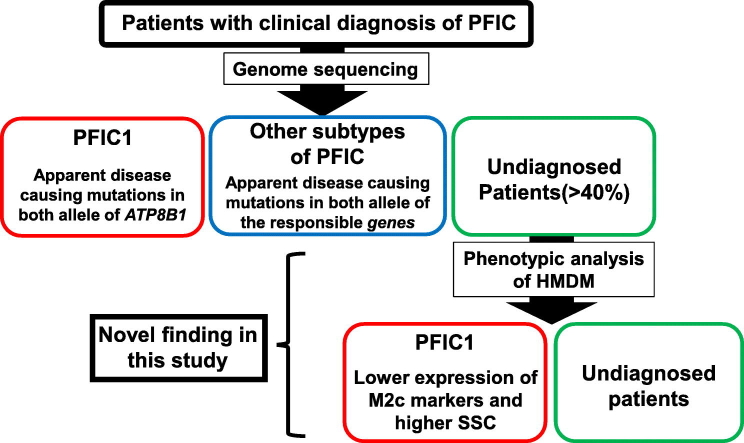
Abstract
Progressive familial intrahepatic cholestasis type 1 (PFIC1), a rare inherited recessive disease resulting from a genetic deficiency in ATP8B1, progresses to liver failure. Because of the difficulty of discriminating PFIC1 from other subtypes of PFIC based on its clinical and histological features and genome sequencing, an alternative method for diagnosing PFIC1 is desirable. Herein, we analyzed human peripheral blood monocyte-derived macrophages (HMDM) and found predominant expression of ATP8B1 in interleukin-10 (IL-10)-induced M2c, a subset of alternatively activated macrophages. SiRNA-mediated depletion of ATP8B1 in IL-10-treated HMDM markedly suppressed the expression of M2c-related surface markers and increased the side scatter (SSC) of M2c, likely via impairment of the IL-10/STAT3 signal transduction pathway. These phenotypic features were confirmed in IL-10-treated HMDM from four PFIC1 patients with disease-causing mutations in both alleles, but not in those from four patients with other subtypes of PFIC. This method identified three PFIC1 patients in a group of PFIC patients undiagnosed by genome sequencing, an identical diagnostic outcome to that achieved by analysis of liver specimens and in vitro mutagenesis studies. In conclusion, ATP8B1 deficiency caused incomplete polarization of HMDM into M2c. Phenotypic analysis of M2c helps to identify PFIC1 patients with no apparent disease-causing mutations in ATP8B1.
Keywords: Pediatric liver disease, Diagnosis, Progressive familial intrahepatic cholestasis
Abbreviations: ACTB, β-actin; BSEP, bile salt export pump; GGT, gamma-glutamyl transferase; HA, hemagglutinin; HBV, hepatitis B virus; HCV, hepatitis C virus; HMDM, human peripheral blood monocyte-derived macrophages; HPBMo, human peripheral blood monocytes; LTx, liver transplantation; PFIC, progressive familial intrahepatic cholestasis; PM, plasma membrane; qPCR, quantitative PCR; siRNA, short interfering RNA; STAT3, signal transducer and activator of transcription 3
Graphical Abstract
Highlights
-
•
ATP8B1, a causal gene of PFIC1, was expressed in IL-10-induced M2c, a subset of alternatively activated macrophages.
-
•
ATP8B1 deficiency caused incomplete polarization of HMDM into M2c, likely via impairment of IL-10/STAT3 signaling.
-
•
Phenotypic analysis of M2c helps to discriminate PFIC1 from other pediatric liver diseases undiagnosed by genomic analysis.
PFIC1, a rare inherited recessive disease resulting from a genetic deficiency in ATP8B1, progresses to liver failure. PFIC1 shares many clinical and histological features with other subtypes of PFIC, but differs in its therapeutic options. Because genome sequencing in patients with a clinical diagnosis of PFIC cannot always identify disease-causing mutations, an alternative method for diagnosing PFIC1 is desirable. We identified expression of ATP8B1 in IL-10-induced M2c, a subset of macrophages, and demonstrated its contribution to normal phenotypic expression of M2c. The phenotypic analysis of M2c helps to discriminate PFIC1 from other pediatric liver diseases undiagnosed by genomic analysis.
1. Introduction
Progressive familial intrahepatic cholestasis (PFIC), an inherited autosomal recessive liver disease, is characterized primarily by persistent intrahepatic cholestasis and jaundice in the first year of life. Its actual prevalence remains unknown, but its estimated incidence varies between 1/50,000 and 1/100,000 births. This disease is classified based on the serum gamma-glutamyl transferase (GGT) level and the causal gene (Davit-Spraul et al., 2009). Patients with PFIC who have normal GGT (normal-GGT PFIC) are termed PFIC type 1, 2, and 4 (PFIC1, PFIC2, and PFIC4) based on whether their genetic deficiency is in ATP8B1, ABCB11, or TJP2, which respectively encode an aminophospholipid flippase expressed in many tissues, a bile salt export pump (BSEP) that mediates biliary excretion of bile acids from hepatocytes, and a component of the epithelial tight junction (Bull et al., 1998, Strautnieks et al., 1998, Sambrotta et al., 2014). However, approximately one-third of individuals with normal-GGT PFIC do not have mutations in ATP8B1 or ABCB11 (Davit-Spraul et al., 2010, Klomp et al., 2004) and mutations in TJP2 seem not to explain all of the remaining patients (Sambrotta et al., 2014). Moreover, in some patients with normal-GGT PFIC, genome sequencing detects a mutation in only one allele of ATP8B1 or ABCB11 or identifies mutations in either gene that are difficult to distinguish as disease-causing mutations or rare normal variants (Chen et al., 2002, Davit-Spraul et al., 2010, Klomp et al., 2004, Liu et al., 2007, Matte et al., 2010). This, together with the fact that individuals with normal-GGT PFIC share many clinical features (Davit-Spraul et al., 2009), makes the determination of the subtypes of PFIC more complex.
Normal-GGT PFIC progresses to severe cholestasis with sustained intractable itching, jaundice, and failure to thrive, resulting in liver failure and death before adulthood (Morotti et al., 2011, Suchy et al., 2014). Although we and other groups have reported that 4-phenylbutyrate improves the intractable itching in patients with PFIC1 (Hasegawa et al., 2014) and the biochemical parameters and liver histology in patients with PFIC2 (Gonzales et al., 2012, Hayashi and Sugiyama, 2007, Hayashi et al., 2005, Naoi et al., 2014), no effective medical therapy for this disease has been established (Morotti et al., 2011, Suchy et al., 2014). Currently, the only curative options for normal-GGT PFIC are surgical procedures including liver transplantation (LTx) (Suchy et al., 2014). However, while LTx solves the immediate problem of liver failure in PFIC1, it is insufficient to overcome an ATP8B1 deficiency because of ongoing steatosis and fibrosis (Hori et al., 2011, Miyagawa-Hayashino et al., 2009). Therefore, to understand the potential of a possible treatment plan, it is of the highest priority to develop a methodology to identify PFIC1 patients correctly at an early phase of their disease course.
The current procedure for diagnosing PFIC1 is to analyze the sequence of ATP8B1 and, if possible, to confirm the hepatic level of ATP8B1 expression in liver biopsy specimens. However, genomic analysis is insufficient to make a definitive diagnosis of PFIC1 except in cases of nonsense, frameshift, and large deletion mutations, which occur in < 40% of patients who carry ATP8B1 mutations in both alleles (Davit-Spraul et al., 2010, Klomp et al., 2004). Studies with liver biopsy specimens can evaluate the impact of the mutation on the expression of ATP8B1, but not on its transport activity, even though the existence of mutations that affect the transport activity of ATP8B1 has been suggested (Folmer et al., 2009, Van Der Velden et al., 2010). In addition, liver biopsy is invasive and involves a high risk of complications such as bleeding, pneumothorax, and pain. Therefore, it is desirable to develop a diagnostic method to evaluate the function of ATP8B1 using specimens that can be collected less invasively.
In the current study, we explored blood cell populations that endogenously expressed ATP8B1 and detected its expression in human peripheral blood monocyte-derived macrophages (HMDM). Of the various subpopulations of HMDM, ATP8B1 was predominantly expressed in M2c, a subset of alternatively activated macrophages that is induced by exposure to interleukin-10 (IL-10) and whose function is related to suppression of immune responses and tissue remodeling (Mantovani et al., 2004). The impact of impaired function of ATP8B1 in M2c was examined by flow cytometric and microscopic analysis using HMDM transfected with siRNA against ATP8B1, and then confirmed using HMDM from PFIC1 patients. Based on the phenotypes identified in this analysis, we tested whether ATP8B1 function was impaired or maintained in patients with a clinical diagnosis of normal-GGT PFIC in whom only one mutant allele of ATP8B1 was detected by genome sequencing (patients with PFIC1-like disease). The results were verified by evaluating expression of ATP8B1 in liver specimens from the patients and by an in vitro mutagenesis study.
2. Materials and Methods
A detailed description of the materials and methods is presented in the Supporting information. All methods used standard techniques and commercially available reagents.
2.1. Sampling of Blood and Liver Specimens from Patients and Control Subjects
This study was approved by the institutional ethics review board at the University of Tokyo and was performed in accordance with the principles stated in the Declaration of Helsinki. Before assessment of subjects, they or their parents (when the subjects were under 18 years of age) provided signed informed consent.
The clinical diagnosis of normal-GGT PFIC was based on the presence of unremitting hepatocellular cholestasis with intractable pruritus, poor growth, jaundice with conjugated hyperbilirubinemia, elevated serum bile acid concentrations, and normal serum GGT level, in the first year of life. Serological, viral, or metabolic markers, imaging, and urine screening were performed to exclude other causes of cholestasis including hepatitis B and C virus (HBV and HCV) infections, inborn errors in bile-acid synthesis, and ductal origin. In patients with a clinical diagnosis of normal-GGT PFIC, ATP8B1 and ABCB11 were analyzed by Sanger sequencing, as described previously (Hasegawa et al., 2014, Naoi et al., 2014). The patients in whom mutations in both alleles of either gene were undetectable by Sanger sequencing were subjected to targeted next-generation sequencing that analyzed genes known to be responsible for neonatal/infantile intrahepatic cholestasis (ATP8B1, ABCB11, ABCB4, TJP2, JAG1, NOTCH2, ABCC2, SLC25A13, HSD3B7, AKR1D1, CYP7B1, VPS33B, BAAT, EPHX1, SLC10A1) (Togawa et al., 2016).
