Abstract
Understanding interactions between above- and belowground components of ecosystems is an important next step in community ecology. These interactions may be fundamental to predicting ecological responses to global change because indirect effects occurring through altered species interactions can outweigh or interact with the direct effects of environmental drivers. In a multiyear field experiment (2010–2015), we tested how experimental addition of a mutualistic leaf endophyte (Epichloë amarillans) associated with American beachgrass (Ammophila breviligulata) interacted with an altered precipitation regime (±30%) to affect the belowground microbial community. Epichloë addition increased host root biomass at the plot scale, but reduced the length of extraradical arbuscular mycorrhizal (AM) fungal hyphae in the soil. Under ambient precipitation alone, the addition of Epichloë increased root biomass per aboveground tiller and reduced the diversity of AM fungi in A. breviligulata roots. Furthermore, with Epichloë added, the diversity of root-associated bacteria declined with higher soil moisture, whereas in its absence, bacterial diversity increased with higher soil moisture. Thus, the aboveground fungal mutualist not only altered the abundance and composition of belowground microbial communities but also affected how belowground communities responded to climate, suggesting that aboveground microbes have potential for cascading influences on community dynamics and ecosystem processes that occur belowground.
Keywords: Epichloë amarillans, bacteria, Ammophila breviligulata, arbuscular mycorrhizal fungi, symbiosis, ecological succession
Symbiotic fungi living inside of a grass species alters the response of soil fungi and bacteria to precipitation.
INTRODUCTION
Studying the interactions between above- and belowground components of ecosystems, including microbial assemblages, is a critical frontier of community ecology (Van der Putten 2012; Philippot et al.2013). Understanding the direction and magnitude of above/belowground interactions may be fundamental to predicting ecological responses to global change because species interactions can create indirect effects that either exacerbate or ameliorate the direct effects of a changing climate, making net outcomes unpredictable (Tylianakis et al.2008; Kivlin, Emery and Rudgers 2013; Singer, Travis and Johst 2013).
Above/belowground interactions between foliar insects and soil microbes are well known to have strong effects on community and ecosystem processes (e.g. Kostenko et al.2012; Van der Putten 2012; A’Bear, Johnson and Jones 2014). However, a potentially important, yet little studied, dimension of integrating above/belowground systems involves interactions between above/belowground microorganisms. Belowground microbial symbionts of plants are recognized to play critical roles in terrestrial ecosystems (Bardgett and van der Putten 2014) such as decomposition (Rousk and Frey 2015), soil stabilization (Fokom et al.2012) and nitrogen cycling (Phillips, Ward and Jones 2014). By contrast, with the exception of foliar pathogens, the significance of aboveground microbial communities in plants has received less attention (Bacon et al.1977; Arnold and Lutzoni 2007; Omacini et al.2012). However, during the past 25 years, culturing, microscopy and sequence analysis of asymptomatic leaves and stems have revealed a ubiquitous and diverse community of bacterial and fungal endophytes that can have diverse ecological effects (Bacon and White 2000; Andrews and Hirano 2012).
One group of aboveground plant microorganisms that can have particularly pervasive belowground effects are the epichloid fungi (family Clavicipitaceae, genus Epichloë) (Leuchtmann et al.2014). These fungi occur systemically in aboveground plant tissues, often conferring protection against abiotic (drought, heat) or biotic (herbivores, foliar pathogens) stressors (Rodriguez et al.2009). Endophyte benefits to host plants can be exceptionally large, resulting in several-fold increases in plant survival, biomass or reproduction (Rudgers, Mattingly and Koslow 2005; Cheplick and Faeth 2009). Strong belowground effects of the Epichloë have been well documented in two systems thus far (Liu et al.2011; Omacini et al.2012). Most work has been on the well-studied forage and turf species, tall fescue grass (Schedonorus arundinaceus). In tall fescue, the presence of the leaf endophyte Epichloë coenophiala suppressed spore abundances of arbuscular mycorrhizal (AM) fungi in field soil (Chu-Chou et al.1992) and reduced AM colonization of roots in both the host plant (Mack and Rudgers 2008) and neighboring plant species (Antunes et al.2008). In tall fescue pastures, endophyte presence also reduced soil microbial biomass and soil respiration (Franzluebbers et al.1999; Franzluebbers and Stuedemann 2005, but see, Van Hecke, Treonis and Kaufman 2005). Mesocosm experiments showed endophyte-mediated suppression of soil archaea, high G+C gram-positive bacteria, Deltaproteobacteria and Planctomycetes in the tall fescue rhizosphere (Jenkins, Franzluebbers and Humayoun 2006), as well as reduced microbial utilization of several substrates (Buyer et al.2011). A recent long-term study, however, reported a higher relative abundance of AM fungi and suppression of Ascomycota in fields with the endophyte present (Rojas et al.2016), suggesting the possibility that above/belowground interactions shift during community succession. Epichloë-mediated changes in soil microbial composition may cause higher soil carbon sequestration (Iqbal et al.2012) and alter nitrogen dynamics in pastures (Franzluebbers and Stuedemann 2005; Bowatte et al.2011). Belowground responses to endophyte presence may derive in part from endophyte effects on the composition of plant root exudates (Novas et al.2011; Guo, McCulley and McNear 2015) or root volatile organic compounds (Rostas, Cripps and Silcock 2015). In addition, endophyte presence in litter or live plants can slow rates of litter decomposition (Lemons, Clay and Rudgers 2005; Omacini et al.2012).
Prior studies have primarily focused on tall fescue–Epichloë interactions due to its economic and agricultural importance; however, in native ecosystems, the potential for above/belowground interactions remains largely unresolved (Cheplick and Faeth 2009; Omacini et al.2012). Interestingly, the few reported effects in native ecosystems thus far show opposite patterns to those in agronomic ecosystems; thus, further exploration of native endophyte symbioses is necessary. For example, surveys of populations with naturally high Epichloë prevalence showed higher abundance and colonization rates of AM fungi for two native host plant species (Novas, Cabral and Godeas 2005; Novas et al.2009). In addition, experimental work in a third native grass species showed that Epichloë increased the abundance of mutualistic AM fungal species and reduced parasitic AM fungal taxa, with net benefits to plant performance (Larimer, Bever and Clay 2012). Finally, in contrast to managed agronomic ecosystems, the effects of aboveground endophytes in native ecosystems may show higher context dependency in their ecological outcomes, i.e. the outcome of their interaction with the plant depends on exogenous, environmental conditions, due to a longer evolutionary history of above/belowground interactions and the absence of artificial selection on the aboveground symbiosis (Cheplick and Faeth 2009).
Understanding the degree of such context-dependency could improve our ability to predict outcomes of above/belowground interactions under future climates. Here, we investigated the influence of an aboveground fungal endophyte symbiosis on belowground microbes in a native dune ecosystem to increase understanding of the importance and prevalence of above/belowground interactions in natural ecosystems. To evaluate the degree of context-dependency, we altered precipitation (±30% ambient) to replicate projected climate changes for Great Lakes dune ecosystems (Emery, Bell-Dereske and Rudgers 2015). Coastal and lacustrine ecosystems are expected to be amongst the most vulnerable to climate change due to their already fragile nature and predicted increases in the intensity of severe weather events, such as storms and droughts, which will accelerate erosion and reduce dune stability (Schlacher et al.2008). Drought may be a particularly important element of climate change for dune plants and microbes in the Great Lakes region. The survival of native dune plants has been shown to be water limited along Lake Michigan (Lichter 2000; Ensign, Webb and Longstaffe 2006), and water was more limiting to plant survival than nutrients in a study of Canadian dunes (Houle 1997). Climate models project increases in evapotranspiration rates and drops in lake levels in the Great Lakes (reviewed in Gronewold et al. 2013), potentially increasing water stress in adjacent dunes. Across 10 general circulation models from the IPCC Fourth Assessment Report (IPCC 2007, www.cccsn.ec.gc.ca/?page = dd-gcm), predicted changes in precipitation for the region ranged from 31% decrease to 19% increase by 2071–2100, compared to baseline data back-projected for each GCM over the period 1971–2000 (Emery, Bell-Dereske and Rudgers 2015; Rudgers et al.2015). The IPCC Fifth Assesement ensemble model predicted a 10%–25% increase in annual preciptiation for the region (IPCC 2014).
Dune ecosystems, particularly early successional dunes, include diverse microbial taxa with a variety of functional roles. Early successional dunes are extremely nitrogen and water limited, and so diazotrophic groups such as Rhizobiales and Burkholderiales, along with drought-tolerant groups such as Acidobacteria, may play particularly important roles in these systems (Dalton et al.2004; Evans and Wallenstein 2014). AM fungi are common plant associates in dunes (Koske and Gemma 1997; Perumal and Maun 1999) as well and may influence plant species composition and soil formation (van der Heijden et al.1998; Bever et al.2010). Additionally, the leaf endophyte Epichloë amarillans can be found in aboveground tissues of the dominant, dune-building grass, Ammophila breviligulata (Drake, White and Belanger in review), and it is especially common in plant material available for dune restoration (Emery, Thompson and Rudgers 2010). Our previous work in this system showed that Epichloë presence increased host plant growth aboveground and reduced the diversity of plant species colonizing the dunes (Emery, Thompson and Rudgers 2010; Emery and Rudgers 2013, 2014; Emery, Bell-Dereske and Rudgers 2015; Rudgers et al.2015). In addition, Epichloë presence in live host plants reduced decomposition rates of litter placed near live plants (Bell-Dereske et al.in press).
In this study, we focused on belowground processes, including soil- and root-associated fungi and bacteria in dunes, which have not received extensive study in Great Lakes dunes. Specifically, we asked: (1) Does aboveground endophyte symbiosis in A. breviligulata affect belowground biomass or the diversity/composition of belowground microbes? (2) Does the amount of growing season precipitation cause context-dependency in the effects of aboveground symbiosis on belowground responses, or does the precipitation regime directly affect the diversity/composition of belowground microbes? To provide new insight into dune soil microbial ecology, we also addressed: (3) Does the diversity and composition of bacteria differ between A. breviligulata roots and the surrounding dune soil matrix?
MATERIALS AND METHODS
Study system
Sand dunes cover much of the Great Lakes shoreline, forming the most extensive freshwater dunes in the world and covering >1000 km2 in Michigan alone (Albert 2000). Great Lakes sand dunes are dominated by Ammophila breviligulata, which stabilizes moving sand during the early stages of dune succession and contributes to early soil carbon enrichment (Olson 1958; Nuñez, Moretti and Simberloff 2011). Additionally, A. breviligulata contributes to biotic engineering of dunes, which can be rapid, altering dune geomorphology within months to years (Godfrey 1977; Lichter 1998a,b). After dunes are stabilized, other plant species colonize and outcompete A. breviligulata, succeeding ultimately to a mixed deciduous-pine forest (Lichter 1998a, 2000).
Study site
The experimental site was located in Leelanau State Park, Leelanau Co., MI, USA (45°10.964΄, –85° 34.578΄). We established the experiment on a large blowout on the leading edge of the second foredune, ∼200 m from the shoreline of Lake Michigan. The blowout was largely devoid of vegetation and showed ongoing sand movement at the time of establishment. The habitat between the first and second dunes was a sparsely vegetated gravel bed.
