SUMMARY
The RASopathy neurofibromatosis type 1 (NF1) is one of the most common autosomal dominant genetic disorders. In NF1 patients, neurological issues may result from damaged myelin, and mice with a neurofibromin gene (Nf1) mutation show white matter (WM) defects including myelin decompaction. Using mouse genetics, we find that altered Nf1 gene-dose in mature oligodendrocytes results in progressive myelin defects and behavioral abnormalities mediated by aberrant Notch activation. Blocking Notch, upstream mitogen-activated protein kinase (MAPK), or nitric oxide signaling rescues myelin defects in hemizygous Nf1 mutants, and pharmacological gamma secretase inhibition rescues aberrant behavior with no effects in wild-type (WT) mice. Concomitant pathway inhibition rescues myelin abnormalities in homozygous mutants. Notch activation is also observed in Nf1+/− mouse brains, and cells containing active Notch are increased in NF1 patient WM. We thus identify Notch as an Nf1 effector regulating myelin structure and behavior in a RASopathy and suggest that inhibition of Notch signaling may be a therapeutic strategy for NF1.
In Brief
López-Juárez et al. find that loss of the RAS-GTP regulator Nf1 in oligodendrocytes leads to myelin and behavioral defects mediated by hyperactive Notch and upstream pathways. Pharmacological inhibition of Notch signaling rescues aberrant behavior in Nf1 mutant mice and may improve neurological manifestations in neurofibromatosis type 1 patients.

INTRODUCTION
The RASopathy neurofibromatosis type 1 (NF1) is one of the most common autosomal dominant genetic disorders. NF1 gene mutation, alone or with subsequent loss of the previously normal somatic allele, can lead to a variety of conditions in NF1 patients, ranging from aesthetic issues such as epidermal hyperpigmentation, to disabling bone malformations and aggressive life-threatening tumors (Ratner and Miller, 2015). NF1-associated neurological abnormalities include learning deficits, delayed acquisition of motor skills, and attention-deficit disorder, with or without hyperactivity; autism manifestations may also be present (Acosta et al., 2006; Garg et al., 2015). Cognitive dysfunction is the most common complication affecting the quality of life of children and adolescents with NF1 (Hyman et al., 2005), many of whom require neuropsychological assessment for educational planning (Acosta et al., 2012).
Significant advances have been made toward understanding how Nf1 mutation impacts neurons, and treatments to ameliorate neuronal abnormalities have been proposed. In animal models, activity of hippocampal interneurons (Cui et al., 2008) and dopaminergic neurons (Diggs-Andrews et al., 2013) are affected by Nf1 mutation. Notably, correlated abnormal behaviors are rescued by treatment with statins (Li et al., 2005) and dopamine re-uptake inhibitors (Brown et al., 2010), respectively. Nevertheless, mixed results in clinical studies (Bearden et al., 2016; van der Vaart et al., 2013, 2016), along with possible memory-associated side effects (Strom et al., 2015), have precluded definitive recommendation of the use of statins in NF1 patients. Therefore, better understanding of molecular mechanisms underlying NF1 neurological issues is crucial to establish successful treatment regimens.
In addition to neuronal defects, 60%–70% of children with NF1 show white matter (WM) abnormalities, including enlarged brain WM tracts, T2 hyperintensities, and altered fractional anisotropy and diffusivity on diffusion tensor imaging (DTI) (Karlsgodt et al., 2012; North, 2000). Myelin produced by mature oligodendrocytes (mOLs) increases nerve impulse velocity; thus, normal brain function requires normal myelin and oligodendrocyte function (Franklin and Gallo, 2014). Indeed, learning and motor skill acquisition correlate with changes in WM and myelin (McKenzie et al., 2014). Nonetheless, research on effects of Nf1 loss in mOLs is limited. We reported nitric oxide (NO)-mediated myelin decompaction >1 year after Nf1 inactivation, correlating with decreased tight junction (TJ) and GAP-junction (GJ) proteins (Mayes et al., 2013). However, what signaling pathways cause these phenotypes and whether loss of Nf1 in oligodendrocytes causes behavioral changes is unknown.
Nf1 is a RAS GTPase-activating protein, so that loss of Nf1 results in increased RAS-mitogen-activated protein kinase (MAPK) signaling (Ratner and Miller, 2015). RAS and Notch pathways can cooperate or antagonize each other in a context-dependent manner. Although Notch acts downstream of Nf1 during differentiation of neural stem cells (Chen et al., 2015), no links between these pathways have been described in mOL. Here, we show genetic and pharmacological evidence indicating that Notch signaling controls progressive and Nf1 gene-dose-dependent myelin defects in the corpus callosum (CC) of PlpCreER;Nf1flox animals. Abnormal behavior in hemizygous mutants is rescued by pharmacological inhibition of Notch. Mechanistically, aberrant Notch activation and myelin defects are rescued by inhibition of NO signaling, revealing NO as a crucial link between RAS and Notch pathways. Increased activation of Notch is also found in Nf1+/− mice and in WM of NF1 patients, suggesting that abnormal Notch signaling is a feature of NF1.
RESULTS
Nf1 Loss in mOLs Causes Nf1 Gene-Dose-Dependent Progressive Myelin Decompaction
We previously reported that myelin decompaction occurs in the optic nerve (ON) 6–12 months after tamoxifen-induced deletion of Nf1 in Plp1-expressing mOLs (Mayes et al., 2013). To study whether Nf1 loss-dependent myelin defects occur in the brain and their progression, we analyzed the CC of adult tamoxifen-treated homozygous (PlpCreER;Nf1flox/flox, henceforth pNf1f/f) and hemizygous (PlpCreER;Nf1flox/+, henceforth pNf1f/+) Nf1 mutant mice and wild-type (WT) animals (Nf1flox or PlpCreER), using electron microscopy (Figures 1A and 1B). Unbiased counting of myelinated axons showed no significant differences in g-ratio of control animals between 1 and 8 months after tamoxifen treatment (Figures S1A and S1C). Significantly decreased g-ratio in Nf1 hemizygous mutants, as compared to WTs, was detected at 1 month with a further decrease 6 months post-tamoxifen, indicating progressive changes in fiber structure. In contrast, in homozygous Nf1 mutants, decreased g-ratio occurred within 1 month post-tamoxifen, and showed slight but significant recovery by 6 months post-tamoxifen (Figures S1A and S1C). At this time point, the density of EGFP reporter-positive recombined cells did not differ between WT and Nf1 mutants (Figures S1F and S1G). Although Nf1 may recombine with different efficiency than EGFP, this result suggests that the trend toward recovery is not due to a depletion of mutant cells. Both parameters determining g-ratio were affected by Nf1 loss in mOLs; pNf1 mutants showed increased myelin thickness due to decompaction at intraperiod lines (number of myelin lamellae remained unchanged, Figures 1B and S1B) and decreased axon diameter (Figure S1E). The latter is likely secondary to disturbed myelin integrity (Cole et al., 1994; Colello et al., 1994), because neurons with axons in the CC do not express the PlpCre allele (Koenning et al., 2012; Mayes et al., 2013).
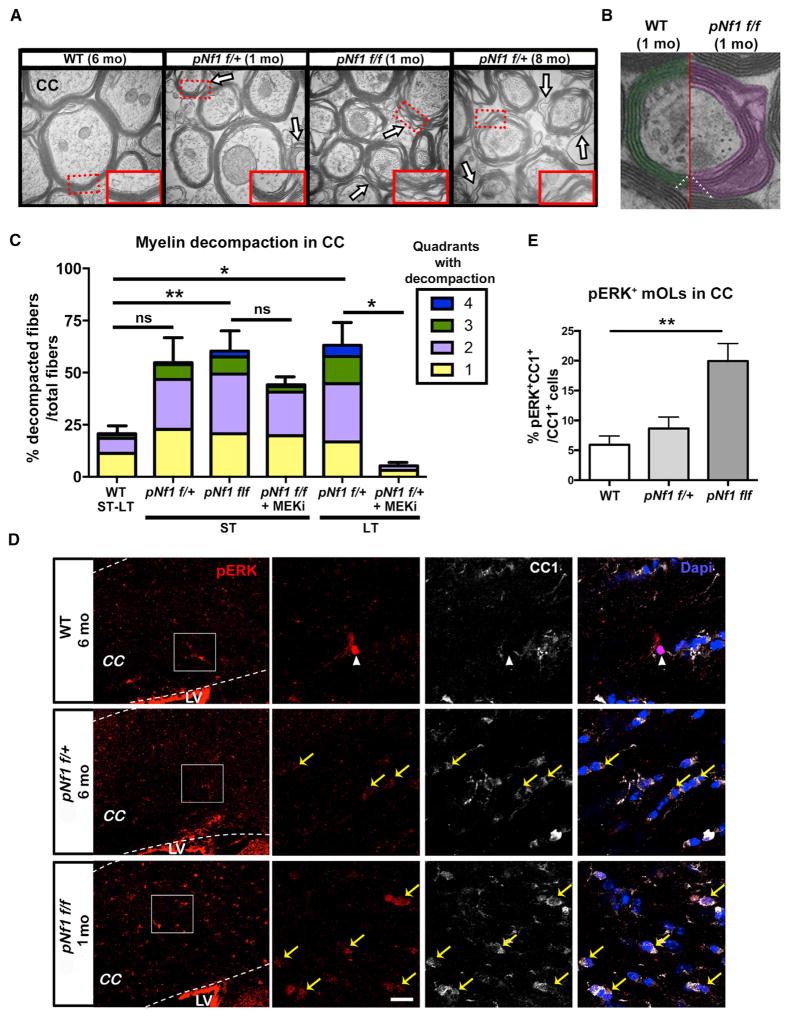
Figure 1. Progressive Myelin Decompaction Regulated by MAPK Signaling in pNf1 Mutants.
