Abstract
Candida albicans is the predominant cause of vulvovaginal candidiasis (VVC). Little is known regarding the genetic diversity of Candida spp. in the vagina or the microvariations in strains over time that may contribute to the development of VVC. This study reports the draft genome sequences of four C. albicans and one C. glabrata strains isolated from women with VVC. An SNP-based whole-genome phylogeny indicates that these isolates are closely related; however, phylogenetic distances between them suggest that there may be genetic adaptations driven by unique host environments. These sequences will facilitate further comparative analyses and ultimately improve our understanding of genetic variation in isolates of Candida spp. that are associated with VVC.
Keywords: Candida albicans, Candida glabrata, single nucleotide polymorphisms (SNP), clinical isolates, yeast infection
Clinical vaginal Candida genomes.
VULVOVAGINAL CANDIDIASIS AND CANDIDA SPP. DIVERSITY
Species of Candida, and most predominantly Candida albicans, are the fungal pathogens responsible for one of the leading causes of vaginal infection in women of reproductive age: vulvovaginal candidiasis (VVC) (Sobel 1985). Though C. albicans is commonly detected in vaginal secretions of women without overt symptoms (Sobel 1988), the mechanisms that trigger the fungal morphologic transition and subsequent proinflammatory immune activation associated with VVC are poorly understood. However, the known virulence factors including phospholipases and proteolytic enzymes that become activated during infection have been well studied (Naglik, Challacombe and Hube 2003; Schaller et al.2003; Lian and Liu 2007; Sobel 2007). Longitudinal karyotyping of C. albicans isolates from women with recurrent VVC suggests that colonization of Candida in the vagina reflects persistence of a single strain that is undergoing genetic microevolution (Vazquez et al.1994). However, comparative genomic analyses using whole-genome sequences for vaginal isolates of C. albicans are currently lacking. It is important to explore genetic variations in genes related to commensalism and/or pathogenicity since these could contribute to phenotypes associated with VVC. A 2014 study conducted at the Broad Institute investigated the genetic and phenotypic variation of 21 clinical C. albicans isolates from different anatomic sites (Hirakawa et al.2015). Their analysis uncovered several types of genetic variation, including aneuploidy, loss of heterozygosity and single nucleotide polymorphism (SNP), which have consequences on general fitness and growth rate of C. albicans. Two of these isolates were collected from vaginal swabs and are, to date, the only published whole-genome sequences for vaginal isolates of C. albicans. A greater appreciation of genomic diversity in C. albicans found in the vagina is important to our understanding of underlying variation that may affect development and treatment of VVC.
CANDIDA ISOLATION AND WHOLE-GENOME SEQUENCING
Vaginal Candida spp. were isolated from vaginal swabs collected from women who experienced VVC (i.e. reported vaginal symptomology characteristic of VVC and were treated with a prescription antimycotic) in a prospective, longitudinal study performed at the University of Alabama, Birmingham, USA (Ravel et al.2013). Prior to use, the swabs collected in this study had been frozen at –80°C in Amies transport medium without glycerol. Amies is often used for collecting, transporting and preserving microbiological specimens and is formulated to maintain the viability of microorganisms without a significant increase in growth, being non-nutritive and phosphate buffered. Nonetheless, Amies could have limited the optimal cultivation of viable Candida spp. Despite these potential limitations, Candida spp. isolates were cultured from frozen vaginal Copan ESwabs after thawing on ice and plating a small aliquot (15 μL) on a yeast extract peptone dextrose (YPD)-rich agar plate. Plates were incubated for 24–48 h at 30°C, and single colonies were speciated using CHROMagar. Four C. albicans and one C. glabrata isolates were obtained from a total of four women. Two of the C. albicans isolates (UAB012_W3 and UAB012_W7) were obtained from the same woman at two separate time points, 4 weeks apart—one during asymptomatic colonization (UAB012_W3) and one during active VVC (UAB012_W7). All other isolates of C. albicans (one from each subject UAB040 and UAB090) and C. glabrata (subject UAB047), alike, were grown from samples collected during a reported episode of VVC. Genomic DNA was extracted using OmniPrep for Fungi DNA from a 3 mL YPD culture of each isolate grown overnight at 30°C. Mate-pair (3 kb) and paired-end (average insert size of 383 bp) libraries were prepared for each isolate using the Illumina TruSeq DNA PCR-Free Library Preparation Kit and an average input of 10.53 μg genomic DNA.
