ABSTRACT
Background: Autoimmune polyendocrine syndrome type-1 (APS-1) is a rare, childhood onset disease caused by mutations in the Autoimmune Regulator gene. The phenotypic expression is highly variable and includes disease manifestations in the oral cavity, including mucocutaneous candidiasis. Increasing evidence suggests a potential role of the skin, oral and gut microbiotas in the pathogenesis of autoimmunity. To date, no information exists regarding the oral microbiota in APS-1.
Objective: To assess the bacterial microbiota of whole saliva in APS-1 patients by using high throughput sequencing.
Design: Whole unstimulated saliva was collected from 10 APS-1 patients and 17 healthy controls and examined by high throughput sequencing of the hypervariable region V1-V2 of 16S rRNA using the 454 GS Junior system. Metastats (http://cbcb.umd.edu/software/metastats) was used to analyse the pyrosequencing reads.
Results: A reduction in the total number of bacterial genera and species was detected in APS-1 compared to healthy controls. The proportion of the major phyla Firmicutes was higher (60% vs 41%, p = 0.002) and Bacteroidetes lower (15% vs 28%, p = 0.007) in APS-1 compared to healthy controls. On the genus level, Streptococcus and Gemella were prevalent in APS-1.
Conclusion: Our findings indicate a significantly altered oral microbiota in APS-1.
KEYWORDS: APS-1, whole saliva, microbiota, bacteria, high throughput sequencing, pyrosequencing
Autoimmune polyendocrine syndrome type-1 (APS-1) or autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy (OMIM 240300) is a rare, monogenic, childhood onset disorder which is clinically defined by the presence of two of the three major disease components: primary adrenal insufficiency, hypoparathyroidism, and chronic mucocutaneous candidiasis (CMC) [1]. However, the clinical picture is highly variable and includes many minor disease components [2]. The Autoimmune Regulator (AIRE) gene is the disease-causing gene [3,4]. AIRE acts as a transcriptional regulator and is almost exclusively expressed in the thymus [5] where it orchestrates the process of negative selection of self-reactive T cells and contributes to the development of regulatory T cells (Tregs) [6,7]. All patients present autoantibodies against autoantigens expressed in the affected tissue [8] and/or against immune mediators such as interferon-omega (ω) and interleukin (IL)-22 [9,10]. Interestingly, circulating autoantibodies and AIRE-mutations can be found before development of clinical APS-1 [11,12] making the role of environmental triggers particularly relevant in the pathogenesis and phenotypic expression.
Increasing evidence indicates that the environment shapes the human immune system and accounts for its heterogeneity among individuals [13]. The oral, gut, and skin microbiotas could play a key role in the pathogenesis of systemic and organ-specific autoimmune diseases [14]. Most APS-1 patients develop disease components affecting the oral cavity; enamel hypoplasia and CMC are both common manifestations [9,15]. Also a Sjögren’s-like syndrome without extractable nuclear antigen autoantibodies has recently been described [15,16]. These oral manifestations probably interfere with the homeostasis of the oral microbiota. Furthermore, autoimmunity against defensins and other antimicrobial substances as observed in APS-1 could change the microbiota [17]. Reduced salivary flow rate changes the oral microbiota [18] and in a study of patients with severe Sjögren’s syndrome Streptococcus salivarius, Neisseria pharyngis, Veillonella species, and Micrococcus mucilaginosus were reduced and the number of Staphylococcus aureus and Candida species were increased compared to healthy controls [19]. Another recent study of primary Sjögren’s syndrome patients with normal salivary flow rate found that the number of bacterial genera and species was lower in patients, and concluded that saliva dysbiosis is a key characteristic of primary Sjögren’s syndrome [20]. Moreover, changes in the oral microbiota are found to be associated with several other diseases including squamous cell carcinoma, atherosclerosis, bacteraemia, and rheumatoid arthritis [21].
In APS-1, no information exists regarding the oral microbiota and only a few studies of the gut microbiota have been reported [17,22,23]. In this study, we characterized the bacterial profile in whole unstimulated saliva of patients by high throughput sequencing, a technique which recovers both cultivated and not-yet-cultivated bacteria, thus giving an in-depth overview of bacteria present.
Materials and methods
Patients and clinical data
A total of 10 APS-1 patients from 6 different families were included. All patients fulfilled the diagnostic criteria of APS-1. They were previously described in the Norwegian cohort [9,24,25] and included in our National Registry of Autoimmune Diseases. Three patients were excluded after an initial quality control. A detailed characterization of the seven remaining patients (five females and two males) is given in Table 1. The mean age was 32.3 years (range 10–64). All participants had their disease onset before the age of 8 years. Three patients presented the three major disease components and the mean number of disease components was five (range 4–7). Enamel hypoplasia was found in all patients. CMC was previously diagnosed in five patients and three had oral candidiasis at the time of sampling (Table 1). Disease-causing AIRE-mutations and autoantibodies against interferon-ω were present in all. All participants gave informed and written consent and the study was approved by The Regional Committee for Medical and Health Research Ethics for Western Norway.
Table 1.
Characteristics of the APS-1 patients. The age at diagnosis for each disease component is written in parentheses. The age of onset denotes the age at which the first APS-1 main component appeared.
