Abstract
Angiogenesis, a prominent feature of pathology, is known to be guided by factors secreted by living cells around a lesion. Although many cells are disrupted in a response to injury, the relevance of degenerating cells in pathological angiogenesis is unclear. Here, we show that the release of lactate dehydrogenase A (LDHA) from degenerating neurons drives central nervous system (CNS) angiogenesis. Silencing neuronal LDHA expression suppressed angiogenesis around experimental autoimmune encephalomyelitis (EAE)- and controlled cortical impact-induced lesions. Extracellular LDHA-mediated angiogenesis was dependent on surface vimentin expression and vascular endothelial growth factor receptor (VEGFR) phosphorylation in vascular endothelial cells. Silencing vimentin expression in vascular endothelial cells prevented angiogenesis around EAE lesions and improved survival in a mouse model of glioblastoma. These results elucidate novel mechanisms that may mediate pathologic angiogenesis and identify a potential molecular target for the treatment of CNS diseases involving angiogenesis.
Keywords: Multiple sclerosis, Cell proliferation, Cancer, Inflammation
Highlights
-
•
Injury leads to the release of intracellular components including LDHA, which promotes angiogenesis in the CNS.
-
•
Cell surface vimentin, which is expressed on vascular endothelial cells, is involved in LHDA-mediated angiogenesis.
Angiogenesis is a prominent feature of many central nervous system (CNS) diseases, and regulates both pathologic progression and wound healing in several diseases. Pathologic CNS is characterized by neuronal damage that leads to the release of intracellular components. However, the effect of damaged cells on angiogenesis has not been clarified. This study revealed that LDHA, which is a known damage marker, promotes CNS-specific angiogenesis. LDHA-mediated angiogenesis depends on vimentin on the surface of vascular endothelial cells. The work described here proposes a novel mechanism by which neurodegeneration drives angiogenesis in the CNS.
1. Introduction
Angiogenesis, the formation of new capillaries from a pre-existing capillary network without the involvement of endothelial precursor cells, is a characteristic of many pathological conditions (Jin and Greenberg, 2005). Because angiogenesis is considered to regulate both pathological progression and wound healing, its modulation is thought to control disease progression (Costa et al., 2007). While most vessels in healthy adults are in a quiescent state, vascular endothelial cells in pathological conditions are actively proliferating (Chung and Ferrara, 2011) through a process that is thought to be affected by environmental stimuli. For example, in the central nervous system (CNS), vascular endothelial cell growth is promoted by vascular endothelial cell growth factor (VEGF) and fibroblast growth factor (FGF), both of which are upregulated in astrocytes, macrophages, and neurons after injury (Vallon et al., 2014). Microglia-derived factors are known to stimulate angiogenesis (Vallon et al., 2014). Most of the research into the molecular dynamics that govern vascular endothelial cell proliferation has been guided by the concept that angiogenic factors are expressed by living cells around lesions under disease conditions (Vallon et al., 2014). In contrast, although cell damage is a key feature of many pathological states, including CNS diseases, the effect of damaged cells around lesions on angiogenesis has not been investigated.
CNS damage causes functional impairment and/or structural dysfunction in nerve cells during disease progression. Neural cell dysfunction in part leads to the disruption of cell membrane, thereby causing the release of intracellular factors into the extracellular space. Recent findings have demonstrated that intracellular factors released from damaged or dying cells, which are recognized as damage-associated molecular patterns (DAMPs), trigger sterile inflammation (Piccinini and Midwood, 2010) and some pro-inflammatory DAMPs have been identified in the CNS (Kigerl et al., 2014, Shichita et al., 2012). Because inflammation and angiogenesis occur simultaneously under pathological conditions (Folkman and Brem, 1992), DAMPs may also influence angiogenesis, although evidence is lacking. It has, however, been reported that molecules identified as proinflammatory DAMP receptors (e.g., toll-like receptors) are also expressed by non-immune cells, including vascular endothelial cells (Stewart et al., 2015). In addition, some pro-inflammatory factors, such as interferon (IFN)-γ and stromal derived factor (SDF)-1, also stimulate vascular endothelial cells, resulting in the regulation of pathological angiogenesis (Belperio et al., 2000). Therefore, we hypothesized that factors released from damaged cells also directly regulate angiogenesis. If this is true, then elucidation of the underlying molecular mechanisms may provide novel concepts and help to promote new therapeutic measures for treating angiogenesis-related diseases.
In the present study, we show that extracellular lactate dehydrogenase A (LDHA), previously known only as a cell damage marker in the circulation, has a pro-angiogenic effect in the adult CNS. LDHA released from degenerating axons drives angiogenesis around the lesions in experimental autoimmune encephalomyelitis (EAE)- and controlled cortical impact (CCI)-induced lesions. Extracellular LDHA interacts with vascular endothelial cells, in a manner dependent on vimentin on the cell surface. Silencing vimentin expression in vascular endothelial cells prevented LDHA-mediated VEGFR2 phosphorylation. In addition, the inhibition of vimentin prevents angiogenesis in response to EAE and prolongs the survival in mouse models of glioblastoma.
2. Results
2.1. Axonal Degeneration Is Synchronized with Angiogenesis in EAE
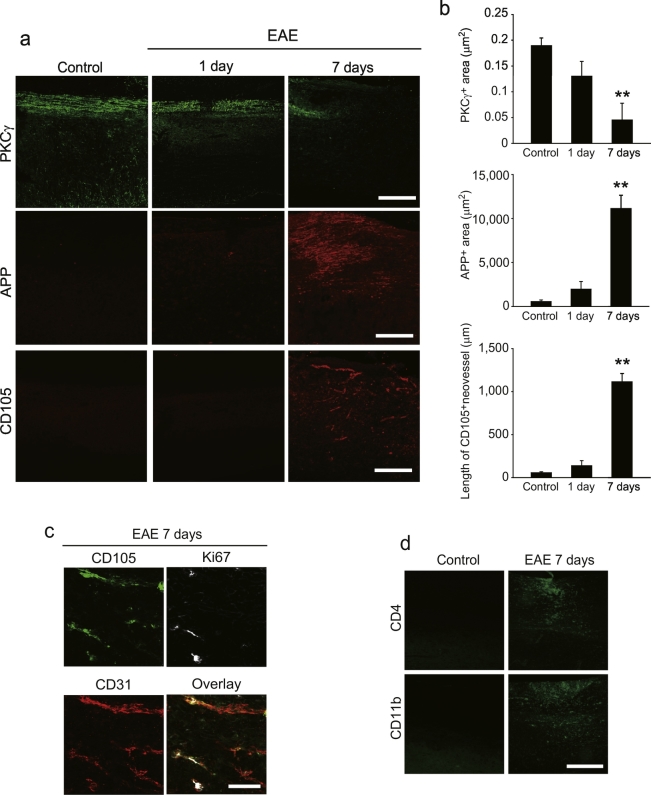
We used a localized model of EAE (Muramatsu et al., 2012) to generate single lesions, characterized by lymphocyte infiltration and angiogenesis. The time course of axonal degeneration and angiogenesis was examined in the spinal cord after EAE induction. Immunohistochemical analyses revealed a reduced intensity of protein kinase C (PKC)γ+ corticospinal tract (CST) labeling around the lesion 7 days after EAE induction (Fig. 1a and b). Here, we also observed perilesional expression of the amyloid precursor protein (APP), which is a marker of axonal damage (Fig. 1a and b). Because we previously reported robust angiogenesis around lesions 1 week after EAE induction (Muramatsu et al., 2012), we explored whether angiogenesis occurs contemporaneously with axonal degeneration. Immunohistochemical analyses demonstrated the presence of CD105+ neovessels around spinal cord lesions 7 days after EAE induction, whereas no signal was detected 1 day after the induction (Fig. 1a and b). These CD105+ areas were also positive for Ki67 and CD31, suggesting that the CD105+ neovessels contain proliferating vascular endothelial cells (Fig. 1c). In addition, we observed lymphocyte accumulation, which is associated with angiogenesis, around the lesions 7 days post-EAE induction (Fig. 1d). These time course data suggest that angiogenesis is synchronized with axonal degeneration in our mouse model of EAE.
Fig. 1.
Neovascularisation synchronises neurodegeneration in EAE.
(a) Representative images of spinal cord sections labeled with PKCγ, APP, or CD105. (b) Quantitative analysis of the fluorescent area of PKCγ or APP around EAE lesions. CD105+ neovessel length around EAE lesions; n = 4–6 for all experiments; error bars represent the s.e.m. **P < 0.01, ANOVA with Tukey's multiple comparison tests. (c) Representative images of spinal cord sections labeled with CD31, Ki67, and CD105. (d) CD4+ and CD11b+ cell accumulation around EAE lesions 7 days after EAE induction. Scale bars for a, d, 200 μm; c, 50 μm.
2.2. LDHA Has Angiogenic Activity In Vitro
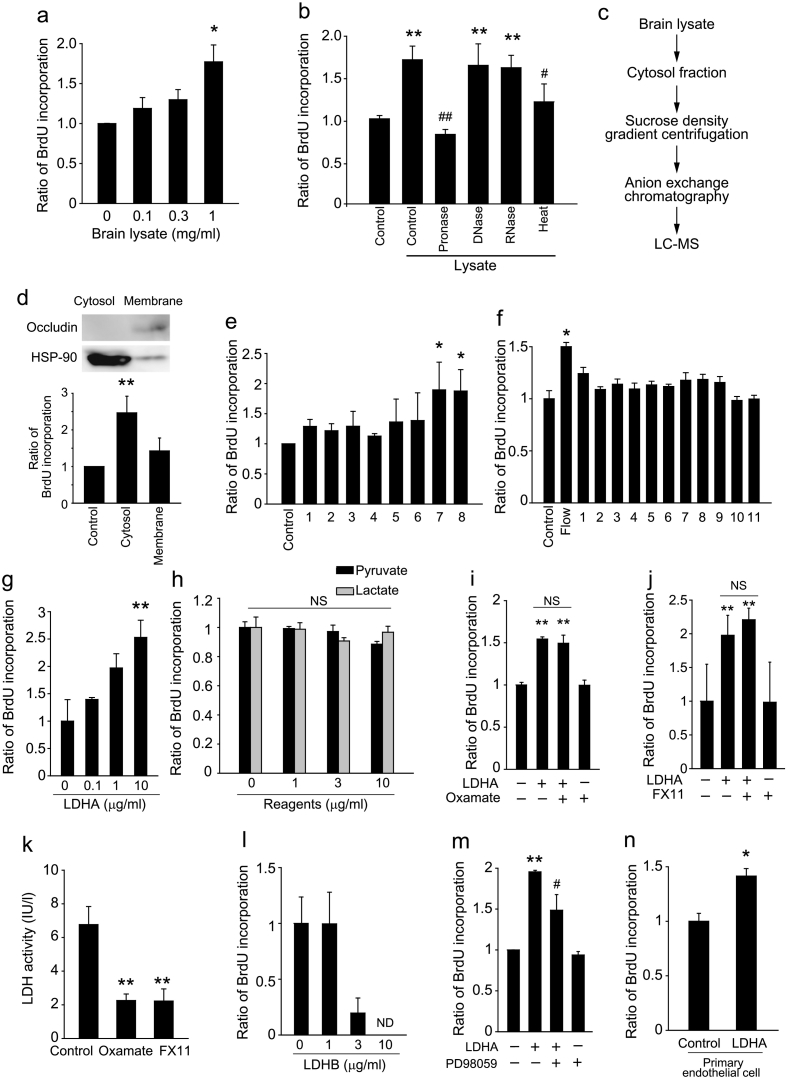
We next investigated the relationship between angiogenesis and axon degeneration. We hypothesized that factors released from degenerating axons stimulate vascular endothelial cell proliferation, because DAMPs promote inflammatory responses (Piccinini and Midwood, 2010). To assess cell proliferation in vitro, we prepared brain lysates from adult mice, as a component of degenerating tissue (Shichita et al., 2012), and used it to treat bEnd.3 vascular endothelial cells (derived from mouse brain tissue). Treatment with the brain lysate increased bromodeoxyuridine (BrdU) incorporation in the cells (Fig. 2a), suggesting that the brain lysate contains a factor(s) that promotes bEnd.3 cell proliferation. We next sought to identify the brain-lysate factor(s) that is responsible for activating the vascular endothelial cell proliferation. Brain lysate that was heated or treated with pronase induced a low degree of BrdU incorporation, whereas robust BrdU incorporation was induced when the brain lysate was treated with DNase or RNase (Fig. 2b). These results indicate that the factor of interest is a heat-susceptible protein. Treatment with fractionated brain lysates revealed that the cytosolic fraction induced cell proliferation (Fig. 2c and d), and sucrose-density gradient centrifugation identified specific subfractions that augmented cell proliferation (Fig. 2e). The active fractions of the final Mono-Q anion exchange chromatography (Fig. 2f) were subjected to liquid chromatography-mass spectrometry (LC-MS) analysis, and 68 proteins were identified in the fraction that induced cell proliferation (Table 1). We then screened these candidate proteins and found that LDHA was able to induce vascular endothelial cell proliferation (Fig. 2g).