(A and B) Electron micrographs of the corpus callosum (CC) of WT, pNf1f/+, and pNf1f/f mice show regions of myelin decompaction (A, arrows; B, purple) at the indicated months (mo) post-tamoxifen treatment. Insets: regions from dotted rectangles (50,000×). Number of myelin wraps (triangles) in WT and pNf1f/f mutants are shown in (B).
(C) The percent of decompacted fibers among total fibers and number of quadrants with decompaction (color code) in the short-term (ST; 1 month) and long-term (LT; 6–10 months) post-tamoxifen is shown. Significantly increased decompaction is found in pNf1f/f mice at ST (n = 5 mice,**p < 0.01), and in pNf1f/+ mice at LT (n = 3 mice,*p < 0.05), as compared to WT (ST-LT, n = 6 mice). Moderate (1–3 quadrants), but not significant (ns), decompaction is observed in pNf1f/+ mice at ST (n = 3 mice). MEKi treatment rescues decompaction in pNf1f/+ mutants (n = 3 mice, *p < 0.05) but does not change decompaction in pNf1f/f mutants (n = 3 mice). Data are presented as the mean + SEM. One-way ANOVA; p = 0.0005, and Tukey’s multiple comparisons test.
(D) Immunostaining of pERK (red) and CC1 (white) in the CC of the indicated genotypes. Magnifications of squared regions (left) depict a pERK+;CC1− cell in WT (nuclear signal, arrowhead), pERK+(faint);CC1+ mOLs (yellow arrows) in pNf1f/+, and pERK+(strong);CC1+ mOLs in pNf1f/f mice. LV, lateral ventricle. Scale bar, 10 μm.
(E) Significantly increased pERK+;CC1+/total CC1+ mOLs are found in pNf1f/f mice as compared to WT (n = 4 mice/genotype, t test;**p = 0.0053, data are the mean + SEM).
See also Figures S1 and S2.
Regions of decompacted myelin were evident in hemizygous and homozygous pNf1 mutants 1 month post-tamoxifen, albeit to different extents (Figures 1A, insets, and 1B). Decompacted fibers among total fibers were quantified. Additionally, severity of decompaction was reflected by the number of quadrants around each axon showing decompaction (Figure 1C, color code). Control animals show baseline myelin “decompaction” (Figure 1C, 1–2 quadrants) in the short- (ST, 1 month) and long-term (LT, 6–9 months) post-tamoxifen treatment, likely indicating cross-sections through paranodal loops and Schmidt-Lanterman incisures. One month post-tamoxifen, pNf1f/+ mutants show decompaction in 1–3 quadrants, but not significant total decompaction, while pNf1f/f show significantly increased total decompacted fibers in 1–4 quadrants, as compared to WTs. Confirming a progressive phenotype, by 6–8 months post-tamoxifen significantly increased decompaction was also detected in hemizygous mice (Figure 1C). Thus, myelin decompaction in the CC is Nf1 gene-dose-dependent and progressive.
Nf1 Regulates Myelin Compaction through RAS-MAPK Pathway Activation
To define the mechanisms responsible for myelin decompaction in the CC of Nf1 mutants, we first analyzed RAS/MAPK pathway activation, which results in phosphorylation and activation of ERK1/2 (pERK). The WT CC showed pERK immunostaining in rare, mainly CC1− cells (Figure 1D). Consistent with activation of MAPK signaling in Nf1 mutants, increased faint pERK signals were observed in CC1+ mOLs of pNf1f/+ mice and strong pERK signals in pNf1f/f mice, at time points when severe decompaction is observable. Quantitative analysis confirmed significantly increased pERK+;CC1+ mOLs in pNf1f/f mice 1 month post-tamoxifen, as compared to controls, while a trend toward increase was detected in pNf1f/+ mice (Figure 1E). Other signal pathways could cause myelin decompaction in pNf1f/+ mutants, and/or modestly increased pERK could cause decompaction in the long-term; if so, decompaction should be sensitive to inhibition of MEK, the ERK kinase. At 9 months post-tamoxifen when myelin decompaction is detectable, we treated Nf1 hemizygous mutants for 21 days with PD0325901, a brain penetrant MEK inhibitor (MEKi, 1.5 mg/kg). At this therapeutically relevant dose (Barrett et al., 2008), MEKi fully rescued myelin decompaction (Figure 1C) and g-ratio (Figure S1D) in pNf1f/+ mice. In fact, myelin was more compacted than WTs, suggesting that basal MAPK signaling contributes to the maintenance of normal compact myelin structure. In homozygous Nf1 mutants, MEKi improved the severity of decompaction (quadrants) and significantly increased g-ratio, but failed to fully rescue these parameters (Figures 1C and S1D), perhaps because the treatment did not abolish abnormal MAPK signaling. In support of this hypothesis, while MEKi significantly decreased pERK+;CC1+ mOLs, they remained significantly increased in pNf1f/f mice as compared to controls (Figures S2A and S2B). Thus, MAPK signaling contributes to the aberrant myelin compaction in Nf1 mutants, but a clinically relevant dose of MEKi is insufficient to rescue cells with complete Nf1 loss of function.
Nf1 Loss Increases Notch Pathway Activity in Glial Cells In Vitro and In Vivo
As in a previous study (Mayes et al., 2013), WT and mutants showed similar MBP expression around CC1+ cells (Figure S2C). We aimed to identify other molecules/pathways downstream ERK activation that influence cell signaling and myelin defects following Nf1 loss. We postulated that transcriptional changes might underlie Nf1-loss effects, as MAPK signaling regulate gene expression (Ishii et al., 2014). Using microarray gene expression analysis of glia-enriched cultures (Bennett et al., 2003), we identified genes with significantly altered expression in Nf1−/− as compared to WT cultures. Interestingly, Hes5, a direct transcriptional target of canonical Notch signaling, was the most upregulated gene in Nf1 mutant cells, and other Notch-related genes (Dll1 and Dll3) were also significantly increased, as validated by qRT-PCR (Figure 2A). Upon ligand binding, a cleaved Notch intracellular domain (NICD) is generated, correlating with Notch activation. NICD detection by western blot increased in Nf1−/− as compared to WT cultures (Figure 2B). To verify these results in vivo in an OL-enriched population, we performed RNA sequencing (RNA-seq) transcriptome analysis using WT and pNf1f/f optic nerves, 1 month post-tamoxifen. We identified significant increases in transcripts encoding the Notch targets Hes5, Cntnap2, and Cttnbp2 (Meier-Stiegen et al., 2010). Transcripts encoding Notch ligands also increased (Dll1, Dlk2, Dner, and Cntn1) or decreased (Thbs1, Postn, and Jag1). Transcripts encoding GJ and TJ proteins expressed in the oligodendrocyte-lineage, including Claudin 11, were also significantly decreased (Figure 2C). To test whether loss of Nf1 in mOLs increases Notch activation in vivo, we analyzed Nf1 mutants carrying the transgene Hes5GFP, a reporter of canonical Notch pathway activation that is not expressed significantly in normal mOLs. WT animals and Nf1 mutants contained, as expected, Hes5GFP+;GFAP+ astrocytes (Figure S3A) and rare Hes5GFP+;CC1+ mOLs in the CC (Figure 2D, insets). Although Hes5GFP+ mOLs were significantly increased in pNf1f/f animals as compared to WT animals (Figures 2E and S4E), they accounted for only 1.41% ± 0.20% of CC1+ cells in the CC, and Hes5GFP signal did not overlap with pERK+CC1+ mOLs (Figure S3B).
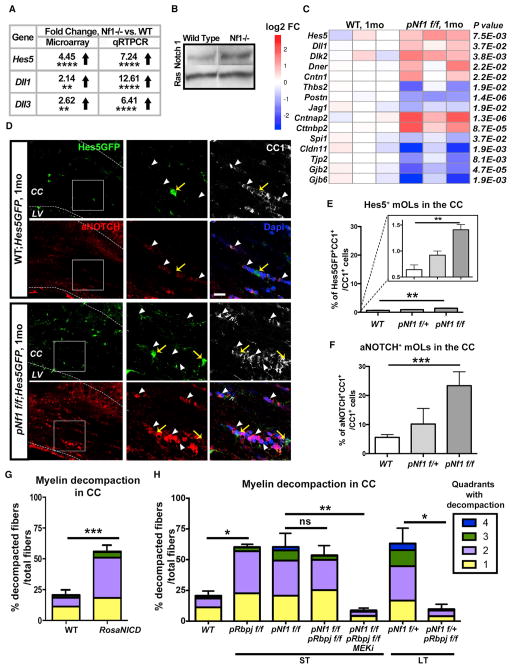
Figure 2. Increased Notch Pathway Activity in mOLs Causes Myelin Decompaction in pNf1 Mutants.
(A) Data from microarray analysis and qRT-PCR validation show increased expression of genes in Nf1−/− glial-enriched cultures (t test, Hes5****p < 0.0001; Dll1**p = 0.0055, Dll3**p = 0.004, n = 3 cultures).
(B) Western blot analysis of Notch1 and Ras10 (loading control) indicates increased activated Notch 1 (97 kD) in Nf1−/− glia-enriched cultures.
(C) RNA-seq analysis of pNf1f/f ON 1 month post-tamoxifen. Heatmap showing gene expression levels (log2 fold change) and p values for genes of canonical and non-canonical Notch ligands (Dll1, Dlk2, Dner, Cntn1, Thbs2, Postn, Jag1), Notch targets (Hes5, Cntnap2, Cttnbp2, Spi1), tight junctions (Cldn11, Tjp2), and GAP junctions (Gjb6, Gjb2). Expression of Mbp, Plp1, Mag, Mog, and Omg did not show significant changes (n = 3/genotype, DESeq normalization method).
