Abstract
Resistance to antifungal drugs is an increasingly significant clinical problem. The most common antifungal resistance encountered is efflux pump-mediated resistance of Candida species to azole drugs. One approach to overcome this resistance is to inhibit the pumps and chemosensitize resistant strains to azole drugs. Drug discovery targeting fungal efflux pumps could thus result in the development of azole-enhancing combination therapy. Heterologous expression of fungal efflux pumps in Saccharomyces cerevisiae provides a versatile system for screening for pump inhibitors. Fungal efflux pumps transport a range of xenobiotics including fluorescent compounds. This enables the use of fluorescence-based detection, as well as growth inhibition assays, in screens to discover compounds targeting efflux-mediated antifungal drug resistance. A variety of medium- and high-throughput screens have been used to identify a number of chemical entities that inhibit fungal efflux pumps.
Keywords: : ABC and MFS transporters, antifungal drug resistance, azole, chemosensitization, heterologous expression, high-throughput screen
Background
Exposure of fungi to antifungal agents provides strong selection for mutations that confer drug resistance. It is evident that there is an impending medical crisis caused by the emergence of microbial drug resistance [1]. Although this problem is most prominent for bacterial pathogens it is also of concern for fungal infections. There are relatively few classes of antifungal agents used in medicine, namely the pyrimidine analog 5-fluorocytosine (5-FC), the polyene antifungals, the azole antifungals and the echinocandins, which restricts clinicians’ choices. While toxicity, pharmacokinetics and drug interactions are important considerations when choosing which antifungal agent to administer, the nature and incidence of antifungal resistance are also important factors [2]. The azole class of antifungals, which includes the imidazoles ketoconazole and clotrimazole and the triazoles fluconazole (FLC), voriconazole, itraconazole, posaconazole and isuvaconazole, are widely used because of their relatively low toxicity and low cost. The resistance of Candida species to FLC, however, has been recognized by the Centers for Disease Control and Prevention as a serious threat to human health [3].
Resistance to 5-FC is associated with mutations in genes encoding the enzymes cytosine deaminase and uracil phosphoribosyltransferase [4–6], which are required for the processing of this pro-drug, and possibly mutations in purine-cytosine permease which is involved in 5-FC uptake [6]. Intrinsic and acquired 5-FC resistance in fungi limits its utility and, as a result, 5-FC is often used in combination with other antifungals.
The polyenes exert their effect by interacting with ergosterol in fungal plasma membranes, forming pores and inducing the production of reactive oxygen species [7]. While some isolates of fungal species (e.g., Candida lusitaniae and Trichosporon beigelii) can show resistance to polyenes, acquired (secondary) resistance to polyene antifungals is rare and may be due to mutations that reduce the ergosterol content of membranes [8], or reduce the effects of reactive oxygen species through increased catalase activity.
The echinocandins are the most recently marketed antifungal drugs, but resistance is already emerging, particularly for the haploid yeast Candida glabrata [9]. Resistance to echinocandins is predominantly associated with point mutations in ‘hot spots’ of the gene encoding the drug target, β-1,3-D-glucan synthase [10]. A clinical concern is the emergence of C. glabrata strains with reduced susceptibilities to both echinocandins and azoles [11]. The emergence of these multiply resistant strains reduces the treatment options for patients significantly.
There are several mechanisms responsible for the azole resistance of fungi [2]. These include mutations in genes involved in ergosterol biosynthesis that provide tolerance to the toxic intermediates that accumulate when sterol 14α-demethylation is inhibited by azoles, overexpression of the drug target lanosterol 14α-demethylase, single or multiple point mutations in the drug target and overexpression of drug efflux pumps. While azole-resistant clinical isolates of the predominant human fungal pathogen, Candida albicans, can possess more than one mechanism of resistance, the most frequent cause of high-level azole resistance is energy-dependent drug efflux [12,13]. Azole antifungals can be effluxed by either major facilitator superfamily (MFS) or ATP-binding cassette (ABC) transporters [14]. In contrast, echinocandins are not substrates of these transporters [15]. Although fungal genomes contain several genes encoding proteins of these two families, it has been shown that in C. albicans the ABC pump Cdr1 is largely responsible for azole resistance [16,17].
The azoles are a tried and tested, well-tolerated and widely used class of antifungal agent. As the main mechanism of high-level azole resistance is energy dependent efflux by membrane bound transporters this opens the possibility of overcoming resistance and salvaging azole use by the inhibition of ABC efflux pumps. This approach to antimicrobial stewardship is analogous to the use of Augmentin for bacterial infections where penicillinase resistance is overcome by the combination of the β-lactamase inhibitor clavulanic acid with the antibacterial Amoxicillin.
Overcoming drug efflux as an adjunct to drug discovery
The concept of overcoming drug resistance by inhibiting efflux pumps is not new – it has been investigated for several decades in relation to the efflux-mediated resistance of tumor cells to chemotherapeutic agents. Ever since the detection of expression of human ABC protein ABCB1 (also known as MDR1 and P-gp) in over 400 human cancers [18], researchers have searched for inhibitors or modulators of ABC transporters. Indeed, three generations of ABCB1 inhibitors have been reported. The first-generation inhibitors included verapamil, cyclosporine A and quinine. Despite potent in vitro activity these compounds showed toxicity in vivo. Second-generation inhibitors were modified to target specific multidrug resistance (MDR) transporters. These compounds, such as valspodar and biricodar had improved bioavailability and less toxicity, but demonstrated off-target effects and lacked significant efficacy in clinical trials [19]. Third-generation MDR modulators were developed using quantitative structure–activity relationship (QSAR) to improve potency and specificity. Drugs such as elacridar, laniquidar and tariquidar inhibit ABCB1 at nanomolar concentrations but, again, definitive demonstration of clinical efficacy is lacking. A problem in the clinical trials of these agents is that tumors can be heterogenous and expression of ABCB1 can vary between patients as well as within tumors [20]. Another important feature of ABCB1 is that it has vital functions required for normal physiology, for instance in maintaining the blood–brain barrier and the renal excretion of metabolites. Thus, ABCB1 inhibitors targeting cancer cells may have unwanted side effects if they impair these functions. Specific inhibitors of fungal efflux proteins are unlikely to have such limitations. Recent attempts to identify human MDR inhibitors have moved away from the scaffolds of first- and second-generation inhibitors and have instead screened compound and natural product libraries or tried to repurpose existing drugs with low toxicity such as tyrosine kinase inhibitors [19].
While it is easy to be discouraged by the lack of success in identifying potent, clinically effective, ABCB1 inhibitors, there are several reasons why targeting fungal efflux pumps may indeed be fruitful. There is a clearer association between the expression of a narrow range of fungal efflux pumps with azole resistance than between ABC pump expression and the chemotherapeutic resistance of human cancers. In addition, fungal ABC proteins have unique features [21], described below, that distinguish them from mammalian ABC transporters. Targeting these features will have the potential to make inhibitors specific and reduce toxicity to the human host.
The approach of inhibiting transporters in order to overcome drug resistance is currently being applied to the bacterial pathogens Pseudomonas aeruginosa, Escherichia coli and Mycobacterium tuberculosis [22,23]. For example, the efflux pump inhibitor timcodar has been shown to increase the potency of the antituberculosis drugs rifampin and isoniazid toward M. tuberculosis in both in vitro and in vivo combination studies even though its precise mechanism of action is unknown [24]. The resistance-nodulation-division family of efflux pumps has received particular attention in P. aeruginosa and E. coli [25]. The structures of relevant efflux pumps, such as AcrB and MexB, have been solved [26–30] and effective efflux pump inhibitors have been identified but have not yet been tested in the clinic. As highlighted in a recent review, potency, obtaining structures of relevant efflux pump inhibitor–target complexes, spectrum of activity, pharmacokinetics and toxicity have all posed significant problems in the development of efflux pump inhibitors for clinical use [31].
Fungal efflux pumps
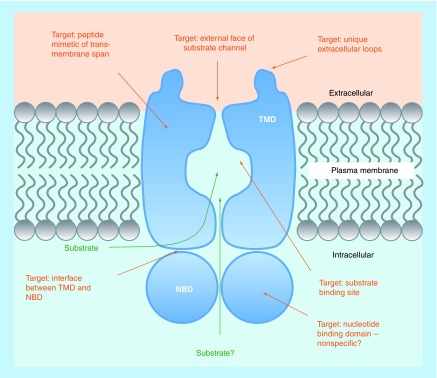
There are two main families of efflux pump proteins in fungi, the ABC proteins and the MFS transporters. ABC proteins contain two types of domain, nucleotide-binding domains (NBDs) and transmembrane domains (TMDs). NBDs are involved in the binding and hydrolysis of ATP which provides the energy for substrate translocation. These domains contain several well-conserved protein motifs including the Walker A motif or P-loop, the Walker B motif and the ABC signature motif or C-loop [21,32]. As these motifs are well-conserved across kingdoms, drugs targeting NBDs are likely to be nonspecific and may demonstrate host toxicity (Figure 1). TMDs comprise six transmembrane spans (TMSs) and are thought to contribute to a substrate channel through the membrane. Most fungal ABC proteins contain two NBDs and two TMDs and are referred to as ‘full-size’ ABC transporters (Figure 1). In these cases the substrate channel is likely formed from all 12 TMSs, or a subset of TMSs [14,32]. It is also likely that ‘half-size’ ABC proteins dimerize to form active transporters. The domain arrangement within full-size ABC proteins varies between transporter sub-class. The order within the pleiotropic drug resistance (PDR) sub-class, which contains most of the transporters involved in fungal drug resistance, is N-NBD1-TMD1-NBD2-TMD2-C [21,33,34]. This domain order is different from that in human ABCB1 which is N-TMD1-NBD1-TMD2-NBD2-C.
Figure 1. . Possible drug targets on fungal ABC efflux pumps.
NBD: Nucleotide-binding domain; TMD: Transmembrane domain.
