Abstract
G protein–coupled receptors (GPCRs) are major regulators of intercellular interactions. They initiate these actions by being activated by a wide variety of natural ligands. Historically, ligands were discovered first, but the advent of molecular biology reversed this trend. Most GPCRs are identified on the basis of their DNA sequences and thus are initially unmatched to known natural ligands. They are termed orphan GPCRs. Discovering their ligands—i.e., “deorphanizing” the GPCRs—gave birth to the field of reverse pharmacology. This review discusses the present status of GPCR deorphanization, presents a few examples of successes and surprises, and highlights difficulties encountered in these efforts.
Keywords: orphan receptors, neuropeptides, reverse pharmacology, phylogeny, lipid mediators
INTRODUCTION
Signaling between cells, which is fundamental to physiology, relies on the transmitter-receptor interaction. Transmitters, as defined in this review, are mostly small molecules. They include biogenic amines, amino acids and derivatives, neuropeptides, chemokines, lipid mediators, nucleotides, polypeptide hormones, olfactory and gustatory molecules, and some other naturally occurring chemicals such as calcium ions and protons. Transmitter-receptor interactions initiate intracellular biochemical pathways that lead to cellular responses and ultimately to physiological changes. Because only a limited number of intracellular biochemical pathways exist, the extracellular recognition process must provide the complexity and diversity that are required to direct the numerous physiological responses of an organism. Yet because this interaction is based on a binary mechanism, diversity must rely on the molecules that can take part. Discovering novel transmitters and receptors is thus of basic interest to biology.
The discovery that the β2-adrenergic receptor and the opsins share a seven-transmembrane (7TM) domain topology (1) led to the concept that receptors that couple to heterotrimeric (αβγ) G proteins form a supergene family, thereafter known as the G protein–coupled receptors (GPCRs) (2). These receptors sense extracellular transmitter molecules, i.e., ligands that are released from one cell to carry a message to another cell. GPCRs are expressed on the plasma membrane of the signal-receiving cell and, when activated by the transmitter, induce intracellular biochemical pathways that lead to a cellular response. Although the 7TM topology has evolved to include receptors that do not couple to heterotrimeric G proteins, and although receptor activation is complex and includes proteins other than G proteins (3–7), one tenet of the basic concept has remained constant over the years: The vast majority of transmitter molecules that have evolved to direct intercellular interactions interact with GPCRs (7TM receptors). This concept positions the GPCRs at the center of signal transmission and endows them with an extraordinary importance in an organism’s life and survival. Consistent with this biological importance, approximately 36% of all currently marketed drugs target approximately 82 GPCRs, making them the most commonly exploited class of drug targets (8).
The GPCRs form the largest family of cell surface receptor molecules. Members of this superfamily were first found by homology screening approaches, such as low-stringency hybridization and degenerate polymerase chain reaction (9, 10). By definition, the receptors cloned by these strategies are in search of their pharmacological identities, in particular of their natural ligands. They are thus termed orphan receptors (11). The quest to unravel these identities began in the second part of the 1980s, once molecular biological approaches (such as those noted above) began to identify orphan receptors.
At that time, there existed approximately 50 transmitters that were potential GPCR ligands, most of which had no identified cognate receptors. Such transmitters had traditionally been discovered on the basis of their ability to regulate physiological responses in organ or tissue preparations. For example, the classical neurotransmitters—e.g., the biogenic amines, acetylcholine, and certain neuropeptides—were discovered this way. Receptors for these molecular entities had been characterized pharmacologically but their molecular structures were unknown. The cloning of the orphan GPCRs offered a solution to this problem. The experimental approach was to express an orphan GPCR in a heterologous cell system and then test it in signal transduction assays with possible transmitters. Some thought that this approach was an unsolvable task. However, insights in interpreting the orphan GPCR tissue expression profiles, as well as random testing with possible ligands, proved successful in matching many of the first identified orphan GPCRs to known transmitters. The first deorphanized GPCRs, the 5HT1A and D2 dopamine receptors, were reported in 1988 (12). This strategy is known as reverse pharmacology (11, 13) because it is directed from a receptor to its natural ligand. Over a five-year span, more than 50 transmitters were matched to orphan GPCRs (14). It is important to note that, because most GPCRs have been matched to transmitters by using this strategy, the vast majority of the GPCRs started indeed as orphan receptors (Table 1).
Table 1.
History of GPCR deorphanizations
| 1986 | GPCRs share sequence similarities; orphan GPCRs can be found by homology |
| 1987 | Report of the first reported orphan GPCR: G-21 |
| 1988 | The first deorphanizations: G-21 as the 5-HT1A receptor and RGB-2 as the dopamine D2 receptor |
| 1989 | Introduction of the PCR-based screening approach for the discovery of orphan GPCRs |
| 1995 | The first novel transmitter as target of an orphan GPCR: orphanin FQ/nociceptin |
| 1990–present | Deorphanized GPCRs |
| Adenosine A1, A2A, A2B, A3 | |
| Adenosine diphosphate | |
| Adrenergic α1A, α1D, α2B, α2C, β1, β3 | |
| Adrenomedullin | |
| Anaphylatoxin C3a, C5a | |
| β-alanine, Angiotensin AT1b | |
| D-amino acids | |
| Apelin, basic L-α amino acids | |
| N-arachidonylglycine | |
| Bile acids | |
| Bradykinin | |
| Bombesin BB1, BB3 | |
| Bovine adrenal medulla peptide 22 | |
| Cannabinoid CB1, CB2 | |
| Calcitonin gene related peptide | |
| Chemokine CCR1, CCR2, CCR3, CCR4, CCR5, CXCR2, CXCR3, CXCR4 | |
| Cholecystokinin CCKa | |
| Cortistatin | |
| Dopamine D1, D2, D3, D4, D5 | |
| Eskine | |
| Estrogen | |
| 5-oxo-ETE | |
| Farnesyl pyrophosphate | |
| Follicle-stimulating hormone | |
| Formyl-peptide FPR2 | |
| Galanin type 2 | |
| Ghrelin | |
| Gonadotropin-releasing hormone | |
| Histamine H2, H3 | |
| α-ketoglutarate | |
| Kynurenic acid | |
| Kisspeptin/metastin | |
| Lactate | |
| Latrotoxin | |
| L-DOPA | |
| Leukotriene B4,C4,D4 | |
| Lysophosphatidic acid | |
| Lysophosphatidylcholine | |
| Lysophosphatidylinositol | |
| Medium- and long-chain fatty acids | |
| Melanin-concentrating hormone | |
| Melanocortin MC1, MC2, MC3, MC4, MC5 | |
| Melatonin MT1, MT2 | |
| 3- and 4-methyl-valeric acid | |
| Motilin | |
| Muscarinic acetylcholine M3, M4, M5 | |
| Neurokinin NK1, NK3 | |
| Neuromedin S, U | |
| Neuropeptides B/W | |
| Neuropeptide FF, AF | |
| Neuropeptide S | |
| Neuropeptide YY1, YY1-like, YY4 | |
| Neurotensin NTR2 | |
| Nicotinic acid | |
| Oleoylethanolamide | |
| Opioid κ, μ | |
| Orexins/hypocretins | |
| Orphanin FQ/nociceptin | |
| Oxysterols | |
| D-phenylalanine | |
| Prokineticins 1/2 | |
| Prolactin-releasing peptide | |
| Prostaglandin DP1, DP2 | |
| Prostanoid EP1, EP2, EP3, EP4, FP, IP1, TP | |
| Protease-activated 2,3 | |
| Proton | |
| Purinoceptor P2Y1, P2Y3, P2Y4, P2Y6, P2Y8 | |
| Psychosine | |
| RFamide-related protein 1,3 | |
| Pyroglutamylated arginine-phenylalanineamide peptide | |
| R-spondins | |
| Relaxin-1, -2, -3 | |
| Serotonin 5-HT1A, 5-HT1B, 5-HT1D, 5-HT1E, 5-HT1F, 5-HT2A, 5-HT2B, 5-HT4, 5-HT5A, 5-HT5B, 5-HT6, 5-HT7 | |
| Sphingosine 1-phosphate | |
| Sphingosylphosphorylcholine | |
| Somatostatin SST1, SST2, SST3, SST4, SST5 | |
| Succinate | |
| Trace amines | |
| Thyrotropin-stimulating hormone | |
| UDP-glucose | |
| Unsaturated long-chain free fatty acids | |
| Uracil nucleotide/cysteinyl leukotrienes | |
| Urotensin II | |
| Vasopressin V1b, V2 | |
| Zn2+ | |
Derived from Reference 14.
Abbreviation: GPCR, G protein–coupled receptor; PCR, polymerase chain reaction.
During the first part of the 1990s, application of the reverse pharmacology strategy matched many GPCRs to known transmitters. Receptors for many neurotransmitters, neuropeptides, hormones, and other first messengers were identified through the use of this approach (14). Moreover, when it was extended to identify receptors that are related on the basis of their primary sequences, the GPCR subfamilies were often found to be more complex than expected (Table 1).
In 1992, the GPCR that binds Δ9-tetrahydrocannabinol, although not an orphan GPCR in the strictest sense, was used as bait to identify anandamide (N-arachidonylethanolamide) (15) as the first novel lipid mediator found by reverse pharmacology. At that time, the number of orphan GPCRs was steadily increasing, as a result of the mining of the database of expressed-sequence-tagged cDNAs (16, 17). There was a paucity of known transmitters to match to all these receptors. Thus, it was proposed that many orphan receptors must bind ligands that had not been characterized because inactive receptors would be expected to have been evolutionarily discarded. This idea was the underlying reason for utilizing orphan GPCRs to isolate their natural ligands and discover novel transmitters (18). The later completion of the human genome (and other genomes) revealed the full extent of GPCR diversity and confirmed the conclusion that the GPCRs outnumbered the then-known potential ligands; thus, many novel transmitters are still to be found.