| Pat. no. | Family no. | Sex | Age | Age of onset | Classic triad | Other manifestations | AIRE-mutations | Autoantibodies | Unstimulated saliva flow rate, saliva pH | Fungal load* | Other oral manifesta-tions |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | I | M | 26 | 4 | HP(4), CMC(9), AI(11) | E(11) | c.967_979del13/ c.967_979del13 |
21OH, IL17, IL22, INF-ω, MAGEB2, PDILT, SCC, TGM4, TH, TPH1 |
Pathological (<0.1 ml/min) pH 5.0 |
Positive. Cheilitis angularis. |
Caries. |
| 2 | I | F | 36 | 4 | HP(4), CMC | E(14), AT(20), V(25) | c.967_979del13/ c.967_979del13 |
21OH, 17OH, AADC, GAD65, IL22, INF-ω, MAGEB2, NALP5, PDILT, SCC, SOX10, TH, TPH1 | Normal (>0.1 ml/min) pH nd. |
Negative. | – |
| 3 | II | F | 10 | 7 | PAI(7), HP(10), CMC | E, M | c.967_979del13/ c.967_979del13 |
21OH, 17OH, AADC, GAD65, IL22, INF-ω, MAGEB2, NALP5, SCC, TH, TPH1 | Normal (>0.1 ml/min) pH 7.0 |
Negative. | – |
| 4 | III | M | 64 | 7 | CMC(7), HP(9), PAI(16) | V(17), Al(21), B12(63), E | c.769C>T/ c.769C>T |
21OH, AADC, IL17, IL22, INF-ω, MAGEB2, SCC, SOX10, TGM4 | Normal (>0.1 ml/min) pH 5.5 |
Positive. Cheilitis angularis. |
Gingivitis. Caries. |
| 5 | IV | F | 24 | 3 | HP(3) | AT(24), E, M | c.967_979del13/ c.967_979del13 |
INF-ω, NALP5 | Normal (>0.1 ml/min) pH 7.0 |
Negative. | Gingivitis. |
| 6 | V | F | 25 | 2 | CMC(2), HP(15) | E(24), Al, E | c.1163_1164insA/ c.1249_1950dupC |
21OH, AADC, IL17, IL22, INF-ω, MAGEB2, NALP5, SOX10 | Normal (>0.1 ml/min) pH 6.0 |
Positive. Cheilitis angularis. |
Mucosal lesions. |
| 7 | VI | F | 41 | 5 | HP(5) | G(19), B12(35), M(39), E | c.934G>A (dominant) mutation) | AADC, GAD65, INF-ω, NALP5, PCA | Pathological (<0.1 ml/min) pH nd. |
Negative. | Caries. |
21OH, 21-hydroxylase; 17OH, 17-α-hydroxylase; AADC, aromatic l-amino acid decarboxylase; Al, alopecia; AT, autoimmune thyroiditis; B12, vitamin-B12 deficiency; CMC, candidiasis; E, enamel hypoplasia; G, hypogonadism; GAD65, glutamic acid decarboxylase 65-kDA isoform; HP, hypoparathyroidism; IL17, interleukin-17; IL22, interleukin-22; INF-ω, interferon-omega; M, malabsorption; MAGEB2, melanoma antigen B2; NALP5, NACHT leucine-rich-repeat protein 5; PAI, primary adrenocortical insufficiency; PCA, parietal cell autoantibodies; PDILT, protein disulphide isomerase-like testis expressed; SCC, side-chain-cleavage enzyme; SOX10, sex determining region Y-box 10; TGM4, transglutaminase 4; TPH1, tryptophan hydroxylase 1; TH, tyrosine hydroxylase; V, vitiligo; YoB, year of birth, nd; not done.
*Standard methods were used for culturing Candida albicans.
Saliva sampling and sample processing
Whole unstimulated saliva was collected from patients (n = 10) and healthy controls (n = 17) with the same age and sex distributions as patients. Saliva samples were collected between 9 a.m. and 11 a.m., and participants were asked not to brush their teeth, eat, or drink for at least 2 h before sampling. No participants were regular smokers or had received antibiotics within the last month before sampling. Saliva pH was measured using a strip test (mColorpHast™ pH Test Strips, MilliporeSigma) and standard methods were used for culturing Candida albicans. Unstimulated saliva flow rate (ml/min) was measured based on a collection period of 15 min (Table 1). Samples were immediately stored at −80°C until analysis.
Sample processing was done as previously described [20,26]. In summary, DNA was extracted from a 250 µl sample volume using the MasterPure™ DNA Purification kit (Epicentre, Illumina Company, Madison, WI) and was dissolved in 45 µl 1 × TE buffer. The 16S rRNA hypervariable region V1-V2 was amplified in three parallel PCRs and sequenced on a 454 GS Junior System (Roche, Branford, CT). Primer sequences and amplifications reactions are given previously [20]. DNA quality and concentrations were assessed with Bioanalyzer 2100 (Agilent, Santa Clara, CA) and Nanodrop 3300 Flurospectrometer (Thermo Scientific, Wilmington, DE), all within the range to perform high throughput sequencing.
Bioinformatics analysis of sequence reads
Bioinformatics analysis of sequence reads was executed as described previously [20]. In brief, raw sequence reads were subjected to a species-level, reference-based taxonomy assignment especially designed for studying the human oral microbial community [20]. The set of 16S rRNA reference sequences previously published by Al-Hebshi et al. [27] and the NCBI 16S rRNA reference sequence set (ftp://ftp.ncbi.nlm.nih.gov/blast/db/16SMicrobial.tar.gz) were combined giving reference sequences representing a total of 1,151 oral and 12,013 non-oral species that were BLASTN-searched for each of the sequence reads. Unassigned reads were then screened for high-quality non-chimeras and subjected to de novo species-level operational taxonomy unit calling for potential novel species. The quantitative insights into microbial ecology pipeline software package version 1.9.1 [28] was used for down-stream analyses, including alpha and beta diversities. A statistical method introduced in Metastats (http://cbcb.umd.edu/software/metastats) was used to reveal significant differences between the microbiota of control saliva and APS-1 saliva. This method employs a false discovery rate to improve specificity in high complexity environments, and handles sparsely sampled features using Fisher’s exact test [29]. p-values ≤ 0.05 were considered significant and Bonferonni Correction for multiple testing was included.
Results
Sequence data
An overview of the sequence read counts in each analysis step is given in Supplemental Table 1.
Composition of the salivary microbiota of APS-1 patients
Five different phyla were detected in APS-1 patients with DNA sequences predominately assigned to the phyla Firmicutes (60%), Bacteroidetes (15%), Proteobacteria (10%), Fusobacteria (8%), and Actinobacteria (6%). Table 2 gives an overview of the significantly different abundances of taxa from control saliva and APS-1 saliva by phyla, genera and species, respectively. In Figure 1, a comparison of the bacterial content of saliva in APS-1 and controls based on the sequencing of the hypervariable 16S rDNA region V1-V2 is presented. Saliva from APS-1 displayed a significantly increased relative abundance of Firmicutes (p = 0.002) and a lower frequency of Bacteroidetes (p = 0.007) compared with controls (Figure 2).
