Summary
Recent studies indicate that even a homogeneous population of cells display heterogeneity in gene expression and response to environmental stimuli. Although promoter structure critically influences the cell-to-cell variation of gene expression in bacteria and lower eukaryotes, it remains unclear what controls the gene expression noise in mammals. Here we report that CTCF decreases cell-to-cell variation of expression by stabilizing enhancer-promoter interaction. We show that CTCF binding sites are interwoven with enhancers within topologically-associated domains (TADs) and a positive correlation is found between CTCF binding and the activity of the associated enhancers. Deletion of CTCF sites compromises enhancer-promoter interactions. Using single-cell flow cytometry and single-molecule RNA-FISH assays, we demonstrate that knocking down of CTCF or deletion of a CTCF binding site results in increased cell-to-cell variation of gene expression, indicating that long-range promoter-enhancer interaction mediated by CTCF plays important roles in controlling the cell-to-cell variation of gene expression in mammalian cells.
In Brief
In this study, Ren G, et al. show CTCF binding sites within TADs stabilize promoterenhancer interactions, which plays an important role in controlling the cell-to-cell variation of gene expression in mammalian cells.
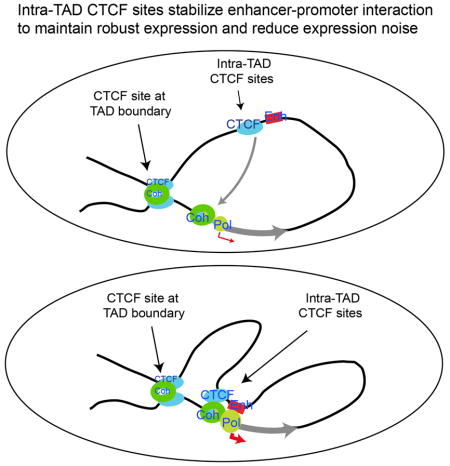
INTRODUCTION
Cell development and differentiation critically depend on precise temporal-spatial control of transcription programs. Increasing evidence indicates substantial cell-to-cell variation of gene expression among a population of the same cells (Sasagawa et al., 2013; Shalek et al., 2014), which is related to heterogeneity in chromatin organization (Jin et al., 2015). Variability of gene expression may result in derailment of normal differentiation programs and lead to phenotypic and disease variations (Aranda-Anzaldo and Dent, 2003; Maamar et al., 2007; Raj et al., 2010; Sharma et al., 2010) as well as differential response to therapeutic treatment of cancers (Yuan et al., 2013). The variation in gene expression in eukaryotic cells may result from numerous mechanisms including fluctuations of upstream regulators (Ji et al., 2013), temporal variations of epigenetic modification states (Metivier et al., 2003), or stochastic bursts of transcription (Larson et al., 2013). Promoter structure is implicated in playing a critical role in controlling the heterogeneity of gene expression in bacteria and yeast (Carey et al., 2013; Murphy et al., 2010). Transcription in mammalian cells is regulated by tens of thousands of enhancers via long-range chromatin interactions. However, due to the lack of understanding of how target genes are regulated by enhancers, it is not clear whether and how long-range chromatin interactions contribute to the heterogeneity of gene expression. In particular, it is unknown whether the insulator binding protein, CTCF, plays a role in controlling expression noise.
RESULTS
To investigate whether CTCF-mediated long-range enhancer-promoter interaction plays a role in controlling gene expression noise, we first analyzed genome-wide chromatin interactions of mouse Th2 cells using a three-enzyme Hi-C protocol (3e Hi-C) that cleaves chromatin with a pool of three 4bp-restriction enzymes (see method section for details, Figure S1A, B, C, and Supplemental Table S1). From the paired-end sequencing data, we identified 81,773 interactions among promoters, enhancers (p300 binding sites) and insulators (CTCF binding sites) in mouse Th2 cells. Among the interactions involving promoters and enhancers, 59–61% of them were detected in two replicate Th2 cell libraries (Figure S1D). Using the 3e Hi-C data, we identified 1,363 TADs in mouse Th2 cells (Figure S1E and data not shown), which exhibited 73–76% overlap with those identified in ES cells (Dixon et al., 2012).
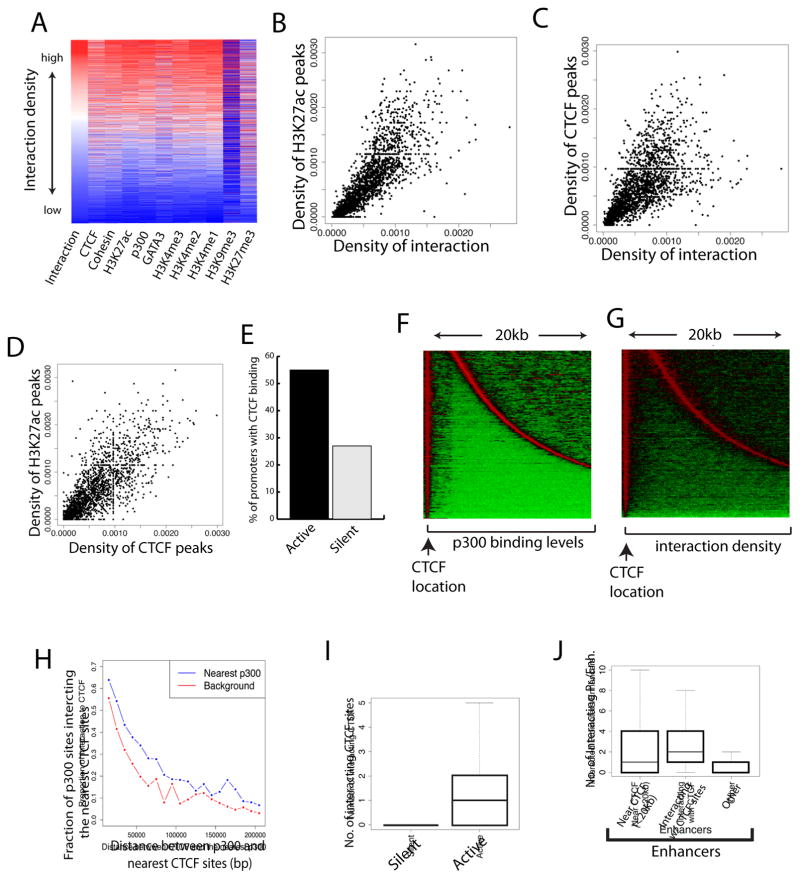
By comparing the long-distance chromatin interactions among the regulatory regions with previously published epigenomic data in mouse Th2 cells (Wei et al., 2011; Wei et al., 2009), we found that the interaction density positively correlates with active marks including H3K27ac, H3K4me1, H3K4me2, and H3K4me3 (Figure 1A). Although previous studies observed an elevated degree of interaction in both H3K4me-marked active domains and PcG-marked repressive domains (Sexton et al., 2012), our identified interacting chromatin regions are positively associated with only active but not repressive marks in Th2 cells (Figure 1A). CTCF, Cohesin (SMC1a), p300, and the Th2-specific transcription factor GATA3 are also positively correlated with the interactivity. The interaction of regulatory elements is positively correlated with the number of active marks and/or transcription factor binding (Figure S1F). The correlation between interaction density and H3K27ac is better illustrated using scatter plot (Figure 1B), suggesting that enhancer activity is positively correlated with interactivity. Pioneering work showed that CTCF mediates chromatin interactions by regulating chromatin domain structure (Dowen et al., 2014; Phillips-Cremins et al., 2013; Tang et al., 2015) and as expected, we observed a positive correlation between the density of CTCF peaks and chromatin interactions (Figure 1C). Interestingly, we also observed a positive correlation between CTCF binding and enhancer activities as indicated by H3K27ac (Figure 1D), suggesting that CTCF binding influences enhancer activity, as suggested by recent data (Sanborn et al., 2015; Yaffe and Tanay, 2011; Zuin et al., 2014).
Figure 1. CTCF binding sites interact with enhancers and promoters and positively correlate with gene activation.
A. Interaction density among regulatory elements positively correlates with CTCF, Cohesin, GATA3, p300 and active histone modifications. The high-confidence interacting regions were sorted based on their interaction density (red: high; blue: low). The binding levels of chromatin proteins and histone modifications determined by ChIP-Seq were plotted as indicated at the bottom of the panel.
B. Scatter plot shows a positive correlation between interaction density and H3K27ac level.
C. Scatter plot shows a positive correlation between interaction density and CTCF binding level.
D. Scatter plot shows a positive correlation between levels of CTCF binding and H3K27ac modification.
E. More active genes are bound by CTCF than silent genes. The fraction of active or inactive promoters (+/− 2kb around TSS) bound by CTCF is plotted.
F. Distribution of the closest p300 site relative to the CTCF sites in the genome. The location of CTCF sites is indicated by the arrow at bottom. The closest p300 sites are found and p300 binding levels are plotted (red: high; green: low). Displayed are the p300 sites located from 3 to 20kb away from the CTCF sites.
G. Interaction density peaks at the CTCF and p300 binding sites. The interaction densities for the chromatin regions described in Panel F above are plotted.
H. p300 sites interact with their neighboring CTCF sites. Plotted are the fractions of p300 sites that interact with the nearest CTCF sites. The background shows the interaction density with the chromatin regions without p300 binding at equal distance.
I. Active promoters exhibit more looping with distal CTCF sites than silent promoters. Genes were separated to active genes (RPKM ≥3) and silent genes (RPKM ≤3). The number of CTCF binding sites (>5kb from TSS), which interact with the promoters, is plotted per promoter on Y-axis.
