Abstract
High-throughput whole genome sequencing has unlocked a multitude of possibilities enabling members of the Neisseria genus to be examined with unprecedented detail, including the human pathogens Neisseria meningitidis and Neisseria gonorrhoeae. To maximise the potential benefit of this for public health, it is becoming increasingly important to ensure that this plethora of data are adequately stored, disseminated and made readily accessible. Investigations facilitating cross-species comparisons as well as the analysis of global datasets will allow differences among and within species and across geographic locations and different times to be identified, improving our understanding of the distinct phenotypes observed. Recent advances in high-throughput platforms that measure the transcriptome, proteome and/or epigenome are also becoming increasingly employed to explore the complexities of Neisseria biology. An integrated approach to the analysis of these is essential to fully understand the impact these may have in the Neisseria genus. This article reviews the current status of some of the tools available for next generation sequence analysis at the dawn of the ‘post-genomic’ era.
Keywords: next-generation sequencing, Neisseria meningitidis, Neisseria gonorrhoeae; Omics analyses
This manuscript discusses how large volumes of genome data available can be organised, analysed and used to best advantage and has a particular focus on the bacterial pathogens: Neisseria meningitidis and Neisseria gonorrhoeae.
INTRODUCTION
In the late 1970s, methods were developed to sequence DNA by chain termination or fragmentation techniques. These revolutionised biology by enabling nucleotide sequences from complete genes to be determined and provided the tools for biologists to examine genetic variation (Sanger, Nicklen and Coulson 1977; Maxam and Gilbert 1980). Around 20 years later, the Sanger sequencing method was used to establish high-quality complete reference genomes for a number of organisms including humans and many important bacterial pathogens. The first pathogens to be sequenced were Haemophilus influenzae and Mycoplasma genitalium (Fleischmann et al.1995; Fraser et al.1995) providing for the first time an insight into the complete genomes of living organisms (Forde and O’Toole 2013). Following this, further improvements in sequencing techniques were made with the use of approaches such as paired-end sequencing and the whole genome shot-gun approach (Roach et al.1995; Weber and Myers 1997). These techniques paved the way for the sequencing of Neisseria meningitidis genomes in 2000, from the isolates Z2491 (serogroup A, accession number: NC_0 03116) and MC58 (serogroup B, NC_0 03112) (Parkhill et al.2000; Tettelin et al.2000). This was followed in 2003 with the genome from N. gonorrhoeae FA1090 (http://www.genome.ou.edu/gono.html, NC_0 02946) and N. lactamica, 20–06, in 2010 (FN995097) (Bennett et al.2010).
In this first phase of genome sequencing, methodologies based on the Sanger sequencing technology dominated; however, these techniques were laborious and time consuming. As a result, sequencing focussed on model organisms with only a few representative isolates from, for example, species of medical importance sequenced. In contrast to conventional sequencing methods, next-generation sequencing (NGS) technologies have the capacity to produce thousands to many millions of sequencing reactions in parallel, without the need for electrophoresis or bacterial cloning and provide a low-cost, high-throughput alternative to conventional Sanger sequencing approaches. The first NGS technology to be released, in 2005, was the pyrosequencing method by 454 Life Sciences (now Roche) with those from the Solexa/Illumina sequencing platform commercialised 1 year later in 2006. In 2007, Sequencing by Oligo Ligation Detection (SOLiD) was released by Applied Biosystems (now Life Technologies) with the Personal Genome Machine (PGM) released in 2010 by Ion Torrent (also now Life Technologies). Initially, these technologies produced short nucleotide (nts) read lengths averaging 35 to 110 nts (Illumina and 454, respectively); however, these have been extended with read lengths of up to 300 nts now possible using the Illumina MiSeq platform. Finally, the Pacific Biosciences (PacBio) sequencing platform PacBio RS II, released in 2011, is able to produce long reads of up to 20 000 nts enabling complete finished bacterial genomes to be obtained relatively rapidly and inexpensively (van Dijk et al.2014).
At the time of writing, the introduction of NGS technology provided the capacity to sequence thousands of bacterial isolates. Current challenges therefore now lie in the handling of the large amount of data generated by these technologies and how these should be organised, analysed and applied to best advantage. This review will explore these issues using the genus Neisseria as an exemplar. The organisation, analysis, application and impact of NGS data in Neisseria research will be discussed with a particular focus on the human pathogens N. meningitidis and N. gonorrhoeae. This review is based on discussions at the Neisseria genomics workshop which took place at the International Pathogenic Neisseria Conference in 2016 in Manchester, UK.
ORGANISATION AND ANALYSIS OF NEISSERIA NGS DATA
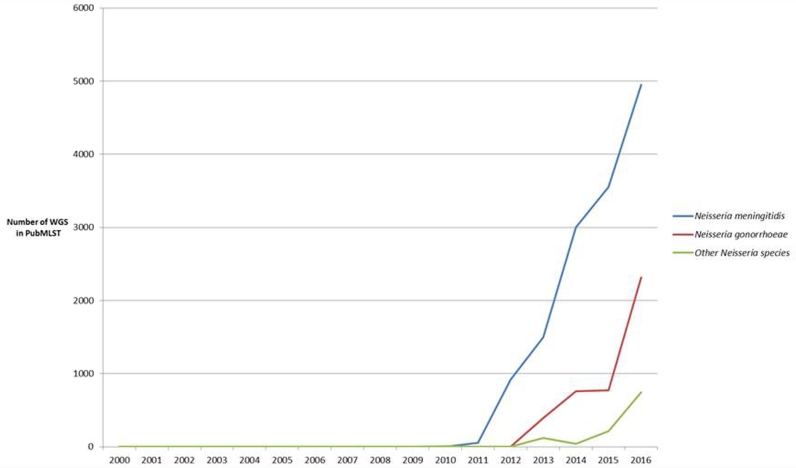
The genus Neisseria includes the human pathogens N. meningitidis and N. gonorrhoeae, which are significant causes of morbidity and mortality worldwide. As a result, isolates of these species have been extensively sequenced since 2000, with NGS generating an abundance of whole genome sequence (WGS) data. At the time of writing, WGS data from N. meningitidis outnumbered any of the other Neisseria species, with WGS from 13 985 meningococci available (Fig. 1), although the availability of WGS from N. gonorrhoeae was rapidly increasing at the time of writing, a consequence of the rise in antimicrobial-resistant gonococci which has provided a renewed incentive to resolve this global health problem (Grad et al.2014; Ezewudo et al.2015; De Silva et al.2016; Didelot et al.2016).
Figure 1.
Cumulative number of assembled WGS data deposited in PubMLST.org/neisseria. Graph depicting cumulative whole genome sequence data available in PubMLST dating from 2000 to the end of 2016. Blue lines depict N. meningitidis WGS data starting with relatively few isolates in 2010 and peaking at 5000 isolates at the end of 2016; red depicts N. gonorrhoeae WGS data peaking at over 2000; green all of the other Neisseria species including N. lactamica, N. subflava, N. polysaccharea, N. cinerea, N. mucosa, N. dentiae, N. musculi, N. oralis, N. bacilliformis and N. elongata.
