Abstract
The S100 protein family is involved in cancer cell invasion and metastasis, but its prognostic value in non-small-cell lung cancer (NSCLC) has not been elucidated. In the present study we investigated the prognostic role of mRNA expression of each individual S100 in NSCLC patients through the Kaplan–Meier plotter (KM plotter) database. Expression of 14 members of the S100 family correlated with overall survival (OS) for all NSCLC patients; 18 members were associated with OS in adenocarcinoma, but none were associated with OS in squamous cell carcinoma. In particular, high mRNA expression level of S100B was associated with better OS in NSCLC patients. The prognostic value of S100 according to smoking status, pathological grades, clinical stages, and chemotherapeutic treatment of NSCLC was further assessed. Although the results should be further verified in clinical trials our findings provide new insights into the prognostic roles of S100 proteins in NSCLC and might promote development of S100-targeted inhibitors for the treatment of NSCLC.
1. Introduction
Lung cancer is the leading cause of cancer-related mortality worldwide and is generally classified as small-cell lung cancer and non-small-cell lung cancer (NSCLC), which is mainly composed of lung adenocarcinoma and lung squamous cell carcinoma. With early and aggressive treatment, the 5-year survival of lung cancer patients is greater than 50%, but once metastatic disease occurs the survival rate drops to 5% [1, 2]. Most patients with NSCLC die due to recurrent disease. Therefore, identification of potential prognostic markers is a matter of great clinical urgency for patients with NSCLC.
The S100 calcium binding protein family, including more than 25 known members the first of which was reported by Moore in 1965, is only expressed in vertebrates and plays a key role in modulating the transmission of various cellular signals [3]. Many studies suggest that the expression of S100 family members is altered in numerous human cancers [4–7] and recent studies have reported that S100 protein may be associated with tumor metastasis [8–10].
In NSCLC, several S100 family members (S100A2, S100A4, S100A7, S100A10, S100A11, and S100A16) have been shown to be related to poor prognosis in different studies [5, 11–13], although the role of the majority of S100 proteins in NSCLC has not been reported.
The Kaplan–Meier (KM) plotter database was generated using gene expression data and survival information downloaded from the Gene Expression Omnibus (GEO) (https://www.ncbi.nlm.nih.gov/geo/). The database contains overall survival (OS) data for 1,926 NSCLC patients [14]. To date, several potential cancer-associated genes have been reported using the KM plotter for breast cancer [15–17], ovarian cancer [18], and gastric cancer [19, 20] in addition to NSCLC. In this study, we assessed the prognostic role of mRNA expression of each individual member of the S100 family in NSCLC patients using the KM plotter database.
2. Materials and Methods
The correlation of individual S100 mRNA expression with OS was analyzed using an online database that was established using gene expression data and survival information of lung cancer patients downloaded from the GEO [14]. Currently, breast cancer [21], ovarian cancer [22], gastric cancer [23], and lung cancer [14] databases have been generated. The database contains a collection of clinical data including histology, stage, grade, gender, and smoking history, and treatment groups include surgery, chemotherapy, and radiotherapy. Briefly, 20 individual members of the S100 family were entered into the database (http://kmplot.com/analysis/index.php?p=service&cancer=lung) to obtain KM survival plots. The requested mRNA expression above or below the median classified the cases into a high expression group and low expression group. These cohorts were compared with a Kaplan–Meier survival plot, and hazard ratio (HR), 95% confidence interval (CI), and log rank p value were determined and displayed on the webpage. A p value < 0.01 was considered statistically significant to reduce the false-positive rate.
