Abstract
Site-directed spin labeling (SDSL) in combination with electron paramagnetic resonance (EPR) spectroscopy is a rapidly expanding powerful biophysical technique to study the structural and dynamic properties of membrane proteins in a native environment. Membrane proteins are responsible for performing important functions in a wide variety of complicated biological systems that are responsible for the survival of living organisms. In this review, a brief introduction of the most popular SDSL EPR techniques and illustrations of recent applications for studying pertinent structural and dynamic properties on membrane proteins will be discussed.
1. Introduction
1.1. Site-Directed Spin Labeling EPR
Earlier biological EPR applications were limited to metalloproteins possessing paramagnetic centers or enzymes with radical cofactors. The absence of unpaired electrons in most biological materials would appear to minimize the application of EPR methods. Molecular biology techniques have been developed to incorporate stable radicals at specific locations on biological systems extending the application of EPR spectroscopy to nearly any biological system. These techniques are called spin labeling. The site-specific introduction of unpaired electrons into biomolecules in the form of spin labels is known as site-directed spin labeling (SDSL) [1, 2]. In SDSL experiments, all native nondisulfide bonded cysteines are removed by replacing them with another amino acid such as an alanine or serine. A unique cysteine residue is then introduced into a recombinant protein using site-directed mutagenesis and subsequently reacted with a sulfhydryl-specific nitroxide reagent to generate a stable spin label side-chain [2–4].
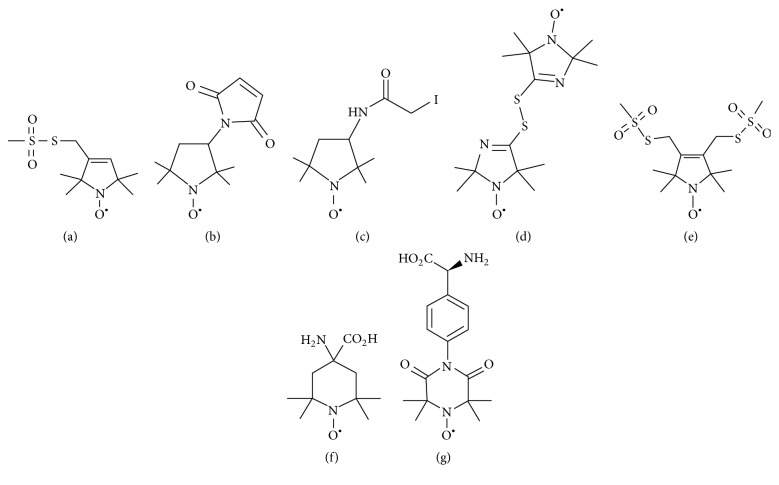
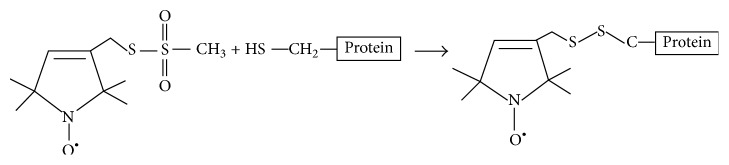
Figure 1 shows the chemical structure of several nitroxide based spin label probes used for EPR spectroscopic studies of biomolecules [5–15]. The spin label probes in Figures 1(a)–1(e) are incorporated using site-directed mutagenesis while spin probes in Figures 1(f)-1(g) are incorporated using solid phase peptide synthesis. A resulting side-chain produced by reaction of the most commonly used spin label, methanethiosulfonate spin label (MTSL), with the cysteine residue of the protein is shown in Figure 2 [8]. We refer a recent book chapter authored by Haugland et al. for details of nitroxide spin labels used for SDSL EPR spectroscopy [6].
Figure 1.
Structure of nitroxide spin labels used in the SDSL EPR study of micromolecules. (a) Methanethiosulfonate spin label (MTSL), (b) maleimide spin label (MSL) N-(1-oxyl-2,2,6,6-tetramethyl-4-piperidinyl) maleimide, (c) iodoacetamide spin label (ISL), (d) bis(1-oxyl-2,2,5,5-tetramethyl-3-imidazolin-4-yl) disulfide (IDSL), (e) bifunctional spin label (BSL), (f) 2,2,6,6-tetramethyl-N-oxyl-4-amino-4-carboxylic acid (TOAC), and (g) 4-(3,3,5,5-tetramethyl-2,6-dioxo-4-oxylpiperazin-1-yl)-l-phenylglycine (TOPP).
Figure 2.
Structure of MTSL (methanethiosulfonate spin label) and the resulting side-chain produced by reaction with a cysteine residue on a protein.
1.2. Origin of Spin Label EPR Spectrum
EPR spectroscopy measures the absorption of microwave radiation corresponding to the energy splitting of an unpaired electron when it is placed in a strong magnetic field. Therefore, an EPR active sample requires the presence of an unpaired electron spin. The simplest EPR active system consists of a single unpaired electron spin residing in a molecular orbital. The electron possesses a magnetic moment and spin quantum number S = 1/2, with magnetic spin components Ms = +1/2 and Ms = −1/2. In the absence of a static magnetic field, these two states are degenerated and have the same energy. However, when an external magnetic field (B0) is applied, the magnetic moment of electron aligns itself either parallel (ms = −1/2) or antiparallel (ms = +1/2) to the field having a specific energy to each alignment. The parallel alignment corresponds to the lower energy state and the antiparallel alignment corresponds to the higher energy state as shown in Figure 3. The energy separation between the lower and the upper state is given by
| (1) |
where ge is the electron's so-called g-factor which varies depending on the electronic configuration of the radical or ion that is similar to a chemical shift parameter in NMR and β is the electron Bohr magneton. The above equation (1) implies that splitting of energy levels is proportional to the magnetic field B0 strength as shown in Figure 3. An unpaired electron spin can flip between the two energy levels by absorbing microwave radiation of energy hν, obeying the fundamental equation of EPR spectroscopy [16]:
| (2) |
where h is Planck's constant and ν is the frequency of the microwave radiation.
Figure 3.
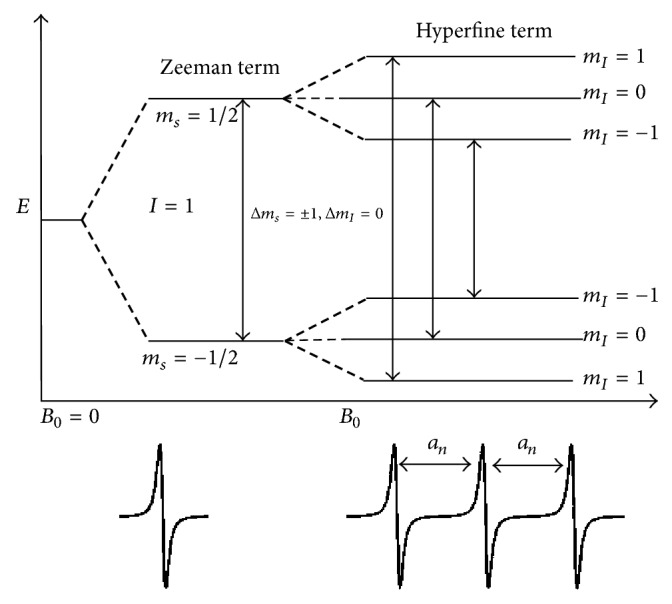
Energy level diagram of a nitroxide spin label in the presence of a magnetic field B0. The lower panel shows the corresponding EPR spectra in the absence and presence of a 14N (I = 1) hyperfine interaction.
In a typical continuous wave- (CW-) EPR experiment, a fixed microwave frequency is applied and the magnetic field B0 is varied. An EPR transition occurs when the energy separation between the two electron spin states matches the constant microwave frequency (2). This phenomenon is known as resonance [16]. In addition to varying B0, the field is also modulated to improve the signal to noise of the spectra. This gives rise to the derivative lineshape typically observed in most EPR spectra. The EPR derivative spectrum is shown in the lower panel of Figure 3. The magnetic field at which this signal appears depends on the g-value, which dictates the slope at which the energy levels for the two spin states change as a function of the magnetic field. In most biological systems, the g-value is anisotropic with an orientation dependence meaning that the effective g-value is different depending on the orientation of the molecule with respect to the applied magnetic field.
If the free electron does not interact with the nearby nuclei, only one line is observed in the EPR spectrum (lower left panel in Figure 3). But for nitroxide spin labels used in most biological studies, the unpaired electron interacts primarily with the nitrogen nucleus (14N, nuclear spin (I) = 1). This interaction is called the hyperfine interaction (A) and depends on the amount of electron spin density on the nucleus, the distance between the electron spin and the nucleus, and the angle between the two with respect to the magnetic field (B0). This hyperfine interaction produces a small change in the allowed energy levels of the electrons and splits the EPR lines into multiple lines depending on the nuclear spin state. The hyperfine splitting due to coupling of an electron to a single 14N nucleus (I = 1) is shown in Figure 3. Other parameters that can influence an EPR spectrum are electron-electron couplings between two sets of spins that can provide valuable distance information in biological systems.