Experimental design
During late May 2010, we established a 2 × 3 factorial field experiment to alter the presence/absence of leaf endophyte symbiosis in A. breviligulata in combination with reduced/ambient/augmented growing season precipitation. Each treatment combination was replicated with 15 2 m × 2 m plots. A full description of the design is reported by Emery, Bell-Dereske and Rudgers (2015). Briefly, precipitation was manipulated using modified Sala rain-out shelters (Yahdjian and Sala 2002). Clear plastic shingles removed ∼30% of ambient rainfall from the reduced rainfall plots, which was collected and added to the augmented rainfall plots after each rain event with watering cans. Both augmented and ambient rainfall plots had mock shelters with shingles oriented upside down to control for any effects on light levels or temperature, without altering the amount of ambient rainfall. To manipulate endophyte presence, we germinated seedlings on 1% water agar and inoculated half with isolates of Epichloë amarillans grown on potato dextrose agar. We used a sterile needle to either wound (sham-inoculate, E- treatment) or insert hyphae into the meristem of each seedling (E+ treatment) (Leuchtmann and Clay 1988). Following inoculation, seedlings were grown in the greenhouse in a 50:50 mix of sterile play sand and Metro-Mix 220 (Sun Gro Horticulture, Agawam, MA). As plants matured, we cloned genotypes by gently separating tillers from the original stock plants. We planted the same set of 12 A. breviligulata genotypes into each E+ plot, and a second set of 12 genotypes into each E- plot. A survey in 2014 showed that mean endophyte frequency was ∼90% in E+ plots and ∼4% in E- plots (David et al. unpublished data).
Response variables
Plot-level measurements
To examine plot-level abiotic conditions, we measured volumetric water content (VWC) at a depth of 40 cm monthly (May–July) in three random locations per plot. We used an M300 soil moisture meter (Aquaterr Instruments & Automation, Costa Mesa, CA, USA). Soil moisture was averaged across the growing season for each year.
To examine how treatments affected plant performance, we counted A. breviligulata tillers per plot each September from 2011–2015. The effects of precipitation and endophyte treatments on A. breviligulata aboveground biomass for 2010–2013 were reported previously by Emery, Bell-Dereske and Rudgers (2015). In the current study, we sampled root biomass during September 2014 using a bulb auger (volume ∼ 695cm3) to collect the tillers and roots from clumps of ∼1–5 tillers. Roots were oven-dried and weighed to calculate per tiller root biomass for each plot. We estimated plot-level root biomass for each year using September tiller counts × per tiller root biomass.
AM fungal root colonization
Fungal colonization of A. breviligulata roots was quantified from composited root samples collected from each plot in July during 2011–2014. Roots were rinsed and placed into 50 ml centrifuge tubes and then soaked in hot 10% KOH for 30 m and stained using the ink (Sheaffer Pen, Shelton, CT) and vinegar method (Vierheilig et al.1998). From each plot, ten 1 cm root sections were mounted on a microscope slide. Using a compound microscope (Leica Microsystems, Wetzlar, Germany) at 200× magnification, the percentage of roots colonized by AM fungal hyphae was recorded using the gridline intercept method (McGonigle et al.1990) with 100 views per slide [(number of views with structures visibly present in roots/total number of views) × 100]. We separately counted coarse AM hyphae, fine AM hyphae (both of which were blue-black and non-septate). Fine AM fungi have been found to more tolerant of extreme environmental conditions than coarse AM fungi (Orchard et al.2016), although their taxonomy is still under debate (Schüßler and Walker 2010)
Extraradical hyphal length
We quantified the length of extraradical AM fungal hyphae in 20 g soil subsamples from each plot collected during 2011–2014. Each subsample was mixed with 500 ml DI water in a 100-ml beaker and stirred at 80% speed for 2 min with a magnetic stir bar. Before solid material settled, the solution was poured through 500 μm and 212 μm sieves to separate sand and large organic material from the hyphal suspension. Residue from the 212 μm filter was rinsed back into a 50-ml beaker using 10 ml of DI water. Twenty drops of 4% Trypan Blue stain was added and left to sit for 45 min. This solution was then filtered through a 38 μm sieve and rinsed with DI water until water ran clear from the sieve. The residue on the 38 μm sieve was rinsed back into a 400 ml beaker using 200 ml of DI water and agitated for 2 min with the stir bar. A 20 ml sample was removed from ∼1 cm below the water surface and drained through a 25 mm glass microanalysis vacuum filter holder fitted with a 0.45 μm mesh nylon membrane. The membrane was then rinsed and dried under vacuum and mounted onto a slide. Hyphal length was estimated using the gridline-intercept method based on 50 fields of view per sample (McGonigle et al. 1990) under a stereomicroscope (Nikon SMZ1500 at 70×). Hyphal lengths were standardized to mm hyphae/g soil based on soil sample mass.
Soil glomalin content
AM fungal spores and extraradical hyphal (ERH) cell walls contain the recalcitrant soil protein glomalin (Wright and Upadhyaya 1996). Glomalin may represent 4%–8% of soil organic carbon in natural ecosystems (Rillig et al.2001), and thus is one measure of ecosystem function (carbon sequestration) provided by mycorrhizal fungi and other soil microbes. Total soil glomalin was estimated by extracting from 1 g soil subsamples per plot during 2010–2014 using the 50 mM sodium citrate buffer and autoclaving method described by Janos, Garamszegi and Beltran (2008). We quantified the Bradford reactive fraction (Bio Rad, Hercules, CA, USA) using bovine serum as a standard. Total soil glomalin has several extractible fractions, and Bradford reactive soil protein (BRSP) has been shown to consistently represent the largest fraction of total soil glomalin (∼90% by volume; Koide and Peoples 2013). Therefore, we used BRSP to operationally define glomalin, keeping in mind that glomalin and BRSP can also be produced by organisms other than AM fungi (Rosier, Hoye and Rillig 2006).
Root and soil collection for microbial diversity and composition
Root and soil samples were collected from each plot in September 2012 for microbial characterization. Roots were collected from three randomly chosen A. breviligulata individuals per plot. Soils were collected from near three plants per plot and homogenized. Roots and soils collected for bacterial extracts were preserved with sucrose lysis buffer (Giovannoni et al.1990) added to saturation. All samples were shipped on dry ice within 24 h of collection. AM fungi root samples to be used in pyrosequencing (details below) were stored at –80°C, and samples for bacterial extraction were stored at –20°C until processing.
454 Pyrosequencing: AM fungi
Freeze-dried root samples were washed with DI water and sterilized with 10% bleach. Samples were disrupted with 0.2 cm3 of 0.1 mm diameter Zirconia Silica beads (BioSpec Products) in a Mixer Mill 300 (Retsch, Haan, Germany). Samples (100 mg) were then extracted using the DNeasy Plant kit (Qiagen, Hilden, Germany) following the manufacturer's protocol. DNA concentration was quantified using a NanoDrop 1000 (Thermo Fisher Scientific, Wilmington, DE) and standardized to 20 ng/μL. Extracted samples were then amplified and sequenced by Mr. DNA (Shallowater, TX). The 28S region of the rDNA was targeted using AM fungal-specific primers. Briefly, PCRs were performed in triplicate 25 μL reactions containing 0.25 mM forward and reverse fusion primer, 0.25 mM dNTP (each), 1x Platinum PCR buffer (Lifetech, Carlsbad, CA), 1.5 mM MgCl2, 1 U Platinum Taq Polymerase (Lifetech, Carlsbad, CA) and 2 μL (∼40 ng) of DNA template. Fusion primers were designed so that the forward primer consisted of the Roche adapter A, followed by a 10 base error-correcting barcode for multiplexing (Hamady et al.2008), and using FLR3 (5΄-TTG AAA GGG AAA CGA TTG AAG T-3΄). The reverse primers included the Roche adapter B, followed by the reverse PCR primer FLR4 (5΄-TAC GTC AAC ATC CTT AAC GAA-3΄) (Gollotte, van Tuinen and Atkinson 2004). The thermal cycler program included an initial 5 min denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s and extension at 72°C for 30 s. A final 7 min extension completed the PCR. PCR amplicons were purified using the Mo-bio Gel Purification Kit (Carlsbad, CA) following the manufacturer's instructions, quantified spectrophotometrically and combined in equimolar concentrations for multiplexed pyrosequencing. Sequencing template was quantitated fluorometrically using a picogreen dye kit, assayed for quality and fragment length on an Agilent Bioanalyzer DNA 1000 chip before library preparation using Roche titanium reagents and titanium procedures. Samples were then sequenced on a Roche 454 FLX titanium instrument (Basel, Switzerland) following the manufacturer's protocols.
454 Pyrosequencing: bacteria
DNA from each of the 90 root-associated (endophytic and surface of the root) and soil samples (0.3 g) was extracted following the cetyltrimethylammonium bromide (CTAB) method described by Mitchell and Takacs-Vesbach (2008), modified to include a bead beating step. Briefly, 0.2 cm3 of 0.1 mm diameter Zirconia Silica beads (BioSpec Products, Bartlesville, OK), 300 μL of 1% CTAB, and 100 μg and 1 mg each of proteinase K and lysozyme, respectively, were added to preserved sample. Samples were incubated with continuous vertical rotation (∼35 rpm) at 37°C for 0.5 h. Sodium dodecyl sulfate was added (final concentration 2%), and samples were returned to the laboratory rotator for 0.5 h at 60°C. Samples were then bead-beaten on a vortexer for 5 min at the medium setting. Nucleic acids were extracted with an equal volume of phenol:chloroform:isoamyl alcohol (25:24:1), followed by an extraction with chloroform and precipitated in 95% ethanol after the addition of 0.1 volume 3 M sodium acetate. Nucleic acids were washed once in 70% ethanol, air-dried and resuspended in 40 μL 10 mM Tris, pH 8.0.
DNA extractions served as template to survey bacterial diversity with barcoded amplicon pyrosequencing of the 16S rDNA gene in each of the 180 samples. The 16S rDNA gene pyrosequencing was performed as described previously (Schwartz et al.2014). PCRs were performed in triplicate as described above for AM fungi. Fusion primers were designed so that the forward primer consisted of the Roche adapter A, followed by a 10 base error-correcting barcode for multiplexing (Hamady et al.2008), and the universal bacterial primer 939F 5΄-TTG ACG GGG GCC CGC ACA AG-3΄. The reverse primers included the Roche adapter B, followed by the reverse PCR primer 1492R 5΄-GTT TAC CTT GTT ACG ACT T-3΄. The thermal cycler program included an initial 5 min denaturation at 95°C, followed by 30 cycles of denaturation at 95°C for 30 s, annealing at 52°C for 30 s and extension at 72°C for 30 s. A final 7 min extension completed the PCR. Sample amplicons were purified, quantified and aggregated as described above. All samples from this study were run on one half region of a sequencing plate, with no more than 96 samples total per region. Pyrosequencing was performed on a Roche 454 FLX instrument (Basel, Switzerland) following manufacturer's protocols at the Molecular Biology Facility in the UNM Biology Department.
Bioinformatics
AM fungal 28S rRNA gene sequencing resulted in 611 624 raw sequences, which then were quality filtered and trimmed to 300bp using fastq_filter in USEARCH8 with default settings (http://drive5.com/usearch/). Sequences were chimera checked, filtered de novo and clustered at 97% similarity into unique operational taxonomic units (OTUs, i.e., DNA sequences or amplicon types) using UPARSE implemented in UEARCH8 (Edgar 2013). USEARCH has been successfully used for the processing and OTU clustering of AM fungal amplicons (Van Geel et al.2014, 2015, 2016; De Beenhouwer et al.2015; Johansen et al.2015). USEARCH8 quality filtering, chimera checking using UCHIME and OTU clustering lead to 44 OTUs and 277 799 reads. Taxonomic affiliation was assigned to OTUs by comparing the representative set of DNA sequences to the MaarjAM database using blastn (Öpik et al.2010). Representative sequences were aligned, and a tree was built in PASTA (Mirarab et al.2015) using RAXML and all other default settings with reference sequences from the online database schuessler.userweb.mwn.de/amphylo/ constructed from Redecker et al. (2013) and Schüßler and Walker (2010). The tree was rooted using Batrachochytrium dendrobatidis as the outgroup. Sequences that did not blast to species in the MaarjAM database (<95% Query coverage and <95% Max identity) but were monophyletic with references sequences in the AM fungal phylogeny were additionally blasted to the NCBI database; OTUs that did not hit AM fungal entries were removed from analysis because they were highly likely to be non-AM fungi. We also removed OTUs with <5 reads total to avoid over splitting (Thiéry et al.2012) and sequencing errors (Dickie 2010). Taxonomic filtering resulted in 34 OTUs and 276 957 reads (Table S1, Supporting Information). We transformed OTU tables using variance stabilizing transformation (VST) in the DeSeq2 package (Love, Huber and Anders 2014) in R (R Core Team 2015) to control for biases in PCR amplification and to avoid biases due to rarefaction (McMurdie and Holmes 2014). The inverse Simpson diversity index was calculated for each sample using the vegan package of R (Oksanen et al.2016). Results were qualitatively the same using a rarefied OTU table [reads = 500 (Table S3, Supporting Information)]. A Bray-Curtis distance matrix was generated from the VST normalized community using Primer V6 (Clarke and Gorley 2006). Weighted and unweighted Unifrac (Lozupone and Knight 2005) distance matrices were generated from the VST normalized community in Quantitative Insights into Microbial Ecology (QIIME). Because results for weighted and unweighted matrices were similar, only results for weighted Unifrac are reported. Importantly, the resulting community matrix was not significantly different than the community matrix produced by quality filtering, chimera checking and OTU clustering using the QIIME pipeline (Caporaso et al.2010). Specifically, the Bray-Curtis matrices were strongly correlated (Mantel: Spearman correlation r = 0.86 P < 0.01), and the ordination structure was significantly correlated (Procrustes correlations r = 0.73 P = 0.001).
