, and Speck S. H. (1999) B cells regulate murine gammaherpesvirus 68 latency. J. Virol. 73, 4651–4661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Speck S. H., and Ganem D. (2010) Viral latency and its regulation: lessons from the gamma-herpesviruses. Cell Host Microbe 8, 100–115 10.1016/j.chom.2010.06.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kenney S. C., and Mertz J. E. (2014) Regulation of the latent-lytic switch in Epstein-Barr virus. Semin. Cancer Biol. 26, 60–68 10.1016/j.semcancer.2014.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tovey M. G., Lenoir G., and Begon-Lours J. (1978) Activation of latent Epstein-Barr virus by antibody to human IgM. Nature 276, 270–272 10.1038/276270a0 [DOI] [PubMed] [Google Scholar]
- 14. Moser J. M., Upton J. W., Gray K. S., and Speck S. H. (2005) Ex vivo stimulation of B cells latently infected with gammaherpesvirus 68 triggers reactivation from latency. J. Virol. 79, 5227–5231 10.1128/JVI.79.8.5227-5231.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takada K. (1984) Cross-linking of cell surface immunoglobulins induces Epstein-Barr virus in Burkitt lymphoma lines. Int. J. Cancer 33, 27–32 10.1002/ijc.2910330106 [DOI] [PubMed] [Google Scholar]
- 16. Daibata M., Humphreys R. E., Takada K., and Sairenji T. (1990) Activation of latent EBV via anti-IgG-triggered, second messenger pathways in the Burkitt's lymphoma cell line Akata. J. Immunol. 144, 4788–4793 [PubMed] [Google Scholar]
- 17. Kati S., Tsao E. H., Günther T., Weidner-Glunde M., Rothämel T., Grundhoff A., Kellam P., and Schulz T. F. (2013) Activation of the B cell antigen receptor triggers reactivation of latent Kaposi's sarcoma-associated herpesvirus in B cells. J. Virol. 87, 8004–8016 10.1128/JVI.00506-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lukac D. M., Renne R., Kirshner J. R., and Ganem D. (1998) Reactivation of Kaposi's sarcoma-associated herpesvirus infection from latency by expression of the ORF 50 transactivator, a homolog of the EBV R protein. Virology 252, 304–312 10.1006/viro.1998.9486 [DOI] [PubMed] [Google Scholar]
- 19. Lukac D. M., Kirshner J. R., and Ganem D. (1999) Transcriptional activation by the product of open reading frame 50 of Kaposi's sarcoma-associated herpesvirus is required for lytic viral reactivation in B cells. J. Virol. 73, 9348–9361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ragoczy T., Heston L., and Miller G. (1998) The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J. Virol. 72, 7978–7984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zalani S., Holley-Guthrie E., and Kenney S. (1996) Epstein-Barr viral latency is disrupted by the immediate-early BRLF1 protein through a cell-specific mechanism. Proc. Natl. Acad. Sci. U.S.A. 93, 9194–9199 10.1073/pnas.93.17.9194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feederle R., Kost M., Baumann M., Janz A., Drouet E., Hammerschmidt W., and Delecluse H. J. (2000) The Epstein-Barr virus lytic program is controlled by the co-operative functions of two transactivators. EMBO J. 19, 3080–3089 10.1093/emboj/19.12.3080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Young R. M., and Staudt L. M. (2013) Targeting pathological B cell receptor signalling in lymphoid malignancies. Nat. Rev. Drug Discov. 12, 229–243 10.1038/nrd3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Iwakiri D., and Takada K. (2004) Phosphatidylinositol 3-kinase is a determinant of responsiveness to B cell antigen receptor-mediated Epstein-Barr virus activation. J. Immunol. 172, 1561–1566 10.4049/jimmunol.172.3.1561 [DOI] [PubMed] [Google Scholar]
- 25. Ma Y., and Hendershot L. M. (2001) The unfolding tale of the unfolded protein response. Cell 107, 827–830 10.1016/S0092-8674(01)00623-7 [DOI] [PubMed] [Google Scholar]