EPR spectroscopy is a very sensitive technique providing up to three orders of magnitude higher sensitivity when compared to nuclear magnetic resonance (NMR) spectroscopy. It can be applied to any size protein without relying on expensive isotopic labels [17]. It is not influenced by the optical properties of the sample. EPR experiments can be performed on a wide range of samples from proteins in solution to highly packed membrane suspensions, tissue samples, ammonium sulfate-precipitated solids, or samples frozen and maintained at cryogenic temperatures [18]. EPR experiments can be conducted at low volume and concentration (~70 nanoliter to several mL sample or even small animals [19, 20]). EPR spectroscopy can answer pertinent structural and dynamic questions related to both solution and membrane bound proteins that are very challenging to be obtained by traditional biophysical methods [21–24]. CW-EPR spectroscopy of spin labeled molecules reveals structural and dynamic information about the motion of the nitroxide side-chain, solvent accessibility, solvent polarity, and intra- or intermolecular distances between two nitroxides or a single nitroxide and another paramagnetic center in the system [3, 8, 15, 22, 25]. The lineshape analysis of the EPR data for a series of spin labeled protein sequences can probe the structural properties of the protein at backbone level spatial resolution [26–29].
1.3. Membrane Proteins
Membrane proteins are responsible for the exchange of signals and physical materials across the membrane and play an essential role in different aspects of cellular activities [30, 31]. Membrane proteins comprise 30% of sequenced genes [32–34]. Mutations in genes and misfolding of membrane proteins are associated with numerous human dysfunctions, disorders, and diseases [35, 36]. Approximately, half of all the FDA approved drugs target membrane proteins [37, 38]. Detailed structural and dynamic information for membrane proteins are vital for understanding intermolecular interactions, protein functions, and regulation [8, 22, 39]. Despite the abundance and clear importance of membrane proteins, very limited knowledge about these systems exists [40, 41]. Membrane proteins can interact with a lipid bilayer in various different fashions or orientations to maintain functional stability. The membrane interacting protein helices may be of varied length or curved in the middle of the membrane bilayer. They may lie flat on membrane surface, cross the membrane at different angles, or form reentrant loops.
X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy are the two most successful and popular biophysical techniques used to probe structural information on protein systems. NMR spectroscopy is also used to obtain dynamic information for a variety of biological systems. Solution NMR can be used to probe the protein structure in a physiologically relevant condition; however, a larger size protein (>~50 kD) is difficult to study using this technique [5, 42–44]. NMR structural studies on membrane proteins are also difficult due to the size of the micelle complex and higher spectral linewidth [45]. X-ray crystallography provides highly resolved structural information but cannot provide detailed dynamic information [46]. In addition, the hydrophobic nature of membrane protein often complicates the process of crystallization, introducing challenges for X-ray crystallographic techniques for studying many membrane proteins [43]. EPR spectroscopy is a rapidly expanding and powerful biophysical technique to resolve these challenges and provides prominent solutions to glean structural and dynamic information on peptides, proteins, macromolecules, and nucleic acids [3, 8, 12, 17, 21–23, 47–50].
2. Application of SDSL EPR Techniques for Studying Membrane Proteins
SDSL in combination with EPR spectroscopy has been widely used to study membrane proteins. This is a very wide topic, which will be discussed in an introductory fashion with recent illustrations in the following sections. For more in-depth information, the following are excellent reviews [8, 12, 15, 22, 24, 54].
2.1. Membrane Protein Dynamics and Topology
The flexible nature of the MTSL nitroxide spin label provides its reorientation motion highly dependent on neighboring side-chains and secondary structure components in its immediate environment and hence can report local structure of the protein. The lineshape of the room temperature EPR spectra reflects the mobility of the spin label side-chain and its relation to protein structures. Spin labeled sites exposed to bulk water show reorientational correlation times of spin label side-chains resulting in very sharp EPR spectral peaks with small linewidths of the central lines. On the contrary, a spin label with very slow motion will be in the rigid limit of the spectrum [55]. In the rigid limit, sample is frozen and the full orientation-dependent parameters are observed. For systems in which the spin label movement falls between these two regions, dynamic properties of the spin label located at the specific site can be described in the terms of a correlation time (τc) [55]. The overall mobility of the nitroxide spin label attached to the protein or peptide is a superposition of the contributions from the motion of the label relative to the peptide backbone, fluctuations of the α-carbon backbone, and the rotational motion of the entire protein or peptide. Under experimental conditions, these motions can be isolated from the EPR spectrum. The inverse linewidth of the central line of the EPR spectrum provides a measure of relative mobility [22, 25, 51, 56]. A plot of the inverse linewidth mobility against the amino acid sequence can produce a periodic data profile, which can be used to predict the local secondary structure of the proteins and peptides [8, 22, 51, 57].
The changes in the spin label mobility can be used to investigate the peptide binding to the membrane [54, 58]. In the solvent phase, a spin labeled peptide or small protein rapidly tumbling leads to an isotropic spectrum with a rotational correlation time of less than nanosecond. However, in a membrane, spin labeled peptides experience restricted mobility, resulting in a broader EPR spectrum with two motional components resulting from the superposition of the signals arising from a free and bound peptide [51, 54, 59–61]. Protein topology in a membrane can be studied with respect to the membrane using nitroxide based SDSL EPR power saturation experiments [8, 60–62]. This method can also be used to identify functional domains in membrane proteins [46]. There are several biologically important membrane proteins such as the prokaryotic potassium channel KcsA, KCNE1, lactose permease protein, integrin β1a, C99 domain of the amyloid precursor protein, bacteriorhodopsin, and KvAP voltage-sensing domain that have been studied in a membrane environment using nitroxide based SDSL CW-EPR spectroscopy to probe the structural topology and dynamic properties [51, 56, 61, 63–67].
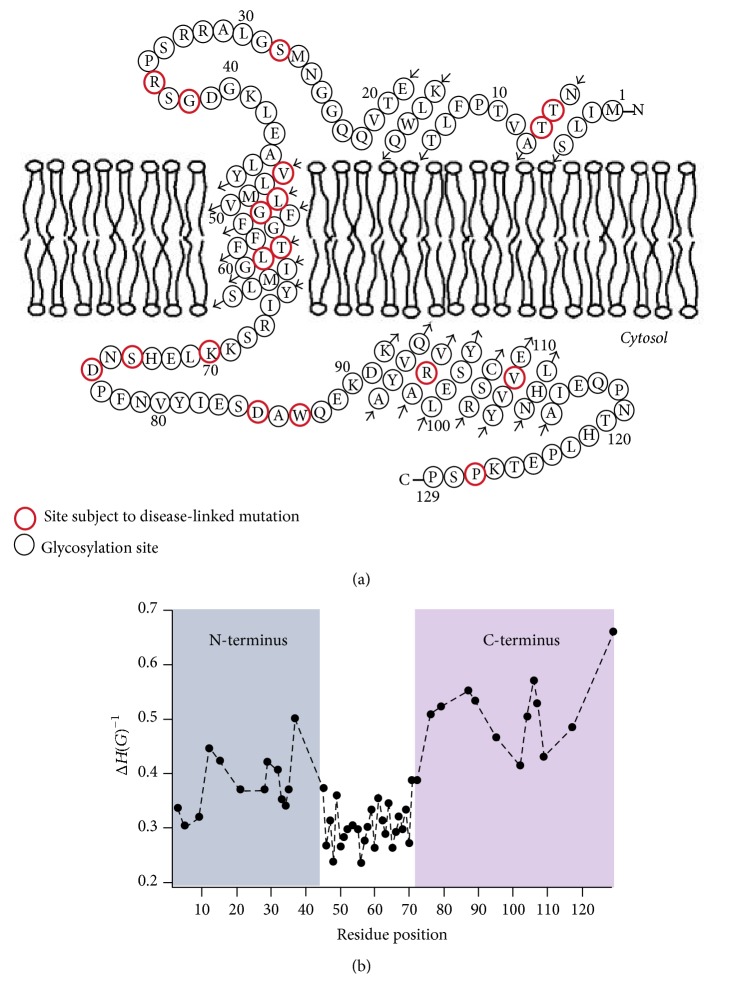
Site-directed spin labeling CW-EPR spectroscopy was recently used to extensively investigate the structural topology and dynamics of KCNE1 in proteoliposomes [51, 68]. KCNE1 is a single pass integral membrane protein which is very important for modulating the functional activities of a voltage gated potassium ion channel (Kv). It is essential for the cardiac action potential that mediates a heartbeat as well as the potassium ion homeostasis in the inner ear. CW-EPR lineshape analysis was performed on 53 sites of spin labeled KCNE1 including all 27 residues of the transmembrane domain (45–71), and 26 residues of the N- and C-termini of KCNE1 in lipid bilayered vesicles to study the nitroxide side-chain motion. The results indicated that the nitroxide spin label side-chains located in the KCNE1 TMD are less mobile when compared to the extracellular region of KCNE1. The EPR data further revealed that the C-terminus of KCNE1 is more mobile when compared to the N-terminus. EPR power saturation data obtained on 41 sites of spin labeled KCNE1 were used to determine the topology of KCNE1 with respect to the 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) lipid bilayers. Also, the data showed that the transmembrane domain is spanning the width of the lipid bilayer, while the extracellular region of KCNE1 is solvent-exposed with some of the portions partially or weakly interacting with the membrane surface. Figure 4 shows the proposed topology and the spin label side-chain mobility of the KCNE1 sequence in lipid bilayers. The CW-EPR data obtained on KCNE1 in various environments (i.e., 1-myristoyl-2-hydroxy-sn-glycero-3-phospho-(1′-rac-glycerol) (sodium salt) (LMPG) micelles, (POPC)/(POPG) liposomes, and POPC/POPG lipodisq nanoparticles) further suggested that the KCNE1 undergoes multiple conformations while interacting with lipid bilayers [51, 53, 68].