Bacterial 16S rRNA gene sequencing resulted in 826 382 raw sequences, which were quality filtered, denoised, screened for PCR errors and chimera checked using AmpliconNoise and Perseus to minimize potential artifacts (Quince et al.2011). The QIIME pipeline was used to analyze alpha and beta diversity of the DNA sequence data (Caporaso et al. 2010). OTUs were identified by the 97% DNA identity criterion using the uclust OTU picker (Edgar 2010) in QIIME. A set of representative DNA sequences was chosen for each unique OTU in QIIME and used for all subsequent analyses. Taxonomic affiliation was assigned to OTUs by comparing the representative DNA sequences to the Green Genes database (gg8.15.13). These DNA sequences were aligned using MUSCLE (Edgar 2004), and a phylogenetic tree necessary for the beta diversity analysis was constructed using FastTree (Price, Dehal and Arkin 2009). We filtered the OTU table to remove samples with <300 reads and OTUs with <5 reads, resulting in 142 total samples (70 root; 72 soil), 8180 OTUs (5003 OTUs in the root, 2120 in the soil) and 807 991 sequences remained. The filtered bacterial OTU table was then normalized using VST as described for the AM fungi data above. To examine the effects of the treatments on root versus soils, filtered OTU tables were separated into root versus soil community, then refiltered to remove samples with <300 reads (removing one root sample and one soil sample) and OTUs with <5 reads within each table (Table S1). The separated files of raw OTU reads were then normalized using VST as described above. Diversity was calculated on the VST normalized data using the inverse Simpson diversity index for each sample using the vegan package of R. Results using this diversity metric were qualitatively similar to a rarefied OTU table [reads = 1000 (Table S4, Supporting Information)]. Bray-Curtis distance matrices were generated from the VST normalized communities using Primer V6 (Clarke and Gorley 2006). Weighted and unweighted Unifrac (Lozupone and Knight 2005) distance matrices were generated from the VST normalized communities in QIIME. Because results for weighted and unweighted matrices were similar, only results for weighted Unifrac are reported. All raw sequence data from this study are available through the NCBI Sequence Read Archive under accession SAMN05354971.
Statistical analyses
Dunes are a highly spatially heterogeneous environment, ranging in depth to the groundwater table, nutrient content, aeolian sand deposition and many other factors. To fully explore our 12 response variables (listed in Table 1) within context of the spatial heterogeneity of the dunes, we compared two types of statistical models. The first set of models (Precipitation Models) used our endophyte (E+/E–) and precipitation (reduced/ambient/augmented) treatments as fixed factors, and included the repeated effect of sampling year when multiple years of data were collected, as well as a fixed effect of the spatial blocking factor within the experiment (three groups of plots). The second set of models (Soil Moisture Model) replaced the fixed effects of the precipitation treatment and spatial blocking factor with the continuous variable of soil moisture (VWC, measured for each plot at 40 cm depth and averaged over multiple sampling dates within each growing season) to better account for spatial heterogeneity among plots in the key belowground measure of water availability.
Table 1.
Response variables tested addressing each hypothesis.
| Hypothesis | Treatment | Sample year(s) | Response variable | Treatment effect size | Test statistic | P | |
|---|---|---|---|---|---|---|---|
| (1) Does aboveground endophyte symbiosis affect | Endophyte | 2011–2015 | Est. per plot root biomass | ↑ 27% | X2 | 3.93 | 0.047 |
| belowground biomass or microbial diversity/composition? | 2011–2014 | Soil ERH length | ↓ 11% | X2 | 4.69 | 0.030 | |
| 2012 | Fine AMF colonization | ↓ 50% | t | −1.95 | 0.052 | ||
| 2013 | Fine AMF colonization | ↑ 35% | t | 1.78 | 0.077 | ||
| Precipitation Models (2A) Does precipitation cause | Precipitation | 2012 | Fine AMF colonization | Augmented ↑ 92% | t | 3.17 | 0.005 |
| context-dependency in the effect of aboveground symbiosis | 2012 | Coarse AMF colonization | Augmented ↓ 26% | t | −2.09 | 0.093 | |
| on belowground microbes, or directly alter microbial | 2014 | Soil ERH length | Augmented ↑ 44% | t | 4.38 | < 0.001 | |
| composition? | Endophyte × precipitation | 2014 | per tiller root biomass | Ambient: E+ ↑ 50% E– | t | 2.33 | 0.022 |
| 2012 | AMF diversity | Ambient: E+ ↓ 25% E– | t | −2.40 | 0.019 | ||
| Soil Moisture Models (2B) Does soil moisture cause | Soil moisture | 2012 | Soil bacterial diversity | r = 0.31 | F | 6.21 | 0.015 |
| context-dependency in the effect of aboveground symbiosis | 2012 | Soil Rhizobiales abundance | r = –0.48 | F | 17.77 | < 0.001 | |
| on belowground microbes, or directly alter microbial | Endophyte × soil moisture | 2012 | Root bacterial diversity | E+ r = –0.40 | F | 5.52 | 0.022 |
| composition? | E– r = 0.20 | ||||||
| 2012 | Root Rhizobiales abundance | E+ r = –0.41 | F | 6.73 | 0.012 | ||
| E– r = 0.24 | |||||||
| 2012 | Soil Actinobacteria abundance | E+ r = 0.26 | F | 12.41 | 0.046 | ||
| E– r = –0.23 | |||||||
| 2012 | Soil Acidobacteria abundance | E+ r = –0.10 | F | 4.14 | 0.001 | ||
| E– r = 0.56 | |||||||
| (3) Does the diversity and composition of bacteria differ | Location | 2012 | Bacterial diversity | Roots 99% > soil | X2 | 52.64 | < 0.001 |
| between A. breviligulata roots and the surrounding dune soil | 2012 | Rhizobiales abundance | Roots 233% < soil | X2 | 57.25 | < 0.001 | |
| matrix? | 2012 | Burkholderiales abundance | Roots 25% < soil | X2 | 21.48 | < 0.001 | |
Effect sizes are presented as the percentage difference between the treatment [i.e. Epichloë present (E+) or altered precipitation (±30%)] and the control [i.e. Epichloë absent (E–) or ambient precipitation]. (2A) When the endophyte × precipitation interaction was significant, we show here results for the precipitation treatment under which the endophyte effect was the strongest. (2B) When the endophyte × soil moisture interaction was significant, Pearson correlation coefficients are shown for each endophyte treatment separately.
Root colonization and soil fungi analyses
Using the Precipitation Model, we analyzed the responses of soil moisture (VWC at 40 cm), percentage of root colonization by fungi, ERH length, glomalin and plot-level estimated root biomass using linear mixed effect repeated measures models in the lme4 package (Bates et al.2015) in R (R Core Team 2015). Because per tiller root biomass was only measured in 2014, we used a general linear model for this variable (one observation per plot). To meet assumptions of Gaussian distributions of errors and homogeneity of variances, we square-root transformed total AM fungal colonization of roots, log-transformed root biomass and fine AM fungi colonization, cube-root-transformed ERH and inverse square-root transformed glomalin estimates.
Community composition analyses: fungi
Using the Precipitation Model, we analyzed the response of the inverse Simpson diversity of the AM fungal community using a linear model in R. AM fungal community structure was analyzed using permutational analysis of variance (PERMANOVA) on the Bray-Curtis and Unifrac weighted (VST normalized matrix) distance matrices using Primer V6 (Clarke and Gorley 2006).
Community composition analyses: bacteria
Using the Precipitation Model with the addition of location of the bacterial community (to address (3) if the bacterial diversity and community differ between roots and soil), we examined responses of inverse Simpson diversity of the bacterial community, along with the relative abundance (in percentage of VST normalized sequences per sample) of key soil functional groups Acidobacteria, Actinobacteria, Rhizobiales and Burkholderiales, using mixed effect models with plot as a random factor using the lme4 package (Bates et al.2015) in R (R Core Team 2015). To meet assumptions of Gaussian distributions of errors and homogeneity of variances, we log-transformed Rhizobiales, Burkholderiales, Actinobacteria and Acidobacteria relative abundance and inverse Simpson diversity. Precipitation Model effects on bacterial community structure were analyzed using PERMANOVA with factors described above plus the location of collection (root vs soil), and all interaction terms with the addition of plot as a random factor. If spatial block was not significant in the full Precipitation Model, it was dropped from the final models.
Community composition analyses: Soil Moisture Model
Using the Soil Moisture Model, we analyzed bacteria and AM fungi community structure using PERMANOVA on the Bray-Curtis and weighted Unifrac (VST normalized matrix) distance matrices using Primer V6. We added the effect of the location of collection (root versus soil) to the Soil Moisture Model for examining the response of the inverse Simpson diversity bacterial community, and relative abundance (in percentage of VST normalized sequences per sample) of Acidobacteria, Actinobacteria, Rhizobiales and Burkholderiales; these models were implemented using linear mixed effect models in the lme4 package (Bates et al.2015) in R (R Core Team 2015). Since soil moisture had significant interactive effects with location of collection (root vs soil) in affecting bacterial community composition and focal bacterial taxonomic groups, we then conducted separate analyses for the root and soil datasets.
RESULTS
Does aboveground endophyte symbiosis in Ammophila breviligulata affect belowground biomass or the diversity/composition of belowground microbes?
Roots
Estimated plot-level root biomass was 27% greater when Epichloë was present compared to endophyte-free plots (Table 1; Fig. 1A), consistent with our previous findings of increased aboveground A. breviligulata biomass when Epichloë was added (Emery, Bell-Dereske and Rudgers 2015).
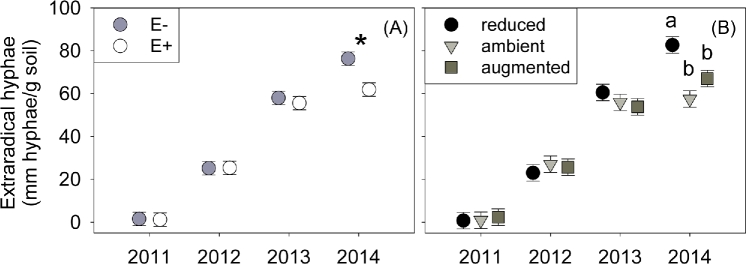
Figure 1.
