Abstract
Bone destruction is a hallmark of myeloma and affects 80% of patients. Myeloma cells trigger bone destruction by activating osteoclasts. We investigated the mechanism by which myeloma cells are regulated to do so. We found that C-reactive protein (CRP), a protein secreted in increased amounts by hepatocytes in response to myeloma-derived cytokines, activated myeloma cells to promote osteoclastogenesis and bone destruction in vivo. In mice bearing human bone grafts and injected with multiple myeloma cells, CRP bound to surface CD32/FcγRII on myeloma cells, which activated p38MAPK-Twist pathway and enhanced the cells’ secretion of osteolytic cytokines. Furthermore, analysis of clinical samples from newly diagnosed myeloma patients revealed a highly positive correlation between the level of serum CRP and the number of osteolytic bone lesions. These findings establish a mechanism by which myeloma cells are activated to promote bone destruction and suggest that CRP may be targeted to prevent or treat myeloma-associated bone disease in patients.
Introduction
More than 80% of patients with multiple myeloma develop bone disease or destruction that causes pathological fractures, severe bone pain, spinal cord compression, and hypercalcemia (1, 2). In healthy adults, bone is a dynamic tissue that is constantly being remodeled by bone-resorbing osteoclasts (OCs) and bone-forming osteoblasts. In patients with myeloma, bone destruction results from increased OC-mediated bone resorption and decreased osteoblast-mediated bone formation. In particular, the resorbed bone that usually occurs in close proximity to myeloma cells is greatly enhanced and rarely heals (3). OCs arise from hematopoietic monocytic precursors. The formation of OCs requires the presence of soluble cytokines such as receptor activator of nuclear factor κ-B ligand (RANKL) and macrophage colony-stimulating factor (M-CSF), which are produced primarily from bone marrow stromal cells. Myeloma cells enhance osteoclastogenesis by producing a number of cytokines such as RANKL, macrophage inflammatory protein (MIP)-1α, and monocyte chemoattractant protein (MCP)-1, which can increase OC differentiation and bone resorption activity (4, 5). However, the mechanism underlying how and what activates myeloma cells to produce these cytokines remains unknown.
C-reactive protein (CRP) is an ancient and highly conserved protein of the pentraxin family. It has five identical subunits forming a planar ring that confers very high stability to the protein. In healthy young adults, the median concentration of CRP is 0.8 mg/L, with a range of 0 to 6 mg/L, but following an acute-phase stimulus, values may increase 10,000-fold, from less than 50 μg/L to more than 500 mg/L (6). Plasma CRP is produced primarily in the liver, synthesized by hepatocytes in response to intermediary inflammatory cytokines such as interleukin (IL)-1β and IL-6. CRP can bind to a variety of ligands, including pneumococcal polysaccharides, membrane phospholipids, apoptotic cells, fibronectin, and ribonuclear particles (6). It also binds to C1q and directly activates the classical complement cascade, or binds to FcγRs, leading to indirect (via classical complement) and direct opsonization (via FcγRs) (7). Through these mechanisms, CRP plays a direct role in a wide range of inflammatory processes and contributes to innate host immunity (8).
Increased levels of CRP are also present in many diseases, including malignancies such as myeloma (9–11), lymphoma (12, 13), and carcinoma (14). High levels of circulating CRP correlate with poor prognosis in myeloma (9, 11) and lymphoma (12). Our previous study showed that CRP enhanced myeloma cell proliferation under stressed conditions and protected myeloma cells from chemotherapy drug-induced apoptosis in a model of human myeloma (15).
Here, through a combination of in vitro, in vivo, and patient studies, we report that CRP has a unique role in myeloma-induced bone disease. Our results show that human CRP binds to CD32/FcγRII on myeloma cells, activates downstream signaling through the mitogen-activated protein kinase (MAPK) p38 and the transcription factor Twist, and promotes the production of osteolytic cytokines from myeloma cells, leading to enhanced myeloma cell-mediated OC differentiation and severe bone resorption in vivo.
Results
CRP promotes lytic bone lesions in myeloma-bearing mice
To determine the effect of CRP on bone destruction under physiological conditions, we injected human recombinant CRP, serum amyloid P component (SAP; an inactive analog of CRP), or PBS (vehicle) into tumor-free SCID or SCID-hu mice, in which the SCID mice were implanted with human bone chips. None caused bone destruction in the implanted human bones in the mice (Figure 1A; left panels).
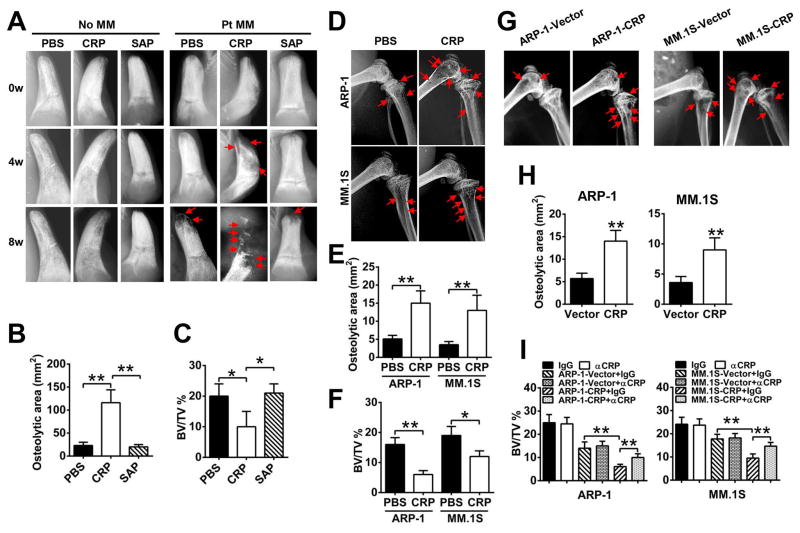
Figure 1. CRP enhances induction of bone lesions in myeloma-bearing mice.
(A) Representative radiographic images of lytic lesions in the implanted human bones of primary myeloma-bearing SCID-hu mice before (0w) or at week 4 (4w) and week 8 (8w) after injection of primary myeloma cells from 1 of 5 patients (Pt MM) and treatment with PBS, CRP (20 μg/mouse) or SAP (20 μg/mouse). Tumor-free mice (no MM) served as controls. (B and C) Histomorphometric analysis of the osteolytic area (B) and bone volume density [(C); assessed as percentage of bone volume over total volume (BV/TV)] in the implanted human bones from the mice described in (A) at 8 weeks. (D) Representative radiographic images of lytic bone lesions in the distal femurs of SCID mice injected with one of two myeloma cell lines (ARP-1 or MM.1S) and treated with PBS or CRP. (n = 10 mice per group). (E and F) Histomorphometric quantitative analysis of the femurs from the mice described in (D), assessing osteolytic area (E) and BV/TV (F). (G and H) Representative radiographic images of bone lesions (G) and histomorphometric quantitative analysis of osteolytic area (H) in the distal femurs of SCID mice (n=10 per group) injected with CRP-expressing or vector-control myeloma cells. Arrows indicate osteolytic lesions in mouse femurs. (I) Micro-CT scanning analysis of BV/TV in the femurs of SCID mice bearing CRP-expressing or vector-control myeloma cells, treated with neutralizing antibodies against CRP or control (IgG). *P < 0.05, **P < 0.01 by student’s t test. Images are representative and data are means ± SD of three independent experiments are shown.
