Abstract
Antibiotic therapies are known to disrupt gastrointestinal (GI) bacterial communities. HIV and pathogenic simian immunodeficiency virus (SIV) infections have also been associated with disrupted GI bacterial communities. We administered a combination antibiotic therapy to six SIV-infected rhesus macaques and collected colon biopsies, stool samples and rectal swabs before and after antibiotics, and evaluated the bacterial communities at each sample site using high-throughput 16S rRNA gene sequencing. The colon mucosa and stool samples displayed different bacterial communities, while the rectal swabs showed a mixture of the mucosal and stool-associated bacteria. Antibiotics disrupted the native bacterial communities at each sample site. The colon mucosa showed depleted abundances of the dominant Helicobacteraceae, while we found depleted abundances of the dominant Ruminococcaceae sp. in the stool. The rectal swabs showed similar trends as the colon mucosa, but were more variable. After the antibiotic treatment, there were increased abundances of similar taxa of facultative anaerobic bacteria, including Lactobacillaceae and Enterobacteriaceae at each sample site.
Keywords: macaque, microbiome, antibiotics, SIV, HIV, rectal swab
Antibiotics alter bacterial communities in the colon, stool and rectal swabs of SIV-infected macaques, leading to a reduction of commensal bacteria and an increase in potentially pathogenic bacteria, including Enterobacteriaceae species.
INTRODUCTION
Antibiotics are widely used in human health and veterinary practices, and are known to disrupt the gastrointestinal (GI) microbiota (Becattini, Taur and Pamer 2016). Simian immunodeficiency virus (SIV) infection of non-human primates (NHP) is the leading animal model to accurately recapitulate the pathogenesis of HIV (Hatziioannou and Evans 2012). Dysbiosis of the intestinal microbiota and translocation of microbes from the colon into the periphery are important processes that drive inflammation in HIV and SIV infections (Brenchley et al.2006; Klase et al.2015), and these processes are likely affected by antibiotic administration. However, the impact of antibiotics on colonic and rectal microbiota in SIV-infected NHP has not been investigated.
We previously reported that oral administration of vancomycin for 7 days followed by a combination of vancomycin and enrofloxacin for 12 days disrupted colonic mucosal bacterial communities of six SIV-infected rhesus macaques treated with antiretroviral therapy (ART), and that a fecal microbiome transplant (FMT) using donor material from a healthy rhesus macaque restored these communities to a pre-antibiotics state. Furthermore, we demonstrated that the FMT was associated with statistically significant increases in peripheral IL-17 and IL-22-producing CD4 + T cells, and statistically significant decreases in activated CD4 + T cells in the jejunal and rectal mucosae (Hensley-McBain et al.2016).
In the previous study, we did not report the bacterial communities detected in the colonic lumen (i.e. stool) or those associated with rectal swabs. Furthermore, the previous study was mostly focused on the effects of FMT on host immunity and did not exclusively evaluate the effects of the antibiotic treatment on the microbiota. Here, we compare the bacterial communities from each site (colon biopsy, stool and rectal swab) before and after antibiotic treatment. We demonstrate that the colonic mucosa and lumen in these macaques had unique bacterial communities, and that the community structure determined from rectal swab samples shared similarities with the mucosal and luminal communities. We further demonstrate that the antibiotic treatment disrupted the normal bacterial communities at each sample site, leading to a reduction in fermentative bacteria and increased abundances of facultative anaerobic bacteria.
MATERIALS AND METHODS
Study animals and antibiotic treatment
Animals were housed and cared for in Association for the Assessment and Accreditation of Laboratory Animal Care international accredited facilities, and all animal procedures were performed according to protocols approved by the Institutional Animal Care and Use Committee of University of Washington (Protocol 4314–01) as previously described (Hensley-McBain et al.2016). Briefly, six male rhesus macaques (Macaca mulatta) were infected intrarectally with SIVMAC239X and started on ART 130 days after infection, which consisted of tenofovir (PMPA; 20 mg kg−1), emtricitabine (FTC; 30 mg kg−1) and raltegravir (50 mg b.i.d). The animals were then treated with an antibiotic regimen consisting of oral vancomycin (15 mg kg−1 b.i.d) for 7 days followed by a combination of oral vancomycin and enrofloxacin (10 mg kg−1 b.i.d.) for 12 additional days. Antibiotics were given with a treat and were monitored to ensure the entire dose was taken. Biopsies of the colon (immersed in RNALater Solution (ThermoFisher Scientific, Waltham, MA)), rectal swabs and stool were collected before and after the antibiotic treatment and were placed at –80°C after collection.
