Abstract
Mammalian Nod-like receptor (NLR) proteins contribute to the regulation and induction of innate and adaptive immunity in mammals, although the function of about half of the currently identified NLR proteins remains poorly characterized. Here we analyzed the function of the primate-specific NLRP11 gene product. We show that NLRP11 is highly expressed in immune cells, including myeloid cells, B cells, and some B cell lymphoma lines. Overexpression of NLRP11 in human cells did not trigger key innate immune signaling pathways, including NF-κB and type I interferon responses. NLRP11 harbors a pyrin domain, which is responsible for inflammasome formation in related NLR proteins. However, NLRP11 did not interact with the inflammasome adaptor protein ASC, and it did not trigger caspase-1 activation. By contrast, expression of NLRP11 specifically repressed NF-κB and type I interferon responses, two key innate immune pathways involved in inflammation. This effect was independent of the pyrin domain and ATPase activity of NLRP11. siRNA-mediated knockdown of NLRP11 in human myeloid THP1 cells validated these findings and revealed enhanced lipopolysaccharide and Sendai virus–induced cytokine and interferon responses, respectively, in cells with reduced NLRP11 expression. In summary, our work identifies a novel role of NLRP11 in the regulation of inflammatory responses in human cells.
Keywords: immunology, inflammasome, innate immunity, NF-kappa B (NF-KB), Nod-like receptor (NLR), interferon, inflammation, lymphoma, signaling, anti-viral, B cell, NLRP11
Introduction
An inflammatory response is triggered upon cell damage and to defend against invading pathogens. In mammals, this is mediated by the activation of cellular pathways cumulating into the release of cytokines, chemokines, and interferons. Downstream, this orchestrates recruitment of effector cells, alerts a systemic response, and restores tissue homeostasis (1). Activation of pattern recognition receptors (PRRs),3 expressed in and on host cells, that respond to pathogen-derived substances, cell damage, and stress initiates this response. On the cellular level, inflammation is mediated by the activation of pro-inflammatory signaling cascades such as the activation of caspase-1, NF-κB, and interferon regulatory factors (IRFs), among others. Chronic or dysfunctional activation of inflammatory signaling can lead to tissue disruption and disease states but also can contribute to malignant transformation, as in the activation of NF-κB in B lymphocytes (2). To control such potential harmful effects of inflammation, inflammatory pathways and their activators are tightly controlled by a network of regulators and feedback loops (3).
In humans, the Toll-like receptor (TLR), NLR, and C-type lectin family members are the most relevant PRRs. Many mammalian NLR proteins have been associated with multiple functions in inflammation and innate and adaptive immune responses (4), and dysfunctions in NLRs are associated with a range of diseases, including Crohn's disease, periodic fever syndromes, and Blau syndrome (5), highlighting their physiologic relevance. NLR proteins have a typical tripartite domain architecture and can be classified functionally according to their N-terminal domain, which is, in most cases, a CARD or PYD (6). Most but not all PYD-containing NLRs can interact with the adaptor molecule ASC to form high-molecular-weight complexes in cells, referred to as inflammasomes, that function as a platform for the activation of caspase-1 and subsequent IL-1β and IL-18 processing. The NLR family member pyrin domain–containing protein 11 (NLRP11, NALP11, NOD17, PYPAF6, PAN10) has the typical tripartite structure of NLRs and contains a PYD effector domain on the amino terminus (6). Many members of the NLR family have been extensively studied in recent years. However, the function of NLRP11 still remains largely elusive. NLRP11 is a primate-specific gene (7) and is highly expressed in oocytes with decreasing expression during oocyte maturation both in humans and in monkeys (8, 9). SNPs in NLRP11 were associated with age at natural menopause (10, 11). NLRP11 might thus have reproduction-related functions such as those reported for the NLRP proteins NLRP2, NLRP5, and NLRP7 (7, 12). Also, a SNP located in the NLRP11 gene is associated with susceptibility to Crohn's disease, an inflammatory bowel disease (13), and a duplication encompassing the NLRP11 gene was identified in patients with juvenile idiopathic arthritis, a rare inflammatory disease in children (14), suggesting further functionalities of NLRP11 in the regulation of inflammatory pathways in humans. These activities might be unrelated to inflammasome functions, as initial molecular analysis showed that NLRP11, besides having a PYD, does not co-localize with the inflammasome adaptor protein ASC and does not induce pro-inflammatory NF-κB signaling upon overexpression in human cells (15). Accordingly, knockdown of NLRP11 in THP1 cells does not affect IL-1β responses induced by heat-killed Acholeplasma laidlawii (16). Overall, the available experimental and genomic data suggest that NLRP11 plays a role in both development and immune regulation, but the molecular details and functions of NLRP11 remain elusive. Here we provide a functional characterization of NLRP11 in human cells, revealing a potential negative regulatory role in NF-κB and IFN pathways and show that NLRP11 is highly expressed in B cell malignancies.
Results
NLRP11 expression analysis
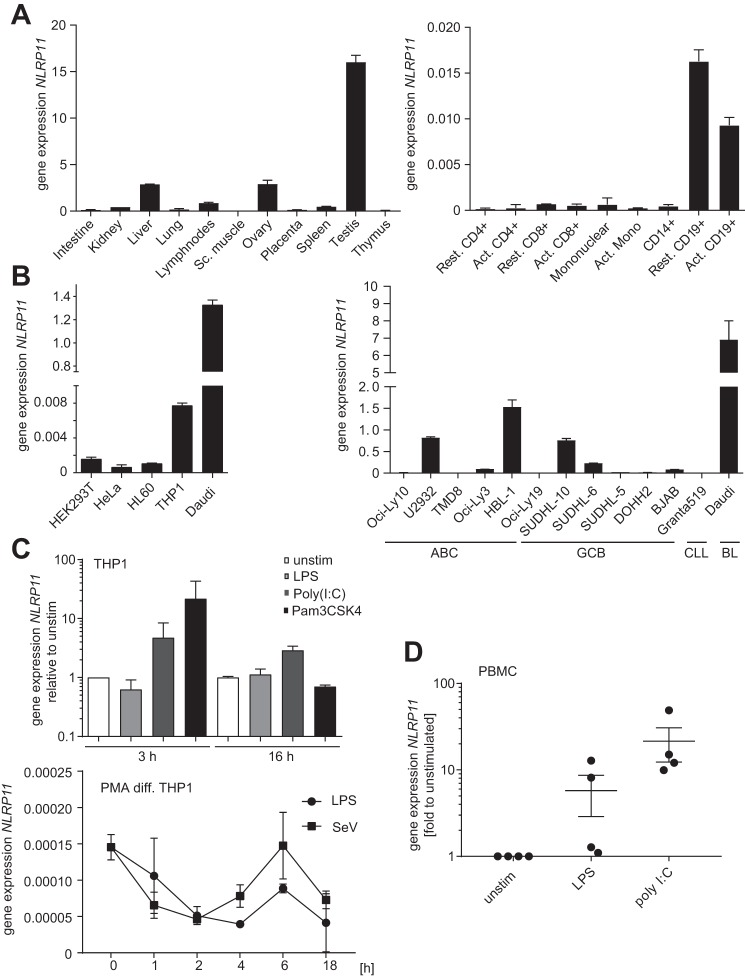
First, we experimentally determined the expression pattern of NLRP11. Quantitative RT-PCR from human tissues revealed high expression of NLRP11 mRNA in the testis, ovary, and liver (Fig. 1A, left panel), in line with expression data deposited at the GTex portal (Fig. S1A). Among the cells of the immune compartment, NLRP11 was highest expressed in B cells (CD19+) (Fig. 1A, right panel). Accordingly, the Burkitt's B cell lymphoma line Daudi expressed the highest levels of NLRP11 mRNA, and robust expression was also seen in myeloid THP1 cells, whereas epithelial cell lines expressed low levels of NLRP11 (Fig. 1B).
