Abstract
We have identified a variant in adenylate cyclase 3 (ADCY3) associated with markedly increased risk of obesity and type 2 diabetes in the Greenlandic population. The variant disrupts a splice-acceptor site and carriers display decreased ADCY3 RNA expression. Additionally, we observe an enrichment of rare ADCY3 loss-of-function variants among type 2 diabetes patients in trans-ethnic cohorts. These findings provide novel information on disease etiology relevant for future treatment strategies.
Identification of homozygous loss-of-function mutations in humans may readily inform about the biological impact of specific genes and point to novel drug targets. We previously identified a loss-of-function variant in TBC1D4 segregating at high frequency in the Greenlandic population displaying a high impact on risk of type 2 diabetes1, confirming the advantage of studying the Greenlandic population due to its extreme demographic history2. Motivated by this, we screened for novel loss-of-function variants in the exome sequencing data from 27 individuals in nine trios that were used to identify the causal TBC1D4 loss-of-function variant1. We identified 46 such variants (Supplementary Table 1) and intersected the location of these variants with loci known to associate with obesity or body mass index (BMI) (Supplementary Fig. 1). One of the variants, which was present in one copy in one of the trio's parents, was situated in a locus where a common non-coding variant has been shown to be associated with BMI in adults and children in genome-wide association studies (GWAS)3,4. The variant (hg19: 2-25050478-C-T, c.2433-1G>A) is predicted to destroy a splice-acceptor site in exon 14 (NM_004036.4) (Fig. 1A) of ADCY3. For this reason, we investigated the specific variant further by genotyping it in two Greenlandic cohorts. This revealed an overall minor allele frequency of 2.3% in the Greenlandic study population (N=4,038, N-heterozygous=172, N-homozygous=7), and a frequency of 3.1% in the Inuit ancestry part of the population. Importantly, the seven homozygous carriers had a 7.3 kg/m2 higher BMI (P=0.00094) compared with the remaining study population (Table 1). Interestingly, we also observed that three of the seven homozygous carriers had type 2 diabetes (P=7.8×10-5, Table 1), while one had impaired fasting glucose and one impaired glucose tolerance. Notably, the association with type 2 diabetes remained significant after adjustment for BMI (P=6.5×10-4), suggesting it is not simply mediated by increased BMI. The effects on BMI and type 2 diabetes were also observed, although with smaller effect sizes, when data were analyzed according to an additive genetic model (Table 1). However, when we compared the recessive and additive models with the full genotype model, we rejected the additive model (BMI: P=0.002; type 2 diabetes: P=0.004), but not the recessive model (BMI: P=0.17; type 2 diabetes: P=0.095). This suggests that the recessive model is appropriate for explaining the effect of the c.2433-1G>A variant.
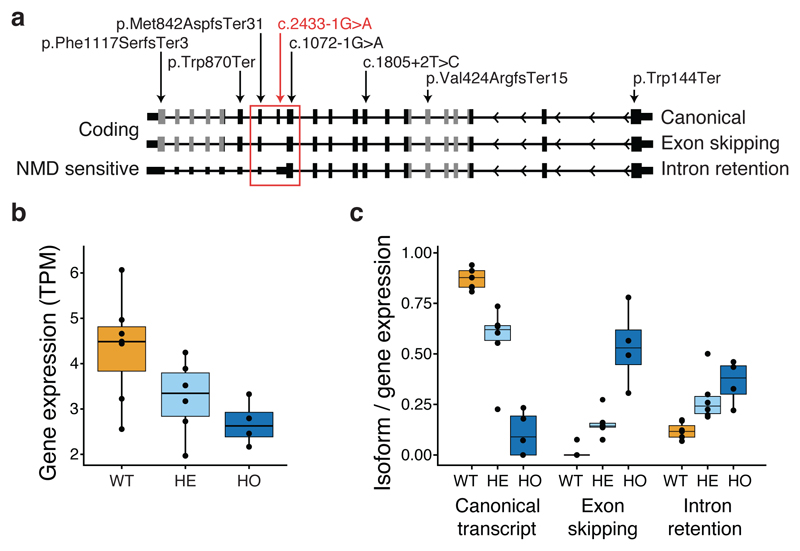
Figure 1. ADCY3 isoforms, observed loss-of-function variants and functional consequences based on RNA sequencing of leukocytes from 17 Greenlandic individuals.
