Abstract
The nephron is the functional unit of the kidney, but the mechanism of nephron formation during human development is unclear. We conducted a detailed analysis of nephron development in humans and mice by immunolabeling, and we compared human and mouse nephron patterning to describe conserved and divergent features. We created protein localization maps that highlight the emerging patterns along the proximal–distal axis of the developing nephron and benchmark expectations for localization of functionally important transcription factors, which revealed unanticipated cellular diversity. Moreover, we identified a novel nephron subdomain marked by Wnt4 expression that we fate-mapped to the proximal mature nephron. Significant conservation was observed between human and mouse patterning. We also determined the time at which markers for mature nephron cell types first emerge—critical data for the renal organoid field. These findings have conceptual implications for the evolutionary processes driving the diversity of mammalian organ systems. Furthermore, these findings provide practical insights beyond those gained with mouse and rat models that will guide in vitro efforts to harness the developmental programs necessary to build human kidney structures.
Keywords: human genetics, nephron, kidney development
Studies predominantly in mouse and rat models have provided a blueprint for mammalian nephrogenesis.1,2 Nephron formation involves a complex series of interactions among nephron progenitor cells (NPCs), overlying interstitial progenitor cells and underlying epithelial cells at the branch tips of the developing ureteric epithelial collecting duct network.1–3 Nephrogenesis within the Six2+/Cited1+ NPC pool4,5 is promoted by Wnt9b/Ctnnd1, Lif, Bmp7, and FAT4 signaling.6–15 Signaling initiates a subset of NPCs to form pretubular aggregates (PTA) beneath the ureteric epithelial branch tips and PTAs activate synthesis of transcriptional regulators (Pax8) and signaling factors (Wnt4 and Fgf8) that are essential for further progression of the nephrogenic program.16–18 In this, Wnt4 is critical for a mesenchymal to epithelial transition that establishes the renal vesicle (RV), the precursor for each nephron.16,19–21
In addition to a classic apical-basal epithelial polarity, the RV displays proximal–distal polarity relative to the adjacent ureteric epithelium. Although careful lineage mapping has not been performed, evidence suggests cells positioned in close contact with the ureteric epithelium generate fates of the distal tubule and connecting segment, whereas the proximal region forms podocytes and parietal epithelium of the renal corpuscle.22,23 The RV undergoes a complex morphogenesis through comma-shaped body and S-shaped body (SSB) stages with a concurrent increase in regional cell complexity along the proximal–distal axis and the formation of a patent–luminal connection between the distal SSB and ureteric epithelial-derived collecting duct network.24
Genetic analysis and in vitro studies have demonstrated that Notch, Bmp, PI3-kinase, Fgf, and Wnt signaling pathways play critical roles in the elaboration of proximal–distal pattern in the RV to SSB transition. Distal cells express Wnt4, and exhibit high levels of Lef1, a transcriptional target and mediator of canonical Wnt signaling.22 Elevating Wnt signaling in vitro leads to an inhibition of proximal and expansion of distal cell identities, consistent with an instructive role for Wnt signaling in promoting distal cell fates.25 Lgr5, a Wnt target, is expressed in a subdomain of the distal SSB, delineating a distal tubule precursor population, and also suggests Wnt dependency in distal identity formation.25,26 Further evidence indicates Fgf818 and appropriate levels of Bmp signaling are also critical.25 At the transcriptional level, distal development is contingent on the activity of Pou3f3 and Lhx1.27,28
The medial segment of the SSB is demarcated by high expression of genes encoding multiple Notch pathway components: the Notch ligands Jag1 and Dll1, Notch receptors Notch1 and Notch2, the Notch pathway modulator Lfng, and Notch transcriptional targets Hes1, Hes5, and HeyL.29–31 Genetic analysis has demonstrated Notch signaling through Notch2 is required for normal development of proximal tubule segments and components of the renal corpuscle.30,31 The transcription factors Irx3 and Hnf1b are both required for medial development.32,33 In the proximal-most region of the nephron, normal podocyte identity development is dependent on the action of several transcriptional regulators, notably Mafb, Tcf21, and Foxc2.34–36 The ongoing function of these factors beyond the SSB stage are unclear, although conditional removal of Tcf21 later in mature podocytes indicates a continuing role in podocyte programs.34 Of note, the precise mapping of distal, medial, and proximal markers to determine potential overlap, has not been performed.
Macroanatomic analyses of the developing human kidney suggest a broadly similar architecture to its murine counterpart.37–40 However, molecular analyses of progenitor compartments comparing the mouse and human kidney have identified distinct regulatory features that may underlie differences in nephron-forming programs.41,42 Here, we performed detailed comparative molecular and cellular analyses to extend an understanding of early nephron patterning in the developing mouse and human kidney. Overall, these studies argue for similar processes at play, although we observed unanticipated cellular diversity in the early epithelializing human nephron. In addition, fate-mapping of cells within a Wnt4+ domain provides a register for the positioning of proximal cell fates within the developing SSB that is likely shared between mouse and man. The complexity of emerging patterns in the human nephron will guide and inform in vitro efforts to recapitulate human nephrogenesis.
Results
Differentiation of NPCs into Early Nephron Structures and Establishment of Transitory Cell Lineages
In the mouse, Six2+/Cited1+ NPCs give rise to the entire nephron.4,5 A similarly positioned population of Six2+/Cited1+ cells is present within the developing human kidney and this population displays a similar transcriptional profile in mouse-human comparisons.40,42 In vitro induction experiments show that a SIX2+-enriched cell population from the human fetal kidney generates nephron-like cell types.43 Here, we focus on the process of nephron formation by the nephron progenitor population comparing week 16–17 human fetal kidneys with the mouse kidney at early (embryo day 15.5 [E15.5]) and late (postnatal day 2 [P2]) stages of development. Tables 1 and 2 summarize the proteins studied, their functional properties, localization and disease association, and overlap comparing human and mouse datasets.
Table 1.
Expression and localization patterns for antibodies and in situ hybridization performed in study
| Kidney Phenotype/Disease | Cap Mesenchyme | Renal Vesicle | SSB Nephron | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Gene Symbol | Gene Name | Protein Type | Mouse | Human | Mouse | Human | Mouse | Human | Mouse | Human | Reference |
| CDH1 | Cadherin 1 | Cell-adhesion protein | − | − | − | − | + | + | + | + | Vestweber et al.47 |
| CITED1 | Cbp/p300-interacting transactivator with Glu/Asp-rich carboxy-terminal domain 1 | Transcription factor | − | − | + | + | − | − | − | − | Boyle et al.64 |
| FOXC2 | Forkhead box C2 | Transcription factor | + | + | + | + | + | + | + | + | Takemoto et al.36 |
| GATA3 | Gata binding protein 3 | Transcription factor | + | + | − | − | − | − | − | + | Grote et al.65 |
| HES1 | Hairy/enhancer of spli homolog 1 | Transcription factor | − | − | + | + | + | + | + | + | Chen and Al-Awqati29 |
| HNF1B | Hnf1 homebox B | Transcription factor | + | + | − | − | + | + | + | + | Heliot et al.33 |
| HOXD11 | Homeobox d11 | Transcription factor | + | − | + | + | unknown | + | unknown | + | Wellik et al.66 |
| JAG1 | Jagged 1 | Notch ligand | + | + | − | − | + | + | + | + | Liu et al.31 |
| KRT8 | Keratin 8 | Cell-adhesion protein | − | − | − | − | − | + | − | + | Chen and Al-Awqati29 |
| LEF1 | Lymphoid enhancer binding factor 1 | Transcription factor | − | − | − | + | + | + | + | + | Mugford et al.22 |
| LHX1 | Lim homeobox gene 1 | Transcription factor | + | − | − | − | + | + | + | + | Kobayashi et al.28 |
| MAFB | v-maf Musculoaponeurotic fibrosarcoma oncogene family, protein B | Transcription factor | + | − | − | − | + | + | + | + | Moriguchi et al.35 |
| PAX2 | Paried box 2 | Transcription factor | + | + | + | + | + | + | + | + | Bouchard et al.17 |
| PAX8 | Paired box 8 | Transcription factor | + | + | − | − | + | + | + | + | Bouchard et al.17 |
| POU3F3 | POU domain, class 3, transcription factor 3 | Transcription factor | + | − | − | − | + | + | + | + | Nakai et al.27 |
| SIX1 | Sine oculis-related homebox 1 | Transcription factor | + | + | − | + | − | + | − | + | Xu et al.53 |
| SIX2 | Sine oculis-related homebox 2 | Transcription factor | + | + | + | + | + | + | − | + | Self et al.44 |
| SOX9 | SRY-box 9 | Transcription factor | + | + | − | − | + | + | + | + | Reginensi et al.49 |
| WNT4 | Wingless-type MMTV integration site family, member 4 | Wnt ligand | + | + | − | − | + | + | + | + | Stark et al.16 |
| WT1 | Wilms tumor protein 1 | Transcription factor | + | + | + | + | + | + | + | + | Armstrong et al.67 |
Table 2.
