Abstract
The morphogen Sonic Hedgehog (SHH) patterns tissues during development by directing cell fates in a concentration-dependent manner. The SHH signal is transmitted across the membrane of target cells by the heptahelical transmembrane protein Smoothened (SMO), which activates the GLI family of transcription factors through a mechanism that is undefined in vertebrates. Using CRISPR-edited null alleles and small molecule inhibitors, we systematically analyzed the epistatic interactions between SMO and three proteins implicated in SMO signaling: the heterotrimeric G-protein subunit GαS, G protein–coupled receptor kinase 2 (GRK2), and the GαS-coupled receptor GPR161. Our experiments uncovered a signaling mechanism that modifies the sensitivity of target cells to SHH and consequently changes the shape of the SHH dose-response curve. In both fibroblasts and spinal neural progenitors, the loss of GPR161, previously implicated as an inhibitor of basal SHH signaling, increased the sensitivity of target cells across the entire spectrum of SHH concentrations. Surprisingly, GRK2, thought to function by antagonizing GPR161, and Gαs, which is activated by GPR161, influenced SHH signaling even in cells lacking GPR161. We propose that the sensitivity of target cells to Hedgehog (Hh) morphogens, and the consequent effects on gene expression and differentiation outcomes, can be controlled by signals from G-protein coupled receptors that converge on Gαs and Protein Kinase A.
Introduction
Secreted ligands of the Hedgehog (Hh) family function as morphogens and pattern tissues, such as the spinal cord, limb bud, and paraxial mesoderm, during development. Activation of the Hh signaling pathway in responsive cells can drive the patterning of spinal neural progenitor subtypes in a manner that depends on both the concentration of the ligand Sonic Hedgehog (SHH) and the duration of SHH exposure (1). The mechanism by which extracellular SHH is transformed into the transcriptional activity that controls target cell identity remains an unresolved question, partly because the mechanisms that convey Hh signals from the cell surface to the nucleus are incompletely understood.
Patched (PTCH) proteins, the transmembrane receptors for Hh ligands, repress the activity of Smoothened (SMO), a Frizzled-family G protein–coupled receptor (GPCR) that transmits the Hh signal across the membrane to the cytoplasm. In the absence of Hh ligands, protein kinase A (PKA) and Suppressor of Fused (SUFU) inhibit the activity of the glioma-associated oncogene family transcription factors GLI2 and GLI3 and promote the proteolysis of GLI3 into a transcriptional repressor fragment (hereafter called GLI3R) (2). Hh ligands inactivate PTCH1, allowing SMO to adopt an active conformation and accumulate in the membrane of the primary cilium (3). Active SMO ultimately antagonizes the inhibitory effect of PKA and SUFU on the GLI proteins. As a result, the formation of GLI3R is blocked and full-length GLI2 and GLI3 are converted to transcriptional activators (hereafter GLI2A and GLI3A) (4–7).
The mechanism by which the Hh signal is transmitted from SMO to GLI2 or GLI3 remains poorly understood in vertebrates. Given the negative role of PKA in Hh signaling in all animals, SMO must somehow antagonize PKA activity or shield GLI proteins from the inhibitory influence of PKA. Several proteins that can influence PKA activity have been found to play a role in signaling at the step between SMO and GLI2 or GLI3. Recent work focused on a key role for the ciliary GPCR GPR161, which has been proposed to function downstream of SMO to repress basal signaling (signaling in the absence of Hh ligands) by promoting the production of GLI3R (8). GPR161 activates the GαS heterotrimeric G-protein, encoded by the Gnas gene, leading to increases in cyclic AMP (cAMP) levels and consequently elevated PKA activity. GPR161 is localized in the ciliary membrane but is cleared from cilia when Hh ligands are received, a step which requires the activity of G protein–coupled receptor kinase 2 (GRK2) (9, 10). Consistent with this model, GRK2 activity has been shown to be required for propogation of the Hh signal in multiple systems (9, 11–17) and Gαs, like GPR161, functions as a negative regulator of the Hh pathway (18–21). In summary, a widely-invoked model for cytoplasmic Hh signaling in vertebrates posits that Hh ligands antagonize GLI3R production by clearing GPR161 from cilia, a step mediated by GRK2, and consequently Gαs activity (8, 9).
However, this model is not fully consistent with neural tube patterning phenotypes in mouse embryos carrying mutations in genes encoding many of these components (8, 13, 18, 22). For example, neural tube patterning in Gpr161-/- embryos is more severely disrupted than in Gli3-/- embryos, suggesting that the resulting phenotype is due to more than a loss of GLI3R (8, 23). Conversely, defects in Gpr161-/- embryos are less severe than those in Gnas-/-, Ptch1-/-, and Sufu-/- embryos, or in embryos lacking PKA activity (8, 18, 22). The epistatic relationship between SMO and GPR161 is also ambiguous. GPR161 is epistatic to SMO when assaying Hh-induced differentiation into NKX6.1-positive neural progenitors, but SMO is epistatic to GPR161 when assaying FOXA2-positive progenitors (8). More generally, the epistatic relationships between SMO, GPR161, GαS, and GRK2 have not been determined.
We set out to test the current model for Hh signaling by SMO using epistasis analysis, quantitative signaling, and differentiation assays in two Hh-responsive cultured cell systems-- cultured fibroblasts and spinal neural progenitor cells (NPCs). NPCs differentiate into specific classes of progenitor subtypes in the neural tube in response to different levels of Hh signaling (24, 25), allowing an assessment of the morphogenetic function of this pathway. Our results reveal that GPR161 regulates Hh signaling in two fundamentally different ways. In addition to its established role in repressing basal, or ligand-independent, Hh signaling, GPR161 plays an equally important role in regulating ligand-driven signaling. We found that GPR161 reduced the sensitivity of target cells to SHH, markedly altering the shape of the SHH dose-response curve. Thus, GPR161 influences neural tube pattering by altering target-cell sensitivity to the SHH morphogen. Unexpectedly, GRK2 retained the ability to promote and Gαs retained the ability to restrain Hh signaling in cells lacking GPR161. We propose that target-cell responses to Hh signaling are influenced through both GPR161-dependent and -independent pathways that are regulated by GRK2 and that converge on GαS and ultimately PKA activity.
Results
GPR161 functions as an attenuator of Hh signaling in NIH/3T3 cells
We used CRISPR-mediated editing to generate loss-of-function mutations in the Gpr161 gene using four different guide RNAs in NIH/3T3 cells, a mouse embryonic fibroblast cell line widely used for the mechanistic analysis of Hh signaling in vitro. In contrast to previous observations in Gpr161-/- mouse embryos, GLI3R abundance was unaltered in all four Gpr161-/- cell lines whether or not the cells were stimulated with SHH (Fig. 1A). The abundance of GLI1, a direct Hh target gene used as a measure of signaling strength, was also very similar between wild-type and Gpr161-/- cells, both in the absence of SHH or in the presence of a saturating concentration of SHH (Fig. 1A).
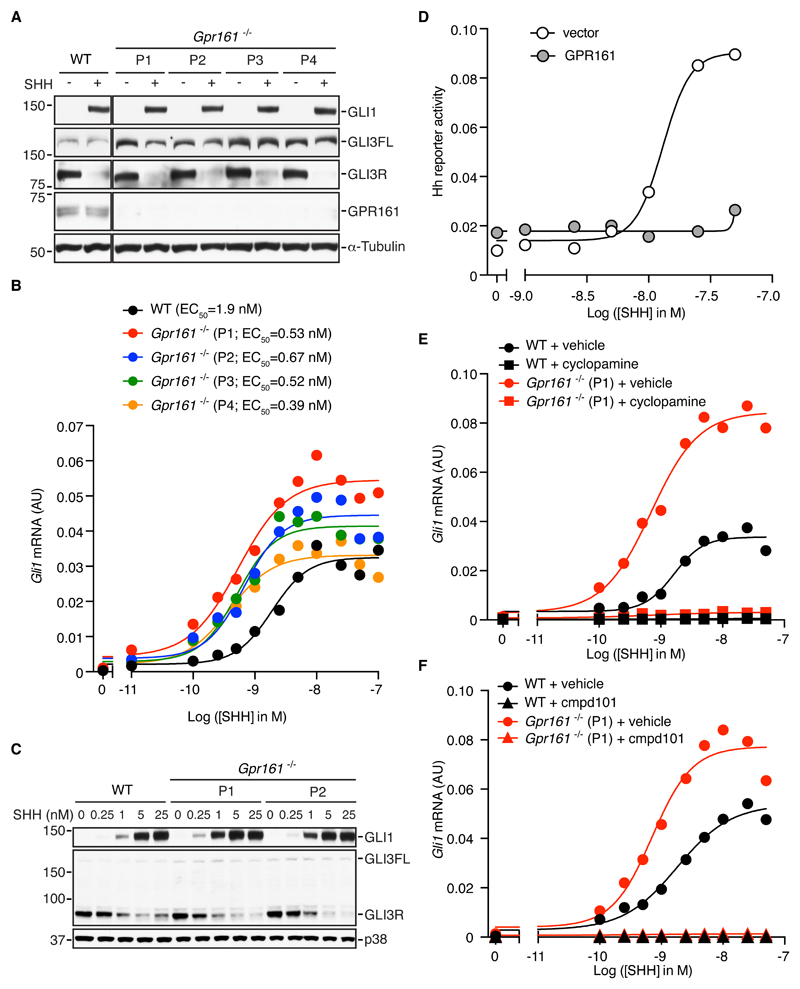
Fig. 1. GPR161 suppresses the potency of Hh ligands in NIH/3T3 cells.
(A) Immunoblot showing the indicated proteins in extracts from SHH-treated wild-type NIH/3T3 cells and four clonal Gpr161-/- NIH/3T3 cell lines (P1-P4) generated using four different guide RNAs. (B) Quantification of Gli1 mRNA relative to Gapdh by qRT-PCR in wild-type (WT) and four independently generated Gpr161-/- NIH/3T3 cell lines after exposure to the indicated range of SHH concentrations. (C) Immunoblot showing the indicated proteins in extracts from wild-type and two different Gpr161-/- NIH/3T3 cell lines (P1 and P2) that were either untreated or treated with increasing concentrations of SHH. (D) SHH-induced activation of a luciferase-based Hh reporter gene in NIH/3T3 cells transiently transfected with an empty vector or a vector carrying a gene encoding GPR161. (E and F) Quantification of Gli1 mRNA relative to Gapdh by qRT-PCR in wild-type or Gpr161-/- NIH/3T3 cells after exposure to the indicated range of SHH concentrations in the presence or absence of small molecules that inhibit SMO (cyclopamine, E) or GRK2 (cmpd101, F). In B, D, E and F each data point represents a mean of three technical replicates. A, B, E and F show representative data from three independent experiments. C and D show representative data from two independent experiments.
Because there was no discernable effect on basal signaling in Gpr161-/- cells, we next asked whether signaling responses were altered in response to SHH. The curve depicting the relationship between Gli1 mRNA and the concentration of SHH was shifted to the left in Gpr161-/- cells compared to wild-type cells, resulting in a ~3-fold to 5-fold increase in the potency of the ligand (Fig. 1B). The concentration of SHH that resulted in half-maximal increase in Gli1 mRNA, a formal measure of potency hereafter denoted EC50, decreased from 1.9 nM in wild-type cells to 0.4-0.7 nM in Gpr161-/- cells (Fig. 1B). In addition, there was a modest increase in the maximum levels of Gli1 mRNA induced by SHH in Gpr161-/- cells (Fig. 1B). The increase in sensitivity to SHH in Gpr161-/- cells is consistent with the neural tube phenotype in Gpr161-/- embryos-- the dorsal expansion of ventral progenitor domains (8). Wild-type and Gpr161-/- cells contained approximately equivalent amounts of GLI3R protein, both at baseline and after exposure to varying doses of SHH (Fig. 1C). GLI3FL levels were modestly increased in cells lacking GPR161 (Fig. 1, A and C), suggesting a defect in the activation of full-length GLI proteins, which are known to be much more unstable in their transcriptionally-active states (5). Over-expression of Gpr161 in NIH/3T3 cells blocked expression of a Hh signaling reporter (Fig. 1D), a phenotype opposite to the enhanced signaling phenotype seen when GPR161 is lost (Fig. 1B).