Figure 1.
Expression pattern of human NLRP11. A–D, qPCR analysis of NLRP11 expression in the indicated human cells and tissue. Data are shown as mean ± S.D. of at least two independently conducted experiments. A, mRNA expression levels of NLRP11 in different human tissues (Multiple Tissue cDNA Panels, Clontech) relative to GAPDH. B, expression of NLRP11 in different human cell lines (left panel) and B cell lymphoma lines (right panel). C, NLRP11 expression measured in total RNA from human THP1 cells. Top panel, undifferentiated THP1 cells were treated with 100 ng/liter LPS, 10 μg/ml poly(I:C), or 100 ng/ml Pam3CSK4 for 3 or 16 h, respectively. Bottom panel, PMA-differentiated (PMA-diff) THP1 cells were treated with LPS or SeV for the indicated time, and NLRP11 expression was measured in total RNA. D, NLRP11 expression measured in total RNA from primary human PBMCs from four donors. Cells were stimulated with 10 ng/ml LPS or 10 μg/ml poly(I:C) for 16 h (mean ± S.E.). ABC, activated B-cell; CLL, chronic lymphocytic leukaemia; BL, Burkitt's lymphoma; GCB, germinal center B-cell–like; unstim, unstimulated.
Analysis of gene chip data showed that, among different tumor entities, NLRP11 was highest expressed in diffuse large B cell lymphoma, hepatocellular carcinoma, and Burkitt's lymphoma (Fig. S1B). The most significant differences in NLRP11 expression were obtained for germinal center B cell lymphomas versus activated B cell lymphoma, glioma versus normal brain astrocytes, and hepatitis B virus-associated hepatocellular carcinoma versus non-diseased liver tissue (Fig. S2A). Analyzing a set of human B cell lymphoma lines revealed a strong heterogeneity in the expression level of NLRP11. Notably, analysis of 2414 gene chip expression sets showed a significant correlation of NLRP11 and NLRP4 expression (Fig. S2B). As NLRP4 and NLRP11 are located next to each other on chromosome 19q13.42 in a bidirectional orientation, this suggests that these two NLRs might share the same promotor.
Next, to analyze whether NLRP11 expression was affected by microbe-associated molecular pattern (MAMP) stimulation, we treated monocytic THP1 cells with the TLR agonists LPS, poly(I:C), and Pam3CSK4. 3 h after treatment, we observed robust induction of NLRP11 expression by TLR3 (poly(I:C)) and TLR1/2 (Pam3CSK4) ligation but not by TLR4 activation by LPS (Fig. 1C). By contrast, activation of PMA macrophage-like differentiated THP1 cells with the TLR4 agonist LPS or with Sendai virus (SeV) provoked a reduction of NLRP11 expression at later time points, with a slight recovery 6 h after activation (Fig. 1C). At later time points, NLRP11 mRNA was consistently less abundant in these cells compared with controls, also upon stimulation with the TLR1/2 ligand Pam3CSK4 (data not shown).
To substantiate the finding that NLRP11 mRNA expression can be induced by MAMPs in monocytes, we used human peripheral blood mononuclear cells (PBMCs) isolated from four different donors and treated these for 16 h with LPS or poly(I:C). Cells from all donors strongly induced NLRP11 expression upon TLR3 ligation, whereas only two donors induced NLRP11 upon TLR4 activation (Fig. 1D). Given the endogenous basal expression of NLRP11 in myeloid cell lines, we used THP1 cells as a model for subsequent analysis of endogenous NLRP11.
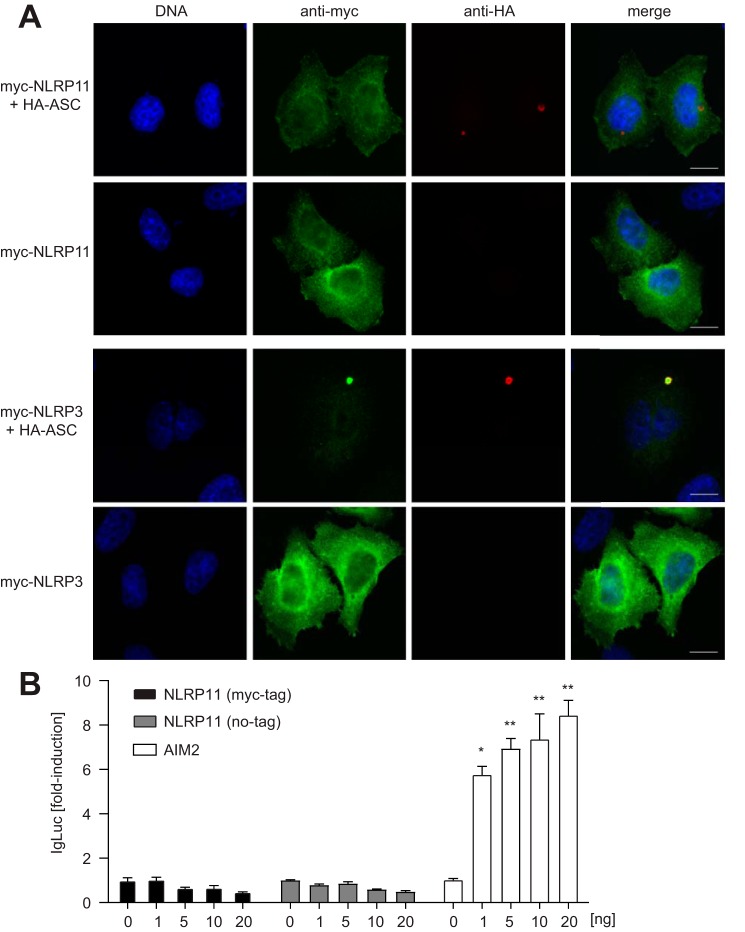
We and others recently showed that certain NLRs exhibit pronounced differences in their subcellular localization, with Nod1 and Nod2 residing at cortical F-actin (17, 18) and NLRC5 shuttling into the nucleus (19). To evaluate the subcellular localization pattern of NLRP11, we transiently transfected HeLa cells with NLRP11, revealing a cytosolic localization pattern with a slight enrichment in endoplasmic reticulum–like structures around the nucleus and some signal at membrane ruffles (Fig. 2A).