First point of detection for commonly used protein markers of mature tubule segments in the human kidney
| Gene Symbol | Gene Name | Stage First Observed | Gudmap Ontology Number |
|---|---|---|---|
| LRP2 | LDL Receptor Related Protein 2 | Proximal segment of SSB | 27764 |
| SLC3A1 | Solute Carrier Family 3 Member 1 | Proximal segment of SSB; early proximal tubule | 27764; 27784 |
| SLC12A1 | Solute Carrier Family 12 Member 1 | Immature loop of Henle ascending limb | 35426 |
| SLC12A3 | Solute Carrier Family 12 Member 3 | Early distal tubule (after capillary loop stage) | 28390 |
| CUBN | Cubilin | Early proximal tubule | 27784 |
| PODXL | Podocalyxin Like | Proximal renal vesicle; visceral epithelium of SSB | 31549; 27766 |
| WT1 | Proximal renal vesicle; visceral epithelium of SSB; proximal segment of SSB | 31549; 27766; 27764 | |
| NPHS2 | Podocin | Visceral epithelium of SSB | 27766 |
| MAFB | Proximal renal vesicle; visceral epithelium of SSB | 31549; 27766 | |
| UMOD | Uromodulin | Early distal tubule (after capillary loop stage) | 28390 |
| AQP1 | Aquaporin 1 | Early proximal tubule (after capillary loop stage) | 27784 |
Proteins detected in figures are summarized and related to data on whether proteins are causative of human or mouse kidney disease/phenotype.
During differentiation, mouse NPCs downregulate expression of transcription factors associated with the NPC-state including Cited1 and Six2; Six2 is required for the self-renewal of NPCs.44 Conversely, early commitment of NPCs is highlighted by the activation of genes encoding other transcriptional regulators such as Pax8, Lef1, and Lhx1, and novel signal components such as the Notch ligand Jag1, Wnt4 and Fgf8.17,22,28,29 Consistent with canonical Wnt signaling triggering nephrogenesis,6,10 activation of the canonical Wnt target Lef1 precedes subsequent expression of Pax8, Fgf8 Wnt4, and Lhx1 in the PTA to RV transition.16–18,28 Thus, Lef1 provides one of the first indicators of initiation of nephrogenesis.22
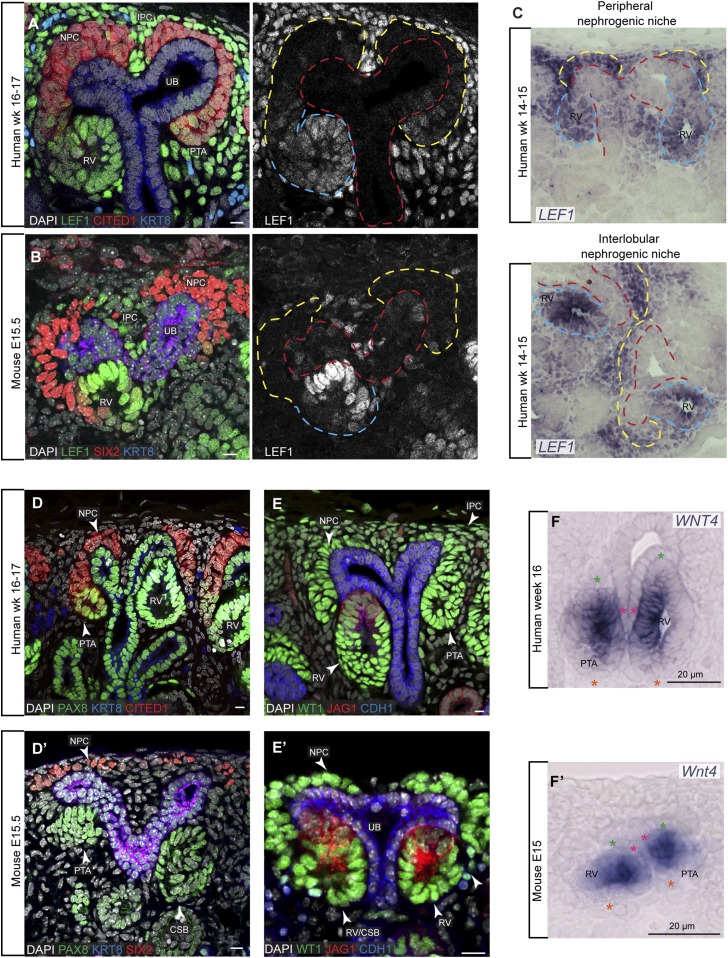
SIX2 and CITED1 downregulation in human NPCs at week 16–17 resembles the E15.5 mouse kidney; however, each protein shows a distinct, human-specific pattern of retention in specific regions of developing nephron intermediates42 (Supplemental Figure 1). CITED1 remains detectable in the proximal PTA, and SIX2 is found in the proximal PTA, RV, and SSB. In the mouse, low Cited1 and Six2 levels were detected in PTAs and the proximal RV, respectively. Lef1, which is only observed in the mouse kidney in conjunction with PTA formation showed a sporadic distribution within human NPCs, close to the PTA transition zone (Figure 1, A and B), consistent with in situ hybridization analysis of LEF1 expression (Figure 1C). To examine whether the “earlier” onset of LEF1 production in human NPCs reflected a relative staging disparity, we immunolabeled Lef1 in the P2 mouse kidney (Supplemental Figure 2). As described by Rumballe and colleagues,45 at P2 the kidney cortex is more densely packed with epithelial structures, fewer NPCs are visible, and structurally recognizable cap mesenchyme populations are infrequent. Commitment to nephron formation is accelerated at P2 relative to E15.5.46 However, as at early stages, most Lef1 was restricted to forming nephrons. Occasional Six2+/Lef1+ cells were observed in the cap mesenchyme clearly distinct from the more extensive SIX2+/LEF1+ population in human NPCs (Supplemental Figure 2). PAX8 was also detected within a similar NPC domain in the human but not the mouse kidney, extending throughout the RV by epithelialization (Figure 1D, Supplemental Figure 3, A, A’, and E). Anti-PAX2 and anti-PAX8 antibodies showed distinct patterns of immunoreactivity (Supplemental Figure 3E). Together these data suggest a temporal and spatial divergence or change in cellular dynamics associated with NPC induction in the human kidney (see Discussion).
Figure 1.
Nephron progenitor induction in human and mouse nephrogenic niches. (A–F) and (D’–F’) Immunofluorescent stains and in situ hybridization on human and mouse kidneys, respectively. Ages and stains as specified on fields. Yellow, red, and cyan dashed lines indicate cap mesenchyme, ureteric bud, and nephrons, respectively. Stars in (F) and (F’) indicate the nephron axes: green, distal; orange, proximal; magenta start indicates ureteric bud. Scale bars on immunofluorescence data indicate 10 µm. For single-channel views see Supplemental Figures 1 and 3. CSB, comma-shaped body; IPC, interstitial progenitor cells; UB, ureteric bud.
Unlike LEF1 and PAX8, WNT4 mRNA and JAG1 were first detected at PTA stages where expression localized to distally located cells (Figure 1, E, E’, F, and F’), a more restricted region at both PTA and RV stages to the mouse.16 JAG1 was absent from early PTAs but present within intracellular vesicles in cells of the late PTAs/early RVs that lay closest to the ureteric bud tip, as in the mouse (Figure 1E, Supplemental Figures 1 and 3C’). By the RV stage, JAG1 localized to the cell surface on the lateral cell of distally located cells (Figure 1E, Supplemental Figure 3C’). At this stage of nephrogenesis, CDH1, a homophilic cell adhesion factor with a broad role in epithelial formation,47 was first evident in the distal RV (Figure 1E, Supplemental Figure 3C’’) as in the mouse.48 However, JAG1 extended beyond distal CDH1 producing cells to medial regions of the RV. At this stage, low levels of the transcriptional regulatory factor SOX9 were evident in a few distal cells of the human RV, a more limited distribution to the mouse at morphologically equivalent stages (Supplemental Figure 1).49 The order of appearance for these proteins/genes during induction was LEF1, followed by PAX8 and WNT4, then JAG1 and LHX1 (data not shown).
In summary, LEF1 and PAX8 activation within morphologically distinct human NPCs likely indicate early inductive signaling not visible in the mouse NPC population. However, the activation of WNT4, LHX1, SOX9, CDH1, and JAG1 and the localization of PAX8 and LEF1 in forming nephrons was quite similar between the two species, as were the parallel morphologic changes accompanying early stages of nephron induction.