Fig. 2.
Extracellular LDHA induces angiogenic properties in vascular endothelial cells.
(a) Concentration dependency of BrdU incorporation in b.End3 cells cultured with brain lysate. (b) BrdU incorporation in b.End3 cells 1 day after stimulation with brain lysate that was heated (98 °C, 10 min) or pretreated with pronase or DNase I, n = 3 each. (c) Strategy used for LDHA identification. (d) BrdU incorporation in b.End3 cells 1 day after stimulation with the indicated fractions of brain lysate. Top panels show fractionation of lysate. (e) BrdU incorporation in b.End3 cells 1 day after stimulation with the indicated sucrose-gradient fractions of brain lysate, n = 4 each. (f) BrdU incorporation in b.End3 cells 1 day after stimulation with each HPLC fraction of brain lysate, n = 3 each. (g) BrdU incorporation in b.End3 cells 1 day after stimulation with recombinant mouse LDHA, n = 4 for each. (h) BrdU incorporation in b.End3 cells with lactate or pyruvate, n = 3 each. (i, j) BrdU incorporation in b.End3 cells pretreated with oxamate (i) or FX11 (j) (inhibitors of LDH enzyme activity) following LDHA stimulation, n = 3 each. (k) LDH activity in b.End3 cells after indicated reagents treatment, n = 4 each. (l) BrdU incorporation in b.End3 cells cultured with LDHB, n = 3 each. (m) BrdU incorporation in b.End3 cells 1 day after stimulation with recombinant mouse LDHA. Cells were pretreated PD98059 (10 μM) for 10 min and then treated with LDHA, n = 4 for each. (n) BrdU incorporation in primary vascular endothelial cells obtained from mouse brain. n = 3–5 for all experiments; error bars represent the s.e.m. *P < 0.05, **P < 0.01 relative to control, #P < 0.05 relative to lysate. ANOVA with Tukey's multiple comparison tests. ND, not detected; NS, not significant.
Table 1.
Brain lysate proteins with a proliferative effect on vascular endothelial cells.
| Rank protein list | Sample | |
|---|---|---|
| 1 | Pkm2 Isoform M1 of Pyruvate kinase isozymes M1/M2 | 318 |
| 2 | Aldoa Fructose-bisphosphate aldolase A | 153 |
| 3 | Beta-s;Hbb-b1 hemoglobin subunit beta-1-like | 136 |
| 4 | Hba-a1;Hba-a2 hemoglobin alpha, adult chain 2 | 121 |
| 5 | Gpi1 Glucose-6-phosphate isomerase | 98 |
| 6 | Pgk1 Phosphoglycerate kinase 1 | 94 |
| 7 | Tkt Transketolase | 86 |
| 8 | Eno1;LOC100503183;LOC100045967;Gm5506 Alpha-enolase | 43 |
| 9 | Ppia Peptidyl-prolyl cis-trans isomerase | 36 |
| 10 | Gstm1 Glutathione S-transferase Mu 1 | 31 |
| 11 | Mdh2 Malate dehydrogenase, mitochondrial | 27 |
| 12 | Me1 NADP-dependent malic enzyme | 25 |
| 13 | Aco2 Aconitate hydratase, mitochondrial | 20 |
| 14 | Tpi1 triosephosphate isomerase | 18 |
| 15 | Ldha L-lactate dehydrogenase A chain | 16 |
| 16 | Park7 Protein DJ-1 | 12 |
| 17 | Sod1 Superoxide dismutase [Cu—Zn] | 12 |
| 18 | Pgd 6-phosphogluconate dehydrogenase, decarboxylating | 11 |
| 19 | Aldh1a1 Retinal dehydrogenase 1 | 11 |
| 20 | Ppid Peptidyl-prolyl cis-trans isomerase D | 10 |
| 21 | Gstp1 Glutathione S-transferase P 1 | 10 |
| 22 | Adssl1 Isoform 1 of Adenylosuccinate synthetase isozyme 1 | 9 |
| 23 | Prdx6 Uncharacterized protein | 9 |
| 24 | Gpd1 Glycerol-3-phosphate dehydrogenase [NAD +], cytoplasm | 8 |
| 25 | LOC100048522 cofilin-1-like | 8 |
| 26 | Prdx5 Isoform Mitochondrial of Peroxiredoxin-5, mitochondrial | 8 |
| 27 | Gsta4 Glutathione S-transferase A4 | 8 |
| 28 | Hrsp12 Ribonuclease UK114 | 8 |
| 29 | Esd S-formylglutathione hydrolase | 7 |
| 30 | Got2 Aspartate aminotransferase, mitochondrial | 7 |
| 31 | Car2 Carbonic anhydrase 2 | 7 |
| 32 | Nme2 Nucleoside diphosphate kinase B | 7 |
| 33 | Aco1 Uncharacterized protein | 7 |
| 34 | Oxct1 Succinyl-CoA:3-ketoacid-coenzyme A transferase 1, mito | 6 |
| 35 | Pfn1 Profilin-1 | 6 |
| 36 | Mt3 Metallothionein-3 | 6 |
| 37 | Akr1a4 Alcohol dehydrogenase [NADP +] | 5 |
| 38 | Asrgl1 L-asparaginase | 5 |
| 39 | Eef1a2 Elongation factor 1-alpha 2 | 5 |
| 40 | Sord Sorbitol dehydrogenase | 4 |
| 41 | Taldo1 Transaldolase | 4 |
| 42 | Nedd8 NEDD8 | 4 |
| 43 | Tppp Tubulin polymerization-promoting protein | 4 |
| 44 | Ass1 Argininosuccinate synthase | 4 |
| 45 | Acot7 cytosolic acyl coenzyme A thioester hydrolase isoform 1 | 3 |
| 46 | Fah Fumarylacetoacetase | 3 |
| 47 | Cryz Quinone oxidoreductase | 3 |
| 48 | Apex1 DNA-(apurinic or apyrimidinic site) lyase | 3 |
| 49 | Adh5 Alcohol dehydrogenase class-3 | 2 |
| 50 | D10Jhu81e;LOC100046684 ES1 protein homolog, mitochondri | 2 |
| 51 | Gm6316 Glyceraldehyde-3-phosphate dehydrogenase | 2 |
| 52 | Mt2 Metallothionein-2 | 2 |
| 53 | Mt1 Metallothionein-1 | 2 |
| 54 | Akr1b3 Aldose reductase | 2 |
| 55 | Cbr1 Carbonyl reductase [NADPH] 1 | 2 |
| 56 | Anxa2 Annexin A2 | 2 |
| 57 | Cfl2 Cofilin-2 | 5 |
| 58 | Mapt 76 kDa protein | 2 |
| 59 | Atp6v1e1 V-type proton ATPase subunit E 1 | 2 |
| 60 | Hagh Isoform 1 of Hydroxyacylglutathione hydrolase, mitochond | 1 |
| 61 | Aldh9a1 4-trimethylaminobutyraldehyde dehydrogenase | 1 |
| 62 | Gstm5 Glutathione S-transferase Mu 5 | 1 |
| 63 | Mif Macrophage migration inhibitory factor | 1 |
| 64 | Dbi acyl-CoA-binding protein isoform 1 | 1 |
| 65 | Nqo1 NAD(P)H dehydrogenase [quinone] 1 | 1 |
| 66 | Hadh Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | 1 |
| 67 | Aars2 Alanyl-tRNA synthetase, mitochondrial | 1 |
| 68 | Asl Argininosuccinate lyase | 1 |
Alignment of identified proteins from the MS analyses in profile mode. We excluded blood-borne proteins from the list.
LDHA forms part of a tetrameric enzyme, and the LDH holoenzyme converts pyruvate to lactate. However, the cell proliferation that was induced by LDHA treatment was not due to increased levels of LDH substrates (Fig. 2h) or greater LDH enzyme activity (Fig. 2i–k). LDHB, an isoenzyme of LDHA, did not enhance BrdU incorporation in bEnd.3 cells (Fig. 2l). LDHA promoted bEnd.3 cell proliferation by a mechanism dependent on p44/42 mitogen-activated protein kinase (MAPK; also known as ERK) activation (Fig. 2m). Moreover, LDHA-mediated cell proliferation was also observed in primary mouse brain vascular endothelial cells (Fig. 2n). Collectively, these data suggested that extracellular LDHA is a novel angiogenic factor that promotes vascular cell proliferation in vitro.
2.3. LDHA Release Promotes Angiogenesis In Vivo
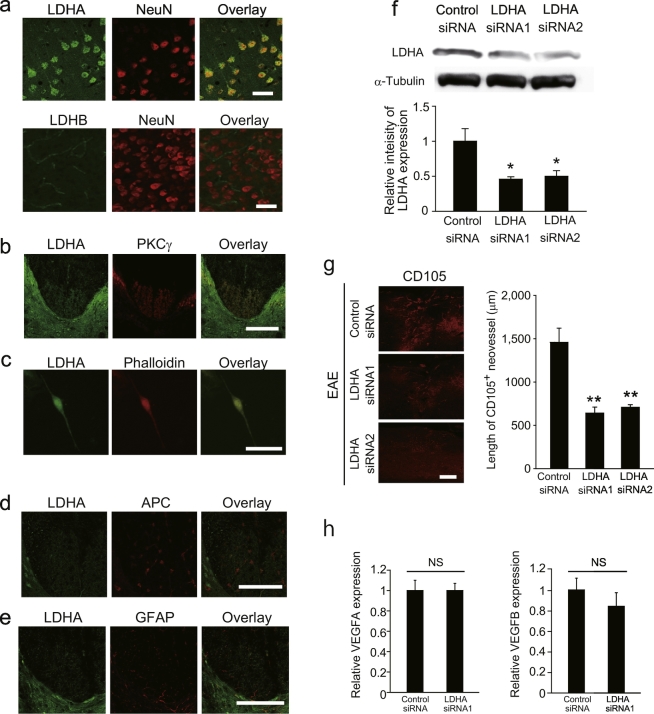
To assess the angiogenic effect of LDHA release during axonal degeneration in vivo, we first examined the expression pattern of LDHA in the adult mouse CNS. Immunohistochemistry revealed that LDHA was expressed in NeuN+ neurons in the adult mouse motor cortex, with particularly an abundant expression in layer V, which includes the soma of CST neurons (Fig. 3a). In contrast, LDHB was not detected in NeuN+ neurons in the motor cortex (Fig. 3a). PKCγ+ CST axons in the dorsal column of the spinal cord also expressed LDHA (Fig. 3b), and immunolabeling of dissociated cortical neuron cultures confirmed LDHA expression in neurites (Fig. 3c). Adenomatous polyposis coli (APC)+ oligodendrocytes and glial fibrillary acidic protein (GFAP)+ astrocytes in the spinal cord expressed very low levels of LDHA (Fig. 3d and e).