We then examined the effects of human CRP on bones in SCID mice xenografted with human myeloma cells. CRP, SAP, or PBS was injected into SCID-hu mice bearing primary human myeloma cells or SCID mice bearing myeloma cell lines. Lytic bone lesions were detected in the implanted human bone chips of SCID-hu mice 8 weeks after tumor inoculation (Figure 1A; right panels), whereas in mice injected with CRP but not SAP or PBS, lytic bone lesions were detected at week 4 after tumor injection. CRP not only accelerated the appearance of bone lesions but also caused more bone destruction, evident by the greater number of lytic bone lesions (Figure 1A; right panels), larger lytic area (Figure 1B), and lower trabecular bone volume (Figure 1C) compared with controls. Similarly, CRP also resulted in more bone lesions in murine femurs of SCID mice bearing human myeloma cells (Figure 1, D–F). The earlier appearance and larger number of bone lesions in CRP-injected myeloma-bearing mice were not due to higher tumor burden because no difference in tumor burden, detected as circulating human IgG secreted by myeloma cells, was detected in myeloma-bearing SCID-hu (Figure S1A) or SCID (Figure S1B) mice injected with or without CRP, indicating that CRP affected not myeloma growth but rather bone destruction in vivo. We injected 20 μg CRP per mouse twice a week continuously for 8 weeks, which maintained CRP concentrations of 3–8 μg/mL in mouse serum (Figure S1, C and D). Mean values of serum CRP in myeloma patients reported for different studies range from 3.8 – 12.0 mg/L, with measured values ranging from 1.5–120 mg/L (9, 11, 16). Therefore, the CRP concentrations in the mice were comparable to those seen in myeloma patients.
To confirm the results, we generated human myeloma cell lines (ARP-1 and MM.1S) that stably produced (Figure S2A) and secreted (Figure S2B) human CRP protein. After myeloma was established in SCID mice, these cells also caused more lytic bone lesions in the femurs of SCID mice as compared with control myeloma cells (Figure 1, G and H). Injection of a neutralizing antibody against human CRP diminished the ability of CRP-producing, but not control, myeloma cells to cause lytic bone lesion in the mice (Figure 1I). Because these CRP-expressing myeloma cells secreted much lower amounts of CRP than those observed in myeloma patients, it is possible that this autocrine CRP, although at low concentrations but constantly secreted, may accumulate on myeloma cells to effectively bind with the surface receptors to activate signaling in myeloma cells. Thus, these results indicate that human CRP is capable of promoting myeloma cell-mediated lytic bone disease in vivo.
CRP enhances OC differentiation and bone resorption activity
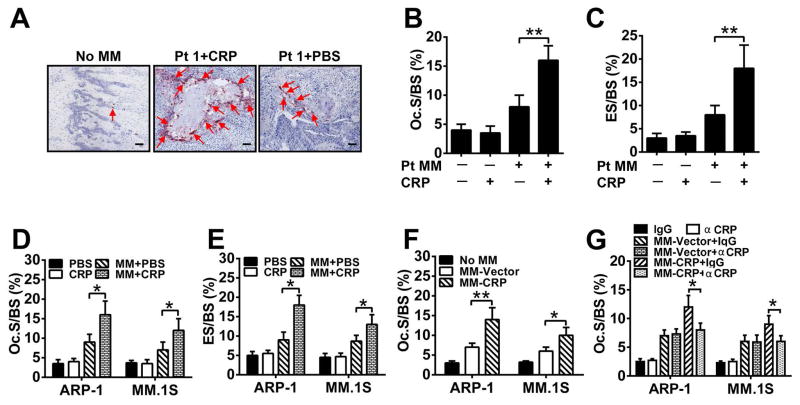
Myeloma manifests as an osteolytic bone disease mainly due to enhanced OC activity (17). To determine whether CRP affected osteoclastogenesis, primary myeloma-bearing mice were sacrificed 8 weeks after tumor injection, and sections of murine femurs or implanted human bone chips were stained for Tartrate-resistant acid phosphatase (TRAP) to identify OCs. CRP injection resulted in significantly more TRAP+ OCs localized on the trabecular bone surface of the bone-tumor interface (Figure 2A, 2B) and more erosion on trabecular bones (Figure 2C) in the implanted human bones of myeloma-bearing SCID-hu mice compared with controls. Similarly, more OCs (Figure 2D) or bone erosion (Figure 2E) were found in the femurs of myeloma-bearing, CRP-injected SCID mice than those injected with PBS, and in mice bearing CRP-producing myeloma cells than those injected with control myeloma cells (Figure 2F). Injection of CRP-neutralizing antibodies but not control IgG abrogated the enhanced bone destruction produced by CRP-producing myeloma cells (Figure 2G).
Figure 2. CRP enhances osteoclastogenesis in myeloma-bearing mice.
(A) Representative images of TRAP+ OCs in human trabecular bones from SCID-hu mice or 8 weeks after inoculation with patient myeloma cells (Pt 1) and treated with CRP or PBS. Arrows indicate OCs in bone sections. No MM, tumor-free mice. Scale bar, 10 μM. (B and C) Histomorphometric quantitative analysis of (B) the number of OCs on the bone surface (Oc. S/BS) and (C) OC-mediated erosion of bone surface, expressed as a percentage of total bone surface (ES/BS), in the implanted human bones of SCID-hu mice that were either tumor-free (−) or bearing primary myeloma cells from one of five patients (Pt MM) and treated with CRP (+) or PBS (−). (D and E) Histomorphometric analysis of (D) Oc. S/BS and (E) ES/BS in the femurs of SCID mice (n = 10 per group) bearing myeloma cells and treated with CRP or PBS. (F) Oc. S/BS in the femurs of SCID mice bearing CRP-expressing (MM-CRP) or vector-control (MM-Vector) cells. (G) Oc. S/BS in the femurs of SCID mice bearing MM-CRP or MM-Vector cells treated with antibodies against CRP (αCRP) or control (IgG). Tumor-free mice (No MM) treated with agents or antibodies served as controls for baseline levels. Data from three independent experiments are shown. *P < 0.05, ** P < 0.01 by student’s t test.
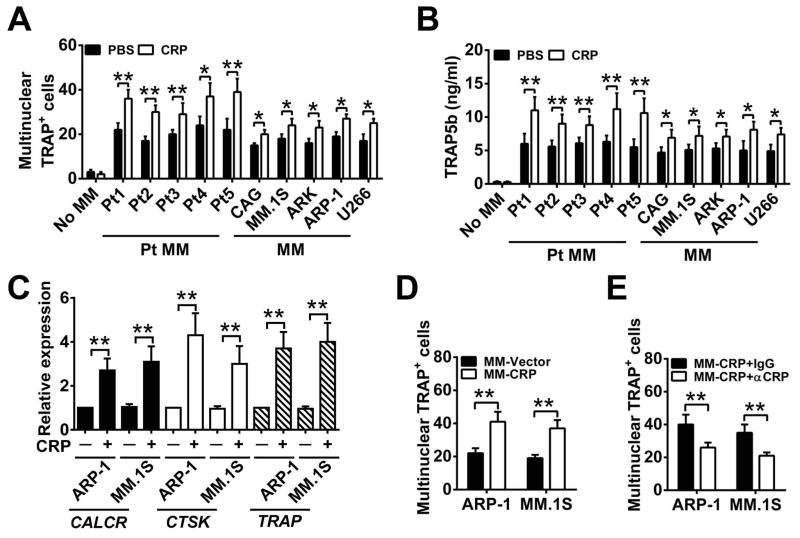
Because the in vivo findings suggested that CRP affected osteoclastogenesis in myeloma-bearing mice, we investigated the role of CRP in OC differentiation in vitro. We observed that adding CRP (5 μg/mL, equal to the average serum levels of CRP in myeloma patients) to cultured OC precursor cells did not induce the formation of mature (multinuclear, TRAP+) OCs (Figure 3A) or increase the levels of TRAP5b (Figure 3B), which is an active component of TRAP, is secreted only by mature OCs, and the level of which reflects OC-mediated bone resorption activity (18). However, coculture of OC precursors with myeloma cells, without addition of RANKL in the medium, induced formation of mature OCs, and addition of CRP (5 μg/mL) to the coculture further and significantly enhanced the generation of mature OCs (Figure 3A, 3B). In line with these results, the expression of OC-associated genes, such as TRAP and those encoding calcitonin receptor (CALCR) and cathepsin K (CTSK) (19), were highly enhanced in OCs cocultured with myeloma cells in the presence of CRP (Figure 3C) as compared with controls. Coculturing OC precursors with CRP-producing human myeloma cells also enhanced mature OC formation (Figure 3D). Once again, neutralizing CRP in the coculture abrogated CRP-induced, myeloma-mediated OC formation (Figure 3E). Together, these results clearly indicate that CRP enhances myeloma cell-mediated osteoclastogenesis in vitro.