The topology of ABC efflux pump domains presents several possible targets for modulating or inhibiting pump function (Figure 1). A unique feature of the PDR ABC transporters of plants and fungi is the presence of two large extracellular loops (ELs) between TMS5 and TMS6 (EL3) and between TMS11 and TMS12 (EL6) [21]. These ELs contain residues and motifs highly conserved among PDR ABC transporters. For example, E704 in the PDRB motif of EL3 is absolutely conserved between 244 PDR ABC proteins [21], and thus likely to be critical for protein folding and/or function. These ELs could represent targets specific for fungal PDR ABC proteins (Figure 1). For example, the unique ELs and the substrate exit channel of the fungal PDR ABC protein C. albicans Cdr1 are thought to contribute to the site of interaction with inhibitory peptide derivative RC21v3 [35]; the isolation of which is discussed below. Another approach to modulating transmembrane protein function is to use mimetics of TMDs to interact and interfere with ABC protein structure and function. It was found that peptide mimics of TMSs 1, 2, 4, 8, 10 and 11 of C. albicans Cdr1 inhibited efflux of the fluorescent substrates Nile Red and rhodamine 6G [36]. The TMS8 peptide was further shown to sensitize azole-resistant C. albicans cells to FLC.
It is thought that interactions between the intracellular loops of the TMDs and the NBDs are important for interdomain cross-talk and coupling between substrate transport and ATP hydrolysis [37]. This opens the possibility of allosteric inhibition and indicates that the interfaces between TMDs and NBDs are possible drug targets (Figure 1).
An obvious ABC protein drug target is the substrate-binding site. An important feature of PDR ABC proteins is their broad substrate specificity, hence the use of the term ‘pleiotropic’ in their classification. This feature has allowed the development of pump function assays based on their ability to efflux fluorescent compounds. However, the broad substrate specificity of the pumps has made it difficult to delimit binding sites and ABCB1 is thought to have multiple discrete drug binding sites [38]. It has been suggested that PDR ABC pumps such as Cdr1 may also have a large binding cavity with discrete binding pockets [32].
MFS proteins are proton antiporters that utilize the electrochemical potential and proton-motive force across membranes to translocate substrates [39]. They do not possess NBDs but do contain TMSs that are thought to form the substrate channel as indicated by the crystal structure of the EmrD multidrug transporter of E. coli [40]. In fungi, there are two subfamilies of MFS transporters which are defined by the number of TMSs within the TMD. The DHA1 subfamily contains 12 TMSs and the DHA2 subfamily 14 TMSs [14,39]. MFS transporters have, in general, a narrower spectrum of substrates than ABC transporters.
Fungi contain many genes encoding transporters, with typically between 14 (Ashbya gossypii) and 54 (Cryptococcus neoformans) ABC genes [21,41]. The number of MFS genes in fungi varies greatly between species with 10 in A. gossypii and 149–174 in C. neoformans [41,42]. Of these transporters, only a subset are involved in drug efflux [43]. Analysis of clinical Candida isolates resistant to azoles has identified that expression of PDR ABC genes orthologous to S. cerevisiae PDR5, and MFS genes orthologous to S. cerevisiae FLR1 are most often associated with azole resistance [13,44–47]. When the expression of efflux pumps in azole-resistant C. albicans clinical isolates was investigated it was found that ABC pumps were more often overexpressed than MFS pumps and that ABC protein Cdr1 (ortholog of S. cerevisiae Pdr5) contributes more than Cdr2 to resistance [16,48]. This was confirmed by disruption of the CDR1 and CDR2 genes in resistant C. albicans isolates [17].
Thus, in terms of targets for overcoming efflux-mediated azole resistance in C. albicans, the ABC protein Cdr1 is a prime candidate. In order to identify Cdr1 inhibitors it is necessary to develop a suitable pump assay and screen libraries of chemical diversity. Pump assays could utilize purified pump protein, but the pumps are membrane proteins and function may be affected by the membrane environment, and reconstitution in membrane vesicles is poorly developed and likely to be technically both demanding and labor intensive. Therefore attention has focused on assays involving screens using intact host cells heterologously expressing efflux pumps. Screening using the target positioned in whole cells as it would appear to drugs applied in vivo has advantages. It allows drugs targeting external features of the efflux pump to be identified, and if compounds that target intracellular components are discovered it immediately indicates that cell permeability is unlikely to be an issue for those compounds.
Heterologous expression of efflux pumps
Screening can be undertaken using the normal fungal host (homologous expression). For example, azole resistant clinical C. albicans isolates have been used to screen for compounds that chemosensitize their growth to FLC, without inhibiting yeast growth directly in the absence of FLC [49]. Such a screening approach has the advantage that hits identified are active against clinical C. albicans isolates. The disadvantage, however, is that clinical isolates are usually not well characterized and may possess multiple mechanisms of resistance [48]. This reduces the likelihood that a compound hitting a single target will have a phenotypic effect and, if hits are found, their targets may need to be identified for the medicinal chemistry optimization of lead compounds. Furthermore, if the target is intracellular, there is the possibility that the compound would be a substrate of a fungal efflux pump – one that was not expressed during the screen.
Heterologous expression of efflux pumps in the model yeast S. cerevisiae provides many advantages for drug screening. S. cerevisiae is a well-studied microorganism and is easily manipulated genetically. The repertoire of ABC and MFS genes involved in efflux-mediated drug resistance is well known [50]. This has enabled the construction of mutants in the laboratory of Andre Goffeau in which the main ABC genes involved in drug efflux have been deleted [51]. These strains are exquisitely sensitive to xenobiotics which can no longer be extruded. In addition, the transcriptional regulator PDR3 was deleted and a gain-of-function mutation (pdr1–3) was introduced in the transcriptional regulator Pdr1 of these strains. This leads to constitutive induction of the PDR5 promoter. A strain in which the PDR5 gene is retained, with homologous constitutive high expression, has been used to screen a large (1.89 × 106) combinatorial D-octapeptide library for efflux pump inhibitors [52]. Another such strain, AD1–8u- and its derivatives, have been used extensively to overexpress heterologous fungal efflux pumps for functional analysis and inhibitor screening [35,53–59]. Inserting the efflux pump gene of interest after the genomic PDR5 promoter leads to constitutive high levels of efflux pump expression. The multiple targets of Pdr1, many of which are involved in membrane protein biosynthesis, are upregulated which results in large amounts of pump protein correctly trafficked to the plasma membrane. For example, when C. albicans Cdr1 was expressed in S. cerevisiae AD1–8u- it accounted for 29% of the plasma membrane protein [56]. Such a high level of correctly localized efflux pump in a strain hypersusceptible to drugs yields a very strong drug resistance phenotype – the efflux pump expressing strain is typically 1000-fold more resistant to azole antifungals than the host strain. The integration of the efflux pump gene as a single copy in the genome, rather than being present at a variable copy number on a plasmid, gives stable strains with consistent levels of efflux gene expression facilitating reproducible drug screens.
This heterologous efflux gene expression system provides a versatile platform that can be used to screen for pump inhibitors based on chemosensitization of growth to FLC, or inhibition of the pumping of fluorescent substrates.
Screening methods to identify inhibitors of fungal efflux pumps
A variety of screening approaches have been used to identify fungal efflux pump inhibitors (Table 1). Although the screens denoted low/medium throughput may have investigated large numbers of compounds, high-throughput screens (HTS) are defined by a significant degree of automation. Both low- and high-throughput screens have included use of C. albicans strains known to overexpress efflux pumps [53,60] or recombinant S. cerevisiae strains hyperexpressing a specific efflux pump from a pathogenic fungus [35,54,55,61–63]. The compound libraries used are varied, but are either large-scale collections of small molecules such as the Molecular Libraries Small Molecule Repository (MLSMR) established by the NIH [64], collections of known drugs such as the Prestwick chemical library [65], collections of natural products and/or extracts [61,66–71], or are members of a family of compounds targeted to a structure of interest, such as a cell surface-targeted peptide library [35].
Table 1. . Primary screening strategies to identify pump inhibitors.
| Assay Approach | Target strain (pump targeted) | Library type (number agents tested) | Basis of screen | Hits identified | Ref. |
|---|---|---|---|---|---|
| Low/medium throughput | Recombinant Saccharomyces cerevisiae overexpressing† specific efflux pump (ScPdr5‡) | Natural product library: culture supernatants from actinomycetes and soil organisms (>10,000) | Chemosensitization to cerulenin and cyclohexamide | Three active extracts including isonitrile and enniatin | [67,68,70,71] |
| Azole-resistant Candida albicans or Candida glabrata clinical isolates overexpressing CaCdr1‡ or CgCdr1‡ | Natural product library: 85,000 extracts from actinomycetes strains (3600) or fungal strains (3500) | Chemosensitization to an azole | Enniatins, beauvericins and 20 milbemycins | [69] | |
| Recombinant S. cerevisiae overexpressing† specific efflux pumps (CaCdr1 or CaMdr1‡) | Natural product library: 10,000 marine extracts from the NCI Open Repository | Chemosensitization to FLC | Two sulfated sterols | [66] | |
| Recombinant S. cerevisiae overexpressing C. albicans efflux pump (CaCdr1 or CaMdr1) | Natural product library: marine algae extracts (>5000) | Chemosensitization to FLC | Capisterones A and B | [61] | |
| Recombinant S. cerevisiae overexpressing§ specific efflux pump (ScPdr5 or CaCdr1) | Targeted small molecule library: combinatorial D-octapeptides (˜1.89 × 106) | Chemosensitization to FLC | KN20; RC21v3 | [35,52] | |
| Azole-resistant C. albicans clinical isolates overexpressing either Mdr1 or Cdr1 and Cdr2 | Targeted library: synthetized analogs of cerulenin (˜30) | Chemosensitization to Brefeldin-A | Three active derivatives | [53] | |
| Azole-resistant C. albicans clinical isolates overexpressing either Mdr1 or Cdr1 and Cdr2 | Targeted library: synthetized derivatives of chalcones (27) | Chemosensitization to FLC | Three active derivatives | [72] | |
| Recombinant S. cerevisiae expressing§C. albicans efflux pump (CaMdr1) | Targeted library: synthetized compounds with squarile core (38) | Chemosensitization to FLC | Five active compounds | [55] | |
| HTS | Recombinant S. cerevisiae expressing§C. albicans efflux pump (CaCdr1 or CaCdr2‡) | Targeted library: Prestwick library of out-of-patent drugs (1200) | Fluorescent substrate (R6G) and flow cytometry | Nine active compounds including clorgyline | [54] |
| Recombinant S. cerevisiae overexpressing† specific efflux pumps (ScPdr5, ScSnq2‡ or CaCdr1) | Targeted library: Phenothiazine derivatives and related compounds (23) | Fluorescent substrate (fluorescein diacetate) microplate format | M961 newly synthesized derivative | [73] | |
| Recombinant S. cerevisiae expressing§C. albicans efflux pumps (multiplex format: CaCdr1, CaCdr2 and CaMdr1) | Small molecule library MLSMR (328,901) | Fluorescent substrate (Nile Red) and flow cytometry | 66 CaCdr2 inhibitors; 21 CaCdr1 inhibitors | [64] | |
| Recombinant S. cerevisiae expressing§C. glabrata efflux pump (CgCdr1) | Small molecule library: combined Prestwick, LOPAC and Tocris libraries (4801) and in-house drug-like compounds (7000) | Fluorescent substrate (R6G) microplate format | 15 active compounds¶ | ||
†Plasmid-based expression of heterologous pump gene.
