Abstract
The identification of genetic factors associated with kidney disease has the potential to provide critical insights into disease mechanisms. Genome-wide association studies have uncovered genomic regions associated with renal function metrics and risk of CKD. UMOD is among the most outstanding loci associated with CKD in the general population, because it has a large effect on eGFR and CKD risk that is consistent across different ethnic groups. The relevance of UMOD for CKD is clear, because the encoded protein, uromodulin (Tamm–Horsfall protein), is exclusively produced by the kidney tubule and has specific biochemical properties that mediate important functions in the kidney and urine. Rare mutations in UMOD are the major cause of autosomal dominant tubulointerstitial kidney disease, a condition that leads to CKD and ESRD. In this brief review, we use the UMOD paradigm to describe how population genetic studies can yield insight into the pathogenesis and prognosis of kidney diseases.
CKD is a global public health burden, affecting as many as 10%–15% of the population worldwide and exceeding 20% in individuals about 60 years old.1 Even in the early stages, CKD is associated with increased prevalence and severity of multiple disorders and adverse outcomes, and it is a major risk factor for accelerated cardiovascular disease and aging.2 Very few pharmacologic interventions have been developed specifically for treating CKD, primarily due to the lack of mechanistic understanding and the lack of biomarkers reflecting the severity of organ damage.
The familial clustering of kidney disorders has long been recognized, and the genetic predisposition to such diseases is emerging as a major source of insights into mechanisms potentially relevant for CKD. In less than a decade, genome-wide studies yielded major breakthroughs in our knowledge of the genetic architecture of kidney diseases, including the role of APOL1 in progression of various nephropathies in individuals of African ancestry,3 the strong signals (HLA-DQA1 and PLA2R1) associated with the risk of membranous nephropathy,4 and the pathogenesis of IgA nephropathy.5 In parallel, the development and implementation of genetic testing, in particular next generation sequencing, allowed to identify the genetic basis of a large number of rare kidney diseases.6 Emerging analyses of the combined effect of rare mutations and modifier gene variants help to explain some phenotype heterogeneity observed in these disorders.7–9
Since 2007, genome-wide association studies (GWASs) have become a very popular tool to uncover genomic regions associated with renal function metrics and CKD risk.10 The success of GWASs was built on their conceptual simplicity and hypothesis-free, unbiased approach.11 Typically, genetic variants uncovered through GWAS are associated with a relatively small increase in disease risk, and their identification requires large study populations.12 The GWASs performed in nephrology were first addressing CKD-defining traits, such as the eGFR on the basis of serum creatinine (eGFRcrea) and the urinary albumin-to-creatinine ratio (UACR), whereas a second group of studies focused on specific CKD etiologies, such as IgA nephropathy or membranous nephropathy. Of note, the latter GWASs have been successful in small- to modest-sized cohorts in identifying common alleles that confer large effects.12
To date, without including studies on diabetic nephropathy, there are >20 GWASs of CKD-defining traits,10,13–32 including one in pediatric subjects25 and two on renal decline.24,30 Of these studies, only a few considered the study of CKD in addition to eGFRcrea or UACR. Most of them limited their analyses to eGFRcrea<60 ml/min per 1.73 m2,10,13,15,16,18,20,22,26 some of them extended their analyses to eGFRcrea<45 ml/min per 1.73 m2,20 and only a few focused their analyses on ESRD.19,23 Studies focusing on CKD on the basis of eGFRcrea did not identify any further genome-wide significant locus other than those already identified by the analysis of eGFRcrea. Taking all studies together, >60 loci are now reported in association with CKD-defining traits, most of them with eGFRcrea in European ancestry individuals.26,29,31
Among all of the loci associated with CKD in the general population, the UMOD one is of particular interest due to its large effect on both eGFR and CKD risk, the consistency of the effect across different ethnic groups, and its relationship with age.33 The relevance of UMOD for CKD is immediate, because the encoded protein, uromodulin (Tamm–Horsfall protein), is exclusively produced by the kidney tubule and has specific biochemical properties that sustain its role in the kidney and urine. Furthermore, rare mutations in UMOD are the major cause of autosomal dominant tubulointerstitial kidney disease (ADTKD), a condition that leads to CKD and ESRD.34 Because UMOD is involved in a continuum between rare mutations and common regulatory variants, physiologically relevant, biochemical investigations of uromodulin are essential to understand its complex role—in the kidney and beyond. This brief review will describe how population genetic studies evidenced risk loci for CKD and related traits and will use the UMOD paradigm to show how this genetic signal may yield pathogenesis and prognostic insights into kidney diseases.
Discovering Genetic Risk Factors for CKD
In recent years, GWASs established themselves as the most powerful tool to detect new genetic variants associated with common diseases. Using this technique, thousands of loci have been associated with several biomarkers and complex disorders. The GWAS technique is rather simple: it consists of running an association test millions of times over the entire genome using linear, logistic, or survival regression models, depending on whether a quantitative, dichotomous, or survival outcome is considered respectively. Models are fitted for the same outcome on one single-nucleotide polymorphism (SNP) at a time. SNPs represent common genetic variations between human subjects and are measured using commercial genotyping arrays. After obtaining a first set of directly genotyped SNPs, the number of SNPs can be expanded via genetic imputation, obtaining a coverage of the genome that is at least ten times larger than the initial set of genotyped SNPs. Imputation is performed using probabilistic methods (usually Markov Chain Monte Carlo) on the basis of large sets of reference fully sequenced genomes, such as those of the 1000 Genomes Consortium or the Haplotype Reference Consortium (HRC).35,36 As an example, a recent GWAS of kidney function traits has been run on nearly 11 million SNPs and insertion/deletion markers.31 Exhaustive overviews of the GWAS method with applications in nephrology have been given previously.11,37
Among the renal function metrics, eGFRcrea has been probably the most studied outcome.12 After uncovering the UMOD locus as associated with eGFRcrea,13 over 60 loci were found in association with this trait in European,13,15,16,17,20,26,31 Asian,22 and mixed ethnic groups.28 Additional results were provided by rare mutation and exome-chip analyses.29,38 The strong transethnic differences in renal function regulation are especially highlighted in the study of albuminuria.32 For the UACR trait, a few but well characterized loci have been identified, including the CUBN gene identified in European, black, and Southern American populations39,40; the RAB38 and HS6ST1 genes identified in diabetic European ancestry subjects27; the BCL2L11 gene identified via genome-wide admixture analysis in Amerindians41; and the HBB gene identified via candidate gene approach but showing genome-wide significance in Africans and Southern Americans.42
In contrast with the successful GWASs of CKD-defining traits, it has been difficult to perform GWASs of clinically defined CKD (i.e., kidney damage or GFR<60 ml/min per 1.73 m2 for at least 3 months).2 Taking into account the heterogeneity of disease etiologies and the small genetic effects that are anticipated for complex diseases, such as diabetes or hypertension, GWASs of clinically defined CKD require large sample sizes that are realistically difficult to obtain.12 To overcome the sample size issue, cross-sectional studies carried out in the general adult population were used.15,22,28 In these studies, CKD 3+ was defined as eGFRcrea<60 ml/min per 1.73 m2 on the basis of a single serum creatinine assessment. No genome-wide significant loci have been identified out of those already reported as significantly associated with eGFRcrea.13,15,20,22,26 Attempts to decrease the eGFRcrea threshold to 45 ml/min per 1.73 m2 did not successfully identified new loci due to lack of power.20 Cross-sectional studies may perform GWASs on CKD stage 3+, but they would miss patients with the most severe cases of CKD stages 4 and 5. Conversely, enrichment for patients with ESRD would then miss the earlier, asymptomatic stages of disease. A recent transethnic analysis showed that most of the eGFRcrea-associated loci are associated with increased risk of CKD defined as eGFRcrea<60 ml/min per 1.73 m2 or incident ESRD.28 Böger et al.43 showed that most of the eGFRcrea-associated loci were also associated with incident CKD or ESRD after 7 years median follow-up. Taken together, these findings support the evidence that, although it is challenging to perform gene discovery analyses on clinically defined CKD, the analysis of CKD-defining traits is providing insight into the biologic pathways underlying CKD.
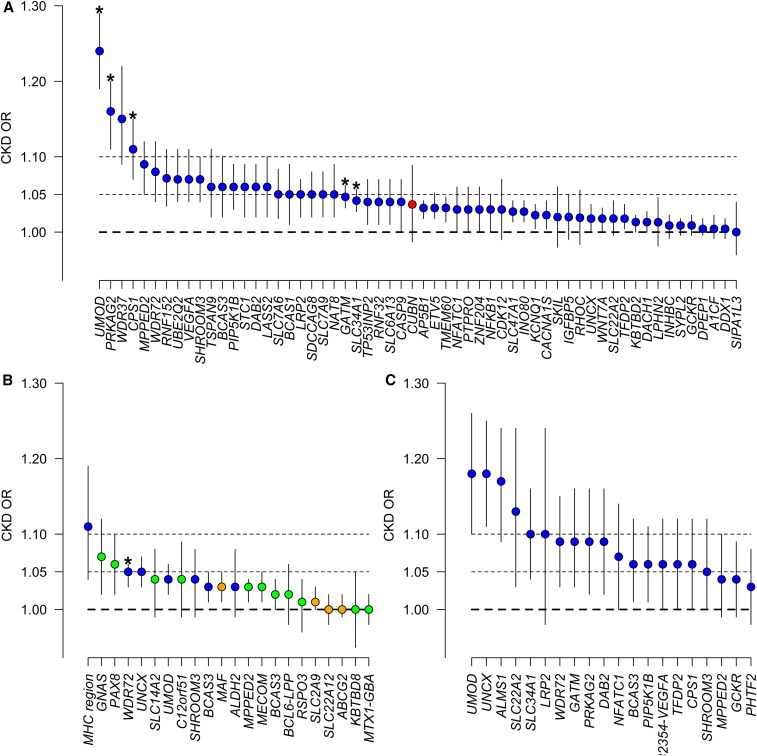
An overview of the effect of different genetic loci on CKD for different ethnicities (Europeans, Asians, and multiethnic) is given in Figure 1. In general, odds ratios (ORs) are small, indicating that each additional copy of the risk allele at each locus confers only a modest increase in the risk of CKD. Just a handful of SNPs would have shown genome-wide significance in a GWAS of CKD, including UMOD, PRKAG2, CPS1, ALMS1-NAT8, and GATM in Europeans (Figure 1A) and WDR72 in Asians (Figure 1B). Among these, loci, such as GATM for instance, reflect the creatinine production pathway11 and probably have little to do with the risk of CKD. Notably, several loci are associated with CKD across ethnicities, including UMOD, SHROOM3, MPPED2, BCAS3, and UNCX. The analysis also indicates that the P value criterion, which drives GWAS discovery, does not reflect effect size but precision. Loci with small but consistent effect across studies will get lower P values than loci with larger but variable effect between populations, maybe because of population-specific features increasing heterogeneity of genetic effects.
Figure 1.