Figure 3. CRP enhances myeloma-induced OC formation and activity in vitro.
(A and B) The number of multinuclear TRAP+ cells (A) and the amount of secreted TRAP5b (B) in cultures of OC precursor cells cultured alone or with either primary myeloma cells isolated from five myeloma patients (Pt MM) or with human myeloma cell lines (MM) in medium without RANKL in the presence of CRP (5 μg/mL) or PBS. (C) Real-time PCR analysis of the expression of CALCR, CTSK, and TRAP in mature OCs cocultured with OC precursors and ARP-1 or MM.1S cells with or without CRP (5 μg/mL). (D) TRAP staining for the number of multinuclear TRAP+ cells in cocultures of OC precursors with vector- or CRP-expressing ARP-1 or MM.1S cells (MM-Vector or MM-CRP) without RANKL. (E) As in (D), in the presence of antibodies against CRP (αCRP; 5 μg/mL) or control IgG. *P < 0.05, **P < 0.01 by student’s t test. Data from four independent experiments are shown.
CRP stimulates the production of OC activators by myeloma cells
Because our results showed that addition of CRP to the transwell coculture of OC precursors and myeloma cells induced OC differentiation (Figure 3), we hypothesized that CRP mediates this effect by upregulating myeloma cell secretion of cytokines that activate OC differentiation. Microarray analysis of gene profiling identified more than 500 genes with 2-fold increased expression in CRP-treated myeloma cells than in PBS-treated cells, including 11 that encode cytokines known to activate osteoclastogenesis: IL-1, MCP-1, IL-11, IL-6, HGF, IL-17, RANKL, M-CSF, BAFF, DcR3, and MIP-1α (Figure S3A). Real-time PCR validated that among these cytokines, RANKL, MCP-1 and MIP-1α were highly expressed in CRP-treated myeloma cells, including patient-derived primary myeloma cells and myeloma cell lines ARP-1 and MM.1S, as compared with control myeloma cells (Figure S3B). Not surprisingly, CRP-expressing ARP-1 and MM.1S cells also expressed a higher level of these cytokines as compared with vector-control myeloma cells (Figure S3C).
To determine the involvement of these cytokines in CRP-enhanced OC differentiation, we added neutralizing antibodies against these cytokines to the coculture of OC precursors and myeloma cells in the presence of CRP. The neutralizing antibodies significantly reduced the number of TRAP+ multinuclear OCs (Figure S4A) and level of TRAP5b (Figure S4B). These results indicate that CRP enhances OC differentiation by upregulating myeloma cell production of OC activators such as RANKL, MCP-1, and MIP-1α.
CRP binds to CD32 on myeloma cells, promotes cytokine production, and induces OC differentiation
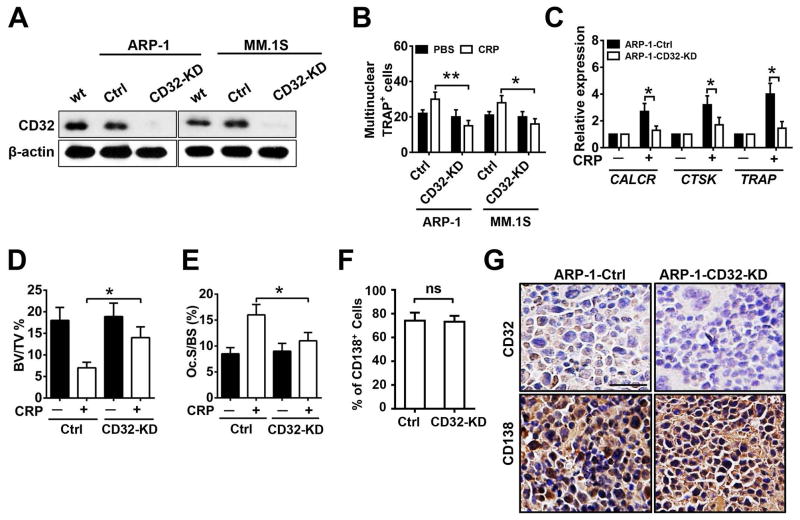
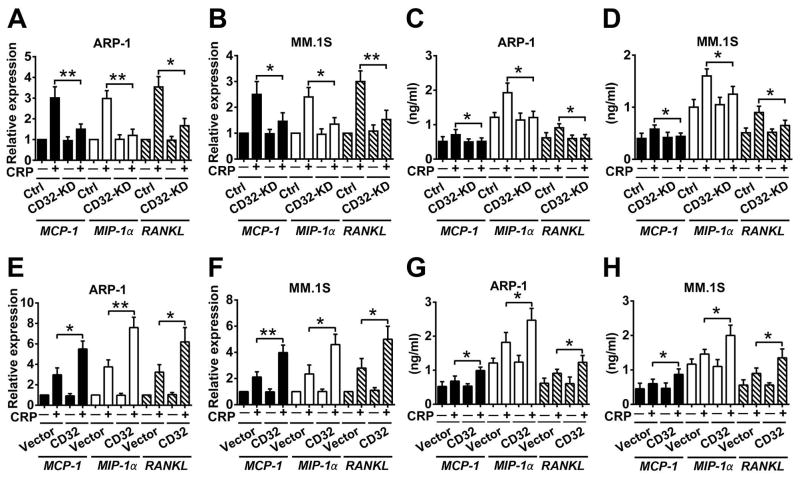
Because CRP has been shown to bind to FcγRs such as CD32 on human cells including myeloma cells (7, 15), we hypothesized that CD32 expressed on myeloma cells may be involved in CRP-induced myeloma-mediated OC differentiation. We generated stable myeloma cell lines (ARP-1 and MM.1S) with significantly reduced expression of CD32 (herein called CD32-KD; Figure 4A). CD32-KD myeloma cells were less able to promote OC differentiation in response to CRP (Figure 4, B and C). Moreover, CD32-KD myeloma cells caused less bone disease in mice injected with CRP (Figure 4, D and E) although similar numbers or percentages of CD32-KD and control MM cells were found in the BM of mice (Figure 4F). As expected, CD32-KD myeloma cells had lower CD32 expression in vivo compared with control cells (Figure 4G). We then added CRP to cultures of CD32-KD or control ARP-1 or MM.1S cells. The CD32-KD myeloma cells had significantly reduced expression (Figure 5, A and B) and secretion (Figure 5, C and D) of MCP-1, MIP-1α and RANKL in response to CRP as compared with control cells.
Figure 4. Knockdown of CD32 in myeloma cells reduces CRP-induced OC differentiation.