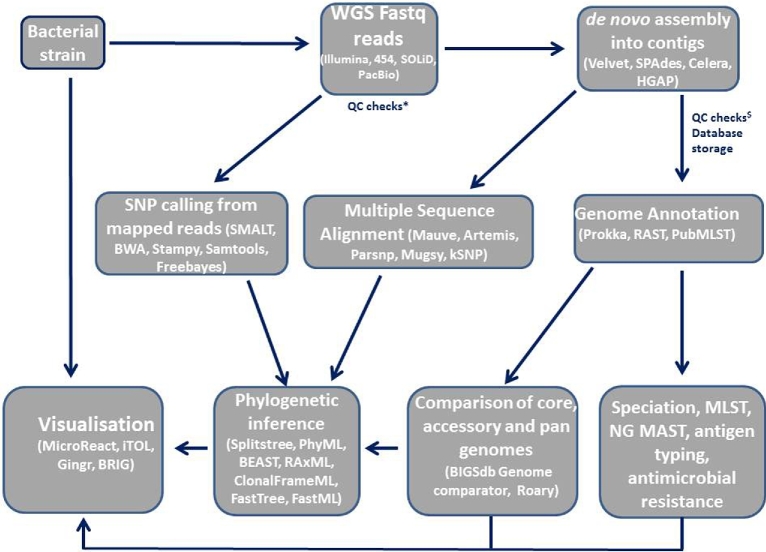
The volume of data generated from WGS can rapidly overwhelm computer storage space with, for example, an average of 150 Mb of data produced per bacterial genome. Such data are therefore rarely stored locally, and are for the most part deposited in the European Nucleotide Archive (ENA) or the National Center for Biotechnology Information (NCBI). A multitude of tools and methodologies are subsequently available for WGS analysis, many of which are not mutually exclusive and are largely dependent on the research question investigated (Fig. 2).
Figure 2.
NGS and analysis pipelines. Most common tools used for the analysis of WGS data. QC checks*: quality control before de novo assembly: poorly identified bases, low quality sequences and/or contaminants such as adaptors. QC checks$: quality control checks after assembly for mixed samples and bacterial contamination.
The Bacterial Isolate Genome Sequence Database (BIGSdb) platform
Once assembled de novo into contiguous stretches of nucleotide sequences (contigs) using assembly software such as Velvet or SPAdes (Zerbino 2010; Bankevich et al.2012), WGS data become more manageable and easy to store with the added benefit that these data can be repeatedly referred to and re-analysed thus generating reproducible and readily accessible data. Neisseria research has benefited from the availability of the PubMLST website (https://pubmlst.org/neisseria) which was initially set up to catalogue the seven multi-locus sequence type (MLST) allelic variants together with isolate provenance metadata (Jolley and Maiden 2013). This has been a continually expanding and well-accessed resource including, at the time of writing, nearly 42 000 isolate records with over 19 000 of these comprising genomes assembled from WGS data and 12 550 sequence types (ST). In addition, isolate records are hyperlinked to raw sequencing data at the ENA.
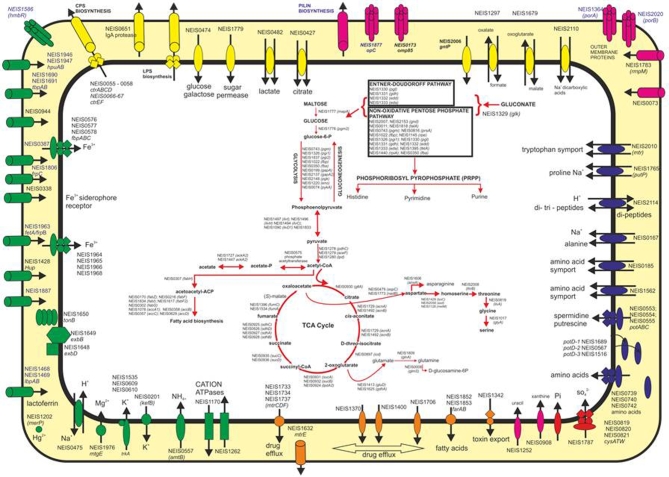
In 2012, PubMLST implemented the Bacterial Isolate Genome Sequence Database (BIGSdb) platform, which was developed to facilitate the flexible storage and exploitation of NGS data (Jolley and Maiden 2010). The platform consists of two kinds of database: (i) a sequence definition database that contains sequences of known alleles for loci as well as allelic profiles for specific schemes such as MLST and (ii) an isolate database that contains isolate provenance and other metadata along with nucleotide sequences associated with that isolate. Sequence definitions have now been established for over 2600 protein-encoding genes, annotated as loci with the NEIS prefix and the majority of these have been organised into schemes dependent on function. Many of these loci are shared across the Neisseria genus permitting WGS data from all Neisseria species to be annotated, including both meningococci and gonococci (Fig. 3).
Figure 3.
NEIS loci defined in PubMLST. Representative figure of a bacterial cell and some of the loci found across Neisseria species. Green structures represent acquisition receptors; pink indicate antigens; orange import/export elements; blue amino acid import/export; yellow sugar import/export; various metabolic pathways are also indicated. Additional loci have been defined in PubMLST. Figure based on the study by Tettelin et al. (2000).
Using gene discovery tools including Prokka and/or RAST (Aziz et al.2008; Seemann 2014), the catalogue of defined loci in PubMLST is continually increased allowing, for example, the entire set of genes found in all isolates from any species to be determined (known as the pan genome). As a result, loci found in mobile genetic elements such as plasmids, the meningococcal disease-associated island as well as secreted toxins associated with maf genomic islands can be annotated in WGS data (Jamet et al.2015; Meyer et al.2016). In addition, locus definitions are not limited to coding sequences, such that any stretch of nucleotide or translated peptide sequence can be defined including intergenic regions, antigenic epitopes, small regulatory RNAs and promoters. To distinguish such features from ORFs, promoter loci in the PubMLST Neisseria database have been assigned the ‘pro’ prefix (for promoter) followed by the corresponding locus suffix for the adjacent gene to differentiate them from coding sequences e.g. proNEIS1635 (mtrR). Point mutations located upstream of the mtrCDE operon and/or the mtrR promoter region upstream of the MtrR transcriptional regulator are associated with increased expression of the MtrCDE efflux pump leading to antimicrobial resistance in gonococci (Hagman and Shafer 1995; Lucas et al.1997; Ohneck et al.2011). Annotation and comparison of this promoter region in combination with loci implicated in antimicrobial resistance therefore allows antimicrobial-resistant genotypes to be determined (Harrison et al.2016). Small non-coding regulatory RNAs (ncRNAs) have also been defined in PubMLST including nrrF (Mellin et al.2010) with plans to include additional ncRNAs in the future as more of these are identified.