The increase in sensitivity to SHH in Gpr161-/- cells in the absence of changes in the abundance of GLI3R (Fig. 1, A and C) or ligand-independent signaling suggested that GPR161 may regulate Hh signling through additional mechanisms that cannot be accommodated by current models. This notion was further reinforced by the epistatic relationship between SMO, GPR161, and GRK2 in NIH/3T3 cells. SMO activity has been shown to be dispensable for some of the increased Hh responses seen in the neural tubes of mouse embryos lacking GPR161, an observation used to place GPR161 downstream of SMO in the pathway (8). However, SHH-stimulated signaling remained dependent on SMO activity in Gpr161-/- NIH/3T3 cells because it could be completely blocked by the SMO antagonist cyclopamine (Fig. 1E). Our epistasis analysis suggests that GPR161 functions in NIH/3T3 cells as a modifier rather than an obligate downstream component of SMO signaling.
Two (non-mutually exclusive) mechanisms have been proposed to explain the positive role of GRK2 in Hh signaling: (1) GRK2 promotes the ciliary localization of SMO and (2) GRK2 induces the clearance of GPR161 from the ciliary membrane, leading to a decrease in Gαs and PKA activity (9). The former model has been questioned by recent studies in zebrafish embryos and fibroblasts showing that SMO accumulates in cilia normally in cells lacking GRK2 activity (9, 17). The latter model is inconsistent with our observation that GPR161 is dispensable for Hh signaling in NIH/3T3 cells (Fig. 1A), whereas GRK2 is essential (17). To establish the epistatic relationship between GRK2 and GPR161, we sought a pharmacological strategy to acutely block the kinase activity of GRK2 and the closely related GRK3. Takeda compound 101 (cmpd101) (26) is a cell-permeable small molecule inhibitor of both GRK2 and GRK3 (GRK2/3) that can inhibit the desensitization of GPCRs (27). Cmpd101 blocked SHH-driven signaling in NIH/3T3 cells with an IC50 of 4.9 µM (fig. S1A), in agreement with the previously reported potency of cmpd101 (IC50 of 3 µM) in inhibiting the desensitization of µ-opioid receptors in intact cell assays (27). Hh signaling strength was evaluated by measuring (1) the mRNA or protein abundance of GLI1 and PTCH1, both of which are encoded by genes that are directly stimulated by SHH signaling, and (2) the abundance of GLI3R protein (fig. S1B). The effect of cmpd101 recapitulated the effect of a genetic deletion of Grk2 in multiple independent clonal cell lines (fig. S1, C to E and table S1). As reported previously for the genetic ablation of Grk2, cmpd101 did not prevent the signal-induced accumulation of SMO in primary cilia (fig. S1F) (9, 17).
If the mechanistic role of GRK2 in Hh signaling is to remove GPR161 from the primary cilium, GRK2 should be dispensable in cells lacking GPR161. However, we found that SHH-induced Gli1 expression was fully inhibited by cmpd101 in Gpr161-/- NIH/3T3 cells, demonstrating that GRK2 must promote Hh signaling in these cells through a mechanism that does not depend exclusively on GPR161 (Fig. 1F). To summarize, our results show that both GRK2 and SMO can function independently of GPR161 in NIH/3T3 cells, a conclusion that is different from the view that SMO and GRK2 promote Hh signaling by antagonizing GPR161 (9). We conclude that GPR161 in NIH/3T3 cells functions as an attenuator of signaling. This should be contrasted with a negative regulator of signaling, such as SUFU, the ablation of which leads to SHH-independent activation of signaling that does not depend on SMO (28).
We also tested whether the ciliary localization or clearance of GPR161 could be affected by GRK2 in NIH/3T3 cells. As previously reported, yellow fluorescent protein (YFP)-tagged GPR161 (GPR161-YFP) stably expressed in NIH/3T3 cells accumulated in the ciliary membrane, and the addition of SHH led to the clearance of GPR161-YFP from cilia (fig. S2, A and B) (8). This clearance could be blocked by a small molecule antagonist of SMO (SANT-1) and by either the genetic ablation of Grk2 or the inhibition of GRK2 kinase activity by cmpd101 (fig. S2, A and B). Endogenous GPR161 was present at levels too low to detect in NIH/3T3 cilia; however, either the genetic or pharmacological antagonism of GRK2 led to a marked increase in endogenous GPR161 in cilia, confirming that GRK2 can reduce the abundance of GPR161 in the cilia of NIH/3T3 cells (fig. S2, C and D).
The central piece of evidence that GPR161 is regulated by SMO is the correlative observation that SHH or agonists of SMO can drive the clearance of GPR161 from the ciliary membrane. However, whether the localization of GPR161 in the ciliary membrane (or its clearance) plays a functional role in Hh signaling remains unknown. Indeed, a C-terminal truncation mutant of GPR161 that cannot be cleared from cilia in response to SHH has no apparent effect on the strength of Hh signaling in cultured cells (9). We also found that the overexpression of Grk2 could drive GPR161 out of cilia, without leading to activation of Hh target genes (fig. S2, E to G). Thus, the ciliary clearance of GPR161 can be uncoupled from the activation of Hh signal transduction.
GRK2 functions at a step between SMO and GαS in Hh signaling
If GRK2 does not influence Hh signaling by controlling SMO localization (9, 17) or by reducing GPR161 in the primary cilium (Fig. 1F), how does it regulate the pathway? To address this question, we performed a comprehensive epistasis analysis to identify the critical step regulated by GRK2 (Fig. 2A). We generated clonal NIH/3T3 cell lines carrying loss-of-function mutations in genes encoding three negative regulators of the pathway (PTCH1, GαS, or SUFU), along with a cell line expressing a constitutively active, oncogenic mutant form of SMO (SMO-W539L, in which Trp539 is mutated to Leu). Each of these cell lines displayed high levels of signaling that did not change in response to SHH stimulation (Fig. 2, B to E and fig. S3, A to G). Pharmacological inhibition of GRK2 kinase activity with cmpd101 or the introduction of loss-of-function mutations in Grk2 reversed the constitutive signaling seen in Ptch1-/- NIH/3T3 cells (Fig. 2B and fig. S3A). Both perturbations also reversed constitutive signaling in Med1-MB cells (fig. S3, H to J), a mouse medulloblastoma cell line that lacks PTCH1 activity and is used to model aspects of Hh-driven human medulloblastomas (29). The IC50 of cmpd101 was very similar between cells that did (4.9 µM) or did not (3.9 µM) contain PTCH1 (fig. S1A and S3H). Therefore, GRK2 must act downstream of PTCH1 in NIH/3T3 cells.
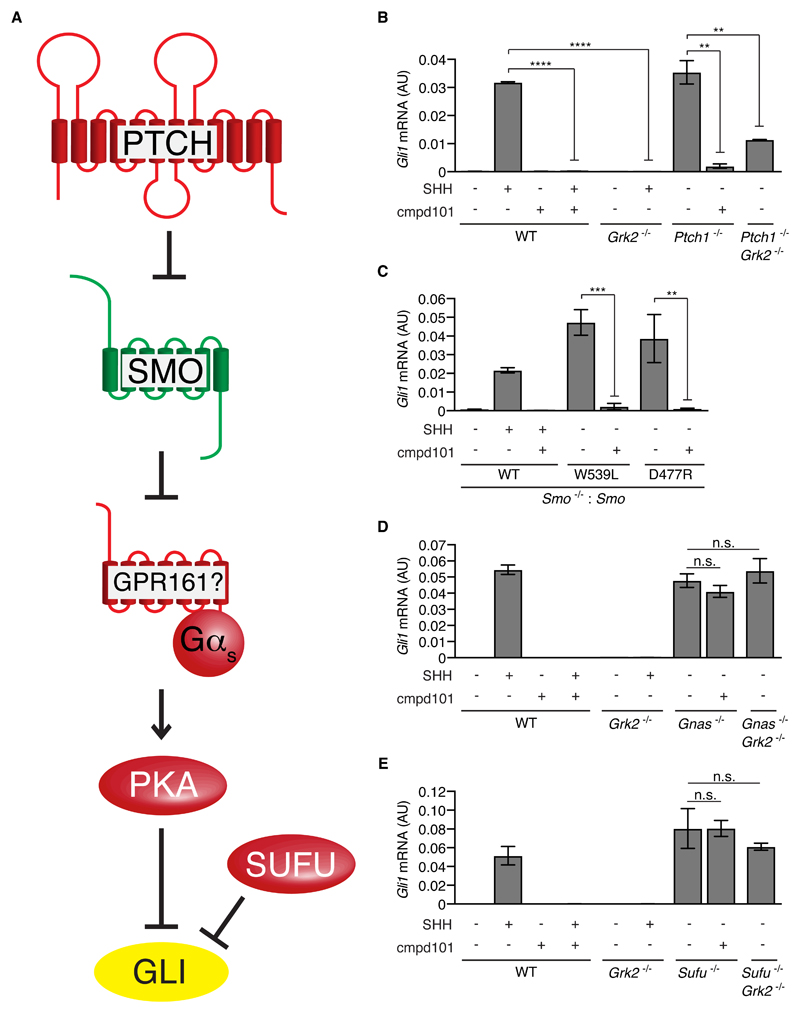
Fig. 2. GRK2 functions at a step between SMO and Gαs in the Hh signaling pathway.
(A) The current model for transduction of Hh signals in vertebrates, with positive regulators in green and negative regulators in red. GLI proteins (yellow) can function as either transcriptional repressors or activators, depending on how they are post-translationally modified and processed. SMO is thought to reduce PKA activity by reducing the amount of GPR161 in primary cilia. Shown to the right of each component are the results from genetic epistasis experiments to establish the order in which the component functions relative to GRK2. (B-E) Quantification of endogenous Gli1 mRNA relative to Gapdh by qRT-PCR in cells of various genotypes that were exposed to the indicated combinations of SHH and the GRK2 inhibitor cmpd101. In B, each data point represents mean±S.D (n=3). In C to E, each data point represents mean±S.D (n=4). Statistical significance was determined by unpaired Welch’s t-test and depicted as follows: P<0.01 (**), P<0.001 (***), P<0.0001 (****), and P>0.05 (n.s., not significant).
With respect to SMO, we have previously shown that the constitutive Hh signaling induced by SMO-W539L requires GRK2 (17). Cmpd101 treatment reduced signaling in cells expressing SMO-W539L (Fig. 2C and fig. S3G). Cmpd101 also blocked signaling in cells carrying SMO mutants (D477G and D477R, in which Asp477 is mutated to Gly or Arg) that are resistant to the clinically-used antagonist vismodegib (Fig. 2C and fig. S3G) (30, 31). These results show that GRK2 functions at the level of SMO or downstream of SMO.
In contrast to the effects on signaling triggered by PTCH1 inactivation or SMO activation, constitutively activated signaling in Gnas-/- and Sufu-/- cells could not be blocked by cmpd101 or by loss-of-function mutations in Grk2 (Fig. 2, D and E and fig. S3, A and B). Taken together, these results suggest that GRK2 is required for signaling at a step in the Hh pathway from SMO to Gαs (the product of Gnas gene) (Fig. 2A). The concordance between the epistatic profile of cmpd101 and the genetic deletion of Grk2 demonstrates that the effects of this small molecule on the Hh pathway are specific.