(A) Western blot analysis for the abundance of CD32 in cultured wild-type (wt) ARP-1 or MM.1S cells and those transfected with non-targeted shRNA (Ctrl) or CD32 shRNA (CD32-KD). β-actin served as loading control. (B) The number of multinuclear TRAP+ cells in cocultures of OC precursors with control or CD32-deficient myeloma cells in the presence of CRP (5 μg/mL) or PBS. (C) Real-time PCR of gene expression in OCs cocultured with control or CD32-deficient ARP-1 myeloma cells and CRP (5 μg/mL) or PBS. (D to G) Histomorphometric analysis of (D) bone volume (BV/TV), (E) the number of OCs present on the bone surface (Oc. S/BS), and immunohistochemcial examination for (F) the percentage of CD138+ cells (myeloma marker) in the bone marrow, and (G) staining for CD32 and CD138 by myeloma cells in bone sections of distal femurs from SCID mice (n=10 per group) injected with Ctrl or CD32-KD ARP-1 cells. Mice receiving no CRP injection served as control in (D) and (E). Scale bar, 50 μM. *P < 0.05, ** P < 0.01 by student’s t test. Data from three independent experiments are shown.
Figure 5. Through myeloma cell CD32, CRP enhances the production of cytokines that mediate OC formation and activation.
(A to D) Real-time PCR analysis of MCP-1, MIP-1α, and RANKL mRNA expression (A, B) and ELISA analysis of secreted protein (C, D) in CD32-knockdown (CD32-KD) ARP-1 or MM.1S cells cultured with CRP (5 μg/mL), compared with those in non-target control shRNA-transfected (Ctrl) cells. (E to H) Analysis of cytokine (MCP-1, MIP-1α, and RANKL) abundance at the mRNA (E, F) and protein (G, H) levels in cultures of vector- or CD32-transfected (CD32) ARP-1 or MM.1S cells cultured with or without CRP (5 μg/mL). *P < 0.05, ** P < 0.01 by student’s t test. Data from three independent experiments are shown.
Because CD32 is involved in various cellular responses, we wanted to exclude the possibility that knockdown of CD32 in myeloma cells affected myeloma cell growth and survival, and thereby compromised their ability to secrete osteolytic cytokines that enhance OC differentiation. Our results showed that knockdown of CD32 did not affect myeloma cell growth (Figure S5A) or survival (Figure S5B) in vitro, or tumor formation and burden in vivo (Figure S5C).
We also generated stable myeloma cell lines with significantly enhanced CD32 expression (Figure S6, A and B) to confirm the role of this molecule. CD32-overexpressing myeloma cells expressed (Figure 5, E and F) and secreted (Figure 5, G and H) more osteolytic cytokines and displayed enhanced ability to promote OC differentiation in vitro (Figure S6, C and D), which resulted in severe bone disease in vivo (Figure S6, E and F) in the presence of CRP. Overexpression of CD32 did not affect myeloma growth and survival in vitro or tumor formation in vivo.
CRP regulates the expression of osteolytic cytokines in myeloma cells through p38 MAPK-Twist signaling
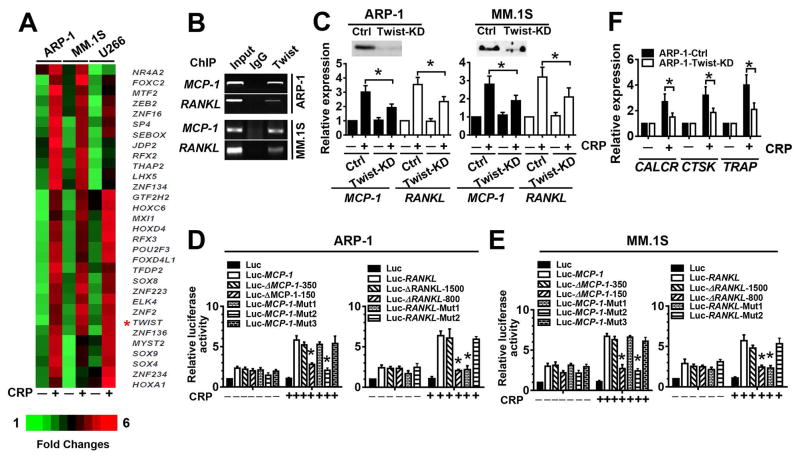
Next, we sought to elucidate the molecular mechanism underlying CRP–CD32-mediated signaling pathways that regulate the production of osteolytic cytokines in myeloma cells. First, we analyzed the expression of transcriptional factors in myeloma cell lines by microarray and identified 31 genes that were at least twofold increase in response to CRP (Figure 6A). We used the software Genomatix (20) to analyze the transcription factor binding sites on the promoter regions of the osteolytic cytokine genes and predicted several potential twist-binding sites. To confirm the results of microarray analysis, we performed a ChIP assay to examine the ability of twist to bind to the promoters of two relevant cytokines (RANKL and MCP-1) genes. We found that twist was enriched on RANKL and MCP-1 promoters in ARP-1 and MM.1S (Figure 6B). To investigate the importance of twist in CRP-induced cytokine production by myeloma cells, we used shRNA to knock down Twist abundance in myeloma cells (Twist-KD, Figure 6C). CRP induction of myeloma cell expression of the osteolytic cytokines MCP-1 and RANKL was significantly reduced in Twist-KD myeloma cells as compared to control cells (Figure 6C). We also cloned the Twist-containing transcription start site of the RANKL and MCP-1 promoters into a luciferase reporter vector. Adding CRP increased the luciferase activity of Luc-RANKL and Luc-MCP-1 in ARP-1 or MM.1S, and truncation of the twist binding sites in the MCP-1 or RANKL gene promoters significantly reduced the activity (Figure 6, D and E), suggesting that the potential twist binding site(s) for RANKL locate at −1500 bp to −800 bp of its start codon, and those for MCP-1 are located at −350 bp to −150 bp of its start codon. Next we performed site-directed mutagenesis on the two putative binding sites for RANKL and three putative binding sites for MCP-1 within each region. Not surprisingly, mutating a single binding site for either RANKL or MCP-1 blocked the transcriptional activation induced by CRP (Figure 6, D and E). Furthermore, CRP-treated twist-KD myeloma cells were less able to induce the expression of OC genes in cocultured OC progenitor cells (Figure 6F), indicating that Twist is crucial to myeloma cell-mediated OC differentiation in response to CRP.
Figure 6. Twist transcriptionally regulates CRP-induced cytokine expression in myeloma cells.
(A) Heat map of the expression of transcriptional factors in myeloma cell lines treated with or without CRP (5 μg/mL) for 24 hours. (B) ChIP assay for the interaction of Twist with the promoters of MCP-1 or RANKL using anti-Twist antibody or rabbit IgG. The input proteins served as controls. (C) Western blot for Twist abundance (top) and real-time PCR for MCP-1 and RANKL mRNAs in Twist-knockdown (KD) myeloma cells and non-targeted shRNA control (Ctrl) cells, cultured with or without CRP. (D and E) Luciferase activity in cultured ARP-1 (D) or MM.1S (E) cells transfected with a luciferase reporter-containing wild-type, truncated mutants, or Twist binding site-mutant RANKL or MCP-1 gene promoters. (F) Real-time PCR for gene expression in OC precursors cocultured with Twist-knockdown or control ARP-1 cells cultured with or without CRP. *P < 0.05, ** P < 0.01 by student’s t test. Data from four independent experiments are shown.
