Abstract
There are significant challenges in identifying receptor-specific functional interactors in vivo. In this issue of Neuron, Ge et al (2018) identify a novel GABAA receptor (GABAAR)-interacting protein, Clptm1 that regulates forward trafficking of GABAARs and inhibitory transmission.
Main text
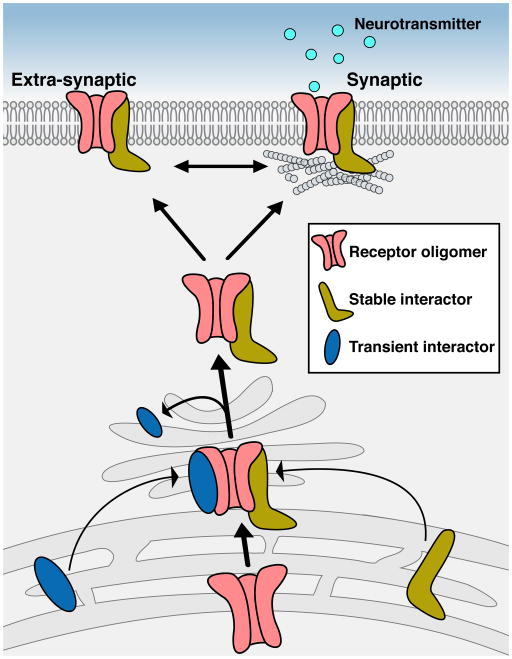
Interactions between neurotransmitter receptors and regulatory proteins control synaptic transmission and plasticity through various pathways. In order to fulfill their function, neurotransmitter receptors first exit the endoplasmic reticulum (ER) and travel to the cell surface. During this process, interactions between the receptor and various binding partners promote and/or regulate receptor assembly and trafficking. Through these interactions, a mature receptor complex is formed, enabling its proper localization and function at the plasma membrane and synapses (Figure 1). Indeed, protein complexes between a neurotransmitter receptor and regulatory, interacting-proteins (‘interactors’) emerge as a common theme in receptor biology. For example, excitatory AMPA and kainate glutamate receptors create a stable complex with their auxiliary proteins TARPs and Neto, respectively, and these interactions are required for proper synaptic transmission (Jackson and Nicoll, 2011; Yan and Tomita, 2012). Recently, GABAARs were found to form a stable tripartite complex with GARLH and Neuroligin-2 (Yamasaki et al., 2017), which is required for synaptic localization of GABAARs, but not forward trafficking (Davenport et al., 2017; Yamasaki et al., 2017).
FIGURE 1. Stable and transient interactors accompany and control neurotransmitter receptors.
During the process of forward trafficking, receptors form complexes with both transient and stable interactors that either limit or facilitate their activity. Transient interactions can support receptor maturation and transport from ER to plasma membrane, whereas stable interactions (such as with auxiliary subunits) can also modulate membrane localization and functional properties. By their nature, functional transient interactors are more challenging to be identified by proteomic methods.
Not only stable, but also transient interactions help shape the receptor landscape on the neuronal membrane (Figure 1). These transient interactions can take place along the various stages of forward receptor trafficking, including complex assembly, ER exit, trafficking, and exocytosis. Uncovering the specific interactors controlling receptor function is critical to our mechanistic understanding of neuronal signaling, yet the biochemical isolation and identification of transient interactors in vivo has proven technically demanding. A major challenge is the fact that transient complexes are (by definition) less abundant, and are therefore harder to detect. Consequently, proteomic approaches to reveal transient interactions may lead to large sets of candidates, with dozens of potential interactors. In some cases, rather than relying solely on classical proteomic approaches, ‘fishing’ for specific transient interactors can be achieved by adopting complementary methods such as high-throughput functional assays. Indeed, taking such a combined approach has led in the past to a functional confirmation of ten AMPA-receptor interactors and identified Porcupine as a transient interactor that modulates AMPAR forward trafficking (Erlenhardt et al., 2016; Schwenk et al., 2012).
A further complicating factor in studying neurotransmitter receptors is heterogeneity in receptor composition. In the case of GABAAR, the receptor assembles as a hetero-pentamer, containing at least one of six α subunits, one of three β subunits and one of ten non-α/β subunits. Various combinations of these subunits give rise to a high degree of GABAAR molecular diversity in the brain. Therefore, identifying specific molecules that control GABAAR trafficking may become a daunting task if trying to account for the many possible subunit combinations.
