Abstract
Chlamydia trachomatis (Ctr) is a bacterial pathogen that causes ocular, urogenital and lymph system infections in humans. It is highly abundant and among its serovars, E, F and D are most prevalent in sexually transmitted disease. However, the number of publicly available genome sequences of the serovars E and F, and thereby our knowledge about the molecular architecture of these serovars, is low. Here we sequenced the genomes of six E and F clinical isolates and one E lab strain, in order to study the genetic variance in these serovars. As observed before, the genomic variation inside the Ctr genomes is very low and the phylogenetic placement in comparison to publicly available genomes is as expected by ompA gene serotyping. However, we observed a large InDel carrying four to five open reading frames in one clinical E sample and in the E lab strain. We have also observed substantial variation on nucleotide and amino acid levels, especially in membrane proteins and secreted proteins. Furthermore, these two groups of proteins are also target for recombination events. One clinical F isolate was genetically heterogeneous and revealed the highest differences on nucleotide level in the pmpE gene.
Keywords: Chlamydia, genome, adhesins, comparative genomics, evolution
Six Chlamydia trachomatis E and F clinical isolates were sequenced and analyzed, which are among the highly abundant and most prevalent serovars in sexually transmitted disease.
BACKGROUND
The human pathogen Chlamydia trachomatis (Ctr) is an obligate intracellular bacterium and the main cause for sexually transmitted diseases worldwide (Bebear and de Barbeyrac 2009) with an increased risk of infertility and ectopic pregnancy when untreated (Paavonen and Eggert-Kruse 1999). It also causes ocular infections up to blindness (Wright, Turner and Taylor 2008). Multiple Ctr strains have been described, which are differentiated based on serotyping of the ompA gene (Yuan, Zhang and Watkins 1989). These are linked to various afflictions, such as the ocular strains A–C, the urogenital strains D–K and the strains L1–L3 causing lymphogranuloma venereum. It has been shown that genetic loci are associated with tissue tropism (Fehlner-Gardiner, Roshick and Carlson 2002; Caldwell, Wood and Crane 2003; Carlson, Hughes and Hogan 2004; Carlson, Porcella and McClarty 2005; Gomes, Nunes and Bruno 2006; Jeffrey, Suchland and Quinn 2010; Andersson, Harris and Seth Smith 2016). The serovars E, F and D are the most abundant among the urogenital strains (Bandea, Kubota and Brown 2001). These strains are less virulent than the L serovars (Almeida, Borges and Ferreira 2012), but they are highly prevalent and therefore a substantial factor in human health (FreundM, Buttlar and Giampaolo 1992; Molano, Meijer and Weiderpass 2005; Frej-Madrzak, Teryks-Wołyniec and Jama-Kmiecik 2015). Asymptomatic infections often remain undetected and are therefore not treated. A prominent example for the link between detection and dispersal is the spread of a novel E serovar in 2006 in Sweden (Ripa and Nilsson 2007; Seth-Smith, Harris and Persson 2009; Unemo, Seth-Smith and Cutcliffe 2010). Although the E and F strains are so abundant, they are not so easy to handle in lab culture like for example the virulent L-strains and consequently our knowledge of their genomic capabilities is still limited. The whole-genome analysis by Harris, Clarke and Seth-Smith (2012) covers only seven E and four F genomes. In order to better understand the pathogenicity of these strains and their natural variability, genome sequences of further representative clinical isolates are highly important. We have sequenced and comparatively analyzed the genomes of seven, mainly clinical, Ctr E and F samples from Germany.