Figure 4.
(a) The proposed topology of the KCNE1 sequence in lipid bilayers. (b) Plot of inverse central EPR resonance linewidth (mI = 0) as a function of residue position of KCNE1 in lipid bilayers (adapted from [51] with permission).
Another recent example of using nitroxide spin labeling CW-EPR spectroscopy is a study on the functional amyloid Obr2A [58]. Obr2A is an isoform of Orb2, having a unique N-terminus domain important for the formation of amyloid-like aggregates and long-term memory in vivo. Soria et al. performed CW-EPR lineshape analysis on several spin labeled sites of Orb2A1-88 in the presence of various lipid concentration to determine the spin label side-chain mobility. The results revealed the increased rigidity of N-terminus of the protein with the increased concentration of lipid vesicles. Their CW-EPR data further revealed that the Orb2A1-88 membrane binding depends on the membrane curvature.
Nitroxide based SDSL CW-EPR spectroscopy at X-band can also be used to study membrane topology of membrane proteins/peptides bound to aligned phospholipid bilayers [69–72]. An excellent example of recent work using this method is the study of phospholamban [72]. McCaffrey et al. applied a bifunctional spin label and X-band EPR spectroscopy on monomeric phospholamban (PLB) to determine the protein structural topology in magnetically aligned bicelles. Phospholamban (PLB) is a single pass integral membrane protein that regulates the cardiac sarcoplasmic reticulum Ca-ATPase (SERCA). The result of this study suggested that the EPR spectra of a bifunctional spin label can be used to accurately determine the orientation and rotational dynamics of an α-helical segment of an integral membrane protein in magnetically aligned bicelles. Recently, the Lorigan lab determined accurate helical tilt angle and dynamics of the AchR M2δ peptide by utilizing the magnetically aligned bicelles EPR technique and multiple TOAC labeled peptide substitutions [71].
2.2. Local Secondary Structure of Membrane Proteins
The assembly, packing, and interaction of membrane proteins with its lipid environment are largely affected by the local secondary structure of membrane proteins. Better information on the local secondary structure is essential for understanding the function, dynamics, and interacting mode of membrane proteins [73, 74]. Nitroxide based SDSL electron spin echo envelope modulation (ESEEM) spectroscopy is a very powerful pulsed EPR spectroscopic technique to measure distances up to 8 Å between a spin label and a single NMR active isotopic nucleus such as deuterium (2H) [75, 76]. It has been used to investigate penetration of water into membranes, localization of proteins, or lipids in lipid membranes [77–81]. Recently SDSL ESEEM spectroscopy has been utilized to directly probe the local secondary structure of membrane proteins/peptides in aqueous as well as lipid membrane environments [52, 79, 80, 82–86]. In this method, a cysteine mutated nitroxide spin label is positioned 2 (i + 2), 3 (i + 3), or 4 (i + 4) residues away from a fully deuterated Leu side-chain (i). The characteristic periodicity of the α-helix (3.6 residue per turn with a pitch of 5.4 Å) and a 310- helix (3.1 residue per turn with a pitch of 6.0 Å) structure gives rise to a unique pattern in the ESEEM spectra. A larger 2H ESEEM peak in the FT frequency domain data is observed for the i + 4 samples, when compared to the i + 3 samples for the α-helix, whereas the opposite pattern is revealed for the 310-helix [52]. For all i + 2 samples, no 2H ESEEM peak in the FT frequency domain data is revealed due to spin labels being too far away from the 2H-labeled side-chain to be detected. These unique patterns provide pertinent local secondary structural information to distinguish between the α-helical and 310-helical structural motifs for protein/peptides using this ESEEM spectroscopic approach with short data acquisition times (~30 min) and small sample concentrations (~100 µM) as well as providing more site-specific secondary structural information compared to other common biophysical approaches such as CD. Figure 5 shows an example of the three-pulse ESEEM data obtained for an amphipathic model peptide, LRL8 [52]. When LRL8 is solubilized in trifluoroethanol (TFE), the peptide adopts an α-helical structure and, alternatively, forms a 310-helical secondary structure when incorporated into liposomes. The inset in Figure 5 shows the spatial relationship between the spin label and the 2H-labeled Leu residue from MD simulations.
Figure 5.
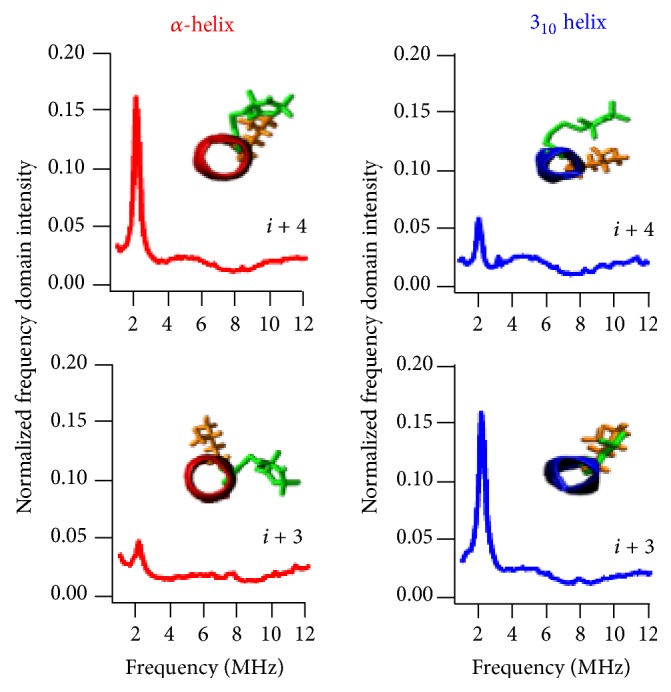
Three-pulse ESEEM FT data of 2H-labeled d10 Leu4 LRL8 peptide in TFE (α-helix) and DPPC liposomes (310-helix) for i + 3 and i + 4 samples (adapted from [52] with permission).
2.3. SDSL Distance Measurement of Membrane Proteins
Distance information can be obtained from two spin labels in terms of either intramolecular distances between sites on the same protein or intermolecular distances between sites on different proteins [18]. The distance is obtained from the magnetic dipolar interactions between the unpaired electrons of two spin labels. The energy of the dipolar interaction is inversely proportional to the cube of the distance (r3). When the distance is less than 20 Å, electron-electron interaction significantly broadens the CW-EPR spectral lineshape. The strength of the interaction is estimated qualitatively from the degree of line broadening using a variety of lineshape analysis techniques and corresponding distance information can be revealed [18, 20, 87–90]. Using dual labeling EPR techniques, distances can be measured to probe secondary, tertiary, and quaternary structures [87]. CW dipolar broadening EPR can provide pertinent structural and functional dynamic information over an intermediate distance range of 8–20 Å [70, 91–93]. Nitroxide based SDSL CW dipolar broadening EPR has been applied to several important biological systems such as bacteriorhodopsin, erythroid β spectrin, AchR M2δ peptide, magainin 2, bacterial K+-translocating protein KtrB, E. coli integral membrane sulfurtransferase, and KCNE1 [53, 70, 90, 93–98].