In this issue of Neuron, overcoming many of the challenges inherent to proteomic-based identification of GABAARs-specific transient interactors, Ge et al. (Ge et al., 2018) applied an optimized proteomic approach to identify Clptm1 as a GABAAR-interacting protein that controls receptor forward trafficking in neurons. To circumvent potential caveats in specificity of antibodies in proteomics, the authors applied a transgenic approach. Using knockin mice that express His-FLAG-YFP-GABAAR γ2, the authors purified the receptor complex through a tandem purification technique with nickel-charged affinity resin and an anti-GFP antibody. This has led to the identification of 39 proteins as well as previously identified associated proteins of GABAAR subunits and neuroligin-2. Next, Ge et al focused on six proteins for further analysis of their interaction in HEK293 cells, and found that three transmembrane proteins (Clptm1, Itm2C and Glgl1) could interact with GABAARs, but not with glycine receptors.
The authors next assessed the functional role of these three proteins and found that only Clptm1 altered GABAARs-mediated currents. This was demonstrated by a substantial reduction in GABA-evoked currents, with no apparent effect on glycine receptor activity. Over-expression of a Myc-tagged Clptm1 resulted in substantial accumulation of Clptm1 and GABAAR in the ER and reduced surface expression of GABAARs, suggesting a role of Clptm1 in intracellular retention. Perhaps most remarkably, knocking down Clptm1 increased GABA-evoked currents in primary cultured neurons, and overexpression and knockdown of clptm1 in vivo reduced and increased miniature inhibitory postsynaptic current (mIPSC) amplitudes, respectively, without altering excitatory miniature excitatory postsynaptic currents (mEPSCs). This set of experiments establishes Cltpm1 as an intracellular molecule for controlling GABAAR forward trafficking, acting likely via ER retention.
Intriguingly, the bi-directional effects of Clptm1 knockdown and overexpression on mIPSCs resemble the process of homeostatic synaptic scaling. In primary neuronal culture preparations, homeostatic synaptic scaling can be defined as a compensatory response in which silencing GABAergic transmission leads to an increase in synaptic GABAAR activity, while silencing overall neuronal firing reduces GABAAR activity. Ge et al. demonstrated that bidirectional manipulation of GABAARs by Clptm1 knockdown and overexpression can either accentuate or occlude homeostatic scaling. This raises the possibility that rather than playing a static role, Clptm1 may participate in the dynamic adjustment of GABAARs number at the synapse.
Overall, the study of Ge et al adds an important step on our way to understanding the cellular machinery that controls inhibitory transmission. It identifies a negative regulator, Clptm1, which acts to trap GABAARs within intracellular compartments and thus limits their expression on the neuronal membrane. Receptor-specific interactors that restrict surface expression highlight the importance of precise and tight control of GABAAR number on the plasma membrane.
Several important questions remain open for future studies. Interestingly, although Clptm1 was originally identified using a tagged GABAAR-γ2, Ge et al detected its interaction (in transfected heterologous cells) with all tested GABAAR subunits (namely γ2, α1, α2, β2, β3). This is surprising, considering the relatively low sequence homology between these different subunits. Future studies can elucidate the precise molecular interaction of Clptm1 with the various subunits of GABAARs. Traditional molecular approaches to characterize interaction domains may be challenging in this case. Typically, deletion mutants or chimeric proteins between functional and non-functional homologous proteins are used for mapping interaction domains. However, applying this approach to study the interaction between two transmembrane proteins is notoriously difficult because of the relatively low tolerance of transmembrane proteins to mutations that affect their structure. With recent advances in cryoelectron microscopy, however, solving the structure of the receptor complex at the atomic level should provide important insights on the interaction between the receptor and its binding partners.
An additional outstanding question concerns the physiological roles that Clptm1 serves by accumulating GABAARs at the ER. GABAARs at the ER may function as a reservoir for surface expression. In such a scenario, Clptm1 or its associated proteins could serve as a dynamic sensor of neuronal activity by releasing GABAARs from the ER upon demand. Indeed, knockdown or overexpression of Clptm1 altered the number of GABAARs at the cell surface, mimicking the effects of homeostatic synaptic scaling in vitro. Future endeavors should reveal whether Clptm1-induced retention of GABAARs plays a physiological role in the dynamic control of inhibitory transmission in vivo. An alternative (though not mutually exclusive) physiological role of Clptm1 might be to delay GABAAR exit from the ER in order to allow proper assembly with other stable interactors. Mature GABAARs form a tri-partite complex with GARLH and Neuroligin2 (Yamasaki et al. 2017), but the underlying mechanisms remain unknown. Interestingly, Clptm1 was identified using a tagged GABAA-γ2 subunit, while GARLH requires the same subunit in order to interact with GABAARs (Martenson et al., 2017; Yamasaki et al., 2017). Thus, Clptm1 may act as a checkpoint for GABAAR complexes upon exiting the ER. Analysis of GABAAR complex assembly and localization in Clptm1 knockout mice can be especially beneficial in revealing such potential roles. Furthermore, complementary proteomic analysis of Clptm1 (as done for GABAAR in the current work by Ge et al) can provide important information on the molecular makeup of GABAAR in complex with Clptm1, and help discover new binding partners involved in Clptm1-induced retention.