Figure 2.
Subcellular localization of human NLRP11 and its role in inflammasome formation. A, indirect immunofluorescence micrograph of HeLa cells transiently transfected for about 24 h with myc-NLRP3, myc-NLRP11, and HA-ASC (as indicated). Scale bars = 10 μm. B, caspase-1 activation assay (iGLuc) in HEK293T cells expressing caspase-1 and ASC (10 ng each) together with the indicted protein. Standard deviation of three experiments conducted in triplicates is shown. *, p < 0.05; **, p < 0.005.
NLRP11 does not induce ASC-mediated NF-κB or IL-1β responses in reconstituted HEK293T cells (15). On the other hand, NLRP11 was recently identified in a short hairpin RNA screen as a candidate that contributes to Mycobacterium tuberculosis–induced IL-1β release (20). Because of these controversial data, we validated a potential role of NLRP11 in forming inflammasomes. To this end, we compared the localization of ectopically expressed NLRP3 and NLRP11 with ASC in HeLa cells. Although NLRP3 was recruited to the single ASC speck per cell, as described before, NLRP11 was not recruited to ASC foci in the cells (Fig. 2A), suggesting that NLRP11 is not involved in inflammasome formation. To corroborate this finding, we tested whether NLRP11 was able to activate caspase-1 using the pro-interleukin (IL)-1β-Gaussia luciferase fusion construct caspase-1 reporter (iGLuc), which measures caspase-1 cleavage (21). Although overexpression of Aim2 resulted in dose-dependent reporter activation, expression of NLRP11 failed to activate caspase-1 in this assay (Fig. 2B). The same results were obtained for an untagged version of NLRP11, ruling out that tagging of the PYD inhibited its function (Fig. 2B).
Taken together, our results revealed that, besides the testis and ovary, NLRP11 is expressed in myeloid cells and to the highest levels in B cells. Different B cell lymphoma lines therefore showed heterogeneous expression of NLRP11. Confirming previous data (15), our data support that NLRP11 is a cytosolic protein that seems not to be involved in inflammasome formation.
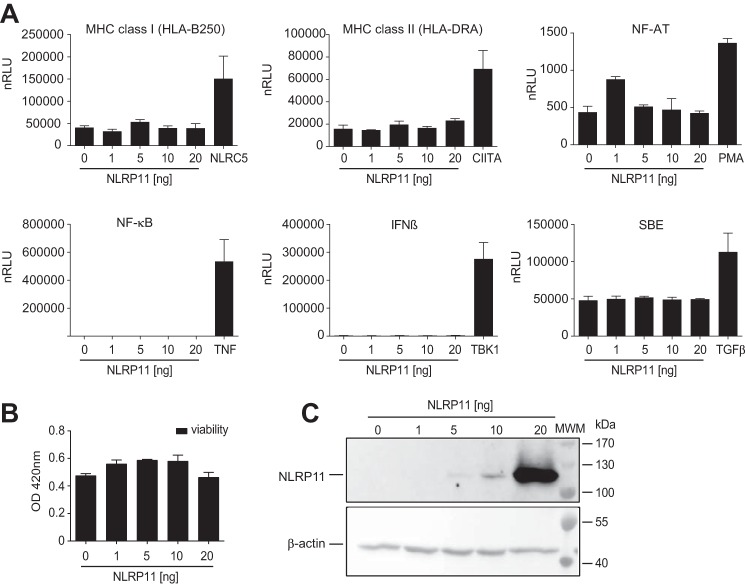
NLRP11 overexpression inhibits TBK1- and MyD88-mediated signaling
NLRP4 was described to affect type I interferon and NF-κB inflammatory responses (22–24). Given the genomic proximity of NLRP11 and NLRP4 and their co-expression, we speculated that NLRP11 might also be involved in inflammatory signaling. To analyze such a potential role of NLRP11, luciferase reporter assays for pathways involved in innate and adaptive immunity were conducted in HEK293T cells. In contrast to other NLRs, such as NLRC5, CIITA, Nod1, and Nod2, which trigger specific cellular signaling pathways upon their overexpression in this system, ectopic expression of NLRP11 did not significantly activate promoters responsive to major histocompatibility complex class I and II, NF-AT, SMAD, IFNβ, or NF-κB (Fig. 3A). By contrast, the corresponding reporter constructs were significantly activated by controls used side by side in each assay (Fig. 3A). Cell viability, as measured by 2,3-Bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide (XTT) assay, was slightly increased by overexpression of NLRP11 (Fig. 3B). Overexpression was validated by immunoblot analysis (Fig. 3C).
Figure 3.
Overexpression of NLRP11 does not trigger pro-inflammatory pathways. A, results from reporter assays with HEK293T cells transfected with reporter plasmids encoding the luciferase gene under the control of different promoters and co-transfected with either 0, 1, 5, 10, or 20 ng of pNLRP11.MYC plasmid. As a control, cells were transfected or stimulated as indicated (plasmid transfection, 5 ng of NLRC5, 5 ng of CIITA, or 20 ng of TBK1 plasmid; treatment, 20 ng/ml TNF, 100 ng/ml TGFβ, or 100 nm PMA). Relative light units normalized to β-galactosidase expression (nRLU) are shown. Error bars represent standard deviation of the mean of representative experiments conducted in triplicates of three independent experiments. B, XTT cell viability assay 16 h after overexpression of NLRP11 in HEK293T cells. Measurement of the optical density (OD) at 450 nm (mean and standard deviation of two independent experiments). C, Western blot from protein lysates from HEK293T cells probed with anti-myc antibody and β-actin as a loading control. **, p < 0.005; ***, p < 0.0005. MWM, molecular weight marker.