Fig. 3.
Axonal LDHA is required for CNS angiogenesis.
(a) Double immunohistochemical labeling for NeuN and LDHA (upper panels) or LDHB (lower panels) in the cerebral cortex. (b) Double immunohistochemical staining for LDHA and PKCγ in the spinal cord. (c) Double immunocytochemical staining for LDHA and phalloidin in cultured cortical neurons. (d, e) Double immunohistochemical staining for LDHA and GFAP (d) or APC (e). (f) LDHA silencing decreases LDHA expression in the motor cortex. Expression of LDHA protein in the motor cortex of mice that underwent transfection of LDHA siRNA into the motor cortex; n = 3 each. (g) Representative images of CD105-labeled spinal cord sections obtained 7 days after EAE induction. Length of CD105+ neovessels around EAE lesions, n = 5 each. (h) Relative expression of Vegfa and Vegfb around EAE lesion with or without Lhda knock down, n = 4 each; all error bars represent the s.e.m. *P < 0.05, **P < 0.01 relative to control, #P < 0.05 relative to lysate. ANOVA with Tukey's multiple comparison tests. Scale bars for a, c, 50 μm; b, d, e, g, 200 μm.
To investigate whether LDHA released from degenerating axons is required for angiogenesis in EAE mice, we injected Ldha siRNA into the motor cortex (Fig. 3f). This procedure facilitated specific knockdown of Ldha in the CST of the spinal cord (Muramatsu et al., 2012). Ldha silencing in the CST inhibited the formation of CD105+ neovessels in the spinal cord after EAE induction (Fig. 3g). We showed that VEGF expression around the lesion was not affected by Ldha knockdown (Fig. 3h). These data suggest that intracellular LDHA, which may be released into the extracellular space from degenerating CST, promotes angiogenesis in a pathological CNS environment.
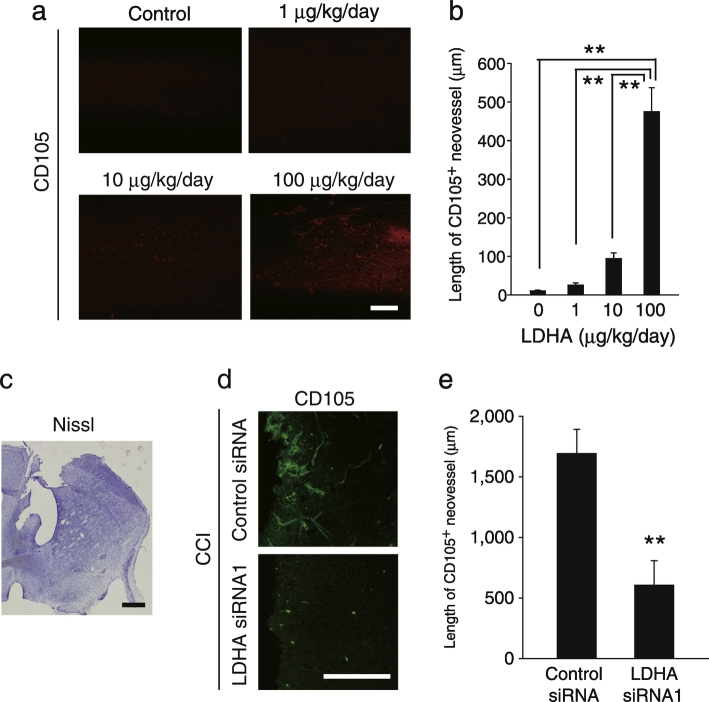
We further investigated the possibility that extracellular LDHA evokes angiogenesis in the adult CNS. Intrathecal administration of recombinant LDHA promoted CD105+ neovessel formation in the spinal cord of mice without EAE induction (Fig. 4a and b). Thus, extracellular LDHA is sufficient to promote angiogenesis in the adult mouse CNS. To further assess the possible angiogenic effect of extracellular LDHA following extensive CNS damage, we employed a CCI model (Fig. 4c). LDHA expression in the brain was knocked down by delivering Ldha siRNA to motor cortex neurons in adult mice (Fig. 3f). Ldha siRNA delivery decreased CD105+ neovessel formation around the CCI-induced lesions (Fig. 4d and e), supporting our hypothesis that extracellular LDHA promotes angiogenesis following CNS injury.
Fig. 4.
LDHA is sufficient to evoke CNS angiogenesis.
(a) Representative images of CD105-labeled spinal cord sections obtained 7 days after LDHA administration. (b) Length of CD105+ neovessels around the LDHA administration site as indicated in a, n = 5 each. (c) Representative image of a Nissl-stained brain section after controlled cortical impact (CCI). (d) Representative image of the CD105-immunolabelled cerebral cortex obtained 7 days after CCI. (e) Length of CD105+ neovessels around CCI lesions as indicated in d; n = 5 each, all error bars represent the s.e.m. **P < 0.01, Student's t-tests. Scale bars, 200 μm.
2.4. LDHA Binds to Surface Vimentin
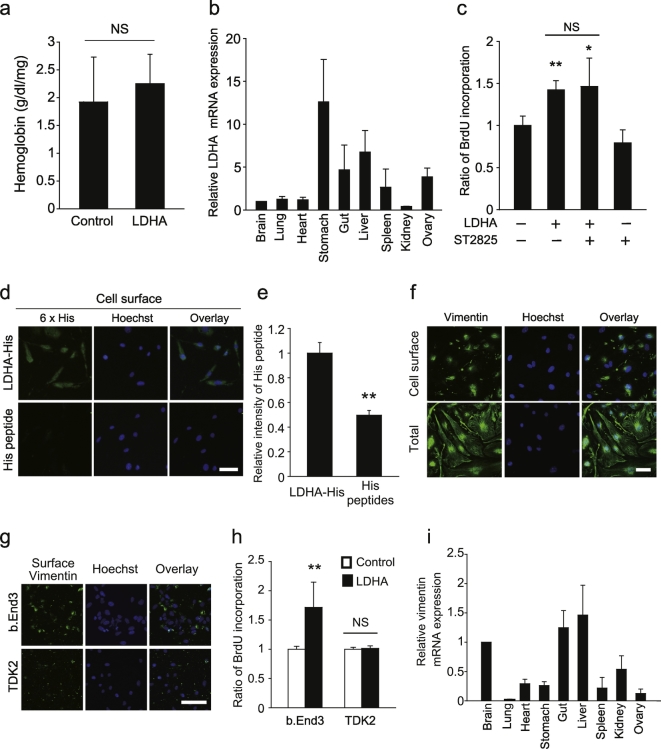
Circulating blood contains LDH, but systemic angiogenesis does not occur in healthy adults, except during pregnancy. Moreover, we found that LDHA administration did not enhance peripheral angiogenesis (Fig. 5a). Therefore, we propose that LDHA-evoked angiogenesis is restricted to the adult mouse CNS. To assess the CNS-specific effects of LDHA on angiogenesis, we analyzed Ldha mRNA expression in different organs. Real-time polymerase chain reaction (PCR) analysis revealed similar Ldha mRNA expression levels in the CNS and peripheral organs (Fig. 5b), suggesting that CNS-specific angiogenesis induced by LDHA is not a consequence of abundant LDHA expression in the CNS.
Fig. 5.
Extracellular LDHA interacts with vimentin on the cell surface.
(a) Matrigel containing LDHA was subcutaneously administered into adult mice. Hemoglobin concentration in the Matrigel 7 days after injection; n = 3 each. (b) LDHA expression is not abundant in the CNS. Relative expression levels of LDHA mRNA in the individual organs of adult mice; n = 3 each. (c) Cells were pretreated with ST2825 (Myd88 inhibitor, 10 μM) for 15 min, followed by LDHA treatment. BrdU incorporation into b.End3 cells was measured 1 d after stimulation with recombinant LDHA; n = 3 for each. (d) Immunocytochemical staining for the His peptide on b.End3 cells. (e) Relative fluorescence intensity indicating His-peptide binding to the cell surface; n = 3 for each. (f) Representative image of surface vimentin expression on non-permeabilised b.End3 cells. (g) Representative image of vimentin on the surfaces of b.End3 and TDK2 cells. (h) BrdU incorporation in b.End3 cells and TDK2 with LDHA; n = 3 for each. (i) Relative expression of vimentin mRNA in the individual organs of adult mice; n = 3 for each, all error bars represent the s.e.m. *P < 0.05, **P < 0.01, Student's t-tests or ANOVA with Tukey's multiple comparison tests. Scale bars, 50 μm.
Further, we questioned whether any LDHA-interacting molecules are highly expressed in the CNS vasculature. To identify molecules that interact with extracellular LDHA, we focused on the involvement of Toll-like receptors (TLRs), which are bound by many DAMPs (Piccinini and Midwood, 2010). Myd88 is a major downstream target of TLRs; however, Myd88 inhibition did not prevent LDHA-mediated proliferation in bEnd.3 cells (Fig. 5c). We then sought to identify molecules that are expressed by vascular endothelial cells and that interact with extracellular LDHA. We assessed the intracellular distribution of exogenous LDHA in cultured vascular endothelial cells. Immunocytochemistry revealed that His-tagged LDHA is located on the surface of bEnd.3 cells (Fig. 5d and e), indicating that the extracellular LDHA-interacting molecule is located on the vascular endothelial cell surface. We prepared membrane fractions from bEnd.3 cells treated with His-tagged LDHA, then purified the His-tagged LDHA. Using MS, we detected abundant peptide fragments of vimentin, an intermediate filament that is reported to be expressed on the surface of several cell types (Zou et al., 2006), in the fraction (Table 2). We also observed vimentin expression on the surface of bEnd.3 cells by immunocytochemistry (Fig. 5f). The relevance of cell surface vimentin in LDHA-mediated bEnd.3 cell proliferation is supported by the low expression levels of surface vimentin in TDK2 cells, a vascular endothelial cell line developed from mouse kidney, which did not show LDHA-mediated proliferation (Fig. 5g and h). Moreover, we observed that Vim mRNA expression in the CNS was not higher than that in peripheral organs (Fig. 5i). Therefore, our data indicate that cell surface vimentin expression in the CNS is key to LDHA-mediated proliferation of vascular endothelial cells.
Table 2.