Developmental Progression from RVs to SSB Nephrons
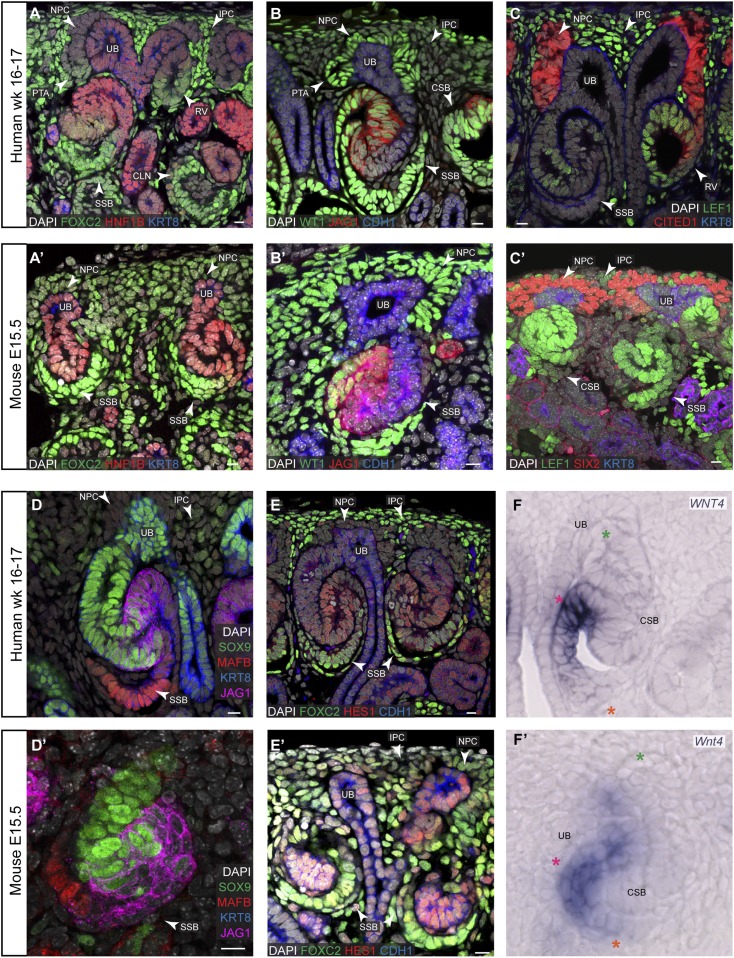
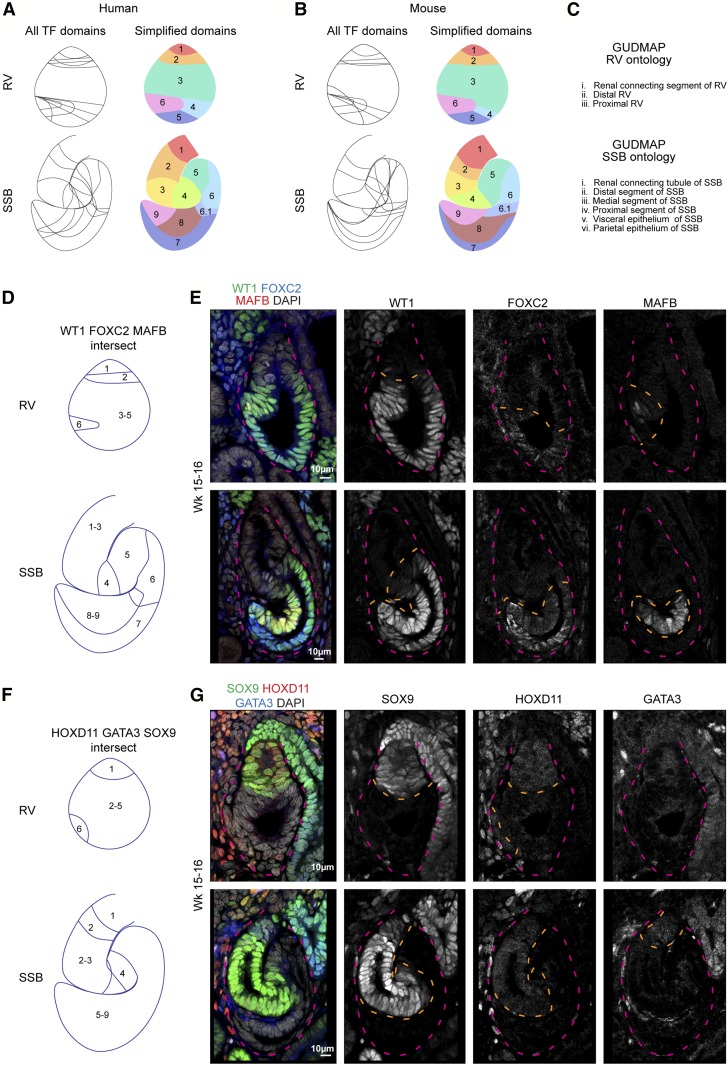
Mouse RVs progress through a series of morphogenetic events that remain poorly understood.2 The ontology for the mouse and human SSB comprises six terms/anatomic domains: renal connecting tubule of SSB, distal segment of SSB, medial segment of SSB, proximal segment of SSB, visceral epithelium of SSB, and parietal epithelium of SSB (www.gudmap.org); although where the boundaries of each domain lie is not clear. Further, there is fluidity in gene expression domains with genes expressed in the RV adopting new patterns within the SSB. Several genes have been shown to identify discrete domains at this stage and loss of their activity results in altered patterning of the nephron. Notable genes, with the normal domains and regions altered on loss-of-function in parentheses, include mutations Hnf1b (distal/medial; loss of proximal/medial nephron regions), Sox9 (distal; none reported), Cdh1 (distal/medial; none reported), Lef1 (distal/medial; none reported), Jag1 (medial; loss of proximal/medial nephron regions), Hes1 (medial; none reported), Wt1 (proximal; none reported), Foxc2 (proximal; loss of podocyte identity), and Mafb (proximal; loss of podocyte identity).25,30,31,33,49–51
As in the mouse SSB, the connecting and distal segments were demarcated by strong labeling of HNF1B, CDH1, SOX9, and PAX8 (Figure 2, A, B, and D, PAX8 in Supplemental Figure 1), the medial and proximal segments by JAG1high, DLL1, HES1, LEF1, but also HNF1B (Figure 2, A–C and E; DLL1 data not shown) and the proximal, parietal, and visceral segments by WT1, MAFB, FOXC2, and JAG1low (Figure 2, A, B, and D).
Figure 2.
Nephron patterning through to the SSB stage in human and mouse kidneys. (A–F) and (A’–F’) Immunofluorescent stains and in situ hybridization on human and mouse kidneys, respectively. Ages and stains as specified on fields. Stars in (F) and (F’) indicate the nephron axes: green, distal; orange, proximal; magenta indicates the ureteric bud. Scale bars indicate 10 µm. CLN, capillary loop stage nephron; CSB, comma-shaped body; IPC, interstitial progenitor cells; UB, ureteric bud.
Rather than sharp boundaries of gene expression, the gradual reduction of gene expression at proximal and distal boundaries generated partially overlapping domains of gene expression, and consequently, a greater potential for cell heterogeneity than is recognized by the current ontology. For example, the region between the distal and the medial domains displayed strong LEF1 labeling and lower but overlapping labeling for SOX9 and JAG1. Similarly, Wnt4/WNT4 transiently demarcates a subset of the proximal segment of the SSB that directly contacts the ureteric epithelium, a region sandwiched between the presumptive renal corpuscle lineages and medial segment; Wnt4/WNT4 expression was rapidly lost after the SSB stage (Figure 2F). Though boundary positions may shift along the proximal distal axis; overall, human and mouse nephrons displayed conserved boundaries of gene activity though transitions varied. As examples, human WT1 extends further into the medial segment than mouse Wt1, whereas human JAG1 shows sharper boundaries than its mouse counterpart (Figure 2, B and D).