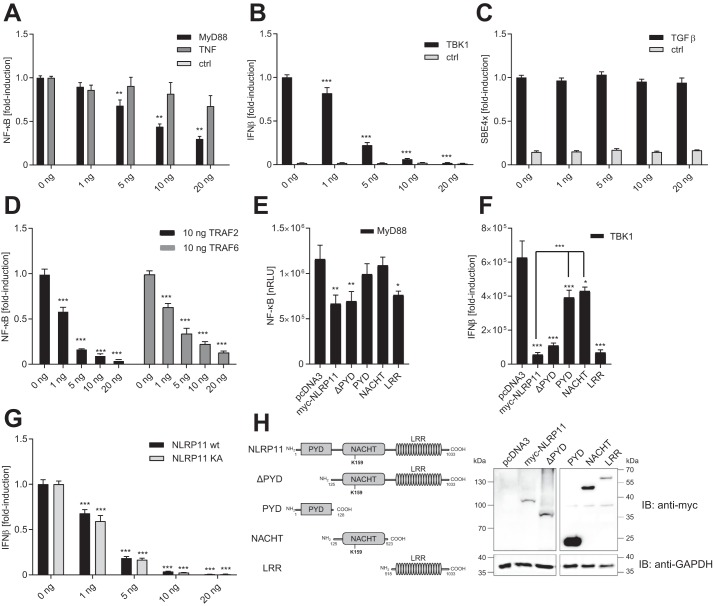
Next, we asked whether NLRP11 might have negative effects on inflammatory signals, as does NLRP4 (23, 25). Using gene reporter assays in HEK293T cells, we found that NLRP11 significantly reduced NF-κB activation downstream of TLRs, induced by MyD88 overexpression already at very low levels (5 ng), where it did not significantly affect TNF-induced NF-κB activation (Fig. 4A). Moreover, NLRP11 overexpression significantly reduced type I interferon reporter (interferon-β promotor) activation induced by TBK1 overexpression (Fig. 4B) in a dose-dependent manner. In contrast, TGFβ-induced SMAD-binding element activation was not affected by NLRP11 overexpression (Fig. 4C), showing specificity of the observed effects. Notably, untagged NLRP11 exhibited comparable activity in repressing both TBK1- and MyD88-induced gene activation, suggesting that the N-terminal myc tag used did not interfere with the functionality of the protein (Fig. S3, A and B).
Figure 4.
NLRP11 overexpression negatively affects innate immune signaling pathways. Gene reporter assays with HEK293T cells transfected with the indicated reporter and NLRP11 expression plasmids. A–D, effect of increasing expression of NLRP11 on inflammatory pathways. A, cells were activated by transfection of 20 ng of MyD88 plasmid or incubation with 20 ng/ml TNF or not treated (control (ctrl)). NF-κB-promoter activity is shown. B, cells were activated by transfection of 20 ng of TBK1 plasmid or empty plasmid (ctrl). IFNβ promoter activity is shown. C, cells were activated with 100 ng/ml TGFβ or not treated (ctrl). Smad-binding element promoter activity is shown. D, cells were activated by transfection of 10 ng of TRAF2 or TRAF6 plasmid. NF-κB promoter activity is shown. E, cells were transfected with the indicated plasmids and activated by transfection of 20 ng of MyD88 plasmid. NF-κB promoter activity is shown. nRLU, normalized relative light units. F, cells were transfected with the indicated plasmids and activated by transfection of 20 ng of TBK1 plasmid or empty plasmid (no TBK1). IFNβ promoter activity is shown. G, effect of the Walker A mutant of NLRP11. Cells were activated by transfection of 20 ng of TBK1 plasmid or empty plasmid (ctrl). IFNβ promoter activity is shown. H, schematic and expression analysis by immunoblot (IB) of the constructs of NLRP11 used in E and F. -Fold reduction relative to control cells transfected with 0 ng of NLRP11 set as 1 (A–D) or normalized relative light units are shown (E–G). Error bars represent standard deviation of the mean of three experiments, each conducted in triplicate. *, p < 0.05; **, p < 0.005; ***, p < 0.0005; n.s., p > 0.99.
Our data above suggest that NLRP11 specifically targets effectors involved in pathways downstream of TLR4 and TBK1. To define these factors, we tested the effect of NLRP11 overexpression on NF-κB responses induced by overexpression of TNF receptor–associated factors (TRAFs), which differentially contribute to both pathways, with TRAF6 mainly contributing to MyD88-dependent responses and TRAF2 mediating TNF-induced responses (26). Surprisingly, NLRP11 inhibited, to virtually the same extent, NF-κB activation induced both by overexpression of TRAF2 and TRAF6 (Fig. 4D), indicating that other, still to be identified proteins downstream of these proteins are the target of NLRP11.
To address which domains of NLRP11 mediate these effects, constructs encoding NLRP11 without the PYD (ΔPYD) and one encoding the PYD (PYD), the NACHT domain (NACHT), and the leucine-rich repeats (LRRs) (LRR) were expressed in HEK293T cells (Fig. 4H), and their effect on MyD88 and TBK1-induced NF-κB and IFN promotor activity was assessed. This revealed that the ΔPYD construct (aa 125–1033) was able to reduce both MyD88-induced NF-κB responses as well as TBK1-induced IFNβ responses to a similar extend as the full-length NLRP11. In contrast, both the PYD construct (aa 1–128) and the NACHT domain (aa 125–523) did not significantly reduce MyD88-induced NF-κB activation and affected TBK1-induced IFNβ promoter activity significantly less than the full-length NLRP11 (Fig. 4, E and F). By contrast, we found that the LRR domain (aa 518–1033) was sufficient for the reduction of both pathways to levels similar to that observed with the full-length protein (Fig. 4, E and F). Expression of the proteins was validated by immunoblotting (Fig. 4H). None of the constructs induced NF-κB or IFNβ promotor activity on its own (data not shown).
Finally, to test whether these effects of NLRP11 expression were dependent on NLRP11 ATPase activity, which affects the function of other NLR proteins, we generated a Walker A mutant of NLRP11 (K159A) (Fig. 4H). Overexpression of NLRP11 K159A protein did affect MyD88- or TBK1-induced responses similar to the wild-type protein (Fig. 4G), suggesting that, for these effects, ATPase functionality is not needed.
Taken together, these experiments showed that NLRP11 expression is able to inhibit NF-κB activation downstream of MyD88 but not TNF and strongly suppressed type I IFN activation induced by TBK1. These effects were mediated by the LRR domain of NLRP11 and did not dependent on the PYD or ATPase activity of NLRP11.