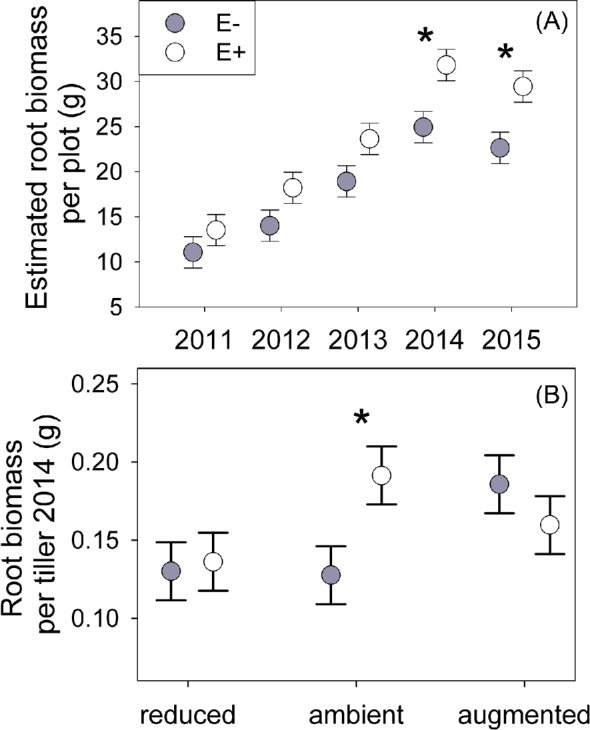
Root biomass (A) estimated per plot (September tiller survey × per tiller root biomass in 2014) and (B) per tiller in 2014 showing treatments with Epichloë (E+, open symbol) versus endophyte-free (E–, filled symbol). For (B) precipitation treatments are 30% reduced (black circle symbol), ambient (gray triangle) or 30% augmented (dark square). ‘*’ indicates P < 0.05 Tukey HSD test. Symbols show means ± s.e.
Fungal abundance
Epichloë presence altered root colonization of A. breviligulata by fungi as well as reducing the abundance of soil ERH by 11% (Table 1). The endophyte-driven reduction in ERH was strongest in 2014, where the presence of Epichloë reduced the ERH by 19% compared to the endophyte-free plots (Fig. 2A). The effects of Epichloë on colonization of fine AM fungal hyphae in roots varied across years (endophyte × year X2 = 8.45; P = 0.038), tending to reduce colonization in 2013 (by 35%) but causing an increase in colonization (50%) during 2012 (Table 1; Fig. 3A). However, Epichloë did not alter hyphal colonization by coarse AM fungal morphotypes (Table S2, Supporting Information). Glomalin concentration, a coarse metric that includes fungi and other microbes, showed no overall response to Epichloë presence (P > 0.65, Table S2).
Figure 2.
Extraradical hyphae in soil (mm hyphae/g soil) from (A) treatments with Epichloë (E+ open symbols) versus endophyte-free (E– filled symbols) and (B) for precipitation treatments: 30% reduced (black circle symbol), ambient (gray triangle) or 30% augmented (dark square). ‘*’ represents P < 0.05. Letters represent Tukey HSD significant differences between means (P < 0.05). Symbols show means ± s.e.
Figure 3.
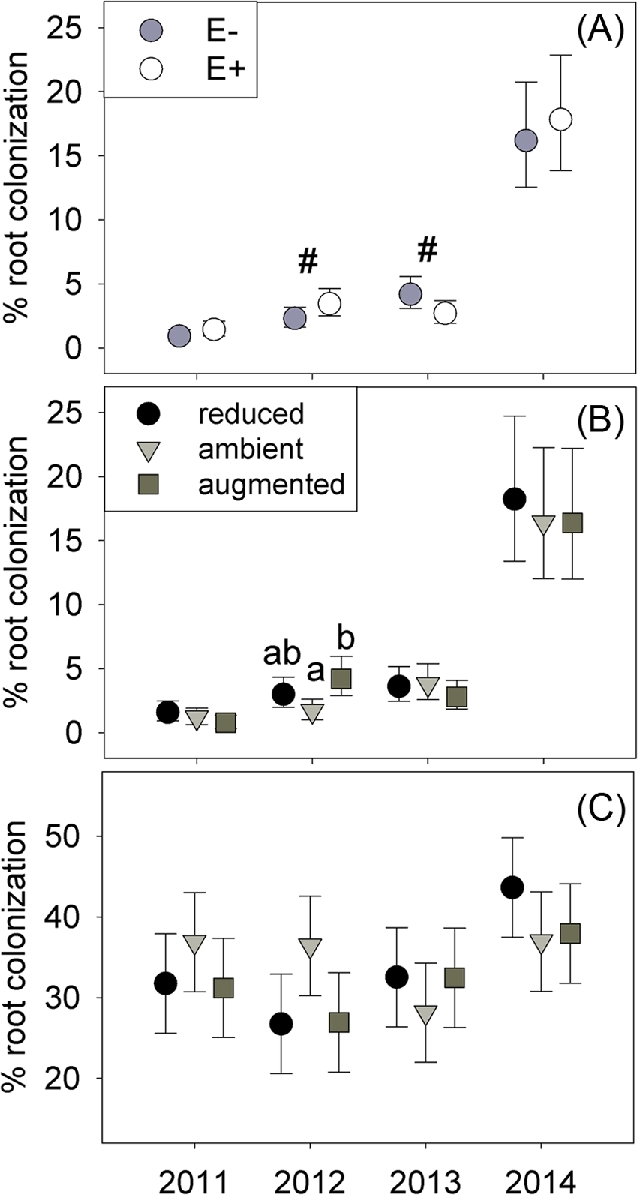
Percentage of root area colonized by hyphae ± 95% CI (not ± s.e.) of (A) fine AM fungal morphotype in plot with Epichloë (E+ open symbols) and endophyte free (E– filled symbols), (B) fine AM fungal morphotype under altered precipitation (C) and coarse AM fungal morphotypes under altered precipitation. For plots (b-c) precipitation treatments: 30% reduced (black circle symbol), ambient (gray triangle) or 30% augmented (dark square). Reported percentage area of roots colonized by the fine AM fungal morphotype are back transformed. Letters represent significant differences between means (P < 0.05, Tukey HSD). ‘#’ shows P < 0.10 in Tukey HSD tests.
Microbial diversity and composition
In contrast to the effects of Epichloë on fungal abundance, there was no main effect on belowground microbial diversity of fungal (Table S3) or bacterial (Table S4) OTUs or community composition (Tables S5 and S6, Supporting Information) despite evidence for context dependency (see Question 2). There was no main endophyte effect on the focal diazotrophic bacteria (Table S7, Supporting Information) or on the focal bacterial phyla (Table S7).
Does the amount of growing season precipitation cause context-dependency in the effects of aboveground symbiosis on belowground responses, or does the precipitation regime directly affect the diversity/composition of belowground microbes?
Direct effects of precipitation on belowground responses
Precipitation directly altered root colonization and soil fungi abundance, but did not affect bacterial or AM fungal diversity or composition (Tables S4–S6). In 2012, A. breviligulata roots from the augmented precipitation treatment had >92% higher colonization by the fine AM fungal morphotype than plots receiving ambient precipitation (precipitation × year X2 = 12.71, P = 0.048; Table 1, Fig. 3B). However, in 2012, there was a trend for increased root colonization of coarse AM fungal morphotypes under ambient precipitation (precipitation × year X2 = 12.30, P = 0.055; Table 1, Fig. 3C). Reduced precipitation increased soil ERH by 44% over ambient precipitation in 2014 but had very little effect in all other years (precipitation × year X2 = 19.71, P = 0.003; Table 1, Fig. 2B).
Belowground context-dependency: roots
The strongest endophyte effects on root biomass occurred under ambient precipitation. Epichloë increased per tiller root biomass during 2014 by 50% compared to endophyte-free plants under ambient precipitation, but had little to no effect on per tiller root biomass under altered precipitation (endophyte × precipitation F2,84 = 2.68, P = 0.075; Table 1; Fig. 1B).
Belowground context-dependency: fungi
The amount of precipitation modified how the endophyte affected AM fungal diversity and glomalin production. Epichloë reduced the diversity of AM fungal VST normalized OTUs by 25% under ambient precipitation, but did not strongly affect diversity under altered precipitation (endophyte × precipitation F2,63 = 3.31, P = 0.043; Table 1; Fig. 4). Consistent with the VST results, endophyte presence similarly reduced AM fungal rarefied diversity (F1,61 = 5.31, P = 0.025; Table S3) with the strongest negative effect under ambient precipitation (Fig. S3, Supporting Information). The interactive effect on glomalin varied with year (endophyte × precipitation × year X2 = 18.13, P = 0.020; Table S2), tending to increase glomalin only under augmented precipitation in year 2012 (when we sampled microbial composition; Fig. S4, Supporting Information). Despite the interactive effects of Epichloë and precipitation on diversity and glomalin, there was no interactive effect on root colonization by AM fungi (Table S2).
Figure 4.
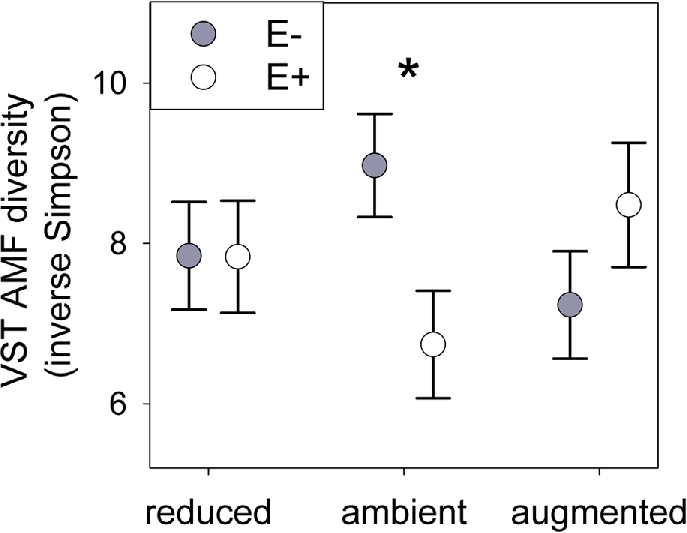
Inverse Simpson diversity of AM fungal sequences from VST normalized root communities from plots with 30% decreased (reduced), ambient or 30% increased precipitation (augmented) with Epichloë (E+ open symbols) and endophyte free (E– filled symbols). ‘*’ represents P < 0.05 Tukey HSD pairwise comparison. Bars show means ± s.e.
Belowground context-dependency: bacteria
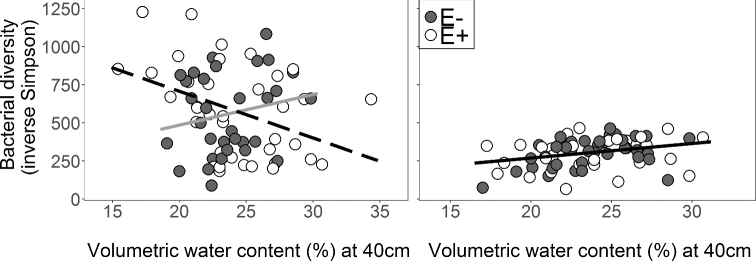
Endophyte presence changed the relationship between bacterial diversity and soil moisture. Specifically, Epichloë presence caused root-associated bacterial diversity to decline with higher soil moisture (endophyte × soil VWC F1,65 = 5.52, P = 0.022). However, in the absence of Epichloë, root-associated bacterial diversity increased with greater soil moisture (slope of E+ (R2 = 0.14) was 252% less than slope of E– (R2 = 0.01); Table 1; Fig. 5A). These effects were likely due to the increased resolution of soil moisture provided by the Soil Moisture Model because our precipitation treatments alone (Precipitation Model) did not alter the effects of Epichloë on root bacterial diversity (Table S4). In contrast to root bacteria, soil bacterial diversity was not affected by Epichloë, and increased with greater soil moisture regardless of Epichloë presence (soil VWC F1,67 = 6.21, P = 0.015, R2 = 0.11; Table 1; Fig. 5B). Endophyte presence did not alter root bacterial, soil bacterial or AM fungal community structure responses to soil moisture, but composition of these three belowground communities did shift with soil moisture (Table S10, Supporting Information).