a. Schematic representation of ADCY3 displaying the three relevant transcript isoforms and their predicted functional consequences annotated to the left (“Coding” or “nonsense mediated decay (NMD) sensitive”). The exons that correspond to the protein domain, Guanylate cyclase, are shown as gray filled boxes, while the rest of the exons are shown as black filled boxes. The red square encapsulates the exons affected by the Greenlandic ADCY3 c.2433-1G>A variant. The locations of the identified loss-of-function variants in ADCY3 in the Greenlandic and trans-ancestry cohorts are shown with arrows in red and black, respectively. Variants were annotated to canonical transcript ADCY3-201 (NM_004036) except c.1072-1G>A, which is annotated to alternative transcript ADCY3-202 (NM_001320613). b. ADCY3 Transcripts Per Million (TPM) normalized gene expression, stratified according to ADCY3 c.2433-1G>A variant genotype groups (WT, wild type; HE, heterozygous; and HO, homozygous). c. ADCY3 transcript isoform fractions for the three quantified isoforms, the canonical isoform, the novel exon skipping and intron retention splice version stratified according to ADCY3 c.2433-1G>A variant genotype groups. Number of individuals in each group in b and c are: WT: 7 GG carriers, HE: 6 GA carriers, and HO: 4 AA carriers. The lower and upper hinges of boxes in b and c correspond to the first and third quartiles of data, respectively, while the middle line is the median and the whiskers extends to the largest and smallest data points no further away than 1.5 times the interquartile range.
Table 1. Association of ADCY3 c.2433-1G>A with obesity and type 2 diabetes-related traits in Greenlandic cohorts.
| Recessive model | Additive model | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Trait | N | β SD | seSD | β | P | β SD | seSD | β | P |
| Type 2 diabetes (cases/ controls) | 301/2,585 | 0.50 | 7.8×10-5 | 0.081 | 0.0014 | ||||
| BMI (kg/m2) | 4,001 | 1.2 | 0.36 | 7.3 | 0.00094 | 0.18 | 0.075 | 1.00 | 0.017 |
| Fat percentage (%) | 2,701 | 1.1 | 0.35 | 8.1 | 0.0024 | 0.18 | 0.078 | 1.56 | 0.024 |
| Fasting plasma glucose (mmol/L) | 3,622 | 0.77 | 0.34 | 0.76 | 0.022 | 0.12 | 0.072 | 0.11 | 0.088 |
| 2-h plasma glucose (mmol/L) | 3,387 | 0.73 | 0.35 | 2.3 | 0.035 | 0.13 | 0.073 | 0.45 | 0.090 |
Results are shown for a recessive and an additive genetic model. βSD and SESD are the effect size and standard error estimated using quantile transformed values of the trait (except for the binary trait type 2 diabetes) and β is the effect size estimated using untransformed values. The P-values were obtained from the analyses of quantile transformed traits (except for the binary trait type 2 diabetes). P-values shown have not been corrected for multiple testing and nominally significant P-values are highlighted in bold.
To further characterize homozygous ADCY3 c.2433-1G>A carriers in the Greenlandic cohorts, we analyzed a number of additional traits related to BMI and type 2 diabetes. The homozygous carriers had an 8.1 percentage points higher body fat percentage (P=0.0024) and a 17 cm larger waist circumference (P=0.0017). In addition, the homozygous carriers had nominally higher levels of fasting and 2-h plasma glucose after an oral glucose tolerance test (P=0.022 and P=0.035, respectively; Table 1). Finally, we also observed nominally significant effects on dyslipidemia and insulin resistance (Supplementary Table 2).
The c.2433-1G>A ADCY3 variant was not observed in sequencing data of up to 138,000 individuals from non-Greenlandic populations collected by the Genome Aggregation Database Consortium5 (gnomAD). To generalize our findings to other populations, we therefore investigated the effect of loss-of-function variants in ADCY3 more generally by analyzing 18,176 samples with exome sequence data generated by the GoT2D, T2DGenes, SIGMA and LuCAMP consortia6,7,8, which are available at the Accelerating Medicines Partnership Type 2 Diabetes Knowledge Portal (AMP-T2D). No homozygous ADCY3 loss-of-function carriers were observed in this dataset, but the analysis included seven predicted ADCY3 loss-of-function variants (Fig. 1A) observed in the heterozygous form in eight individuals, and we observed an enrichment of carriers among type 2 diabetes cases compared with non-diabetic controls (7/8,845 in cases, 1/9,323 in controls, OR 8.6, P=0.044, Supplementary Table 3-5). To further substantiate these findings, exome sequence data from 9,928 Finnish individuals from the METSIM cohort9 were screened for loss-of-function variants in ADCY3; however, none were identified (Markku Laakso, personal communication). Furthermore, we did not find any loss-of-function ADCY3 variants in whole genome sequence data for 3,124 individuals from two Greek isolated populations as part of the HELIC study10. Finally, in up to 138,000 individuals in the gnomAD data, 48 predicted loss-of-function variants were found. All variants had minor allele frequency below 0.007% and only a single homozygous loss-of-function carrier was found for the African-specific ADCY3 c.1072-1G>A variant. Since we cannot obtain phenotypic data for the gnomAD samples, we are unable to evaluate the impact of these variants on obesity and type 2 diabetes.