Depletion of NLRP11 augments NF-κB and IFN signaling in myeloid cells
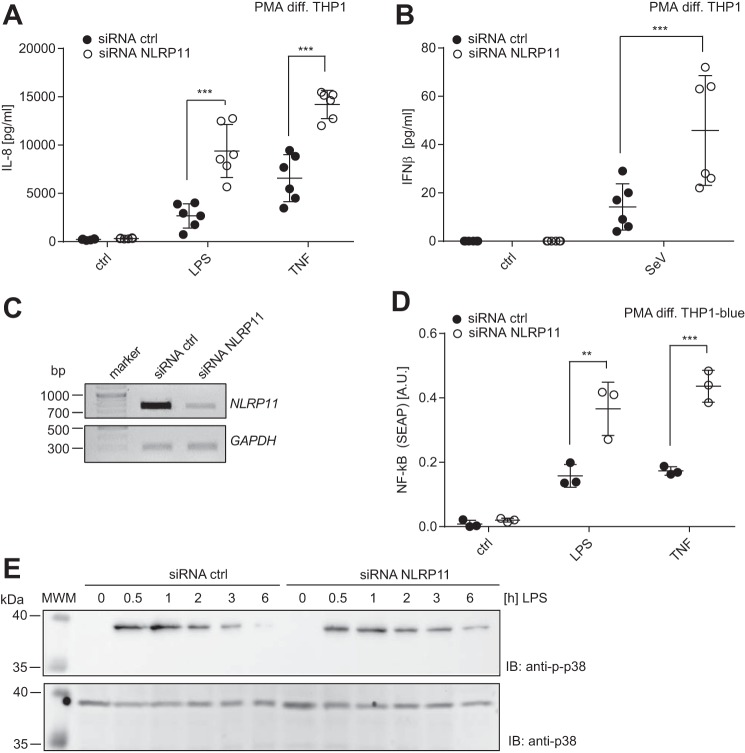
To address the function of endogenous NLRP11, we used differentiated THP1 cells, which express high levels of endogenous NLRP11 (Fig. 1B), and established silencing of NLRP11 expression by siRNA transfection. To this end, macrophage-like differentiated THP1 cells were treated with siRNA, and, after 72 h of silencing with continuous medium exchange, cells were stimulated with LPS, TNF, or SeV. Measurement of released IL-8 and IFNβ by ELISA after 16 h of treatment showed that cells with reduced NLRP11 expression showed significantly augmented IL-8 production upon both TNF and LPS stimulation compared with cells treated under identical conditions with a non-targeting siRNA duplex (Fig. 5A). Similarly, reduced NLRP11 expression correlated with increased type I interferon production upon SeV infection (Fig. 5B). Knockdown of NLRP11 mRNA was validated on the mRNA level by semiquantitative RT-PCR (Fig. 5C). Similar results were obtained using a THP1 reporter line that harbors a secretable alkaline phosphatase (SEAP) under the control of an NF-κB response promotor. Using this line, transfection of the NLRP11-specific siRNA led to increased SEAP secretion compared with cells treated with a non-targeting control siRNA (Fig. 5D). p38 activation in these cells after LPS treatment showed no obvious differences between NLRP11 siRNA and control siRNA treatment (Fig. 5E). Densitometric quantification confirmed no significant change (data not shown). Taken together, these data corroborate the results from the overexpression experiments in HEK293T cells and suggest that NLRP11 acts as a negative regulator of pro-inflammatory signaling in human cells and that NLRP11 attenuates specific signal transduction pathways downstream of TNF-receptor (TNFR) and PRRs.
Figure 5.
NLRP11 silencing enhances pro-inflammatory responses. siRNA-mediated knockdown of NLRP11 in PMA-differentiated (PMA diff.) THP1 cells. Mean + S.D. of two independent experiments, each conducted in biological triplicates (n = 6), is shown. A, IL-8 release from THP1 cells transfected with an NLRP11-specific siRNA or a non-targeting control siRNA (siRNA ctrl) treated for 16 h with LPS or TNF, respectively. B, IFNβ release from THP1 cells transfected with a NLRP11-specific siRNA or a non-targeting control siRNA treated for 16 h with SeV. Ctrl, non-treated cells; n.d., not detectable C, end point PCR with NLRP11 and GAPDH-specific primer sets of a representative experiment from A. D, NF-κB activation in THP1blue reporter cells measured by SEAP secretion upon the indicated siRNA treatment. Cells were stimulated for 16 h with 100 ng/ml LPS or 50 ng/ml TNF, respectively. E, immunoblotting of THP1blue cells treated for the indicated time with 100 ng/ml LPS. Phosphorylation of p38 and total p38 as a loading control are probed. One representative blot of three experiments is shown. ***, p < 0.0005 (two way ANOVA with Sidak's multiple comparisons test).
Discussion
Here we provide the first functional characterization of the human NLRP11 protein, a primate-specific member of the NLR family. Our experimental data and database mining showed that NLRP11 is highly expressed in the testis, ovary, and liver; however, immune cells, predominantly B cells, also express high levels of NLRP11. NLRP11 showed an interesting expression pattern with high expression in some B cell lymphoma lines. However, we were not able to elucidate differences between activated B cell lymphomas and germinal center B cell lines, which both showed remarkably differences between different lines in basal expression. We found that NLRP11 is also expressed in myeloid THP1 cells, as reported by others (16), and we therefore adopted THP1 cells as a model system to evaluate the function of endogenous NLRP11 by siRNA.
Interestingly, the NLRP11 gene is located in the opposite direction, in direct proximity to NLRP4, and co-expression analysis of a curated database showed that the two genes show significant co-expression. NLRP4 was recently identified as a negative regulator of TBK1-mediated type I interferon responses in human cells (23, 25). We observed a similar phenotype for NLRP11 overexpression in identical assays settings, suggesting that the two NLR proteins might have overlapping functions as negative regulators of the interferon pathway. In contrast to our data, a recent report found that overexpression of NLRP11 did not reduce TBK1-driven IFN responses in gene reporter assays in HEK293T cells. However, in that study, the expression of NLRP11 and functionality were not evaluated (9). The nature of this discrepancy awaits independent evaluation.
Overexpression of NLRP11 negatively and specifically affected NF-κB induction downstream of the TLR adaptor protein MyD88 and of TNFR1. Both pathways use a different set of TRAF proteins that appeared to us to be interesting candidates for targets of NLRP11: TLR4/MyD88-triggered NF-κB activation relies on TRAF6, whereas TNFR1-induced responses relies on TRAF2 (for an overview see, Ref. 26). Beside the differential contribution of NLRP11 overexpression on MyD88 versus TNF-induced NF-κB activation, we were surprised that NLRP11 reduced both TRAF2- as well as TRAF6-mediated NF-κB activation to virtually the same extent. The underlying mechanism of the specificity in HEK293T cells thus needs further investigation.
Initial characterization of NLRP11 revealed that NLRP11 does not induce ASC-mediated NF-κB or IL-1β responses in reconstituted HEK293T cells, nor did NLRP11 co-localize with ASC (15). By contrast, NLRP11 was identified in a short hairpin RNA screen as a candidate that contributes to M. tuberculosis–induced IL-1β release. However, no validation was performed (20). Our data are in line with the work by Bertin and co-workers (15), showing that NLRP11 does not associate with ASC in living cells. The function of the PYD of NLRP11 thus remains elusive at present, as it was also dispensable for the observed negative regulatory effect on IFN and NF-κB activation. Notably, the LRR domain of NLRP11 was sufficient for repression of the NF-κB and IFN pathways. Given that LRRs are protein–protein interaction motifs, this raises the possibility that the LRR of NLRP11 acts as an interaction platform for signaling rather than being involved in MAMP sensing.