‡ScPdr5: S. cerevisiae Pdr5; ScSnq2: S. cerevisiae Snq2; CaCdr1: C. albicans Cdr1; CaCdr2 C. albicans Cdr2; CaMdr1: C. albicans Mdr1; CgCdr1: C. glabrata Cdr1.
§Heterologous pump gene integrated into S. cerevisiae genome under the control of the PDR5 promoter.
¶[Cannon et al., unpublished observations].
Screens for inhibitors can be based on yeast growth measurements that demonstrate the reversal of fungal resistance to a pump substrate such as the azoles (chemosensitization) or can utilize the fluorescent properties of certain pump substrates such as rhodamine 6G (R6G) [54,74,75], Nile Red [76] or diS-C3 [77]. However, as noted above, growth chemosensitization assays, unless targeted to counter efflux-mediated resistance, may identify interference with one or more different aspects of resistance, not necessarily involving efflux. A review of antifungal chemosensitizer research [78] noted 24 reports of agents that chemosensitized cells to azoles, but only 13 described the target molecule or pathway in the fungal cell. An HTS of the MLSMR using chemosensitization to the azole FLC and detection of C. albicans growth inhibition with the fluorescent cellular stain Alamar Blue [49,79–81] described follow-up structure–activity relationship (SAR) analysis of three hits. The study identified 3-(3-anisoyl)indazole (derivative 36) as a new small-molecule probe (ML212) showing synergy with FLC, but its mechanism of action has yet to be defined [49]. Another HTS, also probing the MLSMR, was similarly based on identifying azole synergy, but measured biofilm growth inhibition [82]. The hit compound 2-adamantanamine was investigated further and the target was found to be within the ergosterol biosynthesis pathway, although the specific target molecule has not yet been identified [83]. Interestingly, beauvericin, recently described as an inhibitor of C. albicans ABC transporters [84], was identified earlier in a C. parapsilosis growth-based chemosensitization HTS using the azole ketoconazole [85]. In contrast, the use of yeast heterologously expressing a specific target protein, such as a drug efflux pump, allows a more defined screen for inhibitors.
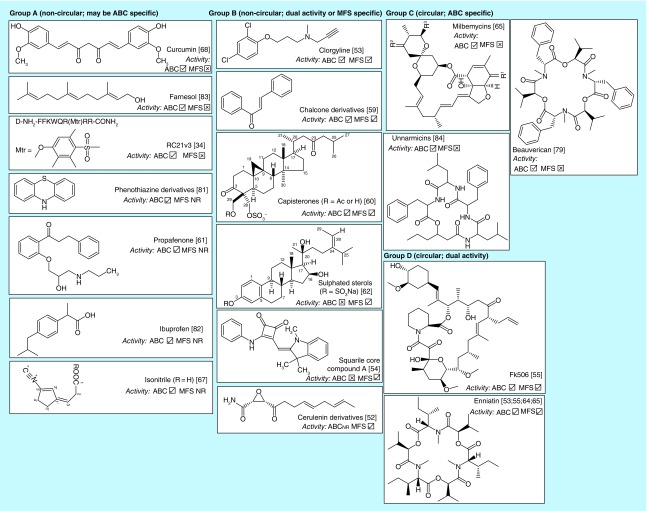
Screens have identified a number of fungal efflux pump inhibitors (Table 1 & Figure 2). Inhibitors identified include isonitrile [71], FK506 [56], enniatins [68], beauvericin [84], milbemycins [69], sulphated sterols [66], capisterones [61], derivatives of cerulenin [53], chalcones [60] and phenothiazine [73,86], ibuprofen [87], farnesol [88], a synthetic peptide [35], compounds with a squarile core [55] and the out-of-patent drug clorgyline [54]. Five of the compounds (RC21v3 [35]; clorgyline [54]; squarile core compounds [55]; beauvericin and unnarmicins [84,89]) were discovered using the heterologous S. cerevisiae AD/pABC3 expression system [56]. This system has been used to express several ABC and MFS efflux pumps, including the human ABC transporter P-gp (HsABCB1) [56] and therefore can also be employed in secondary screens to assess inhibitor specificity. Secondary screening is important because several fungal pump inhibitors (Figure 2) have also been reported as inhibiting P-gp, including curcumin [90] phenothiazine derivatives [57] propafenone [91] chalcones [92] beauvericin [93] and FK506 (tacrolimus) [94].
Figure 2. . Fungal efflux pumps identified through screens.
NR: Not reported.
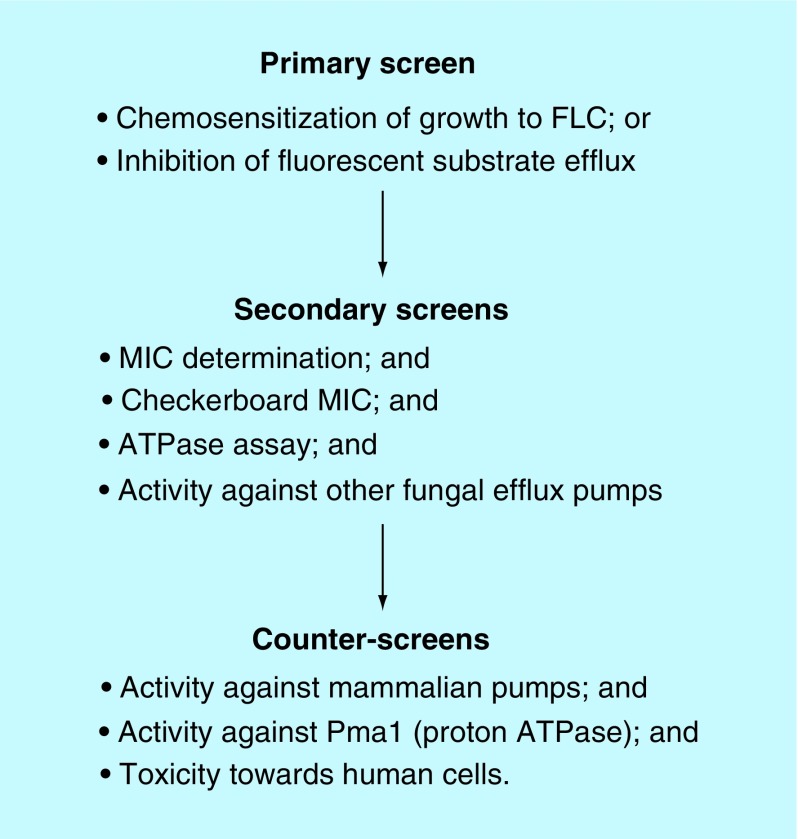
The process involved in either low- or high-throughput screening for specific efflux inhibitors using the pABC3 system is summarized in Figure 3. All hits are subjected to secondary screens or counter-screens as indicated to ensure the required specificity, lack of toxicity, to determine whether the inhibitor is also a pump substrate, and whether it has antifungal activity.
Figure 3. . Screening process using heterologous expression of fungal efflux pumps.
Low-medium throughput primary screens
An agar-based growth assay utilizing heterologous expression of efflux pumps in S. cerevisiae has been used to screen a large (1.89 × 106) combinatorial D-octapeptide library for inhibitors of C. albicans Cdr1 [35]. The octapeptides contained a tri-arginine motif to target the inhibitor molecule to the yeast cell surface, and five additional amino acids with any one of 18 amino acids (glycine and cysteine were excluded) at each position. Initially pools of peptides, with two of the five unknown amino acids identified, were screened, and subsequent deconvolution and optimization steps were applied to identify the most active peptide within resynthesized subpools [35]. The most active component was found to be a D-octapeptide derivative RC21v3. S. cerevisiae cells overexpressing C. albicans Cdr2 or Mdr1 were used as a counter-screen to eliminate chemosensitizers that affected targets other than Cdr1 in the yeast. To increase throughput, the screen used oblong culture dishes on which several compounds or pool samples could be tested. Confirmation that Cdr1 was the target of RC21v3 came from the analysis of suppressor mutants of S. cerevisiae cells expressing Cdr1 that were resistant to RC21v3. Sequencing the CDR1 genes in these strains revealed single amino acid mutations in Cdr1, many within its ELs [35]. Despite the high specificity of RC21v3 for C. albicans Cdr1, and lack of activity against C. albicans Cdr2 or Mdr1, RC21v3 enhanced the activity of FLC against an azole-resistant C. albicans strain in a mouse oral infection model [95].