(D) Immunofluorescence of Hes5GFP (green), aNOTCH (red), and CC1 (white) 1 month post-tamoxifen. In magnification (right) of squared regions: Hes5GFP (yellow arrows) is detected in rare CC1+ (weak) mOLs, aNOTCH (arrowheads) is detected in cytoplasm of mOLs in WT (Hes5GFP) and nuclei of abundant mOLs in pNf1f/f;Hes5GFP mutant (pink, bottom/right). CC, corpus callosum; LV, lateral ventricle. Scale bar, 10 μm.
(E) The percent of Hes5GFP+;CC1+/CC1+ cells significantly increases in pNf1f/f mutants as compared to WT (n = 4 mice/genotype, t test; **p = 0.0013). Inset: y axis enlargement.
(F) The percent of aNOTCH+;CC1+/CC1+ cells significantly increases in pNf1f/f mutants (n = 7 mice) as compared to WT (n = 4 mice, t test; ***p = 0.001). Data are the mean + SEM.
(G and H) Percent of decompacted fibers and severity of decompaction (quadrants) in the indicated genotypes in the short- (1 month) or long-term (6–9 months) post-tamoxifen. Increased decompaction is shown in PlpCre;RosaNICD (G, n = 4 mice, t test, ***p < 0.0001), and pRbpjf/f (H, n = 3 mice, *p < 0.01) as compared to WTs (n = 6 mice). Note that decompaction is not rescued in pNf1f/f;Rbpjf/f (versus pNf1f/f), but it is rescued by additional treatment with MEKi (n = 3, **p < 0.001, one-way ANOVA). Decompaction is fully rescued in pNf1f/+;Rbpjf/f mutants (versus pNf1 f/+, n = 3 mice/genotype, *p < 0.01 one-way ANOVA).
Error bars show ± SEM. See also Figures S3 and S4.
Hes5 is one of many Notch effectors. To test whether other Notch effectors are likely to be activated in mOLs after loss of Nf1, we analyzed NICD immunoreactivity using a specific antibody (aNOTCH). WT mice showed aNOTCH cytoplasmic signals in rare CC1+ mOLs. Significantly increased numbers of aNOTCH+(nuclear);CC1+ mOLs were detected in pNf1f/f animals (Figures 2D and 2F), corroborating Nf1 loss-driven, abnormal, Notch activation. Interestingly, aNOTCH signals were detected in recombinant and non-recombinant cells (Figure S3C), supporting the idea that cell autonomous and non-cell-autonomous effects occur downstream of Nf1 mutation in mOLs (Mayes et al., 2013).
Activation of Notch in mOLs Reduces Claudin 11 Expression and Causes Myelin Decompaction
We investigated whether abnormal Notch activation in mOLs is sufficient to disrupt myelin compaction. We genetically activated Notch signaling in mOLs with the tamoxifen-inducible RosaNICD allele driven by PlpCreER (pNICD). One month post-tamoxifen treatment, myelin decompaction (Figure 2G) and decreased g-ratio (Figure S4A) were found in pNICD animals, as compared to WTs. Decreased Claudin 11 corelates with myelin decompaction in pNf1 mutants (Mayes et al., 2013), and WB analysis showed a significant decrease of this TJ protein in the forebrain of pNICD mice (Figure S4B). Thus, genetic activation of Notch in mOLs mimics phenotypes caused by Nf1 loss.
Notch Genetic Inhibition Rescues Myelin Decompaction in Nf1 Mutants
RBPJ is a transcriptional co-factor required for canonical Notch signaling. To test whether increased canonical Notch signaling regulates phenotypes in Nf1 mutants, we genetically inactivated Notch signaling in mOLs using PlpCreER;Nf1fl/+(or Nf1fl/f);Rbpjfl/fl mice, in which Rbpj and Nf1 are inactivated upon tamoxifen treatment. Unexpectedly, homozygous Rbpj inactivation in mOLs (pRbpjf/f) caused myelin decompaction 1 month post-tamoxifen treatment (Figure 2H). Thus, imbalance in Notch signaling and/or compensatory responses affect myelin compaction in this genetic model. Importantly, hemizygous Nf1 mutants with deletion of Rbpj (pNf1f/+;Rbpjf/f) showed full rescue of myelin decompaction (Figure 2H) and g-ratio (Figure S4C) 6–8 months post-tamoxifen. Inactivation of Rbpj in Nf1 homozygotes rescued g-ratio but did not significantly impact myelin decompaction 1 month post-tamoxifen; however, additional treatment of pNf1f/f;Rbpjf/f mice with MEKi fully rescued compaction to WT levels (Figure 2H). Overall, these data indicate that Notch signaling is crucial for the maintenance of myelin structure in the WT setting and in the context of Nf1 loss.
Gamma Secretase Inhibition Rescues Myelin Defects and Aberrant Behavior in Nf1 Mutants
To test whether Nf1 loss-driven phenotypes can be rescued by pharmacological inhibition of Notch we used the blood-brain barrier-permeable gamma secretase inhibitor (GSI) MRK-003 (Chu et al., 2013). GSI administration to WT animals (weekly for 3 weeks) significantly decreased the number of Hes5GFP+ cells in the CC, confirming drug efficacy (Figure 3A). Importantly, GSI treatment had no effect on myelin compaction in WT animals. In contrast, GSI treatment rescued decompaction (Figure 3B) and g-ratio (Figure S4D) in pNf1f/+ animals 6–10 months post-tamoxifen. In fact, myelin compaction increased over WT levels, similar to the effect of MEKi (Figure 1C). In Nf1 homozygous mutants GSI significantly increased g-ratio (Figure S4D), however, no significant changes in myelin decompaction were observed. These data suggest that GSI treatment normalizes the unbalanced Notch signaling that promotes decompaction in Nf1 mutants.
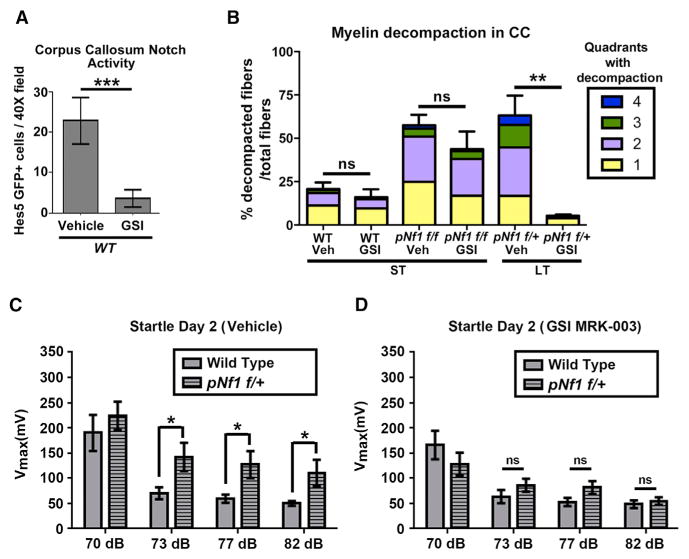
Figure 3. Pharmacological Inhibition of Notch Activation in pNf1 Mutants Rescues Decompaction and Aberrant Behavior.
(A) Number of Hes5GFP+ cells in the CC of vehicle-treated (n = 5 mice) or gamma secretase inhibitor (GSI)-treated (n = 12 mice) WT mice (t test, *p = 0.0001).
(B) The percent of decompacted fibers and quadrants with decompaction does not change after GSI treatment in WT animals (n = 6 vehicle-treated, n = 4 GSI-treated animals, t test, ns, p = 0.4516) and pNf1f/f mutants (n = 5 mice/genotype, ns, p = 0.2768). Decompaction is rescued in pNf1f/+ mice treated with GSI (n = 4 mice, t test, **p = 0.0019) as compared to vehicle-treated pNf1f/+ (n = 3). ST, 1 month post-tamoxifen; LT, 6–10 months post-tamoxifen.
(C and D) Evaluation of the acoustic startle response. (C) pNf1f/+ mutant mice (n = 21 mice) present increased Vmax to the acoustic startle response following the 73 dB, 77 dB, and 82 dB pre-pulse stimuli, as compared to WT mice (n = 19 mice, two-way ANOVA, F[4,152] = 3.05, *p < 0.05). (D) The heightened startle response in pNf1f/+ mutants is abolished after treatment with GSI MRK-003, as no significant difference in Vmax is observed between WT (n = 21 mice) and pNf1 mutants (n = 19 mice, two-way ANOVA, F[4,152] = 2.73).
Error bars show ± SEM. See also Figure S4.
NF1 patients display mutation/loss of 1 NF1 allele and present locomotor, cognitive and attention issues. We therefore tested whether behavior is altered in hemizygous pNf1 mutants. We evaluated sensory gating deficits in Nf1 hemizygous mutant mice (C57BL/6 genetic background), by analyzing the prepulse inhibition of startle response (Vorhees et al., 2011), 9 months post-tamoxifen treatment. Significantly heightened response to startle at 73–82 dB, was found in pNf1 mutants as compared to controls (Figure 3C). Remarkably, all increased startle responses (73–82 dB) were blocked by treatment with GSI for 3 weeks, as compared to littermate WTs (Figure 3D). Thus, hemizygous Nf1 mutation in mOLs causes myelin structural defects and behavioral abnormalities that can be rescued by pharmacological inhibition of Notch signaling.