3. Results
3.1. Prognostic Value of S100 Members in All NSCLC Patients
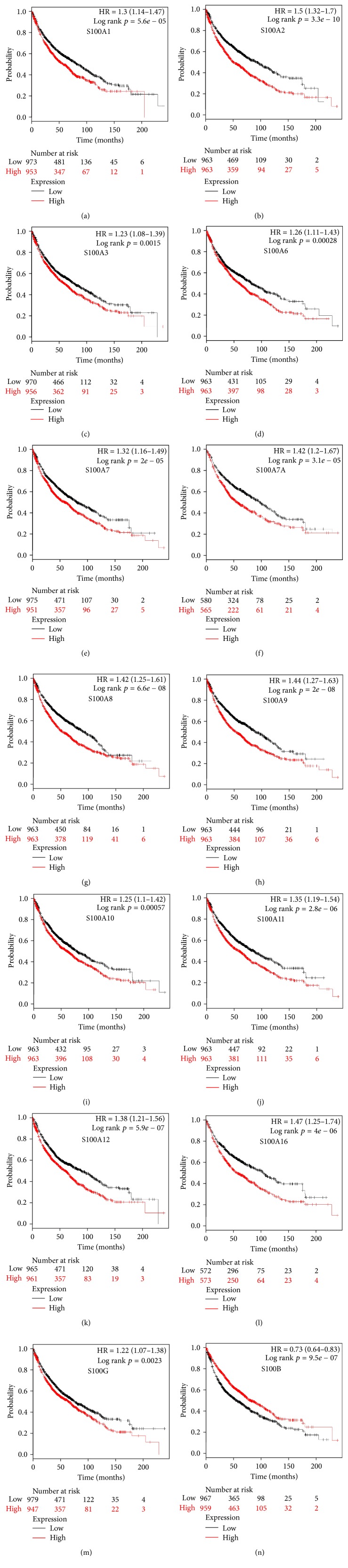
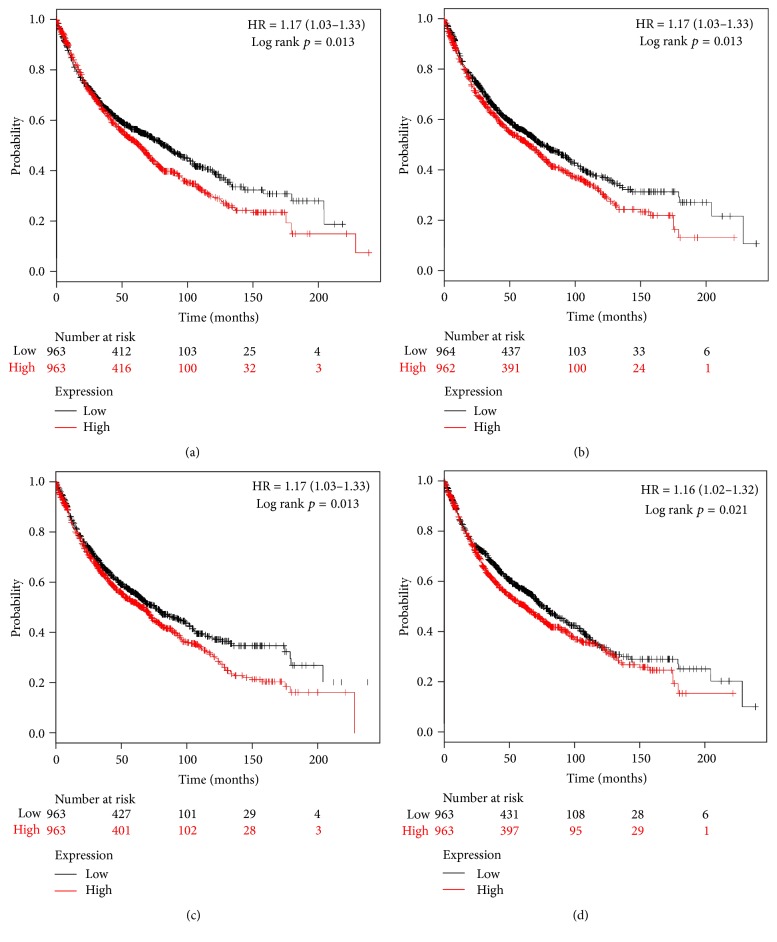
Twenty S100 family members present in NSCLC patients were found in the database (http://kmplot.com), and we determined the prognostic value of mRNA expression of each one individually. Among these 20 S100 members, 14 were significantly associated with prognosis for all NSCLC patients (Figure 1). The survival curves (Figures 1(a)–1(n)) revealed that high mRNA expression of S100B was associated with better prognosis (Figure 1(n), HR = 0.73, 95% CI: 0.64–0.83, and p = 0.0000). The other 13 members were associated with worse prognosis (Figures 1(a)–1(m)). S100A4 (HR = 1.17, 95% CI: 1.03–1.33, and p = 0.0130), S100A13 (HR = 1.17, 95% CI: 1.03–1.33, and p = 0.0130), S100A14 (HR = 1.17, 95% CI: 1.03–1.33, and p = 0.0130), and S100P (HR = 1.16, 95% CI: 1.02–1.32, and p = 0.0210) were modestly associated with poor survival (Figures 2(a)–2(d)), whereas S100A5 (HR = 1.12, 95% CI: 0.98–1.27, and p = 0.0850) and S100Z (HR = 0.90, 95% CI: 0.76–1.06, and p = 0.2100) were not correlated with OS.
Figure 1.
Prognostic value of S100 mRNA expression from the database. (a–n) Survival curves of S100A1 (Affymetrix IDs are valid: 205334_at), S100A2 (204268_at), S100A3 (206027_at), S100A6 (217728_at), S100A7 (205916_at), S100A7A (232170_at), S100A8 (202917_s_at), S100A9 (203535_at), S100A10 (200872_at), S100A11 (200660_at), S100A12 (205863_at), S100A16 (227998_at), S100G (207885_at), and S100B (209686_at) are plotted for all NSCLC patients (n = 1,926). Among them, only S100B mRNA expression was associated with better OS; the other S100 members were associated with worse OS.
Figure 2.
Survival curves for (a) S100A4 (Affymetrix ID is valid: 203186_s_at), (b) S100A13 (202598_at), (c) S10014 (218677_at), and (d) S100P (204351_at) plotted for all NSCLC patients (n = 1,926). All were modestly associated with worse OS.
3.2. Prognostic Values of S100 Members in Different NSCLC Subtypes
The prognostic value of S100 family members was assessed in different intrinsic subtypes of NSCLC, including squamous cell carcinoma and adenocarcinoma. In squamous cell carcinoma, none of high mRNA expression levels of the S100 family members correlated with OS.
In adenocarcinoma patients, S100B mRNA expression level was associated with better OS. S100G (HR = 1.29, 95% CI: 1.03–1.63, and p = 0.0290) and S100Z (HR = 0.91, 95% CI: 0.71–1.16, and p = 0.4300) were not significantly related to prognosis in adenocarcinoma and expression of the other 17 S100 members correlated with worse OS.
3.3. Prognostic Values of S100 Members in NSCLC Patients according to Clinicopathological Features and Treatment
Next, we determined the correlation of S100 with the patients' smoking status, pathological grades, clinical stages, and different chemotherapeutic treatments. As shown in Table 1, high mRNA expression of S100A14 (HR = 1.31, 95% CI: 1.07–1.62, and p = 0.0098) and S100P (HR = 1.38, 95% CI: 1.12–1.7, and p = 0.0022) correlated with worse OS in patients with a history of smoking. High mRNA expression of S100A5 (HR = 2.73, 95% CI: 1.47–5.07, and p = 0.0004), S100A6 (HR = 3.53, 95% CI: 1.87–6.65, and p = 0.0000), S100A13 (HR = 3.32, 95% CI: 1.79–6.16, and p = 0.0001), S100A16 (HR = 5.12, 95% CI: 1.75–14.99, and p = 0.0009), and S100G (HR = 2.93, 95% CI: 1.62–5.31, and p = 0.0002) correlated with worse OS in patients without smoking history. High S100A2, S100A7, S100A8, S100A9, S100A11, and S100A12 mRNA expression was linked to worse OS in patients with and without smoking history. However, only S100B mRNA expression level was associated with better OS in patients with smoking history (HR = 0.71, 95% CI: 0.58–0.87, and p = 0.0012).
Table 1.
