Abstract
Congenital heart defects remain a leading cause of infant mortality in the western world, despite decades of research focusing on cardiovascular development and disease,. With the recent emergence of several high-throughput technologies including RNA sequencing, chromatin immunoprecipitation-coupled sequencing, mass spectrometry-based proteomics analyses, and the numerous variations of these strategies, investigations into cardiac development have been transformed from candidate-based studies into whole-genome, -transcriptome, and -proteome undertakings. In this review, we discuss several reports that have emerged from our lab and others over the last five years that emphasize the versatility of large dataset-based investigations of cardiogenic transcription factors, from phenotypic validations and new gene implications to the identification of novel roles of well-studied transcriptional regulators.
Broadening the understanding of cardiac development and disease
A growing body of work has indicated an association between mutations in several transcription factors (TFs) and congenital heart disease (CHD) (Bruneau, 2013; Prendiville et al., 2014), highlighting the need to elucidate the transcriptional regulation of various aspects of cardiovascular development. High-throughput technologies have dramatically broadened the capacity in which to study regulators of gene networks (see Figure 1 for an overview of the main high-throughput technologies discussed here). High-throughput sequencing of a cell’s transcriptional profile (RNA-seq) allows for comparison of transcriptome-wide expression datasets between wild-type and loss- or gain-of-function conditions. Similarly, coupling high-throughput sequencing to chromatin immunoprecipitation (ChIP-seq) affords global views of DNA-binding proteins’ genomic occupancies. These strategies provide valuable insights into the targets and pathways controlled by transcriptional regulators. Additionally, mass spectrometry-based proteomics furthers understanding of the complex interactions among transcription factors, epigenetic modifiers, and the transcriptional machinery. Application of these strategies to investigations of cardiovascular biology provide a systems-level comprehension of transcriptional regulation not only of normal cardiac development, but of aberrant transcriptional programs that lead to both CHD and idiopathic heart disease.
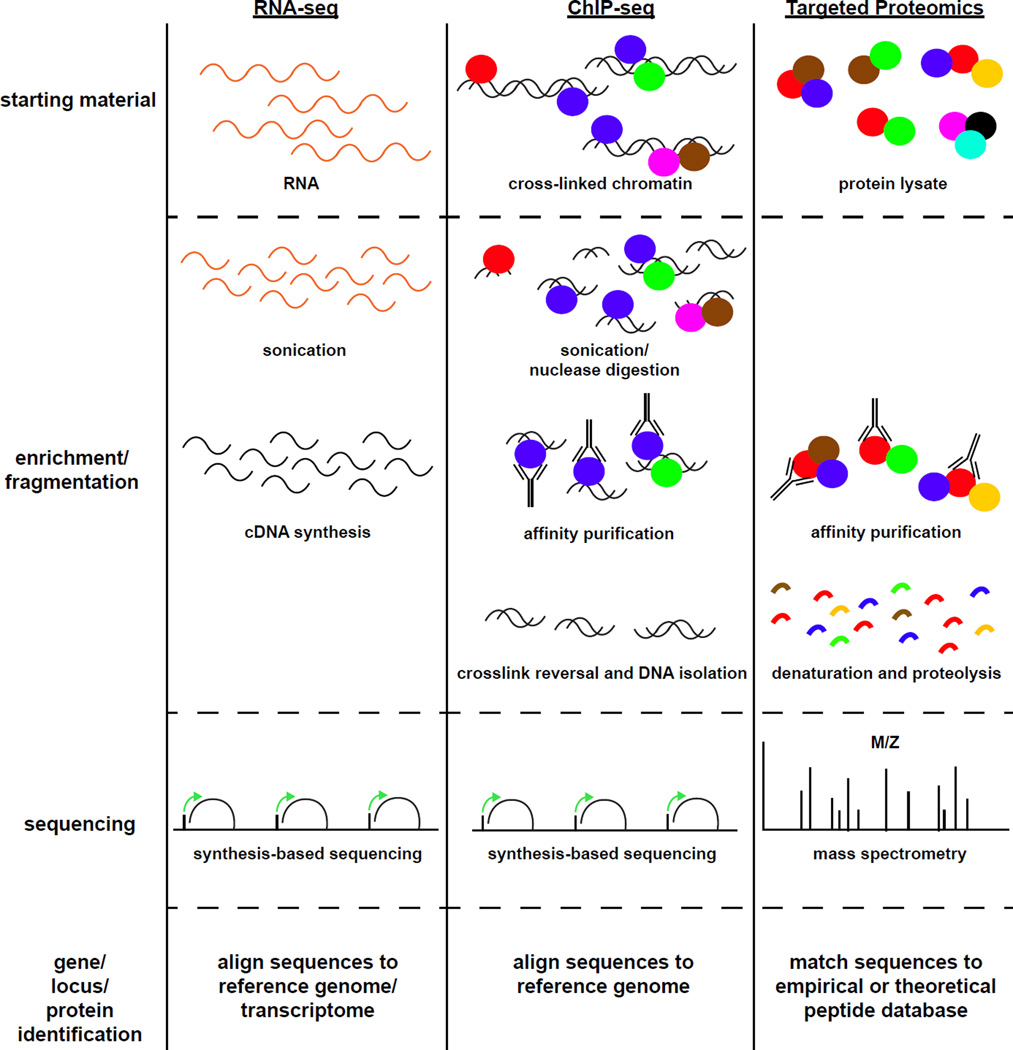
Figure 1. An overview of large-dataset experimental approaches.
RNA-seq, ChIP-seq, and proteomic analyses follow similar workflows. Briefly, starting material to be analyzed is isolated from cells or tissue. Enrichment steps, such as affinity purifications, can be employed to narrow the population of input material analyzed. Molecules of interest are then fragmented, either mechanically or enzymatically, to permit massively parallel sequence analysis. Resulting sequence data is compared to a reference database (either a transcriptome, a genome, or a peptide sequence collection) to attain identities of the molecules of interest.
Transcriptomic Analyses of Cardiac Development
Differential gene expression analysis, in which RNA-seq datasets of different biological states are compared to identify altered levels of gene products, has become the foundation of investigations into TF function. This approach was used to study the role of Mef2c, a key cardiogenic TF (Lin et al., 1997), in outflow tract (OFT) development (Barnes et al., 2016). The murine conditional loss-of-function model Myocyte Enhancer Factor 2C-conditional gene knockout (Mef2c-CKO) recapitulated OFT alignment defects seen in a human CHD patient harboring a mutation in Mef2c (Kodo et al., 2012). Differential RNA-seq gene expression analysis identified teratocarcinoma-derived growth factor 1 (Tdgf1) as a highly down-regulated transcript in Mef2c-CKO OFTs. An essential co-receptor for the Nodal subfamily of TGFβ ligands, Tdgf1 is also mutated in cases of human CHD (Ding et al., 1998; Roessler et al., 2008; Shen and Schier, 2000; Wang et al., 2011). Therefore, the authors’ findings implied that Mef2c regulated OFT response to Nodal signaling during late cardiovascular development. Of future interest is the mechanistic role that Nodal responsiveness plays in properly aligning and connecting the OFT to the vasculature.