Genetic loci for CKD have small effects. ORs for CKD at the loci showing genome-wide significant association with CKD underlying traits. The ORs are reported along with their 95% CIs. The underlying trait that led to uncover each specific locus is identified with a different color: eGFRcrea (blue), albuminuria (red), BUN (green), and uric acid (orange). (A) OR for CKD in the general population of European ancestry at loci reported by the CKDGen Consortium GWAS.26,31 Given that no GWAS of CKD was performed by Gorski et al.31 for these loci and the albuminuria locus cubilin (CUBN),39 ORs for CKD were obtained by a lookup of the GWAS results published in the work by Pattaro et al.26 Given the different genetic imputation platforms (HapMap versus 1000 Genomes), not all loci were available. (B) OR for CKD in the general population of Asian ancestry as reported by the AGEN Consortium.22 *Genome-wide significance for CKD. (C) OR for CKD, defined as eGFRcrea<60 or ESRD, in a general mixed population of European, African, Asian, and Hispanic ancestry.28
Discovery and Associations of the UMOD Locus
The UMOD locus stands out among the loci associated with CKD. In European ancestry individuals, UMOD was the first locus uncovered in association with eGFRcrea, and it was already associated with CKD at a genome-wide significance level.13 CKD association at the UMOD locus is related to the three SNPs in linkage disequilibrium located in the promoter. For these SNPs, the “risk” allele is the common allele, which has a frequency of approximately 80% in Europeans.44 As shown in Figure 1, UMOD shows the largest OR for CKD compared with all other loci in nearly all ethnicities. The OR for CKD is 1.24 (95% confidence interval [95% CI], 1.19 to 1.29) per copy of the UMOD risk allele,26 rising up to 1.35 (95% CI, 1.18 to 1.54) if eGFRcrea<45 ml/min per 1.73 m2 is considered.20 UMOD is, to date, the only locus associated with incident CKD at a genome-wide level of significance, with an OR of about 1.3 per copy of the risk allele, which remained unchanged after baseline eGFRcrea adjustment.43 Furthermore, UMOD is also associated with incident ESRD.43 If all eGFRcrea loci together explain nearly 4% of the trait variance,31 >1% is explained by UMOD alone. Accordingly, UMOD accounts for 25% of the eGFRcrea variability explained by genetic factors.
Recent studies show that the effect of the UMOD eGFRcrea-lowering allele is double in diabetic individuals compared with nonpatients with diabetes (approximately −2.7% versus −1.5% eGFRcrea per copy of the risk allele in patients with diabetes versus nonpatients with diabetes, respectively; P value for difference =0.02).26 However, it is possible that the observed difference by diabetes status in the CKDGen Consortium is confounded by age. In fact, age was shown to interact with UMOD in its association with serum creatinine and CKD.17
In addition to eGFRcrea and CKD, UMOD has been associated with the risk of hypertension and incident cardiovascular disease events in an extreme patient-control study from the Global BPGen Consortium.45 UMOD has also been significantly associated with uric acid levels in an Icelandic cohort17 and a Chinese community–based cohort,46 and it has been associated with reduced risk of kidney stones (OR, 0.88; 95% CI, 0.83 to 0.94) in a combined sample of Icelandic and Dutch subjects.17 Furthermore, the UMOD promoter variants were highly associated with urinary levels of uromodulin (both 24-hour urinary excretion and uromodulin-to-creatinine ratio) in a meta-analysis of 10,884 individuals of European descent.47
Biologic Relevance of the UMOD Locus
The prior knowledge about uromodulin paralleled by genetic evidence involving UMOD in rare kidney diseases and by translational studies has sustained the biologic relevance of the association of UMOD with renal function and CKD.
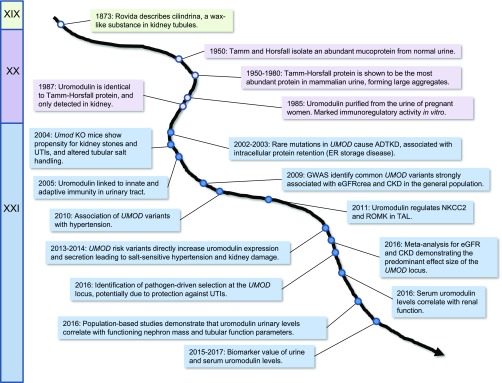
Uromodulin has a long history (Figure 2). First described by Rovida48 as a wax-like material in kidney tubules, the protein was isolated from the urine in 1950 by Tamm and Horsfall49 from the Rockefeller Institute in New York. The Tamm–Horsfall protein, as it was named from there, was found to be the most abundant protein in normal human urine, characterized by a mass of approximately 85 kD, a high carbohydrate content (approximately 30% of the molecular mass), 48 cysteine residues, and a tendency to form aggregates. After the demonstration of an immunoregulatory activity in vitro, the protein was renamed “uromodulin.”50 Uromodulin is exclusively produced in the kidney—by the cells lining the thick ascending limb (TAL) of the loop of Henle.51,52 Studies in uromodulin knockout mice suggested that uromodulin interacts with transporters operating in the TAL to regulate salt transport,53 protects against urinary tract infections (UTIs) via high-affinity binding to the type 1 fimbriae of uropathogenic Escherichia coli,54,55 reduces the propensity to form calcium crystals and kidney stones,56 and plays a role in innate immunity by binding IGs and cytokines and/or activating monocytes and dendritic cells.57,58 In the urine, uromodulin forms a three-dimensional matrix consisting of high molecular weight polymers. The extracellular polymerization is mediated by the proteolytic cleavage of the protein in the apical membrane of TAL cells by the serine protease hepsin.59
Figure 2.
Milestones in the discovery of Tamm–Horsfall/uromodulin protein and the UMOD gene from the XIXth to the XXIst century. ER, endoplasmic reticulum.
In 2002, Hart et al.60 showed that dominant mutations in UMOD are a major cause of ADTKD, a rare disorder often associated with hyperuricemia (due to decreased fractional excretion of urate), gout, and tubulointerstitial damage, invariably leading to CKD and renal failure.34 Most of the UMOD mutations described thus far are missense changes that often replace or delete one of the conserved cysteine residues of the protein, affecting its folding and trafficking to the plasma membrane. This effect has been shown in vitro,61 whereas analysis of patient biopsies has shown typical accumulation of mutant uromodulin in the endoplasmic reticulum, showing that ADTKD-UMOD is, in fact, an endoplasmic reticulum storage disease.62,63
The fact that the top UMOD GWAS variants are in perfect linkage disequilibrium in the promoter of the gene provided an important clue for their biologic relevance.44 Indeed, population studies established that the UMOD promoter variants are strongly associated with urinary levels of uromodulin, each (major) risk allele increasing the urinary excretion of uromodulin in a dose-dependent fashion.47,64 In fact, homozygous carriers of the risk allele had twofold-higher indexed uromodulin levels than the homozygous carriers of the (minor) protective allele.47 Along with eGFR and sodium excretion, the UMOD genotype at rs4293393 was one of the independent predictors of urinary uromodulin levels in a Canadian population.65 Further studies showed that the risk variants exert a robust influence on the activity of the UMOD promoter in vitro reflected by twofold-higher UMOD transcript levels in human kidney biopsies and a higher level of uromodulin in urine.44 The human situation was modeled in a transgenic Umod mouse overexpressing wild-type uromodulin (TgUmodwt): with aging, the kidneys of these mice showed focal lesions and increased expression of damage markers, including lipocalin-2 and Kim-1. These changes, which were not observed in control mice, occurred in absence of renal failure. Similar focal lesions were seen in kidney biopsies from individuals ages >65 years old, and their area was significantly increased in individuals homozygous for UMOD risk variants relative to those carrying protective variants.44 Taken together, these results indicate that increased expression of uromodulin does not lead to renal failure per se. Instead, they support a second hit model, in which high uromodulin production over time predisposes to kidney damage induced by additional comorbidities.44 This model is, in fact, supported by the strong interaction of age with the association of UMOD variants with kidney function.17,20 Other potentially deleterious factors could be the metabolic demand of uromodulin production in TAL cells, the chronic activation of salt reabsorption systems, or the effects of increased uromodulin levels in the lumen of the distal nephron or in the circulation.33
In addition to renal damage and aging lesions, the transgenic Umod mice displayed salt-sensitive hypertension due to activation (phosphorylation) of NKCC2 in the TAL, which was mirrored by increased response to furosemide. An increased response to furosemide was also observed in hypertensive individuals homozygous for the UMOD risk allele.44 The gain-of-function mechanism associated with higher uromodulin expression in this mouse model supports the association of UMOD variants with hypertension44 and completes the link between renal tubular transport of NaCl and BP regulation.66
Prognostic Value: Genotype Versus Biomarker
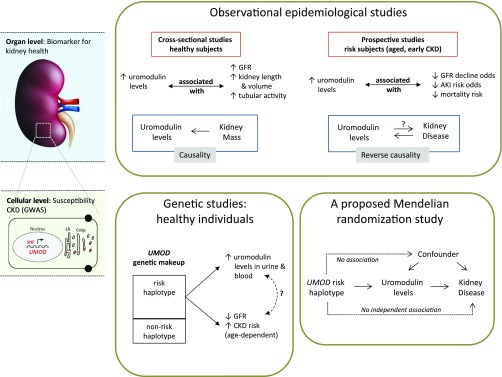
The UMOD locus is associated with eGFR and CKD in the general population. The promoter SNPs constituting the risk haplotype drive increased transcriptional activity and higher levels of uromodulin in kidney and urine and in blood.44,67 Accordingly, one could expect that higher levels of uromodulin would be associated with higher risk of kidney damage in epidemiologic studies. However, recent prospective studies showed that higher levels of uromodulin are protective of kidney damage in at-risk cohorts.67–69 How can we explain these seemingly opposite predictive values of uromodulin levels for the risk of kidney disease (Figure 3)?
Figure 3.
Interpretation of the uromodulin biomarker versus UMOD genotype studies. In observational epidemiologic studies, uromodulin levels largely reflect kidney mass at baseline. Higher levels of uromodulin in urine, representing a higher renal functional reserve, are as such associated with a positive outcome in prospective studies conducted in at-risk individuals. Conversely, decreased levels of uromodulin reflect decreased production and lower functional reserve at baseline. This situation (i.e., when it is the disease [kidney damage] that affects the studied risk factor [uromodulin level]) is typical of reverse causation. In turn, reverse causation represents a bias when evaluating the causality of the UMOD risk haplotype in promoting CKD. Genetic, experimental, and statistical studies in healthy individuals have shown that the UMOD risk haplotype drives the expression of uromodulin in the kidney and its level in urine. The use of analytic methods, such as Mendelian randomization, will provide a valuable opportunity to assess the causality of the UMOD genotype and higher uromodulin levels as a risk factor for CKD. ER, endoplasmic reticulum.
The value of uromodulin as a biomarker originates from its exclusive production by a tubular segment of the kidney, with sizeable levels being detected in normal urine.70,71 Observational studies carried out in the general population showed that urinary uromodulin levels are positively associated with eGFR, markers of tubular transport, and kidney length and volume, suggesting that urinary uromodulin is a biomarker for tubular mass and function.72 A fraction of uromodulin produced in the TAL is released into the circulation.73 Plasma uromodulin concentrations, which are approximately 1000 times lower than urinary levels (nanograms per milliliter versus micrograms per milliliter, respectively), correlate positively with urinary uromodulin excretion and creatinine clearance in patients with CKD.74,75 The levels of uromodulin can thus be considered as biomarkers of kidney tubule function, with higher levels reflecting higher renal functional mass (Figure 3).