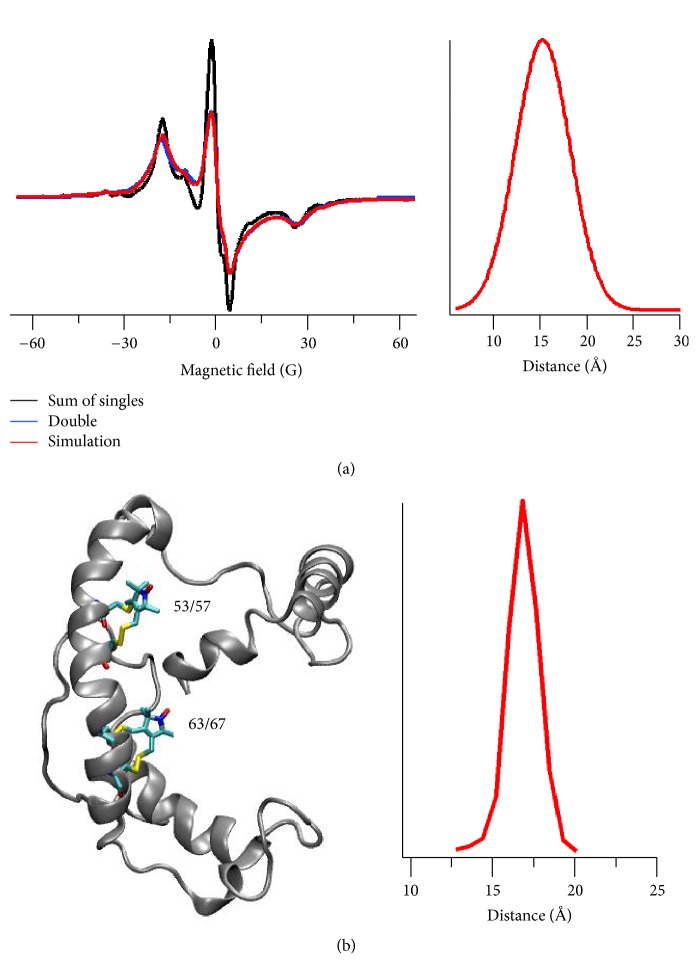
A recent example of using SDSL CW dipolar broadening EPR is the study of an integral membrane protein KCNE1 [53]. Sahu et al. applied CW dipolar broadening EPR technique to measure a distance between two bifunctional spin label (BSLs) attached on the transmembrane domain of KCNE1 in lipid bilayers at room temperature. The experimental result was further validated using a 20 ns molecular dynamics modeling study. Figure 6 shows CW dipolar broadening EPR data on KCNE1 in POPC/POPG lipid bilayers at room temperature. The CW dipolar line broadening EPR data revealed 15 ± 2 Å distance between doubly attached BSLs on KCNE1 (53/57-63/67) which is consistent with molecular dynamics modeling and the solution NMR structure of KCNE1 which yielded a distance of 17 Å. This study demonstrated the utility of investigating the structural and dynamic properties of membrane proteins in physiologically relevant membrane mimetics using BSLs.
Figure 6.
CW dipolar broadening EPR study of KCNE1. (a) CW dipolar broadening EPR spectra of KCNE1 bearing BSLs at sites 53/57 and 63/67 in POPC/POPG liposomes (left panel) and the corresponding distance distribution (right panel) obtained from data analysis by using the Short Distances LabVIEW program. (b) Cartoon representation of KCNE1 bearing two BSLs at sites 53/57 and 63/67 (left panel) and the corresponding distance distribution obtained from 20 ns molecular dynamics trajectory data analysis (right panel) (adapted from [53] with permission).
For longer distances, pulsed EPR double electron-electron resonance (DEER) spectroscopy has been a widely used biophysical technique for measuring distances between 20 and 80 Å [99, 100]. Distance measurements are one of the most popular and rapidly expanding aspects of SDSL EPR spectroscopy. DEER is also known as pulse electron double resonance (PELDOR). In DEER, the measurement of the coupling between the two spins can be performed by monitoring one set of spins while exciting another set of spins with a second microwave frequency that leads to the measurement of distance between them. SDSL DEER spectroscopy has been applied for probing the structure and conformational dynamics of a wide variety of biological systems [9, 99–107]. It can also be used to measure the relative orientation of the spins when the experiment is performed at higher magnetic field of 94 GHz [108]. Although the DEER technique is very popular in the structure biology field, the technical limitations of membrane protein sample preparation in its functional environment introduce challenges in accurate and precise distance measurements. The challenges arise due to much shorter transverse relaxation/phase memory times due to the heterogeneous distribution of spin labeled proteins within the membrane creating local inhomogeneous pockets of high spin concentration and poor DEER modulation in more biologically relevant liposomes when compared to water soluble proteins or membrane proteins in detergent micelles [104]. Additionally, the use of a high effective protein concentration in the liposome samples introduces a strong background contribution causing extreme limits on sensitivity, distance range, and experimental throughput [109]. The protein backbone dynamics and spin label rotameric motions have also a significant contribution to the DEER distance distribution width.
In recent years, impressive studies have been done to overcome these challenges using membrane protein reconstitutions in the presence of unlabeled proteins, bicelles, nanodiscs, lipodisq nanoparticles, a low protein/lipid molar ratio, restricted spin label probes, and Q-band pulse EPR measurements [9, 106, 110–116]. Recently, several labs have utilized DEER distance restraints coupled with molecular dynamics simulations to refine the structure of membrane proteins [56, 106, 117]. These methodological developments have made DEER a powerful and popular structure biology tool to study complicated biological systems such as membrane proteins. SDSL DEER spectroscopy has been applied to study several important membrane protein systems such as bacteriorhodopsin, KCNE1, KCNE3, C99 Amyloid Precursor Protein, KvAP voltage-sensing domain, human dihydroorotate dehydrogenase enzyme (HsDHODH), Influenza A M2 protein, cardiac Na+/Ca2+ exchange (NCX1.1) protein, Na+/Proline Transporter PutP Escherichia coli, tetrameric potassium ion channel KcsA, α-Synuclein, and ABC Transporter MsbA [56, 106, 110–112, 117–124]. SDSL DEER spectroscopy has been recently used to study the oligomerization states of several membrane proteins such as NhaA Na+/H+ antiporter of E. coli, KcsA, M2 transmembrane domain, LptA, and proteorhodopsin [60, 103, 119, 125, 126]. This is a very powerful biophysical technique to determine the oligomeric structure of membrane proteins.
A recent application of SDSL DEER spectroscopy is the conformational study of KCNE3 in proteoliposomes [122]. KCNE3 is a single pass transmembrane protein that modulates a variety of voltage gated ion channels in diverse biological contexts. In epithelial cells, KCNE3 regulates the function of the KCNQ1 potassium ion (K+) channel in a physiologically critical cellular transport process in several organs and whose malfunction causes diseases such as cystic fibrosis (CF), cholera, and pulmonary edema. Kroncke et al. performed SDSL DEER experiments on the protein in different membrane environments to identify a curved α-helical nature of the transmembrane of KCNE3 [122]. Two MTSL spin labels were generated at the ends of the transmembrane domain (TMD) of KCNE3. The dual spin labeled KCNE3 protein was reconstituted into micelles [lyso-myristoylphosphatidylcholine (LMPC)], bicelles (DMPG/DHPC), and 1-palmitoyl-2-oleoyl-phosphatidylcholine (POPC)/1-palmitoyl-2-oleoyl-phosphatidylglycerol (POPG) lipid bilayers. The resulting distances for all three media were statistically identical. These results indicated that the curvature of the TMD is an intrinsic property of KCNE3 that is maintained in lipid bilayers, bicelles, and detergent micelles. The TMD curvature of KCNE3 is important for the kinetics of initial binding of KCNE3 to the KCNQ1 channel.
Another excellent recent application of SDSL DEER spectroscopy is the study of conformational dynamics of a multidrug transporter LmrP from Lactococcus lactis [127]. LmrP is a member of the major facilitator superfamily (MFS). It couples the downhill translocation of protons along their transmembrane gradient to the uphill transport of hydrophobic cytotoxic compounds. The active efflux of diverse cytotoxic compounds through multidrug transporters contributes to bacterial antibiotic resistance. Martens et al. performed DEER distance measurements on the selected spin label pairs of the transporter reconstituted into nanodiscs of different lipid compositions [127]. The resulting DEER data revealed the conformational energy landscape of the transporter modulated by the lipid headgroups. The results further suggested a direct interaction between lipid headgroups and a conserved motif of charged residues that control the conformational equilibrium through an interplay of electrostatic interactions within the protein. Although SDSL EPR has several advantages over the existing biophysical techniques for studying membrane proteins, it needs an incorporation of spin label probes which might not be suitable for all the desired sites on some membrane protein systems due to low spin labeling efficiency.
3. Conclusion
In this review, we discussed some recent applications of nitroxide based SDSL EPR spectroscopic techniques to study important membrane protein systems. SDSL EPR spectroscopy is very popular and growing structure biology technique used to answer pertinent structural and dynamic questions related to biological systems. It can provide important information on complicated biological systems which is very challenging or nearly impossible by using other biophysical techniques.