Similarly, differential gene expression analysis was used to investigate the cardiogenic effects of Forkhead Box C1 (Foxc1) haploinsufficiency (Lambers et al., 2016), which causes ventricular septal defects in Axenfeld-Rieger syndrome patients (Honkanen et al., 2003; Mears et al., 1998). Using an inducible overexpression system in differentiating murine embryonic stem cells (mESCs), the authors identified Foxc1-responsive signaling pathways in addition to genes that are critical to cardiomyocyte differentiation, structure, and function. This analysis complemented a candidate-based assay of Foxc1 depletion effects on established cardiogenic regulators, including T-box transcription factor 1 (Tbx1), the only previously known direct Foxc1 target (Yamagishi et al., 2003), as well as Mef2c, Isl1, and Nkx2.5. Together, these results indicated the importance of Foxc1 in regulating various aspects of cardiomyocyte development, from the exit of pluripotency to building a functionally mature myocardium.
Recently, the transcriptomic dynamics of the differentiation of mesoderm posterior 1 (MESP1)-positive human embryonic stem cells (hESCs) into cardiomyocytes were comprehensively profiled (den Hartogh et al., 2016). In addition to categorizing the expression patterns of known cardiogenic TFs into a concise interaction network, their study provided new preliminary insights into the path from pluripotency to terminal myocardial differentiation. The authors reported a host of potential new transcriptional regulators in various stages of cardiomyocyte development, and their work identified new cell-surface molecules as potential temporal markers of cardiovascular subpopulations during differentiation. Finally, the authors introduced a previously unrecognized role for extracellular matrix (ECM) dynamics in maintaining the appropriate microenvironments as the mesoderm and cardiac precursors develop further down the cardiac lineage. Overall, this RNA-seq based analysis provides the scientific community with a useful resource for profiling transcriptional changes during cardiomyocyte development and opens an array of new research avenues into the differentiation of cardiac cell fates.
Using ChIP-seq to Probe Transcription Factor Occupancy and Epigenetic Dynamics
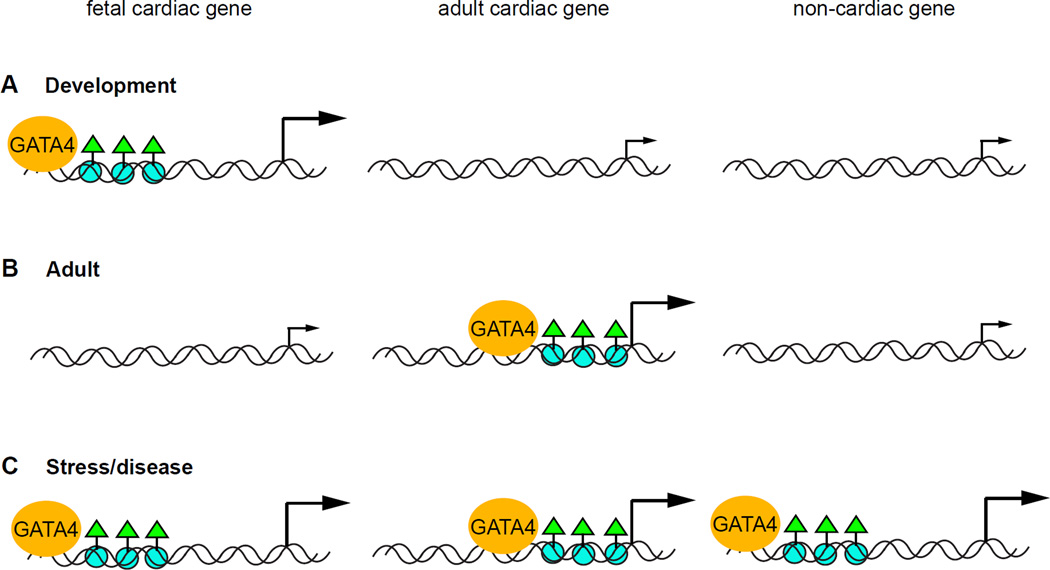
In addition to revealing new transcriptional targets and genetic programs regulated by TFs, high-throughput sequencing approaches have elucidated genome-wide chromatin dynamics as a consequence of TF binding. One study conducted a systematic analysis of chromatin modification changes induced by alterations in Gata4 binding during cardiac development and disease (He et al., 2014). As a pioneer TF in the cardiac cell lineage, Gata4 establishes regions of open chromatin by recruiting histone modifiers such as the histone acetyltransferase (HAT) p300 to its target loci (Cirillo et al., 2002; Dai and Markham, 2001; He et al., 2011; Kuo et al., 1997). The authors correlated Gata4 occupancy with patterns of histone modifications in fetal and adult heart tissue, revealing a fundamental spatial shift in Gata4 binding on its active targets. During development, Gata4 localized to active enhancers at significant distances from transcriptional start sites (TSSs; Figure 2A), whereas Gata4 binding occurred at TSS-proximal sites in adults (Figure 2B). This shift accompanied a change in the enrichment of potential cofactor binding motifs between the fetal and adult heart, suggesting that cofactor availability governs Gata4 binding site choices. Finally, the authors investigated the prevailing model of fetal gene reactivation as the mechanism of stress-induced cardiac hypertrophy (Oka et al., 2007) and extended their promoter/enhancer occupancy analysis to a murine pressure-overload model of hypertrophy. Indeed, Gata4 reestablished occupancy at a subset of its fetal binding sites, supporting the contribution of developmental program reactivation to cardiac hypertrophy. However, a large portion of hypertrophy-associated Gata4 binding sites were never bound in either the fetal or healthy adult heart (Figure 2C), suggesting a greater importance of novel Gata4 binding events in the development of hypertrophic complications.
Figure 2. Dynamic GATA4 genomic occupancy governs epigenetic and physiological states of cardiac biology.
A. In developing cardiomyocytes, GATA4 predominantly binds to distal enhancers of developmental genes and correlates with enrichment of H3K27 acetylation (green triangles on blue circles). Adult cardiac and non-cardiac genes, which are not expressed in developing cardiomyocytes, lack both GATA4 binding and H3K27 acetylation. B. Mature cardiomyocytes display a shift in both GATA4 occupancy and H3K27 acetylation from distal fetal enhancers to proximal adult enhancers. C. During disease states, GATA4 reactivates a subset of its developmental targets, but also binds and activates non-cardiac genes, implying that these novel targets contribute significantly to disease phenotypes.