Proteins which interact with exogenous LDHA on vascular endothelial cells.
| Rank protein list | Sample | |
|---|---|---|
| 1 | Ldha L-lactate dehydrogenase A chain | 249 |
| 2 | Vim Vimentin | 38 |
| 3 | Eif2s3y Putative uncharacterized protein | 33 |
| 4 | Rps19;Rps19-ps6 40S ribosomal protein S19 | 22 |
| 5 | Eif3b Eif3b protein | 15 |
| 6 | Rps4x 40S ribosomal protein S4, X isoform | 14 |
| 7 | Rnf213 LOW QUALITY PROTEIN: RING finger protein | 12 |
| 8 | Eif2s3x Eukaryotic translation initiation factor 2 subuni | 12 |
| 9 | Rpl6 60S ribosomal protein L6 | 12 |
| 10 | Ehd2 EH domain-containing protein 2 | 12 |
| 11 | G3bp1 Ras GTPase-activating protein-binding protein | 11 |
| 12 | Farsa Phenylalanyl-tRNA synthetase alpha chain | 11 |
| 13 | Rps23;Gm15450 40S ribosomal protein S23 | 11 |
| 14 | Ddx3y ATP-dependent RNA helicase DDX3Y | 11 |
| 15 | Hist2h3b;Hist1h3e;Hist1h3c;Hist2h3c2-ps;Hist1h3f;His | 11 |
| 16 | Prps2 Ribose-phosphate pyrophosphokinase 2 | 11 |
| 17 | Tcp1 Isoform 1 of T-complex protein 1 subunit alpha | 10 |
| 18 | Hspd1 Isoform 1 of 60 kDa heat shock protein, mitoch | 10 |
| 19 | Cct3 T-complex protein 1 subunit gamma | 10 |
| 20 | Rps11 40S ribosomal protein S11 | 10 |
| 21 | Hist1h2bj;Hist1h2bn;Hist1h2bf;Hist1h2bl Histone H2B t | 10 |
| 22 | Actc1 Actin, alpha cardiac muscle 1 | 10 |
| 23 | Hist1h1b Histone H1.5 | 10 |
| 24 | Rps10 40S ribosomal protein S10 | 9 |
| 25 | Cad carbamoyl-phosphate synthetase 2, aspartate tran | 9 |
| 26 | Top1 DNA topoisomerase 1 | 9 |
| 27 | Mcm5 DNA replication licensing factor MCM5 | 9 |
| 28 | Twf1 Twinfilin-1 | 9 |
| 29 | Uba52 Ubiquitin-60S ribosomal protein L40 | 9 |
| 30 | Actb Actin, cytoplasmic 1 | 8 |
| 31 | Eif4g2 eukaryotic translation initiation factor 4 gamma | 8 |
| 32 | Cnbp cellular nucleic acid-binding protein isoform 2 | 8 |
| 33 | Hnrnpu Heterogeneous nuclear ribonucleoprotein U | 8 |
| 34 | Lmna Isoform A of Prelamin-A/C | 8 |
| 35 | Pabpc4 Uncharacterized protein | 8 |
| 36 | Sf3b3 Isoform 1 of Splicing factor 3B subunit 3 | 8 |
| 37 | Calm2;Calm1;Calm3 Uncharacterized protein | 8 |
| 38 | Cct8 T-complex protein 1 subunit theta | 7 |
| 39 | Glud1 Glutamate dehydrogenase 1, mitochondrial | 7 |
| 40 | Rpl9 60S ribosomal protein L9 | 7 |
| 41 | Hspg2 basement membrane-specific heparan sulfate p | 7 |
| 42 | Gtpbp1 GTP-binding protein 1 | 7 |
| 43 | Zc3hav1 Uncharacterized protein | 7 |
| 44 | Dhx15 Putative pre-mRNA-splicing factor ATP-depen | 7 |
| 45 | Ifrd1 Interferon-related developmental regulator 1 | 7 |
| 46 | H3f3a;H3f3b Histone H3.3 | 7 |
| 47 | Ssb Lupus La protein homolog | 6 |
| 48 | Strap Serine-threonine kinase receptor-associated pro | 6 |
| 49 | Gm6139 Uncharacterized protein | 6 |
| 50 | Rps16 Rps16 protein | 6 |
| 51 | Hist1h4m;Hist1h4c;Hist1h4k;Hist1h4h;Hist2h4;Hist1h4b | 6 |
| 52 | Ddx6 Probable ATP-dependent RNA helicase DDX6 | 6 |
| 53 | Hist1h1d Histone H1.3 | 6 |
| 54 | Rpl35a;Gm14279;LOC100505045;LOC100505110 60S | 6 |
| 55 | Cct4 T-complex protein 1 subunit delta | 5 |
| 56 | Rps5 40S ribosomal protein S5 | 5 |
| 57 | Rps3 40S ribosomal protein S3 | 5 |
| 58 | Caprin1 caprin-1 isoform c | 5 |
| 59 | Rpl8 60S ribosomal protein L8 | 5 |
| 60 | Drg1 Developmentally-regulated GTP-binding protein 1 | 5 |
| 61 | Rps29;Gm14303 40S ribosomal protein S29 | 5 |
| 62 | Rpl18a 60S ribosomal protein L18a | 5 |
| 63 | Tjp2 Tight junction protein ZO-2 | 5 |
| 64 | [cRAP] K1C9_HUMAN | 5 |
| 65 | Rpl28 60S ribosomal protein L28 | 5 |
| 66 | Dnaja2 DnaJ homolog subfamily A member 2 | 5 |
| 67 | Pak1ip1 p21-activated protein kinase-interacting prote | 5 |
| 68 | Rpl34;Rpl34-ps1;Gm4705 60S ribosomal protein L34 | 5 |
| 69 | Snrpa U1 small nuclear ribonucleoprotein A | 5 |
| 70 | Rpl27 60S ribosomal protein L27 | 5 |
Alignment of identified proteins from the MS analyses in profile mode. Proteins were listed in descending order of difference between LDHA treatment and control.
2.5. Surface Vimentin Expression Drives Angiogenesis via VEGFR2
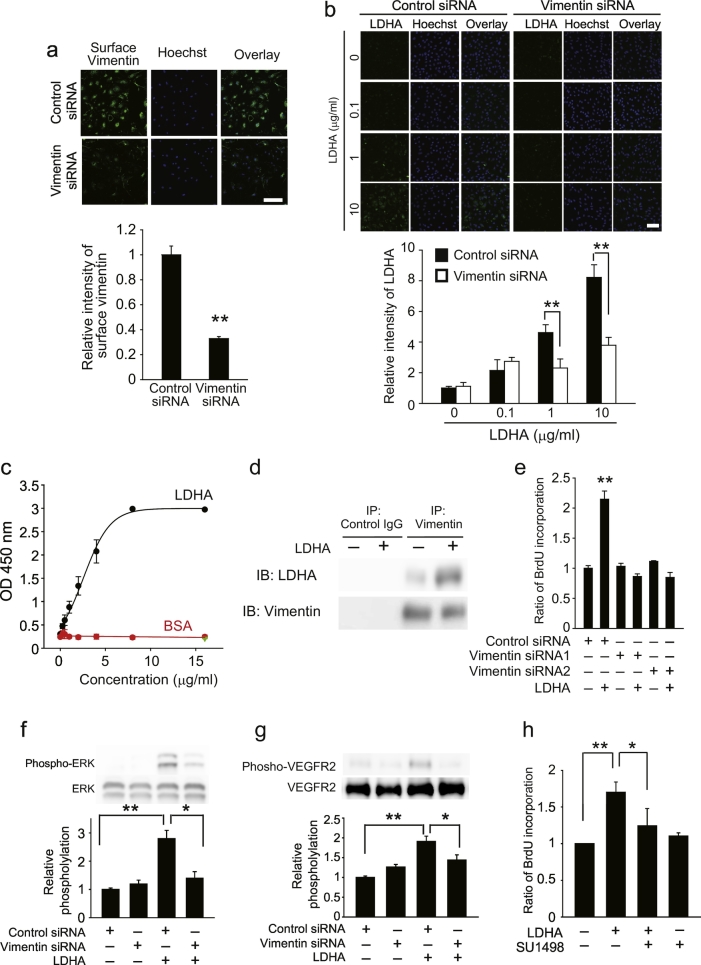
To investigate whether vimentin is involved in LDHA-mediated angiogenesis, we knocked down vimentin in bEnd.3 cells (Fig. 6a), then treated the cells with LDHA. We used cell-based binding assays to evaluate the interaction between LDHA and bEnd.3 cells, and found that exogenous LDHA is expressed on the surface of bEnd.3 cells, and is bound to the cells in a concentration-dependent manner (Fig. 6b). Silencing of Vim in bEnd.3 cells diminished LDHA binding to bEnd.3 cells (Fig. 6b). Direct binding of LDHA to vimentin was detected by ELISA (Fig. 6c). An association between LDHA and vimentin was detected in the bEnd.3 cell lysate after immunoprecipitation with an anti-vimentin antibody (Fig. 6d). We also found that inhibiting vimentin expression abolished LDHA-mediated BrdU incorporation (Fig. 6e).
Fig. 6.
Surface vimentin is involved in LDHA-mediated vascular endothelial cell proliferation.
(a) Upper images show expression of vimentin on the surface of b.End3 cells transfected with vimentin siRNA. Graph shows the quantification of the surface vimentin level shown in images; n = 3 each. (b) Interaction of LDHA with b.End3 cells transfected with vimentin siRNA. Graph shows quantification of the surface vimentin level shown in images; n = 4 each. (c) ELISA indicates direct binding of LDHA and vimentin. n = 5 each. Green symbol indicates the mean in the well which is precoated BSA and then added LDHA into the well. (d) Immunoprecipitation analysis shows that LDHA interact with vimentin. (e) BrdU incorporation in b.End3 cells transfected with vimentin or control siRNA and then stimulated with LDHA. n = 5 each. (f) Representative images of western blotting and quantitative analysis of ERK phosphorylation. n = 3 each. (g) Representative images of western blotting and quantitative analysis of Y1175 site in VEGFR2 phosphorylation. n = 3 each. (h) BrdU incorporation in b.End3 cells pretreated with SU1498 then stimulated with LDHA. n = 3 each, *P < 0.05, **P < 0.01, all error bars represent the s.e.m. **P < 0.05, **P < 0.01, Student's t-tests or ANOVA with Tukey's multiple comparison tests. Scale bars, 50 μm.
We found that LDHA promotes ERK activation in bEnd.3 cells via a vimentin-dependent mechanism (Fig. 6f). If cell surface vimentin functions as transmembrane signal transducer of the LDHA-vimentin complex, its cytoplasmic domain should be required for its activity. However, a topology prediction method, SOSUI (http://bp.nuap.nagoya-u.ac.jp/sosui/), predicted no transmembrane region for vimentin. Therefore, we considered that LDHA-vimentin-mediated intracellular signaling could be required for other proteins that do contain transmembrane regions. Because vascular endothelial cell proliferation is dependent on the phosphorylation of VEGFR-2 at Tyr1175 (Kowanetz and Ferrara, 2006), we asked if Tyr1175 phosphorylation in VEGFR2 is involved in ERK activation and vascular endothelial cell proliferation, and acts in a LDHA-vimentin dependent manner. Western blot analysis revealed that LDHA treatment caused an increase in the phosphorylation of VEGFR2 Y1175 site, and this was reduced by silencing of Vim expression (Fig. 6g). We also found that SU1498 treatment inhibited LDHA-mediated cell proliferation (Fig. 6h). These data suggest that an interaction between LDHA and vimentin causes VEGFR2 phosphorylation, which drives the proliferation in bEnd.3 cells.