From the above, it can be hypothesized that low urinary uromodulin is a marker of poorer tubular health: in at-risk individuals (aged and early CKD), decreased levels of uromodulin reflect decreased production (i.e., decreased renal functional reserve). The latter hypothesis is supported by the studies of Garimella et al.,68 which measured urinary uromodulin levels in 192 participants of the Cardiovascular Health Study over 9 years. Each one SD increase in urinary uromodulin was associated with a 23% lower odds of eGFR decline and a 10% lower risk of mortality. Similarly, lower preoperative urinary uromodulin levels were associated with higher risk of postoperative AKI.69 In these at-risk individuals, decreased levels of uromodulin reflect decreased functional reserve. The latter situation is typical of reverse causation (i.e., when it is the disease [kidney damage] that affects the studied risk factor [uromodulin level]) (Figure 3). Such a situation would then constitute a bias when evaluating the causality of the UMOD genotype and the uromodulin levels in relation to the outcome. The use of analytic methods, such as Mendelian randomization, will provide a valuable opportunity to assess the causality of higher uromodulin levels as a risk factor for CKD.76 Furthermore, even if the UMOD variants show the largest effect size among all known loci for eGFRcrea and CKD, their effect is clearly age dependent and contributing only to a small part of the overall risk of kidney damage, together with many other genetic and environmental factors. Especially regarding age and CKD status, Mendelian randomization studies should take the stratification of study subjects into account to depict a conclusive picture of the causal relation between uromodulin levels and disease risk. Also, one should keep in mind the possibility that the UMOD SNPs may still have additional effects, unrelated to the production of uromodulin, involved in the association with renal disease.
Perspectives
Evolution and Selection
The lead UMOD risk variant (allele T at rs4293393) associated with CKD and hypertension is the ancestral allele, and its prevalence ranges from 70% to 80% in Africans and Europeans to >90% in East Asians. Furthermore, this risk variant directly increases the expression of uromodulin in a dose-dependent fashion, and it causes salt-sensitive hypertension and kidney damage in mice and humans.44 Recent studies suggest that the high prevalence of the ancestral allele could reflect some evolutionary advantage.77 In fact, the ancestral allele shows a global correlation with bacterial diversity (the Human Genome Diversity Project) and the prevalence of antibiotic resistance in Gram-negative uropathogens (i.e., areas with higher prevalence of UTIs). The levels of uromodulin in urine also correlated with urinary leukocytes and nitrites in the population.77 Furthermore, high urinary levels of uromodulin were also associated with lower risk for UTI in older adults from the Cardiovascular Health Study, independent of traditional UTI risk factors.78 Taken together, these studies suggest that the ancestral UMOD allele has been kept at a high frequency due to its protective effect against UTIs, which mainly affect young women and have important consequences in terms of fitness and reproduction. The salt-retaining effect of this variant could also confer an advantage in patients with severe infection, particularly children.79
The persistent high frequencies of the ancestral UMOD allele in areas with stronger selective pressure for its protective effect follow the model of selection on standing variation,80 as observed for the ancestral -3826A allele in UCP1, that has been kept at higher frequencies in populations exposed to low solar radiation.81 Of note, the UMOD situation differs from the ancestral susceptibility model that is applicable when salt retention is the primary motor for evolutionary selection (e.g., variants in the salt-retaining genes CYP3A5 and AGT)77,79 and the selection at the APOL1 locus, where the derived allele rather than the ancestral allele confers a selective advantage against a pathogen.3
Uromodulin as a Target for Treatment
If the causal relation between uromodulin production and the risk of developing CKD is substantiated by MR, the possible benefit of regulating uromodulin production in the TAL by dietary or pharmacologic interventions should be investigated—keeping in mind the age effect associated with the UMOD risk variant.17,44 Another approach could be to modulate the biochemical properties of uromodulin in the urine, which could offer some perspectives for kidney stones, UTIs, and myeloma cast nephropathy.44,82
The opposite effects on BP observed in transgenic and knockout uromodulin mouse models44,83 and the observation of an increased response to furosemide in hypertensive individuals homozygous for the UMOD risk allele44 raise perspectives for a personalized medical approach. For example, the possibility to use a long-acting loop diuretic (torasemide) in patients with resistant hypertension who harbor UMOD promoter risk variants is currently being tested in a prospective study.84
Limitations and Hope for GWAS
The small effect of each individual GWAS locus reflects the polygenic theory that complex diseases are driven by a blend of different pathways, each one responsible of a minimal fraction of the risk of the disease.85,86 This does not mean that genetic findings have little clinical relevance. In fact, as already reported by Köttgen et al.,15 a genetic risk score on the basis of the first 20 loci identified was associated with the doubling of CKD prevalence from about 5% to about 10% in the general population. In addition, regardless of the effect size, each gene identified shed light on a particular pathway. Each of these pathways has the potential to become a therapeutic target to prevent CKD as illustrated in the field of coronary artery disease, where large-scale genetic investigations paved the way to release of targeted therapies into the market.87 In line with such a “blend” theory is the observation that eGFRcrea is not the only phenotype that should be used to uncover genetic loci for CKD: Okada et al.22 showed that loci associated with uric acid and BUN may be associated with CKD at a similar magnitude as eGFRcrea loci. Also, larger meta-GWASs of uromodulin levels may be useful to uncover mechanisms involved in uromodulin production and/or release from TAL cells. In turn, these factors may be viewed as additional traits reflecting renal tubular health.
The fact that all loci reported so far only explain approximately 4% of the total variance of eGFRcrea,31 whereas the genetic heritability of the trait is between 30% and 50%88 means that most of the genetic heritability of eGFRcrea remains unexplained. That pessimistic view could be tempered by growing literature showing that genetic heritability estimates may be inflated89: issues include the design of epidemiologic studies90 and unmodeled genetic and environmental components.91,92
Strategies to expand large genomic studies related to renal function and CKD include larger biobanks with hundreds of thousands of genotyped individuals (e.g., the UK Biobank). The expanded sample size united with improved precision thanks to denser and more precise genetic imputation panels (e.g., the HRC and the 100,000 Genomes Project) will improve the localization of genetic signals associated with a particular phenotype. They will also allow identifying rarer variants (so far, loci uncovered for eGFRcrea were all with a minor allele frequency [MAF] of ≥10% in Europeans). The capacity to go down at least to 1%, hopefully to 0.5%, MAF means getting larger effects and thus, stronger predictive ability. A limitation of previous efforts, such as the CKDGen Consortium, was the analysis of the adult population only. To gain insight into kidney function biology, it would be important to expand efforts, such as those of the PediGFR Consortium.25 Identifying loci associated with renal function before aging-related comorbidities (hypertension, diabetes, and infections) will highlight mechanisms regulating normal renal activity, which may be attractive targets. Before large sequencing studies become available, increased precision in genotyping can be achieved with denser and better imputed genetic maps as illustrated by the discovery of rare variants (MAF<2%) associated with serum creatinine and CKD in an Icelandic cohort.38 By focusing on markers located in the coding regions of the genes, exome chip analysis has the potential to discover rare variants more likely to have a functional value as recently shown by Li et al.29 By exploiting the value of next generation sequencing technologies, future studies should consider fine mapping of top GWAS loci, because it was shown that such loci may carry multiple independent rare variants that substantially increase the amount of explained heritability.93
In conclusion, UMOD has been identified as one of the most outstanding loci associated with renal function parameters and risk of CKD in the general population. Multidisciplinary studies have shown the biologic relevance of the top UMOD variants, which are also associated with hypertension and kidney stones. The fact that UMOD encodes a protein that is exclusively produced by a subset of renal tubular cells and abundantly excreted in the urine offers a unique situation to address the causality between UMOD variants, production of uromodulin, its levels in urine and blood, and risk of disease. These studies should also address the potential of rare variations in UMOD to explain its overall contribution to kidney disease. Examination of public databases, such as the Exome Aggregation Consortium and the Genome Aggregation Database, reveals that UMOD is not mutation intolerant, and there are a fair number of people who have severe UMOD mutations (stop gained, missense involving cysteine residues, or frameshift). Phenotype studies of such individuals may reveal more about the biology of the protein and potential reasons for selection. Considering that the top UMOD variants show pleiotropic effects on multiple phenotypes, a phenome-wide association study approach may uncover additional clinical associations94 and reveal biologic underpinnings involving this fascinating gene.
Disclosures
None.
Acknowledgments
We thank Guido Barbujani, Anthony Bleyer, Murielle Bochud, Kai-Uwe Eckardt, Caroline S. Fox, Barry Freedman, Anna Köttgen, François Madore, Eric Olinger, Belen Ponte, Menno Pruijm, Luca Rampoldi, Rajesh V. Thakker, Stephane Troyanov, Peter Vollenweider, Gérard Waeber, and Sonia Youhanna for fruitful discussions.
O.D. is supported by the Swiss National Centre of Competence in Research Kidney Control of Homeostasis program, Swiss National Science Foundation grant 31003A_169850, and the Rare Disease Initiative Zurich (Radiz), a clinical research priority program of the University of Zurich.