Correlation of S100 member expression with smoking status of NSCLC patients. Expression of S100A2, S100A7, S100A8, S100A9, S100A11, and S100A12 correlated with smoking status of NSCLC patients; S100A5, S100A6, S100A13, S100A16, and S100G were associated worse survival in nonsmoking NSCLC patients; and only S100B, but S100A14 and S100P, was associated with better survival in smoking NSCLC patients.
| S100 family | Affymetrix IDs | Smoking status | HR | 95% CI | p value |
|---|---|---|---|---|---|
| S100A1 | 205334_at | Never smoked | 2.1 | (1.17, 3.77) | 0.0110 |
| Smoked | 1.21 | (0.98, 1.48) | 0.0750 | ||
| S100A2 | 204268_at | Never smoked | 4.1 | (2.1, 8) | 0.0000 |
| Smoked | 1.47 | (1.19, 1.81) | 0.0003 | ||
| S100A3 | 206027_at | Never smoked | 1.91 | (1.07, 3.38) | 0.0250 |
| Smoked | 1.15 | (0.93, 1.41) | 0.2000 | ||
| S100A4 | 203186_s_at | Never smoked | 2.06 | (1.14, 3.7) | 0.0140 |
| Smoked | 1.23 | (1, 1.51) | 0.0500 | ||
| S100A5 | 207763_at | Never smoked | 2.73 | (1.47, 5.07) | 0.0009 |
| Smoked | 1.13 | (0.92, 1.39) | 0.2400 | ||
| S100A6 | 217728_at | Never smoked | 3.53 | (1.87, 6.65) | 0.0000 |
| Smoked | 1.29 | (1.05, 1.59) | 0.0160 | ||
| S100A7 | 205916_at | Never smoked | 4.37 | (2.35, 8.11) | 0.0000 |
| Smoked | 1.34 | (1.09, 1.65) | 0.0053 | ||
| S100A7A | 232170_at | Never smoked | 1.12 | (0.5, 2.5) | 0.7800 |
| Smoked | 1.55 | (1.03, 2.34) | 0.0330 | ||
| S100A8 | 202917_s_at | Never smoked | 2.86 | (1.54, 5.3) | 0.0005 |
| Smoked | 1.76 | (1.42, 2.17) | 0.0000 | ||
| S100A9 | 203535_at | Never smoked | 3.18 | (1.71, 5.9) | 0.0001 |
| Smoked | 1.55 | (1.26, 1.91) | 0.0000 | ||
| S100A10 | 200872_at | Never smoked | 1.66 | (0.93, 2.94) | 0.0810 |
| Smoked | 1.22 | (0.99, 1.5) | 0.0620 | ||
| S100A11 | 200660_at | Never smoked | 4 | (2.09, 7.67) | 0.0000 |
| Smoked | 1.36 | (1.11, 1.68) | 0.0033 | ||
| S100A12 | 205863_at | Never smoked | 2.76 | (1.5, 5.05) | 0.0006 |
| Smoked | 1.37 | (1.11, 1.69) | 0.0028 | ||
| S100A13 | 202598_at | Never smoked | 3.32 | (1.79, 6.16) | 0.0001 |
| Smoked | 1.16 | (0.94, 1.43) | 0.1600 | ||
| S100A14 | 218677_at | Never smoked | 1.8 | (1.02, 3.2) | 0.0410 |
| Smoked | 1.31 | (1.07, 1.62) | 0.0098 | ||
| S100A16 | 227998_at | Never smoked | 5.12 | (1.75, 14.99) | 0.0009 |
| Smoked | 1.29 | (0.85, 1.95) | 0.2300 | ||
| S100B | 209686_at | Never smoked | 0.77 | (0.44, 1.35) | 0.3700 |
| Smoked | 0.71 | (0.58, 0.87) | 0.0012 | ||
| S100G | 207885_at | Never smoked | 2.93 | (1.62, 5.31) | 0.0002 |
| Smoked | 1.25 | (1.02, 1.54) | 0.0340 | ||
| S100P | 204351_at | Never smoked | 1.22 | (0.7, 2.15) | 0.4800 |
| Smoked | 1.38 | (1.12, 1.7) | 0.0022 | ||
| S100Z | 1554876_a_at | Never smoked | 0.82 | (0.37, 1.83) | 0.6300 |
| Smoked | 1.21 | (0.81, 1.82) | 0.3500 |
None of high mRNA expression levels of the S100 family members correlated with OS. The expression of S100P (HR = 1.4, 95% CI: 1.02–1.93, and p = 0.0340) and S100B (HR = 0.47, 95% CI: 0.24–0.93, and p = 0.025) was modestly associated with OS in patients with grade II and III lung cancer, respectively.
High mRNA expression of most of the S100 family members was associated with OS in clinical stage I patients except for S100A14, S100G, and S100Z. Only high RNA expression of S100B was linked to better prognosis (HR = 0.59, 95% CI: 0.45–0.78, and p = 0.0001). High mRNA expression of S100A1 (HR = 1.65, 95% CI: 1.14–2.38, and p = 0.0068), S100A6 (HR = 1.69, 95% CI: 1.16–2.45, and p = 0.0053), and S100B (HR = 0.58, 95% CI: 0.40–0.84, and p = 0.0032) was associated with OS in clinical stage II, and high S100B mRNA expression still correlated with better prognosis in this subgroup. However, none of the S100 mRNAs correlated with OS in clinical stage III patients.
Only S100A11 (HR = 1.89, 95% CI: 1.26–2.84, and p = 0.0018) significantly correlated with survival in patients treated with chemotherapy. High S100A12 (HR = 1.55, 95% CI: 1.11–2.17, and p = 0.0095) and S100G (HR = 1.72, 95% CI: 1.23–2.41, and p = 0.0013) mRNA expression were linked with worse OS in patients who did not undergo chemotherapy.