Notch Activation and Myelin Decompaction Are Due to Increased Nitric Oxide Signaling in Nf1 Mutants
Nf1 loss in oligodendrocytes was reported to result in increased reactive oxygen/nitrogen species in the forebrain, and treatment with the antioxidant N-acetyl cysteine rescued cell-autonomous and non-cell-autonomous defects in the ON of pNf1 mutants (Mayes et al., 2013). To test whether mOL NO contributes to CC myelin abnormalities, Nf1 mutants were treated with a specific inhibitor of NO synthases L-NAME (N-nitroarginine methyl ester, 0.4 mg/kg) for 7 days, starting after severe decompaction is detectable. L-NAME treatment fully rescued myelin compaction (Figure 4A) and g-ratio (Figure S4F) in pNf1f/+ mutants. In pNf1f/f mice, treatment significantly increased g-ratio, but did not rescue compaction to WT levels. Notably, full rescue of de-compaction in the homozygous mutants required inhibiting both Notch and NO signaling (Rbpj deletion and L-NAME treatment) or MAPK and NO signaling (MEKi and L-NAME treatment) (Figures 4A and S4F).
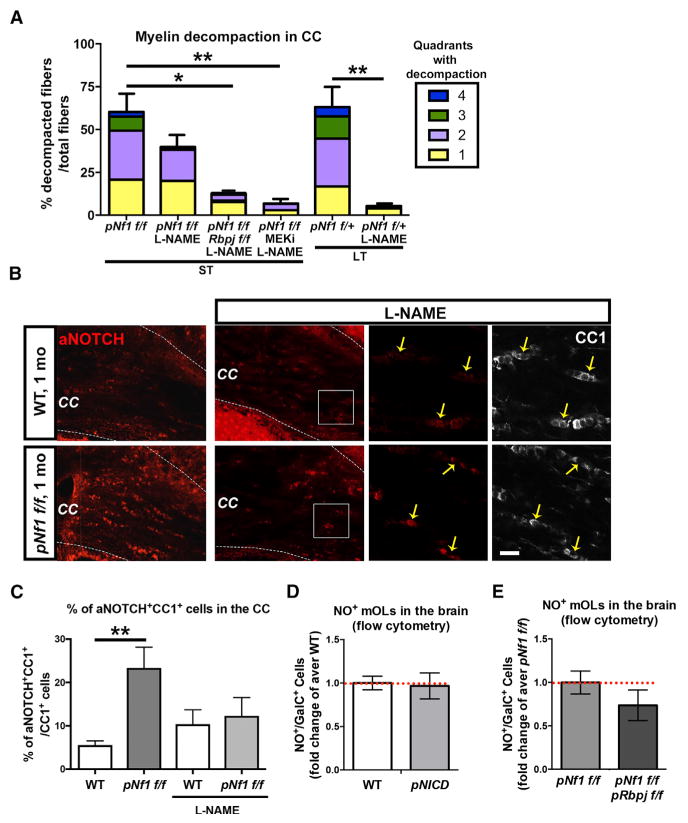
Figure 4. NO Controls Notch Activation and Concomitant Inhibition of Notch/NO or NO/MAPK Signaling Rescues Decompaction in pNf1f/f Mutants.
(A) Decompacted fibers (%) and quadrants with decompaction in mice treated with L-NAME (7 days) and MEKi (21 days). L-NAME treatment rescues myelin compaction in the long-term (LT) pNf1f/+ mice (n = 4 mice/treatment, **p < 0.01). Note that % of decompacted fibers does not significantly change in L-NAME-treated pNf1f/f mice (ST, short-term), but decompaction is fully rescued in L-NAME-treated pNf1f/f;Rbpjf/f (n = 3 mice, *p < 0.05), or in L-NAME;MEKi-treated pNf1f/f (n = 3 mice, **p < 0.01), as compared to pNf1f/f mice.
(B) Immunostaining of aNOTCH (red) and CC1 (white) in the CC of vehicle- or L-NAME-treated WTs and pNf1f/f mutants. Note the lower number of aNOTCH+;CC1+ cells in L-NAME-treated pNf1f/f mutants, as compared to untreated mutants. Scale bar, 10 μm.
(C) WT and pNf1f/f mice show no significantly different numbers of aNOTCH+;CC1+ mOLs (yellow arrows in B) after L-NAME treatment (n = 3 mice/genotype, t test, p = 0.9721). Untreated animals (from Figure 2F) are shown for comparison.
(D) Flow cytometry analysis of forebrain cells indicates that the number of GalC+ mOLs showing NO signals does not change in PlpCreER;RosaNICD (pNICD) mice, as compared to WT animals (n = 3 mice/genotype, t test, p = 0.86).
(E) Flow cytometry analysis of forebrain cells indicates that the number of GalC+ mOLs showing NO signals does not change in pNf1f/f;pRbpjf/f mice as compared to pNf1f/f mutants (n = 3 mice/genotype, t test, p = 0.29).
Error bars show ± SEM. See also Figure S4.
We therefore tested whether elevated NO levels drive Notch activation (Charles et al., 2010; Jeon et al., 2014) in Nf1 mutants or whether Notch activation is an independent Nf1-driven signal in mOLs. L-NAME treatment for 7 days rescued the abnormally increased number of aNotch+;CC1+/CC1+ mOLs in the CC of pNf1f/f mutants, as compared to L-NAME-treated WT mice (Figures 4B and 4C). Thus, abnormally increased Notch signaling in mOL is reduced by NOS inhibition. Flow cytometry analysis of forebrain showed that neither NO nor superoxide signals changed in Hes5GFP+ cells of pNf1f/f mutants, versus Hes5+ cells in WT animals (Figure S4E). Demonstrating that Notch does not drive NO, overexpression of NICD in mOLs (PlpCreER;RosaNICD) did not alter numbers of NO+GalC+ cells, and NO+ cells remained unchanged in Nf1 mutants with genetic inactivation of canonical Notch signaling as compared to pNf1f/f mutants (Figures 4D and 4E). A linear MAPK/NO/Notch pathway downstream Nf1 loss might explain why single agent treatments fully rescue myelin decompaction in hemizygous pNf1 mutants. However, combined inhibition of MAPK, NO, and Notch signaling (Figures 2H and 4A) is necessary to normalize effects in homozygous mutants plausibly because of a more robust pathway activation; durable feedback loops may also explain this finding.
Nf1 Loss and Oligodendrocyte Numbers
Nf1 loss in mOLs was found to increase numbers of Olig2+ cells in the CC 4 days after tamoxifen treatment (Mayes et al., 2013). To test if this increase is durable, we quantified CC Olig2+ cells 1 month post-tamoxifen. Olig2+ cells were not significantly increased in pNf1f/f mutants versus WT (Figures S6A and S6B). As Olig2 labels all oligodendrocyte lineage cells, we identified by flow cytometry-specific forebrain cell populations using the markers PDGFRα, O4, and GalC. mOLs (PDGFRα− GalC+ or O4−GalC+) did not show altered numbers in pNf1 mutants. Therefore, overall changes in numbers of oligodendrocytes are unlikely to account for our findings. We note that maturing OLs (PDGFRα+GalC+) were significantly increased in pNf1f/f animals (Figure S6C), and within the Hes5+ population, O4+GalC+ cells were also significantly increased (Figure S6D). Non-cell-autonomous effects on oligodendrocyte progenitors and/or re-expression of immature OL markers in GalC+ mOL might explain these phenotypes.
Nf1+/−Mice Show Increased Notch Activity, and NF1 Patients Show Hyperactivated Notch
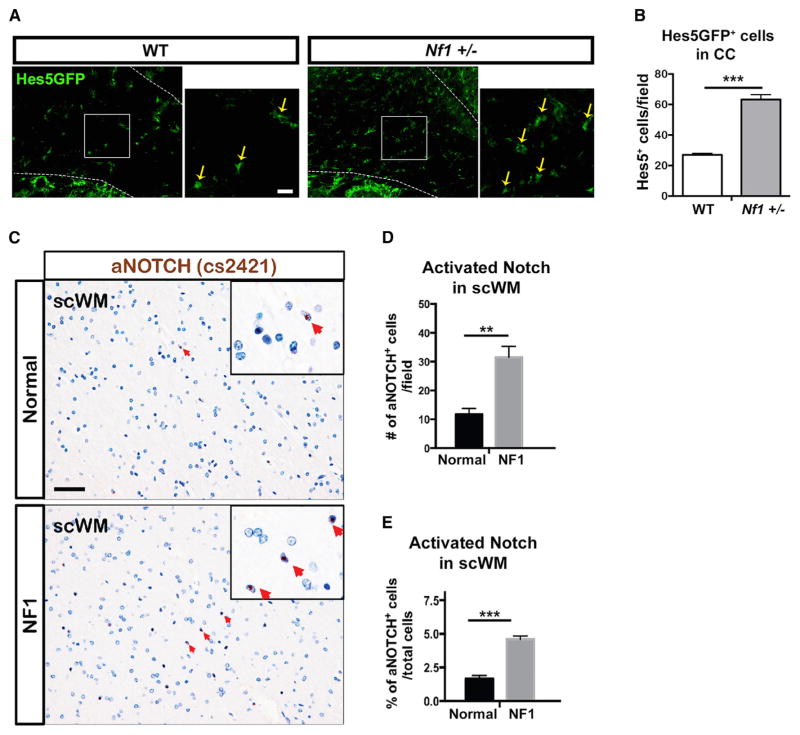
To determine whether control of myelin compaction through Notch signaling might be relevant to NF1 patients, we first evaluated Notch activation in the brain of Nf1+/−mice, which mimic NF1 patients genotype with germline mutation in one Nf1 allele in all brain cells. Notch activation was elevated in Nf1+/−mutants, as in the Notch-activity reporter mouse Nf1+/−;Hes5GFP, numbers of Hes5GFP+ cells in CC were significantly increased, as compared to WT animals (Figures 5A and 5B).