DEORPHANIZING GPCRs TO DISCOVER NOVEL TRANSMITTERS
The concept that an orphan receptor could be used as bait to identify a novel transmitter required the application of two technologies (18, 19): (a) preparing and purifying active compounds from tissue extracts and (b) monitoring receptor activation. Because of a lack of prior knowledge about the structure or activity of the transmitter, the success of this approach relies on the ability to carry out repetitive assays.
Regarding the purification of active compounds, we recognized that GPCRs are activated by a plethora of ligands (18, 20); most of these are small molecules, and a few are larger polypeptides (Table 2). Ligands of GPCRs include the following: GABA and glutamate, acetylcholine, biogenic amines, neuropeptides, chemokines, lipid mediators, nucleotides, polypeptide hormones, olfactory and gustatory molecules, and other naturally occurring chemicals such as calcium and hydrogen ions (protons). Thus, the ligand of an orphan GPCR can exhibit a spectrum of chemical compositions; it may be hydrophilic or hydrophobic and small or large. Because GPCRs whose sequences are related likely bind transmitters from the same chemical class, phylogenetic analyses of orphan receptors can help direct purification strategies. Another consideration is that GPCRs initiate responses that may be endocrine, paracrine, or exocrine in nature. Consequently, the transmitter may not be synthesized in the same tissue where the orphan receptor is expressed.
Table 2.
The GPCRs of the Rhodopsin family and their transmitters
| Ligand type | Phylogenetic cluster | Ligands | Number of paired receptors | Number of orphan receptors | Names of orphan receptors |
|---|---|---|---|---|---|
| Light | Opsin | Photon | 9 | 3 | GPR21, GPR45, GPR52 |
| Amine | Biogenic amine | Acetylcholine, serotonin, adrenaline, noradrenaline, dopamine, histamine, tyramine | 37 | 12 | GPR88, GPR152, GPR153, GPR160, GPR162, and seven TAARs |
| Melatonin | Melatonin | 2 | 4 | GPR50, GPR61, GPR62, GPR135 | |
| Peptide | Peptide | Endothelins, prokineticins, orexins, TRH, CCK neurokinins, neuromedins, NPY, NPAF-FF, GRP, PrRP, vasopressin, oxytocin, GnRH, neurotensin, NPS, motilin, ghrelin, apelin, QRFP | 37 | 5 | BRS-3, GPR37, GPR37L1, GPR150, GPR175 |
| SOG | Galanin, kisspeptin, opioids, somatostatins, urotensins, MCH, NPB-NPW | 18 | 1 | GPR151 | |
| MECA | MSH, AgRP | 5 | |||
| MAS-related | Various peptides | 4 | 8 | GPR19, GPR101, GPR161, MAS1L, MRGPRF, MRGPRE, MRGPRG, MRGPRX3 | |
| Chemokine | Angiotensin, bradykinin, formylpeptide, XCL relaxin, anaphylatoxin, CCL, CXCL, CXCL3 | 34 | 7 | CCRL2, GPR1, GPR15, GPR20, GPR25, GPR32, GPR182 | |
| Glycoprotein | LH, FSH, TSH, relaxin, R-spondins | 8 | 2 | GPR26, GPR78 | |
| Purine | Thrombin | 4 | |||
| Lipid | MECA | Cannabinoids, LPA, S1P, estrogen | 17 | 3 | GPR3, GPR6, GPR12 |
| Purine | Leukotrienes, LPI, LPS, NA-glycine, free fatty acids, PAF | 15 | 1 | GPR31 | |
| Prostaglandin | Prostaglandins | 9 | |||
| SOG | Leukotrienes | 2 | |||
| Nucleotide/nucleoside | Purine | ADP, ATP, UDP, UTP, UDP-glucose | 12 | 5 | GPR27, GPR85, GPR171, GPR173, GPR174 |
| MECA | Adenosine | 4 | |||
| Amino acid | MAS-related | β-alanine, D-amino acids | 2 | ||
| Proton | Purine | H+ | 4 | ||
| Olfactory | Olfactory | Olfactants | few | ~380 |
Quantifying receptor reactivity is also necessary. An orphan GPCR is typically expressed in a heterologous cell line in which its activation is recorded by one or more biochemical assays. Numerous intracellular signaling pathways can be assessed (21), but one hurdle remains. When investigators transfect a GPCR cDNA into a heterologous cell, the heterologous receptor is typically expressed under the regulation of a strong promoter, which directs the synthesis of high, often unnatural, amounts of the receptor protein. Although this approach can ensure the generation of readily detectable intracellular responses, it can also induce unexpected biochemical reactions. Activation of a highly expressed GPCR may spill into a signaling pathway that is not its canonical one (22). This secondary activation can result in the “discovery” of a low-affinity ligand, especially if it is present in high amounts; however, this ligand is not the true, primary one for that receptor. Artifactual interactions constitute another caveat of monitoring overexpressed receptor proteins. All GPCRs are hydrophobic molecules that have the potential, at least to a certain extent, to interact with one another. Such dimerization, including heterodimerization (23), can be an issue when deorphanizing GPCRs because an orphan GPCR coupled to a known GPCR can mistakenly lead one to conclude that the ligand of the known GPCR is also the ligand of the orphan GPCR. These caveats are particularly important when synthetic ligands are matched to orphan GPCRs because the cutoff of an acceptable affinity is defined by the investigator.
The deorphanization of GPCRs requires the application of numerous repetitive assays, many examples of which exist (e.g., 24, 25). Because screening large libraries of synthetic compounds is the mainstay of the pharmaceutical industry, recent years have seen the development of high-throughput technologies, many of which can be used or adapted for monitoring GPCR reactivity.
IMPACT OF THE DEORPHANIZATION OF GPCRs
The Salient Successes: The Novel Neuropeptides
The first attempt at finding a novel transmitter through an orphan GPCR led to the discovery of a neuropeptide (26, 27). The orphan GPCR ORL-1 (opioid receptor-like 1) (28) had been cloned through its homology to the opioid receptors (29). Its activation was monitored by quantifying decreases in intracellular cAMP levels. Because phylogenic analyses classified ORL-1 as a peptidergic GPCR and because ORL-1 is expressed in the central nervous system, peptidic brain tissue extracts were prepared, purified, and fractionated. Fractions were tested for their abilities to inhibit adenylyl cyclase activity in cells that were stably transfected with ORL-1. A peptide with 17 amino acids was ultimately isolated and named orphanin FQ or nociceptin (OFQ/N) (26, 27). Its structural similarities to the opioid peptides made it of immediate interest, yet it was proven not to bind to opioid receptors (30).
The second successful attempt at discovering novel transmitters through orphan GPCRs involved the screening of more than 50 different orphan GPCRs by assaying their ability to induce intracellular calcium release when subjected to peptidic extracts. Two peaks of activities were detected when the receptor HFGAN72 was challenged by purified extract fractions. This led to the characterization of two sequentially related peptides, orexin A and B (31). These peptides were identified independently as hypocretins by use of an RNA-subtraction approach (32). The discovery of orexins was rapidly followed by that of two other novel peptides, prolactin-releasing peptide and apelin, as the natural ligands of the orphan GPCRs GPR10 and APJ, respectively (33, 34). That discovery, in turn, was followed by the discovery of ghrelin as the endogenous ligand of the growth hormone secretagogue receptor (35). In the first decade of the millennium, kisspeptin/metastin was identified as the ligand of GPR54 (36); the prokineticins PK1 and PK2 were identified as the ligands of two highly related receptors, GPR73a and GPR73b (37, 38); and neuropeptide B and neuropeptide W were identified as the ligands of the primary sequence–related GPR7 and GPR8 (39–41). Neuropeptide S was later shown to be the natural ligand of GPR154 (42, 43), and neuromedin S was identified as that of the FM-4/TGR-1 receptor (44). In addition, relaxin-3 [also matched to the orphan LGR7 (45)] was found to be the ligand of GPCR142 (46). Finally, the R-spondins (a family of secreted proteins that activate Wnt/β-catenin signaling) were identified as endogenous ligands of LGR4, LGR5, and LGR6 (47, 48).
Discovery of each novel neuropeptide has resulted in a flurry of subsequent studies. Over the years, much has been learned from each of these new systems, although much still remains to be discovered. Studies have investigated the pharmacological profiles of those neuropeptides and the anatomical pathways of these newly recognized peptide/receptor systems. Altogether, they have helped provide information about a large number of physiological processes, ranging from higher brain function and peripheral nervous system–mediated organ control to immunological and endocrine functions. It would require much more space than is available here to enumerate all the roles that have been attributed to the new neuropeptides, so we encourage readers to consult recent reviews that discuss their biology (49–61). Notably, ghrelin has been shown to be a major regulator of energy homeostasis and growth hormone release (54, 62, 63). Furthermore, discovery of the orexin/hypocretin and kisspeptin/GPR54 neuropeptide systems has helped reveal the long-sought basis of particular genetic disorders: The former is critically involved in the etiology of narcolepsy (64, 65), and mutations in the latter reveal its role in the regulation of gonadotropins and in the induction of puberty (66).
All of these novel neuropeptide systems have been the focus of drug discovery efforts by the pharmaceutical industry. With the availability of synthetic agonists and antagonists, an understanding of their physiological functions and potential therapeutic applications is expected to substantially increase in the coming years. In fact, drugs targeting the OFQ/N system (50) and the orexin/hypocretin system (67, 68) have already entered clinical trials. Of note, these were the earliest discoveries related to deorphanization, and the development cycles for drugs, especially for ones directed at new targets, often require more than 10 years.
