Abstract
Enterohemorrhagic Escherichia coli (EHEC) is a food-borne pathogen that assembles a type III secretion system (T3SS) on its surface. The last portion of the T3SS, called the ‘translocon’, is composed of a filament and a pore complex that is inserted into the membrane of intestinal epithelial cells. The genes encoding the translocon (espADB) are part of the LEE4 operon. Their expression is regulated by a complex post-transcriptional mechanism that involves the processing of LEE4 mRNA by the essential endoribonuclease RNase E. Here, we report the construction of an EHEC strain (TEA028-rne) in which RNase E can be induced by adding IPTG to the culture medium. EHEC cells deficient in RNase E displayed an abnormal morphology and slower growth, in agreement with published observations in E. coli K-12. Under those conditions, EspA and EspB were produced at higher concentrations, and protein secretion still occurred. These results indicate that RNase E negatively regulates translocon protein synthesis and demonstrate the utility of E. coli strain TEA028-rne as a tool for investigating the influence of this ribonuclease on EHEC gene expression in vitro.
Keywords: enterohemorrhagic Escherichia coli; ribonuclease E; type III secretion system; posttranscriptional regulation; LEE4 operon, EHEC
Type III secretion system components are negatively regulated by ribonuclease E in enterohemorrhagic Escherichia coli.
INTRODUCTION
The pathogen enterohemorrhagic Escherichia coli (EHEC) O157:H7 employs a type III secretion system (T3SS) to attach to intestinal cells and to deliver effector proteins that disrupt cell function (Jarvis and Kaper 1996; Stevens and Frankel 2014). The terminal portion of the T3SS, called the ‘translocon’, encompasses a long filament that is composed of EspA subunits and ends in a complex of the EspB and EspD proteins (Knutton et al.1998; Luo and Donnenberg 2011). This complex is thought to form a pore in the membrane of eukaryotic cells to allow the passage of effector proteins into the cytoplasm (Ide et al.2001; Shaw et al.2001).
The espADB genes are part of the LEE4 operon, which also contains genes encoding the regulator of secretion SepL (Deng et al.2005, 2015), chaperones CesD2 and L0017 (Neves et al.2003; Su et al.2008), the T3SS needle component EscF (Wilson et al.2001) and the effector protein EspF (Nougayrède and Donnenberg 2004). The LEE4 operon, whose regulation is complex and has only partially been elucidated, is required for bacterial attachment to the recto-anal junction in the bovine reservoir (Naylor et al.2005) and initial adhesion to cultured cells through the EspA filaments (Sharma et al.2016). The LEE4 transcript undergoes post-transcriptional processing by the endoribonuclease RNase E near the 3΄ end of sepL, the first gene of LEE4, to generate an espADB mRNA product that has a monophosphorylated 5΄ end and an AU-rich leader region (Lodato and Kaper 2009). These features would ordinarily render espADB mRNA highly susceptible to further degradation by RNase E, but the binding of ribosomes to a mini-open reading frame located in the leader region increases transcript stability (Lodato et al.2012). Processing of the primary transcript appears to be important for determining the stoichiometry of the EspADB proteins and the regulator SepL. Recently, it was proposed that RNase E is a positive post-transcriptional regulator of LEE4 (Connolly, Finlay and Roe 2015). However, the role of RNase E as a regulator of LEE4 is rather unclear because of its overlapping contributions to the processing and degradation of LEE4 mRNA and its possible influence on the stability of protein-coding mRNAs or small RNAs (sRNAs) that may regulate LEE4 gene expression.
In this study, we examined the effect of RNase E on the production of the translocon proteins EspA and EspB. To do so, we constructed an EHEC strain in which RNase E levels can be manipulated by varying the concentration of an inducer in the culture medium.
MATERIALS AND METHODS
Strains and growth conditions
The strains used in this work are described in Table I. Bacteria were grown in Luria-Bertani (LB) medium or Dulbecco's Modified Eagle's Medium (DMEM) low glucose (HyClone, GE Healthcare), which were supplemented with antibiotics and IPTG when required. Antibiotics were added at the following concentrations: ampicillin (100 μg mL−1), tetracycline (6 μg mL−1), chloramphenicol (25 μg mL−1). When grown in DMEM, the strains were inoculated from overnight cultures in LB at a ratio of 1/500 and then incubated statically for 15 h at 37°C. CFU quantification was done on LB agar supplemented with antibiotics and 100 μM IPTG. To generate P1 lysates, R agar and R-top agar were prepared according to Miller (1992). The R agar composition was 1% tryptone, 0.1% yeast extract, 1.2% agar, 0.8% NaCl; CaCl2 and glucose were added after autoclaving (final concentrations of 2 mM and 0.1%, respectively). R-top agar had the same composition as R agar except that it contained 0.8% agar.