WGS data deposited in PubMLST are automatically annotated for any of the defined loci, updating isolate records with allele numbers. New alleles are automatically identified if they exhibit ≥98% sequence identity over ≥98% total length with previously identified variants. This results in a continually expanding catalogue of the diversity of all loci found in Neisseria. Gene-based comparisons can then be undertaken using the Genome Comparator tool within BIGSdb, which compares isolates using any number of loci predefined in the database or a reference genome. Through the use of this gene-by-gene approach to genome analysis, any combination of genes of interest can be compared between isolates ranging from the comparison of single loci to analyses of whole genomes resulting in a genome-wide MLST profile i.e. wgMLST (Maiden et al.2013). The benefit of this gene-by-gene approach being that the functional significance of genetic diversity can be rapidly deduced and compared between datasets.
WGS analysis tools
PubMLST is particularly useful for the gene-by-gene annotation, comparison and analysis of WGS data; however, additional tools are available that facilitate the analysis of larger datasets including several thousand isolates. Genome alignment and phylogenetic representation can become computationally intensive in the Neisseria genome where genomic rearrangements, duplications, deletions and insertions frequently occur resulting in a genome formed of conserved regions interspersed with highly diverse regions (Hao et al.2011). Tools such as Gubbins (Genealogies Unbiased By recomBinations In Nucleotide Sequences) can be used to identify, and exclude from phylogenetic analysis, loci subject to recombination effectively resulting in the analysis of the core non-recombinant genome which can be further analysed using tools such as ClonalFrameML (Croucher et al.2015). Alternative tools including single nucleotide polymorphism (SNP) typing methods are also available. Read-based mapping algorithms carry out assembly-free analyses using a finished reference genome and a sensitive read mapper such as BWA or Smalt followed by a variant caller such as samtools/bcftools and variant filter methods (Li and Durbin 2009; Li et al.2009). These methods require read data that are not always available and can be substantially larger than assembled genomes. In addition, read mapping can be sensitive to contaminants, misalign repetitive sequences and introduce bias in phylogenetic reconstructions. This form of analysis is also dependent on the selection and availability of an appropriate reference genome.
K-mer matching can be used to identify and/or cluster homologous genomic sequence data and has recently been extended to detect SNPs within large datasets. This has been achieved through the development of the software kSNP which identifies odd-length k-mers that match all but the central position (Gardner, Slezak and Hall 2015). The location of the putative SNPs can then be determined by mapping matched k-mers back to a reference genome. kSNP therefore avoids the use of whole-genome alignments and has the added benefit of accepting both assembled or short read data as input files.
Although pattern-based approaches such as that employed by kSNP have been found to closely reconstruct phylogenetic trees and are superior in this respect to other alignment-free methods, accurate phylogenetic inference can only be reliably obtained through the generation of maximum-likelihood distances computed from multiple sequence alignments (Hohl and Ragan 2007). Methods that combine whole-genome alignment with read-mapping may overcome problems associated with sensitivity and accuracy, whilst still allowing the rapid analysis of large datasets: the software Parsnp has been specifically designed for this purpose (Treangen et al.2014). Parsnp identifies maximal-unique matches among assembled genomes to both recruit similar genomes and anchor the multiple alignments (Treangen et al.2014). Core-genome alignments, variant calls and a SNP tree are then generated which can be visually explored using the accompanying software Gingr. For example, Parsnp was recently used to analyse WGS data from the bacterial pathogen, Streptococcus agalactiae. In this study, maximum-likelihood trees were generated based on concatenated SNPs from aligned core genomes revealing the occurrence of recombination events between ST23 and ST17 S. agalactiae isolates and the emergence of a serotype IV ST-425 lineage (Campisi et al.2016).
The principle underlying all of these methods is that the core genome, which includes genes present in all isolates from one species, will predominantly contain essential genes vertically inherited from the common ancestor and with a high signal-to-noise ratio. These will thus allow accurate representations of WGS data while avoiding difficulties with aligning and interpreting the evolution of complex and diverse genomic regions. Using a combination of methods and tools including the BIGSdb Genome Comparator and Roary (Page et al.2015), core genomes have been characterised and defined in PubMLST at the Neisseria genus level (Neisseria genus cgMLST) (Bennett et al.2010), the species level for the meningococcus (N. meningitidis cgMLST) (Bratcher et al.2014) and the gonococcus (N. gonorrhoeae cgMLST) (Harrison et al.2017) as well as the lineage-specific level (Harrison et al.2015). The genus-wide availability of loci in PubMLST combined with multiple core genome catalogues will ultimately allow genomic differences between species to be identified. For example, 1605 core loci have been characterised in the meningococcus with a total of 1653 core loci defined in gonococci. Comparison of loci from these core datasets has revealed that 1395 of these were in common to both species leaving 210 loci unique to meningococci and 258 loci distinct to gonococci (Harrison, O.B. unpublished). Further analyses of these loci may unravel some of the fundamental differences observed in both species including the distinct pathologies caused.
APPLICATIONS AND IMPLICATIONS OF NGS DATA FOR NEISSERIA
Effective and robust diagnostics using WGS data
Major benefits of WGS data in the Neisseria community have included N. meningitidis outbreak detection (Mulhall et al.2016), N. meningitidis and N. gonorrhoeae isolate characterisation and disease surveillance (Lucidarme et al.2015; Jacobsson et al.2016; Stefanelli et al.2016), and Neisseria taxonomy using ribosomal MLST (Bennett et al.2012, 2014; Bennett, Jolley and Maiden 2013). A major achievement in particular has been the publicly available online database containing WGS from invasive meningococcal isolates originating from England and Wales and dating from 2010 to the present providing data to the Neisseria community in almost real-time (MRF-MGL). In addition, there is the possibility in the near future for antimicrobial resistance profiles to be detected in WGS data from N. gonorrhoeae (Harrison et al.2016; Abrams and Trees 2017; Eyre et al.2017). WGS also enables species identification through quantitative assessment of genome-wide similarity between a new isolate and a curated multispecies reference collection, using statistics such as average nucleotide identity (Richter and Rossello-Mora 2009). Faster algorithms such as MASH allow this approach to be extended to much larger reference collections (Ondov et al.2016). In addition, WGS data have recently been used to detect novel genetic mutations which may have led to the dissemination of new invasive meningococci or antimicrobial-resistant gonococci (Grad et al.2014; Taha et al.2016; Toh et al.2017). Using WGS data, there is also the potential to combine pathogen and patient genomic sequence data to better understand patient response and susceptibility to infection (Davila et al.2010; Renner et al.2012).