The Surprises: The Unexpected Transmitters
A so-called unexpected transmitter can be defined as a naturally occurring molecule that was not expected to exert its action through a specific receptor. Uridine is the major form of pyrimidine nucleoside taken up by the brain. It is phosphorylated to nucleotides, which are used for RNA synthesis as well as for the synthesis of membrane constituents and glycosylation. In addition, uracil nucleotides may be released from neuronal and glial cells, and GPCRs that recognize UTP and UDP (P2Y2, P2Y4, P2Y6) have been identified. A surprise was the discovery that the orphan GPCR KIAA0001 is activated not by UTP or UDP, but instead by UDP-glucose at an affinity in the 100-nM range (69). UDP-glucose was previously thought only to be a glucosyl donor in the biosynthesis of carbohydrates and not to be a transmitter. Consequently, KIAA0001 became P2Y14, whose physiological agonist is UDP-glucose.
Other surprises as unexpected transmitters are succinate and α-ketoglutarate, which activate the orphan GPCRs GPR91 and GPR99, respectively, with affinities in the 25–70-μM range (70). Succinic acid was identified as the agonist of GPR91 through its isolation and purification from renal extracts. Succinate had been previously shown to have a role in the reabsorption of phosphate and glucose in the proximal tubule and to stimulate gluconeogenesis (70). In GPR91-deficient mice, succinate was unable to induce an increase in blood pressure, a response that occurred in wild-type mice, thus implying that this activity of succinate relies on GPR91 activation. This was a surprising result because succinate is a tricarboxylic acid cycle intermediate that is not expected to be secreted from cells or to act extracellularly. Succinate, which can be released upon mechanical stress or cell death, acts through GPR91 to govern retinal angiogenesis and to participate in the ability of retinal ganglion neurons to act as sensors of ischemic stress (71).
The Promiscuous GPCRs
Several transmitters found via the deorphanization of GPCRs have led to a revision of ideas regarding pharmacological specificity. GPCRs are traditionally expected to exhibit specificity for one particular natural ligand (72), as a result of evolutionary processes that aim at diversifying the regulation of intercellular interactions. The one ligand–one receptor rule was known not to be absolute, as exemplified by the numerous receptors for catecholamines, acetylcholine, histamine, and serotonin. On the one hand, a GPCR may bind more than one naturally occurring transmitter (e.g., norepinephrine and epinephrine bind to multiple α1- and α2-adrenergic receptors and to β 1-adrenergic receptors), but such transmitters also share structural similarities and are often part of the same synthesis pathway (e.g., the neuropeptides synthesized from the same precursor) (73). On the other hand, a natural ligand may activate more than one GPCR, but those receptors generally share a higher degree of sequence homology such that they are grouped into a subfamily.
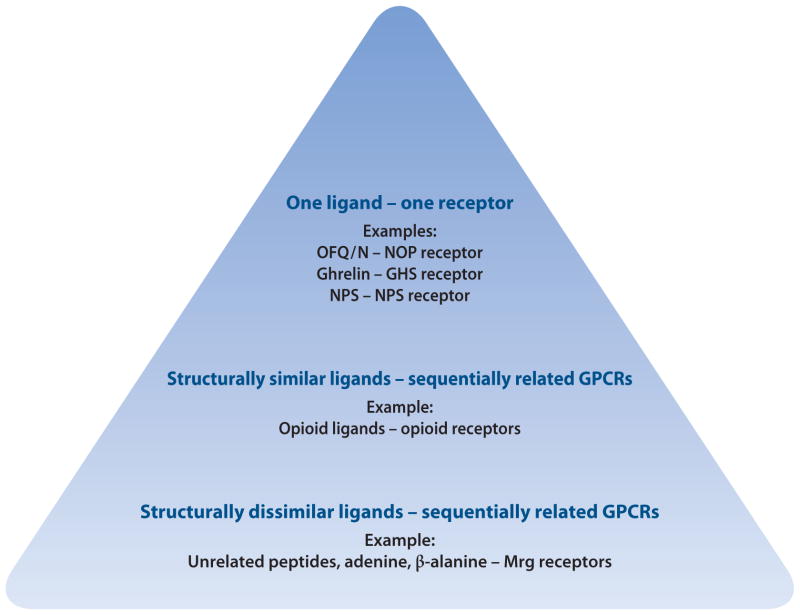
Such a relationship does not occur for the family of the MAS-related GPCRs (also known as Mrgs or sensory neuron–specific receptors). This family of orphan GPCRs, which is predominantly expressed in dorsal root ganglia, may have a role in nociception. A variable number of Mrgs exist in humans, rats, and mice, making it difficult to classify the receptors on the basis of orthologous relationships (74). Being part of a subfamily, the Mrgs might be expected to bind similar transmitters. Instead, physiological agonists for the Mrgs include structurally diverse transmitters: RFamide peptides for some mouse Mrgs (75); BAM22 (76) and cortistatin (77) for two human Mrgs; adenine (78) for a rat Mrg; and β-alanine (79) for an Mrg found in humans, rats, and mice. These transmitters were generally identified by random matching, which revealed specificity and efficiency in their activation of particular Mrgs. For example, the RFamides and BAM22 peptides have affinities in the low-nanomolar range (76, 80), as expected for peptides binding to GPCRs. In this respect, it is remarkable that one of the Mrg receptors, MrgA3, is activated by chloroquine, an agent that induces an itch response. Mice lacking a cluster of Mrg genes, including MrgA3, have significant deficits in chloroquine-induced itch, suggesting that some Mrg receptors may act as itch receptors (81) (Figure 1). These results indicate that some GPCR subfamilies may have broad pharmacological profiles, which may result from the organism’s need to have responses that can be considered pleiotropic, such as the one caused by itch.
Figure 1.
Evolution of the pharmacological specificity concept. Abbreviations: GHS, growth hormone secretagogue; GPCR, G protein–coupled receptor; OFQ/N, orphanin FQ or nociceptin; NOP, OFQ/N receptor; NPS, neuropeptide S.
WHERE DOES DEORPHANIZATION STAND NOW?
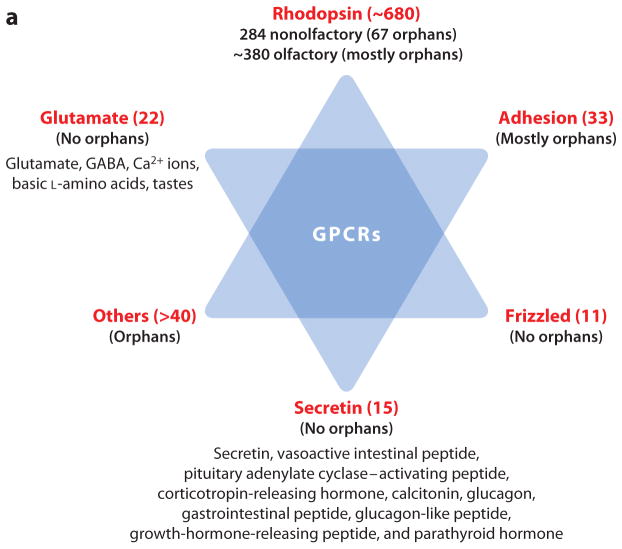
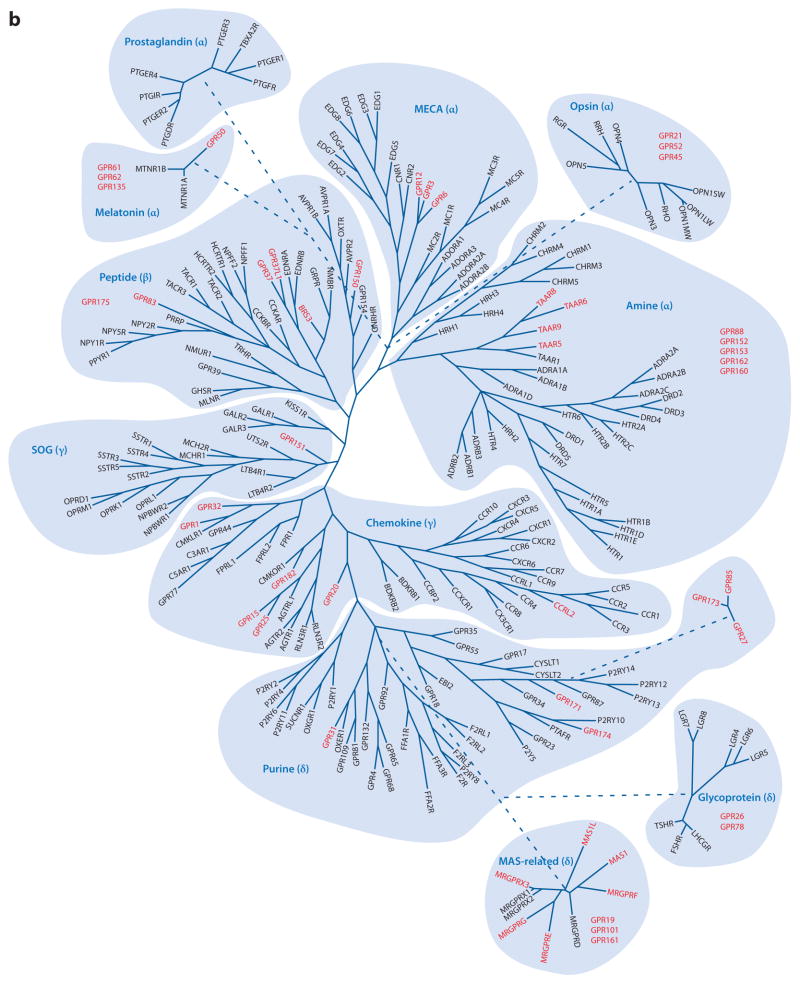
The human genome contains approximately 800 GPCR genes (and approximately 600 GPCR pseudogenes), more than 400 of which are classified as chemosensory because they recognize signals of external origin such as odors, pheromones, and tastes. On the basis of their nucleotide and inferred amino acid sequences, the GPCRs can be classified according to the GRAFS system (82) (Figure 2a), named on the basis of its five main classes: Glutamate, Rhodopsin, Adhesion, Frizzled, and Secretin. The Rhodopsin family can be further divided into four groups: the α-group, the β-group, the γ-group, and the δ-group (Figure 2b).