Peripheral blood samples from patients with normal-GGT PFIC and age-matched controls including healthy individuals, obese individuals, and those with pancreatitis, HBV, HCV, Alagille syndrome, citrullinemia, and biliary atresia were collected in EDTA-2 K-coated blood sampling tubes (Becton Dickinson, Tokyo, Japan) for isolation of human peripheral blood monocytes (HPBMo) and their subsequent differentiation into human monocyte-derived macrophages (HMDM). In each assay except for Fig. 1c in which interindividual variability was examined in HMDM, the monocytes pooled from more than three control subjects were differentiated into HMDM and then used to identify blood cell populations expressing ATP8B1 and to examine the impact of ATP8B1 in HMDM, and employed as control cells to evaluate HMDM from each patient with normal-GGT PFIC.
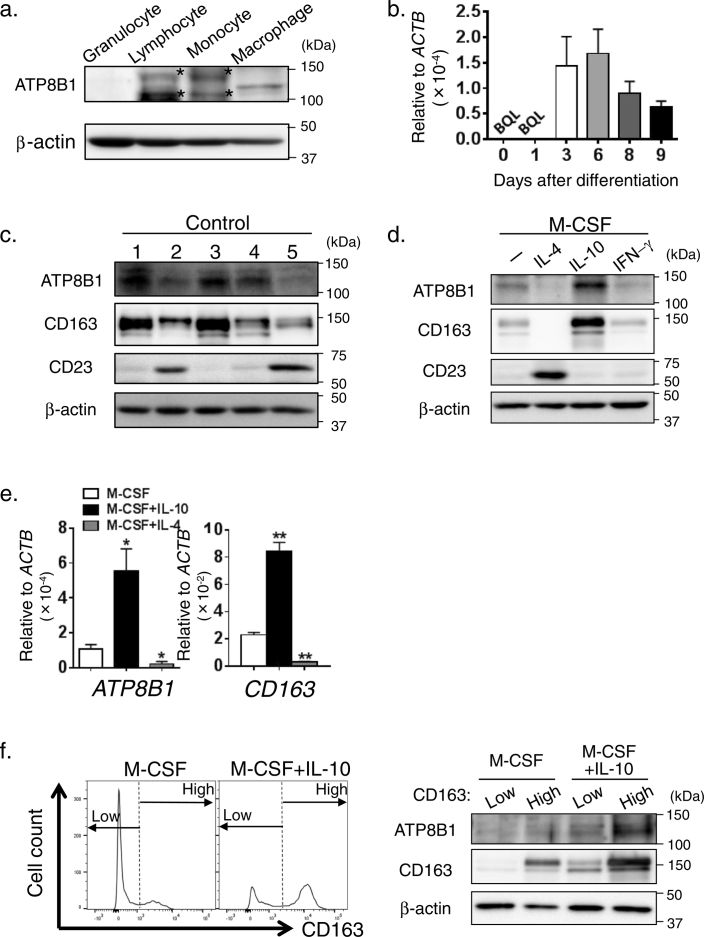
Fig. 1.
Expression of ATP8B1 in M2c.
(a) Expression of ATP8B1 protein in HMDM. Granulocytes, lymphocytes, and monocytes were prepared from human peripheral blood. HPBMo were differentiated into HMDM by culture for 9 days in RPMI/M-CSF. Cell lysates of these cells were analyzed by immunoblotting. *, nonspecific bands. (b) Expressions of ATP8B1 mRNA in HMDM. During differentiation of HPBMo into HMDM, RNA was prepared and subjected to qPCR analysis. The expression of ATP8B1 in each sample was normalized to that of β-actin (ACTB). Each bar represents the mean ± SEM of triplicate determinations. BQL, below the limit of quantitation. (c–f) HPBMo were seeded, differentiated into HMDM by culture for 9 days in RPMI/M-CSF (c), and then cultured for 6 days in RPMI/M-CSF with or without IFN-γ (40 ng/ml) (d), IL-4 (20 ng/ml) (d, e), and IL-10 (40 ng/ml) (d–f) to elicit polarization into M1, M2a, and M2c, respectively. (c) Variation between donors in the abundance of ATP8B1 in HMDM. The HMDM were lysed and analyzed by immunoblotting. 1; citrullinemia, 2; HBV, 3; Alagille syndrome, 4; biliary atresia, 5; healthy. (d) Expression of ATP8B1 in each macrophage subset. Macrophages treated with or without each cytokine were lysed and analyzed by immunoblotting. The polarization of HMDM into each macrophage subset was confirmed by the presence of CD23 (M2a marker) and CD163 (M2c marker). (e) Expression of ATP8B1 mRNA in each macrophage subset. RNA was prepared from macrophages treated with or without IL-4 (20 ng/ml) and IL-10 (40 ng/ml) and subjected to qPCR analysis. The expression of ATP8B1 and CD163 in each reaction was normalized to that of ACTB. Each bar represents the mean ± SEM of triplicate determinations. *, P < 0.05, **, P < 0.01 (f) Presence of ATP8B1 in M2c. Nonpolarized and IL-10-treated HMDM were stained with FITC-labeled anti-CD163 antibody and sorted by flow cytometry into two populations with high or low levels of expression of CD163 as shown in the histograms (left). The sorted cells were lysed and analyzed by immunoblotting (right). In (a–f), a representative result of at least two independent experiments is shown.
A liver sample from the patients with normal-GGT PFIC and the age-matched control subjects (those with HBV, HCV, or Alagille syndrome) was obtained by biopsy for diagnosis or at the time of LTx, snap-frozen in liquid nitrogen, and stored at − 80 °C. The membrane fractions from these specimens were prepared as described previously (Hasegawa et al., 2014, Naoi et al., 2014).
2.2. Reagents and Antibodies
Human Ab serum (14-490E) was obtained from Lonza (Basel, Switzerland). Human macrophage colony-stimulating factor (M-CSF; 300–25), interleukin-10 (IL-10; 571,002), interleukin-4 (IL-4; 574,002), and interferon gamma (IFN-γ; 57,202) were purchased from Peprotech (Rocky Hill, NJ) or BioLegend (San Diego, CA). The sequence of the siRNA against human ATP8B1 (siATP8B1, Cosmo Bio, Tokyo, Japan) was as follows: sense 5′-CAGCCTCTGCTATGTAGAAdTdT-3′ and antisense 5′-TTCTACATAGCAGAGGCTGdTdT-3′. Negative control siRNA (S20C-0600; Cosmo Bio) was employed as control siRNA (siControl). Antibodies against ATP8B1 were raised in rabbits using an oligopeptide (WPSESDKIQKHRKRLKAEEQ). The other primary antibodies used are listed in Supporting Table 1. AlexaFluor-labeled secondary antibodies were purchased from Molecular Probes (Life Technologies, Carlsbad, CA). All other chemicals were of analytical grade.
2.3. Preparation of HMDM and their Polarization into Subsets
HPBMo were obtained using a monocyte enrichment cocktail (StemCell Technologies, Vancouver, BC, Canada) according to the manufacturer's instructions, seeded on Repcell dishes (CellSeed, Tokyo, Japan) for flow cytometric analysis or 12-well plates for the other analyses, and differentiated into HMDM by culturing for 8–11 days in RPMI 1640 (Thermo Fisher Scientific, Waltham, MA) containing 10% human serum, 1% penicillin-streptomycin, and 10 ng/ml human M-CSF (RPMI/M-CSF). Then, the HMDM were either maintained for 6 days in RPMI/M-CSF (nonpolarized HMDM), or cultured in RPMI/M-CSF containing 40 ng/ml human IFN-γ, 20 ng/ml IL-4, or 40 ng/ml IL-10 to elicit polarization into M1, M2a, and M2c subsets, respectively. The cells were cultured at 37 °C in an atmosphere of 5% CO2 in air at 95% humidity. The resulting cells were used to prepare RNA and cell lysate, and for immunocytochemistry and flow cytometric analysis as described in the Supporting information.
2.4. In Vitro Mutagenesis Study
To characterize the ATP8B1 mutations identified in patients with PFIC1 and PFIC1-like disease, CHO-K1 cells (ATCC CCL-61) were transfected with the indicated vector using XtremeGeneHP (Roche) according to the manufacturer's instructions, and cultured in Ham's F12 nutrient mixture (Thermo Fisher Scientific) supplemented with 10% fetal bovine serum at 37 °C in an atmosphere of 5% CO2 in air at 95% humidity. At 48 h after the transfection, the cells were subjected to cell surface biotinylation, immunocytochemistry, and the flippase assay as described in the Supporting information.
2.5. Statistical Analysis
Data are presented as means ± standard error of the mean (SEM), unless otherwise indicated. The differences between two variables and multiple variables were assessed at the 95% confidence level using Student's t-tests and analysis of variance with a post-hoc Dunnett's test, respectively. The data were analyzed using Prism software (v. 6; GraphPad Software, La Jolla, CA).
3. Results
3.1. Expression of ATP8B1 in M2c
HPBMo were cultured in RPMI/M-CSF to differentiate into HMDM. After 9 days of culture, the cells appeared as a mixture of round and spindle shaped cells, morphologically characteristic of HMDM (Supporting Fig. 1a) (Kittan et al., 2013, Rey-Giraud et al., 2012) that were stained with antibody against 25F9, a human mature macrophage marker (Pilling et al., 2009), but not with antibody against CD93, a human monocyte marker (Pilling et al., 2009), indicating that the monocytes was correctly differentiated into HMDM (Supporting Fig. 1b). Immunoblot analysis showed that ATP8B1 was expressed on HMDM, but not on granulocytes, lymphocytes, or monocytes (Fig. 1a). By qPCR, ATP8B1 expression appeared after 3 days of culture of HPBMo in RPMI/M-CSF and was maintained in the HMDM (Fig. 1b).