Table 2.
Differences in abundances of taxa in APS-1 and control saliva. Significant (p ≤ 0.05) differences in relative abundance of taxa from control saliva and APS-1 saliva as estimated by Metastats http://cbcb.umd.edu/software/metastats. Differences significant after Bonferonni Correction for multiple testing are marked with *.
| Taxon | APS-1 saliva (n = 7) | Control saliva (n = 17) | Metastat p-value |
|---|---|---|---|
| Phyla | |||
| Firmicutes | 0.65118 ± 0.02551 | 0.41118 ± 0.02572 | 0.002 |
| Bacteroidetes | 0.11271 ± 0.03162 | 0.26009 ± 0.03845 | 0.007 |
| Genera | |||
| Streptococcus | 0.53448 ± 0.08523 | 0.18016 ± 0.01963 | 0.0001* |
| Gemella | 0.01467 ± 0.00023 | 0.00253 ± 0.00062 | 0.036 |
| Prevotella | 0.05995 ± 0.01021 | 0.17357 ± 0.02970 | 0.001 |
| Veillonella | 0.04049 ± 0.00925 | 0.16254 ± 0.02760 | 0.0001* |
| Neissera | 0.02065 ± 0.00965 | 0.05508 ± 0.01269 | 0.029 |
| Actinomyces | 0.00148 ± 0.00055 | 0.00673 ± 0.00159 | 0.002 |
| Megashaera | 0.00000 | 0.00631 ± 0.00287 | 0.027 |
| Lachnospiraceae_ [G-3] | 0.00059 ± 0.00033 | 0.00417 ± 0.00097 | 0.001 |
| Lachnoanaerobaculum | 0.00024 ± 0.00019 | 0.00123 ± 0.00033 | 0.008 |
| Eubacterium_[XIVa][G-1] | 0.00025 ± 0.00018 | 0.00109 ± 0.00031 | 0.019 |
| Ruminococcaceae_[G-3] | 0.00002 ± 0.00776 | 0.00119 ± 0.00448 | 0.016 |
| Bacteroides | 0.00000 | 0.00049 ± 0.00019 | 0.012 |
| Veillonella | 0.00000 | 0.00053 ± 0.00025 | 0.031 |
| Bacteroidetes_[G-3] | 0.00000 | 0.00032 ± 0.00010 | 0.002 |
| Peptostreptococcaceae_[XI][G-4] | 0.00000 | 0.00023 ± 0.00011 | 0.034 |
| Mitsuokella | 0.00000 | 0.00033 ± 0.00016 | 0.038 |
| Desulfovibrio | 0.00000 | 0.00405 ± 0.00040 | 0.018 |
| Catonella | 0.00000 | 0.00015 ± 0.00013 | 0.048 |
| Species | |||
| Streptococcus sp._str._C300 | 0.12958 ± 0.03003 | 0.03065 ± 0.00618 | 0.001 |
| Streptococcus multispecies_spp24_2 | 0.11555 ± 0.05022 | 0.00877 ± 0.00349 | 0.031 |
| Fusobacterium nucleatum_ss_animalis | 0.02401 ± 0.01178 | 0.00083 ± 0.00038 | 0.046 |
| Gemella haemolysans | 0.01308 ± 0.00599 | 0.00132 ± 0.00047 | 0.047 |
| Capnocytophaga ochracea | 0.00112 ± 0.00035 | 0.00004 ± 0.00002 | 0.023 |
| Veillonella parvula_group | 0.03273 ± 0.00653 | 0.10745 ± 0.01904 | 0.0003* |
| Prevotella melaninogenica | 0.02205 ± 0.00876 | 0.09752 ± 0.01762 | 0.0002* |
| Veillonella atypica | 0.00776 ± 0.00389 | 0.05509 ± 0.01369 | 0.001 |
| Neisseria flavescens/subflava | 0.01723 ± 0.00873 | 0.04995 ± 0.01204 | 0.023 |
| Porphyromonas gingivalis | 0.00000 | 0.02849 ± 0.01465 | 0.048 |
| Prevotella pallens | 0.00059 ± 0.00036 | 0.02308 ± 0.00617 | 0.0004* |
| Campylobacter concisus | 0.00133 ± 0.00083 | 0.01390 ± 0.00382 | 0.001 |
| Prevotella salivae | 0.00054 ± 0.00024 | 0.00848 ± 0.00293 | 0.006 |
| Prevotella veroralis_nov_95.28% | 0.00000 | 0.00723 ± 0.00287 | 0.010 |
| Solobacterium moorei | 0.00251 ± 0.00121 | 0.00638 ± 0.00154 | 0.045 |
| Megasphare micronuciformis | 0.00000 | 0.00631 ± 0.00287 | 0.023 |
| Atopobium parvulum | 0.00099 ± 0.00026 | 0.00579 ± 0.00223 | 0.028 |
| Lachnospiraceae_[G-3] sp._oral_taxon_100_nov_83.29% | 0.00049 ± 0.00032 | 0.00342 ± 0.00082 | 0.001 |
| Actinomyces sp._Oral_Taxon_180 | 0.00043 ± 0.00022 | 0.00328 ± 0.00103 | 0.006 |