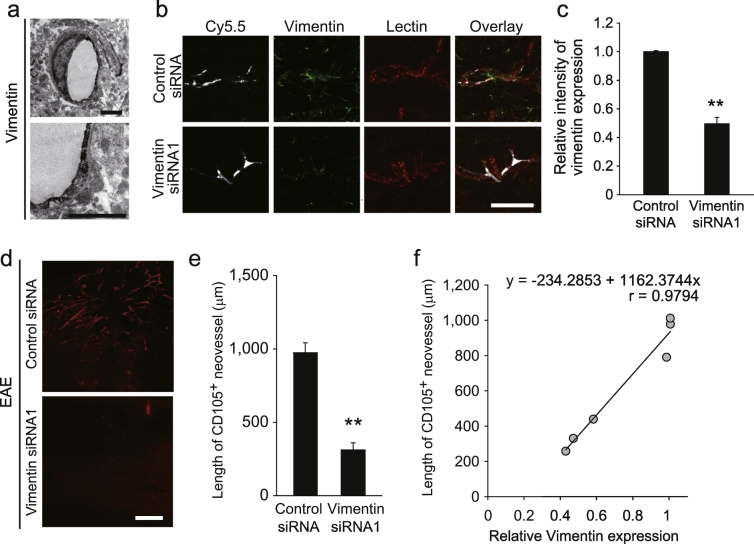
To determine whether vimentin expression on the surface of vascular endothelial cells is required for neurodegeneration-mediated angiogenesis during CNS pathology, we examined vimentin expression on the extraluminal vasculature surface in the spinal cord of adult mice using immunoelectron microscopy (Fig. 7a). We selectively knocked down Vim expression in CD31+ vascular endothelial cells in the mouse spinal cord (Fig. 7b and c) and evaluated the formation of CD105+ neovessels around the EAE lesions. Mice in which vimentin expression was inhibited did not exhibit robust angiogenesis around the EAE lesions, as compared with control mice (Fig. 7d and e). We also detected a correlation between vimentin intensity and CD105+ neovessel length (Fig. 7f). These results indicate that vimentin may be involved in neurodegeneration-mediated angiogenesis in the adult CNS.
Fig. 7.
Vascular endothelial cell vimentin is associated with neurodegeneration-related angiogenesis.
(a) Representative immuno-electron microscopy images of surface vimentin on vascular endothelial cells in the spinal cord. (b) Representative image of a spinal cord section injected with CD31-targeted liposomes containing Cy5.5 dye and the indicated oligonucleotides. Scale bars, 50 μm. (c) The graph shows the relative intensity of vimentin expression in Cy5.5+ CD31+ double-positive cells; n = 3 each, error bars represent the s.e.m. (d) Representative images of the CD105-immunolabelled spinal cord sections. (e) Length of CD105+ neovessels around EAE lesions as indicated in e. n = 5 each. (f) Correlation of vimentin expression and CD105+ neovessel length. **P < 0.01, Student's t-tests. All error bars represent the s.e.m. **P < 0.05, **P < 0.01, Student's t-tests or log-rank tests. Scale bars for a, 2 μm; b, d 50 μm, e, 200 μm.
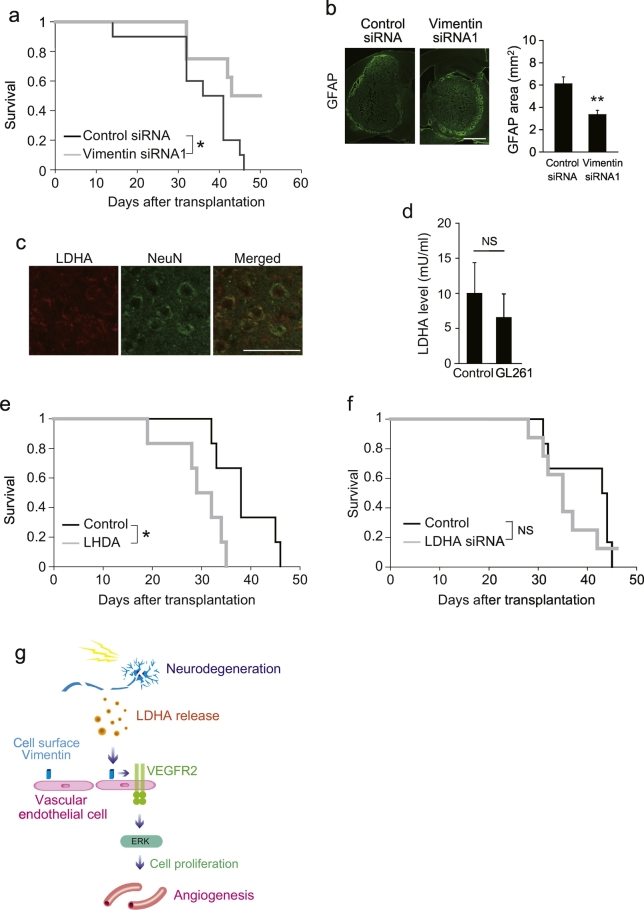
Finally, we sought to determine whether vimentin on vascular endothelial cells is involved in angiogenesis-associated disease progression at the behavioral level. Neurodegeneration and neovascularization are known hallmarks of malignant glioma, and neovascularization is associated with tumor development (Watkins and Sontheimer, 2013). We evaluated the survival of mice that received glioblastoma cell transplants into the brain in addition to the selective knockdown of vimentin in CD31+ vascular endothelial cells. Silencing Vim expression in vascular endothelial cells extended the survival of these mice, compared with control mice (Fig. 8a). Consistent with improved survival, silencing Vim expression inhibited the expansion of GL261 cells in the brain (Fig. 8b). Although we detected LDHA expression in NeuN+ cells around the transplanted GL261 cells (Fig. 8c), LDHA levels in the cerebrospinal fluid of these mice were not different compared to those in the cerebrospinal fluid of intact mice (Fig. 8d). This suggests that LDHA functions in a restricted area, rather than throughout the brain. Moreover, exogenous LDHA treatment shortened the survival of mice that underwent GL261 cell transplantation (Fig. 8e). In contrast, transplantation of GL261 cells in which LDHA was knocked down, did not affect survival (Fig. 8f). This suggests that the function of LDHA in survival is not dependent on a non-cell autonomous effect on glioblastoma cells. Collectively, these data indicate that vimentin expressed by vascular endothelial cells may affect mortality in CNS diseases.
Fig. 8.
LDHA-vimentin interaction is involved in survival of glioblastoma model mice.
(a) Kaplan − Meier survival curve of the glioblastoma mice treated with CD31 antibodies conjugated with liposomes containing vimentin siRNA or control siRNA. n = 10 for control siRNA, n = 8 for vimentin siRNA. (b) Effect of vimentin in vascular endotheilal cell on the growth of GL261 tumors in xenograft model. The graph shows the area of GL261 cells which are visualized by GFAP staining; n = 6 for control, 7 for vimentin siRNA. (c) Double immunohistochemical labeling for NeuN and LDHA around GL261 cells. (d) LDHA level in the cerebrospinal fluid (CSF) after GL261 transplantation. CSF was collected 30 days after cell transplantation; n = 4 each. (e) Kaplan − Meier survival curve of the glioblastoma mice treated with recombinant LDHA. n = 6 for each. (f) Kaplan − Meier survival curve of the glioblastoma mice transplanted with GL261 cell after LDHA knockdown. n = 6 for control, 8 for LDHA siRNA. (g) Schematic model of the role of neurodegeneration in central nervous system (CNS) angiogenesis, Student's t-tests. All error bars represent the s.e.m. **P < 0.05, **P < 0.01, Student's t-tests or log-rank tests. Scale bars for b, 1 mm; c, 50 μm.
3. Discussion
The results of this study demonstrate that a factor released from damaged cells can trigger angiogenesis in the CNS (Fig. 8g). The function of DAMPs has mainly been investigated in the context of inflammation, but our findings reveal a novel pro-angiogenic role for DAMPs. Among the abundant molecules produced inside cells, we found that LDHA, normally localized to the cytoplasm and passively released from damaged neurons, is a novel CNS-specific angiogenic factor. Although it has been reported that LDH can bind to cell membranes (Muronetz et al., 1996, Dym et al., 2000), the function of extracellular LDH was previously unknown. Our finding is the first demonstration that extracellular LDHA exerts biochemical activity related to vascular endothelial cell proliferation in the CNS. Vascular endothelial cell proliferation is only one of the processes involved in vascular rebuilding under pathological CNS conditions, and the role of extracellular LDHA on other angiogenesis mechanisms, such as proliferation and recruitment of perivascular cells (e.g., pericytes, smooth muscle cells) (Potente et al., 2011) remains unclear. It has been reported that LDH levels in the cerebrospinal fluid (CSF) are elevated within a day after brain injury (Maas, 1997) and that vascular endothelial cell proliferation is induced within 48 h after ischemic brain damage (Landis, 1994). In contrast, pericyte accumulation around the vessels is observed 2 weeks after injury (Kokovay et al., 2006). Given the timing of changes in LDH levels and angiogenic cellular responses, it is conceivable that damaged cell-derived factors play a strong supportive role in early angiogenesis, such as in vascular endothelial cell proliferation, at the time when there is an abundant release of intracellular components around the lesion site.
It has been reported that intracellular LDH promotes peripheral angiogenesis via a glycolysis-dependent mechanism, which mediates the activation of VEGF signaling (Koukourakis et al., 2003) and/or modulation of endothelial cell–extracellular matrix interactions (Sudhakaran et al., 2009). In contrast, we observed that exogenous LDHA treatment does not promote peripheral angiogenesis. Therefore, the angiogenic mechanisms induced by intra- and extracellular LDHA may be different. In this context, we found that extracellular LDHA mediates vascular endothelial cell proliferation and is required for vimentin expression on the surface of vascular endothelial cells. The role of vimentin in pathological angiogenesis is supported by reports that vimentin-null mice show a significant retardation in wound healing (Eckes et al., 2000), which is assisted by angiogenesis (Battegay, 1995). However, we should note that a lack of vimentin expression has no marked effect on the peripheral organs of mice (Colucci-Guyon et al., 1994). Because organ formation is strongly dependent on vascular development, we propose that the function of vimentin differs under normal and pathological conditions, and that vimentin strongly supports tissue repair processes, such as pathological angiogenesis. One possible reason for this difference may be a change in the localization of vimentin. We showed that vimentin is externalized during platelet activation in patients with myocardial ischemia (Podor et al., 2002). In addition, stimulation of human macrophages activates the phosphorylation of vimentin, leading to its expression on the cell surface (Mor-Vaknin et al., 2002). These reports prompted us to consider that vimentin localization in vascular endothelial cells may also change, such that cell surface vimentin expression increases under pathological CNS conditions, resulting in enhanced LDHA-mediated cell proliferation. We found that LDHA treatment promoted CD105+ neovessel formation in the intact mouse spinal cord, indicating that the level of cell surface vimentin on normal vascular endothelial cells in the CNS is sufficient to evoke LDHA-mediated angiogenesis. This is supported by a previous report that peptides that bind to cell surface vimentin promote human umbilical endothelial cell proliferation (Glaser-Gabay et al., 2011). However, the difference in CD105+ neovessel formation in response to EAE in mice after control or Ldha siRNA treatment was larger than that caused by LDHA administration in the spinal cord of control mice. This finding suggests that, under pathological CNS conditions, vascular endothelial cells are more responsive to extracellular LDHA.
Regarding the mechanism by which the extracellular LDHA-vimentin interaction regulates intracellular signaling, we found that LDHA promotes VEGFR phosphorylation, which in turn is dependent on vimentin expression. Although it remains to be determined whether other proteins are present in the protein complex that promotes vascular endothelial cell proliferation, our data suggest that VEGFR phosphorylation participates in propagating the signals to vascular endothelial cells. Although the mechanism by which cell surface vimentin stimulates VEGFR phosphorylation is open for question, one possibility is VEGFR transactivation. It has been reported that VEGFR activation is induced by extracellular Sphingosine 1-phosphate (S1P) without the release of endogenous VEGF, indicating that VEGFR transactivation occurs in a VEGF-independent manner. For S1P-mediated VEGFR activation, S1P receptor-coupled G protein signaling is partially involved in VEGFR transactivation (Spiegel and Milstien, 2003). However, considering that cell surface vimentin may lack a cytoplasmic domain, the mechanism by which it activates VEGFR may be different. In the present study, we did not investigate other mechanisms, such as a direct interaction between the extracellular domains of vimentin and VEGFR or an involvement of other proteins between vimentin and VEGFR, therefore further studies will be required to uncover the mechanism that underlies this surface vimentin-mediated VEGFR activation.