Mapping Transcription Factors to the Developing Human Nephron Reveals Additional Cellular Complexity
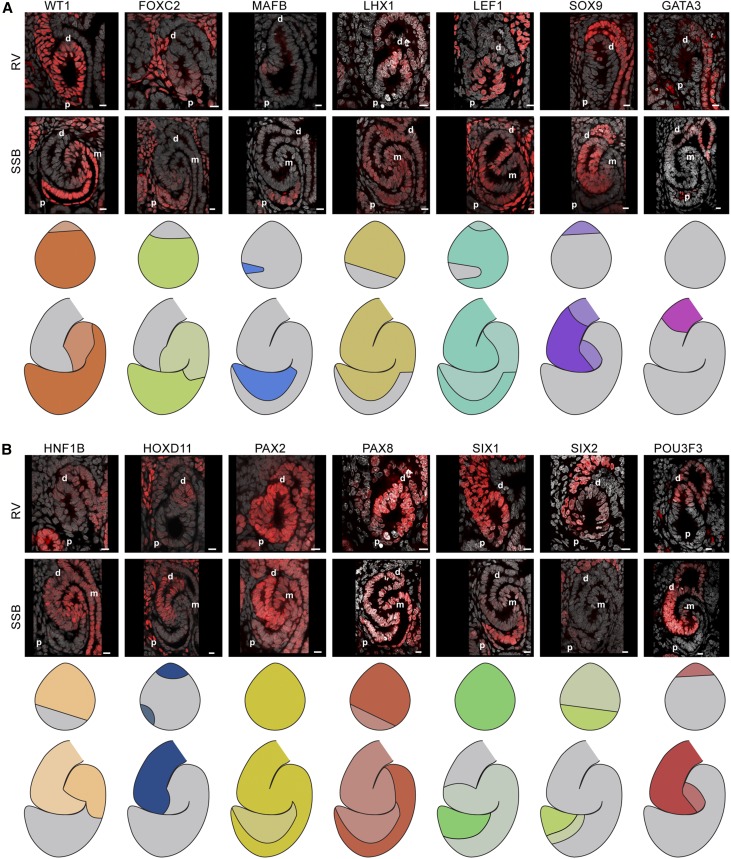
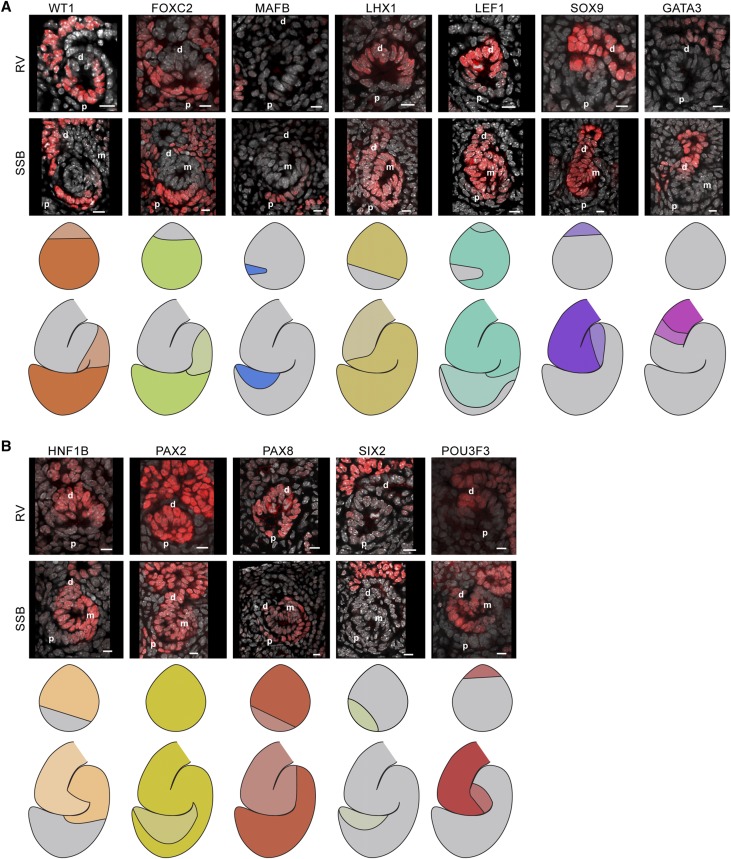
To better define the domains in the human SSB and relate the molecular organization between the RV and SSB, we performed immunostaining for 14 transcription factors present in the mouse and human RV and SSB (WT1, FOXC2, MAFB, LHX1, LEF1, SOX9, GATA3, HNF1B, HOXD11, PAX2, PAX8, SIX1, SIX2, and POU3F3) and mapped their localization to nephron models extending the current mouse-focused understanding of individual factors to a high-resolution synthesis of the data (Figures 3 and 4).17,22,27,28,33,35,41,44,49,50,52–54
Figure 3.
Transcription factor maps in the human renal vesicle and SSB nephron. (A and B) Single-channel immunofluorescent stains with DAPI showing transcription factor localization patterns in renal vesicles and SSB nephrons. Nephron model schematics indicate where the transcription factor is present; a two-level color scheme used to indicate strong and weak detection where applicable. Scale bars indicate 10 µm. Proximal (p), medial (m), and distal (d) segments indicated on fields.
Figure 4.
Transcription factor maps in the mouse renal vesicle and SSB nephron. (A and B) Single-channel immunofluorescent stains with DAPI showing transcription factor localization patterns in renal vesicles and SSB nephrons. Nephron model schematics indicate where the transcription factor is present; a two-level color scheme used to indicate strong and weak detection where applicable. Scale bars indicate 10 µm. Proximal (p), medial (m), and distal (d) segments indicated on fields.
Simplified comparative domain maps made on the basis of the markers above were compiled for the mouse and human RV (Figures 3A and 4A) and SSB (Figures 3B and 4B). These maps predict additional molecular diversity beyond those defined by current working ontologies (Figure 5C). The human RV model indicated the presence of at least six domains (Figure 5A): two distal domains (1–2), a large medial (3), and three proximal (4–6) domains. The SSB model suggested nine domains (Figure 5A); domains 4 and 6 were intersected by multiple protein domains so additional molecular heterogeneity is expected within these cell populations. The mouse models displayed analogous domains (Figure 5B). To test whether the predicted cellular diversity from these models was reflected by actual molecular diversity, we costained human RVs and SSBs for WT1, FOXC2, and MAFB, which are predicted to divide the RV into regions 1, 2, 3–5, and 6, and distinguish between domains 1–3, 4, 5, 6, 7, and 8–9 in the SSB (Figure 5D). Consistent with the model, this analysis detected predicted domains on the basis of the presence, absence, and levels of these three factors (Figure 5E). Further, the transcription factors HOXD11, GATA3, and SOX9 separated the RVs into domains 1, 2–5, and 6 and 1, 2, 2–3, 4, and 5–9 in the SSB (Figure 5G), as predicted (Figure 5F). In summary, there is clearly greater molecular and cellular heterogeneity in the early stages of nephrogenesis than previously documented. Further, the conservation in the observed heterogeneity between the mouse and human suggests a functional relevance to the emerging pattern of the mammalian nephron.
Figure 5.
Diversity in the human and mouse renal vesicle and SSB nephron. (A–C) Predicted cellular diversity in the human and mouse RV and SSB and current ontological terms. (D and E) Predicted and tested subdomains identified by detection of WT1, FOXC2, and MAFB in human nephrons. (F and G) Predicted and tested subdomains identified by detection of SOX9, GATA3, and HOXD11 in human nephrons. Dashed magenta line outlines nephrons. Dashed orange line indicates region where the transcription factor is detected.
Delineating the Emergence of Mature Nephron Segment Markers in the Early Nephron
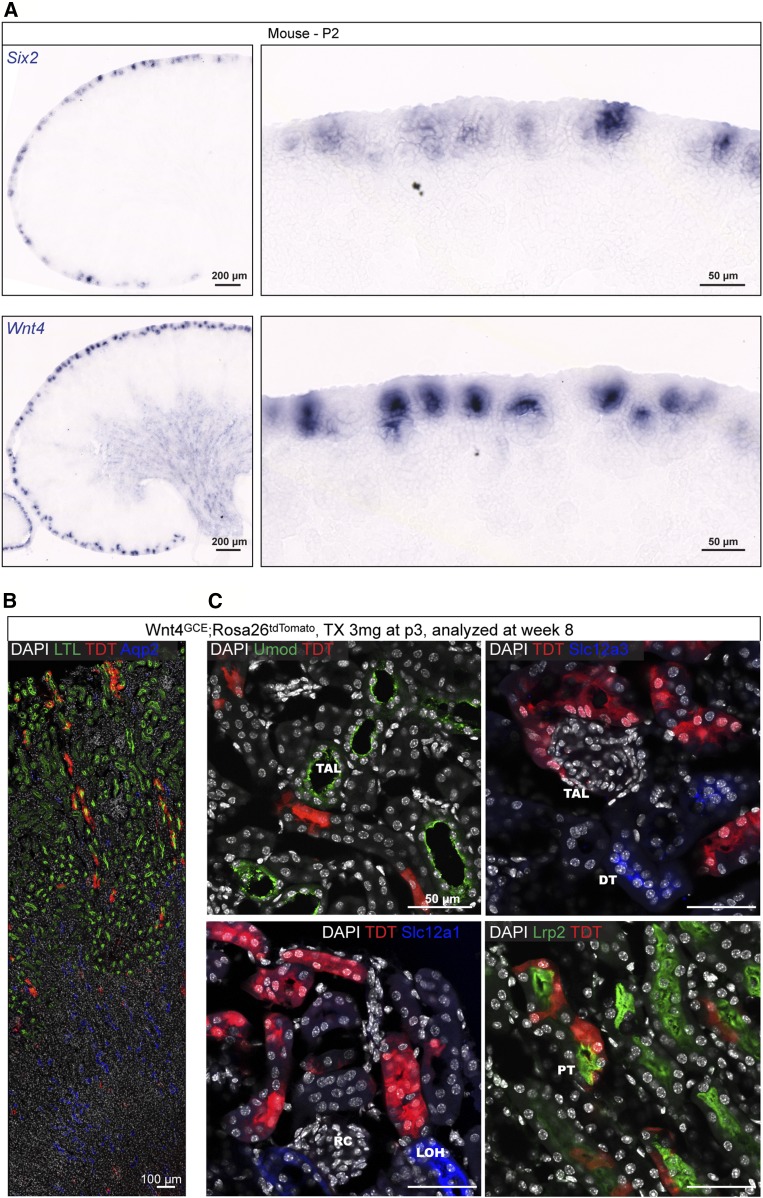
Domain 6 in the SSB closely aligns with the domain of Wnt4/WNT4 expression (Figure 2F). To delineate this precursor/mature-segment relationship, we performed Wnt4-CRE–mediated fate-mapping in the mouse kidney. At P2, NPC populations are depleted and the remaining Six2-expressing cells are found within epithelializing structures (Figure 6A). Wnt4 is at this point expressed in some PTA stages, but predominantly in RVs and SSBs (Figure 6A). Therefore, injection of tamoxifen at P3 into neonatal Wnt4CreERT2;Rosa26td-Tomato mice4,55 results in stable activation of td-Tomato in descendants of domain 6. Analysis of kidneys in adult mice at week 8 showed td-Tomato+ cells generated exclusively LTL and Lrp2+ proximal tubule cells in the kidney cortex (Figure 6, B and C)56,57; labeled cells were negative for renal corpuscle markers and for Umod, Slc12a1, and Slc12a3, which demarcate the ascending loop of Henle and the distal convoluted tubule.58–60
Figure 6.