In another insightful investigation into Gata4 function, the tissue-specific activities of Gata4-responsive enhancers in the atrioventricular canal (AVC) and the chamber myocardium were analyzed (Stefanovic et al., 2014). In this study, the authors combined transgenic reporter activity analysis with whole-genome assessments of TF chromatin binding and histone modification patterns. This strategy enabled the identification of regulatory elements that behave as switches, modulating Gata4-mediated transcription based on the expression of tissue-specific cofactors. In the AVC, Gata4 occupancy was correlated with that of the HAT p300, as well as with the enrichment of the active-enhancer histone H3 lysine 27 acetylation (H3K27ac) mark on enhancers of several genes involved in AVC identity (e.g. Tbx2, Tbx3;(Habets et al., 2002; Hoogaars et al., 2004) . In the chamber myocardium, these same regulatory elements lacked H3K27ac but the colocalization of Gata4 and histone deacetylases (HDACs) was observed. The opposite was apparent on Gata4-bound elements of genes that are important for chamber formation, such as Nppa and Hey2 (Houweling et al., 2005; Leimeister et al., 1999; Nakagawa et al., 1999). These sites were enriched for H3K27ac in the chamber myocardium but lacked the active-enhancer mark in the AVC. The authors built on this tissue-specific chromatin occupancy by integrating signaling pathway activity into the choice of transcriptional activation or repression. In both in vitro and in vivo experiments, they demonstrated that Bmp signaling, which is involved in AVC development and functionally interacts with both Gata4 and HATs (Dai and Markham, 2001; Kawamura et al., 2005; Ma et al., 2005; Moskowitz et al., 2011; Ross et al., 2006), synergized with Gata4 occupancy at the AVC enhancers to stimulate transcriptional activity. Conversely, in the chamber myocardium, inactivation of the AVC enhancers corresponded to active Notch signaling and occupancy of the Notch transcriptional effector Hey2. Intriguingly, the in vitro activation of a Gata4-responsive enhancer construct by Gata4 and Bmp2 was completely suppressed by Hey1/2 activity, highlighting the importance of the interplay of signaling pathways in determining regulatory element states.
Collectively, these mechanistic studies of Gata4 function highlight the versatility of high-throughput sequencing approaches for gaining new functional insights into well-studied developmental TFs, as well as for elucidating the causal mechanisms of disease states.
Combining Transcriptome and Chromatin Dynamics of Cardiogenic Transcription Factors
A widely used strategy to probe the functions of transcriptional regulators combines whole-transcriptome profiling with the high-throughput sequencing of TF-bound genomic regions. This workflow allows for genome-wide identification of directly activated and directly repressed TF targets. Applying this strategy to the Hippo-pathway transcriptional coactivator Yes-associated protein (YAP), the phosphoinositol-3-kinase catalytic subunit Pi3kcb was identified as a downstream regulator of cardiomyocyte proliferation and survival (Lin et al., 2015). YAP localized to an intron in the Pi3kcb locus through an interaction with the DNA-binding TF TEA Domain Family Member 1 (TEAD1). Furthermore, altering the expression of YAP both in vitro and in vivo positively correlated with changes in Pi3kcb expression. Mechanistically, the authors showed that Pi3kcb mediated the regulation of AKT signaling by YAP in order to positively regulate cardiomyocyte proliferation in both healthy mice and murine models of myocardial infarction. In summary, through a combination of high-throughput sequencing technologies, an enzymatic signaling pathway was identified as a new key regulator of cardiomyocyte growth in development and disease.
Using a similar strategy, a functional link between Hedgehog signaling and Tbx5-mediated transcriptional regulation during second heart field (SHF) specification was revealed (Hoffmann et al., 2014), confirming their previously identified genetic interaction (Xie et al., 2012). The authors overlaid SHF-specific ChIP-seq data for Glioblastoma 3 (Gli3), a major transcriptional effector of the Hedgehog pathway, with a differential gene expression analysis of Sonic Hedgehog (Shh)-deficient SHF tissue. This approach led to the discovery that Foxf1a and Foxf2 are direct transcriptional targets of Hedgehog signaling in the posterior SHF. A genetic interaction between the two Forkhead box TFs confirmed their requirement for atrial septation, as double heterozygous mice phenocopied a loss of Shh signaling in the SHF. The authors then confirmed that Tbx5 and Gli3 co-occupied the same regulatory region of the Foxf1a locus, and demonstrated synergistic activity of Tbx5 and Gli3 on the genomic element in transient transcriptional assays. Altogether, this study utilized a combination of large dataset analysis to define an SHF-specific transcriptional regulatory network responsible for atrial septation. By identifying Foxf1a as an integrator of Hedgehog and Tbx5-mediated transcriptional regulation, the authors provided a mechanistic basis for the observed genetic interaction of the Hedgehog pathway and Tbx5 function. They likewise identified Foxf1a and Foxf2 as new players in the development of the atrial septum and as novel candidate genes for atrial septal defect (ASD)-based CHDs.
A combination of RNA-seq and ChIP-seq was also used to investigate the role of Sox9 in the development of the cardiac cushion mesenchyme, which gives rise to the valves of the heart (Garside et al., 2015). Sox9, a TF that is essential for proper murine valve formation and is mutated in human conditions that affect heart development, is expressed in numerous proliferating embryonic tissue types (Akiyama et al., 2002; Lincoln et al., 2007; Montero et al., 2002; Rockich et al., 2013; Trowe et al., 2010; Wagner et al., 1994). A comparison of genomic occupancy in cushions versus limb buds that was overlaid with a differential gene expression analysis identified both common and tissue-specific Sox9 functions. Although Sox9 localized to numerous cell-cycle regulator genes in both tissues, reinforcing its general role in proliferation, the authors found that Sox9 also directly regulated a host of well-established valve-associated TFs, including Twist, Hand2, and Mecom/Evi1, as well as ECM components that are critical for valve formation. Furthermore, a significant portion of direct targets was upregulated upon loss of Sox9 function, indicating the dual mechanistic roles of Sox9 as both a transcriptional repressor and activator. In summary, this study utilized a combination of large dataset approaches to the understanding of Sox9 function in valvulogenesis as well as to extend the roles of this critical TF to tissue-specific cell fate decisions.
Using a combination of a genomic binding analysis and whole transcriptome profiling via microarray, mechanistic insights into cardiac arrhythmia-associated non-coding variants were revealed (van den Boogaard et al., 2012). The authors began with a systematic ChIP-seq-based assessment of in vivo chromatin binding for TFs involved in the development of the cardiac conduction system, including the transcriptional repressor Tbx3. This analysis identified loci shared between Tbx3 and its relative Tbx5. However, the associated genes, highly enriched for ion channels and gap junctions, were activated by Tbx5 in non-conduction cardiomyocytes but were repressed by Tbx3 in the conduction system. These findings are in line with previous reports that Tbx3-mediated transcriptional repression is critical for the proper differentiation of the conduction system myocardium (Bakker et al., 2008; Hoogaars et al., 2007; Horsthuis et al., 2009). The authors demonstrated that a single nucleotide polymorphism present in a conserved Tbx3/Tbx5-bound enhancer of Scn5a, a sodium channel that is essential for rapid cardiomyocyte depolarization (Papadatos et al., 2002), reduced Tbx3/Tbx5 binding affinity and, consequently, altered Scn5a expression. The authors hypothesized that this enhancer plays an important role in distinguishing myocardial subpopulations by preferentially binding distinct TFs in different tissues. Furthermore, in combination with other non-coding variants, perturbation of this conserved regulatory sequence could significantly affect the proper expression of functional proteins that participate in cardiac conduction. Hence, these results offer insights into a mechanistic link between a disease phenotype and an associated genomic non-coding variant.