Finally, the proteomic analysis by Ge et al identified 39 proteins and extensively studied 6 of them. It is common for proteomic studies to generate relatively large lists of potentially interesting targets. Indeed, different proteomic approaches have been successful in identifying additional sets of GABAAR-associated proteins that may serve important roles in the cellular and physiological regulation of these receptors (Heller et al., 2012; Nakamura et al., 2016). Considering the many functions that GABAAR-associated proteins can serve in receptor maturation, trafficking, membrane localization and biophysical properties, further analysis of individual interactors and their in vivo functions should provide important insights. Currently, sifting through large amounts of candidates to identify the most promising ones remains labor intensive. Establishing high-throughput pipelines to systematically evaluate the functional contribution of all proteomic hits is therefore of high priority to the field and should bring us closer to a complete mechanistic understanding of a receptor’s life cycle in the living neuron, and the consequences for brain function.
Acknowledgments
S.T. is supported by NIH/NIMH MH115705.
References
- Davenport EC, Pendolino V, Kontou G, McGee TP, Sheehan DF, Lopez-Domenech G, Farrant M, Kittler JT. An Essential Role for the Tetraspanin LHFPL4 in the Cell-Type-Specific Targeting and Clustering of Synaptic GABAA Receptors. Cell reports. 2017;21:70–83. doi: 10.1016/j.celrep.2017.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erlenhardt N, Yu H, Abiraman K, Yamasaki T, Wadiche JI, Tomita S, Bredt DS. Porcupine Controls Hippocampal AMPAR Levels, Composition, and Synaptic Transmission. Cell reports. 2016;14:782–794. doi: 10.1016/j.celrep.2015.12.078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge Y, Kang Y, Cassidy RM, Moon K, Lewis R, Wong ROL, Foster LJ, Craig AM. Clptm1 Limits Forward Trafficking of GABAA Receptors to Scale Inhibitory Synaptic Strength. Neuron. 2018 doi: 10.1016/j.neuron.2017.12.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heller EA, Zhang W, Selimi F, Earnheart JC, Slimak MA, Santos-Torres J, Ibanez-Tallon I, Aoki C, Chait BT, Heintz N. The biochemical anatomy of cortical inhibitory synapses. PloS one. 2012;7:e39572. doi: 10.1371/journal.pone.0039572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson AC, Nicoll RA. The Expanding Social Network of Ionotropic Glutamate Receptors: TARPs and Other Transmembrane Auxiliary Subunits. Neuron. 2011;70:178–199. doi: 10.1016/j.neuron.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martenson JS, Yamasaki T, Chaudhury NH, Albrecht D, Tomita S. Assembly rules for GABAA receptor complexes in the brain. eLife. 2017:6. doi: 10.7554/eLife.27443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura Y, Morrow DH, Modgil A, Huyghe D, Deeb TZ, Lumb MJ, Davies PA, Moss SJ. Proteomic Characterization of Inhibitory Synapses Using a Novel pHluorin-tagged gamma-Aminobutyric Acid Receptor, Type A (GABAA), alpha2 Subunit Knock-in Mouse. J Biol Chem. 2016;291:12394–12407. doi: 10.1074/jbc.M116.724443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwenk J, Harmel N, Brechet A, Zolles G, Berkefeld H, Muller CS, Bildl W, Baehrens D, Huber B, Kulik A, et al. High-resolution proteomics unravel architecture and molecular diversity of native AMPA receptor complexes. Neuron. 2012;74:621–633. doi: 10.1016/j.neuron.2012.03.034. [DOI] [PubMed] [Google Scholar]
- Yamasaki T, Hoyos-Ramirez E, Martenson JS, Morimoto-Tomita M, Tomita S. GARLH family proteins stabilize GABAA receptors at synapses. Neuron. 2017;93:1138–1152. doi: 10.1016/j.neuron.2017.02.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan D, Tomita S. Defined criteria for auxiliary subunits of glutamate receptors. J Physiol. 2012;590:21–31. doi: 10.1113/jphysiol.2011.213868. [DOI] [PMC free article] [PubMed] [Google Scholar]