| Actinomyces odontolyticus | 0.00047 ± 0.00015 | 0.00247 ± 0.00062 | 0.001 |
| Prevotella multispecies_sppn1_2_nov_82.34% | 0.00027 ± 0.00019 | 0.00158 ± 0.00579 | 0.026 |
| Lachnoanaerobaculum orale | 0.00024 ± 0.00019 | 0.00123 ± 0.00332 | 0.008 |
| Eubacterium_[XIVa][G-1] saburreum | 0.00025 ± 0.00018 | 0.00109 ± 0.00313 | 0.016 |
| Tannerella forsythia_nov_96.52% | 0.00000 | 0.00095 ± 0.00311 | 0.002 |
| Aggregatibacter aphrophilus | 0.00000 | 0.00080 ± 0.00035 | 0.018 |
| Porphyromonas gingivalis_nov_96.46% | 0.00000 | 0.00079 ± 0.00037 | 0.026 |
| Leptotrichia sp._oral_taxon_219_nov_83.33% | 0.00021 ± 0.00019 | 0.00077 ± 0.00020 | 0.039 |
| Fretibacterium fastidiosum | 0.00000 | 0.00074 ± 0.00033 | 0.019 |
| Ruminococcaceae_[G-3] sp._oral_taxon_366_nov_78.83% | 0.00002 ± 0.00002 | 0.00054 ± 0.00015 | 0.001 |
| Veillonella sp._oral_taxon_780_nov_82.14% | 0.00000 | 0.00053 ± 0.00025 | 0.029 |
| Mogibacterium neglectum | 0.00010 ± 0.00008 | 0.00051 ± 0.00190 | 0.044 |
| Haemophilus parainfluenzae_nov_96.89% | 0.00000 | 0.00043 ± 0.00026 | 0.018 |
| Desulfovibrio sp._oral_taxon_040 | 0.00000 | 0.00040 ± 0.00040 | 0.018 |
| Mitsuokella sp._Oral_Taxon_H31_nov_78.38% | 0.00000 | 0.00033 ± 0.00016 | 0.035 |
| Streptococcus constellatus_nov_80.22% | 0.00000 | 0.00032 ± 0.00013 | 0.008 |
| Porphyromonas gingivalis_nov_91.05% | 0.00004 ± 0.00004 | 0.00032 ± 0.00011 | 0.019 |
| Bacteroidetes_[G-3] sp._oral_taxon_280_nov_92.66% | 0.00000 | 0.00031 ± 0.00010 | 0.015 |
| Alloprevotella rava_nov_86.47% | 0.00000 | 0.00029 ± 0.00009 | 0.002 |
| Fretibacterium sp._oral_taxon_359 | 0.00000 | 0.00025 ± 0.00016 | 0.029 |
| Prevotella sp._oral_taxon_515_nov_80.56% | 0.00000 | 0.00024 ± 0.00013 | 0.018 |
| Prevotella sp._oral_taxon_515_nov_78.25% | 0.00000 | 0.00023 ± 0.00016 | 0.029 |
| Peptostreptococcaceae_[XI][G-4] sp._oral_taxon_369 | 0.00000 | 0.00022 ± 0.00011 | 0.031 |
| Catonella sp._oral_taxon_164_nov_95.73% | 0.00000 | 0.00015 ± 0.00014 | 0.048 |
Figure 1.
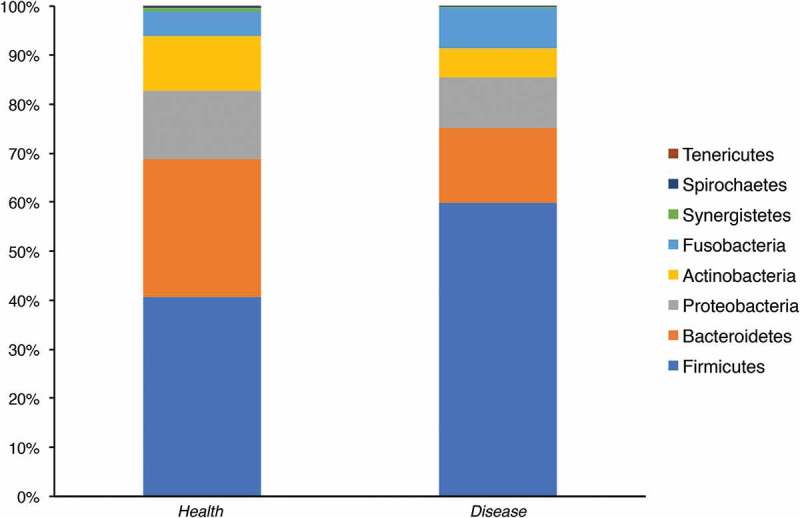
Bacterial phyla detected in APS-1 patients (n = 7) and control saliva (n = 17). Comparison of microbiota in APS-1 and healthy saliva determined by sequencing the hypervariable 16S rDNA region V1-V2. Relative abundance of the different phyla in control and APS-1 samples is shown.
Figure 2.
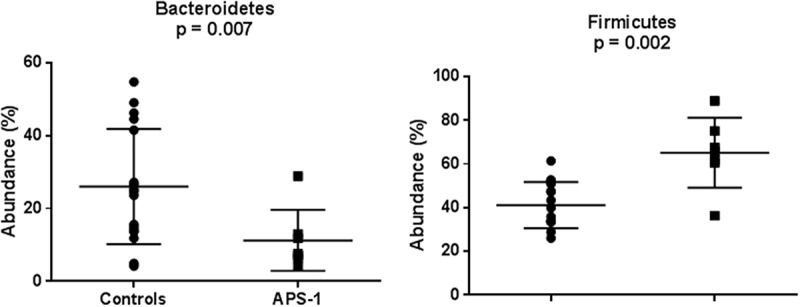
The relative abundances of the phyla Bacteroidetes and Firmicutes in saliva of healthy controls (n = 17) and APS-1 patients (n = 7) illustrated with boxplots. The lines indicate means and standard deviations.
A total of 64 bacterial genera were detected in APS-1 and 90 in healthy controls. Figure 3 gives the relative abundances of the 18 major genera found in APS-1 samples and controls. Moreover, a total of 18 genera showed significant differences in abundances (Table 2). Metastats analyses showed that the genera Streptococcus and Gemella were significantly higher in patients than controls (p = 0.0001 and 0.036, respectively; Figure 4) whereas Prevotella and Veillonella were significantly higher in controls (p = 0.001 and <0.001, respectively; Figure 4). Among the genera with significant difference in abundance, eight genera were absent from whole saliva of APS-1 patients (Table 2).