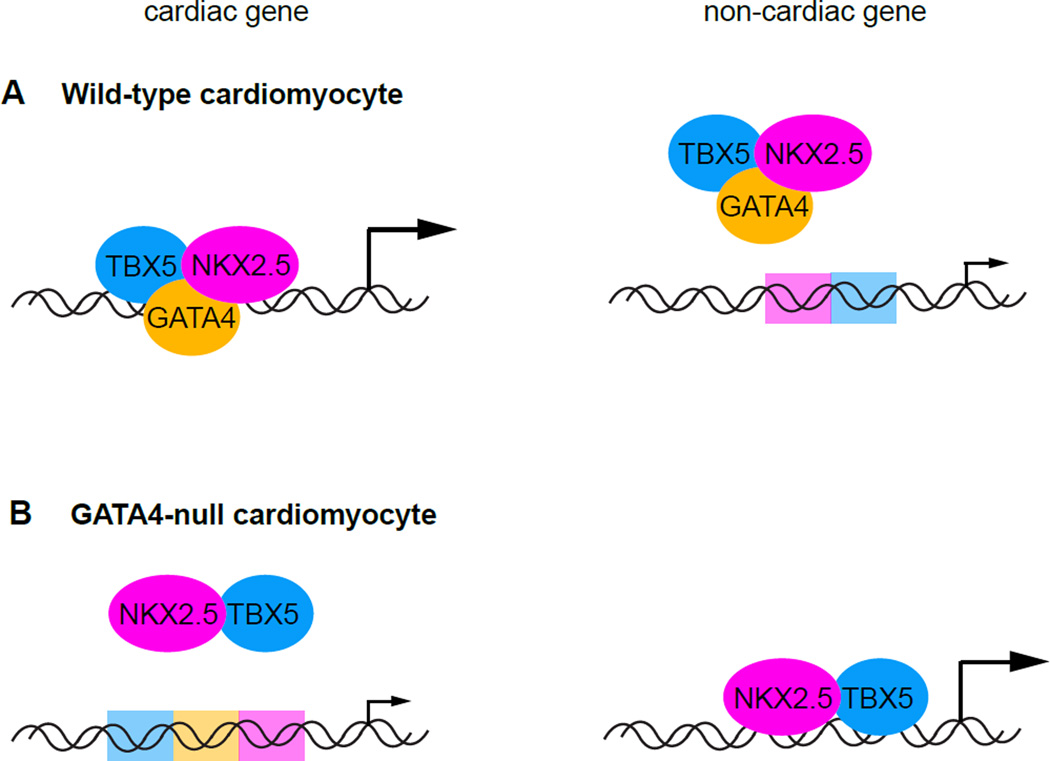
In addition to using large dataset platforms as initial screens to identify novel cardiogenic factors, technologies including RNA-seq and ChIP-seq have been applied directly to the molecular mechanisms of transcriptional regulation in cardiac development. ChIP-seq variations have been used to probe the interactions between the components of the well-studied Nkx2.5-Tbx5-Gata4 triumvirate of cardiogenic TFs (Luna-Zurita et al., 2016) (Figure 3). The investigation began with the transcriptomic profiling of Nkx2.5 and Tbx5 single- or double-knockout mESCs that differentiated along the cardiac lineage. Consequently, the analysis revealed an unexpectedly complex and dynamic relationship among genes regulated by each TF, implying a complicated temporal pattern of genomic co-occupancy. Extending their analysis to include Gata4, the authors performed a comprehensive ChIP-exo-based survey of genomic binding for all three TFs during cardiomyocyte differentiation and found that their genomic occupancy results reflected the findings of their transcriptomic profiling. The relationships between binding positions proved to be temporally complex and dynamic, with examples of different occupancy combinations running the spectrum between all and none. Intriguingly, the loss of a single factor significantly altered the binding patterns of the others beyond the expected concomitant loss at co-bound loci (Figure 3B). Specifically, the absence of one factor often led to a strengthening of the binding of the other factors at ectopic sites, i.e. sites not normally bound in a wild-type situation (Figure 3A). This finding has surprising implications for the etiologies of CHDs caused by TF mutations, as disease phenotypes may not be the lone direct consequences of TF loss- or gain-of-function. Physiological complications might arise partially due to ectopically activated or repressed genetic programs as the binding partners of the mutated TFs are redistributed throughout the genome. Consequently, this study will likely have significant future impacts on the clinical views of CHD mechanisms and treatments.
Figure 3. Relative binding site orientations influence activities of heterotypic transcription factor complexes.
A. The TBX5-GATA4-NKX2.5 complex binds to regulatory sequences in particular arrangements (e.g. proper combinations, spacing, head/tail orientations). Non-optimal binding sites are not occupied by the complex. B. Loss of complex members (e.g. GATA4-null cardiomyocytes) can redistribute remaining members to ectopic sites in the genome, leading to inappropriate gene expression and potentially exacerbating loss-of-function phenotypes.
Proteomics Analysis of Transcription Factor Coregulators
In addition to identifying downstream target genes for a given TF, significant progress has been made in elucidating the molecular mechanisms of gene regulation during cardiac development. Mass spectrometry has been used to identify the TF co-regulators involved in both cardiac chamber formation and epicardial maturation. For example, immunoisolation of Tcf21-containing transcriptional complexes revealed the transcriptional repression capabilities of this crucial epicardium-associated TF (Tandon et al., 2013). The results of this mechanistic investigation were substantiated by a differential RNA-seq gene expression analysis, which identified a host of proepicardium-expressed genes whose expression inappropriately persisted following the loss of Tcf21 function. Furthermore, the perdurance of these genes in the epicardial lineage coincided with the retention of proepicardium-like cellular phenotypes including persistent migratory behavior and failure to undergo a proper mesenchymal-to-epithelial transition. Overall, this work identified transcriptional repression as a key role of Tcf21 during the transition of the proepicardial organ to a mature epicardium.
The transcriptional mechanisms employed by Tbx20, a T-box TF expressed throughout cardiac development in species ranging from flies to humans (Ahn et al., 2000; Brown et al., 2003; Griffin et al., 2000; Iio et al., 2001; Meins et al., 2000), were studied using a similar approach (Kaltenbrun et al., 2013). The results of the proteomics analysis revealed that the Nucleosome Remodeling and Deacetylase (NuRD) complex, a chromatin-modifying transcriptional repression complex, is physically associated with Tbx20. The recruitment of NuRD activity to Tbx20 transcriptional targets is mediated by the interaction of Groucho/ transducin-like Enhancer of split (TLE) corepressors and an evolutionarily conserved Engrailed Homology 1 (Eh1) domain (Chen et al., 1999) in Tbx20, exemplifying a combinatorial specificity in the selection of regulatory mechanisms by DNA-binding TFs. After in vivo confirmation of the interaction of Tbx20 and TLE3 in murine embryonic hearts, the authors demonstrated the functional relevance of the Tbx20-TLE interaction via RNA-seq-based differential gene expression analysis. A mutation in the Eh1 domain diminished Tbx20 overexpression phenotypes in Xenopus embryos, concomitant with an upregulation of biologically relevant genes that are repressed by wild-type Tbx20. Interestingly, several of these genes play roles in cardiomyocyte differentiation. Given its persistent expression throughout early cardiac development, these findings suggest that Tbx20 might serve as a transcriptional switch, repressing cardiac genes until temporally regulated transcriptional coactivators displace Tbx20-associated corepressors during subsequent stages of differentiation.