The abundance of ATP8B1 in HMDM varied between individuals (Fig. 1c). HMDM induced by treatment with M-CSF comprise several subsets of macrophages (Chen et al., 2015, Kittan et al., 2013, Rey-Giraud et al., 2012). To identify the subset expressing ATP8B1, HMDM were maintained in RPMI/M-CSF alone or polarized into M1, M2a, and M2c subsets by the addition of IFN-γ, IL-4, or IL-10, respectively. The polarization of HMDM into M1, M2a, and M2c was confirmed by expression of CD80 (Supporting Fig. 1c), CD23, and CD163 (Fig. 1d), markers of human M1, M2a, and M2c, respectively (Rey-Giraud et al., 2012). Compared with its expression in nonpolarized HMDM, the expression of ATP8B1 was increased by treatment with IL-10 and was abrogated by treatment with IFN-γ and IL-4 (Fig. 1d), indicating that ATP8B1 was exclusively expressed in M2c. qPCR analysis suggested that transcriptional regulation was predominantly responsible for the increase and decrease of ATP8B1 expression in HMDM induced by IL-10 and IL-4 treatment, respectively (Fig. 1e). The expression of ATP8B1 in M2c was confirmed by sorting of nonpolarized and IL-10-treated HMDM based on low and high expression of CD163, a marker of human M2c, and subsequent immunoblot analysis (Fig. 1f). In both types of macrophages, the expression of ATP8B1 was higher in the CD163-positive subset. The lower band in blots for CD163 (Figs. 1c, d, and f) was nonspecific because there was no significant difference in intensity between HMDM with low and high expression of CD163 (Fig. 1f).
3.2. ATP8B1-Deficient M2c Show Decreased Expression of M2c Markers and Increased Side Scatter (SSC)
To examine the phenotypic and morphological impact of ATP8B1 in M2c, HMDM were transfected with siControl or siATP8B1 and then cultured in RPMI/M-CSF with or without IL-10 (Fig. 2a, b). No effect of ATP8B1 deficiency on differentiation of HPBMo into HMDM was detected by flow cytometric analysis, which showed that > 95% of both siControl- and siATP8B1-transfected cells identified from forward scatter- and SSC-based gating were positive for CD68, a macrophage marker, and negative for CD93, a monocyte marker (Supporting Fig. 2a). The mean fluorescence intensity (MFI) of CD163, CD14, and CD16, surface markers of M2c (Rey-Giraud et al., 2012, Zizzo et al., 2012), and the number of cells positive for each marker were higher in IL-10-treated HMDM than in nonpolarized HMDM (Fig. 2c). Both the MFI and the percent of cells positive for CD163 and CD14, but not for CD16, were markedly inhibited in both IL-10-treated and -untreated HMDM after ATP8B1 depletion (Fig. 2c), which is consistent with the positive correlation between the abundance of ATP8B1 and CD163 in nonpolarized HMDM in individuals (Fig. 1c). The influence of ATP8B1 on these M2c markers was considered to originate at the mRNA level (Supporting Fig. 3).
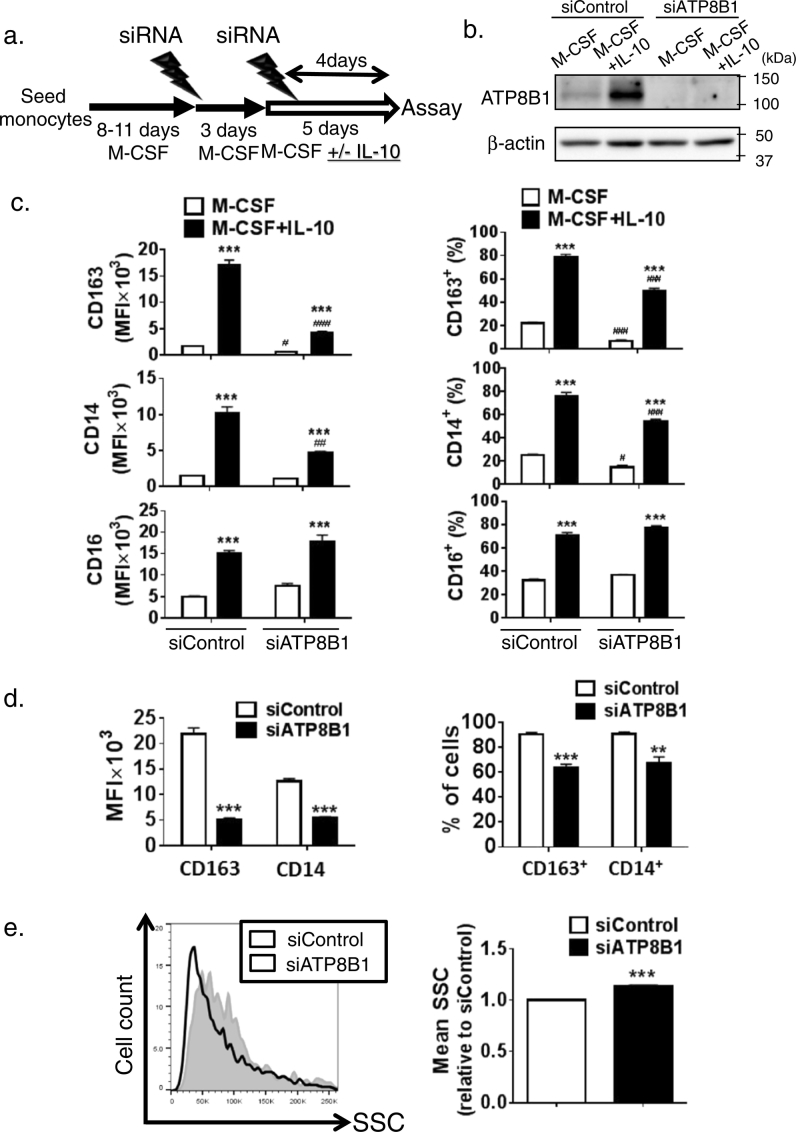
Fig. 2.
Depletion of ATP8B1 decreases expression of M2c markers.
HPBMo were seeded, differentiated into HMDM by culture for 8 to 11 days in RPMI/M-CSF, and then transfected with siControl or siATP8B1. Four days later, a second siRNA transfection was performed. During the last five days, the cells were cultured in RPMI/M-CSF with or without IL-10 (40 ng/ml), which induced polarization into M2c. (a) Schematic diagram illustrating the culture procedure. (b) Confirmation of ATP8B1 depletion. The cells were lysed and analyzed by immunoblotting. (c) Lower expression of M2c markers, CD163 and CD14, by ATP8B1 suppression. The cells were stained with fluorochrome-labeled antibodies against CD163, CD14, and CD16, subjected to flow cytometry, and the results were analyzed using FlowJo software (v. 10). Expression of each marker in the cells, which was gated according to FSC/SSC properties as described in Supporting Fig. 2a, was determined as MFI (left) and as percentages of CD163+, CD14+, and CD16+ cells (right). Data are shown as means ± SEM of triplicate determinations (c). *, P < 0.05, **, P < 0.01, ***, P < 0.001 versus nonpolarized HMDM; #, P < 0.05, ###, P < 0.001 versus siControl. (d, e) Expression of CD163 and CD14 and mean value of SSC in CD16+ cells from IL-10-treated HMDM. FlowJo software (v. 10) was used to gate the CD16+ cells and determine the MFI of CD163 and CD14 (d, left) and the percentages of CD163+ and CD14+ cells (d, right). The CD16+ cells are plotted on a histogram with SSC (e, left) and the mean value of SSC is calculated (e, right). Data shown in (d, e, right) are means ± SEM of triplicate determinations. **, P < 0.01, ***, P < 0.001. In (b–e), a representative result of four independent experiments is shown.
Based on these findings, CD16+ cells were gated from IL-10-treated HMDM and used to analyze ATP8B1 function in M2c further. In this subset, 90% of siControl-transfected cells and 65% of siATP8B1-transfected cells were positive for CD163 and CD14 (Fig. 2d, right), indicating that ATP8B1 deficiency caused a 77% and 56% decrease in the MFIs of CD163 and CD14, respectively (Fig. 2d, left). M2c, CD163+ CD14+ CD16+ cells, had lower SSC (internal complexity) than the other subsets comprising the IL-10-treated HMDM (Supporting Fig. 4). The mean SSC of ATP8B1-deficient CD16+ cells gated from IL-10-treated HMDM was 13% higher than that of control CD16+ cells (Fig. 2e). Together, these results suggest that ATP8B1 deficiency causes incomplete polarization of HMDM into M2c.
3.3. Impaired IL-10/STAT3 Signaling in ATP8B1-Deficient HMDM
IL-10, a potent immunosuppressive cytokine, limits proinflammatory responses, controls inflammatory disease progression, and prevents excessive tissue disruption because of inflammation (Murray, 2006, Ouyang et al., 2011). In HMDM, its anti-inflammatory action is initiated by binding to the IL-10 receptor (IL-10R) and is predominantly mediated by a transcription factor, signal transducer and activator of transcription 3 (STAT3) (Williams et al., 2004). After the binding of IL-10 to IL-10R, STAT3 is activated via phosphorylation, which causes its translocation from the cytosol to the nucleus, where it binds to the IFN-γ activated sequence in target promoters, and thereby facilitates transcription of anti-inflammatory genes (Murray, 2006, Ouyang et al., 2011).
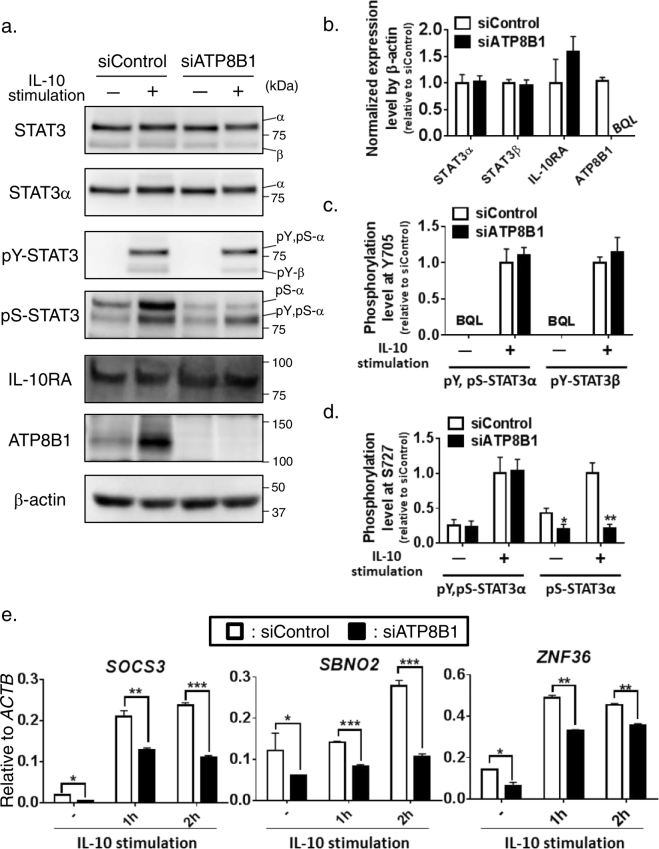
To understand the mechanism underlying the incomplete IL-10-driven polarization of ATP8B1-deficient HMDM into M2c, the influence of ATP8B1 deficiency on the IL-10/STAT3 signal transduction pathway was explored. IL-10 stimulus elicits phosphorylation of STAT3α, the major isoform of STAT3 in HMDM, at tyrosine 705 and serine 727 and generates two phosphoforms of STAT3α: pY, pS-STAT3α, which is phosphorylated at both tyrosine 705 and serine 727, a canonical activated form of STAT3 (Wen et al., 1995), and pS-STAT3α, phosphorylated only at serine 727, a noncanonical activated form of STAT3 that is required for its optimal activation (Fig. 3a) (Liu et al., 2003, Waitkus et al., 2014, Zhu et al., 2015). Suppression of ATP8B1 in HMDM had no influence on the expression of IL-10R, STAT3α, and STAT3β, an alternative splicing variant of STAT3 that lacks the transactivation domain (Fig. 3a, b). However, ATP8B1 depletion attenuated the IL-10-driven formation of pS-STAT3α, but not of pY, pS-STAT3α (Fig. 3a, c, and d). The amount of pS-STAT3 was 54% lower in the steady state and 80% lower 60 min after IL-10 stimulation in HMDM transfected with siATP8B1 than in those transfected by siControl (Fig. 3d). Consistent with this finding, ATP8B1 suppression markedly reduced the mRNA expression of the STAT3-regulated genes, SOCS3, ZNF36, and SBNO2 (El Kasmi et al., 2007, Hutchins et al., 2012, Schaljo et al., 2009), both in the steady state and following IL-10 stimulus (Fig. 3e).