Table I.
Strains used in this study.
| Strain | Description | Source or reference |
|---|---|---|
| TEA028 | Derivative of EHEC serotype O157:H7 strain EDL933, ΔgalETKM::tetA | Ho and Waldor (2007) |
| TEA028-rne | Derivative of TEA028 into which the IPTG-inducible, HA- and His6-tagged rne gene from E. coli RNE102 has been transduced. This strain also produces the LacIQ repressor from a plasmid | This study |
| EDL933 ΔgalU | Derivative of EDL933 in which a kanamycin resistance marker has been inserted into the galU gene | This study |
| EDL933ΔLEE | Derivative of EDL933 in which the LEE pathogenicity island encoding the T3SS has been deleted | Pósfai et al. (1997) |
| RNE102 | Derivative of E. coli Keio BW25113. The rne gene is HA- and His6-tagged and transcribed from a Plac promoter. Thus, IPTG is required for rne transcription. An ampicillin resistance marker is located upstream Plac | This study |
| RNE100 | Derivative of E. coli Keio BW25113. The rne gene is HA- and His6-tagged and transcribed from the rne promoter | This study |
| MC4100 | Non-pathogenic E. coli K-12 strain used as a positive control for P1 transduction experiments | Casadaban (1976) |
Determination of generation times
LB medium was inoculated with overnight cultures of the strains TEA028 and TEA028-rne at a ratio of 1/500 and dispensed into a multiwell plate. In the case of strain TEA028-rne, the LB medium was supplemented with appropriate antibiotics and with IPTG at various concentrations (500, 100, 1 and 0.1 μM). The multiwell plates were incubated at 37°C with agitation in a BioTek Epoch 2 plate reader, and cell growth was followed by measuring optical density at 600 nm at 15 min intervals. Generation times were calculated as reported by Hall et al. (2014), using the software GrowthRates 2.1. Only growth curves with correlation coefficients >0.995 were considered in the growth rate analysis.
Construction of plasmid pLacIQ and EDL933 galU::kan strain
The lacI gene was amplified from EHEC genomic DNA by PCR (AccuPrime Taq DNA Polymerase, Thermo Fisher Scientific), using the forward primer f-prlacIq (5΄-CCCAAGCTTGTGCAAAA CCTTTCGCGGTATG-3΄) and the reverse primer r-prlacIq (5΄- CGCGGATCCTCACATTAATTGCGTTGCGCTC-3΄). In the forward and reverse primers, underlined sequences correspond to HindIII and BamHI restriction sites, respectively. The bold ‘T’ nucleotide in the forward primer introduced a single nucleotide change in the lacI promoter to enhance the expression of the lacI gene. The amplified DNA fragment was cloned between the HindIII and BamHI restriction sites of plasmid pACYC184rrnB (Lodato and Kaper 2009) to generate plasmid pLacIQ, which was then introduced into the TEA028-rne strain. The cloned lacIQ gene sequence was confirmed by Sanger sequencing. The EDL933 galU::kan strain was constructed by the gene doctoring method described by Lee et al (2009). The kanamycin resistance gene (kan) was amplified with primers f-GalU and r-GalU, using plasmid pDOC-K as the template. The sequences of primers were f-GalU (5΄-CCGGAATTCGCCGTTATCCCCGTTGCGGGATTAGGAACCAGGATGTTGCCGGCGGACCGGTCAATTGGCTGGAG-3΄) and r-GalU (5΄-CCGCTCGAGCCAGGCTTTAAATTCCGTGCCAAGGGTGTTATGACGAATACCGTAAATATCCTCCTTAGTTCC-3΄). In the f-GalU and r-GalU primers, the underlined sequences are EcoRI and XhoI restriction sites, respectively, while the sequences in italics are homologous to the galU gene. The PCR fragment was cloned between the EcoRI and XhoI sites of plasmid pDOC-C, and the resulting plasmid was then introduced into the EDL933 strain. Next, the strain was transformed with plasmid pACBSCE, which carries the genes encoding the λ-Red proteins to facilitate recombination and the gene for the restriction enzyme I-SceI. The recombinants were able to grow in the presence of kanamycin but not in minimal medium with galactose as the carbon source. PCR, restriction enzyme digests and DNA ligation were done according to the manufacturers’ recommendations.