Multiple-Platform Approaches to Studying Transcriptional Control of Cardiac Development
Combining the three major aspects of TF function, transcriptome dynamics, genome occupancy, and the cofactor interactome offers a powerful strategy for in-depth investigations into transcriptional regulation. A combination of differential gene expression analysis, ChIP-seq studies, and proteomics was used to provide intriguing insights into the function of Isl1 during the differentiation of cardiac progenitors (Wang et al., 2016). Using an ESC differentiation model to generate the cardiac progenitors, the authors identified direct transcriptional targets of Isl1. The onset of Isl1 occupancy correlated strongly with a switch from inactive- to active-enhancer epigenetic modifications, suggesting Isl1 recruits chromatin-modifying enzymes to stimulate target gene transcription. The knockdown of Isl1 led to a loss of cardiomyocyte terminal differentiation, a marked reduction in Isl1 target gene expression, and the maintenance of inactive-enhancer chromatin marks. The authors identified Isl1 transcriptional coregulators via a proteomics screen and focused on JMJD3, a histone demethylase previously implicated in mesoderm specification and cardiac lineage differentiation (Ohtani et al., 2013). Partial depletion of JMJD3 impaired ESC cardiomyocyte differentiation, the expression of Isl1 transcriptional targets, and the clearance of the H3K27me3 repressive mark from Isl1-bound enhancers. Re-ChIP experiments indicated that Isl1 and JMJD3 co-occupied the same enhancer elements of Mef2c and MyoD, two critical regulators of cardiomyocyte differentiation previously identified as Isl1 transcriptional targets (Dodou et al., 2004; Kwon et al., 2009; Takeuchi et al., 2005). Finally, the authors demonstrated that Isl1 knockdown prevented JMJD3 occupancy at representative Isl1 target enhancers. Overall, these results greatly expand the current understanding of the molecular mechanisms by which an already well-studied TF regulates aspects of cardiac development.
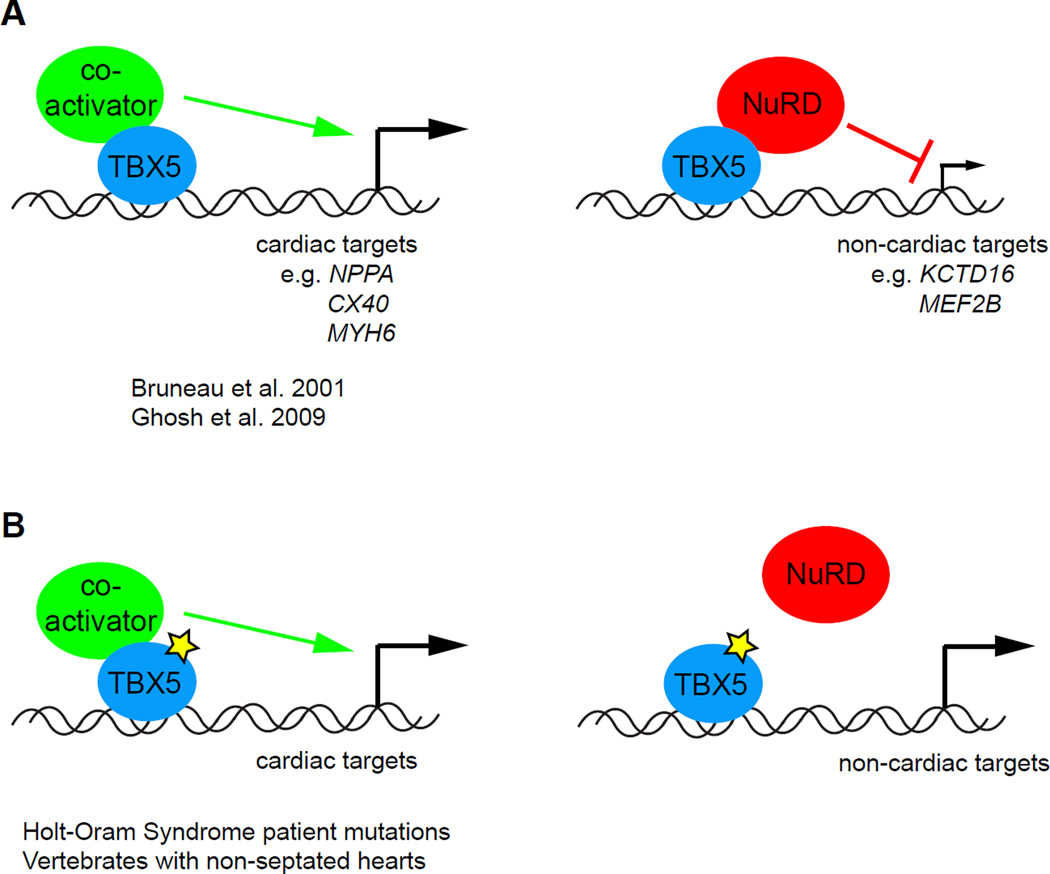
The molecular mechanisms employed by the widely-studied cardiogenic T-box TF Tbx5 were investigated in a comprehensive analysis utilizing multiple large dataset platforms (Waldron et al., 2016). Following a proteomics analysis of Tbx5 transcriptional coregulators, the authors discovered and biochemically verified a physical interaction between Tbx5 and the NuRD transcriptional repression complex. This physical interaction was functionally validated in vivo via a genetic interaction between Tbx5 and the NuRD component Mta1, with compound heterozygous mice displaying synergistic, severe cardiac abnormalities including atrioventricular septal defects. The authors biochemically mapped the NuRD-interaction domain (NID) to a previously uncharacterized region of Tbx5 and identified human CHD patient mutations that fall in this region (Heinritz et al., 2005). After identifying the direct targets of Tbx5-mediated transcriptional repression via overlaying differential gene expression analysis with previously reported ChIP-seq data (He et al., 2011), the authors demonstrated that the human CHD mutations in the Tbx5 NID not only disrupted the biochemical interaction but impaired the ability of Tbx5 to properly repress a subset of its targets (Figure 4). Finally, the authors undertook a phylogenetic analysis to highlight the strict evolutionary conservation of the Tbx5 NID sequence. In doing so, they revealed a potential causal relationship between the Tbx5-NuRD interaction and the evolution of atrial septation. Although the NID amino acid sequence is strictly conserved among vertebrates with septated hearts, those with unseptated hearts (e.g. zebrafish) bear NID substitutions that are identical to human CHD mutations. In addition to establishing Tbx5 as a repressor of gene programs incongruent with cardiomyocyte development, this work reveals the etiology of certain cases of human CHD and proposes a molecular mechanism for the evolution of cardiac chamber septation, namely the concurrent evolution of the interaction between Tbx5 and the NuRD repression complex.
Figure 4. TBX5 promotes cardiac septation via NuRD-mediated transcriptional repression.
A. During cardiac development, TBX5 activates transcription of relevant downstream targets, including cardiomyocyte structural genes. At non-cardiac targets, TBX5 recruits the NuRD chromatin remodeling complex to inhibit lineage-inappropriate gene expression. Together, these functions allow for proper development and septation of the heart. B. Disruption of the TBX5-NuRD interaction (yellow star), either through disease mutations or evolutionary mechanisms, derepresses targets of TBX5-mediated repression, leading to congenital heart defects such as AVSDs.