Figure 3.
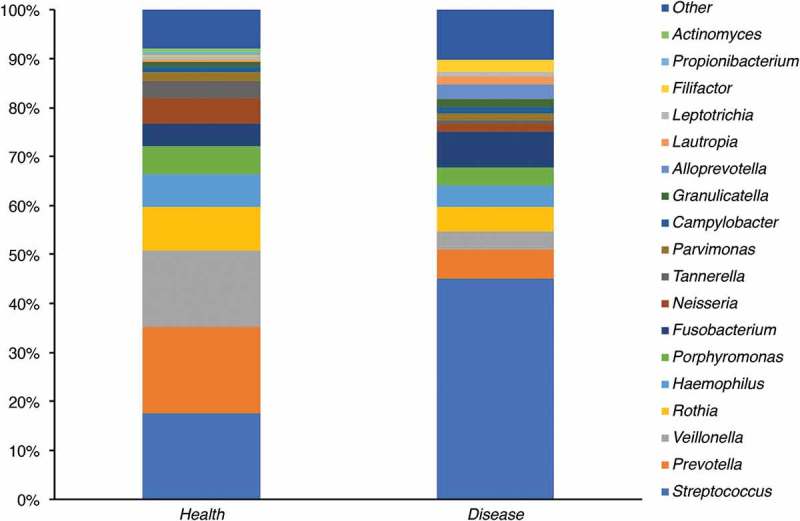
Bacterial genera detected in saliva from APS-1 patients (n = 7) and controls (n = 17). Groups designated as ‘Other’ represents minor groups classified. The Y-axis represents relative abundance. An increase in the genus Streptococcus in APS-1 saliva relative to control saliva is demonstrated.
Figure 4.
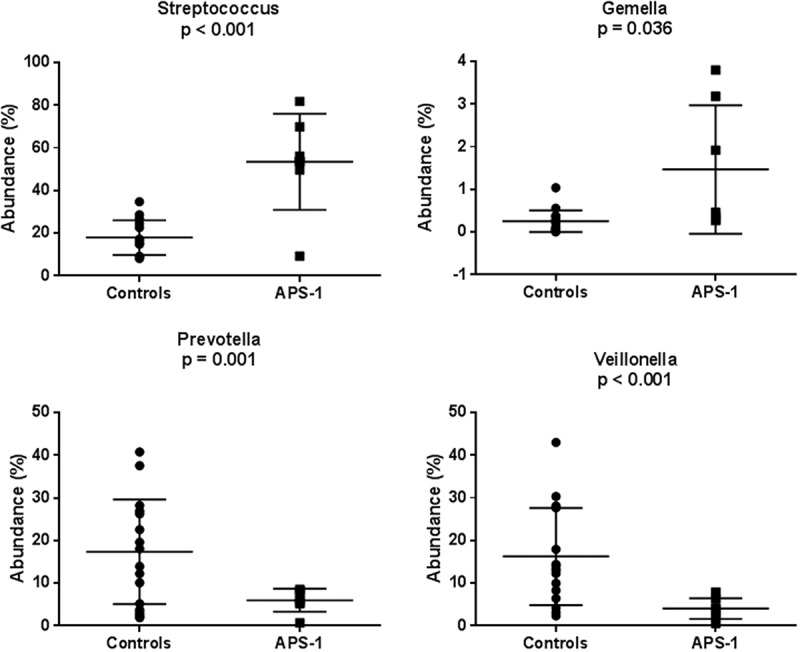
The relative abundances of the genera Streptococcus, Gemella, Prevotella, and Veillonella in saliva of healthy controls (n = 17) and APS-1 patients (n = 7) illustrated with boxplots. The lines indicate means and standard deviations.
In APS-1 patients, the sequencing data revealed the most abundant species to be Streptococcus sp. str. C300, Streptococcus multispecies spp.24_2, Streptococcus mitis, Streptococcus infantis, and Haemophilus parainfluenzae. Among these, only Streptococcus sp. str. C300 and Streptococcus multispecies spp.24_2 were increased compared to controls (p = 0.001, 0.032, respectively). The other species found more abundant in APS-1 patients were Fusobacterium nucleatum subsp. animalis, Gemella haemolysans, Ruminococcaceae [G-3] sp. Oral taxon 366, and Capnocytophaga ochracea (p = 0.046, 0.047, <0.001, 0.002, respectively). The Veillonella parvula group and Prevotella melaninogenica were most abundant in controls. Table 2 shows all species with significant differences in abundance compared to control saliva and APS-1 saliva as estimated by Metastats.
Species richness and diversity
Samples from healthy controls had a higher species richness and diversity than APS-1 samples. Figure 5(a) illustrates that the average species richness is higher in healthy subjects based on the observed species rarefaction curves Figure 5(b,c) gives the estimated species richness evaluated on the Chao1 matrix and the rarefaction curves based on the Shannon index, respectively showing an overall difference in alpha diversity, although the latter shows no difference between health and APS-1. However, the Shannon index measures both richness and abundance, hence the evenness of the community. Figure 5(c) therefore indicates that there is no significant difference between APS-1 and controls in terms of species evenness. Finally, Figure 5(d) gives a PCoA 3D plot of all samples with the distances calculated using weighed normalized UniFrac matrix, and clearly indicates that the two groups have distinct beta diversity. Taken together, these results show a richer diversity in controls compared to APS-1.
Figure 5.