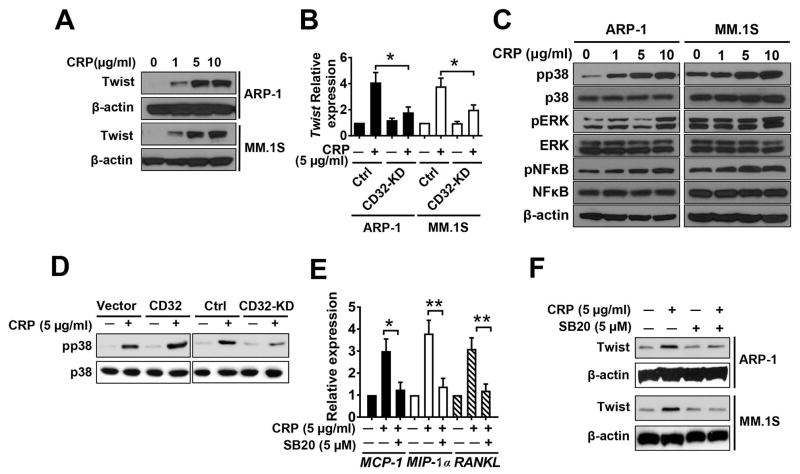
To further examine the effect of CRP on twist expression, we treated ARP-1 or MM.1S cells with various concentrations (1–10 μg/mL) of CRP and assessed twist expression by western blot. CRP enhanced twist expression in a dose-dependent manner (Figure 7A). In contrast, Twist expression was reduced in CRP-treated CD32-KD myeloma cells (Figure 7B). To further investigate the potential downstream targets of Twist, we added various concentrations (1–10 μg/mL) of CRP to cultures of myeloma cells and found that CRP enhanced phosphorylation of p38 MAPK but not ERK or NF-κB in a dose-dependent manner (Figure 7C). CRP promoted the levels of phosphorylated p38 MAPK in control and in CD32-overexpressing myeloma cells, but not in CD32-KD myeloma cells (Figure 7D), indicating that CRP activates the p38 MAPK pathway in myeloma cells through surface CD32. To examine the importance of p38 MAPK in CRP-induced cytokine production in myeloma cells, we added the p38 MAPK-specific inhibitor SB202190 to cultures of myeloma cells along with CRP. SB202190 significantly reduced the expression of osteolytic cytokine mRNAs (Figure 7E) and reduced CRP-induced Twist protein abundance (Figure 7F). These results clearly showed that CRP binds to CD32 and activates p38 MAPK-Twist signaling pathways, leading to increased production of osteolytic cytokines by myeloma cells.
Figure 7. Crosslinking CD32 by CRP activates p38 MAPK-Twist in myeloma cells.
Cells. (A) Western blot for Twist abundance in ARP-1 or MM.1S cells treated with various doses of CRP for 24 hours. (B) Real-time PCR for TWIST mRNA levels in CD32-knockdown (CD32-KD) or control myeloma cells cultured with or without CRP. (C) Western blot analysis for dose-dependent effects of CRP on the phosphorylation (p) of p38 MAPK, ERK, and NF-κB in myeloma cells. (D) Western blot analysis for effects of CRP on the phosphorylation of p38 MAPK in CD32-overexpressing (CD32) or knockdown (CD32-KD) ARP-1 cells compared with the respective controls. (E) Real-time PCR analysis of the expression of MCP-1, MIP-1α and RANKL in ARP-1 cells cultured with or without CRP and the p38 MAPK inhibitor SB202190 (SB20). (F) Western blot analysis for the effect of SB20 on CRP-induced Twist abundance. *P < 0.05, ** P < 0.01 by student’s t test. Data from three independent experiments are shown.
Serum CRP levels significantly and positively correlate with bone lesions in patients with newly diagnosed myeloma
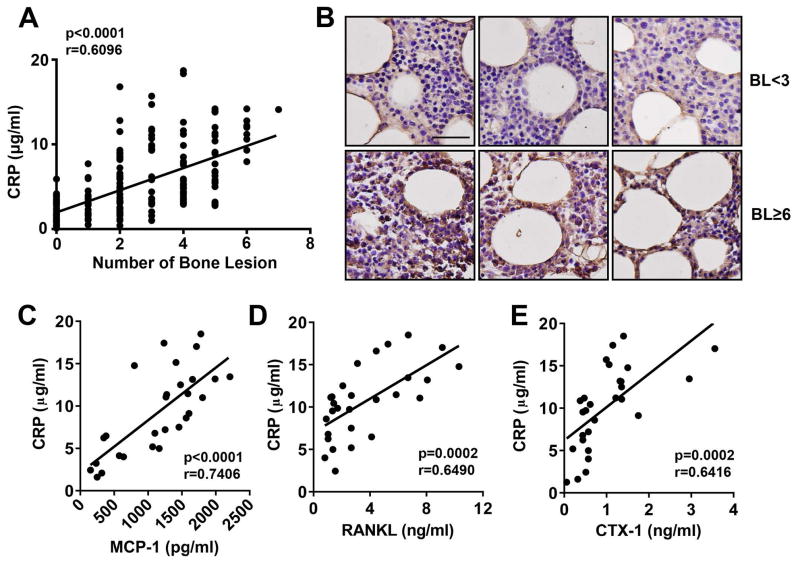
Finally, we examined the relationship between serum CRP and development of bone disease in patients with myeloma. In newly diagnosed myeloma patients, serum CRP was quantified by ELISA, and bone disease and the number of bone lesions were detected by X-ray and MRI. As shown in Figure 8A, serum CRP level significantly and positively correlated with the number of bone lesions (n = 244; r = 0.6096; P < 0.0001). We examined bone marrow biopsies from myeloma patients with high (≥ 6 bone lesions) or low (< 3 bone lesions) numbers of bone lesions, and found that more CRP protein was present in the bone marrow myeloma cells of patients with more bone disease (Figure 8B). Moreover, positive correlations were also found between the levels of serum RANKL (Figure 8C) or MCP-1 (Figure 8D) and CRP level. We quantified serum CTx-1, which reflects the bone resorption activity of OCs, and observed that CTx-1 and CRP level (Figure 8E) also positively correlated. These results are clinically significant because they suggest that high levels of CRP may be a key causative factor in the development of the bone disease observed in 80% of patients with myeloma.
Figure 8. Relationship between serum CRP level and osteolytic bone lesion number in patients with newly diagnosed myeloma.
(A) Correlation between serum CRP level and the number of lytic bone lesions in 244 patients with newly diagnosed myeloma. CRP was measured by ELISA, and bone lesions were detected by radiography (x-ray or MRI). (B) Representative images of immunohistochemical staining for CRP accumulation in the bone marrow of myeloma patients (3 per group) with low (BL < 3) or high (BL ≥ 6) numbers of bone lesions. Scale bar, 50 μM. (C–E) Correlations between the level of circulating CRP and (C) MCP-1, (D) RANKL, and (E) CTx-1 in 28 randomly selected patients with newly diagnosed myeloma, calculated by Pearson’s correlation coefficient analysis.
Discussion
Myeloma is accompanied by bone disease in the vast majority of patients (1, 2). Myeloma cells disrupt the delicate balance between bone formation and resorption, leading to debilitating osteolytic bone lesions (3). Although it is well established that myeloma cells enhance OC differentiation and activity via secreting osteolytic cytokines (17), the mechanism underlying how myeloma cells are regulated and activated to secrete these cytokines remains elusive. In this study we clearly showed that human CRP, which is secreted in elevated amounts by hepatocytes in response to cytokines (IL-1 or IL-6) derived from myeloma cells in patients (6), may be responsible, at least in part, for activating myeloma cells to promote osteoclastogenesis and inducing bone destruction in vivo. CRP binds with surface CD32/FcγRII, activates p38 MAPK-twist pathways, and stimulates the secretion of osteolytic cytokines by myeloma cells. Interestingly, our previous study showed that CRP at a much higher concentration (25 μg/mL) activated ERK and NF-κB but inhibited p38 MAPK (15). Our clinical analyses examining the relationship between the level of serum CRP and the number of osteolytic bone lesions in newly diagnosed patients support this conclusion. Thus, this study reveals a previously unknown mechanism that explains how myeloma cells are activated to promote osteoclastogenesis and induce bone destruction in human myeloma. Our study also suggests that CRP may be a therapeutic target for preventing or treating bone disease in patients with myeloma.