4. Discussion
In this study, we investigated the expression level of each individual S100 family member and its prognostic value in NSCLC. Among them, 14 members were significantly associated with prognosis, but only S100B was significantly associated with better prognosis. However, the molecular mechanisms by which S100 proteins contribute to disease aggression are not understood. Many studies have suggested that the expression of S100 protein in NSCLC is related to prognosis. High expression of S100A4 has been observed in NSCLC and was associated with differentiation and metastasis of tumor cells [11]; however, our results showed that S100A4 was not significantly related to OS in NSCLC.
Among the 14 S100 family members mentioned above, S100A1, S100A3, S100A7A, S100A12, and S100G display increased expression in several cancers [8, 24–26]. Our results suggested that high mRNA expression of these members was significantly associated with worse prognosis in all NSCLC patients. However, the roles of S100A1, S100A3, S100A7A, S100A12, S10016, and S100G proteins in lung cancer are rarely reported. In this study, we selectively discuss the other eight members.
High protein expression of S100A2 has been confirmed in the early stage of NSCLC [27] and is a prognostic marker associated with poor survival and a high risk of metastasis [4, 28]. Furthermore, S100A2 is considered a novel transcriptional target of p53 homologues, playing a pivotal role in regulating the cell cycle and triggering apoptotic programmed cell death in response to DNA damage or stress [29–31]. Epidermal growth factor receptor (EGFR) signaling is the main regulatory pathway for S100A2 transcription in human keratinocytes and other epithelial cells [32]. In addition, S100A2 induced epithelial-mesenchymal transition (EMT), increased invasive capability, loosened colony morphology in soft agar, and enhanced Akt phosphorylation in A549 lung cancer cells to promote tumorigenic actions and tumor growth [33]. Our data suggested that high mRNA expression of S100A2 was associated with worse OS in patients with stage I NSCLC.
The expression and prognostic role of S100A6 have been identified in thyroid [34], colorectal [35], and osteosarcoma [36] cancers. In NSCLC, S100A6 is associated with cell proliferation [37, 38]. Furthermore, high S100A6 protein level could lead to cell apoptosis by facilitating the apoptotic action of p53 [39]. Consistent with these findings, increased mRNA expression of S100A6 was significantly associated with poor prognosis in NSCLC patients in our study.
Expression of S100A7 mRNA and protein is increased mainly in squamous cell carcinoma tissues and breast cancer [40–42]. S100A7 was demonstrated to be involved in cancer growth and metastasis through modulation of the tumor microenvironment [43, 44]. Knockdown of S100A7 attenuated lung cancer growth by disruption of nuclear factor-κB activity, and S100A7 was reported as a potential diagnostic marker in lung squamous cell carcinoma [45]. Overexpression of S100A7 was associated with poor survival in SCC cells [45]. In this study, high mRNA expression of S100A7 was associated with worse OS in adenocarcinoma, but not in squamous cell carcinoma.
S100A10 forms a heterotetramer with annexin A2, which acts as a receptor of plasminogen and is involved in the conversion of plasminogen to plasmin [46]. Plasmin contributes to degradation of the basement membrane and ECM [46, 47]. S100A10 was overexpressed in renal cell carcinoma, anaplastic thyroid carcinoma, gallbladder, and colorectal cancer [48–51]. Decreased expression of S100A10 in HT1080 fibrosarcoma cells and colorectal cancer cells weakened their invasiveness and metastatic potential, suggesting that S100A10 contributes to cancer cell invasiveness [52, 53]. Katono et al. reported that S100A10 was highly expressed in lung adenocarcinomas and suggested that S100A10 may enhance the invasiveness of tumor cells by increasing plasmin production [13]. We reported a similar result, with high S100A10 expression related to poorer prognosis in NSCLC patients.
Upregulation of S100A11 plays a major role in lung cancer progression [54]. S100A11 protein was selectively expressed in NSCLC and displayed a particularly prominent effect in KRAS-mutated lung adenocarcinomas [5, 54]. It was reported that the A549 and LTEP-a-2 cell lines, which have lost S100A11 expression, show a marked suppression in tumor growth, and S100A11 knockdown also significantly inhibited tumor growth in vivo [54]. Zagryazhskaya et al. reported that S100A11 might be involved in regulation of chemoresistance of NSCLC cells [55]. We showed that high mRNA expression of S100A11 was associated with worse OS in NSCLC patients treated with chemotherapy and those with early-stage disease.
The protein heterodimer of S100A8 and S100A9 has been implicated in tumor development and progression [56]. The S100A8/A9 heterodimer has been shown to promote accumulation of myeloid-derived suppressor cells (MDSCs) in the primary tumor and their recruitment to premetastatic lungs [57, 58]. MDSCs function to induce expansion and local accumulation of regulatory T cells and suppress the antitumor immune response of natural killer cells [58, 59]. Eisenblaetter et al. reported increased S100A8/A9 levels in premetastatic lung tissue and accumulation of MDSC-like monocytes [60]. The authors proposed S100A8/A9 as a potent imaging biomarker for tumor-mediated immune remodeling [60]. In the present study, high expression levels of S100A8 and S100A9 were negative prognostic markers in NSCLC patients.
S100B is mainly found in Schwann cells of the peripheral nervous system and extraneuronally in melanocytes, adipocytes, and chondrocytes [61, 62] and is implicated in regulating enzyme activities, cell growth, and differentiation [63, 64]. S100B plays a prognostic role in the majority of brain metastases of melanoma [65]. Similarly, elevated serum S100B levels in NSCLC might be associated with the development of brain metastasis [66, 67]. S100B overexpression could contribute to cancer progression by interfering with p53 activity [64, 66]. These results imply that S100B may be a predictor of poor prognosis in lung cancer. However, contrary to our expectation, high expression of S100B in lung cancer was associated with better OS in this study. There are two possible reasons for this discrepant result: first, we obtained this result using the retrospective Kaplan–Meier plotter database and second study of S100B has mainly focused on in vitro systems and a future clinical trial is needed to validate the findings.