Whole-genome sequencing was performed on 1 lane of Illumina HiSeq2000 at the Institute for Genome Sciences (IGS), Genomics Resource Center, resulting in appreciable coverage per genome—averaging ∼250× and ∼290× coverage for mate-pair and paired-end libraries, respectively (Table 1). On average, 39.3 million sequence reads were generated from each mate-pair library and 44.9 million sequence reads from each paired-end library. The sequence reads generated for each strain were assembled into a draft genome using default parameters in ALLPATHS-LG (Butler et al.2008), a high-quality, short-read de novo genome assembler. Insert size and standard deviation of insert length were estimated using Velvet (Zerbino and Birney 2008). Average genome size for C. albicans isolates is 15.4 Mb, while the C. glabrata isolate is 12.3 Mb. Relevant assembly statistics can be found in Table 1. Genome assemblies for the four C. albicans isolates contained 321 contigs per genome, on average. The assembly for the one C. glabrata genome resulted in just 48 contigs. While there are presently eight other published genome sequences for C. glabrata isolates, this is the first of an isolate collected from the vagina. Structural and functional annotation predictions were performed using a custom Eukaryotic annotation pipeline, as previously described (Chibucos et al.2015), using C. albicans SC5314 (15.5 Mb) as reference genome (van het Hoog et al.2007). The average number of gene models predicted in the four C. albicans isolates is 5445 and 5843 using the ab initio gene prediction tools, GeneMark-ES and Augustus, respectively. Considering % completeness of these genomes, these gene prediction models obtained using the Core Eukaryotic Genes Mapping Approach (CEGMA) (Parra, Bradnam and Korf. 2007) (Table 1) are in line with the 6218 ORFs listed for C. albicans SC5314 in the Candida Genome Database (http://www.candidagenome.org). The relatively higher CEGMA % completeness for C. glabrata UAB047 (96.77%) compared to the other C. albicans isolates is most likely due to greater quality in genome assembly and C. glabrata being a slightly smaller and less complex genome. The genome sequence for C. glabrata UAB047 produced just 48 contigs (N90 = 19), whereas the number of contigs for the C. albicans genomes ranged from 226 to 430 (Table 1).
Table 1.
Whole-genome sequencing and assembly statistics.
| Strain | UAB012_W3 | UAB012_W7 | UAB040 | UAB090 | UAB047 |
|---|---|---|---|---|---|
| Taxa | C. albicans | C. albicans | C. albicans | C. albicans | C. glabrata |
| State at collection | Healthy | VVC | VVC | VVC | VVC |
| Genome Sequencing: Illumina HiSeq2000 | |||||
| Mate-pair library | |||||
| Avg library fragment length | 342 | 387 | 295 | 383 | 337 |
| Insert length | 3339 | 3167 | 2893 | 2952 | 3258 |
| Total reads | 39 059 084 | 38 081 146 | 37 682 010 | 30 562 766 | 50 780 034 |
| Read length | 101 | 101 | 101 | 101 | 101 |
| Coverage | 250 | 243 | 241 | 195 | 325 |
| Paired-end library | |||||
| Avg library fragment length | 393 | 352 | 393 | 358 | 421 |
| Total reads | 49 341 596 | 53 111 316 | 57 339 790 | 23 586 682 | 41 287 118 |
| Read length | 101 | 101 | 101 | 101 | 101 |
| Coverage | 315 | 340 | 367 | 151 | 264 |
| Genome assembly: ALLPATHS-LG de novo assembly | |||||
| Genome size (bp) | 15 270 022 | 15 645 509 | 14 987 076 | 15 527 721 | 12 254 043 |
| Number of contigs | 309 | 320 | 226 | 430 | 48 |
| Mean length | 49 417 | 48 892 | 66 314 | 36 110 | 255 292 |
| Max length | 336 679 | 445 608 | 935 413 | 363 854 | 1424 620 |
| N50 | 43 | 39 | 31 | 59 | 6 |
| N50 length | 129 269 | 136 759 | 143 362 | 74 258 | 695 543 |
| N90 | 136 | 133 | 106 | 207 | 15 |
| N90 length | 31 054 | 33 048 | 44 817 | 21 413 | 360 839 |
| Genome annotation: custom eukaryotic pipeline | |||||
| CEGMA % completeness | |||||
| Complete core eukaryotic genes | 86.29% | 90.73% | 92.34% | 90.32% | 96.77% |
| Partial core eukaryotic genes | 90.73% | 93.55% | 95.97% | 93.95% | 96.77% |
| Predicted gene models | |||||
| GeneMark-ES | 5514 | 5443 | 5323 | 5501 | 4858 |
| Augustus | 5755 | 5850 | 5862 | 5906 | 4541 |
| Predicted non-coding RNA genes | |||||