To investigate the functional impact of the ADCY3 c.2433-1G>A variant, we performed deep RNA sequencing in leukocytes from 17 Greenlandic individuals (7 GG carriers, 6 GA carriers and 4 AA carriers). The RNA sequencing data confirmed that ADCY3 was expressed in the wild type GG carriers and that exon 14 (NM_004036.4) of ADCY3 was expressed and spliced in its canonical form (Supplementary Fig. 2A). The inclusion of exon 14 in the mature mRNA was further confirmed by RNA sequence data from adipose tissue of a healthy Caucasian female donor11. Importantly, we found that the overall RNA expression level of ADCY3 was severely decreased in the homozygous AA carriers, while the heterozygous GA carriers showed an intermediate expression level (Fig. 1B). The RNA sequencing data further confirmed that the predicted disruption of exon 14 splice-acceptor site by variant c.2433-1G>A has an impact on the molecular phenotype. Specifically, the data predict that two novel ADCY3 isoforms are expressed in the variant carriers: one transcript isoform where exon 14 is skipped and an alternative splice-acceptor site at exon 15 is used and one transcript isoform in which the intron between exon 13 and exon 14 is retained (Supplementary Fig. 2B). We quantified these alternative splicing events by comparing the expression of the three predicted ADCY3 isoforms using isoform fraction (IF) (Fig. 1C) and the percentage of spliced in (PSI) at the relevant splice sites (Supplementary Fig. 2B). Both analyses demonstrated that the homozygous AA carriers had severely affected splice pattern and mainly used intron retention (median IF 0.38, median PSI 75%) or exon skipping (IF 0.53, PSI 24%), while the wild type GG carriers had the canonical splice pattern (IF 0.88, PSI 87%). In all analyses, heterozygous carriers showed an intermediate level of alternative splicing (Fig. 1C; Supplementary Fig. 2B & C). Importantly, we predict the isoform with an intron retention to be sensitive to nonsense mediated decay due to a premature stop codon12 (Fig. 1A). This predicted degradation naturally would lead to further reduction of ADCY3 protein levels.
ADCY3 encodes an adenylate cyclase with a wide tissue distribution showing high levels in subcutaneous and visceral adipose tissue, intermediate levels in brain and rather low levels in pancreas and skeletal muscle in GTEx Project data. ADCY3 catalyzes the synthesis of cAMP from ATP. cAMP is an essential second-messenger in intracellular signaling downstream of key metabolic mediators such glucagon-like peptide 1, ghrelin and α-melanocyte stimulating hormone13, and cAMP signaling has been linked to control of adipose tissue development and function, as well as insulin secretion in beta cells14. In addition, mouse models have indicated that ADCY3 plays an important role in the regulation of adiposity and glucose homeostasis. Hence, in mice, Adcy3 haplo-insufficiency causes impaired insulin sensitivity and dyslipidaemia15, Adcy3 gain-of-function protects from diet induced obesity16, and Adcy3 knock-out mice show increased fat mass, hyperphagia, depression-like phenotypes, and leptin resistance17,18. Possibly, leptin resistance occurs through disrupted cAMP signaling in primary cilia in hypothalamus affecting downstream signaling and morphology of the neurons17,18. Interestingly, previously described syndromic forms of obesity, including Bardet-Biedl and Alström syndrome, are caused by altered function of primary cilia, and are besides from obesity characterized by diabetes19. Furthermore, common variation in ADCY5, a gene in the same family as ADCY3, is known to be associated with fasting plasma glucose levels and risk of type 2 diabetes20.