J. Enhancers near or interacting with CTCF sites are more interactive. The p300-bound enhancers are separated to three categories: (1) with at least one CTCF site within a distance of 20 kb; (2) interacting with a CTCF site; and (3) others. The number of interacting promoters or enhancers is plotted for each category (Y-axis).
The mammalian genomes are organized into topologically-associated domains (TADs) and sub-functional domains (Dixon et al., 2012; Duan et al., 2010; Lieberman-Aiden et al., 2009; Nora et al., 2012; Sexton et al., 2012; Tanizawa et al., 2010). Previous studies suggested that CTCF binding is enriched at the domain boundary regions and CTCF is established as a key player for the maintenance of chromatin domain boundaries (Cuddapah et al., 2009; Dixon et al., 2012; Dowen et al., 2014; Nora et al., 2012; Phillips-Cremins et al., 2013; Schwartz et al., 2012; Sofueva et al., 2013; Tang et al., 2015; Zuin et al., 2014). However, ChIP-Seq data (Barski et al., 2007; Kim et al., 2007) indicate that only a minor fraction of CTCF binding sites are located to boundary regions (Dixon et al., 2012). We found that the vast majority (80%) of CTCF sites are interspersed with enhancers in active chromatin domains in Th2 cells, raising the possibility that CTCF may have other important functions in addition to the well-established insulator/boundary function. Indeed, CTCF and its interaction partner, Cohesin, have been previously suggested to contribute to gene regulation by mechanisms involving long-distance chromatin interactions (Faure et al., 2012; Kagey et al., 2010; Parelho et al., 2008; Rubio et al., 2008; Seitan et al., 2013). To test whether CTCF is generally associated with gene activation, we examined its enrichment at promoters of active and silent genes in Th2 cells. The analysis revealed that while only 27% of silent promoters were bound by CTCF, 53% of active promoters were bound by CTCF in Th2 cells (Figure 1E).
To investigate the relationship between the binding sites of CTCF and p300, an enhancer binding protein, we identified CTCF binding sites and searched for their closest p300 binding sites in the genome. The analysis indicated that about 70% of CTCF binding sites are located next to at least one p300 binding site within a distance of 20 kb (Figure 1F). Interestingly, strong chromatin interactions were detected at the CTCF and p300 binding sites (Figure 1G). Furthermore, the CTCF sites exhibited significantly higher interactions with their neighboring p300 sites compared to the control regions of same distance but without p300 binding (Figure 1H). To test whether the CTCF sites, which are interspersed with enhancers, interact with promoters during gene activation, we examined the CTCF sites that are looped to the promoter regions of active and silent genes. The analysis revealed that active promoters exhibit significantly higher interaction with CTCF sites than silent promoters (Figure 1I). Remarkably, enhancers with a neighboring CTCF site exhibited a greater tendency to interact with promoters than those without neighboring CTCF sites (Figure 1J). Furthermore, the enhancers that interacted with CTCF sites also exhibited significantly higher interactions with promoters and/or other enhancers than those without interacting CTCF sites (Figure 1J). These results suggest that CTCF binding sites interact with their neighboring enhancers and facilitate the functional interaction between enhancers and promoters.
To characterize the genes that may be regulated by long-distance enhancer-promoter interactions, we identified the top 650 genes that exhibit the most enhancer-promoter interactions (Supplemental Table S1). Interestingly, the genes with the most long-distance interactions are highly enriched in pathways and categories related to immune function and T-cell activation (Figure S2A). In contrast, the highest expressed genes in Th2 cells are significantly enriched in housekeeping functions (Supplemental Table S1, Figure S2B). These results indicate that lineage-specific genes are most likely regulated by long-distance enhancer-promoter interactions, while house-keeping genes are mainly regulated by proximal promoter elements.
To test whether CTCF regulates expression of the tissue-specific genes that are associated with long-distance enhancer-promoter interactions, we knocked down CTCF expression by about 60% using shRNAs in a mouse T cell line, EL4 cells (Figure S2C, D). The CTCF knockdown resulted in 819 down-regulated and 879 up-regulated genes, respectively (Supplemental Table S1). Interestingly, the top ten enriched gene categories in down-regulated genes included leukocyte activation, phosphoprotein and T cell activation (Figure S2E), which are involved in immune functions. The down-regulation of several cell specific genes including Cd5, Thy-1, and Gata3 was confirmed by qPCR analysis (Figure S2F). In contrast, the up-regulated genes were enriched in gene categories including ribosome and ribosome proteins (Figure S2G), which belong to house-keeping genes and may not involve CTCF-mediated enhancer-promoter interactions.
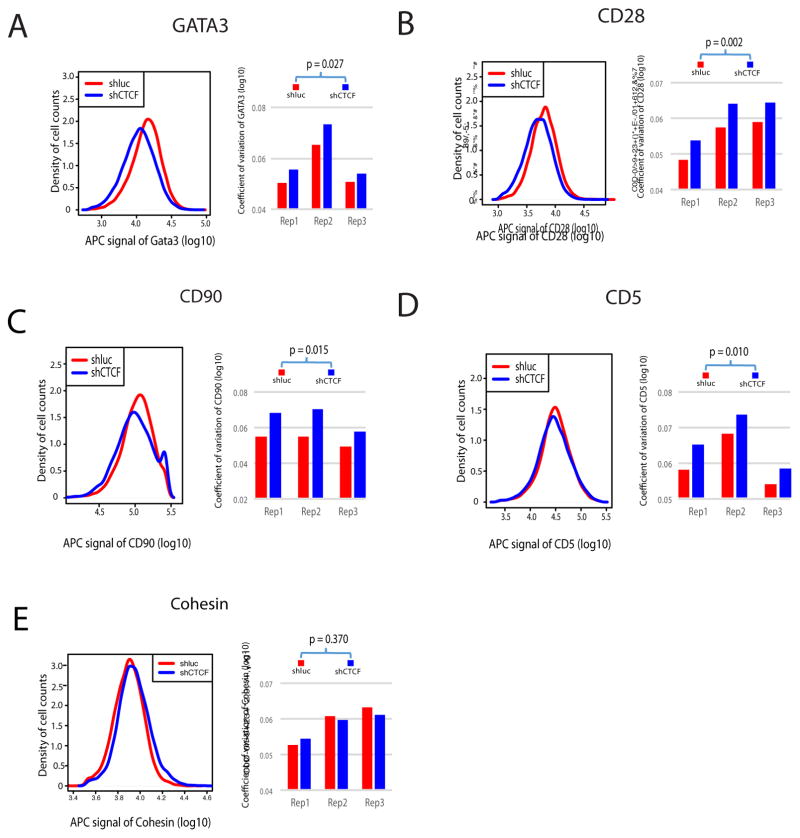
Although these results support the notion that CTCF contributes to expression of the lineage-specific genes by mediating the interaction between enhancers and promoters, only 71 of the 650 most interactive genes were significant down-regulated while the vast majority of them displayed only modest changes or no change in expression upon CTCF knockdown, raising the question what roles the CTCF-mediated enhancer-promoter interaction plays in the expression of these genes. Since promoter elements are critical determinants of expression noise in bacteria and lower eukaryotic cells (Carey et al., 2013; Murphy et al., 2010), we reasoned that CTCF-mediated enhancer-promoter interaction may contribute to the control of expression noise and help to reduce cell-to-cell variation of expression in mammalian cells. To test this hypothesis, we examined the expression of several genes using fluorescence-activated cell sorting (FACS) that monitors gene expression in each single cell. Knockdown of CTCF only modestly decreased the expression of GATA3 (Figure 2A, left panel). To measure the cell-to-cell variation of GATA3 expression, the variance of expression and coefficient of variation (CV) were calculated, which exhibited significant increases in three replicates (Figure 2A, right panel). Among the six CTCF-bound T cell-specific genes analyzed, we found four (GATA3, CD90, CD28, CD5) displayed statistically significant increase in variance while two (CD3e and STAT6) did not show significant changes (Figure 2A, B, C, D; Figure S2H and data not shown; Supplemental Table S1). Two ubiquitously expressed genes (Cohesin and Condensin) did not show significant changes either (Figure 2E and data not shown). These results indicate that CTCF contributes to reduced expression variability.
Figure 2. Knocking down of CTCF results in increased cell-to-cell variation.
FACS analysis revealed increased cell-to-cell variation of expression of GATA3 (A), CD28 (B), CD90 (C), and CD5 (D). No significantly changed variation of expression was detected for Cohesin (E). Left panels show the distribution of gene expression with the x-axis indicating the expression level and y-axis indicating the cell density. Right panels are bar plots for the coefficient of variation that measures the expression variation of cells from individual replicate. APC signal were log10 transformed. Replicates for KD cells and control cells were paired based on experiment date. P-value was obtained by paired t-test.