Finally, we provide experimental evidence for the function of endogenous NLRP11 in myeloid cells. siRNA-mediated knockdown of endogenous NLRP11 in macrophage-like THP1 cells confirmed our results obtained upon overexpression of NLRP11 in HEK293T cells, as reduced NLRP11 levels increased both LPS/TLR4- and TNF/TNFR1-mediated IL-8 secretion and NF-κB activation in macrophage-like THP1 cells. Moreover, we observed augmented SeV-induced type I interferon production in THP1 cells upon reduction of NLRP11 expression. Of note, p38 activation downstream of TLR4 was not strongly affected by NLRP11 knockdown.
Similar to what we propose for NLRP11, NLRP4 (23) and NLRP14 (27) have also both been described as negative regulators of IFN responses. Interestingly, NLRP4 and NLRP11 are organized in an antiparallel manner in the genome, and their co-expression suggests the use of a common promotor or shared promotor elements. This implies that these two NLRs might have overlapping functions. Given that NLRP11 evolved later and is a primate-specific NLR might indicate that NLRP11 has additional functions compared with NLRP4. Notably, database mining revealed significant co-expression of the two proteins in several cells and tissue. Moreover, NLRP11 expression was profoundly altered in diffuse large B-cell lymphoma (DLBCL), glioma, and hepatocellular carcinoma cohorts, although this does not show causality. Still, inflammatory signaling pathways contribute to B cell lymphoma transformation, and thus it is tempting to speculate that NLRP11 and NLRP4 might contribute to cancer. Unfortunately, technical issues prevented us from studying depletion of NLRP11 in B cells. Further development of suitable knockdown approaches in B cells will facilitate analysis of the function of NLRP11 in B cells.
Another interesting aspect is the high basal expression of NLRP11 in the reproductive system. This is shared with other NLR proteins, notably NLRP14. For NLRP14, it was shown that it acts as a repressor for intercellular DNA sensing during oocyte fertilization (27). These NLRPs and others might act at the interface of immunity and reproduction (12) but also have additional roles in immunity in the adult animal.
In summary, we show here that NLRP11 is expressed in a panel of immune cells, notably myeloid cells and B cells, and functions as a negative regulator of inflammatory responses. The differential expression of NLRP11 in B cell lymphoma lines and cancer entities is an interesting point that warrants future research to elucidate whether NLRP11 might be involved in the malignant transformation process.
Experimental procedures
Cells and cell culture
HEK293T and HeLa cells were grown at 37 °C with 5% CO2 in Dulbecco's modified Eagle's medium containing 10% heat-inactivated fetal calf serum and penicillin–streptomycin. THP-1 cells were maintained in RPMI 1640 (Thermo Fisher) containing 10% heat-inactivated fetal calf serum and penicillin–streptomycin. THP-1blue cells (thp-sp, InvivoGen) were maintained in RPMI 1640 (Thermo Fisher) containing 10% heat-inactivated fetal calf serum, penicillin–streptomycin, and zeocin. Cells were continuously tested for mycoplasma contamination by PCR. PBMCs were isolated from whole blood obtained from two different donors by using Ficoll density gradient centrifugation.
Plasmids and reagents
The coding sequence of human NLRP11, corresponding to NM_145007.3, was cloned into a pCDNA3.1 backbone fused to a 5′ located myc tag. Plasmids encoding truncations NLRP10 were generated by PCR cloning in the same vector. The NLRP11 Walker A mutation (K159A) was generated by QuikChange site-directed mutagenesis and cloned into the backbone vector described above. The iGluc plasmid was a kind gift from Veit Hornung (Ludwig Maximilians University of Munich). All plasmids were validated by DNA sequencing.
Plasmids encoding human MyD88 and TBK1 were a kind gift from Alexander Weber and Kate Fitzgerald, respectively. The gene reporter plasmids used are described in Ref. 28. The expression plasmids encoding NLRC5 and CIITA are described in Ref. 29. High molecular weight (HMW) poly(I:C), LPS (ultrapure lipopolysaccharide, Escherichia coli 0111:B4), PMA (phorbol 12-myristate 13-acetate), Pam3CSK4, and recombinantly expressed TNF were obtained from InvivoGen. TGFβ was purchased from eBioscience.
Gene reporter assays in HEK293T cells
HEK293T cells were plated at a density of 30.000 cells/well of a 96-well plate and transiently transfected using X-tremeGENE 9 DNA transfection reagent (Roche) with 8.6 ng of β-galactosidase, 13 ng of the respective luciferase reporter constructs, and the indicated amounts of plasmids encoding NLRP11, TRAF2, TRAF6, MyD88, TBK1, and pcDNA was added up to constant total DNA levels. 24 h after transfection, cells were lysed in 100 μl of lysis buffer containing 25 mm Tris (pH 8.0), 8 mm MgCl2, 1% Triton X-100, and 15% glycerol per well. 50 μl of the cell lysates was transferred to a white, non-transparent 96-well plate, and luciferase activity was measured as luminescence in a multiplate reader (Enspire, PerkinElmer Life Sciences) upon automatic dispension of 100 μl of luciferase substrate solution containing 1.3 μm ATP and 770 ng/ml d-luciferin (Sigma).The residual 50 μl of cell lysate was supplemented with 100 μl of 1 mg/ml O-nitrophenyl-β-d-galactopyranoside in 60 mm Na2HPO4, 40 mm NaH2PO4, 10 mm KCl, and 1 mm MgSO4 (pH 7.0) per well and incubated at 37 °C for 30 min, and absorption was measured at 405 nm (620 nm reference) as β-galactosidase activity. Luciferase activity was normalized to β-galactosidase activity. The iGluc assay for caspase-1 activation was performed as described recently, after optimization of plasmids concentration for optimal readout (21).
siRNA treatment of THP1 cells
Knockdown of NLRP11 was performed using Hiperfect transfection of PMA-differentiated THP1 or THP1blue cells with the siRNA NLRP11_6 (Qiagen, CACGACCTTGCAGCTGTCGAA) and a non-targeting control siRNA (All-Star negative control, Qiagen) as described in Ref. 28.
RNA extraction and PCR
RNA was extracted from the cells using the RNeasy Plus Mini Kit from Qiagen according to the protocol of the manufacturer. Concentration and purity of the total RNA was measured optically (NanoPhotometer P360, Implen). cDNA synthesis was performed using the iScript cDNA synthesis kit (Bio-Rad). For screening NLRP11 expression in different human tissues and immune cells, cDNA from the Multiple Tissue cDNA Panels (Clontech) was used.