Figure 5. Notch Signaling Increases in Nf1+/− mice, and aNotch+ Cells Are Increased in NF1 Patient WM.
(A and B) The number of Hes5GFP+ cells in the CC (A) of Nf1+/− mice is significantly increased (B), as compared to WT animals (n = 3 mice/genotype, t test, ***p = 0.0004).
(C) Immunodetection of NICD (aNOTCH) with a specific antibody (Cell Signaling, cs2421) in the subcortical WM (scWM) of NF1 patient brains (n = 2) and normal human brains (n = 2). Representative aNOTCH nuclear signals are shown with red arrows (insets). Scale bars, 50 μm.
(D) Quantification of total aNOTCH+ cells per 20× field suggests increased Notch activity in the scWM of NF1 patients, as compared to normal brain (n = 2 NF1 patients, n = 2 normal brains, 2 technical replicates/condition, t test, **p = 0.0042).
(E) Quantification of aNOTCH+ cells as percent of total cells per 20× field supports increased Notch activity in the scWM of NF1 patients, as compared to normal brain (n = 2 NF1 patients, n = 2 normal brains, 2 technical replicates/condition, t test, ***p = 0.0014). Error and significance of the graphs reflect only experimental variability.
Error bars show ± SEM. See also Figure S5.
Finally, we used two antibodies to detect cells with activated Notch+ (aNOTCH or NICD) in brain sections from previously described adult NF1 patients (Nordlund et al., 1995). Both antibodies (ab8925 and cs2421) detected higher number of NICD+ cells in the subcortical WM of NF1 patients (n = 2), as compared to normal adult brain (n = 2) (Figures 5C and S5A). Quantification of two technical replicates (reflecting experimental variability, not the variability of the biological process), showed significantly increased number of aNOTCH+ cells (Figure 5D). To address the issue of precisely matching brain regions in the limited specimens available, we normalized the percentage of aNOTCH+ cells to the total number of cells per field and found similar results (Figure 5E). Overall, our data are consistent with the idea that abnormal Notch signaling downstream of Nf1 loss and NO increase contribute to NF1 patient pathology.
DISCUSSION
The study of WM abnormalities that correlate with motor, sensory, or cognitive changes in pathological situations may help to identify mechanisms relevant to normal brain function. We focused on brain pathology in NF1, a common genetic disorder in which patients show WM disorganization on brain imaging, and neurological deficits. We show that Notch signaling, along with MAPK and NO, are aberrantly activated after Nf1 loss in mature oligodendrocytes, causing myelin decompaction and changing animal behavior.
Patient mutations in genes throughout the RAS-MAPK pathway (Rasopathy genes) compromise brain function (Rauen, 2013). Nf1 is a well-studied Rasopathy gene that acts as a regulator of RAS-MAPK signaling (Donovan et al., 2002; Ratner and Miller, 2015). Increasing evidence confirms a role for MAPK activation in NF1 brain. For example, abnormally enlarged CC resulting from Nf1 loss is rescued by perinatal MEK inhibition (Wang et al., 2012). In oligodendrocyte progenitors with Nf1 mutation, basal and FGF-stimulated RAS-GTP are increased, proliferation increases, and differentiation is aberrant, although myelination occurs (Bennett et al., 2003). In contrast to these developmental studies, our analyses ensue after deletion of Nf1 in mature cells. Our results are encouraging as they support the idea that alterations in NF1 brain may be susceptible both to prevention and to therapy.
Myelin decompaction resulting from Nf1 loss in oligodendrocytes accounts for pathological increases in myelin thickness, as numbers of myelin lamellae did not significantly change. This result differs from studies in which the RAS effector proteins ERK1/2 regulate myelin thickness by increasing numbers of myelin wraps (Ishii et al., 2013), as assessed using gain and loss-of-function models. In our model, after Nf1 loss, RAS, MEK, and ERK proteins are present at normal levels; therefore, extrinsic stimuli activate transient signaling through MEK/ERK. The transience of the RAS signal after Nf1 loss likely accounts for the discrepancy with studies of constitutive activation or complete loss of ERK. Other RAS effector pathways activated by loss of Nf1 may also contribute to decompaction after homozygous Nf1 loss, although this seems less likely given that MEK inhibition alone blocks decompaction after hemizygous Nf1 loss.
Increased Notch signaling following Nf1 loss was demonstrated by microarray/RNA-seq analyses and corroborated by in vivo detection of NICD in CC1+ mOLs. Cell-autonomous roles of oligodendrocyte Notch in controlling myelin compaction are supported by our genetic gain- (NICD) and loss-of-function (Rbpj) models. Furthermore, although GSI can have effects on other substrates, our finding that GSI rescues aberrant myelin decompaction is consistent with our detection of active nuclear NICD in oligodendrocytes and the genetic data indicating that Notch blockade via Rbpj loss reverses effects of Nf1 loss. The correlation between rescued decompaction and GSI rescue of abnormal startle response in pNf1 mutants suggests that myelin compaction is relevant to normal behavior. Importantly, GSI treatment did not affect WT myelin structure or behavior in our study. It will be of great interest to define a direct link between abnormal activation of Notch with decompaction and behavior in animal models and whether increased activation of Notch in NF1 patients is linked to their neurological issues.
Notch, via Hes5, regulates many processes in immature oligodendrocyte lineage cells (Liu et al., 2006; Watkins et al., 2008). We found that Hes5 transcript is increased in a Nf1 mutant glial precursor-enriched population in vitro, and NICD and Hes1/5 increases were recently reported downstream of Nf1 inactivation in neural stem cells (Chen et al., 2015). However, it is likely that Notch effector-driven cell-autonomous effects on mOL do not require Hes5, in that CC1+Hes5+ are rare (albeit increased) in Nf1 mutants and do not show detectable ERK activation. It remains possible that in Nf1 mutants, Hes5+ cells and/or an increased population of O4+GalC+ maturing oligodendrocytes, contribute to abnormal myelin compaction. However, the broad expression of mOL NotchICD in the absence of Hes5 strongly suggests the pathological relevance of alternate Notch effector proteins in this setting.
We previously reported that Nf1 mutation in oligodendrocytes causes small but significant increase in NO levels in the forebrain (Mayes et al., 2013). Here, we extend the results to CC oligodendrocytes and demonstrate that increased NO is upstream of Notch-mediated myelin decompaction. In oligodendrocytes, NO is protective at low levels, while at high levels causes ERK1/2-mediated toxicity (Li et al., 2011) and downregulation of myelin genes preceding cell death (Jana and Pahan, 2013). Also at low concentrations, NO induces cGMP-mediated morphological changes (Garthwaite et al., 2015). As no increased mOL cell death was found in pNf1 mice (Mayes et al., 2013), it is probable that NO does not accumulate to lethal levels after Nf1 loss. Yet, myelin structural defects develop in an Nf1 gene-dose-dependent manner. We propose that the slow progression of myelin decompaction in hemizygous Nf1 mutants is caused by modest increased MAPK and downstream signals that causes cumulative damage, while biallelic loss of Nf1 causes higher levels of pathway activation and more rapid decompaction. Other Nf1 gene-dose-dependent phenotypes have been reported. For example, Nf1 hemizygous oligodendrocyte precursors are expanded in vitro, to a lesser extent than Nf1 homozygous mutant cells (Bennett et al., 2003). Only Nf1−/− sympathetic neurons develop neurites independent of growth factor (Vogel et al., 1995), but Nf1+/− mice show defects in Morris water maze tests and dopamine-based learning (Anastasaki et al., 2015).
It is well known that RAS-GTP signaling can act up or downstream of Notch and have a cooperative or antagonistic outcomes (Sundaram, 2005). In fact, multilineage differentiation of neural stem cells is controlled by Nf1 via RAF/MEK pathway and Notch activation (Chen et al., 2015), however, the link between these signal pathways is unclear. We find that increased active Notch is rescued by L-NAME treatment, suggesting that NO is a mediator between RAS-Nocth signaling in Nf1 mutants. Thus, a linear MAPK/NO/Notch pathway might drive myelin de-compaction and behavioral phenotypes in hemizygous mutants (Figure 6A). In support of this idea, single agent treatment with MEKi (Figure 1C), GSI (Figure 3B), or L-NAME (Figure 4A) fully rescue myelin decompaction, and GSI alone rescues behavioral abnormalities. Similar pathway interactions have been reported in PDGF-induced gliomas, in which NO/cGMP/PKG (Charles et al., 2010) or NO/ID4/Jagged1 (Jeon et al., 2014) signaling drive Notch activation and tumor growth.