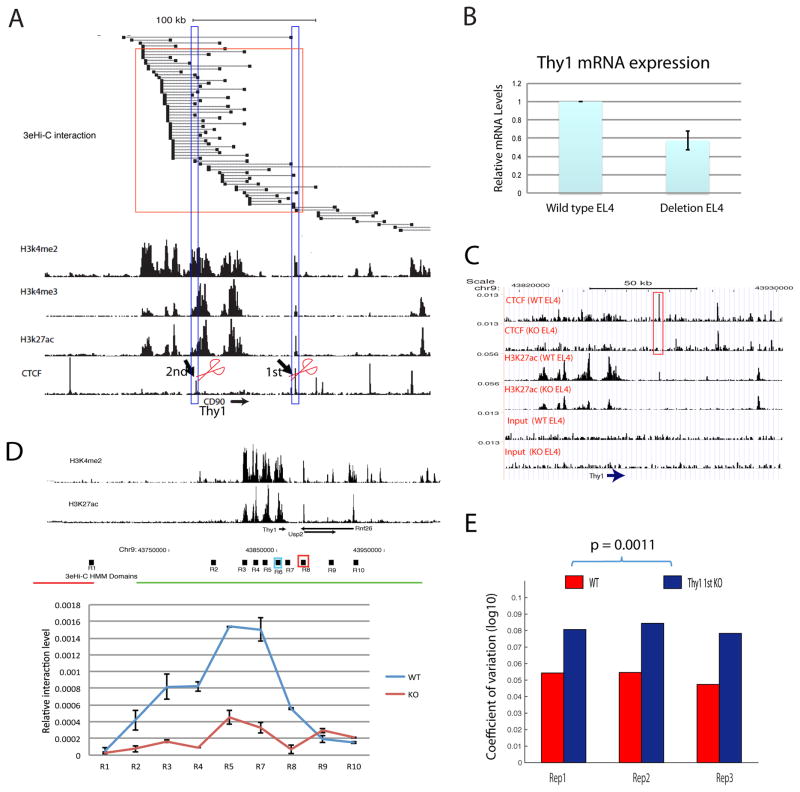
The observed increase in cell-to-cell variation of expression in CTCF knockdown cells may be caused by heterogeneous CTCF knockdown efficiency across different cells and/or indirect effects of CTCF knockdown in the cells. To rule out these possibilities, we examined the cell-to-cell variation of gene expression after deleting specific CTCF binding sites using CRISPR-CAS9. The Thy1 gene, which encodes a T cell-specific cell surface marker CD90, exhibits extensive interaction among its promoter and potential enhancers marked by H3K27ac and H3K4me2 and is bound by CTCF at several sites (Figure 3A). Our 3e Hi-C data revealed that the 1st and 2nd CTCF binding sites directly interact with the Thy1 promoter and enhancers (Figure 3A, high-lighted in the blue rectangles). Deletion of the 1st CTCF binding site (Figure S3A) resulted in decreased Thy1 expression by 40% (Figure 3B). Our data show that the deletion abolished CTCF binding to this site and severely compromised H3K27ac across the entire Thy1 gene region but not other nearby regions (Figure 3C). Furthermore, our analysis of chromatin interaction using 3C assays with the Thy1 promoter as an anchor point revealed that the interactions between the Thy1 promoter (R6 region) with the CTCF binding region and enhancer regions are substantially decreased (Figure 3D, R3, R4, R5, R7, R8 regions). Interestingly, there were no significant changes in interaction between Thy1 promoter region with the nearby gene promoter regions R9, and R10 (Figure 3D), suggesting CTCF binding to this site specifically mediates the Thy1 promoter-enhancer interaction. Furthermore, there was no significant change in interaction between R6 region and R1 region that is located in a neighboring TAD boundary (Figure 3D), suggesting that deletion of this CTCF binding site did not disrupt the TAD structure. Finally, we found a significantly higher cell-to-cell variation of gene expression in the CRISPR knockout cells using FACS (Figure 3E, Supplemental Table S2). These results indicate that CTCF binding to the 1st site (R8 region) of the Thy1 gene stabilizes the interaction between the Thy1 promoter and its enhancers and thus reduces the cell-to-cell variation of Thy1 expression.
Figure 3. CTCF binding sites at the Thy1 locus contribute to the functional interaction between Thy1 promoter and its enhancers and the expression noise control. CD90 protein is encoded by the Thy1 gene.
A. The chromatin interactions surrounding the Thy1 gene locus are shown in the upper panel and H3K4me2, H3K4me3, H3K27ac and CTCF ChIP-Seq signals are shown in the lower panels. The red rectangle highlights the interactions between the CTCF binding site and Thy1 promoter and enhancers. The CTCF binding sites high-lighted by scissors were deleted separately in EL4 cells using CRISPR/CAS9.
B. Deletion of the 1st CTCF binding site decreased Thy1 mRNA levels. Total RNAs isolated from the wild type or CTCF site deletion EL4 clones were analyzed by quantitative reverse-transcription PCR and normalized to GAPDH.
C. The 1st CTCF site deletion abolished CTCF binding and compromised the H3K27ac modification at the Thy1 gene locus. The genome browser images show the ChIP-Seq data for CTCF binding, H3K27ac and chromatin input signals in wild type and CRISPR deletion EL4 cells. The high-lighted CTCF peak indicates the location of CRISPR deletion.
D. The 1st CTCF site deletion compromised the enhancer-promoter interaction in the Thy1 gene locus. Top panel shows the H3K4me2 and H3K27ac ChIP-Seq signals. The red and green horizontal lines below the ChIP-Seq tracks indicate the different TADs called by HMM. The bottom panel shows the relative chromatin interaction intensity of the Thy1 promoter with various enhancer regions indicated above the top panel (R1 to R10) by 3C analysis. The blue rectangle marked as R6 is the anchor site for the 3C analysis. The red rectangle marked R8 region is the deleted 1st CTCF site. Data show average of two independent experiments and are represented as mean ± SEM. WT: control cells; KO: CRISPR/CAS9 deletion cells.
E. Deletion of the 1st CTCF site resulted in increased cell-to-cell variation of expression of CD90 protein encoded by the Thy1 gene as measured by FACS assay.
To test if CTCF contributes to the promoter-enhancer interaction of other genes, we deleted CTCF binding sites in the Cd5 and Runx3 gene loci using CRISPR-CAS9 (Figure S3B, C, Figure S4A, B). CRISPR/CAS9-mediated deletion of the CTCF binding sites modestly reduced expression of CD5 and Runx3 at mRNA levels (Figure S3D, S4C). Our 3C assays using R4 region (Figure S3E, left panel) or R7 region (Figure S3E, right panel) of the Cd5 gene as an anchor point revealed that the interaction between the Cd5 gene promoter with the CTCF binding region and enhancer regions are significantly decreased in the CRISPR deletion cells. Similarly, deletion of the CTCF binding site at the Runx3 gene locus also compromised the interaction between CTCF binding site, promoter and enhancers (Figure S4D). However, no significant changes in interaction were observed between the CTCF binding site and Runx3 promoter with a potential regulatory region (R1 region) located in a neighboring TAD (Figure S4D), suggesting that the TAD structure is not disrupted by deletion of the CTCF binding site. Together, these results indicate that CTCF binding stabilizes the interaction between the promoter and its enhancers.
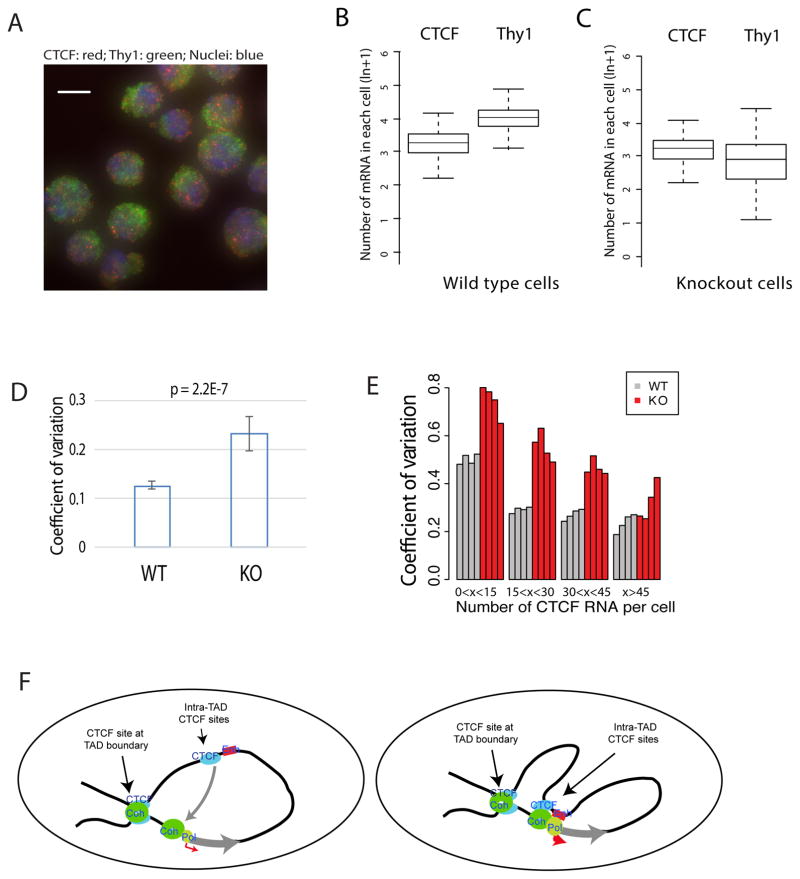
In order to directly measure the number of mRNA molecules in single cells, we employed single molecule RNA-FISH assays (Figure 4A). The number of CTCF and Thy1 mRNAs was counted in each of about 2000 individual cells of wild type and the CRISPR deletion cells (Figures 4B, C; Supplemental Table S2). CTCF and Thy1 exhibited a mean value of about 25 and 48 molecules per cell, respectively, in wild type cells (Figure 4B). The number of Thy1 mRNA decreased to a mean value of 27 molecules in the CRISPR knockout cells (Figure 4C). A significantly higher cell-to-cell variation in expression was observed in the knockout cells (Figure 4D). Similarly, we observed that deletion of the CTCF binding site of the Cd5 gene locus led to significantly increased cell-to-cell variation of CD5 expression using the RNA-FISH assay (Figure S3F, Supplemental Table S2).