A benefit of WGS can be through the provision of comprehensive information on bacterial populations, including the analysis of complex genomic regions that previously required several independent PCR- and sequencing-based assays (Dolan Thomas et al.2011; Wang et al.2011, 2012). This in turn can, for example, accelerate the time required for species identification and improve clinical diagnosis including the provision of subtyping information (Jolley et al.2012), particularly in clinical specimens where viable cultures could not be obtained (Joseph et al.2016). Indeed, the use of molecular-based approaches to identify both meningococci and gonococci has been increasingly employed with detection of gonorrhoea in the UK now routinely performed using nucleic acid amplification tests, while over half of the invasive meningococcal disease (IMD) cases in the UK between 2009 and 2010 were diagnosed solely by PCR (Heinsbroek et al.2013; Mohammed et al.2015). A substantial benefit of whole genome sequencing will therefore be in the ability to improve diagnosis in large numbers of isolates with low cost per sample while also providing genomic sequence data for additional analyses (Jolley and Maiden 2013; Maiden and Harrison 2016), all of which prior to WGS would have been cost-prohibitive.
An additional application of WGS in clinical diagnostics is through the enhanced surveillance of N. meningitidis capsular groups associated with IMD. Conjugate polysaccharide vaccines have successfully reduced the burden of meningitis and septicaemia globally. Such vaccines can, however, lead to the emergence of capsule replacement, as seen with Streptococcus pneumoniae (Wyres et al.2013). In addition, through immune selection, meningococci with capsule types included in a vaccine formulation may acquire capsule genes from non-vaccine isolates giving rise to new variants through capsule switching. The continued surveillance of serogroups associated with IMD is therefore essential to monitor such events which may not be apparent using conventional seroagglutination and PCR methods (Mustapha et al.2016). Serogroup B meningococcal disease is the only major disease-associated capsular group of meningococci for which a polysaccharide conjugate vaccine is unavailable; therefore, multicomponent protein-based vaccines that include outer membrane proteins have been developed (Giuliani et al.2006; Bambini et al.2009) with WGS data facilitating the assessment of antigen diversity among broad N. meningitidis isolate collections (Brehony et al.2015). Two such vaccines, Bexsero® and Trumenba® have recently been licensed for use in the UK and the USA, and post-implementation surveillance of IMD meningococci, including characterisation of vaccine antigens, is essential to assess the effectiveness of such vaccines. A Bexsero Antigen Sequence Type (BAST) scheme, based on deduced peptide sequence variants from the antigen targets included in Bexsero (fHbp, PorA, NHBA and NadA) has been implemented in the PubMLST database allowing vaccine antigen variants to be characterised (Brehony et al.2016). Results from this study, which included invasive meningococci isolated in Great Britain and Ireland, showed that BASTs were strongly associated with serogroup and clonal complex and demonstrated a significant increase in BAST-2 associated with the increased prevalence of serogroup W clonal complex 11 meningococci.
Recommendations and use of nomenclature
The effective use of WGS data for public health purposes requires the continued development of standardised software tools and effective data sharing networks to ensure that clear and robust results can be reported and compared globally. The practice of grouping and naming organisms on the basis of genotypic and phenotypic similarity facilitates communication and decision making. A principle of biological systematics is that biological names reflect evolutionary lineages and are therefore applied to clades (i.e. monophyletic groups). Due to the clonal reproduction of bacteria, clades can be identified even within species, although genetic recombination mediated by horizontal gene transfer means that clonal relationships do not necessarily describe the origins of the bacterial genomes over longer periods of time (Spratt 2004).
During outbreaks, pathogenic isolates tend to be grouped together on the assumption of sharing a recent common ancestor, representing one or more virulent strains circulating in the affected community. The complexity of genetic changes present, however, particularly when genetic differences occur in genes present only in some isolates (the accessory genome), demonstrates the challenges of interpreting genetic variation among isolates recovered from a single outbreak (Bergholz et al.2015). Genomic changes can occur even during a single case of IMD in one patient although evidence for concomitant within-host evolution of invasive phenotypes has not been detected so far (Omer et al.2011; Klughammer et al.2017; Lees et al.2017). This therefore introduces particular issues such as what should be the lowest cut-off value in genomic changes for transmission to be accurately detected (Hatherell et al.2016)? In addition, WGS analyses should always consider the possibility that patients may be co-infected with two or more distinct genotypes, particularly in the case of N. gonorrhoeae (Martin and Ison 2003). Such narrowly defined outbreak isolates will, however, only be relevant for a short time, as they will either become extinct or undergo further diversification.
Phylogeny-based definitions can account for the diversification of an outbreak strain over time, for example, the ‘Hajj clone’ from the Saudi Arabian meningococcal outbreak of 2000 (Lucidarme et al.2015; Mustapha et al.2015; Retchless et al.2016), but most isolates will not belong to a clade with a definable origin. A generalisable and stable nomenclature that facilitates communication may be best achieved by using an operational definition based on simple genomic similarity measures, which can serve as a surrogate for phylogenetic relationships among closely related strains (CDC unpublished data). A widely useful nomenclature scheme will need to include multiple ranks of within-species diversity to allow the rapid communication of genomic similarity for a variety of purposes, including short-term outbreak investigation, long-term surveillance programs and evolutionary studies. The PubMLST database currently performs genome clustering using a single-linkage algorithm based on cgMLST profiles. This precomputes groups of similar genomes based on allelic mismatch thresholds of 200, 100, 50 or 25 loci to at least one other member of the group. This clustering does not produce a stable nomenclature, due to the transient nature of groups which can merge as data are added, but will readily facilitate the identification of similar isolates in outbreaks or for wider epidemiological purposes. For a stable nomenclature, especially required for long-term surveillance, it would be possible to cluster based on thresholds of allelic differences to defined central cgMLST genotypes in the same way that standard MLST-based clonal complexes are defined for Neisseria.
FUTURE PROSPECTS
Challenges and prospects of functional genomic data integration
The collection of approaches referred to as ‘omics’ spans an increasing variety of fields such as genomics (the quantitative study of protein coding genes, regulatory elements and non-coding sequences), transcriptomics (RNA and gene expression), proteomics (protein abundance) and epigenomics (DNA methylation patterns). Ongoing advances in high-throughput sequencing methods facilitate such analyses, and will further our understanding of Neisseria infection and biology. For example, distinct base modifications implicated in epigenetic modifications can be determined in assembled WGS data using single molecule, real-time (SMRT) genome sequencing technology. Using SMRT, the recognition sites for three key N. meningitidis N(6)-adenine DNA methyltransferases (ModA11, ModA12 and ModD1) were identified unravelling global methylation patterns which can result in phase-variable changes in meningococcal gene expression (Seib et al.2015).