In humans, common variants in the ADCY3 locus have been associated with higher BMI3,4 and total as well as truncal fat mass21. Thus, the phenotype observed in Greenlandic homozygous loss-of-function carriers, characterized by truncal adiposity, insulin resistance, dyslipidemia and type 2 diabetes, is in accordance with and elaborates on the phenotype observed for GWAS-identified variants. Our findings for loss-of-function carriers implicate ADCY3 as the causal transcript in the reported GWAS-identified locus, and coherent evidence from genetic and biological studies suggest that pharmacological modulation of this target may possibly be a valid future therapy for obesity and type 2 diabetes.
In conclusion, we identified an ADCY3 loss-of-function variant in Greenlandic Inuit, which increases adiposity and risk of type 2 diabetes in homozygous carriers, and to a lesser extent in heterozygous carriers. Concomitantly, we detected decreased ADCY3 RNA expression levels in homozygous carriers, and again to a lesser extent in heterozygous carriers. Furthermore, we show that the variant disrupts a splice-acceptor site, triggering exon skipping or intron retention, where the latter is predicted to confer nonsense-mediated decay. The association with type 2 diabetes was substantiated in trans-ethnic heterozygous carriers of rare ADCY3 variants underlining the possible role of ADCY3 as a future target for prevention and treatment of obesity and type 2 diabetes.
Online methods
Study samples
The Greenlandic samples are from two different cohorts: B99 (N=1,401) and Inuit Health in Transition (IHIT) (N=3,115), which were collected in Greenland as a part of general population health surveys conducted in 1999-2001 and 2005-2010, respectively22,23. Two hundred and ninety-five individuals overlapped between the two cohorts and were assigned to the B99 dataset. Clinical characteristics of the participants are shown in Supplementary Table 6.
Ethical considerations
The study has received ethics approval from the Commission for Scientific Research in Greenland (project 2011-13, ref. no. 2011-056978; and project 2013-13, ref.no. 2013-090702) and was conducted in accordance with the ethical standards of the Declaration of Helsinki, second revision. All participants gave written consent after being informed about the study both orally and in writing.
Phenotypic data
Height and weight were measured wearing light indoor clothes. Waist circumference was measured midway between the rib cage and the iliac crest and hip circumference at its maximum. Weight was measured on a standard electronic clinical scale. Fat percentage was calculated from bioimpedance measurements (Tanita TBF-300MA) in IHIT participants. Intraabdominal adipose tissue was assessed by ultrasonography, which is considered a valid and reproducible method compared with CT and MRI24, using a 3.5 MHz transducer with the participant in supine position and at the end of a normal expiration. Tests for intra- and inter-observer variation were performed in IHIT and were in the range of 1.9-5.6% 25. In IHIT, all participants underwent an oral glucose tolerance test (OGTT). In B99, participants above 24 years of age had fasting blood samples taken and participants above 35 years underwent an OGTT. At the baseline health examination, venous blood samples were drawn after an overnight fast of at least 8 hours. After this, participants received a standard 75-g OGTT, with blood samples drawn 2 hours after the glucose intake. Only fasting venous plasma glucose was measured in participants with known diabetes. Fasting and 2-hours (2-h) plasma glucose values were analyzed with the Hitachi 912 system (Roche Diagnostics, Mannheim, Germany). Fasting and 2-h serum insulin levels were analyzed by an immunoassay method excluding des-31,32 split products and intact proinsulin (AutoDELFIA; Perkin Elmer, Waltham, MA). Indices of insulin sensitivity and insulin secretion were derived from plasma glucose and serum insulin measures from the OGTT. Hepatic insulin sensitivity was estimated by the homeostasis model assessment (HOMA-IR = fasting glucose [mmol/L] × (fasting insulin [pmol/L]/6.945) / 22.5) 26 and by a peripheral insulin sensitivity index (ISI0,120) calculated as: ISI0,120 = ( ((75000 + (fasting glucose[mmol/L] × 18 – 2-h glucose × 18) × 0.19 × weight [kg])/120) / ((fasting glucose[mmol/L] × 18 + 2-h glucose[mmol/L] × 18) / 2) ) / log((fasting insulin[pmol/L]/6.945 + 2-h insulin[pmol/L]/6.945)/2) 27. Analyses of quantitative glycemic traits were performed excluding individuals with previously diagnosed diabetes. Individuals taking lipid-lowering medication were removed in analysis of fasting serum lipids. Dichotomous glycaemia variables were constructed to test for association with type 2 diabetes classified according to the World Health Organization criteria28.