Figure 4. Single-molecule RNA-FISH shows increased cell-to-cell variation of Thy1 mRNA in the CTCF site-deleted cells.
A. Typical images of RNA-FISH for detecting CTCF mRNA (red), Thy1 mRNA (green) and DNA (blue).
B. Box plots showing the numbers of CTCF and Thy1 mRNA molecules per cell in wild type EL4 cells. The box plot is from one representative of 4 replicates.
C. Box plots showing the numbers of CTCF and Thy1 mRNA molecules per cell in the 1st CTCF site-deleted EL4 cells. The box plot is from one representative of 12 replicates.
D. Deletion of the 1st CTCF site results in increased coefficient of variation in the number of Thy1 mRNAs per cell. The bar plot shows the distribution of CVs of 4 replicates for WT and 12 replicates for KO. Data represented as mean ± SEM. P-value was obtained by t-test.
E. The variation of Thy1 mRNA per cell caused by deletion of the 1st CTCF site is related to the number of CTCF mRNA in the cell. On the X-axis, the cells are sorted to four groups according to the number of CTCF mRNAs per cell (0–14; 15–29; 30–44; and >44). Y-axis indicates the coefficient of variation of Thy1 mRNA. Grey bars indicate the wild type EL4 cells and red bars indicate the CTCF site deletion EL4 cells. Each bar represents one replicate.
F. CTCF and Cohesin organize chromatin to large domains (left panel) and facilitate long-distance enhancer-promoter interaction to decrease fluctuation of expression and maintain robustness of expression (right panel).
The change in cell-to-cell variation of Thy1 expression appears to be related to the number of CTCF mRNA per cell. Higher variation of Thy1 expression was observed in the cells with fewer CTCF mRNA molecules than the cells with more CTCF mRNAs when the 1st CTCF binding site was deleted (Figure 4E), suggesting that higher CTCF expression may compensate for the deletion of the CTCF site possibly through other CTCF binding sites in the region. To test this hypothesis, we deleted the 2nd CTCF binding site upstream of the Thy1 gene (Figure 3A, 2nd, Figure S4E). Deletion of this CTCF binding site resulted in about 40% reduction of Thy1 mRNA level (Figure S4F). Furthermore, significantly higher cell-to-cell variation of Thy1 mRNA expression was observed in the deletion cells as compared to control cells by RNA-FISH assays (Figure S4G, Supplemental Table S2). At protein levels, higher variation of CD90 was also observed in the CTCF site deletion cells by FACS assays (Figure S4H). Together, our results indicate that multiple CTCF binding sites may contribute to the stability of promoter-enhancer interaction and thus reduce gene expression noise.
DISCUSTION
Gene expression is stochastic in nature (Blake et al., 2003; Coulon et al., 2013; Elliott et al., 1995) and transcriptional fluctuations have been suggested to contribute to probabilistic differentiation and evolution (Eldar and Elowitz, 2010). Indeed, the fluctuation of Nanog expression in ES cells appears to be a critical factor in influencing the decision for self-renewal or differentiation (Kalmar et al., 2009). However, uncontrolled fluctuation of gene expression may be deleterious to normal development and differentiation programs and associated with disease progression (Capp, 2005; Yuan et al., 2013). Our data reveal a molecular mechanism for reducing expression heterogeneity: enhancer-promoter interactions mediated by CTCF. CTCF and Cohesin maintain topologically associated domains and subdomains by mediating stable chromatin loop formation (Faure et al., 2012; Kagey et al., 2010; Parelho et al., 2008; Rubio et al., 2008; Seitan et al., 2013), which is favored by a pair of convergent CTCF binding sites (Guo et al., 2015; Rao et al., 2014; Tang et al., 2015) and may be formed by CTCF/Cohesin-mediated loop extrusion (Nichols and Corces, 2015; Sanborn et al., 2015). We show that CTCF binding sites are interwoven with enhancer elements and facilitate enhancer-promoter interactions. We propose that “quasi stable” loops may form by the interaction of Cohesin with CTCF that binds to the CTCF sites with either convergent or divergent motifs near enhancers, possibly through Cohesin-mediated loop extrusion (Nichols and Corces, 2015), which brings the enhancers to the proximity of their target promoters and stabilizes the enhancer-promoter interaction and thus decreases the fluctuation of the promoter activity (Figure 4F). This interaction may be further stabilized by a positive feedback loop by RNA, and CTCF-interacting protein YY1 (Sigova et al., 2015). Our study provides an alternative explanation for those recent findings that targeted degradation of CTCF/Cohesin have very mild effect on the overall transcriptional changes (Nora et al., 2017). In addition to its function in maintaining intact chromatin domain structures, our results suggest that genetic mutations of CTCF may also affect disease progression by changed ability of mediating enhancer-promoter interaction and thus controlling the fluctuation of transcription.
STAR & Method
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| anti-mouse CD4 APC | eBioscience | Cat# 17-0042-82 |
| anti-mouse CD62L Pacific Blue | Biolegend | Cat# 104424 |
| anti-mouse CD44 APC-Cy7 | BD bioscience | Cat# 560568 |
| anti-mouse CD25 PE | eBioscience | Cat# 12-0251-82 |
| Hamster anti-mouse CD3e APC | BD bioscience | Cat# 553066 |
| anti-mouse CD5 APC | eBioscience | Cat# 17-0051-82 |
| anti-mouse CD28 APC | eBioscience | Cat# 17-0281-82 |
| anti-mouse CD44 APC | Biolegend | Cat# 100412 |
| anti-mouse CD90.2 APC | eBioscience | Cat# 17-0902-82 |
| Alexa Fluor® 647 Mouse anti-GATA3 | BD bioscience | Cat# 560068 |
| Recombinant Murine IL-4 | Peprotech | Cat# 214-14 |
| Recombinant Murine IL-2 | Peprotech | Cat# 212-12 |
| anti-murine IFN-γ | Peprotech | Cat# 500-P119 |
| anti-murine IL-12 | Peprotech | Cat# 500-M59 |
| H3K4me1 | Abcam | Cat# ab8895 |
| H3K4me2 | Abcam | Cat# ab32356 |
| H3K27ac | Abcam | Cat# ab4729 |
| Gata3 | BD biosciences | Cat# 558686 |
| Stat6 | Santa Cruz Biotechnology | Cat# sc-374021 |
| Cohesin (SMC1a) | Bethyl | Cat# A300-055A |
| CTCF | Millipore | Cat# 07-729 |
| RNA Polymerase II | Abcam | Cat# ab5408 |
| Bacterial and Virus Strains | ||
| One Shot™ Stbl3™ Chemically Competent E. coli | Thermo Fisher Scientific | Cat# C737303 |
| Biological Samples | ||
| C57BL/6 mice | Jackson Laboratory | Stock No: 000664 Black 6 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Ampicillin | Sigma-Aldrich | Cat#A1593 |
| Puromycin | Sigma-Aldrich | Cat# P8833-10MG |
| Critical Commercial Assays | ||
| Prolong Gold Mountant with DAPI | ThermoFisher Scientific | P36941 |
| Deposited Data | ||
| Raw data files for ChIP sequencing | NCBI database (GSE66343) | |
| Raw data files for bulk RNA-seq sequencing | NCBI database (GSE66343) | |
| Raw data files for 3e Hi-C sequencing | NCBI database (GSE66343) | |
| FACS data files | See supplemental Tables S2 | |
| Experimental Models: Cell Lines | ||
| Mouse lymphoma: EL4 cells | ATCC | TIB-39 |
| Experimental Models: Organisms/Strains | ||
| Oligonucleotides | ||
| Supplemental table S1 Sheet 3 for the list of Oligos used in this study | ||
| Recombinant DNA | ||
| pSpCas9(BB)-2A-Puro (PX459) | Cong et al., 2013 | Addgene Plasmid #48139 |
| RNAi-Ready pSIREN-RetroQ-ZsGreen1 Vector | Clontech Laboratories | Cat# 632455 |
| Software and Algorithms | ||
| Bowtie 2 | Langmead, et al., 2012 | http://bowtie-bio.sourceforge.net/bowtie2/index.shtml |
| DVAID | Huang da et al., 2009 | https://david.ncifcrf.gov/ |
| R | R Core Team. 2017 | www.R-project.org |
| EdgeR | Robinson et al., 2010 | http://bioconductor.org/packages/release/bioc/html/edgeR.html |
| FlowJo X software | FlowJo, LLC | https://www.flowjo.com/ |
| CellProfiler | Carpenter et al., 2006 | http://cellprofiler.org/ |
| IDL software | http://www.larsonlab.net/ | |
| RNA-fish Oligo | http://www.oligo.net/ | |
| Fiji | https://fiji.sc/ | |
| Micromanager | https://micro-manager.org/ | |
| Other | ||
| Amaxa® Cell Line Nucleofector® Kit L | Lonza | VCA-1005 |
| Foxp3 staining kit | eBioscience | Cat# 00-5523-00 |
| PureLink™ HiPure Plasmid Filter Midiprep Kit | Thermo Fisher Scientific | Cat# K210014 |
| End-It™ DNA End-Repair Kit | Epicentre | Cat# ER81050 |
Contact for Reagent and Resource Sharing
Further information and requests for reagents may be directed to, and will be fulfilled by the corresponding author Keji Zhao (zhaok@nhlbi.nih.gov).