Fig. 3.
Influence of ATP8B1 depletion on IL-10/STAT3 signal transduction pathway in HMDM.
HMDM deficient for ATP8B1 were prepared as described in Fig. 2. The HMDM were stimulated with or without IL-10 (40 ng/ml) for 60 min and subjected to prepare whole cell lysates (a–d) or RNA (e). (a–c) Tyrosine and serine phosphorylation of STAT3. The prepared cell lysates (5 μg) were analyzed by immunoblotting (a). Expression level of each protein (b) and phosphorylation level of STAT3 (c, d) was quantified as described in the Supporting information. Data shown are means ± SEM of three independent experiments. α, STAT3α; β, STAT3β; pY,pS-α, pY,pS-STAT3α; pY-β, pY-STAT3β; pS-α, pS-STAT3α; BQL, below the limit of quantitation. (e) mRNA expression of STAT3-regulated genes. The isolated RNA was subjected to qPCR analysis. The expression of SOCS3, SBNO2, and ZNF36 in each reaction was normalized to that of ACTB. Data shown are means ± SEM of triplicate determinations. In (a, e), a representative result of three independent experiments is shown. *, P < 0.05, **, P < 0.01, ***, P < 0.001 versus siControl.
3.4. Determination of the Subtype of PFIC Using Liver Specimens and an in vitro Mutagenesis Study
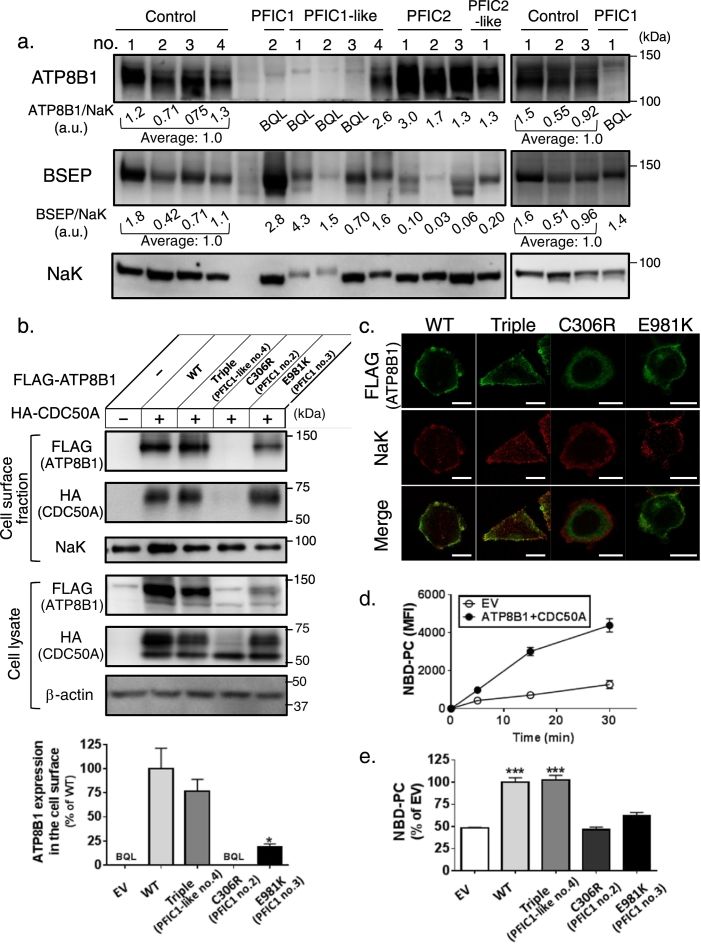
Sequencing of the genes responsible for neonatal/infantile intrahepatic cholestasis (ATP8B1, ABCB11, ABCB4, TJP2, JAG1, NOTCH2, ABCC2, SLC25A13, HSD3B7, AKR1D1, CYP7B1, VPS33B, BAAT, EPHX1, SLC10A1) in 12 individuals with a clinical diagnosis of normal-GGT PFIC (Hasegawa et al., 2014, Naoi et al., 2014, Togawa et al., 2016) identified mutations in both alleles of ATP8B1 (PFIC1) in four patients and in one allele (PFIC1-like) in four patients; and mutations in both alleles of ABCB11 (which encodes BSEP) (PFIC2) in three patients and in one allele (PFIC2-like) in one patient (Table 1). The patient with PFIC1-like and PFIC2-like disease had no mutations in other relevant genes. The genetic diagnosis of PFIC1 and PFIC2 was further supported by much lower expression of ATP8B1 and BSEP in membrane fractions from liver specimens of patients with PFIC1 and PFIC2, respectively, than in control subjects (Fig. 4a). Although liver specimens were unavailable for two patients (no.3 and 4) with PFIC1, their hepatic expression of ATP8B1 would be extremely low because they clearly had disease-causing mutations (frameshift mutation and nonsense mutation) or a missense mutation, c.2941G > A (p.E981K) that could decrease exogenous ATP8B1WT–FLAG expression by about 80% in CHO-K1 cells expressing HA-CDC50A, which forms a complex with ATP8B1 and assists its correct trafficking to the plasma membrane (PM) (Fig. 4b) (Paulusma et al., 2008).
Table 1.
Analysis of genome sequencing, liver specimens, and in vitro mutagenesis in normal-GGT PFIC patients.
| Disease | Current age | Gender | Genome |
Liver |
In vitro Mutagenesis |
|||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
ATP8B1 |
ABCB11 |
ATP8B1 | BSEP | Alelle 1 of ATP8B1 |
||||||||
| Allele 1 | Allele 2 | Allele 1 | Allele 2 | Expression | Localization | Function | ||||||
| PFIC1 | no.1 | 3y | M | c.2124_2125insGAGCTACAGCTATTGAAGGC (p.K709fsX41) | exon 2–6 del | NF | BQL | + | ND | |||
| no.2 | 4y | F | c.916 T > C (p.C306R) |
c.2854C > T (p.R952X) |
NF | BQL | + | BQL | ER | ↓↓ | ||
| no.3a | 10y | F | c.2941G > A (p.E981K) |
c.1371del (p.G457fsX6) |
NF | NA | ↓ | PM and intracellular | ↓↓ | |||
| no.4 | 10y | F | c.727del (p.L243fsX28) |
c.2854C > T (p.R952X) |
NF | NA | ND | |||||
| PFIC1-like | no.1b | 5y | M | c.3033-34del (p.L1011fsX18) |
NF | NF | BQL | + | ND | |||
| no.2b | 10y | M | c.1585-87del (p.F529del) |
NF | NF | BQL | + | ND | ||||
| no.3c | 8y | M | c.3579_3589del (p.R1193fsX39) |
NF | NF | BQL | + | ND | ||||
| no.4 | 17y | M | c.234C > G (p.H78Q) c.1729A > G (p.I577V) c.2021 T > C (p.M674 T) |
NF | NF | + | + | + | PM | + | ||
| PFIC2 | no.1 | 3y | F | NF | c.386G > A (p.C129Y) |
c.1460G > A (p.R487H) |
+ | ↓↓ | ND | |||
| no.2 | 2y | F | NF | c.386G > A (p.C129Y) |
c.386G > A (p.C129Y) |
+ | ↓↓ | ND | ||||
| no.3d | 3y | F | NF | c.3692G > A (p.R1231Q) |
c.3692G > A (p.R1231Q) |
+ | ↓↓ | ND | ||||
| PFIC2-like | no.1 | 3y | F | NF | c.386G > A (p.C129Y) |
NF | + | ↓ | ND | |||
BQL, below quantification limit; ER, endoplasmic reticulum; F, female; M, male; NA, not available; ND, not done; NF, not found; PM, plasma membrane; +, normal; ↓, decreased; ↓↓, severely decreased.
Numakura C et al.,Pediatr Int. 2011 Feb;53(1):107–10.
Mochizuki K et al., Pediatr Surg Int. 2012 Jan; 28(1):51–4.
Fig. 4.
Diagnosis of PFIC subtype using liver specimens and in vitro mutagenesis study in the patients with normal-GGT PFIC.