Conventional deep transcriptome sequencing (RNA-seq) as well as transcriptome analyses using tiling arrays have already enriched our understanding of meningococcal and gonococcal transcriptome dynamics and organisation. In particular, these technologies have allowed the genome-wide identification of small ncRNAs in meningococci and gonococci demonstrating their important contribution in Neisseria physiology (Mellin et al.2010; Huis in 't Veld et al.2011; Wachter and Hill 2015), including the regulation of large metabolic adaptations (Pannekoek et al.2017), survival in human blood (Del Tordello et al.2012) as well as during infection in vivo (Fagnocchi et al.2015; McClure et al.2015) and enhancement of pilin variation in both meningococci and gonococci (Cahoon and Seifert 2013; Tan et al.2015). The still expanding array of high-throughput sequencing-based technologies (reviewed in Barquist and Vogel 2015) is adding novel data on Neisseria genome architecture and gene expression regulation at an unprecedented detail and with increasing pace. These technologies include differential RNA-seq (dRNA-seq) (Sharma et al.2010) and Term-seq (Dar et al.2016) which allow the genome-wide identification down to the single nucleotide level of transcript boundaries and operons as well as the reliable identification of novel transcripts including trans-acting ncRNAs and cis-regulatory RNA elements such as riboswitches or attenuators. Accordingly, using dRNA-seq, Heidrich et al. (2017) mapped 1625 transcriptional start sites in the genome from N. meningitidis strain 8013 along with 64 ncRNAs and revealed an unexpected paucity of classical σ70-type promoters. Furthermore, Zhang et al. (2013) demonstrated that meningococci possessed the most streamlined CRISPR/Cas system characterised to date, thus limiting natural transformation—the primary source of genetic variation in this species.
The specific interactions between transcripts and nucleic acid-binding proteins such as Hfq can be studied on a genome-wide basis under various experimental conditions with new sequencing-based technologies such as RIP-seq (Sittka et al.2008), CLIP-seq (Holmqvist et al.2016) or Grad-seq (Smirnov et al.2016), which allow transcriptome-wide mapping of Neisseria RNA-binding protein target sites to rapidly describe functional RNA landscapes. High-throughput sequencing has been further used to modify classical genome-wide transposon mutagenesis approaches in order to determine gene function in various experimental conditions. For example, Capel et al. (2016) were the first to use transposon insertion site sequencing (Tn-seq) to comprehensively identify 228 protein-coding genes as well as 33 ncRNA candidates required for meningococcal colonisation of human cells. A recently developed sequencing technology is dual RNA-seq (Aprianto et al.2016; Westermann et al.2016) which allows the simultaneous profiling of all RNA classes in both bacteria and the eukaryotic host cell, and thus promises to deliver novel insights into Neisseria infection biology. Given the high genetic diversity of the Neisseria, however, it remains to be shown whether the results obtained for one particular genotype will be representative of the entire species. In fact, it is known that the high genetic diversity observed might not be entirely neutral (Buckee et al.2008) and may result in phenotypic differences at the transcriptomic (Joseph et al.2010) and likely metabolic level therefore affecting virulence in a strain-dependent manner (Schoen et al.2014). In support of this hypothesis, a recent systems biology analysis, comparing the transcriptomes from a carried and invasive-associated meningococcus in conditions mimicking infection, indicated that inactivating mutations in amino acid metabolism genes were buffered at the transcriptional level. Consequently, while both meningococci were able to grow in human blood, they showed significant differences in the expression of numerous virulence-associated determinants suggesting that meningococcal virulence was linked to transcriptional buffering of cryptic genetic variation in metabolic genes (Ampattu et al.2017).
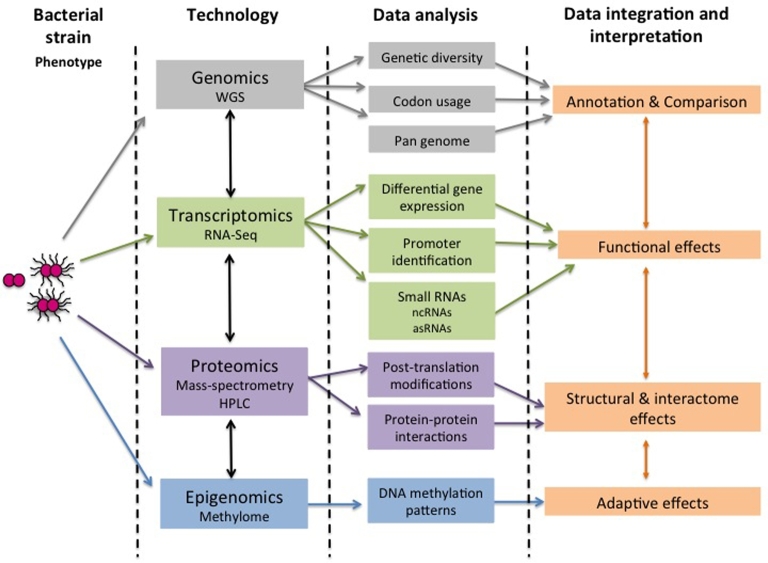
All these genome-wide analyses may generate more data than is analysed and published; however, these data are not lost but can be used to improve functional genome annotation and provide the raw material for integrative analyses that in turn can guide functional and molecular studies. The feasibility and power of combining genomic data with computational simulation to predict phenotype from genotype has already been demonstrated for a number of model species such as Mycoplasma genitalium (Karr et al.2012) or Escherichia coli (Carrera et al.2014). Dedicated databases and software exist for the storage and analysis of Neisseria WGS from a large number of strains such as BIGSdb (Jolley and Maiden 2010) or NeMeSys (Rusniok et al.2009). Future planned developments of BIGSdb will allow the arbitrary storage and querying of any number of sparsely-populated fields facilitating the integration of complete phenotypic and transcriptomic data with existing genome and provenance information. In addition, a computational genome-scale metabolic model has already been established and experimentally verified for N. meningitidis MC58 (Baart et al.2007; Mendum et al.2011). Given that such genome-scale models are able to highlight strain-specific adaptations to different nutritional environments corresponding to bacterial pathotypes (Monk et al.2013), it is anticipated that computational model-based analyses of the huge omics datasets becoming available in the near future will be key in understanding the impact of genetic variation in both meningococcal and gonococcal phenotypes. Integration of the large and heterogeneous omics datasets and making these accessible for comprehensive computational analyses will therefore be instrumental in enriching our understanding of Neisseria biology (Fig. 4).
Figure 4.
Omics platforms. Flow diagram depicting available Omics approaches some of which have been used in Neisseria research. Possible ways in which these approaches can be associated are also indicated.
CONCLUSIONS
From the elucidation of the structure of DNA to the publication of the first bacterial genome, our fascination with DNA and the genome it encodes shows no sign of diminishing. The impact genetic differences may have in a bacterial population ranging from the ability to successfully avoid the immune response to becoming antimicrobial resistant has become more carefully considered and understood. It is only through the comparison and analysis of large-scale bacterial populations, however, that we will be able to fully appreciate the consequences of these. Future challenges now lie in establishing ways in which the genome can be linked with the transcriptome, proteome and epigenome, ultimately resulting in a connection between the genotype and the phenotype and unravelling the complexity of bacterial infections.
Acknowledgments
Disclaimer: The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the CDC.