Construction of Escherichia coli strain RNE102
The Plac-rneΔ337 allele of CJ1832 (Jain, Deana and Belasco 2002) was transferred to E. coli strain BW25113 by P1 transduction in the presence of 10 μM IPTG and selection for ampicillin resistance. The rne gene of the transductant was then modified further by allelic exchange (Edwards, Keller and Schifferli 1998) so as to add carboxy-terminal HA and hexahistidine tags (GYPYDVPDYAGHHHHHH) to RNase E. The resulting recombinant strain, RNE102, encodes epitope-tagged RNase E under the control of Plac. Its genotype was verified by chromosomal DNA sequencing and immunoblotting.
P1 transduction
The protocol of Ho and Waldor (2007) was followed with minor modifications. Cells from overnight cultures of recipient strains (E. coli K-12 strain MC 4100, EHEC TEA028, EHEC galU::kan) in LB broth (0.5 mL) were pelleted and resuspended in 200 μl of MC buffer (5 mM MgSO4, 50 mM CaCl2). The cells were infected with 50–100 μl of a P1 lysate (∼3 × 109 PFU mL−1) and incubated for 15 min at 37°C. LB medium containing 10 mM sodium citrate (2 mL) was added, and the cells were incubated for 1 h at room temperature, pelleted, resuspended in 100 μl of 1M sodium citrate and plated on LB agar supplemented with ampicillin (100 μg mL−1) and IPTG (300 μM).
P1 lysate production
The procedure described by Miller (1992) was followed. Briefly, a 1:100 dilution of an overnight culture of E. coli strain RNE102 was prepared in LB medium supplemented with 100 μM IPTG and 5 mM CaCl2. The bacterial subculture was incubated at 37°C without agitation for ∼6 h and then for 1 h at 37°C with agitation. An aliquot (1 mL) of the subculture was collected and infected with 100 μl of a P1 stock. After incubation for 20 min at 37°C, 2.5 mL of R-top agar was added and the infected cells were plated on R agar plates and incubated overnight at 37°C. The soft agar layer was scraped and washed with 1 mL of LB medium. The recovered liquid was treated with chloroform (100 μl), vortexed and spun in a centrifuge to eliminate cellular debris. The supernatant containing the P1 lysate was stored at 4°C.
Preparation of protein extracts and western blotting
Cells from bacterial cultures were collected by centrifugation (15 000 × g for 5 min), resuspended in water and boiled for 5 min. The cell extracts were mixed with 2.5% SDS and 4 M urea (final concentrations), and then total protein was quantified by the DC Protein Assay kit (Bio-RAD). The cell extracts were mixed with urea to disrupt RNase E aggregates that otherwise are not well resolved by PAGE, and aliquots containing the same amount of total protein were mixed with Laemmli Buffer (Bio-RAD) containing β-mercaptoethanol (5%) and subjected to electrophoresis on an SDS-10% polyacryalamide gel. Proteins were transferred to a PVDF membrane (Immobilon P-Millipore) by a semidry method. The membrane was blocked with chemiluminescence blocker (Bløk, Millipore) or Tropix I-Block for 30 min and then incubated overnight at 4°C with primary antibodies. The blot was washed with PBS containing 0.05% Tween 20 (3 × 5 min) and then incubated for 1–3 h with the secondary antibody. After washing again, the membrane was incubated for 5 min with a chemiluminescent substrate (Clarity Western ECL substrate, Bio-RAD), and then scanned in a C-DiGit Blot Scanner (LiCor). Alternatively, the membrane was incubated with Pierce 1-Step Ultra TMB Blotting Solution (Thermo Scientific). The antibodies used for RNase E detection were anti-6X His tag (GeneTex) and anti-mouse IgG (HRP) (GeneTex), whereas those for GroEL detection were anti-GroEL mouse monoclonal (GeneTex) and anti-mouse IgG (HRP) (GeneTex) antibodies. The EspA and EspB proteins were detected with polyclonal antibodies (Lodato and Kaper 2009) and EasyBlot anti-rabbit IgG (HRP) (GeneTex).
Protein precipitation from supernatants
Supernatants from bacterial cultures were collected after centrifugation (14 000 × g for 10 min at 4°C). The proteins they contained were precipitated from equivalent volumes (2.6 mL) of the culture supernatants by addition of deoxycholic acid (2.2% final concentration) and trichloroacetic acid (TCA, 10% final concentration), incubation on ice for 30 min and subsequent centrifugation for 10 min at 15 000 rpm and 4°C. Bovine serum albumin (BSA) was added to each sample prior to TCA addition as a control for the efficiency of precipitation. Pellets were washed with chilled acetone at –20°C for 1 h on ice with occasional vortexing, and the samples were then centrifuged (15 000 × g for 10 min at 4°C). The resulting pellets were air-dried, mixed with sample buffer and analyzed by PAGE. Proteins were stained with lumitein (Biotium) following the manufacturer's recommendations.