(a) Expression of ATP8B1 and BSEP in livers of patients with normal-GGT PFIC. Membrane fractions were prepared from liver specimens of the patients, and 5 μg (left panel) and 20 μg (right panel) of the specimens were subjected to immunoblotting. The signal intensity of ATP8B1 and BSEP relative to that of NaK is presented below each panel. Control 1; HBV, Control 2; HCV, Control 3; HCV, Control 4; Alagille syndrome. a.u., arbitrary unit; BQL, below the limit of quantitation; NaK, Na+, K+-ATPase α1 subunit. (b–d) Characterization of missense mutations of ATP8B1 in patients with PFIC1 and PFIC1-like disease. CHO-K1 cells were transfected with pShuttle-ATP8B1WT–FLAG, ATP8B1Triple–FLAG, ATP8B1C306R–FLAG, or ATP8B1E981K–FLAG together with pShuttle-HA–CDC50A and subjected to cell surface biotinylation (b), immunocytochemistry (c), and a flippase assay (d, e). CHO-K1 cells transfected with the corresponding empty vector were employed as control cells. A representative result of at least two independent experiments is shown. (b) Protein expression. Upper, the cells were biotinylated, lysed, precipitated with streptavidin, and then analyzed by immunoblotting. Lower, quantification of the amount of the mutated ATP8B1 in the cell surface fraction. The band intensity shown in the upper panel was quantified as described in the Supporting information. Each bar represents the mean ± SEM of triplicate determinations. *, P < 0.05. (c) Cellular localization. The cells were subjected to immunocytochemistry and analyzed by confocal immunofluorescence microscopy. Yellow in the merged images indicates colocalization of FLAG and NaK. Scale bar: 10 μm. (d, e) Determination of flippase activity. The cells were incubated with NBD-PC at 15 °C for the indicated time (d) or 15 min (e) and then with fatty acid-free BSA on ice for 5 min. The residual fluorescence intensity associated with the cells was determined by flow cytometry. Each bar represents the mean ± SEM of triplicate determinations. ***, P < 0.001. BQL, below the limit of quantitation; EV, empty vector; WT, wild type.
The abundance of ATP8B1 in the membrane fractions from liver varied between patients with PFIC1-like disease (Fig. 4a). Expression of ATP8B1 was below the quantification limit in PFIC1-like patients 1, 2, and 3, but normal in PFIC1-like patient 4, who possessed three missense mutations in one allele of ATP8B1, each of which has been reported to occur at an allele frequency of about 5% in East Asian populations (http://gnomad.broadinstitute.org/). ATP8B1Triple–FLAG, a mutated form of ATP8B1–FLAG incorporating these three mutations, showed equivalent cell surface expression and similar cellular localization to ATP8B1WT–FLAG in CHO-K1 cells (Fig. 4b, c). The intrinsic molecular function of ATP8B1 is to translocate aminophospholipids such as phosphatidylcholine from the outer leaflet to the inner leaflet of biological membranes (Paulusma et al., 2008, Takatsu et al., 2014). Incorporation into the inner leaflet of the PM of nitrobenzoxadiazole-labeled phosphatidylcholine (NBD-PC), which cannot be extracted with fatty acid-free BSA, was linear for up to 15 min in ATP8B1WT–FLAG-transfected CHO-K1 cells (Fig. 4d). Its level at 15 min was unaffected by the three mutations harbored by PFIC1-like patient 4, but was decreased to an equal degree to that in empty-vector-transfected cells after the introduction of c.916 T > C (p.C306R) and c.2941G > A (p.E981K) mutations (Fig. 4e), which were harbored in PFIC1 patients 2 and 3 (Table 1). Overall, these results indicated that PFIC1-like patients 1, 2, and 3, but not patients 4, suffered from PFIC1.
3.5. Diagnosis of PFIC1 in Patients With Normal-GGT PFIC Using Phenotypic Characteristics of M2c
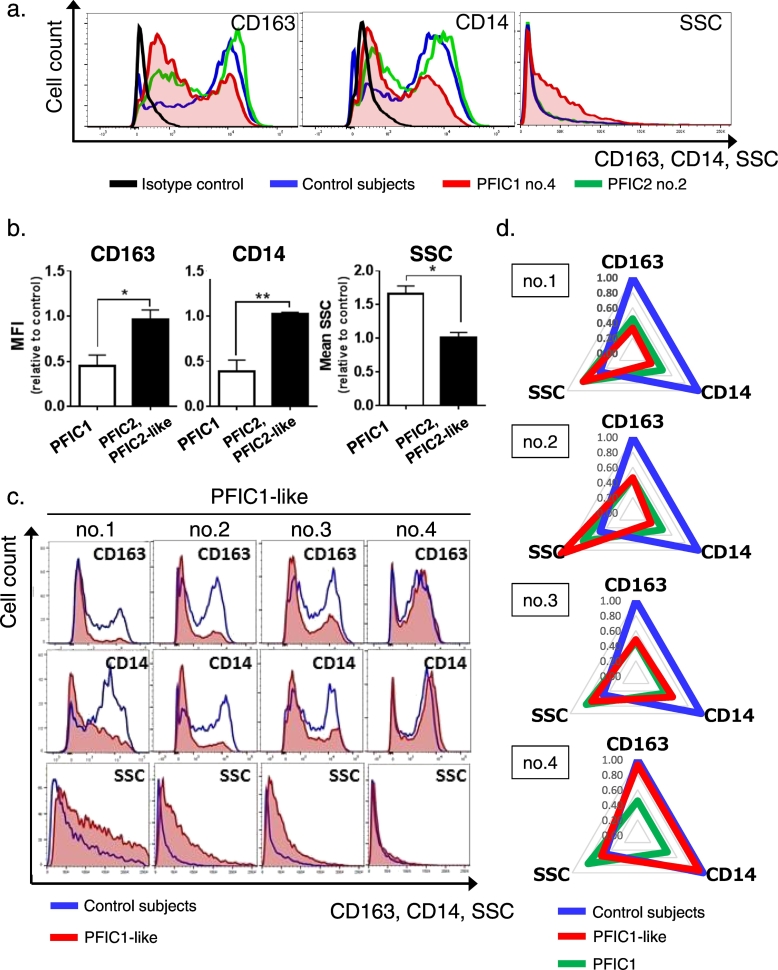
Based on the finding that siRNA-mediated suppression of ATP8B1 caused incomplete polarization of HMDM into M2c (Fig. 2), HPBMo from patients with normal-GGT PFIC were differentiated into HMDM and their polarization to M2c were facilitated by IL-10 stimulation. Flow cytometric analysis of HPBMo based on surface expression of CD14 and CD16, which divided HPBMo into three functionally distinct populations, showed no significant difference between PFIC1 patients and control subjects (Supporting Fig. 5). Correct differentiation of HPBMo into HMDM in the patients with PFIC1 was confirmed by their staining for CD68, but not for CD93 (Supporting Fig. 2b). In all four patients with PFIC1, IL-10-treated HMDM showed decreased expression of CD163 and CD14 and increased SSC (Fig. 5a), compared with cells from control subjects, which were prepared from the HPBMo of at least three individuals to minimize interindividual variability (Fig. 1c). The IL-10-treated HMDM from patients with PFIC2 and PFIC2-like disease, who in the early stages of disease present with clinical symptom similar to those of PFIC1 (Davit-Spraul et al., 2009), showed an almost identical phenotypic and morphological pattern to those of the control subjects. The MFI of CD163 and CD14 and the mean value of SSC in IL-10-treated HMDM calculated relative to the values in control subjects, were 61 ± 8% (P < 0.05) and 68 ± 6% (P < 0.01) lower and 57 ± 8% (P < 0.01) higher, respectively, in patients with PFIC1 than in patients with PFIC2 and PFIC2-like disease (Fig. 5b). The IL-10-treated HMDM from PFIC1-like patients 1, 2, and 3, but not patient 4, showed the same phenotypic and morphological pattern as those from the patients with PFIC1 (Fig. 5c, d), indicating that the same diagnosis could be made using M2c as was made based on the amount of ATP8B1 in liver and the in vitro mutagenesis study.
Fig. 5.
Identifying PFIC1 patients using phenotypic and morphological characteristic of M2c from patients with normal-GGT PFIC.
HPBMo were prepared from patients with normal-GGT PFIC. The obtained HPBMo were seeded, differentiated into HMDM by culture for 4 to 10 days in RPMI/M-CSF, and then cultured for 6 days in RPMI/M-CSF with IL-10 (40 ng/ml) to elicit polarization into M2c. The IL-10-treated HMDM were stained with fluorochrome-labeled antibodies against CD163 and CD14, subjected to flow cytometry, and the results were analyzed as described in Fig. 2c using FlowJo software (v. 10). Seven independent experiments were performed to analyze 12 patients (n = 4 PFIC1 patients; n = 4 PFIC2 and PFIC2-like patients; n = 4 PFIC1-like patients). In each experiment, HPBMo from more than three age-matched control subjects were pooled to minimize interindividual variability and employed as control cells. (a, b) Phenotypic and morphological differences in IL-10-treated HMDM of PFIC1 patients from patients with other PFIC subtypes. Graphs show cell surface expression of CD163 and CD14, and SSC of IL-10-treated HMDM on the horizontal axis (a) (control subjects, blue line; PFIC1 patient no.4, red filled line; PFIC2 patient no.2, green line; isotype control, black line). A representative result of four PFIC1 patients and four PFIC2 and PFIC2-like disease patients is shown. The MFI of CD163 and CD14 and the mean value of SSC in the cells from each patient with PFIC1, PFIC2, and PFIC2-like disease were determined and expressed relative to those of the cells from age-matched control subjects analyzed simultaneously (b). Each bar represents mean ± standard deviation (n = 4 PFIC1 patients; n = 4 PFIC2 and PFIC2-like patients). *, P < 0.05; **, P < 0.01. (c, d) Discrimination of patients with PFIC1 from patients with PFIC1-like disease. Graphs in (c) show cell surface expression of CD163 and CD14, and SSC of IL-10-treated HMDM on the horizontal axis (control subjects, blue line; PFIC1-like patients, red filled line). Each patient with PFIC1-like disease was analyzed in independent experiments. Their mean values were quantified as relative to those of the cells from the age-matched control subjects in each experiment and are plotted on a radar chart (d) (control subjects, blue line; PFIC1-like patients, red line; PFIC1 patients, green line). Data for PFIC1 patients are those shown in (b).
4. Discussion
PFIC1 shares many clinical and liver pathological features with other subtypes of normal-GGT PFIC, but differs in the available therapeutic options (Davit-Spraul et al., 2009). Patients with PFIC1 develop graft steatosis with or without fibrosis as well as severe diarrhea after LTx and sometimes need re-transplantation (Hori et al., 2011, Miyagawa-Hayashino et al., 2009), whereas it is the only curative option for the disease-related liver cirrhosis in individuals with other subtypes of normal-GGT PFIC. Therefore, to identify the preferred first-line therapy for these patients accurately, it is of the highest priority to discriminate PFIC1 correctly from other types of normal-GGT PFIC at an early phase of the disease.