Acknowledgments
Researchers of this work were generously supported by a National Institute of Health Grant R01 GM108026 and a National Science Foundation Award CHE- 1305664.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
References
- 1.Stone T. J., Buckman T., Nordio P. L., McConnell H. M. Spin-labeled biomolecules. Proceedings of the National Acadamy of Sciences of the United States of America. 1965;54(4):1010–1017. doi: 10.1073/pnas.54.4.1010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cornish V. W., Benson D. R., Altenbach C. A., Hideg K., Hubbell W. L., Schultz P. G. Site-specific incorporation of biophysical probes into proteins. Proceedings of the National Acadamy of Sciences of the United States of America. 1994;91(8):2910–2914. doi: 10.1073/pnas.91.8.2910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klare J. P., Steinhoff H.-J. Spin labeling EPR. Photosynthesis Research. 2009;102(2):377–390. doi: 10.1007/s11120-009-9490-7. [DOI] [PubMed] [Google Scholar]
- 4.Steinhoff H.-J. Multi-frequency EPR spectroscopy studies of the structure and conformational changes of site-directed spin labelled membrane proteins. Supramolecular Structure and Function 8. 2005:157–177. doi: 10.1007/0-306-48662-8_10. doi: 10.1007/0-306-48662-8_10. [DOI] [Google Scholar]
- 5.Klare J. P. A Versatile Tool to Study Protein-Protein Interaction. InTech; 2012. Site-directed spin labelingand electron paramagnetic resonance (EPR) spectroscopy: [DOI] [Google Scholar]
- 6.Haugland M. M., Anderson E. A., Lovett J. E. Tuning the properties of nitroxide spin labels for use in electron paramagnetic resonance spectroscopy through chemical modification of the nitroxide framework. In: Chechik V., Murphy D. M., editors. Electron Paramagnetic Resonance. 2017. pp. 1–34. [DOI] [Google Scholar]
- 7.Balo A. R., Feyrer H., Ernst O. P. Toward precise interpretation of DEER-based distance distributions: insights from structural characterization of V1 spin-labeled side chains. Biochemistry. 2016;55(37):5256–5263. doi: 10.1021/acs.biochem.6b00608. [DOI] [PubMed] [Google Scholar]
- 8.Sahu I. D., McCarrick R. M., Lorigan G. A. Use of electron paramagnetic resonance to solve biochemical problems. Biochemistry. 2013;52(35):5967–5984. doi: 10.1021/bi400834a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sahu I. D., McCarrick R. M., Troxel K. R., et al. DEER EPR measurements for membrane protein structures via bifunctional spin labels and lipodisq nanoparticles. Biochemistry. 2013;52(38):6627–6632. doi: 10.1021/bi4009984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fleissner M. R., Bridges M. D., Brooks E. K., et al. Structure and dynamics of a conformationally constrained nitroxide side chain and applications in EPR spectroscopy. Proceedings of the National Acadamy of Sciences of the United States of America. 2011;108(39):16241–16246. doi: 10.1073/pnas.1111420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schreier S., Bozelli Jr J. C., Marín N., Vieira R. F. F., Nakaie C. R. The spin label amino acid TOAC and its uses in studies of peptides: chemical, physicochemical, spectroscopic, and conformational aspects. Biophysical Reviews. 2012;4(1):45–66. doi: 10.1007/s12551-011-0064-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbell W. L., López C. J., Altenbach C., Yang Z. Technological advances in site-directed spin labeling of proteins. Current Opinion in Structural Biology. 2013;23(5):725–733. doi: 10.1016/j.sbi.2013.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stoller S., Sicoli G., Baranova T. Y., Bennati M., Diederichsen U. TOPP: a novel nitroxide-labeled amino acid for EPR distance measurements. Angewandte Chemie International Edition. 2011;50(41):9743–9746. doi: 10.1002/anie.201103315. [DOI] [PubMed] [Google Scholar]
- 14.Fielding A. J., Concilio M. G., Heaven G., Hollas M. A. New developments in spin labels for pulsed dipolar EPR. Molecules. 2014;19(10):16998–17025. doi: 10.3390/molecules191016998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Roser P., Schmidt M. J., Drescher M., Summerer D. Site-directed spin labeling of proteins for distance measurements: In vitro and in cells. Organic & Biomolecular Chemistry. 2016;14(24):5468–5476. doi: 10.1039/c6ob00473c. [DOI] [PubMed] [Google Scholar]
- 16.Weil J. A., Bolton J. R. Electron paramagnetic resonance: Elementary Theory and Practical Applications. Hoboken, NJ, USA: Wiley-Interscience; 2007. [Google Scholar]
- 17.Berliner L. J. From spin-labeled proteins to in vivo EPR applications. European Biophysics Journal. 2010;39(4):579–588. doi: 10.1007/s00249-009-0534-x. [DOI] [PubMed] [Google Scholar]
- 18.Hustedt E. J., Beth A. H. Nitroxide spin-spin interactions: applications to protein structure and dynamics. Annual Review of Biophysics. 1999;28:129–153. doi: 10.1146/annurev.biophys.28.1.129. [DOI] [PubMed] [Google Scholar]
- 19.Liu K. J., Gast P., Moussavi M., et al. Lithium phthalocyanine: a probe for electron paramagnetic resonance oximetry in viable biological systems. Proceedings of the National Acadamy of Sciences of the United States of America. 1993;90(12):5438–5442. doi: 10.1073/pnas.90.12.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hustedt E. J., Smirnov A. I., Laub C. F., Cobb C. E., Beth A. H. Molecular distances from dipolar coupled spin-labels: the global analysis of multifrequency continuous wave electron paramagnetic resonance data. Biophysical Journal. 1997;72(4):1861–1877. doi: 10.1016/S0006-3495(97)78832-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hubbell W. L., Gross A., Langen R., Lietzow M. A. Recent advances in site-directed spin labeling of proteins. Current Opinion in Structural Biology. 1998;8(5):649–656. doi: 10.1016/S0959-440X(98)80158-9. [DOI] [PubMed] [Google Scholar]
- 22.Klug C. S., Feix J. B. Methods and applications of site-directed spin labeling EPR spectroscopy. Methods in Cell Biology. 2008;84:617–658. doi: 10.1016/S0091-679X(07)84020-9. [DOI] [PubMed] [Google Scholar]
- 23.Hubbell W. L., Mchaourab H. S., Altenbach C., Lietzow M. A. Watching proteins move using site-directed spin labeling. Structure. 1996;4(7):779–783. doi: 10.1016/S0969-2126(96)00085-8. [DOI] [PubMed] [Google Scholar]
- 24.Bordignon E., Steinhoff H. J. Steinhoff, Membrane protein structure and dynamics studied by site-directed spin-labeling ESR. ESR Spectroscopy in Membrane Biophysics. 2007;27:129–164. doi: 10.1007/978-0-387-49367-1. [DOI] [Google Scholar]
- 25.Sahu I. D., Lorigan G. A. Biophysical EPR studies applied to membrane proteins. Journal of Physical Chemistry & Biophysics. 2015;5(6):p. 188. doi: 10.4172/2161-0398.1000188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jeschke G., Bender A., Schweikardt T., Panek G., Decker H., Paulsen H. Localization of the N-terminal domain in light-harvesting chlorophyll a/b protein by EPR measurements. The Journal of Biological Chemistry. 2005;280(19):18623–18630. doi: 10.1074/jbc.M501171200. [DOI] [PubMed] [Google Scholar]
- 27.Mchaourab H. S., Perozo E. Determination of protein folds and conformational dynamics using spin-labeling EPR spectroscopy. In: Berliner L., Eaton G., Eaton S., editors. Biological Magnetic Resonance. Vol. 19. Boston, MA, USA: Springer; 2002. pp. 185–247. (Biological Magnetic Resonance). [DOI] [Google Scholar]
- 28.Perozo E., Marien Cortes D., Cuello L. G. Three-dimensional architecture and gating mechanism of a K+ channel studied by EPR spectroscopy. Nature Structural & Molecular Biology. 1998;5(6):459–469. doi: 10.1038/nsb0698-459. [DOI] [PubMed] [Google Scholar]
- 29.Vásquez V., Sotomayor M., Marien Cortes D., Roux B., Schulten K., Perozo E. Three-dimensional architecture of membrane-embedded MscS in the closed conformation. Journal of Molecular Biology. 2008;378(1):55–70. doi: 10.1016/j.jmb.2007.10.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Congreve M., Marshall F. The impact of GPCR structures on pharmacology and structure-based drug design. British Journal of Pharmacology. 2010;159(5):986–996. doi: 10.1111/j.1476-5381.2009.00476.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baker M. Structural biology: the gatekeepers revealed. Nature. 2010;465(7299):823–826. doi: 10.1038/465823a. [DOI] [PubMed] [Google Scholar]