Identification and selection of loss-of function variants to study
To identify the novel loss-of-function variants in the Greenlandic population we used the exome data from 27 Greenlandic individuals in nine trios presented previously1. In this dataset, we identified 46 loss-of-function variants, which were not present in dbSNP (version 138 and below) and that were not present in NHLBI Exome Sequencing Project (ESP) (Supplementary Table 1). We then used the GWAS catalogue (downloaded January 2013) to assign known associations to the loci with the loss-of-function variants using associations with P-values of less than 10-7. Finally, we selected the variants with known associations to obesity or BMI for further study. This selection procedure left us with one predicted loss-of-function variant, ADCY3 c.2433-1G>A (Supplementary Fig. 1). This variant is located in ADCY3 and was present in a single parent in one of the nine trios in heterozygous form.
Genotype data
Subsequently, we genotyped ADCY3 c.2433-1G>A in the two Greenlandic cohorts, IHIT and B99, using KASP Genotyping Assay (LGC Genomics, Hoddlesdon, UK). Genotyping call rate was 99.2% and no mismatches were observed in 362 samples genotyped in duplicate. For the association testing, we used quality filtered MetaboChip genotype data to base our genetic similarity matrix estimates on. The genotyping and quality filtering of the MetaboChip data has been described in detail earlier29. The filtered MetaboChip dataset contains data from 2,791 individuals from IHIT and 1,336 individuals from B99 and we have ADCY3 c.2433-1G>A genotyping data for 4,038 of these individuals, 2,779 from IHIT and 1,259 from B99.
Association testing and replication
Greenlandic cohorts
To test for association between the ADCY3 variant and the different phenotypes of interest we used a linear mixed model, implemented in the software GEMMA30, in order to account for relatedness and admixture1. For each phenotype, GEMMA was applied to data from the subset of individuals across the two cohorts with information available about that specific phenotype. The genetic similarity matrix required as input to GEMMA was estimated from MetaboChip genotype data1. For each phenotype, we performed two association tests: a test where we assumed a recessive effect model and a test where we assumed an additive effect model. All tests were performed with sex, age and cohort included as covariates. An additional analysis also included BMI as covariate. Before all tests for association with quantitative traits, we quantile transformed the phenotype data to a normal distribution separately for each sex, which resulted in the results reported as βSD, seSD and P. However, to get effect sizes in the units the traits were measured in, we also performed the analyses without transforming the phenotype data, which resulted in the results reported as β. For the binary trait, type 2 diabetes, we did not perform any transformation and the test results are reported as β and P.
To assess the appropriateness of the recessive effect model and the additive effect model for BMI and type 2 diabetes we compared each of these effects models against a full genotype model, where each of the three genotypes have an independent effect. To do this, we used the same linear model framework as we did for the association tests. These additional tests were only performed in the IHIT cohort because there were no homozygous carriers in B99, which in practice means the tests cannot be performed if B99 is included.
Accelerating Medicines Partnership Type 2 Diabetes Knowledge Portal (AMP-T2D)
Data from AMP-T2D generated as part of the GoT2D, T2DGenes, SIGMA and LuCAMP consortia6,7,8 were queried for association between a burden of loss-of-function variants in ADCY3 and type 2 diabetes. The data contained 18,869 samples with exome sequence data of which 18,176 samples were informative for type 2 diabetes. Samples originated from five ancestries (6,356 Europeans, 5,722 Hispanic, 2,199 South Asians, 2,158 East Asians and 1,741 African Americans). Variants annotated as stop-gained, frameshift or in a splice adapter/donor site were considered loss-of-function. Burden association analyses were performed with each individual coded as carrying a loss-of-function variant or not. We used logistic regression and adjusted for principal components 1-4 as well as age and sex by including these as covariates. We also analyzed all other genes with at least five loss-of-function variants with at least one mutation carrier for each variant. Only loss-of-function variants with a minor allele frequency (MAF) <5% were used. This analysis showed that there is no general inflation in the test statistic (Supplementary Fig. 3).
Greek isolated populations
As part of the HELIC study10, 1,642 samples from the Pomak villages in Northern Greece and 1,482 samples from the Mylopotamos villages in Crete were sequenced at an average depth of 18.6x and 22.5x, respectively using the HiSeqX platform. Adapter-ligated libraries were amplified by 6 cycles of PCR and subjected to DNA sequencing according to manufacturer’s instructions. GVCFs were created for 200 equally sized chunks using GATK HaplotypeCaller v3.5, combined into batches of 150 samples using GATK CombineGVCFs and called using GATK GenotypeGVCFs. Variant-level QC was performed using GATK VQSR v.3.5.