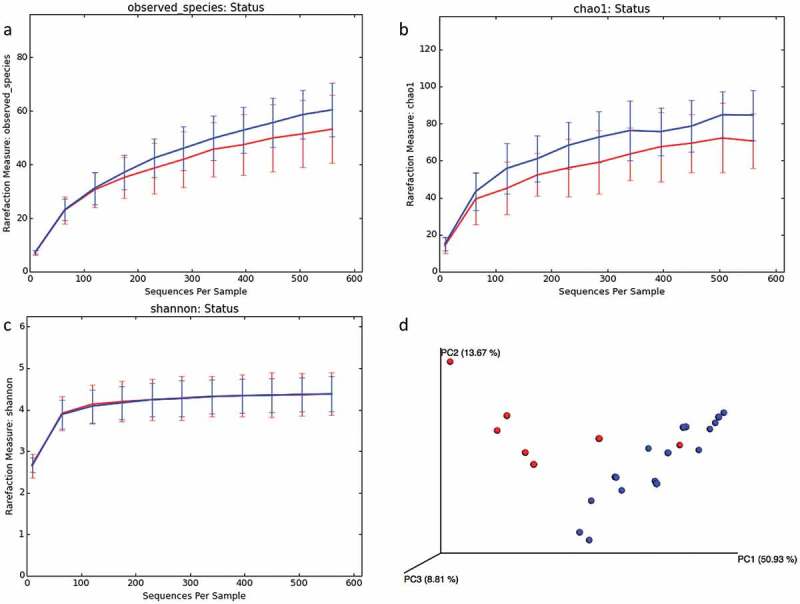
Comparison of microbial diversity in saliva samples of APS-1 patients (n = 7) and health samples (n = 17). Average rarefaction curves of APS-1 and health samples reported as (a) observed species, (b) Chao1 estimator, (c) Shannon index, (d) PCoA 3D plots of all samples with distances calculated using weighed normalized UniFrac matrix. Blue curves and dots represent health samples; and red represents APS-1.
Discussion
Increasing evidence suggests a potential role of the skin, oral, and gut microbiotas in the pathogenesis of autoimmunity [14]. Using high throughput sequencing and a comprehensive bioinformatics approach to analyse next generation 16S rDNA pyrosequencing reads, we here present hitherto undescribed significant differences in the composition of the salivary microbiota comparing APS-1 patients and healthy controls, indicating a possible contribution in pathogenesis and clinical expression.
The most striking differences were a higher portion of Firmicutes and a reduction of Bacteroidetes in APS-1 patients. Similar findings were recently described in primary Sjögrens syndrome [20] and a reduced species diversity and an altered ratio between Firmicutes and Bacteroidetes were described in the intestinal microbiota in several autoimmune diseases [30–33]. However, Firmicutes represents a phylum where most bacteria have a Gram-positive cell wall including the oral genera Streptococcus, Lactobacillus, Selenomonas, Clostridium, and Eubacterium. The phylum Bacteroidetes is composed of Gram-negative bacteria. On a genus level, Streptococcus and Gemella were increased in APS-1. However, a reduction in the total number of bacterial genera was seen. Overall, these novel findings indicate a significant altered oral microbiota in APS-1.
This is the first report on the oral microbiota in APS-1, and only a few studies have investigated the gut microbiota in patients [17,22,23]. A comparative analysis of the intestinal microbiota of APS-1 patients with gastrointestinal manifestations showed significant enrichment of segmented filamentous bacteria [17], which are Gram-positive commensal bacteria with the potential to adhere to epithelial cells and induce T helper (Th) 17 responses. Another study reported that APS-1 patients develop early and sustained responses to gut microbial antigens reminiscent to Crohn’s disease [23] linked to defects in Tregs. Finally, products from commensal bacteria have the potential to indirectly regulate thymic Aire expression in mice [22]. Based on the above, this indicates that the microbiota contributes in shaping immunity in APS-1 although the molecular mechanisms are incompletely characterized.
Factors known to directly affect the immune system, and consequently, the risk of autoimmunity, such as genetics, gender, and diet may also exert their effects by modulating microbiota profiles and functions. Using animal models of experimental colitis [34] and arthritis [35] it was shown that Gram-negative bacteria, possibly through the TLR2/IL-10 axis, reduced inflammation [34], whereas Gram-positive bacteria contributed to a more severe disease [35]. In a mouse model of Sjögren’s disease, depletion of the intestinal microbiome worsened the ocular response to desiccation, while the overall severity of disease correlated with intestinal microbiome diversity [36]. Two recent characterizations of the oral microbiota in primary Sjögren’s syndrome demonstrated a significant shift in the oral microbiota of patients and reduced numbers of genera [20,37], suggesting a role of the oral microbiota in the pathogenesis. Interestingly, these findings are comparable to what we currently describe in APS-1.
CMC caused by C. albicans is the most common and earliest manifestation of APS-1 [2]. The clinical course varies from periodical to chronic and usually affects the oral mucosa [1,2,9]. To date, neutralizing autoantibodies against the Th17 cytokines IL-17A, IL-17F, and IL-22 are suggested to explain the impairment in mucosal immunity in APS-1 patients [10,38]. However, an important line of defence against oral CMC is the oral microbiota that prevents infections by their interplay with immune cells, nutrients, metabolic products and by secreting antagonistic molecules, which together balance local inflammatory responses [39]. C. albicans can form biofilms with many oral bacteria, including streptococci [40], which have synergistic or antagonistic influences on C. albicans. Noteworthy, recent work has highlighted the critical role of metabolic products from specific gut microbiota such as lactobacilli in priming IL-22 dependent mucosal immune responses by innate lymphoid cells via the aryl hydrocarbon receptor, which is fundamental for protection against uncontrolled local Candida expansion [41]. Further, a shift in salivary microbiota has been linked to the risk of oral cancer in selected groups of patients [42]. We found significantly increased abundance of streptococci in APS-1 saliva compared to healthy controls and several streptococci were among the most abundant species in APS-1 saliva. To speculate, an altered microbiota may change the profile of immune regulatory metabolic products, and thus, contribute in altering immunity in APS-1 patients. Still it remains unclear whether an altered microbiota causes disease manifestations or the altered microbiota is an effect of disease components.
The obvious drawbacks of this study are the low number of patients included and the heterogeneity within this group. Initially 10 patients were included but technical issues with the sequencing made us exclude 3 patients. We plan to include more patients trying to correlate the different taxa to clinical manifestations of APS-1 and calculate absolute microbial abundances using a quantitative polymerase chain reactions assay. The same approach could be used to characterize the skin and the gut microbiotas in APS-1.