- 32.Landreh M., Robinson C. V. A new window into the molecular physiology of membrane proteins. The Journal of Physiology. 2015;593(2):355–362. doi: 10.1113/jphysiol.2014.283150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moraes I., Evans G., Sanchez-Weatherby J., Newstead S., Stewart P. D. S. Membrane protein structure determination—the next generation. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2014;1838(1):78–87. doi: 10.1016/j.bbamem.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wallin E., von Heijne G. Genome-wide analysis of integral membrane proteins from eubacterial, archaean, and eukaryotic organisms. Protein Science. 1998;7(4):1029–1038. doi: 10.1002/pro.5560070420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Conn P. M., Ulloa-Aguirre A., Ito J., Janovick J. A. G protein-coupled receptor trafficking in health and disease: lessons learned to prepare for therapeutic mutant rescue in vivo. Pharmacological Reviews. 2007;59(3):225–250. doi: 10.1124/pr.59.3.2. [DOI] [PubMed] [Google Scholar]
- 36.Cheung J. C., Deber C. M. Misfolding of the cystic fibrosis transmembrane conductance regulator and disease. Biochemistry. 2008;47(6):1465–1473. doi: 10.1021/bi702209s. [DOI] [PubMed] [Google Scholar]
- 37.von Heijne G. The membrane protein universe: what's out there and why bother? Journal of Internal Medicine. 2007;261(6):543–557. doi: 10.1111/j.1365-2796.2007.01792.x. [DOI] [PubMed] [Google Scholar]
- 38.Overington J. P., Al-Lazikani B., Hopkins A. L. How many drug targets are there? Nature Reviews Drug Discovery. 2006;5(12):993–996. doi: 10.1038/nrd2199. [DOI] [PubMed] [Google Scholar]
- 39.Engel A., Gaub H. E. Structure and mechanics of membrane proteins. Annual Review of Biochemistry. 2008;77:127–148. doi: 10.1146/annurev.biochem.77.062706.154450. [DOI] [PubMed] [Google Scholar]
- 40.Das B. B., Park S. H., Opella S. J. Membrane protein structure from rotational diffusion. Biochimica et Biophysica Acta (BBA)—Biomembranes. 2015;1848(1):229–245. doi: 10.1016/j.bbamem.2014.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kang H. J., Lee C., Drew D. Breaking the barriers in membrane protein crystallography. The International Journal of Biochemistry & Cell Biology. 2013;45(3):636–644. doi: 10.1016/j.biocel.2012.12.018. [DOI] [PubMed] [Google Scholar]
- 42.Wüthrich K. NMR studies of structure and function of biological macromolecules (Nobel Lecture) Journal of Biomolecular NMR. 2003;27:13–39. doi: 10.1023/B:BIRE.0000007689.27614.7c. [DOI] [PubMed] [Google Scholar]
- 43.Acharya K. R., Lloyd M. D. The advantages and limitations of protein crystal structures. Trends in Pharmacological Sciences. 2005;26(1):10–14. doi: 10.1016/j.tips.2004.10.011. [DOI] [PubMed] [Google Scholar]
- 44.Schiemann O., Prisner T. F. Long-range distance determinations in biomacromolecules by EPR spectroscopy. Quarterly Reviews of Biophysics. 2007;40(1):1–53. doi: 10.1017/S003358350700460X. [DOI] [PubMed] [Google Scholar]
- 45.Torres J., Stevens T. J., Samsó M. Membrane proteins: the 'Wild West' of structural biology. Trends in Biochemical Sciences. 2003;28(3):137–144. doi: 10.1016/S0968-0004(03)00026-4. [DOI] [PubMed] [Google Scholar]
- 46.Columbus L., Hubbell W. L. A new spin on protein dynamics. Trends in Biochemical Sciences. 2002;27(6):288–295. doi: 10.1016/S0968-0004(02)02095-9. [DOI] [PubMed] [Google Scholar]
- 47.Fanucci G. E., Cafiso D. S. Recent advances and applications of site-directed spin labeling. Current Opinion in Structural Biology. 2006;16(5):644–653. doi: 10.1016/j.sbi.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 48.Qin P. Z., Dieckmann T. Application of NMR and EPR methods to the study of RNA. Current Opinion in Structural Biology. 2004;14(3):350–359. doi: 10.1016/j.sbi.2004.04.002. [DOI] [PubMed] [Google Scholar]
- 49.Speicher D. W. Characterization of protein primary structure. Developments in Biological Standardization. 1998;96:27–28. [PubMed] [Google Scholar]
- 50.Claxton D. P., Kazmier K., Mishra S., McHaourab H. S. Navigating membrane protein structure, dynamics, and energy landscapes using spin labeling and EPR spectroscopy. Methods in Enzymology. 2015;564:349–387. doi: 10.1016/bs.mie.2015.07.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sahu I. D., Craig A. F., Dunagan M. M., et al. Probing structural dynamics and topology of the kcne1 membrane protein in lipid bilayers via site-directed spin labeling and electron paramagnetic resonance spectroscopy. Biochemistry. 2015;54(41):6402–6412. doi: 10.1021/acs.biochem.5b00505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bottorf L., Rafferty S., Sahu I. D., McCarrick R. M., Lorigan G. A. Utilizing electron spin echo envelope modulation to distinguish between the local secondary structures of an α-helix and an amphipathic 310-helical peptide. The Journal of Physical Chemistry B. 2017;121(14):2961–2967. doi: 10.1021/acs.jpcb.7b00626. [DOI] [PubMed] [Google Scholar]
- 53.Sahu I. D., Craig A. F., Dunagum M. M., McCarrick R. M., Lorigan G. A. Characterization of bifunctional spin labels for investigating the structural and dynamic properties of membrane proteins using EPR spectroscopy. The Journal of Physical Chemistry B. 2017;121(39):9185–9195. doi: 10.1021/acs.jpcb.7b07631. [DOI] [PubMed] [Google Scholar]
- 54.Klug C. S., Feix J. B. SDSL: A survey of biological applications. Biological Magnetic Resonance. 2004;24:269–308. doi: 10.1007/b111471. [DOI] [Google Scholar]
- 55.Stoll S., Schweiger A. Easyspin: simulating cw ESR spectra. Biological Magnetic Resonance. 2007;27:299–321. [Google Scholar]
- 56.Li Q., Wanderling S., Sompornpisut P., Perozo E. Structural basis of lipid-driven conformational transitions in the KvAP voltage-sensing domain. Nature Structural & Molecular Biology. 2014;21(2):160–166. doi: 10.1038/nsmb.2747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cuello L. G., Cortes D. M., Perozo E. Molecular architecture of the KvAP voltage-dependent K+ channel in a lipid bilayer. Science. 2004;306(5695):491–495. doi: 10.1126/science.1101373. [DOI] [PubMed] [Google Scholar]
- 58.Soria M. A., Cervantes S. A., Bajakian T. H., Siemer A. B. The functional amyloid Orb2A binds to lipid membranes. Biophysical Journal. 2017;113(1):37–47. doi: 10.1016/j.bpj.2017.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Victor K. G., Cafiso D. S. Location and dynamics of basic peptides at the membrane interface: electron paramagnetic resonance spectroscopy of tetramethyl-piperidine-N-oxyl-4-amino-4-carboxylic acid-labeled peptides. Biophysical Journal. 2001;81(4):2241–2250. doi: 10.1016/S0006-3495(01)75871-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kim S. S., Upshur M. A., Saotome K., et al. Cholesterol-dependent conformational exchange of the C-terminal domain of the influenza A M2 protein. Biochemistry. 2015;54(49):7157–7167. doi: 10.1021/acs.biochem.5b01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yu L., Wang W., Ling S., et al. CW-EPR studies revealed different motional properties and oligomeric states of the integrin β1a transmembrane domain in detergent micelles or liposomes. Scientific Reports. 2015;5, article no. 7848 doi: 10.1038/srep07848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Altenbach C., Greenhalgh D. A., Khorana H. G., Hubbell W. L. A collision gradient method to determine the immersion depth of nitroxides in lipid bilayers: application to spin-labeled mutants of bacteriorhodopsin. Proceedings of the National Acadamy of Sciences of the United States of America. 1994;91(5):1667–1671. doi: 10.1073/pnas.91.5.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cortes D. M., Cuello L. G., Perozo E. Molecular architecture of full-length KcsA role of cytoplasmic domains in ion permeation and activation gating. The Journal of General Physiology. 2001;117(2):165–180. doi: 10.1085/jgp.117.2.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voss J., He M. M., Hubbell W. L., Ronald Kaback H. Site-directed spin labeling demonstrates that transmembrane domain XII in the lactose permease of Escherichia coli is an α-helix. Biochemistry. 1996;35(39):12915–12918. doi: 10.1021/bi9608774. [DOI] [PubMed] [Google Scholar]
- 65.Song Y., Hustedt E. J., Brandon S., Sanders C. R. Competition between homodimerization and cholesterol binding to the C99 domain of the amyloid precursor protein. Biochemistry. 2013;52(30):5051–5064. doi: 10.1021/bi400735x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perozo E., Hubbell W. L. Transmembrane voltage control in liposomes-the use of bacteriorhodopsin as a light-driven current source. Biophysical Journal. 1993;64:A222–A222. doi: 10.1021/bi00090a025. [DOI] [PubMed] [Google Scholar]
- 67.Hubbell W. L., Altenbach C. Investigation of structure and dynamics in membrane proteins using site-directed spin labeling. Current Opinion in Structural Biology. 1994;4(4):566–573. doi: 10.1016/S0959-440X(94)90219-4. [DOI] [Google Scholar]
- 68.Sahu I. D., Zhang R., Dunagan M. M., Craig A. F., Lorigan G. A. Characterization of KCNE1 inside lipodisq nanoparticles for EPR spectroscopic studies of membrane proteins. The Journal of Physical Chemistry B. 2017;121(21):5312–5321. doi: 10.1021/acs.jpcb.7b01705. [DOI] [PubMed] [Google Scholar]