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
References
- 1.Levin A, Tonelli M, Bonventre J, Coresh J, Donner JA, Fogo AB, Fox CS, Gansevoort RT, Heerspink HJL, Jardine M, Kasiske B, Köttgen A, Kretzler M, Levey AS, Luyckx VA, Mehta R, Moe O, Obrador G, Pannu N, Parikh CR, Perkovic V, Pollock C, Stenvinkel P, Tuttle KR, Wheeler DC, Eckardt KU; ISN Global Kidney Health Summit participants : Global kidney health 2017 and beyond: A roadmap for closing gaps in care, research, and policy [published online ahead of print April 20, 2017]. Lancet 10.1016/S0140-6736(17)30788-2 [DOI] [PubMed]
- 2.Eckardt KU, Coresh J, Devuyst O, Johnson RJ, Köttgen A, Levey AS, Levin A: Evolving importance of kidney disease: From subspecialty to global health burden. Lancet 382: 158–169, 2013 [DOI] [PubMed] [Google Scholar]
- 3.Genovese G, Friedman DJ, Ross MD, Lecordier L, Uzureau P, Freedman BI, Bowden DW, Langefeld CD, Oleksyk TK, Uscinski Knob AL, Bernhardy AJ, Hicks PJ, Nelson GW, Vanhollebeke B, Winkler CA, Kopp JB, Pays E, Pollak MR: Association of trypanolytic ApoL1 variants with kidney disease in African Americans. Science 329: 841–845, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanescu HC, Arcos-Burgos M, Medlar A, Bockenhauer D, Kottgen A, Dragomirescu L, Voinescu C, Patel N, Pearce K, Hubank M, Stephens HA, Laundy V, Padmanabhan S, Zawadzka A, Hofstra JM, Coenen MJ, den Heijer M, Kiemeney LA, Bacq-Daian D, Stengel B, Powis SH, Brenchley P, Feehally J, Rees AJ, Debiec H, Wetzels JF, Ronco P, Mathieson PW, Kleta R: Risk HLA-DQA1 and PLA(2)R1 alleles in idiopathic membranous nephropathy. N Engl J Med 364: 616–626, 2011 [DOI] [PubMed] [Google Scholar]
- 5.Kiryluk K, Li Y, Scolari F, Sanna-Cherchi S, Choi M, Verbitsky M, Fasel D, Lata S, Prakash S, Shapiro S, Fischman C, Snyder HJ, Appel G, Izzi C, Viola BF, Dallera N, Del Vecchio L, Barlassina C, Salvi E, Bertinetto FE, Amoroso A, Savoldi S, Rocchietti M, Amore A, Peruzzi L, Coppo R, Salvadori M, Ravani P, Magistroni R, Ghiggeri GM, Caridi G, Bodria M, Lugani F, Allegri L, Delsante M, Maiorana M, Magnano A, Frasca G, Boer E, Boscutti G, Ponticelli C, Mignani R, Marcantoni C, Di Landro D, Santoro D, Pani A, Polci R, Feriozzi S, Chicca S, Galliani M, Gigante M, Gesualdo L, Zamboli P, Battaglia GG, Garozzo M, Maixnerová D, Tesar V, Eitner F, Rauen T, Floege J, Kovacs T, Nagy J, Mucha K, Pączek L, Zaniew M, Mizerska-Wasiak M, Roszkowska-Blaim M, Pawlaczyk K, Gale D, Barratt J, Thibaudin L, Berthoux F, Canaud G, Boland A, Metzger M, Panzer U, Suzuki H, Goto S, Narita I, Caliskan Y, Xie J, Hou P, Chen N, Zhang H, Wyatt RJ, Novak J, Julian BA, Feehally J, Stengel B, Cusi D, Lifton RP, Gharavi AG: Discovery of new risk loci for IgA nephropathy implicates genes involved in immunity against intestinal pathogens. Nat Genet 46: 1187–1196, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Devuyst O, Knoers NV, Remuzzi G, Schaefer F; Board of the Working Group for Inherited Kidney Diseases of the European Renal Association and European Dialysis and Transplant Association : Rare inherited kidney diseases: Challenges, opportunities, and perspectives. Lancet 383: 1844–1859, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis EE, Zhang Q, Liu Q, Diplas BH, Davey LM, Hartley J, Stoetzel C, Szymanska K, Ramaswami G, Logan CV, Muzny DM, Young AC, Wheeler DA, Cruz P, Morgan M, Lewis LR, Cherukuri P, Maskeri B, Hansen NF, Mullikin JC, Blakesley RW, Bouffard GG, Gyapay G, Rieger S, Tönshoff B, Kern I, Soliman NA, Neuhaus TJ, Swoboda KJ, Kayserili H, Gallagher TE, Lewis RA, Bergmann C, Otto EA, Saunier S, Scambler PJ, Beales PL, Gleeson JG, Maher ER, Attié-Bitach T, Dollfus H, Johnson CA, Green ED, Gibbs RA, Hildebrandt F, Pierce EA, Katsanis N; NISC Comparative Sequencing Program : TTC21B contributes both causal and modifying alleles across the ciliopathy spectrum. Nat Genet 43: 189–196, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tory K, Menyhárd DK, Woerner S, Nevo F, Gribouval O, Kerti A, Stráner P, Arrondel C, Huynh Cong E, Tulassay T, Mollet G, Perczel A, Antignac C: Mutation-dependent recessive inheritance of NPHS2-associated steroid-resistant nephrotic syndrome. Nat Genet 46: 299–304, 2014 [DOI] [PubMed] [Google Scholar]
- 9.Bullich G, Trujillano D, Santín S, Ossowski S, Mendizábal S, Fraga G, Madrid Á, Ariceta G, Ballarín J, Torra R, Estivill X, Ars E: Targeted next-generation sequencing in steroid-resistant nephrotic syndrome: Mutations in multiple glomerular genes may influence disease severity. Eur J Hum Genet 23: 1192–1199, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hwang SJ, Yang Q, Meigs JB, Pearce EN, Fox CS: A genome-wide association for kidney function and endocrine-related traits in the NHLBI’s Framingham Heart Study. BMC Med Genet 8[Suppl 1]: S10, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.O’Seaghdha CM, Fox CS: Genome-wide association studies of chronic kidney disease: What have we learned? Nat Rev Nephrol 8: 89–99, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wuttke M, Köttgen A: Insights into kidney diseases from genome-wide association studies. Nat Rev Nephrol 12: 549–562, 2016 [DOI] [PubMed] [Google Scholar]
- 13.Köttgen A, Glazer NL, Dehghan A, Hwang SJ, Katz R, Li M, Yang Q, Gudnason V, Launer LJ, Harris TB, Smith AV, Arking DE, Astor BC, Boerwinkle E, Ehret GB, Ruczinski I, Scharpf RB, Chen YD, de Boer IH, Haritunians T, Lumley T, Sarnak M, Siscovick D, Benjamin EJ, Levy D, Upadhyay A, Aulchenko YS, Hofman A, Rivadeneira F, Uitterlinden AG, van Duijn CM, Chasman DI, Paré G, Ridker PM, Kao WH, Witteman JC, Coresh J, Shlipak MG, Fox CS: Multiple loci associated with indices of renal function and chronic kidney disease. Nat Genet 41: 712–717, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pattaro C, De Grandi A, Vitart V, Hayward C, Franke A, Aulchenko YS, Johansson A, Wild SH, Melville SA, Isaacs A, Polasek O, Ellinghaus D, Kolcic I, Nöthlings U, Zgaga L, Zemunik T, Gnewuch C, Schreiber S, Campbell S, Hastie N, Boban M, Meitinger T, Oostra BA, Riegler P, Minelli C, Wright AF, Campbell H, van Duijn CM, Gyllensten U, Wilson JF, Krawczak M, Rudan I, Pramstaller PP; EUROSPAN consortium : A meta-analysis of genome-wide data from five European isolates reveals an association of COL22A1, SYT1, and GABRR2 with serum creatinine level. BMC Med Genet 11: 41, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Köttgen A, Pattaro C, Böger CA, Fuchsberger C, Olden M, Glazer NL, Parsa A, Gao X, Yang Q, Smith AV, O’Connell JR, Li M, Schmidt H, Tanaka T, Isaacs A, Ketkar S, Hwang SJ, Johnson AD, Dehghan A, Teumer A, Paré G, Atkinson EJ, Zeller T, Lohman K, Cornelis MC, Probst-Hensch NM, Kronenberg F, Tönjes A, Hayward C, Aspelund T, Eiriksdottir G, Launer LJ, Harris TB, Rampersaud E, Mitchell BD, Arking DE, Boerwinkle E, Struchalin M, Cavalieri M, Singleton A, Giallauria F, Metter J, de Boer IH, Haritunians T, Lumley T, Siscovick D, Psaty BM, Zillikens MC, Oostra BA, Feitosa M, Province M, de Andrade M, Turner ST, Schillert A, Ziegler A, Wild PS, Schnabel RB, Wilde S, Munzel TF, Leak TS, Illig T, Klopp N, Meisinger C, Wichmann HE, Koenig W, Zgaga L, Zemunik T, Kolcic I, Minelli C, Hu FB, Johansson A, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Schreiber S, Aulchenko YS, Felix JF, Rivadeneira F, Uitterlinden AG, Hofman A, Imboden M, Nitsch D, Brandstätter A, Kollerits B, Kedenko L, Mägi R, Stumvoll M, Kovacs P, Boban M, Campbell S, Endlich K, Völzke H, Kroemer HK, Nauck M, Völker U, Polasek O, Vitart V, Badola S, Parker AN, Ridker PM, Kardia SL, Blankenberg S, Liu Y, Curhan GC, Franke A, Rochat T, Paulweber B, Prokopenko I, Wang W, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Shlipak MG, van Duijn CM, Borecki I, Krämer BK, Rudan I, Gyllensten U, Wilson JF, Witteman JC, Pramstaller PP, Rettig R, Hastie N, Chasman DI, Kao WH, Heid IM, Fox CS: New loci associated with kidney function and chronic kidney disease. Nat Genet 42: 376–384, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chambers JC, Zhang W, Lord GM, van der Harst P, Lawlor DA, Sehmi JS, Gale DP, Wass MN, Ahmadi KR, Bakker SJ, Beckmann J, Bilo HJ, Bochud M, Brown MJ, Caulfield MJ, Connell JM, Cook HT, Cotlarciuc I, Davey Smith G, de Silva R, Deng G, Devuyst O, Dikkeschei LD, Dimkovic N, Dockrell M, Dominiczak A, Ebrahim S, Eggermann T, Farrall M, Ferrucci L, Floege J, Forouhi NG, Gansevoort RT, Han X, Hedblad B, Homan van der Heide JJ, Hepkema BG, Hernandez-Fuentes M, Hypponen E, Johnson T, de Jong PE, Kleefstra N, Lagou V, Lapsley M, Li Y, Loos RJ, Luan J, Luttropp K, Maréchal C, Melander O, Munroe PB, Nordfors L, Parsa A, Peltonen L, Penninx BW, Perucha E, Pouta A, Prokopenko I, Roderick PJ, Ruokonen A, Samani NJ, Sanna S, Schalling M, Schlessinger D, Schlieper G, Seelen MA, Shuldiner AR, Sjögren M, Smit JH, Snieder H, Soranzo N, Spector TD, Stenvinkel P, Sternberg MJ, Swaminathan R, Tanaka T, Ubink-Veltmaat LJ, Uda M, Vollenweider P, Wallace C, Waterworth D, Zerres K, Waeber G, Wareham NJ, Maxwell PH, McCarthy MI, Jarvelin MR, Mooser V, Abecasis GR, Lightstone L, Scott J, Navis G, Elliott P, Kooner JS: Genetic loci influencing kidney function and chronic kidney disease. Nat Genet 42: 373–375, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gudbjartsson DF, Holm H, Indridason OS, Thorleifsson G, Edvardsson V, Sulem P, de Vegt F, d’Ancona FC, den Heijer M, Wetzels JF, Franzson L, Rafnar T, Kristjansson