CRP is a sensitive systemic marker of inflammation and tissue damage (6). Its level in the serum is increased in patients with infection, inflammatory disease, necrosis, or malignancy, including myeloma, lymphoma, and carcinoma, and high levels of circulating CRP correlate with poor prognosis in patients with myeloma or lymphoma (9–13). We hypothesized that CRP may not be just a surrogate for tumor burden in myeloma but also plays a central role in myeloma growth, survival, and drug resistance. Indeed, we reported that adding human CRP to myeloma cultures at levels seen in patients with myeloma or other tumors promoted tumor cell proliferation under stressed conditions and protected tumor cells from apoptosis induced by chemotherapy, IL-6 withdrawal, or serum deprivation in vitro and in vivo (15). The results of this study further showed that CRP may be an important player in myeloma-mediated bone destruction in human myeloma. As inflammation has long been hypothesized to be linked to cancer (21), our studies suggest a possible link between inflammation or its products and the pathogenesis and bone destruction in myeloma.
Twist is a basic helix-loop-helix (bHLH) transcriptional factor essential in embryological morphogenesis, and it belongs to the HLH superfamily of proteins, which are involved in a variety of regulatory processes in organisms (22). Initially identified in Drosophila where it is involved in mesodermal patterning and morphogenetic movement (23), Twist is a highly conserved protein and a master regulator of morphogenesis. Previous studies have also shown that Twist is important in bone development and is expressed in primary osteoblastic cells (24) and preosteoblasts (25). In situ analysis of murine embryonic development provided evidence that Twist abundance is reduced during endochondral and intramembranous fetal bone development (26). Twist proteins inhibit skeletal development through direct and indirect inhibition of Runx2 (27, 28), a key regulator of osteogenic differentiation. Finally, expression of TWIST1 and TWIST2 in mesenchymal cells may promote adipogenesis (29). Together, these observations suggest that Twist proteins are important to normal bone marrow stromal cell function. However, Twist also is essential to tumor metastasis. Ectopic expression of Twist results in loss of E-cadherin-mediated cell-cell adhesion, activation of mesenchymal markers, and induction of cell motility; it contributes to metastasis by promoting an epithelial-mesenchymal transition. In human breast cancers, high TWIST expression correlates with the presence of invasive lobular carcinoma (30). The role of Twist in bone destruction in myeloma is unclear. Our results indicate that Twist mediates CRP-induced upregulation of MCP-1, RANKL, and MIP-1α expression in myeloma cells, leading to an enhanced osteoclast-mediated bone resorption. Further studies are needed to elucidate its effect on pathogenesis of myeloma-associated bone disease.
Materials and Methods
Primary myeloma cells and myeloma cell lines
This study was approved by the Institutional Review Board at the University of Texas MD Anderson Cancer Center, and all patients provided written informed consent. Bone marrow aspirates were obtained from newly diagnosed patients with myeloma as part of their routine clinical work-up. Patient CD138+ myeloma cells were isolated from the aspirates by magnetic bead sorting (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany). The myeloma cell lines ARP-1 and ARK were kindly provided by the Arkansas Cancer Research Center, Little Rock, AR. Other cell lines were purchased from the American Type Culture Collection (Rockville, MD). All myeloma cells were cultured in RPMI-1640 medium supplemented with 10% FBS (Life Technologies, Carlsbad, CA).
Antibodies and reagents
Neutralizing antibodies against CRP, MIP-1α, MCP-1, RANKL, and control IgG, and recombinant human CRP, M-CSF, RANKL, and SAP were purchased from R&D Systems (Minneapolis, MN). The p38 MAPK-specific inhibitor was purchased from Axon Medchem BV (Groningen, Netherlands). Except where specified, antibodies for western blotting analysis were purchased from Cell Signaling Technology, MA.
In vitro OC formation and function assays
Human OCs were generated from peripheral blood, CD14 antibody–coated magnetic bead purified (Miltenyi Biotec) monocytes of healthy blood donors, and cultured in α-MEM medium supplemented with 10% of fetal bovine serum (FBS) with 25 ng/ml M-CSF (R&D Systems) for 7 days to obtain the precursors of osteoclasts as we previously reported (5). The precursors were then co-cultured with myeloma cells in medium without or with a low dose of 10 ng/mL RANKL for an additional 7 days to induce mature osteoclast formation. To examine the effects of myeloma cells and CRP on OC differentiation, OC precursors (1 × 105 cells/mL) were seeded in the wells and cocultured with myeloma cells (0.5 × 106 cells/mL) in transwell inserts for 7 days, in OC medium without the addition of RANKL, in the presence or absence of CRP. Cells were fixed with 4% formaldehyde and stained for TRAP by using the Leukocyte Acid Phosphatase Kit (Sigma). At the end of 2-week culture, culture supernatants were collected and used for quantifying by ELISA (CrossLaps; Biosciences Diagnostics) secreted TRAP5b.
Generation of knockdown and overexpression myeloma cells
The shRNAs specific for CD32 or twist were purchased from Santa Cruz Technology (Dallas, TX) according to the manufacturer’s instructions. After lentiviral infection, transduced cells were harvested to examine CD32 or twist expression or used for experiments. The cells with CD32 or twist knockdown were drug-selected. CRP and CD32 overexpression was achieved using the retroviral expression vector pBABEhybro, according to the manufacturer’s instructions.
Western blotting analysis
Cells were harvested and lysed with lysis buffer (50 mM Tris, pH 7.5, 140 mM NaCl, 5 mM EDTA, 5 mM NaN3, 1% Triton-X-100, 1% NP-40, 1 × protease inhibitor cocktail). Cell lysates were subjected to SDS-PAGE, transferred to a polyvinylidene difluoride membrane, and immunoblotted with antibodies against CRP (R&D Systems) and phosphorylated or non-phosphorylated kinases p38 (Cell Signaling Technology) at 4°C overnight. Antibodies were diluted according to the manufacture’s recommendations. The membrane was stripped and reprobed with anti-β-actin antibodies (Sigma) to ensure equal protein loading. Secondary antibodies conjugated to horseradish peroxidase were used for detection and followed by enhanced chemiluminescence (Pierce Biotechnology) and autoradiography.
Real-time PCR
Total RNA was isolated with an RNeasy kit (Qiagen). An aliquot of 1 μg total RNA was subjected to reverse transcription with SuperScript II (Invitrogen) RT PCR kit, and 1 ng of the final cDNA was applied to real-time PCR amplification with SYBRGreen using StepOnePlus real-time PCR system (ABI). The primers used are listed in table S1.
Myeloma cell apoptosis
The fraction of apoptotic cells was determined by staining with fluoroscein isothiocyanate (FITC)-conjugated annexin-V and propidium iodide (PI), and analyzed by flow cytometry (BD LSRFortessa, BD Biosciences) according to manufacturer’s instructions.
Mouse models, radiographic, histology, and bone histomorphometry
CB.17 SCID mice were purchased from Harlan (Indianapolis, IN). All mice were maintained in American Association of Laboratory Animal Care–accredited facilities, and the studies were approved by the Institutional Animal Care and Use Committee of the University of Texas MD Anderson Cancer Center. Six- to eight-week-old mice were injected intravenously with (1 × 106 cells per mouse) human myeloma cell lines ARP-1 and MM.1S, and myeloma-bearing mice were then treated with CRP (20 μg/mouse; twice a week for 6–8 weeks) or vehicle (PBS). Serum was collected from mice daily during the treatment and tested for myeloma-secreted M-proteins (human Ig) or their light chains by ELISA.
SCID-hu hosts were developed as described previously (31). Fetal bones (10-mm pieces, purchased from Advanced Bioscience Resources Inc) were implanted subcutaneously and allowed to become vascularized (6–10 weeks). Purified myeloma cells (≥ 106 cells per mouse) were injected into implanted human bones to establish myeloma (4–6 weeks, n=5 per group). After 7 days, mice were treated with CRP or SAP (20 μg/mouse) or equal volume of PBS twice a week for 8 weeks. Serum was obtained from the mice weekly and assayed by ELISA for the human M-component.