DNA extraction and sequencing
Genomic DNA was extracted from the colon biopsies, rectal swabs and stool using the Omni Bead Ruptor (Omni International, Kennesaw, GA) with 2.8-mm ceramic beads, followed by purification using the RNA/DNA All-Prep Kit (Qiagen, Germantown, MA). We used 5 ng of DNA to generate 460 bp amplicons by targeting the V3-V4 hypervariable regions of the 16S rRNA gene. To achieve this, we utilized the previously described 341F-805R primer pair targeting the V3-V4 (Table 1) region of the bacterial 16S rRNA gene (Herlemann et al.2011; Klindworth et al.2013). PCR conditions were as follows: 95°C for 3 min followed by 25 cycles of 95°C for 30 s, 55°C for 30 s and 72°C for 30 s, followed by a final extension at 72°C for 5 min. Nextera XT dual index adaptors were incorporated by performing 12 PCR cycles using a FailSafe PCR system, cleaned using 1.1X AMPure XP Beads (Beckman Coulter, Brea, CA) and quantified using DNA High Sensitivity Qubit (ThermoFisher). The samples were then mixed in equimolar ratios. These final libraries were then loaded onto a 600-cycle Illumina MiSeq Kit and sequenced using an Illumina MiSeq (Illumina, San Diego, CA) at 2 pM concentration with 5% PhiX control and sequenced using Nextera sequencing read and index primers (Hensley-McBain et al.2016).
Table 1.
Primers used in this study.
| Name | Sequence (5΄→3΄) | Purpose | Reference |
|---|---|---|---|
| 341F | CCTACGGGNGGCWGCAG | Illumina 16S rRNA gene sequencing | Klindworth et al. (2013) |
| 805R | GACTACHVGGGTATCTAATCC | Illumina 16S rRNA gene sequencing | Klindworth et al. (2013) |
| 8F | AGAGTTTGATCCTGGCTCAG | Cloning whole 16S rRNA genes | Galkiewicz and Kellogg (2008) |
| 1542R | AAGGAGGTGATCCAGCCGCA | Cloning whole 16S rRNA genes | Galkiewicz and Kellogg (2008) |
| 1055yF | ATGGYTGTCGTCAGCT | Total bacterial 16S qPCR | Ritalahti et al. (2006) |
| 1392R | ACGGGCGGTGTGTAC | Total bacterial 16S qPCR | Dionisi et al. (2003) |
| 16STaq1115 | FAM-CAACGAGCGCAACCC-BHQ1 | Total bacterial 16S qPCR | Dionisi et al. (2003) |
| HelicoF | ACCAAGGCWATGACGGGTATC | Helicobacter-specific 16S qPCR | Huijsdens et al. (2004) |
| HelicoR | CGGAGTTAGCCGGTGCTTATT | Helicobacter-specific 16S qPCR | Huijsdens et al. (2004) |
| HelicoTaq | FAM-AACCTTCATCCTCCACGCGGC-BHQ1 | Helicobacter-specific 16S qPCR | Huijsdens et al. (2004) |
Analysis of 16S rRNA gene sequencing data
Paired-end reads were joined using the PANDAseq assembler (Masella et al.2012) and the resulting reads were analyzed using the QIIME V1.9.0 pipeline (Caporaso et al.2010). We used the split_libraries_fastq.py script to demultiplex and quality filter the sequence reads using a quality score cutoff of 20. After quality filtering, we obtained 3214 697 total reads with an average of 91 848 reads per sample and a median sequence length of 443 base pairs. We clustered sequences into operational taxonomic units (OTUs) with an open-reference approach at 97% similarity using the UCLUST algorithm and assigned taxonomy to each OTU based using the SILVA database (Pruesse et al.2007), as implemented by the pick_otus.py script. This resulted in an average of ∼45 000 OTUs per sample. We identified potentially chimeric sequences using the ChimeraSlayer algorithm as implemented in the identify_chimeric_seqs.py script and removed those sequences from the analysis. To further analyze the species within the Helicobacteraceae family, we pulled all sequences assigned to the family Helicobacteraceae and searched these against the NCBI 16S rRNA gene database using the BLAST algorithm. We subsampled 40 000 sequences per sample from the resulting OTU tables and calculated the Shannon diversity, and performed principal coordinate analysis (PCoA) based on Bray-Curtis distances using the vegan and ape packages in R. Wilcoxon signed-rank tests were performed using Graphpad Prism. Linear discriminant analysis effect size (LEfSe) analysis was performed using online tools (Segata et al.2011). All sequence data from this study have been uploaded to the NCBI SRA under BioProject Accession PRJNA385206.