Statistical analysis
The experiments were each repeated at least twice. Graphs and statistical analysis were performed with GraphPad Prism 7.01 software. Colony forming units and generation times were analyzed by one-way ANOVA corrected for Dunnett's multiple comparisons test. Cell lengths were analyzed by non-parametric ANOVA (Kruskal-Wallis test) corrected for Dunnett's multiple comparisons test.
RESULTS AND DISCUSSION
RNase E is an essential enzyme in Escherichia coli. Therefore, to investigate its influence on translocon protein synthesis in EHEC, we used P1 transduction to place the RNase E gene (rne) under the control of an IPTG-inducible Plac promoter. Although P1 transduction is a common genetic manipulation in E. coli K-12 strains, this method is hampered in EHEC, presumably because the O antigen shields the P1 receptor. Ho and Waldor (2007) reported that chromosomal markers can be moved effectively among EHEC strains that carry deletions of galactose utilization genes (ΔgalETKM or ΔgalU). Thus, we decided to use their method to generate an EHEC strain in which RNase E synthesis is inducible. To this end, we produced P1 lysates of the E. coli K-12 strain RNE102 (Table I and Materials and Methods section), which carries an rne gene under the control of a Plac promoter and a nearby ampicillin resistance marker. Hence, transcription of the rne gene in this strain can be controlled by adjusting the concentration of IPTG in the culture medium, and the proximity of the antibiotic resistance marker facilitates selection after P1 transduction. Two EHEC gal– strains, TEA028 (ΔgalETKM) and EDL933 ΔgalU (Table I), were used as recipients for transduction from RNE102. We were unable to transduce the ampicillin resistance marker to the EHEC ΔgalU strain, and only one ampicillin-resistant colony was obtained by transduction into the TEA028 strain. By contrast, using the same P1 lysates to transduce E. coli K-12 strain MC4100 resulted in more than a hundred ampicillin-resistant colonies. Therefore, P1 transduction of EHEC gal mutants was possible in our hands but was less efficient than previously reported (Ho and Waldor 2007).
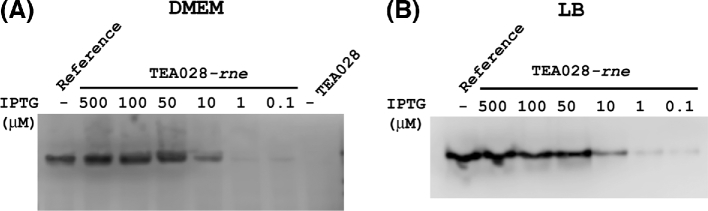
To achieve tighter control of RNase E synthesis in the P1 transductant of TEA028, we introduced a plasmid that overproduces the lac repressor (pLacIQ). Hereafter, we will refer to the resulting plasmid-bearing strain as TEA028-rne. As expected, the concentration of RNase E in TEA028-rne varied depending on the concentration of IPTG in the culture medium. RNase E was scarce at IPTG concentrations ≤1 μM but present at levels that resembled the reference strain at 50–500 μM IPTG (Fig. 1).
Figure 1.
RNase E synthesis in the RNase E-inducible strain TEA028-rne. In the EHEC strain TEA028-rne, transcription of the rne gene is under the control of an IPTG-inducible promoter. Bacteria were grown in (A) DMEM or (B) LB medium supplemented with various concentrations of IPTG, as indicated. The reference is an E. coli K-12 strain in which RNase E is expressed under the control of its own regulatory region (E. coli K-12 strain RNE100). In both TEA028-rne and the reference strain, RNase E bears a C-terminal hexahistidine (6xHis) tag. Cellular proteins were subjected to SDS-PAGE and western blot analysis with antibodies against the 6xHis tag. The parental strain (TEA028), in which RNase E lacks a 6xHis tag and thus is undetectable with these antibodies, served as a negative control. The immunoblot in panel A was also stained with Ponceau S (not shown) to show that equal amounts of total protein had been loaded in each lane.