Low-to-medium-throughput growth chemosensization-based screens are also useful when a screen is based on a lead compound with activity that may be improved by derivatization, or in screens of a particular family of related compounds. Cerulenin is a substrate of several antifungal efflux pumps [96,97] and screening of approximately 30 synthesized analogs identified three as inhibitors of C. albicans MFS transporter Mdr1p [53]. Derivatives of an oxathiolone-fused chalcone with antifungal activity [72] were shown to have ABC and MFS pump chemosensitizing activities [60] and a screen of structurally related low molecular mass compounds with a cyclobutene-dione (squarile) core identified inhibitors of the C. albicans Mdr1p efflux pump [55]. In the last example, overexpression of C. albicans Mdr1 in S. cerevisiae was used as the primary screen and S. cerevisiae overexpressing C. albicans Cdr1 was used as a counter-screen to eliminate compounds that affected ABC pumps as well as the targeted MFS efflux pump Mdr1.
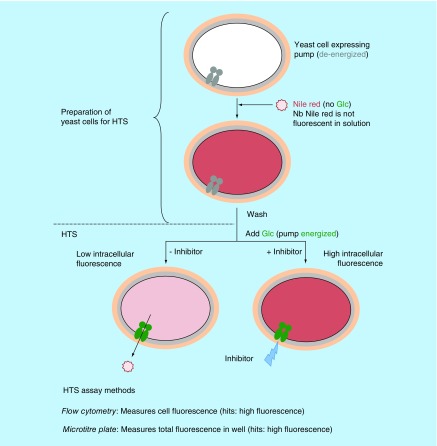
Although assays using growth measures of chemosensitization have identified inhibitors of efflux pumps, assays using fluorescent substrates are more readily applicable to HTS, as automated systems for assay readouts have been developed. A fluorescent substrate is used as a surrogate for the target antifungal drug with inhibition of the efflux pump causing retention of the fluorescent substrate within the yeast cells (Figure 4). Reported fluorescent substrates of fungal efflux pumps include the mitochondrial stains R6G and rhodamine-123 (R123) [58], the lipid stain Nile Red [76,98] and the potentiometric probe 3,3′-dipropylthiacarbocyanine iodide (diS-C3(3)) [60,99]. Automation and selection of suitable pump substrate(s), control strains, control inhibitors and compound libraries are critical for HTS. Phenotypic screens which use whole cells, such as the yeast screens described here, have the strategic advantage that toxic side effects can be monitored within the assay system [100]. Multiplexing (including more than one yeast strain in the assay, each expressing different pumps) further increases throughput, by allowing simultaneous testing of several targets. Care must be taken to incorporate appropriate secondary screens, as fluorescent substrates of efflux may respond differently to inhibition than the antifungal targeted. For example, FLC does not inhibit the efflux of R6G by C. albicans Cdr1. Additionally, such assays are only applicable to yeast cells, not hyphal cells, which aggregate, cannot be held in suspension, and have variable intracellular volumes. Systems used for HTS include: the HyperCyt® HT flow cytometry platform [100–102]; or a combination of multidrop delivery systems, liquid handling workstations and fluorimeters using 384 microtiter plates to reduce reagent volumes [103]. Our group has used both systems in HTS screens for inhibitors of C. albicans efflux pumps [54,76] and the C. glabrata Cdr1 efflux pump [cannon et al., unpublished observations].
Figure 4. . Whole-cell fluorescence-based fungal efflux pump assays for high-throughput screens.
HTS: High-throughput screen
High-throughput primary screens using recombinant S. cerevisiae strains
Flow cytometry-based HTS assays
Flow cytometry has been used to screen the Prestwick chemical library, using the fluorescent substrate R6G and individual recombinant yeast strains expressing either C. albicans ABC transporter Cdr1 or Cdr2 [54]. Yeast cells were preloaded with R6G and distributed into 384-well microtiter plates which were configured with control wells for negative, unblocked, transporter activity controls (low fluorescence) and positive, blocked (transporter inhibited by enniatin B) controls (high fluorescence), respectively. The AD/pABC3 empty cloning cassette control strain was also used as a positive labeling control (high fluorescence), and acted as a further counter screen to identify any off-target effects of the test compounds against fungal cells. Test compounds, stored as dimethyl sulfoxide stocks, were added to the test wells and, after incubation, the cell-associated fluorescence was measured with the HyperCyt® system to determine pump activity. Specialized software (IDLeQuery) was used to analyze the data. A fluorescence reading with a fivefold change in median fluorescence signal, relative to the negative controls without added inhibitor, defined hits. Of the nine hits obtained, only clorgyline showed a >90% inhibition of both Cdr1 and Cdr2. The broad-spectrum efflux pump inhibitory activities of clorgyline were confirmed in secondary assays [54].
The flow cytometry assay was developed further as a multiplex assay in which three ‘sentinel’ strains (expressing C. albicans Cdr1, Cdr2 or Mdr1), each tagged with a different fluorphore, were included in each well. The fluorescent substrate was Nile Red, which is effluxed by all three pumps (R6G is not a substrate of MFS pumps) and has the additional advantage that it is only fluorescent when cell associated [76]. This screen was applied to the MLSMR (329,018 compounds). A total of 357 compounds from 40 clusters of related compounds were cherry-picked for further analysis using strains expressing individual pumps and also for dose-response determinations. The single point data from the cherry-pick analysis have been uploaded to PubChem (AIDs 588522, 588518, 588520 and 588517). Hits were analyzed both for activity against all efflux pumps (broad-spectrum) and for specific activity against C. albicans ABC transporters Cdr1 and Cdr2 or the MDR transporter Mdr1. Several broad-spectrum hits were confirmed but dose response curves did not show any compounds with inhibition at submicromolar concentrations [63]. Interestingly, at least three of the hits (SID #s 4254537; 26660549 and 26661268) were also identified in a related HTS screen (AID1979) which used growth chemosensitization of a C. albicans strain to FLC as the basis of the HTS.
Spectrofluorimetric-based assay
Microplate spectrofluorometers have been used to measure the R6G effluxed from yeast cells, preloaded under energy-depleted conditions, into the culture supernatant following filtration to remove the yeast [16]. It might be assumed that filtration would be necessary because the remaining intracellular R6G fluorescence would overwhelm or cancel out the increasing supernatant signal. Such a filtration step would preclude development of a fluorimeter-based automated HTD assay. In contrast, we postulated that the fluorescent substrate Nile Red, which is only fluorescent when cell associated, would be compatible with fluorimeter plate-reader based HTS (Figure 4), and our preliminary experiments have confirmed this. We have also found that the signal from R6G effluxed from energized yeast cells into the assay supernatant can, in fact, be detected in wells still containing the cells. This possibly reflects quenching of the intracellular fluorescence compared with that from the supernatant. Further experiments were undertaken to confirm the utility of both substrates for a spectrofluorometric-based HTS and to optimize assay conditions [Cannon et al., unpublished observations]. Strains expressing either C. albicans Cdr1 or C. glabrata Cdr1, pre-loaded with R6G, were stable on ice for some hours allowing time for distribution into multiple 384-well microtiter plates. The optimal glucose assay concentration was found to be 20 mM. Following optimization, the R6G efflux signal:background (S:B) ratio (comparing assays in the presence or absence of glucose) was consistently ≥3.0. In a preliminary automatic dispensing experiment using a multidrop apparatus, satisfactory Z factor values [104] of between 0.68 and 0.78 were obtained (36 replicates). Using these conditions, we completed a pilot screen with the S. cerevisiae strain expressing C. glabrata Cdr1 [Cardno et al., unpublished observations]. The screen comprised 40 × 384-well library plates containing 12,401 compounds, consisting of 4801 known drugs and a further 7600 in-house library compounds. Each compound was dispensed into 384 well microtiter plates, and each plate also contained replicate wells of the control inhibitor beauvericin (Figure 2). A suspension of yeast cells, pre-loaded with R6G under de-energized conditions, was seeded directly onto compounds before the addition of glucose to start efflux pump activity. Plates were incubated at room temperature for 1 h, and efflux pump activity was measured by quantifying fluorescence on an Envision plate reader. Fifteen hits were obtained, but efflux inhibitory activity remains to be confirmed in secondary assays.
Secondary assays to confirm & characterize screen hits
We have developed routine assays of fungal efflux pump functions and demonstrated that these assays can be used to assess the effectiveness of inhibitors [35,54–56]. These assays include whole-cell-based assays (agarose disk chemosensitization; liquid MIC chemosensitization; R6G efflux) and an in vitro assay of pump activity (oligomycin-sensitive ATPase activity in plasma membrane preparations) (Figure 3). The assays are complementary: the growth-based chemosensitization assays demonstrate both direct and indirect effects of compounds tested over a long time scale (24 h); inhibition of efflux of R6G from whole cells into filtered supernatants measures direct inhibition of efflux over a short time scale (minutes) and the in vitro oligomycin-sensitive ATPase activity determines inhibitory activity at the molecular level. The pH dependence and oligomycin sensitivity of the ATPase activity measured in this assay differentiates it from the activity of the plasma membrane H+-ATPase Pma1 present in the membrane preparations.
Whole cell assays
Agarose diffusion chemosensitization assay: chemosensitization to antifungals can be demonstrated by agarose diffusion, using either S. cerevisiae strains expressing fungal efflux pumps or resistant clinical isolates. Inert sterile filter paper disks containing each hit compound are placed at suitable distances from each other on a lawn of the yeast cells inoculated on agarose containing sub-MIC concentrations of the azole [56,58]. A zone of growth inhibition around the disk indicates chemosensitization which can then be confirmed in liquid assays as described below.