Figure 2.
(a) The G protein–coupled receptors (GPCRs) classified according to the GRAFS system (named on the basis of its five main classes: Glutamate, Rhodopsin, Adhesion, Frizzled, and Secretin). The numbers indicate the approximate numbers of receptors in the different classes. The ligands of the Glutamate and Secretin receptors are also indicated. (b) Schematic representation of the phylogenetic relationships of the human Rhodopsin GPCRs. The phylogenetic relationships are based on those discussed in Reference 83. Orphan GPCRs are shown in red. Some GPCR subgroups, analyzed as separate groups in Reference 83, have been placed according to their phylogenetic positions as described in Reference 85. Encircled areas outline phylogenetic clusters. Receptors with ambiguous relationships to other members are indicated by a dotted line.
Approximately 67 orphan GPCRs share structural relationships with the Rhodopsin families, 32 of which fall into the phylogenetic trees (Figure 1). In addition, 17 exhibit enough similarities to known GPCRs to be assigned to the different tree classes but not to particular branches. Analyses of the sequences of these 49 GPCRs lead us to propose putative types of ligands for them (Table 2). The other orphan GPCRs are only distantly related. Additional highly divergent orphan GPCRs are only distantly related to the Rhodopsin family (83, 85) (Figure 2).
Discrepancies Between the Phylogenetic Clusters and the Ligand Class
The common way to initiate a search for a novel transmitter is for the experimenter to choose a purification strategy that depends on the chemical nature of the expected ligand. GPCRs that are related by sequence are thought to bind ligands from the same chemical class; consequently, receptor phylogenetic analyses can help direct the purification strategy. However, the deorphanization of the GPCRs that bind lipid mediators is an example that shows how GPCRs that bind one chemical type of transmitter may not be closely related phylogenetically.
Lysophosphatidic acid (LPA) is a phospholipid that acts as a lipid mediator. Its first receptor was the orphan GPCR Edg-2, now renamed LPA1 (86). The discovery of LPA1 led to a search for other Edg-like receptors, and seven more were found. Edg-4 and Edg-7 are LPA receptors (LPA2 and LPA3), and Edg-1 (which has 33% identity with Edg-2) binds the structurally similar lipid sphingosine 1-phosphate, often termed S1P. On the basis of their sequences, the GPCRs closest to the Edg receptors are the cannabinoid receptors; those two groups represent a cluster of GPCRs activated by lipid mediators. However, another orphan, GPR23, was shown to also bind LPA (and is termed LPA4) (87, 88). GPR23 shares only 10% amino acid identity with LPA1 and is closely related to the purinergic P2Y receptors. P2Y5, the receptor most closely related to GPR23, was shown to bind purines but is also activated by LPA. In addition, the orphan GPR92, which is related to GPR23 and P2Y5, is also activated by LPA (and is termed LPA6). LPA6 is activated by uncharacteristically high concentrations of LPA (EC50 in the low micromolar range), which provides an example of the difficulty in defining the extent of the non-Edg-like LPA receptor cluster. GPR23, LPA5, and LPA6 therefore form such a cluster that is phylogenetically separated from the originally identified LPA receptors. Receptors that bind lipid mediators are found in at least three different phylogenetic clusters among the Rhodopsin receptors. It is possible that the ancestral Rhodopsin receptors were promiscuous in binding both lipids and other ligands and that the differences found for LPA receptors, for example, result from evolutionary subspecification. This is in line with the ability of GPR23 to bind LPA and purines or in line with GPR17, a dual uracil-nucleotide and cysteinyl-leukotriene receptor that is intermediate in its phylogenetic position between P2Y and CysLT receptors (89).
Another example of discrepancy between the phylogenetic clustering and the chemical nature of the ligand is found among the GPCRs that bind peptides. The Secretin receptors are activated by peptides but are most closely related to the Adhesion receptor family, as shown by the long evolutionary relationship among the common ancestors of these families. Yet the Adhesion receptors do not bind peptides (90). Moreover, the majority of peptide binding receptors are found among the Rhodopsin receptors in three different phylogenetic clusters: the peptide, the MECA family, and the SOG family. The MECA family includes GPCRs that bind peptide (melanin-concentrating hormone), lipids (cannabinoid and LPA), and purine ligands (adenosine). Such evolutionary “flip-flopping” between ligand classes highlights the difficulty of interpreting phylogenetic analyses.
The Reluctant Deorphanizations: One Example, Bombesin Receptor Subtype-3
A large group of GPCRs have not yet been deorphanized. Excluding the olfactory receptors, this group is composed of 53 receptors in the Rhodopsin family. Undoubtedly, many of these have been subjected to (as-yet unsuccessful) attempts at deorphanization. Below we discuss bombesin receptor subtype-3 (BRS-3) as an example of the difficulties that one can encounter in the deorphanization process.
The mammalian bombesin receptor family is composed of three GPCRs: neuromedin B receptor (NMB-R, or BB1), whose endogenous ligand is neuromedin B (NMB); gastrin-releasing peptide receptor (GRP-R, or BB2), which binds gastrin-releasing peptide (GRP); and the orphan GPCR BRS-3 (or BB3) (91). BRS-3 does not interact with high affinity with NMB, GRP, or any of the naturally occurring bombesin-like peptides (92). BRS-3-deficient mice develop late-onset mild obesity, partially attributed to a reduced metabolic rate (93). These mice also have hyperphagia, hypertension, insulin resistance, leptin resistance, and defects in glucose metabolism. Although the mechanism by which the lack of BRS-3 contributes to these metabolic syndromes is largely unknown, the potential role of BRS-3 in the regulation of energy homeostasis makes it a new target for therapeutic interventions related to obesity and diabetes. Furthermore, BRS-3 is expressed predominantly in the hypothalamus, which regulates homeostasis. Therefore, BRS-3 agonists have been synthesized and used to demonstrate that activation of BRS-3 leads to coupling with the Gq class of heterotrimeric G proteins and to calcium signaling (91).
Several groups, including ours, have attempted to identify the natural ligand of BRS-3. On the basis of its sequence, we expected that its ligand would be a peptide. The availability of synthetic agonists can assist in studies by being able to show that BRS-3 is expressed on cell surface membranes, that it can be activated, and that its activation can be monitored. Brain, as well as peripheral tissue, extracts prepared from different species have been tested. Because of the possible role of BRS-3 in the regulation of feeding, tissues from fed and starved animals have been studied. The possibility that BRS-3 would form dimers in order to be active has also been investigated, in particular by coexpressing BRS-3 with the BB1 or BB2 receptor. Unfortunately, though, the natural ligand of BRS-3 has thus far not been identified.
Some Caveats
In view of the difficulties that are encountered in deorphanizing GPCRs, it is not surprising that some efforts to identify ligands of orphan GPCRs led to claims of ligand-receptor pairings that could not be replicated and that remain controversial. Probably the earliest example was the claim that dynorphin A might be the endogenous ligand of ORL-1 (94), only to be corrected a few months later by the isolation and publication of the OFQ/N structure Another example was the pairing of obestatin with GPR39 (95). Obestatin is predicted to be a product of the processing of the ghrelin precursor protein, and GPR39 is structurally related to the receptors for ghrelin, motilin, and neuromedin U. However, subsequent reports did not verify the interaction of obestatin with GPR39 (96–101), and the original authors later reported that they were unable to reproduce their initial findings (102), even though another report also claimed that obestatin interacts with GPR39 (103). Other data then indicated that Zn2+ ions activate GPR39, and one study isolated Zn2+ from tissue extracts (98, 104). As indicated above for BRS-3, GPR39 is predicted to be a peptide receptor, making it difficult to imagine how it could evolve to be activated by an inorganic heavy metal ion. However, an olfactory receptor uses copper as a cofactor to recognize the volatile odorant (methylthio)methanethiol (105), indicating that metal ions may act as cofactors in the activation of GPCRs.
The pairing of the head activator peptide with GPR37 is more elusive (106). Head activator, a protein originally isolated from hydra, was claimed to exist in mammals (107), although a precursor for head activator has not been found in mammalian genomes. As a final example, Arg8-vasotocin, the teleost fish homolog of vasopressin, was proposed as a ligand for GPR154 (108), even though GPR154 is a receptor for neuropeptide S (42, 43). These examples highlight some of the obstacles and pitfalls in the search for the natural ligands of orphan GPCRs.
CONCLUSIONS AND PERSPECTIVE
Tremendous efforts have been directed at deorphanizing GPCRs. Striking successes have been achieved, and greater understanding of many physiological responses has resulted from this success. Many GPCRs remain to be deorphanized, and the number of successful deorphanizations has drastically decreased in recent years. BRS-3 provides an example of the problems that one encounters during GPCR deorphanization.