Concluding Remarks
The establishment of genome, transcriptome, and proteome-wide analytical technologies has revolutionized molecular investigations into a wide array of biological questions. Research into CHD, a leading cause of infant mortality in the developed world, has already benefited tremendously in a short time from these large dataset strategies. As the technologies are standardized and optimized, costs are reduced, and large dataset platforms become more accessible to a greater number of researchers, insights into cardiovascular development and disease states will increase by orders of magnitude. Exciting future research prospects will move beyond investigations of a single or limited number of TFs and explore the roles of higher-order chromatin structures in cardiovascular biology. Pioneering work has already implicated chromatin looping and domain architecture in lineage specification, cardiac development, and disease, using chromatin conformation capture (3C) technologies coupled with high-throughput sequencing analysis (Aguirre et al., 2015; Caputo et al., 2015; Dowen et al., 2014; Korostowski et al., 2011; Sergeeva et al., 2016). More widespread use of these strategies, in conjunction with large-scale proteomics analysis, could provide a wealth of information on spatiotemporal genome dynamics, as well as the compositions of protein complexes that mediate them. These efforts will, in turn, vastly improve our understanding of the factors and mechanisms at play during both normal cardiac development and cardiovascular disease states, including CHD.
Outstanding Questions Box.
How does combinatorial transcription factor binding regulate higher-order chromatin structure?
How does higher-order chromatin architecture govern proper cardiovascular development?
To what extent does ectopic transcription factor binding contribute to phenotypes of congenital and idiopathic heart disease?
Trends Box.
Large dataset platforms allow for systems-based investigations into transcription factor function during cardiovascular development.
Combining large dataset approaches provides comprehensive views of transcription factor function, revealing many transcriptional regulators to be both activators and repressors.
Transcription factor binding to both regulatory sequences and co-regulators is heavily context-dependent, relying on genomic element orientation and spacing, development-dictated co-expression, and chromatin landscapes.
Upon mutation of cardiogenic transcription factors, redistribution of their co-regulators to ectopic genomic binding sites may contribute significantly to disease etiologies.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguirre LA, Alonso ME, Badía-Careaga C, Rollán I, Arias C, Fernández-Miñán A, López-Jiménez E, Aránega A, Gómez-Skarmeta JL, Franco D. Long-range regulatory interactions at the 4q25 atrial fibrillation risk locus involve PITX2c and ENPEP. BMC biology. 2015;13:1. doi: 10.1186/s12915-015-0138-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn D-g, Ruvinsky I, Oates AC, Silver LM, Ho RK. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Akiyama H, Chaboissier M-C, Martin JF, Schedl A, de Crombrugghe B. The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6. Genes Dev. 2002;16:2813–2828. doi: 10.1101/gad.1017802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bakker ML, Boukens BJ, Mommersteeg MT, Brons JF, Wakker V, Moorman AF, Christoffels VM. Transcription factor Tbx3 is required for the specification of the atrioventricular conduction system. Circ Res. 2008;102:1340–1349. doi: 10.1161/CIRCRESAHA.107.169565. [DOI] [PubMed] [Google Scholar]
- Barnes RM, Harris IS, Jaehnig EJ, Sauls K, Sinha T, Rojas A, Schachterle W, McCulley DJ, Norris RA, Black BL. MEF2C regulates outflow tract alignment and transcriptional control of Tdgf1. Development. 2016;143:774–779. doi: 10.1242/dev.126383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown D, Binder O, Pagratis M, Parr BA, Conlon FL. Developmental expression of the Xenopus laevis Tbx20 orthologue. Development genes and evolution. 2003;212:604–607. doi: 10.1007/s00427-002-0276-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bruneau BG. Signaling and transcriptional networks in heart development and regeneration. Cold Spring Harb Perspect Biol. 2013;5:a008292. doi: 10.1101/cshperspect.a008292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caputo L, Witzel HR, Kolovos P, Cheedipudi S, Looso M, Mylona A, van IJcken WF, Laugwitz K-L, Evans SM, Braun T. The Isl1/Ldb1 complex orchestrates genome-wide chromatin organization to instruct differentiation of multipotent cardiac progenitors. Cell stem cell. 2015;17:287–299. doi: 10.1016/j.stem.2015.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Dai YS, Markham BE. p300 Functions as a coactivator of transcription factor GATA-4. J Biol Chem. 2001;276:37178–37185. doi: 10.1074/jbc.M103731200. [DOI] [PubMed] [Google Scholar]
- den Hartogh SC, Wolstencroft K, Mummery CL, Passier R. A comprehensive gene expression analysis at sequential stages of in vitro cardiac differentiation from isolated MESP1-expressing-mesoderm progenitors. Sci Rep. 2016;6:19386. doi: 10.1038/srep19386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding J, Yang L, Yan YT, Chen A, Desai N, Wynshaw-Boris A, Shen MM. Cripto is required for correct orientation of the anterior-posterior axis in the mouse embryo. Nature. 1998;395:702–707. doi: 10.1038/27215. [DOI] [PubMed] [Google Scholar]
- Dodou E, Verzi MP, Anderson JP, Xu S-M, Black BL. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Development. 2004;131:3931–3942. doi: 10.1242/dev.01256. [DOI] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garside VC, Cullum R, Alder O, Lu DY, Vander Werff R, Bilenky M, Zhao Y, Jones SJ, Marra MA, Underhill TM, et al. SOX9 modulates the expression of key transcription factors required for heart valve development. Development. 2015;142:4340–4350. doi: 10.1242/dev.125252. [DOI] [PubMed] [Google Scholar]
- Griffin K, Stoller J, Gibson M, Chen S, Yelon D, Stainier D, Kimelman D. A conserved role for H15-related T-box transcription factors in zebrafish and Drosophila heart formation. Dev Biol. 2000;218:235–247. doi: 10.1006/dbio.1999.9571. [DOI] [PubMed] [Google Scholar]