FUNDING
OBH and MCJM are funded by the Wellcome Trust (087622/Z/08/2) and the Oxford Martin School. KAJ and JEB are funded by Wellcome Trust Biomedical Resource Grant (104992). CS is funded by the German Research Foundation (DFG) grant (SCHO1322/1). XW and ACR are supported by the CDC.
Conflict of interest. None declared.
REFERENCES
- Abrams AJ, Trees DL. Genomic sequencing of Neisseria gonorrhoeae to respond to the urgent threat of antimicrobial-resistant gonorrhoea. Pathog Dis 2017, 75 DOI: 10.1093/femspd/ftx041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ampattu BJ, Hagmann L, Liang C et al. . Transcriptomic buffering of cryptic genetic variation contributes to meningococcal virulence. BMC Genomics 2017, 18:282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aprianto R, Slager J, Holsappel S et al. . Time-resolved dual RNA-seq reveals extensive rewiring of lung epithelial and pneumococcal transcriptomes during early infection. Genome Biol 2016;17:198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aziz RK, Bartels D, Best AA et al. . The RAST Server: rapid annotations using subsystems technology. BMC Genomics 2008;9:75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baart GJ, Zomer B, de Haan A et al. . Modeling Neisseria meningitidis metabolism: from genome to metabolic fluxes. Genome Biol 2007;8:R136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bambini S, Muzzi A, Olcen P et al. . Distribution and genetic variability of three vaccine components in a panel of strains representative of the diversity of serogroup B meningococcus. Vaccine 2009;27:2794–803. [DOI] [PubMed] [Google Scholar]
- Bankevich A, Nurk S, Antipov D et al. . SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 2012;19:455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barquist L, Vogel J. Accelerating discovery and functional analysis of small RNAs with new technologies. Annu Rev Genet 2015;49:367–94. [DOI] [PubMed] [Google Scholar]
- Bennett JS, Bentley SD, Vernikos GS et al. . Independent evolution of the core and accessory gene sets in the genus Neisseria: insights gained from the genome of Neisseria lactamica isolate 020-06. BMC Genomics 2010;11:652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Jolley KA, Earle SG et al. . A genomic approach to bacterial taxonomy: an examination and proposed reclassification of species within the genus Neisseria. Microbiology 2012;158:1570–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Jolley KA, Maiden MC. Genome sequence analyses show that Neisseria oralis is the same species as ‘Neisseria mucosa var. heidelbergensis’. Int J Syst Evol Microbiol 2013;63:3920–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett JS, Watkins ER, Jolley KA et al. . Identifying Neisseria species using the 50S ribosomal protein L6 (rplF) gene. J Clin Microbiol 2014;52:1375–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergholz TM, den Bakker HC, Katz LS et al. . Determination of evolutionary relationships of outbreak-associated Listeria monocytogenes strains of serotypes 1/2a and 1/2b by whole-genome sequencing. Appl Environ Microb 2015;82:928–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratcher HB, Corton C, Jolley KA et al. . A gene-by-gene population genomics platform: de novo assembly, annotation and genealogical analysis of 108 representative Neisseria meningitidis genomes. BMC Genomics 2014;15:1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehony C, Hill DM, Lucidarme J et al. . Meningococcal vaccine antigen diversity in global databases. Euro Surveill 2015;20:30084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehony C, Rodrigues CM, Borrow R et al. . Distribution of Bexsero® Antigen Sequence Types (BASTs) in invasive meningococcal disease isolates: Implications for immunisation. Vaccine 2016;34:4690–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckee CO, Jolley K, Recker M et al. . Role of selection in the emergence of lineages and the evolution of virulence in Neisseria meningitidis. P Natl Acad Sci USA 2008;105:15082–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cahoon LA, Seifert HS. Transcription of a cis-acting, noncoding, small RNA is required for pilin antigenic variation in Neisseria gonorrhoeae. PLoS Pathog 2013;9:e1003074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi E, Rinaudo CD, Donati C et al. . Serotype IV Streptococcus agalactiae ST-452 has arisen from large genomic recombination events between CC23 and the hypervirulent CC17 lineages. Sci Rep 2016;6:29799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capel E, Zomer AL, Nussbaumer T et al. . Comprehensive identification of meningococcal genes and small noncoding RNAs required for host cell colonization. Mbio 2016;7:e01173–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrera J, Estrela R, Luo J et al. . An integrative, multi-scale, genome-wide model reveals the phenotypic landscape of Escherichia coli. Mol Syst Biol 2014;10:735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croucher NJ, Page AJ, Connor TR et al. . Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res 2015;43:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar D, Shamir M, Mellin JR et al. . Term-seq reveals abundant ribo-regulation of antibiotics resistance in bacteria. Science 2016;352:aad9822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davila S, Wright VJ, Khor CC et al. . Genome-wide association study identifies variants in the CFH region associated with host susceptibility to meningococcal disease. Nat Genet 2010;42:772–6. [DOI] [PubMed] [Google Scholar]
- De Silva D, Peters J, Cole K et al. . Whole-genome sequencing to determine transmission of Neisseria gonorrhoeae: an observational study. Lancet Infect Dis 2016;16:1295–1303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Tordello E, Bottini S, Muzzi A et al. . Analysis of the regulated transcriptome of Neisseria meningitidis in human blood using a tiling array. J Bacteriol 2012;194:6217–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didelot X, Dordel J, Whittles LK et al. . Genomic analysis and comparison of two gonorrhoea outbreaks. Mbio 2016;7:e00525–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolan Thomas J, Hatcher CP, Satterfield DA et al. . sodC-based real-time PCR for detection of Neisseria meningitidis. PLoS One 2011;6:e19361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyre DW, De Silva D, Cole K et al. . WGS to predict antibiotic MICs for Neisseria gonorrhoeae. J Antimicrob Chemoth 2017. DOI: 10.1093/jac/dkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezewudo MN, Joseph SJ, Castillo-Ramirez S et al. . Population structure of Neisseria gonorrhoeae based on whole genome data and its relationship with antibiotic resistance. PeerJ 2015;3:e806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fagnocchi L, Bottini S, Golfieri G et al. . Global transcriptome analysis reveals small RNAs affecting Neisseria meningitidis bacteremia. PLoS One 2015;10:e0126325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleischmann RD, Adams MD, White O et al. . Whole-genome random sequencing and assembly of Haemophilus influenzae RD. Science 1995;269:496–512. [DOI] [PubMed] [Google Scholar]
- Forde BM, O’Toole PW. Next-generation sequencing technologies and their impact on microbial genomics. Brief Funct Genomics 2013;12:440–53. [DOI] [PubMed] [Google Scholar]
- Fraser CM, Gocayne JD, White O et al. . The minimal gene complement of Mycoplasma genitalium. Science 1995;270:397–403. [DOI] [PubMed] [Google Scholar]
- Gardner SN, Slezak T, Hall BG. kSNP3.0: SNP detection and phylogenetic analysis of genomes without genome alignment or reference genome. Bioinformatics 2015;31:2877–8. [DOI] [PubMed] [Google Scholar]