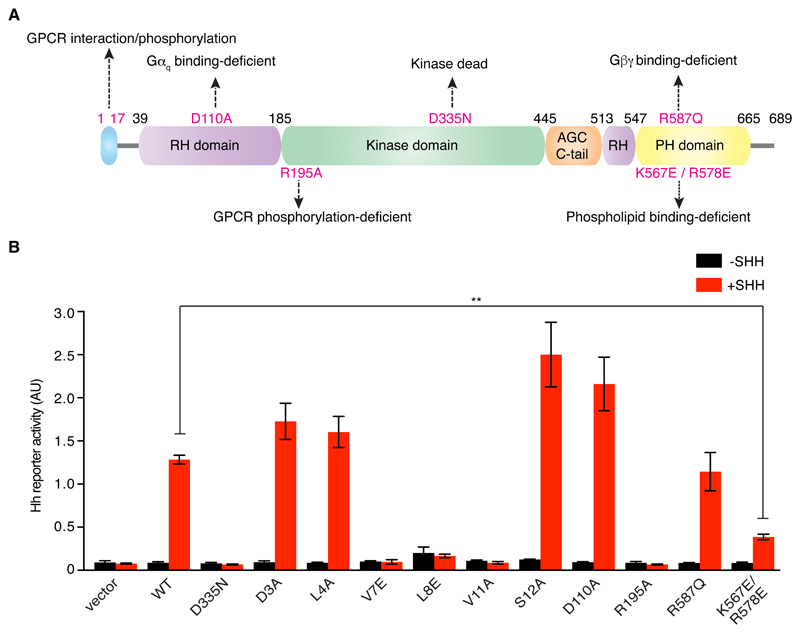
To understand how GRK2 may function in NIH/3T3 cells, we took advantage of previous studies that have mapped critical amino acid residues in GRK2 that are required for kinase activity, membrane recruitment, interaction with GPCRs, and binding to G protein subunits (32–35) (Fig. 3A). We generated mutant GRK2 variants that each specifically affect one aspect of GRK2 function and tested their ability to rescue the Hh phenotype observed in Grk2-/- cells. Mutations in residues on one face of an N-terminal helix that are known to mediate the interaction of GRK2 with GPCRs failed to rescue Hh reporter expression in Grk2-/- cells (Fig. 3B) (35). In addition, GRK2 function in Hh signaling depended on residues that mediate membrane recruitment through anionic phospholipid binding (Lys567 and Arg578), but not on residues implicated in binding to the Gβγ complex (Arg587) or to Gαq (Asp110) (Fig. 3, A and B). Taken together with the epistasis analysis above, these data suggest that GRK2 may function in Hh signaling by regulating the activity or abundance of a GPCR other than GPR161 in NIH/3T3 cells. These mutagenesis experiments are also consistent with the possibility that GRK2 regulates SMO itself (because SMO is a GPCR); however, in this latter scenario, GRK2 would have to regulate SMO activity by a mechanism other than altering its ciliary localization, which has been excluded by two independent studies (9, 17).
Fig. 3. Mapping the residues that are critical for GRK2 function in Hh signaling.
(A) Domain structure of GRK2 [adapted from (61)]. RH – regulator of G-protein signaling homology; AGC C-tail – C-terminal extension of the kinase domain reminiscent of Protein Kinases A, G, and C; PH - pleckstrin homology. Amino acid residues mutated in our analysis are indicated, along with their previously established effects on GRK2 function. (B) Activation of a luciferase-based Hh reporter gene in Grk2-/- NIH/3T3 cells in response to SHH after transient transfection with an empty vector or a vector carrying genes encoding the indicated mutant forms of GRK2-GFP. Each data point represents mean±S.D (n=3). Statistical significance was determined by unpaired Welch’s t-test and depicted as follows: P<0.01 (**).
GRK2 and GRK3 are essential for Hh responses in Neural Progenitor Cells
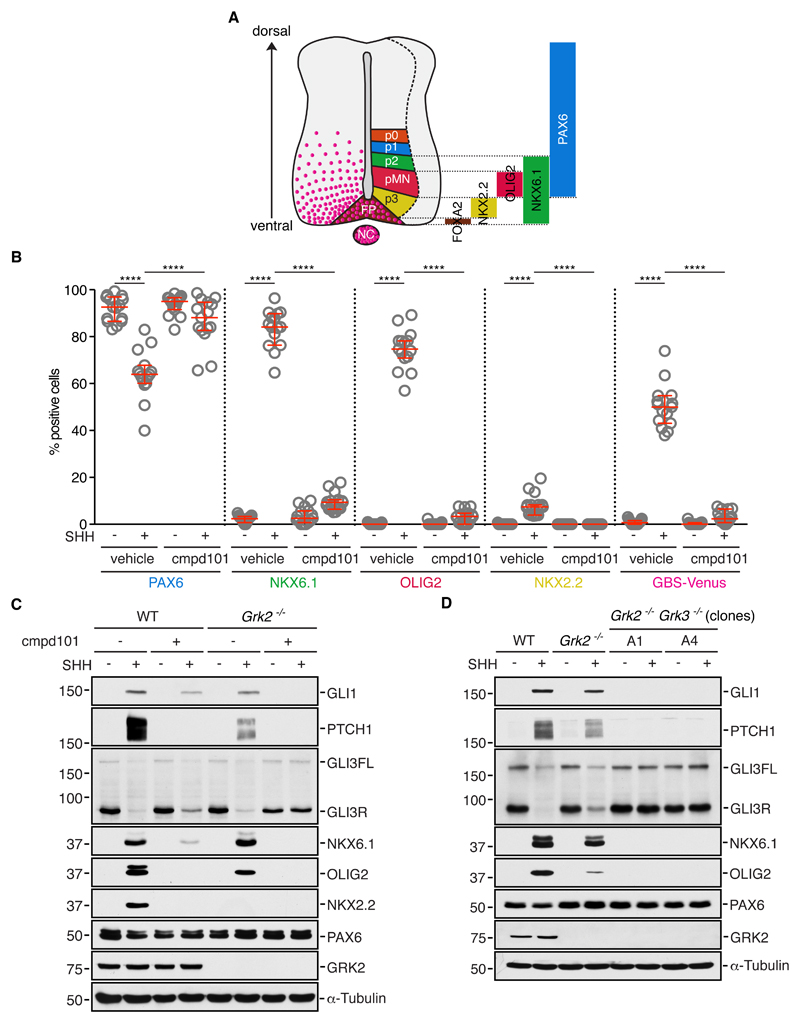
Mouse embryos with loss-of-function mutations in Gpr161 have a defect in patterning of the ventral neural tube, where neural identity is determined by a gradient of SHH ligand secreted by the notochord and the floorplate (8, 36) (Fig. 4A). This well-established morphogenetic activity of Hh ligands can be recapitulated in spinal cord neural progenitor cells (NPCs) derived from embryonic stem cells. As in NIH/3T3 cells, Hh signaling activity can be assessed in NPCs by measuring the induction of GLI1 and PTCH1. However, NPCs also afford a more physiological readout of Hh signaling strength—the adoption of different cell fates that can be assayed by the expression of genes encoding transcription factors that define progenitor identity (Fig. 4A) (25). These transcription factors can be divided into two groups: Class I transcription factors that are expressed in the absence of Hh (PAX6) and Class II transcription factors whose expression is driven by increasing amounts of Hh signaling (low – NKX6.1, medium – OLIG2, high – NKX2.2, and highest – FOXA2). These transcription factors that define neural identity in the spinal cord can be assessed by (1) measuring their mRNA or protein abundance in bulk NPC cultures or (2) counting the number of individual cells positive for each transcription factor after immunostaining. In addition, we took advantage of NPCs produced from a mouse embryonic stem cell line (mESC) harbouring the fluorescent marker Venus under the control of a Hh-responsive promoter carrying multiple GLI binding sites (GBS) from the FOXA2 locus (hereafter called GBS-Venus). Given that GPR161 depletion had no effect on basal signaling in NIH/3T3 cells, we reassessed the roles of GPR161 and GRK2 in NPCs.
Fig. 4. GRK2 and GRK3 activity is required for spinal neural cell fates that are dependent on all levels of Hh signaling.
(A) A schematic of the progenitor domains within the embryonic spinal cord (adapted from (36)). NC -notochord; FP - floorplate; pMN - motor neuron progenitors; p0, p1, p2, p3 - ventral interneuron progenitors. The progenitor domains are a product of a high-to-low SHH gradient (pink) along the ventral-to-dorsal axis. The bars on the right represent the transcription factors present in each progenitor domain. (B) Neural progenitor cells (NPCs) carrying the fluorescent Hh reporter GBS-Venus were left untreated or treated with SHH, cmpd101, or SHH+cmpd101 followed by immunofluorescence staining to count the percentage of cells positive for transcription factors that define ventral progenitor sub-types summarized in (A). Each data point represents the data from one image of a NPC colony (see fig. S4A) consisting of 100-200 cells, and each condition is represented by 15 different colonies. The experiment was repeated twice. Medians with interquartile ranges are shown with statistical significance determined by the Mann-Whitney non-parametric ANOVA test and depicted as follows: P<0.0001 (****). (C and D) Immunoblots (representative of two independent experiments) showing the indicated proteins in wild-type (WT), Grk2-/- (C), or Grk2-/- Grk3-/- double-null NPCs (D) treated with the indicated combinations of SHH and cmpd101. Two Grk2-/- Grk3-/- clonal NPC lines (A1 and A4) are shown in (D) and an independent Grk2-/- NPC cell line is shown in fig. S4C.
When cultured without SHH, NPCs contained low levels of GLI1 and PTCH1 and high levels of PAX6 (Fig. 4, B and C and fig. S4A). Upon SHH addition, GLI1 and PTCH1 levels increased (Fig. 4C) concomitant with a decrease in PAX6 (Fig. 4, B and C and fig. S4A). A majority of NPCs differentiated into NKX6.1-positive and OLIG2-positive progenitors, and a smaller fraction into NKX2.2-positive progenitors (Fig. 4, B and C and fig. S4A).
Because both Grk2 and Grk3 are expressed in NPCs (fig. S4D), we tested the dual inhibitor cmpd101 these cells. Cmpd101 blocked all readouts of Hh signaling activity, including target gene induction, as measured by PTCH1, GLI1 (Fig. 4C) and GBS-Venus expression (Fig. 4B), and differentiation of progenitor subtypes driven by low (NKX6.1), medium (OLIG2), and high (NKX2.2) levels of signaling (Fig. 4, B and C and fig. S4A). Thus, GRK2/3 activity is required to mediate all responses to SHH in NPCs.
Unlike NIH/3T3 cells, which only express Grk2, NPCs express both Grk2 and Grk3 (fig. S4D). NPCs carrying loss-of-function mutations in Grk2 alone responded to SHH (Fig. 4C and fig. S4B,C) and remained sensitive to cmpd101 (Fig. 4C and fig. S4A), suggesting that GRK2 and GRK3 play redundant roles. Indeed, loss-of-function mutations in both Grk2 and Grk3 (Grk2-/- Grk3-/- double null cells) resulted in a near-complete block in Hh-induced differentiation into ventral cell types, analogous to the effect seen with the dual inhibitor cmpd101 (Fig. 4D). GRK2 and GRK3 were functionally redundant because GRK3 could support Hh signaling in NIH/3T3 cells lacking GRK2 (fig. S4E). Redundancy between the paralogous Grk2 and Grk3 genes in mice provides a likely explanation for why Grk2-/- mice display a much milder Hh signaling phenotype compared to zebrafish grk2-/- mutants; the zebrafish genome contains a single gene (named grk2) that encodes a protein most similar to mammalian GRK3 (17, 37). In summary, GRK2 and GRK3 are together required in NPCs for Hh signaling and for Hh-driven differentiation, raising the important question of whether they regulate the pathway through GPR161 in NPCs.