- Habets PE, Moorman AF, Clout DE, van Roon MA, Lingbeek M, van Lohuizen M, Campione M, Christoffels VM. Cooperative action of Tbx2 and Nkx2.5 inhibits ANF expression in the atrioventricular canal: implications for cardiac chamber formation. Genes Dev. 2002;16:1234–1246. doi: 10.1101/gad.222902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Gu F, Hu Y, Ma Q, Ye LY, Akiyama JA, Visel A, Pennacchio LA, Pu WT. Dynamic GATA4 enhancers shape the chromatin landscape central to heart development and disease. Nat Commun. 2014;5:4907. doi: 10.1038/ncomms5907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He A, Kong SW, Ma Q, Pu WT. Co-occupancy by multiple cardiac transcription factors identifies transcriptional enhancers active in heart. Proc Natl Acad Sci U S A. 2011;108:5632–5637. doi: 10.1073/pnas.1016959108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinritz W, Lin S, Moschik A, Froster UG. The human TBX5 gene mutation database. Human mutation. 2005;26:397. doi: 10.1002/humu.9375. [DOI] [PubMed] [Google Scholar]
- Hoffmann AD, Yang XH, Burnicka-Turek O, Bosman JD, Ren X, Steimle JD, Vokes SA, McMahon AP, Kalinichenko VV, Moskowitz IP. Foxf genes integrate tbx5 and hedgehog pathways in the second heart field for cardiac septation. PLoS Genet. 2014;10:e1004604. doi: 10.1371/journal.pgen.1004604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honkanen RA, Nishimura DY, Swiderski RE, Bennett SR, Hong S, Kwon YH, Stone EM, Sheffield VC, Alward WL. A family with Axenfeld-Rieger syndrome and Peters Anomaly caused by a point mutation (Phe112Ser) in the FOXC1 gene. Am J Ophthalmol. 2003;135:368–375. doi: 10.1016/s0002-9394(02)02061-5. [DOI] [PubMed] [Google Scholar]
- Hoogaars WM, Engel A, Brons JF, Verkerk AO, de Lange FJ, Wong LE, Bakker ML, Clout DE, Wakker V, Barnett P. Tbx3 controls the sinoatrial node gene program and imposes pacemaker function on the atria. Genes Dev. 2007;21:1098–1112. doi: 10.1101/gad.416007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoogaars WM, Tessari A, Moorman AF, de Boer PA, Hagoort J, Soufan AT, Campione M, Christoffels VM. The transcriptional repressor Tbx3 delineates the developing central conduction system of the heart. Cardiovasc Res. 2004;62:489–499. doi: 10.1016/j.cardiores.2004.01.030. [DOI] [PubMed] [Google Scholar]
- Horsthuis T, Buermans HP, Brons JF, Verkerk AO, Bakker ML, Wakker V, Clout DE, Moorman AF, AC’t Hoen P, Christoffels VM. Gene expression profiling of the forming atrioventricular node using a novel tbx3-based node-specific transgenic reporter. Circ Res. 2009;105:61–69. doi: 10.1161/CIRCRESAHA.108.192443. [DOI] [PubMed] [Google Scholar]
- Houweling AC, van Borren MM, Moorman AF, Christoffels VM. Expression and regulation of the atrial natriuretic factor encoding gene Nppa during development and disease. Cardiovasc Res. 2005;67:583–593. doi: 10.1016/j.cardiores.2005.06.013. [DOI] [PubMed] [Google Scholar]
- Iio A, Koide M, Hidaka K, Morisaki T. Expression pattern of novel chick T-box gene, Tbx20. Development genes and evolution. 2001;211:559–562. doi: 10.1007/s00427-001-0187-y. [DOI] [PubMed] [Google Scholar]
- Kaltenbrun E, Greco TM, Slagle CE, Kennedy LM, Li T, Cristea IM, Conlon FL. A Gro/TLE-NuRD corepressor complex facilitates Tbx20-dependent transcriptional repression. J Proteome Res. 2013;12:5395–5409. doi: 10.1021/pr400818c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawamura T, Ono K, Morimoto T, Wada H, Hirai M, Hidaka K, Morisaki T, Heike T, Nakahata T, Kita T. Acetylation of GATA-4 is involved in the differentiation of embryonic stem cells into cardiac myocytes. Journal of Biological Chemistry. 2005;280:19682–19688. doi: 10.1074/jbc.M412428200. [DOI] [PubMed] [Google Scholar]
- Kodo K, Nishizawa T, Furutani M, Arai S, Ishihara K, Oda M, Makino S, Fukuda K, Takahashi T, Matsuoka R, et al. Genetic analysis of essential cardiac transcription factors in 256 patients with non-syndromic congenital heart defects. Circ J. 2012;76:1703–1711. doi: 10.1253/circj.cj-11-1389. [DOI] [PubMed] [Google Scholar]
- Korostowski L, Raval A, Breuer G, Engel N. Enhancer-driven chromatin interactions during development promote escape from silencing by a long non-coding RNA. Epigenetics & chromatin. 2011;4:1. doi: 10.1186/1756-8935-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuo CT, Morrisey EE, Anandappa R, Sigrist K, Lu MM, Parmacek MS, Soudais C, Leiden JM. GATA4 transcription factor is required for ventral morphogenesis and heart tube formation. Genes Dev. 1997;11:1048–1060. doi: 10.1101/gad.11.8.1048. [DOI] [PubMed] [Google Scholar]
- Kwon C, Qian L, Cheng P, Nigam V, Arnold J, Srivastava D. A regulatory pathway involving Notchl/β-catenin/Isl1 determines cardiac progenitor cell fate. Nature cell biology. 2009;11:951–957. doi: 10.1038/ncb1906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambers E, Arnone B, Fatima A, Qin G, Wasserstrom JA, Kume T. Foxc1 Regulates Early Cardiomyogenesis and Functional Properties of Embryonic Stem Cell Derived Cardiomyocytes. Stem Cells. 2016 doi: 10.1002/stem.2301. [DOI] [PubMed] [Google Scholar]
- Leimeister C, Externbrink A, Klamt B, Gessler M. Hey genes: a novel subfamily of hairy- and Enhancer of split related genes specifically expressed during mouse embryogenesis. Mech Dev. 1999;85:173–177. doi: 10.1016/s0925-4773(99)00080-5. [DOI] [PubMed] [Google Scholar]
- Lin Q, Schwarz J, Bucana C, Olson EN. Control of mouse cardiac morphogenesis and myogenesis by transcription factor MEF2C. Science. 1997;276:1404–1407. doi: 10.1126/science.276.5317.1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z, Zhou P, von Gise A, Gu F, Ma Q, Chen J, Guo H, van Gorp PR, Wang DZ, Pu WT. Pi3kcb links Hippo-YAP and PI3K-AKT signaling pathways to promote cardiomyocyte proliferation and survival. Circ Res. 2015;116:35–45. doi: 10.1161/CIRCRESAHA.115.304457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lincoln J, Kist R, Scherer G, Yutzey KE. Sox9 is required for precursor cell expansion and extracellular matrix organization during mouse heart valve development. Dev Biol. 2007;305:120–132. doi: 10.1016/j.ydbio.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luna-Zurita L, Stirnimann CU, Glatt S, Kaynak BL, Thomas S, Baudin F, Samee MA, He D, Small EM, Mileikovsky M, et al. Complex Interdependence Regulates Heterotypic Transcription Factor Distribution and Coordinates Cardiogenesis. Cell. 2016;164:999–1014. doi: 10.1016/j.cell.2016.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma L, Lu M-F, Schwartz RJ, Martin JF. Bmp2 is essential for cardiac cushion epithelial-mesenchymal transition and myocardial patterning. Development. 2005;132:5601–5611. doi: 10.1242/dev.02156. [DOI] [PubMed] [Google Scholar]
- Mears AJ, Jordan T, Mirzayans F, Dubois S, Kume T, Parlee M, Ritch R, Koop B, Kuo WL, Collins C, et al. Mutations of the forkhead/winged-helix gene, FKHL7, in patients with Axenfeld-Rieger anomaly. Am J Hum Genet. 1998;63:1316–1328. doi: 10.1086/302109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meins M, Henderson DJ, Bhattacharya SS, Sowden JC. Characterization of the human TBX20 gene, a new member of the T-Box gene family closely related to the Drosophila H15 gene. Genomics. 2000;67:317–332. doi: 10.1006/geno.2000.6249. [DOI] [PubMed] [Google Scholar]