Tobacco smoke contains many classes of carcinogens. Nicotine, one of the carcinogens, is the addictive and most active component of tobacco smoke. Although it is not directly involved in tumorigenesis, it has been shown to promote tumor growth and metastasis [68, 69]. In vivo, nicotine can be converted to cotinine, which has tumor-promoting effects in the lung such as inducing abnormal cell formation and proliferation, promoting tumor growth, and inhibiting apoptosis [68, 70]. Furthermore, nicotine genotoxicity was described through activation of cell surface nicotinic acetylcholine receptors (nAChRs), which led to increased levels of reactive oxygen species (ROS) [71, 72]. At present, there are no reports suggesting a direct correlation between nicotine and S100 proteins in NSCLC. Recently, nicotine was reported to promote NSCLC growth and metastasis by inducing the secretion of stem cell factor [73]. Cigarette smoke induces airway inflammation by downregulating S100A8/A9 [74]. In our study, expression of S100A2, S100A7, S100A8, S100A9, S100A11, and S100A12 but not that of S100A1, S100A3, S100A4, S100A7A, S100A10, and S100Z correlated with smoking status of NSCLC patients; S100A5, S100A6, S100A13, S100A16, and S100G were associated with worse survival in nonsmoking NSCLC patients; and only S100B, but S100A14 and S100P, was associated with better survival in smoking NSCLC patients.
5. Conclusions
In the present study, the prognostic value of mRNA expression of 20 members of the S100 family in NSCLC patients was assessed using the KM plotter database. Among them, 14 members were associated with prognosis in all NSCLC patients. However, only increased S100B mRNA expression was significantly associated with better OS in all NSCLC patients. The prognostic value of the S100 protein should be further evaluated in clinical studies. These results will be helpful for investigating the relationship between proteins and disease and their roles in different signaling pathways. Our study provides new insights into the prognostic functions of S100 proteins in NSCLC and might promote development of S100 targeted inhibitors for the treatment of NSCLC.
Acknowledgments
The study was supported by grants from the National Natural Science Foundation of China (nos. 81460018, 81560154, and 81760168) and Jiangxi Provincial Science Technology Foundation of China (no. 20151BBG70073).
Conflicts of Interest
The authors declare that no conflicts of interest exist.
References
- 1.Goldstraw P., Crowley J., Chansky K., et al. The IASLC lung cancer staging project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. Journal of Thoracic Oncology. 2007;2(8):706–714. doi: 10.1097/JTO.0b013e31812f3c1a. [DOI] [PubMed] [Google Scholar]
- 2.Rami-Porta R., Bolejack V., Giroux D. J., et al. The IASLC lung cancer staging project: The new database to inform the eighth edition of the TNM classification of lung cancer. Journal of Thoracic Oncology. 2014;9(11):1618–1624. doi: 10.1097/JTO.0000000000000334. [DOI] [PubMed] [Google Scholar]
- 3.Moore B. W. A soluble protein characteristic of the nervous system. Biochemical & Biophysical Research Communications. 1965;19(6):p. 739. doi: 10.1016/0006-291X(65)90320-7. [DOI] [PubMed] [Google Scholar]
- 4.Wang T., Liang Y., Thakur A., et al. Expression and clinicopathological significance of S100 calcium binding protein A2 in lung cancer patients of Chinese Han ethnicity. Clinica Chimica Acta. 2017;464:118–122. doi: 10.1016/j.cca.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 5.Woo T., Okudela K., Mitsui H., et al. Up-regulation of S100A11 in lung adenocarcinoma-its potential relationship with cancer progression. PLoS ONE. 2015;10(11) doi: 10.1371/journal.pone.0142642.e0142642 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moravkova P., Kohoutova D., Rejchrt S., Cyrany J., Bures J. Role of S100 Proteins in Colorectal Carcinogenesis. Gastroenterology Research and Practice. 2016;2016 doi: 10.1155/2016/2632703.2632703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cross S. S., Hamdy F. C., Deloulme J. C., Rehman I. Expression of S100 proteins in normal human tissues and common cancers using tissue microarrays: S100A6, S100A8, S100A9 and S100A11 are all overexpressed in common cancers. Histopathology. 2005;46(3):256–269. doi: 10.1111/j.1365-2559.2005.02097.x. [DOI] [PubMed] [Google Scholar]
- 8.Tian T., Liu Z., Chen H., Cui Z. S100A1 promotes cell proliferation and migration and is associated with lymph node metastasis in ovarian cancer. Discovery Medicine. 2017;23(127, article no. 3) [PubMed] [Google Scholar]
- 9.Chen A., Wang L., Li B., et al. Reduction in Migratory Phenotype in a Metastasized Breast Cancer Cell Line via Downregulation of S100A4 and GRM3. Scientific Reports. 2017;7(1) doi: 10.1038/s41598-017-03811-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fei F., Qu J., Zhang M., Li Y., Zhang S. S100A4 in cancer progression and metastasis: A systematic review. Oncotarget . doi: 10.18632/oncotarget.18016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen N., Sato D., Saiki Y., Sunamura M., Fukushige S., Horii A. S100A4 is frequently overexpressed in lung cancer cells and promotes cell growth and cell motility. Biochemical and Biophysical Research Communications. 2014;447(3):459–464. doi: 10.1016/j.bbrc.2014.04.025. [DOI] [PubMed] [Google Scholar]
- 12.Saito K., Kobayashi M., Nagashio R., et al. S100A16 is a prognostic marker for lung adenocarcinomas. Asian Pacific Journal of Cancer Prevention. 2015;16(16):7039–7044. doi: 10.7314/APJCP.2015.16.16.7039. [DOI] [PubMed] [Google Scholar]