GPR161 inhibits low-level Hh responses and attenuates high-level Hh responses in NPCs
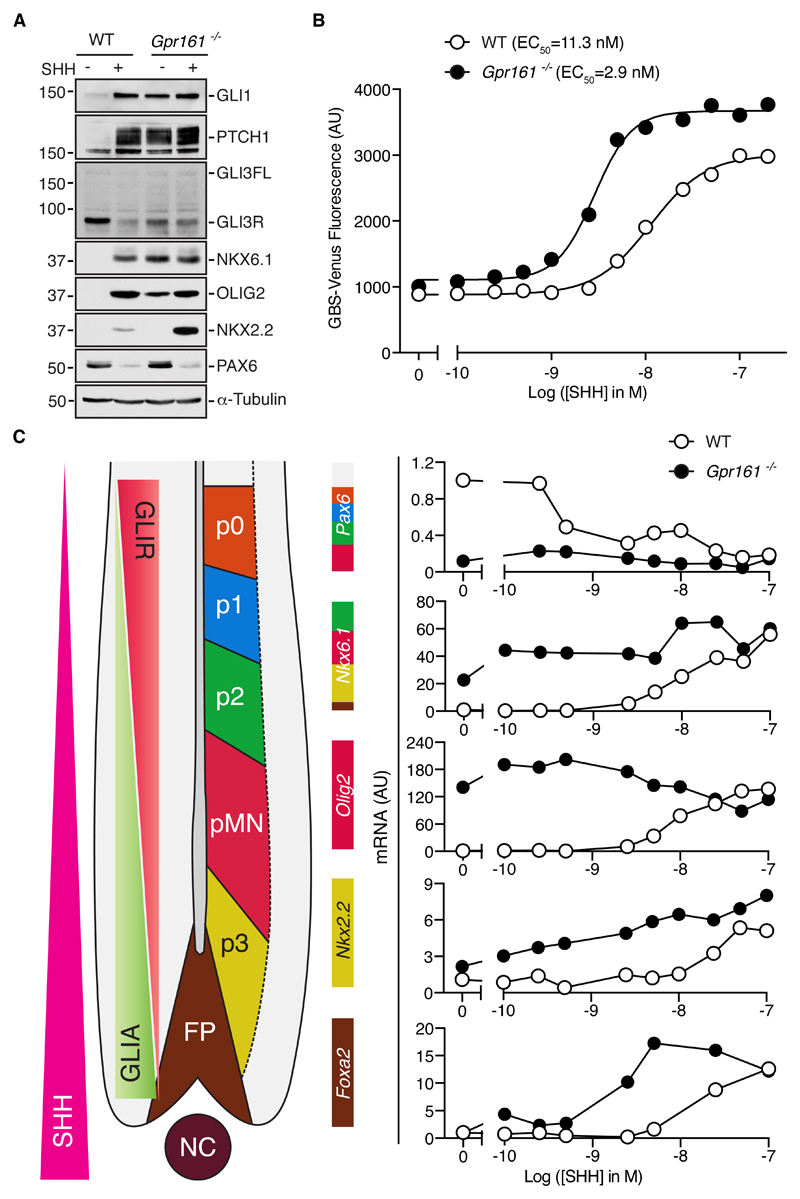
Because GPR161 was originally shown to be a negative regulator of Hh signaling in the neural tube (8), we generated Gpr161-/- NPCs (fig. S5A) and tested their differentiation in response to SHH. Consistent with the phenotype observed in the neural tube of Gpr161-/- mouse embryos (8), Hh signaling was modestly increased in Gpr161-/- NPCs in the absence of Hh ligands (Fig. 5A). This finding was in contrast to the absence of basal Hh pathway activation in NIH/3T3 cells lacking GPR161 (Fig. 1A), highlighting its tissue or cell-type specificity. The stronger effect of GPR161 loss on basal Hh signaling in NPCs was not caused by the higher expression of Gpr161 in NPC cells compared to NIH/3T3 cells (fig. S5B).
Fig. 5. GPR161 suppresses low-level Hh responses and attenuates high-level Hh responses in neural progenitor cells.
(A) Hh signaling was assessed (n=3 independent experiments) using immunoblots of extracts from wild-type and Gpr161-/- NPCs left untreated or treated with SHH. (B) Activation of the GBS-Venus reporter in wild-type (WT) or Gpr161-/- NPCs treated with increasing concentrations of SHH. Each data point represents median reporter fluorescence calculated from 10,000 cells. (C) A schematic (left) of the progenitor domains within the embryonic spinal cord along with opposing gradients of GLIR and GLIA proposed to establish the spatial pattern of neural subtypes. Pax6, Nkx6.1, Olig2, Nkx2.2, and Foxa2 mRNAs were quantified by qRT-PCR (normalized to Gapdh) in wild-type and Gpr161-/- NPCs treated with increasing concentrations of SHH. Immediately to the left of each graph, the domains in the neural tube where each transcription factor is present is depicted as a color code based on the diagram to the left. Each data point represents a mean of three technical replicates. The experiments in B and C were repeated twice.
In unstimulated Gpr161-/- NPCs, we observed an increase in the abundances of GLI1 and PTCH1 and a reduction in GLI3R (Fig. 5A). GLI3 processing into the GLI3R fragment depends on direct PKA-mediated phosphorylation; hence, loss of GPR161, a GαS-coupled receptor, could prevent GLI3R production by decreasing cAMP levels and PKA activity (8, 38). A predicted consequence of GLI3R loss is the de-repression of markers responsive to low-level Hh signals (23, 39). Indeed, the analysis of markers of ventral neural progenitor subtypes revealed low-level activation of Hh signaling. Most NPCs lacking Gpr161 contained NKX6.1, driven by the lowest levels of Hh signaling (Fig. 5A and fig. S5C), whereas wild-type cells differentiated predominantly into PAX6-positive NPCs. However, a smaller fraction were positive for OLIG2, the production of which is dependent on intermediate-level Hh signaling, and very few were positive for NKX2.2, which requires the high levels of Hh signaling (Fig. 5A and fig. S6A). The basal level of the GBS-Venus reporter fluorescence, which is driven by GLI1 binding sites from the Foxa2 promoter and responsive to the highest levels of Hh signaling, was not increased in Gpr161-/- NPCs compared to wild-type NPCs (Fig. 5B). However, analogous to Gpr161-/- NIH/3T3 cells, Gpr161-/- NPCs were 3- to 4-fold more sensitive to SHH (EC50=2.9 nM) compared to wild-type cells (EC50=11.3 nM) (Fig. 5B).
To clearly understand the effects of GPR161 loss on Hh-driven neural progenitor specification, we simultaneously measured mRNA abundances for a panel of five markers known to be driven by various levels of Hh signaling in NPC cultures treated with increasing doses of SHH, allowing us to construct detailed SHH dose-response curves for each of the markers in both wild-type and Gpr161-/- NPCs (Fig. 5C). The loss of GPR161 led to the SHH-independent expression of Nkx6.1 and Olig2, progenitor subtype markers repressed by GLI3R, which in turn is inhibited by low-level Hh signals (Fig. 5C). In contrast, Nkx2.2 and Foxa2, markers that depend on high-level Hh signals and GLI2 activator function (GLI2A), were not induced in untreated Gpr161-/- NPCs (Fig. 5C). Instead, reminiscent of the effect of GPR161 loss in NIH/3T3 cells, the SHH dose-response curves for the induction of NKX2.2 and FOXA2 were shifted to the left (Fig. 5C). Thus, the loss of GPR161 sensitized cells to Hh ligands, consistent with the ventralized patterning observed in the neural tube in Gpr161-/- embryos (8).
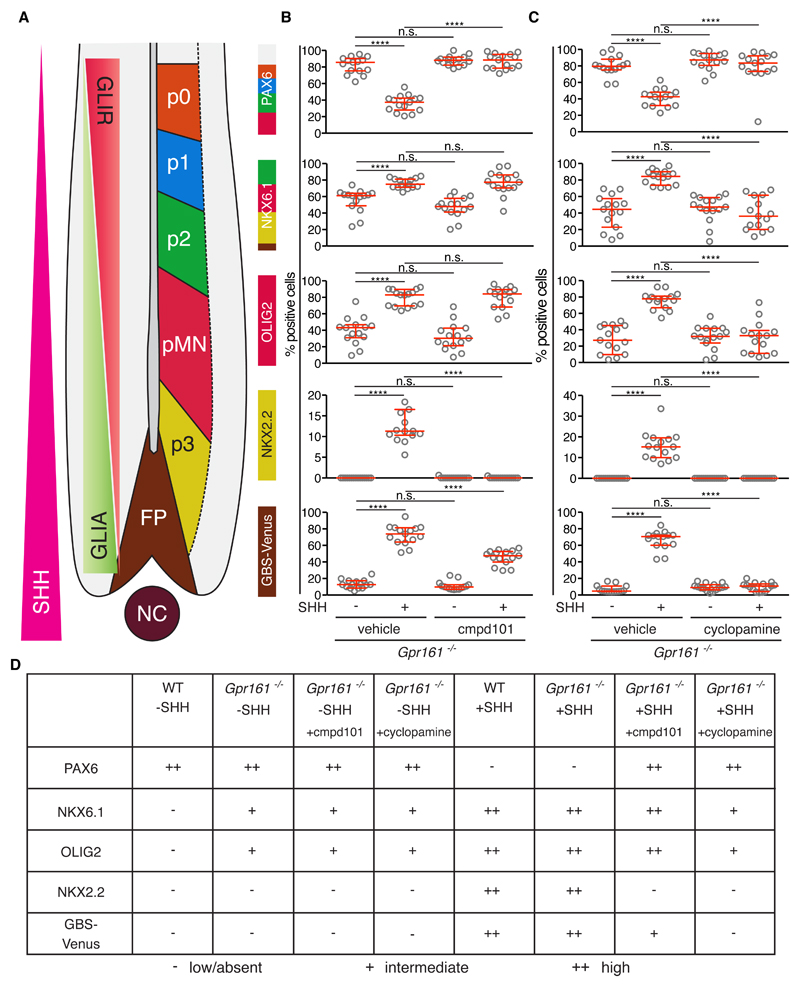
Is the activity of either GRK2/3 or SMO required for Hh signaling in the absence of GPR161 in NPCs, like it is in NIH/3T3 cells? GPR161 is considered epistatic to both SMO and GRK2, predicting that neither should be required for signaling in Gpr161-/- cells (8, 9). In agreement with this view, the SHH-independent, constitutive signaling activity seen in Gpr161-/- NPCs was largely unaffected by cmpd101 and cyclopamine. In the absence of SHH, the increase in NKX6.1- and OLIG2-positive NPCs could not be prevented by pharmacologically inhibiting either SMO or GRK2 (Fig. 6, A to D and fig. S6, A and B). This is consistent with the hypothesis that differentiation into NKX6.1- or OLIG2-positive progenitor subtypes was likely to be driven by the loss of GLI3R in Gpr161-/- NPCs (Fig. 5A). The increased expression of both Nkx6.1 and Olig2 observed in response to SHH in Gpr161-/- cells could be prevented by SMO inhibition with cyclopamine, but not by cmpd101, perhaps because GRK2/3 activity could not be completely inhibited by cmpd101 or because it was not required under these conditions.
Fig. 6. GRK2 and SMO are required for high-level Hh responses in Gpr161-/- NPCs.
(A) A schematic of the progenitor domains, with reciprocal GLIR and GLIA domains, as previously shown in Fig. 5C. (B and C) The percentage of cells containing progenitor subtype markers (NKX6.1, OLIG2, NKX2.2, and GBS-Venus) with or without SHH exposure in Gpr161-/- NPCs assessed at 48 h after inhibition of GRK2/3 with cmpd101 (B) or inhibition of SMO with cyclopamine (C). Each data point represents the data from one image of a NPC colony (see fig. S6A) consisting of 100-200 cells, and each condition is represented by 15 different colonies. The experiment was repeated twice. Medians with interquartile ranges are shown with statistical significance determined by the Mann-Whitney non-parametric ANOVA test and depicted as follows: P<0.0001 (****) and and P>0.05 (ns, not significant). (D) Summary of the effect of GRK2 and SMO inhibitors on the differentiation of wild-type (WT) and Gpr161-/- NPCs.
In striking contrast, both cmpd101 and cyclopamine inhibited SHH-induced, high-level Hh signaling in Gpr161-/- NPCs, resulting in a marked decrease in the specification of NKX2.2- and GBS-Venus–positive cells (Fig. 6, A to D and fig. S6, A and B). Thus, GRK2/3 and SMO must play roles independent of GPR161 in transducing high-level Hh signals, which are presumably mediated predominantly by GLI2A. Taken together, our analysis in cultured NPCs resolves a paradoxical observation in the developing neural tube of mouse embryos: the induction of NKX6.1- and OLIG2-positive progenitors in Gpr161-/- was independent on SMO activity, but the induction of FOXA2 (requiring high level of Hh signaling) remained dependent on SMO activity (8).
In summary, GPR161 and GRK2 appear to be regulating low- and high-level Hh responses in NPCs through apparently distinct mechanisms. GPR161 suppresses low-level Hh responses but only attenuates high-level Hh responses. Although GRK2 activity is essential for all Hh responses, it functions through GPR161 only to regulate low-level (but not high-level) Hh signaling. Thus, low- and high-level responses to the SHH morphogen may be mediated by distinct molecular pathways.