Fate mapping of SSB nephron Wnt4 expression to an adult nephron segment. (A) In situ hybridization on P2 mouse kidneys for Six2 and Wnt4. (B and C) td-Tomato+ cells in a week 8 mouse kidney post fate-mapping from P3. Immunofluorescent stains as stated on fields. DT, distal tubule; LOH, loop of Henle; PT, proximal tubule; RC, renal corpuscle; TAL, thin ascending limb of the loop of Henle.
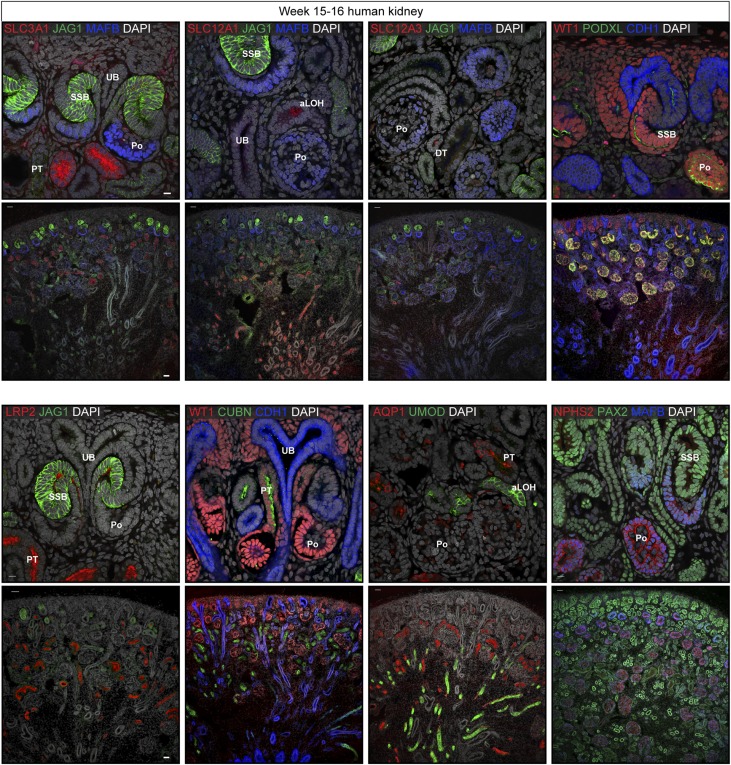
The observation that distinct transcriptional domains exist within the SSB begs the question, when are functional proteins mediating physiologic actions of mature nephron segments first detected? Temporally, mature nephron segments first emerge between week 10 and 11 of human development.40 However, how their emergence reflects patterning within the SSB or later capillary loop stage nephron (CLS) that forms from elongation of the SSB is unclear. To address this question, we examined the distribution of well characterized proteins that have been used as markers of mature nephron identities in pluripotent stem cell-derived organoid models of kidney development; from proximal to distal: the renal corpuscle and podocyte markers PODXL, NPHS2, WT1, and MAFB; proximal tubule markers SLC3A1, LRP2, CUBN, and AQP1; ascending loop of Henle markers SLC12A1 and UMOD; and distal convoluted tubule marker SLC12A3. LRP2, CUBN, PODXL, MAFB, WT1, and NPHS2 were first detected at the SSB stage (Figure 7). SLC3A1 was first detected at low levels in the late SSB, whereas AQP1, UMOD, and SLC12A1 emerged in CLS nephrons. UMOD and SLC12a3 were first detected after the CLS in more elongated loops of Henle and distal tubules, respectively (Figure 7, Tables 1 and 2). These data are consistent with the general view of a proximal to distal progression in the local production of key proteins underlying regional nephron functions. However, each of these proteins are present at markedly elevated levels in functional nephrons (compare high and low power fields in Figure 7).
Figure 7.
Activation of mature cell-lineage markers in the early development nephron. Immunofluorescent stains for markers of the distal, proximal, loop of Henle, and renal corpuscle domains of the nephron. Stains as specified on fields. Tissue from a week 5–16 human kidney. Scale bar is 10 µm and 50 µm in higher and lower magnification fields, respectively. aLOH, ascending loop of Henle; DT, distal tubule; Po, podocytes; PT, proximal tubule; UB, ureteric bud.
Discussion
We report a detailed characterization of the anatomic and molecular patterning of the human nephron from induction to SSB formation. The findings complement recent descriptions of human kidney development and comparative studies of the mouse and human nephrogenic niche.40,42 The data lead to four important findings. First, inductive programs are demonstrably active in NPCs in the human nephrogenic niche. Second, there is significantly greater cellular diversity in developing nephrons with strong conservation between the human and mouse kidney. Third, fate tracing to link emerging patterns with mature structures demonstrates that a Wnt4+ population localized to the proximal SSB comprises proximal tubule precursors in the mouse, and likely the human kidney. Finally, the analysis of segment specific markers of mature nephron cell-types connects patterning with the emerging regional anatomy of the functional mammalian nephron.
Early Inductive Responses Initiate in the Human Cap Mesenchyme
In the mouse, Lef1 is first detected in the PTA and is thought to be a direct response to canonical Wnt/β-catenin signaling from the ureteric bud.6,22 This is considered a key event in nephron induction, followed by the activation of Wnt target genes Pax8, Wnt4, and Lhx1, each essential for nephron formation and epithelialization.16,17,19–21,28 In the human niche, LEF1 is detected in NPCs nearest to the ureteric tip and in NPCs connecting to developing PTAs and RVs, whereas in the mouse LEF1 has a later onset in already formed RVs. Thus, human NPCs display overt Wnt-driven commitment to nephrogenesis at an earlier stage the mouse. Further, the human niche appears to form a continuum of LEF1+/PAX8+ NPCs extending to coalescing PTAs and RVs, a population not readily detected in either the E15.5 or P2 mouse kidney. A clear physical separation between NPCs and already committed progeny may not occur until the RV stage and it is only after incorporation into the PTA and RV that WNT4 and LHX1 turn on, as in the E15.5 and P2 mouse kidney.
One explanation for the differences here may be the distinct kinetics of mouse and human nephrogenesis. Previous estimates suggest that the transition from PTA to SSB can takes up to 3–8 days in humans, but <24 hours in mice.40 Consequently, a more protracted process in the human kidney could lend increased temporal resolution, enabling distinct stages in the nephrogenic program to be more readily distinguished. Interestingly, the temporal order to the NPC induction with LEF1, likely reporting elevated Wnt signaling, followed by PAX8, WNT4, and LHX1 is consistent with genetic studies in the mouse that have placed Pax8 upstream of Wnt416 and Wnt4 upstream of Lhx1.10
Cellular Diversity and Patterning of the Early Nephron
Current mouse and human kidney anatomic ontologies (www.gudmap.org) propose three terms for the RV and six terms for the SSB. The mapping of 14 transcription factors, chosen for their readily identifiable nuclear organization and direct relevance to patterning events themselves, identified at least six domains in the RV, dividing the RV into three distal and medial domains, and three proximal domains. Domain six corresponded to the region where active NPC recruitment was occurring displaying the highest levels of SIX2 presumably reflecting the recent recruitment from SIX2+ NPCs and the perdurance of SIX2 mRNA and protein in recruited cells. MAFB+ cells first emerged in this RV domain. In the SSB, we detected nine domains with additional diversity detected in the distal and medial segments, each comprising three domains. Of note, domain six in SSBs faithfully recapitulated an expression domain of WNT4 in the SSB (Figure 2F). This region of the SSB is in close proximity to the ureteric epithelium (Figure 2, D and F), where it would be predicted to receive high levels of collecting duct–derived ligands such as WNT9B and LIF.6,15 Importantly, costaining with selected proximal (WT1, FOXC2, MAFB) and distal (HOXD11, GATA3, SOX9) protein marker sets validated predictions from the composite models on the basis of the distribution of single factors.