In conclusion, the oral microbiota in APS-1 patients is altered compared to healthy controls and seems to be a characteristic of the syndrome. In general, the skin, oral, and intestinal microbiotas in APS-1 patients should be further investigated to reveal their potentially contribution in pathogenesis and phenotypic expression of the syndrome.
Supplementary Material
Acknowledgments
The authors would like to thank the DNA Sequencing Facility, Department of Biochemistry, University of Cambridge for sequencing services (http://www.bioc.cam.ac.uk/dnasequencing).
Biographies
Øyvind Bruserud is MD and PhD-fellow at the University of Bergen with research interests in autoimmune polyendocrine syndrome type 1.
Huma Siddiqui is a molecular microbiologist with experience in genomics and metagenomics. At the Department of Oral Biology, Faculty of Dentistry, University of Oslo, she has been researching the genomics of oral bacteria.
Mihaela Cuida Marthinussen is DMD and associate professor in the field of clinical dentistry at the University of Bergen. Her main research interest is use of saliva as a diagnostic tool.
Tsute Chen is an associate research investigator at The Forsyth Institute, Cambridge, MA, USA and an instructor of the Department of Oral Medicine, Infection and Immunity, Harvard School of Dental Medicine, Boston, MA, USA. Tsute Chen’s research interests include studies of human oral microbiome in both taxonomy and genomics. Tsute Chen is one of the authors of the ‘Human Oral Microbiome Database’ (http://www.homd.org) and has been developing bioinformatics software tools for analysing the NGS genomic and transcriptomic data.
Roland Jonsson is DMD, PhD and professor at the University of Bergen and head of Broegelmann Research Laboratory. He has a particular interest in Sjögren’s syndrome.
Bergithe Eikeland Oftedal is an immunologist working in the field of immunogenetics. She has a particular interest in the development of T cells and how this shapes the immune repertoire. She is currently a visiting scientist at the University of Oxford.
Ingar Olsen is professor emeritus and guest researcher at Department of Oral Biology, Faculty of Dentistry, University of Oslo. Senior Research Investigator, Department of Microbiology, Forsyth Institute, Cambridge, MA. DDS from the Faculty of Dentistry, University of Oslo in 1966. Dr Odont. In 1976. Professor in oral microbiology 1988. Dean for research 2002-2008. Previously, main supervisor of more than 20 PhD students.
Eystein Husebye is professor of endocrinology at the University of Bergen with research interest in autoimmune endocrine diseases, in particular Addison’s disease and autoimmune polyendocrine syndrome type 1.
Bøe Wolff is a senior researcher in the field of molecular biology and immunology at the University of Bergen. Her main research interest is to understand background mechanisms for autoimmune diseases.
Funding Statement
This study was supported by grants from the University of Bergen, the Bergen Research Foundation, Stiftelsen KG Jebsen, the Regional Health Authorities of Western Norway (grant no 977878), The Norwegian Research Council (grant no 213704) and the Novo Nordisk Foundation (grant no NNF14OC0011005). IO was funded by the European Commission (FP7-HEALTH-306029 ‘TRIGGER’). RJ was funded by EU H2020 contract HarmonicSS (contract nr: H2020-SC1-2016-RTD/731944) and the Broegelmann Foundation. Grant from Research Council of Norway (250030) to BEO
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1]. Perheentupa J. Autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. J Clin Endocrinol Metab. 2006;91:2843–9. [DOI] [PubMed] [Google Scholar]
- [2]. Husebye ES, Perheentupa J, Rautemaa R, et al. Clinical manifestations and management of patients with autoimmune polyendocrine syndrome type I. J Intern Med. 2009;265:514–529. [DOI] [PubMed] [Google Scholar]
- [3]. Nagamine K, Peterson P, Scott HS, et al. Positional cloning of the APECED gene. Nat Genet. 1997;17:393–398. [DOI] [PubMed] [Google Scholar]
- [4]. Consortium F-GA. An autoimmune disease, APECED, caused by mutations in a novel gene featuring two PHD-type zinc-finger domains. Nat Genet. 1997;17:399–403. [DOI] [PubMed] [Google Scholar]
- [5]. Pitkänen J, Vähämurto P, Krohn K, et al. Subcellular localization of the autoimmune regulator protein. Characterization of nuclear targeting and transcriptional activation domain. J Biol Chem. 2001;276:19597–19602. [DOI] [PubMed] [Google Scholar]
- [6]. Anderson MS, Venanzi ES, Klein L, et al. Projection of an immunological self shadow within the thymus by the aire protein. Science. 2002;298:1395–1401. [DOI] [PubMed] [Google Scholar]
- [7]. Yang S, Fujikado N, Kolodin D, et al. Immune tolerance. Regulatory T cells generated early in life play a distinct role in maintaining self-tolerance. Science. 2015;348:589–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8]. Söderbergh A, Gustafsson J, Ekwall O, et al. Autoantibodies linked to autoimmune polyendocrine syndrome type I are prevalent in Down syndrome. Acta Paediatr. 2006;95:1657–1660. [DOI] [PubMed] [Google Scholar]
- [9]. Bruserud O, Oftedal BE, Landegren N, et al. A longitudinal follow-up of autoimmune polyendocrine syndrome type 1. J Clin Endocrinol Metab. 2016;101:2975–2983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10]. Kisand K, Wolff ASB, Podkrajsek KT, et al. Chronic mucocutaneous candidiasis in APECED or thymoma patients correlates with autoimmunity to Th17-associated cytokines. J Exp Med. 2010;207:299–308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11]. Meager A, Visvalingam K, Peterson P, et al. Anti-interferon autoantibodies in autoimmune polyendocrinopathy syndrome type 1. PLoS Med. 2006;3:e289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12]. Wolff AS, Sarkadi AK, Maródi L, et al. Anti-cytokine autoantibodies preceding onset of autoimmune polyendocrine syndrome type I features in early childhood. J Clin Immunol. 2013;33:1341–1348. [DOI] [PubMed] [Google Scholar]