Figure 5.
Linear regressions of soil moisture at 40 cm versus (A) root bacterial and (B) soil bacterial inverse Simpson diversity from VST normalized communities. In plots with the A. breviligulata–Epichloë symbiosis, there was a negative correlation between soil moisture and bacterial OTU diversity (E+ open symbols and dashed line, y = –30.68 × x + 1321.01, R2 = 0.14). In plots without endophytes, there was a weak positive correlation between soil moisture and bacterial diversity (E– filled symbols and solid gray line y = 20.18 × x + 81.64, R2 = 0.01). Soil moisture was positively correlated with soil bacterial diversity (black line y = 9.78 × x + 70.83, R2 = 0.11).
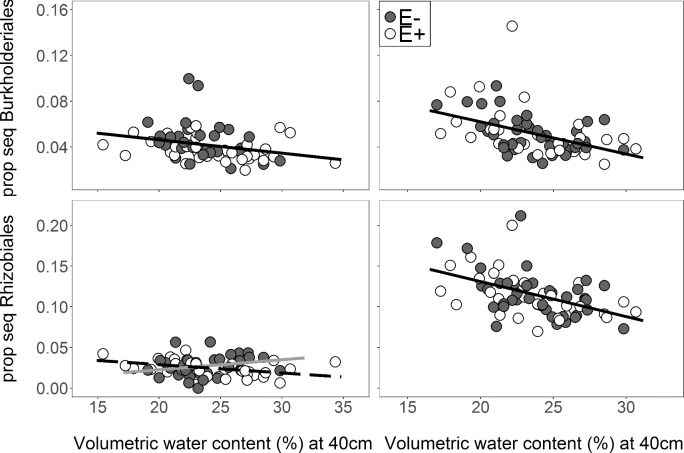
Endophyte presence also altered the responses of diazotrophic and specific soil bacteria phyla to soil moisture. For putative diazotrophs in roots, Epichloë presence caused the relative abundance of Rhizobiales to decline with soil moisture; however, when the endophyte was absent, they weakly increased with higher soil moisture (Table 1; Fig. 6A). The relative abundances of three additional groups (soil Rhizobiales, root Burkholderiales and soil Burkholderiales) decreased with increasing soil moisture (Table 1), but the endophyte did not alter these relationships (endophyte × soil VWC P > 0.15; Fig. 6; Table S9, Supporting Information).
Figure 6.
Linear regressions of soil moisture at 40 cm versus relative abundance of VST normalized sequences composed of (A) Burkholderiales in samples from roots, (B) Burkholderiales from soils, (C) Rhizobiales from roots and (D) Rhizobiales from soils associated with A. breviligulata. Endophyte presence (E+ open symbols and E– filled symbols) did not affect Burkholderiales abundance, but there was a positive correlation between soil moisture and the proportion of sequences composed of Burkholderiales (root: y = –0.0012 × x + 0.069, R2 = 0.07 and soil: y = –0.0028 × x + 0.12, R2 = 0.19). Soil moisture was negatively correlated with proportion of sequences composed of Rhizobiales (E+ white dots and dashed line y = 0.0010 × x + 0.049, R2 = 0.17); however, there was a positive correlation between soil moisture when the endophyte was absent (E– grey dots and solid gray line y = 0.0012 × x – 0.0016, R2 = 0.025). Soil Rhizobiales decreased with increasing soil moisture (black line: y = –0.0043 × x + 0.22, R2 = 0.22).
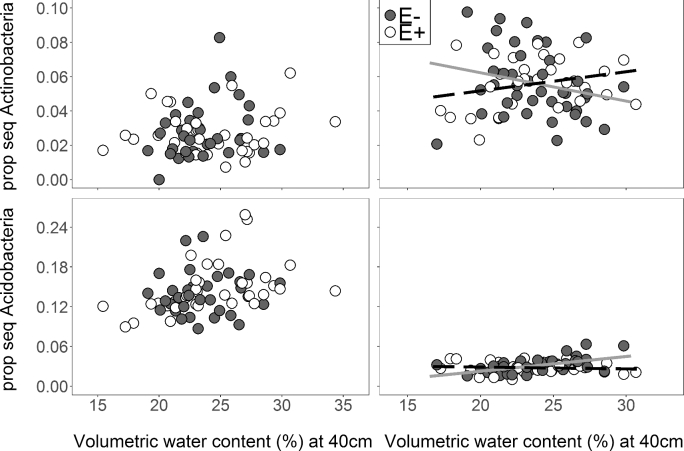
Interestingly, endophyte presence altered how soil Actinobacteria and Acidobacteria responded to soil moisture (Table 1), but did not affect the response of these phyla when they resided in roots (endophyte × soil VWC P > 0.15; Fig. 7A and C). Soil Actinobacteria tended to increase with higher soil moisture when the endophyte was present, but tended to decrease when the endophyte was absent (Fig. 7B). In contrast, soil Acidobacteria increased in relative abundance with increasing soil moisture only when the endophyte was absent (Table 1; Fig. 7D; Table S9).
Figure 7.
Linear regressions of soil moisture at 40 cm versus relative abundance of VST normalized sequences composed of (A) Actinobacteria in samples from roots, (B) Actinobacteria in samples from soils, (C) Acidobacteria from roots and (D) Acidobacteria from soils associated with A. breviligulata. In plots with the A. breviligulata-Epichloë symbiosis, there was a weak positive correlation between soil moisture and proportion of sequences composed of Actinobacteria (E+ open symbols and dashed line, y = 0.0011 × x + 0.029, R2 = 0.035). In plots without endophytes, there was a weak positive correlation between soil moisture and the abundance of Actinobacteria (E– filled symbols and solid gray line y = –0.0017 × x + 0.095, R2 = 0.031). There was no correlation between soil moisture and Acidobacterial abundance when the Epichloë symbiosis was present (E+ open symbols and dashed line, y = 0.00027 × x + 0.034, R2 < 0.001). When the endophyte is absent, there is a positive correlation between soil moisture and the portion of sequences composed of Acidobacteria (E– filled symbols and solid gray line y = 0.0022×x – 0.022, R2 = 0.30).
Does the diversity and composition of bacteria differ between Ammophila breviligulata roots and the surrounding dune soil matrix?
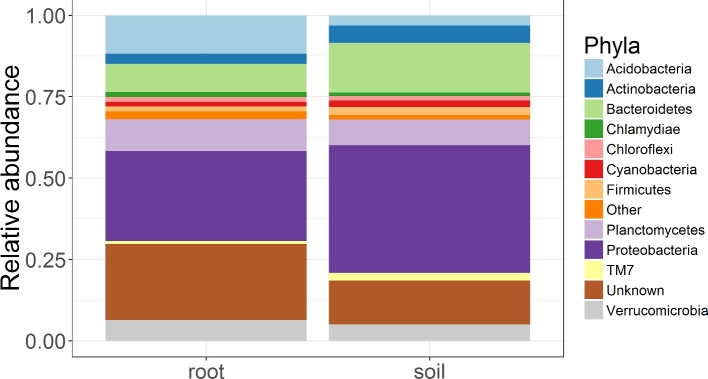
In the nutrient poor soil of dunes, proximity to A. breviligulata roots strongly altered the composition and diversity of the bacterial community (Figs 8 and 9). VST normalized root-associated bacteria were ∼99% more diverse than the soil bacterial community (Table 1; Fig. 9A), and rarefied diversity was 9-fold higher in roots than in soil (Table 1; Fig. S5, Supporting Information). Rhizobiales were more abundant in soils, where that clade made up 10% of the sequences, than in the roots of A. breviligulata where Rhizobiales constituted just 3% of total sequences (Table 1; Fig. 6C and D). Burkholderiales was only slightly more abundant in the soils, representing 5% of total sequences in the soil versus 4% of total sequences in the roots (Table 1; Fig. 6A and B).
Figure 8.
Relative abundance (VST) of reads in each of the dominant bacterial phyla for communities associated with A. breviligulata roots versus the surrounding soil matrix.
Figure 9.
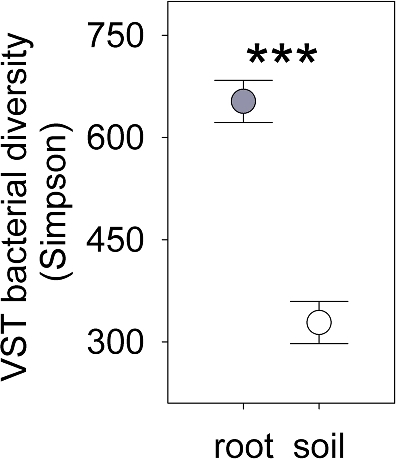
Bacterial inverse Simpson diversity in A. breviligulata from VST normalized communities in roots (filled symbols) and the surrounding soils (open symbols). ‘***’ represents P < 0.001 significance. Bars show means ± s.e.
DISCUSSION
Leaf Epichloë increased belowground biomass while reducing AM fungal diversity and ERH
Epichloë presence in the aboveground tissue of A. breviligulata increased root biomass nearly 30%, and reduced AM fungal diversity and ERH production. The tradeoff between root biomass and ERH production has been found in other systems (De Deyn et al.2009), with plants that invest more in roots possibly having reduced need for ERH to acquire nutrients. To our knowledge, ours is the first study that has used AM-specific primers to examine whether Epichloë affects root fungal composition in the field. Here, Epichloë reduced the diversity of the AM fungal community. In contrast, Rojas et al. (2016) showed that Epichloë presence increased the relative abundance of AM fungal sequences when they examined the soil fungal community via more general ITS fungal primers, which do not yield good resolution of AM fungi and may not amplify AM fungal sequences when they are not abundant in the sample (reviewed in Lindahl et al. 2013 and Tedersoo et al.2015). The reduction in AM fungal diversity reported here could influence plant succession through shared hyphal networks or altered access to symbiont partners (van der Heijden et al.1998; Enkhtuya, Poschl and Vosatka 2005).
An endophyte-mediated shift in soil fungi could also influence dune ecosystem processes. Here, the presence of Epichloë reduced the hyphal length of ERH in the soil consistently across precipitation treatments and years. Reduced ERH may contribute to slower decomposition in the dunes and help explain our prior observation that Epichloë presence in live A. breviligulata plants reduced the rate of decomposition of litter buried in our plots (Bell-Dereske et al.in press). Endophyte-caused reductions in soil fungi may similarly slow decomposition rates in tall fescue pastures (e.g. Siegrist et al.2010) and also explain their higher levels of carbon sequestration (Iqbal et al.2012). In contrast to our result for ERH, a long-term field study on the tall fescue–Epichloë coenophiala symbiosis showed no endophyte effect on soil fungal biomass; however, Ascomycota strongly declined with endophyte-presence (Rojas et al.2016). Although we did not sequence Ascomycota, we found no significant shifts in root colonization by dark septate endophyte morphotypes, common in this fungal clade. Also, in contrast to our results, a study of Bromus auleticus–E. pampeana suggested that Epichloë addition increased the diversity of soil fungal cultures, specifically phosphorus-solubilizing, rhizospheric fungi (Arrieta et al.2015). Furthermore, a previous study on tall fescue found endophyte-mediated increases in the activity of the fungal community as measured by respiration (Casas et al.2011). Thus, our results suggest that the belowground effects of Epichloë are not easily generalizable across host species or ecosystems. Characterizing effects across a greater diversity of systems may help in formulating a predictive framework for the magnitude and direction of effects.