GENOME RECONSTRUCTION
We sequenced 8875 105 paired-end sequence reads (Table 1; Table S1; Supplementary methods) from six Ctr clinical samples and one Ctr lab strain (E DK-20). These were assembled into one closed chromosome and one plasmid for each sample. We compared the closed genomes to one E (E 150), one F (F SW4) and one D (UW-3/CX) reference sequence which are publicly available (Fig. S1; Table S2, Supporting Information). Eight regions have particularly high SNP densities (Fig. 1A; Table S3, Supporting Information). Only six SNPs and one deletion were found in only four of the plasmid sequences (Ctr E 32931, 8873, DK-20 and F 6068) (Table S4, Supporting Information). The deletion and two SNPs have been observed in smaller and larger fractions of the sequenced plasmids (Table S4), so the distribution is not always homogeneous. From the seven samples investigated in this study, six showed evidence for single infection but Ctr F 6068 consisted of a heterogeneous population. Its reconstructed genome represents the major component but for 70 positions we see SNPs with around 21% coverage, meaning that this sample had a subpopulation of this percentage (Table S5, Supporting Information).
Table 1.
Overview on samples and genomes.
| Data set | Strain name/ isolate | Serotype | Country | City | Source | Year of isolation | Clinical manifestation | Nr passages | Length of chromosome in bp | Length of plasmid in bp | CDS | Accession |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CtrE-103 | 103 | E | GER | Female | 1992 | 14 | 1043 019 | 7502 | 970 | CP015294, CP015295 | ||
| CtrE-160 | 160 | E | GER | Female | 1995 | 11 | 1043 007 | 7502 | 968 | CP015296, CP015297 | ||
| CtrE-32 921 | 32921 | E | GER | Stadtroda | Vagina, 20-year-old female | 2003 | 4 | 1048 917 | 7502 | 970 | CP015302, CP015303 | |
| CtrE-547 | 547 | E | GER | Female | 1991 | 12 | 1043 003 | 7502 | 963 | CP015298, CP015299 | ||
| CtrE-8873 | 8873 | E | GER | Jena | Female | 1998 | Urethritis | 9 | 1042 717 | 7493 | 970 | CP015300, CP015301 |
| CtrE-DK-20 | DK-20 | E | GB | London | Institute of Ophthalmology | 1977 | Unknown | 1048 033 | 7502 | 967 | CP015304, CP015305 | |
| Reference strain (Treharne, Darougar and Jones 1977) | ||||||||||||
| CtrF-6068 | 6068 | F | GER | Jena | Urethra, 19-year-old female | 1997 | Urethritis | 10 | 1042 738 | 7493 | 969 | CP015306, CP015307 |
Overview about C. trachomatis sample names, sample metadata, sequencing characteristics and genome summary.
Figure 1.
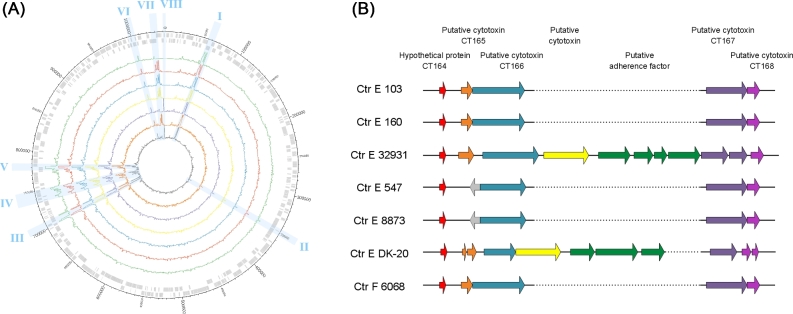
SNP density and the large insert. (A) This plot shows the SNP densities over the genome sequences of all samples. On the two outermost circles, the genes are plotted in gray, first the forward then the reverse ones. Starting from the outside the genomes are E 103 green, E 160 red, E 32931 blue, E 547 yellow, E 8873 purple, E DK-20 orange and F 6068 gray. The light-blue areas indicate regions with a high SNP density affecting the following proteins. I: Three hypothetical proteins CT_049, CT_050 and CT_051; II: The hypothetical protein CT_310 and the V-type ATP synthase subunit E CT_311; III: Hypothetical protein CT_619 and hypothetical protein CT_622; IV: The intergenic region between formyltetrahydrofolate synthetase CT_649, recombinase RecA CT_650 and a hypothetical protein CT_651; V: major outer membrane protein CT_681; VI: membrane protein CT_852; VII: outer membrane protein PmpE and PmpF, CT_869 and CT_870; VIII: outer membrane protein PmpH CT_872 and hypothetical protein CT_873. (B) Scheme of the region surrounding the large insert plus the predicted annotation of the genes and their homologs in C. trachomatis D/UW-3/CX (CT numbers). Whereas CT_164 is present in all seven samples, the ORF representing CT_165 shows changes in E DK-20 and E 32 931, as well as in E 547 and E 8873, where it is replaced by an ORF on the other strand. CT_166 is also affected by the insert in E 32931 and DK-20. The first ORF of the insert is predicted to be a cytotoxin, followed by three or four putative adherence factors. The insert in E DK-20 is around 900 bp shorter and in E DK-20 and E 32931 either CT_167 or CT_168 is disturbed.