Estimation of admixture proportions, ancestral allele frequencies and relatedness
For the Greenlanders, all admixture proportions were obtained from ref. 1. To estimate the ancestral Inuit allele frequencies for the ADCY3 variant we performed maximum likelihood estimation using the likelihood model from ADMIXTURE31 with the admixture proportions fixed and including only genotype data from the variant of interest. To achieve the maximum likelihood estimates we applied an EM-algorithm. We estimated relatedness using relateAdmix32 based on the estimated admixture proportions. None of the seven homozygous carriers are closely related (2nd-degree or closer), however one pair may be first cousins with an estimated kinship coefficient of 0.08.
RNA sequencing analysis
RNA sequencing was performed on leukocytes from 17 Greenlandic individuals (7 GG carriers, 6 GA carriers and 4 AA carriers). The total RNA was extracted from 2.5 mL of peripheral blood with the PAXGene Blood miRNA Kit (Qiagen) according to the manufacturer's protocol, and subjected to on-column DNase I treatment with RNase-free Dnase (Qiagen). The RNA quality and purity were checked using Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit) and NanoDrop™, respectively.
The RNA sequencing library was prepared following the instructions of the TruSeq RNA Sample Prep Kit v2 (Illumina): For mRNA isolation and fragmentation 200 ng of total RNA was purified by oligo-dT beads before fragmentation with Elute, Prime, Fragment Mix. First-strand cDNA was generated using First Strand Mix and SuperScript II (Invitrogen) reverse transcription master mix (reaction condition: 25°C for 10 min; 42°C for 50 min; 70°C for 15 min). The second-strand was synthesized by adding Second Strand Master Mix (16°C for 1h). The fragmented cDNA was end-repaired and purified with Ampure XP Beads (AGENCOURT). A-Tailing Mix was added and incubated (37°C for 30 min). For the adapter ligation, Adenylate 3'Ends DNA, RNA Index Adapter and Ligation Mix, were mixed and incubated (30°C for 10 min). End-repaired DNA was purified with Ampure XP Beads (AGENCOURT). Several rounds of PCR amplification with PCR Primer Cocktail and PCR Master Mix were performed to enrich the cDNA fragments. The PCR products were purified with Ampure XP Beads (AGENCOURT). The average molecule length was measured using the Agilent 2100 bioanalyzer instrument (Agilent DNA 1000 Reagents), and by real-time quantitative PCR (TaqMan Probe). The qualified libraries were amplified on cBot to generate the cluster on the flowcell (TruSeq PE Cluster Kit V3–cBot–HS, Illumina). The amplified flow cell was sequenced paired-end on the HiSeq 4000 System (TruSeq SBS KIT-HS V3, Illumina).
The obtained RNA-seq libraries, as well as one RNA-seq library from adipose tissue of a healthy Caucasian female11 were processed by Trimmomatic v 0.32 33 trimming the reads using HEADCROP:11 LEADING:22 SLIDINGWINDOW:4:22 MINLEN:25. Quality of all libraries were checked with FastQC, before and after trimming. Trimmed reads were mapped un-stranded to the human reference genome (hg19) using Hisat2 v2.0.1-beta34 with splice site information from GENCODE v19 35 and set to annotate properly paired reads as those with a minimum insert size from 0 to 1000 nucleotides. Sashimi plots36 were generated from the IGV genome browser37. PSI was calculated as rexon:intron / (rexon:intron + rexon:exon) where r is the number of reads indicating the different splice patterns. rexon:intron was obtained by extracting relevant GENCODE v19 35 exon:intron boundaries +/- 3 nucleotides. Reads mapping to these regions were quantified using featureCounts from the R package Rsubread38 specifying countChimericFragments=FALSE and minOverlap=6, ensuring that reads map across the exon boundary. rexon:exon was calculated by running featureCounts with the juncCounts=TRUE parameter. Plots were made using ggplot2. For transcript quantification, we constructed a gtf file containing three versions of GENCODE v19 transcript ENST00000260600.5, the longest transcript for ADCY3. The corresponding genomic sequences were extracted using Cufflinks gffread tool39 and a Salmon index was build using Salmon v0.8.2 40. Untrimmed fastq files were used to quantify the resulting transcripts using Salmon v0.8.2 0. Calculations of isoform fractions and prediction of functional consequences, were done with the R package IsoformSwitchAnalyzeR12 using standard parameters except the filtering where geneExpressionCutoff = 1, and isoformExpressionCutoff = 0.5, and keepIsoformInAllConditions=TRUE, supplied to the preFilter() function. Hmmer scan v.3.1 41 was used to predict pfamA protein domains as described in the IsoformSwitchAnalyzeR vignette and integrated into the analysis by the analyzePFAM() function. The functional consequences of isoform switches was predicted by IsoformSwitchAnalyzeR’s analyzeSwitchConsequences() function.