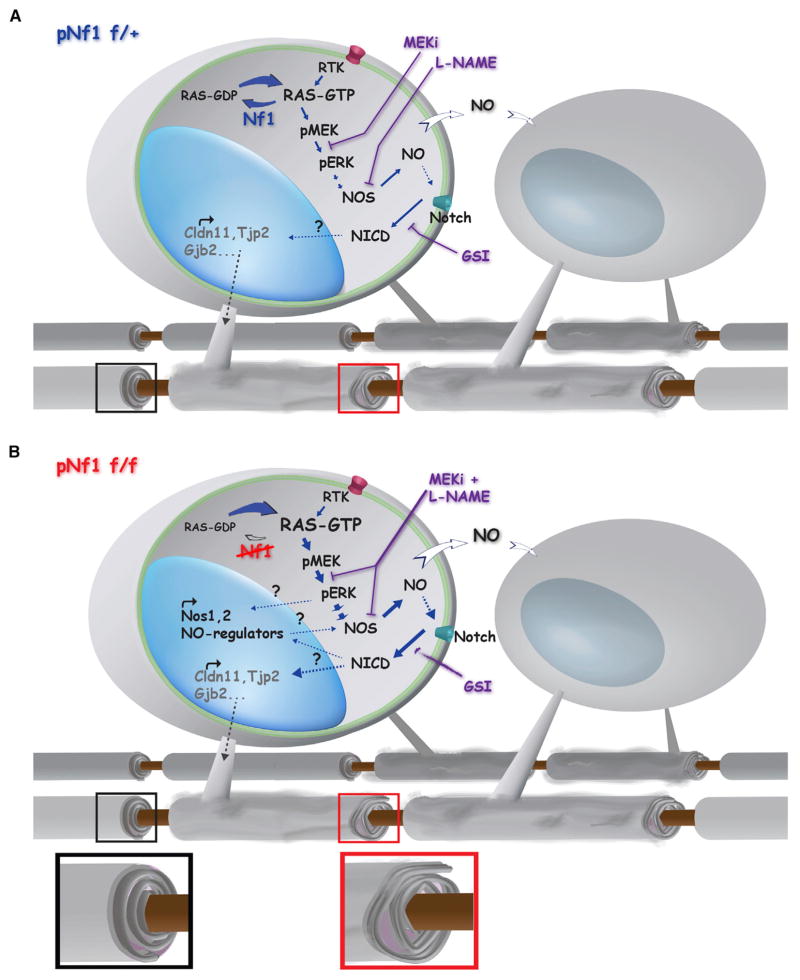
Figure 6. Model of Nf1 Loss-Driven Myelin Decompaction.
Increased MAPK/NO/Notch signaling may mediate myelin decompaction (gray-dotted arrows and insets), by downregulation of TJ/GJ proteins involved in myelin compaction such as Claudin 11 (Cldn11), ZO-2 (Tjp2), and Connexin 26 (Gjb2) in pNf1 mutants.
(A) Decreased Nf1 (blue) results in increased RAS-GTP in mature oligodendrocytes, leading to hyperactive MAPK signaling (RTK →RAS →pMEK →pERK). Production of NO increases in response to hyperactive MAPK pathway (NOS, dashed arrow). Subsequently, NO promotes directly or indirectly (dotted line) Notch cleavage and translocation of NICD to the nucleus, where it regulates gene expression. Furthermore, diffusion of NO from Nf1 mutant oligodendrocytes (white arrows) might affect nearby oligodendrocytes increasing the number of decompacted fibers (bottom). Either pharmacological treatment alone (MEKi, L-NAME or GSI) rescues decompaction (purple lines).
(B) Absent Nf1 (red) results in accumulation of RAS-GTP and stronger induction (thick arrows) of MAPK signaling and NO production. It is possible that long-lasting transcriptional and/or post-transcriptional changes (dotted lines with question marks) in molecules controlling NO levels (for example NOS1-2) might contribute to increases in NO. Subsequently, NO promotes Notch activation and regulation of myelin genes. Combination of inhibitors (MEKi and L-NAME, purple fused lines) rescues decompaction, while single agent treatments (for example GSI, fading purple arrow) improves severity but does not rescue decompaction. Note that genetic inactivation of Notch (Figure 5) combined with MEKi or L-NAME, also rescues compaction in this setting.
A linear MAPK/NO/Notch pathway, however, does not easily explain why blocking each pathway alone does not rescue decompaction in homozygous mutants. A parsimonious explanation is that the more robust activation of RAS in pNf1f/f mutants requires longer treatments with single drugs, higher doses, or combinations of drugs, to fully diminish the effects of RAS over activation (Figure 6B). Alternatively, increased MAPK, NO, and Notch signaling might cause durable transcriptional or posttranscriptional changes that feedback and potentiate the effects of the entire pathway. In support of this idea, NOS1-3 proteins are increased in pNf1 homozygous mutants (Mayes et al., 2013). Given that oligodendrocytes express NOS1 (Yao et al., 2012) and possibly NOS2 (Boullerne and Benjamins, 2006), transcriptional regulation of these and other genes involved in the pathway is conceivable. The specific Notch effector(s) controlling myelin compaction remain unknown. Myelin proteins regulating compaction, including Claudin 11, might be involved. We found that genetic activation of Notch decreases Claudin 11. Of note, mice with loss of the Plp1 and Cldn11 show impaired motor behavior and myelin decompaction, similar to those found in pNf1 mutants (Chow et al., 2005). Regulation of Cldn11 by Notch has not been reported, and in silico analysis do not show binding sites for CSL/Rbpj (http://genome.ucsc.edu). However, a binding site for PU.1, a transcription factor regulated by Notch (Chen et al., 2008) and downregulated in pNf1 mutants, is upstream of Cldn11. Additional downregulation of TJ/GJ genes (Tjp2, Gjb6, Gjb2) may also contribute to abnormal myelin and behavior (Figure 6).
Most myelinated fibers in Nf1 mutants showed decompaction, even though deletion of Nf1 occurs in a smaller number of mOLs (Koenning et al., 2012; Mayes et al., 2013). This result suggests that a diffusible messenger signals to non-recombinant cells. Diverse paracrine roles of NO have been widely reported in the brain (Calabrese et al., 2007), thus, NO could mediate effects to neighbor cells in pNf1 mutants (Figure 6). In fact, several non-cell-autonomous defects are detected after deletion of Nf1 in mOLs (Mayes et al., 2013).
In summary, activation of Notch signaling, downstream of MAPK and NO, results in myelin decompaction and behavioral changes in Nf1 hemizygous mutants. It is believed that most cells in NF1 patient brains contain one normal NF1 allele and one mutant/lost allele of the NF1 gene. However, there are brain regions with especially poor organization that may represent regions of biallelic NF1 loss, based on DTI (Ferraz-Filho et al., 2012). If biallelic NF1 mutations are present in oligodendrocytes in NF1 patient brains and cause WM alteration (Ferraz-Filho et al., 2012; Karlsgodt et al., 2012), drug combinations might be needed therapeutically. Importantly, our findings that NO/Notch signaling is perturbed in Nf1+/− mice, and that NICD increases in NF1 patients raise the intriguing possibility that the pathways we identified in mouse mOLs will be relevant to NF1 patients. Based on these studies, drugs that block the Nf1 loss-dependent increased NO and subsequent Notch activation should be considered to treat disease manifestations in NF1.
EXPERIMENTAL PROCEDURES
Mouse Strains
All mouse studies were approved by the Cincinnati Children’s Hospital Research Foundation Institutional Animal Care and Use Review Committee (IACUC). The generation and genotyping of mice carrying Nf1fl/fl [Nf1tm1Par/J] (Zhu et al., 2002), PlpCre [B6.Cg-Tg(Plp1-Cre/ERT)3Pop/J] (Doerflinger et al., 2003), Hes5-GFP [Tg1(Hes5GFP)Gsat/Mmmh] (Gong et al., 2003), Rbpjfl/fl [RBP-Jf/f] (Han et al., 2002), RosaNICD [Gt(ROSA)26Sortm1(Notch1)DAM/J] (Murtaugh et al., 2003), and Nf1+/− (Brannan et al., 1994) alleles, are described in previous studies. Mice were maintained on the C57BL/6 background. For behavioral studies, only adult female mice were used. In all other studies, we used adult mice of both sexes. Mice were housed in a temperature- and humidity-controlled vivarium on a 12 hr light-dark cycle with free access to food and water.
Pre-clinical Therapeutics
Gamma secretase inhibitor (GSI,MRK-003; Merck) was made fresh weekly and dosed at 300 mg/kg in 0.5% methocel by oral gavage (Lewis et al., 2007; Sparey et al., 2005). For pathology, we dosed mice once weekly for 4 weeks and sacrificed 6 hr after the last dose (n = 5 doses). For flow analysis, we dosed mice once weekly for 1 week and sacrificed mice 6 hr after the last dose (n = 2 doses). Fresh solution of L-NAME (L-NG-nitroarginine methyl ester, 100 μM in 1× PBS; Sigma-Aldrich) was administered daily at 0.4 mg/kg. For pathology and flow analysis, mice were injected intraperitoneally (i.p.) daily for 7 days and sacrificed 6 hr after the last dose. MEK inhibitor (PD0325901; Pfizer) was made fresh weekly and dosed at 1.5 mg/kg/day in 0.5% methocel/0.2% Tween 80, by oral gavage. For pathology, we dosed mice every day for 3 weeks. For flow analysis, we dosed mice every day for 7 days and sacrificed 6 hr after the last dose.
Behavior
Behavioral assessments were completed on pNf1fl/+ mice (F12, backcrossed onto C57BL/6) mice. Acoustic startle response, with prepulse inhibition, was assessed as described (Vorhees et al., 2011). White noise background of 70 dB, prepulses of 73, 77, and 82 dB, and a mixed frequency startle stimulus of 120 dB were used. Animals received 100 trials per day with equal numbers of trials using a Latin square design that was duplicated. Animals were tested on 2 consecutive days and the data from the second day were analyzed, because animals that are tested on a single day for the acoustic startle response tend to have greater variability that can mask group differences.
Human Brain Tissues
Brain specimens were described previously (Nordlund et al., 1995). Briefly, NFI brains were obtained from patients diagnosed with NF1 based on NIH guidelines. None of these patients showed any evidence of neurological disease other than NF1. Control/normal brains were obtained from adult patients with no neurological diseases reported, although a history of depression was reported in one. All brains were harvested within 8 hr after death. Brains were sliced and placed in 4% paraformaldehyde/0.1 M phosphate buffer for 72 hr and prepared for paraffin embedding and storage.