| Predicted tRNA | 111 | 118 | 131 | 111 | 207 |
| Predicted rRNA | 0 | 0 | 0 | 0 | 0 |
WHOLE-GENOME PHYLOGENY OF CANDIDA ALBICANS FROM VARIOUS BODY SITES
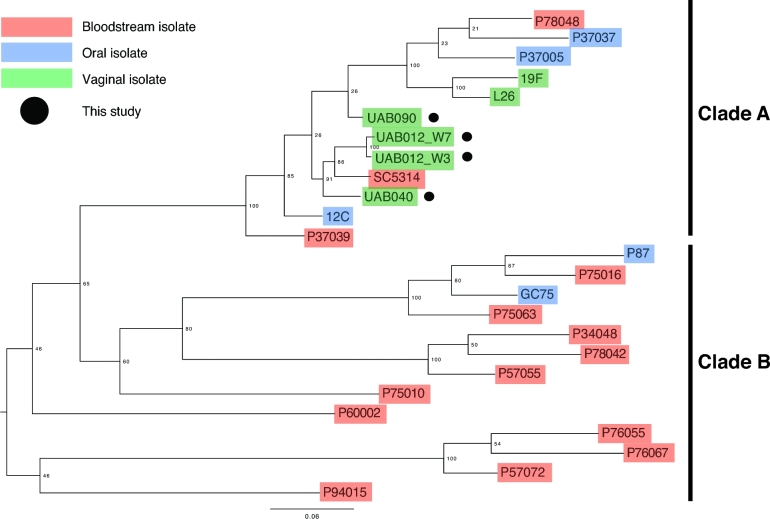
An SNP-based whole-genome phylogeny of 25 clinical C. albicans isolates (four isolates sequenced as part of this study and 21 sequenced at the Broad Institute (Hirakawa et al.2015)) was generated using the open-source In Silico Genotyper (ISG) (Sahl et al.2015) (Fig. 1). The ISG pipeline uses MUMMER v.3.22 (Delcher, Salzberg and Phillippy 2003) to identify SNPs relative to the C. albicans SC5314 reference genome. A total of 17 374 conserved SNP sites identified across all 25 C. albicans genomes (i.e. a base call was available for each of the 25 genomes) were concatenated and used to generate a maximum-likelihood phylogeny with 100 bootstrap values using RAxML v.7.2.8 software (Stamatakis 2014). The phylogenetic tree was midpoint rooted and labeled in FigTree v.1.4.2 (http://tree.bio.ed.ac.uk/software/figtree/). ISG has been previously used to resolve the population structure for Rhizopus strains, another diploid fungal pathogen of the order Mucorales (Chibucos et al.2016). First, we show that there are two major clades in this phylogeny that primarily distinguish mucosal isolates (clade A: oral and vaginal) from bloodstream isolates (clade B) (Fig. 1). Clusters of closely related genomes observed in the tree are comparable to those reported by Hirakawa et al., who inferred phylogenetic relationships of 21 C. albicans strains using a total of 112 223 parsimony informative SNP positions (Wilgenbusch and Swofford 2003; Hirakawa et al.2015). Their maximum parsimony phylogeny is based on total SNPs, whereas our maximum-likelihood phylogeny is only based on conserved SNP sites. Interestingly, vaginal isolates map closest to one another on the tree, providing evidence for phylogenetic relatedness among isolates occupying this niche. Of note, we observed clustering of the isolates sequenced in this study. Although these vaginal isolates were obtained from women in a limited geographical area, we do not expect a strong geographical influence on the genetic relatedness of these Candida spp. Pinto de Andrade et al. (2000) used PCR fingerprinting of vaginal C. albicans isolates from Portugal, Angola, Madagascar and Germany, and found a very weak correlation between vaginal C. albicans genotype and origin of the isolates. Furthermore, C. albicans UAB012_W3 and UAB012_W7 which were isolated from the same women at two different time points are highly similar, indicating that an isolate present prior to VVC was responsible for the symptomatic infection. It is also possible that VVC in UAB012 is associated with the acquisition of a new C. albicans strain that was not isolated in culture. However, since others have shown that VVC is most often associated with persistence of a single C. albicans strain (Vazquez et al.1994), our current hypothesis is that changing conditions within the vaginal environment, including the microbiota, may be associated with differences in C. albicans colonization (UAB012_W3) and virulence (UAB012_W7). The phylogenetic clustering of vaginal isolates from different women may be an indication of genetic adaptation to the uniquely acidic (pH< 4) and highly dynamic vaginal environment. More work is needed to explore genetic microvariations in Candida spp. genomes within the vagina, as these could be important contributors to virulence mechanisms and antifungal resistance. Further investigation of the genetic relatedness of mucosal C. albicans isolates and how they differ from bloodstream isolates could provide insight into mechanisms of pathogenesis and/or host adaptation that can be exploited with therapeutic targets.
Figure 1.
SNP-based whole-genome maximum-likelihood phylogenic tree generated using 17 374 conserved SNP sites identified with the ISG (Sahl et al.2015) from genome sequences from clinical C. albicans isolates from various body sites (red: bloodstream, blue: oral, green: vaginal). The four clinical C. albicans genomes sequenced in this study are noted with a black circle, and all others are from Hirakawa et al. (2015).