Experimental Method and Subject Details
Cell culture
EL4 cells were cultured in DMEM (Gibco Invitrogen) supplemented with 50 IU/mL penicillin and 50 μg/mL streptomycin (Gibco Invitrogen) and 10% heat inactivated calf serum (Sigma, USA). Cultures were maintained by replacement of fresh medium every 3 days, and cell density was kept between 1 × 105 and 1 × 106 cells/ml. For generation of Th2 cells, naïve CD4+ T cells were isolated from the lymph nodes of C57BL/6 mice from the Jackson Laboratory (JAX) by staining with Pacific Blue-anti-CD62L, APC-anti-CD4, APC-Cy7- anti-CD44 and PE-anti-CD25 and sorting for CD4+CD25-CD62LhiCD44low population by FACSAria (BD Biosciences) as described before (Hu et al., 2013). For Th2 differentiation, the cytokine and antibody cocktail containing IL-4 (5000 U/ml), IL-2 (100 U/ml), anti-IFNγ (10 μg/ml) and anti-IL-12 (10 μg/ml) was added to the cell culture and two rounds polarizations were performed (Hu et al., 2013).
Antibodies, ChIP-Seq and RNA-Seq assays
The following antibodies were used for ChIP-Seq analysis: H3K27ac (ab4729, Abcam); Cohesin (SMC1) (A300-055A, Bethyl), CTCF (07-729, Millipore), RNA Polymerase II (ab5408, Abcam). For generating ChIP-Seq libraries, chromatin extracts were prepared by sonication of cells following crosslinking with 1% formaldehyde for 10 minutes at room temperature. The ChIP was performed with 1 μg specific antibodies as described (Barski et al., 2007) and ChIP-Seq libraries were prepared using indexing primers as described (Hu et al., 2013). RNA-Seq libraries using polyA RNA isolated from both the WT and KO cells were prepared as described (Chepelev et al., 2009).
Gene Expression Analysis
Total RNAs from the knockdown, knock out and control cells were extracted with miRNeasy Micro Kit (Qiagen). cDNA was synthesized by using oligo (dT)20, and SuperScript™ III Reverse Transcriptase (Invitrogen) according to the manufacturers’ instructions. RT-qPCR samples were mixed with the following Taqman probes mixture (Applied Biosystems) and run on Lightcycle 96 (Roche): Gapdh: Mm03302249_g1; CTCF: Mm00484027_m1; CD5: Mm00432417_m1; Cohesin (SMC1a): Mm00490624_m1; Runx3: Mm00490666_m1; Thy1: Mm00493681_m1. Results were normalized to Gapdh gene mRNA level.
Analysis of expression variation in wild type and CTCF knockdown EL4 by FACS
EL4 cells were infected with shCTCF retroviral particles packaged in GP2-293 cells. After five days of infection, half of the cells were sorted for GFP+ cells to check knockdown efficiency by using RT-qPCR and/or RNA-seq analysis; the other half was used for expression variation analysis by Flow Cytometry. The immunostaining was done using 0.5 μg specific antibodies for 2 × 106 cells. For surface proteins Cd3e, Cd5, Cd28, Cd44, and Cd90 (BD Biosciences or eBioscience), cells were stained in PBS containing 0.3% BSA at 4°C for 30 minutes (eBioscience). For Gata3 (BD Biosciences), and STAT6 (Miltenyi), intracellular staining was done according to the manufacturers’ instructions. For Cohesin (SMC1a), and CTCF, cells were permeabilized with the transcription factor staining buffer reagent (eBioscience, 00-5523-00), incubated with the primary antibody at 25°C for 30 minutes, then stained with the secondary antibody (TRITC conjunction goat anti rabbit IgG) at 25°C for 30 minutes. Data were acquired on a FACSCanto II flow cytometer (Becton Dickinson) and analyzed using FlowJo X software. The same cell surface protein staining and analytical procedures were performed for the Thy1 2nd CTCF binding site deletion and control cells.
The following CTCF shRNA target sequences were used for the knockdown experiments:
mouse CTCF- shRNA 7: 5′-GGTGCAATTGAGAACATTATA
mouse CTCF- shRNA 6: 5′-TGGACGATACCCAGATCATAA
3e Hi-C assay
The three enzyme Hi-C (3e Hi-C) was performed similar to in situ Hi-C (Rao et al., 2014) with modifications as described below. In vitro differentiated murine Th2 cells were cross-linked with 1% formaldehyde for 10 minutes at 25°C. 10 × 106 cells were lysed in 10 ml lysis buffer (10 mM Tris-HCl pH8.0, 10 mM NaCl, 0.2% NP40; 10 μl protease inhibitors (Sigma)) with rotation at 4°C for 60 minutes. The cells were then treated with 400 μl 1X NEB cutsmart buffer with 0.1% SDS at 65°C for 10 minutes, followed by addition of 44 μl 10% Triton X-100 to quench SDS. Chromatin was subsequently digested with 20 Units CviQ I (NEB), and 20 Units CviA II (NEB) at 25°C for 20 minutes, then with 20 Units Bfa I (NEB) at 37°C for 20 minutes. The reaction was stopped by washing the cells twice with 600 μl wash buffer (10mM NaCl, 1mM EDTA, 0.1% triton-100). The DNA ends were blunted and labeled with biotin by Klenow enzyme in the presence of dCTP, dGTP, dTTP, biotin-14-dATP, followed by ligation using T4 DNA ligase. After reverse crosslinking, the samples were treated with T4 DNA polymerase to remove biotin labels at the DNA ends. Then, DNA was fragmented to 300–500 bp by sonication with a Bioruptor sonicator (Diagenode UCD-200). Next, The DNA was end-repaired, followed by A-addition as described previously (Lieberman-Aiden et al., 2009). The remaining biotinylated DNA fragments were then captured using Dynabeads MyOne Streptavin C1 Beads (Invitrogen) by incubating for 30 minutes at 25°C with rotation. The DNA on beads was ligated to the Illumina Paired End Adaptors. Following PCR-amplification of the libraries, DNA fragments from 300 to 700 bp were purified from 2% E-gel and sequenced on Hi-Seq 2500.
3C assay
For each sample, 2 to 5 × 106 cells were cross-linked with 1% formaldehyde for 10 min at 25°C. The reaction was quenched by the addition of 125 mM glycine for 5 minutes at 25°C. Cross-linked cells were washed with PBS and resuspended in 10 ml lysis buffer (10 mM Tris-HCl, pH 8.0, 10 mM NaCl, 0.2% NP-40 and proteinase inhibitors) and lysed with a Dounce homogenizer. Following Bgl II (NEB) digestion for Thy1 and Runx3 gene loci, and Hind III (NEB) digestion for CD5 gene locus, 3C ligation was performed as previously described (Lieberman-Aiden et al., 2009). The 3C interactions at the three Thy1, CD5, Runx3 loci were analyzed by quantitative real-time PCR using custom designed probes as previously described (Xu et al., 2011). The amount of DNA in the qPCR reactions was normalized across 3C libraries for one using a custom Taqman probe directed against the ACTB gene locus. The sequences of primers and probes used in the 3C assays are listed in the supplemental table S1 oligo list.
Deletion of CTCF binding sites by CRISPR/Cas9
Genome Editing was carried out by CRISPR/Cas9 system (Cong et al., 2013). The CRISPR/Cas9 plasmid (PX459) pairs were nucleofected to EL4 cells by using Amaxa® Cell Line Nucleofector® Kit L according to the manufacturer’s instructions (Lonza). One plasmid expressing PmaxGFP® which include in the Kit L was co-transfected. After 24 hours, the cells were selected in fresh medium containing 4 μg/ml puromycin (Sigma). After 72 hours, limiting dilution was performed and selection was continued for 8–12 days with DMEM medium containing 0.5 μg/ml puromycin. Individual clones were picked and genomic DNA was extracted following the manufacturer’s instructions (Redextract-N-AMP tissue kit, SIGMA). Genotyping was done by using locus-specific PCR primers under the following PCR conditions: 98°C for 30 s; 35 cycles (98°C for 10 s, 60°C for 30 s, 72°C for 35 s); 72°C for 5 min; hold at 4°C. PCR products were run on 2% agarose gel. Genotyping primers and CRISPR/cas9 sgRNA target sequences for Thy1 1st CTCF site, Thy1 2nd CTCF site, CD5 gene CTCF site, and Runx3 gene CTCF site are listed in the supplemental S1 oligo list part. Positive clones were expanded for further experiments.
RNA-FISH assay
RNA-FISH Probe Set Design
Thy1 and CTCF probes were designed from mRNA sequence with Oligo (Rychlik, 2007) and ordered from Biosearch Technologies. 48 probes were designed each for Thy1, CD5, and CTCF mRNA (Supplement Table 2). The probe sets were singly labeled with Quasar570 and Quasar670, respectively.