The most formidable challenge is ligand availability. Extremely limited quantities of a ligand can render the search fruitless. An example is the isolation of urotensin II–related peptide (109), which required the use of 400 rat brains as starting material. Another hurdle exists if the location of the orphan GPCR does not coincide with the site of synthesis of its ligand. The discovery of ghrelin (35) is an example: The receptor is in the brain, but its ligand is produced in the stomach. Limited availability of the ligand may also occur if the expression of the ligand is tightly regulated—for example, if its expression is low (and essentially nondetectable) under resting conditions but significantly increased in response to environmental or homeostatic factors, such as development, food restriction, reproductive status, circadian events, injury, or infection.
Another challenge is functionality of the orphan GPCR. An example is the requirement of an orphan GPCR to form functional heterodimers, as occurs for the GABAB receptor (110). Although the heterodimeric GABAB receptor may be an exception, relative to the large number of GPCRs that are functional by themselves, this possibility cannot be excluded, especially when studying orphan GPCRs. Additional accessory proteins may also be required; for example, the calcitonin/adrenomedullin GPCR changes its ligand selectivity and gains functionality by association with receptor activity-modifying proteins (RAMPs) (111). Although this form of heterodimerization has thus far been observed only among class B GPCRs, potentially as-yet undiscovered RAMPs of unrelated structures exist and are required by certain orphan GPCRs to form functional receptors. It is also possible that certain orphan GPCRs are not activated by ligands but instead act to modulate the activity of other GPCRs, as occurs for the interaction between the melatonin 1 receptor and GPR50, a related orphan GPCR (112).
A requirement of more than one ligand molecule to simultaneously bind to a GPCR might also explain unsuccessful attempts to discover natural ligands for certain orphan GPCRs. Some GPCRs bind dimeric ligands (glycoprotein hormones) and ligand-gated ion channels, e.g., the N-methyl-D-aspartate receptors, and are examples of multiple ligands modulating receptor protein activity. Moreover, secretory vesicles often contain neuropeptides together with a small-molecule transmitter, such as biogenic amines, glutamate, or nucleotides (e.g., ATP). Owing to the large number of possible combinations of such constituents along with “traditional” neuropeptide and small-molecule neurotransmitters, it will be difficult to include all such possibilities in a screening protocol. To date, there has been no identification of a ligand for an orphan receptor that would fit one of the scenarios just outlined, but each suggests starting points for new experimental approaches.
Finally, we believe it is important to consider the evolutionary perspective. The Rhodopsin GPCR family has undergone large expansion during vertebrate evolution; this expansion coincides with development of so-called higher brain functions. A characteristic of many of the Rhodopsin receptor genes is their lack of introns, a property that contrasts with those of the other GPCR families. Evolutionary events, driven by RNA-based mechanisms instead of classical gene duplications, may be responsible for the lack of introns (113), allowing for the appearance of chimeric proteins that contain sequences originating from different receptors. This might contribute to the discrepancies between phylogenetic clusters and ligand preferences and help explain the ligand promiscuity found for several GPCRs. Such relatively rapid evolution may also create greater opportunities for new GPCRs to act in noncanonical ways. In this respect, the lack of a specific ligand removes the evolutionary constraint essential to receptor function and may speed its evolution. Therefore, it is possible that some orphan GPCRs do not act via the typical transmitter-receptor binary model.
The experience described above for BRS-3 reflects some of the problems encountered in GPCR deorphanization. It is thus far unique in one aspect: Even though the natural agonist of BRS-3 in not known, a drug targeted to this receptor entered clinical trials (114). The pharmacological properties of an orally active, potent, and selective BRS-3 agonist has been tested in a double-blind, randomized, placebo-controlled study in healthy and obese male volunteers. A single dose transiently increased blood pressure and led to penile erection, feelings of hot or cold, and/or jitteriness but not to changes in body temperature, heart rate, plasma glucose levels, or feelings of hunger or satiety. These effects could be attributed to BRS-3 activation. To our knowledge, BRS-3 is the only orphan GPCR that has led to the development of a drug administered to humans. Thus, even if orphan receptors cannot be deorphanized, using them as therapeutic targets may still be possible. This is a testament to the importance of orphan GPCRs in pharmaceutical research.
SUMMARY POINTS.
GPCR deorphanization dates from 1988, when the first two orphan GPCRs were identified as the 5-HT1A and D2 dopamine receptors.
Researchers initially deorphanized orphan GPCRs by matching them to known neurotransmitters.
In 1995, GPCR deorphanization gained a new dimension, when an orphan GPCR was used as a target to discover a novel neuropeptide, OFQ/N.
GPCR deorphanization has led to the discovery of a dozen new families of neuropeptides—including the orexin/hypocretin family, the kisspeptin family, and the ghrelin family—and thereby has enriched understanding of physiological responses such as metabolism and reproduction and disease states such as narcolepsy.
GPCR deorphanization has revealed that some previously known biochemical entities display unexpected intercellular signaling roles. Examples include UDP-glucose, succinate, and α-ketoglutarate.
The results of GPCR deorphanization have extended the concept of specificity among classes of GPCR ligands.
More than 800 human GPCRs exist, half of which are chemosensory receptors. The Rhodopsin GPCR family includes approximately 67 GPCRs that await deorphanization.
Acknowledgments
In view of the extent of this subject, we are unable to cover it entirely. We apologize to our many colleagues whose work was not rightly recognized or was omitted. This work was supported by National Institutes of Health grants MH60231 and DA024746, an Established Investigator Award from the National Alliance for Research on Schizophrenia and Depression (NARSAD), and a Tourette Syndrome Association award to O.C.; by a National Institute of Health grant (MH093521) to R.K.R.; and by grants from the Swedish Research Council and the Novo Nordisk Foundation to R.F. and H.B.S.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.Dixon RA, Kobilka BK, Strader DJ, Benovic JL, Dohlman HG, et al. Cloning of the gene and cDNA for mammalian β-adrenergic receptor and homology with rhodopsin. Nature. 1986;321:75–79. doi: 10.1038/321075a0. [DOI] [PubMed] [Google Scholar]
- 2.Dohlman HG, Caron MG, Lefkowitz RJ. A family of receptors coupled to guanine nucleotide regulatory proteins. Biochemistry. 1987;26:2657–64. doi: 10.1021/bi00384a001. [DOI] [PubMed] [Google Scholar]
- 3.Bockaert J, Pin JP. Molecular tinkering of G protein–coupled receptors: an evolutionary success. EMBO J. 1999;18:1723–29. doi: 10.1093/emboj/18.7.1723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Angers S, Salahpour A, Bouvier M. Dimerization: an emerging concept for G protein–coupled receptor ontogeny and function. Annu Rev Pharmacol Toxicol. 2002;42:409–35. doi: 10.1146/annurev.pharmtox.42.091701.082314. [DOI] [PubMed] [Google Scholar]
- 5.Pierce KL, Premont RT, Lefkowitz RJ. Seven-transmembrane receptors. Nat Rev Mol Cell Biol. 2002;3:639–50. doi: 10.1038/nrm908. [DOI] [PubMed] [Google Scholar]
- 6.Kristiansen K. Molecular mechanisms of ligand binding, signaling, and regulation within the superfamily of G-protein-coupled receptors: molecular modeling and mutagenesis approaches to receptor structure and function. Pharmacol Ther. 2004;103:21–80. doi: 10.1016/j.pharmthera.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 7.Reiter E, Ahn S, Shukla AK, Lefkowitz RJ. Molecular mechanism of β-arrestin-biased agonism at seven-transmembrane receptors. Annu Rev Pharmacol Toxicol. 2012;52:179–97. doi: 10.1146/annurev.pharmtox.010909.105800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rask-Andersen M, Almen MS, Schiöth HB. Trends in the exploitation of novel drug targets. Nat Rev Drug Discov. 2011;10:579–90. doi: 10.1038/nrd3478. [DOI] [PubMed] [Google Scholar]