K, Bjornsdottir US, Eyjolfsson GI, Kiemeney LA, Kong A, Palsson R, Thorsteinsdottir U, Stefansson K: Association of variants at UMOD with chronic kidney disease and kidney stones-role of age and comorbid diseases. PLoS Genet 6: e1001039, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liu CT, Garnaas MK, Tin A, Kottgen A, Franceschini N, Peralta CA, de Boer IH, Lu X, Atkinson E, Ding J, Nalls M, Shriner D, Coresh J, Kutlar A, Bibbins-Domingo K, Siscovick D, Akylbekova E, Wyatt S, Astor B, Mychaleckjy J, Li M, Reilly MP, Townsend RR, Adeyemo A, Zonderman AB, de Andrade M, Turner ST, Mosley TH, Harris TB, Rotimi CN, Liu Y, Kardia SL, Evans MK, Shlipak MG, Kramer H, Flessner MF, Dreisbach AW, Goessling W, Cupples LA, Kao WL, Fox CS; CKDGen Consortium : Genetic association for renal traits among participants of African ancestry reveals new loci for renal function. PLoS Genet 7: e1002264, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kao WH, Klag MJ, Meoni LA, Reich D, Berthier-Schaad Y, Li M, Coresh J, Patterson N, Tandon A, Powe NR, Fink NE, Sadler JH, Weir MR, Abboud HE, Adler SG, Divers J, Iyengar SK, Freedman BI, Kimmel PL, Knowler WC, Kohn OF, Kramp K, Leehey DJ, Nicholas SB, Pahl MV, Schelling JR, Sedor JR, Thornley-Brown D, Winkler CA, Smith MW, Parekh RS, Family Investigation of Nephropathy and Diabetes Research Group: MYH9 is associated with nondiabetic end-stage renal disease in African Americans. Nat Genet 40: 1185–1192, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pattaro C, Köttgen A, Teumer A, Garnaas M, Böger CA, Fuchsberger C, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa M, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson Å, Tönjes A, Dehghan A, Chouraki V, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Kollerits B, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu FB, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Koenig W, Illig T, Döring A, Wichmann HE, Kolcic I, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Endlich K, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Giulianini F, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Metzger M, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki I, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman JC, Hayward C, Ridker P, Parsa A, Bochud M, Heid IM, Goessling W, Chasman DI, Kao WH, Fox CS; CARDIoGRAM Consortium; ICBP Consortium; CARe Consortium; Wellcome Trust Case Control Consortium 2 (WTCCC2) : Genome-wide association and functional follow-up reveals new loci for kidney function. PLoS Genet 8: e1002584, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chasman DI, Fuchsberger C, Pattaro C, Teumer A, Böger CA, Endlich K, Olden M, Chen MH, Tin A, Taliun D, Li M, Gao X, Gorski M, Yang Q, Hundertmark C, Foster MC, O’Seaghdha CM, Glazer N, Isaacs A, Liu CT, Smith AV, O’Connell JR, Struchalin M, Tanaka T, Li G, Johnson AD, Gierman HJ, Feitosa MF, Hwang SJ, Atkinson EJ, Lohman K, Cornelis MC, Johansson A, Tönjes A, Dehghan A, Lambert JC, Holliday EG, Sorice R, Kutalik Z, Lehtimäki T, Esko T, Deshmukh H, Ulivi S, Chu AY, Murgia F, Trompet S, Imboden M, Coassin S, Pistis G, Harris TB, Launer LJ, Aspelund T, Eiriksdottir G, Mitchell BD, Boerwinkle E, Schmidt H, Cavalieri M, Rao M, Hu F, Demirkan A, Oostra BA, de Andrade M, Turner ST, Ding J, Andrews JS, Freedman BI, Giulianini F, Koenig W, Illig T, Meisinger C, Gieger C, Zgaga L, Zemunik T, Boban M, Minelli C, Wheeler HE, Igl W, Zaboli G, Wild SH, Wright AF, Campbell H, Ellinghaus D, Nöthlings U, Jacobs G, Biffar R, Ernst F, Homuth G, Kroemer HK, Nauck M, Stracke S, Völker U, Völzke H, Kovacs P, Stumvoll M, Mägi R, Hofman A, Uitterlinden AG, Rivadeneira F, Aulchenko YS, Polasek O, Hastie N, Vitart V, Helmer C, Wang JJ, Stengel B, Ruggiero D, Bergmann S, Kähönen M, Viikari J, Nikopensius T, Province M, Ketkar S, Colhoun H, Doney A, Robino A, Krämer BK, Portas L, Ford I, Buckley BM, Adam M, Thun GA, Paulweber B, Haun M, Sala C, Mitchell P, Ciullo M, Kim SK, Vollenweider P, Raitakari O, Metspalu A, Palmer C, Gasparini P, Pirastu M, Jukema JW, Probst-Hensch NM, Kronenberg F, Toniolo D, Gudnason V, Shuldiner AR, Coresh J, Schmidt R, Ferrucci L, Siscovick DS, van Duijn CM, Borecki IB, Kardia SL, Liu Y, Curhan GC, Rudan I, Gyllensten U, Wilson JF, Franke A, Pramstaller PP, Rettig R, Prokopenko I, Witteman J, Hayward C, Ridker PM, Parsa A, Bochud M, Heid IM, Kao WH, Fox CS, Köttgen A; CARDIoGRAM Consortium; ICBP Consortium; CARe Consortium; WTCCC2 : Integration of genome-wide association studies with biological knowledge identifies six novel genes related to kidney function. Hum Mol Genet 21: 5329–5343, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Okada Y, Sim X, Go MJ, Wu JY, Gu D, Takeuchi F, Takahashi A, Maeda S, Tsunoda T, Chen P, Lim SC, Wong TY, Liu J, Young TL, Aung T, Seielstad M, Teo YY, Kim YJ, Lee JY, Han BG, Kang D, Chen CH, Tsai FJ, Chang LC, Fann SJ, Mei H, Rao DC, Hixson JE, Chen S, Katsuya T, Isono M, Ogihara T, Chambers JC, Zhang W, Kooner JS, Albrecht E, Yamamoto K, Kubo M, Nakamura Y, Kamatani N, Kato N, He J, Chen YT, Cho YS, Tai ES, Tanaka T; KidneyGen Consortium; CKDGen Consortium; GUGC consortium : Meta-analysis identifies multiple loci associated with kidney function-related traits in east Asian populations. Nat Genet 44: 904–909, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamada Y, Nishida T, Ichihara S, Kato K, Fujimaki T, Oguri M, Horibe H, Yoshida T, Watanabe S, Satoh K, Aoyagi Y, Fukuda M, Sawabe M: Identification of chromosome 3q28 and ALPK1 as susceptibility loci for chronic kidney disease in Japanese individuals by a genome-wide association study. J Med Genet 50: 410–418, 2013 [DOI] [PubMed] [Google Scholar]
- 24.Gorski M, Tin A, Garnaas M, McMahon GM, Chu AY, Tayo BO, Pattaro C, Teumer A, Chasman DI, Chalmers J, Hamet P, Tremblay J, Woodward M, Aspelund T, Eiriksdottir G, Gudnason V, Harris TB, Launer LJ, Smith AV, Mitchell BD, O’Connell JR, Shuldiner AR, Coresh J, Li M, Freudenberger P, Hofer E, Schmidt H, Schmidt R, Holliday EG, Mitchell P, Wang JJ, de Boer IH, Li G, Siscovick DS, Kutalik Z, Corre T, Vollenweider P, Waeber G, Gupta J, Kanetsky PA, Hwang SJ, Olden M, Yang Q, de Andrade M, Atkinson EJ, Kardia SL, Turner ST, Stafford JM, Ding J, Liu Y, Barlassina C, Cusi D, Salvi E, Staessen JA, Ridker PM, Grallert H, Meisinger C, Müller-Nurasyid M, Krämer BK, Kramer H, Rosas SE, Nolte IM, Penninx BW, Snieder H, Fabiola Del Greco M, Franke A, Nöthlings U, Lieb W, Bakker SJ, Gansevoort RT, van der Harst P, Dehghan A, Franco OH, Hofman A, Rivadeneira F, Sedaghat S, Uitterlinden AG, Coassin S, Haun M, Kollerits B, Kronenberg F, Paulweber B, Aumann N, Endlich K, Pietzner M, Völker U, Rettig R, Chouraki V, Helmer C, Lambert JC, Metzger M, Stengel B, Lehtimäki T, Lyytikäinen LP, Raitakari O, Johnson A, Parsa A, Bochud M, Heid IM, Goessling W, Köttgen A, Kao WH, Fox CS, Böger CA: Genome-wide association study of kidney function decline in individuals of European descent. Kidney Int 87: 1017–1029, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuttke M, Wong CS, Wühl E, Epting D, Luo L, Hoppmann A, Doyon A, Li Y, Sözeri B, Thurn D, Helmstädter M, Huber TB, Blydt-Hansen TD, Kramer-Zucker A, Mehls O, Melk A, Querfeld U, Furth SL, Warady BA, Schaefer F, Köttgen A; CKDGen Consortium : Genetic loci associated with renal function measures and chronic kidney disease in children: The pediatric investigation for genetic factors linked with renal progression consortium. Nephrol Dial Transplant 31: 262–269, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pattaro C, Teumer A, Gorski M, Chu AY, Li M, Mijatovic V, Garnaas M, Tin A, Sorice R, Li Y, Taliun D, Olden M, Foster M, Yang Q, Chen MH, Pers TH, Johnson AD, Ko YA, Fuchsberger C, Tayo B, Nalls M, Feitosa MF, Isaacs A, Dehghan A, d’Adamo P, Adeyemo A, Dieffenbach AK, Zonderman AB, Nolte IM, van der Most PJ, Wright AF, Shuldiner AR, Morrison AC, Hofman A, Smith AV, Dreisbach AW, Franke A, Uitterlinden AG, Metspalu A, Tonjes A, Lupo A, Robino A, Johansson Å, Demirkan A, Kollerits B, Freedman BI, Ponte B, Oostra BA, Paulweber B, Krämer BK, Mitchell BD, Buckley BM, Peralta CA, Hayward C, Helmer C, Rotimi CN, Shaffer CM, Müller C, Sala C, van Duijn CM, Saint-Pierre A, Ackermann D, Shriner D, Ruggiero D, Toniolo D, Lu Y, Cusi D, Czamara D, Ellinghaus D, Siscovick DS, Ruderfer D, Gieger C, Grallert H, Rochtchina E, Atkinson EJ, Holliday EG, Boerwinkle E, Salvi E, Bottinger EP, Murgia F, Rivadeneira F, Ernst F, Kronenberg F, Hu FB, Navis GJ, Curhan GC, Ehret GB, Homuth G, Coassin S, Thun GA, Pistis G, Gambaro G, Malerba G, Montgomery GW, Eiriksdottir G, Jacobs G, Li G, Wichmann HE, Campbell H, Schmidt H, Wallaschofski H, Völzke H, Brenner H, Kroemer HK, Kramer H, Lin H, Leach IM, Ford I, Guessous I, Rudan I, Prokopenko I, Borecki I, Heid IM, Kolcic I, Persico I, Jukema JW, Wilson JF, Felix JF, Divers J, Lambert JC, Stafford JM, Gaspoz JM, Smith JA, Faul JD, Wang JJ, Ding J, Hirschhorn JN, Attia J, Whitfield JB, Chalmers J, Viikari J, Coresh J, Denny JC, Karjalainen J, Fernandes JK, Endlich K, Butterbach K, Keene KL, Lohman K, Portas L, Launer LJ, Lyytikäinen LP, Yengo L, Franke L, Ferrucci L, Rose LM, Kedenko L, Rao M, Struchalin M, Kleber ME, Cavalieri M, Haun M, Cornelis MC, Ciullo M, Pirastu M, de Andrade M, McEvoy MA, Woodward M, Adam M, Cocca M, Nauck M, Imboden M, Waldenberger M, Pruijm M, Metzger M, Stumvoll M, Evans MK, Sale