Despite detecting an influence of Epichloë on AM fungi and ERH, we found that most microbial responses were insensitive to the main effect of endophyte presence. That belowground fungi were more sensitive than bacteria is in line with prior work showing higher sensitivity of fungi than bacteria to Epichloë in tall fescue pastures (Rojas et al.2016). Additionally, Great Lakes dunes have a spatially heterogeneous abiotic environment (Lichter 1998a,b; Ensign, Webb and Longstaffe 2006). Our results suggest that this heterogeneity is the primary driver of both root and soil microbial diversity and composition because spatial blocking effects tended to outweigh the biotic influence of Epichloë presence in leaves for many microbial responses variables. Factors that could be structuring the microbial community are likely to be scale dependent, with global trends driven by soil pH (Fierer and Jackson 2006) and temperature (Zhou et al.2016), and local patterns reflective of variable soil moisture, nutrients or salinity (Van Horn et al.2013; Okie et al.2015).
Limited context-dependency in fungal community responses
There were few fungal responses to the precipitation treatment, with the exception of the coarse metric of soil glomalin. Additionally, both glomalin and fine AM fungal hyphae showed year-to-year variability in responses to Epichloë, which may also indicate that the climate context alters above/belowground interactions. For example, Epichloë tended to increase colonization by fine AM fungal hyphae in 2012, but reduced the abundance of fine hyphae in 2013. The fine hyphae (previously categorized as Glomus tenue; Schüßler and Walker 2010) have been suggested to be drought resistant (Staddon, Gregersen and Jakobsen 2004). Our site experienced increased drought during August–September 2012 (Fig. S6, Supporting Information). Interestingly, our augmented precipitation treatment also increased fine AM fungi in 2012, which may indicate that Epichloë and precipitation addition helped to alleviate the effects of extreme drought on fine AM fungi. However, the effects of fine AM fungi on host plant responses to climate are poorly understood due to their low detection in environmental samples and difficulties in culturing (Orchard et al.2016). In the tall fescue–E. coenophiala system, endophyte presence reduced soil glomalin (Buyer et al.2011); however, unlike our system, Epichloë also reduced total root colonization and spores of AM fungi (Chu-Chou et al.1992; Mack and Rudgers 2008). Interannual variability in climate could underlie differences among years, but a longer time series would be needed to resolve such an influence in our system.
Epichloë causes context-dependent responses of bacteria to soil moisture
To our knowledge, ours is the first study to report that a foliar endophyte alters how belowground bacterial diversity responds to an abiotic gradient. Resolving such relationships is important for refining predictions on how plant–microbe interactions will change under future environmental conditions. Specifically, root-associated bacterial diversity decreased in wetter soils only when Epichloë was present (Fig. 5A). The abundance of root-associated Rhizobiales showed the same negative relationship with soil moisture when the endophyte was present. However, with Epichloë absent, root bacterial diversity and Rhizobiales abundance increased with soil moisture, similar to the overall positive effect of soil moisture on soil bacterial diversity. Previous research has shown that Actinobacteria are likely copiotrophic and sensitive to changes in soil moisture, whereas Acidobacteria are more oligotrophic and resilient to changes in moisture (Fierer, Bradford and Jackson 2007; Evans and Wallenstein 2014). Consistent with this past work, dune soil Actinobacteria tended to increase in relative abundance with soil moisture when the endophyte was present and showed greater sensitivity to soil moisture than soil Acidobacteria. The effect of Epichloë on bacterial abundance was similarly context dependent on soil type in tall fescue pastures, where endophyte presence reduced the abundance of more phyla of bacteria in clay loam soils than in loamy sand (Jenkins, Franzluebbers and Humayoun 2006).
It remains unclear why Epichloë alters the responsiveness of bacterial diversity to soil moisture (or soil texture). Growth and feeding strategies seem to be phylogenetically conserved in some of the dominant soil phyla (Fierer, Bradford and Jackson 2007), which leads to somewhat predictable shifts in the community composition in response to changes in soil moisture (Evans, Wallenstein and Burke 2014). It is possible that by altering plant root characteristics, such as root exudates (Franzluebbers and Hill 2005; Guo, McCulley and McNear 2015) or root biomass, Epichloë shifts limitations on bacterial diversity from carbon-based resource availability to water limitation, increasing bacterial responsiveness to soil moisture. Adding carbon to E-plots could provide a direct test of this hypothesis. Alternatively, Epichloë presence also widened the range of soil moistures observed across plots (Fig. 5), possibly making it easier to detect an influence of soil moisture on bacterial diversity. Prior work has shown that Epichloë can promote host tolerance of drought (Malinowski and Belesky 2000), including our past work on A. breviligulata (Emery, Thompson and Rudgers 2010). Previous studies have additionally suggested that Epichloë can alter plant water relations in ways that retain soil moisture for longer periods of time (Elmi and West 1995; Kannadan and Rudgers 2008). Thus, plots with Epichloë could have an expanded range of soil moisture values. In support of this hypothesis, the coefficient of variation in soil moisture for E+ plots was 42% higher (CV = 16%) than in endophyte-free plots (CV = 11%), and differences in the range of soil moistures observed were not due to imbalance in the sample sizes among treatments.
Since roots had vastly higher microbial diversity and richness than soils, these context-dependent endophyte effects on precipitation could strongly influence the total diversity of bacterial species in dune ecosystems. Even a relatively small reduction in diazotrophic bacteria (i.e. Rhizobiales) in response to soil moisture could affect plant succession because dune soils are so nitrogen poor (Lichter 1998b, 2000). In contrast, we found no effect of Epichloë on the Burkholderiales, for which both root and soil communities showed a negative relationship with soil moisture. Despite the responsiveness of bacterial diversity, and specifically of diazotrophs, in our system, we have not detected significant shifts in total N, nitrate or ammonium in dune soils, based on ion resin exchange membranes placed in plots during the 2013 growing season (data not shown). Future investigations of N process rates or extracellular enzyme activities could be useful for resolving the N cycle in this system.
Ammophila breviligulata roots harbor islands of bacterial biodiversity
In many plants, roots selectively filter microbial communities, constraining microbial diversity relative to that of the surrounding soil matrix (Wang, Yang and Falcão Salles 2016). Contrary to this general pattern, roots of A. breviligulata act more as islands of microbial biodiversity than as a selective filter. Perhaps root exudates from A. breviligulata provide much needed resources for bacteria, explaining the elevated diversity of bacteria compared to that of the soil. On the other hand, resource inputs from root exudates could lead to antagonistic interactions among bacterial species, increasing the diversity of the root community (Czárán, Hoekstra and Pagie 2002; Schlatter et al.2015). In more productive ecosystems, the root and rhizosphere typically harbor lower bacterial diversity than surrounding soils (reviewed in Faure, Vereecke and Leveau 2008), an effect that grows stronger with a longer time of interaction with plant roots (Shi et al.2015). Although most research on the selective effect of roots on bacterial communities has focused on few well-studied plants, such as Arabidopsis thaliana (Bulgarelli et al.2013), barley and rice (Bulgarelli et al.2015; Edwards et al.2015), studies of wild species have found a similar selective effect of the host root on bacterial communities (Dean et al.2015; Nuccio et al.2016).
Roots of A. breviligulata harbored higher relative abundances of Burkholderiales than of Rhizobiales, suggesting that roots may selectively favor this group of diazotrophic bacteria. In contrast to the diversity pattern of the whole bacterial community, soils actually had higher proportions of both diazotrophic clades than did roots. Prior work suggested that members of Burkholderiales inhabit the rhizosheaths of grasses, such as Ammophila, that grow in extremely nutrient poor soils (Wullstein, Bruening and Bollen 1979; Wullstein 1991; Bergmann et al.2009). Diazotrophic bacteria in the root sheaths of A. breviligulata may be an important, but unresolved, part of the nitrogen cycle in nutrient-poor dune ecosystems. For example, the roots of the sister species A. arenaria hosted the diazotrophic bacterial species Burkholderia tropicalis in the Pacific Northwest (Dalton et al.2004). Although we did not directly examine levels of nitrogen fixation or nif gene expression, our detection of Burkholderiales suggests that root-associated taxa are present in North American dunes as well. In addition, Rhizobiales made up a significant fraction of the soil bacterial community (∼10%), suggesting that free-living diazotrophs could make important contributions to the nitrogen cycle in dune soils.
CONCLUSION
An aboveground fungal endophyte reduced AM fungal diversity and abundance and altered how root-associated bacteria responded to soil moisture. Most belowground responses to the aboveground endophyte varied among years, demonstrating context-dependency that may be caused by interannual variation in drought. Within the spatially heterogeneous, low nutrient and high disturbance ecosystem of Great Lakes dunes, plant roots acted as an important resource for belowground microbes, strongly increasing microbial diversity relative to that in the soil. Our work highlights the importance of examining aboveground microbes as factors that influence belowground microbes and sheds new light on above/belowground microbial interactions.
Supplementary Material
Supplementary data are available at FEMSEC online.
Acknowledgments
We thank A. J. Davitt, Kerri Crawford, Isaiah Reynolds, Shelly Gleason, Tamara Milton, Brandon McCormick, Caitlin Rhodes, Catherine Fargen, Emily Moehlman, Alex Bryant, Joy Uwimbabazi, Brad Gottshall, Jeff Masters and Keera Lowe for assistance with field work and experimental set-up. Thanks to David Janos for help with glomalin analyses, Shelley MacNeil for the bacterial laboratory work, and Valérie Huguet for assistance with fungal laboratory work. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSEC online.
FUNDING
This work has been funded by National Science Foundation Graduate Research Fellowship (NSFGRFP) #0940902 and the George Melendez Wright Climate Change Fellowship to LBD and National Science Foundation Division of Environmental Biology (NSFDEB) #0918267 and #1145588 to JAR and SME and National Science Foundation Office of Polar Programs (NSFOPP) #1142102 to CTV. Research reported in this publication was supported by the National Institute of General Medical Sciences of the National Institutes of Health under Award Number P30GM110907 to the Center for Evolutionary and Theoretical Immunology at UNM.
Conflict of interest. None declared.