Quantitative polymerase chain reaction
We used quantitative polymerase chain reaction (qPCR) to quantify total bacterial 16S rRNA genes at each sample site. We prepared triplicate 10 μL reactions of each sample and standard containing 1X SsoAdvanced Universal Probes Supermix (BioRad, Hercules, CA), and 300 nM each of forward primer, 1055yF (Ritalahti et al.2006); reverse primer; 1392R, and TaqMan probe, 16STaq1115 (Dionisi et al.2003) (Table 1). The TaqMan probe carried a 5΄ FAM dye and a 3΄ BHQ1 quencher. To generate a standard curve, we PCR amplified the entire 16S rRNA gene of Lactobacillus rhamnosus ATCC53103 using the universal bacterial primers 8F and 1542R and cloned this amplicon into a plasmid using the TOPO TA Cloning Kit (ThermoFisher Scientific). We then serially diluted this plasmid such that final standard curve ranged from 7.1–7.1 × 106 16S rRNA gene copies per microliter. Reactions were initially denatured at 95°C for 2 min followed by 40 cycles of 95°C for 10 s, 56°C for 20 s and 68°C for 20 s. We performed qPCR using a BioRad CFX96 Real-Time PCR Detection System (BioRad, Hercules, CA). We performed Helicobacter genus specific qPCR using previously described primers (Table 1) and qPCR methods (Huijsdens et al.2004). For this, we prepared a qPCR standard as described above, but instead used the PCR-amplified 16S rRNA gene from Helicobacter pylori in the TOPO-TA reaction. For the colon biopsy and stool samples, we normalized copy numbers to the amount of input biomass. As this information was not available for rectal swabs, we reported total copies detected.
RESULTS
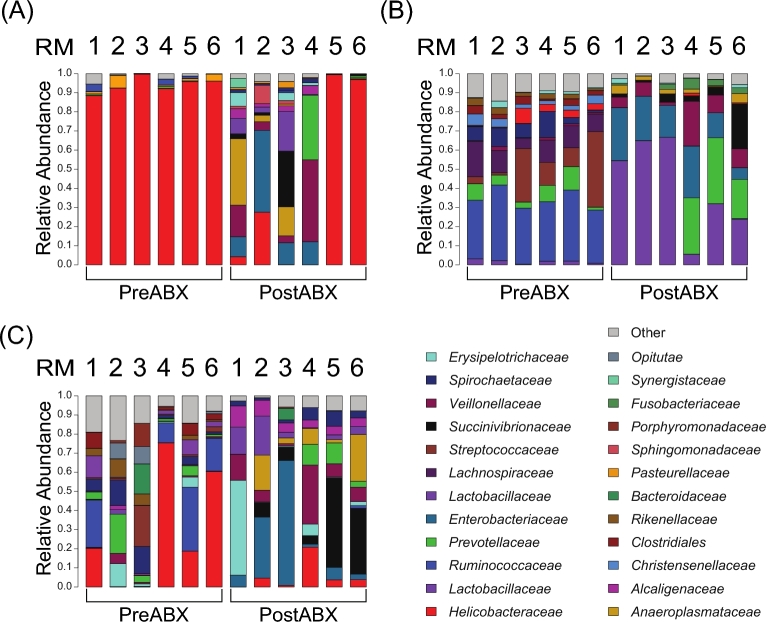
Prior to the antibiotic treatment, the colon biopsies (Fig. 1A) were dominated by bacteria from the family Helicobacteraceae, on average representing 94.1 ± 3.9% of the total community. This high abundance of Helicobacteraceae in the colonic mucosa was unexpected, as mucosal bacterial communities in humans often display a greater degree of diversity (Stearns et al.2011; Dillon et al.2014). Thus, we further evaluated the bacterial species within the family Helicobacteraceae across each sample site and found that it was composed mostly of Helicobacter fennalliae (53.2%) and H. macacae (45.7%). We also detected a variety of other Helicobacter species as well as several Campylobacter species, but these totaled <1% of the Helicobacteraceae species (Table 2).
Figure 1.
Bacterial community composition was altered by antibiotic treatment. Before the antibiotic treatment (PreABX), the colon biospies (A) were dominated by Helicobacteraceae while the stool communities (B) were composed of Ruminococcaceae, Lachnospiraceae and other anaerobic bacteria. The rectal swab (C) communities represented a mixture of the bacteria from the mucosa and stool. After the antibiotic treatment (PostABX), there was an expansion of Enterobacteriaceae, Lactobacillaceae and other facultative anaerobic bacterial phylotypes at all three sites. However, each site displayed unique bacterial communities.
Table 2.
Species detected within the family Helicobacteraceae.