- 13.Katono K., Sato Y., Jiang S.-X., et al. Clinicopathological significance of S100A10 expression in lung adenocarcinomas. Asian Pacific Journal of Cancer Prevention. 2016;17(1):289–294. doi: 10.7314/APJCP.2016.17.1.289. [DOI] [PubMed] [Google Scholar]
- 14.Győrffy B., Surowiak P., Budczies J., Lánczky A. Online survival analysis software to assess the prognostic value of biomarkers using transcriptomic data in non-small-cell lung cancer. PLoS ONE. 2013;8(12) doi: 10.1371/journal.pone.0082241.e82241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tilghman S. L., Townley I., Zhong Q., et al. Proteomic signatures of acquired letrozole resistance in breast cancer: Suppressed estrogen signaling and increased cell motility and invasiveness. Molecular & Cellular Proteomics. 2013;12(9):2440–2455. doi: 10.1074/mcp.M112.023861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adam M. A. New prognostic factors in breast cancer. Advances in Clinical & Experimental Medicine Official Organ Wroclaw Medical University. 2013;22(1):p. 5. [PubMed] [Google Scholar]
- 17.MacIejczyk A., Jagoda E., Wysocka T., et al. ABCC2 (MRP2, cMOAT) localized in the nuclear envelope of breast carcinoma cells correlates with poor clinical outcome. Pathology & Oncology Research. 2012;18(2):331–342. doi: 10.1007/s12253-011-9449-9. [DOI] [PubMed] [Google Scholar]
- 18.Ma Y. M., Zhao S. Prognostic values of aldehyde dehydrogenase 1 isoenzymes in ovarian cancer. Oncotargets & Therapy. 2016;9(1):p. 1981. doi: 10.2147/OTT.S101063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kai L., Guo X., Wang Z., et al. The prognostic roles of ALDH1 isoenzymes in gastric cancer. Oncotargets & Therapy. 2016;9(1):p. 3405. doi: 10.2147/OTT.S102314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xia P., Xu X.-Y. Prognostic significance of CD44 in human colon cancer and gastric cancer: Evidence from bioinformatic analyses. Oncotarget . 2016;7(29):45538–45546. doi: 10.18632/oncotarget.9998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gyorffy B., Lanczky A., Eklund A. C., et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Research and Treatment. 2010;123(3):725–731. doi: 10.1007/s10549-009-0674-9. [DOI] [PubMed] [Google Scholar]
- 22.Gyorffy B., Lánczky A., Szállási Z. Implementing an online tool for genomewide validation of survival-associated biomarkers in ovarian-cancer using microarray data from 1287 patients. Endocrine-Related Cancer. 2012;19(2):197–208. doi: 10.1530/erc-11-0329. [DOI] [PubMed] [Google Scholar]
- 23.Szász A. M., Lánczky A., Nagy Á., et al. Cross-validation of survival associated biomarkers in gastric cancer using transcriptomic data of 1,065 patients. Oncotarget . 2016;7(31):49322–49333. doi: 10.18632/oncotarget.10337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mckiernan E., Mcdermott E. W., Evoy D., et al. The role of S100 genes in breast cancer progression. Tumour Biology the Journal of the International Society for Oncodevelopmental Biology & Medicine. 2011;32(3):441–450. doi: 10.1007/s13277-010-0137-2. [DOI] [PubMed] [Google Scholar]
- 25.Sviatoha V., Tani E., Kleina R., Sperga M., Skoog L. Immunohistochemical analysis of the S100A1, S100B, CD44 and Bcl-2 antigens and the rate of cell proliferation assessed by Ki-67 antibody in benign and malignant melanocytic tumours. Melanoma Research. 2010;20(2):118–125. doi: 10.1097/CMR.0b013e3283350554. [DOI] [PubMed] [Google Scholar]
- 26.Kang M. S100A3 suppression inhibits in vitro and in vivo tumor growth and invasion of human castration-resistant prostate cancer cells. Urology. 2015;85(1):273–e9. doi: 10.1016/j.urology.2014.09.018. [DOI] [PubMed] [Google Scholar]
- 27.Feng G., Xu X., Youssef E. M., Lotan R. Diminished expression of S100A2, a putative tumor suppressor, at early stage of human lung carcinogenesis. Cancer Research. 2001;61(21):7999–8004. [PubMed] [Google Scholar]
- 28.Bulk E., Sargin B., Krug U., et al. S100A2 induces metastasis in non-small cell lung cancer. Clinical Cancer Research. 2009;15(1):22–29. doi: 10.1158/1078-0432.CCR-08-0953. [DOI] [PubMed] [Google Scholar]
- 29.Van Dieck J., Brandt T., Teufel D. P., Veprintsev D. B., Joerger A. C., Fersht A. R. Molecular basis of S100 proteins interacting with the p53 homologs p63 and p73. Oncogene. 2010;29(14):2024–2035. doi: 10.1038/onc.2009.490. [DOI] [PubMed] [Google Scholar]
- 30.van Dieck J., Fernandez-Fernandez M. R., Veprintsev D. B., Fersht A. R. Modulation of the oligomerization state of p53 by differential binding of proteins of the S100 family to p53 monomers and tetramers. The Journal of Biological Chemistry. 2009;284(20):13804–13811. doi: 10.1074/jbc.M901351200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kirschner R. D., Sänger K., Müller G. A., Engeland K. Transcriptional activation of the tumor suppressor and differentiation gene S100A2 by a novel p63-binding site. Nucleic Acids Research. 2008;36(9):2969–2980. doi: 10.1093/nar/gkn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deshpande R., Woods T. L., Fu J., Zhang T., Stoll S. W., Elder J. T. Biochemical characterization of S100A2 in human keratinocytes: Subcellular localization, dimerization, and oxidative cross-linking. Journal of Investigative Dermatology. 2000;115(3):477–485. doi: 10.1046/j.1523-1747.2000.00078.x. [DOI] [PubMed] [Google Scholar]