Statistical Analysis
The minimal number of animals per statistically significant experiment was three. Comparison between two groups used Student’s t tests with a significance cutoff of p < 0.05. Comparison of three or more groups used the one-way ANOVA, followed by Tukey post hoc test with a significance cutoff of p < 0.05. For unbiased counting of myelinated axons, mice were randomized and treated in a blinded manner. For behavior, acoustic startle was analyzed by mixed linear Two-way ANOVA with factors of genotype and trial. Further comparisons averaged across interval, day, and trial for genotype differences were compared by (two-tailed) t test for independent samples.
Supplementary Material
Highlights.
Experimental oligodendrocyte Nf1 inactivation causes myelin and behavioral defects
Hyper-active Notch in oligodendrocytes is necessary and sufficient to disrupt myelin
Pharmacological inhibition of Notch normalizes myelin and behavior in Nf1 mutants
Nf1+/− mouse and patient brains suggest Notch hyper-activation in NF1 pathogenesis
Acknowledgments
Shyra J. Miller and Nisha Schuler provided analyzed microarray data for cultured cells. Debra A. Mayes provided input into early phases of the project. Bethany Bresnen edited figures. We thank Brian Popko (U. of Chicago), Luis F. Parada (UTSW), and Tasuku Honjo (Kyoto-U) for mouse lines, Marie Dominique Fillipi and Monica DeLay for assistance with flow cytometry, Merck for MRK-003, and Kevin Shannon (UCSF) for PD0325901. This work was supported by a grant from the DOD program on Neurofibromatosis W81-WH-10-10116 and NIH R01 NS091037 (to N.R.). The Cincinnati Children’s Hospital Research Foundation Flow and Pathology Cores provided support for these studies (NIH P30 DK0909710551).
Footnotes
ACCESSION NUMBERS
The accession number for the RNA-seq data reported in this paper is GEO: GSE96738.
Supplemental Information includes Supplemental Experimental Procedures and six figures and can be found with this article online at http://dx.doi.org/10.1016/j.celrep.2017.03.073.
AUTHOR CONTRIBUTIONS
A.L.-J. and H.E.T. conducted most experiments and analyzed data. J.W.P conducted animal work and drug dosing. M.R.B. performed microarray and western blot. T.A.R. assisted with perfusions. G.C. prepared blocks and sections for electron microscopy. S.H.S. assisted with statistical analyses. S.H.S. and M.B. evaluated g-ratios and myelin decompaction in electron micrographs. M.T.W. and C.V.V. analyzed behavior. A.L.-J. wrote the manuscript. N.R. conceived and directed the project, contributed to data analysis and interpretation, and edited the manuscript.
References
- Acosta MT, Gioia GA, Silva AJ. Neurofibromatosis type 1: new insights into neurocognitive issues. Curr Neurol Neurosci Rep. 2006;6:136–143. doi: 10.1007/s11910-996-0036-5. [DOI] [PubMed] [Google Scholar]
- Acosta MT, Bearden CE, Castellanos FX, Cutting L, Elgersma Y, Gioia G, Gutmann DH, Lee YS, Legius E, Muenke M, et al. The Learning Disabilities Network (LeaDNet): using neurofibromatosis type 1 (NF1) as a paradigm for translational research. Am J Med Genet A. 2012;158A:2225–2232. doi: 10.1002/ajmg.a.35535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastasaki C, Woo AS, Messiaen LM, Gutmann DH. Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum Mol Genet. 2015;24:3518–3528. doi: 10.1093/hmg/ddv103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett SD, Bridges AJ, Dudley DT, Saltiel AR, Fergus JH, Flamme CM, Delaney AM, Kaufman M, LePage S, Leopold WR, et al. The discovery of the benzhydroxamate MEK inhibitors CI-1040 and PD 0325901. Bioorg Med Chem Lett. 2008;18:6501–6504. doi: 10.1016/j.bmcl.2008.10.054. [DOI] [PubMed] [Google Scholar]
- Bearden CE, Hellemann GS, Rosser T, Montojo C, Jonas R, Enrique N, Pacheco L, Hussain SA, Wu JY, Ho JS, et al. A randomized placebo-controlled lovastatin trial for neurobehavioral function in neurofibromatosis I. Ann Clin Transl Neurol. 2016;3:266–279. doi: 10.1002/acn3.288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett MR, Rizvi TA, Karyala S, McKinnon RD, Ratner N. Aberrant growth and differentiation of oligodendrocyte progenitors in neurofibromatosis type 1 mutants. J Neurosci. 2003;23:7207–7217. doi: 10.1523/JNEUROSCI.23-18-07207.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boullerne AI, Benjamins JA. Nitric oxide synthase expression and nitric oxide toxicity in oligodendrocytes. Antioxid Redox Signal. 2006;8:967–980. doi: 10.1089/ars.2006.8.967. [DOI] [PubMed] [Google Scholar]
- Brannan CI, Perkins AS, Vogel KS, Ratner N, Nordlund ML, Reid SW, Buchberg AM, Jenkins NA, Parada LF, Copeland NG. Targeted disruption of the neurofibromatosis type-1 gene leads to developmental abnormalities in heart and various neural crest-derived tissues. Genes Dev. 1994;8:1019–1029. doi: 10.1101/gad.8.9.1019. [DOI] [PubMed] [Google Scholar]
- Brown JA, Emnett RJ, White CR, Yuede CM, Conyers SB, O’Malley KL, Wozniak DF, Gutmann DH. Reduced striatal dopamine underlies the attention system dysfunction in neurofibromatosis-1 mutant mice. Hum Mol Genet. 2010;19:4515–4528. doi: 10.1093/hmg/ddq382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese V, Mancuso C, Calvani M, Rizzarelli E, Butterfield DA, Stella AM. Nitric oxide in the central nervous system: neuroprotection versus neurotoxicity. Nat Rev Neurosci. 2007;8:766–775. doi: 10.1038/nrn2214. [DOI] [PubMed] [Google Scholar]
- Charles N, Ozawa T, Squatrito M, Bleau AM, Brennan CW, Hambardzumyan D, Holland EC. Perivascular nitric oxide activates notch signaling and promotes stem-like character in PDGF-induced glioma cells. Cell Stem Cell. 2010;6:141–152. doi: 10.1016/j.stem.2010.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PM, Yen CC, Wang WS, Lin YJ, Chu CJ, Chiou TJ, Liu JH, Yang MH. Down-regulation of Notch-1 expression decreases PU. 1-mediated myeloid differentiation signaling in acute myeloid leukemia. Int J Oncol. 2008;32:1335–1341. doi: 10.3892/ijo_32_6_1335. [DOI] [PubMed] [Google Scholar]
- Chen YH, Gianino SM, Gutmann DH. Neurofibromatosis-1 regulation of neural stem cell proliferation and multilineage differentiation operates through distinct RAS effector pathways. Genes Dev. 2015;29:1677–1682. doi: 10.1101/gad.261677.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow E, Mottahedeh J, Prins M, Ridder W, Nusinowitz S, Bronstein JM. Disrupted compaction of CNS myelin in an OSP/Claudin-11 and PLP/DM20 double knockout mouse. Mol Cell Neurosci. 2005;29:405–413. doi: 10.1016/j.mcn.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Chu Q, Orr BA, Semenkow S, Bar EE, Eberhart CG. Prolonged inhibition of glioblastoma xenograft initiation and clonogenic growth following in vivo Notch blockade. Clin Cancer Res. 2013;19:3224–3233. doi: 10.1158/1078-0432.CCR-12-2119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole JS, Messing A, Trojanowski JQ, Lee VM. Modulation of axon diameter and neurofilaments by hypomyelinating Schwann cells in transgenic mice. J Neurosci. 1994;14:6956–6966. doi: 10.1523/JNEUROSCI.14-11-06956.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colello RJ, Pott U, Schwab ME. The role of oligodendrocytes and myelin on axon maturation in the developing rat retinofugal pathway. J Neurosci. 1994;14:2594–2605. doi: 10.1523/JNEUROSCI.14-05-02594.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui Y, Costa RM, Murphy GG, Elgersma Y, Zhu Y, Gutmann DH, Parada LF, Mody I, Silva AJ. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell. 2008;135:549–560. doi: 10.1016/j.cell.2008.09.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diggs-Andrews KA, Tokuda K, Izumi Y, Zorumski CF, Wozniak DF, Gutmann DH. Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann Neurol. 2013;73:309–315. doi: 10.1002/ana.23793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doerflinger NH, Macklin WB, Popko B. Inducible site-specific recombination in myelinating cells. Genesis. 2003;35:63–72. doi: 10.1002/gene.10154. [DOI] [PubMed] [Google Scholar]
- Donovan S, Shannon KM, Bollag G. GTPase activating proteins: critical regulators of intracellular signaling. Biochim Biophys Acta. 2002;1602:23–45. doi: 10.1016/s0304-419x(01)00041-5. [DOI] [PubMed] [Google Scholar]
- Ferraz-Filho JR, da Rocha AJ, Muniz MP, Souza AS, Goloni-Bertollo EM, Pavarino-Bertelli EC. Diffusion tensor MR imaging in neurofibromatosis type 1: expanding the knowledge of microstructural brain abnormalities. Pediatr Radiol. 2012;42:449–454. doi: 10.1007/s00247-011-2274-1. [DOI] [PubMed] [Google Scholar]
- Franklin RJ, Gallo V. The translational biology of remyelination: past, present, and future. Glia. 2014;62:1905–1915. doi: 10.1002/glia.22622. [DOI] [PubMed] [Google Scholar]