SUMMARY
Expanding our archive of published Candida spp. genomes will support comparative analyses to further our understanding of Candida spp. commensalism and pathogenesis. With respect to VVC, comparative Candida genomics will improve our understanding of genetic and phenotypic variations that take place within the vagina over time in response to a highly dynamic vaginal environment. These genomes will facilitate understanding the mechanisms of commensalism and pathogenesis, which ultimately could be targeted with new therapeutics.
FUNGAL GENOMIC ACCESSIONS
These Whole Genome Shotgun projects have been deposited at DDBJ/ENA/GenBank (http://www.ncbi.nlm.nih.gov/genbank/) under the accessions NETM00000000, NETN00000000, NETO00000000, NETQ00000000, NETP00000000 corresponding to strains Candida albicans UAB012_W3, UAB012_W7, UAB040 and UAB090, and C. glabrata UAB047, respectively. The versions described in this paper are the first versions: NETM01000000, NETN01000000, NETO01000000, NETQ01000000 and NETP01000000.
Acknowledgments
We thank Dr Tracy Hazen for guidance on using the In Silico Genotyper.
FUNDING
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under Award Number F31AI116023 and UH2AI083264.
Conflict of interest. None declared.
REFERENCES
- Butler J, MacCallum I, Kleber M et al. ALLPATHS: de novo assembly of whole-genome shotgun microreads. Genome Res 2008;18:810–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibucos MC, Etienne KA, Orvis J et al. The genome sequence of four isolates from the family Lichtheimiaceae. Pathog Dis 2015;73, DOI: 10.1093/femspd/ftv024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chibucos MC, Soliman S, Gebremariam T et al. An integrated genomic and transcriptomic survey of mucormycosis-causing fungi. Nat Commun 2016;7:12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delcher AL, Salzberg SL, Phillippy AM. Using MUMmer to identify similar regions in large sequence sets. Curr Protoc Bioinformatics 2003, Chapter 10, Unit 10.3. [DOI] [PubMed] [Google Scholar]
- Hirakawa MP, Martinez DA, Sakthikumar S et al. Genetic and phenotypic intra-species variation in Candida albicans. Genome Res 2015;25:413–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian CH, Liu WD. Differential expression of Candida albicans secreted aspartyl proteinase in human vulvovaginal candidiasis. Mycoses 2007;50:383–90. [DOI] [PubMed] [Google Scholar]
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol R 2003;67:400–28, table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G, Bradnam K, Korf I. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 2007;23:1061–7. [DOI] [PubMed] [Google Scholar]
- Pinto de Andrade M, Schonian G, Forche A et al. Assessment of genetic relatedness of vaginal isolates of Candida albicans from different geographical origins. Int J Med Microbiol 2000;290:97–104. [DOI] [PubMed] [Google Scholar]
- Ravel J, Brotman RM, Gajer P et al. Daily temporal dynamics of vaginal microbiota before, during and after episodes of bacterial vaginosis. Microbiome 2013;1:29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahl JW, Beckstrom-Sternberg SM, Babic-Sternberg J et al. The In Silico Genotyper (ISG): an open-source pipeline to rapidly identify and annotate nucleotide variants for comparative genomics applications. BioRx IV 2015, DOI: 10.1101/015578. [DOI] [Google Scholar]
- Schaller M, Bein M, Korting HC et al. The secreted aspartyl proteinases Sap1 and Sap2 cause tissue damage in an in vitro model of vaginal candidiasis based on reconstituted human vaginal epithelium. Infect Immun 2003;71: 3227–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobel JD. Epidemiology and pathogenesis of recurrent vulvovaginal candidiasis. Am J Obst Gynecol 1985;152:924–35. [DOI] [PubMed] [Google Scholar]
- Sobel JD. Pathogenesis and epidemiology of vulvovaginal candidiasis. Ann N Y Acad Sci 1988;544:547–57. [DOI] [PubMed] [Google Scholar]
- Sobel JD. Vulvovaginal candidosis. Lancet 2007;369:1961–71. [DOI] [PubMed] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 2014;30:1312–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van het Hoog M, Rast TJ, Martchenko M et al. Assembly of the Candida albicans genome into sixteen supercontigs aligned on the eight chromosomes. Genome Biol 2007;8:R52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez JA, Sobel JD, Demitriou R et al. Karyotyping of Candida albicans isolates obtained longitudinally in women with recurrent vulvovaginal candidiasis. J Infect Dis 1994;170:1566–9. [DOI] [PubMed] [Google Scholar]
- Wilgenbusch JC, Swofford D. Inferring evolutionary trees with PAUP*. Curr Protoc Bioinformatics 2003, Chapter 6, Unit 6.4. [DOI] [PubMed] [Google Scholar]
- Zerbino DR, Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res 2008;18:821–9. [DOI] [PMC free article] [PubMed] [Google Scholar]