- 33.Naz S., Ranganathan P., Bodapati P., Shastry A. H., Mishra L. N., Kondaiah P. Regulation of S100A2 expression by TGF-β-induced MEK/ERK signalling and its role in cell migration/invasion. Biochemical Journal. 2012;447(1):81–91. doi: 10.1042/bj20120014. [DOI] [PubMed] [Google Scholar]
- 34.Ito Y., Yoshida H., Tomoda C., et al. Expression of S100A2 and S100A6 in thyroid carcinomas. Histopathology. 2005;46(5):569–575. doi: 10.1111/j.1365-2559.2005.02137.x. [DOI] [PubMed] [Google Scholar]
- 35.Komatsu K., Kobune-Fujiwara Y., Andoh A., et al. Increased expression of S100A6 at the invading fronts of the primary lesion and liver metastasis in patients with colorectal adenocarcinoma. British Journal of Cancer. 2000;83(6):769–774. doi: 10.1054/bjoc.2000.1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luu H. H., Zhou L., Haydon R. C., et al. Increased expression of S100A6 is associated with decreased metastasis and inhibition of cell migration and anchorage independent growth in human osteosarcoma. Cancer Letters. 2005;229(1):135–148. doi: 10.1016/j.canlet.2005.02.015. [DOI] [PubMed] [Google Scholar]
- 37.Jian Z., Tomizawa Y., Yanagitani N., Iijima H., Sano T., Nakajima T. Papillary adenocarcinoma of the lung is a more advanced adenocarcinoma than bronchioloalveolar carcinoma that is composed of two distinct histological subtypes. Pathology International. 2005;55(10):619–625. doi: 10.1111/j.1440-1827.2005.01879.x. [DOI] [PubMed] [Google Scholar]
- 38.Ishii A., Suzuki M., Satomi K., et al. Increased cytoplasmic S100A6 expression is associated with pulmonary adenocarcinoma progression. Pathology International. 2009;59(9):623–630. doi: 10.1111/j.1440-1827.2009.02417.x. [DOI] [PubMed] [Google Scholar]
- 39.Słomnicki Ł. P., Nawrot B., Leśniak W. S100A6 binds p53 and affects its activity. The International Journal of Biochemistry & Cell Biology. 2009;41(4):784–790. doi: 10.1016/j.biocel.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 40.Nasser M. W., Qamri Z., Deol Y. S., et al. S100A7 enhances mammary tumorigenesis through upregulation of inflammatory pathways. Cancer Research. 2012;72(3):604–615. doi: 10.1158/0008-5472.CAN-11-0669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tripathi S. C., Matta A., Kaur J., et al. Nuclear S100A7 is associated with poor prognosis in head and neck cancer. PLoS ONE. 2010;5(8) doi: 10.1371/journal.pone.0011939.e11939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Barbieri M. R., Andrade C. D. C., Silva W. A., et al. Expression of human protein S100A7 (psoriasin), preparation of antibody and application to human larynx squamous cell carcinoma. BMC Research Notes. 2011;4, article no. 494 doi: 10.1186/1756-0500-4-494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang H., Wang Y., Chen Y., et al. Identification and validation of S100A7 associated with lung squamous cell carcinoma metastasis to brain. Lung Cancer. 2007;57(1):37–45. doi: 10.1016/j.lungcan.2007.02.020. [DOI] [PubMed] [Google Scholar]
- 44.Nasser M. W., Wani N. A., Ahirwar D. K., et al. RAGE mediates S100A7-induced breast cancer growth and metastasis by modulating the tumor microenvironment. Cancer Research. 2015;75(6):974–985. doi: 10.1158/0008-5472.CAN-14-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu G., Wu Q., Liu G., Song X., Zhang J. Knockdown of S100A7 reduces lung squamous cell carcinoma cell growth in vitro and in vivo. International Journal of Clinical and Experimental Pathology. 2014;7(11):8279–8289. [PMC free article] [PubMed] [Google Scholar]
- 46.Hedhli N., Falcone D. J., Huang B., et al. The Annexin A2/S100A10 System in Health and Disease: Emerging Paradigms. Journal of Biomedicine & Biotechnology. 2012;2012(2) doi: 10.1155/2012/406273.406273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu Y., Myrvang H. K., Dekker L. V. Annexin A2 complexes with S100 proteins: Structure, function and pharmacological manipulation. British Journal of Pharmacology. 2015;172(7):1664–1676. doi: 10.1111/bph.12978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Domoto T., Miyama Y., Suzuki H., et al. Evaluation of S100A10, annexin II and B-FABP expression as markers for renal cell carcinoma. Cancer Science. 2007;98(1):77–82. doi: 10.1111/j.1349-7006.2006.00355.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ito Y., Arai K., Nozawa R., et al. S100A10 expression in thyroid neoplasms originating from the follicular epithelium: Contribution to the aggressive characteristic of anaplastic carcinoma. Anticancer Reseach. 2007;27(4 C):2679–2683. [PubMed] [Google Scholar]
- 50.Tan Y., Ma S.-Y., Wang F.-Q., et al. Proteomic-based analysis for identification of potential serum biomarkers in gallbladder cancer. Oncology Reports. 2011;26(4):853–859. doi: 10.3892/or.2011.1353. [DOI] [PubMed] [Google Scholar]
- 51.Shang J., Zhang Z., Song W., et al. S100A10 as a novel biomarker in colorectal cancer. Tumour Biology the Journal of the International Society for Oncodevelopmental Biology & Medicine. 2013;34(6):3785–3790. doi: 10.1007/s13277-013-0962-1. [DOI] [PubMed] [Google Scholar]
- 52.Choi K.-S., Fogg D. K., Yoon C.-S., Waisman D. M. p11 regulates extracellular plasmin production and invasiveness of HT1080 fibrosarcoma cells. The FASEB Journal. 2003;17(2):235–246. doi: 10.1096/fj.02-0697com. [DOI] [PubMed] [Google Scholar]
- 53.Zhang L., Fogg D. K., Waisman D. M. RNA Interference-mediated Silencing of the S100A10 Gene Attenuates Plasmin Generation and Invasiveness of Colo 222 Colorectal Cancer Cells. The Journal of Biological Chemistry. 2004;279(3):2053–2062. doi: 10.1074/jbc.M310357200. [DOI] [PubMed] [Google Scholar]