GRK2 and GRK3 are required for Hh signaling through a GαS-dependent pathway
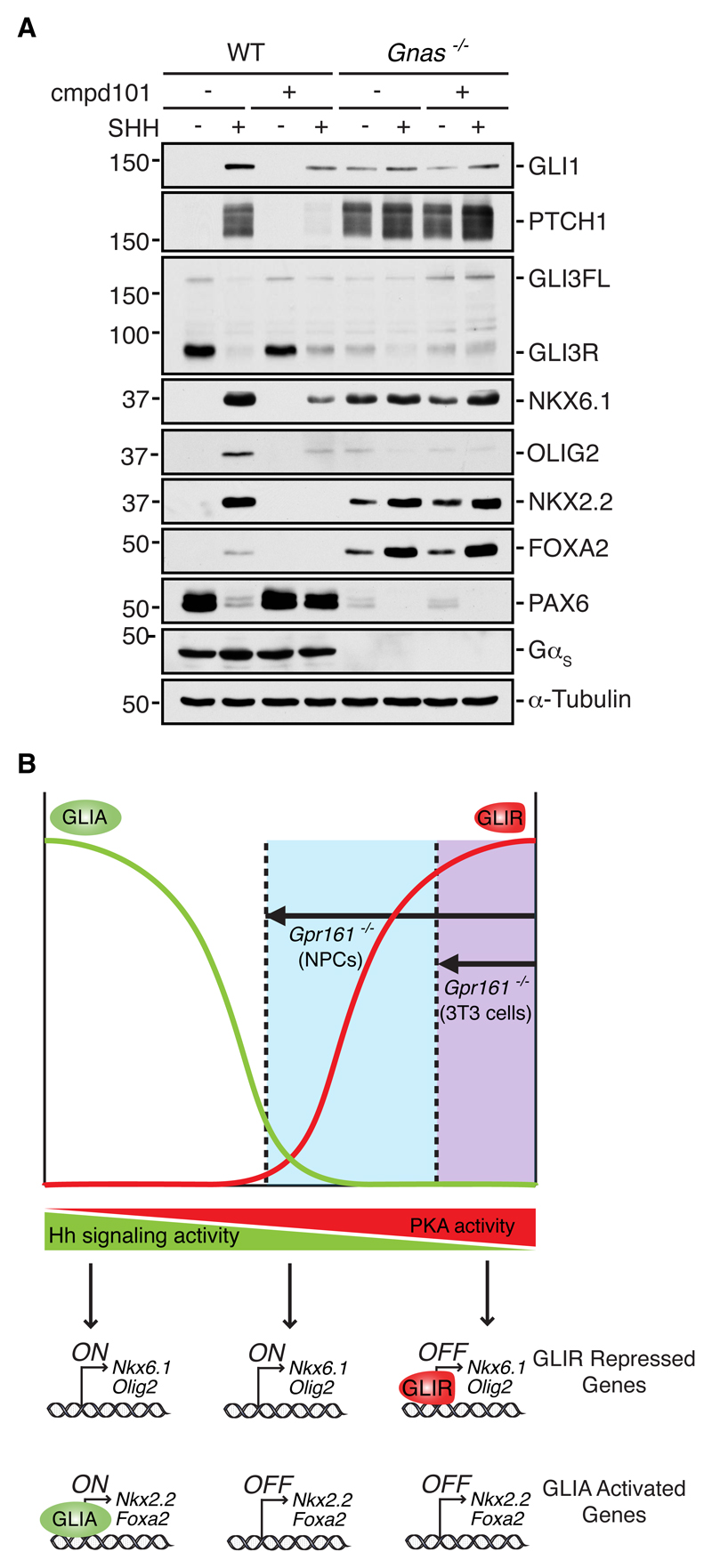
Our work in NIH/3T3 cells demonstrated that GRK2 was dispensable for Hh signaling in the absence of GαS (Fig. 2D) and also provided evidence that an alternative GPCR (other than GPR161) may mediate the effect of GRK2 on Hh signaling (Fig. 1A). Thus, high-level, GRK2-mediated responses in NPCs, which do not depend on GPR161, may also be regulated by an alternative GαS-coupled GPCR. Elimination of GαS from NPCs increased the abundance of both NKX2.2 and FOXA2, which require the highest levels of Hh signaling (Fig. 7A and fig. S7) and were not induced by the loss of Gpr161 (Fig. 5, A and B). Cmpd101 did not inhibit Hh signaling in cells lacking GαS (Fig. 7A). Therefore, GRK2 appears to function upstream of GαS in NPCs, as it does in NIH/3T3 cells, raising the possibility that Hh signaling can be regulated by GαS-coupled GPCRs other than GPR161. The higher activation of Hh signaling observed in both mouse embryos and cultured cells depleted of GαS also suggests the involvement of alternative GαS-coupled GPCRs.
Fig. 7. GαS negatively regulates all levels of Hh signaling in NPCs.
(A) Immunoblot (representative of three independent experiments) showing the effect of GRK2 and GRK3 inhibition (by cmpd101) on SHH-induced responses in wild-type (WT) and Gnas-/- NPCs. FOXA2 abundance is driven by the highest level of Hh signaling during neural tube development and hence can be considered a marker of maximal Hh signaling in this system. (B) A model for the different consequences of GPR161 loss in NIH/3T3 cells and NPCs. Decreasing PKA activity drives increasing levels of signaling, first by preventing GLIR formation and subsequently by allowing formation of GLIA. We speculate that the decrease in PKA activity (arrows) produced by the loss of GPR161 is greater in NPCs compared to NIH/3T3 cells (upper panel). In NPCs only, this decrease in PKA activity drops below the threshold required for the biogenesis of GLI3R and consequently allows for the adoption of cell fates (lower panel; NKX6.1- and OLIG2-positive cells) repressed by GLI3R in the basal state. In both NPCs and NIH/3T3 cells, the lowered PKA activity sensitizes cells to SHH-induced responses, shifting the SHH dose-response curve to the left.
Discussion
We used genetic and pharmacological approaches in two Hh-responsive cell culture systems to dissect the epistatic relationships between GRK2, GPR161, and GαS in Hh signaling. Our results highlight the importance of distinguishing between negative regulators and attenuators of signaling pathways. Negative regulators, such as SUFU, PTCH1, and GαS, keep the pathway off in the absence of ligand. Their loss leads to constitutive activation of the pathway in the absence of ligand, and they must be antagonized to allow signal propagation to the nucleus. In contrast, attenuators are not essential to signal transduction, but may function as components of control systems (such as feedback loops) that dampen signaling by limiting ligand potency or efficacy. Our results show that GPR161 functions as an attenuator of Hh signaling in NIH/3T3 cells and of high-level Hh signals in NPCs. In these contexts, the loss of GPR161 does not lead to spontaneous activation of the pathway, but instead increases the sensitivity to SHH, with signaling remaining dependent on SMO. However, consistent with previous reports, GPR161 functions as a bona fide negative regulator of lower-level Hh responses in NPCs. On the other hand, GRK2 and GαS were clearly positive and negative regulators, respectively, of signaling across the full range of Hh responses in both fibroblasts and NPCs, suggesting both GPR161-dependent and -independent roles for these regulators. Because SHH is a morphogen, the shape of the SHH dose-response curve is predicted to have an effect on tissue patterning. Genes like Gpr161 may regulate patterning in a unique way, functioning by shaping the sensitivity to morphogens rather than by acting as simple positive or negative regulators of signal propagation.
How could GPR161 have distinct effects on Hh signaling in NPCs compared to NIH/3T3 cells? We favor a model that builds on the previous observation that Hh responsiveness is exquisitely sensitive to the amount of cellular PKA activity (5). Increasing PKA activity leads to a progressive loss in cellular responsiveness to SHH (Fig. 7B). PKA directly phosphorylates and regulates the transcriptional activity of the bifunctional GLI proteins, which can exist either as transcriptional repressors (GLIR) or activators (GLIA). PKA blocks GLI transcriptional activity in a graded manner using a multi-site phosphorylation code: phosphorylation at a group of four residues promotes the proteolytic conversion of GLIs into GLIRs; however, the phosphorylation of an additional set of two residues is required to fully block GLIA (6). SHH can independently prevent GLIR formation and promote GLIA formation by controlling this phosphorylation code.
Given that cellular PKA activity is a central determinant of signaling strength, we propose that the balance between the activity of GαS-coupled GPCRs (which would raise cAMP levels and hence PKA activity) and the activity of Gαi-coupled GPCRs (which would reduce cAMP levels and hence PKA activity) may set the sensitivity of target cells to Hh ligands in a tissue- or cell-type specific manner. Thus, the different effects of GPR161 on Hh signaling in NIH/3T3 cells and NPCs may be explained by cell-type–specific effects on PKA activity (Fig. 7B). In NPCs (but not in NIH/3T3 cells) the decline in PKA activity when GPR161 is eliminated drops below the threshold for GLI3R production, leading to activation of differentiation programs that depend on low- and medium-level Hh responses (Fig. 7B). However, in both NPCs and NIH/3T3 cells, reduced PKA activity caused by GPR161 loss results in increased sensitivity to Hh ligands, manifested as a leftward shift of the SHH dose-response curve.
This model can also explain how the loss of GPR161 in NPCs can have apparently distinct effects on high- and low-level Hh responses. The spontaneous differentiation phenotype of Gpr161-/- NPCs is likely to be explained by the loss of GLI3R in these cells in the absence of any Hh ligands (8) (Fig. 7B). The production of NKX6.1 and OLIG2, the transcription factors found in NPCs lacking GPR161, is known to be repressed by GLI3R in the neural tube (25, 39, 40). However, the loss of GPR161 may not be sufficient to fully activate GLI2A, which drives the production of NKX2.2 and FOXA2 in response to high-level Hh signals in the ventral-most domains of the neural tube (Fig. 7B). These results are consistent with the observation that neural tube patterning defects in mouse embryos lacking GPR161 are more severe than those lacking GLI3 but less severe than those lacking other negative regulators like PKA and GαS (18, 19, 21, 22).
In comparison to GPR161, GRK2 regulates the production of transcription factors that depend on both GLI3R and GLI2A for their expression. The effect of GRK2 on GLI3R-regulated genes depends on GPR161 because GRK2 activity is dispensable for the production of NKX6.1 and OLIG2 in Gpr161-/- NPCs (Fig. 6, A and B). However, even in Gpr161-/- NPCs, GRK2 is still necessary for SHH to induce transcription factors that depend on GLI2A (Fig. 6, B and D and fig. S6A), demonstrating a GPR161-independent role for GRK2 in regulating high-level Hh responses. Although the mechanism by which GRK2/3 regulate GPR161-independent signaling remains to be determined, it may involve either (1) the regulation of an alternate GαS-coupled GPCR or (2) regulation of high-level signaling by SMO itself, which does not couple to GαS.
A key unresolved question is whether GαS-coupled GPCRs, including GPR161, are directly regulated by Hh ligands or whether they simply set the basal PKA activity level in cells that then modulates signaling strength or sensitivity. It has been challenging to observe changes in PKA enzymatic activity or cAMP levels in response to Hh ligands, though it is possible that such changes are restricted to a microdomain or to the cilium (41, 42). A direct regulatory role has been suggested based on the observation that GPR161 is cleared from the ciliary membrane in response to SHH or SMO agonists (8, 9). However, our observations in NIH/3T3 cells show that ciliary trafficking changes, even when correlated with signal activation, may not play a functional role. Other components that could be regulated by SMO include Gαi proteins, which reduce PKA activity and can be directly activated by SMO (43–47), GRK2, or the GLI-SUFU complex. NPCs lacking Gαs, despite displaying very high levels of Hh target gene transcription, remain responsive to SHH, suggesting that SMO regulates a process that is at least partially independent of GαS and hence of the GPCRs that couple to GαS (Fig. 7A).
We conclude with a few comments about the therapeutic relevance of our findings. SMO antagonists are FDA-approved drugs used in advanced basal cell cancers. As with many other protein-targeted therapies, resistance has been observed in the clinic, predominantly caused by mutations in SMO itself that prevent drug binding or drug activity (31, 48, 49). Thus, Hh inhibitors that act downstream of SMO are of interest (50). GRK2/3 inhibitors, which have been explored largely for their potential utility in heart failure, should be effective in blocking signaling mediated by drug-resistant mutants of SMO (fig. S3G).
Materials and Methods
Cell Culture
Flp-In-3T3 (a sub-line of NIH/3T3 cells) and 293FT cell lines were purchased from Life Technologies. The Med1-MB cell line was a kind gift from Dr. Matthew P. Scott, Stanford University. Stable cell lines expressing SMO mutants in the background of Smo-/- mouse embryonic fibroblasts (MEFs) were previously described (51). Stable Flp-In-3T3 cell lines expressing tagged GRK2 (C-terminal GFP- or 1D4 tag), PTCH1 (C-terminal APEX2-1D4 tag, GαS (internal GFP tag), GPR161 (C-terminal YFP-FLAG tag, and SUFU (C-terminal APEX-1D4 tag) were generated as previously described (52). All the above mentioned cell lines were cultured in Dulbecco's Modified Eagle Medium (DMEM) containing high glucose (Thermo Scientific) and supplemented with 10% fetal bovine serum (FBS) (Atlanta Biologicals), 1 mM sodium pyruvate (Gibco), 2 mM L-Glutamine (Gemini Biosciences), 1x MEM non-essential amino acids solution (Gibco), penicillin (40 U/ml), and streptomycin (40 µg/ml) (Gemini Biosciences) in a humidified atmosphere containing 5% CO2 at 37 °C. To induce ciliation, cells were grown to confluence in DMEM containing 10% FBS and then switched to medium containing 0.5% FBS for 24 h. Treatment with Hh pathway agonists (25 nM SHH and 200 nM SAG), antagonists (200 nM SANT-1, 5 µM cyclopamine, and 1 µM vismodegib), and GRK2/3 inhibitor (25 µM cmpd101) was done for either 4 h following serum starvation (immunofluorescence staining) or included during the 24 h serum starvation step [qRT-PCR (except for Fig. S1A and S3H, in which treatment was 4 h), Western Blotting, and reporter assays].