The organization predicted here can be further tested through single-cell RNA analysis and should serve as a useful template for relational mapping of these datasets and for exploring the in vivo relevance of nephron patterning processes in in vitro kidney organoid models of mouse and human nephrogenesis. Establishing direct evidence to link domains of emerging cell diversity to the distinct anatomy of the mature nephron is challenging, especially where coexpression of more than one factor, or levels of factor activity, complicate genetic approaches. However, we were able to show that the SSB-specific Wnt4 domain corresponds with cells fated to give rise to proximal tubule cells. Similar studies will only be possible in the human kidney in organoid models, and these will only be relevant to normal development if normal nephrogenic programs are shown to occur in vitro.
Nephron Patterning and Mature Kidney Markers
Analysis of the onset of detectable levels of several key markers of mature cell types in the adult nephron suggest a proximal to distal hierarchy in nephron maturation, which is consistent with distal identities forming last during kidney development.40 Delayed development of distal identities compared with proximal ones may reflect a primary need to generate a proximal filtration device before any other functions are necessary, and a secondary requirement for recovery of low molecular weight compounds (e.g., glucose, sodium), as would be critical to organismal fitness. Mature distal cell types have not been reported in pluripotent stem cell-derived renal organoids,61–63 also suggestive of a late developmental time for the maturation of these fates or the absence of environmental cues directing their development.
Concise Methods
The protocols as relating to human kidney material, animal husbandry, in situ hybridization, immunolabeling, and microscopy are as described previously in this series of articles.40,42 Specific details pertinent to this study are described here.
Animal Care and Embryo Collection
All animal work was reviewed and institutionally approved by Institutional Animal Care and Use Committees at the University of Southern California and performed according to institutional guidelines. Wnt4GCE mice were generated as described previously4 and were mated with Rosa26tdToamto mice (B6.Cg-Gt(ROSA)26Sortm14(CAG-td-Tomato)Hze/J)55 obtained from Jackson Laboratories. Timed matings were set up to recover neonates at the appropriate age. Tamoxifen was injected at P3 and kidneys collected at week 8. Three experimental animals were sectioned and stained for this analysis.
Antibody-Directed Analyses
Immunofluorescence staining was performed as outlined previously40 with the following primary antibodies: PAX2 (AF3364, 1:500; R&D Systems; 901001, 1:500; Biolegend), PAX8 (189249, 1:1000; Abcam), HES1 (11988, 1:300; Cell Signaling), SALL1 (PP-K9814–00, 1:500; R&D Systems), WT1 (ab89901, 1:1000; Abcam), FOXC2 (AF6989, 1:500; R&D Systems), LHX1 (sc-19341, 1:300; Santa Cruz Biotechnology), LEF1 (2230, 1:300; Cell Signaling; sc-8591, 1:100; Santa Cruz Biotechnology), SOX9 (ab185230, 1:1000; Abcam), GATA3 (AF2605, 1:300; R&D Systems), HNF1B (sc-22840, 1:300; Santa Cruz Biotechnology), HOXD11 (ab60715, 1:300; Abcam), POU3F3 (PA5–64311, 1:1000; Thermo Fisher Scientific), SIX1 (12891, 1:1000; Cell Signaling), JAG1 (AF599, 1:300; R&D Systems), CUBN (sc-20607, 1:500; Santa Cruz Biotechnology), AQP1 (ab168387, 1:1000; Abcam), AQP2 (sc-9882, 1:300; Santa Cruz Biotechnology), SLC3A1 (HPA038360, 1:500; Sigma), CALB1 (C9848, 1:300; Sigma), MAFB (sc-10022, 1:300; Santa Cruz Biotechnology), NPHS2 (ab50339, 1:10,000; Abcam), LRP2 (MBS690201, 1:1000; MyBioSource), SLC12A1 (HPA018107, 1:1000; Sigma), UMOD (AF5144 and AF5175, 1:500; R&D Systems), SLC12A3 (HPA028748, 1:300; Sigma), SIX2 (SAB1401533, 1:500; Sigma Aldrich), SIX2 (MBS610128, 1:1000; MyBioSource), CITED1 (ab55467, 1:300; Abcam), KRT8 (troma-1, 1:50; DSHB), β-laminin (sc-33709, 1:300; Santa Cruz Biotechnology), and CDH1 (610182, 1:300; BD Transduction Laboratories). Secondary antibodies were purchased from Molecular Probes (Thermo Fisher Scientific) and used at a 1:1000 dilution.
Sample Numbers Analyzed
The number (n) of independent human fetal kidneys analyzed for each antibody were as follows: CITED1 (n=6), LEF1 (n=6), KRT8 (n=6), SIX2 (n=6), PAX8 (n=3), WT1 (n=6), JAG1 (n=6), CDH1 (n=6), FOXC2 (n=6), HNF1B (n=6), HES1 (n=3), SOX9 (n=4), MAFB (n=5), LHX1 (n=3), GATA3 (n=4), HOXD11 (n=3), PAX2 (n=4), SIX1 (n=5), SIX2 (n=5), POU3F3 (n=3), SLC3A1 (n=3), SLC12A1 (n=4), SLC12A3 (n=4), PODXL (n=3), LRP2 (n=4), CUBN (n=4), UMOD (n=3), AQP1 (n=4), NPHS2 (n=4), CALB1 (n=4), and SALL1 (n=3).
In Situ Hybridization and Confocal Imaging
In situ hybridization on frozen sections of mouse and human kidney samples followed previously published procedures40,42 (https://www.gudmap.org/Research/Protocols/McMahon.html). Imaging of immunofluorescent and in situ hybridization signals was performed as described previously.40,42
Nephron Models
To generate nephron models of transcription factor patterns, kidneys were immunostained for each transcription factor. Schematized anatomic models were generated for nephrons representing two-dimensional sections through the mid-line of RVs or SSB nephrons. Immunofluorescent stains were considered, and localization patterns binned into visually distinguishable levels of protein intensity. Each antibody stain was detectable within a range, and was binned into three categories, thereby likely reducing actual complexity: absent, detected, or detected at a high level (gray, medium intensity color, intense color). Each factor map is underpinned by examining greater than five RV and SSB stages, together with intermediate stages, to produce maps representing the location of specific sets of markers. Sample numbers are indicated elsewhere in the Concise Methods section. To generate intersection maps of protein localization patterns, semitransparent maps were superimposed in Illustrator (Adobe) (Figure 4, A and B). On merging, larger and smaller domains became apparent, highlighted by the distribution of the various markers. Domains which were intersected multiple times in analysis of adjacent sections were simplified into a single broader domain (e.g., domain 4).
Disclosures
None.
Supplementary Material
Acknowledgments
We thank all members of the McMahon laboratory for helpful discussion. We thank Dr. Rachel Steward and Dr. Melissa Wilson for their help providing tissue samples and with institutional review board approval processes. N.O.L., T.T., and A.P.M. planned experiments and analyzed data. N.O.L. and T.T. assembled the figures. N.O.L., T.T., R.K.P., J.G., and E.R. collected data. M.E.T. and B.G. provided embryonic and fetal kidneys. N.O.L. and A.P.M. wrote the manuscript, incorporating input from all authors.
“We thank Hongsuda Tangmunarunkit and Laura Pearlman for their work integrating the imaging data into the GenitoUrinary Development Molecular Anatomy Project (GUDMAP) database.”
Work in A.P.M.’s laboratory was supported by grants from the National Institutes of Health (DK107350, DK094526, DK110792) and the California Institute for Regenerative Medicine (LA1-06536).
Footnotes
Published online ahead of print. Publication date available at www.jasn.org.
See related editorials, “The Era of Human Developmental Nephrology,” and “Evolution and Kidney Development: A Rosetta Stone for Nephrology,” on pages 705–706 and 706–709, respectively.
This article contains supplemental material online at http://jasn.asnjournals.org/lookup/suppl/doi:10.1681/ASN.2017091036/-/DCSupplemental.