Checkerboard liquid chemosensitization assay: if hit compounds target the fungal efflux pumps then they will chemosensitize S. cerevisiae cells expressing the pumps to FLC and other azoles (when present in the medium at sub-MIC levels) [35,54,55]. Initially the minimal inhibitory concentrations (MICs) of the azoles and the hit compounds for the pump-expressing S. cerevisiae strain are determined. Then microtiter plates containing the azole and hit compound in a checkerboard format (the azole is diluted twofold across the columns of a separate plate before transfer to the experimental plate containing twofold dilutions of the hit compound down the rows) are inoculated with the yeast strain. Cell growth is monitored at 24 and 48 h. The effect of sub-MIC concentrations of hit compound on the azole MIC, and vice versa, can confirm additivity or synergy between the azole and pump inhibitor. After 48 h incubation, samples from wells with no visible growth can be spotted on agar plates with no azole to test for cell viability, indicating whether the azole/inhibitor combination is fungicidal or whether the fungistatic property of the azole is retained. These assays can be used to confirm activity against azole-resistant clinical isolates.
Confirmation of efflux inhibition: efflux detected in the HTS may reflect induced permeability of the yeast cells rather than inhibition of the transporter, therefore all inhibitors should be checked in an assay based on detection of rhodamine efflux into filtered supernatants [16] or centifuged supernatants [58] (this cannot be done with the substrate Nile Red as it is nonfluorescent in solution). Cells preloaded with R6G or R123 are distributed to microplate wells and appropriate dilutions of the putative pump inhibitor and controls added. Glucose is then added to the cells to start efflux and at a predetermined appropriate time the cell suspension is transferred to a glass fiber filter plate and supernatants are collected into a receiver plate by application of a vacuum. R6G fluorescence can then be determined in a spectrofluorometer using excitation and the emission wavelengths of 485 and 520 nm, respectively, and the effectiveness of the test inhibitor determined.
Counter-screen: it is important to ensure that inhibitors of fungal efflux pumps do not affect important mammalian transporters such as ABCB1. The chemosensitization and efflux assays described above can be carried out using S. cerevisiae strains expressing mammalian pumps.
In vitro assays using purified plasma membrane preparations
ATPase assay: if a hit compound is indeed targeting the ABC fungal efflux pump it should inhibit the ATPase activity of the pump. Plasma membranes can be obtained from S. cerevisiae with minimal mitochondrial contamination [105] in which ABC pump-related ATPase activity can be distinguished from the plasma membrane proton pump (Pma1p) ATPase activity due to its oligomycin sensitivity.
Toxicity assays: secondary assays to ensure hit compounds are not toxic to human cells are needed before animal toxicity and efficacy evaluations can be undertaken. Red blood cell lysis is a useful indicator of direct effects on human cell membranes; release of hemoglobin can be determined by absorbance at 540 nm. Two other absorbance peaks for oxidized heme, 405 and 576 nm, can also be used to measure hemolysis if needed to overcome interference from the absorbance of the test compound. Measurements for a nonhemolyzed control or for 100% hemolysis can be determined in the presence of buffer only or buffer containing a detergent such as Triton X-100, respectively. The polyene antifungal amphotericin B provides a reference hemolysis value [35]. Cultured human cells, such as HEp2 cells, can be used to flag other toxicity issues. For example, cell viability in the presence or absence of test compounds can be determined using live-dead fluorescent stains and confocal microscopy [35]. Again toxicity can be evaluated relative to the antifungal amphotericin B.
This panel of secondary- and counter-assays can be carried out with hits and optimized leads as a prelude to animal and clinical testing of efflux pump inhibitors. Activity in animal models of fungal infection has been demonstrated for some efflux pump inhibitors, such as the milbemycins [106] and the octapeptide derivative RC21v3 [95].
Future perspective
Invasive fungal infections represent an emerging global health challenge which is accentuated by drug resistance. There is a pressing need to develop novel treatment strategies in order to overcome drug-resistant fungal infections.
Drug resistance is an inevitable consequence of microbial adaptation and evolution in response to selective pressure. This means there is a constant need to develop new antifungal agents, but also to devise ways of minimizing or overcoming the emergence of antifungal resistance.
The development of potent specific pump inhibitors that target efflux pumps responsible for xenobiotic efflux would benefit from atomic-level pump structures that could guide drug design. There are currently no crystal structures for fungal PDR-type ABC proteins which differ significantly from members of other ABC subfamilies.
Heterologous expression of efflux pumps in S. cerevisiae and the study of suppressor mutants resistant to potential pump inhibitors can confirm that the inhibitor target is the efflux pump. The suppressor mutants can also indicate molecular features within the target that may contribute to susceptibility and the likelihood of resistance to the inhibitor developing.
The combination of drugs targeting different molecules, that include drug efflux pumps, should also chemosensitize susceptible cells to antifungals and reduce the frequency with which drug-resistant isolates emerge.
The value of specific versus broad-spectrum pump inhibitors remains a key consideration. The specificity of efflux pump inhibitors can be determined easily using a panel of S. cerevisiae strains expressing individual efflux pumps from a range of organisms. Further research is needed to determine the extent to which specific pump inhibitors can overcome the clinical drug resistance of fungal infections.
Executive summary.
Background
Fungi pose a serious threat to human health and there is a limited number of classes of antifungal agents available.
Drug resistance of fungal pathogens is an increasing clinical problem.
An alternative to discovering new drug targets is to overcome known resistance mechanisms for otherwise efficacious antifungal drugs.
Efflux pumps as drug targets
Efflux pumps are responsible for high-level resistance of fungi to drugs such as the azole antifungals. Until it becomes possible to design inhibitors in this class that are not substrates of drug efflux pumps, the pumps have potential as secondary sites for chemotherapeutic intervention.
Efflux pumps are druggable targets, as inhibitors have been identified.
There are several sites on ABC efflux pumps that can be targeted.
Drug efflux pumps are often localized in the plasma membrane and present extracellular targets. This means that drugs targeting extracellular portions of the pumps will not need to penetrate the cell membrane, will not be subject to cellular detoxification or efflux.
There are multiple efflux pump genes in fungal genomes. The efficacy of pump inhibitors in overcoming drug resistance depends on the contribution of different pumps to the resistance phenotype and the specificity of inhibitors to these pumps.
Efflux pumps play vital roles in human cells. It is important that fungal pump inhibitors do not affect these vital host functions.
Advantages of using Saccharomyces cerevisiae to express fungal efflux pump targets
Saccharomyces cerevisiae expresses fungal efflux pumps at high levels. S. cerevisiae can be easily genetically manipulated to delete endogenous efflux pumps. This means that heterologous efflux pumps can be studied in the absence of other pumps and the high expression level gives a strong phenotype facilitating inhibitor screening in vitro and with whole cells.
S. cerevisiae is a robust micro-organism that is easily cultured and integrating heterologous pump genes at a chromosomal locus provides very stable strains. Both haploid and diploid strains of S. cerevisiae can be generated. Haploid strains are better hosts for genetic manipulations and gene expression, whereas diploid strains are better for modelling allele polymorphisms.
Panels of S. cerevisiae strains each expressing different efflux pumps enables valuable secondary screens to ensure novel antifungals are not substrates of efflux pumps.
Saccharomyces cerevisiae suppressor mutants can be used to identify the molecular target of chemosensitizers.
Outcomes of screens for fungal efflux pump inhibitors
A variety of medium-throughput and high-throughput screens have been used to identify fungal efflux pump inhibitors.
A variety of chemical compounds and peptides have been identified that inhibit efflux pumps in vitro.
Some of these inhibitors are very specific and only inhibit a narrow range of efflux pumps. Others have broad specificities and inhibit both ABC and MFS transporters.
One specific ABC pump inhibitor, modified D-peptide RC21v3, has been shown to be effective in a mouse model of oral candidiasis caused by a fluconazole-resistant clinical isolate by acting synergistically with fluconazole to reduce symptoms and fungal burden.
No fungal efflux pump inhibitors have entered clinical trials.
Footnotes
Financial & competing interests disclosure
RD Cannon acknowledges funding from the NIH (R01-DE016855-01; R03-MH087406-1A1) and the Royal Society of New Zealand Marsden Fund (UOO1305). LA Sklar acknowledges funding from the NIH (U54 MH074425/MH086490). TS Cardno was funded by the Health Research Council of New Zealand's International Investment Opportunities Fund (IIOF 09_04). BC Monk was supported by the Health Research Council of New Zealand. AR Holmes was supported by the University of Otago. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Michael CA, Dominey-Howes D, Labbate M. The antimicrobial resistance crisis: causes, consequences, and management. Front. Public Health. 2014;2:145. doi: 10.3389/fpubh.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prasad R, Shah AH, Rawal MK. Antifungals: mechanism of action and drug resistance. Adv. Exp. Med. Biol. 2016;892:327–349. doi: 10.1007/978-3-319-25304-6_14. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States, 2013. www.cdc.gov/drugresistance/threat-report-2013
- 4.Charlier C, El Sissy C, Bachelier-Bassi S, et al. Acquired flucytosine resistance during combination therapy with caspofungin and flucytosine for Candida glabrata cystitis. Antimicrob. Agents Chemother. 2015;60(1):662–665. doi: 10.1128/AAC.02265-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dodgson AR, Dodgson KJ, Pujol C, Pfaller MA, Soll DR. Clade-specific flucytosine resistance is due to a single nucleotide change in the FUR1 gene of Candida albicans . Antimicrob. Agents Chemother. 2004;48(6):2223–2227. doi: 10.1128/AAC.48.6.2223-2227.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hope WW, Tabernero L, Denning DW, Anderson MJ. Molecular mechanisms of primary resistance to flucytosine in Candida albicans . Antimicrob. Agents Chemother. 2004;48(11):4377–4386. doi: 10.1128/AAC.48.11.4377-4386.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mesa-Arango AC, Trevijano-Contador N, Roman E, et al. The production of reactive oxygen species is a universal action mechanism of Amphotericin B against pathogenic yeasts and contributes to the fungicidal effect of this drug. Antimicrob. Agents Chemother. 2014;58(11):6627–6638. doi: 10.1128/AAC.03570-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jensen RH, Astvad KM, Silva LV, et al. Stepwise emergence of azole, echinocandin and amphotericin B multidrug resistance in vivo in Candida albicans orchestrated by multiple genetic alterations. J. Antimicrob. Chemother. 2015;70(9):2551–2555. doi: 10.1093/jac/dkv140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arendrup MC, Perlin DS. Echinocandin resistance: an emerging clinical problem? Curr. Opin. Infect. Dis. 2014;27(6):484–492. doi: 10.1097/QCO.0000000000000111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlin DS. Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 2007;10(3):121–130. doi: 10.1016/j.drup.2007.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pfaller MA, Castanheira M, Lockhart SR, Ahlquist AM, Messer SA, Jones RN. Frequency of decreased susceptibility and resistance to echinocandins among fluconazole-resistant bloodstream isolates of Candida glabrata . J. Clin. Microbiol. 2012;50(4):1199–1203. doi: 10.1128/JCM.06112-11. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Report of multiresistant fungal clinical isolates.