The current study showed using siRNA-mediated ATP8B1 suppression that ATP8B1 contributes to IL-10-driven polarization of HMDM into M2c, and explored the usefulness of this finding for the diagnosis of PFIC1 using HMDM from patients with PFIC1, PFIC2, and undiagnosed normal-GGT PFIC. This method resulted in the correct diagnosis of all the tested patients with PFIC1 who had disease-causing mutations in both alleles of ATP8B1 (Fig. 4 and Table 1), because IL-10-treated HMDM from patients with PFIC1 showed significantly lower expression of typical M2c surface markers and higher SSC than those from control subjects. HMDM from patients with PFIC2 and PFIC2-like disease, other subtypes of normal-GGT PFIC, showed an almost identical phenotypic and morphological pattern to controls (Fig. 5a, b).
The results of this analysis in four patients with PFIC1-like disease who harbored only one mutant allele of ATP8B1 and no mutations in the other genes responsible for neonatal/infantile intrahepatic cholestasis, meant that three of four patients (patients 1 to 3) were diagnosed with PFIC1 (Fig. 5c, d). These three patients, who showed no ATP8B1 expression in liver specimens (Fig. 4a), presented with the typical clinical symptoms of PFIC1, including cholestasis with normal GGT, intractable itching, diarrhea/steatorrhea, failure to thrive, and mild intellectual disability. The histology of their liver specimens showed mild fibrosis with hepatocellular and canalicular cholestasis lacking giant cell transformation, which is compatible with PFIC1 (Table 2) (Davit-Spraul et al., 2010, Pawlikowska et al., 2010, Suchy et al., 2014). Furthermore, an electron microscopic analysis showed Byler's bile in bile canaliculus of these patients, which is highly suggestive of PFIC1 (Suchy et al., 2014). The other disease-causing mutation in ATP8B1 present in these patients may be outside the sequenced regions in areas such as transcriptional regulatory elements or untranslated regions. The other patient with PFIC-like disease (patient 4) showed similar liver histology to that of PFIC1 and presented with clinical symptoms similar to those of PFIC1 since 6 months old, including cholestasis with normal GGT, failure to thrive, severe pruritus, severe intellectual disability, and sensorineural deafness (Table 2) (Davit-Spraul et al., 2010, Pawlikowska et al., 2010, Suchy et al., 2014). However, in contrast to the situation with PFIC1, his serum bilirubin level returned to normal at 19 months old and his serum transaminases were elevated only episodically. These clinical symptoms that differed from those of PFIC1 were consistent with the normal ATP8B1 expression in his liver specimen (Fig. 4a), and the lack of a significant impact of the three mutations he possessed on the expression, cellular localization, and transport activity of ATP8B1 (Fig. 4b–d). Genetic diagnosis of this patient by whole-exome sequencing is under way. Together, these results demonstrated that the phenotypic and morphological analysis of IL-10-treated HMDM made it possible to assess the functional deficiency of ATP8B1 in individuals with a clinical diagnosis of normal-GGT PFIC.
Table 2.
Clinical and biochemical features and liver histology in PFIC1 and PFIC1-like patients.
| Features | Diagnostic findings of PFIC1 [% in PFIC1] | PFIC1 |
PFIC1-like |
||||||
|---|---|---|---|---|---|---|---|---|---|
| no.1 | no.2 | no.3d | no.4 | no.1e | no.2e | no.3f | no.4 | ||
| Presenting symptoms | |||||||||
| Age of onset | Before 1 year | 2 months | 4 months | 2 months | 2 months | 2 months | 3 months | 3 months | 6 months |
| Cholestasis | + [100%] | + | + | + | + | + | + | + | + |
| Jaundice | + [73%]a | + | + | + | + | + | + | + | + (resolved at 1y7m) |
| Intractable itching | + [100%]b | + | + | + | + | + | + | + | + |
| Diarrhea | + [61%]a | − | + | − | − | + | + | + | − |
| Steatorrhea | + [NA] | − | + | − | − | + | + | + | − |
| Sensorineural deafness | + [31%]a | − | − | − | − | − | − | − | + |
| Failure to thrive | + [90%]a | + | + | + | + | + | + | + | + |
| Short stature | + [NA] | + | + | + | + | + | + | + | + |
| Weight-for-height | > Normal [NA] | Normal | > Normal | > Normal | > Normal | > Normal | > Normal | Normal | > Normal |
| Mental retardation | + [NA] | − | + | + | + | + | + | − | ++ |
| Pneumonia | + [13%]a | − | − | − | − | − | − | + | − |
| Rickets | + [46%]a | − | + | + | + | + | − | − | − |
| Pancreatitis | + [12%]a | − | − | − | − | − | − | − | − |
| Serum assay | |||||||||
| AST, ALT | < 2 × Normalc [NA] | < 2 × Normal | > 2 × Normal | < 2 × Normal | < 2 × Normal | < 2 × Normal | < 2 × Normal | < 2 × Normal | > 2 × Normal |
| GGT | Low to normal [NA] | Normal | Normal | Low | Low | Low | Low | Low | Low |
| Total bilirubin | Elevated [NA] | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated (returned to normal at 1y7m) |
| Total bile acid | Elevated [NA] | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated | Elevated |
| Liver histology | |||||||||
| Hepatocellular and canalicular cholestasis | +c [NA] | + | + | + | + | + | + | + | + |
| Giant transformation of hepatocytes | No or fewc [NA] | Few | No | No | No | No | No | No | No |
| Fibrosis | +c [NA] | − | + | + | + | + | + | + | + |
| Surgical procedure (age) | PEBD (4 months) | LDLTx (2 years 4 months) | PEBD (3 years 6 months) | PEBD (1 years 3 months) | − | − | PIBD (1 year 6 months) | − | |
LDLTx, living donor liver transplantation; NA, not available; PEBD, percutaneous external biliary drainage; PIBD, partial internal biliary diversion.
Numakura C et al., Pediatr Int. 2011 Feb; 53(1):107–10.
Mochizuki K et al., Pediatr Surg Int. 2012 Jan;28(1):51–4.
IL-10/STAT3 signaling is initiated by binding of IL-10 to the IL-10R and subsequent tyrosine phosphorylation of the IL-10R by Janus kinase 1. This in turn results in STAT3, an obligate transcription factor for IL-10 signaling, docking with IL-10R and becoming activated by phosphorylation (Williams et al., 2004). A functional deficiency of ATP8B1 in HMDM attenuated the IL-10/STAT3 signal transduction pathway by decreasing pS-STAT3α (Fig. 3), which is required for its optimal activation (Liu et al., 2003, Waitkus et al., 2014, Zhu et al., 2015), and resulted in the incomplete IL-10-driven polarization of HMDM into M2c (Fig. 2, Fig. 5). ATP8B1, a member of the P4 subfamily of P-type adenosine triphosphatases, is expressed on the PM and translocates aminophospholipids from the outer leaflet to the inner leaflet, thereby contributing to making the PM a rigid, liquid-ordered membrane (Paulusma et al., 2008, Takatsu et al., 2014). Although the mechanism underlying the regulation of pS-STAT3α by ATP8B1 is unclear, a functional deficiency of ATP8B1 may disrupt the well-organized aminophospholipid asymmetry of the PM (Paulusma et al., 2009) and affect tethering to the PM of the kinases responsible for generation of pS-STAT3α. This concept is supported by the observation that ATP8B1 depletion inactivates protein kinase Cζ, whose activation depends on recruitment to the PM (Frankenberg et al., 2008). Various serine kinases, including JNK, ERK, mTOR, and several forms of protein kinase C, are implicated in the phosphorylation of STAT3 at serine 727, but their contribution differs in each cell type and with different stimuli (Aziz et al., 2007, Decker and Kovarik, 2000). Identification of the kinases that mediate formation of pS-STAT3α in HMDM stimulated with IL-10 will clarify the molecular mechanism underpinning the IL-10-driven polarization of HMDM into M2c and the molecular role of ATP8B1 in this process.
Treatment of HMDM with IL-10 to elicit their polarization into M2c, suppresses fluid phase pinocytosis and mannose receptor-mediated uptake (Montaner et al., 1999) and induces Mer tyrosine kinase, a member of the TAM subfamily of receptors that recognizes phosphatidylserine through protein S and Gas6. This enables HMDM to clear preferentially and efficiently early apoptotic cells derived from various processes including normal homeostasis, tissue turnover, and immune responses against pathogens (Xu et al., 2006, Zizzo et al., 2012). Therefore, M2c suppress persistent apoptosis and the accumulation of secondary necrotic cells, which are highly inflammatory because their autolysis releases cytotoxic, proinflammatory and immunogenic molecules including damage-associated molecular patterns (Silva, 2010). The anti-inflammatory actions of M2c are amplified and prolonged by their uptake of apoptotic cells and subsequent IL-10 secretion, generating a positive feedback loop of M2c homeostasis (Xu et al., 2006, Zizzo et al., 2012). ATP8B1 deficiency in HMDM resulted in incomplete polarization of HMDM into M2c (Fig. 2, Fig. 3, Fig. 5), which might prevent the resolution of inflammation and contribute, at least in part, to the pathogenesis of the clinical features of PFIC1, including steatohepatitis following LTx (Hori et al., 2011, Miyagawa-Hayashino et al., 2009), pancreatitis (Pawlikowska et al., 2010), and atherosclerosis (Nagasaka et al., 2005), in which cholesterol-laden apoptotic foam cells progress to secondary necrosis, causing inflammation and plaque instability. This could be supported by a recent study using THP-1-derived Mφ, which found that ATP8B1 depletion prevents LPS-induced internalization of TLR4 and may thereby induce a chronic inflammatory response (van der Mark et al., 2017). However, the pro-inflammatory (M1) or anti-inflammatory (M2) phenotypes generated in HMDM in response to pathophysiologic stimuli are distinct from THP-1-derived Mφ in which no induction of cell surface markers characteristic of M2a and M2c by stimulation with IL-4 or IL-10, respectively, was observed (Shiratori et al., 2017). Further studies to clarify the roles of ATP8B1 in M2c and the functions of M2c in liver physiology and pathophysiology should pave the way to understanding the pathogenic mechanism of progressive cholestatic liver failure and the extrahepatic manifestations of PFIC1, and allow the development of new therapies for PFIC1.
In conclusion, our present study showed that ATP8B1 deficiency caused incomplete polarization of HMDM into M2c via impairment of the IL-10/STAT3 signal transduction pathway. Genome sequencing and liver histological analysis, both of which are currently employed to diagnose PFIC1, are insufficient for that purpose because of the shared features of liver histology and the difficulties in identifying disease-causing mutations. Phenotypic and morphological analyses of IL-10-driven HMDM help to discriminate PFIC1 patients from individuals who have a clinical diagnosis of normal-GGT PFIC and no apparent disease-causing mutations in ATP8B1. The application of these findings as an alternative diagnostic method for PFIC1 must be validated by future studies including larger numbers of patients with neonatal/infantile cholestasis than was possible in this study. If confirmed, this method may assist with identifying the optimal treatment plan in patients with normal-GGT PFIC and in understanding the actual prevalence and clinical course of PFIC1.