MM, Kähönen M, Boban M, Bochud M, Rheinberger M, Verweij N, Bouatia-Naji N, Martin NG, Hastie N, Probst-Hensch N, Soranzo N, Devuyst O, Raitakari O, Gottesman O, Franco OH, Polasek O, Gasparini P, Munroe PB, Ridker PM, Mitchell P, Muntner P, Meisinger C, Smit JH, Kovacs P, Wild PS, Froguel P, Rettig R, Mägi R, Biffar R, Schmidt R, Middelberg RP, Carroll RJ, Penninx BW, Scott RJ, Katz R, Sedaghat S, Wild SH, Kardia SL, Ulivi S, Hwang SJ, Enroth S, Kloiber S, Trompet S, Stengel B, Hancock SJ, Turner ST, Rosas SE, Stracke S, Harris TB, Zeller T, Zemunik T, Lehtimäki T, Illig T, Aspelund T, Nikopensius T, Esko T, Tanaka T, Gyllensten U, Völker U, Emilsson V, Vitart V, Aalto V, Gudnason V, Chouraki V, Chen WM, Igl W, März W, Koenig W, Lieb W, Loos RJ, Liu Y, Snieder H, Pramstaller PP, Parsa A, O’Connell JR, Susztak K, Hamet P, Tremblay J, de Boer IH, Böger CA, Goessling W, Chasman DI, Köttgen A, Kao WH, Fox CS; ICBP Consortium; AGEN Consortium; CARDIOGRAM; CHARGe-Heart Failure Group; ECHOGen Consortium : Genetic associations at 53 loci highlight cell types and biological pathways relevant for kidney function. Nat Commun 7: 10023, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Teumer A, Tin A, Sorice R, Gorski M, Yeo NC, Chu AY, Li M, Li Y, Mijatovic V, Ko YA, Taliun D, Luciani A, Chen MH, Yang Q, Foster MC, Olden M, Hiraki LT, Tayo BO, Fuchsberger C, Dieffenbach AK, Shuldiner AR, Smith AV, Zappa AM, Lupo A, Kollerits B, Ponte B, Stengel B, Krämer BK, Paulweber B, Mitchell BD, Hayward C, Helmer C, Meisinger C, Gieger C, Shaffer CM, Müller C, Langenberg C, Ackermann D, Siscovick D, Boerwinkle E, Kronenberg F, Ehret GB, Homuth G, Waeber G, Navis G, Gambaro G, Malerba G, Eiriksdottir G, Li G, Wichmann HE, Grallert H, Wallaschofski H, Völzke H, Brenner H, Kramer H, Mateo Leach I, Rudan I, Hillege HL, Beckmann JS, Lambert JC, Luan J, Zhao JH, Chalmers J, Coresh J, Denny JC, Butterbach K, Launer LJ, Ferrucci L, Kedenko L, Haun M, Metzger M, Woodward M, Hoffman MJ, Nauck M, Waldenberger M, Pruijm M, Bochud M, Rheinberger M, Verweij N, Wareham NJ, Endlich N, Soranzo N, Polasek O, van der Harst P, Pramstaller PP, Vollenweider P, Wild PS, Gansevoort RT, Rettig R, Biffar R, Carroll RJ, Katz R, Loos RJ, Hwang SJ, Coassin S, Bergmann S, Rosas SE, Stracke S, Harris TB, Corre T, Zeller T, Illig T, Aspelund T, Tanaka T, Lendeckel U, Völker U, Gudnason V, Chouraki V, Koenig W, Kutalik Z, O’Connell JR, Parsa A, Heid IM, Paterson AD, de Boer IH, Devuyst O, Lazar J, Endlich K, Susztak K, Tremblay J, Hamet P, Jacob HJ, Böger CA, Fox CS, Pattaro C, Köttgen A; DCCT/EDIC : Genome-wide association studies identify genetic loci associated with albuminuria in diabetes. Diabetes 65: 803–817, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahajan A, Rodan AR, Le TH, Gaulton KJ, Haessler J, Stilp AM, Kamatani Y, Zhu G, Sofer T, Puri S, Schellinger JN, Chu PL, Cechova S, van Zuydam N, Arnlov J, Flessner MF, Giedraitis V, Heath AC, Kubo M, Larsson A, Lindgren CM, Madden PAF, Montgomery GW, Papanicolaou GJ, Reiner AP, Sundström J, Thornton TA, Lind L, Ingelsson E, Cai J, Martin NG, Kooperberg C, Matsuda K, Whitfield JB, Okada Y, Laurie CC, Morris AP, Franceschini N; SUMMIT Consortium; BioBank Japan Project : Trans-ethnic fine mapping highlights kidney-function genes linked to salt sensitivity. Am J Hum Genet 99: 636–646, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li M, Li Y, Weeks O, Mijatovic V, Teumer A, Huffman JE, Tromp G, Fuchsberger C, Gorski M, Lyytikäinen LP, Nutile T, Sedaghat S, Sorice R, Tin A, Yang Q, Ahluwalia TS, Arking DE, Bihlmeyer NA, Böger CA, Carroll RJ, Chasman DI, Cornelis MC, Dehghan A, Faul JD, Feitosa MF, Gambaro G, Gasparini P, Giulianini F, Heid I, Huang J, Imboden M, Jackson AU, Jeff J, Jhun MA, Katz R, Kifley A, Kilpeläinen TO, Kumar A, Laakso M, Li-Gao R, Lohman K, Lu Y, Mägi R, Malerba G, Mihailov E, Mohlke KL, Mook-Kanamori DO, Robino A, Ruderfer D, Salvi E, Schick UM, Schulz CA, Smith AV, Smith JA, Traglia M, Yerges-Armstrong LM, Zhao W, Goodarzi MO, Kraja AT, Liu C, Wessel J, Boerwinkle E, Borecki IB, Bork-Jensen J, Bottinger EP, Braga D, Brandslund I, Brody JA, Campbell A, Carey DJ, Christensen C, Coresh J, Crook E, Curhan GC, Cusi D, de Boer IH, de Vries AP, Denny JC, Devuyst O, Dreisbach AW, Endlich K, Esko T, Franco OH, Fulop T, Gerhard GS, Glümer C, Gottesman O, Grarup N, Gudnason V, Hansen T, Harris TB, Hayward C, Hocking L, Hofman A, Hu FB, Husemoen LL, Jackson RD, Jørgensen T, Jørgensen ME, Kähönen M, Kardia SL, König W, Kooperberg C, Kriebel J, Launer LJ, Lauritzen T, Lehtimäki T, Levy D, Linksted P, Linneberg A, Liu Y, Loos RJ, Lupo A, Meisinger C, Melander O, Metspalu A, Mitchell P, Nauck M, Nürnberg P, Orho-Melander M, Parsa A, Pedersen O, Peters A, Peters U, Polasek O, Porteous D, Probst-Hensch NM, Psaty BM, Qi L, Raitakari OT, Reiner AP, Rettig R, Ridker PM, Rivadeneira F, Rossouw JE, Schmidt F, Siscovick D, Soranzo N, Strauch K, Toniolo D, Turner ST, Uitterlinden AG, Ulivi S, Velayutham D, Völker U, Völzke H, Waldenberger M, Wang JJ, Weir DR, Witte D, Kuivaniemi H, Fox CS, Franceschini N, Goessling W, Köttgen A, Chu AY; CHARGE Glycemic-T2D Working Group; CHARGE Blood Pressure Working Group : SOS2 and ACP1 loci identified through large-scale exome chip analysis regulate kidney development and function. J Am Soc Nephrol 28: 981–994, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Parsa A, Kanetsky PA, Xiao R, Gupta J, Mitra N, Limou S, Xie D, Xu H, Anderson AH, Ojo A, Kusek JW, Lora CM, Hamm LL, He J, Sandholm N, Jeff J, Raj DE, Böger CA, Bottinger E, Salimi S, Parekh RS, Adler SG, Langefeld CD, Bowden DW, Groop PH, Forsblom C, Freedman BI, Lipkowitz M, Fox CS, Winkler CA, Feldman HI; FIND Consortium; and the Chronic Renal Insufficiency Cohort (CRIC) Study Investigators : Genome-wide association of CKD progression: The Chronic Renal Insufficiency Cohort study. J Am Soc Nephrol 28: 923–934, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gorski M, van der Most PJ, Teumer A, Chu AY, Li M, Mijatovic V, Nolte IM, Cocca M, Taliun D, Gomez F, Li Y, Tayo B, Tin A, Feitosa MF, Aspelund T, Attia J, Biffar R, Bochud M, Boerwinkle E, Borecki I, Bottinger EP, Chen MH, Chouraki V, Ciullo M, Coresh J, Cornelis MC, Curhan GC, d’Adamo AP, Dehghan A, Dengler L, Ding J, Eiriksdottir G, Endlich K, Enroth S, Esko T, Franco OH, Gasparini P, Gieger C, Girotto G, Gottesman O, Gudnason V, Gyllensten U, Hancock SJ, Harris TB, Helmer C, Höllerer S, Hofer E, Hofman A, Holliday EG, Homuth G, Hu FB, Huth C, Hutri-Kähönen N, Hwang SJ, Imboden M, Johansson Å, Kähönen M, König W, Kramer H, Krämer BK, Kumar A, Kutalik Z, Lambert JC, Launer LJ, Lehtimäki T, de Borst M, Navis G, Swertz M, Liu Y, Lohman K, Loos RJF, Lu Y, Lyytikäinen LP, McEvoy MA, Meisinger C, Meitinger T, Metspalu A, Metzger M, Mihailov E, Mitchell P, Nauck M, Oldehinkel AJ, Olden M, Wjh Penninx B, Pistis G, Pramstaller PP, Probst-Hensch N, Raitakari OT, Rettig R, Ridker PM, Rivadeneira F, Robino A, Rosas SE, Ruderfer D, Ruggiero D, Saba Y, Sala C, Schmidt H, Schmidt R, Scott RJ, Sedaghat S, Smith AV, Sorice R, Stengel B, Stracke S, Strauch K, Toniolo D, Uitterlinden AG, Ulivi S, Viikari JS, Völker U, Vollenweider P, Völzke H, Vuckovic D, Waldenberger M, Jin Wang J, Yang Q, Chasman DI, Tromp G, Snieder H, Heid IM, Fox CS, Köttgen A, Pattaro C, Böger CA, Fuchsberger C: 1000 Genomes-based meta-analysis identifies 10 novel loci for kidney function. Sci Rep 7: 45040, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pattaro C: Genome-wide association studies of albuminuria: Towards genetic stratification in diabetes? [published online ahead of print September 16, 2017]. J Nephrol 10.1007/s40620-017-0437-3 [DOI] [PubMed] [Google Scholar]
- 33.Devuyst O, Olinger E, Rampoldi L: Uromodulin: From physiology to rare and complex kidney disorders. Nat Rev Nephrol 13: 525–544, 2017 [DOI] [PubMed] [Google Scholar]
- 34.Eckardt KU, Alper SL, Antignac C, Bleyer AJ, Chauveau D, Dahan K, Deltas C, Hosking A, Kmoch S, Rampoldi L, Wiesener M, Wolf MT, Devuyst O; Kidney Disease: Improving Global Outcomes : Autosomal dominant tubulointerstitial kidney disease: Diagnosis, classification, and management--A KDIGO consensus report. Kidney Int 88: 676–683, 2015 [DOI] [PubMed] [Google Scholar]
- 35.1000 Genomes Project Consortium; Auton A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini JL, McCarthy S, McVean GA, Abecasis GR: A global reference for human genetic variation. Nature 526: 68–74, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCarthy S, Das S, Kretzschmar W, Delaneau O, Wood AR, Teumer A, Kang HM, Fuchsberger C, Danecek P, Sharp K, Luo Y, Sidore C, Kwong A, Timpson N, Koskinen S, Vrieze S, Scott LJ, Zhang H, Mahajan A, Veldink J, Peters U, Pato C, van Duijn CM, Gillies CE, Gandin I, Mezzavilla M, Gilly A, Cocca M, Traglia M, Angius A, Barrett JC, Boomsma D, Branham K, Breen G, Brummett CM, Busonero F, Campbell H, Chan A, Chen S, Chew E, Collins FS, Corbin LJ, Smith GD, Dedoussis G, Dorr M, Farmaki AE, Ferrucci L, Forer L, Fraser RM, Gabriel S, Levy S, Groop L, Harrison T, Hattersley A, Holmen OL, Hveem K, Kretzler M, Lee JC, McGue M, Meitinger T, Melzer D, Min JL, Mohlke KL, Vincent JB, Nauck M, Nickerson D, Palotie A, Pato M, Pirastu N, McInnis M, Richards JB, Sala C, Salomaa V, Schlessinger D, Schoenherr S, Slagboom PE, Small K, Spector T, Stambolian D, Tuke M, Tuomilehto J, Van den Berg LH, Van Rheenen W, Volker U, Wijmenga C, Toniolo D, Zeggini E, Gasparini P, Sampson MG, Wilson JF, Frayling T, de Bakker PI, Swertz MA, McCarroll S, Kooperberg C, Dekker A, Altshuler D, Willer C, Iacono W, Ripatti S, Soranzo N, Walter K, Swaroop A, Cucca F, Anderson CA, Myers RM, Boehnke M, McCarthy MI, Durbin R, Haplotype Reference Consortium: A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet 48: 1279–83, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li Y, Köttgen A: Genetic investigations of kidney disease: Core curriculum 2013. Am J Kidney Dis 61: 832–844, 2013 [DOI] [PubMed] [Google Scholar]