- 12.Liu JY, Shi C, Wang Y, Li WJ, Zhao Y, Xiang MJ. Mechanisms of azole resistance in Candida albicans clinical isolates from Shanghai, China. Res. Microbiol. 2015;166(3):153–161. doi: 10.1016/j.resmic.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 13.Mane A, Vidhate P, Kusro C, et al. Molecular mechanisms associated with fluconazole resistance in clinical Candida albicans isolates from India. Mycoses. 2016;59(2):93–100. doi: 10.1111/myc.12439. [DOI] [PubMed] [Google Scholar]
- 14.Cannon RD, Lamping E, Holmes AR, et al. Efflux-mediated antifungal drug resistance. Clin. Microbiol. Rev. 2009;22(2):291–321. doi: 10.1128/CMR.00051-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Major review of efflux-mediated fungal drug resistance.
- 15.Niimi K, Maki K, Ikeda F, et al. Overexpression of Candida albicans CDR1, CDR2, or MDR1 does not produce significant changes in echinocandin susceptibility. Antimicrob. Agents Chemother. 2006;50(4):1148–1155. doi: 10.1128/AAC.50.4.1148-1155.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holmes AR, Lin YH, Niimi K, et al. ABC transporter Cdr1p contributes more than Cdr2p does to fluconazole efflux in fluconazole-resistant Candida albicans clinical isolates. Antimicrob. Agents Chemother. 2008;52(11):3851–3862. doi: 10.1128/AAC.00463-08. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Evidence that Cdr1 is the most important efflux pump in Candida albicans
- 17.Tsao S, Rahkhoodaee F, Raymond M. Relative contributions of the Candida albicans ABC transporters Cdr1p and Cdr2p to clinical azole resistance. Antimicrob. Agents Chemother. 2009;53(4):1344–1352. doi: 10.1128/AAC.00926-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Goldstein LJ, Galski H, Fojo A, et al. Expression of a multidrug resistance gene in human cancers. J. Natl Cancer Inst. 1989;81(2):116–124. doi: 10.1093/jnci/81.2.116. [DOI] [PubMed] [Google Scholar]
- 19.Kathawala RJ, Gupta P, Ashby CR, Jr., Chen ZS. The modulation of ABC transporter-mediated multidrug resistance in cancer: a review of the past decade. Drug Resist. Updat. 2015;18:1–17. doi: 10.1016/j.drup.2014.11.002. [DOI] [PubMed] [Google Scholar]
- 20.Gottesman MM, Pastan IH. The role of multidrug resistance efflux pumps in cancer: revisiting a JNCI publication exploring expression of the MDR1 (P-glycoprotein) gene. J. Natl Cancer Inst. 2015;107(9):djv222. doi: 10.1093/jnci/djv222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lamping E, Baret PV, Holmes AR, Monk BC, Goffeau A, Cannon RD. Fungal PDR transporters: phylogeny, topology, motifs and function. Fungal Genet. Biol. 2010;47(2):127–142. doi: 10.1016/j.fgb.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aparna V, Dineshkumar K, Mohanalakshmi N, Velmurugan D, Hopper W. Identification of natural compound inhibitors for multidrug efflux pumps of Escherichia coli and Pseudomonas aeruginosa using in silico high-throughput virtual screening and in vitro validation. PLoS ONE. 2014;9(7):e101840. doi: 10.1371/journal.pone.0101840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gupta S, Cohen KA, Winglee K, Maiga M, Diarra B, Bishai WR. Efflux inhibition with verapamil potentiates bedaquiline in Mycobacterium tuberculosis . Antimicrob. Agents Chemother. 2014;58(1):574–576. doi: 10.1128/AAC.01462-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grossman TH, Shoen CM, Jones SM, Jones PL, Cynamon MH, Locher CP. The efflux pump inhibitor timcodar improves the potency of antimycobacterial agents. Antimicrob. Agents Chemother. 2015;59(3):1534–1541. doi: 10.1128/AAC.04271-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lomovskaya O, Zgurskaya HI, Totrov M, Watkins WJ. Waltzing transporters and ‘the dance macabre’ between humans and bacteria. Nat. Rev. Drug Discov. 2007;6(1):56–65. doi: 10.1038/nrd2200. [DOI] [PubMed] [Google Scholar]
- 26.Murakami S, Nakashima R, Yamashita E, Matsumoto T, Yamaguchi A. Crystal structures of a multidrug transporter reveal a functionally rotating mechanism. Nature. 2006;443(7108):173–179. doi: 10.1038/nature05076. [DOI] [PubMed] [Google Scholar]
- 27.Nakashima R, Sakurai K, Yamasaki S, et al. Structural basis for the inhibition of bacterial multidrug exporters. Nature. 2013;500(7460):102–106. doi: 10.1038/nature12300. [DOI] [PubMed] [Google Scholar]
- 28.Nakashima R, Sakurai K, Yamasaki S, Nishino K, Yamaguchi A. Structures of the multidrug exporter AcrB reveal a proximal multisite drug-binding pocket. Nature. 2011;480(7378):565–569. doi: 10.1038/nature10641. [DOI] [PubMed] [Google Scholar]
- 29.Sennhauser G, Amstutz P, Briand C, Storchenegger O, Grutter MG. Drug export pathway of multidrug exporter AcrB revealed by DARPin inhibitors. PLoS Biol. 2007;5(1):e7. doi: 10.1371/journal.pbio.0050007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sennhauser G, Bukowska MA, Briand C, Grutter MG. Crystal structure of the multidrug exporter MexB from Pseudomonas aeruginosa . J. Mol. Biol. 2009;389(1):134–145. doi: 10.1016/j.jmb.2009.04.001. [DOI] [PubMed] [Google Scholar]
- 31.Opperman TJ, Nguyen ST. Recent advances toward a molecular mechanism of efflux pump inhibition. Front. Microbiol. 2015;6:421. doi: 10.3389/fmicb.2015.00421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Prasad R, Goffeau A. Yeast ATP-binding cassette transporters conferring multidrug resistance. Annu. Rev. Microbiol. 2012;66:39–63. doi: 10.1146/annurev-micro-092611-150111. [DOI] [PubMed] [Google Scholar]; •• Review of ABC efflux pumps in yeast.
- 33.Cannon RD, Holmes AR. Learning the ABC of oral fungal drug resistance. Mol. Oral. Microbiol. 2015;30(6):425–437. doi: 10.1111/omi.12109. [DOI] [PubMed] [Google Scholar]
- 34.Prasad R, Rawal MK, Shah AH. Candida efflux ATPases and antiporters in clinical drug resistance. Adv. Exp. Med. Biol. 2016;892:351–376. doi: 10.1007/978-3-319-25304-6_15. [DOI] [PubMed] [Google Scholar]
- 35.Niimi K, Harding DR, Holmes AR, et al. Specific interactions between the Candida albicans ABC transporter Cdr1p ectodomain and a D-octapeptide derivative inhibitor. Mol. Microbiol. 2012;85(4):747–767. doi: 10.1111/j.1365-2958.2012.08140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maurya IK, Thota CK, Verma SD, et al. Rationally designed transmembrane peptide mimics of the multidrug transporter protein Cdr1 act as antagonists to selectively block drug efflux and chemosensitize azole-resistant clinical isolates of Candida albicans . J. Biol. Chem. 2013;288(23):16775–16787. doi: 10.1074/jbc.M113.467159. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Shah AH, Rawal MK, Dhamgaye S, Komath SS, Saxena AK, Prasad R. Mutational analysis of intracellular loops identify cross talk with nucleotide binding domains of yeast ABC transporter Cdr1p. Sci. Rep. 2015;5:11211. doi: 10.1038/srep11211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Safa AR. Identification and characterization of the binding sites of P-glycoprotein for multidrug resistance-related drugs and modulators. Curr. Med. Chem. Anticancer Agents. 2004;4(1):1–17. doi: 10.2174/1568011043482142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sa-Correia I, dos Santos SC, Teixeira MC, Cabrito TR, Mira NP. Drug:H+ antiporters in chemical stress response in yeast. Trends Microbiol. 2009;17(1):22–31. doi: 10.1016/j.tim.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 40.Yin Y, He X, Szewczyk P, Nguyen T, Chang G. Structure of the multidrug transporter EmrD from Escherichia coli . Science. 2006;312(5774):741–744. doi: 10.1126/science.1125629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gbelska Y, Krijger JJ, Breunig KD. Evolution of gene families: the multidrug resistance transporter genes in five related yeast species. FEMS Yeast Res. 2006;6(3):345–355. doi: 10.1111/j.1567-1364.2006.00058.x. [DOI] [PubMed] [Google Scholar]
- 42.Janbon G, Ormerod KL, Paulet D, et al. Analysis of the genome and transcriptome of Cryptococcus neoformans var. grubii reveals complex RNA expression and microevolution leading to virulence attenuation. PLoS Genet. 2014;10(4):e1004261. doi: 10.1371/journal.pgen.1004261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Costa C, Dias PJ, Sa-Correia I, Teixeira MC. MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front. Physiol. 2014;5:197. doi: 10.3389/fphys.2014.00197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maebashi K, Niimi M, Kudoh M, et al. Mechanisms of fluconazole resistance in Candida albicans isolates from Japanese AIDS patients. J. Antimicrob. Chemother. 2001;47(5):527–536. doi: 10.1093/jac/47.5.527. [DOI] [PubMed] [Google Scholar]
- 45.Sanglard D, Ischer F, Calabrese D, Majcherczyk PA, Bille J. The ATP binding cassette transporter gene CgCDR1 from Candida glabrata is involved in the resistance of clinical isolates to azole antifungal agents. Antimicrob. Agents Chemother. 1999;43(11):2753–2765. doi: 10.1128/aac.43.11.2753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanglard D, Kuchler K, Ischer F, Pagani JL, Monod M, Bille J. Mechanisms of resistance to azole antifungal agents in Candida albicans isolates from AIDS patients involve specific multidrug transporters. Antimicrob. Agents Chemother. 1995;39(11):2378–2386. doi: 10.1128/aac.39.11.2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.White TC. Increased mRNA levels of ERG16, CDR, and MDR1 correlate with increases in azole resistance in Candida albicans isolates from a patient infected with human immunodeficiency virus. Antimicrob. Agents Chemother. 1997;41(7):1482–1487. doi: 10.1128/aac.41.7.1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.White TC, Holleman S, Dy F, Mirels LF, Stevens DA. Resistance mechanisms in clinical isolates of Candida albicans . Antimicrob. Agents Chemother. 2002;46(6):1704–1713. doi: 10.1128/AAC.46.6.1704-1713.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Youngsaye W, Hartland CL, Morgan BJ, et al. ML212: a small-molecule probe for investigating fluconazole resistance mechanisms in Candida albicans . Beilstein J. Org. Chem. 2013;9:1501–1507. doi: 10.3762/bjoc.9.171. [DOI] [PMC free article] [PubMed] [Google Scholar]; • High-throughput screening (HTS) of MLSMR based on chemosensitization to azole.