Regarding LDH subtypes, we observed that LDHA promoted bEnd.3 cell proliferation, while LDHB did not. This reciprocal effect between LDHA and LDHB may confuse the angiogenic outcome of damaged cells. Our study shows that neurodegeneration promotes angiogenesis, which is consistent with the fact that the neurons express abundant LDHA, but not LDHB. These observations suggest that the angiogenic function of damaged cells is affected by the balance between LDHA and LDHB expression.
In our glioblastoma experiments, silencing LDHA in glioblastoma cells did not affect the survival of recipient mice. This observation suggests that LDHA in transplanted cells is not involved in disease progression in the glioblastoma model. In contrast, we found that LDHA administration shortened survival compared with controls, and silencing vimentin in vascular endothelial cells extended the survival of recipient mice. These data support the idea that an interaction between extracellular LDHA and vimentin in vascular endothelial cells is involved in disease progression in the glioblastoma model.
The findings of this study identify a novel concept that a molecule released from damaged neurons can control CNS angiogenesis. Previous research into neurovascular interactions has focused on the intriguing parallels between neuronal and vasculature networks: nerves and vessels track alongside each other to reach their targets throughout the body. The formation of these networks has been studied based on the idea that the same guidance cues mediate development of both networks (Quaegebeur et al., 2011), or that one system mediates the development of the other (Makita et al., 2008). These systems are thought to be conserved in terms of both developmental and post-injury repair processes, and the networks markedly change at the structural and/or functional level after injury. However, with the exception of neuronal rewiring (Muramatsu et al., 2012), neurovascular interactions that occur after injury have not been described. Our findings reveal unexpected neurovascular interactions in the injured adult CNS that may be relevant to our understanding of neuronal damage, which is a hallmark of many CNS disorders.
4. Materials and Methods
4.1. Animals
C57BL/6J mice were obtained from Charles River Japan or Japan SLC (Japan). All experimental procedures were approved by the Institutional Animal Care and Use Committee of Osaka University (Approval No. 24-067-041).
4.2. Vascular Endothelial Cell Culture and Cell Proliferation Assay
TDK2 and bEnd.3 cells were obtained from the Japanese Cancer Research Resources Bank and American Type Culture Collection, respectively. Cells were maintained in Dulbecco's Modified Eagle's Medium (DMEM; Sigma-Aldrich) supplemented with 10% (vol/vol) heat-inactivated fetal bovine serum (FBS; Gibco). GL261 cells were maintained as previously described (Ono et al., 2014).
Primary cultures of brain vascular endothelial cells were prepared from cerebral cortices obtained from 3-week-old mice. The cerebral cortices were digested in a mixture of 1 mg/ml collagenase type 2 (Worthington Biochemical Corp) and 6.7 μg/ml DNase (Sigma-Aldrich) in DMEM for 90 min at 37 °C. The cell pellet was separated by centrifugation in 20% bovine serum albumin (BSA)-DMEM (1000 × g, 10 ms), then incubated for another 45 min at 37 °C with a mixture of 1 mg/ml collagenase-dispase (Roche) and 6.7 μg/ml DNase in DMEM at 37 °C. Brain capillaries were isolated on a 33% continuous Percoll gradient (GE Healthcare) and plated on culture dishes coated with 0.1 mg/ml collagen type IV (Sigma-Aldrich) and 0.1 mg/ml fibronectin (Sigma-Aldrich). All cultures were maintained in DMEM supplemented with 10% (vol/vol) heat-inactivated FBS and puromycin (1 μg/ml).
To assess proliferation, cells were plated at a density of 1 × 104 cells/ml in DMEM supplemented with 10% FBS. BrdU was added to cultures after 2 h (bEnd.3 cells) or 24 h (primary vascular endothelial cells). Thereafter, cell proliferation was estimated by measuring BrdU incorporation into newly synthesized cellular DNA using the Cell Proliferation Enzyme-Linked Immunosorbent and BrdU (colorimetric) Kit (Roche), according to the manufacturer's instructions. To inhibit ERK activation and LDH enzymic activity, cells were treated with the following reagents: PD98059 (10 μM, Sigma-Aldrich), Oxamate (100 mM) FX11 (10 μM, Calbiochem).
4.3. LDH Activity Assay
LDH activity in the culture was measured using QuantiChrom™ Lactate Dehydrogenase Kit (BioAssay Systems) according to the manufactory's instruction.
4.4. Brain Lysate Preparation and MS Analysis
The mice were transcardially perfused with ice-cold saline. The brains were removed and homogenized in 1 mM Tris/EGTA (pH 7.5) and centrifuged at 13,000 × g for 10 min. The supernatant was collected and used as brain lysate. For digestion experiments, brain lysate was incubated with pronase (1 U/ml, Roche), DNase I (100 μg/ml, Sigma-Aldrich), or RNase (100 μg/ml, Roche) at 37 °C for 1 h. For sucrose-density gradient centrifugation, the brain lysate was ultracentrifuged at 100,000 × g for 1 h. The supernatant was layered on a 10–40% (w/w) linear sucrose gradient in DMEM and centrifuged at 100,000 × g for 16 h. Sucrose was depleted from each sucrose gradient fraction by ultrafiltration using an Amicon Ultra-0.5 centrifugal filter unit (0.5 ml, 3 K, Millipore). We collected fractions 7 and 8, which strongly induced cell proliferation. These fractions were applied to an anionic column (MonoQ, GE Healthcare), and the eluate was condensed by ultrafiltration with an Amicon Ultra-0.5 centrifugal filter unit followed by trypsin treatment and nano-LC-MS/MS, using a Synapt G2 HDMS mass spectrometer (Waters).
4.5. Cell Lysate Preparation and MS Analysis
bEnd.3 cells were lysed in phosphate-buffered saline (PBS) containing 1% Triton X-100 and protease inhibitors at 4 °C. The 8000 × g-non-precipitable and 100,000 × g-precipitable cell lysate fractions were incubated with His-tagged recombinant LDHA (USCN) or a 6 × His-tag peptide (Abcam) for 2 h at 4 °C. Proteins retained on His Mag Sepharose Ni magnetic beads (GE Healthcare) were identified by nano-LC-MS/MS. LC-MS analysis was performed after trypsin treatment using a Synapt G2 HDMS mass spectrometer.
4.6. Localized EAE Mouse Model
Adult female C57BL/6J mice (6–8-weeks-old) were subcutaneously immunized with an emulsion of 50 μg myelin oligodendrocyte glycoprotein (MOG)35–55 (Sigma-Aldrich) in complete Freund's adjuvant (Difco) containing 500 μg Mycobacterium tuberculosis H37Ra (Difco) on day 0. This immunization protocol alone rarely induces disseminated disease; the few mice in which dissemination occurred after this protocol were excluded from subsequent experiments. After 3 weeks post-immunization, we performed a dorsal laminectomy at vertebra Th10 and injected 1 μl of a cytokine mixture comprised of 250 ng recombinant mouse tumor necrosis factor α (TNF-α; R&D Systems) and 150 U recombinant mouse interferon γ (IFN-γ; R&D Systems) dissolved in PBS over a 3-min period (CST coordinate: 0.5 mm depth at the midline) into the dorsal column of the thoracic spinal cord. All mice received an intravenous injection of 200 ng pertussis toxin (List Biological Laboratories) in 100 μl PBS at 48 h after injection of TNF-α and IFN-γ. To conduct pharmacological administration into the spinal cord, the cannula connected to Alzet osmotic pump (model no. 1007D; Alzet Corp) was placed under the dura at the thoracic spinal cord. The pump was implanted subcutaneously dorsally. The pumps were filled with recombinant mouse LHDA (0, 1, 10, 100 μg/kg/day). The pump was implanted subcutaneously in the back of mice.
4.7. CCI Injuries
The mouse scalp was retracted, and a 4-mm-diameter circular craniotomy was performed on the left side, using a drill, with the center 0 mm anteroposterior and 2 mm lateral to the bregma. Brain injury was induced by unilateral CCI over the somatosensory cortex using a pneumatic impact device (AmScien Instruments) with a 3-mm-diameter tip. The impact was made at a depth of 1.0 mm and at a speed 4.0–4.5 m/s (Miyake et al., 2015).
4.8. Matrigel Plug Assay
Mice were subcutaneously injected with 200 μl BD Matrigel Matrix Growth Factor Reduced (BD Biosciences) containing 10 μg/ml mouse recombinant LDHA (USCN). Matrigel plugs were removed after 7 days and homogenized in 100 μl deionized water. After centrifugation, the supernatant was collected, and the hemoglobin concentration was measured with the Hemoglobin Colorimetric Assay Kit (Cayman Chemical), according to the manufacturer's instructions.
4.9. siRNA Preparation and Transfection
Mouse Ldha siRNA and Vim siRNA were purchased from Applied Biosystems (Silencer siRNA). The sense and antisense strands of the siRNAs were as follows: Ldha siRNA1, 5′-CAGUGGAUAUCUUGACCUATT-3′ (sense) and 5′-UAGGUCAAGAUAUCCACUGGA-3′ (antisense); Ldha siRNA2, 5′-GUUCAUCAUUCCCAACAUUTT-3′ (sense) and 5′-AAUGUUGGGAAUGAUGAACTT-3′ (antisense); Vim siRNA1, 5′-GGUUGACACCCACUCAAAAtt-3′ (sense) and 5′-UUUUGAGUGGGUGUCAACCag-3′ (antisense). Vim siRNA2 was purchased from Santa Cruz (cat no. sc-29523). Cortical neuron transfection with Ldha siRNA was performed using a Nucleofector transfection device (Amaxa), according to the manufacturer's protocols. Transfection of Vim siRNA into bEnd.3 cells was performed using Lipofectamine RNAiMAX (Invitrogen). The cells were cultured for 72 h after transfection prior to immunocytochemical analysis.
For siRNA transfection into the motor cortex, aliquots of stock solution were mixed with the i-Fect transfection reagent (Neuromics). The mice received intracortical injections of siRNA (2 μg/μl; coordinates from the bregma: 2.0 mm posterior/2.0 mm lateral, 2.0 mm posterior/2.5 mm lateral, 2.5 mm posterior/2.0 mm lateral, and 2.0 mm posterior/2.5 mm lateral, all at a depth of 0.8 mm into the cortex). The siRNAs were delivered as either lipid-encapsulated Ldha-specific siRNA or i-Fect-encapsulated non-targeting double-stranded RNA (dsRNA; control mismatch siRNA) 3 days before EAE induction (Muramatsu et al., 2012). Silencer™ Select Negative Control No. 1 siRNA was used for control siRNA. Injection of siRNA or the transfection reagents alone into the motor cortex did not cause any behavioral changes as assessed in an open-field test (data not shown).
To knockdown vimentin expression in vascular endothelial cells in vivo, liposome-encapsulated (GLYCOPIPO) Vim siRNA and Cy5.5 fluorescent dye was synthesized by Katayama Chemical Industries. The liposome was further labeled with an anti-CD31 antibody (Abcam) using an antibody labeling kit (Sumisho Pharma International) according to the manufacturer's instructions. The antibody-conjugated liposome was injected into the exposed thoracic spinal cord (at Th10) over a 2-ms period (CST coordinates: 1 mm lateral, 0.5 mm depth). Liposome targeting of vascular endothelial cells was confirmed in spinal cord sections stained with DyLight 594-labeled Lycopersicon esculentum (Tomato) Lectin (Vector Laboratories). Liposome injection into the spinal cord did not result in any behavioral changes (data not shown).