- 38.Sveinbjornsson G, Mikaelsdottir E, Palsson R, Indridason OS, Holm H, Jonasdottir A, Helgason A, Sigurdsson S, Jonasdottir A, Sigurdsson A, Eyjolfsson GI, Sigurdardottir O, Magnusson OT, Kong A, Masson G, Sulem P, Olafsson I, Thorsteinsdottir U, Gudbjartsson DF, Stefansson K: Rare mutations associating with serum creatinine and chronic kidney disease. Hum Mol Genet 23: 6935–6943, 2014 [DOI] [PubMed] [Google Scholar]
- 39.Böger CA, Chen MH, Tin A, Olden M, Köttgen A, de Boer IH, Fuchsberger C, O’Seaghdha CM, Pattaro C, Teumer A, Liu CT, Glazer NL, Li M, O’Connell JR, Tanaka T, Peralta CA, Kutalik Z, Luan J, Zhao JH, Hwang SJ, Akylbekova E, Kramer H, van der Harst P, Smith AV, Lohman K, de Andrade M, Hayward C, Kollerits B, Tönjes A, Aspelund T, Ingelsson E, Eiriksdottir G, Launer LJ, Harris TB, Shuldiner AR, Mitchell BD, Arking DE, Franceschini N, Boerwinkle E, Egan J, Hernandez D, Reilly M, Townsend RR, Lumley T, Siscovick DS, Psaty BM, Kestenbaum B, Haritunians T, Bergmann S, Vollenweider P, Waeber G, Mooser V, Waterworth D, Johnson AD, Florez JC, Meigs JB, Lu X, Turner ST, Atkinson EJ, Leak TS, Aasarød K, Skorpen F, Syvänen AC, Illig T, Baumert J, Koenig W, Krämer BK, Devuyst O, Mychaleckyj JC, Minelli C, Bakker SJ, Kedenko L, Paulweber B, Coassin S, Endlich K, Kroemer HK, Biffar R, Stracke S, Völzke H, Stumvoll M, Mägi R, Campbell H, Vitart V, Hastie ND, Gudnason V, Kardia SL, Liu Y, Polasek O, Curhan G, Kronenberg F, Prokopenko I, Rudan I, Arnlöv J, Hallan S, Navis G, Parsa A, Ferrucci L, Coresh J, Shlipak MG, Bull SB, Paterson NJ, Wichmann HE, Wareham NJ, Loos RJ, Rotter JI, Pramstaller PP, Cupples LA, Beckmann JS, Yang Q, Heid IM, Rettig R, Dreisbach AW, Bochud M, Fox CS, Kao WH; CKDGen Consortium : CUBN is a gene locus for albuminuria. J Am Soc Nephrol 22: 555–570, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kramer HJ, Stilp AM, Laurie CC, Reiner AP, Lash J, Daviglus ML, Rosas SE, Ricardo AC, Tayo BO, Flessner MF, Kerr KF, Peralta C, Durazo-Arvizu R, Conomos M, Thornton T, Rotter J, Taylor KD, Cai J, Eckfeldt J, Chen H, Papanicolau G, Franceschini N: African ancestry-specific alleles and kidney disease risk in Hispanics/Latinos. J Am Soc Nephrol 28: 915–922, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brown LA, Sofer T, Stilp AM, Baier LJ, Kramer HJ, Masindova I, Levy D, Hanson RL, Moncrieft AE, Redline S, Rosas SE, Lash JP, Cai J, Laurie CC, Browning S, Thornton T, Franceschini N: Admixture mapping identifies an amerindian ancestry locus associated with albuminuria in Hispanics in the United States. J Am Soc Nephrol 28: 2211–2220, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Naik RP, Derebail VK, Grams ME, Franceschini N, Auer PL, Peloso GM, Young BA, Lettre G, Peralta CA, Katz R, Hyacinth HI, Quarells RC, Grove ML, Bick AG, Fontanillas P, Rich SS, Smith JD, Boerwinkle E, Rosamond WD, Ito K, Lanzkron S, Coresh J, Correa A, Sarto GE, Key NS, Jacobs DR, Kathiresan S, Bibbins-Domingo K, Kshirsagar AV, Wilson JG, Reiner AP: Association of sickle cell trait with chronic kidney disease and albuminuria in African Americans. JAMA 312: 2115–2125, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Böger CA, Gorski M, Li M, Hoffmann MM, Huang C, Yang Q, Teumer A, Krane V, O’Seaghdha CM, Kutalik Z, Wichmann HE, Haak T, Boes E, Coassin S, Coresh J, Kollerits B, Haun M, Paulweber B, Köttgen A, Li G, Shlipak MG, Powe N, Hwang SJ, Dehghan A, Rivadeneira F, Uitterlinden A, Hofman A, Beckmann JS, Krämer BK, Witteman J, Bochud M, Siscovick D, Rettig R, Kronenberg F, Wanner C, Thadhani RI, Heid IM, Fox CS, Kao WH; CKDGen Consortium : Association of eGFR-related loci identified by GWAS with incident CKD and ESRD. PLoS Genet 7: e1002292, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trudu M, Janas S, Lanzani C, Debaix H, Schaeffer C, Ikehata M, Citterio L, Demaretz S, Trevisani F, Ristagno G, Glaudemans B, Laghmani K, Dell’Antonio G, Loffing J, Rastaldi MP, Manunta P, Devuyst O, Rampoldi L; SKIPOGH team : Common noncoding UMOD gene variants induce salt-sensitive hypertension and kidney damage by increasing uromodulin expression. Nat Med 19: 1655–1660, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Padmanabhan S, Melander O, Johnson T, Di Blasio AM, Lee WK, Gentilini D, Hastie CE, Menni C, Monti MC, Delles C, Laing S, Corso B, Navis G, Kwakernaak AJ, van der Harst P, Bochud M, Maillard M, Burnier M, Hedner T, Kjeldsen S, Wahlstrand B, Sjögren M, Fava C, Montagnana M, Danese E, Torffvit O, Hedblad B, Snieder H, Connell JM, Brown M, Samani NJ, Farrall M, Cesana G, Mancia G, Signorini S, Grassi G, Eyheramendy S, Wichmann HE, Laan M, Strachan DP, Sever P, Shields DC, Stanton A, Vollenweider P, Teumer A, Völzke H, Rettig R, Newton-Cheh C, Arora P, Zhang F, Soranzo N, Spector TD, Lucas G, Kathiresan S, Siscovick DS, Luan J, Loos RJ, Wareham NJ, Penninx BW, Nolte IM, McBride M, Miller WH, Nicklin SA, Baker AH, Graham D, McDonald RA, Pell JP, Sattar N, Welsh P, Munroe P, Caulfield MJ, Zanchetti A, Dominiczak AF; Global BPgen Consortium : Genome-wide association study of blood pressure extremes identifies variant near UMOD associated with hypertension. PLoS Genet 6: e1001177, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han J, Liu Y, Rao F, Nievergelt CM, O’Connor DT, Wang X, Liu L, Bu D, Liang Y, Wang F, Zhang L, Zhang H, Chen Y, Wang H: Common genetic variants of the human uromodulin gene regulate transcription and predict plasma uric acid levels. Kidney Int 83: 733–740, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Olden M, Corre T, Hayward C, Toniolo D, Ulivi S, Gasparini P, Pistis G, Hwang SJ, Bergmann S, Campbell H, Cocca M, Gandin I, Girotto G, Glaudemans B, Hastie ND, Loffing J, Polasek O, Rampoldi L, Rudan I, Sala C, Traglia M, Vollenweider P, Vuckovic D, Youhanna S, Weber J, Wright AF, Kutalik Z, Bochud M, Fox CS, Devuyst O: Common variants in UMOD associate with urinary uromodulin levels: A meta-analysis. J Am Soc Nephrol 25: 1869–1882, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rovida CL: Conclusione degli studi intorno all’origine istologica dei cilindri dell’urina. Riv Clin Bologna 2a: 303–306, 1873 [Google Scholar]
- 49.Tamm I, Horsfall FL Jr: Characterization and separation of an inhibitor of viral hemagglutination present in urine. Proc Soc Exp Biol Med 74: 106–108, 1950 [PubMed] [Google Scholar]
- 50.Muchmore AV, Decker JM: Uromodulin: A unique 85-kilodalton immunosuppressive glycoprotein isolated from urine of pregnant women. Science 229: 479–481, 1985 [DOI] [PubMed] [Google Scholar]
- 51.McKenzie JK, McQueen EG: Immunofluorescent localization of Tamm-Horsfall mucoprotein in human kidney. J Clin Pathol 22: 334–339, 1969 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pennica D, Kohr WJ, Kuang WJ, Glaister D, Aggarwal BB, Chen EY, Goeddel DV: Identification of human uromodulin as the Tamm-Horsfall urinary glycoprotein. Science 236: 83–88, 1987 [DOI] [PubMed] [Google Scholar]
- 53.Mutig K, Kahl T, Saritas T, Godes M, Persson P, Bates J, Raffi H, Rampoldi L, Uchida S, Hille C, Dosche C, Kumar S, Castañeda-Bueno M, Gamba G, Bachmann S: Activation of the bumetanide-sensitive Na+,K+,2Cl- cotransporter (NKCC2) is facilitated by Tamm-Horsfall protein in a chloride-sensitive manner. J Biol Chem 286: 30200–30210, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Serafini-Cessi F, Monti A, Cavallone D: N-Glycans carried by Tamm-Horsfall glycoprotein have a crucial role in the defense against urinary tract diseases. Glycoconj J 22: 383–394, 2005 [DOI] [PubMed] [Google Scholar]
- 55.Bates JM, Raffi HM, Prasadan K, Mascarenhas R, Laszik Z, Maeda N, Hultgren SJ, Kumar S: Tamm-Horsfall protein knockout mice are more prone to urinary tract infection: Rapid communication. Kidney Int 65: 791–797, 2004 [DOI] [PubMed] [Google Scholar]
- 56.Mo L, Huang HY, Zhu XH, Shapiro E, Hasty DL, Wu XR: Tamm-Horsfall protein is a critical renal defense factor protecting against calcium oxalate crystal formation. Kidney Int 66: 1159–1166, 2004 [DOI] [PubMed] [Google Scholar]