CHROMOSOMAL INSERTION/DELETION (InDel)
We compared the reconstructed genomes not only in between each other but also to the publicly available Ctr genomes (Table S2). We identified a large InDel (around 5 kb long) in Ctr E DK-20 and E 32931. In Ctr E 32931, the region is 5913 nucleotides long and consists of five open reading frames (ORFs), whereas the region in Ctr E DK-20 is, with 5041 nucleotides and four ORFs, slightly shorter (Fig. 1B). The InDel is in both cases located between the putative cytotoxins CT_166 and CT_167. We observed variation in the predicted gene structure in close proximity to the InDel. There is a frameshift predicted in E 32931 in CT_167 and also in E DK-20 in CT_165 and CT_168. We found the complete E 32931 InDel with 100% identity only in Ctr E SotonE8. With a high similarity for the E 32931 InDel and 100% identity for the E DK-20 InDel, we find it also in the in vitro-generated strains Ctr RC-L2(s)/3, RC-J(s)/122, RC-F/69, RC-F(s)/342 and RC-F(s)/852 (Jeffrey, Suchland and Eriksen 2013). With only a few mismatches the InDels were also found in Ctr serotypes H and J, which are also infecting the urogenital tract (Carlson, Hughes and Hogan 2004), annotated as cytotoxin genes in UW-36 and UW-4. The 3΄ part of around 1720 nucleotides is also present in A, B and C serotypes (A2497, A/363, A/HAR-13, A/7249, A/5291, B/Jali20/OT, B/TZ1A828/OT and C/TW-3) and it is also described as cytotoxin genes in AP2, Har-13 and TW-3 (Carlson, Hughes and Hogan 2004). Regions homologous to the InDel were also found in Chlamydia suis and C. muridarum with an average amino acid sequence identity of 70%.
PHYLOGENETIC RECONSTRUCTION
The maximum-likelihood phylogenetic tree of the ompA gene shows a clear separation of the E and F serovars and the D reference (Fig. S2, Supporting Information). As expected, the reconstructed samples are closely located to their respective reference genomes. We also used the complete genome sequences of our samples together with 69 public available genomes including the 50 genomes investigated by Harris, Clarke and Seth-Smith (2012) for phylogenetic reconstruction (Fig. S3, Supporting Information). All E and F serovars are in their respective subtrees, which is consistent with the ompA phylogeny and therefore we can exclude recombination in ompA for the seven reconstructed genomes. Interestingly, the only E serovar, in which we found the large InDel (Ctr E SotonE8), is the nearest neighbor to Ctr E 32931. This could be an indication that their common ancestor might also have carried this InDel if it was not acquired by recombination.
RECOMBINATION
Based on the SNPs, we observed significant evidence for recombination within the seven reconstructed genomes and the three reference genomes (E-150, F-SW4 and D/UW-3/CX). The EqualAngle tree reconstruction shows connections in between the serovar E cluster and E 8873 as well as the F serovars (Fig. S4, Supporting Information). Recombination events were found inside the newly sequenced E and F serovars (Fig. S5; Table S6, Supporting Information).