References
- 1.Saxen L: Organogenesis of the Kidney, Cambridge, Cambridge University Press, 1987 [Google Scholar]
- 2.McMahon AP: Development of the mammalian kidney. Curr Top Dev Biol 117: 31–64, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Costantini F, Kopan R: Patterning a complex organ: Branching morphogenesis and nephron segmentation in kidney development. Dev Cell 18: 698–712, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kobayashi A, Valerius MT, Mugford JW, Carroll TJ, Self M, Oliver G, McMahon AP: Six2 defines and regulates a multipotent self-renewing nephron progenitor population throughout mammalian kidney development. Cell Stem Cell 3: 169–181, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Boyle S, Misfeldt A, Chandler KJ, Deal KK, Southard-Smith EM, Mortlock DP, Baldwin HS, de Caestecker M: Fate mapping using Cited1-CreERT2 mice demonstrates that the cap mesenchyme contains self-renewing progenitor cells and gives rise exclusively to nephronic epithelia. Dev Biol 313: 234–245, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Carroll TJ, Park JS, Hayashi S, Majumdar A, McMahon AP: Wnt9b plays a central role in the regulation of mesenchymal to epithelial transitions underlying organogenesis of the mammalian urogenital system. Dev Cell 9: 283–292, 2005 [DOI] [PubMed] [Google Scholar]
- 7.Karner CM, Das A, Ma Z, Self M, Chen C, Lum L, Oliver G, Carroll TJ: Canonical Wnt9b signaling balances progenitor cell expansion and differentiation during kidney development. Development 138: 1247–1257, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brown AC, Muthukrishnan SD, Guay JA, Adams DC, Schafer DA, Fetting JL, Oxburgh L: Role for compartmentalization in nephron progenitor differentiation. Proc Natl Acad Sci U S A 110: 4640–4645, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies JA, Garrod DR: Induction of early stages of kidney tubule differentiation by lithium ions. Dev Biol 167: 50–60, 1995 [DOI] [PubMed] [Google Scholar]
- 10.Park J-S, Valerius MT, McMahon AP: Wnt/beta-catenin signaling regulates nephron induction during mouse kidney development. Development 134: 2533–2539, 2007 [DOI] [PubMed] [Google Scholar]
- 11.Kuure S, Popsueva A, Jakobson M, Sainio K, Sariola H: Glycogen synthase kinase-3 inactivation and stabilization of beta-catenin induce nephron differentiation in isolated mouse and rat kidney mesenchymes. J Am Soc Nephrol 18: 1130–1139, 2007 [DOI] [PubMed] [Google Scholar]
- 12.Mao Y, Francis-West P, Irvine KD: Fat4/Dchs1 signaling between stromal and cap mesenchyme cells influences nephrogenesis and ureteric bud branching. Development 142: 2574–2585, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Badouel C, Zander MA, Liscio N, Bagherie-Lachidan M, Sopko R, Coyaud E, Raught B, Miller FD, McNeill H: Fat1 interacts with Fat4 to regulate neural tube closure, neural progenitor proliferation and apical constriction during mouse brain development. Development 142: 2781–2791, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Das A, Tanigawa S, Karner CM, Xin M, Lum L, Chen C, Olson EN, Perantoni AO, Carroll TJ: Stromal–epithelial crosstalk regulates kidney progenitor cell differentiation. Nat Cell Biol 15: 1035–1044, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barasch J, Yang J, Ware CB, Taga T, Yoshida K, Erdjument-Bromage H, Tempst P, Parravicini E, Malach S, Aranoff T, Oliver JA: Mesenchymal to epithelial conversion in rat metanephros is induced by LIF. Cell 99: 377–386, 1999 [DOI] [PubMed] [Google Scholar]
- 16.Stark K, Vainio S, Vassileva G, McMahon AP: Epithelial transformation of metanephric mesenchyme in the developing kidney regulated by Wnt-4. Nature 372: 679–683, 1994 [DOI] [PubMed] [Google Scholar]
- 17.Bouchard M, Souabni A, Mandler M, Neubüser A, Busslinger M: Nephric lineage specification by Pax2 and Pax8. Genes Dev 16: 2958–2970, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grieshammer U, Cebrián C, Ilagan R, Meyers E, Herzlinger D, Martin GR: FGF8 is required for cell survival at distinct stages of nephrogenesis and for regulation of gene expression in nascent nephrons. Development 132: 3847–3857, 2005 [DOI] [PubMed] [Google Scholar]
- 19.Kispert A, Vainio S, McMahon AP: Wnt-4 is a mesenchymal signal for epithelial transformation of metanephric mesenchyme in the developing kidney. Development 125: 4225–4234, 1998 [DOI] [PubMed] [Google Scholar]
- 20.Burn SF, Webb A, Berry RL, Davies JA, Ferrer-Vaquer A, Hadjantonakis AK, Hastie ND, Hohenstein P: Calcium/NFAT signalling promotes early nephrogenesis. Dev Biol 352: 288–298, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tanigawa S, Wang H, Yang Y, Sharma N, Tarasova N, Ajima R, Yamaguchi TP, Rodriguez LG, Perantoni AO: Wnt4 induces nephronic tubules in metanephric mesenchyme by a non-canonical mechanism. Dev Biol 352: 58–69, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mugford JW, Yu J, Kobayashi A, McMahon AP: High-resolution gene expression analysis of the developing mouse kidney defines novel cellular compartments within the nephron progenitor population. Dev Biol 333: 312–323, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Georgas K, Rumballe B, Valerius MT, Chiu HS, Thiagarajan RD, Lesieur E, Aronow BJ, Brunskill EW, Combes AN, Tang D, Taylor D, Grimmond SM, Potter SS, McMahon AP, Little MH: Analysis of early nephron patterning reveals a role for distal RV proliferation in fusion to the ureteric tip via a cap mesenchyme-derived connecting segment. Dev Biol 332: 273–286, 2009 [DOI] [PubMed] [Google Scholar]
- 24.Kao RM, Vasilyev A, Miyawaki A, Drummond IA, McMahon AP: Invasion of distal nephron precursors associates with tubular interconnection during nephrogenesis. J Am Soc Nephrol 23: 1682–1690, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindström NO, Lawrence ML, Burn SF, Johansson JA, Bakker ER, Ridgway RA, Chang C-H, Karolak MJ, Oxburgh L, Headon DJ, Sansom OJ, Smits R, Davies JA, Hohenstein P: Integrated β-catenin, BMP, PTEN, and Notch signalling patterns the nephron. eLife 3: e04000, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barker N, Rookmaaker MB, Kujala P, Ng A, Leushacke M, Snippert H, van de Wetering M, Tan S, Van Es JH, Huch M, Poulsom R, Verhaar MC, Peters PJ, Clevers H: Lgr5(+ve) stem/progenitor cells contribute to nephron formation during kidney development. Cell Reports 2: 540–552, 2012 [DOI] [PubMed] [Google Scholar]
- 27.Nakai S, Sugitani Y, Sato H, Ito S, Miura Y, Ogawa M, Nishi M, Jishage K, Minowa O, Noda T: Crucial roles of Brn1 in distal tubule formation and function in mouse kidney. Development 130: 4751–4759, 2003 [DOI] [PubMed] [Google Scholar]
- 28.Kobayashi A, Kwan K-M, Carroll TJ, McMahon AP, Mendelsohn CL, Behringer RR: Distinct and sequential tissue-specific activities of the LIM-class homeobox gene Lim1 for tubular morphogenesis during kidney development. Development 132: 2809–2823, 2005 [DOI] [PubMed] [Google Scholar]
- 29.Chen L, Al-Awqati Q: Segmental expression of Notch and Hairy genes in nephrogenesis. Am J Physiol Renal Physiol 288: F939–F952, 2005 [DOI] [PubMed] [Google Scholar]
- 30.Cheng H-T, Kim M, Valerius MT, Surendran K, Schuster-Gossler K, Gossler A, McMahon AP, Kopan R: Notch2, but not Notch1, is required for proximal fate acquisition in the mammalian nephron. Development 134: 801–811, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu Z, Chen S, Boyle S, Zhu Y, Zhang A, Piwnica-Worms DR, Ilagan MXG, Kopan R: The extracellular domain of Notch2 increases its cell-surface abundance and ligand responsiveness during kidney development. Dev Cell 25: 585–598, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Reggiani L, Raciti D, Airik R, Kispert A, Brändli AW: The prepattern transcription factor Irx3 directs nephron segment identity. Genes Dev 21: 2358–2370, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heliot C, Desgrange A, Buisson I, Prunskaite-Hyyryläinen R, Shan J, Vainio S, Umbhauer M, Cereghini S: HNF1B controls proximal-intermediate nephron segment identity in vertebrates by regulating Notch signalling components and Irx1/2. Development 140: 873–885, 2013 [DOI] [PubMed] [Google Scholar]