- 9.Bunzow JR, Van Tol HH, Grandy DK, Albert P, Salon J, et al. Cloning and expression of a rat D2 dopamine receptor cDNA. Nature. 1988;336:783–87. doi: 10.1038/336783a0. Demonstrates that an orphan GPCR, RGB-2, is the D2 dopamine receptor. [DOI] [PubMed] [Google Scholar]
- 10.Libert F, Parmentier M, Lefort A, Dinsart C, Van Sande J, et al. Selective amplification and cloning of four members of the G protein-coupled receptor family. Science. 1989;244:569–72. doi: 10.1126/science.2541503. [DOI] [PubMed] [Google Scholar]
- 11.Libert F, Schiffmann SN, Lefort A, Parmentier M, Gerard C, et al. The orphan receptor cDNA RDC7 encodes an A1 adenosine receptor. EMBO J. 1991;10:1677–82. doi: 10.1002/j.1460-2075.1991.tb07691.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fargin A, Raymond JR, Lohse MJ, Kobilka BK, Caron MG, Lefkowitz RJ. The genomic clone G-21 which resembles a β-adrenergic receptor sequence encodes the 5-HT1A receptor. Nature. 1988;335:358–60. doi: 10.1038/335358a0. Demonstrates that the orphan GPCR G-21 is the 5HT1A receptor. [DOI] [PubMed] [Google Scholar]
- 13.Mills A, Duggan MJ. Orphan seven transmembrane domain receptors: reversing pharmacology. Trends Biotechnol. 1994;12:47–49. doi: 10.1016/0167-7799(94)90099-X. [DOI] [PubMed] [Google Scholar]
- 14.Civelli O, Saito Y, Wang Z, Nothacker HP, Reinscheid RK. Orphan GPCRs and their ligands. Pharmacol Ther. 2006;110:525–32. doi: 10.1016/j.pharmthera.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 15.Devane WA, Hanus L, Breuer A, Pertwee RG, Stevenson LA, et al. Isolation and structure of a brain constituent that binds to the cannabinoid receptor. Science. 1992;258:1946–49. doi: 10.1126/science.1470919. [DOI] [PubMed] [Google Scholar]
- 16.Marchese A, Docherty JM, Nguyen T, Heiber M, Cheng R, et al. Cloning of human genes encoding novel G protein–coupled receptors. Genomics. 1994;23:609–18. doi: 10.1006/geno.1994.1549. [DOI] [PubMed] [Google Scholar]
- 17.O’Dowd BF, Nguyen T, Marchese A, Cheng R, Lynch KR, et al. Discovery of three novel G-protein-coupled receptor genes. Genomics. 1998;47:310–13. doi: 10.1006/geno.1998.5095. [DOI] [PubMed] [Google Scholar]
- 18.Civelli O. Functional genomics: the search for novel neurotransmitters and neuropeptides. FEBS Lett. 1998;430:55–58. doi: 10.1016/s0014-5793(98)00524-9. [DOI] [PubMed] [Google Scholar]
- 19.Civelli O, Nothacker HP, Reinscheid R. Reverse physiology: discovery of the novel neuropeptide, orphanin FQ/nociceptin. Crit Rev Neurobiol. 1998;12:163–76. doi: 10.1615/critrevneurobiol.v12.i3.10. [DOI] [PubMed] [Google Scholar]
- 20.Civelli O, Nothacker HP, Saito Y, Wang Z, Lin SH, Reinscheid RK. Novel neurotransmitters as natural ligands of orphan G-protein-coupled receptors. Trends Neurosci. 2001;24:230–37. doi: 10.1016/s0166-2236(00)01763-x. [DOI] [PubMed] [Google Scholar]
- 21.Ozawa A, Lindberg I, Roth B, Kroeze WK. Deorphanization of novel peptides and their receptors. AAPS J. 2010;12:378–84. doi: 10.1208/s12248-010-9198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu YF, Civelli O, Zhou QY, Albert PR. Cholera toxin-sensitive 3′,5′-cyclic adenosine monophosphate and calcium signals of the human dopamine-D1 receptor: selective potentiation by protein kinase A. Mol Endocrinol. 1992;6(11):1815–24. doi: 10.1210/mend.6.11.1282671. [DOI] [PubMed] [Google Scholar]
- 23.Ferre S, Baler R, Bouvier M, Caron MG, Devi LA, et al. Building a new conceptual framework for receptor heteromers. Nat Chem Biol. 2009;5:131–34. doi: 10.1038/nchembio0309-131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eglen RM, Reisine T. New insights into GPCR function: implications for HTS. Methods Mol Biol. 2009;552:1–13. doi: 10.1007/978-1-60327-317-6_1. [DOI] [PubMed] [Google Scholar]
- 25.Wess G, Urmann M, Sickenberger B. Medicinal chemistry: challenges and opportunities. Angew Chem Int Ed. 2001;40:3341–50. [PubMed] [Google Scholar]
- 26.Meunier JC, Mollereau C, Toll L, Suaudeau C, Moisand C, et al. Isolation and structure of the endogenous agonist of opioid receptor-like ORL1 receptor. Nature. 1995;377:532–35. doi: 10.1038/377532a0. Discover OFQ/N, the first new neuropeptide discovered by GPCR deorphanization. [DOI] [PubMed] [Google Scholar]
- 27.Reinscheid RK, Nothacker HP, Bourson A, Ardati A, Henningsen RA, et al. Orphanin FQ: a neuropeptide that activates an opioidlike G protein-coupled receptor. Science. 1995;270:792–94. doi: 10.1126/science.270.5237.792. Discover OFQ/N, the first new neuropeptide discovered by GPCR deorphanization. [DOI] [PubMed] [Google Scholar]
- 28.Mollereau C, Parmentier M, Mailleux P, Butour JL, Moisand C, et al. ORL1, a novel member of the opioid receptor family: cloning, functional expression and localization. FEBS Lett. 1994;341:33–38. doi: 10.1016/0014-5793(94)80235-1. [DOI] [PubMed] [Google Scholar]
- 29.Henderson G, McKnight AT. The orphan opioid receptor and its endogenous ligand—nociceptin/orphanin FQ. Trends Pharmacol Sci. 1997;18:293–300. [PubMed] [Google Scholar]
- 30.Reinscheid RK, Higelin J, Henningsen RA, Monsma FJ, Jr, Civelli O. Structures that delineate orphanin FQ and dynorphin A pharmacological selectivities. J Biol Chem. 1998;273:1490–95. doi: 10.1074/jbc.273.3.1490. [DOI] [PubMed] [Google Scholar]
- 31.Sakurai T, Amemiya A, Ishii M, Matsuzaki I, Chemelli RM, et al. Orexins and orexin receptors: a family of hypothalamic neuropeptides and G protein-coupled receptors that regulate feeding behavior. Cell. 1998;92:573–85. doi: 10.1016/s0092-8674(00)80949-6. [DOI] [PubMed] [Google Scholar]
- 32.de Lecea L, Kilduff TS, Peyron C, Gao X, Foye PE, et al. The hypocretins: hypothalamus-specific peptides with neuroexcitatory activity. Proc Natl Acad Sci USA. 1998;95:322–27. doi: 10.1073/pnas.95.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinuma S, Habata Y, Fujii R, Kawamata Y, Hosoya M, et al. A prolactin-releasing peptide in the brain. Nature. 1998;393:272–76. doi: 10.1038/30515. [DOI] [PubMed] [Google Scholar]
- 34.Tatemoto K, Hosoya M, Habata Y, Fujii R, Kakegawa T, et al. Isolation and characterization of a novel endogenous peptide ligand for the human APJ receptor. Biochem Biophys Res Commun. 1998;251:471–76. doi: 10.1006/bbrc.1998.9489. [DOI] [PubMed] [Google Scholar]
- 35.Kojima M, Hosoda H, Date Y, Nakazato M, Matsuo H, Kangawa K. Ghrelin is a growth-hormone-releasing acylated peptide from stomach. Nature. 1999;402:656–60. doi: 10.1038/45230. Presents how a GPCR deorphanization leads to the discovery of ghrelin. [DOI] [PubMed] [Google Scholar]
- 36.Ohtaki T, Shintani Y, Honda S, Matsumoto H, Hori A, et al. Metastasis suppressor gene KiSS-1 encodes peptide ligand of a G-protein-coupled receptor. Nature. 2001;411:613–17. doi: 10.1038/35079135. [DOI] [PubMed] [Google Scholar]
- 37.Lin DC, Bullock CM, Ehlert FJ, Chen JL, Tian H, Zhou QY. Identification and molecular characterization of two closely related G protein-coupled receptors activated by prokineticins/endocrine gland vascular endothelial growth factor. J Biol Chem. 2002;277:19276–80. doi: 10.1074/jbc.M202139200. [DOI] [PubMed] [Google Scholar]
- 38.Masuda Y, Takatsu Y, Terao Y, Kumano S, Ishibashi Y, et al. Isolation and identification of EG-VEGF/prokineticins as cognate ligands for two orphan G-protein-coupled receptors. Biochem Biophys Res Commun. 2002;293:396–402. doi: 10.1016/S0006-291X(02)00239-5. [DOI] [PubMed] [Google Scholar]
- 39.Fujii R, Yoshida H, Fukusumi S, Habata Y, Hosoya M, et al. Identification of a neuropeptide modified with bromine as an endogenous ligand for GPR7. J Biol Chem. 2002;277:34010–16. doi: 10.1074/jbc.M205883200. [DOI] [PubMed] [Google Scholar]
- 40.Shimomura Y, Harada M, Goto M, Sugo T, Matsumoto Y, et al. Identification of neuropeptide W as the endogenous ligand for orphan G-protein-coupled receptors GPR7 and GPR8. J Biol Chem. 2002;277:35826–32. doi: 10.1074/jbc.M205337200. [DOI] [PubMed] [Google Scholar]
- 41.Brezillon S, Lannoy V, Franssen JD, Le Poul E, Dupriez V, et al. Identification of natural ligands for the orphan G protein-coupled receptors GPR7 and GPR8. J Biol Chem. 2003;278:776–83. doi: 10.1074/jbc.M206396200. [DOI] [PubMed] [Google Scholar]