RNA-FISH sample preparation and hybridization
CTCF binding site deletion cells (Thy1 1st, Thy1 2nd, and CD5 binding sites) and control cells were cultured in normal DMEM as described above. For fixing, cells were washed two times with PBS, and then resuspended at 1×106/ml using 0.5 ml DMEM without FBS, subsequently incubated at 25°C for five minutes without moving plate. Then, 0.5 ml 4% paraformaldehyde was added to each well and samples were kept on ice for 10 minutes, and followed by washing samples with PBS two times. Next, 2 ml 70% ethanol were added to samples and were stored at 4°C for next step. The standard Biosearch Technologies protocol was then used as described previously (Raj et al., 2008). Briefly, sample coverslips were incubated with 1ml wash buffer (10% Deionized formamide, 2 X SSC in water) for 5 minutes at 25°C. Coverslips were depleted of excess wash buffer by holding coverslip edges upright against a kimwipe. A drop of hybridization solution was placed on a petri dish lined with parafilm (50 ul of 10% dextran sulfate, 10% Deionized formamide, 2 X SSC in water, 100 nM RNA-FISH each probe per sample). Coverslips were then placed by facing the sample sides down on hybridization solution. A kimwipe soaked with 1ml of water was placed inside the petri dish, and the petri dish edges were sealed with parafilm. The coverslips were incubated in a light tight incubator at 37°C for 4 hours. Samples were then placed back in 12 well dishes and washed twice with 1ml wash buffer for 30 minutes each at 37°C. Samples were rinsed with 2 X SSC and placed in 1 X PBS. Coverslips were air dried for 5 minutes and mounted on glass coverslips in Invitrogen Prolong Gold Mountant with DAPI P36941.
Imaging
Coverslips were imaged with a custom-built microscope equipped with a Hamamatsu ORCA-FLASH4.0 CMOS camera C11440, Lumencor Spectra X light source, Zeiss Objective Plan-Apochromat 40X/1.4 Oil DIC M27 420762-9900-000 and a custom quad band pass filter set for DAPI, FITC, CY3, CY5 designed Chroma Part No: 280948, 268285. Components were controlled through Micromanager (Edelstein et al., 2010). The Quasar570 probeset was excited with the Green 550/15 light source. The Quasar670 probeset was excited with the Red 640/30 light source. Z planes spanning 16 microns at 0.5micron intervals were acquired for each field of view. 25 fields were acquired for each sample.
RNA-FISH analysis
Maximum intensity projections were generated in Imagej/FIJI (Schindelin et al., 2012) and corrected for non-uniform field illumination. Cells were then segmented into cell and nucleus using CellProfiler (Carpenter et al., 2006). RNA spots were then identified as previously described (Lenstra et al., 2015; Thompson et al., 2002) by using custom IDL software. IDL software can be downloaded from: http://www.larsonlab.net/ After counted signal molecule number, get the natural log-transformed values for each single cell, the coefficients of variations were calculated using the following equation:
σ = standard deviation; μ = mean.
Data Analysis
Hi-C data and mapping
The paired-end reads of Hi-C libraries were mapped to the mouse reference genome (mm9) using bowtie (Langmead and Salzberg, 2012) and the PETs with the mapping quality >10 were kept, then the 3′-end of unmapped reads were iteratively trimmed by 5bp and realign the reads to the mouse genome until the reads are as short as 25bp, which could significantly improve the mappability (Imakaev et al., 2012). The PETs information was provided in supplemental table S1.
Correlation between biological repeats
The genome was split into the same sized bins. For selected bin i, the interaction density (number of reads linked the two bins) between the bin i and j were calculated. The bin j was one of the 50 closest bins around the bin i due to the high proximate of the interaction. Then the two interaction matrices were used to calculate the correlation between two libraries. The sizes of bins were set to 1Kb, 2Kb, 4Kb, 5Kb, 10Kb, 20Kb and 50Kb.
Topological domain and its boundary
The topological domains were identified based established method (Dixon et al., 2012). In short, we split the genome into 40Kb bins and calculated the upstream and downstream interaction bias in 2Mb range to obtain the directionality index (DI) of each bin. Then we implemented the Hidden Markov Model (HMM) on the directionality index to infer the topological domains and their boundaries. Two domain boundaries in two different libraries were considered shared only if the topological domain boundaries in two Hi-C libraries directly overlapped with each other.
Generation of interaction heatmap
To produce heatmaps, the genome was divided into 1Mb bins (later, 100Kb, 10Kb, 5Kb, 1Kb and so on) and each interaction was binned according to the location of both ends to produce the matrix M. It is assumed that roughly equal numbers of Hi-C reads should originate from each bin of equal size in the genome since 3e Hi-C is an unbiased assay covering the whole genome. However, different number of reads were observed in each bin due to the systematic bias including the number of restriction enzyme recognition sites, GC content and sequence uniqueness. We normalized the 3e Hi-C interaction matrix using the observed number of interactions divided by the expected number of reads. To calculate the expected Hi-C reads between two given regions, we used the following equation: . Where N is the total number of reads in the 3e Hi-C library and np and nq are total number of reads in region p and q. This formulation assumes that bin region has a uniform probability of interacting with any other region in the genome.
ChIP-Seq and peak calling
We produced the ChIP-Seq data for H3K27ac, H3K4me2, H3K4me3, CTCF, Cohesin (SMC1a) in Th2 cell and H3K27ac, H3K4me2, H3K4me3, CTCF, RNA Polymerase II data in EL4 cells. Then we collected the other histone modification and transcription factor ChIP-Seq datasets from a variety of publically available databases. The reads were mapped to the mouse genome mm9 using bowetie2 and the reads with the mapping quality >10 were kept. The peaks were called using MACS and the peaks with a p-value <1e-9 were kept.
RNA-Seq and cell-specific genes
The reads from RNA-Seq libraries were mapped to the mouse genome (mm9) using bowtie2 (Langmead and Salzberg, 2012). The gene expression level was measured by RPKM (Reads Per Kilobase per million mapped reads) and number of reads in a gene. The cell specific genes between ES cells and Th2 cells were identified using EdgeR (FDR < 0.05; Fold change > 1.5 or < 2/3) (Robinson et al., 2010).
Analysis of interaction around active epigenetic marks and transcription factor binding sites
We sorted the histone modification peaks based on their p-values from MACS and grouped them into 200 equal sized bins. We plotted the heatmap covering a region spanning 2Kb upstream and downstream of the summit of the peak (80 non-overlapping 50-bp window) using the reads density. The density was indicated by color, with green to red denoting low to high. Then we plotted the heatmap of interaction density based on the sorted and binned peaks (Figure 1A). We selected 9 active markers (H3K27ac, H3K4me1, H3K4me2, H3K4me3, p300, Gata3, Stat6, CTCF and Cohesin (SMC1a) to investigate whether more active markers could lead to high interaction. The analyses showed that regions with more markers have higher interaction density with other regions than those with fewer active markers (Figure S1F), indicating that very active regions involve more complex chromatin-chromatin interactions.
Identifying promoter-enhancer interaction
We treated regions +/− 1kb surrounding TSS of refSeq gene (mm9) as promoters. The regions +/− 1kb surrounding the summit of p300 binding sites were treated as the enhancers. We counted the all the PETs that linked any two of these regions (promoter-promoter, promoter-enhancers and enhancer-enhancer). Similar as the general chromatin interaction, the number of PETs that linked two regions should follow a hyper-geometric distribution in null hypothesis. We assumed there are N PETs linked any two different regions p and q, the number of PETs with one end located in region p and q are np and nq, respectively. We get the formula:
in which Ipq is the observed number of PETs linked region p and q. Multiple testing correction was performed and the interactions between two regions with FDR>0.05 and 2 PETs were kept for further analysis. Our method is similar to GotHiC method (Mifsud et al., 2017) except that we focused on interaction calls among regulatory regions.
Identifying the interactions between regulatory elements
We merged the binding sites of 9 markers (p300, H3K4me1, H3K4me2, H3K4me3, Gata3, Stat6, H3k27ac, CTCF and Cohesin (SMC1a) in Th2 to get a pool of regulatory elements. To determine whether two regulatory elements interacted with each other in the cell nucleus, a simple method is to count the number of PETs that linked the two regulatory element regions. Then we tested weather these interactions using the hyper-geometric distribution as above and only keep the interaction linked by at least 2 PETs.
GO Enrichment analysis
To examine whether particular gene categories/pathways were enriched in certain gene lists, the GO enrichment analysis were performed using DVAID (Huang da et al., 2009). The GO categories with FDR<0.05 were consider as significant.
The calculated of Coefficient of variation
In order to calculate the Coefficient of variations for RNA-fish data and FACS data, the counts of mRNA copies were natural-log transformed with a pseudo count of one to avoid infinite value for each single cell for every RNA-fish data, and the values for the APC signal were log10 transformed for each single cell for all the FACS data. Then CV was calculated by standard deviation (σ) dividing by mean (μ) for each sample. P-value were calculated by paired t-test for FACS data.
Quantification and Statistical Analysis
Three independent experiments in flow cytometry data shown in Figure 2. Data shows average of two independent experiments and error bars indicate S.E.M. in Figure 3B, C, D and Figure 4B, C, D. Two independent batches of samples were used for data reported in Figure S2 E, and Figure S2 G. Data show average of two independent experiments and are represented as mean ± SEM for Figure S2 H, Figure S3 C, D, E, Figure S4 C, D, and figure S5 B, C, D. *indicates p < 0.05, **indicates p < 0.01.
Data Availability
Data Resources
All softwares used in this study are listed in the Key Resources Table, all the data in this manuscript have been deposited in the NCBI database (GEO: GSE66343) and can be accessed: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=mpqdykumjpgpbin&acc=GSE66343.
Supplementary Material
Supplemental Figure S1. Related to Figure 1. Three enzyme Hi-C (3e Hi-C) is a reliable method to detect genome wide chromatin interaction.