Data availability
The Greenlandic exome sequencing and RNA sequencing data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) under accession number EGAS00001002727. Trans-ethnic data analyzed in the project are available at http://www.type2diabetesgenetics.org/gene/geneInfo/ADCY3 upon registration at the website. Repositories of genetic variation and gene expression were queried at http://gnomad.broadinstitute.org/ and https://www.gtexportal.org/, respectively.
Supplementary Material
Acknowledgements
We gratefully acknowledge the participants in the Greenlandic health surveys. We thank Dr. Flannick, Broad Institute, US for help with obtaining the AMP-T2D exome sequencing data. The Novo Nordisk Foundation Center for Basic Metabolic Research is an independent Research Center at the University of Copenhagen partially funded by an unrestricted donation from the Novo Nordisk Foundation (www.metabol.ku.dk). This project was also funded by the Danish Council for Independent Research (Sapere Aude DFF-Ung Eliteforsker grant 11-120909 from FSS to I.M.; DFF-4090-00244 from FNU to I.M.; Sapere Aude DFF-Forskningsleder 6108-00038B from FNU to R.A., DFF-4181-00383 to A.A.), the Steno Diabetes Center Copenhagen (www.steno.dk), Simon Fougner Hartmanns Familiefond to N.G., the Lundbeck foundation (R215-2015-4174) to A.A., the Novo Nordisk Foundation (NNF15OC0017918 to N.G.; NNF16OC0019986 to N.G.; NNFCC0018486 to T.H.) and the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation programme (grant agreement no. 638173) to R.A.. The Greenlandic health surveys (IHIT and B99) were supported by Karen Elise Jensen’s Foundation, the Department of Health in Greenland, NunaFonden, Medical Research Council of Denmark, Medical Research Council of Greenland, and the Commission for Scientific Research in Greenland.
Footnotes
URLs
Accelerating Medicines Partnership Type 2 Diabetes Knowledge Portal: http://www.type2diabetesgenetics.org/
Genome Aggregation Database (gnomAD): http://gnomad.broadinstitute.org/
GTEx Portal: https://www.gtexportal.org/
FastQC: http://www.bioinformatics.babraham.ac.uk/projects/fastqc/
Contributions
T.H. and A.A. conceived and headed the project. I.M. and A.A. designed the statistical setup for the association testing, while T.H., N.G, M.K.A. and O.P. designed the experimental setup for the DNA extraction, genotyping and sequencing. M.E.J. and P.B. were PIs of the population studies in Greenland, and together with C.V.L.L., and I.K.D.-P. they provided the Greenlandic samples, collected and defined the phenotypes and provided context of these samples. I.M., N.G., E.J. and A.A. performed the association analyses. T.K. and Y.M. designed the experimental setup for RNA extraction and sequencing. A.G, D.S, G.D. and E.Z. performed the loss-of-function analysis in the Greek cohorts. K.V.S., M.D. and R.A. performed the RNA sequence analysis. N.G., I.M., M.K.A., A.A. and T.H. wrote the majority of the manuscript with input from all authors. All authors approved the final version of the manuscript.
Competing Financial Interests Statement
The authors report no competing financial interests.