- Garg S, Plasschaert E, Descheemaeker MJ, Huson S, Borghgraef M, Vogels A, Evans DG, Legius E, Green J. Autism spectrum disorder profile in neurofibromatosis type I. J Autism Dev Disord. 2015;45:1649–1657. doi: 10.1007/s10803-014-2321-5. [DOI] [PubMed] [Google Scholar]
- Garthwaite G, Hampden-Smith K, Wilson GW, Goodwin DA, Garthwaite J. Nitric oxide targets oligodendrocytes and promotes their morphological differentiation. Glia. 2015;63:383–399. doi: 10.1002/glia.22759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong S, Zheng C, Doughty ML, Losos K, Didkovsky N, Schambra UB, Nowak NJ, Joyner A, Leblanc G, Hatten ME, Heintz N. A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature. 2003;425:917–925. doi: 10.1038/nature02033. [DOI] [PubMed] [Google Scholar]
- Han H, Tanigaki K, Yamamoto N, Kuroda K, Yoshimoto M, Nakahata T, Ikuta K, Honjo T. Inducible gene knockout of transcription factor recombination signal binding protein-J reveals its essential role in T versus B lineage decision. Int Immunol. 2002;14:637–645. doi: 10.1093/intimm/dxf030. [DOI] [PubMed] [Google Scholar]
- Hyman SL, Shores A, North KN. The nature and frequency of cognitive deficits in children with neurofibromatosis type 1. Neurology. 2005;65:1037–1044. doi: 10.1212/01.wnl.0000179303.72345.ce. [DOI] [PubMed] [Google Scholar]
- Ishii A, Furusho M, Bansal R. Sustained activation of ERK1/2 MAPK in oligodendrocytes and schwann cells enhances myelin growth and stimulates oligodendrocyte progenitor expansion. J Neurosci. 2013;33:175–186. doi: 10.1523/JNEUROSCI.4403-12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii A, Furusho M, Dupree JL, Bansal R. Role of ERK1/2 MAPK signaling in the maintenance of myelin and axonal integrity in the adult CNS. J Neurosci. 2014;34:16031–16045. doi: 10.1523/JNEUROSCI.3360-14.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jana M, Pahan K. Down-regulation of myelin gene expression in human oligodendrocytes by nitric oxide: implications for demyelination in multiple sclerosis. J Clin Cell Immunol. 2013:4. doi: 10.4172/2155-9899.1000157. http://dx.doi.org/10.4172/2155-9899.1000157. [DOI] [PMC free article] [PubMed]
- Jeon HM, Kim SH, Jin X, Park JB, Kim SH, Joshi K, Nakano I, Kim H. Crosstalk between glioma-initiating cells and endothelial cells drives tumor progression. Cancer Res. 2014;74:4482–4492. doi: 10.1158/0008-5472.CAN-13-1597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlsgodt KH, Rosser T, Lutkenhoff ES, Cannon TD, Silva A, Bearden CE. Alterations in white matter microstructure in neurofibro-matosis-1. PLoS ONE. 2012;7:e47854. doi: 10.1371/journal.pone.0047854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koenning M, Jackson S, Hay CM, Faux C, Kilpatrick TJ, Willingham M, Emery B. Myelin gene regulatory factor is required for maintenance of myelin and mature oligodendrocyte identity in the adult CNS. J Neurosci. 2012;32:12528–12542. doi: 10.1523/JNEUROSCI.1069-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis HD, Leveridge M, Strack PR, Haldon CD, O’neil J, Kim H, Madin A, Hannam JC, Look AT, Kohl N, et al. Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of notch signaling. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]
- Li W, Cui Y, Kushner SA, Brown RA, Jentsch JD, Frankland PW, Cannon TD, Silva AJ. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr Biol. 2005;15:1961–1967. doi: 10.1016/j.cub.2005.09.043. [DOI] [PubMed] [Google Scholar]
- Li S, Vana AC, Ribeiro R, Zhang Y. Distinct role of nitric oxide and peroxynitrite in mediating oligodendrocyte toxicity in culture and in experimental autoimmune encephalomyelitis. Neuroscience. 2011;184:107–119. doi: 10.1016/j.neuroscience.2011.04.007. [DOI] [PubMed] [Google Scholar]
- Liu A, Li J, Marin-Husstege M, Kageyama R, Fan Y, Gelinas C, Casaccia-Bonnefil P. A molecular insight of Hes5-dependent inhibition of myelin gene expression: old partners and new players. EMBO J. 2006;25:4833–4842. doi: 10.1038/sj.emboj.7601352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayes DA, Rizvi TA, Titus-Mitchell H, Oberst R, Ciraolo GM, Vorhees CV, Robinson AP, Miller SD, Cancelas JA, Stemmer-Rachamimov AO, Ratner N. Nf1 loss and Ras hyperactivation in oligodendrocytes induce NOS-driven defects in myelin and vasculature. Cell Rep. 2013;4:1197–1212. doi: 10.1016/j.celrep.2013.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenzie IA, Ohayon D, Li H, de Faria JP, Emery B, Tohyama K, Richardson WD. Motor skill learning requires active central myelination. Science. 2014;346:318–322. doi: 10.1126/science.1254960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meier-Stiegen F, Schwanbeck R, Bernoth K, Martini S, Hieronymus T, Ruau D, Zenke M, Just U. Activated Notch1 target genes during embryonic cell differentiation depend on the cellular context and include lineage determinants and inhibitors. PLoS ONE. 2010;5:e11481. doi: 10.1371/journal.pone.0011481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murtaugh LC, Stanger BZ, Kwan KM, Melton DA. Notch signaling controls multiple steps of pancreatic differentiation. Proc Natl Acad Sci USA. 2003;100:14920–14925. doi: 10.1073/pnas.2436557100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordlund ML, Rizvi TA, Brannan CI, Ratner N. Neurofibromin expression and astrogliosis in neurofibromatosis (type 1) brains. J Neuropathol Exp Neurol. 1995;54:588–600. doi: 10.1097/00005072-199507000-00013. [DOI] [PubMed] [Google Scholar]
- North K. Neurofibromatosis type 1. Am J Med Genet. 2000;97:119–127. doi: 10.1002/1096-8628(200022)97:2<119::aid-ajmg3>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- Ratner N, Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer. 2015;15:290–301. doi: 10.1038/nrc3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rauen KA. The RASopathies. Annu Rev Genomics Hum Genet. 2013;14:355–369. doi: 10.1146/annurev-genom-091212-153523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparey T, Beher D, Best J, Biba M, Castro JL, Clarke E, Hannam J, Harrison T, Lewis H, Madin A, et al. Cyclic sulfamide gamma-secretase inhibitors. Bioorg Med Chem Lett. 2005;15:4212–4216. doi: 10.1016/j.bmcl.2005.06.084. [DOI] [PubMed] [Google Scholar]
- Strom BL, Schinnar R, Karlawish J, Hennessy S, Teal V, Bilker WB. Statin therapy and risk of acute memory impairment. JAMA Intern Med. 2015;175:1399–1405. doi: 10.1001/jamainternmed.2015.2092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaram MV. The love-hate relationship between Ras and Notch. Genes Dev. 2005;19:1825–1839. doi: 10.1101/gad.1330605. [DOI] [PubMed] [Google Scholar]
- van der Vaart T, Plasschaert E, Rietman AB, Renard M, Oostenbrink R, Vogels A, de Wit MC, Descheemaeker MJ, Vergouwe Y, Catsman-Berrevoets CE, et al. Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): a randomised, placebo-controlled trial. Lancet Neurol. 2013;12:1076–1083. doi: 10.1016/S1474-4422(13)70227-8. [DOI] [PubMed] [Google Scholar]
- van der Vaart T, Rietman AB, Plasschaert E, Legius E, Elgersma Y, Moll HA NF1-SIMCODA Study Group. Behavioral and cognitive outcomes for clinical trials in children with neurofibromatosis type 1. Neurology. 2016;86:154–160. doi: 10.1212/WNL.0000000000002118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel KS, Brannan CI, Jenkins NA, Copeland NG, Parada LF. Loss of neurofibromin results in neurotrophin-independent survival of embryonic sensory and sympathetic neurons. Cell. 1995;82:733–742. doi: 10.1016/0092-8674(95)90470-0. [DOI] [PubMed] [Google Scholar]
- Vorhees CV, Morford LR, Graham DL, Skelton MR, Williams MT. Effects of periadolescent fluoxetine and paroxetine on elevated plus-maze, acoustic startle, and swimming immobility in rats while on and off-drug. Behav Brain Funct. 2011;7:41. doi: 10.1186/1744-9081-7-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Kim E, Wang X, Novitch BG, Yoshikawa K, Chang LS, Zhu Y. ERK inhibition rescues defects in fate specification of Nf1-deficient neural progenitors and brain abnormalities. Cell. 2012;150:816–830. doi: 10.1016/j.cell.2012.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins TA, Emery B, Mulinyawe S, Barres BA. Distinct stages of myelination regulated by gamma-secretase and astrocytes in a rapidly myelinating CNS coculture system. Neuron. 2008;60:555–569. doi: 10.1016/j.neuron.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao SY, Natarajan C, Sriram S. nNOS mediated mitochondrial injury in LPS stimulated oligodendrocytes. Mitochondrion. 2012;12:336–344. doi: 10.1016/j.mito.2012.01.002. [DOI] [PubMed] [Google Scholar]
- Zhu Y, Ghosh P, Charnay P, Burns DK, Parada LF. Neurofibromas in NF1: Schwann cell origin and role of tumor environment. Science. 2002;296:920–922. doi: 10.1126/science.1068452. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.