Funding Sources
This work was supported by Practical Research Project for Rare/Intractable Diseases of Japan Agency for Medical Research and Development, AMED, and Astellas Foundation for Research on Metabolic Disorders, to H.H.. Funding agencies played no role in study design, data collection, data analysis, interpretation or writing of the report.
Conflict of Interest
The authors declare no conflicts of interest associated with this study.
Author Contributions
H.H. conceived the study, designed and performed the experiments, collected and analyzed the data, and wrote the manuscript. S.N. designed and performed the experiments, collected and analyzed the data. Y.H. performed in vitro mutagenesis study and analyzed the data. T.T. performed targeted next-generation sequencing and analyzed the data. Y.H., H.K., M.S., K.M., S.W., D.A., S.N., K.M., A.F., M.K., and A.I. provided biological samples and clinical information of PFIC patients. M.S. provided biological samples and clinical information of PFIC patients and revised the manuscript for intellectual content. H.N. K.B., and H.K. revised the manuscript for intellectual content. All authors approved the manuscript before submission.
Acknowledgments
Acknowledgments
We thank Dr. K. Hirano (Osaka University Graduate School of Medicine) for collaboration on the early stages of this work and Dr. T Isojima (Graduate School of Medicine, University of Tokyo), Dr. H. Takikawa (Teikyo University School of Medicine), Dr. Y. Inomata and Dr. Y. Ohya (Kumamoto University), Dr. H. Egawa (Tokyo Women's Medical University), Dr. H. Okajima and Dr. S. Okamoto (Kyoto University Graduate School of Medicine), Dr. T. Yorifuji (Osaka City General Hospital) for providing human blood samples on the early stages of this work. We also thank the families who participated in this study.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2017.10.007.
Appendix A. Supplementary data
References
- Aziz M.H., Manoharan H.T., Church D.R., Dreckschmidt N.E., Zhong W., Oberley T.D., Wilding G., Verma A.K. Protein kinase Cepsilon interacts with signal transducers and activators of transcription 3 (Stat3), phosphorylates Stat3Ser727, and regulates its constitutive activation in prostate cancer. Cancer Res. 2007;67:8828–8838. doi: 10.1158/0008-5472.CAN-07-1604. [DOI] [PubMed] [Google Scholar]
- Bull L.N., Van Eijk M.J., Pawlikowska L., Deyoung J.A., Juijn J.A., Liao M., Klomp L.W., Lomri N., Berger R., Scharschmidt B.F., Knisely A.S., Houwen R.H., Freimer N.B. A gene encoding a P-type ATPase mutated in two forms of hereditary cholestasis. Nat. Genet. 1998;18:219–224. doi: 10.1038/ng0398-219. [DOI] [PubMed] [Google Scholar]
- Chen H.L., Chang P.S., Hsu H.C., Ni Y.H., Hsu H.Y., Lee J.H., Jeng Y.M., Shau W.Y., Chang M.H. FIC1 and BSEP defects in Taiwanese patients with chronic intrahepatic cholestasis with low gamma-glutamyltranspeptidase levels. J. Pediatr. 2002;140:119–124. doi: 10.1067/mpd.2002.119993. [DOI] [PubMed] [Google Scholar]
- Chen S., So E.C., Strome S.E., Zhang X. Impact of detachment methods on M2 macrophage phenotype and function. J. Immunol. Methods. 2015;426:56–61. doi: 10.1016/j.jim.2015.08.001. [DOI] [PubMed] [Google Scholar]
- Davit-Spraul A., Gonzales E., Baussan C., Jacquemin E. Progressive familial intrahepatic cholestasis. Orphanet J. Rare Dis. 2009;4:1. doi: 10.1186/1750-1172-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davit-Spraul A., Fabre M., Branchereau S., Baussan C., Gonzales E., Stieger B., Bernard O., Jacquemin E. ATP8B1 and ABCB11 analysis in 62 children with normal gamma-glutamyl transferase progressive familial intrahepatic cholestasis (PFIC): phenotypic differences between PFIC1 and PFIC2 and natural history. Hepatology. 2010;51:1645–1655. doi: 10.1002/hep.23539. [DOI] [PubMed] [Google Scholar]
- Decker T., Kovarik P. Serine phosphorylation of STATs. Oncogene. 2000;19:2628–2637. doi: 10.1038/sj.onc.1203481. [DOI] [PubMed] [Google Scholar]
- El Kasmi K.C., Smith A.M., Williams L., Neale G., Panopoulos A.D., Watowich S.S., Hacker H., Foxwell B.M., Murray P.J. Cutting edge: a transcriptional repressor and corepressor induced by the STAT3-regulated anti-inflammatory signaling pathway. J. Immunol. 2007;179:7215–7219. doi: 10.4049/jimmunol.179.11.7215. [DOI] [PubMed] [Google Scholar]
- Folmer D.E., Van Der Mark V.A., Ho-Mok K.S., Oude Elferink R.P., Paulusma C.C. Differential effects of progressive familial intrahepatic cholestasis type 1 and benign recurrent intrahepatic cholestasis type 1 mutations on canalicular localization of ATP8B1. Hepatology. 2009;50:1597–1605. doi: 10.1002/hep.23158. [DOI] [PubMed] [Google Scholar]
- Frankenberg T., Miloh T., Chen F.Y., Ananthanarayanan M., Sun A.Q., Balasubramaniyan N., Arias I., Setchell K.D., Suchy F.J., Shneider B.L. The membrane protein ATPase class I type 8B member 1 signals through protein kinase C zeta to activate the farnesoid X receptor. Hepatology. 2008;48:1896–1905. doi: 10.1002/hep.22431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzales E., Grosse B., Cassio D., Davit-Spraul A., Fabre M., Jacquemin E. Successful mutation-specific chaperone therapy with 4-phenylbutyrate in a child with progressive familial intrahepatic cholestasis type 2. J. Hepatol. 2012;57:695–698. doi: 10.1016/j.jhep.2012.04.017. [DOI] [PubMed] [Google Scholar]
- Hasegawa Y., Hayashi H., Naoi S., Kondou H., Bessho K., Igarashi K., Hanada K., Nakao K., Kimura T., Konishi A., Nagasaka H., Miyoshi Y., Ozono K., Kusuhara H. Intractable itch relieved by 4-phenylbutyrate therapy in patients with progressive familial intrahepatic cholestasis type 1. Orphanet J Rare Dis. 2014;9:89. doi: 10.1186/1750-1172-9-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Sugiyama Y. 4-phenylbutyrate enhances the cell surface expression and the transport capacity of wild-type and mutated bile salt export pumps. Hepatology. 2007;45:1506–1516. doi: 10.1002/hep.21630. [DOI] [PubMed] [Google Scholar]
- Hayashi H., Takada T., Suzuki H., Akita H., Sugiyama Y. Two common PFIC2 mutations are associated with the impaired membrane trafficking of BSEP/ABCB11. Hepatology. 2005;41:916–924. doi: 10.1002/hep.20627. [DOI] [PubMed] [Google Scholar]
- Hori T., Egawa H., Takada Y., Ueda M., Oike F., Ogura Y., Sakamoto S., Kasahara M., Ogawa K., Miyagawa-Hayashino A., Yonekawa Y., Yorifuji T., Watanabe K., Doi H., Nguyen J.H., Chen F., Baine A.M., Gardner L.B., Uemoto S. Progressive familial intrahepatic cholestasis: a single-center experience of living-donor liver transplantation during two decades in Japan. Clin. Transpl. 2011;25:776–785. doi: 10.1111/j.1399-0012.2010.01368.x. [DOI] [PubMed] [Google Scholar]
- Hutchins A.P., Poulain S., Miranda-Saavedra D. Genome-wide analysis of STAT3 binding in vivo predicts effectors of the anti-inflammatory response in macrophages. Blood. 2012;119:e110–9. doi: 10.1182/blood-2011-09-381483. [DOI] [PubMed] [Google Scholar]
- Kittan N.A., Allen R.M., Dhaliwal A., Cavassani K.A., Schaller M., Gallagher K.A., Carson W.F.T., Mukherjee S., Grembecka J., Cierpicki T., Jarai G., Westwick J., Kunkel S.L., Hogaboam C.M. Cytokine induced phenotypic and epigenetic signatures are key to establishing specific macrophage phenotypes. PLoS One. 2013;8 doi: 10.1371/journal.pone.0078045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klomp L.W., Vargas J.C., Van Mil S.W., Pawlikowska L., Strautnieks S.S., Van Eijk M.J., Juijn J.A., Pabon-Pena C., Smith L.B., Deyoung J.A., Byrne J.A., Gombert J., Van Der Brugge G., Berger R., Jankowska I., Pawlowska J., Villa E., Knisely A.S., Thompson R.J., Freimer N.B., Houwen R.H., Bull L.N. Characterization of mutations in ATP8B1 associated with hereditary cholestasis. Hepatology. 2004;40:27–38. doi: 10.1002/hep.20285. [DOI] [PubMed] [Google Scholar]
- Liu H., Ma Y., Cole S.M., Zander C., Chen K.H., Karras J., Pope R.M. Serine phosphorylation of STAT3 is essential for Mcl-1 expression and macrophage survival. Blood. 2003;102:344–352. doi: 10.1182/blood-2002-11-3396. [DOI] [PubMed] [Google Scholar]
- Liu C., Aronow B.J., Jegga A.G., Wang N., Miethke A., Mourya R., Bezerra J.A. Novel resequencing chip customized to diagnose mutations in patients with inherited syndromes of intrahepatic cholestasis. Gastroenterology. 2007;132:119–126. doi: 10.1053/j.gastro.2006.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantovani A., Sica A., Sozzani S., Allavena P., Vecchi A., Locati M. The chemokine system in diverse forms of macrophage activation and polarization. Trends Immunol. 2004;25:677–686. doi: 10.1016/j.it.2004.09.015. [DOI] [PubMed] [Google Scholar]