References
- 1.Moltke I, et al. Nature. 2014;512:190–3. doi: 10.1038/nature13425. [DOI] [PubMed] [Google Scholar]
- 2.Pedersen CT, et al. Genetics. 2017;205:787–801. doi: 10.1534/genetics.116.193821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Speliotes EK, et al. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Warrington NM, et al. Int J Epidemiol. 2015;44:700–12. doi: 10.1093/ije/dyv077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lek M, et al. Nature. 2016;536:285–91. doi: 10.1038/nature19057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fuchsberger C, et al. Nature. 2016;536:41–7. [Google Scholar]
- 7.The Sigma Type Diabetes Consortium et al. JAMA. 2014;311:2305–14. [Google Scholar]
- 8.Lohmueller KE, et al. Am J Hum Genet. 2013;93:1072–86. doi: 10.1016/j.ajhg.2013.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laakso M, et al. J Lipid Res. 2017;58:481–493. doi: 10.1194/jlr.O072629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Panoutsopoulou K, et al. Nat Commun. 2014;5:5345. doi: 10.1038/ncomms6345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu Y, et al. PLoS One. 2013;8:e66883. doi: 10.1371/journal.pone.0066883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vitting-Seerup K, Sandelin A. Mol Cancer Res. 2017;15:1206–1220. doi: 10.1158/1541-7786.MCR-16-0459. [DOI] [PubMed] [Google Scholar]
- 13.Xu TR, Yang Y, Ward R, Gao L, Liu Y. Cell Signal. 2013;25:2413–23. doi: 10.1016/j.cellsig.2013.07.025. [DOI] [PubMed] [Google Scholar]
- 14.Yang H, et al. J Mol Endocrinol. 2016;57:R93–R108. doi: 10.1530/JME-15-0316. [DOI] [PubMed] [Google Scholar]
- 15.Tong T, et al. Sci Rep. 2016;6:34179. doi: 10.1038/srep34179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pitman JL, et al. PLoS One. 2014;9:e110226. doi: 10.1371/journal.pone.0110226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, et al. PLoS One. 2009;4:e6979. doi: 10.1371/journal.pone.0006979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chen X, et al. Biol Psychiatry. 2016;80:836–848. doi: 10.1016/j.biopsych.2015.12.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaisse C, Reiter JF, Berbari NF. Cold Spring Harb Perspect Biol. 2017;9 doi: 10.1101/cshperspect.a028217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dupuis J, et al. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tachmazidou I, et al. Am J Hum Genet. 2017;100:865–884. doi: 10.1016/j.ajhg.2017.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bjerregaard P, et al. Int J Circumpolar Health. 2003;62(Suppl 1):3–79. doi: 10.3402/ijch.v62i0.18212. [DOI] [PubMed] [Google Scholar]
- 23.Bjerregaard P. 2011 http://www.si-folkesundhed.dk/upload/inuit_health_in_transition_Greenland_methods_5_2nd_revision.pdf.
- 24.Philipsen A, et al. PLoS One. 2015;10:e0123062. doi: 10.1371/journal.pone.0123062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jørgensen ME, et al. Diabetes Care. 2013;36:2988–94. doi: 10.2337/dc12-2703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matthews DR, et al. Diabetologia. 1985;28:412–9. doi: 10.1007/BF00280883. [DOI] [PubMed] [Google Scholar]
- 27.Gutt M, et al. Diabetes Res Clin Pract. 2000;47:177–84. doi: 10.1016/s0168-8227(99)00116-3. [DOI] [PubMed] [Google Scholar]
- 28.World Health Organization Study Group. World Health Organization; Geneva: 1999. [Google Scholar]
- 29.Andersen MK, et al. PLoS Genet. 2016;12:e1006119. doi: 10.1371/journal.pgen.1006119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou X, Stephens M. Nat Genet. 2012;44:821–4. doi: 10.1038/ng.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Alexander DH, Novembre J, Lange K. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Moltke I, Albrechtsen A. Bioinformatics. 2014;30:1027–28. doi: 10.1093/bioinformatics/btt652. [DOI] [PubMed] [Google Scholar]
- 33.Bolger AM, Lohse M, Usadel B. Bioinformatics. 2014;30:2114–20. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim D, Langmead B, Salzberg SL. Nat Methods. 2015;12:357–60. doi: 10.1038/nmeth.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harrow J, et al. Genome Res. 2012;22:1760–74. doi: 10.1101/gr.135350.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz Y, et al. Bioinformatics. 2015;31:2400–2. doi: 10.1093/bioinformatics/btv034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Robinson JT, et al. Nat Biotechnol. 2011;29:24–6. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao Y, Smyth GK, Shi W. Bioinformatics. 2014;30:923–30. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 39.Trapnell C, et al. Nat Biotechnol. 2010;28:511–5. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Nat Methods. 2017;14:417–419. doi: 10.1038/nmeth.4197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Punta M, et al. Nucleic Acids Res. 2012;40:D290–301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The Greenlandic exome sequencing and RNA sequencing data have been deposited at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega/home) under accession number EGAS00001002727. Trans-ethnic data analyzed in the project are available at http://www.type2diabetesgenetics.org/gene/geneInfo/ADCY3 upon registration at the website. Repositories of genetic variation and gene expression were queried at http://gnomad.broadinstitute.org/ and https://www.gtexportal.org/, respectively.