To quantify NLRP11 mRNA expression in B cell lymphoma, cDNA from different diffuse large B cell lymphoma cell lines, including both active B cell and germinal center types, was obtained upon RNA isolation with the RNeasy kit (Qiagen), DNA digestion using the Turbo DNA-free kit (Thermo Fisher), and reverse transcription using the high-capacity RNA-to-cDNA kit (Thermo Fisher).
qPCR was performed using iQ SYBR Green Supermix (Bio-Rad) in a CFX cycler and the following primers: NLRP11, ACGAGCCCACATGCCAAATA (forward) and GTTTTCTCAGACTCCCGCCA (reverse); GAPDH, ATG CCA GTG AGC TTC CCG TTC AG and GGT ATC GTG GAA GGA CTC ATG AC (reverse). For end-point PCR the following primes were used: NLRP11 GTTCACCTCACTGCTCACGA (forward) and NLRP11CGCTTCAGGACAGTACACGT (reverse). The specificity of the amplification was assured by sequencing of the qPCR amplification products (GATC) and validating that they corresponded to the NLRP11 sequence NM_145007.3. Semiquantitative RT-PCR was performed using Taq polymerase (Promega) on cDNA.
Indirect immunofluorescence
HeLa cells were transfected with the indicated plasmids using Lipofectamine 2000, fixed in 4% paraformaldehyde in phosphate-buffered saline, and permeabilized with 0.5% Triton X-100 for 5 min. The cells were incubated in 5% fetal calf serum in phosphate-buffered saline. The primary antibodies were rabbit anti-HA (Y11, Santa Cruz Biotechnology, sc-805) and mouse anti-myc (9E10, Santa Cruz Biotechnology). The secondary antibodies were Alexa 488–conjugated goat anti-mouse IgG and Alexa 546–conjugated goat anti-rabbit IgG (Molecular Probes). DNA was stained with Hoechst 33258 (Sigma). The images were acquired on a Leica DMi8 microscope using the HC PL APO ×63/1.40 oil objective and processed using ImageJ and the Leica LasX software.
Cell viability assay and immunoblotting
Cell viability was assessed in cells 16 h after transfection by the XTT kit from Roche according to the instructions of the manufacturer. Western blotting was performed as described in Ref. 28. Antibodies used were as follows: anti-myc (Santa Cruz Biotechnology, 9E10), anti-GAPDH (Santa Cruz Biotechnology, FL-335), anti-p-p38 (Cell Signaling Technology, 9216), and anti-p38 (Cell Signaling Technology, 9212).
Detection of cytokines and QUANTI-Blue assay
IL-8 (CXCL8) and IFNβ were quantified by ELISA (Duoset DY208 and DY814–05, Bio-techne) according to the instructions of the manufacturer. To measure SEAP activity in the supernatants of THP1blue cells, 20 μl of supernatant was incubated with 200 μl of QUANTI-Blue (InvivoGen, rep-qb) and A620 was detected after 2 h.
Data analysis and graphics
Data were analyzed and graphed using Excel and GraphPad Prism 7.0. The figures were generated using Adobe Illustrator CS6. Statistical differences were calculated by two-way analysis of variance using Dunnett's multiple comparisons test.
Author contributions
K. E. and Y. P. data curation; K. E., E. B., and T. A. K. formal analysis; K. E., I. K., and A. S. validation; K. E., E. B., I. K., A. S., Y. P., and Y. C. G. investigation; K. E., E. B., A. S., Y. P., Y. C. G., and A. N. R. W. methodology; K. E., I. K., A. S., A. N. R. W., and T. A. K. writing-review and editing; E. B., I. K., and T. A. K. visualization; A. N. R. W. resources; T. A. K. conceptualization; T. A. K. supervision; T. A. K. funding acquisition; T. A. K. writing-original draft; T. A. K. project administration.
Supplementary Material
Acknowledgment
We thank Axel Lorentz for critical reading of the manuscript.
This work was supported by funds from the University of Hohenheim. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S3.
- PRR
- pattern-recognition receptor
- PBMC
- peripheral blood mononuclear cells
- PYD
- pyrin domain
- IL
- interleukin
- IFN
- interferon
- LPS
- lipopolysaccharide
- LRR
- leucine-rich repeats
- MAMP
- microbe-associated molecular pattern
- PMA
- phorbol 12-myristate 13-acetate
- SeV
- Sendai virus
- TLR
- Toll-like receptor
- XTT
- 2,3-Bis-(2-methoxy-4-nitro-5-sulfophenyl)-2H-tetrazolium-5-carboxanilide
- TNF
- tumor necrosis factor
- TNFR
- tumor necrosis factor receptor
- TRAF
- TNF receptor–associated factor
- aa
- amino acids
- SEAP
- secretable alkaline phosphatase
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- TGF
- transforming growth factor
- NLR
- Nod-like receptor.