4.10. Histology and Immunohistochemistry
Mice were transcardially perfused with 4% paraformaldehyde (PFA) in PBS. Spinal cord and brain tissues were post-fixed with 4% PFA in PBS at 4 °C overnight following immersion in PBS containing 30% sucrose. For free-floating immunohistochemistry, 30-μm-thick sections were obtained at 300-μm intervals and permeabilized in PBS containing 0.1% Triton X-100 and 10% goat serum for 1 h at room temperature. The sections were then incubated with primary antibodies overnight at 4 °C and then with a fluorescent-labeled secondary antibody for 3 h at room temperature. Sections were mounted on uncoated slides (Matsunami Glass), air-dried, and sealed with coverslips. Immunofluorescence images were captured with an Olympus FluoView FV1200 microscope and a BX60 fluorescence microscope equipped with a cooled CCD camera DP80 (Olympus).
The primary antibodies used were as follows: rabbit anti-mouse LDHA (1:100, PAB370Mu01; USCN), rabbit anti-mouse PKC-γ (1:100, sc-211; Santa Cruz Biotechnology, RRID:AB_632234), rabbit anti-human PKCγ (1:100, LS-C91497; LSBio, RRID:AB_2171760), rabbit anti-human LDHB (ab75167), rabbit anti-mouse NeuN (1:100, MAB377; Covance, RRID:AB_2298772), rat anti-mouse CD4 (1:100, 550,278; BD Biosciences, RRID:AB_393574), rabbit anti-mouse Iba1 (1:100, sc-98468; Santa Cruz Biotechnology), mouse anti-mouse GFAP (1:100, G3893; Sigma-Aldrich), mouse anti-APC (1:100, OP80; Calbiochem, RRID:AB_2224389), rabbit anti-mouse CD31 (1:100, ab28364; Abcam, RRID:AB_726362), rat anti-mouse CD105 (1:100, 550,546; BD Biosciences, RRID:AB_2277916), rabbit anti-amyloid β (A4) precursor protein (APP) (1:100, A8718, Sigma) and mouse anti-mouse Ki67 (1: 200, 556,003; BD Biosciences, RRID:AB_396287). As secondary antibodies, we used an Alexa Fluor 488- or 568-conjugated goat anti-rabbit IgG antibody, a goat anti-rat IgG antibody, and a goat anti-mouse IgG antibody (Invitrogen). For Ki67 labeling, the sections were pretreated with HistoVT One (Nacalai Tesque), according to the manufacturer's instructions.
To estimate neovessel lengths, we measured the lengths of CD105+ capillaries around the EAE lesions. To evaluate angiogenesis in EAE, the length of the vessels in grey matter was measured in sections around the targeted EAE lesion (for EAE, segment Th10 immediately rostral to the lesion center; for CCI, lesion edge through the entire rostral-caudal extent of the lesion). The means were calculated from measurements made with 3–7 sections. For Nissl staining, sections were stained with 1% cresyl violet for 10 min.
4.11. Primary Cortical Neuron Culture
Primary cortical neuron cultures were obtained from the cerebral cortex of postnatal day 1 mice. Cerebral cortices were dissociated by treatment with 0.25% trypsin in PBS for 15 min at 37 °C and the trypsin was neutralized with DMEM containing 10% FBS, followed by filtration through a 70-μm pore size membrane filter. Cells were plated in poly-L-lysine-coated plastic culture dishes and maintained in DMEM supplemented with 10% FBS.
4.12. Quantitative Real-Time PCR
Total RNA was extracted from tissues using the TRIzol Reagent (Invitrogen), and cDNA was synthesized with a High Fidelity PrimeScript II RT-PCR Kit (TaKaRa), according to the manufacturers' instructions. Quantitative PCR was performed using an ABI ViiA7 real-time PCR system (Applied Biosystems). PCR reactions were performed using Power SYBR Green PCR Master Mix (Applied Biosystems) with 200 nM primers and 10 ng cDNA. The PCR conditions used included 1 cycle at 95 °C for 10 s, which was followed by 40 cycles of 95 °C/15 s, and 60 °C/60 s. Amplicon specificity was monitored by melting curve analysis. The following primers were used: Ldha forward 5′-TGCGTGCTGGAGCCACT-3′, Ldha reverse 5′-GCGAGGAGAAGCAGCGTG-3′, Vim forward 5′-CGAGTACCGGAGACAGGTGC, Vim reverse 5′-GCGCTCCAGGGACTCGTTA-3′, Vegfa forward 5′-CCTGGTGGACATCTTCCAGGAGTACC-3′, Vegfa reverse 5′-GAAGCTCATCTCTCCTATGTGCTGGC-3′, Vegfb forward 5′- TCTCGCCATCTTTTATCTCCCAG-3′, Vegfb reverse 5′-CAGAACCCAAATCCCGTTATTG-3′, Gapdh forward 5′-TCACCACCATGGAGAAGGC-3′, and Gapdh reverse 5′-GCTAAGCAGTTGGTGGTGCA-3′.
4.13. Western Blot Analysis
Samples were homogenized in lysis buffer (50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% Triton X-100, 0.1% sodium dodecyl sulfate [SDS]) containing protease inhibitor (Roche). In tissue samples, mice were perfused with ice-cold PBS before organs were isolated for analysis. The lysates were clarified by centrifugation at 13,000 × g at 4 °C for 20 min, and the supernatants were collected and normalized in terms of protein concentrations. Proteins were separated by SDS-polyacrylamide gel electrophoresis and transferred to Immobilon-P membranes (Millipore). After blocking with PBS containing 5% skim milk and 0.05% Tween 20, the membranes were incubated with appropriate primary antibodies.
A fluorescence-conjugated secondary antibody and an Electrogenerated Chemiluminescence System (GE Healthcare) were used for signal detection. The membrane was exposed to an LAS-3000 imaging system (Fujifilm) according to the manufacturer's specifications. The protein bands were quantified using ImageJ software (National Institutes of Health, NIH). The primary antibodies used were as follows: mouse α-tubulin (1:1000, sc-5286; Santa Cruz Biotechnology, RRID:AB_628411), rabbit anti-LDHA (1:1000, PAB370Mu01; USCN), rabbit anti-heat shock protein 90 (1:1000, ab191302; Abcam), rabbit anti-occludin (1:1000, ab31721; Abcam), rabbit anti-phospho-p44/42 MAPK (1:1000, 9101; Cell Signaling Technology), and rabbit anti-p44/42 MAPK (1:1000, 9102, Cell Signaling Technology). Horseradish peroxidase-conjugated anti-mouse IgG and anti-rabbit IgG were used as secondary antibodies.
4.14. Ligand-Binding Assay
bEnd.3 cells were incubated for 30 min with 1 μg/ml LDHA-His and 6 × His peptides in DMEM supplemented with 10% FBS. The cells were washed in PBS and fixed with 4% PFA for 30 min at room temperature. To detect LDHA binding, the cells were labeled with an anti-LDHA antibody without prior permeabilization. Nuclei were stained with Hoechst 33,342.
4.15. Immunocytochemistry
After culturing, the cells were washed in PBS and fixed for 30 min at room temperature with 4% PFA. Cells were incubated with PBS containing 10% goat serum, with or without permeabilization for 1 h at room temperature, followed by overnight incubation with primary antibodies at 4 °C. After washing with PBS, cells were incubated with a fluorescently labeled secondary antibody for 1 h at room temperature. To visualize neurites, cells were also stained with and Rhodamine Phalloidin (Invitrogen) for 1 h at room temperature. Nuclear staining was performed with Hoechst 33342 (Sigma-Aldrich). Immunofluorescence images were captured with an Olympus microscope. Fluorescence intensity was measured by ImageJ software (NIH). The primary antibodies were as follows: chicken anti-vimentin (1:250, AB5733; Millipore, RRID:AB_11212377) and rabbit anti-LDHA (1:100, PAB370Mu01, USCN).
4.16. Elisa
ELISA was performed using 96-well microplates (Thermo Fisher Scientific) that were coated with 2 μg/ml of BSA or vimentin. The wells were incubated with 5% (w/v) BSA-PBS 1 h at room temperature. The wells then incubated with recombinant mouse LDHA (160–0 ng/ml) or BSA (160–0 ng/ml) diluted in PBST. Two hours after incubation, the wells were washed and anti-LDHA antibody was added. One hour after incubation, the wells were incubated with HRP-conjugated secondary antibodies. Protein bindings were detected by using substrate reagent, and stop solution (R & D Systems). Absorbance was measured at 450 nm.
4.17. Immunoprecipitation
The culture of bEnd.3 cells were stimulates with recombinant mouse LDHA (1 μg/ml) in binding buffer (PBS containg 25 mM BSA) for 5 min. Cell lysis was then carried out using a lysis buffer (1% Triton X-100, 150 mM NaCl, 20 mM Hepes, pH 7.5, complete protease inhibitors (Roche Applied Science), and 1 mM phenylmethylsulfonyl fluoride), and immunoprecipitation was performed by treatment with 10 μg of the corresponding antibody and protein G-Sepharose beads overnight under centrifugation at 4 °C. After the protein precipitates were washed five times, the proteins were separated by SDS-PAGE followed by immunoblotting.
4.18. Immunoelectron Microscopy
Mice were transcardially perfused with 4% PFA in 0.1 M phosphate buffer (PB). Spinal cord tissue was removed and fixed overnight in PB containing 30% (w/v) sucrose. The samples were cut into 10-μm-thick sections using a microtome (HM550; Thermo Fisher Scientific). The sections were treated with a 50 mM glycine solution and then treated with 0.25% saponin solution. The sections were blocked in 0.1 M PB containing 0.005% saponin and 10% BSA for 30 min at room temperature. The sections were then incubated with a chicken anti-vimentin antibody (1:150, Millipore, RRID:AB_11212377), followed by labeling overnight at 4 °C with a biotin-conjugated anti-chicken IgY secondary antibody (1:200, Abcam). The reaction products were visualized using 50 mM Tris-HCl buffer (pH 7.4) containing 0.02% diaminobenzidine tetrahydrochloride and 0.01% H2O2. The sections were incubated with an avidin-biotin complex from the ABC Kit (Vector Laboratories) and were re-fixed in 0.1 M PB containing 1% glutaraldehyde (Koyama et al., 2013). All electron micrographs were obtained using a transmission electron microscope (H-7650, Hitachi).
4.19. Glioblastoma Model
The mouse glioblastoma model was established as previously described (Benedetti et al., 2000). Briefly, GL261 cells were transplanted into the brains of 8-week-old C57BL/6j female mice. A total of 2 × 104 cells was suspended in 2 μl of DMEM containing 10% FBS and injected stereotaxically into the left striatum at a depth of 3 mm (coordinates with respect to bregma: 0.2 mm posterior and 2.5 mm left). To inhibit vimentin expression, a 28-gauge stainless steel cannula attached to a plastic pedestal (Brain Infusion Kit 3; ALZET) was introduced through a burr hole in the skull and into the left striatum (coordinates from bregma: 0.2 mm posterior, 2.5 mm left, and 3 mm depth from the skull surface) at 1 week post-cell transplantation. The cannula was cemented to the skull using cyanoacrylate and connected via plastic tubing to a subcutaneously implanted ALZET osmotic pump (Model 1007D, ALZET). The pumps were filled with CD31 antibody (Abcam)-conjugated liposomes encapsulating control siRNA (saline), or CD31 antibody (Abcam)-conjugated liposomes encapsulating vimentin siRNA. Control and vimentin siRNA delivery was performed at 9.5 μg siRNA per kg per day (dissolved in saline), over 1 week. The pump was implanted subcutaneously in the back of mice.
For LDHA treatment, the cannula connected to Alzet osmotic pump (model no. 1007D; Alzet Corp) was placed under the dura at the thoracic spinal cord. The pump was implanted subcutaneously dorsally. The pumps were filled with recombinant mouse LHDA (10 μg/kg/day). LDHA treatment was started from 1 week after cell transplantation and continued 2 weeks.