| Accession no. | Number of sequences | Fraction of Helicobacteraceae sequences | Nearest BLAST hit species |
|---|---|---|---|
| NG_042881 | 581 758 | 0.532471384 | Helicobacter fennelliae |
| NG_042884 | 499 625 | 0.457296703 | Helicobacter macacae |
| NR_146694 | 5270 | 0.004823525 | Helicobacter canicola |
| NR_135861 | 3064 | 0.002804418 | Helicobacter himalayensis |
| NR_044761 | 789 | 0.000722156 | Helicobacter pylori |
| NR_116595 | 334 | 0.000305703 | Helicobacter pullorum |
| NG_042880 | 304 | 0.000278245 | Helicobacter fennelliae |
| NR_115104 | 268 | 0.000245295 | Helicobacter canadensis |
| NR_025941 | 225 | 0.000205938 | Helicobacter cinaedi |
| NR_074476 | 144 | 0.0001318 | Helicobacter cetorum |
| NR_114557 | 124 | 0.000113495 | Helicobacter winghamensis |
| NR_024836 | 100 | 9.1528E-05 | Helicobacter ganmani |
| NR_026074 | 98 | 8.96974E-05 | Helicobacter rodentium |
| NR_043799 | 93 | 8.5121E-05 | Helicobacter brantae |
| NR_118528 | 80 | 7.32224E-05 | Campylobacter upsaliensis |
| NR_025124 | 77 | 7.04765E-05 | Helicobacter aurati |
| NR_044169 | 45 | 4.11876E-05 | Helicobacter suis |
| NR_029182 | 32 | 2.9289E-05 | Helicobacter bilis |
| NR_025939 | 31 | 2.83737E-05 | Helicobacter muridarum |
| NR_118526 | 27 | 2.47126E-05 | Campylobacter showae |
| NR_041825 | 18 | 1.6475E-05 | Helicobacter marmotae |
| NR_118513 | 16 | 1.46445E-05 | Campylobacter curvus |
| NR_028019 | 9 | 8.23752E-06 | Helicobacter pametensis |
| NR_043052 | 7 | 6.40696E-06 | Helicobacter canis |
| NR_043108 | 6 | 5.49168E-06 | Helicobacter cholecystus |
| NR_115105 | 6 | 5.49168E-06 | Helicobacter equorum |
| NR_025952 | 4 | 3.66112E-06 | Helicobacter hepaticus |
| NR_025935 | 3 | 2.74584E-06 | Helicobacter felis |
| NR_118516 | 3 | 2.74584E-06 | Campylobacter gracilis |
| NG_042882 | 2 | 1.83056E-06 | Helicobacter mastomyrinus |
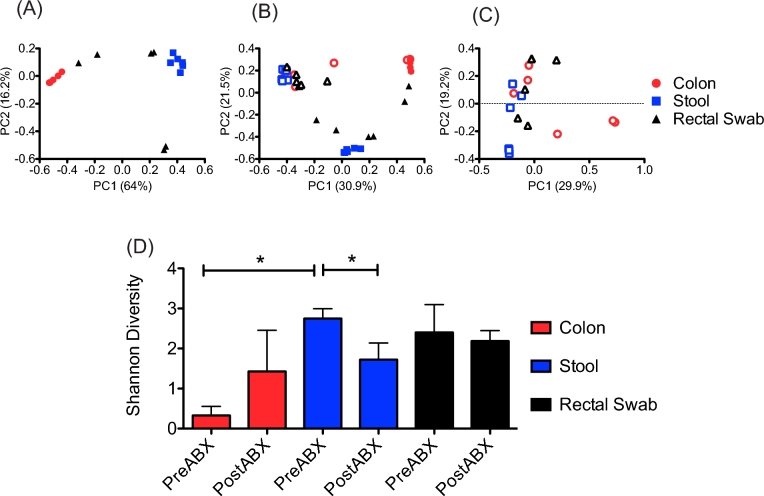
In contrast, the stool bacterial community (Fig. 1B) showed a more even distribution of Ruminococcaceae (32.6 ± 4.7%), Streptococcaceae (15.7 ± 15%), Lachnospiraceae (11.3 ± 4.3%) and Prevotellaceae (6.6 ± 4.0%), among a variety of other bacterial families. The bacteria detected on the rectal swabs (Fig. 1C) were similar to those from the colon biopsy and stool samples, including Helicobacteraceae, Ruminococcaceae and Prevotellaceae. However, the rectal swab communities for each animal were distinct from the corresponding biopsy and stool samples, and the rectal swabs were more variable between the animals. Indeed, PCoA of the bacterial abundance and diversity in the samples taken before the antibiotic treatment revealed that the rectal swab samples appeared to share features with the mucosal and fecal communities (Fig. 2A). This grouping of the rectal swab samples also emphasized that the rectum is a unique compartment.
Figure 2.
Antibiotics altered bacterial community diversity. PCoA of the samples taken before antibiotic treatment demonstrated that the mucosal (red circles) and stool-associated (blue squares) communities clustered separately, while those from the rectal swabs (black triangles) formed an intermediate grouping (A). Comparing all of the samples from before (closed symbols) and after (open symbols) the antibiotic treatment demonstrated that there was a greater degree of similarity between the bacterial communities at each site after the antibiotic treatment (B). This trend was further demonstrated by PCoA of samples taken after the antibiotic treatment, which did not show distinctive clustering (C). The antibiotic treatment led to increased species diversity in the colon mucosa, but decreased diversity in the stool-associated communities, while rectal swabs showed a minor, but not statistically significant, decrease in diversity. A Wilcoxon signed-rank test was used to determine statistical significance * = P < 0.05 (D).