- Giuliani MM, Adu-Bobie J, Comanducci M et al. . A universal vaccine for serogroup B meningococcus. P Natl Acad Sci USA 2006;103:10834–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grad YH, Kirkcaldy RD, Trees D et al. . Genomic epidemiology of Neisseria gonorrhoeae with reduced susceptibility to cefixime in the USA: a retrospective observational study. Lancet Infect Dis 2014;14:220–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagman KE, Shafer WM. Transcriptional control of the mtr efflux system of Neisseria gonorrhoeae. J Bacteriol 1995;177:4162–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hao W, Ma JH, Warren K et al. . Extensive genomic variation within clonal complexes of Neisseria meningitidis. Genome Biol Evol 2011;3:1406–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Bray JE, Maiden MC et al. . Genomic analysis of the evolution and global spread of hyper-invasive meningococcal lineage 5. EBioMedicine 2015;2:234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Clemence M, Dillard JP et al. . Genomic analyses of Neisseria gonorrhoeae reveal an association of the gonococcal genetic island with antimicrobial resistance. J Infect 2016;73:578–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison OB, Cole K, Peters J et al. . Genomic analysis of urogenital and rectal Neisseria meningitidis isolates reveals encapsulated hyperinvasive meningococci and coincident multidrug-resistant gonococci. Sex Transm Infect 2017. DOI: 10.1136/sextrans-2016-052781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatherell HA, Colijn C, Stagg HR et al. . Interpreting whole genome sequencing for investigating tuberculosis transmission: a systematic review. BMC Med 2016;14:21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidrich N, Bauriedl S, Barquist L et al. . The primary transcriptome of Neisseria meningitidis and its interaction with the RNA chaperone Hfq. Nucleic Acids Res 2017;45:6147–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinsbroek E, Ladhani S, Gray S et al. . Added value of PCR-testing for confirmation of invasive meningococcal disease in England. J Infect 2013;67:385–90. [DOI] [PubMed] [Google Scholar]
- Hohl M, Ragan MA. Is multiple-sequence alignment required for accurate inference of phylogeny? Syst Biol 2007;56:206–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Wright PR, Li L et al. . Global RNA recognition patterns of post-transcriptional regulators Hfq and CsrA revealed by UV crosslinking in vivo. EMBO J 2016;35:991–1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huis in 't Veld RA, Willemsen AM, van Kampen AH et al. . Deep sequencing whole transcriptome exploration of the sigmaE regulon in Neisseria meningitidis. PLoS One 2011;6:e29002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobsson S, Golparian D, Cole M et al. . WGS analysis and molecular resistance mechanisms of azithromycin-resistant (MIC > 2 mg/L) Neisseria gonorrhoeae isolates in Europe from 2009 to 2014. J Antimicrob Chemoth 2016;71:3109–16. [DOI] [PubMed] [Google Scholar]
- Jamet A, Jousset AB, Euphrasie D et al. . A new family of secreted toxins in pathogenic Neisseria species. PLoS Pathog 2015;11:e1004592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 2010;11:595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Maiden MC. Automated extraction of typing information for bacterial pathogens from whole genome sequence data: Neisseria meningitidis as an exemplar. Euro Surveill 2013;18:20379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jolley KA, Bliss CM, Bennett JS et al. . Ribosomal Multi-Locus Sequence Typing: universal characterization of bacteria from domain to strain. Microbiology 2012;158:1005–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B, Schneiker-Bekel S, Schramm-Gluck A et al. . Comparative genome biology of a serogroup B carriage and disease strain supports a polygenic nature of meningococcal virulence. J Bacteriol 2010;192:5363–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph SJ, Li B, Petit Iii RA et al. . The single-species metagenome: subtyping Staphylococcus aureus core genome sequences from shotgun metagenomic data. PeerJ 2016;4:e2571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr JR, Sanghvi JC, Macklin DN et al. . A whole-cell computational model predicts phenotype from genotype. Cell 2012;150:389–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klughammer J, Dittrich M, Blom J et al. . Comparative genome sequencing reveals within-host genetic changes in Neisseria meningitidis during invasive disease. PLoS One 2017;12: e0169892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lees JA, Kremer PH, Manso AS et al. . Large scale genomic analysis shows no evidence for pathogen adaptation between the blood and cerebrospinal fluid niches during bacterial meningitis. Microbial Genomics 2017;3:e000103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009;25:1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A et al. . The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009;25:2078–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas CE, Balthazar JT, Hagman KE et al. . The MtrR repressor binds the DNA sequence between the mtrR and mtrC genes of Neisseria gonorrhoeae. J Bacteriol 1997;179:4123–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucidarme J, Hill DM, Bratcher HB et al. . Genomic resolution of an aggressive, widespread, diverse and expanding meningococcal serogroup B, C and W lineage. J Infect 2015;71:544–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure R, Nudel K, Massari P et al. . The gonococcal transcriptome during infection of the lower genital tract in women. PLoS One 2015;10:e0133982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, Harrison OB. The population and functional genomics of the Neisseria revealed with gene-by-gene approaches. J Clin Microbiol 2016;54:1949–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiden MC, Jansen van Rensburg MJ, Bray JE et al. . MLST revisited: the gene-by-gene approach to bacterial genomics. Nat Rev Microbiol 2013;11:728–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin IM, Ison CA. Detection of mixed infection of Neisseria gonorrhoeae. Sex Transm Infect 2003;79:56–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol 1980;65:499–560. [DOI] [PubMed] [Google Scholar]
- Mellin JR, McClure R, Lopez D et al. . Role of Hfq in iron-dependent and -independent gene regulation in Neisseria meningitidis. Microbiology 2010;156:2316–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendum TA, Newcombe J, Mannan AA et al. . Interrogation of global mutagenesis data with a genome scale model of Neisseria meningitidis to assess gene fitness in vitro and in sera. Genome Biol 2011;12:R127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer J, Brissac T, Frapy E et al. . Characterization of MDAPhi, a temperate filamentous bacteriophage of Neisseria meningitidis. Microbiology 2016;162:268–82. [DOI] [PubMed] [Google Scholar]
- Mohammed H, Ison CA, Obi C et al. . Frequency and correlates of culture-positive infection with Neisseria gonorrhoeae in England: a review of sentinel surveillance data. Sex Transm Infect 2015;91:287–93. [DOI] [PubMed] [Google Scholar]
- Monk JM, Charusanti P, Aziz RK et al. . Genome-scale metabolic reconstructions of multiple Escherichia coli strains highlight strain-specific adaptations to nutritional environments. Proceedings of the National Academy of Sciences U S A 2013;110:20338–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MRF-MGL MRF Meningococcus Genome Library. Meningitis Research Foundation http://www.meningitis.org/research/genome.