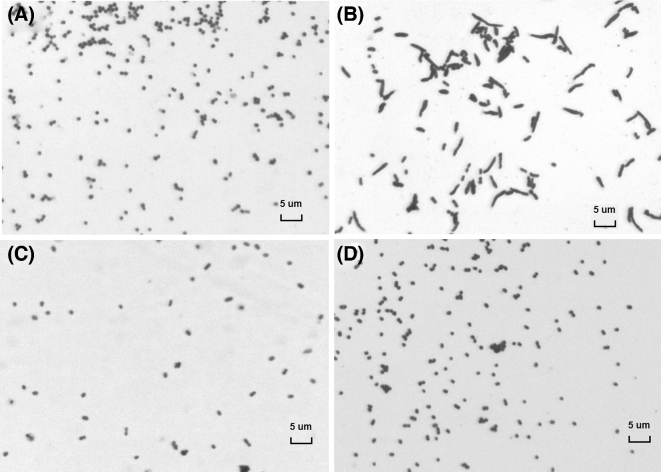
We then examined the effect of RNase E levels on cell shape. Cells deficient in RNase E displayed an altered morphology characterized by filamentation (Fig. 2 and Figs S1 and S2, Supporting Information). A similar growth defect has been described for E. coli K-12 strains at low RNase E concentrations (Goldblum and Apririon 1981; Cam et al.1996; Jain, Deana and Belasco 2002). The formation of filaments has been attributed to the altered decay of ftsA and ftsZ mRNA, which encode two proteins essential for cell division. The resulting reduction in the ratio of FtsZ/FtsA proteins affects septum formation (Tamura et al.2006).
Figure 2.
RNase E depletion causes abnormal morphology of EHEC cells. (A) TEA028; (B, C, D) TEA028-rne. Bacteria were grown overnight in LB medium either in the absence of IPTG (A) or in the presence of 0.1 μM (B), 100 μM (C) or 500 μM (D) IPTG. Smears were stained with crystal violet. At the lowest IPTG concentration (0.1 μM), TEA028-rne cells grew abnormally as filaments. Magnification = ×1000. Bar: 5 μM.
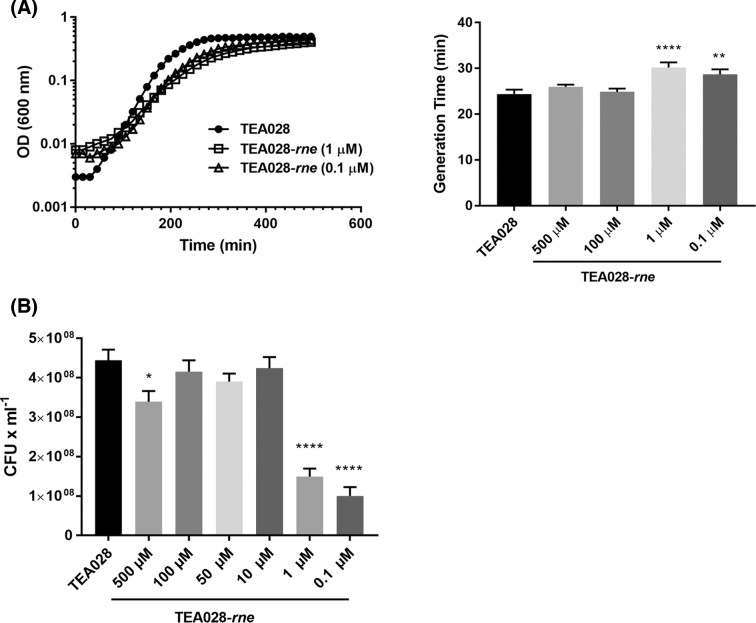
Depletion of RNase E was observed not only to alter cell morphology but also to slow cell growth in rich media (Fig. 3A). In addition, CFU yields were reduced at low concentrations of IPTG in the defined medium DMEM (Fig. 3B), which is normally used to grow EHEC because it stimulates the expression of virulence genes (Tatsuno et al.2000). The growth of TEA028-rne was significantly retarded at IPTG concentrations (0.1 or 1 μM) that cause RNase E to be produced at levels well below its concentration in the reference strain. Diminished cell growth has also been reported for E. coli K-12 cells in which RNase E has been depleted (Jain, Deana and Belasco 2002).
Figure 3.
IPTG-dependent growth of the RNase E-inducible strain TEA028-rne in various culture media. (A) Bacteria were grown in LB media supplemented with IPTG as indicated. Representative growth curves of TEA028 (control) and TEA028-rne (grown in the presence of 1 or 0.1 μM IPTG) are shown on the left. Growth curves for TEA028-rne at 100 or 500 μM IPTG, which resemble that for TEA028, are omitted for clarity. (B) Bacteria were grown in DMEM medium supplemented with IPTG as indicated. Colony forming units (CFU) were measured after overnight growth under static conditions. Means and standard errors are indicated in each graph. In panel A: **adjusted p-value versus TEA028 = 0.0098; ****adjusted p-value versus TEA028 = 0.0001. In panel B: *adjusted p-value versus TEA028 = 0.018; ****adjusted p-value versus TEA028 = 0.0001.