REFERENCES
- A’Bear AD, Johnson SN, Jones TH. Putting the ‘upstairs–downstairs’ into ecosystem service: What can aboveground–belowground ecology tell us? Biol Control 2014;75:97–107. [Google Scholar]
- Albert D. Borne of the Wind: An Introduction to the Ecology of Michigan Sand Dunes. Lansing, MI: Michigan Biological Features Inventory, 2000. [Google Scholar]
- Andrews JH, Hirano SS. Microbial Ecology of Leaves. New York: Springer, 2012. [Google Scholar]
- Antunes PM, Miller J, Carvalho LM et al. Even after death the endophytic fungus of Schedonorus phoenix reduces the arbuscular mycorrhizas of other plants. Funct Ecol 2008;22:912–8. [Google Scholar]
- Arnold AE, Lutzoni F. Diversity and host range of foliar fungal endophytes: are tropical leaves biodiversity hotspots? Ecology 2007;88:541–9. [DOI] [PubMed] [Google Scholar]
- Arrieta AM, Iannone LJ, Scervino JM et al. A foliar endophyte increases the diversity of phosphorus-solubilizing rhizospheric fungi and mycorrhizal colonization in the wild grass Bromus auleticus. Fungal Ecol 2015;17:146–54. [Google Scholar]
- Bacon CW, Porter JK, Robbins JD et al. Epichloë typhina from toxic tall fescue grasses. Appl Environ Microb 1977;34:576–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacon CW, White JF. Microbial Endophytes. New York: Marcel Dekker, Inc., 2000. [Google Scholar]
- Bardgett RD, van der Putten WH. Belowground biodiversity and ecosystem functioning. Nature 2014;515:505–11. [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, Bolker B et al. Fitting linear mixed-effects models using lme4. J Stat Softw 2015;67:48. [Google Scholar]
- Bell-Dereske L, Gao X, Masiello CA et al. Plant-fungal symbiosis affects litter decomposition during primary succession. Oikos in press. [Google Scholar]
- Bergmann D, Zehfus M, Zierer L et al. Grass rhizosheaths: Associated bacterial communities and potential for nitrogen fixation. West N Am Naturalist 2009;69:105–14. [Google Scholar]
- Bever JD, Dickie IA, Facelli E et al. Rooting theories of plant community ecology in microbial interactions. Trends Ecol Evol 2010;25:468–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowatte S, Barrett B, Luscombe C et al. Effect of grass species and fungal endophyte on soil nitrification potential. N Z J Agr Res 2011;54:275–84. [Google Scholar]
- Bulgarelli D, Garrido-Oter R, Münch Philipp C et al. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015;17:392–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulgarelli D, Schlaeppi K, Spaepen S et al. Structure and functions of the bacterial microbiota of plants. Annu Rev Plant Biol 2013;64:807–38. [DOI] [PubMed] [Google Scholar]
- Buyer JS, Zuberer DA, Nichols KA et al. Soil microbial community function, structure, and glomalin in response to tall fescue endophyte infection. Plant Soil 2011;339:401–12. [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. QIIME allows analysis of high-throughput community sequencing data. Nat Meth 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casas C, Omacini M, Montecchia M et al. Soil microbial community responses to the fungal endophyte Neotyphodium in Italian ryegrass. Plant Soil 2011;340:347–55. [Google Scholar]
- Cheplick GP, Faeth SH. Ecology and Evolution of the Grass-endophyte Symbiosis. New York: Oxford University Press, 2009. [Google Scholar]
- Chu-Chou M, Guo B, An ZQ et al. Suppression of mycorrhizal fungi in fescue by the Acremonium coenophialum endophyte. Soil Biol Biochem 1992;24:633–7. [Google Scholar]
- Clarke K, Gorley R. PRIMER v6: User Manual/Tutorial. Plymouth: PRIMER-E, 2006, 192. [Google Scholar]
- Czárán TL, Hoekstra RF, Pagie L. Chemical warfare between microbes promotes biodiversity. P Natl Acad Sci USA 2002;99:786–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalton DA, Kramer S, Azios N et al. Endophytic nitrogen fixation in dune grasses (Ammophila arenaria and Elymus mollis) from Oregon. FEMS Microbiol Ecol 2004;49:469–79. [DOI] [PubMed] [Google Scholar]
- De Beenhouwer M, Van Geel M, Ceulemans T et al. Changing soil characteristics alter the arbuscular mycorrhizal fungi communities of Arabica coffee (Coffea arabica) in Ethiopia across a management intensity gradient. Soil Biol Biochem 2015;91:133–9. [Google Scholar]
- De Deyn GB, Biere A, van der Putten WH et al. Chemical defense, mycorrhizal colonization and growth responses in Plantago lanceolata L. Oecologia 2009;160:433–42. [DOI] [PubMed] [Google Scholar]
- Dean SL, Farrer EC, Porras-Alfaro A et al. Assembly of root-associated bacteria communities: interactions between abiotic and biotic factors. Environ Microbiol Rep 2015;7:102–10. [DOI] [PubMed] [Google Scholar]
- Dickie IA. Insidious effects of sequencing errors on perceived diversity in molecular surveys. New Phytol 2010;188:916–8. [DOI] [PubMed] [Google Scholar]
- Drake I, White JF Jr, Belanger FC. Identification of the fungal endophyte of Ammophila breviligulata (American beach grass) as Epichloe amarillans Mycotaxon. in review. [DOI] [PMC free article] [PubMed]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 2004;32:1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010;26:2460–1. [DOI] [PubMed] [Google Scholar]
- Edgar RC. UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 2013;10:996–8. [DOI] [PubMed] [Google Scholar]
- Edwards J, Johnson C, Santos-Medellín C et al. Structure, variation, and assembly of the root-associated microbiomes of rice. P Natl Acad Sci USA 2015;112:E911–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elmi AA, West CP. Endophyte infection effects on stomatal conductance, osmotic adjustment and drought recovery of tall fescue. New Phytol 1995;131:61–67. [DOI] [PubMed] [Google Scholar]
- Emery SM, Bell-Dereske L, Rudgers JA. Fungal symbiosis and precipitation alter traits and dune building by the ecosystem engineer, Ammophila breviligulata. Ecology 2015;96:927–35. [DOI] [PubMed] [Google Scholar]
- Emery SM, Rudgers JA. Impacts of simulated climate change and fungal symbionts on survival and growth of a foundation species in sand dunes. Oecologia 2013;173:1601–12. [DOI] [PubMed] [Google Scholar]
- Emery SM, Rudgers JA. Biotic and abiotic predictors of ecosystem engineering traits of the dune building grass, Ammophila breviligulata. Ecosphere 2014;5:art87. [Google Scholar]
- Emery SM, Thompson D, Rudgers JA. Variation in endophyte symbiosis, herbivory and drought tolerance of Ammophila breviligulata populations in the Great Lakes region. Am Midl Nat 2010;163:186–96. [Google Scholar]
- Enkhtuya B, Poschl M, Vosatka M. Native grass facilitates mycorrhizal colonisation and P uptake of tree seedlings in two anthropogenic substrates. Water Air Soil Poll 2005;166:217–36. [Google Scholar]
- Ensign KL, Webb EA, Longstaffe FJ. Microenvironmental and seasonal variations in soil water content of the unsaturated zone of a sand dune system at Pinery Provincial Park, Ontario, Canada. Geoderma 2006;136:788–802. [Google Scholar]
- Evans SE, Wallenstein MD. Climate change alters ecological strategies of soil bacteria. Ecol Lett 2014;17:155–64. [DOI] [PubMed] [Google Scholar]
- Evans SE, Wallenstein MD, Burke IC. Is bacterial moisture niche a good predictor of shifts in community composition under long-term drought? Ecology 2014;95:110–22. [DOI] [PubMed] [Google Scholar]
- Faure D, Vereecke D, Leveau JHJ. Molecular communication in the rhizosphere. Plant Soil 2008;321:279–303. [Google Scholar]
- Fierer N, Jackson RB. The diversity and biogeography of soil bacterial communities. P Natl Acad Sci USA 2006;103:626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fierer N, Bradford MA, Jackson RB. Toward an ecological classification of soil bacteria. Ecology 2007;88:1354–64. [DOI] [PubMed] [Google Scholar]
- Fokom R, Adamou S, Teugwa MC et al. Glomalin related soil protein, carbon, nitrogen and soil aggregate stability as affected by land use variation in the humid forest zone of south Cameroon. Soil Till Res 2012;120:69–75. [Google Scholar]
- Franzluebbers AJ, Hill NS. Soil carbon, nitrogen, and ergot alkaloids with short- and long-term exposure to endophyte-infected and endophyte-free tall fescue. Soil Sci Soc Am J 2005;69:404–12. [Google Scholar]
- Franzluebbers AJ, Nazih N, Stuedemann JA et al. Soil carbon and nitrogen pools under low- and high-endophyte-infected tall fescue. Soil Sci Soc Am J 1999;63:1687–94. [Google Scholar]
- Franzluebbers AJ, Stuedemann JA. Soil carbon and nitrogen pools in response to tall fescue endophyte infection, fertilization, and cultivar. Soil Sci Soc Am J 2005;69:396–403. [Google Scholar]
- Giovannoni SJ, DeLong EF, Schmidt TM et al. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microb 1990;56:2572–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey PJ. Climate, plant response and development of dunes on barrier beaches along the U.S. east coast. Int J Biometeorol 1977;21:203–16. [Google Scholar]
- Gollotte A, van Tuinen D, Atkinson D. Diversity of arbuscular mycorrhizal fungi colonising roots of the grass species Agrostis capillaris and Lolium perenne in a field experiment. Mycorrhiza 2004;14:111–7. [DOI] [PubMed] [Google Scholar]
- Gronewold A, Fortin V, Lofgren B et al. Coasts, water levels, and climate change: A Great Lakes perspective. Climatic Change 2013;120:697–711. [Google Scholar]
- Guo J, McCulley RL, McNear DH. Tall fescue cultivar and fungal endophyte combinations influence plant growth and root exudate composition. Front Plant Sci 2015;6:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamady M, Walker JJ, Harris JK et al. Error-correcting barcoded primers for pyrosequencing hundreds of samples in multiplex. Nat Methods 2008;5:235–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houle G. Interactions between resources and abiotic conditions control plant performance on subarctic coastal dunes. Am J Bot 1997;84:1729–37. [PubMed] [Google Scholar]
- IPCC. Climate Change 2007: Synthesis Report. Contribution of Working Groups I, II and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change. Geneva, Switzerland, 2007. [Google Scholar]
- IPCC. Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part B: Regional Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change. Cambridge, United Kingdom and New York, NY, USA, 2014. [Google Scholar]
- Iqbal J, Siegrist JA, Nelson JA et al. Fungal endophyte infection increases carbon sequestration potential of southeastern USA tall fescue stands. Soil Biol Biochem 2012;44:81–92. [Google Scholar]
- Janos DP, Garamszegi S, Beltran B. Glomalin extraction and measurement. Soil Biol Biochem 2008;40:728–39. [Google Scholar]
- Jenkins MB, Franzluebbers AJ, Humayoun SB. Assessing short-term responses of prokaryotic communities in bulk and rhizosphere soils to tall fescue endophyte infection. Plant Soil 2006;289:309–20. [Google Scholar]
- Johansen RB, Vestberg M, Burns BR et al. A coastal sand dune in New Zealand reveals high arbuscular mycorrhizal fungal diversity. Symbiosis 2015;66:111–21. [Google Scholar]
- Kannadan S, Rudgers JA. Endophyte symbiosis benefits a rare grass under low water availability. Funct Ecol 2008;22:706–13. [Google Scholar]
- Kivlin SN, Emery SM, Rudgers JA. Fungal symbionts alter plant responses to global change. Am J Bot 2013;100:1445–57. [DOI] [PubMed] [Google Scholar]
- Koide RT, Peoples MS. Behavior of Bradford-reactive substances is consistent with predictions for glomalin. Appl Soil Ecol 2013;63:8–14. [Google Scholar]
- Koske RE, Gemma JN. Mycorrhizae and succession in plantings of beachgrass in sand dunes. Am J Bot 1997;84:118–30. [Google Scholar]
- Kostenko O, van de Voorde TFJ, Mulder PPJ et al. Legacy effects of aboveground–belowground interactions. Ecol Lett 2012;15:813–21. [DOI] [PubMed] [Google Scholar]
- Larimer AL, Bever JD, Clay K. Consequences of simultaneous interactions of fungal endophytes and arbuscular mycorrhizal fungi with a shared host grass. Oikos 2012;121:2090–6. [Google Scholar]
- Lemons A, Clay K, Rudgers JA. Connecting plant–microbial interactions above and belowground: a fungal endophyte affects decomposition. Oecologia 2005;145:595–604. [DOI] [PubMed] [Google Scholar]
- Leuchtmann A, Bacon CW, Schardl CL et al. Nomenclatural realignment of Neotyphodium species with genus Epichloë. Mycologia 2014;106:202–15. [DOI] [PubMed] [Google Scholar]
- Leuchtmann A, Clay K. Experimental infection of host grasses and sedges with Atkinsonella hypoxylon and Balansia cyperi (Balansiae, Clavicipitaceae). Mycologia 1988;80:291–7. [Google Scholar]
- Lichter J. Primary succession and forest development on coastal Lake Michigan sand dunes. Ecol Monogr 1998a;68:487–510. [Google Scholar]
- Lichter J. Rates of weathering and chemical depletion in soils across a chronosequence of Lake Michigan sand dunes. Geoderma 1998b;85:255–82. [Google Scholar]
- Lichter J. Colonization constraints during primary succession on coastal Lake Michigan sand dunes. J Ecol 2000;88:825–39. [Google Scholar]
- Lindahl BD, Nilsson RH, Tedersoo L et al. Fungal community analysis by high-throughput sequencing of amplified markers – a user's guide. New Phytol 2013;199:288–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Parsons AJ, Xue H et al. Competition between foliar Neotyphodium lolii endophytes and mycorrhizal Glomus spp. fungi in Lolium perenne depends on resource supply and host carbohydrate content. Funct Ecol 2011;25:910–20. [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microb 2005;71:8228–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGonigle TP, Miller MH, Evans DG et al. A new method which gives an objective measure of colonization of roots by vesicular—arbuscular mycorrhizal fungi. New Phytol 1990;115:495–501. [DOI] [PubMed] [Google Scholar]
- Mack KML, Rudgers JA. Balancing multiple mutualists: asymmetric interactions among plants, arbuscular mycorrhizal fungi, and fungal endophytes. Oikos 2008;117:310–20. [Google Scholar]
- McMurdie PJ, Holmes S. Waste not, want not: Why rarefying microbiome data is inadmissible. PLoS Comput Biol 2014;10:e1003531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malinowski DP, Belesky DP. Adaptations of endophyte-infected cool-season grasses to environmental stresses: mechanisms of drought and mineral stress tolerance. Crop Sci 2000;40:923–40. [Google Scholar]
- Mirarab S, Nguyen N, Guo S et al. PASTA: Ultra-large multiple sequence alignment for nucleotide and amino-acid sequences. J Comput Biol 2015;22:377–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell KR, Takacs-Vesbach CD. A comparison of methods for total community DNA preservation and extraction from various thermal environments. J Ind Microbiol Biot 2008;35:1139–47. [DOI] [PubMed] [Google Scholar]
- Novas MV, Cabral D, Godeas AM. Interaction between grass endophytes and mycorrhizas in Bromus setifolius from Patagonia, Argentina. Symbiosis 2005;40:23–30 [Google Scholar]
- Novas MV, Iannone LJ, Godeas AM et al. Evidence for leaf endophyte regulation of root symbionts: effect of Neotyphodium endophytes on the pre-infective state of mycorrhizal fungi. Symbiosis 2011;55:19–28. [Google Scholar]
- Novas VM, Iannone L, Godeas A et al. Positive association between mycorrhiza and foliar endophytes in Poa bonariensis, a native grass. Mycol Prog 2009;8:75–81. [Google Scholar]
- Nuccio EE, Anderson-Furgeson J, Estera KY et al. Climate and edaphic controllers influence rhizosphere community assembly for a wild annual grass. Ecology 2016;97:1307–18. [DOI] [PubMed] [Google Scholar]
- Nuñez MA, Moretti A, Simberloff D. Propagule pressure hypothesis not supported by an 80-year experiment on woody species invasion. Oikos 2011;120:1311–16. [Google Scholar]
- Okie JG, Van Horn DJ, Storch D et al. Niche and metabolic principles explain patterns of diversity and distribution: theory and a case study with soil bacterial communities. P Roy Soc B: Biol Sci 2015;282:20142630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oksanen J, Blanchet FG, Kindt R et al. vegan: Community ecology package. R package version 2.3-5, 2016, https://CRAN.R-project.org/package=vegan.