Supplemental Figure S2. Related to Figure 2. Knockdown of CTCF results in decreased expression of some T lineage-specific genes and increased cell-to-cell variation of expression in mouse EL4 cells.
Supplemental Figure S3. Related to Figure 3 and Figure 4. CTCF binding site at the Cd5 gene locus contributes to the functional interaction between Cd5 promoter with its enhancers and the expression noise control.
Supplemental Figure S4. Related to Figure 3 and Figure 4. CTCF binding sites at the Runx3, and Thy1 gene loci contribute to the functional interaction between promoters with their enhancers and noise control of gene expression.
Supplemental Table S1. Related to Figure 1 and Figure 2. Statistics of 3e Hi-C libraries, public data used the study, the highest expressed genes in Th2 cells, list of Oligonucleotides used in the study, the most interactive genes in Th2 cells, down-regulated and up-regulated genes in CTCF knockdown EL4 cells.
Supplemental Table S2. Related to Figure 3 and Figure 4. FACS data of CD5, CD90, Gata3 and CD28 in CTCF knockdown and control EL4 cells, FACS data of CD90 in CRISPR deletion cells, single-cell RNA-FISH data in CRISPR deletion EL4 cells.
Highlights.
3e Hi-C is a robust technique to measure genome-wide chromatin interactions.
CTCF binding correlates with enhancer activity and enhancer-promoter interaction.
CTCF stabilizes enhancer-promoter interaction and maintains robust gene expression.
Deletion of CTCF binding sites increases cell-to-cell variation of gene expression.
Acknowledgments
We thank Dr. Benjamin Stanton for critical reading of the manuscript and Timothy Zhou for setting up the WashU genome browser in lab for visualizing 3e Hi-C data. PX459 was a gift from Dr. Kai Ge. The next generation sequencing of the libraries was performed by the NHLBI DNA Sequencing and Genomics Core facility; cell sorting was performed by the NHLBI Flow Cytometry Core facility. The work was supported by Division of Intramural Research, National Heart, Lung and Blood Institute, National Institutes of Health.
Footnotes
Author Contribution
“G. R., K.C., and J. R. performed the experiments and W.J., G.H. and J.R. analyzed the data. D.R.L. and K.Z. supervised the study. K.Z. conceived the study and wrote the manuscript with inputs from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aranda-Anzaldo A, Dent MA. Developmental noise, ageing and cancer. Mechanisms of ageing and development. 2003;124:711–720. doi: 10.1016/s0047-6374(03)00089-7. [DOI] [PubMed] [Google Scholar]
- Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- Blake WJ, M KA, Cantor CR, Collins JJ. Noise in eukaryotic gene expression. Nature. 2003;422:633–637. doi: 10.1038/nature01546. [DOI] [PubMed] [Google Scholar]
- Capp JP. Stochastic gene expression, disruption of tissue averaging effects and cancer as a disease of development. BioEssays: news and reviews in molecular, cellular and developmental biology. 2005;27:1277–1285. doi: 10.1002/bies.20326. [DOI] [PubMed] [Google Scholar]
- Carey LB, van Dijk D, Sloot PM, Kaandorp JA, Segal E. Promoter sequence determines the relationship between expression level and noise. PLoS biology. 2013;11:e1001528. doi: 10.1371/journal.pbio.1001528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpenter AE, Jones TR, Lamprecht MR, Clarke C, Kang IH, Friman O, Guertin DA, Chang JH, Lindquist RA, Moffat J, et al. CellProfiler: image analysis software for identifying and quantifying cell phenotypes. Genome biology. 2006;7:R100. doi: 10.1186/gb-2006-7-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chepelev I, Wei G, Tang Q, Zhao K. Detection of single nucleotide variations in expressed exons of the human genome using RNA-Seq. Nucleic acids research. 2009;37:e106. doi: 10.1093/nar/gkp507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulon A, Chow CC, Singer RH, Larson DR. Eukaryotic transcriptional dynamics: from single molecules to cell populations. Nat Rev Genet. 2013;14:572–584. doi: 10.1038/nrg3484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuddapah S, Jothi R, Schones DE, Roh TY, Cui K, Zhao K. Global analysis of the insulator binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome research. 2009;19:24–32. doi: 10.1101/gr.082800.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, Hu M, Liu JS, Ren B. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485:376–380. doi: 10.1038/nature11082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowen JM, Fan ZP, Hnisz D, Ren G, Abraham BJ, Zhang LN, Weintraub AS, Schuijers J, Lee TI, Zhao K, et al. Control of cell identity genes occurs in insulated neighborhoods in Mammalian chromosomes. Cell. 2014;159:374–387. doi: 10.1016/j.cell.2014.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Z, Andronescu M, Schutz K, McIlwain S, Kim YJ, Lee C, Shendure J, Fields S, Blau CA, Noble WS. A three-dimensional model of the yeast genome. Nature. 2010;465:363–367. doi: 10.1038/nature08973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edelstein A, Amodaj N, Hoover K, Vale R, Stuurman N. Computer control of microscopes using micro Manager. Current protocols in molecular biology/edited by Frederick M Ausubel [et al] 2010;Chapter 14(Unit14):20. doi: 10.1002/0471142727.mb1420s92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eldar A, Elowitz MB. Functional roles for noise in genetic circuits. Nature. 2010;467:167–173. doi: 10.1038/nature09326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott JI, Festenstein R, Tolaini M, Kioussis D. Random activation of a transgene under the control of a hybrid hCD2 locus control region/Ig enhancer regulatory element. The EMBO journal. 1995;14:575–584. doi: 10.1002/j.1460-2075.1995.tb07033.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faure AJ, Schmidt D, Watt S, Schwalie PC, Wilson MD, Xu H, Ramsay RG, Odom DT, Flicek P. Cohesin regulates tissue-specific expression by stabilizing highly occupied cis-regulatory modules. Genome research. 2012;22:2163–2175. doi: 10.1101/gr.136507.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Xu Q, Canzio D, Shou J, Li J, Gorkin DU, Jung I, Wu H, Zhai Y, Tang Y, et al. CRISPR Inversion of CTCF Sites Alters Genome Topology and Enhancer/Promoter Function. Cell. 2015;162:900–910. doi: 10.1016/j.cell.2015.07.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cui K, Northrup D, Liu C, Wang C, Tang Q, Ge K, Levens D, Crane-Robinson C, Zhao K. H2A. Z facilitates access of active and repressive complexes to chromatin in embryonic stem cell self-renewal and differentiation. Cell stem cell. 2013;12:180–192. doi: 10.1016/j.stem.2012.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Imakaev M, Fudenberg G, McCord RP, Naumova N, Goloborodko A, Lajoie BR, Dekker J, Mirny LA. Iterative correction of Hi-C data reveals hallmarks of chromosome organization. Nat Methods. 2012;9:999–1003. doi: 10.1038/nmeth.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji N, Middelkoop TC, Mentink RA, Betist MC, Tonegawa S, Mooijman D, Korswagen HC, van Oudenaarden A. Feedback control of gene expression variability in the Caenorhabditis elegans Wnt pathway. Cell. 2013;155:869–880. doi: 10.1016/j.cell.2013.09.060. [DOI] [PubMed] [Google Scholar]
- Jin W, Tang Q, Wan M, Cui K, Zhang Y, Ren G, Ni B, Sklar J, Przytycka TM, Childs R, et al. Genome-wide detection of DNase I hypersensitive sites in single cells and FFPE tissue samples. Nature. 2015;528:142–146. doi: 10.1038/nature15740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalmar T, Lim C, Hayward P, Munoz-Descalzo S, Nichols J, Garcia-Ojalvo J, Martinez Arias A. Regulated fluctuations in nanog expression mediate cell fate decisions in embryonic stem cells. PLoS biology. 2009;7:e1000149. doi: 10.1371/journal.pbio.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Abdullaev ZK, Smith AD, Ching KA, Loukinov DI, Green RD, Zhang MQ, Lobanenkov VV, Ren B. Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell. 2007;128:1231–1245. doi: 10.1016/j.cell.2006.12.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson DR, Fritzsch C, Sun L, Meng X, Lawrence DS, Singer RH. Direct observation of frequency modulated transcription in single cells using light activation. Elife. 2013;2:e00750. doi: 10.7554/eLife.00750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenstra TL, Coulon A, Chow CC, Larson DR. Single-Molecule Imaging Reveals a Switch between Spurious and Functional ncRNA Transcription. Molecular cell. 2015;60:597–610. doi: 10.1016/j.molcel.2015.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science. 2009;326:289–293. doi: 10.1126/science.1181369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maamar H, Raj A, Dubnau D. Noise in gene expression determines cell fate in Bacillus subtilis. Science. 2007;317:526–529. doi: 10.1126/science.1140818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metivier R, Penot G, Hubner MR, Reid G, Brand H, Kos M, Gannon F. Estrogen receptor-alpha directs ordered, cyclical, and combinatorial recruitment of cofactors on a natural target promoter. Cell. 2003;115:751–763. doi: 10.1016/s0092-8674(03)00934-6. [DOI] [PubMed] [Google Scholar]