EVOLUTIONARY SELECTION
We investigated the genes with high Ka/Ks ratios in between the seven genomes and the three reference genomes, which are thought to be those who evolve under positive selection. After ranking and statistical testing (Tables S7 and S8, Supporting Information), the top-ranking ratios were CT_868, CT_867 (Misaghi, Balsara and Catic 2006) and CT_694 (Bullock, Hower and Fields 2012), which are all experimentally verified type III secreted, effector proteins. CT_089 (Fields and Hackstadt 2000) and CT_116 (Subtil, Delevoye and Balañá 2005) are predicted to be secreted by the type III secretion system of Ctr. CT_198 with its function for transmembrane transport is involved in transport or possibly presented outside Chlamydia, so the high Ka/Ks ratio is very plausible. The endonuclease CT_157 has also been seen as highly polymorphic in Kari, Whitmire and Carlson (2008), and the authors suspect that it is involved in pathogenicity of Ctr strains. The hypothetical protein CT_105 is likely to be a type III substrate of Ctr (da Cunha, Milho and Almeida 2014). In contrast, the high ratios in the hypothetical proteins CT_168 and CT_244 point toward possible candidates for being secreted effectors, membrane proteins or being involved in transport to the endosome or host cell. The Ka/Ks ratio in the pmp gene family is below 1 for pmpD and pmpI, and also for pmpA and pmpB and around 1 for pmpC (Table S9, Supporting Information).
DISCUSSION
We sequenced and reconstructed the complete genomes of seven Ctr E and F strains, six of them from clinical samples and one from a lab strain (E DK-20). We observed highest diversity at loci coding for hypothetical proteins, as well as ompA and the pmpE and pmpF genes. These loci are in agreement with increased SNP and homoplasy density regions found within other Ctr serovars (Harris, Clarke and Seth-Smith 2012). Besides hypothetical proteins, mainly membrane proteins and secreted proteins show high numbers of SNPs. This strengthens the assumption of higher evolutionary variability of genes involved in interactions with the host. The phylogenetic placement agrees overall with the tree previously presented (Harris, Clarke and Seth-Smith 2012). Compared to the phylogenetic tree of the ompA gene, the whole-genome-based tree indicates that especially the lab strain Ctr E DK-20 has a higher genetic distance to the other E serovars. Ctr E 8873 has diverged most early from the other E serovars. We found evidence for several recombination events, covering genes with diverse functions including several membrane-related ones. Similar to all other E and F strains, the subpopulation in F 6068 differs most in the pmpE gene which indicates that the genomic variation is focused on this particular membrane protein. Co-infections of Ctr serovars (Bandea, Kubota and Brown 2001; Jurstrand, Falk and Fredlund 2001; Lee, Park and Kim 2006; Gharsallah, Frikha-Gargouri and Sellami 2012; Zhang, Zhao and Wang 2012; Rodriguez-Dominguez, Gonzalez-Alba and Puerta 2015; Gallo Vaulet, Entrocassi and Portu 2016) are the prerequisite for recombination. In the F 6068 sample, we could detect a co-infection. However, the genetic difference between the two genotypes was small. A large genomic InDel of about 5 kb in two samples seems to originate from the last common ancestor of Chlamydia suis, C. muridarum and Ctr. It might have been acquired via recombination (Jeffrey, Suchland and Eriksen 2013). The full-length InDel is only present in two samples from this study (E DK-20 and E 32931) and in Ctr E SotonE8, in the H and J serovars and in five in vitro artificially generated strains (Jeffrey, Suchland and Eriksen 2013). Small parts of the InDel have also been previously described (Carlson, Hughes and Hogan 2004; Unemo, Seth-Smith and Cutcliffe 2010). It encodes a large cytotoxin gene in C. muridarum, whereas in the reconstructed E serovars it is split into four or five ORFs, predicted to be cytotoxin genes and one adherence factor. High Ka/Ks ratios indicate that selection favors changes in the amino acid sequences. As expected, the genes with the highest Ka/Ks ratios are mainly type III secreted proteins (known and predicted), hypothetical or involved in the transport to the endosome or host. Statistical evidence strengthens the assumption of a more variable secretome, compared to the majority of proteins. In summary, our study demonstrates a substantial genomic variation in the abundant Ctr E and F strains. These loci and genes may have high impact on the pathogenicity of Ctr, and will be relevant for the development of novel diagnostic tools and vaccines.