Neural progenitor differentiation
A stable GBS-Venus reporter line (DVI2) was generated using HM1 mESCs. The 8xGBS-H2B::Venus Shh reporter was cloned into the HPRT targeting vector, pSKB1 (53). The HPRT-8xGBS-H2B::Venus construct was integrated into the HPRT locus of HM1 cells using nucleofection. Positive clones were selected for 8 days using hypoxanthine aminopterin thymidine (HAT medium) (Sigma-Aldrich). Single colonies were expanded on feeder layers in ES cell medium with leukemia inhibitory factor (LIF) and characterized by PCR. To prepare the dishes for mESC maintenance, feeders were plated onto dishes coated with 0.1% gelatin (Sigma-Aldrich) overnight. The mESCs were cultured with feeders in mESC media (DMEM containing high glucose, 15% Optima FBS (Atlanta Biologicals), 1x MEM non-essential amino acids, 1% penicillin/streptomycin, 2 mM L-glutamine, 1% EmbryoMax nucleosides (Millipore), 55 µM 2-mercaptoethanol (Gibco), and 1000 U/ml ESGRO LIF (Millipore)). The mESCs were differentiated into spinal neural progenitors using a previously described protocol with minor modifications (54). Briefly, for this differentiation, the feeders were first removed from the mESCs. This was achieved by lifting the cells off the maintenance plates with 0.25% Trypsin/EDTA and incubating the cells on 10cm tissue culture plates for two short successive periods (20 min each). To induce neural differentiation, the cells were plated onto either gelatin-coated glass coverslips (12mm diameter, placed in a 24-well plate) at a density of 24x103 cells/coverslip or onto gelatin-coated CellBIND plates (Corning) at a density of 60x103 cells/6-well or 100 x103 cells/10 cm plate. For the differentiation, cells were plated in N2B27 media (Dulbecco’s Modified Eagle’s Medium F12 (Gibco) and Neurobasal Medium (Gibco) (1:1 ratio) supplemented with N-2 Supplement (Gibco), B-27 Supplement (Gibco), 1% penicillin/streptomycin (Gemini Bio-Products), 2mM L-glutamine (Gemini Bio-Products), 40 µg/ml Bovine Serum Albumin (Sigma), and 55 µM 2-mercaptoethanol (Gibco) with varying components. On Day 0 (plating day) and Day 1, cells were cultured in N2B27 with 10 ng/ml bFGF (R&D). On Day 2, the cells were cultured in N2B27 with 10 ng/ml bFGF (R&D) and 5 µM CHIR99021 (Axon). On Day 3, the cells were cultured in N2B27 containing the following components: retinoic acid (RA, 100 nM, Sigma-Aldrich), RA+SHH (25 nM), RA+cmpd101 (5 µM), RA+SHH+cmpd101, RA+cyclopamine (5 µM), RA+SHH+cyclopamine, RA+SANT-1 (200 nM), or RA+SHH+SANT-1. On Day 4, the cell culture media was replenished with fresh media containing the same components. On Day 5 the cells were rinsed with PBS and either fixed with 4% PFA for further analysis using immunohistochemistry or the protein was extracted for Western blot analysis.
Constructs
Mouse Grk2 and Grk3 were tagged with a C-terminal GFP or 1D4 and cloned into the pEF5/FRT/V5-DEST vector (Life Technologies). Grk2 mutants were generated by site-directed mutagenesis. Mouse Gpr161 was tagged with a dual YFP-FLAG tag and cloned into the pEF5/FRT/V5-DEST vector. Mouse Ptch1 and Sufu were tagged with a dual APEX2-1D4 tag and cloned into the pEF5/FRT/V5-DEST vector. Rat Gnas with an internal GFP tag (55) was cloned into the pEF5/FRT/V5-DEST vector.
Reagents and Antibodies
Recombinant SHH was generated as previously described (56). The following chemicals were used: SAG (Enzo Life Sciences), SANT-1 (EMD Millipore), cyclopamine (LC Labs), vismodegib (LC Labs), cmpd101 (HelloBio), and puromycin (Sigma-Aldrich). The following primary antibodies were used: mouse anti-GRK2 (sc-13143; Santa Cruz; 1:500), mouse anti-GLI1 (2643; Cell Signaling; 1:500), goat anti-GLI3 (AF3690; R&D; 1:200), rabbit anti-p38 (ab7952; Abcam; 1:2000), mouse anti-GαS (ac-135914; Santa Cruz; 1:200), goat anti-GFP (600-101-215; Rockland; 1:500), rabbit anti-GFP (NB600-308; Novus Biologicals; 1:5000), mouse anti-1D4 (The University of British Columbia; 1:5000), mouse anti-α-Tubulin (T6199; Sigma-Aldrich; 1:10000), mouse anti-acetylated-Tubulin (T6793; Sigma-Aldrich; 1:10000), mouse anti-NKX2.2 (74.5A5, Developmental Studies Hybridoma Bank; 1:100), mouse anti-NKX6.1 (F55A10, Developmental Studies Hybridoma Bank; 1:100), rabbit anti-PAX6 (AB2237; EMD Millipore; 1:1000), and mouse anti-FOXA2 (4C7; Developmental Studies Hybridoma Bank; 1:100). Rabbit polyclonal antibodies recognizing PTCH1, SMO, and SUFU have been described previously (5, 57). Rabbit anti-GPR161 (1:150) and guinea pig anti-OLIG2 (1:20000) antibodies have been described previously (8, 58). Secondary antibodies conjugated to horseradish peroxidase or Alexa Fluor dyes were obtained from Jackson Laboratories and Thermo Fisher Scientific, respectively. All other reagents and chemicals were of the highest quality available.
CRISPR/Cas9-mediated gene editing in 3T3 and NPCs
Flp-In-3T3 Grk2-/-, Ptch1-/-, Gnas-/-, Sufu-/-, Gpr161-/-, Ptch1-/- Grk2-/-, Gnas-/- Grk2-/-, and Sufu-/- Grk2-/- cells were generated by CRISPR-Cas9 gene editing technology as previously described (59). Briefly, guide RNA sequences targeting Grk2 (5’-ctggaacacgtcccctcgg-3’), Gnas (5’-gaccgaggaccagcgcaacg-3’) and Gpr161 (5’- ggtgactagctccatccgga-3’; pool 1, 5’-gtctgccgcccctatttggt-3’; pool 2, 5’-gatgaccaaccaaatagggg-3’; pool 3, 5’-ctacggtttcatcttccggg-3’; pool 4) were cloned into lentiCRISPR v2 plasmid (Addgene#52961). Lentivirus was generated in 293FT cells by transfecting 3,000,000 cells with 8 µg lentiCRISPR v2 plasmid, 4 µg pCMV-VSV-G (60) plasmid (Addgene#8454), 4 µg psPAX2 packaging plasmid (Addgene#12260), and 48 µl of 1 mg/ml polyethylenimine (Polysciences). The culture medium was changed after 14 h post transfection and the lentivirus was collected after 48 h by a brief spin at 400xg followed by a filtration through 0.45 µm low-protein binding membrane (Pall Corporation). Flp-In-3T3 cells were transduced with the lentivirus and selected with puromycin containing DMEM (2 µg/ml) for ten days. Single cells were sorted into a 96-well plate using FACSAria II (BD Biosciences). Multiple single cell–derived clones (except for Gpr161, for which pooled knockout cell lines were used) were analyzed by Western blotting to obtain clones with complete depletion of the gene product. To generate Ptch1-/- and Sufu-/- cells, guide RNA sequences targeting Ptch1 (5’-agctaatctcgagaccaacg-3’) and Sufu (5’-gcggcgacactctccgtaga-3’) were cloned into pX330-U6-Chimeric_BB-CBh-hSpCas9 (PX330; Addgene#42230). Flp-In-3T3 cells were transiently transfected with the CRISPR plasmid along with a control GFP plasmid using X-tremeGENE 9 (Roche), and GFP positive single cells were sorted into a 96-well plate using FACSAria II. Single cell derived null clones were obtained by western blotting. Knockout of Grk2 in the background of Ptch1-/-, Gnas-/-, and Sufu-/- was achieved using the lentiCRISPR v2 plasmid expressing the guide RNA targeting Grk2 with the exception of Gnas-/- cells in which a blasticidin resisitance cassette containing lentiCRISPR v2 plasmid was used (kindly provided by Dr. Henry Ho, UC Davis). Gpr161-/-, Grk2-/-, Grk2-/- Grk3-/-, and Gnas-/- NPCs were generated using CRISPR-Cas9 gene editing technology. Briefly, guide RNA sequences targeting Gpr161 (5’-ggtgactagctccatccgga-3’), Grk2 (5’-ctggaacacgtcccctcgg-3’), Grk3 (dual guides - 5’-attctgtcagtggaagaacg-3’ and 5’- gctgtctctcgttagcactg-3’), and Gnas (5’-cgttaaacccattaacatgc-3’ for clone C1; dual guides 5-gttgtcccgccccaactatc-3' and 5'-gtggtgtagcgagcgaactc-3' for clones C3, C6 and D5) were cloned into the plasmid PX459 (Addgene #48139). These guide constructs were electroporated into GBS-Venus mESCs using the Lonza nucleofection system (Nucleofector 2b Device using the program A-023 and Lonza Cell Nucleofector Kit #VAPH-1001). mESCs were cultured under feeder-free conditions in 2i media (Dulbecco’s Modified Eagle’s Medium F12 (Gibco) and Neurobasal Medium (Gibco) (1:1 ratio) supplemented with N-2 Supplement (Gibco), B-27 Supplement (Gibco), 1% penicillin/streptomycin (Gemini Bio-Products), 2mM L-glutamine (Gemini Bio-Products), 40 µg/ml Bovine Serum Albumin (Sigma), 55 µM 2-mercaptoethanol (Gibco), 5 µM CHIR99021 (Axon), 1 µM PD 98059 (Axon), and 1000 U/ml ESGRO LIF (Millipore)). Antibiotic selection was performed 24 h after nucleofection, in 2i media containing 1.5 µg/ml puromycin for 48 h. Approximately 1 week after selection, individual mESC colonies were manually picked, expanded, and the genomic DNA was collected using QuickExtract DNA Extraction Solution (Epicentre). The region surrounding the guide target was PCR amplified and sequenced to determine if non-homologous end joining resulted in a nonsense or frameshift mutation.
Real-Time Quantitative Reverse Transcription PCR (Real-Time qRT-PCR)
Real-Time qRT-PCR in Flp-In 3T3 cells was performed using the Power SYBR Green Cells-to-CT Kit (Thermo Fisher Scientific) on a QuantStudio 5 Real-Time PCR System (Applied Biosystems) with custom designed primers for Gli1 (forward primer: 5’-ccaagccaactttatgtcaggg-3’ and reverse primer: 5’-agcccgcttctttgttaatttga-3’), Grk2 (forward primer: 5’-cgatacttctacttgttccc-3’ and reverse primer: 5’-tctggatcactatcacactg-3’), Grk3 (forward primer: 5’-ctggacaacgaagaggatagg-3’ and reverse primer: 5’-tgtagccgtcacctgtttc-3’), Gpr161 (forward primer: 5’-ctcacgcttggggtcattg-3’ and reverse primer: 5’-gagccagatgtagacgagagc-3’), and Gapdh (forward primer: 5’-agtggcaaagtggagatt-3’ and reverse primer: 5’-gtggagtcatactggaaca-3’). For NPCs, RNA extraction and cDNA synthesis was done using TRIzol reagent (Life Technologies) and iScript Reverse Transcription Supermix (Biorad) following the manufacturer’s instructions. The following primers were used for Real-Time qRT-PCR: Nkx6.1 (forward primer: 5’-cccggagtgatgcagagt-3’ and reverse primer: 5’-gaacgtgggtctggtgtgtt-3’), Olig2 (forward primer: 5’-agaccgagccaacaccag-3’ and reverse primer: 5’-aagctctcgaatgatccttcttt-3’), Nkx2.2 (forward primer: 5’-cagcctcatccgtctcac-3’ and 5’-tcacctccatacctttctcc-3’), Foxa2 (forward primer: 5’-ggagtgtactccaggcctatta-3’ and 5’-ctccactcagcctctcatttc-3’), and Pax6 (forward primer: 5’-acccggcagaagatcgtag-3’ and 5’-tttgcatctgcatgggtct-3’). Transcript levels relative to gapdh were calculated using the ΔCt method.