References
- 1. Medzhitov R. (2008) Origin and physiological roles of inflammation. Nature 454, 428–435 10.1038/nature07201 [DOI] [PubMed] [Google Scholar]
- 2. Sasaki Y., and Iwai K. (2016) Roles of the NF-κB pathway in B-lymphocyte biology. Curr. Top. Microbiol. Immunol 393, 177–209 [DOI] [PubMed] [Google Scholar]
- 3. Afonina I. S., Zhong Z., Karin M., and Beyaert R. (2017) Limiting inflammation: the negative regulation of NF-κB and the NLRP3 inflammasome. Nat. Immunol. 18, 861–869 10.1038/ni.3772 [DOI] [PubMed] [Google Scholar]
- 4. Franchi L., Warner N., Viani K., and Nuñez G. (2009) Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 227, 106–128 10.1111/j.1600-065X.2008.00734.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zhong Y., Kinio A., and Saleh M. (2013) Functions of NOD-like receptors in human diseases. Front. Immunol. 4, 333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Meunier E., and Broz P. (2017) Evolutionary convergence and divergence in NLR function and structure. Trends Immunol. 38, 744–757 10.1016/j.it.2017.04.005 [DOI] [PubMed] [Google Scholar]
- 7. Tian X., Pascal G., and Monget P. (2009) Evolution and functional divergence of NLRP genes in mammalian reproductive systems. BMC Evol. Biol. 9, 202 10.1186/1471-2148-9-202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. McDaniel P., and Wu X. (2009) Identification of oocyte-selective NLRP genes in rhesus macaque monkeys (Macaca mulatta). Mol. Reprod. Dev. 76, 151–159 10.1002/mrd.20937 [DOI] [PubMed] [Google Scholar]
- 9. Zhang L., Mo J., Swanson K. V., Wen H., Petrucelli A., Gregory S. M., Zhang Z., Schneider M., Jiang Y., Fitzgerald K. A., Ouyang S., Liu Z. J., Damania B., Shu H. B., Duncan J. A., and Ting J. P. (2014) NLRC3, a member of the NLR family of proteins, is a negative regulator of innate immune signaling induced by the DNA sensor STING. Immunity 40, 329–341 10.1016/j.immuni.2014.01.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Shen C., Delahanty R. J., Gao Y. T., Lu W., Xiang Y. B., Zheng Y., Cai Q., Zheng W., Shu X. O., and Long J. (2013) Evaluating GWAS-identified SNPs for age at natural menopause among Chinese women. PLoS ONE 8, e58766 10.1371/journal.pone.0058766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Stolk L., Perry J. R., Chasman D. I., He C., Mangino M., Sulem P., Barbalic M., Broer L., Byrne E. M., Ernst F., Esko T., Franceschini N., Gudbjartsson D. F., Hottenga J. J., Kraft P., et al. (2012) Meta-analyses identify 13 loci associated with age at menopause and highlight DNA repair and immune pathways. Nat. Genet. 44, 260–268 10.1038/ng.1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Gorp H., Kuchmiy A., Van Hauwermeiren F., and Lamkanfi M. (2014) NOD-like receptors interfacing the immune and reproductive systems. FEBS J. 281, 4568–4582 10.1111/febs.13014 [DOI] [PubMed] [Google Scholar]
- 13. Cummings J. R., Cooney R. M., Clarke G., Beckly J., Geremia A., Pathan S., Hancock L., Guo C., Cardon L. R., and Jewell D. P. (2010) The genetics of NOD-like receptors in Crohn's disease. Tissue Antigens 76, 48–56 [DOI] [PubMed] [Google Scholar]
- 14. Tadaki H., Saitsu H., Nishimura-Tadaki A., Imagawa T., Kikuchi M., Hara R., Kaneko U., Kishi T., Miyamae T., Miyake N., Doi H., Tsurusaki Y., Sakai H., Yokota S., and Matsumoto N. (2011) De novo 19q13.42 duplications involving NLRP gene cluster in a patient with systemic-onset juvenile idiopathic arthritis. J. Hum. Genet. 56, 343–347 10.1038/jhg.2011.16 [DOI] [PubMed] [Google Scholar]
- 15. Grenier J. M., Wang L., Manji G. A., Huang W. J., Al-Garawi A., Kelly R., Carlson A., Merriam S., Lora J. M., Briskin M., DiStefano P. S., and Bertin J. (2002) Functional screening of five PYPAF family members identifies PYPAF5 as a novel regulator of NF-κB and caspase-1. FEBS Lett. 530, 73–78 10.1016/S0014-5793(02)03416-6 [DOI] [PubMed] [Google Scholar]
- 16. Khare S., Dorfleutner A., Bryan N. B., Yun C., Radian A. D., de Almeida L., Rojanasakul Y., and Stehlik C. (2012) An NLRP7-containing inflammasome mediates recognition of microbial lipopeptides in human macrophages. Immunity 36, 464–476 10.1016/j.immuni.2012.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kufer T. A., Kremmer E., Banks D. J., and Philpott D. J. (2006) Role for Erbin in bacterial activation of Nod2. Infect. Immun. 74, 3115–3124 10.1128/IAI.00035-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kufer T. A., Kremmer E., Adam A. C., Philpott D. J., and Sansonetti P. J. (2008) The pattern-recognition molecule Nod1 is localized at the plasma membrane at sites of bacterial interaction. Cell. Microbiol. 10, 477–486 [DOI] [PubMed] [Google Scholar]
- 19. Neerincx A., Rodriguez G. M., Steimle V., and Kufer T. A. (2012) NLRC5 controls basal MHC class I gene expression in an MHC enhanceosome-dependent manner. J. Immunol. 188, 4940–4950 10.4049/jimmunol.1103136 [DOI] [PubMed] [Google Scholar]
- 20. Mishra B. B., Moura-Alves P., Sonawane A., Hacohen N., Griffiths G., Moita L. F., and Anes E. (2010) Mycobacterium tuberculosis protein ESAT-6 is a potent activator of the NLRP3/ASC inflammasome. Cell. Microbiol. 12, 1046–1063 10.1111/j.1462-5822.2010.01450.x [DOI] [PubMed] [Google Scholar]
- 21. Bartok E., Bauernfeind F., Khaminets M. G., Jakobs C., Monks B., Fitzgerald K. A., Latz E., and Hornung V. (2013) iGLuc: a luciferase-based inflammasome and protease activity reporter. Nat. Methods 10, 147–154 10.1038/nmeth.2327 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhu L., Hu Z., Liu J., Gao J., and Lin B. (2015) Gene expression profile analysis identifies metastasis and chemoresistance-associated genes in epithelial ovarian carcinoma cells. Med. Oncol. 32, 426 10.1007/s12032-014-0426-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Cui J., Li Y., Zhu L., Liu D., Songyang Z., Wang H. Y., and Wang R. F. (2012) NLRP4 negatively regulates type I interferon signaling by targeting the kinase TBK1 for degradation via the ubiquitin ligase DTX4. Nat. Immunol. 13, 387–395 10.1038/ni.2239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lian Y. G., Zhao H. Y., Wang S. J., Xu Q. L., and Xia X. J. (2017) NLRP4 is an essential negative regulator of fructose-induced cardiac injury in vitro and in vivo. Biomed. Pharmacother. 91, 590–601 10.1016/j.biopha.2017.04.120 [DOI] [PubMed] [Google Scholar]
- 25. Charoenthongtrakul S., Gao L., and Harhaj E. W. (2012) The NLRP4-DTX4 axis: a key suppressor of TBK1 and innate antiviral signaling. Cell Mol. Immunol. 9, 431–433 10.1038/cmi.2012.49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Xie P. (2013) TRAF molecules in cell signaling and in human diseases. J. Mol. Signal 8, 7 10.1186/1750-2187-8-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Abe T., Lee A., Sitharam R., Kesner J., Rabadan R., and Shapira S. D. (2017) Germ-cell-specific inflammasome component NLRP14 negatively regulates cytosolic nucleic acid sensing to promote fertilization. Immunity 46, 621–634 10.1016/j.immuni.2017.03.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neerincx A., Lautz K., Menning M., Kremmer E., Zigrino P., Hösel M., Büning H., Schwarzenbacher R., and Kufer T. A. (2010) A role for the human nucleotide-binding domain, leucine-rich repeat-containing family member NLRC5 in antiviral responses. J. Biol. Chem. 285, 26223–26232 10.1074/jbc.M110.109736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Neerincx A., Jakobshagen K., Utermohlen O., Buning H., Steimle V., and Kufer T. A. (2014) The N-terminal domain of NLRC5 confers transcriptional activity for MHC class I and II gene expression. J. Immunol. 193, 3090–3100 10.4049/jimmunol.1401065 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.