- 42.Xu YL, Reinscheid RK, Huitron-Resendiz S, Clark SD, Wang Z, et al. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron. 2004;43:487–97. doi: 10.1016/j.neuron.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 43.Sato S, Shintani Y, Miyajima N, Yoshimura K. World Patent No. WO 02/345 A1 Novel G protein-coupled receptor protein and DNA thereof. 2002
- 44.Mori K, Miyazato M, Ida T, Murakami N, Serino R, et al. Identification of neuromedin S and its possible role in the mammalian circadian oscillator system. EMBO J. 2005;24:325–35. doi: 10.1038/sj.emboj.7600526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sudo S, Kumagai J, Nishi S, Layfield S, Ferraro T, et al. H3 relaxin is a specific ligand for LGR7 and activates the receptor by interacting with both the ectodomain and the exoloop 2. J Biol Chem. 2003;278:7855–62. doi: 10.1074/jbc.M212457200. [DOI] [PubMed] [Google Scholar]
- 46.Liu C, Chen J, Sutton S, Roland B, Kuei C, et al. Identification of relaxin-3/INSL7 as a ligand for GPCR142. J Biol Chem. 2003;278:50765–70. doi: 10.1074/jbc.M308996200. [DOI] [PubMed] [Google Scholar]
- 47.de Lau W, Barker N, Low TY, Koo BK, Li VSW, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–97. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 48.Carmon KS, Gong X, Lin QS, Thomas A, Liu QY. R-spondins function as ligands of the orphan receptors LGR4 and LGR5 to regulate Wnt/β-catenin signaling. Proc Natl Acad Sci USA. 2011;108:11452–7. doi: 10.1073/pnas.1106083108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Civelli O. The orphanin FQ/nociceptin (OFQ/N) system. Results Probl Cell Differ. 2008;46:1–25. doi: 10.1007/400_2007_057. [DOI] [PubMed] [Google Scholar]
- 50.Lambert DG. The nociceptin/orphanin FQ receptor: a target with broad therapeutic potential. Nat Rev Drug Discov. 2008;7:694–710. doi: 10.1038/nrd2572. [DOI] [PubMed] [Google Scholar]
- 51.Matsuki T, Sakurai T. Orexins and orexin receptors: from molecules to integrative physiology. Results Probl Cell Differ. 2008;46:27–55. doi: 10.1007/400_2007_047. [DOI] [PubMed] [Google Scholar]
- 52.Willie JT, Chemelli RM, Sinton CM, Yanagisawa M. To eat or to sleep? Orexin in the regulation of feeding and wakefulness. Annu Rev Neurosci. 2001;24:429–58. doi: 10.1146/annurev.neuro.24.1.429. [DOI] [PubMed] [Google Scholar]
- 53.Lin SH. Prolactin-releasing peptide. Results Probl Cell Differ. 2008;46:57–88. doi: 10.1007/400_2007_048. [DOI] [PubMed] [Google Scholar]
- 54.Kojima M, Kangawa K. Structure and function of ghrelin. Results Probl Cell Differ. 2008;46:89–115. doi: 10.1007/400_2007_049. [DOI] [PubMed] [Google Scholar]
- 55.Colledge WH. GPR54 and kisspeptins. Results Probl Cell Differ. 2008;46:117–43. doi: 10.1007/400_2007_050. [DOI] [PubMed] [Google Scholar]
- 56.Colledge WH. Kisspeptins and GnRH neuronal signalling. Trends Endocrinol Metab. 2009;20:115–21. doi: 10.1016/j.tem.2008.10.005. [DOI] [PubMed] [Google Scholar]
- 57.Reinscheid RK. Neuropeptide S: anatomy, pharmacology, genetics and physiological functions. Results Probl Cell Differ. 2008;46:145–58. doi: 10.1007/400_2007_051. [DOI] [PubMed] [Google Scholar]
- 58.Zhou QY, Meidan R. Biological function of prokineticins. Results Probl Cell Differ. 2008;46:181–99. doi: 10.1007/400_2007_053. [DOI] [PubMed] [Google Scholar]
- 59.Mori K, Miyazato M, Kangawa K. Neuromedin S: discovery and functions. Results Probl Cell Differ. 2008;46:201–12. doi: 10.1007/400_2007_054. [DOI] [PubMed] [Google Scholar]
- 60.Liu C, Lovenberg TW. Relaxin-3, INSL5, and their receptors. Results Probl Cell Differ. 2008;46:213–37. doi: 10.1007/400_2007_055. [DOI] [PubMed] [Google Scholar]
- 61.Hondo M, Ishii M, Sakurai T. The NPB/NPW neuropeptide system and its role in regulating energy homeostasis, pain, and emotion. Results Probl Cell Differ. 2008;46:239–56. doi: 10.1007/400_2007_056. [DOI] [PubMed] [Google Scholar]
- 62.Kojima M, Kangawa K. Ghrelin: from gene to physiological function. Results Probl Cell Differ. 2010;50:185–205. doi: 10.1007/400_2009_28. [DOI] [PubMed] [Google Scholar]
- 63.Kojima M, Kangawa K. Ghrelin: more than endogenous growth hormone secretagogue. Ann N Y Acad Sci. 2010;1200:140–48. doi: 10.1111/j.1749-6632.2010.05516.x. [DOI] [PubMed] [Google Scholar]
- 64.Lin L, Faraco J, Li R, Kadotani H, Rogers W, et al. The sleep disorder canine narcolepsy is caused by a mutation in the hypocretin (orexin) receptor 2 gene. Cell. 1999;98:365–76. doi: 10.1016/s0092-8674(00)81965-0. Shows that narcolepsy is caused by a mutation of a deorphanized GPCR (hypocretin/orexin receptor 2) in dogs. [DOI] [PubMed] [Google Scholar]
- 65.Chemelli RM, Willie JT, Sinton CM, Elmquist JK, Scammell T, et al. Narcolepsy in orexin knockout mice: molecular genetics of sleep regulation. Cell. 1999;98:437–51. doi: 10.1016/s0092-8674(00)81973-x. Shows that ablation of a deorphanized GPCR system (orexin/hypocretin) causes narcolepsy. [DOI] [PubMed] [Google Scholar]
- 66.de Roux N, Genin E, Carel JC, Matsuda F, Chaussain JL, Milgrom E. Hypogonadotropic hypogonadism due to loss of function of the KiSS1-derived peptide receptor GPR54. Proc Natl Acad Sci USA. 2003;100:10972–76. doi: 10.1073/pnas.1834399100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Coleman PJ, Renger JJ. Orexin receptor antagonists: a review of promising compounds patented since 2006. Expert Opin Ther Pat. 2010;20:307–24. doi: 10.1517/13543770903567085. [DOI] [PubMed] [Google Scholar]
- 68.Scammell TE, Winrow CJ. Orexin receptors: pharmacology and therapeutic opportunities. Annu Rev Pharmacol Toxicol. 2011;51:243–66. doi: 10.1146/annurev-pharmtox-010510-100528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chambers JK, Macdonald LE, Sarau HM, Ames RS, Freeman K, et al. A G protein-coupled receptor for UDP-glucose. J Biol Chem. 2000;275:10767–71. doi: 10.1074/jbc.275.15.10767. [DOI] [PubMed] [Google Scholar]
- 70.He W, Miao FJ, Lin DC, Schwandner RT, Wang Z, et al. Citric acid cycle intermediates as ligands for orphan G-protein-coupled receptors. Nature. 2004;429:188–93. doi: 10.1038/nature02488. [DOI] [PubMed] [Google Scholar]
- 71.Sapieha P, Sirinyan M, Hamel D, Zaniolo K, Joyal JS, et al. The succinate receptor GPR91 in neurons has a major role in retinal angiogenesis. Nat Med. 2008;14:1067–76. doi: 10.1038/nm.1873. [DOI] [PubMed] [Google Scholar]
- 72.Goldstein A. Interactions of narcotic antagonists with receptor sites. Adv Biochem Psychopharmacol. 1973;8:471–81. [PubMed] [Google Scholar]
- 73.Douglass J, Civelli O, Herbert E. Polyprotein gene expression: generation of diversity of neuroendocrine peptides. Annu Rev Biochem. 1984;53:665–715. doi: 10.1146/annurev.bi.53.070184.003313. [DOI] [PubMed] [Google Scholar]
- 74.Zylka MJ, Dong X, Southwell AL, Anderson DJ. Atypical expansion in mice of the sensory neuron-specific Mrg G protein-coupled receptor family. Proc Natl Acad Sci USA. 2003;100:10043–48. doi: 10.1073/pnas.1732949100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Dong X, Han S, Zylka MJ, Simon MI, Anderson DJ. A diverse family of GPCRs expressed in specific subsets of nociceptive sensory neurons. Cell. 2001;106:619–32. doi: 10.1016/s0092-8674(01)00483-4. [DOI] [PubMed] [Google Scholar]
- 76.Lembo PM, Grazzini E, Groblewski T, O’Donnell D, Roy MO, et al. Proenkephalin A gene products activate a new family of sensory neuron–specific GPCRs. Nat Neurosci. 2002;5:201–9. doi: 10.1038/nn815. [DOI] [PubMed] [Google Scholar]
- 77.Robas N, Mead E, Fidock M. MrgX2 is a high potency cortistatin receptor expressed in dorsal root ganglion. J Biol Chem. 2003;278:44400–4. doi: 10.1074/jbc.M302456200. [DOI] [PubMed] [Google Scholar]
- 78.Bender E, Buist A, Jurzak M, Langlois X, Baggerman G, et al. Characterization of an orphan G protein-coupled receptor localized in the dorsal root ganglia reveals adenine as a signaling molecule. Proc Natl Acad Sci USA. 2002;99:8573–78. doi: 10.1073/pnas.122016499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Shinohara T, Harada M, Ogi K, Maruyama M, Fujii R, et al. Identification of a G protein-coupled receptor specifically responsive to β-alanine. J Biol Chem. 2004;279:23559–64. doi: 10.1074/jbc.M314240200. [DOI] [PubMed] [Google Scholar]