- 26. Patil C., and Walter P. (2001) Intracellular signaling from the endoplasmic reticulum to the nucleus: The unfolded protein response in yeast and mammals. Curr. Opin. Cell Biol. 13, 349–355 10.1016/S0955-0674(00)00219-2 [DOI] [PubMed] [Google Scholar]
- 27. Rutkowski D. T., and Kaufman R. J. (2004) A trip to the ER: Coping with stress. Trends Cell Biol. 14, 20–28 10.1016/j.tcb.2003.11.001 [DOI] [PubMed] [Google Scholar]
- 28. Hetz C. (2012) The unfolded protein response: Controlling cell fate decisions under ER stress and beyond. Nat. Rev. Mol. Cell Biol. 13, 89–102 [DOI] [PubMed] [Google Scholar]
- 29. Gorman A. M., Healy S. J., Jäger R., and Samali A. (2012) Stress management at the ER: Regulators of ER stress-induced apoptosis. Pharmacol. Ther. 134, 306–316 10.1016/j.pharmthera.2012.02.003 [DOI] [PubMed] [Google Scholar]
- 30. Thastrup O., Cullen P. J., Drøbak B. K., Hanley M. R., and Dawson A. P. (1990) Thapsigargin, a tumor promoter, discharges intracellular Ca2+ stores by specific inhibition of the endoplasmic reticulum Ca2(+)-ATPase. Proc. Natl. Acad. Sci. U.S.A. 87, 2466–2470 10.1073/pnas.87.7.2466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Taylor G. M., Raghuwanshi S. K., Rowe D. T., Wadowsky R. M., and Rosendorff A. (2011) Endoplasmic reticulum stress causes EBV lytic replication. Blood 118, 5528–5539 10.1182/blood-2011-04-347112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Leung H. J., Duran E. M., Kurtoglu M., Andreansky S., Lampidis T. J., and Mesri E. A. (2012) Activation of the unfolded protein response by 2-deoxy-d-glucose inhibits Kaposi's sarcoma-associated herpesvirus replication and gene expression. Antimicrob. Agents Chemother. 56, 5794–5803 10.1128/AAC.01126-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Skalet A. H., Isler J. A., King L. B., Harding H. P., Ron D., and Monroe J. G. (2005) Rapid B cell receptor-induced unfolded protein response in nonsecretory B cells correlates with pro- versus antiapoptotic cell fate. J. Biol. Chem. 280, 39762–39771 10.1074/jbc.M502640200 [DOI] [PubMed] [Google Scholar]
- 34. Krysov S., Steele A. J., Coelho V., Linley A., Sanchez Hidalgo M., Carter M., Potter K. N., Kennedy B., Duncombe A. S., Ashton-Key M., Forconi F., Stevenson F. K., and Packham G. (2014) Stimulation of surface IgM of chronic lymphocytic leukemia cells induces an unfolded protein response dependent on BTK and SYK. Blood 124, 3101–3109 10.1182/blood-2014-04-567198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Forrest J. C., and Speck S. H. (2008) Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade. J. Virol. 82, 7688–7699 10.1128/JVI.02689-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Liang X., Paden C. R., Morales F. M., Powers R. P., Jacob J., and Speck S. H. (2011) Murine gamma-herpesvirus immortalization of fetal liver-derived B cells requires both the viral cyclin D homolog and latency-associated nuclear antigen. PLoS Pathog. 7, e1002220 10.1371/journal.ppat.1002220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shirley C. M., Chen J., Shamay M., Li H., Zahnow C. A., Hayward S. D., and Ambinder R. F. (2011) Bortezomib induction of C/EBPβ mediates Epstein-Barr virus lytic activation in Burkitt lymphoma. Blood 117, 6297–6303 10.1182/blood-2011-01-332379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yan B. C., Adachi T., and Tsubata T. (2008) ER stress is involved in B cell antigen receptor ligation-induced apoptosis. Biochem. Biophys. Res. Commun. 365, 143–148 10.1016/j.bbrc.2007.10.137 [DOI] [PubMed] [Google Scholar]
- 39. Urano F., Wang X., Bertolotti A., Zhang Y., Chung P., Harding H. P., and Ron D. (2000) Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666 10.1126/science.287.5453.664 [DOI] [PubMed] [Google Scholar]