- 69.Inbaraj J. J., Cardon T. B., Laryukhin M., Grosser S. M., Lorigan G. A. Determining the topology of integral membrane peptides using EPR spectroscopy. Journal of the American Chemical Society. 2006;128(29):9549–9554. doi: 10.1021/ja0622204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sahu I. D., Hustedt E. J., Ghimire H., Inbaraj J. J., McCarrick R. M., Lorigan G. A. CW dipolar broadening EPR spectroscopy and mechanically aligned bilayers used to measure distance and relative orientation between two TOAC spin labels on an antimicrobial peptide. Journal of Magnetic Resonance. 2014;249:72–79. doi: 10.1016/j.jmr.2014.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sahu I. D., Mayo D. J., Subbaraman N., Inbaraj J. J., McCarrick R. M., Lorigan G. A. Probing topology and dynamics of the second transmembrane domain (M2δ) of the acetyl choline receptor using magnetically aligned lipid bilayers (bicelles) and EPR spectroscopy. Chemistry and Physics of Lipids. 2017;206:9–15. doi: 10.1016/j.chemphyslip.2017.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.McCaffrey J. E., James Z. M., Svensson B., Binder B. P., Thomas D. D. A bifunctional spin label reports the structural topology of phospholamban in magnetically-aligned bicelles. Journal of Magnetic Resonance. 2016;262:50–56. doi: 10.1016/j.jmr.2015.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kubota T., Lacroix J. J., Bezanilla F., Correa A. M. Probing α-3(10) transitions in a voltage-sensing S4 helix. Biophysical Journal. 2014;107(5):1117–1128. doi: 10.1016/j.bpj.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yu X., Lorigan G. A. Secondary structure, backbone dynamics, and structural topology of phospholamban and its phosphorylated and Arg9Cys-mutated forms in phospholipid bilayers utilizing 13C and 15N solid-state NMR spectroscopy. The Journal of Physical Chemistry B. 2014;118(8):2124–2133. doi: 10.1021/jp500316s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Cieslak J. A., Focia P. J., Gross A. Electron spin-echo envelope modulation (ESEEM) reveals water and phosphate interactions with the KcsA potassium channel. Biochemistry. 2010;49(7):1486–1494. doi: 10.1021/bi9016523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Lorigan G. A., Britt R. D., Kim J. H., Hille R. Electron spin echo envelope modulation spectroscopy of the molybdenum center of xanthine oxidase. BBA—Bioenergetics. 1994;1185(3):284–294. doi: 10.1016/0005-2728(94)90243-7. [DOI] [PubMed] [Google Scholar]
- 77.Bartucci R., Guzzi R., Esmann M., Marsh D. Water penetration profile at the protein-lipid interface in Na,K-ATPase membranes. Biophysical Journal. 2014;107(6):1375–1382. doi: 10.1016/j.bpj.2014.07.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Carmieli R., Papo N., Zimmermann H., Potapov A., Shai Y., Goldfarb D. Utilizing ESEEM spectroscopy to locate the position of specific regions of membrane-active peptides within model membranes. Biophysical Journal. 2006;90(2):492–505. doi: 10.1529/biophysj.105.062992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Liu L., Sahu I. D., Mayo D. J., et al. Enhancement of electron spin echo envelope modulation spectroscopic methods to investigate the secondary structure of membrane proteins. The Journal of Physical Chemistry B. 2012;116(36):11041–11045. doi: 10.1021/jp304669b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mayo D., Zhou A., Sahu I., et al. Probing the structure of membrane proteins with electron spin echo envelope modulation spectroscopy. Protein Science. 2011;20(7):1100–1104. doi: 10.1002/pro.656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Milov A. D., Samoilova R. I., Tsvetkov Y. D., et al. Structure of self-aggregated alamethicin in ePC membranes detected by pulsed electron-electron double resonance and electron spin echo envelope modulation spectroscopies. Biophysical Journal. 2009;96(8):3197–3209. doi: 10.1016/j.bpj.2009.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Zhou A., Abu-Baker S., Sahu I. D., et al. Determining α-helical and β-sheet secondary structures via pulsed electron spin resonance spectroscopy. Biochemistry. 2012;51(38):7417–7419. doi: 10.1021/bi3010736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Liu L., Mayo D. J., Sahu I. D., et al. Determining the secondary structure of membrane proteins and peptides via electron spin echo envelope modulation (ESEEM) spectroscopy. Methods in Enzymology. 2015;564:289–313. doi: 10.1016/bs.mie.2015.06.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zhang R., Sahu I. D., Gibson K. R., et al. Development of electron spin echo envelope modulation spectroscopy to probe the secondary structure of recombinant membrane proteins in a lipid bilayer. Protein Science. 2015;24:1707–1713. doi: 10.1002/pro.2795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Liu L., Hess J., Sahu I. D., FitzGerald P. G., McCarrick R. M., Lorigan G. A. Probing the local secondary structure of human vimentin with electron spin echo envelope modulation (ESEEM) spectroscopy. The Journal of Physical Chemistry B. 2016;120(48):12321–12326. doi: 10.1021/acs.jpcb.6b10054. [DOI] [PubMed] [Google Scholar]
- 86.Liu L., Sahu I. D., McCarrick R. M., Lorigan G. A. Probing the secondary structure of membrane peptides using 2H-Labeled d10-leucine via site-directed spin-labeling and electron spin echo envelope modulation spectroscopy. The Journal of Physical Chemistry B. 2016;120(4):633–640. doi: 10.1021/acs.jpcb.5b09040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Hustedt E. J., Stein R. A., Sethaphong L., Brandon S., Zhou Z., DeSensi S. C. Dipolar coupling between nitroxide spin labels: the development and application of a tether-in-a-cone model. Biophysical Journal. 2006;90(1):340–356. doi: 10.1529/biophysj.105.068544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Banham J. E., Baker C. M., Ceola S., et al. Distance measurements in the borderline region of applicability of CW EPR and DEER: a model study on a homologous series of spin-labelled peptides. Journal of Magnetic Resonance. 2008;191(2):202–218. doi: 10.1016/j.jmr.2007.11.023. [DOI] [PubMed] [Google Scholar]
- 89.Rabenstein M. D., Shin Y.-K. Determination of the distance between two spin labels attached to a macromolecule. Proceedings of the National Acadamy of Sciences of the United States of America. 1995;92(18):8239–8243. doi: 10.1073/pnas.92.18.8239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Czogalla A., Pieciul A., Jezierski A., Sikorski A. F. Attaching a spin to a protein—site-directed spin labeling in structural biology. Acta Biochimica Polonica. 2007;54(2):235–244. [PubMed] [Google Scholar]
- 91.Chiang Y.-W., Zheng T.-Y., Kao C.-J., Horng J.-C. Determination of interspin distance distributions by cw-ESR is a single linear inverse problem. Biophysical Journal. 2009;97(3):930–936. doi: 10.1016/j.bpj.2009.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kittell A. W., Hustedt E. J., Hyde J. S. Inter-spin distance determination using L-band (1-2 GHz) non-adiabatic rapid sweep electron paramagnetic resonance (NARS EPR) Journal of Magnetic Resonance. 2012;221:51–56. doi: 10.1016/j.jmr.2012.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ghimire H., Hustedt E. J., Sahu I. D., et al. Distance measurements on a dual-labeled toac achr m2δ peptide in mechanically aligned DMPC bilayers via dipolar broadening CW-EPR spectroscopy. The Journal of Physical Chemistry B. 2012;116(12):3866–3873. doi: 10.1021/jp212272d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Czogalla A., Jaszewski A. R., Diakowski W., Bok E., Jezierski A., Sikorski A. F. Structural insight into an ankyrin-sensitive lipid-binding site of erythroid β-spectrin. Molecular Membrane Biology. 2007;24(3):215–224. doi: 10.1080/09687860601102427. [DOI] [PubMed] [Google Scholar]
- 95.Essen L.-O., Siegert R., Lehmann W. D., Oesterhelt D. Lipid patches in membrane protein oligomers: crystal structure of the bacteriorhodopsin-lipid complex. Proceedings of the National Acadamy of Sciences of the United States of America. 1998;95(20):11673–11678. doi: 10.1073/pnas.95.20.11673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ling S., Wang W., Yu L., et al. Structure of an E. coli integral membrane sulfurtransferase and its structural transition upon SCN- binding defined by EPR-based hybrid method. Scientific Reports. 2016;6 doi: 10.1038/srep20025.20025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hänelt I., Wunnicke D., Müller-Trimbusch M., et al. Membrane region M2C2 in subunit KtrB of the K+ uptake system KtrAB from Vibrio alginolyticus forms a flexible gate controlling K + flux: An electron paramagnetic resonance study. The Journal of Biological Chemistry. 2010;285(36):28210–28219. doi: 10.1074/jbc.M110.139311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Steinhoff H.-J. Inter- and intra-molecular distances determined by EPR spectroscopy and site-directed spin labeling reveal protein-protein and protein-oligonucleotide interaction. biological chemistry. 2004;385(10):913–920. doi: 10.1515/BC.2004.119. [DOI] [PubMed] [Google Scholar]