Following the antibiotic treatment, there were dramatic shifts in the bacterial communities detected in all three of the sample sites. Interestingly, the colon biopsy bacterial communities from RM5 and RM6 did not show a response to the antibiotic treatment, and instead retained high abundances of Helicobacteraceae. However, the stool and rectal swab samples from these animals did show altered bacterial community composition after the antibiotic treatment (Fig. 1). A further interesting trend revealed by PCoA of the samples from before and after the antibiotic treatment was that the bacterial communities from all three of the sample sites were more similar after the antibiotic treatment than they had been before the antibiotic treatment, as demonstrated by clustering of the post-antibiotic samples in PCoA plots (Fig. 2B). PCoA of the samples taken after the antibiotic treatment confirmed this trend (Fig. 2C) demonstrated by less distinctive clustering of the samples based on the sampling site. This indicated that the antibiotic treatment led to increased abundances of similar bacteria without regard to the biogeographic location. We also found that the antibiotic treatments altered bacterial species diversity at each sample site. The colon biopsy samples showed significantly increased diversity after the antibiotic treatment, while that of the stool samples was significantly decreased. There was no significant change in the species diversity of the rectal swabs after the antibiotic treatment (Fig. 2C).
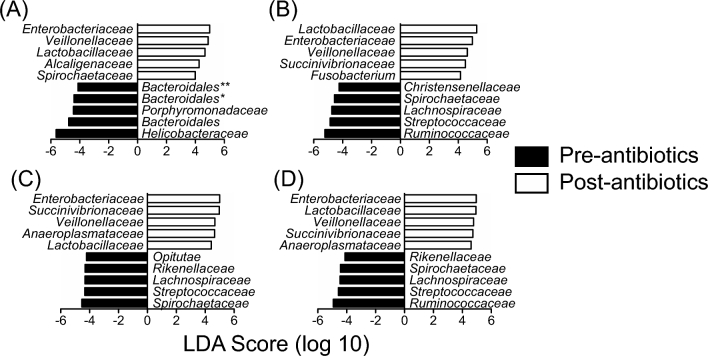
We used LEfSe analysis to determine which bacterial families were most differentially abundant before and after the antibiotic treatment at each sample site as well as across all of the sample sites (Fig. 3). Because the colon biopsies from RM5 and 6 did not show a response to the antibiotics, we omitted these samples from the analysis and only performed LEfSe analysis using animals that responded to the antibiotic treatment. The LEfSe analysis indicated that, across all sample sites, Enterobacteriaceae, Lactobacillaceae, Veillonellaceae and Succinivibrionaceae were most significantly expanded while Ruminococcaceae, Streptococcaceae, Lachnospiraceae and Treponema were most significantly depleted. Helicobacteraceae was identified as being significantly depleted after the antibiotic treatment only when the colon biopsy samples were analyzed independently by LEfSe analysis.
Figure 3.
Facultative anaerobic bacteria were enriched after antibiotic treatment. LEfSe analysis of the bacterial communities of the colon biopsies (A), stool (B), rectal swabs (C) and across all sites (D) before and after the antibiotic treatment demonstrated that facultative anaerobic bacterial taxa, including Enterobacteriaceae and Lactobacillaceae, were significantly expanded after the antibiotic treatment, while many fermentative obligate anaerobes such as Ruminococcaceae were depleted.
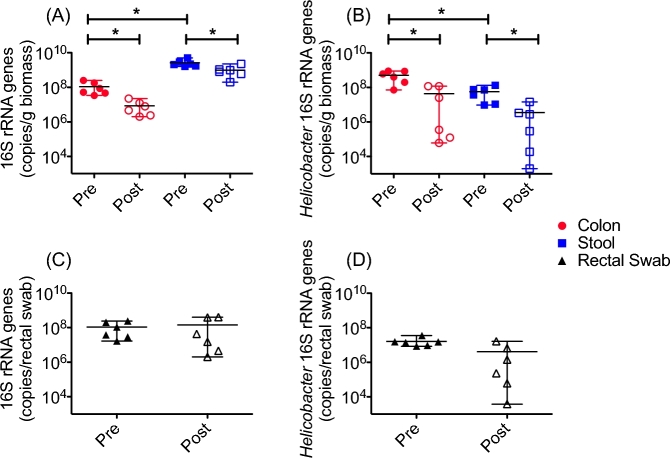
Finally, we estimated the total bacterial load and Helicobacter load at each sample site before and after the antibiotic treatment using qPCR targeting either all bacterial 16S rRNA genes or only Helicobacter 16S rRNA genes (Fig. 4). We found that the stool bacterial communities had significantly higher total copy numbers than did those from the colon biopsies and that the bacterial load at these sites was significantly decreased after the antibiotic treatment (Fig. 4A). In accordance with the high-throughput sequencing data, the colon mucosa showed significantly higher copies of Helicobacter 16S rRNA genes than did the stool, and these were also significantly decreased after the antibiotic treatment (Fig. 4B). Because there was no way to measure the amount of biomass collected on each rectal swab, we were unable to compare these communities with those from the colon biopsies or stool in terms of total bacterial load. Interestingly, the total 16S rRNA gene copies detected from the rectal swabs were similar to that of the colon mucosa before the antibiotic treatment. However, after the antibiotic treatment, the rectal swab samples did not show a significant reduction in the total bacterial load, although some rectal swab samples did show reduced bacterial loads after the antibiotic treatment (Fig. 4C). We also observed similar trends for the Helicobacter 16S rRNA genes (Fig. 4D).