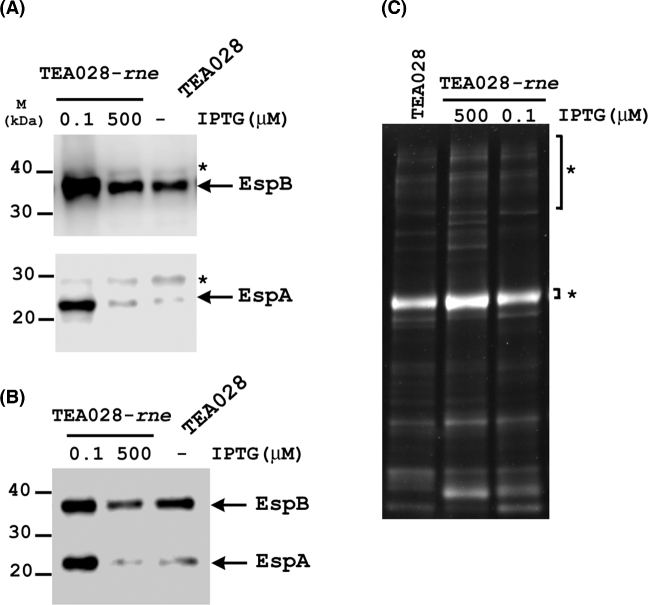
Next, we investigated the effect of RNase E levels on the production of two translocon proteins (EspA and EspB) and on protein secretion. A lower than normal concentration of RNase E (0.1 μM IPTG) increased EspB and especially EspA levels in both cell pellets and culture supernatants (Fig. 4A and B). Other proteins could also be detected in the culture supernatants at low RNase E levels (Fig. 4C), indicating that secretion still occurs. By contrast, the cytoplasmic protein GroEL was undetectable in the culture supernatants (Fig. S3, Supporting Information), indicating that the presence of translocon proteins in the supernatants was mainly due to secretion and not to cell lysis. As expected, protein levels were significantly reduced in supernatants of an EHEC strain lacking type III secretion (Fig. S4, Supporting Information). The increase of EspA and EspB levels under conditions of RNase E deficiency implies either that the degradation of espADB mRNA is RNase E-dependent or that processing of the primary LEE4 transcript in the sepL coding region negatively affects the production of translocon proteins. Contradicting the latter proposition is the previous observation that production of EspA was not noticeably affected by a deletion of the RNase E processing site in sepL (Lodato and Kaper 2009). However, that earlier deletion was made in a plasmid-borne fragment of the LEE4 operon in which only the sepL, espA and espD genes were present. Ascertaining whether or not LEE4 mRNA processing affects the production of translocon proteins in the context of the entire operon will require the more difficult approach of deleting the RNase E processing site in the chromosomal sepL gene.
Figure 4.
IPTG-dependent production and secretion of EspA and EspB by the RNase E-inducible strain TEA028-rne. TEA028-rne and its parental strain (TEA028) were grown statically overnight in DMEM supplemented with IPTG at the indicated concentrations. Cellular proteins from (A) bacterial pellets or (B) supernatants were subjected to SDS-PAGE and western blotting with antisera raised against EspA or EspB. Arrows indicate the EspA and EspB proteins, while asterisks indicate non-specific proteins that cross-reacted with the antisera. In panel C, secreted proteins in equivalent volumes of culture supernatants were precipitated and examined by SDS-PAGE and staining with lumitein. Asterisks indicate BSA, which was added to each supernatant as an internal control for the efficiency of protein precipitation.
Besides affecting mRNA lifetimes directly, RNase E can influence gene expression indirectly by degrading sRNAs that act post-transcriptionally to regulate specific mRNAs (Waters and Storz 2009). In recent years, various sRNAs that regulate virulence have been discovered in EHEC (Bhatt et al.2016; Hücker et al.2017), and some of them have been found to exert either a positive (Gruber and Sperandio 2015) or negative (Gruber and Sperandio 2014) effect on LEE4 gene expression. However, in most cases the role of RNase E has not yet been defined. In principle, the balance of direct and indirect effects of RNase E on LEE4 may be important for producing the translocon components in the correct ratio and for their timely and efficient assembly during infection.
It remains to be determined whether cells with low RNase E content are able to attach to eukaryotic cells and form the characteristic pedestals that are the hallmark of EHEC infections (Knutton et al.1989). Of note is that RNase E processes another polycistronic transcript, LEE5, which encodes proteins required for the intimate attachment of EHEC to host cells (Gruber and Sperandio 2014). Therefore, processing of EHEC transcripts by RNase E or other nucleases appears to be of broad significance.