- 57.Säemann MD, Weichhart T, Zeyda M, Staffler G, Schunn M, Stuhlmeier KM, Sobanov Y, Stulnig TM, Akira S, von Gabain A, von Ahsen U, Hörl WH, Zlabinger GJ: Tamm-Horsfall glycoprotein links innate immune cell activation with adaptive immunity via a Toll-like receptor-4-dependent mechanism. J Clin Invest 115: 468–475, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Darisipudi MN, Thomasova D, Mulay SR, Brech D, Noessner E, Liapis H, Anders HJ: Uromodulin triggers IL-1β-dependent innate immunity via the NLRP3 inflammasome. J Am Soc Nephrol 23: 1783–1789, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Brunati M, Perucca S, Han L, Cattaneo A, Consolato F, Andolfo A, Schaeffer C, Olinger E, Peng J, Santambrogio S, Perrier R, Li S, Bokhove M, Bachi A, Hummler E, Devuyst O, Wu Q, Jovine L, Rampoldi L: The serine protease hepsin mediates urinary secretion and polymerisation of Zona Pellucida domain protein uromodulin. eLife 4: e08887, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hart TC, Gorry MC, Hart PS, Woodard AS, Shihabi Z, Sandhu J, Shirts B, Xu L, Zhu H, Barmada MM, Bleyer AJ: Mutations of the UMOD gene are responsible for medullary cystic kidney disease 2 and familial juvenile hyperuricaemic nephropathy. J Med Genet 39: 882–892, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bernascone I, Vavassori S, Di Pentima A, Santambrogio S, Lamorte G, Amoroso A, Scolari F, Ghiggeri GM, Casari G, Polishchuk R, Rampoldi L: Defective intracellular trafficking of uromodulin mutant isoforms. Traffic 7: 1567–1579, 2006 [DOI] [PubMed] [Google Scholar]
- 62.Dahan K, Devuyst O, Smaers M, Vertommen D, Loute G, Poux JM, Viron B, Jacquot C, Gagnadoux MF, Chauveau D, Büchler M, Cochat P, Cosyns JP, Mougenot B, Rider MH, Antignac C, Verellen-Dumoulin C, Pirson Y: A cluster of mutations in the UMOD gene causes familial juvenile hyperuricemic nephropathy with abnormal expression of uromodulin. J Am Soc Nephrol 14: 2883–2893, 2003 [DOI] [PubMed] [Google Scholar]
- 63.Nasr SH, Lucia JP, Galgano SJ, Markowitz GS, D’Agati VD: Uromodulin storage disease. Kidney Int 73: 971–976, 2008 [DOI] [PubMed] [Google Scholar]
- 64.Köttgen A, Hwang SJ, Larson MG, Van Eyk JE, Fu Q, Benjamin EJ, Dehghan A, Glazer NL, Kao WH, Harris TB, Gudnason V, Shlipak MG, Yang Q, Coresh J, Levy D, Fox CS: Uromodulin levels associate with a common UMOD variant and risk for incident CKD. J Am Soc Nephrol 21: 337–344, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Troyanov S, Delmas-Frenette C, Bollée G, Youhanna S, Bruat V, Awadalla P, Devuyst O, Madore F: Clinical,genetic, and urinary factors associated with uromodulin excretion. Clin J Am Soc Nephrol 11: 62–69, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lifton RP: Genetic dissection of human blood pressure variation: Common pathways from rare phenotypes. Harvey Lect 100: 71–101, 2004–2005 [PubMed] [Google Scholar]
- 67.Delgado GE, Kleber ME, Scharnagl H, Krämer BK, März W, Scherberich JE: Serum uromodulin and mortality risk in patients undergoing coronary angiography. J Am Soc Nephrol 28: 2201–2210, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Garimella PS, Biggs ML, Katz R, Ix JH, Bennett MR, Devarajan P, Kestenbaum BR, Siscovick DS, Jensen MK, Shlipak MG, Chaves PH, Sarnak MJ: Urinary uromodulin, kidney function, and cardiovascular disease in elderly adults. Kidney Int 88: 1126–1134, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Garimella PS, Jaber BL, Tighiouart H, Liangos O, Bennett MR, Devarajan P, El-Achkar TM, Sarnak MJ: Association of preoperative urinary uromodulin with AKI after cardiac surgery. Clin J Am Soc Nephrol 12: 10–18, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Youhanna S, Weber J, Beaujean V, Glaudemans B, Sobek J, Devuyst O: Determination of uromodulin in human urine: Influence of storage and processing. Nephrol Dial Transplant 29: 136–145, 2014 [DOI] [PubMed] [Google Scholar]
- 71.Hammond TG, Moes S, Youhanna S, Jennings P, Devuyst O, Odermatt A, Jenö P: Development and characterization of a pseudo multiple reaction monitoring method for the quantification of human uromodulin in urine. Bioanalysis 8: 1279–1296, 2016 [DOI] [PubMed] [Google Scholar]
- 72.Pruijm M, Ponte B, Ackermann D, Paccaud F, Guessous I, Ehret G, Pechère-Bertschi A, Vogt B, Mohaupt MG, Martin PY, Youhanna SC, Nägele N, Vollenweider P, Waeber G, Burnier M, Devuyst O, Bochud M: Associations of urinary uromodulin with clinical characteristics and markers of tubular function in the general population. Clin J Am Soc Nephrol 11: 70–80, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Scherberich JE, Gruber R, Nockher WA, Christensen EI, Schmitt H, Herbst V, Block M, Kaden J, Schlumberger W: Serum uromodulin-a marker of kidney function and renal parenchymal integrity [published online ahead of print February 16, 2017]. Nephrol Dial Transplant 10.1093/ndt/gfw422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Thornley C, Dawnay A, Cattell WR: Human Tamm-Horsfall glycoprotein: Urinary and plasma levels in normal subjects and patients with renal disease determined by a fully validated radioimmunoassay. Clin Sci (Lond) 68: 529–535, 1985 [DOI] [PubMed] [Google Scholar]
- 75.Steubl D, Block M, Herbst V, Nockher WA, Schlumberger W, Satanovskij R, Angermann S, Hasenau AL, Stecher L, Heemann U, Renders L, Scherberich J: Plasma uromodulin correlates with kidney function and identifies early stages in chronic kidney disease patients. Medicine (Baltimore) 95: e3011, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sekula P, Del Greco M F, Pattaro C, Köttgen A: Mendelian randomization as an approach to assess causality using observational data. J Am Soc Nephrol 27: 3253–3265, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ghirotto S, Tassi F, Barbujani G, Pattini L, Hayward C, Vollenweider P, Bochud M, Rampoldi L, Devuyst O: The uromodulin gene locus shows evidence of pathogen adaptation through human evolution. J Am Soc Nephrol 27: 2983–2996, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Garimella PS, Bartz TM, Ix JH, Chonchol M, Shlipak MG, Devarajan P, Bennett MR, Sarnak MJ: Urinary uromodulin and risk of urinary tract infections: The Cardiovascular Health Study. Am J Kidney Dis 69: 744–751, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rossier BC, Bochud M, Devuyst O: The hypertension pandemic: An evolutionary perspective. Physiology (Bethesda) 32: 112–125, 2017 [DOI] [PubMed] [Google Scholar]
- 80.Przeworski M, Coop G, Wall JD: The signature of positive selection on standing genetic variation. Evolution 59: 2312–2323, 2005 [PubMed] [Google Scholar]
- 81.Hancock AM, Clark VJ, Qian Y, Di Rienzo A: Population genetic analysis of the uncoupling proteins supports a role for UCP3 in human cold resistance. Mol Biol Evol 28: 601–614, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ying WZ, Allen CE, Curtis LM, Aaron KJ, Sanders PW: Mechanism and prevention of acute kidney injury from cast nephropathy in a rodent model. J Clin Invest 122: 1777–1785, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Graham LA, Padmanabhan S, Fraser NJ, Kumar S, Bates JM, Raffi HS, Welsh P, Beattie W, Hao S, Leh S, Hultstrom M, Ferreri NR, Dominiczak AF, Graham D, McBride MW: Validation of uromodulin as a candidate gene for human essential hypertension. Hypertension 63: 551–558, 2014 [DOI] [PubMed] [Google Scholar]
- 84.Dominiczak A, Delles C, Padmanabhan S: Genomics and precision medicine for clinicians and scientists in hypertension. Hypertension 69: e10–e13, 2017 [DOI] [PubMed] [Google Scholar]
- 85.Khera AV, Kathiresan S: Is coronary atherosclerosis one disease or many? Setting realistic expectations for precision medicine. Circulation 135: 1005–1007, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.McCarthy MI: Painting a new picture of personalised medicine for diabetes. Diabetologia 60: 793–799, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Khera AV, Kathiresan S: Genetics of coronary artery disease: Discovery, biology and clinical translation. Nat Rev Genet 18: 331–344, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pattaro C, Aulchenko YS, Isaacs A, Vitart V, Hayward C, Franklin CS, Polasek O, Kolcic I, Biloglav Z, Campbell S, Hastie N, Lauc G, Meitinger T, Oostra BA, Gyllensten U, Wilson JF, Pichler I, Hicks AA, Campbell H, Wright AF, Rudan I, van Duijn CM, Riegler P, Marroni F, Pramstaller PP; EUROSPAN Consortium : Genome-wide linkage analysis of serum creatinine in three isolated European populations. Kidney Int 76: 297–306, 2009 [DOI] [PubMed] [Google Scholar]
- 89.Vinkhuyzen AA, Wray NR, Yang J, Goddard ME, Visscher PM: Estimation and partition of heritability in human populations using whole-genome analysis methods. Annu Rev Genet 47: 75–95, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Noce D, Gögele M, Schwienbacher C, Caprioli G, De Grandi A, Foco L, Platzgummer S, Pramstaller PP, Pattaro C: Sequential recruitment of study participants may inflate genetic heritability estimates. Hum Genet 136: 743–757, 2017 [DOI] [PubMed] [Google Scholar]
- 91.Zuk O, Hechter E, Sunyaev SR, Lander ES: The mystery of missing heritability: Genetic interactions create phantom heritability. Proc Natl Acad Sci U S A 109: 1193–1198, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu C, Dupuis J, Larson MG, Cupples LA, Ordovas JM, Vasan RS, Meigs JB, Jacques PF, Levy D: Revisiting heritability accounting for shared environmental effects and maternal inheritance. Hum Genet 134: 169–179, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Sanna S, Li B, Mulas A, Sidore C, Kang HM, Jackson AU, Piras MG, Usala G, Maninchedda G, Sassu A, Serra F, Palmas MA, Wood WH 3rd, Njølstad I, Laakso M, Hveem K, Tuomilehto J, Lakka TA, Rauramaa R, Boehnke M, Cucca F, Uda M, Schlessinger D, Nagaraja R, Abecasis GR: Fine mapping of five loci associated with low-density lipoprotein cholesterol detects variants that double the explained heritability. PLoS Genet 7: e1002198, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bush WS, Oetjens MT, Crawford DC: Unravelling the human genome-phenome relationship using phenome-wide association studies. Nat Rev Genet 17: 129–145, 2016 [DOI] [PubMed] [Google Scholar]