SUPPLEMENTARY DATA
Supplementary data are available at FEMSPD online.
Conflict of Interest. None declared.
Supplementary data are available at FEMSPD online.
REFERENCES
- Almeida F, Borges V, Ferreira R et al. Polymorphisms in inc proteins and differential expression of inc genes among Chlamydia trachomatis strains correlate with invasiveness and tropism of lymphogranuloma venereum isolates. J Bacteriol 2012;194:6574–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson P, Harris SR, Seth Smith HM et al. Chlamydia trachomatis from Australian Aboriginal people with trachoma are polyphyletic composed of multiple distinctive lineages. Nat Commun 2016;7:10688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandea CI, Kubota K, Brown TM et al. Typing of Chlamydia trachomatis strains from urine samples by amplification and sequencing the major outer membrane protein gene (omp1). Sex Transm Infect 2001;77:419–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bebear C, de Barbeyrac B. Genital Chlamydia trachomatis infections. Clin Microbiol Infect 2009;15:4–10. [DOI] [PubMed] [Google Scholar]
- Bullock HD, Hower S, Fields KA. Domain analyses reveal that Chlamydia trachomatis CT694 protein belongs to the membrane-localized family of type III effector proteins. J Biol Chem 2012;287:28078–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caldwell HD, Wood H, Crane D et al. Polymorphisms in Chlamydia trachomatis tryptophan synthase genes differentiate between genital and ocular isolates. J Clin Invest 2003;111:1757–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Hughes S, Hogan D et al. Polymorphisms in the Chlamydia trachomatis cytotoxin locus associated with ocular and genital isolates. Infect Immun 2004;72:7063–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlson JH, Porcella SF, McClarty G et al. Comparative genomic analysis of Chlamydia trachomatis oculotropic and genitotropic strains. Infect Immun 2005;73:6407–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- da Cunha M, Milho C, Almeida F et al. Identification of type III secretion substrates of Chlamydia trachomatis using Yersinia enterocolitica as a heterologous system. BMC Microbiol 2014;14:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fehlner-Gardiner C, Roshick C, Carlson JH et al. Molecular basis defining human Chlamydia trachomatis tissue tropism. A possible role for tryptophan synthase. J Biol Chem 2002;277:26893–903. [DOI] [PubMed] [Google Scholar]
- Fields KA, Hackstadt T. Evidence for the secretion of Chlamydia trachomatis CopN by a type III secretion mechanism. Mol Microbiol 2000;38:1048–60. [DOI] [PubMed] [Google Scholar]
- Frej-Madrzak M, Teryks-Wołyniec D, Jama-Kmiecik A et al. Diagnosing Chlamydia trachomatis urinary tract infections—preliminary report. Adv Clin Exp Med 2015;24:441–5. [DOI] [PubMed] [Google Scholar]
- Freund KM, Buttlar CA, Giampaolo C et al. The use of cervical cytology to identify women at risk for chlamydial infection. Am J Prev Med 1992;8:292–7. [PubMed] [Google Scholar]
- Gallo Vaulet L, Entrocassi C, Portu AI et al. High frequency of Chlamydia trachomatis mixed infections detected by microarray assay in South American samples. PLoS One 2016;11:e0153511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gharsallah H, Frikha-Gargouri O, Sellami H et al. Chlamydia trachomatis genovar distribution in clinical urogenital specimens from Tunisian patients: high prevalence of C. trachomatis genovar E and mixed infections. BMC Infect Dis 2012;12:333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes JP, Nunes A, Bruno WJ et al. Polymorphisms in the nine polymorphic membrane proteins of Chlamydia trachomatis across all serovars: evidence for serovar Da recombination and correlation with tissue tropism. J Bacteriol 2006;188:275–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris SR, Clarke IN, Seth-Smith