Figure 4.
Antibiotics decreased total bacterial and Helicobacter load in colon mucsoa and stool. qPCR of total 16S rRNA genes revealed that the stool (blue squares) had a significantly higher bacterial load than did the colon mucosa (red circles), and that the total bacterial load before the antibiotic treatment (closed symbols) was significantly greater than the bacterial load after the antibiotic treatment (open symbols) in both compartments (A). In contrast, the colon mucosa showed a significantly higher load of Helicobacter 16S rRNA genes than did the stool, and the antibiotic treatment reduced the Helicobacter load at both sites (B). The rectal swab samples (black triangles) showed reduction of both total bacterial (C) and Helicobacter(D) 16S rRNA genes after the antibiotic treatment, but this did not reach statistical significance. A Wilcoxon signed-rank test was used to determine statistical significance * = P < 0.05.
DISCUSSION
Here, we reported on the effects of a combination antibiotic treatment on mucosal and luminal bacterial communities in six ART-treated, SIV-infected male rhesus macaques. As has been previously reported in healthy and SIV-infected macaques, the colonic mucosa of these macaques was heavily dominated by Helicobacteraceae (Yasuda et al.2015), which we determined to be H. fennalliae and H. macacae. Several Helicobacter spp. have been isolated from the colon mucosa of rhesus macaques, including H. cinaedi (Fox et al.2001) and H. macacae (Lertpiriyapong et al.2014). While Helicobacter spp. are often associated with GI inflammation, the animals in this study did not display overt colonic inflammation, suggesting that the Helicobacteraceae sp. in these animals was not pathogenic. By comparison, the stool bacterial communities were more diverse and composed mostly of bacteria from the phyla Firmicutes and Bacteroides. This distinct compartmentalization of the mucosa and stool is in contrast to humans, which typically display similar bacterial taxa in these compartments (Stearns et al.2011; Lozupone et al.2013; Dillon et al.2014). Helicobacter spp. are microaerophillic (Fox 2001), while the majority of the stool-associated bacteria, such as Prevotellaceae and Ruminococcaceae, are typically obligate anaerobes. This indicates that the mucosa possibly has a higher oxygen content than the lumen, which may be derived from oxygen flowing to host cells at the mucosal surface, as has been previously suggested (Yasuda et al.2015). Indeed, oxygen gradients in the colonic mucosa have previously been linked to differential bacterial community composition when comparing mucosal and luminal communities (Albenberg et al.2014; De Weirdt and Van de Wiele 2015). Such environmental factors combined with the ability of Helicobacter spp. to efficiently colonize intestinal mucosa (Kao, Sheu and Wu2016) may help explain the high abundance of Helicobacteraceae in the mucosa of these animals. However, the specific factors contributing to this require further investigation.
Some of the bacterial communities detected on the rectal swabs were more similar to the colon mucosa, while others were more similar to the stool communities, and this was likely influenced by the capture of stool on the rectal swab. Rectal swabs are often used to approximate colon mucosal bacterial communities in lieu of biopsies due to the non-invasive nature of sample collection. While this approach appears to work well in humans (Budding et al.2014), the differences between mucosal and luminal bacterial communities in these macaques indicates that complete clearing of stool from the rectal mucosa (e.g. by enema) is necessary to ensure that rectal swabs accurately reflect the colon mucosal bacterial communities. These data also indicate that rectal bacterial communities may truly be unique from those of the colon mucosa and stool. Thus, the rectum may be an important factor to consider in the context of transmission of certain viruses, such as HIV, as mucosal bacterial communities have been shown to affect processes related to mucosal homeostasis, such as wound healing rates (Zevin et al.2016).
The antibiotic treatment disrupted the bacterial community structure of all three compartments, and significantly decreased bacterial load in the colon and stool. Intriguingly, the composition of the colon biopsy communities from RM5 and RM6 showed no response to the antibiotics, while the stool bacterial communities from these animals showed a response similar to the other animals. However, there was a decrease in the total bacterial load in the colonic mucosa of these animals. One potential explanation for this phenomenon is that the Helicobacteraceae sp. residing in the colon of these animals was resistant to the antibiotics used. Vancomycin acts by inhibiting cell wall synthesis and is thus ineffective against most Gram-negative bacteria, including Proteobacteria. Indeed, vancomycin is often used in growth media to select for Helicobacter spp. (Fox et al.2001). Enrofloxacin is a fluoroquinolone antibiotic that acts by inhibiting DNA synthesis, but a variety of mechanisms, such as target site modification target site blockade, or increased efflux can reduce their efficacy (Redgrave et al.2014). This may suggest that the vancomycin may have had no effect on the colonic Helicobacteraceae spp. and that those from RM5/6 were resistant to enrofloxacin. However, this study lacked the temporal resolution to attribute the observed disruption of microbiota to one specific antibiotic. Further investigation of the dynamics of intestinal bacterial communities during combination antibiotic therapies and assessment of antibiotic resistance genes will be valuable to understand these mechanisms.