- 80.Han SK, Dong X, Hwang JI, Zylka MJ, Anderson DJ, Simon MI. Orphan G protein-coupled receptors MrgA1 and MrgC11 are distinctively activated by RF-amide-related peptides through the Gαq/11 pathway. Proc Natl Acad Sci USA. 2002;99:14740–45. doi: 10.1073/pnas.192565799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Liu Q, Tang Z, Surdenikova L, Kim S, Patel KN, et al. Sensory neuron-specific GPCR Mrgprs are itch receptors mediating chloroquine-induced pruritus. Cell. 2009;139:1353–65. doi: 10.1016/j.cell.2009.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Fredriksson R, Lagerstrom MC, Lundin LG, Schiöth HB. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Mol Pharmacol. 2003;63:1256–72. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- 83.Gloriam DE, Fredriksson R, Schiöth HB. The G protein-coupled receptor subset of the rat genome. BMC Genomics. 2007;8:338. doi: 10.1186/1471-2164-8-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lagerstrom MC, Schiöth HB. Structural diversity of G protein-coupled receptors and significance for drug discovery. Nat Rev Drug Discov. 2008;7:339–57. doi: 10.1038/nrd2518. [DOI] [PubMed] [Google Scholar]
- 85.Bjarnadottir TK, Gloriam DE, Hellstrand SH, Kristiansson H, Fredriksson R, Schiöth HB. Comprehensive repertoire and phylogenetic analysis of the G protein-coupled receptors in human and mouse. Genomics. 2006;88:263–73. doi: 10.1016/j.ygeno.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 86.Choi JW, Herr DR, Noguchi K, Yung YC, Lee CW, et al. LPA receptors: subtypes and biological actions. Annu Rev Pharmacol Toxicol. 2010;50:157–86. doi: 10.1146/annurev.pharmtox.010909.105753. [DOI] [PubMed] [Google Scholar]
- 87.Bradshaw HB, Lee SH, McHugh D. Orphan endogenous lipids and orphan GPCRs: a good match. Prostaglandins Other Lipid Mediat. 2009;89:131–34. doi: 10.1016/j.prostaglandins.2009.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Yanagida K, Ishii S. Non-Edg family LPA receptors: the cutting edge of LPA research. J Biochem. 2011;150:223–32. doi: 10.1093/jb/mvr087. [DOI] [PubMed] [Google Scholar]
- 89.Ciana P, Fumagalli M, Trincavelli ML, Verderio C, Rosa P, et al. The orphan receptor GPR17 identified as a new dual uracil nucleotides/cysteinyl-leukotrienes receptor. EMBO J. 2006;25:4615–27. doi: 10.1038/sj.emboj.7601341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Nordstrom KJ, Lagerstrom MC, Waller LM, Fredriksson R, Schiöth HB. The Secretin GPCRs descended from the family of Adhesion GPCRs. Mol Biol Evol. 2009;26:71–84. doi: 10.1093/molbev/msn228. [DOI] [PubMed] [Google Scholar]
- 91.Jensen RT, Battey JF, Spindel ER, Benya RV. International union of pharmacology. LXVIII Mammalian bombesin receptors: nomenclature, distribution, pharmacology, signaling, and functions in normal and disease states. Pharmacol Rev. 2008;60:1–42. doi: 10.1124/pr.107.07108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Mantey SA, Weber HC, Sainz E, Akeson M, Ryan RR, et al. Discovery of a high affinity radioligand for the human orphan receptor, bombesin receptor subtype 3, which demonstrates that it has a unique pharmacology compared with other mammalian bombesin receptors. J Biol Chem. 1997;272:26062–71. doi: 10.1074/jbc.272.41.26062. [DOI] [PubMed] [Google Scholar]
- 93.Ohki-Hamazaki H, Watase K, Yamamoto K, Ogura H, Yamano M, et al. Mice lacking bombesin receptor subtype-3 develop metabolic defects and obesity. Nature. 1997;390:165–69. doi: 10.1038/36568. [DOI] [PubMed] [Google Scholar]
- 94.Zhang S, Yu L. Identification of dynorphins as endogenous ligands for an opioid receptor-like orphan receptor. J Biol Chem. 1995;270:22772–76. doi: 10.1074/jbc.270.39.22772. [DOI] [PubMed] [Google Scholar]
- 95.Zhang JV, Ren PG, Avsian-Kretchmer O, Luo CW, Rauch R, et al. Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science. 2005;310:996–99. doi: 10.1126/science.1117255. [DOI] [PubMed] [Google Scholar]
- 96.Chartrel N, Alvear-Perez R, Leprince J, Iturrioz X, Reaux-Le Goazigo A, et al. Comment on “Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science. 2007;315:766. doi: 10.1126/science.1135047. [DOI] [PubMed] [Google Scholar]
- 97.Gourcerol G, St-Pierre DH, Tache Y. Lack of obestatin effects on food intake: Should obestatin be renamed ghrelin-associated peptide (GAP)? Regul Pept. 2007;141:1–7. doi: 10.1016/j.regpep.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 98.Holst B, Egerod KL, Schild E, Vickers SP, Cheetham S, et al. GPR39 signaling is stimulated by zinc ions but not by obestatin. Endocrinology. 2007;148:13–20. doi: 10.1210/en.2006-0933. [DOI] [PubMed] [Google Scholar]
- 99.Lauwers E, Landuyt B, Arckens L, Schoofs L, Luyten W. Obestatin does not activate orphan G protein-coupled receptor GPR39. Biochem Biophys Res Commun. 2006;351:21–25. doi: 10.1016/j.bbrc.2006.09.141. [DOI] [PubMed] [Google Scholar]
- 100.Nogueiras R, Pfluger P, Tovar S, Arnold M, Mitchell S, et al. Effects of obestatin on energy balance and growth hormone secretion in rodents. Endocrinology. 2007;148:21–26. doi: 10.1210/en.2006-0915. [DOI] [PubMed] [Google Scholar]
- 101.Seoane LM, Al-Massadi O, Pazos Y, Pagotto U, Casanueva FF. Central obestatin administration does not modify either spontaneous or ghrelin-induced food intake in rats. J Endocrinol Investig. 2006;29:RC13–15. doi: 10.1007/BF03344174. [DOI] [PubMed] [Google Scholar]
- 102.Zhang JV, Klein C, Ren PG, Kass S, Ver Donck L, et al. Response to comment on “Obestatin, a peptide encoded by the ghrelin gene, opposes ghrelin’s effects on food intake. Science. 2007;315:766. [Google Scholar]
- 103.Zhang JV, Jahr H, Luo CW, Klein C, Van Kolen K, et al. Obestatin induction of early-response gene expression in gastrointestinal and adipose tissues and the mediatory role of G protein-coupled receptor, GPR39. Mol Endocrinol. 2008;22:1464–75. doi: 10.1210/me.2007-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Yasuda SI, Miyazaki T, Munechika K, Yamashita M, Ikeda Y, Kamizono A. Isolation of Zn2+ as an endogenous agonist of GPR39 from fetal bovine serum. J Recept Signal Transduct Res. 2007;27:235–46. doi: 10.1080/10799890701506147. [DOI] [PubMed] [Google Scholar]
- 105.Duan X, Block E, Li Z, Connelly T, Zhang J, et al. Crucial role of copper in detection of metal-coordinating odorants. Proc Natl Acad Sci USA. 2012;109:3492–97. doi: 10.1073/pnas.1111297109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Rezgaoui M, Susens U, Ignatov A, Gelderblom M, Glassmeier G, et al. The neuropeptide head activator is a high-affinity ligand for the orphan G-protein-coupled receptor GPR37. J Cell Sci. 2006;119:542–49. doi: 10.1242/jcs.02766. [DOI] [PubMed] [Google Scholar]
- 107.Bodenmüller H, Schaller HC. Conserved amino acid sequence of a neuropeptide, the head activator, from coelenterates to humans. Nature. 1981;293:579–80. doi: 10.1038/293579a0. [DOI] [PubMed] [Google Scholar]
- 108.Usui M, Aoshima H, Yamamoto Y, Luziga C, Mamba K. Characterization and distribution of an arginine vasotocin receptor in mouse. J Vet Med Sci. 2006;68:655–61. doi: 10.1292/jvms.68.655. [DOI] [PubMed] [Google Scholar]
- 109.Sugo T, Murakami Y, Shimomura Y, Haradam M, Abe M, et al. Identification of urotensin II-related peptide as the urotensin II-immunoreactive molecule in the rat brain. Biochem Biophys Res Commun. 2003;310:860–68. doi: 10.1016/j.bbrc.2003.09.102. [DOI] [PubMed] [Google Scholar]
- 110.Kaupmann K, Huggel K, Heid J, Flor PJ, Bischoff S, et al. Expression cloning of GABAB receptors uncovers similarity to metabotropic glutamate receptors. Nature. 1997;386:239–46. doi: 10.1038/386239a0. [DOI] [PubMed] [Google Scholar]
- 111.McLatchie LM, Fraser NJ, Main MJ, Wise A, Brown J, et al. RAMPs regulate the transport and ligand specificity of the calcitonin-receptor-like receptor. Nature. 1998;393:333–39. doi: 10.1038/30666. [DOI] [PubMed] [Google Scholar]
- 112.Levoye A, Dam J, Ayoub MA, Guilllaume JL, Couturier C, et al. The orphan GPR50 receptor specifically inhibits MT1 melatonin receptor function through heterodimerization. EMBO J. 2006;25:3012–23. doi: 10.1038/sj.emboj.7601193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Fridmanis D, Fredriksson R, Kapa I, Schiöth HB, Klovins J. Formation of new genes explains lower intron density in mammalian Rhodopsin G protein-coupled receptors. Mol Phylogenet Evol. 2007;43:864–80. doi: 10.1016/j.ympev.2006.11.007. [DOI] [PubMed] [Google Scholar]
- 114.Reitman M, Dishy V, Moreau A, Denney WS, Liu C, et al. Pharmacokinetics and pharmaco-dynamics of MK-5046, a bombesin receptor subtype-3 (BRS-3) agonist, in healthy patients. J Clin Pharmacol. 2012;52:1306–16. doi: 10.1177/0091270011419854. [DOI] [PMC free article] [PubMed] [Google Scholar]