- [13]. Brodin P, Jojic V, Gao T, et al. Variation in the human immune system is largely driven by non-heritable influences. Cell. 2015;160:37–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14]. Shamriz O, Mizrahi H, Werbner M, et al. Microbiota at the crossroads of autoimmunity. Autoimmun Rev. 2016;15:859–869. [DOI] [PubMed] [Google Scholar]
- [15]. Ferre EM, Rose SR, Rosenzweig SD, et al. Redefined clinical features and diagnostic criteria in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy. JCI Insight. 2016;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16]. Oftedal BE, Marthinussen MC, Erichsen MM, et al. Impaired salivary gland activity in patients with autoimmune polyendocrine syndrome type I. Autoimmunity. 2017;50:211–222. [DOI] [PubMed] [Google Scholar]
- [17]. Dobeš J, Neuwirth A, Dobešová M, et al. Gastrointestinal autoimmunity associated with loss of central tolerance to enteric alpha-defensins. Gastroenterology. 2015;149:139–150. [DOI] [PubMed] [Google Scholar]
- [18]. AlmståhI IA, Wikström M, Stenberg I, et al. Oral microbiota associated with hyposalivation of different origins. Oral Microbiol Immunol. 2003;18:1–8. [DOI] [PubMed] [Google Scholar]
- [19]. MacFarlane TW. The oral ecology of patients with severe Sjogren’s’ syndrome. Microbios. 1984;41:99–106. [PubMed] [Google Scholar]
- [20]. Siddiqui H, Chen T, Aliko A, et al. Microbiological and bioinformatics analysis of primary Sjogren’s syndrome patients with normal salivation. J Oral Microbiol. 2016;8:31119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21]. He J, Li Y, Cao Y, et al. The oral microbiome diversity and its relation to human diseases. Folia Microbiol (Praha). 2015;60:69–80. [DOI] [PubMed] [Google Scholar]
- [22]. Nakajima A, Negishi N, Tsurui H, et al. Commensal bacteria regulate thymic Aire expression. PLoS One. 2014;9:e105904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23]. Hetemäki I, Jarva H, Kluger N, et al. Anticommensal responses are associated with regulatory T cell defect in autoimmune polyendocrinopathy-candidiasis-ectodermal dystrophy patients. J Immunol. 2016;196:2955–2964. [DOI] [PubMed] [Google Scholar]
- [24]. Wolff AS, Erichsen MM, Meager A, et al. Autoimmune polyendocrine syndrome type 1 in Norway: phenotypic variation, autoantibodies, and novel mutations in the autoimmune regulator gene. J Clin Endocrinol Metab. 2007;92:595–603. [DOI] [PubMed] [Google Scholar]
- [25]. Myhre AG, Halonen M, Eskelin P, et al. Autoimmune polyendocrine syndrome type 1 (APS I) in Norway. Clin Endocrinol (Oxf). 2001;54:211–217. [DOI] [PubMed] [Google Scholar]
- [26]. Siddiqui H, Nederbragt AJ, Lagesen K, et al. Assessing diversity of the female urine microbiota by high throughput sequencing of 16S rDNA amplicons. BMC Microbiol. 2011;11:244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27]. Al-Hebshi NN, Nasher AT, Idris AM, et al. Robust species taxonomy assignment algorithm for 16S rRNA NGS reads: application to oral carcinoma samples. J Oral Microbiol. 2015;7:28934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28]. Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29]. White JR, Nagarajan N, Pop M. Statistical methods for detecting differentially abundant features in clinical metanogenic samples. PLoS Comput Biol. 2009;5:e1000352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30]. Giongo A, Gano KA, Crabb DB, et al. Toward defining the autoimmune microbiome for type 1 diabetes. Isme J. 2011;5:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Bhagava P, Moury EM. Gut microbiome and multiple sclerosis. Curr Neurol Neurosci Rep. 2014;492. [DOI] [PubMed] [Google Scholar]
- [32]. Chen J, Wright K, Davis JM, et al. An expression of rare lineage intestinal micobes characterizes rheumatoid arthritis. Genome Med. 2016;8:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Hevia A, Milani C, López P, et al. Intestinal dysbiosis associated with systemic lupus erythematosus. MBio. 2014;30(5):e01548–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34]. Oppong GO, Rapsinski GJ, Tursi SA, et al. Biofilm-associated bacterial amyloids dampen inflammation in the gut: oral treatment with curli fibres reduces the severity of hapten-induced colitis in mice. NPJ Biofilms Microbiomes. 2015;1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35]. Kohashi O, Kohashi Y, Takahashi T, et al. Reverse effect of gram-positive bacteria vs. gram-negative bacteria on adjuvant-induced arthritis in germfree rats. Microbiol Immunol. 1985;29:487–497. [DOI] [PubMed] [Google Scholar]
- [36]. de Paiva CS, Jones DB, Stern ME, et al. Altered mucosal microbiome diversity and disease severity in Sjogren syndrome. Sci Rep. 2016;6:23561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37]. Li M, Zou Y, Jiang Q, et al. A preliminary study of the oral microbiota in Chinese patients with Sjogren’s syndrome. Arch Oral Biol. 2016;70:143–148. [DOI] [PubMed] [Google Scholar]
- [38]. Puel A, Döffinger R, Natividad A, et al. Autoantibodies against IL-17A, IL-17F, and IL-22 in patients with chronic mucocutaneous candidiasis and autoimmune polyendocrine syndrome type I. J Exp Med. 2010;207:291–297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39]. Salvatori O, Puri S, Tati S, et al. Innate immunity and saliva in Candida albicans-mediated oral diseases. J Dent Res. 2016;95:365–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40]. Harriott MM, Noverr MC. Importance of Candida-bacterial polymicrobial biofilms in disease. Trends Microbiol. 2011;19:557–563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41]. Zelante T, Iannitti RG, Cunha C, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39:372–385. [DOI] [PubMed] [Google Scholar]
- [42]. Furquim CP, Soares GM, Ribeiro LL, et al. The salivary microbiome and oral cancer risk: A pilot study in fanconi anemia. J Dent Res. 2017;96(3):292–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