Western Blotting
Whole cell extracts from Flp-In-3T3 and NPCs were prepared in RIPA lysis buffer (50 mM Tris-HCl pH-8.0, 150 mM NaCl, 2% NP-40, 0.25% Deoxycholate, 0.1% SDS, 1 mM NaF, 1mM DTT, 10% glycerol, 1x SIGMAFAST protease inhibitor cocktail (Sigma-Aldrich), and 1x PhosSTOP (Roche)). Samples were resuspended in Laemmli buffer, incubated for 30 min at room temperature and subjected to SDS-PAGE. The resolved proteins were transferred onto a nitrocellulose membrane (Bio-Rad) using a wet electroblotting system (Bio-Rad) followed by immunoblotting.
Hh reporter assay
Hh reporter assay was performed as described previously (17). Briefly, wild-type or Grk2-/- NIH/3T3 cells were seeded in 96-well plates and transfected after 24 h using X-tremeGENE 9 (Roche) following manufacturer’s instructions. The transfection mix consisted of 32 ng firefly luciferase reporter driven by an 8xGli-responsive promoter, 8 ng of a Renilla luciferase reporter driven by a constitutive TK promoter (Promega) and 1 ng of vector control or GRK2/GRK3 construct or 10 ng of GPR161. 48 h post transfection, confluent cells were serum starved for 24 h in the presence of SHH (25 nM) or SAG (100 nM). Reporter activity was measured using the Dual-Luciferase Reporter kit (Promega) and read on a Synergy H1 Hybrid Multi-Mode Microplate Reader (BioTek). The GLI luciferase to Renilla luciferase ratio is reported as “Hh reporter activity”.
Immunofluorescence staining and Image quantifications
Immunofluorescence staining of YFP-tagged GPR161 and endogenous SMO in Flp-In-3T3 cells was performed as described previously (52). Ciliary GPR161 and SMO quantifications were done as previously described (52). For immunofluorescence staining of NPCs, cells were rinsed once with PBS and fixed in 4% PFA for 10 min at room temperature. After blocking for 30 min at room temperature in antibody blocking solution (1% Horse serum and 0.1% Triton X-100 in PBS), primary antibodies were applied overnight at 4 °C. Coverslips were rinsed with PBS-T (0.1% Triton X-100 in PBS), secondary antibodies were applied at room temperature for 1 hour, rinsed with PBS-T, and then mounted in ProLong Diamond Antifade Mountant (Life Technologies). Fluorescent images were collected on a Leica TCS SP5 confocal imaging system equipped with a 40x oil immersion objective and captured using the Leica Application Suite X (LASX) software. In each experiment, coverslips from each condition were grown, collected, and processed together to ensure uniformity across fixation and staining times. While collecting images, the gain, offset, and laser power settings on the microscope were held constant for each antibody. 15 images were taken per condition. To ensure full representation, z-stacks were acquired and counts were performed on compressed images. Cell counts were collected using the NIH ImageJ software suite with Cell Counter plugin. In total, over 1500 cells were analyzed per condition and each experiment was independently conducted at least twice. Representative images were processed equally using Adobe Photoshop and Adobe Illustrator.
Quantification and statistical analysis
In all data panels, representative data from two-four independent experiments is shown. The statistical significance of differences between the Gli1 mRNA levels or Hh reporter activity between two samples was determined by an unpaired Welch’s t-test, with a post Bonferroni correction applied to correct for multiple comparisions (grouped by graph). Statistical analysis for differences in ciliary fluorescence intensity (NIH/3T3 cells) or neural differentiation marker positivity (in NPCs) between two groups was evaluated using the Mann-Whitney non-parametric ANOVA test, with a Bonferroni correction applied to correct for multiple comparisions (grouped by graph). The statistical tests were performed in consultation with an expert statistician in the department of Biomedical Data Science in Stanford University School of Medicine.
Supplementary Material
Acknowledgements
We are grateful to Dr. Swetha Mohan, Department of Neurology, UC San Francisco, for help with the generation of Gnas-/- NIH/3T3 cell lines. We thank Dr. Andres Lebensohn, Dr. Broder Schmidt, and Dr. Ramin Dubey for comments on the manuscript. We thank Dr. Alex McMillan, PhD (Department of Biomedical Data Science, Stanford University School of Medicine) for expert advice on statistical analysis.
Funding: GVP was supported by a postdoctoral fellowship from the American Heart Association; JHK by the Stanford School of Medicine Dean’s postdoctoral fellowship and Jump Start Award for Excellence in Research; MG by the MaxDelbrück Center for Molecular Medicine in the Helmholtz Association; AS by the HFSP LTF (LT000401/2014-L) and the People Programme (Marie Curie Actions) of the European Union's Seventh Framework Programme FP7-2013 under REA grant agreement n° 624973. Work in RRs lab was supported by the National Institutes of Health grants (GM105448, GM112988, GM118082) to RR. Work in JB’s lab was supported by the BBSRC (grant reference BB/J015539/1) and the Francis Crick Institute, which receives its funding from Cancer Research UK (FC001051), the UK Medical Research Council (FC001051), and the Wellcome Trust (FC001051; WT098326MA). Work in PWI’s lab was supported by the Toh Kian Chui foundation.
Footnotes
Author Contributions: Conceptualization, GVP, JHK, JB, PWI and RR; Methodology, GVP, JHK, BBP, AS, JB, and RR; Validation, GVP and JHK; Formal Analysis, GVP; Investigation, GVP and JHK; Resources, BBP, AS, MG, RS, GL, and PWI; Writing – Original Draft, GVP, JHK, and RR; Writing – Review & Editing, GVP, JHK, PWI, AS, MG, JB, and RR; Visualization, GVP, and JHK; Supervision, JB and RR; Project Administration, GVP; Funding Acquisition, JB and RR.
Competing Interests: PWI owns a small amount of stock shares in Curis, Inc., a biotechnology company with interests in pharmacologically targeting Hedgehog signaling.
References
- 1.Dessaud E, Yang LL, Hill K, Cox B, Ulloa F, Ribeiro A, Mynett A, Novitch BG, Briscoe J. Interpretation of the sonic hedgehog morphogen gradient by a temporal adaptation mechanism. Nature. 2007;450:717–720. doi: 10.1038/nature06347. [DOI] [PubMed] [Google Scholar]
- 2.Wang B, Fallon JF, Beachy PA. Hedgehog-regulated processing of Gli3 produces an anterior/posterior repressor gradient in the developing vertebrate limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 3.Corbit KC, Aanstad P, Singla V, Norman AR, Stainier DYR, Reiter JF. Vertebrate Smoothened functions at the primary cilium. Nature. 2005;437:1018–1021. doi: 10.1038/nature04117. [DOI] [PubMed] [Google Scholar]
- 4.Hui C-C, Angers S. Gli Proteins in Development and Disease. Annu Rev Cell Dev Biol. 2011;27:513–537. doi: 10.1146/annurev-cellbio-092910-154048. [DOI] [PubMed] [Google Scholar]
- 5.Humke EW, Dorn KV, Milenkovic L, Scott MP, Rohatgi R. The output of Hedgehog signaling is controlled by the dynamic association between Suppressor of Fused and the Gli proteins. Genes Dev. 2010;24:670–682. doi: 10.1101/gad.1902910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Niewiadomski P, Kong JH, Ahrends R, Ma Y, Humke EW, Khan S, Teruel MN, Novitch BG, Rohatgi R. Gli Protein Activity Is Controlled by Multisite Phosphorylation in Vertebrate Hedgehog Signaling. Cell Reports. 2014;6:168–181. doi: 10.1016/j.celrep.2013.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: recruitment to cilia and dissociation of SuFu–Gli protein complexes. J Cell Biol. 2010;191:415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S, Wen X, Ratti N, Loktev A, Rangell L, Scales SJ, Jackson PK. The Ciliary G-Protein-Coupled Receptor Gpr161 Negatively Regulates the Sonic Hedgehog Pathway via cAMP Signaling. Cell. 2013;152:210–223. doi: 10.1016/j.cell.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 9.Pal K, Hwang S-H, Somatilaka B, Badgandi H, Jackson PK, DeFea K, Mukhopadhyay S. Smoothened determines β-arrestin–mediated removal of the G protein–coupled receptor Gpr161 from the primary cilium. J Cell Biol. 2016;212:861–875. doi: 10.1083/jcb.201506132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pitcher JA, Freedman NJ, Lefkowitz RJ. G Protein–Coupled Receptor Kinases. 2003;67:653–692. doi: 10.1146/annurev.biochem.67.1.653. [DOI] [PubMed] [Google Scholar]
- 11.Chen W, Ren X-R, Nelson CD, Barak LS, Chen JK, Beachy PA, de Sauvage F, Lefkowitz RJ. Activity-Dependent Internalization of Smoothened Mediated by ß-Arrestin 2 and GRK2. Science. 2004;306:2257–2260. doi: 10.1126/science.1104135. [DOI] [PubMed] [Google Scholar]
- 12.Meloni AR, Fralish GB, Kelly P, Salahpour A, Chen JK, Wechsler-Reya RJ, Lefkowitz RJ, Caron MG. Smoothened signal transduction is promoted by G protein-coupled receptor kinase 2. Molecular and Cellular Biology. 2006;26:7550–7560. doi: 10.1128/MCB.00546-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Philipp M, Fralish GB, Meloni AR, Chen W, MacInnes AW, Barak LS, Caron MG. Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol Biol Cell. 2008;19:5478–5489. doi: 10.1091/mbc.E08-05-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Li S, Tong C, Zhao Y, Wang B, Liu Y, Jia J, Jiang J. G protein-coupled receptor kinase 2 promotes high-level Hedgehog signaling by regulating the active state of Smo through kinase-dependent and kinase-independent mechanisms in Drosophila. Genes Dev. 2010;24:2054–2067. doi: 10.1101/gad.1948710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y, Sasai N, Ma G, Yue T, Jia J, Briscoe J, Jiang J. Sonic Hedgehog Dependent Phosphorylation by CK1α and GRK2 Is Required for Ciliary Accumulation and Activation of Smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Evron T, Daigle TL, Caron MG. GRK2: multiple roles beyond G protein-coupled receptor desensitization. Trends in Pharmacological Sciences. 2012;33:154–164. doi: 10.1016/j.tips.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhao Z, Lee RTH, Pusapati GV, Iyu A, Rohatgi R, Ingham PW. An essential role for Grk2 in Hedgehog signalling downstream of Smoothened. EMBO Rep. 2016;17:739–752. doi: 10.15252/embr.201541532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Regard JB, Malhotra D, Gvozdenovic-Jeremic J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS, Yang Y. Activation of Hedgehog signaling by loss of GNAS causes heterotopic ossification. Nat Med. 2013;19:1505–1512. doi: 10.1038/nm.3314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.He X, Zhang L, Chen Y, Remke M, Shih D, Lu F, Wang H, Deng Y, Yu Y, Xia Y, Wu X, et al. The G protein α subunit Gαs is a tumor suppressor in Sonic hedgehog–driven medulloblastoma. Nat Med. 2014;20:1035–1042. doi: 10.1038/nm.3666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iglesias-Bartolome R, Torres D, Marone R, Feng X, Martin D, Simaan M, Chen M, Weinstein LS, Taylor SS, Molinolo AA, Gutkind JS. Inactivation of a G[alpha]s-PKA tumour suppressor pathway in skin stem cells initiates basal-cell carcinogenesis. Nat Cell Biol. 2015;17:793–803. doi: 10.1038/ncb3164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hwang S-H, Mukhopadhyay S. G-protein-coupled receptors and localized signaling in the primary cilium during ventral neural tube patterning. Birth Defects Research Part A: Clinical and Molecular Teratology. 2015;103:12–19. doi: 10.1002/bdra.23267. [DOI] [PubMed] [Google Scholar]