- 54.Hao J., Wang K., Yue Y., et al. Selective expression of S100A11 in lung cancer and its role in regulating proliferation of adenocarcinomas cells. Molecular and Cellular Biochemistry. 2012;359(1-2):323–332. doi: 10.1007/s11010-011-1026-8. [DOI] [PubMed] [Google Scholar]
- 55.Zagryazhskaya A., Surova O., Akbar N. S., et al. Tudor staphylococcal nuclease drives chemoresistance of non-small cell lung carcinoma cells by regulating S100A1. Oncotarget. 2015;6(14):12156–12173. doi: 10.18632/oncotarget.8805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gebhardt C., Breitenbach U., Tuckermann J. P., Dittrich B. T., Richter K. H., Angel P. Calgranulins S100A8 and S100A9 are negatively regulated by glucocorticoids in a c-Fos-dependent manner and overexpressed throughout skin carcinogenesis. Oncogene. 2002;21(27):4266–4276. doi: 10.1038/sj.onc.1205521. [DOI] [PubMed] [Google Scholar]
- 57.Burke M., Choksawangkarn W., Edwards N., Ostrand-Rosenberg S., Fenselau C. Exosomes from myeloid-derived suppressor cells carry biologically active proteins. Journal of Proteome Research. 2014;13(2):836–843. doi: 10.1021/pr400879c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Srivastava M. K., Andersson Å., Zhu L., et al. Myeloid suppressor cells and immune modulation in lung cancer. Immunotherapy. 2012;4(3):291–304. doi: 10.2217/imt.11.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindau D., Gielen P., Kroesen M., Wesseling P., Adema G. J. The immunosuppressive tumour network: myeloid-derived suppressor cells, regulatory T cells and natural killer T cells. The Journal of Immunology. 2013;138(2):105–115. doi: 10.1111/imm.12036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Eisenblaetter M., Flores-Borja F., Lee J. J., et al. Visualization of tumor-immune interaction - Target-specific imaging of S100A8/A9 reveals pre-metastatic niche establishment. Theranostics. 2017;7(9):2392–2401. doi: 10.7150/thno.17138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sorci G., Riuzzi F., Arcuri C., et al. The many faces of S100B protein: when an extracellular factor inactivates its own receptor and activates another one. Italian Journal of Anatomy and Embryology. 2010;115(1-2):147–151. [PubMed] [Google Scholar]
- 62.Heizmann C. W., Fritz G., Schäfer B. W. S100 proteins: structure, functions and pathology. Frontiers in Bioscience. 2002;7:d1356–d1368. doi: 10.2741/heizmann. [DOI] [PubMed] [Google Scholar]
- 63.Sorci G., Bianchi R., Riuzzi F. S100B Protein, A Damage-Associated Molecular Pattern Protein in the Brain and Heart, and Beyond. Cardiovascular Psychiatry & Neurology. 2010;2010(2090-0163):p. 13. doi: 10.1155/2010/656481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin J., Yang Q., Wilder P. T., Carrier F., Weber D. J. The calcium-binding protein S100B down-regulates p53 and apoptosis in malignant melanoma. The Journal of Biological Chemistry. 2010;285(35):27487–27498. doi: 10.1074/jbc.M110.155382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bouwhuis M. G., Suciu S., Kruit W., et al. Prognostic value of serial blood S100B determinations in stage IIB-III melanoma patients: A corollary study to EORTC trial 18952. European Journal of Cancer. 2011;47(3):361–368. doi: 10.1016/j.ejca.2010.10.005. [DOI] [PubMed] [Google Scholar]
- 66.Pang X., Min J., Liu L., Liu Y., Ma N., Zhang H. S100B protein as a possible participant in the brain metastasis of NSCLC. Medical Oncology. 2012;29(4):2626–2632. doi: 10.1007/s12032-012-0169-0. [DOI] [PubMed] [Google Scholar]
- 67.Jiang W., Jia Q., Liu L., et al. S100B promotes the proliferation, migration and invasion of specific brain metastatic lung adenocarcinoma cell line. Cell Biochemistry & Function. 2011;29(7):582–588. doi: 10.1002/cbf.1791. [DOI] [PubMed] [Google Scholar]
- 68.Grando S. A. Connections of nicotine to cancer. Nature Reviews Cancer. 2014;14(6):419–429. doi: 10.1038/nrc3725. [DOI] [PubMed] [Google Scholar]
- 69.Schaal C., Chellappan S. P. Nicotine-mediated cell proliferation and tumor progression in smoking-related cancers. Molecular Cancer Research. 2014;12(1):14–23. doi: 10.1158/1541-7786.MCR-13-0541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Nakada T., Kiyotani K., Iwano S., et al. Lung tumorigenesis promoted by anti-apoptotic effects of cotinine, a nicotine metabolite through activation of PI3K/Akt pathway. Journal of Toxicological Sciences. 2012;37(3):555–563. doi: 10.2131/jts.37.555. [DOI] [PubMed] [Google Scholar]
- 71.Ginzkey C., Friehs G., Koehler C., Hackenberg S., Hagen R., Kleinsasser N. H. Assessment of nicotine-induced DNA damage in a genotoxicological test battery. Mutation Research - Genetic Toxicology and Environmental Mutagenesis. 2013;751(1):34–39. doi: 10.1016/j.mrgentox.2012.11.004. [DOI] [PubMed] [Google Scholar]
- 72.Ginzkey C., Stueber T., Friehs G., et al. Analysis of nicotine-induced DNA damage in cells of the human respiratory tract. Toxicology Letters. 2012;208(1):23–29. doi: 10.1016/j.toxlet.2011.09.029. [DOI] [PubMed] [Google Scholar]
- 73.Perumal D., Pillai S., Nguyen J., Schaal C., Coppola D., Chellappan S. P. Nicotinic acetylcholine receptors induce c-Kit ligand/Stem Cell Factor and promote stemness in an ARRB1/β-arrestin-1 dependent manner in NSCLC. Oncotarget . 2014;5(21):10486–10502. doi: 10.18632/oncotarget.2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen M., Wang T., Shen Y., et al. Knockout of RAGE ameliorates mainstream cigarette smoke-induced airway inflammation in mice. International Immunopharmacology. 2017;50:230–235. doi: 10.1016/j.intimp.2017.06.018. [DOI] [PubMed] [Google Scholar]