- Olson JS. Rates of succession and soil changes on Southern Lake Michigan sand dunes. Botanical Gazette 1958;119:125–70. [Google Scholar]
- Omacini M, Semmartin M, Pérez LI et al. Grass–endophyte symbiosis: a neglected aboveground interaction with multiple belowground consequences. Appl Soil Ecol 2012;61:273–9. [Google Scholar]
- Öpik M, Vanatoa A, Vanatoa E et al. The online database MaarjAM reveals global and ecosystemic distribution patterns in arbuscular mycorrhizal fungi (Glomeromycota). New Phytol 2010;188:223–41. [DOI] [PubMed] [Google Scholar]
- Orchard S, Standish RJ, Nicol D et al. The response of fine root endophyte (Glomus tenue) to waterlogging is dependent on host plant species and soil type. Plant Soil 2016;403:305–15. [Google Scholar]
- Perumal M. The role of mycorrhizal fungi in growth enhancement of dune plants following burial in sand. Funct Ecol 1999;13:560–6. [Google Scholar]
- Philippot L, Raaijmakers JM, Lemanceau P et al. Going back to the roots: the microbial ecology of the rhizosphere. Nat Rev Microbiol 2013;11:789–99. [DOI] [PubMed] [Google Scholar]
- Phillips LA, Ward V, Jones MD. Ectomycorrhizal fungi contribute to soil organic matter cycling in sub-boreal forests. ISME J 2014;8:699–713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26:1641–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quince C, Lanzen A, Davenport RJ et al. Removing noise from pyrosequenced amplicons. BMC Bioinformatics 2011;12:1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redecker D, Schüßler A, Stockinger H et al. An evidence-based consensus for the classification of arbuscular mycorrhizal fungi (Glomeromycota). Mycorrhiza 2013;23:515–31. [DOI] [PubMed] [Google Scholar]
- Rillig MC, Wright SF, Nichols KA et al. Large contribution of arbuscular mycorrhizal fungi to soil carbon pools in tropical forest soils. Plant Soil 2001;233:167–77. [Google Scholar]
- Rodriguez RJ, White JF, Arnold AE et al. Fungal endophytes: diversity and functional roles. New Phytol 2009;182:314–30. [DOI] [PubMed] [Google Scholar]
- Rojas X, Guo J, Leff JW et al. Infection with a shoot-specific fungal endophyte (Epichloë) alters tall fescue soil microbial communities. Microbial Ecol 2016;72:197–206. [DOI] [PubMed] [Google Scholar]
- Rosier CL, Hoye AT, Rillig MC. Glomalin-related soil protein: Assessment of current detection and quantification tools. Soil Biol Biochem 2006;38:2205–11. [Google Scholar]
- Rostas M, Cripps MG, Silcock P. Aboveground endophyte affects root volatile emission and host plant selection of a belowground insect. Oecologia 2015;177:487–97. [DOI] [PubMed] [Google Scholar]
- Rousk J, Frey SD. Revisiting the hypothesis that fungal-to-bacterial dominance characterizes turnover of soil organic matter and nutrients. Ecol Monogr 2015;85:457–72. [Google Scholar]
- Rudgers J, Mattingly W, Koslow J. Mutualistic fungus promotes plant invasion into diverse communities. Oecologia 2005;144:463–71. [DOI] [PubMed] [Google Scholar]
- Rudgers JA, Bell-Dereske L, Crawford KM et al. Fungal symbiont effects on dune plant diversity depend on precipitation. J Ecol 2015;103:219–30. [Google Scholar]
- Schlacher TA, Schoeman DS, Dugan J et al. Sandy beach ecosystems: key features, sampling issues, management challenges and climate change impacts. Marine Ecol 2008;29:70–90. [Google Scholar]
- Schlatter DC, Bakker MG, Bradeen JM et al. Plant community richness and microbial interactions structure bacterial communities in soil. Ecology 2015;96:134–42. [DOI] [PubMed] [Google Scholar]
- Schüßler A, Walker C. The Glomeromycota: a species list with new families and new genera. Gloucester, UK: 2010. [Google Scholar]
- Schwartz E, Van Horn DJ, Buelow HN et al. Characterization of growing bacterial populations in McMurdo Dry Valley soils through stable isotope probing with 18O-water. FEMS Microbiol Ecol 2014;89:415–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi S, Nuccio E, Herman DJ et al. Successional trajectories of rhizosphere bacterial communities over consecutive seasons. mBio 2015;6:e00746-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegrist JA, McCulley RL, Bush LP et al. Alkaloids may not be responsible for endophyte-associated reductions in tall fescue decomposition rates. Funct Ecol 2010;24:460–8. [Google Scholar]
- Singer A, Travis JMJ, Johst K. Interspecific interactions affect species and community responses to climate shifts. Oikos 2013;122:358–66. [Google Scholar]
- Staddon PL, Gregersen R, Jakobsen I. The response of two Glomus mycorrhizal fungi and a fine endophyte to elevated atmospheric CO2, soil warming and drought. Global Change Biology 2004;10:1909–21. [Google Scholar]
- Team RC R: A Language and Environment for Statistical Computing. Vienna, Austria: R Foundation for Statistical Computing, 2015. [Google Scholar]
- Tedersoo L, Anslan S, Bahram M et al. Shotgun metagenomes and multiple primer pair-barcode combinations of amplicons reveal biases in metabarcoding analyses of fungi. MycoKeys 2015;10:1–43. [Google Scholar]
- Thiéry O, Moora M, Vasar M et al. Inter- and intrasporal nuclear ribosomal gene sequence variation within one isolate of arbuscular mycorrhizal fungus, Diversispora sp. Symbiosis 2012;58:135–47. [Google Scholar]
- Tylianakis JM, Didham RK, Bascompte J et al. Global change and species interactions in terrestrial ecosystems. Ecol Lett 2008;11:1351–63. [DOI] [PubMed] [Google Scholar]
- van der Heijden MGA, Klironomos JN, Ursic M et al. Mycorrhizal fungal diversity determines plant biodiversity, ecosystem variability and productivity. Nature 1998;396:69–72. [Google Scholar]
- Van der Putten WH. Climate change, aboveground-belowground interactions, and species' range shifts. Ann Rev Ecol Evol Syst 2012;43:365–83. [Google Scholar]
- Van Geel M, Busschaert P, Honnay O et al. Evaluation of six primer pairs targeting the nuclear rRNA operon for characterization of arbuscular mycorrhizal fungal (AMF) communities using 454 pyrosequencing. J Microbiol Meth 2014;106:93–100. [DOI] [PubMed] [Google Scholar]
- van Geel M, Ceustermans A, van Hemelrijck W et al. Decrease in diversity and changes in community composition of arbuscular mycorrhizal fungi in roots of apple trees with increasing orchard management intensity across a regional scale. Mol Ecol 2015;24:941–52. [DOI] [PubMed] [Google Scholar]
- Van Geel M, De Beenhouwer M, Ceulemans T et al. Application of slow-release phosphorus fertilizers increases arbuscular mycorrhizal fungal diversity in the roots of apple trees. Plant Soil 2016;402:291–301. [Google Scholar]
- Van Hecke MM, Treonis AM, Kaufman JR. How does the fungal endophyte Neotyphodium coenophialum affect tall fescue (Festuca arundinacea) rhizodeposition and soil microorganisms? Plant Soil 2005;275:101–9. [Google Scholar]
- Van Horn DJ, Van Horn ML, Barrett JE et al. Factors controlling soil microbial biomass and bacterial diversity and community composition in a cold desert ecosystem: role of geographic scale. PLoS One 2013;8:e66103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vierheilig H, Coughlan AP, Wyss U et al. Ink and vinegar, a simple staining technique for arbuscular-mycorrhizal fungi. Appl Environ Microb 1998;64:5004–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Yang P, Falcão Salles J. Distribution of root-associated bacterial communities along a salt-marsh primary succession. Front Plant Sci 2016;6:1188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright SF, Upadhyaya A. Extraction of an abundant and unusual protein from soil and comparison with hyphal protein of arbuscular mycorrhizal fungi. Soil Sci 1996;161:575–86. [Google Scholar]
- Wullstein LH. Variation in N2 fixation (C2H2 reduction) associated with rhizosheaths of Indian Ricegrass (Stipa hymenoides). Am Midl Nat 1991;126:76–81. [Google Scholar]
- Wullstein LH, Bruening ML, Bollen WB. Nitrogen fixation associated with sand grain root sheaths (rhizosheaths) of certain xeric grasses. Physiol Plantarum 1979;46:1–4. [Google Scholar]
- Yahdjian L, Sala O. A rainout shelter design for intercepting different amounts of rainfall. Oecologia 2002;133:95–101. [DOI] [PubMed] [Google Scholar]
- Zhou J, Deng Y, Shen L et al. Temperature mediates continental-scale diversity of microbes in forest soils. Nat Commun 2016;7:12083. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSEC online.