To measure the tumor size, brain sections were obtained 30 days after cell transplantation. GL261 cells were visualized by anti-GFAP staining (Newcomb and Zagzag, 2009). GFAP+ areas were measured in each brain sections and average GFAP+ areas were calculated per samples.
4.20. Statistical Analysis
Data are presented as mean ± s.e.m. Statistics were analyzed using either the unpaired Student's t-test, repeated measures analysis of variance (ANOVA), or one-way ANOVA followed by Scheffe's test or Tukey's test. P < 0.05 was considered significant.
Author Contributions
H.L. and N.M. performed in vivo experiments. R.M. wrote the manuscript. T.H. performed in vitro experiments. M.H. conducted glioblastoma experiments. Y.K. performed the immunoelectron microscopy experiments. M.K. helped real time PCR analysis. K·O and M.S. contribute to glioblastoma experiments. T.Y. provided the direction of immunoprecipitation experiments.
Conflict of Interest
The authors declare that they have no conflict of interest.
Acknowledgments
We are grateful to Prof. Akihiko Yoshimura, Dr. Takashi Shichita, and Prof. Seiji Takashima, Prof. Akira Kikuchi and Dr. Shogo Tanabe for technical advice. This work was supported by a Grant-in-Aid for Scientific Research (B) (Grant No. 16H04672 to R.M.) from the Japan Society Promotion Science. We also thank the Osaka University Program for the Support of Networking among Present and Future Researchers and the Center of Medical Research and Education, Graduate School of Medicine, Osaka University for technical support.
References
- Battegay E.J. Angiogenesis: mechanistic insights, neovascular diseases, and therapeutic prospects. J. Mol. Med. 1995:333–346. doi: 10.1007/BF00192885. [DOI] [PubMed] [Google Scholar]
- Belperio J.A., Micheal K.P., Arenberg D.A., Addision C.L., Ehlert J.E., Burdick M.D., Strieter R.M., Portier M.M., Dunia I. CXC chemokines in angiogenesis. J. Leukoc. Biol. 2000;68:1–8. [PubMed] [Google Scholar]
- Benedetti S., Pirola B., Pollo B., Magrassi L. Gene therapy of experimental brain tumors using neural progenitor cells. Nat. Med. 2000;6:447–450. doi: 10.1038/74710. [DOI] [PubMed] [Google Scholar]
- Chung A.S., Ferrara N. Developmental and pathological angiogenesis. Annu. Rev. Cell Dev. Biol. 2011;27:563–584. doi: 10.1146/annurev-cellbio-092910-154002. [DOI] [PubMed] [Google Scholar]
- Colucci-Guyon E., Portier M.M., Dunia I., Paulin D., Pournin S., Babinet C. Mice lacking vimentin develop and reproduce without an obvious phenotype. Cell. 1994;79:679–694. doi: 10.1016/0092-8674(94)90553-3. [DOI] [PubMed] [Google Scholar]
- Costa C., Incio J., Soares R. Angiogenesis and chronic inflammation: cause or consequence? Angiogenesis. 2007;10:149–166. doi: 10.1007/s10456-007-9074-0. [DOI] [PubMed] [Google Scholar]
- Dym O., Pratt E.A., Ho C., Eisenberg D. The crystal structure of D-lactate dehydrogenase, a peripheral membrane respiratory enzyme. Proc. Natl. Acad. Sci. U. S. A. 2000;97:9413–9418. doi: 10.1073/pnas.97.17.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eckes B., Colucci-Guyon E., Smola H., Nodder S., Babinet C., Krieg T., Martin P. Impaired wound healing in embryonic and adult mice lacking vimentin. J. Cell Sci. 2000;113:2455–2462. doi: 10.1242/jcs.113.13.2455. [DOI] [PubMed] [Google Scholar]
- Folkman J., Brem H. Inflammation: Basic Principles and Clinical Correlates. Second Edition. 1992. Angiogenesis and inflammation. [Google Scholar]
- Glaser-Gabay L., Raiter A., Battler A., Hardy B. Endothelial cell surface vimentin binding peptide induces angiogenesis under hypoxic/ischemic conditions. Microvasc. Res. 2011;82:221–226. doi: 10.1016/j.mvr.2011.07.006. [DOI] [PubMed] [Google Scholar]
- Jin K., Greenberg D.A. From angiogenesis to neuropathology. Nature. 2005;438(7070):954–959. doi: 10.1038/nature04481. [DOI] [PubMed] [Google Scholar]
- Kigerl K.A., de Rivero Vaccari J.P., Dietrich W.D., Popovich P.G., Keane R.W. Pattern recognition receptors and central nervous system repair. Exp. Neurol. 2014;258:5–16. doi: 10.1016/j.expneurol.2014.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kokovay E., Li L., Cunningham L.A. Angiogenic recruitment of pericytes from bone marrow after stroke. J. Cereb. Blood Flow Metab. 2006;26:545–555. doi: 10.1038/sj.jcbfm.9600214. [DOI] [PubMed] [Google Scholar]
- Koukourakis M.I., Giatromanolaki A., Sivridis E., Bougioukas G., Didilis V., Gatter K.C., Harris A.L., Tumour and Angiogenesis Research Group Lactate dehydrogenase-5 (LDH-5) overexpression in non-small-cell lung cancer tissues is linked to tumour hypoxia, angiogenic factor production and poor prognosis. Br. J. Cancer. 2003;89:877–885. doi: 10.1038/sj.bjc.6601205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kowanetz M., Ferrara N. Vascular endothelial growth factor signaling pathways: therapeutic perspective. Clin. Cancer Res. 2006;12:5018–5022. doi: 10.1158/1078-0432.CCR-06-1520. [DOI] [PubMed] [Google Scholar]
- Koyama Y., Nishida T., Tohyama M. Establishment of an optimised protocol for a Golgi-electron microscopy method based on a Golgi-Cox staining procedure with a commercial kit. J. Neurosci. Methods. 2013;218(1):103–109. doi: 10.1016/j.jneumeth.2013.05.004. [DOI] [PubMed] [Google Scholar]
- Landis D.M. The early reactions of non-neuronal cells to brain injury. Annu. Rev. Neurosci. 1994;17:133–151. doi: 10.1146/annurev.ne.17.030194.001025. [DOI] [PubMed] [Google Scholar]
- Maas A.I. Cerebrospinal fluid enzymes in acute brain injury. 1. Dynamics of changes in CSF enzyme activity after acute experimental brain injury. J. Neurol. Neurosurg. Psychiatry. 1997;40:655–665. doi: 10.1136/jnnp.40.7.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Makita T., Sucov H.M., Gariepy C.E., Yanagisawa M., Ginty D.D. Endothelins are vascular-derived axonal guidance cues for developing sympathetic neurons. Nature. 2008;452:759–763. doi: 10.1038/nature06859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyake S., Muramatsu R., Hamaguchi M., Yamashita T. Prolyl hydroxylase regulates axonal rewiring and motor recovery after traumatic brain injury. Cell Death Dis. 2015;6:e1638. doi: 10.1038/cddis.2015.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mor-Vaknin N., Punturieri A., Sitwala K., Markovitz D.M. Vimentin is secreted by activated macrophages. Nat. Cell Biol. 2002;5:59–63. doi: 10.1038/ncb898. [DOI] [PubMed] [Google Scholar]
- Muramatsu R., Takahashi C., Miyake S., Fujimura H., Mochizuki H., Yamashita T. Angiogenesis induced by CNS inflammation promotes neuronal remodeling through vessel-derived prostacyclin. Nat. Med. 2012;18:1658–1664. doi: 10.1038/nm.2943. [DOI] [PubMed] [Google Scholar]
- Muronetz V.I., Shcherbatova N.A., Nagradova N.K. Interaction of NAD-dependent dehydrogenases with human erythrocyte membranes. Appl. Biochem. Biotechnol. 1996;61:39–46. doi: 10.1007/BF02785686. [DOI] [PubMed] [Google Scholar]
- Newcomb W.E., Zagzag D. The murine GL261 glioma experimental model to assess novel brain tumor treatment. CNS Cancer. 2009:227–241. [Google Scholar]
- Ono K., Suzuki H., Higa M., Tabata K., Sawada M. Glutamate release from astrocyte cell-line GL261 via alterations in the intracellular ion environment. J. Neural Transm. 2014;121:245–257. doi: 10.1007/s00702-013-1096-8. [DOI] [PubMed] [Google Scholar]
- Piccinini A.M., Midwood K.S. DAMPening inflammation by modulating TLR signalling. Mediat. Inflamm. 2010:pii:672395. doi: 10.1155/2010/672395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Podor T.J., Singh D., Chindemi P., Foulon D.M., McKelvie R., Weitz J.I., Austin R., Boudreau G., Davies R. Vimentin exposed on activated platelets and platelet microparticles localizes vitronectin and plasminogen activator inhibitor complexes on their surface. J. Biol. Chem. 2002;277:7529–7539. doi: 10.1074/jbc.M109675200. [DOI] [PubMed] [Google Scholar]
- Potente M., Gerhardt H., Carmeliet P. Basic and therapeutic aspects of angiogenesis. Cell. 2011;146:873–887. doi: 10.1016/j.cell.2011.08.039. [DOI] [PubMed] [Google Scholar]
- Quaegebeur A., Lange C., Carmeliet P. The neurovascular link in health and disease: molecular mechanisms and therapeutic implications. Neuron. 2011;71:406–424. doi: 10.1016/j.neuron.2011.07.013. [DOI] [PubMed] [Google Scholar]
- Shichita T., Hasegawa E., Kimura A., Morita R., Sakaguchi R., Takada I., Sekiya T., Ooboshi H., Kitazono T., Yanagawa T., Ishii T., Takahashi H., Mori S., Nishibori M., Kuroda K., Akira S., Miyake K., Yoshimura A. Peroxiredoxin family proteins are key initiators of post-ischemic inflammation in the brain. Nat. Med. 2012;18:911–917. doi: 10.1038/nm.2749. [DOI] [PubMed] [Google Scholar]
- Spiegel S., Milstien S. Sphingosine-1-phosphate: an enigmatic signalling lipid. Nat. Rev. Mol. Cell Biol. 2003;4:397–407. doi: 10.1038/nrm1103. [DOI] [PubMed] [Google Scholar]
- Stewart E.A., Wei R., Branch M.J., Sidney L.E., Amoaku W.M. Expression of toll-like receptors in human retinal and choroidal vascular endothelial cells. Exp. Eye Res. 2015;138:114–123. doi: 10.1016/j.exer.2015.06.012. [DOI] [PubMed] [Google Scholar]
- Sudhakaran P.R., Viji R.I., Kiran M.S., Sameer Kumar V.B. Endothelial cell–laminin interaction: modulation of LDH expression involves α6β4 integrin–FAK–p38MAPK pathway. Glycoconj. J. 2009;26:697–704. doi: 10.1007/s10719-008-9188-7. [DOI] [PubMed] [Google Scholar]
- Vallon M., Chang J., Zhang H., Kuo C.J. Developmental and pathological angiogenesis in the central nervous system. Cell. Mol. Life Sci. 2014;71:3489–3506. doi: 10.1007/s00018-014-1625-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watkins S., Sontheimer H. Unique biology of gliomas: challenges and opportunities. Trends Neurosci. 2013;35:546–556. doi: 10.1016/j.tins.2012.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Y., He L., Huang S.H. Identification of a surface protein on human brain microvascular endothelial cells as vimentin interacting with Escherichia coli invasion protein IbeA. Biochem. Biophys. Res. Commun. 2006;351:625–630. doi: 10.1016/j.bbrc.2006.10.091. [DOI] [PubMed] [Google Scholar]