- 50.De Hertogh B, Carvajal E, Talla E, Dujon B, Baret P, Goffeau A. Phylogenetic classification of transporters and other membrane proteins from Saccharomyces cerevisiae . Funct. Integr. Genomics. 2002;2(4–5):154–170. doi: 10.1007/s10142-002-0060-8. [DOI] [PubMed] [Google Scholar]
- 51.Decottignies A, Grant AM, Nichols JW, de Wet H, McIntosh DB, Goffeau A. ATPase and multidrug transport activities of the overexpressed yeast ABC protein Yor1p. J. Biol. Chem. 1998;273(20):12612–12622. doi: 10.1074/jbc.273.20.12612. [DOI] [PubMed] [Google Scholar]; • Construction of versatile S. cerevisiae strain for HTS.
- 52.Niimi K, Harding DR, Parshot R, et al. Chemosensitization of fluconazole resistance in Saccharomyces cerevisiae and pathogenic fungi by a D-octapeptide derivative. Antimicrob. Agents Chemother. 2004;48(4):1256–1271. doi: 10.1128/AAC.48.4.1256-1271.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Diwischek F, Morschhauser J, Holzgrabe U. Cerulenin analogues as inhibitors of efflux pumps in drug-resistant Candida albicans . Arch. Pharm. (Weinheim) 2009;342(3):150–164. doi: 10.1002/ardp.200800160. [DOI] [PubMed] [Google Scholar]
- 54.Holmes AR, Keniya MV, Ivnitski-Steele I, et al. The monoamine oxidase A inhibitor clorgyline is a broad-spectrum inhibitor of fungal ABC and MFS transporter efflux pump activities which reverses the azole resistance of Candida albicans and Candida glabrata clinical isolates. Antimicrob. Agents Chemother. 2012;56(3):1508–1515. doi: 10.1128/AAC.05706-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Keniya MV, Fleischer E, Klinger A, Cannon RD, Monk BC. Inhibitors of the Candida albicans major facilitator superfamily transporter Mdr1p responsible for fluconazole resistance. PLoS ONE. 2015;10(5):e0126350. doi: 10.1371/journal.pone.0126350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lamping E, Monk BC, Niimi K, et al. Characterization of three classes of membrane proteins involved in fungal azole resistance by functional hyperexpression in Saccharomyces cerevisiae . Eukaryot. Cell. 2007;6(7):1150–1165. doi: 10.1128/EC.00091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Maki N, Dey S. Biochemical and pharmacological properties of an allosteric modulator site of the human P-glycoprotein (ABCB1) Biochem. Pharmacol. 2006;72(2):145–155. doi: 10.1016/j.bcp.2006.04.008. [DOI] [PubMed] [Google Scholar]
- 58.Nakamura K, Niimi M, Niimi K, et al. Functional expression of Candida albicans drug efflux pump Cdr1p in a Saccharomyces cerevisiae strain deficient in membrane transporters. Antimicrob. Agents Chemother. 2001;45(12):3366–3374. doi: 10.1128/AAC.45.12.3366-3374.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shukla S, Saini P, Smriti, Jha S, Ambudkar SV, Prasad R. Functional characterization of Candida albicans ABC transporter Cdr1p. Eukaryot. Cell. 2003;2(6):1361–1375. doi: 10.1128/EC.2.6.1361-1375.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lacka I, Konieczny MT, Bulakowska A, et al. Chemosensitization of multidrug resistant Candida albicans by the oxathiolone fused chalcone derivatives. Front. Microbiol. 2015;6:783. doi: 10.3389/fmicb.2015.00783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Li XC, Jacob MR, Ding Y, et al. Capisterones A and B, which enhance fluconazole activity in Saccharomyces cerevisiae, from the marine green alga Penicillus capitatus . J. Nat. Prod. 2006;69(4):542–546. doi: 10.1021/np050396y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schuetzer-Muehlbauer M, Willinger B, Egner R, Ecker G, Kuchler K. Reversal of antifungal resistance mediated by ABC efflux pumps from Candida albicans functionally expressed in yeast. Int. J. Antimicrob. Agents. 2003;22(3):291–300. doi: 10.1016/s0924-8579(03)00213-9. [DOI] [PubMed] [Google Scholar]
- 63.Cannon R, Sklar LA, Lindsley C. Phenotypic HTS multiplex for anti-fungal efflux pump inhibitors. PubChem Bioassay Screening Summary; PubChem AID 485335. 2010. http://pubchem.ncbi.nlm.nih.gov/assay/assay.cgi?aid=485335
- 64.NIH Molecular Libraries Small Molecule Repository Home Page. https://mlsmr.evotec.com/MLSMR_HomePage/index.html
- 65.Prestwick Chemical Library®. www.prestwickchemical.com/prestwick-chemical-library.html 220 Boulevard Gonthier d'Andernach Parc d'innnovation, 67400 ILLKIRCH, France.
- 66.Digirolamo JA, Li XC, Jacob MR, Clark AM, Ferreira D. Reversal of fluconazole resistance by sulfated sterols from the marine sponge Topsentia sp . J. Nat. Prod. 2009;72(8):1524–1528. doi: 10.1021/np900177m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Hiraga K, Wanigasekera A, Sugi H, Hamanaka N, Oda K. A novel screening for inhibitors of a pleiotropic drug resistant pump, Pdr5, in Saccharomyces cerevisiae . Biosci. Biotechnol. Biochem. 2001;65(7):1589–1595. doi: 10.1271/bbb.65.1589. [DOI] [PubMed] [Google Scholar]; • Screening in S. cerevisiae using plasmid-based target expression.
- 68.Hiraga K, Yamamoto S, Fukuda H, Hamanaka N, Oda K. Enniatin has a new function as an inhibitor of Pdr5p, one of the ABC transporters in Saccharomyces cerevisiae . Biochem. Biophys. Res. Commun. 2005;328(4):1119–1125. doi: 10.1016/j.bbrc.2005.01.075. [DOI] [PubMed] [Google Scholar]
- 69.Lee MD, Galazzo JL, Staley AL, et al. Microbial fermentation-derived inhibitors of efflux-pump-mediated drug resistance. Farmaco. 2001;56(1–2):81–85. doi: 10.1016/s0014-827x(01)01002-3. [DOI] [PubMed] [Google Scholar]; • Early use of homologous pump overexpression in inhibitor screens.
- 70.Wanigasekera A, Hiraga K, Hamanaka N, Oda K. Purification and some properties of an inhibitor for a yeast pleiotropic drug resistant pump from Kitasatospora sp. E-420. Biosci. Biotechnol. Biochem. 2001;65(10):2353–2357. doi: 10.1271/bbb.65.2353. [DOI] [PubMed] [Google Scholar]
- 71.Yamamoto S, Hiraga K, Abiko A, Hamanaka N, Oda K. A new function of isonitrile as an inhibitor of the Pdr5p multidrug ABC transporter in Saccharomyces cerevisiae . Biochem. Biophys. Res. Commun. 2005;330(2):622–628. doi: 10.1016/j.bbrc.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 72.Lacka I, Konieczny MT, Bulakowska A, Rzymowski T, Milewski S. Antifungal action of the oxathiolone-fused chalcone derivative. Mycoses. 2011;54(5):e407–e414. doi: 10.1111/j.1439-0507.2010.01936.x. [DOI] [PubMed] [Google Scholar]
- 73.Kolaczkowski M, Kolaczkowska A, Motohashi N, Michalak K. New high-throughput screening assay to reveal similarities and differences in inhibitory sensitivities of multidrug ATP-binding cassette transporters. Antimicrob. Agents Chemother. 2009;53(4):1516–1527. doi: 10.1128/AAC.00956-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Sharma M, Manoharlal R, Shukla S, et al. Curcumin modulates efflux mediated by yeast ABC multidrug transporters and is synergistic with antifungals. Antimicrob. Agents Chemother. 2009;53(8):3256–3265. doi: 10.1128/AAC.01497-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Singh P, Kaur J, Yadav B, Komath SS. Design, synthesis and evaluations of acridone derivatives using Candida albicans – search for MDR modulators led to the identification of an anti-candidiasis agent. Bioorg. Med. Chem. 2009;17(11):3973–3979. doi: 10.1016/j.bmc.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 76.Ivnitski-Steele I, Holmes AR, Lamping E, Monk BC, Cannon RD, Sklar LA. Identification of Nile red as a fluorescent substrate of the Candida albicans ATP-binding cassette transporters Cdr1p and Cdr2p and the major facilitator superfamily transporter Mdr1p. Anal. Biochem. 2009;394(1):87–91. doi: 10.1016/j.ab.2009.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Fluorescent substrate of multiple efflux pumps.