While the construction of RNase E-inducible strains has been described for E. coli K-12 (Jain, Deana and Belasco 2002), to our knowledge this is the first report of such a modification of the rne gene on the chromosome of a Gram-negative pathogen. Few studies have examined the role of RNase E in pathogenesis. In Yersinia pseudotuberculosis, overproducing a truncated form of RNase E that lacked the non-catalytic C-terminal domain was found to impair type III secretion and the infection of macrophage-like cells in vitro (Yang, Jain and Schesser 2008). In addition, a similar RNase E truncation in Salmonella enterica serovar Typhimurium attenuated infection of insect larvae (Viegas et al.2013). The TEA028-rne strain described here is expected to prove useful for examining the role of RNase E in the expression of the many EHEC virulence genes in vitro, such as those required for Shiga-toxin synthesis or adhesion to eukaryotic cells (Torres, Zhou and Kaper 2005; Mellies and Lorenzen 2014).
Supplementary Material
Supplementary data are available at FEMSLE online.
Acknowledgments
We thank Dr Immo Hansen for the use of the light microscope and Mrs. Kristina Gonzales for help with the microscope software.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSLE online.
FUNDING
This work was supported by start-up funds from New Mexico State University and by an Institutional Development Award (IDeA) [P20GM103451] and a research project grant [R01GM035769 (JGB)] from the National Institute of General Medical Sciences of the National Institutes of Health.
Conflict of interest. None declared.
REFERENCES
- Bhatt S, Egan M, Jenkins V et al. . The tip of the iceberg: on the roles of regulatory small RNAs in the virulence of enterohemorrhagic and enteropathogenic Escherichia coli. Front Cell Infect Mi 2016;6:105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cam K, Rome G, Krisch HM et al. . RNase E processing of essential cell division genes mRNA in Escherichia coli. Nucleic Acids Res 1996;24:3065–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban MJ. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol 1976;104:541–55. [DOI] [PubMed] [Google Scholar]
- Connolly JP, Finlay BB, Roe AJ. From ingestion to colonization: the influence of the host environment on regulation of the LEE encoded type III secretion system in enterohaemorrhagic Escherichia coli. Front Microbiol 2015;6:568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Li Y, Hardwidge PR et al. . Regulation of type III secretion hierarchy of translocators and effectors in attaching and effacing bacterial pathogens. Infect Immun 2005;73:2135–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng W, Yu HB, Li Y et al. . SepD/SepL-dependent secretion signals of the type III secretion system translocator proteins in enteropathogenic Escherichia coli. J Bacteriol 2015;197:1263–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards RA, Keller LH, Schifferli DM. Improved allelic exchange vectors and their use to analyze 987P fimbria gene expression. Gene 1998;207:149–57. [DOI] [PubMed] [Google Scholar]
- Goldblum K, Apririon D. Inactivation of the ribonucleic acid-processing enzyme ribonuclease E blocks cell division. J Bacteriol 1981;146:128–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CC, Sperandio V. Posttranscriptional control of microbe-induced rearrangement of host cell actin. MBio 2014;5:e01025–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruber CC, Sperandio V. Global analysis of posttranscriptional regulation by GlmY and GlmZ in enterohemorrhagic Escherichia coli O157:H7. Infect Immun 2015;83:1286–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall BG, Acar H, Nandipati A et al. . Growth rates made easy. Mol Biol Evol 2014;31:232–8. [DOI] [PubMed] [Google Scholar]
- Hücker SM, Simon S, Scherer S et al. . Transcriptional and translational regulation by RNA thermometers, riboswitches and the sRNA DsrA in Escherichia coli O157:H7 Sakai under combined cold and osmotic stress adaptation. FEMS Microbiol Lett 2017;364, DOI: 10.1093/femsle/fnw262. [DOI] [PubMed] [Google Scholar]
- Ho TD, Waldor MK. Enterohemorrhagic Escherichia coli O157:H7 gal mutants are sensitive to bacteriophage P1 and defective in intestinal colonization. Infect Immun 2007;75:1661–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ide T, Laarmann S, Greune L et al. . Characterization of translocation pores inserted into plasma membranes by type III-secreted Esp proteins of enteropathogenic Escherichia coli. Cell Microbiol 2001;3:669–79. [DOI] [PubMed] [Google Scholar]
- Jain C, Deana A, Belasco JG. Consequences of RNase E scarcity in Escherichia coli. Mol Microbiol 2002;43:1053–64. [DOI] [PubMed] [Google Scholar]