HM et al. Whole-genome analysis of diverse Chlamydia trachomatis strains identifies phylogenetic relationships masked by current clinical typing. Nat Genet 2012;44:413–9, S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Eriksen SG et al. Genomic and phenotypic characterization of in vitro-generated Chlamydia trachomatis recombinants. BMC Microbiol 2013;13:142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffrey BM, Suchland RJ, Quinn KL et al. Genome sequencing of recent clinical Chlamydia trachomatis strains identifies loci associated with tissue tropism and regions of apparent recombination. Infect Immun 2010;78:2544–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurstrand M, Falk L, Fredlund H et al. Characterization of Chlamydia trachomatis omp1 genotypes among sexually transmitted disease patients in Sweden. J Clin Microbiol 2001;39:3915–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kari L, Whitmire WM, Carlson JH et al. Pathogenic diversity among Chlamydia trachomatis ocular strains in nonhuman primates is affected by subtle genomic variations. J Infect Dis 2008;197:449–56. [DOI] [PubMed] [Google Scholar]
- Lee G, Park J, Kim B et al. OmpA genotyping of Chlamydia trachomatis from Korean female sex workers. J Infect 2006;52:451–4. [DOI] [PubMed] [Google Scholar]
- Misaghi S, Balsara ZR, Catic A et al. Chlamydia trachomatis-derived deubiquitinating enzymes in mammalian cells during infection. Mol Microbiol 2006;61:142–50. [DOI] [PubMed] [Google Scholar]
- Molano M, Meijer CJ, Weiderpass E et al. The natural course of Chlamydia trachomatis infection in asymptomatic Colombian women: a 5-year follow-up study. J Infect Dis 2005;191:907–16. [DOI] [PubMed] [Google Scholar]
- Paavonen J, Eggert-Kruse W. Chlamydia trachomatis: impact on human reproduction. Hum Reprod Update 1999;5:433–47. [DOI] [PubMed] [Google Scholar]
- Ripa T, Nilsson PA. A Chlamydia trachomatis strain with a 377-bp deletion in the cryptic plasmid causing false-negative nucleic acid amplification tests. Sex Transm Dis 2007;34:255–6. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Dominguez M, Gonzalez-Alba JM, Puerta T et al. High prevalence of co-infections by invasive and non-invasive Chlamydia trachomatis genotypes during the lymphogranuloma venereum outbreak in Spain. PLoS One 2015;10:e0126145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seth-Smith HM, Harris SR, Persson K et al. Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 2009;10:239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subtil A, Delevoye C, Balañá ME et al. A directed screen for chlamydial proteins secreted by a type III mechanism identifies a translocated protein and numerous other new candidates. Mol Microbiol 2005;56:1636–47. [DOI] [PubMed] [Google Scholar]
- Treharne JD, Darougar S, Jones BR. Modification of the microimmunofluorescence test to provide a routine serodiagnostic test for chlamydial infection. J Clin Pathol 1977;30:510–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unemo M, Seth-Smith HM, Cutcliffe LT et al. The Swedish new variant of Chlamydia trachomatis: genome sequence, morphology, cell tropism and phenotypic characterization. Microbiology 2010;156:1394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright HR, Turner A, Taylor HR. Trachoma. Lancet 2008;371:1945–54. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Zhang YX, Watkins NG et al. Nucleotide and deduced amino acid sequences for the four variable domains of the major outer membrane proteins of the 15 Chlamydia trachomatis serovars. Infect Immun 1989;57:1040–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JJ, Zhao GL, Wang F et al. Molecular epidemiology of genital Chlamydia trachomatis infection in Shenzhen, China. Sex Transm Infect 2012;88:272–7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data are available at FEMSPD online.