At all of the sample sites, the antibiotic treatment led to increased abundances of similar bacterial phylotypes, especially Enterobacteriaceae and Lactobacillaceae. Multidrug-resistant Enterobacteriaceae are a major emerging public health threat (Savard and Perl 2012) and populations of these bacteria have been shown to increase following antibiotic treatment in a variety of settings, possibly by occupying metabolic niches that were previously occupied by bacterial groups that were reduced by the antibiotics (Ng et al.2013; Keeney et al.2014; Lankelma et al.2017). Indeed, antibiotics have previously been linked to altered bacterial respiration and redox states, and expansion of enteric bacteria through metabolism of free monosaccharides liberated by the host or microbiota (Ng et al.2013; Dwyer et al.2014; Lobritz et al.2015; Faber et al.2016). The specific changes to microbiome function associated with antibiotic treatment in these animals are the subject of ongoing metagenomic studies.
Colonic microbiota play a key role in maintaining homeostais of the mucosal immune system (Belkaid and Hand 2014), and disruption of the microbiome by antibiotic treatment may also influence immunity. Indeed, current research suggests that antibiotics can induce mucosal inflammation (Knoop et al.2016, 2017). Similarly, HIV infection is associated with disruption of colon mucosal microbiota and immunity independent of antibiotics (Dillon et al.2014; Somsouk et al.2015). Thus, antibiotic treatments may exacerbate these symptoms and further disrupt mucosal homeostasis in HIV-infected individuals, and further investigation of the possible effects of antibiotics on mucosal immunity in HIV infection is warranted to aid in the development of improved therapeutics. Finally, these data show that antibiotic use must be minimized in studies designed to evaluate the effects of colonic and rectal bacteria in the context of SIV infection models.
Acknowledgements
We would like to thank all animal technicians and veterinary staff of the Washington National Primate Research Center. We would also like to thank the laboratory of Dr Nina Salama for their gift of genomic DNA from Helicobacter pylori.
FUNDING
This work was supported by the National Institutes of Health [K22AI098440] and by start-up funds to N. Klatt from the University of Washington Department of Pharmaceutics and the Washington National Primate Research Center.
Conflicts of interest. None declared.
REFERENCES
- Albenberg L, Esipova TV, Judge CP et al. . Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 2014;147:1055–63.e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becattini S, Taur Y, Pamer EG. Antibiotic-induced changes in the intestinal microbiota and disease. Trends Mol Med 2016;22:458–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belkaid Y, Hand TW. Role of the microbiota in immunity and inflammation. Cell 2014;157:121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenchley JM, Price DA, Schacker TW. et al.Microbial translocation is a cause of systemic immune activation in chronic HIV infection. Nat Med 2006;12:1365–71. [DOI] [PubMed] [Google Scholar]
- Budding AE, Grasman ME, Eck A et al. . Rectal swabs for analysis of the intestinal microbiota. PLoS One 2014;9:e101344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caporaso JG, Kuczynski J, Stombaugh J et al. . QIIME allows analysis of high-throughput community sequencing data. Nat Methods 2010;7:335–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Weirdt R, Van de Wiele T. Micromanagement in the gut: microenvironmental factors govern colon mucosal biofilm structure and functionality. NPJ Biofilms Microbiomes 2015;1:15026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillon SM, Lee EJ, Kotter CV et al. . An altered intestinal mucosal microbiome in HIV-1 infection is associated with mucosal and systemic immune activation and endotoxemia. Mucosal Immunol 2014;7:983–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dionisi HM, Harms G, Layton AC et al. . Power analysis for real-time PCR quantification of genes in activated sludge and analysis of the variability introduced by DNA extraction. Appl Environ Microb 2003;69:6597–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dwyer DJ, Belenky PA, Yang JH et al. . Antibiotics induce redox-related physiological alterations as part of their lethality. P Natl Acad Sci USA 2014;111:E2100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faber F, Tran L, Byndloss MX et al. . Host-mediated sugar oxidation promotes post-antibiotic pathogen expansion. Nature 2016;534:697–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG. The non-H pylori helicobacters: their expanding role in gastrointestinal and systemic diseases. Gut 2001;50:273–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Handt L, Sheppard BJ et al. . Isolation of Helicobacter cinaedi from the colon, liver, and mesenteric lymph node of a rhesus monkey with chronic colitis and hepatitis. J Clin Microbiol 2001;39:1580–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galkiewicz JP, Kellogg CA. Cross-kingdom amplification using bacteria-specific