- 77.Szczepaniak J, Lukaszewicz M, Krasowska A. Detection of inhibitors of Candida albicans Cdr transporters using a diS-C3(3) fluorescence. Front. Microbiol. 2015;6:176. doi: 10.3389/fmicb.2015.00176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Campbell BC, Chan KL, Kim JH. Chemosensitization as a means to augment commercial antifungal agents. Front. Microbiol. 2012;3:79. doi: 10.3389/fmicb.2012.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hartland CL, Youngsaye W, Dockendorff C, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD: 2010. Identification of small molecules that selectively inhibit fluconazole-resistant Candida albicans in the presence of fluconazole but not in its absence – Probe 3. [PubMed] [Google Scholar]
- 80.Hartland CL, Youngsaye W, Morgan B, et al. Probe Reports from the NIH Molecular Libraries Program. Bethesda, MD, USA: 2010. Identification of small molecules that selectively inhibit fluconazole-resistant Candida albicans in the presence of fluconazole but not in its absence - Probe 2. [PubMed] [Google Scholar]
- 81.Youngsaye W, Vincent B, Hartland CL, et al. Piperazinyl quinolines as chemosensitizers to increase fluconazole susceptibility of Candida albicans clinical isolates. Bioorg. Med. Chem. Lett. 2011;21(18):5502–5505. doi: 10.1016/j.bmcl.2011.06.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.LaFleur MD, Lucumi E, Napper AD, Diamond SL, Lewis K. Novel high-throughput screen against Candida albicans identifies antifungal potentiators and agents effective against biofilms. J. Antimicrob. Chemother. 2011;66(4):820–826. doi: 10.1093/jac/dkq530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lafleur MD, Sun L, Lister I, et al. Potentiation of azole antifungals by 2-adamantanamine. Antimicrob. Agents Chemother. 2013;57(8):3585–3592. doi: 10.1128/AAC.00294-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Tanabe K, Lamping E, Nagi M, et al. Chimeras of Candida albicans Cdr1p and Cdr2p reveal features of pleiotropic drug resistance transporter structure and function. Mol. Microbiol. 2011;82(2):416–433. doi: 10.1111/j.1365-2958.2011.07820.x. [DOI] [PubMed] [Google Scholar]
- 85.Zhang L, Yan K, Zhang Y, et al. High-throughput synergy screening identifies microbial metabolites as combination agents for the treatment of fungal infections. Proc. Natl Acad. Sci. USA. 2007;104(11):4606–4611. doi: 10.1073/pnas.0609370104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kolaczkowski M, Michalak K, Motohashi N. Phenothiazines as potent modulators of yeast multidrug resistance. Int. J. Antimicrob. Agents. 2003;22(3):279–283. doi: 10.1016/s0924-8579(03)00214-0. [DOI] [PubMed] [Google Scholar]
- 87.Ricardo E, Costa-de-Oliveira S, Dias AS, Guerra J, Rodrigues AG, Pina-Vaz C. Ibuprofen reverts antifungal resistance on Candida albicans showing overexpression of CDR genes. FEMS Yeast Res. 2009;9(4):618–625. doi: 10.1111/j.1567-1364.2009.00504.x. [DOI] [PubMed] [Google Scholar]
- 88.Sharma M, Prasad R. The quorum-sensing molecule farnesol is a modulator of drug efflux mediated by ABC multidrug transporters and synergizes with drugs in Candida albicans . Antimicrob. Agents Chemother. 2011;55(10):4834–4843. doi: 10.1128/AAC.00344-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tanabe K, Lamping E, Adachi K, et al. Inhibition of fungal ABC transporters by unnarmicin A and unnarmicin C, novel cyclic peptides from marine bacterium. Biochem. Biophys. Res. Commun. 2007;364(4):990–995. doi: 10.1016/j.bbrc.2007.10.110. [DOI] [PubMed] [Google Scholar]
- 90.Limtrakul P, Anuchapreeda S, Buddhasukh D. Modulation of human multidrug-resistance MDR-1 gene by natural curcuminoids. BMC Cancer. 2004;4:13. doi: 10.1186/1471-2407-4-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Schmid D, Ecker G, Kopp S, Hitzler M, Chiba P. Structure–activity relationship studies of propafenone analogs based on P-glycoprotein ATPase activity measurements. Biochem. Pharmacol. 1999;58(9):1447–1456. doi: 10.1016/s0006-2952(99)00229-4. [DOI] [PubMed] [Google Scholar]
- 92.Liu XL, Tee HW, Go ML. Functionalized chalcones as selective inhibitors of P-glycoprotein and breast cancer resistance protein. Bioorg. Med. Chem. 2008;16(1):171–180. doi: 10.1016/j.bmc.2007.10.006. [DOI] [PubMed] [Google Scholar]
- 93.Dornetshuber R, Heffeter P, Sulyok M, et al. Interactions between ABC-transport proteins and the secondary Fusarium metabolites enniatin and beauvericin. Mol. Nutr. Food Res. 2009;53(7):904–920. doi: 10.1002/mnfr.200800384. [DOI] [PubMed] [Google Scholar]
- 94.Suzuki K, Saito K, Tsujimura S, et al. Tacrolimus, a calcineurin inhibitor, overcomes treatment unresponsiveness mediated by P-glycoprotein on lymphocytes in refractory rheumatoid arthritis. J. Rheumatol. 2010;37(3):512–520. doi: 10.3899/jrheum.090048. [DOI] [PubMed] [Google Scholar]
- 95.Hayama K, Ishibashi H, Ishijima SA, et al. A D-octapeptide drug efflux pump inhibitor acts synergistically with azoles in a murine oral candidiasis infection model. FEMS Microbiol. Lett. 2012;328(2):130–137. doi: 10.1111/j.1574-6968.2011.02490.x. [DOI] [PubMed] [Google Scholar]; •• In vivo efficacy of fungal efflux pump inhibitor.
- 96.Sanglard D, Ischer F, Monod M, Bille J. Susceptibilities of Candida albicans multidrug transporter mutants to various antifungal agents and other metabolic inhibitors. Antimicrob. Agents Chemother. 1996;40(10):2300–2305. doi: 10.1128/aac.40.10.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wirsching S, Moran GP, Sullivan DJ, Coleman DC, Morschhauser J. MDR1-mediated drug resistance in Candida dubliniensis . Antimicrob. Agents Chemother. 2001;45(12):3416–3421. doi: 10.1128/AAC.45.12.3416-3421.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Vachova L, Stovicek V, Hlavacek O, et al. Flo11p, drug efflux pumps, and the extracellular matrix cooperate to form biofilm yeast colonies. J. Cell Biol. 2011;194(5):679–687. doi: 10.1083/jcb.201103129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gaskova D, Cadek R, Chaloupka R, Vacata V, Gebel J, Sigler K. Monitoring the kinetics and performance of yeast membrane ABC transporters by diS-C3(3) fluorescence. Int. J. Biochem. Cell Biol. 2002;34(8):931–937. doi: 10.1016/s1357-2725(02)00013-4. [DOI] [PubMed] [Google Scholar]
- 100.Edwards BS, Sklar LA. Flow cytometry: impact on early drug discovery. J. Biomol. Screen. 2015;20(6):689–707. doi: 10.1177/1087057115578273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Tegos GP, Evangelisti AM, Strouse JJ, Ursu O, Bologa C, Sklar LA. A high throughput flow cytometric assay platform targeting transporter inhibition. Drug Discov. Today Technol. 2014;12:e95–e103. doi: 10.1016/j.ddtec.2014.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Use of flow cytometry in HTS for pump inhibitors.
- 102.Young SM, Bologa C, Prossnitz ER, Oprea TI, Sklar LA, Edwards BS. High-throughput screening with HyperCyt flow cytometry to detect small molecule formylpeptide receptor ligands. J. Biomol. Screen. 2005;10(4):374–382. doi: 10.1177/1087057105274532. [DOI] [PubMed] [Google Scholar]
- 103.Sandberg M, Maattanen A, Peltonen J, Vuorela PM, Fallarero A. Automating a 96-well microtitre plate model for Staphylococcus aureus biofilms: an approach to screening of natural antimicrobial compounds. Int. J. Antimicrob. Agents. 2008;32(3):233–240. doi: 10.1016/j.ijantimicag.2008.04.022. [DOI] [PubMed] [Google Scholar]
- 104.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4(2):67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 105.Goffeau A, Dufour JP. Plasma membrane ATPase from the yeast Saccharomyces cerevisiae . Methods Enzymol. 1988;157:528–533. doi: 10.1016/0076-6879(88)57101-x. [DOI] [PubMed] [Google Scholar]; • Method for isolating yeast plama membranes for secondary screens.
- 106.Silva LV, Sanguinetti M, Vandeputte P, Torelli R, Rochat B, Sanglard D. Milbemycins: more than efflux inhibitors for fungal pathogens. Antimicrob. Agents Chemother. 2013;57(2):873–86. doi: 10.1128/AAC.02040-12. [DOI] [PMC free article] [PubMed] [Google Scholar]