- Jarvis KG, Kaper JB. Secretion of extracellular proteins by enterohemorrhagic Escherichia coli via a putative type III secretion system. Infect Immun 1996;64:4826–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Baldwin T, Williams PH et al. . Actin accumulation at sites of bacterial adhesion to tissue culture cells: basis of a new diagnostic test for enteropathogenic and enterohemorrhagic Escherichia coli. Infect Immun 1989;57:1290–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutton S, Rosenshine I, Pallen MJ et al. . A novel EspA-associated surface organelle of enteropathogenic Escherichia coli involved in protein translocation into epithelial cells. EMBO J 1998;17:2166–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DJ, Bingle LE, Heurlier K et al. . Gene doctoring: a method for recombineering in laboratory and pathogenic Escherichia coli strains. BMC Microbiol 2009;9:252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato PB, Kaper JB. Post-transcriptional processing of the LEE4 operon in enterohaemorrhagic Escherichia coli. Mol Microbiol 2009;71:273–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodato PB, Hsieh PK, Belasco JG et al. . The ribosome binding site of a mini-ORF protects a T3SS mRNA from degradation by RNase E. Mol Microbiol 2012;86:1167–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo W, Donnenberg MS. Interactions and predicted host membrane topology of the enteropathogenic Escherichia coli translocator protein EspB. J Bacteriol 2011;193:2972–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellies JL, Lorenzen E. Enterohemorrhagic Escherichia coli virulence gene regulation. Microbiol Spectr 2014;2:EHEC-0004–2013. [DOI] [PubMed] [Google Scholar]
- Miller JH. A Short Course in Bacterial Genetics: A Laboratory Manual and Handbook for Escherichia coli and Related Bacteria. USA: Cold Spring Harbor Laboratory Press, 1992. [Google Scholar]
- Naylor SW, Roe AJ, Nart P et al. . Escherichia coli O157: H7 forms attaching and effacing lesions at the terminal rectum of cattle and colonization requires the LEE4 operon. Microbiology 2005;151:2773–81. [DOI] [PubMed] [Google Scholar]
- Neves BC, Mundy R, Petrovska L et al. . CesD2 of enteropathogenic Escherichia coli is a second chaperone for the type III secretion translocator protein EspD. Infect Immun 2003;71:2130–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrède JP, Donnenberg MS. Enteropathogenic Escherichia coli EspF is targeted to mitochondria and is required to initiate the mitochondrial death pathway. Cell Microbiol 2004;6:1097–111. [DOI] [PubMed] [Google Scholar]
- Pósfai G, Koob MD, Kirkpatrick HA et al. . Versatile insertion plasmids for targeted genome manipulations in bacteria: isolation, deletion, and rescue of the pathogenicity island LEE of the Escherichia coli O157:H7 genome. J Bacteriol 1997;179:4426–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma VK, Kudva IT, Bearson BL et al. . Contributions of EspA filaments and curli fimbriae in cellular adherence and biofilm formation of enterohemorrhagic Escherichia coli O157:H7. PLoS One 2016;11:e0149745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw RK, Daniell S, Ebel F et al. . Esp A filament-mediated protein translocation into red blood cells. 2001;3:213–22. [DOI] [PubMed] [Google Scholar]
- Stevens MP, Frankel GM. The locus of enterocyte effacement and associated virulence factors of enterohemorrhagic Escherichia coli. Microbiol Spectr 2014;2:EHEC-0007–2013. [DOI] [PubMed] [Google Scholar]
- Su MS, Kao HC, Lin CN et al. . Gene l0017 encodes a second chaperone for EspA of enterohaemorrhagic Escherichia coli O157:H7. Microbiology 2008;154:1094–103. [DOI] [PubMed] [Google Scholar]
- Tamura M, Lee K, Miller CA et al. . RNase E maintenance of proper FtsZ/FtsA ratio required for nonfilamentous growth of Escherichia coli cells but not for colony-forming ability. J Bacteriol 2006;188:5145–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuno I, Kimura H, Okutani A et al. . Isolation and characterization of mini-Tn5Km2 insertion mutants of enterohemorrhagic Escherichia coli O157:H7 deficient in adherence to Caco-2 cells. Infect Immun 2000;68:5943–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres AG, Zhou X, Kaper JB. Adherence of diarrheagenic Escherichia coli strains to epithelial cells. Infect Immun 2005;73:18–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Mil-Homens D, Fialho AM et al. . The virulence of Salmonella enterica serovar Typhimurium in the insect model Galleria mellonella is impaired by mutations in RNase E and RNase III. Appl Environ Microb 2013;79:6124–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters LS, Storz G. Regulatory RNAs in bacteria. Cell 2009;136:615–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson RK, Shaw RK, Daniell S et al. . Role of EscF, a putative needle complex protein in the type III protein translocation system of enteropathogenic Escherichia coli. Cell Microbiol 2001;3:753–62. [DOI] [PubMed] [Google Scholar]
- Yang J, Jain C, Schesser K. RNase E regulates the Yersinia type 3 secretion system. J Bacteriol 2008;190:3774–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSLE online.