- 40. Zhang Y., Liu R., Ni M., Gill P., and Lee A. S. (2010) Cell surface relocalization of the endoplasmic reticulum chaperone and unfolded protein response regulator GRP78/BiP. J. Biol. Chem. 285, 15065–15075 10.1074/jbc.M109.087445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bhende P. M., Dickerson S. J., Sun X., Feng W. H., and Kenney S. C. (2007) X-box-binding protein 1 activates lytic Epstein-Barr virus gene expression in combination with protein kinase D. J. Virol. 81, 7363–7370 10.1128/JVI.00154-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Dalton-Griffin L., Wilson S. J., and Kellam P. (2009) X-box binding protein 1 contributes to induction of the Kaposi's sarcoma-associated herpesvirus lytic cycle under hypoxic conditions. J. Virol. 83, 7202–7209 10.1128/JVI.00076-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Bettigole S. E., and Glimcher L. H. (2015) Endoplasmic reticulum stress in immunity. Annu. Rev. Immunol. 33, 107–138 10.1146/annurev-immunol-032414-112116 [DOI] [PubMed] [Google Scholar]
- 44. Aragon I. V., Barrington R. A., Jackowski S., Mori K., and Brewer J. W. (2012) The specialized unfolded protein response of B lymphocytes: ATF6α-independent development of antibody-secreting B cells. Mol. Immunol. 51, 347–355 10.1016/j.molimm.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Amon W., and Farrell P. J. (2005) Reactivation of Epstein-Barr virus from latency. Rev. Med. Virol. 15, 149–156 10.1002/rmv.456 [DOI] [PubMed] [Google Scholar]
- 46. Chang P. J., Hung C. H., Wang S. S., Tsai P. H., Shih Y. J., Chen L. Y., Huang H. Y., Wei L. H., Yen J. B., Lin C. L., and Chen L. W. (2014) Identification and characterization of two novel spliced genes located in the orf47-orf46-orf45 gene locus of Kaposi's sarcoma-associated herpesvirus. J. Virol. 88, 10092–10109 10.1128/JVI.01445-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nishitoh H. (2012) CHOP is a multifunctional transcription factor in the ER stress response. J. Biochem. 151, 217–219 10.1093/jb/mvr143 [DOI] [PubMed] [Google Scholar]
- 48. Merksamer P. I., and Papa F. R. (2010) The UPR and cell fate at a glance. J. Cell Sci. 123, 1003–1006 10.1242/jcs.035832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Matar C. G., Rangaswamy U. S., Wakeman B. S., Iwakoshi N., and Speck S. H. (2014) Murine gammaherpesvirus 68 reactivation from B cells requires IRF4 but not XBP-1. J. Virol. 88, 11600–11610 10.1128/JVI.01876-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Qian Z., Xuan B., Chapa T. J., Gualberto N., and Yu D. (2012) Murine cytomegalovirus targets transcription factor ATF4 to exploit the unfolded-protein response. J. Virol. 86, 6712–6723 10.1128/JVI.00200-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee D. Y., and Sugden B. (2008) The LMP1 oncogene of EBV activates PERK and the unfolded protein response to drive its own synthesis. Blood 111, 2280–2289 10.1182/blood-2007-07-100032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Liu S., Pavlova I. V., Virgin H. W. 4th, and Speck S. H. (2000) Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol. 74, 2029–2037 10.1128/JVI.74.4.2029-2037.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stahl J. A., Paden C. R., Chavan S. S., MacLeod V., Edmondson R. D., Speck S. H., and Forrest J. C. (2012) Amplification of JNK signaling is necessary to complete the murine gammaherpesvirus 68 lytic replication cycle. J. Virol. 86, 13253–13262 10.1128/JVI.01432-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Tan L., Zhang C., Dematos J., Kuang L., Jung J. U., and Liang X. (2016) CD95 Signaling inhibits B cell receptor-mediated gammaherpesvirus replication in apoptosis-resistant B lymphoma cells. J. Virol. 90, 9782–9796 10.1128/JVI.00668-16 [DOI] [PMC free article] [PubMed] [Google Scholar]