- 34.Maezawa Y, Onay T, Scott RP, Keir LS, Dimke H, Li C, Eremina V, Maezawa Y, Jeansson M, Shan J, Binnie M, Lewin M, Ghosh A, Miner JH, Vainio SJ, Quaggin SE: Loss of the podocyte-expressed transcription factor Tcf21/Pod1 results in podocyte differentiation defects and FSGS. J Am Soc Nephrol 25: 2459–2470, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Moriguchi T, Hamada M, Morito N, Terunuma T, Hasegawa K, Zhang C, Yokomizo T, Esaki R, Kuroda E, Yoh K, Kudo T, Nagata M, Greaves DR, Engel JD, Yamamoto M, Takahashi S: MafB is essential for renal development and F4/80 expression in macrophages. Mol Cell Biol 26: 5715–5727, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Takemoto M, He L, Norlin J, Patrakka J, Xiao Z, Petrova T, Bondjers C, Asp J, Wallgard E, Sun Y, Samuelsson T, Mostad P, Lundin S, Miura N, Sado Y, Alitalo K, Quaggin SE, Tryggvason K, Betsholtz C: Large-scale identification of genes implicated in kidney glomerulus development and function. EMBO J 25: 1160–1174, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Peter K: Untersuchungen über Bau und Entwicklung der Niere, Jena, Verlag von G. Fischer, 1927 [Google Scholar]
- 38.Potter EL: Normal and Abnormal Development of the Kidney, Chicago, Year Book Medical Publishers, Inc., 1972 [Google Scholar]
- 39.Oliver J: Nephrons and Kidneys: A Quantitative Study of Development and Evolutionary Mammalian Renal Architectonics, New York, Harper & Row, 1968 [Google Scholar]
- 40.Lindström NO, McMahon JA, Guo J, Tran T, Guo Q, Rutledge E, Parvez RK, Saribekyan G, Schuler RE, Liao C, Kim AD, Abdelhalim A, Ruffins SW, Thornton ME, Basking L, Grubbs B, Kesselman C, McMahon A: Conserved and divergent features of human and mouse kidney organogenesis. J Am Soc Nephrol 29: 785–805, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O’Brien LL, Guo Q, Lee Y, Tran T, Benazet JD, Whitney PH, Valouev A, McMahon AP: Differential regulation of mouse and human nephron progenitors by the Six family of transcriptional regulators. Development 143: 595–608, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lindström NO, Guo J, Kim AD, Tran T, Guo Q, De Sena Brandine G, Ransick A, Parvez R, Thornton ME, Basking L, Grubbs B, McMahon JA, Smith AD, McMahon AP: Conserved and divergent features of mesenchymal progenitor cell types within the cortical nephrogenic niche of the mouse and human kidney. J Am Soc Nephrol 29: 806–824, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li Z, Araoka T, Wu J, Liao HK, Li M, Lazo M, Zhou B, Sui Y, Wu M-Z, Tamura I, Xia Y, Beyret E, Matsusaka T, Pastan I, Rodriguez Esteban C, Guillen I, Guillen P, Campistol JM, Izpisua Belmonte JC: 3D culture supports long-term expansion of mouse and human nephrogenic progenitors. Cell Stem Cell 19: 516–529, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Self M, Lagutin OV, Bowling B, Hendrix J, Cai Y, Dressler GR, Oliver G: Six2 is required for suppression of nephrogenesis and progenitor renewal in the developing kidney. EMBO J 25: 5214–5228, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thiagarajan RD, Georgas KM, Rumballe BA, Lesieur E, Chiu HS, Taylor D, Tang DTP, Grimmond SM, Little MH: Identification of anchor genes during kidney development defines ontological relationships, molecular subcompartments and regulatory pathways. PLoS One 6: e17286, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Short KM, Combes AN, Lefevre J, Ju AL, Georgas KM, Lamberton T, Cairncross O, Rumballe BA, McMahon AP, Hamilton NA, Smyth IM, Little MH: Global quantification of tissue dynamics in the developing mouse kidney. Dev Cell 29: 188–202, 2014 [DOI] [PubMed] [Google Scholar]
- 47.Vestweber D, Kemler R, Ekblom P: Cell-adhesion molecule uvomorulin during kidney development. Dev Biol 112: 213–221, 1985 [DOI] [PubMed] [Google Scholar]
- 48.Cho EA, Patterson LT, Brookhiser WT, Mah S, Kintner C, Dressler GR: Differential expression and function of cadherin-6 during renal epithelium development. Development 125: 803–812, 1998. 9449663 [Google Scholar]
- 49.Reginensi A, Clarkson M, Neirijnck Y, Lu B, Ohyama T, Groves AK, Sock E, Wegner M, Costantini F, Chaboissier MC, Schedl A: SOX9 controls epithelial branching by activating RET effector genes during kidney development. Hum Mol Genet 20: 1143–1153, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kume T, Deng K, Hogan BL: Murine forkhead/winged helix genes Foxc1 (Mf1) and Foxc2 (Mfh1) are required for the early organogenesis of the kidney and urinary tract. Development 127: 1387–1395, 2000 [DOI] [PubMed] [Google Scholar]
- 51.Motojima M, Ogiwara S, Matsusaka T, Kim SY, Sagawa N, Abe K, Ohtsuka M: Conditional knockout of Foxc2 gene in kidney: Efficient generation of conditional alleles of single-exon gene by double-selection system. Mamm Genome 27: 62–69, 2016 [DOI] [PubMed] [Google Scholar]
- 52.Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JBL: The expression of the Wilms’ tumour gene, WT1, in the developing mammalian embryo. Mech Dev 40: 85–97, 1993 [DOI] [PubMed] [Google Scholar]
- 53.Xu P-X, Zheng W, Huang L, Maire P, Laclef C, Silvius D: Six1 is required for the early organogenesis of mammalian kidney. Development 130: 3085–3094, 2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kaku Y, Taguchi A, Tanigawa S, Haque F, Sakuma T, Yamamoto T, Nishinakamura R: PAX2 is dispensable for in vitro nephron formation from human induced pluripotent stem cells. Sci Rep 7: 4554, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H: A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci 13: 133–140, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moestrup SK, Birn H, Fischer PB, Petersen CM, Verroust PJ, Sim RB, Christensen EI, Nexø E: Megalin-mediated endocytosis of transcobalamin-vitamin-B12 complexes suggests a role of the receptor in vitamin-B12 homeostasis. Proc Natl Acad Sci U S A 93: 8612–8617, 1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laitinen L, Virtanen I, Saxén L: Changes in the glycosylation pattern during embryonic development of mouse kidney as revealed with lectin conjugates. J Histochem Cytochem 35: 55–65, 1987 [DOI] [PubMed] [Google Scholar]
- 58.Gamba G, Miyanoshita A, Lombardi M, Lytton J, Lee WS, Hediger MA, Hebert SC: Molecular cloning, primary structure, and characterization of two members of the mammalian electroneutral sodium-(potassium)-chloride cotransporter family expressed in kidney. J Biol Chem 269: 17713–17722, 1994 [PubMed] [Google Scholar]
- 59.Lytle C, Xu JC, Biemesderfer D, Forbush B: Distribution and diversity of Na-K-Cl cotransport proteins: A study with monoclonal antibodies. Am J Physiol 269: C1496–C1505, 1995 [DOI] [PubMed] [Google Scholar]
- 60.Pennica D, Kohr WJ, Kuang WJ, Glaister D, Aggarwal BB, Chen EY, Goeddel DV: Identification of human uromodulin as the Tamm-Horsfall urinary glycoprotein. Science 236: 83–88, 1987 [DOI] [PubMed] [Google Scholar]
- 61.Taguchi A, Kaku Y, Ohmori T, Sharmin S, Ogawa M, Sasaki H, Nishinakamura R: Redefining the in vivo origin of metanephric nephron progenitors enables generation of complex kidney structures from pluripotent stem cells. Cell Stem Cell 14: 53–67, 2014 [DOI] [PubMed] [Google Scholar]
- 62.Takasato M, Er PX, Chiu HS, Maier B, Baillie GJ, Ferguson C, Parton RG, Wolvetang EJ, Roost MS, Chuva de Sousa Lopes SM, Little MH: Kidney organoids from human iPS cells contain multiple lineages and model human nephrogenesis. Nature 526: 564–568, 2015 [DOI] [PubMed] [Google Scholar]
- 63.Morizane R, Lam AQ, Freedman BS, Kishi S, Valerius MT, Bonventre JV: Nephron organoids derived from human pluripotent stem cells model kidney development and injury. Nat Biotechnol 33: 1193–1200, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Boyle S, Shioda T, Perantoni AO, De Caestecker M: Cited1 and Cited2 are differentially expressed in the developing kidney but are not required for nephrogenesis. Dev Dyn 236: 2321–2330, 2007 [DOI] [PubMed] [Google Scholar]
- 65.Grote D, Boualia SK, Souabni A, Merkel C, Chi X, Costantini F, Carroll T, Bouchard M: Gata3 acts downstream of β-catenin signaling to prevent ectopic metanephric kidney induction. PLoS Genet 4: 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wellik DM, Hawkes PJ, Capecchi MR: Hox11 paralogous genes are essential for metanephric kidney induction. Genes Dev 16: 1423–1432, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Armstrong JF, Pritchard-Jones K, Bickmore WA, Hastie ND, Bard JBL: The expression of the Wilms' tumour gene, WT1, in the developing mammalian embryo. Mech Dev 40: 85–97, 1992 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.