- Mulhall RM, Brehony C, O’Connor L et al. . Resolution of a protracted serogroup B meningococcal outbreak with whole genome sequencing shows inter species genetic transfer. J Clin Microbiol 2016;54:2891–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha MM, Marsh JW, Krauland MG et al. . Genomic epidemiology of hypervirulent serogroup W, ST-11 Neisseria meningitidis. EBioMedicine 2015;2:1447–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mustapha MM, Marsh JW, Krauland MG et al. . Genomic investigation reveals highly conserved, mosaic, recombination events associated with capsular switching among invasive Neisseria meningitidis serogroup W sequence type (ST)-11 strains. Geno Biol Evol 2016;8:2065–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohneck EA, Zalucki YM, Johnson PJ et al. . A novel mechanism of high-level, broad-spectrum antibiotic resistance caused by a single base pair change in Neisseria gonorrhoeae. Mbio 2011;2:e00187–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Omer H, Rose G, Jolley KA et al. . Genotypic and phenotypic modifications of Neisseria meningitidis after an accidental human passage. PLos One 2011;6:-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ondov BD, Treangen TJ, Melsted P et al. . Mash: fast genome and metagenome distance estimation using MinHash. Genome Biol 2016;17:132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page AJ, Cummins CA, Hunt M et al. . Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015;31:3691–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pannekoek Y, Huis In 't Veld RA, Schipper K et al. . Neisseria meningitidis uses sibling small regulatory RNAs to switch from cataplerotic to anaplerotic metabolism. Mbio 2017;8:pii:e02293-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parkhill J, Achtman M, James KD et al. . Complete DNA sequence of a serogroup A strain of Neisseria meningitidis Z2491. Nature 2000;404:502–6. [DOI] [PubMed] [Google Scholar]
- Renner P, Roger T, Bochud PY et al. . A functional microsatellite of the macrophage migration inhibitory factor gene associated with meningococcal disease. FASEB J 2012;26:907–16. [DOI] [PubMed] [Google Scholar]
- Retchless AC, Hu F, Ouedraogo AS et al. . The establishment and diversification of epidemic-associated serogroup W meningococcus in the African Meningitis Belt, 1994 to 2012. mSphere 2016;1:e00201–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richter M, Rossello-Mora R. Shifting the genomic gold standard for the prokaryotic species definition. P Natl Acad Sci USA 2009;106:19126–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roach JC, Boysen C, Wang K et al. . Pairwise end sequencing: a unified approach to genomic mapping and sequencing. Genomics 1995;26:345–53. [DOI] [PubMed] [Google Scholar]
- Rusniok C, Vallenet D, Floquet S et al. . NeMeSys: a biological resource for narrowing the gap between sequence and function in the human pathogen Neisseria meningitidis. Genome Biol 2009;10:R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. P Natl Acad Sci USA 1977;74:5463–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoen C, Kischkies L, Elias J et al. . Metabolism and virulence in Neisseria meningitidis. Front Cell Infect Microbiol 2014;4:114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics 2014;30:2068–9. [DOI] [PubMed] [Google Scholar]
- Seib KL, Jen FE, Tan A et al. . Specificity of the ModA11, ModA12 and ModD1 epigenetic regulator N6-adenine DNA methyltransferases of Neisseria meningitidis. Nucleic Acids Res 2015;43:4150–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma CM, Hoffmann S, Darfeuille F et al. . The primary transcriptome of the major human pathogen Helicobacter pylori. Nature 2010;464:250–5. [DOI] [PubMed] [Google Scholar]
- Sittka A, Lucchini S, Papenfort K et al. . Deep sequencing analysis of small noncoding RNA and mRNA targets of the global post-transcriptional regulator, Hfq. PLoS Genet 2008;4:e1000163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Forstner KU, Holmqvist E et al. . Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. P Natl Acad Sci USA 2016;113:11591–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spratt BG. Exploring the concept of clonality in bacteria. Methods Mol Biol 2004;266:323–52. [DOI] [PubMed] [Google Scholar]
- Stefanelli P, Fazio C, Neri A et al. . Genome-based study of a spatio-temporal cluster of invasive meningococcal disease due to Neisseria meningitidis serogroup C, clonal complex 11. J Infect 2016;73:136–44. [DOI] [PubMed] [Google Scholar]
- Taha MK, Claus H, Lappann M et al. . Evolutionary events associated with an outbreak of meningococcal disease in men who have sex with men. PLoS One 2016;11:e0154047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan FY, Wormann ME, Loh E et al. . Characterization of a novel antisense RNA in the major pilin locus of Neisseria meningitidis influencing antigenic variation. J Bacteriol 2015;197:1757–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tettelin H, Saunders NJ, Heidelberg J et al. . Complete genome sequence of Neisseria meningitidis serogroup B strain MC58. Science 2000;287:1809–15. [DOI] [PubMed] [Google Scholar]
- Toh E, Gangaiah D, Batteiger BE et al. . Neisseria meningitidis ST11 complex isolates associated with nongonococcal urethritis, Indiana, USA, 2015-2016. Emerg Infect Dis 2017;23:336–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Treangen TJ, Ondov BD, Koren S et al. . The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol 2014;15:524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Dijk EL, Auger H, Jaszczyszyn Y et al. . Ten years of next-generation sequencing technology. Trends Genet 2014;30:418–26. [DOI] [PubMed] [Google Scholar]
- Wachter J, Hill SA. Small transcriptome analysis indicates that the enzyme RppH influences both the quality and quantity of sRNAs in Neisseria gonorrhoeae. FEMS Microbiol Lett 2015;362:1–7. DOI: 10.1093/femsle/fnu059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Cohn A, Comanducci M et al. . Prevalence and genetic diversity of candidate vaccine antigens among invasive Neisseria meningitidis isolates in the United States. Vaccine 2011;29:4739–44. [DOI] [PubMed] [Google Scholar]
- Wang X, Theodore MJ, Mair R et al. . Clinical validation of multiplex real-time PCR assays for detection of bacterial meningitis pathogens. J Clin Microbiol 2012;50:702–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber JL, Myers EW. Human whole-genome shotgun sequencing. Genome Res 1997;7:401–9. [DOI] [PubMed] [Google Scholar]
- Westermann AJ, Forstner KU, Amman F et al. . Dual RNA-seq unveils noncoding RNA functions in host-pathogen interactions. Nature 2016;529:496–501. [DOI] [PubMed] [Google Scholar]
- Wyres KL, Lambertsen LM, Croucher NJ et al. . Pneumococcal capsular switching: a historical perspective. J Infect Dis 2013;207:439–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino D. Using the velvet de novo assembler for short-read sequencing technologies. Curr Proto Bioinformat 2010;11:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Heidrich N, Ampattu BJ et al. . Processing-independent CRISPR RNAs limit natural transformation in Neisseria meningitidis. Mol Cell 2013;50:488–503. [DOI] [PMC free article] [PubMed] [Google Scholar]