Mice were x-rayed weekly starting at 1 to 2 weeks after tumor injection. To measure the size of lytic bone lesions, radiographs were scanned with a Faxitron X-ray cabinet (Faxitron X-ray). The total number of osteolytic lesions or area was quantified using Image J software (National Institutes of Health). For histological and bone histomorphometric analyses, mice were killed and mouse femurs were fixed in 10% neutral-buffered formalin for 18 hours. To detect mature OCs, a leukocyte acid phosphatase staining kit (Sigma) was used to stain sections with TRAP according to the manufacturer’s instructions. The number of TRAP-positive, multinuclear (> 3) OCs per millimeter of bone at the bone-tumor interface was calculated using a computerized image analysis system.
Immunohistochemistry
Formalin-fixed, paraffin-embedded sections of SCID mouse femurs were deparaffinized with xylene and rehydrated to water through a graded alcohol series. Endogenous peroxidase activity was quenched with 3% hydrogen peroxide. Expression of CD138 and CD32 was detected using specific antibodies (R&D Systems and LifeSpan BioSciences, Inc). Signal was detected using secondary biotinylated antibodies and streptavidin/horseradish peroxidase. Chromagen 3,3-diaminobenzidine/H2O2 (DAKO) was used, and slides were counterstained with hematoxylin. All slides were observed under light microscopy, and images were captured with a SPOT RT camera (Diagnostic Instruments, Burlingame, CA).
Chromatin Immunoprecipitation (ChIP) Assays
ChIP analysis with anti-twist was performed using a commercially available kit (Upstate Biotechnology, Lake Placid, NY, USA). Immunoprecipitates and total chromatin input were reverse cross-linked, DNA was isolated, and 1 μl DNA was used for PCR with primers specific for the gene promoter region for RANKL and for MCP-1. The primer sequences were as follows: (1) RANKL forward, CTTGGACCTCCAGAAAGACAG; reverse ACTCTTATAAACCGCTTGGAGAG; (2) MCP-1 forward, CTGCTAGGCTTCTATGATGCTAC; reverse, TCACTGCTGAGACCAAATGAG.
Transfection and luciferase assay
Cells at 50% confluence were transfected in 12-well petri dishes using Lipofectamine (Gibco-BRL, Grand Island, NY). The total amount of DNA was adjusted to 1.0 μg by adding empty vector. We determined luciferase activity by using a luciferase assay system (Promega, Madison, WI). As a reference plasmid to normalize transfection efficiency, 25 ng pRL-CMV plasmid (Promega) was cotransfected in all experiments.
Microarray analysis
We performed the microarray analysis as described previously (32). At least a 2-fold difference between control myeloma cells and CRP-treated myeloma cells was required. Microarray data has been deposited into Gene Expression Omnibus (GEO) database (GSE103697).
Statistical analysis
All data are shown as mean ± standard deviation. Student’s t test was used to compare various experimental groups; significance was set at an α level of 0.05.
Supplementary Material
Figure S1. Injection of CRP does not affect tumor growth in vivo.
Figure S2. Generation of CRP-expressing myeloma cells.
Figure S3. CRP upregulates myeloma cell production of cytokines important for OC activation.
Figure S4. Antibodies specific for OC-activating cytokines inhibit CRP-induced OC activation.
Figure S5. Knockdown or overexpression of CD32 does not affect myeloma cell proliferation or survival.
Figure S6. Overexpression of CD32 in myeloma cells enhances CRP-induced OC differentiation.
Table S1. Primers used for real-time PCR.
Acknowledgments
We thank Cassandra Talerico, PhD, Dept. of Cancer Biology, Cleveland Clinic, for helpful editorial assistance, and the MD Anderson Myeloma Tissue Bank for providing patient samples.
Funding: This work was supported by grants from NCI R01s (CA190863 and CA193362 to J.Y. and CA138398, CA200539, CA211073, and CA214811 to Q.Y.), ACS Research Scholar Grant (127337-RSG-15-069-01-TBG), grants from UTMDACC IRG-Basic Research, LRF, ASH, and National Natural Science Foundation of China Grant No. 81470356 to J.Y., as well as LLS TRP (6469-15) and MMRF Senior Investigator Award to Q.Y. This work was also supported by NIH/NCI under award number P30CA016672 and used the Small Animal Imaging Facility for radiography and Research Histopathology Facility for histological studies.
Footnotes
Competing interests: The authors declare no competing financial interests.
Data and material availability: Microarray data has been deposited into Gene Expression Omnibus (GEO) database (GSE103697).
Author contributions: Conceptualization, Q.Y. and J.Y.; Methodology, J.Y, Z.L., H.L., J. He, J.Y., Q.W. and E.D.; Microarray analysis, W.M. and E.D.; Providing samples and critical suggestions, P.L., J.D., R.Z.O., and J. Hou; Statistical consultation, Z.Y.; Writing, J.Y., H.L., and Q.Y; Funding Acquisition, Q.Y. and J.Y.
References and Notes
- 1.Kyle RA, Rajkumar SV. Multiple myeloma. N Engl J Med. 2004;351:1860–1873. doi: 10.1056/NEJMra041875. [DOI] [PubMed] [Google Scholar]
- 2.Tian E, Zhan F, Walker R, Rasmussen E, Ma Y, Barlogie B, Shaughnessy JD., Jr The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483–2494. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 3.Giuliani N, Rizzoli V, Roodman GD. Multiple myeloma bone disease: Pathophysiology of osteoblast inhibition. Blood. 2006;108:3992–3996. doi: 10.1182/blood-2006-05-026112. [DOI] [PubMed] [Google Scholar]
- 4.Roodman GD. Biology of osteoclast activation in cancer. J Clin Oncol. 2001;19:3562–3571. doi: 10.1200/JCO.2001.19.15.3562. [DOI] [PubMed] [Google Scholar]
- 5.He J, Liu Z, Zheng Y, Qian J, Li H, Lu Y, Xu J, Hong B, Zhang M, Lin P, Cai Z, Orlowski RZ, Kwak LW, Yi Q, Yang J. p38 MAPK in myeloma cells regulates osteoclast and osteoblast activity and induces bone destruction. Cancer Res. 2012;72:6393–6402. doi: 10.1158/0008-5472.CAN-12-2664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pepys MB, Hirschfield GM. C-reactive protein: a critical update. J Clin Invest. 2003;111:1805–1812. doi: 10.1172/JCI18921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein MP, Mold C, Du Clos TW. C-reactive protein binding to murine leukocytes requires Fc gamma receptors. J Immunol. 2000;164:1514–1520. doi: 10.4049/jimmunol.164.3.1514. [DOI] [PubMed] [Google Scholar]
- 8.Stein MP, Edberg JC, Kimberly RP, Mangan EK, Bharadwaj D, Mold C, Du Clos TW. C-reactive protein binding to FcgammaRIIa on human monocytes and neutrophils is allele-specific. J Clin Invest. 2000;105:369–376. doi: 10.1172/JCI7817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bataille R, Boccadoro M, Klein B, Durie B, Pileri A. C-reactive protein and beta-2 microglobulin produce a simple and powerful myeloma staging system. Blood. 1992;80:733–737. [PubMed] [Google Scholar]