- Matte U., Mourya R., Miethke A., Liu C., Kauffmann G., Moyer K., Zhang K., Bezerra J.A. Analysis of gene mutations in children with cholestasis of undefined etiology. J. Pediatr. Gastroenterol. Nutr. 2010;51:488–493. doi: 10.1097/MPG.0b013e3181dffe8f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa-Hayashino A., Egawa H., Yorifuji T., Hasegawa M., Haga H., Tsuruyama T., Wen M.C., Sumazaki R., Manabe T., Uemoto S. Allograft steatohepatitis in progressive familial intrahepatic cholestasis type 1 after living donor liver transplantation. Liver Transpl. 2009;15:610–618. doi: 10.1002/lt.21686. [DOI] [PubMed] [Google Scholar]
- Montaner L.J., Da Silva R.P., Sun J., Sutterwala S., Hollinshead M., Vaux D., Gordon S. Type 1 and type 2 cytokine regulation of macrophage endocytosis: differential activation by IL-4/IL-13 as opposed to IFN-gamma or IL-10. J. Immunol. 1999;162:4606–4613. [PubMed] [Google Scholar]
- Morotti R.A., Suchy F.J., Magid M.S. Progressive familial intrahepatic cholestasis (PFIC) type 1, 2, and 3: a review of the liver pathology findings. Semin. Liver Dis. 2011;31:3–10. doi: 10.1055/s-0031-1272831. [DOI] [PubMed] [Google Scholar]
- Murray P.J. Understanding and exploiting the endogenous interleukin-10/STAT3-mediated anti-inflammatory response. Curr. Opin. Pharmacol. 2006;6:379–386. doi: 10.1016/j.coph.2006.01.010. [DOI] [PubMed] [Google Scholar]
- Nagasaka H., Yorifuji T., Egawa H., Yanai H., Fujisawa T., Kosugiyama K., Matsui A., Hasegawa M., Okada T., Takayanagi M., Chiba H., Kobayashi K. Evaluation of risk for atherosclerosis in Alagille syndrome and progressive familial intrahepatic cholestasis: two congenital cholestatic diseases with different lipoprotein metabolisms. J. Pediatr. 2005;146:329–335. doi: 10.1016/j.jpeds.2004.10.047. [DOI] [PubMed] [Google Scholar]
- Naoi S., Hayashi H., Inoue T., Tanikawa K., Igarashi K., Nagasaka H., Kage M., Takikawa H., Sugiyama Y., Inui A., Nagai T., Kusuhara H. Improved liver function and relieved pruritus after 4-phenylbutyrate therapy in a patient with progressive familial intrahepatic cholestasis type 2. J. Pediatr. 2014;164(1219–1227) doi: 10.1016/j.jpeds.2013.12.032. [DOI] [PubMed] [Google Scholar]
- Ouyang W., Rutz S., Crellin N.K., Valdez P.A., Hymowitz S.G. Regulation and functions of the IL-10 family of cytokines in inflammation and disease. Annu. Rev. Immunol. 2011;29:71–109. doi: 10.1146/annurev-immunol-031210-101312. [DOI] [PubMed] [Google Scholar]
- Paulusma C.C., Folmer D.E., Ho-Mok K.S., De Waart D.R., Hilarius P.M., Verhoeven A.J., Oude Elferink R.P. ATP8B1 requires an accessory protein for endoplasmic reticulum exit and plasma membrane lipid flippase activity. Hepatology. 2008;47:268–278. doi: 10.1002/hep.21950. [DOI] [PubMed] [Google Scholar]
- Paulusma C.C., De Waart D.R., Kunne C., Mok K.S., Elferink R.P. Activity of the bile salt export pump (ABCB11) is critically dependent on canalicular membrane cholesterol content. J. Biol. Chem. 2009;284:9947–9954. doi: 10.1074/jbc.M808667200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlikowska L., Strautnieks S., Jankowska I., Czubkowski P., Emerick K., Antoniou A., Wanty C., Fischler B., Jacquemin E., Wali S., Blanchard S., Nielsen I.M., Bourke B., McQuaid S., Lacaille F., Byrne J.A., Van Eerde A.M., Kolho K.L., Klomp L., Houwen R., Bacchetti P., Lobritto S., Hupertz V., McClean P., Mieli-Vergani G., Shneider B., Nemeth A., Sokal E., Freimer N.B., Knisely A.S., Rosenthal P., Whitington P.F., Pawlowska J., Thompson R.J., Bull L.N. Differences in presentation and progression between severe FIC1 and BSEP deficiencies. J. Hepatol. 2010;53:170–178. doi: 10.1016/j.jhep.2010.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilling D., Fan T., Huang D., Kaul B., Gomer R.H. Identification of markers that distinguish monocyte-derived fibrocytes from monocytes, macrophages, and fibroblasts. PLoS One. 2009;4 doi: 10.1371/journal.pone.0007475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rey-Giraud F., Hafner M., Ries C.H. In vitro generation of monocyte-derived macrophages under serum-free conditions improves their tumor promoting functions. PLoS One. 2012;7:e42656. doi: 10.1371/journal.pone.0042656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrotta M., Strautnieks S., Papouli E., Rushton P., Clark B.E., Parry D.A., Logan C.V., Newbury L.J., Kamath B.M., Ling S., Grammatikopoulos T., Wagner B.E., Magee J.C., Sokol R.J., Mieli-Vergani G., University of Washington Center for Mendelian, G, Smith J.D., Johnson C.A., Mcclean P., Simpson M.A., Knisely A.S., Bull L.N., Thompson R.J. Mutations in TJP2 cause progressive cholestatic liver disease. Nat. Genet. 2014;46:326–328. doi: 10.1038/ng.2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaljo B., Kratochvill F., Gratz N., Sadzak I., Sauer I., Hammer M., Vogl C., Strobl B., Muller M., Blackshear P.J., Poli V., Lang R., Murray P.J., Kovarik P. Tristetraprolin is required for full anti-inflammatory response of murine macrophages to IL-10. J. Immunol. 2009;183:1197–1206. doi: 10.4049/jimmunol.0803883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiratori H., Feinweber C., Luckhardt S., Linke B., Resch E., Geisslinger G., Weigert A., Parnham M.J. THP-1 and human peripheral blood mononuclear cell-derived macrophages differ in their capacity to polarize in vitro. Mol. Immunol. 2017;88:58–68. doi: 10.1016/j.molimm.2017.05.027. [DOI] [PubMed] [Google Scholar]
- Silva M.T. Secondary necrosis: the natural outcome of the complete apoptotic program. FEBS Lett. 2010;584:4491–4499. doi: 10.1016/j.febslet.2010.10.046. [DOI] [PubMed] [Google Scholar]
- Strautnieks S.S., Bull L.N., Knisely A.S., Kocoshis S.A., Dahl N., Arnell H., Sokal E., Dahan K., Childs S., Ling V., Tanner M.S., Kagalwalla A.F., Nemeth A., Pawlowska J., Baker A., Mieli-Vergani G., Freimer N.B., Gardiner R.M., Thompson R.J. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat. Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- Suchy F.J., Sundaram S., Shneider B. Liver Disease in Children. 4th Edition. 2014. 13. Familial hepatocellular cholestasis; pp. 199–215. [Google Scholar]
- Takatsu H., Tanaka G., Segawa K., Suzuki J., Nagata S., Nakayama K., Shin H.W. Phospholipid flippase activities and substrate specificities of human type IV P-type ATPases localized to the plasma membrane. J. Biol. Chem. 2014;289:33543–33556. doi: 10.1074/jbc.M114.593012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Togawa T., Sugiura T., Ito K., Endo T., Aoyama K., Ohashi K., Negishi Y., Kudo T., Ito R., Kikuchi A., Arai-Ichinoi N., Kure S., Saitoh S. Molecular genetic dissection and neonatal/infantile intrahepatic cholestasis using targeted next-generation sequencing. J. Pediatr. 2016;171(171–177) doi: 10.1016/j.jpeds.2016.01.006. [DOI] [PubMed] [Google Scholar]
- Van Der Mark V.A., Ghiboub M., Marsman C., Zhao J., Van Dijk R., Hiralall J.K., Ho-Mok K.S., Castricum Z., De Jonge W.J., Oude Elferink R.P., Paulusma C.C. Phospholipid flippases attenuate LPS-induced TLR4 signaling by mediating endocytic retrieval of Toll-like receptor 4. Cell. Mol. Life Sci. 2017;74:715–730. doi: 10.1007/s00018-016-2360-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Der Velden L.M., Stapelbroek J.M., Krieger E., Van Den Berghe P.V., Berger R., Verhulst P.M., Holthuis J.C., Houwen R.H., Klomp L.W., Van De Graaf S.F. Folding defects in P-type ATP 8B1 associated with hereditary cholestasis are ameliorated by 4-phenylbutyrate. Hepatology. 2010;51:286–296. doi: 10.1002/hep.23268. [DOI] [PubMed] [Google Scholar]
- Waitkus M.S., Chandrasekharan U.M., Willard B., Tee T.L., Hsieh J.K., Przybycin C.G., Rini B.I., Dicorleto P.E. Signal integration and gene induction by a functionally distinct STAT3 phosphoform. Mol. Cell. Biol. 2014;34:1800–1811. doi: 10.1128/MCB.00034-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen, Z., Zhong, Z. & Darnell, J. E., JR. 1995. Maximal activation of transcription by Stat1 and Stat3 requires both tyrosine and serine phosphorylation. Cell, 82, 241–50. [DOI] [PubMed]
- Williams L., Bradley L., Smith A., Foxwell B. Signal transducer and activator of transcription 3 is the dominant mediator of the anti-inflammatory effects of IL-10 in human macrophages. J. Immunol. 2004;172:567–576. doi: 10.4049/jimmunol.172.1.567. [DOI] [PubMed] [Google Scholar]
- Xu W., Roos A., Schlagwein N., Woltman A.M., Daha M.R., Van Kooten C. IL-10-producing macrophages preferentially clear early apoptotic cells. Blood. 2006;107:4930–4937. doi: 10.1182/blood-2005-10-4144. [DOI] [PubMed] [Google Scholar]
- Zhu Y.P., Brown J.R., Sag D., Zhang L., Suttles J. Adenosine 5′-monophosphate-activated protein kinase regulates IL-10-mediated anti-inflammatory signaling pathways in macrophages. J. Immunol. 2015;194:584–594. doi: 10.4049/jimmunol.1401024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zizzo G., Hilliard B.A., Monestier M., Cohen P.L. Efficient clearance of early apoptotic cells by human macrophages requires M2c polarization and MerTK induction. J. Immunol. 2012;189:3508–3520. doi: 10.4049/jimmunol.1200662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.