- Montero J, Giron B, Arrechedera H, Cheng Y-C, Scotting P, Chimal-Monroy J, Garcia-Porrero J, Hurle J. Expression of Sox8, Sox9 and Sox10 in the developing valves and autonomic nerves of the embryonic heart. Mech Dev. 2002;118:199–202. doi: 10.1016/s0925-4773(02)00249-6. [DOI] [PubMed] [Google Scholar]
- Moskowitz IP, Wang J, Peterson MA, Pu WT, Mackinnon AC, Oxburgh L, Chu GC, Sarkar M, Berul C, Smoot L. Transcription factor genes Smad4 and Gata4 cooperatively regulate cardiac valve development. Proceedings of the National Academy of Sciences. 2011;108:4006–4011. doi: 10.1073/pnas.1019025108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa O, Nakagawa M, Richardson JA, Olson EN, Srivastava D. HRT1, HRT2, and HRT3: a new subclass of bHLH transcription factors marking specific cardiac, somitic, and pharyngeal arch segments. Dev Biol. 1999;216:72–84. doi: 10.1006/dbio.1999.9454. [DOI] [PubMed] [Google Scholar]
- Ohtani K, Zhao C, Dobreva G, Manavski Y, Kluge B, Braun T, Rieger MA, Zeiher AM, Dimmeler S. Jmjd3 controls mesodermal and cardiovascular differentiation of embryonic stem cells. Circ Res. 2013;113:856–862. doi: 10.1161/CIRCRESAHA.113.302035. [DOI] [PubMed] [Google Scholar]
- Oka T, Xu J, Molkentin JD. Re-employment of developmental transcription factors in adult heart disease. Semin Cell Dev Biol. 2007;18:117–131. doi: 10.1016/j.semcdb.2006.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadatos GA, Wallerstein PM, Head CE, Ratcliff R, Brady PA, Benndorf K, Saumarez RC, Trezise AE, Huang CL-H, Vandenberg JI. Slowed conduction and ventricular tachycardia after targeted disruption of the cardiac sodium channel gene Scn5a. Proceedings of the National Academy of Sciences. 2002;99:6210–6215. doi: 10.1073/pnas.082121299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendiville T, Jay PY, Pu WT. Insights into the genetic structure of congenital heart disease from human and murine studies on monogenic disorders. Cold Spring Harb Perspect Med. 2014:4. doi: 10.1101/cshperspect.a013946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rockich BE, Hrycaj SM, Shih HP, Nagy MS, Ferguson MA, Kopp JL, Sander M, Wellik DM, Spence JR. Sox9 plays multiple roles in the lung epithelium during branching morphogenesis. Proceedings of the National Academy of Sciences. 2013;110:E4456–E4464. doi: 10.1073/pnas.1311847110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roessler E, Ouspenskaia MV, Karkera JD, Velez JI, Kantipong A, Lacbawan F, Bowers P, Belmont JW, Towbin JA, Goldmuntz E, et al. Reduced NODAL signaling strength via mutation of several pathway members including FOXH1 is linked to human heart defects and holoprosencephaly. Am J Hum Genet. 2008;83:18–29. doi: 10.1016/j.ajhg.2008.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S, Cheung E, Petrakis TG, Howell M, Kraus WL, Hill CS. Smads orchestrate specific histone modifications and chromatin remodeling to activate transcription. The EMBO journal. 2006;25:4490–4502. doi: 10.1038/sj.emboj.7601332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sergeeva IA, Hooijkaas IB, Ruijter JM, van der Made I, de Groot NE, van de Werken HJ, Creemers EE, Christoffels VM. Identification of a regulatory domain controlling the Nppa-Nppb gene cluster during heart development and stress. Development, dev. 2016:132019. doi: 10.1242/dev.132019. [DOI] [PubMed] [Google Scholar]
- Shen MM, Schier AF. The EGF-CFC gene family in vertebrate development. Trends Genet. 2000;16:303–309. doi: 10.1016/s0168-9525(00)02006-0. [DOI] [PubMed] [Google Scholar]
- Stefanovic S, Barnett P, van Duijvenboden K, Weber D, Gessler M, Christoffels VM. GATA-dependent regulatory switches establish atrioventricular canal specificity during heart development. Nat Commun. 2014;5:3680. doi: 10.1038/ncomms4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi JK, Mileikovskaia M, Koshiba-Takeuchi K, Heidt AB, Mori AD, Arruda EP, Gertsenstein M, Georges R, Davidson L, Mo R. Tbx20 dose-dependently regulates transcription factor networks required for mouse heart and motoneuron development. Development. 2005;132:2463–2474. doi: 10.1242/dev.01827. [DOI] [PubMed] [Google Scholar]
- Tandon P, Miteva YV, Kuchenbrod LM, Cristea IM, Conlon FL. Tcf21 regulates the specification and maturation of proepicardial cells. Development. 2013;140:2409–2421. doi: 10.1242/dev.093385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowe M-O, Shah S, Petry M, Airik R, Schuster-Gossler K, Kist R, Kispert A. Loss of Sox9 in the periotic mesenchyme affects mesenchymal expansion and differentiation, and epithelial morphogenesis during cochlea development in the mouse. Dev Biol. 2010;342:51–62. doi: 10.1016/j.ydbio.2010.03.014. [DOI] [PubMed] [Google Scholar]
- van den Boogaard M, Wong LY, Tessadori F, Bakker ML, Dreizehnter LK, Wakker V, Bezzina CR, t Hoen PA, Bakkers J, Barnett P, et al. Genetic variation in T-box binding element functionally affects SCN5A/SCN10A enhancer. J Clin Invest. 2012;122:2519–2530. doi: 10.1172/JCI62613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner T, Wirth J, Meyer J, Zabel B, Held M, Zimmer J, Pasantes J, Bricarelli FD, Keutel J, Hustert E. Autosomal sex reversal and campomelic dysplasia are caused by mutations in and around the SRY-related gene SOX9. Cell. 1994;79:1111–1120. doi: 10.1016/0092-8674(94)90041-8. [DOI] [PubMed] [Google Scholar]
- Waldron L, Steimle JD, Greco TM, Gomez NC, Dorr KM, Kweon J, Temple B, Yang XH, Wilczewski CM, Davis IJ, et al. The Cardiac TBX5 Interactome Reveals a Chromatin Remodeling Network Essential for Cardiac Septation. Dev Cell. 2016;36:262–275. doi: 10.1016/j.devcel.2016.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Yan J, Peng Z, Wang J, Liu S, Xie X, Ma X. Teratocarcinoma-derived growth factor 1 (TDGF1) sequence variants in patients with congenital heart defect. Int J Cardiol. 2011;146:225–227. doi: 10.1016/j.ijcard.2009.08.046. [DOI] [PubMed] [Google Scholar]
- Wang Y, Li Y, Guo C, Lu Q, Wang W, Jia Z, Chen P, Ma K, Reinberg D, Zhou C. ISL1 and JMJD3 synergistically control cardiac differentiation of embryonic stem cells. Nucleic Acids Res. 2016 doi: 10.1093/nar/gkw301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Hoffmann AD, Burnicka-Turek O, Friedland-Little JM, Zhang K, Moskowitz IP. Tbx5-hedgehog molecular networks are essential in the second heart field for atrial septation. Dev Cell. 2012;23:280–291. doi: 10.1016/j.devcel.2012.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi H, Maeda J, Hu T, McAnally J, Conway SJ, Kume T, Meyers EN, Yamagishi C, Srivastava D. Tbx1 is regulated by tissue-specific forkhead proteins through a common Sonic hedgehog-responsive enhancer. Genes Dev. 2003;17:269–281. doi: 10.1101/gad.1048903. [DOI] [PMC free article] [PubMed] [Google Scholar]