- 10.Ludwig H, Durie BG, Bolejack V, Turesson I, Kyle RA, Blade J, Fonseca R, Dimopoulos M, Shimizu K, San Miguel J, Westin J, Harousseau JL, Beksac M, Boccadoro M, Palumbo A, Barlogie B, Shustik C, Cavo M, Greipp PR, Joshua D, Attal M, Sonneveld P, Crowley J. Myeloma in patients younger than age 50 years presents with more favorable features and shows better survival: an analysis of 10 549 patients from the International Myeloma Working Group. Blood. 2008;111:4039–4047. doi: 10.1182/blood-2007-03-081018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tienhaara A, Pulkki K, Mattila K, Irjala K, Pelliniemi TT. Serum immunoreactive interleukin-6 and C-reactive protein levels in patients with multiple myeloma at diagnosis. Br J Haematol. 1994;86:391–393. doi: 10.1111/j.1365-2141.1994.tb04748.x. [DOI] [PubMed] [Google Scholar]
- 12.Legouffe E, Rodriguez C, Picot MC, Richard B, Klein B, Rossi JF, Commes T. C-reactive protein serum level is a valuable and simple prognostic marker in non Hodgkin’s lymphoma. Leuk Lymphoma. 1998;31:351–357. doi: 10.3109/10428199809059228. [DOI] [PubMed] [Google Scholar]
- 13.Pedersen LM, Bergmann OJ. Urinary albumin excretion and its relationship to C-reactive protein and proinflammatory cytokines in patients with cancer and febrile neutropenia. Scand J Infect Dis. 2003;35:491–494. doi: 10.1080/00365540310012415. [DOI] [PubMed] [Google Scholar]
- 14.Reichle A, Bross K, Vogt T, Bataille F, Wild P, Berand A, Krause SW, Andreesen R. Pioglitazone and rofecoxib combined with angiostatically scheduled trofosfamide in the treatment of far-advanced melanoma and soft tissue sarcoma. Cancer. 2004;101:2247–2256. doi: 10.1002/cncr.20574. [DOI] [PubMed] [Google Scholar]
- 15.Yang J, Wezeman M, Zhang X, Lin P, Wang M, Qian J, Wan B, Kwak LW, Yu L, Yi Q. Human C-reactive protein binds activating Fcgamma receptors and protects myeloma tumor cells from apoptosis. Cancer Cell. 2007;12:252–265. doi: 10.1016/j.ccr.2007.08.008. [DOI] [PubMed] [Google Scholar]
- 16.Barlogie B, Tricot GJ, van Rhee F, Angtuaco E, Walker R, Epstein J, Shaughnessy JD, Jagannath S, Bolejack V, Gurley J, Hoering A, Vesole D, Desikan R, Siegel D, Mehta J, Singhal S, Munshi NC, Dhodapkar M, Jenkins B, Attal M, Harousseau JL, Crowley J. Long-term outcome results of the first tandem autotransplant trial for multiple myeloma. Br J Haematol. 2006;135:158–164. doi: 10.1111/j.1365-2141.2006.06271.x. [DOI] [PubMed] [Google Scholar]
- 17.Roodman GD. Treatment strategies for bone disease. Bone Marrow Transplant. 2007;40:1139–1146. doi: 10.1038/sj.bmt.1705802. [DOI] [PubMed] [Google Scholar]
- 18.Alatalo SL, Halleen JM, Hentunen TA, Monkkonen J, Vaananen HK. Rapid screening method for osteoclast differentiation in vitro that measures tartrate-resistant acid phosphatase 5b activity secreted into the culture medium. Clin Chem. 2000;46:1751–1754. [PubMed] [Google Scholar]
- 19.Asagiri M, Takayanagi H. The molecular understanding of osteoclast differentiation. Bone. 2007;40:251–264. doi: 10.1016/j.bone.2006.09.023. [DOI] [PubMed] [Google Scholar]
- 20.Scherf M, Klingenhoff A, Frech K, Quandt K, Schneider R, Grote K, Frisch M, Gailus-Durner V, Seidel A, Brack-Werner R, Werner T. First pass annotation of promoters on human chromosome 22. Genome Res. 2001;11:333–340. doi: 10.1101/gr.154601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jan YN, Jan LY. Functional gene cassettes in development. Proc Natl Acad Sci U S A. 1993;90:8305–8307. doi: 10.1073/pnas.90.18.8305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thisse B, el Messal M, Perrin-Schmitt F. The twist gene: isolation of a Drosophila zygotic gene necessary for the establishment of dorsoventral pattern. Nucleic Acids Res. 1987;15:3439–3453. doi: 10.1093/nar/15.8.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murray SS, Glackin CA, Winters KA, Gazit D, Kahn AJ, Murray EJ. Expression of helix-loop-helix regulatory genes during differentiation of mouse osteoblastic cells. J Bone Miner Res. 1992;7:1131–1138. doi: 10.1002/jbmr.5650071004. [DOI] [PubMed] [Google Scholar]
- 25.Rice DP, Aberg T, Chan Y, Tang Z, Kettunen PJ, Pakarinen L, Maxson RE, Thesleff I. Integration of FGF and TWIST in calvarial bone and suture development. Development. 2000;127:1845–1855. doi: 10.1242/dev.127.9.1845. [DOI] [PubMed] [Google Scholar]
- 26.Alborzi A, Mac K, Glackin CA, Murray SS, Zernik JH. Endochondral and intramembranous fetal bone development: osteoblastic cell proliferation, and expression of alkaline phosphatase, m-twist, and histone H4. J Craniofac Genet Dev Biol. 1996;16:94–106. [PubMed] [Google Scholar]
- 27.Bialek P, Kern B, Yang X, Schrock M, Sosic D, Hong N, Wu H, Yu K, Ornitz DM, Olson EN, Justice MJ, Karsenty G. A twist code determines the onset of osteoblast differentiation. Dev Cell. 2004;6:423–435. doi: 10.1016/s1534-5807(04)00058-9. [DOI] [PubMed] [Google Scholar]
- 28.Isenmann S, Arthur A, Zannettino AC, Turner JL, Shi S, Glackin CA, Gronthos S. TWIST family of basic helix-loop-helix transcription factors mediate human mesenchymal stem cell growth and commitment. Stem Cells. 2009;27:2457–2468. doi: 10.1002/stem.181. [DOI] [PubMed] [Google Scholar]
- 29.Miraoui H, Marie PJ. Pivotal role of Twist in skeletal biology and pathology. Gene. 2010;468:1–7. doi: 10.1016/j.gene.2010.07.013. [DOI] [PubMed] [Google Scholar]
- 30.Yang J, Mani SA, Donaher JL, Ramaswamy S, Itzykson RA, Come C, Savagner P, Gitelman I, Richardson A, Weinberg RA. Twist, a master regulator of morphogenesis, plays an essential role in tumor metastasis. Cell. 2004;117:927–939. doi: 10.1016/j.cell.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 31.Yaccoby S, Barlogie B, Epstein J. Primary myeloma cells growing in SCID-hu mice: a model for studying the biology and treatment of myeloma and its manifestations. Blood. 1998;92:2908–2913. [PubMed] [Google Scholar]
- 32.Ma W, Wang M, Wang ZQ, Sun L, Graber D, Matthews J, Champlin R, Yi Q, Orlowski RZ, Kwak LW, Weber DM, Thomas SK, Shah J, Kornblau S, Davis RE. Effect of long-term storage in TRIzol on microarray-based gene expression profiling. Cancer Epidemiol Biomarkers Prev. 2010;19:2445–2452. doi: 10.1158/1055-9965.EPI-10-0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Injection of CRP does not affect tumor growth in vivo.
Figure S2. Generation of CRP-expressing myeloma cells.
Figure S3. CRP upregulates myeloma cell production of cytokines important for OC activation.
Figure S4. Antibodies specific for OC-activating cytokines inhibit CRP-induced OC activation.
Figure S5. Knockdown or overexpression of CD32 does not affect myeloma cell proliferation or survival.
Figure S6. Overexpression of CD32 in myeloma cells enhances CRP-induced OC differentiation.
Table S1. Primers used for real-time PCR.