- Mifsud B, Martincorena I, Darbo E, Sugar R, Schoenfelder S, Fraser P, Luscombe NM. GOTHiC, a probabilistic model to resolve complex biases and to identify real interactions in Hi-C data. PLoS One. 2017;12:e0174744. doi: 10.1371/journal.pone.0174744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy KF, Adams RM, Wang X, Balazsi G, Collins JJ. Tuning and controlling gene expression noise in synthetic gene networks. Nucleic acids research. 2010;38:2712–2726. doi: 10.1093/nar/gkq091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols MH, Corces VG. A CTCF Code for 3D Genome Architecture. Cell. 2015;162:703–705. doi: 10.1016/j.cell.2015.07.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Goloborodko A, Valton AL, Gibcus JH, Uebersohn A, Abdennur N, Dekker J, Mirny LA, Bruneau BG. Targeted Degradation of CTCF Decouples Local Insulation of Chromosome Domains from Genomic Compartmentalization. Cell. 2017;169:930–944. e922. doi: 10.1016/j.cell.2017.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nora EP, Lajoie BR, Schulz EG, Giorgetti L, Okamoto I, Servant N, Piolot T, van Berkum NL, Meisig J, Sedat J, et al. Spatial partitioning of the regulatory landscape of the X-inactivation centre. Nature. 2012;485:381–385. doi: 10.1038/nature11049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parelho V, Hadjur S, Spivakov M, Leleu M, Sauer S, Gregson HC, Jarmuz A, Canzonetta C, Webster Z, Nesterova T, et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Phillips-Cremins JE, Sauria ME, Sanyal A, Gerasimova TI, Lajoie BR, Bell JS, Ong CT, Hookway TA, Guo C, Sun Y, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153:1281–1295. doi: 10.1016/j.cell.2013.04.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, Rifkin SA, Andersen E, van Oudenaarden A. Variability in gene expression underlies incomplete penetrance. Nature. 2010;463:913–918. doi: 10.1038/nature08781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raj A, van den Bogaard P, Rifkin SA, van Oudenaarden A, Tyagi S. Imaging individual mRNA molecules using multiple singly labeled probes. Nat Methods. 2008;5:877–879. doi: 10.1038/nmeth.1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rao SS, Huntley MH, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, Sanborn AL, Machol I, Omer AD, Lander ES, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159:1665–1680. doi: 10.1016/j.cell.2014.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc Natl Acad Sci U S A. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik W. OLIGO 7 primer analysis software. Methods in molecular biology. 2007;402:35–60. doi: 10.1007/978-1-59745-528-2_2. [DOI] [PubMed] [Google Scholar]
- Sanborn AL, Rao SS, Huang SC, Durand NC, Huntley MH, Jewett AI, Bochkov ID, Chinnappan D, Cutkosky A, Li J, et al. Chromatin extrusion explains key features of loop and domain formation in wild-type and engineered genomes. Proc Natl Acad Sci U S A. 2015;112:E6456–6465. doi: 10.1073/pnas.1518552112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasagawa Y, Nikaido I, Hayashi T, Danno H, Uno KD, Imai T, Ueda HR. Quartz-Seq: a highly reproducible and sensitive single-cell RNA sequencing method, reveals non-genetic gene-expression heterogeneity. Genome biology. 2013;14:R31. doi: 10.1186/gb-2013-14-4-r31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nat Methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz YB, Linder-Basso D, Kharchenko PV, Tolstorukov MY, Kim M, Li HB, Gorchakov AA, Minoda A, Shanower G, Alekseyenko AA, et al. Nature and function of insulator protein binding sites in the Drosophila genome. Genome research. 2012;22:2188–2198. doi: 10.1101/gr.138156.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seitan VC, Faure AJ, Zhan Y, McCord RP, Lajoie BR, Ing-Simmons E, Lenhard B, Giorgetti L, Heard E, Fisher AG, et al. Cohesin-based chromatin interactions enable regulated gene expression within preexisting architectural compartments. Genome research. 2013;23:2066–2077. doi: 10.1101/gr.161620.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, Parrinello H, Tanay A, Cavalli G. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148:458–472. doi: 10.1016/j.cell.2012.01.010. [DOI] [PubMed] [Google Scholar]
- Shalek AK, Satija R, Shuga J, Trombetta JJ, Gennert D, Lu D, Chen P, Gertner RS, Gaublomme JT, Yosef N, et al. Single-cell RNA-seq reveals dynamic paracrine control of cellular variation. Nature. 2014;510:363–369. doi: 10.1038/nature13437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma SV, Lee DY, Li B, Quinlan MP, Takahashi F, Maheswaran S, McDermott U, Azizian N, Zou L, Fischbach MA, et al. A chromatinmediated reversible drug-tolerant state in cancer cell subpopulations. Cell. 2010;141:69–80. doi: 10.1016/j.cell.2010.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigova AA, Abraham BJ, Ji X, Molinie B, Hannett NM, Guo YE, Jangi M, Giallourakis CC, Sharp PA, Young RA. Transcription factor trapping by RNA in gene regulatory elements. Science. 2015;350:978–981. doi: 10.1126/science.aad3346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sofueva S, Yaffe E, Chan WC, Georgopoulou D, Vietri Rudan M, Mira-Bontenbal H, Pollard SM, Schroth GP, Tanay A, Hadjur S. Cohesin-mediated interactions organize chromosomal domain architecture. The EMBO journal. 2013;32:3119–3129. doi: 10.1038/emboj.2013.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang Z, Luo OJ, Li X, Zheng M, Zhu JJ, Szalaj P, Trzaskoma P, Magalska A, Wlodarczyk J, Ruszczycki B, et al. CTCF-Mediated Human 3D Genome Architecture Reveals Chromatin Topology for Transcription. Cell. 2015;163:1611–1627. doi: 10.1016/j.cell.2015.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanizawa H, Iwasaki O, Tanaka A, Capizzi JR, Wickramasinghe P, Lee M, Fu Z, Noma K. Mapping of long-range associations throughout the fission yeast genome reveals global genome organization linked to transcriptional regulation. Nucleic acids research. 2010;38:8164–8177. doi: 10.1093/nar/gkq955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson RE, Larson DR, Webb WW. Precise nanometer localization analysis for individual fluorescent probes. Biophysical journal. 2002;82:2775–2783. doi: 10.1016/S0006-3495(02)75618-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Abraham BJ, Yagi R, Jothi R, Cui K, Sharma S, Narlikar L, Northrup DL, Tang Q, Paul WE, et al. Genome-wide analyses of transcription factor GATA3-mediated gene regulation in distinct T cell types. Immunity. 2011;35:299–311. doi: 10.1016/j.immuni.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei G, Wei L, Zhu J, Zang C, Hu-Li J, Yao Z, Cui K, Kanno Y, Roh TY, Watford WT, et al. Global mapping of H3K4me3 and H3K27me3 reveals specificity and plasticity in lineage fate determination of differentiating CD4+ T cells. Immunity. 2009;30:155–167. doi: 10.1016/j.immuni.2008.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z, Wei G, Chepelev I, Zhao K, Felsenfeld G. Mapping of INS promoter interactions reveals its role in long-range regulation of SYT8 transcription. Nature structural & molecular biology. 2011;18:372–378. doi: 10.1038/nsmb.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaffe E, Tanay A. Probabilistic modeling of Hi-C contact maps eliminates systematic biases to characterize global chromosomal architecture. Nat Genet. 2011;43:1059–1065. doi: 10.1038/ng.947. [DOI] [PubMed] [Google Scholar]
- Yuan P, Ito K, Perez-Lorenzo R, Del Guzzo C, Lee JH, Shen CH, Bosenberg MW, McMahon M, Cantley LC, Zheng B. Phenformin enhances the therapeutic benefit of BRAF(V600E) inhibition in melanoma. Proc Natl Acad Sci U S A. 2013;110:18226–18231. doi: 10.1073/pnas.1317577110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuin J, Dixon JR, van der Reijden MI, Ye Z, Kolovos P, Brouwer RW, van de Corput MP, van de Werken HJ, Knoch TA, van IWF, et al. Cohesin and CTCF differentially affect chromatin architecture and gene expression in human cells. Proc Natl Acad Sci U S A. 2014;111:996–1001. doi: 10.1073/pnas.1317788111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Related to Figure 1. Three enzyme Hi-C (3e Hi-C) is a reliable method to detect genome wide chromatin interaction.
Supplemental Figure S2. Related to Figure 2. Knockdown of CTCF results in decreased expression of some T lineage-specific genes and increased cell-to-cell variation of expression in mouse EL4 cells.
Supplemental Figure S3. Related to Figure 3 and Figure 4. CTCF binding site at the Cd5 gene locus contributes to the functional interaction between Cd5 promoter with its enhancers and the expression noise control.
Supplemental Figure S4. Related to Figure 3 and Figure 4. CTCF binding sites at the Runx3, and Thy1 gene loci contribute to the functional interaction between promoters with their enhancers and noise control of gene expression.
Supplemental Table S1. Related to Figure 1 and Figure 2. Statistics of 3e Hi-C libraries, public data used the study, the highest expressed genes in Th2 cells, list of Oligonucleotides used in the study, the most interactive genes in Th2 cells, down-regulated and up-regulated genes in CTCF knockdown EL4 cells.
Supplemental Table S2. Related to Figure 3 and Figure 4. FACS data of CD5, CD90, Gata3 and CD28 in CTCF knockdown and control EL4 cells, FACS data of CD90 in CRISPR deletion cells, single-cell RNA-FISH data in CRISPR deletion EL4 cells.
Data Availability Statement
Data Resources
All softwares used in this study are listed in the Key Resources Table, all the data in this manuscript have been deposited in the NCBI database (GEO: GSE66343) and can be accessed: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=mpqdykumjpgpbin&acc=GSE66343.