- 22.Tuson M, He M, Anderson KV. Protein kinase A acts at the basal body of the primary cilium to prevent Gli2 activation and ventralization of the mouse neural tube. Development. 2011;138:4921–4930. doi: 10.1242/dev.070805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Persson M, Stamataki D, te Welscher P, Andersson E, Böse J, Rüther U, Ericson J, Briscoe J. Dorsal-ventral patterning of the spinal cord requires Gli3 transcriptional repressor activity. Genes Dev. 2002;16:2865–2878. doi: 10.1101/gad.243402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessell TM. Neuronal specification in the spinal cord: inductive signals and transcriptional codes. Nat Rev Genet. 2000;1:20–29. doi: 10.1038/35049541. [DOI] [PubMed] [Google Scholar]
- 25.Cohen M, Briscoe J, Blassberg R. Morphogen interpretation: the transcriptional logic of neural tube patterning. Current Opinion in Genetics & Development. 2013;23:423–428. doi: 10.1016/j.gde.2013.04.003. [DOI] [PubMed] [Google Scholar]
- 26.Thal DM, Yeow RY, Schoenau C, Huber J, Tesmer JJG. Molecular Mechanism of Selectivity among G Protein-Coupled Receptor Kinase 2 Inhibitors. Mol Pharmacol. 2011;80:294–303. doi: 10.1124/mol.111.071522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lowe JD, Sanderson HS, Cooke AE, Ostovar M, Tsisanova E, Withey SL, Chavkin C, Husbands SM, Kelly E, Henderson G, Bailey CP. Role of G Protein-Coupled Receptor Kinases 2 and 3 in -Opioid Receptor Desensitization and Internalization. Mol Pharmacol. 2015;88:347–356. doi: 10.1124/mol.115.098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Svärd J, Henricson KH, Persson-Lek M, Rozell B, Lauth M, Bergström Å, Ericson J, Toftgård R, Teglund S. Genetic Elimination of Suppressor of Fused Reveals an Essential Repressor Function in the Mammalian Hedgehog Signaling Pathway. Developmental Cell. 2006;10:187–197. doi: 10.1016/j.devcel.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 29.Gephart MGH, Su YS, Bandara S, Tsai F-C, Hong J, Conley N, Rayburn H, Milenkovic L, Meyer T, Scott MP. Neuropilin-2 contributes to tumorigenicity in a mouse model of Hedgehog pathway medulloblastoma. J Neurooncol. 2013;115:161–168. doi: 10.1007/s11060-013-1216-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dijkgraaf GJP, Alicke B, Weinmann L, Januario T, West K, Modrusan Z, Burdick D, Goldsmith R, Robarge K, Sutherlin D, Scales SJ, et al. Small Molecule Inhibition of GDC-0449 Refractory Smoothened Mutants and Downstream Mechanisms of Drug Resistance. Cancer Research. 2011;71:435–444. doi: 10.1158/0008-5472.CAN-10-2876. [DOI] [PubMed] [Google Scholar]
- 31.Yauch RL, Dijkgraaf GJP, Alicke B, Januario T, Ahn CP, Holcomb T, Pujara K, Stinson J, Callahan CA, Tang T, Bazan JF, et al. Smoothened Mutation Confers Resistance to a Hedgehog Pathway Inhibitor in Medulloblastoma. Science. 2009;326:572–574. doi: 10.1126/science.1179386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pao CS, Barker BL, Benovic JL. Role of the Amino Terminus of G Protein-Coupled Receptor Kinase 2 in Receptor Phosphorylation. Biochemistry. 2009;48:7325–7333. doi: 10.1021/bi900408g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gurevich EV, Tesmer JJG, Mushegian A, Gurevich VV. G protein-coupled receptor kinases: More than just kinases and not only for GPCRs. Pharmacology & Therapeutics. 2012;133:40–69. doi: 10.1016/j.pharmthera.2011.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sterne-Marr R, Tesmer JJG, Day PW, Stracquatanio RP, Cilente J-AE, O'Connor KE, Pronin AN, Benovic JL, Wedegaertner PB. G protein-coupled receptor Kinase 2/G alpha q/11 interaction. A novel surface on a regulator of G protein signaling homology domain for binding G alpha subunits. Journal of Biological Chemistry. 2003;278:6050–6058. doi: 10.1074/jbc.M208787200. [DOI] [PubMed] [Google Scholar]
- 35.Beautrait A, Michalski KR, Lopez TS, Mannix KM, McDonald DJ, Cutter AR, Medina CB, Hebert AM, Francis CJ, Bouvier M, Tesmer JJG, et al. Mapping the putative G protein-coupled receptor (GPCR) docking site on GPCR kinase 2: insights from intact cell phosphorylation and recruitment assays. J Biol Chem. 2014;289:25262–25275. doi: 10.1074/jbc.M114.593178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ribes V, Briscoe J. Establishing and Interpreting Graded Sonic Hedgehog Signaling during Vertebrate Neural Tube Patterning: The Role of Negative Feedback. Cold Spring Harb Perspect Biol. 2009;1:a002014–a002014. doi: 10.1101/cshperspect.a002014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Philipp M, Fralish GB, Meloni AR, Chen W, MacInnes AW, Barak LS, Caron MG. Smoothened signaling in vertebrates is facilitated by a G protein-coupled receptor kinase. Mol Biol Cell. 2008;19:5478–5489. doi: 10.1091/mbc.E08-05-0448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang B, Fallon JF, Beachy PA. Hedgehog-Regulated Processing of Gli3 Produces an Anterior/Posterior Repressor Gradient in the Developing Vertebrate Limb. Cell. 2000;100:423–434. doi: 10.1016/s0092-8674(00)80678-9. [DOI] [PubMed] [Google Scholar]
- 39.Oosterveen T, Kurdija S, Alekseenko Z, Uhde CW, Bergsland M, Sandberg M, Andersson E, Dias JM, Muhr J, Ericson J. Mechanistic Differences in the Transcriptional Interpretation of Local and Long-Range Shh Morphogen Signaling. Developmental Cell. 2012;23:1006–1019. doi: 10.1016/j.devcel.2012.09.015. [DOI] [PubMed] [Google Scholar]
- 40.Junker JP, Peterson KA, Nishi Y, Mao J, McMahon AP, van Oudenaarden A. A Predictive Model of Bifunctional Transcription Factor Signaling during Embryonic Tissue Patterning. Developmental Cell. 2014;31:448–460. doi: 10.1016/j.devcel.2014.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Moore BS, Stepanchick AN, Tewson PH, Hartle CM, Zhang J, Quinn AM, Hughes TE, Mirshahi T. Cilia have high cAMP levels that are inhibited by Sonic Hedgehog-regulated calcium dynamics. Proc Natl Acad Sci USA. 2016;113:13069–13074. doi: 10.1073/pnas.1602393113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vuolo L, Herrera A, Torroba B, Menendez A, Pons S. Ciliary adenylyl cyclases control the Hedgehog pathway. J Cell Sci. 2015;128:2928–2937. doi: 10.1242/jcs.172635. [DOI] [PubMed] [Google Scholar]
- 43.DeCamp DL, Thompson TM, de Sauvage FJ, Lerner MR. Smoothened activates Galphai-mediated signaling in frog melanophores. Journal of Biological Chemistry. 2000;275:26322–26327. doi: 10.1074/jbc.M004055200. [DOI] [PubMed] [Google Scholar]
- 44.Riobo NA, Saucy B, DiLizio C, Manning DR. Activation of heterotrimeric G proteins by Smoothened. Proc Natl Acad Sci USA. 2006;103:12607–12612. doi: 10.1073/pnas.0600880103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ogden SK, Fei DL, Schilling NS, Ahmed YF, Hwa J, Robbins DJ. G protein G|[agr]|i functions immediately downstream of Smoothened in Hedgehog signalling. Nature. 2008;456:967–970. doi: 10.1038/nature07459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barzi M, Kostrz D, Menendez A, Pons S. Sonic Hedgehog-induced proliferation requires specific Gα inhibitory proteins. J Biol Chem. 2011;286:8067–8074. doi: 10.1074/jbc.M110.178772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Shen F, Cheng L, Douglas AE, Riobo NA, Manning DR. Smoothened Is a Fully Competent Activator of the Heterotrimeric G Protein Gi. Mol Pharmacol. 2013;83:691–697. doi: 10.1124/mol.112.082511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sharpe HJ, Pau G, Dijkgraaf GJ, Basset-Seguin N, Modrusan Z, Januario T, Tsui V, Durham AB, Dlugosz AA, Haverty PM, Bourgon R, et al. Genomic Analysis of Smoothened Inhibitor Resistance in Basal Cell Carcinoma. Cancer Cell. 2015;27:327–341. doi: 10.1016/j.ccell.2015.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Atwood SX, Sarin KY, Whitson RJ, Li JR, Kim G, Rezaee M, Ally MS, Kim J, Yao C, Chang ALS, Oro AE, et al. Smoothened Variants Explain the Majority of Drug Resistance in Basal Cell Carcinoma. Cancer Cell. 2015;27:342–353. doi: 10.1016/j.ccell.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kim J, Aftab BT, Tang JY, Kim D, Lee AH, Rezaee M, Kim J, Chen B, King EM, Borodovsky A, Riggins GJ, et al. Itraconazole and Arsenic Trioxide Inhibit Hedgehog Pathway Activation and Tumor Growth Associated with Acquired Resistance to Smoothened Antagonists. Cancer Cell. 2013;23:23–34. doi: 10.1016/j.ccr.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Byrne EFX, Sircar R, Miller PS, Hedger G, Luchetti G, Nachtergaele S, Tully MD, Mydock-McGrane L, Covey DF, Rambo RP, Sansom MSP, et al. Structural basis of Smoothened regulation by its extracellular domains. Nature Publishing Group. 2016;535:517–522. doi: 10.1038/nature18934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Pusapati GV, Hughes CE, Dorn KV, Zhang D, Sugianto P, Aravind L, Rohatgi R. EFCAB7 and IQCE regulate hedgehog signaling by tethering the EVC-EVC2 complex to the base of primary cilia. Developmental Cell. 2014;28:483–496. doi: 10.1016/j.devcel.2014.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc Natl Acad Sci USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gouti M, Tsakiridis A, Wymeersch FJ, Huang Y, Kleinjung J, Wilson V, Briscoe J. In Vitro Generation of Neuromesodermal Progenitors Reveals Distinct Roles for Wnt Signalling in the Specification of Spinal Cord and Paraxial Mesoderm Identity. PLoS Biol. 2014;12:e1001937. doi: 10.1371/journal.pbio.1001937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J-Z, Rasenick MM. Real-Time Visualization of a Fluorescent Gαs: Dissociation of the Activated G Protein from Plasma Membrane. Mol Pharmacol. 2002;61:352–359. doi: 10.1124/mol.61.2.352. [DOI] [PubMed] [Google Scholar]
- 56.Luchetti G, Sircar R, Kong JH, Nachtergaele S, Sagner A, Byrne EF, Covey DF, Siebold C, Rohatgi R, Pan D. Cholesterol activates the G-protein coupled receptor Smoothened to promote Hedgehog signaling. eLife Sciences. 2016;5:e20304. doi: 10.7554/eLife.20304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rohatgi R, Milenkovic L, Scott MP. Patched1 Regulates Hedgehog Signaling at the Primary Cilium. Science. 2007;317:372–376. doi: 10.1126/science.1139740. [DOI] [PubMed] [Google Scholar]
- 58.Novitch BG, Wichterle H, Jessell TM, Sockanathan S. A Requirement for Retinoic Acid-Mediated Transcriptional Activation in Ventral Neural Patterning and Motor Neuron Specification. Neuron. 2003;40:81–95. doi: 10.1016/j.neuron.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 59.Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Stewart SA, Dykxhoorn DM, Palliser D, Mizuno H, Yu EY, An DS, Sabatini DM, Chen ISY, Hahn WC, Sharp PA, Weinberg RA, et al. Lentivirus-delivered stable gene silencing by RNAi in primary cells. RNA. 2003;9:493–501. doi: 10.1261/rna.2192803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Homan KT, Tesmer JJ. Structural insights into G protein-coupled receptor kinase function. Current Opinion in Cell Biology. 2014;27:25–31. doi: 10.1016/j.ceb.2013.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.