primers: complications for studies of coral microbial ecology. Appl Environ Microb 2008;74:7828–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Evans DT. Animal models for HIV/AIDS research. Nat Rev Microbiol 2012;10:852–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hensley-McBain T, Zevin AS, Manuzak J et al. . Effects of fecal microbial transplantation on microbiome and immunity in simian immunodeficiency virus-infected macaques. J Virol 2016;90:4981–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlemann DP, Labrenz M, Jurgens K et al. . Transitions in bacterial communities along the 2000 km salinity gradient of the Baltic Sea. ISME J 2011;5:1571–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huijsdens XW, Linskens RK, Koppes J et al. . Detection of Helicobacter species DNA by quantitative PCR in the gastrointestinal tract of healthy individuals and of patients with inflammatory bowel disease. FEMS Immunol Med Mic 2004;41:79–84. [DOI] [PubMed] [Google Scholar]
- Kao CY, Sheu BS, Wu JJ. Helicobacter pylori infection: An overview of bacterial virulence factors and pathogenesis. Biomed J 2016;39:14–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keeney KM, Yurist-Doutsch S, Arrieta MC et al. . Effects of antibiotics on human microbiota and subsequent disease. Annu Rev Microbiol 2014;68:217–35. [DOI] [PubMed] [Google Scholar]
- Klase Z, Ortiz A, Deleage C et al. . Dysbiotic bacteria translocate in progressive SIV infection. Mucosal Immunol 2015;8:1009–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klindworth A, Pruesse E, Schweer T et al. . Evaluation of general 16S ribosomal RNA gene PCR primers for classical and next-generation sequencing-based diversity studies. Nucleic Acids Res 2013;41:e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, Gustafsson JK, McDonald KG et al. . Antibiotics promote the sampling of luminal antigens and bacteria via colonic goblet cell associated antigen passages. Gut Microbes 2017;8:400–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop KA, McDonald KG, Kulkarni DH et al. . Antibiotics promote inflammation through the translocation of native commensal colonic bacteria. Gut 2016;65:1100–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankelma JM, Cranendonk DR, Belzer C et al. . Antibiotic-induced gut microbiota disruption during human endotoxemia: a randomised controlled study. Gut 2017;66:1623–30. [DOI] [PubMed] [Google Scholar]
- Lertpiriyapong K, Handt L, Feng Y et al. . Pathogenic properties of enterohepatic Helicobacter spp. isolated from rhesus macaques with intestinal adenocarcinoma. J Med Microbiol 2014;63:1004–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lobritz MA, Belenky P, Porter CB et al. . Antibiotic efficacy is linked to bacterial cellular respiration. P Natl Acad Sci USA 2015;112:8173–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Li M, Campbell TB et al. . Alterations in the gut microbiota associated with HIV-1 infection. Cell Host Microbe 2013;14:329–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masella AP, Bartram AK, Truszkowski JM et al. . PANDAseq: paired-end assembler for illumina sequences. BMC Bioinformatics 2012;13:31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng KM, Ferreyra JA, Higginbottom SK et al. . Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens. Nature 2013;502:96–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruesse E, Quast C, Knittel K et al. . SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 2007;35:7188–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redgrave LS, Sutton SB, Webber MA et al. . Fluoroquinolone resistance: mechanisms, impact on bacteria, and role in evolutionary success. Trends Microbiol 2014;22:438–45. [DOI] [PubMed] [Google Scholar]
- Ritalahti KM, Amos BK, Sung Y et al. . Quantitative PCR targeting 16S rRNA and reductive dehalogenase genes simultaneously monitors multiple Dehalococcoides strains. Appl Environ Microb 2006;72:2765–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savard P, Perl TM. A call for action: managing the emergence of multidrug-resistant Enterobacteriaceae in the acute care settings. Curr Opin Infect Dis 2012;25:371–7. [DOI] [PubMed] [Google Scholar]
- Segata N, Izard J, Waldron L et al. . Metagenomic biomarker discovery and explanation. Genome Biol 2011;12:R60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Somsouk M, Estes JD, Deleage C et al. . Gut epithelial barrier and systemic inflammation during chronic HIV infection. AIDS 2015;29:43–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stearns JC, Lynch MD, Senadheera DB et al. . Bacterial biogeography of the human digestive tract. Sci Rep 2011;1:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasuda K, Oh K, Ren B et al. . Biogeography of the intestinal mucosal and lumenal microbiome in the rhesus macaque. Cell Host Microbe 2015;17:385–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zevin AS, Xie IY, Birse K et al. . Microbiome composition and function drives wound-healing impairment in the female genital tract. PLoS Pathog 2016;12:e1005889. [DOI] [PMC free article] [PubMed] [Google Scholar]