- 99.Jeschke G., Polyhach Y. Distance measurements on spin-labelled biomacromolecules by pulsed electron paramagnetic resonance. Physical Chemistry Chemical Physics. 2007;9(16):1895–1910. doi: 10.1039/b614920k. [DOI] [PubMed] [Google Scholar]
- 100.Borbat P. P., Mchaourab H. S., Freed J. H. Protein structure determination using long-distance constraints from double-quantum coherence ESR: study of T4 lysozyme. Journal of the American Chemical Society. 2002;124(19):5304–5314. doi: 10.1021/ja020040y. [DOI] [PubMed] [Google Scholar]
- 101.Schiemann O., Piton N., Mu Y., Stock G., Engels J. W., Prisner T. F. A PELDOR-based nanometer distance ruler for oligonucleotides. Journal of the American Chemical Society. 2004;126(18):5722–5729. doi: 10.1021/ja0393877. [DOI] [PubMed] [Google Scholar]
- 102.Milov A. D., Tsvetkov Y. D., Formaggio F., Crisma M., Toniolo C., Raap J. Self-assembling properties of membrane-modifying peptides studied by PELDOR and CW-ESR spectroscopies. Journal of the American Chemical Society. 2000;122(16):3843–3848. doi: 10.1021/ja993870t. [DOI] [PubMed] [Google Scholar]
- 103.Hilger D., Jung H., Padan E., et al. Assessing oligomerization of membrane proteins by four-pulse DEER: pH-dependent dimerization of NhaA Na+/H+ antiporter of E. coli. Biophysical Journal. 2005;89(2):1328–1338. doi: 10.1529/biophysj.105.062232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Jeschke G. DEER distance measurements on proteins. Annual Review of Physical Chemistry. 2012;63:419–446. doi: 10.1146/annurev-physchem-032511-143716. [DOI] [PubMed] [Google Scholar]
- 105.Banham J. E., Timmel C. R., Abbott R. J. M., Lea S. M., Jeschke G. The characterization of weak protein-protein interactions: evidence from DEER for the trimerization of a von willebrand factor a domain in solution. Angewandte Chemie International Edition. 2006;45(7):1058–1061. doi: 10.1002/anie.200503720. [DOI] [PubMed] [Google Scholar]
- 106.Sahu I. D., Kroncke B. M., Zhang R., et al. Structural investigation of the transmembrane domain of KCNE1 in proteoliposomes. Biochemistry. 2014;53(40):6392–6401. doi: 10.1021/bi500943p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Meyer V., Swanson M. A., Clouston L. J., et al. Room-temperature distance measurements of immobilized Spin-labeled Protein by DEER/PELDOR. Biophysical Journal. 2015;108(5):1213–1219. doi: 10.1016/j.bpj.2015.01.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Reginsson G. W., Hunter R. I., Cruickshank P. A. S., et al. W-band PELDOR with 1 kW microwave power: Molecular geometry, flexibility and exchange coupling. Journal of Magnetic Resonance. 2012;216:175–182. doi: 10.1016/j.jmr.2012.01.019. [DOI] [PubMed] [Google Scholar]
- 109.McHaourab H. S., Steed P. R., Kazmier K. Toward the fourth dimension of membrane protein structure: Insight into dynamics from spin-labeling EPR spectroscopy. Structure. 2011;19(11):1549–1561. doi: 10.1016/j.str.2011.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zou P., Bortolus M., Mchaourab H. S. Conformational cycle of the ABC Transporter MsbA in liposomes: detailed analysis using double electron-electron resonance spectroscopy. Journal of Molecular Biology. 2009;393(3):586–597. doi: 10.1016/j.jmb.2009.08.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Endeward B., Butterwick J. A., MacKinnon R., Prisner T. F. Pulsed electron-electron double-resonance determination of spin-label distances and orientations on the tetrameric potassium ion channel KcsA. Journal of the American Chemical Society. 2009;131(42):15246–15250. doi: 10.1021/ja904808n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Georgieva E. R., Ramlall T. F., Borbat P. P., Freed J. H., Eliezer D. Membrane-bound α-synuclein forms an extended helix: Long-distance pulsed ESR measurements using vesicles, bicelles, and rodlike micelles. Journal of the American Chemical Society. 2008;130(39):12856–12857. doi: 10.1021/ja804517m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Zou P., Mchaourab H. S. Increased sensitivity and extended range of distance measurements in Spin-labeled membrane proteins: Q-band double electron-electron resonance and nanoscale bilayers. Biophysical Journal. 2010;98(6):L18–L20. doi: 10.1016/j.bpj.2009.12.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Xu Q., Ellena J. F., Kim M., Cafiso D. S. Substrate-dependent unfolding of the energy coupling motif of a membrane transport protein determined by double electron-electron resonance. Biochemistry. 2006;45(36):10847–10854. doi: 10.1021/bi061051x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Polyhach Y., Bordignon E., Tschaggelar R., Gandra S., Godt A., Jeschke G. High sensitivity and versatility of the DEER experiment on nitroxide radical pairs at Q-band frequencies. Physical Chemistry Chemical Physics. 2012;14(30):10762–10773. doi: 10.1039/c2cp41520h. [DOI] [PubMed] [Google Scholar]
- 116.Cunningham T. F., Putterman M. R., Desai A., Horne W. S., Saxena S. The double-histidine Cu2+-binding motif: a highly rigid, site-specific spin probe for electron spin resonance distance measurements. Angewandte Chemie International Edition. 2015;54(21):6330–6334. doi: 10.1002/anie.201501968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Jao C. C., Hegde B. G., Chenb J., Haworth I. S., Langen R. Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement. Proceedings of the National Acadamy of Sciences of the United States of America. 2008;105(50):19666–19671. doi: 10.1073/pnas.0807826105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Vicente E. F., Sahu I. D., Costa-Filho A. J., Cilli E. M., Lorigan G. A. Conformational changes of the Hs DHODH N-terminal Microdomain via DEER Spectroscopy. The Journal of Physical Chemistry B. 2015;119(28):8693–8697. doi: 10.1021/acs.jpcb.5b01706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Georgieva E. R., Borbat P. P., Norman H. D., Freed J. H. Mechanism of influenza A M2 transmembrane domain assembly in lipid membranes. Scientific Reports. 2015;5 doi: 10.1038/srep11757.11757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Dixit M., Kim S., Matthews G. F., et al. Structural arrangement of the intracellular Ca2+ binding domains of the cardiac Na+/Ca2+ exchanger (NCX1.1): effects of Ca2+ binding. The Journal of Biological Chemistry. 2013;288(6):4194–4207. doi: 10.1074/jbc.M112.423293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Hilger D., Polyhach Y., Jung H., Jeschke G. Backbone structure of transmembrane domain IX of the Na+ /proline transporter PutP of Escherichia coli. Biophysical Journal. 2009;96(1):217–225. doi: 10.1016/j.bpj.2008.09.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kroncke B. M., Van Horn W. D., Smith J., et al. Structural basis for KCNE3 modulation of potassium recycling in epithelia. Science Advances. 2016;2(9):e1501228–e1501228. doi: 10.1126/sciadv.1501228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Barrett P. J., Song Y., Van Horn W. D., et al. The amyloid precursor protein has a flexible transmembrane domain and binds cholesterol. Science. 2012;336(6085):1168–1171. doi: 10.1126/science.1219988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Mullen A., Hall J., Diegel J., Hassan I., Fey A., MacMillan F. Membrane transporters studied by EPR spectroscopy: structure determination and elucidation of functional dynamics. Biochemical Society Transactions. 2016;44(3):905–915. doi: 10.1042/BST20160024. [DOI] [PubMed] [Google Scholar]
- 125.Dalmas O., Hyde H. C., Hulse R. E., Perozo E. Symmetry-constrained analysis of pulsed Double Electron-Electron Resonance (DEER) spectroscopy reveals the dynamic nature of the KcsA activation gate. Journal of the American Chemical Society. 2012;134(39):16360–16369. doi: 10.1021/ja3069038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Merten J. A., Schultz K. M., Klug C. S. Concentration-dependent oligomerization and oligomeric arrangement of LptA. Protein Science. 2012;21(2):211–218. doi: 10.1002/pro.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Martens C., Stein R. A., Masureel M., et al. Lipids modulate the conformational dynamics of a secondary multidrug transporter. Nature Structural & Molecular Biology. 2016;23(8):744–751. doi: 10.1038/nsmb.3262. [DOI] [PMC free article] [PubMed] [Google Scholar]